Abstract
The cell wall composition and autolytic properties of passage-selected glycopeptide-intermediate Staphylococcus aureus (GISA) isolates and their parent strains were studied in order to investigate the mechanism of decreased vancomycin susceptibility. GISA had relatively modest changes in peptidoglycan composition involving peptidoglycan interpeptide bridges and somewhat decreased cross-linking compared to that of parent strains. The cell wall phosphorus content of GISA strains was lower than that of susceptible parent strains, indicating somewhat lower wall teichoic acid levels in the GISA strains. Similar to whole cells, isolated crude cell walls retaining autolytic activity of GISA had drastically reduced autolytic activity compared to that of parent strains, and this arose early in the development of the GISA phenotype. This was due to an alteration in the autolytic enzymes of GISA as revealed by normal susceptibility of GISA-purified cell walls to parental strain autolysin extract and lower activity and altered peptidoglycan hydrolase activity profiles in GISA autolysin extracts compared to those of parent strains. Northern blot analysis indicated that expression of atl, the major autolysin gene, was significantly downregulated in a GISA strain compared to that of its parent strain. In contrast to whole cells, which showed decreased lysostaphin susceptibility, purified cell walls of GISA showed increased susceptibility to lysostaphin. We suggest that in our GISA strains, decreased autolytic activity is involved in the tolerance of vancomycin and the activities of endogenous autolysins are important in conferring sensitivity to lysostaphin on whole cells.
For several decades the glycopeptide antibiotic vancomycin remained the drug to which Staphylococcus aureus was uniformly susceptible. However, since 1997 clinical isolates with reduced susceptibility to glycopeptide antibiotics (glycopeptide-intermediate S. aureus [GISA]) have arisen during long-term vancomycin therapy in various countries around the world (22).
Although they have been areas of active investigation, the biochemistry and genetics of vancomycin resistance are not fully understood in either clinically- or laboratory-derived GISA. Studies of the cell walls of GISA have been a focus of investigation. A spectrum of changes in peptidoglycan composition in different GISA strains has been described previously (5). These changes in peptidoglycan composition include a decrease in the proportion of highly cross-linked muropeptide species and a concomitant increase in the representation of muropeptide monomers and dimers in muramidase digests of isolated peptidoglycan and, hence, a decrease in cross-linking (5, 20, 47). It has been proposed that GISA strains have an increased number of d-Ala-d-Ala residues in their cell walls that form false targets that sequester vancomycin in the cell wall, preventing it from reaching its target d-Ala-d-Ala in membrane-anchored lipid disaccharide-pentapeptide peptidoglycan precursors (21, 47).
Cui and associates have emphasized the importance of a thickened cell wall in vancomycin resistance, finding it to be a common feature of clinical GISA strains (9, 10). Evidence has been provided that cell wall synthesis is activated in strain Mu50 (19).
It is generally held that multiple mutations occur during the development of GISA (21, 38, 48). Insertional inactivation of the mutS gene, which is involved in methyl-directed DNA mismatch repair, increases the frequency of mutation of S. aureus to vancomycin resistance (35, 45). However, we found the mutS gene to be intact in a variety of clinical GISA strains, laboratory GISA strains, and their parent strains (34). Avison et al. (1) carried out an in silico comparison of the genome sequence of GISA strain Mu50 and those of vancomycin-susceptible strains. Several loss-of-function mutations in genes involved in cell wall biosynthesis and intermediary metabolism were identified.
A series of well-characterized, passage-selected GISA from various vancomycin-susceptible parent strains was previously reported (38). SmaI chromosomal restriction fragment length pulsed-field gel electrophoresis studies have confirmed the close relationship between the parent and GISA strains within each set of strains (17). However, some loss of phage typeability was noted in GISA strains compared to that of their parent strains. In an effort to further investigate the mechanism of vancomycin resistance in S. aureus, we report here on the cell wall composition and associated properties of three sets of vancomycin-susceptible parent and GISA strains. A variety of alterations were observed in peptidoglycan composition, and the strains were markedly defective in autolytic activity.
MATERIALS AND METHODS
Strains and growth conditions.
The following three sets of strains were used in this study, and the vancomycin MICs, in micrograms per milliliter, for these strains are given in parentheses (38): BB270 (2), BB270V5 (6), BB270V15 (16), 13136p−m+ (2), 13136p−m+V5 (6), 13136p−m+V20 (20), COL (3), and COLV10 (8). The strains are all methicillin resistant. Strain BB270 is a heterogeneous methicillin-resistant transductant in an 8325 background (16). Strains 13136p−m+ and COL are heterogeneous and homogeneous methicillin-resistant strains, respectively, that were originally isolated in a clinical setting in the 1960s. Strain COL has been studied extensively (47), and its genome has been sequenced (www.tigr.org). The strains were maintained as previously described (38). Cultures were grown in tryptic soy broth (TSB) (Difco Laboratories, Detroit, Mich.) at 37°C with shaking at 250 rpm.
Preparation of cell walls.
Cell walls for determination of peptidoglycan composition were prepared by breaking cells with glass beads in a Bead-Beater (Biospec Products, Bartlesville, Okla.) and treatment with sodium dodecyl sulfate and trypsin as described by Roos et al. (44) and Stranden et al. (50). Teichoic acid was removed by suspending cell walls in 40% (vol/vol) hydrofluoric acid. Crude cell walls retaining autolytic activity were prepared as described by Gustafson et al. (16). Purified cell walls, which do not retain autolytic activity, were prepared as described by Qoronfleh and Wilkinson (40). Both purified and crude cell walls retain teichoic acid covalently linked to peptidoglycan.
Analysis of peptidoglycan composition.
Peptidoglycan was digested to muropeptides by using Streptomyces globisporus muramidase, and the products were analyzed by reverse-phase high-performance liquid chromatography (HPLC) as described by Roos et al. (44) and Stranden et al. (50).
