Abstract
Arabidopsis cryptochrome 2 (CRY2) is a blue light receptor that mediates light inhibition of hypocotyl elongation and long-day promotion of floral initiation. CRY2 is known to undergo blue light-dependent phosphorylation, which is believed to serve regulatory roles in the function of CRY2. We report here a biochemical and genetics study of CRY2 phosphorylation. Using mass spectrometry analysis, we identified at least three serine residues in the CCE domain of CRY2 (S588, S599, and S605) that undergo blue light-dependent phosphorylation in Arabidopsis seedlings. A study of serine-substitution mutations of the CCE domain of CRY2 demonstrates that CRY2 contains two types of phosphorylation in the CCE domain, one in the serine cluster that cause electrophoretic mobility upshift and the other outside of the serine cluster that do not seem to cause mobility upshift. We showed that mutations in the serine residues within and outside of the serine cluster diminished blue light-dependent CRY2 phosphorylation, degradation, and physiological activities. These results support the hypothesis that blue light-dependent phosphorylation of the CCE domain determines the photosensitivity of Arabidopsis CRY2.
Keywords: light regulation, light signaling, physiology of plant growth, Arabidopsis
Short Summary
This study identified, for the first time, light-dependent phosphorylation sites of a cryptochrome protein purified from plants. Using the transgenic approach, it also demonstrated a number of interesting biochemical and functional features of the blue light-dependent phosphorylation of a plant cryptochrome photoreceptor.
INTRODUCTION
Cryptochromes are photolyase-related blue light receptors that regulate photomorphogenesis in plants and the circadian clock in plants and animals (Cashmore, 2003; Chaves et al., 2011; Lin and Shalitin, 2003; Sancar, 2003). Arabidopsis genome encodes two cryptochromes, cryptochrome 1 (CRY1, 681 amino acids) and cryptochrome 2 (CRY2, 621 amino acids), which mediate over lapping blue light responses, such as inhibition of hypocotyl elongation and promotion of floral initiation (Ahmad and Cashmore, 1993; Guo et al., 1998; Lin et al., 1998). Although the mechanism of initial photochemistry of cryptochromes remains unclear, the signaling mechanism of plant cryptochromes is believed to involve light regulation of transcription and protein stability. For example, photoexcited CRY2 interacts with the bHLH transcription factors CIBs (CRY-Interacting BHLHs) to directly modulate transcription in Arabidopsis, and CRY2 also interacts with SPA1 (Suppressor of Phytochrome A 1) and COP1 (Constitutive Photomorphogenesis 1) to alter stability of transcription factors required for the light-dependent transcriptional changes and photomorphogenesis (Liu et al., 2011a; Liu et al., 2011b; Liu et al., 2008; Liu et al., 2013; Meng et al., 2013; Wang et al., 2001; Yang et al., 2001; Zuo et al., 2011).
Cryptochromes are characterized by two domains, the PHR (Photolyase Homologous Region) domain (about 500 residues) that is evolutionarily conserved in DNA photolyases and cryptochromes, and the CCE (CRY C-terminal Extension) domain (also referred to as CCT domain) that is absent in photolyases (Lin and Shalitin, 2003). The CCE domains of cryptochromes in plants and animals share little sequence similarity, but they are intrinsically disordered yet functionally indispensible for cryptochromes in both evolutionary branches (Busza et al., 2004; Chaves et al., 2006; Partch et al., 2005; Rosato et al., 2001; Yang et al., 2000; Yu et al., 2007b; Zhu et al., 2003). Cryptochromes are phosphoproteins and protein phosphorylation is believed to play important roles in the function and regulation of cryptochromes in both plants and animals (Lamia et al., 2009; Shalitin et al., 2002; Shalitin et al., 2003). Arabidopsis cryptochromes appear to be phosphorylated in multiple residues of at least the CCE domain (Harada et al., 2005; Shalitin et al., 2002; Yu et al., 2007b). Phosphorylation of the CCE domain correlates with the function of CRY2 (Sang et al., 2005; Shalitin et al., 2002; Yang et al., 2000). For example, the GUS (β-glucuronidase)-CCT2 fusion protein, which contains the CCE domain without the PHR domain of CRY2, is constitutively phosphorylated, constitutively dimerized, and constitutively active in plants. Interestingly, the GFP (Green Fluorescent Protein)-CRY2-N563 fusion protein (GFP-N563), which contains the N-terminal 563 residues but misses the serine-rich C-terminal half (49 residues) of the CCE domain of CRY2, remained photo-phosphorylated without mobility upshift (Yu et al., 2007b), implying that CRY2 undergoes two types of photo-phosphorylation, one type of CRY2 phosphorylation causes mobility upshift whereas the other type does not cause mobility upshift. It has been reported recently that Casein kinase1 CK1.3 and CK1.4 phosphorylate CRY2 at serine-587 (S587) and threonine-603 (T603) to affect the function and regulation of CRY2 (Tan et al., 2013). These observations are consistent with the hypothesis that the blue light-induced phosphorylation determines the function and regulation of CRY2 (Shalitin et al., 2002). However, the in vivo phosphorylation sites of cryptochromes have not been identified in nay plant cryptochromes, and how protein phosphorylation at different residues of plant cryptochromes affects their function and regulation remains unclear. To address these questions, we investigated the phosphorylatable residues of CRY2 by mass spectrometry analysis of CRY2 purified from plants, and we also systematically analyzed serine-substitution mutants of CRY2 in transgenic Arabidopsis. Results of these experiments demonstrate that CRY2 undergoes blue light-dependent phosphorylation in multiple serine residues of the CCE domain, including S598, S599, and S605, and that the combined phosphorylation pattern of CRY2 determines the photosensitivity of the physiological activities and blue light regulation of CRY2.
RESULTS
Identification of blue light-dependent phosphorylation sites of CRY2 by mass spectrometry
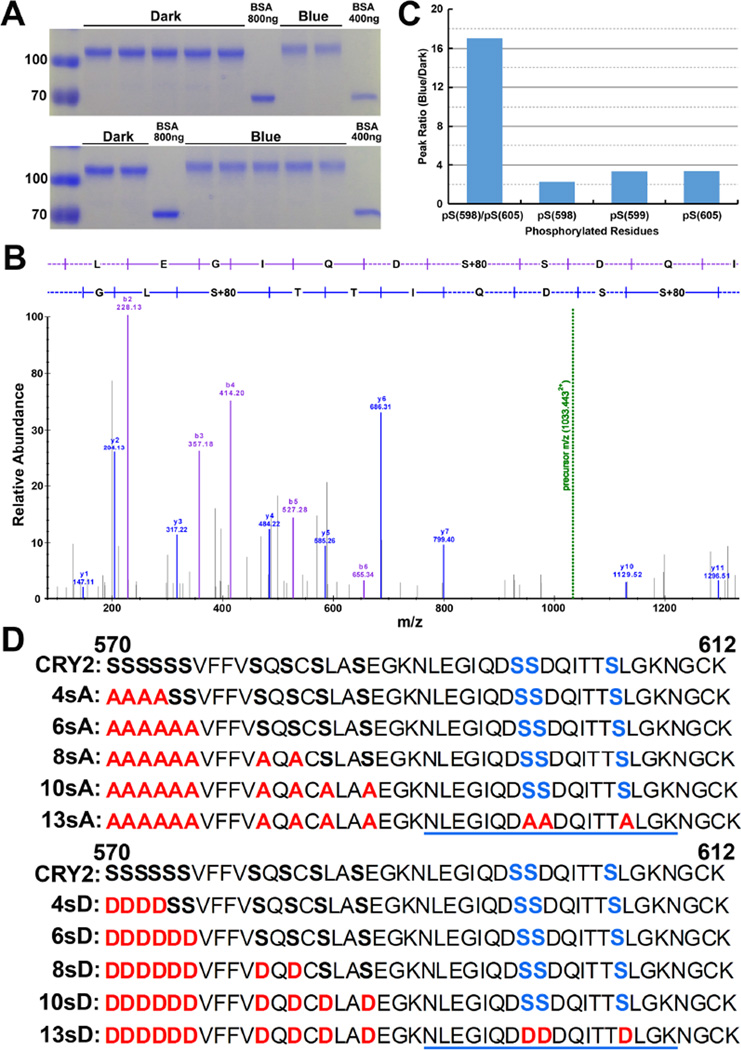
To identify the blue light-induced phosphorylation sites of CRY2 in vivo, we purified and analyzed the GFP-CRY2 recombinant protein from the transgenic Arabidopsis plants (Yu et al., 2009). We have previously shown that GFP-CRY2 is functionally indistinguishable from the endogenous CRY2 protein in all the photophysiological and photobiochemical properties examined (Yu et al., 2009). The GFP-CRY2 protein was purified from 7-day-old etiolated seedlings without (D) or with a brief blue light treatment (B30, 50 µmol m−2 s−1 for 30 min), using the GFP-trap affinity chromatography (Fig. 1A)(Rothbauer et al., 2008). Similar to the endogenous CRY2, the GFP-CRY2 recombinant proteins in etiolated seedlings exposed to blue light exhibit the electrophoreticmobility upshift and reduced level of protein, which are due to blue light-dependent CRY2 phosphorylation, ubiquitination, and degradation (Fig. 1A) (Lin et al., 1998; Shalitin et al., 2002; Yu et al., 2007a). We analyzed the light-dependent phosphorylation of the GFP-CRY2 protein purified from plants using label-free quantitative mass spectrometry (see Methods). This MS experiment detected a phosphopeptide of CRY2, NLEGIQDSSDQITTSLGK (Spectral FDR<0.05), which is monophosphorylated or diphosphorylated at three residues S598, S599, and S605 (Fig. 1B–D, Fig. S1–3, Table S1). This is the first time that the blue light-dependent phosphorylation sites of a cryptochrome are determined directly from the cryptochrome protein purified from plants. Although additional phosphorylation sites of CRY2 were also identified in this experiment (Liu, 2015), we focused on the analysis of phosphopeptide NLEGIQDSSDQITTSLGK in the present report. Fig. 1C shows four different phosphorylation events detected in this phosphopeptide of CRY2 that exhibited clear blue light responsiveness, among which the S598/S605 diphosphorylation exhibits a markedly higher blue light responsiveness than that of the monophosphorylation events in any neighboring serine residue of this phosphopeptide. This result appears to suggest that the blue light-dependent monophospohorylation facilitates further phosphorylation of the neighboring serine residues in this region of CRY2.
