Abstract
Yeast cells modulate their protein synthesis capacity in response to physiological needs through the transcriptional control of ribosomal protein (RP) genes. Here we demonstrate that the transcription factor Sfp1, previously shown to play a role in the control of cell size, regulates RP gene expression in response to nutrients and stress. Under optimal growth conditions, Sfp1 is localized to the nucleus, bound to the promoters of RP genes, and helps promote RP gene expression. In response to inhibition of target of rapamycin (TOR) signaling, stress, or changes in nutrient availability, Sfp1 is released from RP gene promoters and leaves the nucleus, and RP gene transcription is down-regulated. Additionally, cells lacking Sfp1 fail to appropriately modulate RP gene expression in response to environmental cues. We conclude that Sfp1 integrates information from nutrient- and stress-responsive signaling pathways to help control RP gene expression.
The size of cells, organs, and organisms is influenced by genetic and environmental factors that act to coordinate cell growth and cell division (1). Recent studies in yeast, Drosophila, and mammalian cells indicate that the evolutionarily conserved target of rapamycin (TOR) pathway plays an important role in modulating cell growth and cell size (2). For example, defects in the Drosophila TOR pathway result both in cells reduced in size at all stages of the cell cycle and in a smaller organism (3, 4). TOR signaling influences cell growth by regulating transcription, translation, and ribosome biogenesis, which collectively modulate cell mass by influencing protein synthesis (5).
Studies in budding yeast have provided important insights into the physiological role and regulation of the TOR pathway and its connection to cell size and growth control, revealing that TOR regulates cell growth in response to nutrient availability (5, 6). The TOR pathway is active in favorable nutrient conditions and promotes cell growth. When nutrients become limiting (or cells are treated with rapamycin), the TOR pathway is inactivated, leading to the inhibition of protein synthesis, changes in gene transcription, cell cycle arrest in G1, and up-regulation of ubiquitination and autophagy (5). The regulation of TOR by nutrient availability may be evolutionarily conserved, given that it has been reported that the mammalian TOR pathway is responsive to amino acid levels (7, 8).
A major output of the TOR pathway is the modulation of gene transcription (9), which has been well characterized in budding yeast by microarray analysis (10, 11). By an uncharacterized mechanism, TOR promotes the transcription of ribosomal protein (RP) and ribosome biogenesis genes in favorable nutrient conditions (12), thereby promoting cell growth. The TOR pathway also acts in favorable nutrient conditions to repress transcription of starvation-specific genes by sequestering several stress- and nutrient-responsive transcription factors (Msn2, Msn4, Rtg1, Rtg3, Gln3, and Gat1) in the cytoplasm (13, 14). Regulation of these factors is mediated by different branches of the TOR pathway. TOR inhibits nuclear accumulation of Gln3 and Gat1 by regulating binding of the inhibitor Tap42 to the PP2A and Sit4 phosphatases, which act on Gln3 and Gat1 (13). In contrast, the Rtg2 and Mks1 proteins mediate TOR regulation of Rtg1 and Rtg3 (15). Finally, TOR promotes cytosolic accumulation of the Msn2/4 proteins by regulating their interaction with a cytosolic anchor belonging to the 14-3-3 protein family (13). When TOR is inactivated by nutrient limitation or by treatment of cells with rapamycin, the transcription factors Gln3, Gat1, Rtg1, Rtg3, Msn2, and Msn4 translocate to the nucleus and activate transcription of target genes involved in nutrient utilization and the stress response (13, 14). Also in response to TOR inactivation, transcription of RP and ribosome biogenesis genes is down-regulated (12). The downstream effectors of TOR important for the modulation of ribosome production are not known, but the Tap42/PP2A and Rtg branches of the TOR pathway are not required (16). Additionally, the transcription factor target(s) for the TOR pathway relevant for the control of ribosome production has not been identified.
We have identified a transcription factor target of the TOR pathway important for the modulation of RP gene transcription: the zinc finger-containing transcription factor Sfp1. Sfp1 is an activator of RP gene expression that is localized to the nucleus in exponentially dividing cells. In response to inactivation of the TOR pathway, cell stress, or nutrient limitation, Sfp1 relocalizes to the cytoplasm, and RP genes are down-regulated. We have also found that Sfp1 localization is controlled by the nutrient-responsive cAMP/protein kinase A (PKA) pathway. Moreover, cells lacking Sfp1 are unable to properly regulate RP gene expression in response to environmental conditions. These observations suggest that Sfp1 plays an important role in modulating cell growth and RP gene expression in response to environmental cues.
Materials and Methods
Strains. Strains genotypes are provided in Table 1, which is published as supporting information on the PNAS web site.
GFP-Tagged Yeast Strains and Microscopic Imaging. Strains expressing GFP-tagged proteins are described in ref. 17. Each strain produces a single C-terminal GFP-tagged protein expressed from its chromosomal locus under the control of its native promoter. Strains expressing known or predicted site-specific DNA-binding proteins (GFP-tagged) (Table 2, which is published as supporting information on the PNAS web site) were screened for changes in subcellular localization upon treatment with 100 nM rapamycin by fluorescence microscopy. Aliquots of strains grown to mid-logarithmic phase in synthetic dextrose medium were analyzed in 96-well glass-bottom microscope slides (BD Falcon, BD Biosciences) pretreated with Con A (50 μg·ml-1) to ensure cell adhesion. Cells were analyzed by fluorescence microscopy with a digital imaging-capable Nikon TE200/300 inverted microscope and a Roper Q57 charge-coupled device camera with an oil-immersed objective at ×100 magnification. To monitor changes in subcellular localization of the tagged proteins upon rapamycin treatment, a specific field in each well was selected and its absolute position saved. By using a script in metamorph Version 4.6r8 imaging software (Universal Imaging, Downingtown, PA), fluorescence microscopy images of the selected fields were taken as a function of time after rapamycin treatment, and the stage was automatically advanced between wells on the 96-well slide, generating a picture of the same live cells every 4-5 min.