Measurement of purified cell wall phosphorus levels.
Total phosphorus was measured by the method of Fiske and SubbaRow as described by Leloir and Gardini (29).
Lysostaphin susceptibility of purified cell walls.
Purified cell walls were resuspended in 0.05 M Tris-HCl, pH 7.5, to an optical density at 620 nm (OD620) of 0.5 to 0.6, and lysostaphin (Sigma) was added to a final concentration of 5 μg ml−1. The OD620 of the suspension was measured at intervals during incubation at 37°C.
Autolysis of crude cell walls.
Crude cell walls were resuspended in 0.05 M KH2PO4-K2HPO4 buffer (pH 7.2) to an OD580 of 0.4 to 0.6. The suspension was incubated at 37°C, and the OD580 was measured at intervals (40).
Digestion of purified cell walls by autolysin extract.
An autolysin extract was prepared by freeze-thaw of exponential-phase cells in 0.01 M KH2PO4-K2HPO4 buffer (pH 7.2) as described by Qoronfleh and Wilkinson (40). Purified cell walls were resuspended in autolysin extract diluted 1:1 at an OD580 of 0.4 to 0.6. The suspension was incubated at 37°C, and the OD580 was measured at intervals.
Peptidoglycan hydrolase profiles by zymography.
Zymographic analyses of peptidoglycan hydrolase profiles were performed as described by Mani et al. (31). An overnight culture was used to inoculate 100 ml of TSB medium and was allowed to grow to an OD580 of 0.7 at 37°C with shaking. The cells were centrifuged (13,380 × g, 4°C, 10 min), washed once with cold water and 0.01 M phosphate buffer (pH 7.0), and resuspended in 1 ml of the buffer. Freeze-thaw autolysin extract was prepared by subjecting the cell suspension to three freeze-thaw cycles (−80°C for 1 h and then 37°C for 10 min). The suspension was centrifuged (13,380 × g, 10 min), and the supernatant was used as the freeze-thaw autolysin extract. Extracellular autolysins were obtained by centrifuging 15 ml of overnight culture (13,380 × g, 4°C, 10 min). The supernatant was then filter-concentrated 100-fold with a Centricon 3 concentrator (Millipore, Billerica, Mass.) (18). Cytoplasmic autolysins were obtained from 100 ml of culture grown to an OD580 of 0.7 in TSB at 37°C. Cells were harvested and broken in a Mini Bead-Beater (Biospec Products) using glass beads (16). For electrophoresis, 10 μg of protein of the various autolysin-containing fractions was electrophoresed in a sodium dodecyl sulfate-polyacrylamide gel (10%) that included 0.2% (wt/vol) Micrococcus luteus cells (Sigma) or heat-killed and lyophilized S. aureus cells. Following electrophoresis the gel was incubated at 37°C in renaturation buffer (25 mM Tris-HCl [pH 8.0] containing 1% Triton X-100) overnight. The gel was stained with 1% methylene blue in 0.01% KOH and was destained in deionized water. Peptidoglycan hydrolases were observed as clearings in the gel.
Northern blot analysis of atl expression.
Overnight-grown culture was inoculated in 20 ml of TSB and incubated at 37°C with shaking to an OD580 of 0.7. Two milliliters of sample was then taken and immediately centrifuged (13,380 × g, 4°C, 5 min). Total RNA was isolated from the pellet by using the QIAGEN RNeasy kit (QIAGEN) according to the manufacturer's instructions. Five micrograms of total RNA and formamide-bromophenol dye were mixed and incubated at 65°C for 5 min followed by quick chilling on ice. Denatured total RNA was separated by formaldehyde-denatured agarose gel electrophoresis in 1× morpholinepropanesulfonic acid buffer. After evaluating the quality of the RNA by ethidium bromide staining, the agarose gel was subjected to 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) transfer to Immobilon-Ny+ transfer membrane (Millipore) followed by UV cross-linking in a Stratalinker UV Cross-linker (Stratagene, La Jolla, Calif.). The transferred membrane was hybridized with radiolabeled randomly primed DNA, which was synthesized in the presence of [α-32P]dCTP (ICN Pharmaceuticals, Costa Mesa, Calif.), at 68°C for 18 h. The atl gene-specific PCR product was used as template, and radiolabeled probe was synthesized using the Prime-a-Gene labeling system (Promega, Madison, Wis.) in the presence of [α-32P]dCTP and used to probe the membrane. The hybridized membrane was subjected to low- and moderate-stringency washes prior to autoradiography. The size of the atl transcript was determined by comparison to an RNA ladder (Invitrogen) run on the same gel.
RESULTS
Modest changes in peptidoglycan composition in GISA strains.
Peptidoglycan preparations from the various strains were digested with muramidase, which cleaves after N-acetyl-muramic acid residues, leaving peptide cross-bridges intact in the various disaccharide peptide units generated through the activity of the enzyme. The muropeptides generated were analyzed by HPLC, and peaks were identified and designated using previously described nomenclature (44, 50). The proportions of muropeptide monomers, dimers, trimers, and oligomers are summarized in Table 1.
TABLE 1.