Fig.1. Blue light-dependent phosphorylation of the C-terminal tail of CRY2.
A). The Coomassie Blue-stained gel showing the GFP-CRY2 fusion protein purified from plants. The bands shown are excised, and subject to proteolysis for MS analysis. BSA was included as the loading control. (B). A representative tandem mass spectrum of the NLEGIQDpSSDQITTpSLGK peptide (m/z 1033.4427). Fragments matched within 10ppm are annotated as either the B-ions (purple) or Y-ions (blue) series. The green dashed line indicates precursor m/z. (C). The abundance ratio of the indicated phosphopeptide was calculated by determining the label-free MS1 peak area of each phosphopeptide under the blue light and dark conditions. All phosphopeptide quantities were normalized to the amount of unmodified CRY2 detected in each condition (see Supplemental Table 1). (D). The Amino acid sequence of the C-terminal tail of CRY2 (residues 570 to 612). Sequences of the serine-substitution mutations containing the alanine (A) or aspartic acid (D) replacements in the indicated mutations are highlighted in red, the phosphopeptide and the residues identified by the MS analysis (S598, S599, and S605) are underlined and highlighted in blue, respectively.
Expression and subcellular localization of the serine-substitution CRY2 mutants in transgenic Arabidopsis plants
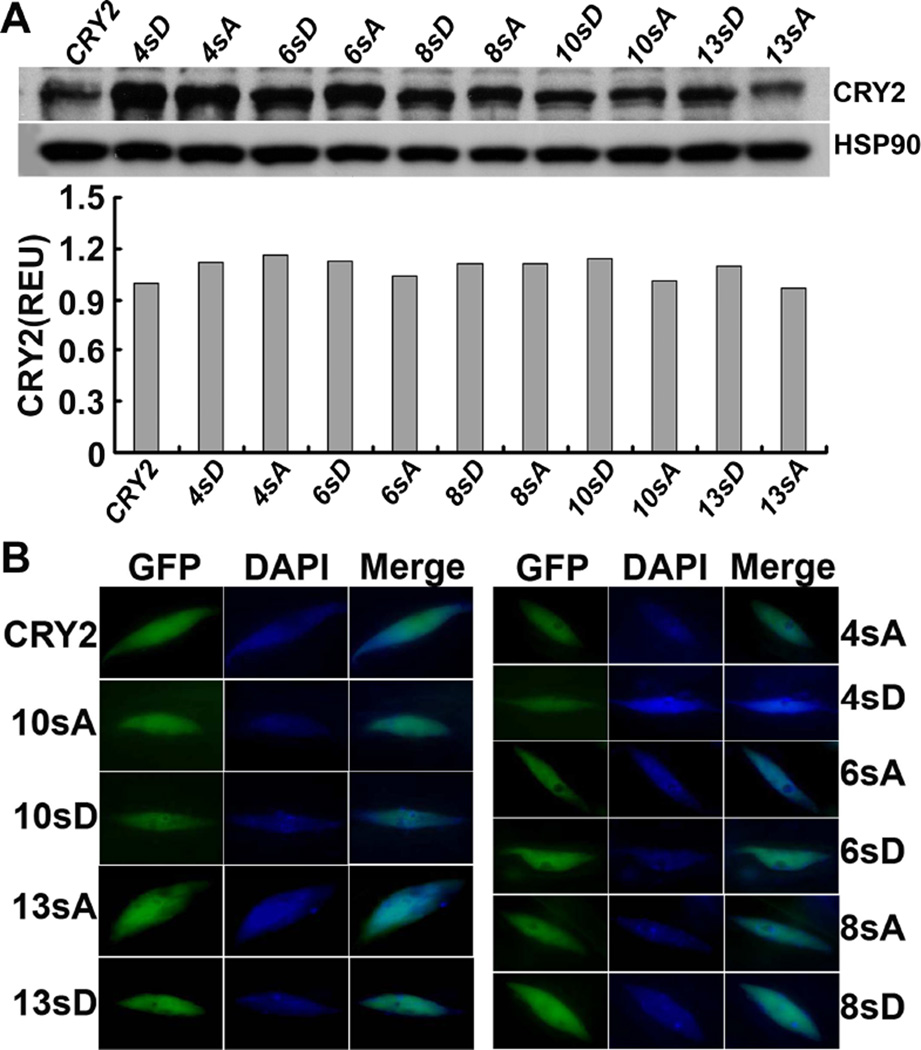
To further investigate photo-phosphorylation of CRY2, we performed a systematic analysis of the serine-rich C-terminal tail of the CCE domain of CRY2. This 43-residue C-terminal tail of CRY2 (from residues 570 to 612) contains 13 serine residues (30.2%), including a 6-residue serine cluster (S570 to S575) and the three phosphorylatable serine residues (S598, S599, and S605) identified in our MS experiment (Fig. 1). We replaced serine residues with non-phosphomimetic alanines (S-to-A) or phosphomimetic aspartic acids (S-to-D) by site-specific mutagenesis, using the plasmid template encoding the GFP-CRY2 recombinant protein (Fig. 1D). And we prepared transgenic plants expressing the serine-substitution mutants or the wild-type GFP-CRY2 control in the cry1cry2 double mutant background (Yu et al., 2009). For simplicity, the transgenic lines expressing the wild-type GFP-CRY2 control and the GFP-CRY2 recombinant protein are referred to as the “wild-type CRY2”, whereas the transgenic lines expressing the serine-substitution GFP-CRY2 mutants and the respective GFP-CRY2 mutant proteins are referred to as xsA or xsD, in which × indicates the number of serine residues within the 43-residue C-terminal tail of CRY2 that are substituted by A or D, respectively. For example, 4sA or 4sD refer to the GFP-CRY2 mutant proteins, of which 4 out of 6 serine residues of the serine cluster are replaced by alanines or aspartates, respectively. Similarly, 13sA or 13sD refer to the mutations, of which all 13 serine residues between residues 570 to 612 of the C-terminal tail of CRY2 are substituted by alanines or aspartates, respectively. The sequences of all 10 serine-substitution mutations of CRY2 analyzed in this report are shown in Fig. 1D. To ensure that the phenotypic change of the serine-substitution mutants of CRY2 is not due to a lower level of mutant protein expression, we selected transgenic lines that express GFP-CRY2 mutant proteins at the level slightly higher or comparable to that of the wild-type GFP-CRY2 control (Fig. 2A). Fig. 2 also shows that all CRY2 mutant proteins examined located in the nucleus of Arabidopsis cells as the endogenous CRY2 or the wild-type GFP-CRY2 (Yu et al., 2007a), suggesting that the mutant CRY2 proteins are not denatured. This result may not be surprising, because CCE is an unstructured domain of cryptochromes and mutations in this domain may not drastically disrupt the overall structure of the CRY2 holoprotein (Li, 2012; Partch et al., 2005).