cDNA Microarrays. Congenic wild-type (BY4741, MATa) and sfp1Δ strains were grown to early logarithmic phase (OD600 ≈ 0.3) in yeast extract/peptone/dextrose medium at 30°C. Cells were treated with rapamycin (100 nM) or the drug carrier (90% ethanol/10% Tween 20) for 30 min or 60 min, harvested, and flash-frozen in liquid nitrogen. Microarray analysis was performed as described in ref. 18. Total and poly(A) RNA was isolated, and poly(A) RNA was reverse-transcribed by incorporating amino-allyl dUTP (Sigma). The resulting cDNAs were labeled by using monofunctional reactive Cy3 and Cy5 dyes (Amersham Pharmacia) in the presence of sodium bicarbonate. The standard deviation from the mean expression ratio of 1 was calculated by using all of the individual gene expression measurements in a given experiment. This value provides an indication of the number and magnitude of changes occurring in that experiment. Data presented are the average of three independent microarray experiments. (The complete data set used to create Fig. 4A can be downloaded as Table 3, which is published as supporting information on the PNAS web site.)
Fig. 4.
Sfp1 is a direct regulator of RP genes and is required for appropriate down-regulation of RP gene expression in response to stress. (A) Microarray analysis comparing expression of RP genes in the following lanes: a, wild-type strain treated with rapamycin (Cy5 red) versus an untreated wild-type strain (Cy3 green) (1 h after addition of 100 nM rapamycin); b, sfp1Δ strain treated with rapamycin (Cy5 red) versus untreated sfp1Δ (Cy3 green) (1 h); c, sfp1Δ (Cy5 red) versus wild type (Cy3 green); d, sfp1Δ treated with rapamycin (Cy5 red) versus wild type treated with rapamycin (Cy3 green) (1 h). Data presented are the average of three independent microarray experiments. When examining the behavior of genes from various functional groups (47, 48) in the microarray experiments, RPs were the most significantly regulated functional group in lanes a, b, and c with P < 7.19 × 10-106, 1.13 × 10-25, and 5.9 × 10-104, respectively. (B) Quantitative RT-PCR validation of microarray data. Expression levels of several RP mRNAs relative to ACT1 transcript levels were measured in wild-type and sfp1Δ strains, untreated or treated with 100 nM rapamycin (rap) for 1 h. (Upper) The fold repression of RP genes in response to rapamycin in both wild-type and sfp1Δ strains. (Lower) The comparison of RP gene expression in the sfp1Δ strain versus wild type, either under optimal growth conditions or after treatment with rapamycin. Values are the averages of three independent experiments; error bars show SEM. (C) Yeast cells lacking Sfp1 are defective in RP regulation in response to oxidative stress. Expression levels of several RP mRNAs relative to the ACT1 transcript were measured by quantitative RT-PCR in wild-type and sfp1Δ strains untreated or treated with 0.4 mM H2O2 for 60 min. (Upper) The fold repression of RP genes in response to H2O2 in both wild-type and sfp1Δ strains. (Lower) The comparison of RP gene expression in the sfp1Δ strain versus wild type, either under optimal growth conditions or after treatment with H2O2. Values are the averages of three independent experiments; error bars show SEM. (D) Chromatin immunoprecipitation analysis of Sfp1-HA3 in cells treated or untreated with 100 nM rapamycin. Sfp1 binding for the indicated promoter DNA relative to ACT1 DNA is represented by (immunoprecipitated DNA/input DNA)/(ACT1 immunoprecipitated DNA/ACT1 input DNA). Values are the averages of three independent experiments; error bars show standard deviations.
Chromatin Immunoprecipitation. The chromatin immunoprecipitation protocol was similar to that described in ref. 19. Cells expressing Sfp1-HA3 were grown in yeast extract/peptone/dextrose medium at 30°C, treated or not treated with rapamycin (100 nM) for 30 min or 60 min, fixed with 1% formaldehyde for 15 min at room temperature, and harvested. To obtain DNA, the cell pellet was processed and immunoprecipitated with a monoclonal anti-hemagglutinin antibody (Roche Applied Science, clone no. 12CA5), and copurifying DNA was purified by phenol-chloroform and chloroform extraction. The DNA was then quantified by using an Opticon real-time PCR machine (MJ Research, Waltham, MA) as described in ref. 20. At least three independent extracts were analyzed for each strain and growth condition. We observed binding of Sfp1 to the promoters of the following genes: RPL1B, RPS31, RPL23B, RPS9A, RPL27B, RPS0B, RPL12A, RPL13A, RPL11A, RPL8B, RPS26A, RPL2B, RPS22B, RPS8A, RPL5, RPS5, RPP0, RPL34B, and RPL4B.