Muropeptide composition of GISA and parent strains
| Strain | Composition (%)
|
|||
|---|---|---|---|---|
| Monomer | Dimer | Trimer | Oligomer | |
| BB270 | 5.7 | 9.3 | 11.5 | 72.3 |
| BB270V5 | 8.0 | 10.6 | 12.7 | 68.0 |
| BB270V15 | 9.5 | 12.1 | 13.2 | 64.5 |
| 13136p−m+ | 7.1 | 11.6 | 11.5 | 66.8 |
| 13136p−m+V5 | 7.2 | 11.4 | 12.5 | 69.0 |
| 13136p−m+V20 | 8.4 | 11.5 | 13.3 | 65.4 |
| COL | 6.0 | 9.1 | 10.6 | 73.2 |
| COLV10 | 7.1 | 9.0 | 10.2 | 72.5 |
The HPLC chromatograms of strains BB270, BB270V5, and BB270V15 are shown in Fig. 1a to c. Strain BB270 had a typical muropeptide profile similar to those of previously reported vancomycin-susceptible strains (5, 20, 47). Strain BB270V5 showed slightly decreased cross-linking, whereas cross-linking in strain BB270Vl5 was significantly decreased, as judged by an increase in the size of the monomer peaks and a decrease in the size of the oligomer peaks. The primary peaks for all of these strains were M4 (monomer unit with five glycine residues) and D3 (dimer equivalent to M4). In strains BB270V5 and BB270V15 there were clear increases in peaks M2 (monomer unit with a single glycine residue instead of five), M10 (monomer unit with an l-alanine residue linked to l-lysine replacing the first of the five glycine residues), and D10 (a dimer equivalent to M10). In contrast to strain Mu50 (20), there was not a large peak of M9 corresponding to a disaccharide pentapeptide monomer with a pentaglycine bridge containing d-glutamate instead of d-isoglutamine in the stem peptide.
FIG. 1.
HPLC muropeptide elution profiles of mutanolysin-digested peptidoglycan from GISA and parent strains. (a) BB270; (b) BB270V5; (c) BB270V15; (d) 13136p−m+; (e) 13136p−m+V5; (f) 13136p−m+V20; (g) COL; (h) COLV10.
The muropeptide profiles of strains 13136p−m+, 13136p−m+V5, and 13136p−m+V20 are shown in Fig. 1d to f. Similar to BB270 GISA strains, there was an increase in muropeptides containing an l-alanine residue in the interpeptide bridge (M10 and D10) in 13136p−m+V5 and 13136p−m+V20. However, an increase in M2 in these strains similar to that of the BB270 series was not seen. Again, no large M9 peak was seen in this series of strains.
The muropeptide profiles of strains COL and COLV10 are shown in Fig. 1g and h. The muropeptide profile of strain COL is consistent with published reports on this strain (47). Compared to strains BB270 and 13136p−m+, COL has a higher proportion of peak M1 monomer units lacking glycine residues. A major decrease in peptidoglycan cross-linking was not observed in strain COL V10, unlike the independently isolated COL GISA derivative COL-VM50 (47). A striking aspect of the muropeptide profile of strain COLV10 was an increased amount of peaks representing muropeptides with no glycine residues (M1, M1*) or a reduced number of glycine residues: M2, a single glycine residue, and M3, three glycine residues.
Lower teichoic acid content of GISA-purified cell walls.
The purified cell wall phosphorus content of the various strains was determined as a measure of wall teichoic acid content (Table 2). The GISA strains showed a decreased phosphorus content of 9% in strain BB270V15, 35% in strain 13136 p−m+V20, and 24% in strain COLV10 compared to those of their respective parent strains. Growth in the presence of vancomycin had no effect on the teichoic acid content of strains COL and COLV10.
TABLE 2.
Phosphorus content of purified cell walls of GISA and parent strains
| Strain | Phosphorus (μmol mg−1 of purified cell walls)a |
|---|---|
| BB270 | 0.91 ± 0.09 |
| BB270V5 | 0.90 ± 0.10 |
| BB270V15 | 0.83 ± 0.04 |
| 13136p−m+ | 0.79 ± 0.06 |
| 13136p−m+V5 | 0.73 ± 0.02 |
| 13136p−m+V20 | 0.51 ± 0.08 |
| COL | 0.58 ± 0.08 |
| COLV10 | 0.44 ± 0.01 |
| COL grown with vancomycin | 0.60 ± 0.04 |
| COLV10 grown with vancomycin | 0.44 ± 0.07 |
Values are ± standard errors of the means. They are the results of duplicate determinations on three independent batches of purified cell walls.
Increased lysostaphin susceptibility of GISA-purified cell walls.
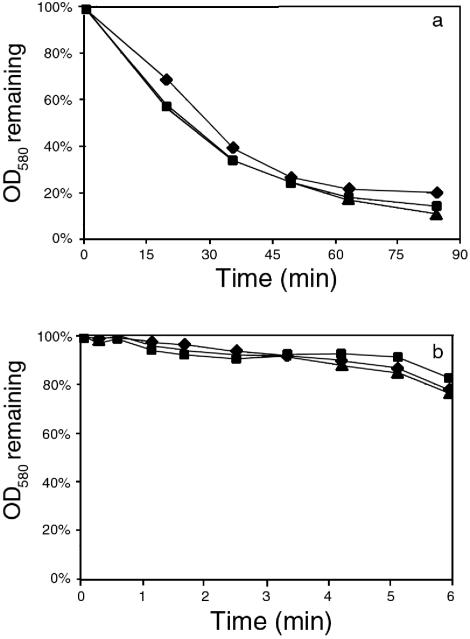
Boyle-Vavra et al. (3) and Pfeltz et al. (38) have reported that intact cells of GISA are more resistant to lysostaphin than are susceptible strains. Hence, we determined the susceptibility of purified cell walls to lysostaphin (Fig. 2). Surprisingly, the purified cell walls of the GISA derivatives of BB270 and 13136p−m+ were more susceptible to lysostaphin than were the purified cell walls of their respective parent strains. In contrast, the intact GISA cells showed much lower lysostaphin susceptibility than their parent strains (data not shown). This was also in contrast to the decreased autolysis of crude cell walls from the GISA strains (see below). There was little difference in lysostaphin susceptibility between COL and COLV10. Growth in the presence of vancomycin did not result in major changes in the lysostaphin susceptibility of the purified cell walls of COLV10.
FIG. 2.