Fig.2. Analyses of protein expression and subcellular localization of the serine-substitution mutants of CRY2 in the transgenic plants.
(A) Immunblots showing expression of the fusion proteins of GFP to the wild-type CRY2 (CRY2) or the serine-substitution mutants of CRY2 (4sD, 6sA and 6sD, 8sA and 8sD, 10sA and 10sD, 13sA and 13sD) in transgenic seedlings. 6-day-old transgenic seedlings were grown in continuous blue light (60µmol m−2 s−1), proteins were extracted and fractioned by 10% SDS-PAGE, blotted, probed with the anti-CRY2 antibody (CRY2), stripped, and reprobed with anti-HSP90 antibody (HSP90). Relative levels of CRY2 expression of the indicated transgenic lines are estimated by normalization of the CRY2 signals with the HSP90 signals and shown below. (B) Fluorescence images showing nuclear distribution of GFP-CRY2 and the serine-substitution mutants of CRY2. The GFP and DAPI fluorescence images were taken from the nuclei isolated from 6-day-old seedlings of the indicated genotypes.
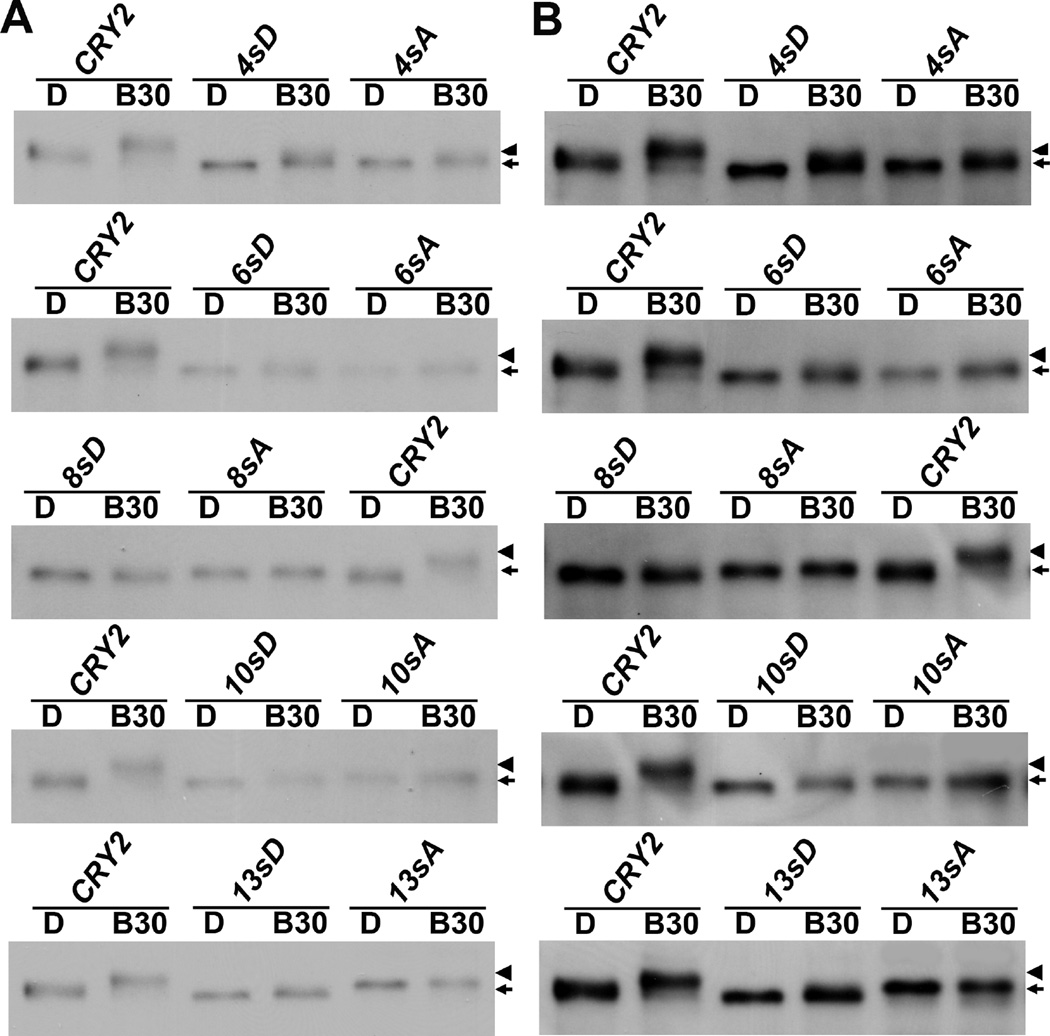
Mutations in the serine cluster abolished the phosphorylation events detectable by the electrophoretic mobility upshift assay
To examine the blue light-dependent phosphorylation of the serine-substitution mutants of CRY2 in the transgenic plants, etiolated seedlings without blue light treatment (D) or treated with blue light (B30, 60 µmol m−2 s−1 for 30 min) were analyzed by immunoblot probed with the anti-CRY2 antibody (Fig. 3). We compared the blue light-dependent phosphorylation of different GFP-CRY2 recombinant proteins by the electrophoresis mobility shift assay (Shalitin et al., 2002), in which both the weak and the strong exposures of the same immunoblot are included to help distinguishing subtle difference in electrophoretic mobility of CRY2 in SDS-PAGE gels (Fig. 3). We have previously shown that both the endogenous CRY2 protein and the wild-type GFP-CRY2 recombinant proteins undergo blue light-dependent phosphorylation and exhibit electrophoretic mobility upshift (Shalitin et al., 2002; Yu et al., 2009; Yu et al., 2007b; Zuo et al., 2011). As expected, the wild-type GFP-CRY2 control exhibited the mobility upshift or protein phosphorylation in response to blue light (Fig. 3, CRY2, B30). In contrast, no blue light-dependent mobility upshift was observed in any of the 10 serine substitution mutants of CRY2, except 4sA/D that might retain some residual mobility upshift (Fig. 3).
Fig.3. Loss of blue light-induced phosphorylation of the serine-substitution mutations of CRY2.
(A) Immunoblots showing the blue light-dependent mobility upshift of the wild-type GFP-CRY2 and loss of mobility upshift of the serine-substitution mutants. 6-day-old seedlings were grown in dark (D) and transferred to blue light (60 µmol m−2 s−1) for 30min (B30). Protein samples were extracted and fractioned by 10% SDS-PAGE gels, blotted, probed with anti-CRY2 antibody. Arrowhead indicates mobility upshifted or phosphorylated CRY2 band, and arrow indicates mostly the unphosphorylated CRY2.
The results shown in Fig. 3 revealed two interesting aspects of CRY2 phosphorylation. First, most CRY2 mutants examined, except 13sA/D, are not mutated in the phosphorylatable S598, S599, and S605 residues, but none of them showed a clear discernable electrophoretic mobility upshift (Fig. 3). This result, together with the facts that all those mutants contain mutations in the serine cluster (S570–S575), that proteins phosphorylated in serine clusters are prone to electrophoretic mobility upshift (Hereld et al., 1994), and that protein phosphorylation do not always cause electrophoretic mobility upshift (Dephoure et al., 2013; Peck, 2006), suggests that blue light induces two type of phosphorylation in the CRY2 protein: one occurs in the serine cluster that causes mobility upshift, whereas the other type occurs in residues outside of the serine cluster that does not cause mobility upshift (prediction #1). In consistent with this proposition, the GFP-N563 fusion protein that lacks all 13 serine residues examined in this report remains phosphorylated by 32P without electrophoretic mobility upshift, although the phosphorylation activity of GFP-N563 is markedly weaker than that of the wild-type GFP-CRY2 control (Yu et al., 2007b). Secondly, similar to the non-phosphomimetic S-to-A mutants, the phosphomimetic S-to-D mutants of CRY2 also lost blue light-dependent mobility upshift (Fig. 3). This observation is in contrast to other phosphoproteins, such as PIF3. The phosphomimetic S-to-D mutations of PIF3 mimic both mobility upshift and functional activity of the phosphorylated PIF3 protein (Ni et al., 2013). Because it is well known that phosphomimetic mutations do not always reproduce the changes caused by protein phosphorylation (Dephoure et al., 2013; Peck, 2006), we predict that both the non-phosphomimetic S-to-A mutations and the phosphomimetic S-to-D mutations of CRY2 result in loss-of-function phenotype in not only mobility upshift but also the function of CRY2 (prediction #2).