Quantitative RT-PCR. Congenic wild-type (BY4741, MATa) and sfp1Δ strains were grown to early logarithmic phase (OD600 ≈ 0.3) in yeast extract/peptone/dextrose medium at 30°C. Cells were treated with rapamycin (100 nM) or the drug carrier (90% ethanol/10% Tween 20) for 60 min or untreated or treated with 0.4 mM H2O2 for 60 min, harvested, and flash-frozen in liquid nitrogen. Total RNA was isolated as described above, and mRNA was reverse-transcribed by using gene-specific primers. As a control for genomic DNA contamination, a fraction of each sample was processed without any oligo present. The resulting cDNA was then quantified by using an Opticon real-time PCR machine (MJ Research) as described in ref. 20. In each sample, levels of mRNA expression were normalized to the expression of the ACT1 transcript. Three independent extracts were analyzed for each strain and growth condition, and the mean and SEM were calculated.
Results
The TOR Pathway Controls Sfp1 Subcellular Localization. To explore how the TOR signaling pathway modulates changes in gene regulation, we carried out a screen to identify new transcription factors involved in the regulation of the transcriptional response to rapamycin treatment. Because the activity of many transcription factors in the TOR pathway is controlled through regulation of their subcellular localization (13, 14), we assayed changes in transcription factor localization as an indicator of changes in transcription factor activity. We used a collection of ≈250 strains (Table 2), each expressing a single characterized or predicted site-specific DNA-binding protein (Incyte, Wilmington, DE) (21) tagged with GFP at its C terminus and expressed from its native chromosomal locus under the control of its native promoter (17). These strains were treated with rapamycin, and fluorescence micrographs of live cells were captured as a function of time. To facilitate the detection of changes in localization, each time-course was recorded by using a single field of cells.
As expected, this screen identified the transcription factors previously known to act downstream of TOR: Msn2/4, Gln3, Gat1, Rtg1, and Rtg3. These factors are all localized to the cytoplasm in untreated cells and relocalize to the nucleus in the presence of rapamycin (data not shown). The screen also identified a transcription factor whose regulation had not been previously studied, Sfp1. Surprisingly, Sfp1 localization is regulated in a manner opposite to that of the known TOR downstream transcriptional effectors in that it localizes mainly to the nucleus in dividing cells grown in standard laboratory conditions and relocalizes to the cytoplasm when cells are treated with rapamycin (Fig. 1A). The decrease in Sfp1 nuclear concentration is through regulation of nucleocytoplasmic trafficking, because Sfp1 protein levels are not significantly reduced during the time it is relocalized (data not shown). The effect of rapamycin on Sfp1 localization is mediated through the TOR pathway because in a strain containing a rapamycin-resistant mutation in Tor1 Sfp1 is localized to the nucleus and is not relocalized to the cytoplasm when cells are treated with rapamycin (Fig. 1B).
Fig. 1.
The TOR pathway controls Sfp1 subcellular localization. (A) Time course of Sfp1-GFP localization after TOR pathway inactivation. Cells were grown in synthetic dextrose medium, treated with 100 nM rapamycin, and analyzed by fluorescence microscopy. Images of the same live cells were captured every 4-5 min. (B) Rapamycin induces Sfp1 relocalization to the cytoplasm through inhibition of TOR signaling. Sfp1-GFP localization in rapamycin-resistant (TOR1-1) and wild-type strains untreated or treated with rapamycin (45 min after addition of 100 nM rapamycin).
Bioinformatics Analysis and the Identification of Candidate Target Genes for Sfp1. Sfp1 is a zinc-finger transcription factor that was recently shown to play a role in the control of cell size in budding yeast (22, 23). Cells lacking SFP1 are small and have defects in the transcription of genes involved in ribosome biogenesis and those that encode RPs (23). Because cells containing mutations in genes involved in ribosome biogenesis are also small, it was previously suggested (23) that the reduced cell size of sfp1Δ mutants may be the result of a defect in expression of ribosome biogenesis genes. However, further studies have been unable to demonstrate the binding of Sfp1 to the promoters of ribosome biogenesis genes (24). Large-scale studies indicated that Sfp1 may bind the promoters of a large variety of genes, including some of the RP genes. Thus, it remains unclear what genes Sfp1 directly regulates.
To formulate a hypothesis regarding the direct targets of Sfp1, we reasoned that the expression of bona fide targets of Sfp1 should be coordinately controlled. We therefore devised a bioinformatics approach to examine which subset of the genes potentially bound by Sfp1 are also consistently coregulated in relevant conditions and could therefore be its key targets. To perform this analysis we combined information from two genome-wide datasets. A large-scale study of yeast transcription factor binding in the genome, assayed by using chromatin immunoprecipitation followed by detection on a microarray containing intergenic sequences (25), provided a set of target genes that are potentially directly regulated by each of those transcription factors. A compendium of 1,006 previously published microarray experiments (26, 27) provided gene expression data for a variety of potentially relevant biological conditions.