Lysostaphin susceptibilities of purified cell walls from GISA and parent strains. (a) BB270 series. (♦) BB270, (▪) BB270V5, (▴) BB270V15. (b) 13136p−m+ series. (♦) 13136p−m+, (▪) 13136p−m+V5, (▴) 13136p−m+V20. (c) COL series. COL grown with (⋄) and without (♦) vancomycin and COLV10 grown with (□) and without (▪) vancomycin.
Decreased autolysis of GISA crude cell walls.
Pfeltz et al. (38) reported that GISA strains, in general, had decreased whole-cell autolytic activity compared to that of their vancomycin-susceptible parent strains. To investigate this further, the autolysis of isolated crude cell walls that retain autolytic activity was studied. Crude cell walls from both strains BB270V5 and BB270V15 had drastically reduced autolytic activity compared to that of parent strain crude cell walls (Fig. 3a). Clearly a reduction in autolytic activity had arisen early (BB270V5) in the development of reduced vancomycin susceptibility. A similar pattern was also seen with the 13136p−m+ series of strains (Fig. 3b). Strain COLV10 showed lower autolytic activity than strain COL (Fig. 3c), and growth in the presence of vancomycin led to a further decrease in autolysis. Sieradzki and Tomasz (47) have previously reported that growth of a strain COL GISA derivative in the presence of vancomycin leads to decreased autolytic activity.
FIG. 3.
Autolysis of GISA and parent strain crude cell walls. (a) BB270 series. (♦) BB270, (▪) BB270V5, (▴) BB270V15. (b) 13136p−m+ series. (♦) 13136p−m+, (▪) 13136p−m+V5, (▴) 13136p−m+V20. (c) COL series. COL grown with (⋄) and without (♦) vancomycin and COLV10 grown with (□) and without (▪) vancomycin.
In an effort to try to determine whether the decreased rate of autolysis of GISA crude cell walls was due to a change in the composition and structure of the cell wall or a change in the cell's autolytic enzymes, the susceptibility of purified cell walls to a freeze-thaw autolysin extract was investigated. In Fig. 4a the susceptibilities of purified cell walls from strains BB270, BB270V5, and BB270V15 to freeze-thaw autolysin extract from strain BB270 are shown. The purified cell walls from each strain showed a similar susceptibility to strain BB270 autolysin extract. This indicates that the decreased autolytic activity of BB270V5 and BB270V15 cells and crude cell walls is not due to altered cell wall structure but is due to altered autolytic enzymes in the GISA strains. When GISA strain BB270V15 was used as the source of the freeze-thaw autolysin extract, the purified cell walls from the BB270 series of strains showed similar low rates of autolysis, in contrast to when the BB270 freeze-thaw extract was used as the source of autolysins (Fig. 4b). Similar findings were obtained with the 13136p−m+ series of strains (data not shown). Interestingly, the protein concentration of the freeze-thaw extract from GISA cells was significantly lower than that of the extract from the same amount of parent strain cells. These findings suggested that there is a smaller amount or less activity of cell wall-associated autolysin in the GISA strains.
FIG. 4.
Digestion of GISA- and parent strain-purified cell walls by freeze-thaw autolysin extract from strain BB270 (a) and strain BB270V15 (b). (♦) BB270, (▪) BB270V5, (▴) BB270V15.
Altered peptidoglycan hydrolase profile of GISA freeze-thaw, cytoplasmic, and extracellular fractions.
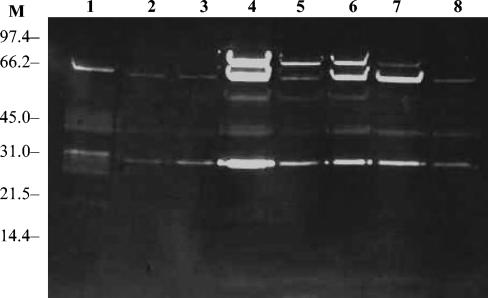
The peptidoglycan hydrolase profiles of freeze-thaw autolysin extracts from the parent and GISA strains with M. luteus cells as substrate are shown in Fig. 5. The three parent strains BB270, 13136p−m+, and COL showed typical S. aureus peptidoglycan hydrolase activity profiles with multiple bands of activity and major bands of activity at approximately 28 and 63 kDa. The major difference in the GISA strain freeze-thaw extracts was that the bands were less intense, even though equivalent amounts of proteins were loaded on the gels. In addition, some changes in the number of bands observed were noted.
FIG. 5.
Peptidoglycan hydrolase profiles of freeze-thaw autolysin extracts from GISA and parent strains against M. luteus cells. Lanes: M, molecular mass markers in kilodaltons; 1, BB270; 2, BB270V5; 3, BB270V15; 4, 13136p−m+; 5, 13136p−m+V5; 6, 13136p−m+V20; 7, COL; 8, COLV10.
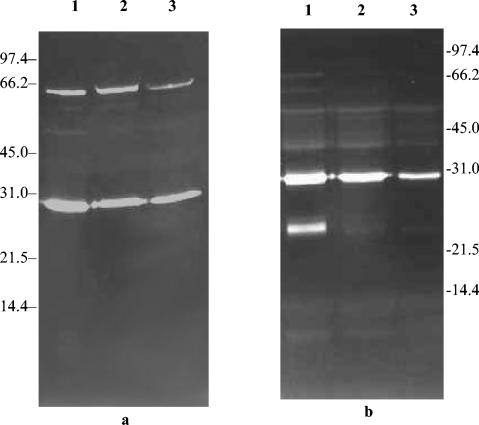
These results suggested that smaller amounts of autolysin were becoming associated with the GISA strain cell walls. Accordingly, the peptidoglycan hydrolase profiles of the cytoplasmic and extracellular fractions of the 13136p−m+ series of strains were determined (Fig. 6). In the cytoplasmic fraction the same two major bands were present in each of the strains (Fig. 6a). The intensity of the bands was somewhat lower in strains 13136p−m+V5 and 13136p−m+V20 than in strain 13136p−m+. The most striking aspect of extracellular autolysin profiles (Fig. 6b) was the apparent absence of the ∼25-kDa major band that is prominent in the parent strain. Other bands were less intense in the GISA strains. This suggests that less autolysin may have been produced in the GISA strains.
