Abstract
Ganglion cells (GCs), the retinal output neurons, receive synaptic inputs from bipolar and amacrine cells in the inner plexiform layer (IPL) and send information to the brain nuclei via the optic nerve. Although GCs constitute less than 1% of the total retinal cells, they occur in numerous types and are the first neurons formed during retinal development. Using Brn3a and Brn3b mutant mice in which the alkaline phosphatase gene was knocked-in (Badea et al. [Neuron] 2009;61:852–864; Badea and Nathans [Vision Res] 2011;51:269–279), we studied the general effects after gene removal on the retinal neuropil together with the consequences of lack of development of large numbers of GCs onto the remaining retinal neurons of the same class. We analyzed the morphology, number, and general architecture of various neuronal types presynaptic to GCs, searching for changes secondary to the decrement in the number of their postsynaptic partners, as well as the morphology and distribution of retinal astrocytes, for their strong topographical relation to GCs. We found that, despite GC losses, retinal organization in Brn3 null mice is remarkably similar to that of wild-type controls.
INDEXING TERMS: Brn3 transcription factors, ganglion cells, inner plexiform layer, mosaics, synapses, RRIDs: AB_94166, AB_2314052, AB_10000340, AB_2079751, AB_399431, AB_2533912, AB_2278725, AB_477035, AB_477345, AB_2492226, AB_390204, AB_10013783
Retinal ganglion cells (GCs) are the only output neurons of the retina, carrying visual information to retinorecipient centers of the brain. They integrate and process data collected in the inner plexiform layer (IPL) from amacrine and bipolar cells. During retinal development, GCs are the first neurons that exit the cell cycle. In the mouse, they start migrating toward their final location as early as embryonic day 11.5 (E11.5). Although their absolute number accounts for less than 1% of the total cells of the adult mouse retina, GCs occur in a large variety of types—21 morphologically distinct GC types have been described for the mouse, as for most mammalian retinas. Differences in number, shape, size, and distribution over the retinal surface, molecular determinants, central projections, and physiological properties distinguish the various cell types (Roska and Werblin, 2001; Rockhill et al., 2002; Wässle, 2004; Badea and Nathans, 2011; Masland, 2012a). It is believed that each one of these features is determined during development by a combination of extrinsic factors, which include growth factors, signaling molecules and hormones, and the strict control of intrinsic components, such as transcription factors, adhesion and intracellular signaling molecules, etc. (Siegert et al., 2009).
Math5 and Brn3b are the master regulators for the specification of GCs, being expressed as early as E11.5 in the mouse (Brown et al., 1998). This coincides with an age when retinal precursors differentiate into GCs. Brn3a and Brn3c are transcription factors highly specific for GCs in the retina. They are strictly related to Brn3b in both structure and function, as their DNA binding motif shares 95% similarity (Liu et al., 2000).
Various mouse models carrying mutations in the Brn3 genes have been generated (Erkman et al., 1996; Gan et al., 1996; Xiang et al., 1996, 1997). Brn3b null animals display 70% reduction in the number of all GCs, with the remaining ones showing intra- and extraretinal axon guidance defects (Badea et al., 2009a). The effects of Brn3a deletion were initially impossible to study because mice homozygous for a null mutation die at birth, probably due to severe defects in the dorsal root and trigeminal ganglia (Xiang et al., 1996). In contrast, Brn3c null mice do not show any significant retinal defect (Xiang et al., 1997; Badea and Nathans, 2011). Using a Cre-dependent conditional knockout (KO) approach coupled with expression of a histochemical reporter, alkaline phosphatase (AP), Brn3aCKOAP/KO and Brn3bCKOAP/KO mice were generated in which it was possible to test the effects of removing each of the Brn3 genes on the GCs and on the entire retina. This approach showed that ablation of Brn3a causes about a 30% decrease in the number of GCs and major stratification defects of their dendrites in the IPL (Badea et al., 2009a; Shi et al., 2013). Comparisons between the Brn3aCKOAP/KO and Brn3bCKOAP/KO strains revealed how different combinations of Brn3 transcription factors contribute to generate specific attributes of GC types.
The present study provides a systematic examination of the retina of the Brn3aCKOAP/KO and Brn3bCKOAP/KO mice described above, analyzed from the perspective of the input neurons to GCs, with an investigation into whether they had undergone structural rearrangements due to major changes in the number and morphology of their postsynaptic partners. Using specific immunostaining, quantitative neuroanatomy, and electron microscopy, we investigated potential changes and reorganization in the number, architecture, and networks established by bipolar and amacrine cells, the physiological presynaptic partners of GCs, also giving an account of the overall synaptic contacts established by these cells in the IPL. Potentially propagated effects to the outer retina organization and to the astrocytic network were studied as well. The analysis was carried out in parallel for Brn3aAP/KO and Brn3bAP/KO mice, with the expectation of differences reflecting strain-specific abnormalities in GCs. Instead, we found that the fine structure of the retina distal to GCs is remarkably similar in the two mutant strains and in their wild-type controls.