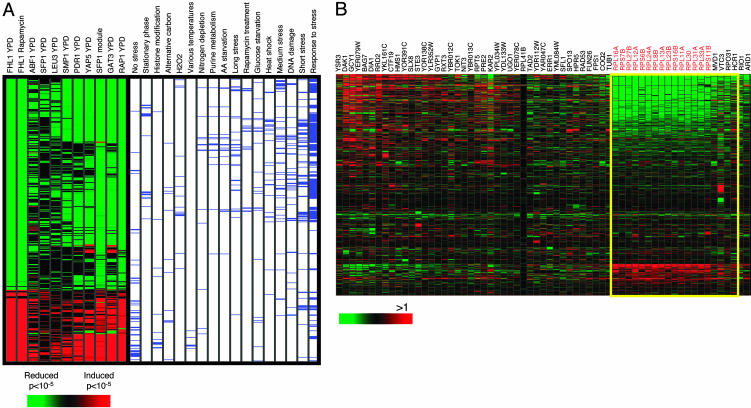
We applied a two-step method (described in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site) to identify target genes of Sfp1. In the first step, we associated each transcription factor with a set of microarrays and biological conditions in which a significant subset of its targets is coordinately regulated. We used a statistical Kolmogorov-Smirnov test to determine whether a subset of the candidate target genes of a given transcription factor is differentially regulated (e.g., up-regulated or down-regulated) in a coordinated way in a given microarray experiment. We also annotated each microarray experiment with a biological description (e.g., heat shock or cell cycle), and then used a hypergeometric test to identify biological conditions that are significantly enriched in those microarrays for which we detected significant coordination in the expression of the candidate target genes of transcription factors. By using this approach we found that a significant fraction of the genes bound by Sfp1 in the large-scale chromatin immunoprecipitation experiment were down-regulated in microarray experiments in which cells were treated with heat shock or oxidative stress, grown to stationary phase, or subjected to nutrient deprivation (Fig. 2A). Interestingly, clustering transcription factors based on the patterns of their target regulation revealed that candidate target genes of known transcriptional regulators of RP gene expression, Abf1 and Rap1, as well as a putative regulator, Fhl1, exhibited similar behavior.
Fig. 2.
Bioinformatics analysis reveals candidate Sfp1 target genes. (A) A significant fraction of potential Sfp1 target genes are coordinately repressed in stress and nutrient deprivation. (Left) A transcription factor-microarray matrix is shown, where red and green elements indicate a significant test for induction and repression of the candidate transcription factor target genes in a given microarray, respectively. The intensity of each entry corresponds to the -log10(P value) of the statistical test (black elements indicate a insignificant result). The matrix is the subcluster of the full analysis on 106 transcription factors and indicates the similar behavior of candidate target gene sets for Sfp1, Rap1, Abf1, Fhl1, and several additional factors. The matrix is filtered to show only microarrays with at least one significant test for the targets of those factors. (Right) The biological conditions associated with each microarray are shown as blue elements. Only annotations that were significant for candidate target gene sets of at least one factor in the Sfp1 cluster (P < 0.01, after a false discovery rate statistical correction for multiple hypotheses) are shown. Significant annotations for Sfp1 include response to stress (P < 1.24 × 10-8), amino acid (AA) starvation (P < 2.14 × 10-6), nitrogen depletion (P < 8.13 × 10-5), stationary phase (P < 4.17 × 10-8), and response to glucose starvation (P < 2.4 × 10-4). The targets of Sfp1 were also significantly repressed in many additional individual experiments, including treatment with hydrogen peroxide, DTT, MMS, and rapamycin. (B) RP genes underlie the coordinated expression pattern of Sfp1 targets. The matrix shows the expression pattern of all 63 potential Sfp1 target genes, highlighting (yellow) those targets identified by our methods to significantly (P < 10-25) underlie the coordinated changes identified in A. Among the 18 detected bona fide target genes, 14 encode RPs (red highlight). When iterating the analysis in A with this refined set of targets, the resulting profile is even more prominent (Sfp1 module column in A).
In the second step, we derived the bona fide set of targets for the transcription factor by identifying genes that underlie the coherent expression behavior. To this end, we performed a third statistical test that identified the subset of candidate target genes whose expression changes are consistent with the overall pattern of significant changes discovered in the first step. Indeed, the candidate Sfp1 target genes that underlie its associated coherent expression behavior consist almost exclusively of RP genes (P < 1.6 × 10-11 for enrichment of RP genes) (Fig. 2B). These observations suggest that Sfp1 may be a direct regulator of RP gene expression.
Sfp1 Links Physiological Changes to Regulation of RP Gene Expression. Both rapamycin treatment and stress conditions cause repression of RP genes (10, 28, 29), raising the possibility that Sfp1 may be regulated in response to stress and nutrient availability and link physiological changes to the regulation of RP genes. If this hypothesis is correct, we expect that Sfp1 localization will also change in response to stress and growth conditions known to cause down-regulation in RP gene expression, including (i) treatment with osmotic stress, DTT, hydrogen peroxide, or methyl methanesulfonate (MMS) (28, 29); (ii) a block in the secretory pathway caused by tunicamycin addition (30); (iii) entry into stationary phase (29, 31); and (iv) glucose starvation (32, 33). Indeed, treatment of cells with osmotic stress, DTT, hydrogen peroxide, MMS, tunicamycin, or entry into stationary phase all cause Sfp1 to be relocalized to the cytoplasm (Fig. 3). Furthermore, Sfp1 is relocalized to the cytoplasm in response to glucose starvation (Fig. 3B) and rapidly reenters the nucleus when glucose is added back to the medium, a condition known to induce rapid induction of RP gene transcription (32, 33). This rapid nuclear accumulation of Sfp1 further confirms that subcellular localization is regulated. In each of these stresses or nutrient changes, the timescale of Sfp1 relocalization matches the kinetics of changes in RP gene expression (12, 29-33). Therefore, Sfp1 is localized to the nucleus under conditions in which RP genes are actively transcribed and is redistributed to the cytoplasm in response to signals that cause down-regulation of RP gene transcription.
Fig. 3.