FIG. 6.
Peptidoglycan hydrolase profiles of the cytoplasmic (a) and extracellular (b) fractions of GISA and parent strains against M. luteus cells. Molecular mass markers are indicated in kilodaltons on both sides of gels. Lanes: 1, 13136p−m+; 2, 13136p−m+V5; 3, 13136p−m+V20.
Similar peptidoglycan hydrolase profiles of parent strain freeze-thaw extracts on parent and GISA strain cells as substrates.
Boyle-Vavra et al. (4) reported that GISA strains that showed reduced autolysis had poor peptidoglycan hydrolase activity in various fractions against GISA strains but not against a vancomycin-susceptible strain or against M. luteus cells. This low level of activity of GISA autolysin preparations was attributed to a change in the cell wall of the GISA strains. In contrast, we found that freeze-thaw autolysin extract from GISA parent strains had more activity against M. luteus cells in determination of peptidoglycan hydrolase profiles than the extract from GISA strains (Fig. 5). Accordingly, we compared the activity of freeze-thaw autolysin extract from strains 13136p−m+ and 13136p−m+V20 to that of heat-killed 13136p−m+ and 13136p−m+V20 cells. Freeze-thaw autolysin extract from 13136p−m+ had more activity against both susceptible parent (13136p−m+) and GISA (13136p−m+V20) heat-killed cells than the extract from 13136p−m+V20 (Fig. 7). However, freeze-thaw autolysin extract of 13136p-m+V20 had poor activity, and some peptidoglycan hydrolase bands were missing against both susceptible (13136p−m+) and resistant (13136p−m+V20) heat-killed cells (Fig. 7). This crisscross experiment indicates that the poor peptidoglycan hydrolase activity of GISA strains is due to the reduced activity of autolysin extracts rather than alterations in the cell wall composition of GISA strains.
FIG. 7.
Peptidoglycan hydrolase profiles of freeze-thaw autolysin extracts of parent strain 13136p−m+ and GISA strain 13136p−m+V20 against 13136p−m+ (a) and 13136p−m+V20 (b) cells. Molecular mass markers are indicated in kilodaltons on both sides of gels. (a) Lanes: 1, 13136p−m+; 2, 13136p−m+V20 extract against 13136p−m+ cells. (b) Lanes: 1, 13136p−m+; 2, 13136p−m+V20 extract against 13136p−m+V20 cells.
Decreased atl transcription in a GISA strain.
Northern blot analysis was used to analyze the expression level of the major, bifunctional autolysin gene atl in GISA. Total RNA was isolated from both a parent (13136p−m+) and GISA (13136p−m+ V20 strain) and was hybridized with an atl-specific probe. The hybridization signal was weak in the GISA strain, whereas the parent strain showed a higher intensity of signal (Fig. 8). This indicates decreased transcription of atl in this GISA strain compared to that of its parent strain.
FIG. 8.

Northern blot analysis of atl expression in parent strain 13136p−m+ and its GISA derivative 13136p−m+V20. The corresponding ethidium bromide-stained gel is shown underneath the blot.
DISCUSSION
In BB270 GISA strains there was a decrease in peptidoglycan cross-linking compared to that in the parent strain. The decreases in cross-linking appeared to be less striking than those observed in a COL-derived GISA strain described by Sieradzki and Tomasz (47, 48). In strains BB270V5 and BB270V15 there were increases in monomer units with a single glycine residue. There was also an increase in muropeptides containing l-alanine attached to the epsilon amino group of l-lysine in the BB270 GISAs and the 13136p−m+ GISA that were not present in their parent strains. A somewhat different pattern was noted for GISA strain COLV10. Compared to that of the parent strain COL, there was an increase in muropeptides with no or a reduced number of glycine residues. No major changes in peptidoglycan cross-linking were noted for the GISA strains COLV10, and 13136p−m+V5, or 13136p−m+V20. In none of our GISA strains was there large amounts of peptidoglycan muropeptides with a glutamate rather than an isoglutamine residue in the stem peptide, in contrast to clinical GISA strain Mu50 (20). However, Boyle-Vavra et al. (5) have shown that this muropeptide is not specific to GISA because it is also found in glycopeptide-susceptible strains. Other peptidoglycan modifications noted in different GISA strains include an increase in cross-linking (33) and an increase in the chain length distribution of the polysaccharide backbone of peptidoglycan (27, 46). Overall, the changes observed in peptidoglycan composition in our GISA strains appear to be relatively modest. Clearly, as pointed out by Boyle-Vavra (5), a spectrum of changes occur in the peptidoglycan composition of various GISA strains.
As revealed by phosphorus levels in purified cell walls, the GISA strains had less teichoic acid than the parent strains, and this was presumably compensated for by an increase in the proportion of peptidoglycan in the walls. Jenni and Berger-Bächi (26) reported variation in the levels of wall teichoic acid in a number of S. aureus strains. Teichoic acid comprises part of the S. aureus phage receptor site (7). Gustafson et al. (17) have reported a decrease in the number of phage types to which the BB270, 13136 p−m+, and COL GISA strains were susceptible compared to the numbers for their parent strains. This might involve decreased wall teichoic acid levels and the severe autolytic deficiency in the GISA strains (see below), which may interfere with phage lysis.