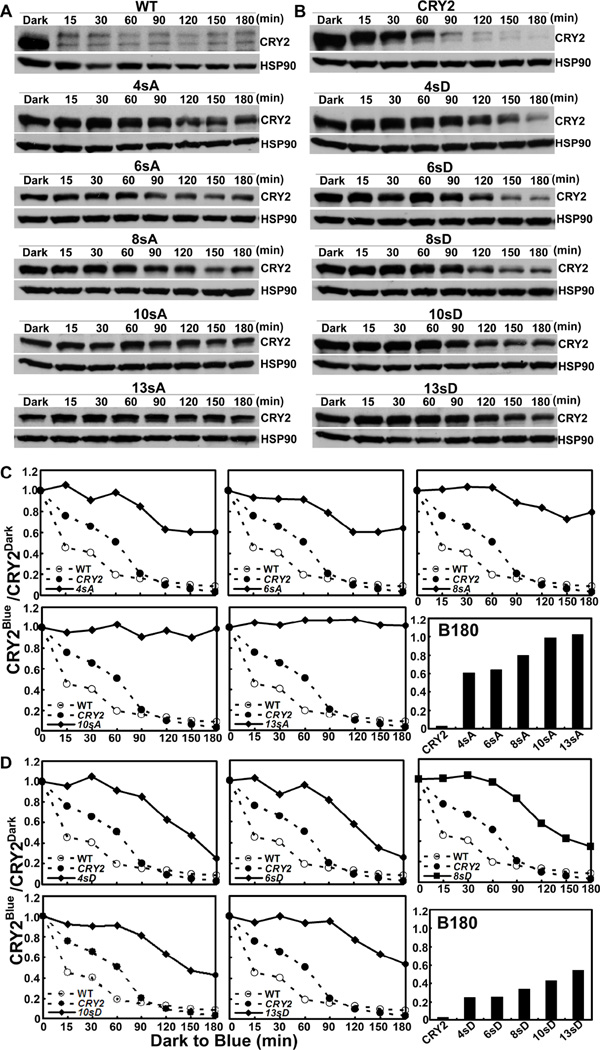
The phosphorylation-deficient CRY2 mutants are impaired in blue light-dependent proteolysis
We tested the above two predictions first by analyses of how the serine-substitution mutations affects blue light- and phosphorylation-dependent degradation of CRY2 in vivo. We have shown previously that CRY2 undergoes blue light-dependent proteolysis that is dependent on the blue light-induced phosphorylation and ubiquitination of the CRY2 protein (Shalitin et al., 2002; Yu et al., 2007a; Yu et al., 2009; Zuo et al., 2012). And we compared the kinetics of the blue light-dependent degradation of the mutant and the wild-type GFP-CRY2 proteins in etiolated seedlings exposed to blue light (Fig. 4). Fig. 4 shows that, the endogenous CRY2 (WT) and the wild-type GFP-CRY2 control (CRY2) undergo blue light-dependent degradation, whereas the CRY2 mutant proteins containing progressively increased serine-substitution mutations were progressively more stable in blue light (Fig. 4). For example, the degradation of the 4sA/D and 6sA/D mutants was detectable after a prolonged exposure to blue light, although it was slower than the wild-type GFP-CRY2 (Fig. 4A, 4C). The CRY2 mutants that contain more serine substitutions, such as 8sA and 10sA, exhibited further decreased rates of proteolysisin comparison to that of 4sA and 6sA (Fig. 4A, 4C), and the 13sA mutant mutated in all 13 serine residues of the C-terminal tail of CRY2 exhibited no apparent proteolysis in response to blue light (Fig. 4A, 4C). This progressively decreased blue light sensitivity of the serine-substitution mutants, which is more apparent with longer blue light exposure (Fig. 4C, B180), argues that most mutants except 13sA may remain weakly phosphorylated, ubiquitinated, and degraded in response to blue light. In other words, as we predicted (prediction #1), both the phosphorylation in the serine cluster detectable by mobility upshift assay and the phosphorylation in other residues undetectable by the mobility upshift assay contribute to the photobiochemical activities of CRY2. Similar to the non-phosphomimetic S-to-A mutations, the phosphomimetic S-to-D mutants of CRY2 also exhibited progressive impairment of the blue light-induced proteolysis of CRY2, although the effects of S-to-D mutations on the blue light-dependent proteolysis are generally weaker than that of the S-to-A mutations (Fig. 4B, 4D). This result is consistent with our second prediction (prediction #2) that both the non-phosphomimetic S-to-A mutation and the phosphomimetic S-to-D mutants of CRY2 cause loss-of-function phenotypes.
Fig.4. The serine-substitution mutations reduce or abolish blue light-dependent degradation of CRY2.
(A–B) Immunoblot showing expression of the serine-substitution mutants of CRY2 in etiolated seedlings exposed to blue light. 6-day-old seedlings grown in dark were transferred to blue light (60 µmol m−2 s−1) for the indicated time (min). Proteins were extracted, fractioned by 10% SDS-PAGE gels, blotted, probed with anti-CRY2 (CRY2), stripped, and reprobed with anti-HSP90 antibody (HSP90) are shown.
(C–D) The relative level of CRY2 proteins calculated by normalization of the CRY2 signals in blue light-treated samples to that of the etiolated seedlings, and presented as CRY2Blue (signal intensity of CRY2 in blue-light treated seedlings)/CRY2Dark (signal intensity of CRY2 in seedlings before blue light treatment).
The phosphorylation-deficient CRY2 mutants are impaired in the blue light-dependent hypocotyl inhibition response
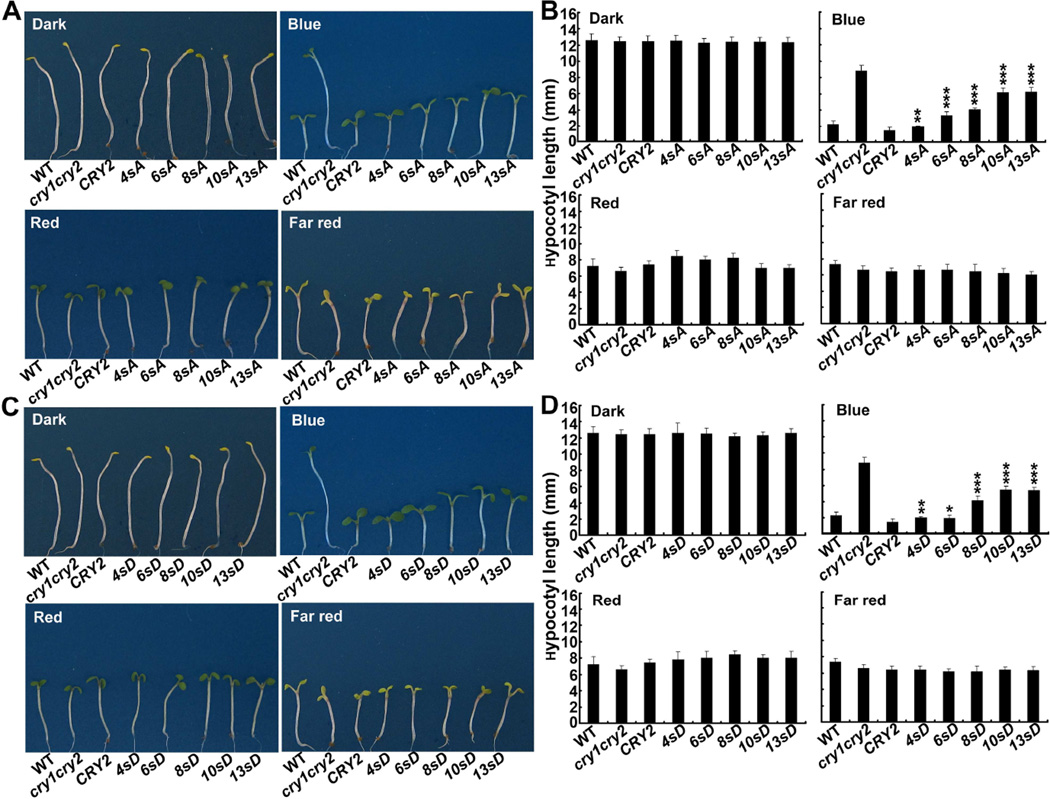
The observation that progressive increase of the number of serine-substitution mutations aggravates defects in the blue light-dependent CRY2 degradation predicts that many or all serine residues in the serine-rich C-terminal tail of CRY2 are phosphorylatable to contribute to the photosensitivity of CRY2. In consistent with this hypothesis, we found that the progressively increased serine substitutions of CRY2 also resulted in progressively decreased activity of the CRY2 mutants to rescue the blue light-specific long hypocotyl phenotype of the cry1cry2 mutant parent (Fig. 5–7). Figure 5 shows a clear trend that the transgenic seedlings expressing the CRY2 mutants containing progressively increased serine substitutions exhibited the phenotype progressively more similar to of that the cry1cry2 mutant parents (Fig. 5B, Blue). For example, the seedlings expressing the CRY2 mutant that contain 4 serine-substitution mutations (4sA) are slightly taller (p<0.01) than that of the seedlings expressing the wild-type GFP-CRY2 control. In contrast, the seedlings expressing the CRY2 mutants containing 6 or more S-to-A mutations (6sA, 8sA, 10sA, and 13sA) were progressively and significantly taller than that expressing the wild-type GFP-CRY2 control (p<0.001). No such difference was found in seedlings grown in darkness, or red light, or far-red light, indicating the phenotype difference is indeed due to the mutation of CRY2. Similar to the S-to-A mutants, the phosphomimetic S-to-D mutants of CRY2 also exhibited progressively reduced activity mediating blue light inhibition of hypocotyl elongation (Fig. 5C–D).