Cytosolic localization of Sfp1 correlates with conditions of RP gene down-regulation. (A) Sfp1-GFP localization under different stress conditions: 0.4 mM H2O2 (30 min after addition); 0.5 M NaCl (15 min after addition); 2 mM DTT (60 min after addition); 0.1% MMS (45 min after addition); 2.5 μg/ml tunicamycin (3.5 h after treatment). Images of the same field of cells were captured every 5 min after addition of the stress reagent; only one representative time point is shown. (B) Sfp1-GFP localization under different nutrient availability conditions: cells exponentially growing; cells grown to stationary phase; cells transferred to medium containing ethanol as a carbon source (10 min after transfer); and cells 10 min after refeeding of glucose (to cells in no glucose).
Yeast Cells Lacking Sfp1 Do Not Properly Regulate RP Gene Expression in Stress Conditions. If Sfp1 is a regulator of RP gene transcription, we expect that sfp1 mutant cells will be defective in RP gene transcription and will have defects in the regulation of RP genes that occurs in response to stress. Indeed, as has been shown previously, we find that cells lacking SFP1 have reduced levels of RP gene transcription under normal growth conditions (23), suggesting that Sfp1 is an activator of RP gene expression (Fig. 4 A, lane c; B Lower; and C Lower). To test whether sfp1 mutant cells show defects in regulation of RP gene transcription in response to stress, we treated wild-type and sfp1Δ cells with rapamycin and analyzed gene expression by whole-genome microarray analysis and by reverse transcription followed by quantitative PCR. Rapamycin treatment of wild-type cells causes dramatic down-regulation of RP gene transcription (10-12) (Fig. 4 A, lane a, and B, WT/WT+rap). In contrast, we find that cells lacking SFP1 do not appropriately down-regulate RP gene transcription when treated with rapamycin (Fig. 4 A, lanes b and d, and B, Δsfp1/Δsfp1+rap and Δsfp1+rap/WT+rap). Rapamycin still triggers some reduction of RP gene expression in the sfp1 mutant strain (Fig. 4 A, lane b, and B, Δsfp1/Δsfp1+rap), so it is likely that there is an additional mechanism for down-regulation of these genes, but deletion of SFP1 clearly abrogates wild-type patterns of RP gene regulation. Similar results were obtained with wild-type and sfp1Δ cells treated with 0.4 mM H2O2 (Fig. 4C), indicating that sfp1Δ cells are defective in RP regulation in response to stresses other than rapamycin. These results indicate that Sfp1 is required for proper regulation of RP gene expression in stress conditions.
Regulation of Sfp1 Binding to RP Gene Promoters. To test whether Sfp1 is a direct regulator of RP gene transcription, we used chromatin immunoprecipitation experiments to determine whether Sfp1 binds to the promoters of RP genes. We observe binding of Sfp1 to the promoters of 19 of 23 RP genes tested (e.g., Fig. 4D; see Materials and Methods). In contrast, we were unable to detect binding of Sfp1 to the promoters of genes involved in ribosome biogenesis or to the SSB2, YEF3, and IMD4 genes whose expression is highly dependent on Sfp1 (Fig. 4D). Furthermore, binding of Sfp1 to RP gene promoters is regulated: Sfp1 binding is disrupted in response to rapamycin treatment. It is not known whether binding of Sfp1 to RP gene promoters occurs through direct interaction with DNA or through interaction with other proteins. We conclude that Sfp1 directly regulates RP gene transcription and that binding of Sfp1 to RP promoters is nutrient- and stress-sensitive.
Role of the TOR and PKA Pathways in Regulating Sfp1 Localization. To further investigate how the TOR signaling pathway controls Sfp1 activity, we examined Sfp1 localization in strains with mutations in downstream effectors of the TOR pathway. Recent studies demonstrate that down-regulation of RP gene expression in response to rapamycin treatment does not require the TOR effectors Sit4 or Tap42 (16). Accordingly, mutation of SIT4 or PP2A-1 does not affect rapamycin-induced relocalization of Sfp1 to the cytoplasm (data not shown). Similarly, mutation of RTG2 or MKS1 had no effect on Sfp1 relocalization upon rapamycin treatment (data not shown). These observations suggest that downstream effectors of TOR other than the well characterized Sit4/Tap42 and Rtg1/Rtg2/Rtg3 branches mediate the regulation of RP gene expression and Sfp1 localization.
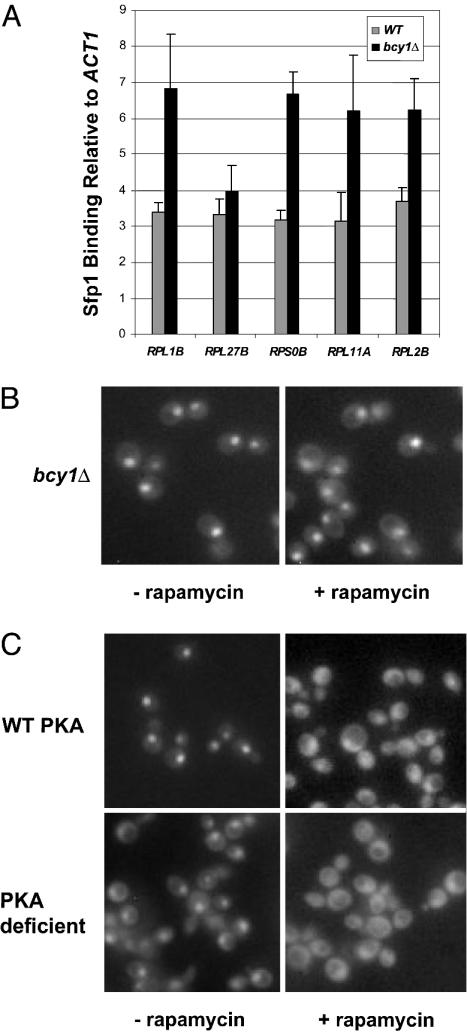
The cAMP/PKA pathway has also been shown to influence RP gene transcription in response to nutrient and carbon availability (34-36). cAMP causes activation of PKA by binding to the regulatory subunit Bcy1, triggering release and activation of the PKA catalytic subunits (37). Constitutively high PKA activity (in a bcy1Δ strain) results in an ≈2-fold increase in transcription of RP genes (34). Accordingly, we find that ≈2-fold more Sfp1 is bound to the promoters of RP genes in the bcy1Δ strain than in the wild-type strain (Fig. 5A).