The significance of altered cell wall teichoic acid levels in the context of decreased vancomycin susceptibility and autolysin is not yet clear. Peschel et al. (37) have described a mutant deficient in d-alanine esters in wall teichoic acid and lipoteichoic acid. The mutant showed increased vancomycin and lysostaphin susceptibility but decreased autolysis compared to that of the wild type. In contrast to our findings of decreased teichoic acid levels in the walls of our GISA strains, Sieradzki and Tomasz (49) found somewhat higher teichoic acid levels in some clinical GISA strains compared to that of their isogenic parent strain. These authors have proposed that a key component of vancomycin resistance is an alteration in teichoic acid structure or biosynthesis.
The BB270 and 13136p−m+ GISA strains in this study were highly deficient in autolytic activity, and this appeared to develop early as revealed by strains BB270V5 and 13136p−m+V5. Autolysis deficiency has been noted in other laboratory strains (47) and in clinical GISA isolates (4, 49). The well-known clinical GISA strain Mu50 remains a perplexing exception to this common finding (23), in that strain Mu50 appears to show increased autolytic activity compared to that of unrelated S. aureus isolates.
Our GISA strains showed low susceptibility to lysostaphin (38), as has been found previously for other laboratory strains (11) and a clinical isolate (3). To our surprise, purified cell walls of our GISA strains did not show decreased susceptibility to lysostaphin but rather showed increased susceptibility compared to that of their respective parent strains. Climo et al. (8) described some lysostaphin-resistant, oxacillin-resistant S. aureus strains. Lysostaphin resistance was associated with a change in the interpeptide cross-bridge from a pentaglycine to a single glycine residue, as revealed by a large increase in the M2 peak in the mutants. We saw no large increase in the M2 peak in our strains. In the coagulase-negative staphylococcal species Staphylococcus simulans and Staphylococcus capitis, incorporation of serine residues into the third and fifth position of the interpeptide bridge, through the activities of the lif and epr genes in S. simulans and S. capitis, respectively, is believed to be responsible for the natural low susceptibility to lysostaphin of these species (12, 53). We noted no such changes in our strains. Rather, we propose that in intact cells the cell's autolysins participate in the lysis seen upon lysostaphin treatment and that the defective autolysis of GISA strains is mainly responsible for the poor susceptibility of intact cells to lysostaphin. In support of this, Pfeltz et al. (38) have shown that intact cells of the atl knockout mutant SH108 are more lysostaphin-resistant than atl intact strains. However, strain SH108 does not show decreased vancomycin susceptibility (MIC, 1 μg ml−1) (38). Also, cells that have received a brief heat shock, which is unlikely to cause major changes in peptidoglycan composition, show decreased autolysis (39) and decreased susceptibility to lysostaphin (unpublished observations).
We have made some investigations of the mechanism of autolysin deficiency. It appears that changes in the amount or activity of the autolytic enzymes are mainly responsible for the autolysin deficiency, even though some changes in peptidoglycan composition were evident in the walls of the GISA strains. Freeze-thaw autolysin extract of the GISA strains had less intense bands in peptidoglycan hydrolase profile gels than did parent strain autolysin extracts when equal amounts of protein were loaded, and some differences in the bands present were noted. The freeze-thaw autolysin extract is believed to represent cell wall-associated autolytic enzymes that are released from the walls (24). Significantly, parent strain freeze-thaw autolysin was equally active on purified cell walls from the parent or GISA strains. GISA autolysin extract had similarly poor activity on purified cell walls from parent or GISA strains. In contrast to the findings of Boyle-Vavra et al. (4) and Sieradzki and Tomasz (49), we did not find that autolysis-deficient GISA strains were resistant to lysis by autolysin-containing fractions from susceptible strains, as judged by lysis of GISA purified cell walls, or heat-killed cells in peptidoglycan hydrolase determinations.
Several autolysin genes are present in S. aureus. atl is the gene that encodes the major S. aureus autolysin that undergoes posttranslation processing to yield a 63-kDa amidase and a 53.5-kDa glucosaminidase (13, 36). Other autolysin genes include lytM, which encodes glycylglycine endopeptidase (41), and lytN, which possibly encodes muramidase activity (51). Boyle-Vavra et al. (4) have shown that atl was not mutated in several laboratory and clinical GISA strains that were examined. Assays of autolysin activity suggest decreased expression of autolysin(s) in GISA strains. Our Northern blot analysis suggests that the expression of atl is significantly reduced in GISA strain 13136p−m+V20. In contrast, Mongodin et al. (32) found that several cell wall-associated autolysin genes, including atl, were upregulated in two vancomycin-resistant strains selected by passage in the presence of vancomycin of two clinical GISA strains, one of which was Mu50, which has been reported to have increased autolytic activity. However, Kuroda et al. (28) and Utaida et al. (54) found that atl was downregulated in S. aureus exposed to cell wall-active antibiotics.
The regulation of autolysin activity and expression in S. aureus is complex. Two two-component regulatory systems, LytSR (18) and ArlRS (14), are negative regulators of autolysin activity. LytSR controls lrgA and lrgB, which are downstream from lytSR and negatively regulate autolysin activity. Ingavale et al. (25) have recently described a novel regulator of autolytic activity, known as Rat, that decreased the expression of lytSR, lrgAB, and arlRS and negatively regulated the expression of lytM and lytN but not atl. Other genes potentially involved in autolysis regulation include sarA (15), scdA (6), cidAB (42), and sspA, which encodes the major serine protease in S. aureus (43). Further work will be necessary to understand the mechanism of downregulation of autolysin in GISA strains.