Fig.5. The serine substitution mutations reduce the activity of CRY2 to mediate blue light inhibition of hypocotyl elongation.
Representative seedlings (A, C) and hypocotyl lengths (B, D) of 6-day-old transgenic seedlings of indicated genotypes. Seedlings were grown in dark or continuous blue (25µmol m−2 s−1), red (15µmol m−2 s−1), and far red (5µmol m−2 s−1) light for 6 days, and hypocotyl lengths of indicated genotypes were measured (n ≥ 20). Hypocotyl lengths of seedlings expressing the serine-substitution mutant CRY2 that are significantly longer than the control seedlings expressing the wild-type CRY2 transgene are indicated *(p≤0.05), **(p≤0.01), or ***(p≤0.001), respectively (Turkey’s LSD Test).
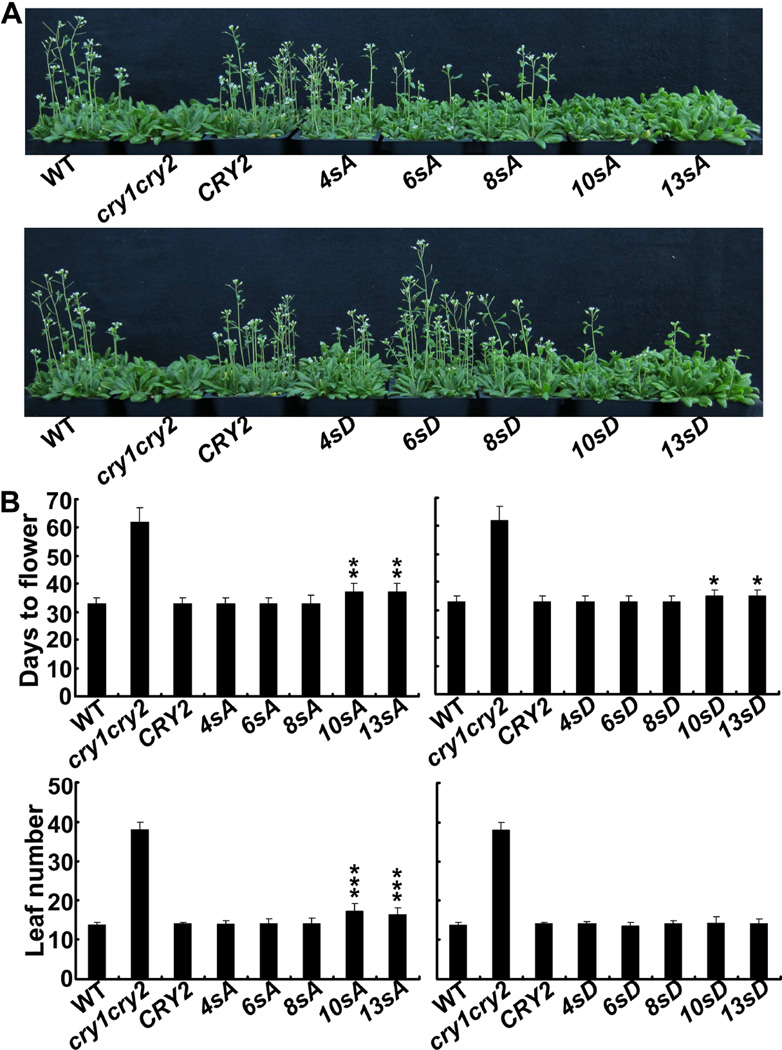
Fig.7. The serine substitution mutations do not cause significant change of the activity of CRY2 promoting floral initiation.
(A) Images of 35-day-old plants grown in long day condition.
(B) Days to flowering and rosette leaf number at flowering (± SD) of the respective genotypes are shown (≥20). Transgenic lines expressing the serine-substitution mutant CRY2 that showed significantly delayed flowering are indicated by*(p≤0.05), **(p≤0.01), or ***(p≤0.001), respectively (Turkey’s LSD Test).
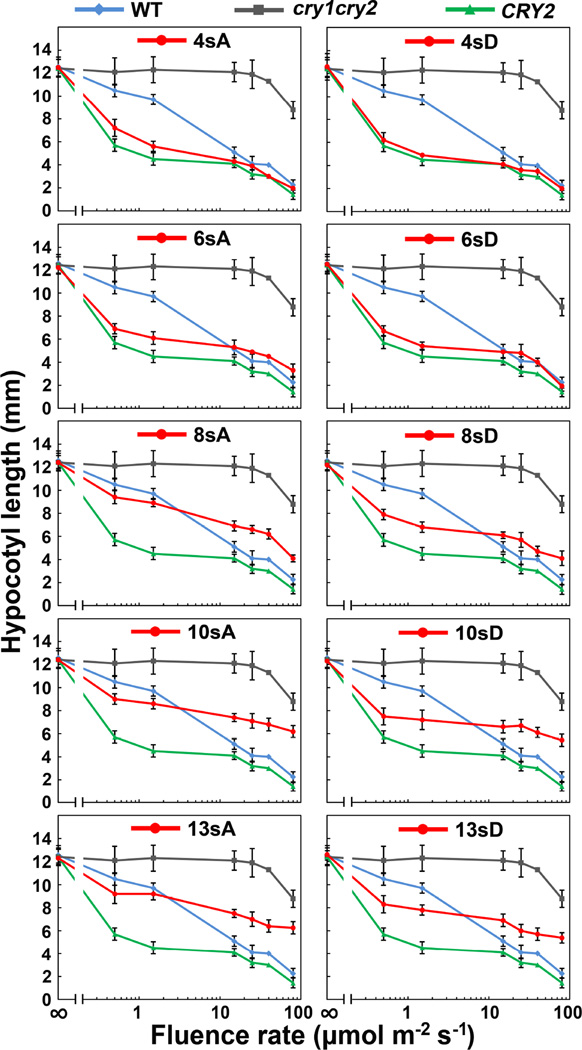
To confirm this observation, we analyzed the fluence rate response of the seedlings expressing the serine-substitution CRY2 mutants (Fig. 6). Fig. 6 shows that the cry1cry2 mutant exhibited no blue light inhibition of hypocotyl elongation in seedlings grown under most light intensities tested, except the highest fluence rates that is likely due to the actions of phytochromes (Franklin et al., 2003). The transgenic seedlings expressing the 4sA/D and 6sA/D mutant proteins exhibited either similar or slightly reduced hypocotyl lengths in comparison to the seedlings expressing the wild-type GFP-CRY2 control when grown in different fluence rates of blue light tested (Fig. 5, 4sA, 6sA, 4sD, 6sD), indicating that those CRY2 mutants retain substantial physiological activity. In contrast, transgenic seedlings expressing the mutant CRY2 proteins that contain more than 6 serine substitutions (8sA, 10sA, 13sA, 8sD, 10sD, and 13sD) need over 100 folds higher fluence rate of blue light to achieve the similar extent of hypocotyl inhibition (Fig. 6). This difference is not due to lower levels of mutant CRY2 expression, because the steady-state level of those mutant CRY2 proteins are higher or comparable to that of the wild-type GFP-CRY2 control (Fig. 2A), and the mutant CRY2 proteins are more stable than the wild-type GFP-CRY2 control in etiolated seedlings exposed to blue light (Fig. 4). Therefore, as we predicted, the progressively increased serine substitutions progressively reduced photosensitivity and activity of CRY2.
Fig.6. Fluence rate-response of the hypocotyl inhibition response of transgenic seedlings expressing the serine-substitution mutants of CRY2.
6-day-old seedlings of indicated genotypes were grown under continuous blue light with fluence rate of 0, 0.5, 1.5, 15, 25, 40 or 80 µmol m−2 s−1, respectively. The fluence rates are shown in the log scale; hypocotyl lengths and standard deviations (n ≥ 20) are shown.