Fig. 5.
The TOR and PKA pathways control subcellular localization of Sfp1. (A) Chromatin immunoprecipitation analysis of Sfp1-HA3 in a strain containing constitutively active PKA (bcy1Δ) and in a wild-type strain. Sfp1 binding for the indicated promoter DNA relative to ACT1 DNA is represented by (immunoprecipitated DNA/input DNA)/(ACT1 immunoprecipitated DNA/ACT1 input DNA). Values are the averages of three independent experiments; error bars show standard deviations. (B) Sfp1-GFP localization in a bcy1Δ strain untreated or treated with rapamycin (45 min after addition of 100 nM rapamycin). (C) Sfp1-GFP localization in PKA-deficient (tpk1Δ tpk2Δ tpk3Δ msn2Δ msn4Δ) and PKA wild-type (msn2Δ msn4Δ) strains untreated or treated with rapamycin (45 min after addition of 100 nM rapamycin).
Several studies suggest that there may be interactions between the TOR and PKA pathways (13, 38). Moreover, it has been proposed that TOR functions through PKA to modulate RP gene transcription (39). Constitutively high PKA activity (e.g., bcy1 strain) inhibits rapamycin-induced down-regulation of RP gene transcription (39). Accordingly, we see that Sfp1 remains localized to the nucleus in a bcy1 strain treated with rapamycin (Fig. 5B). These observations are consistent with a model in which Sfp1 is regulated by the branch of the TOR pathway containing PKA. However, Sfp1 is still partially localized to the nucleus in a strain lacking PKA activity (Fig. 5C) and moves to the cytoplasm when cells are treated with rapamycin, indicating that TOR can maintain Sfp1 in the nucleus independently of PKA.
Although the PKA catalytic subunits, encoded by TPK1, TPK2, and TPK3, are redundant in most cases, they sometimes show different substrate specificities and functions (40-42). To determine whether a specific PKA subunit controls Sfp1 nuclear localization, we generated strains containing a single PKA catalytic subunit and lacking the regulatory subunit Bcy1. In all three mutant strains, constitutive activity of the single catalytic subunit reduced Sfp1 relocalization in response to rapamycin (Fig. 7, which is published as supporting information on the PNAS web site), indicating that the catalytic subunits of PKA act redundantly to promote nuclear localization of Sfp1.
It has been proposed that the PKA pathway is an integral part of the stress response (43). We therefore tested whether the PKA pathway is involved in the regulation of Sfp1 localization in response to environmental insults. We find that a strain lacking PKA activity is still able to relocalize Sfp1 in response to osmotic and oxidative stress (Fig. 8, which is published as supporting information on the PNAS web site), indicating that these stress stimuli can be transmitted to Sfp1 independently of PKA.
Discussion
Cells have evolved mechanisms to coordinately regulate the synthesis of RPs in response to nutrient availability and other changes in environmental conditions that alter the demand for protein biosynthetic capacity (36). This regulation is extremely important for the economy of the cell, because the synthesis of RPs is a major consumer of the resources of the cell. In yeast, this regulation is effected largely at the level of transcription, where expression of genes encoding RPs accounts for ≈50% of total RNA polymerase II transcription (36). Genome-wide expression analyses have shown that coordinate repression of RP genes is a common response to all environmental stresses tested (28, 29) and to nutrient starvation and entry into stationary phase (10, 28, 29, 31-33). Interestingly, however, the kinetics and amplitude of the down-regulation are different in each stress condition. Remarkably, for each condition we find that the kinetics of Sfp1 relocalization to the cytoplasm matches the kinetics of RP repression. We propose that one mechanism by which nutrient starvation and environmental stresses modulate transcription of RP genes is by regulating Sfp1 activity. In support of this model, our data demonstrate that cells lacking Sfp1 do not properly regulate RP mRNA levels in response to environmental conditions. We speculate that cells lacking Sfp1 may not appropriately communicate the signal for down-regulation to the RP gene promoters. For example, it may be that Sfp1 is required to establish/maintain chromatin in a state that permits down-regulation of the RP genes. Alternatively, if Sfp1 interacts with other transcription factors bound to the promoters of RP genes, it may be required for these factors to receive the down-regulation signal. Our observation that significant regulation of RP gene expression occurs in the absence of Sfp1 in response to rapamycin treatment supports the idea that other transcription factors are likely to be targets of the TOR pathway. Although the transcriptional regulators Abf1, Rap1, and Fhl1 have been implicated in RP gene expression (25, 36), there is no evidence that they are targets of the TOR pathway.