Studies of various clinical and laboratory GISA have revealed alterations in cell walls and cell wall-associated properties of such strains, although the changes are not uniform among different strains and in some cases are even contradictory. Hanaki et al. (19) reported activated peptidoglycan synthesis in a GISA, whereas Sieradzki and Tomasz (49) reported a decreased rate of peptidoglycan biosynthesis. We report decreased wall teichoic acid levels, whereas Sieradzki and Tomasz (49) found increased teichoic acid levels in their GISA strains. However, from this paper and those of Boyle-Vavra et al. (4) and Sieradzki and Tomasz (49), evidence is accumulating that decreased autolysis is an important aspect of the GISA phenotype.
Although the exact role of autolysins in the killing of bacteria by cell wall-active antibiotics has been a subject of debate for more than 30 years (2, 30), it is clear that autolysin deficiency leads to tolerance to multiple cell wall-active antibiotics (52). The minimum bactericidal concentrations of vancomycin for the GISA strains used in this study ranged from 16 to 64 μg ml−1 compared to minimum bactericidal concentrations of 8 μg ml−1 for strains BB270 and COL (38). A significant decrease in autolytic activity early in the development of vancomycin resistance such as that found in our strains and in clinical strains studied by Boyle-Vavra et al. (4) and Sieradzki and Tomasz (49) may allow the cell to tolerate vancomycin that might otherwise cause cell killing in autolysis-sufficient bacteria. This might be an important step in the development of higher levels of vancomycin resistance.
Acknowledgments
This work was supported by grants AI 49964 and AI 43970 to B.J.W. and R.K.J.
REFERENCES
- 1.Avison, M. B., P. M. Bennett, R. A. Howe, and T. R. Walsh. 2002. Preliminary analysis of the genetic basis for vancomycin resistance in Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 49:255-260. [DOI] [PubMed] [Google Scholar]
- 2.Bayles, K. W. 2003. Are the molecular strategies that control apoptosis conserved in bacteria? Trends Microbiol. 11:306-311. [DOI] [PubMed] [Google Scholar]
- 3.Boyle-Vavra, S., R. B. Carey, and R. S. Daum. 2001. Development of vancomycin and lysostaphin resistance in a methicillin-resistant Staphylococcus aureus isolate. J. Antimicrob. Chemother. 48:617-625. [DOI] [PubMed] [Google Scholar]
- 4.Boyle-Vavra, S., M. Challapalli, and R. S. Daum. 2003. Resistance to autolysis in vancomycin-selected Staphylococcus aureus isolates precedes vancomycin-intermediate resistance. Antimicrob. Agents Chemother. 47:2036-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle-Vavra, S., H. Labischinski, C. C. Ebert, K. Ehlert, and R. S. Daum. 2001. A spectrum of changes in peptidoglycan composition of glycopeptide-intermediate clinical Staphylococcus aureus isolates. Antimicrob. Agents Chemother. 45:280-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunskill, E. W., B. L. de Jonge, and K. W. Bayles. 1997. Staphylococcus aureus scdA gene: a novel locus that affects cell division and morphogenesis. Microbiology 143:2877-2882. [DOI] [PubMed] [Google Scholar]
- 7.Chatterjee, A. N. 1969. Use of bacteriophage-resistant mutants to study the nature of the bacteriophage receptor site of Staphylococcus aureus. J. Bacteriol. 98:519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Climo, M. W., K. Ehlert, and G. L. Archer. 2001. Mechanism and suppression of lysostaphin resistance in oxacillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui, L., X. Ma, K. Sato, K. Okuma, F. C. Tenover, E. M. Mamizuka, C. G. Gemmell, M. N. Kim, M. C. Ploy, N. E. Solh, V. Ferraz, and K. Hiramatsu. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J. Clin. Microbiol. 41:5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui, L., H. Murakami, K. Kuwahara-Arai, H. Hanaki, and K. Hiramatsu. 2000. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expression by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 44:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daum, R. S., S. Gupta, R. Sabbath, and W. M. Milewski. 1992. Characterization of Staphylococcus aureus isolates with decreased susceptibility to vancomycin and teicoplanin: isolation and purification of a constitutively produced protein associated with decreased susceptibility. J. Infect. Dis. 166:1066-1072. [DOI] [PubMed] [Google Scholar]
- 12.Ehlert, K., M. Tschierske, C. Mori, W. Schröder, and B. Berger-Bächi. 2000. Site-specific serine incorporation by Lif and Epr into positions 3 and 5 of the staphylococcal interpeptide bridge. J. Bacteriol. 182:2635-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foster, S. J. 1995. Molecular characterization and functional analysis of the major autolysin of Staphylococcus aureus 8325-4. J. Bacteriol. 177:5723-5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto, D. F., and K. W. Bayles. 1998. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J. Bacteriol. 180:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafson, J., B. Berger-Bächi, A. Strassle, and B. J. Wilkinson. 1992. Autolysis of methicillin-resistant and -susceptible Staphylococcus aureus. Antimicrob. Agents Chemother. 36:566-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustafson, J. E., F. G. O'Brien, G. W. Coombs, M. J. Malkowski, W. B. Grubb, R. F. Pfeltz, and B. J. Wilkinson. 2003. Alterations in phage-typing patterns in vancomycin-intermediate Staphylococcus aureus. J. Med. Microbiol. 52:1-4. [DOI] [PubMed] [Google Scholar]
- 18.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanaki, H., K. Kuwahara-Arai, S. Boyle-Vavra, R. S. Daum, H. Labischinski, and K. Hiramatsu. 1998. Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. J. Antimicrob. Chemother. 42:199-209. [DOI] [PubMed] [Google Scholar]
- 20.Hanaki, H., H. Labischinski, Y. Inaba, N. Kondo, H. Murakami, and K. Hiramatsu. 1998. Increase in glutamine-non-amidated muropeptides in the peptidoglycan of vancomycin-resistant Staphylococcus aureus strain Mu50. J. Antimicrob. Chemother. 42:315-320. [DOI] [PubMed] [Google Scholar]
- 21.Hiramatsu, K. 1998. Vancomycin resistance in staphylococci. Drug Resist. Updates 1:135-150. [DOI] [PubMed] [Google Scholar]