In addition to mediating blue light inhibition of hypocotyl elongation, CRY2 also mediates long-day promotion of floral initiation (Guo et al., 1998). Fig. 7 demonstrates that most S-to-A mutants tested, except 10sA and 13sA, were able to rescue the late-flowering phenotype of the cry1cry2 parent (Fig. 7). The transgenic plants expressing the 10sA and 13sA mutants showed statistically significant delay in flowering (p<0.01), demonstrating again that increased serine substitutions of CRY2 aggravates the impairment of CRY2 activity to promote floral initiation in long-day photoperiods. The S-to-D mutation had relatively weaker effect on the CRY2 activity than the S-to-A mutations. The transgenic plants expressing 10sD and 13sD mutants exhibited only slightly (p<0.05) delayed flowering measured by “Days to flower”, but showed no significant difference in “leaf number” at flowering (Fig. 7). It is not clear why the serine-substitution mutations of CRY2 have more profound defects in mediating blue light inhibition of hypocotyl elongation than that in promoting floral initiation. However, given that CRY2 interacts with multiple proteins to exert different functions in plant development (Liu et al., 2011a; Liu et al., 2011b; Liu et al., 2008; Liu et al., 2013; Meng et al., 2013; Wang et al., 2001; Yang et al., 2001; Zuo et al., 2011), it is conceivable that different CRY2-interacting proteins regulating hypocotyl growth or floral initiation may be differentially affected by blue light-dependent serine phosphorylation or the serine substitution mutations. This hypothesis remains to be tested.
DISCUSSIONS
In the present study, we investigated blue light-dependent phosphorylation of the GFP-CRY2 protein purified from plants by MS analyses, and identified at least three in vivo blue light-dependent phosphorylation sites (S598, S599, and S605) of CRY2. Further experiments are needed to elucidate all in vivo phosphorylation sites of CRY2. In addition, our analyses of the transgenic plants expressing the serine-substitution mutants of CRY2 showed that progressively increased serine substitutions resulted in progressive impairment of the regulation and physiological activities of CRY2 in response to blue light. These results are consistent with the hypothesis that the blue light-dependent phosphorylation of CRY2 determines the photosensitivity of this photoreceptor. It is noteworthy that the serine-substitution mutations may also result in structure perturbations to affect the photosensitivity of CRY2. For example, in yeast cells, the serine-substitution mutant of CRY2 showed progressively reduced activity in the blue light-dependent interaction with the CRY2-signaling proteins CIB1 and SPA1 (Fig. S5). Because yeast cells do not have the bona fide CRY2 kinases, whereas CRY2 may not have the autophosphorylation activity (Ozgur and Sancar, 2006; Yu et al., 2009), the defect of CRY2 to interact with CIB1 and SPA1 in yeast cells may result from conformation changes of the CRY2 mutants. However, because the CCE domain of cryptochromes is intrinsically unstructured (Partch et al., 2005), that all serine-substitution mutants of CRY2 expressed normally and located correctly in the nucleus in transgenic plants (Fig. 2), and that all mutants examined retained partial activities in the transgenic plants (Fig. 5–7), the reduced photosensitivity of serine-substitution mutants of CRY2 in plants are more likely due to the reduced phosphorylation of those mutants.
Our study also discovered two previously unknown aspects of the blue light-dependent photophosphorylation of CRY2. First, our results argue that CRY2 undergoes two types of phosphorylation, the phosphorylation in the 6-residue serine cluster that causes electrophoretic mobility upshift and the phosphorylation in other residues (such as S598, S599, and S605) that do not result in electrophoretic mobility upshift. Additional studies are needed to verify this phenomenon and to elucidate the underling mechanistic implication of the two types of phosphorylation to the function and regulation of CRY2. Second, in contrast to the expectation that the phosphomimetic S-to-D mutations might cause gain-of-function effects to mimic the changes of protein phosphorylation, the S-to-D mutations of CRY2 showed loss-of-function effect similar to that of the non-phosphomimetic S-to-A mutations in all phenotypes analyzed in our study, including light-dependent mobility upshift, protein stability, and physiological activities. It is well-known that phosphomimetic mutation do not always mimic or reproduce the changes introduced by protein phosphorylation (Dephoure et al., 2013). For example, phosphomimetic S-to-D mutations of the mammalian mCRY1 caused loss-of-function phenotype with respect to the transcription suppression activity (Sanada et al., 2004). Whether or not a phosphomimetic mutation imitates the changes introduced by protein phosphorylation is dependent on the exact structural and functional roles of the respective phosphorylation event. For example, a phosphomimetic mutation may incline to reproduce the changes introduced by phosphorylation if the native protein is phosphorylated to produce a modified residue specifically recognized by another protein. On the other hand, a phosphomimetic mutation may be less likely to mimic the changes introduced by phosphorylation if the native protein is phosphorylated to establish a charge-dependent structural change. It has been previously proposed that blue light triggers separation of the PHR domain and CCE domain to activate cryptochromes (Yang et al., 2000; Yu et al., 2007b), and that phosphorylation of the CCE domain may facilitate such domain separation by a charge-dependent electrostatic repelling mechanism (Yu et al., 2007b). According to this hypothesis, the number of negative charges introduced by light-dependent phosphorylation is important to the light-dependent structural change of cryptochromes. However, the negative charge carried by an aspartate (ca. −1 charge per residue) is at least 50% less than that introduced by phosphorylation (ca. −1.5 to −2 charges per residue) at pH7.2 estimated for the Arabidopsis nuclear compartment (Shen et al.). This charge difference may explain the loss-of-function instead of gain-of-function phenotype of the pphosphomimetic S-to-D mutations of CRY2. It would be informative to test whether the same phenomenon is true for CRY1. In this regard, it is particularly interesting that the truncated CRY1 or CRY2 recombinant proteins composed of the PHR/CNT domain in the absence of most (GFP-N563 of CRY2) or all (CNT1 of CRY1) of the CCE/CCT sequences (He et al., 2015; Yu et al., 2007b), or composed of the CCE/CCT domain in the absence of the PHR sequences (GUS-CCT1 and GUS-CCT2)(Yang et al., 2000), or composed of a short sequence from each of the two domains (NC80) (Yu et al., 2007b) are all reported to be active in the transgenic plants. These seemingly perplex observations may be explained by the phosphorylation-dependent domain separation model given that different CRY-signaling complexes interact with either or both domains of cryptochromes (Liu et al., 2011a; Liu et al., 2011b; Liu et al., 2008; Liu et al., 2013; Meng et al., 2013; Wang et al., 2001; Yang et al., 2001; Zuo et al., 2011). This hypothesis may be further tested by elucidation of the specific structural elements of the PHR or CCE domain that interact with the known or presently unknown CRY-signaling protein or protein complexes.
MATERIALS AND METHODS
Plant Materials
Transgenic plants expressing the GFP-CRY2 fusion protein containing site-specific serine-substitution mutations of CRY2 were prepared in the cry1cry2 mutant background by the floral dip transformation method as described (Yu et al., 2007a). The control transgenic plants expressing the wild-type GFP-CRY2 fusion protein in the cry1cry2 mutant background was described previously (Yu et al., 2007a). To investigate the response of hypocotyl of transgenic plants under different light conditions, seeds were surface sterilized for 15min in 20% bleach, and rinsed with sterile, distilled water for 3 to 4 times. About 50 seeds were placed on the MS agar medium. After being incubated in the dark at 4°C for 4 days, seeds were exposed to white light for 12 hours before transferring to temperature controlled growth chambers, and grown under continuous blue, red, far red light or in the dark at 22–24°C. After 6 days, the hypocotyl lengths of more than 20 seedlings were measured manually (Zhao et al., 2007) and the standard deviations are calculated. To measure flowering time of the transgenic plants, seeds were sown in soil and placed in the cold room for 4 days, and then placed in the long-day growth chamber. The days to flowering and the total leaf number at flowering were counted as described (Mockler et al., 2003).