Sfp1 senses and responds to all environmental insults we tested. To do so, Sfp1 has to integrate information from the major pathways that regulate RP gene expression in response to nutrients and external stresses: the PKA and TOR pathways (12, 34, 35). We show that the TOR and PKA pathways contribute to maintaining Sfp1 in the nucleus, thus keeping RP gene expression turned on in favorable nutrient conditions. It has been proposed that TOR functions through PKA to modulate RP gene transcription (39). Although our data are consistent with this model, we also demonstrate that there is a branch of the TOR pathway that controls localization of Sfp1 independently of PKA. We envision that the TOR and PKA pathways function redundantly to maintain Sfp1 in the nucleus (Fig. 6). It is not clear how other stimuli regulate Sfp1 localization, but osmotic and oxidative stress signals do not appear to require PKA.
Fig. 6.
Model for regulation of Sfp1 by the PKA and TOR pathways (see text for details).
Several studies indicate that TOR and PKA act together to control the subcellular localization of a number of proteins involved in modulating cell growth in response to changes in nutrients and stress. In addition to promoting nuclear accumulation of Sfp1, the TOR and PKA pathways promote cytoplasmic accumulation (and thus inhibition) of the kinase Rim15, a positive regulator of entry into stationary phase (44), and the transcription factors Msn2/4 (13, 45, 46). Thus, when conditions are favorable and nutrients are available, the TOR and PKA pathways inhibit transcription of stress and stationary phase programs and induce RP gene expression, promoting cell growth. Under unfavorable growth conditions, inactivation of TOR and PKA will induce cytosolic localization of Sfp1 and nuclear accumulation and activation of Rim15 and Msn2/4, causing down-regulation of RP genes, induction of stress genes, and entry into stationary phase. The regulation of protein activity through control of subcellular localization is particularly well suited for responding to changing environmental conditions, because it is rapid, reversible, and does not require new protein synthesis.
The TOR pathway in both mammalian cells and Drosophila has an important role in controlling cell size through effects on cell growth, in part through regulation of translation (1, 2). Inactivation of TOR or key downstream effectors in these systems results in small cell size. The observation that yeast cells lacking Sfp1 are unusually small (23), coupled with our finding that Sfp1 is a target of the TOR pathway, suggests that the TOR pathway in yeast also has a role in modulating cell size. Although the downstream effectors of TOR relevant for cell-size control differ in Drosophila, mammalian, and yeast cells, they are all involved in the regulation of protein synthesis, reinforcing previous studies that suggest cell size is modulated in part through the control of translational capacity.
Supplementary Material
Acknowledgments
We thank Jose Luis Riechmann (California Institute of Technology, Pasadena) for the list of candidate yeast transcription factors; Gustav Ammerer (University of Vienna, Vienna), Stephen Garrett (University of Medicine and Dentistry of New Jersey, Newark), and Vladimir Denic (University of California, San Francisco) for yeast strains; Mike Springer, Jonathan Raser, and Andrew Capaldi for help with experimental design and analysis; and Ted Powers, Vladimir Denic, and members of the O'Shea laboratory for comments on the manuscript. This work was supported by National Institutes of Health Grant GM59034 (to E.K.O.) and by the Howard Hughes Medical Institute and the David and Lucile Packard Foundation.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: TOR, target of rapamycin; RP, ribosomal protein; PKA, protein kinase A; MMS, methyl methanesulfonate.
See accompanying Biography on page 14312.
References
- 1.Saucedo, L. J. & Edgar, B. A. (2002) Curr. Opin. Genet. Dev. 12, 565-571. [DOI] [PubMed] [Google Scholar]
- 2.Jacinto, E. & Hall, M. N. (2003) Nat. Rev. Mol. Cell. Biol. 4, 117-126. [DOI] [PubMed] [Google Scholar]
- 3.Zhang, H., Stallock, J. P., Ng, J. C., Reinhard, C. & Neufeld, T. P. (2000) Genes Dev. 14, 2712-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oldham, S., Montagne, J., Radimerski, T., Thomas, G. & Hafen, E. (2000) Genes Dev. 14, 2689-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crespo, J. L. & Hall, M. N. (2002) Microbiol. Mol. Biol. Rev. 66, 579-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rohde, J., Heitman, J. & Cardenas, M. E. (2001) J. Biol. Chem. 276, 9583-9586. [DOI] [PubMed] [Google Scholar]
- 7.Hara, K., Yonezawa, K., Weng, Q. P., Kozlowski, M. T., Belham, C. & Avruch, J. (1998) J. Biol. Chem. 273, 14484-14494. [DOI] [PubMed] [Google Scholar]
- 8.Kim, D. H., Sarbassov, D. D., Ali, S. M., King, J. E., Latek, R. R., Erdjument-Bromage, H., Tempst, P. & Sabatini, D. M. (2002) Cell 110, 163-175. [DOI] [PubMed] [Google Scholar]
- 9.Powers, T., Dilova, I., Chen, C. Y. & Wedaman, K. (2004) Curr. Top Microbiol. Immunol. 279, 39-51. [DOI] [PubMed] [Google Scholar]