- 22.Hiramatsu, K. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect. Dis. 1:147-155. [DOI] [PubMed] [Google Scholar]
- 23.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 24.Huff, E., C. S. Silverman, N. J. Adams, and W. S. Awkard. 1970. Extracellular cell wall lytic enzyme from Staphylococcus aureus: purification and partial characterization. J. Bacteriol. 103:761-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingavale, S. S., W. Van Wauel, and A. L. Cheung. 2003. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1456. [DOI] [PubMed] [Google Scholar]
- 26.Jenni, R., and B. Berger-Bächi. 1998. Teichoic acid content in different lineages of Staphylococcus aureus NCTC8325. Arch. Microbiol. 170:171-178. [DOI] [PubMed] [Google Scholar]
- 27.Komatsuzawa, H., K. Ohta, S. Yamada, K. Ehlert, H. Labischinski, J. Kajimura, T. Fujiwara, and M. Sugai. 2002. Increased glycan chain length distribution and decreased susceptibility to moenomycin in a vancomycin-resistant Staphylococcus aureus mutant. Antimicrob. Agents Chemother. 46:75-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 29.Leloir, L. T., and C. E. Gardini. 1957. Characterization of phosphorus compounds by acid lability. Methods Enzymol. 3:840-850. [Google Scholar]
- 30.Lewis, K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64:503-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mani, N., P. Tobin, and R. K. Jayaswal. 1993. Isolation and characterization of autolysis-defective mutants of Staphylococcus aureus created by Tn917-lacZ mutagenesis. J. Bacteriol. 185:1493-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreira, B., S. Boyle-Vavra, B. L. M. de Jonge, and R. S. Daum. 1997. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 41:1788-1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muthaiyan, A., R. K. Jayaswal, and B. J. Wilkinson. 2004. Intact mutS gene in laboratory-derived and clinical glycopeptide-intermediate Staphylococcus aureus strains. Antimicrob. Agents Chemother. 48:623-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Neill, A. J., and I. Chopra. 2002. Insertional inactivation of mutS in Staphylococcus aureus reveals potential for elevated mutation frequencies, although the prevalence of mutation in clinical isolates is low. J. Antimicrob. Chemother. 50:161-169. [DOI] [PubMed] [Google Scholar]
- 36.Oshida, T., M. Sugai, H. Komatsuzawa, Y.-M. Hong, H. Suginaka, and A. Tomasz. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-β-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. USA 92:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peschel, A., C. Vuong, M. Otto, and F. Götz. 2003. The d-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44:2845-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfeltz, R. F., V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal, and B. J. Wilkinson. 2000. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 44:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qoronfleh, M. W., J. E. Gustafson, and B. J. Wilkinson. 1998. Conditions that induce Staphylococcus aureus heat shock proteins also inhibit autolysis. FEMS Microbiol. Lett. 166:103-107. [DOI] [PubMed] [Google Scholar]
- 40.Qoronfleh, M. W., and B. J. Wilkinson. 1986. Effects of growth of methicillin-resistant and -susceptible Staphylococcus aureus in the presence of β-lactams on peptidoglycan structure and susceptibility to lytic enzymes. Antimicrob. Agents Chemother. 29:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramadurai, L., and R. K. Jayaswal. 1997. Molecular cloning, sequencing, and expression of lytM, a unique autolytic gene of Staphylococcus aureus. J. Bacteriol. 179:3625-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, K. C., B. A. Firek, J. B. Nelson, S. J. Jang, T. G. Patton, and K. W. Bayles. 2003. The Staphylococcus aureus cidAB operon: evaluation of its role in regulation of murein hydrolase activity and penicillin tolerance. J. Bacteriol. 185:2635-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of Staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roos, M., E. Pittenauer, E. Schmid, M. Beyer, B. Reinike, G. Allmaier, and H. Labischinski. 1998. Improved high-performance liquid chromatographic separation of peptidoglycan isolated from various Staphylococcus aureus strains for mass spectrometric characterization. J. Chromatogr. B 705:183-192. [DOI] [PubMed] [Google Scholar]
- 45.Schaaff, F., A. Reipert, and G. Bierbaum. 2002. An elevated mutation frequency favors development of vancomycin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 46:3540-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 47.Sieradzki, K., and A. Tomasz. 1997. Inhibition of cell wall turnover and autolysis by vancomycin in a highly vancomycin-resistant mutant of Staphylococcus aureus. J. Bacteriol. 179:2557-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sieradzki, K., and A. Tomasz. 1999. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J. Bacteriol. 181:7566-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sieradzki, K., and A. Tomasz. 2003. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J. Bacteriol. 185:7103-7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stranden, A. M., K. Ehlert, H. Labischinski, and B. Berger-Bächi. 1997. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 179:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugai, M., T. Fujiwara, H. Komatsuzawa, and H. Suginaka. 1998. Identification and molecular characterization of a gene homologous to epr (endopeptidase resistance gene) in Staphylococcus aureus. J. Bacteriol. 97:837-847. [DOI] [PubMed] [Google Scholar]
- 52.Tomasz, A., A. Albino, and E. Zanati. 1970. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227:138-140. [DOI] [PubMed] [Google Scholar]
- 53.Tschierske, M., K. Ehlert, A. M. Stranden, and B. Berger-Bächi. 1997. Lif, the lysostaphin immunity factor, complements FemB in staphylococcal peptidoglycan interpeptide bridge formation. FEMS Microbiol. Lett. 153:261-264. [DOI] [PubMed] [Google Scholar]
- 54.Utaida, S., P. M. Dunman, D. Macapagal, E. Murphy, S. J. Projan, V. K. Singh, R. K. Jayaswal, and B. J. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell wall stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]