Mass Spectrometry analysis
The GFP-CRY2 fusion protein was purified by the GFP-Trap method as described (Rothbauer et al., 2008). The GFP-CRY2 proein fractionated by the gels shown in Fig. 1A were excised, digested in the excised gel slices using trypsin, and subject to MS analysis (Kaiser and Wohlschlegel, 2005; Kelstrup et al., 2012). The peptide digests were desalted and fractionated online using a 75 µM inner diameter fritted fused silica capillary column with a 5 µM pulled electrospray tip packed in-house with 17 cm of Luna C18(2) 3 µM reversed phase particles (Phenomonex). The gradient was delivered via an easy-nLC 1000 ultra high pressure liquid chromatography (UHPLC) system (Thermo Scientific). MS/MS spectra were collected on a Q-Exactive mass spectrometer (Thermo Scientific)(Kelstrup et al., 2012; Michalski et al., 2011). Data analysis was performed using the ProLuCID, and DTASelect2 algorithms as implemented in the Integrated Proteomics Pipeline - IP2 (Integrated Proteomics Applications, Inc., San Diego, CA)(Cociorva et al., 2007; Tabb et al., 2002; Xu et al., 2006). Phosphopeptides were identified using a differential modification search that considered a mass shift of +79.9663 on serines, threonines and tyrosines. All peptide-spectrum-matches were evaluated by DTASelect2 and filtered to achieve a maximum false detection rate of 5% using a decoy database approach (Elias and Gygi, 2007). Phosphosite localization confidence was assessed using the phosphoRS algorithm and a set of in-house scripts with all reported phosphopeptides possessing a localization probability score of greater than 99%(Taus et al., 2011). Individual spectra were examined within the ProteoWizard suite’s See MS (Kessner et al., 2008). Label-free quantitation of the peptides was performed using the Skyline software package (MacLean et al., 2010). Peak integration was carried out through manual interrogation of the data (MacLean et al., 2010; Schilling et al., 2012). For each peptide, peak areas were summed for three isotopic peaks (M, M+1, M+2) to serve as the peptide’s quantitative measure. For each dataset, phosphopeptide abundances were normalized to the amount of unmodified CRY2 detected in the sample. Unmodified CRY2 abundance was determined as the average of the integrated peak areas for identified CRY2 peptides which were uniquely mapping, had no missed tryptic cleavage sites, and showed no evidence of phosphorylation.
Site-Specific Mutagenesis
The transgenes encoding the GFP-CRY2 fusion protein (Yu et al., 2007b) was used as the template to prepare all mutants described in this study. Amino acid substitutions of serine (S) to alanine (A) or serine (S) to aspartic acid (D) were introduced into CRY2 coding sequence by the site-directed mutagenesis. The QuickChange II Site-specific mutagenesis Kit (Stratagene, Inc) was used to prepare individual and combined mutants, according to the manufacturer’s instructions. All mutants were confirmed by DNA sequencing. Site-mutated fragments were cloned into pEGAD vector and fused to the C-terminus of GFP sequence to generate the following mutations of CRY2: 4sA (S570A, S571A, S572A, S573A), 6sA (S570A, S571A, S572A, S573A, S574A, S575A), 8sA (S570A, S571A, S572A, S573A, S574A, S757A, S580A, S582A), 10sA (S570A, S571A, S572A, S573A, S574A, S575A, S580A, S582A, S584A, S587A) 13sA (S570A, S571A, S572A, S573A, S574A, S575A, S580A, S582A, S584A, S587A, S598A, S599A, S605A). Similar method was used to prepare the 4sD (S570D, S571D, S572D, S573D), 6sD (S570D, S571D, S572D, S573D, S574D, S575D), 8sD (S570D, S571D, S572D, S573D, S574D, S575D, S580D, S582D), 10sD (S570D, S571D, S572D, S573D, S574D, S575D, S580D, S582D, S584D, S587D) and 13sD (S570D, S571D, S572D, S573D, S574D, S757D, S580D, S582D, S584D, S587D, S598D, S599D, S605D) mutant constructs. The recombinant CRY2 transgenes that expressing the wild-type or mutant GFP-CRY2 fusion proteins were transformed to the cry1cry2 double mutant (Col accession).
The primers (F: forward, R: reverse) used in the site-specific mutagenesis of CRY2 coding sequence are the following:
4sA-F; GTTTTCGACTGCTGAAGCTGCTGCTGCTTCGAGTGTGTTTTTCG
4sA-R; CGAAAAACACACTCGAAGCAGCAGCAGCTTCAGCAGTCGAAAAC
4sD-F; GTTTTCGACTGCTGAAGATGATGATGATTCGAGTGTGTTTTTCG
4sD-R; CGAAAAACACACTCGAATCATCATCATCTTCAGCAGTCGAAAAC
6sA-F; GCTGAAGCTGCTGCTGCTGCGGCTGTGTTTTTCGTTTCGC
6sA-R; GCGAAACGAAAAACACAGCCGCAGCAGCAGCAGCTTCAGC
6sD-F; GCTGAAGATGATGATGATGATGATGTGTTTTTCGTTTCGC
6sD-R; GCGAAACGAAAAACACATCATCATCATCATCATCTTCAGC
8sA-F, GTGTTTTTCGTTGCGCAGGCTTGCTCGTTGGCATC
8sA-R, GATGCCAACGAGCAAGCCTGCGCAACGAAAAACAC
8sD-F, GTGTTTTTCGTTGACCAGGATTGCTCGTTGGCATC
8sD-R, GATGCCAACGAGCAATCCTGGTCAACGAAAAACAC
10sA-F, GTTGCGCAGGCTTGCGCGTTGGCAGCAGAAGGGAAGAATC
10sA-R, GATTCTTCCCTTCTGCTGCCAACGCGCAAGCCTGCGCAAC
10sD-F, GTTGACCAGGATTGCGACTTGGCAGATGAAGGGAAGAATC
10sD-R, GATTCTTCCCTTCATCTGCCAAGTCGCAATCCTGGTCAAC
13sA-F, GATCAGATTACTACAGCTTTGGGAAAAAATGG
13sA-R, CCATTTTTTCCCAAAGCTGTAGTAATCTGATC
13sD-F, GATCAGATTACTACAGATTTGGGAAAAAATGG
13sD-R, CCATTTTTTCCCAAATCTGTAGTAATCTGATC
Yeast two hybrid assays
The coding sequence of CRY2 was cloned into bait vector pBridge, and CIB1 and SPA1 were cloned into prey vector pGADT7 as reported previously (Liu et al., 2011a; Liu et al., 2008; Zuo et al., 2011). The mutated fragments of CRY2 was amplified from transgenes encoding 4sA, 6sA, 8sA, 10sA, and 13sA proteins generated by the sense (TGTATCGCCGGAATTCATGAAGATGGACAAAAAGACTA) and antisense (TTGGCTGCAGGTCGACTTTGCAACCATTTTTTCCCAA) primers (with the EcoR I and Sal I restriction sites), sub-cloned into the EcoR I and Sal I sites of bait vector pBridge using the Clontech Infusion PCR cloningkit (Clontech), and confirmed by DNA sequencing. The bait and the prey plasmids were co-transformed into the yeast strain AH109. The protein-protein interaction was analyzed using the histidine auxotrophy assay, by which yeast colonies were grown in duplicate onto His− and His+ plates. One duplicate was grown under blue light (25 molem−2 s−1), at 28°C, for 2–3 days; the second duplicate was wrapped in aluminum foil to block the light, and grown in the same condition. Liquid assays were as previously described (Zuo et al., 2011).
Protein analyses
To analyze blue light-induced CRY2 degradation, seedlings were grown on MS medium for 6 days in the dark, and exposed to blue light (60 µmole m−2 s−1) for indicated time. Total proteins were extracted, fractionated in 10% SDS-PAGE gels, and transferred to nitrocellulose membranes for immunoblots. The blots were probed by anti-CRY2 antibody or anti-HSP90 antibody for the loading control. The immunoblot signals were quantified using ImageJ (http://rsb.info.nih.gov/ij/), the relative level of CRY2 is calculated by normalization of the “blue light” signal with the dark control as described (Yu et al., 2007b).
To investigate the blue-light induced CRY2 phosphorylation, seedlings were grown on MS solid medium for 6 days in the dark, and then exposed to blue light (60 µmol m−2 s−1) for 30 min. To show the fine resolution of migration alterations resulting from protein phosphorylation, low voltage (120v) was used and the gel was run for about 4 hours before transferring to nitrocellulose membranes for immunoblots. The blot was probed with anti-CRY2 antibody to show the CRY2 level, and the mobility upshift of CRY2 indicates phosphorylation.