- 10.Hardwick, J. S., Kuruvilla, F. G., Tong, J. K., Shamji, A. F. & Schreiber, S. L. (1999) Proc. Natl. Acad. Sci. USA 96, 14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cardenas, M. E., Cutler, N. S., Lorenz, M. C., Di Como, C. J. & Heitman, J. (1999) Genes Dev. 13, 3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powers, T. & Walter, P. (1999) Mol. Biol. Cell 10, 987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck, T. & Hall, M. N. (1999) Nature 402, 689-692. [DOI] [PubMed] [Google Scholar]
- 14.Komeili, A., Wedaman, K. P., O'Shea, E. K. & Powers, T. (2000) J. Cell Biol. 151, 863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dilova, I., Chen, C. Y. & Powers, T. (2002) Curr. Biol. 12, 389-395. [DOI] [PubMed] [Google Scholar]
- 16.Duvel, K., Santhanam, A., Garrett, S., Schneper, L. & Broach, J. R. (2003) Mol. Cell 11, 1467-1478. [DOI] [PubMed] [Google Scholar]
- 17.Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S. & O'Shea, E. K. (2003) Nature 425, 686-691. [DOI] [PubMed] [Google Scholar]
- 18.Carroll, A. S., Bishop, A. C., DeRisi, J. L., Shokat, K. M. & O'Shea, E. K. (2001) Proc. Natl. Acad. Sci. USA 98, 12578-12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strahl-Bolsinger, S., Hecht, A., Luo, K. & Grunstein, M. (1997) Genes Dev. 11, 83-93. [DOI] [PubMed] [Google Scholar]
- 20.Steger, D. J., Haswell, E. S., Miller, A. L., Wente, S. R. & O'Shea, E. K. (2003) Science 299, 114-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., Adam, L., Pineda, O., Ratcliffe, O. J., Samaha, R. R., et al. (2000) Science 290, 2105-2110. [DOI] [PubMed] [Google Scholar]
- 22.Blumberg, H. & Silver, P. (1991) Gene 107, 101-110. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen, P., Nishikawa, J. L., Breitkreutz, B. J. & Tyers, M. (2002) Science 297, 395-400. [DOI] [PubMed] [Google Scholar]
- 24.Fingerman, I., Nagaraj, V., Norris, D. & Vershon, A. K. (2003) Eukaryotic Cell 2, 1061-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, T. I., Rinaldi, N. J., Robert, F., Odom, D. T., Bar-Joseph, Z., Gerber, G. K., Hannett, N. M., Harbison, C. T., Thompson, C. M., Simon, I., et al. (2002) Science 298, 799-804. [DOI] [PubMed] [Google Scholar]
- 26.Bar-Joseph, Z., Gerber, G. K., Lee, T. I., Rinaldi, N. J., Yoo, J. Y., Robert, F., Gordon, D. B., Fraenkel, E., Jaakkola, T. S., Young, R. A. & Gifford, D. K. (2003) Nat. Biotechnol. 21, 1337-1342. [DOI] [PubMed] [Google Scholar]
- 27.Ihmels, J., Levy, R. & Barkai, N. (2004) Nat. Biotechnol. 22, 86-92. [DOI] [PubMed] [Google Scholar]
- 28.Causton, H. C., Ren, B., Koh, S. S., Harbison, C. T., Kanin, E., Jennings, E. G., Lee, T. I., True, H. L., Lander, E. S. & Young, R. A. (2001) Mol. Biol. Cell 12, 323-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., Botstein, D. & Brown, P. O. (2000) Mol. Biol. Cell 11, 4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuta, K. & Warner, J. R. (1994) Mol. Cell. Biol. 14, 2493-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ju, Q. & Warner, J. R. (1994) Yeast 10, 151-157. [DOI] [PubMed] [Google Scholar]
- 32.Kief, D. R. & Warner, J. R. (1981) Mol. Cell. Biol. 1, 1007-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herruer, M. H., Mager, W. H., Woudt, L. P., Nieuwint, R. T., Wassenaar, G. M., Groeneveld, P. & Planta, R. J. (1987) Nucleic Acids Res. 15, 10133-10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein, C. & Struhl, K. (1994) Mol. Cell. Biol. 14, 1920-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuman-Silberberg, F. S., Bhattacharya, S. & Broach, J. R. (1995) Mol. Cell. Biol. 15, 3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Warner, J. R. (1999) Trends Biochem. Sci. 24, 437-440. [DOI] [PubMed] [Google Scholar]
- 37.Broach, J. R. (1991) Curr. Opin. Genet. Dev. 1, 370-377. [DOI] [PubMed] [Google Scholar]
- 38.Cutler, N. S., Pan, X., Heitman, J. & Cardenas, M. E. (2001) Mol. Biol. Cell 12, 4103-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmelzle, T., Beck, T., Martin, D. E. & Hall, M. N. (2004) Mol. Cell. Biol. 24, 338-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robertson, L. S. & Fink, G. R. (1998) Proc. Natl. Acad. Sci. USA 95, 13783-13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan, X. & Heitman, J. (1999) Mol. Cell. Biol. 19, 4874-4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson, L. S., Causton, H. C., Young, R. A. & Fink, G. R. (2000) Proc. Natl. Acad. Sci. USA 97, 5984-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, A., Ward, M. P. & Garrett, S. (1998) EMBO J. 17, 3556-3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reinders, A., Burckert, N., Boller, T., Wiemken, A. & De Virgilio, C. (1998) Genes Dev. 12, 2943-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedruzzi, I., Dubouloz, F., Cameroni, E., Wanke, V., Roosen, J., Winderickx, J. & De Virgilio, C. (2003) Mol. Cell 12, 1607-1613. [DOI] [PubMed] [Google Scholar]
- 46.Gorner, W., Durchschlag, E., Martinez-Pastor, M. T., Estruch, F., Ammerer, G., Hamilton, B., Ruis, H. & Schuller, C. (1998) Genes Dev. 12, 586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., Davis, A. P., Dolinski, K., Dwight, S. S., Eppig, J. T., et al. (2000) Nat. Genet. 25, 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. (2002) Nucleic Acids Res. 30, 42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.