Supplementary Material
Acknowledgments
This work is supported in part by National Institute of Health (GM56265 to CL, GM089778 to JAW), research funds from Fujian Agriculture and Forestry University (to BFPC), the Jilin University (research support to LSPBG), the MOA Transgenic Research Grant (2010ZX08010-002 to BL), and National Natural Science Foundation of China (31171176 to XY and 31422041 to BL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366:162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. doi: 10.1126/science.1096973. [DOI] [PubMed] [Google Scholar]
- Cashmore AR. Cryptochromes: enabling plants and animals to determine circadian time. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annual Review of Plant Biology. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- Chaves I, Yagita K, Barnhoorn S, Okamura H, van der Horst GT, Tamanini F. Functional evolution of the photolyase/cryptochrome protein family: importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol Cell Biol. 2006;26:1743–1753. doi: 10.1128/MCB.26.5.1743-1753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociorva D, D LT, Yates JR. Validation of tandem mass spectrometry database search results using DTASelect. Current protocols in bioinformatics/editoral board, Andreas D. Baxevanis … [et al. 2007;Chapter 13(Unit 13):14. doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Gould KL, Gygi SP, Kellogg DR. Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Molecular Biology of the Cell. 2013;24:535–542. doi: 10.1091/mbc.E12-09-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC. Mutant Analyses Define Multiple Roles for Phytochrome C in Arabidopsis Photomorphogenesis. The Plant Cell Online. 2003;15:1981–1989. doi: 10.1105/tpc.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279:1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- He S-B, Wang W-X, Zhang J-Y, Xu F, Lian H-L, Li L, Yang H-Q. The CNT1 Domain of Arabidopsis CRY1 alone Is Sufficient to Mediate Blue-light Inhibition of Hypocotyl Elongation. Molecular Plant. 2015 doi: 10.1016/j.molp.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Hereld D, Vaughan R, Kim JY, Borleis J, Devreotes P. Localization of ligand-induced phosphorylation sites to serine clusters in the C-terminal domain of the Dictyostelium cAMP receptor, cAR1. Journal of Biological Chemistry. 1994;269:7036–7044. [PubMed] [Google Scholar]
- Kaiser P, Wohlschlegel J. Identification of ubiquitination sites and determination of ubiquitin-chain architectures by mass spectrometry. Methods Enzymol. 2005;399:266–277. doi: 10.1016/S0076-6879(05)99018-6. [DOI] [PubMed] [Google Scholar]
- Kelstrup CD, Young C, Lavallee R, Nielsen ML, Olsen JV. Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole orbitrap mass spectrometer. J Proteome Res. 2012;11:3487–3497. doi: 10.1021/pr3000249. [DOI] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Analytical Chemistry. Hunan University; 2012. The Biochemical Study of Arabidopsis Blue Light Receptor CRY2 Photoactivation Mechanism and function. [Google Scholar]
- Lin C, Shalitin D. Cryptochrome Structure and Signal transduction. Annu. Rev. Plant Biol. 2003;54:469–496. doi: 10.1146/annurev.arplant.54.110901.160901. [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci U S A. 1998;95:2686–2690. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Zuo Z, Liu H, Liu X, Lin C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes & development. 2011a;25:1029–1034. doi: 10.1101/gad.2025011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends in plant science. 2011b;16:684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yu X, Li K, Klejnot J, Yang H, Lisiero D, Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- Liu Q. PhD Thesis. College of Plant Science: Jilin University; 2015. A study of the CIK kinases that phosphorylate photoexcited cryptochromes in Arabidopsis. [Google Scholar]
- Liu Y, Li X, Li K, Liu H, Lin C. Multiple bHLH Proteins form Heterodimers to Mediate CRY2-Dependent Regulation of Flowering-Time in <italic>Arabidopsis</italic>. PLoS Genet. 2013;9:e1003861. doi: 10.1371/journal.pgen.1003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Li H, Wang Q, Liu B, Lin C. Blue-Light, Dependent Interaction between Cryptochrome2 and CIB1 Regulates Transcription and Leaf Senescence in Soybean. The Plant Cell Online. 2013;25:4405–4420. doi: 10.1105/tpc.113.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalski A, Damoc E, Hauschild JP, Lange O, Wieghaus A, Makarov A, Nagaraj N, Cox J, Mann M, Horning S. Mass spectrometry-based proteomics using Q 20 Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Molecular & cellular proteomics : MCP. 2011;10 doi: 10.1074/mcp.M111.011015. M111 011015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockler T, Yang H, Yu X, Parikh D, Cheng YC, Dolan S, Lin C. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci U S A. 2003;100:2140–2145. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Chalkley RJ, Pham TND, Guan S, Maltby DA, Burlingame AL, Wang Z-Y, Quail PH. Multisite Light-Induced Phosphorylation of the Transcription Factor PIF3 Is Necessary for Both Its Rapid Degradation and Concomitant Negative Feedback Modulation of Photoreceptor phyB Levels in Arabidopsis. The Plant Cell Online. 2013;25:2679–2698. doi: 10.1105/tpc.113.112342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Sancar A. Analysis of autophosphorylating kinase activities of Arabidopsis and human cryptochromes. Biochemistry. 2006;45:13369–13374. doi: 10.1021/bi061556n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partch CL, Clarkson MW, Ozgur S, Lee AL, Sancar A. Role of structural plasticity in signal transduction by the cryptochrome blue-light photoreceptor. Biochemistry. 2005;44:3795–3805. doi: 10.1021/bi047545g. [DOI] [PubMed] [Google Scholar]
- Peck SC. Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. The Plant Journal. 2006;45:512–522. doi: 10.1111/j.1365-313X.2005.02613.x. [DOI] [PubMed] [Google Scholar]
- Rosato E, Codd V, Mazzotta G, Piccin A, Zordan M, Costa R, Kyriacou CP. Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of CRY. Curr Biol. 2001;11:909–917. doi: 10.1016/s0960-9822(01)00259-7. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A Versatile Nanotrap for Biochemical and Functional Studies with Fluorescent Fusion Proteins. Molecular & Cellular Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Sanada K, Harada Y, Sakai M, Todo T, Fukada Y. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes Cells. 2004;9:697–708. doi: 10.1111/j.1356-9597.2004.00758.x. [DOI] [PubMed] [Google Scholar]
- Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ. N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell. 2005;17:1569–1584. doi: 10.1105/tpc.104.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Molecular & cellular proteomics : MCP. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature. 2002;417:763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- Shalitin D, Yu X, Maymon M, Mockler T, Lin C. Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell. 2003;15:2421–2429. doi: 10.1105/tpc.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zeng Y, Zhuang X, Sun L, Yao X, Pimpl P, Jiang L. Organelle pH in the Arabidopsis Endomembrane System. Molecular Plant. 6:1419–1437. doi: 10.1093/mp/sst079. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S-T, Dai C, Liu H-T, Xue H-W. Arabidopsis Casein Kinase1 Proteins CK1.3 and CK1.4 Phosphorylate Cryptochrome2 to Regulate Blue Light Signaling. The Plant Cell Online. 2013;25:2618–2632. doi: 10.1105/tpc.113.114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taus T, Kocher T, Pichler P, Paschke C, Schmidt A, Henrich C, Mechtler K. Universal and confident phosphorylation site localization using phosphoRS. J Proteome Res. 2011;10:5354–5362. doi: 10.1021/pr200611n. [DOI] [PubMed] [Google Scholar]
- Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294:154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- Xu T, Venable JD, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR., III ProLuCID, A Fast and Sensitive Tandem Mass Spectra-based Protein Identification Programs. Molecular & Cellular Proteomics. 2006;5:S174. [Google Scholar]
- Yang H-Q, Wu Y-J, Tang R-H, Liu D, Liu Y, Cashmore AR. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103:815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- Yang HQ, Tang RH, Cashmore AR. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell. 2001;13:2573–2587. doi: 10.1105/tpc.010367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C. Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell. 2007a;19:3146–3156. doi: 10.1105/tpc.107.053017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C. Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. Plant Cell. 2009;21:118–130. doi: 10.1105/tpc.108.061663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Shalitin D, Liu X, Maymon M, Klejnot J, Yang H, Lopez J, Zhao X, Bendehakkalu KT, Lin C. Derepression of the NC80 motif is critical for the photoactivation of Arabidopsis CRY2. Proc Natl Acad Sci U S A. 2007b;104:7289–7294. doi: 10.1073/pnas.0701912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu X, Foo E, Symons GM, Lopez J, Bendehakkalu KT, Xiang J, Weller JL, Liu X, Reid JB, et al. A study of Gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Phy. 2007;145:106–118. doi: 10.1104/pp.107.099838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Conte F, Green CB. Nuclear localization and transcriptional repression are confined to separable domains in the circadian protein CRYPTOCHROME. Curr Biol. 2003;13:1653–1658. doi: 10.1016/j.cub.2003.08.033. [DOI] [PubMed] [Google Scholar]
- Zuo Z, Liu H, Liu B, Liu X, Lin C. Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Current biology. 2011;21:841–847. doi: 10.1016/j.cub.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z-C, Meng Y-Y, Yu X-H, Zhang Z-L, Feng D-S, Sun S-F, Liu B, Lin C-T. A Study of the Blue-Light-Dependent Phosphorylation, Degradation, and Photobody Formation of Arabidopsis CRY2. Molecular Plant. 2012;5:726–733. doi: 10.1093/mp/sss007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.