MATERIALS AND METHODS
Mouse lines
All experimental procedures were in accordance with the National Eye Institute Animal Care and Use Committee (Animal Study Protocol NEI-640) and with the Italian and European laws regulating the experimental use of animals for research. All mouse lines used in this study were previously characterized: retinal specific Cre expression was achieved using the Pax6α:Cre line (Marquardt et al., 2001); conditional knock-in reporter alleles were Brn3aCKOAP and Brn3bCKOAP mice (Badea et al., 2009a, 2012; Badea and Nathans, 2011); and conventional KO alleles for Brn3a and Brn3b were Brn3aKO (Xiang et al., 1996); and Brn3bKO (Gan et al., 1996). All lines were maintained on a mixed C57Bl6/SV129 background. To generate retinal specific ablation of Brn3a or Brn3b, Pax6α:Cre; Brn3aWT/KO or Pax6α:Cre; Brn3bWT/KO males were crossed with Brn3aCKOAP/CKOAP or Brn3bCKOAP/CKOAP females. Resulting offspring are either Pax6α:Cre; Brn3aCKOAP/WT (Brn3a heterozygote) or Pax6α: Cre; Brn3aCKOAP/KO (Brn3a KO) and Pax6α:Cre; Brn3bCKOAP/WT (Brn3b heterozygote) or Pax6α:Cre; Brn3bCKOAP/KO (Brn3b KO). In these offspring, the Brn3 gene encoded by the conditional allele is replaced by AP specifically at the level of the retina (Brn3aAP). Brn3 heterozygotes (Pax6α:Cre; Brn3aCKOAP/WT, Pax6α:Cre; Brn3bCKOAP/WT), which carry a normal Brn3 allele on the pair chromosome, are phenotypically wild type (WT). In Pax6α:Cre Brn3aCKOAP/KO; and Pax6α:Cre Brn3bCKOAP/KO; mice, the second Brn3 allele is constitutively lost, thus generating a null genotype in the expression domain of the Pax6α:Cre transgene (Marquardt et al., 2001; Badea et al., 2009a). AP expression and full KO phenotypes are achieved in the entire retina with the exception of a V-shaped stripe with dorsoventral orientation (Marquardt et al., 2001). Unless otherwise specified, all the studies illustrated here were restricted to retinal regions with full Cre recombination and therefore complete Brn3a or Brn3b ablation.
Tissue preparation, histology, and immunohistochemistry
For immunohistochemistry (ICCH), adult animals, between 3 and 6 months old, were anesthetized with ketamine and xylazine before sacrifice, and the eyes were enucleated, fixed for 1 hour in 4% paraformaldehyde (PFA) in 0.1 M sodium–phosphate buffer (PB), and then transferred to 0.1 M PB. A reference nasal mark was made on the eyes prior to enucleation, with a small cut in the same location, keeping the orientation identical for all samples. For ICCH experiments on vertical sections, whole eyes were infiltrated gradually in 30% sucrose in 0.1M PB, then embedded in OCT TissueTek (Sakura, Alphen aan den Rijn, NL), frozen at −80 °C, and stored until use. Vertical sections of 12 µm were cut at with cryostat at −20 °C and collected on glass slides. Retinal sections were then washed 3 × 15 minutes in 0.01 M phosphate-buffered saline (PBS), blocked in a solution containing 5% serum of the species in which the secondary antibody was raised, and 0.3% Triton X-100, for 2 hours at room temperature. Then the sections were incubated overnight at 4 °C in the primary antibody mixture containing 1% serum, 0.1% Triton X-100, followed by multiple PBS rinses and incubation with the secondary antibody mixture, diluted as above. Secondary antibodies were: Alexa 488 anti-mouse and anti-rabbit (Life Technologies, Monza, Italy); and Rhodamine Red X anti-mouse, anti-rabbit, and anti-goat (Jackson ImmunoResearch Europe, Newmarket, UK), all diluted 1:1,000. For nuclear counterstaining, the sections were incubated for 2–5 minutes in Hoechst 33258 (Sigma-Aldrich, Milan, Italy), in YOYO or in ethidium homodimer 1 (both from Life Technologies), all diluted 1:1,000. The slides were mounted in Vectashield (H1000; Vector, Burlingame, CA) and cover-slipped. Images were obtained on a Leica TCS SL confocal microscope (Leica Microsystems, Milan, Italy) equipped with an argon and a helium/neon laser, using a 40×/1.25 HCX PL APO oil objective. Additional images were obtained with a Zeiss Imager.Z2 microscope equipped with an Apotome 2 device (Zeiss, Milan, Italy), using a 40×/0.75 EC Plan Neofluar air, a 40×/1.25 Plan Neofluar oil, and a 63×/1.4 Plan Apochromat oil objective. Brightness and contrast of images were adjusted with Adobe Photoshop (San Jose, CA). Image analysis was done with MetamorphV® software (v. 5.0r1 Meta-morph, Universal Imaging, Downingtown, PA) or with the Zeiss software ZENV®PRO 2012.
As a rule, cryostat retinal sections from Pax6α:Cre; Brn3CKOAP/WT and Pax6α:Cre; Brn3CKOAP/KO mice were collected on the same slide, to ensure comparisons of matching retinal locations and eccentricities and to minimize handling differences during the ICCH procedures, which followed standardized protocols. Microscope acquisition parameters determining resolution and thickness of synthetic focus images were kept constant for KO and WT specimens used for comparisons; all measurements were repeated at least 3 times for each sample studied, on more than 3 biological replicates (four images per section–two at peripheral and two at central locations, always avoiding the area of incomplete recombination, for a minimum of three sections per retina/mouse.
For whole-mount ICCH, the retinas were isolated from eye cups, the vitreous was removed, and four cuts were made to delimitate the four quadrants. After extensive washes in PBS, the retinas were blocked overnight at 4 °C in a solution containing 0.5% Triton X-100 and 5% serum of the donor species of the secondary antibody. The specimens were then incubated for 3–5 days at 4 °C with the primary antibodies against choline acetyltransferase (ChAT), tyrosine hydroxylase (TH), RNA binding protein with multiple splicing (RBPMS), and glial fibrillary acidic protein (GFAP) antibodies to label cholinergic, dopaminergic amacrine cells, GCs and astrocytes, respectively. Primary antibodies (used at the concentrations shown in Table 1) were diluted with 5% serum and 0.3% Triton X-100. After three washes of 30 minutes each in PBS, the retinas were incubated in the secondary antibody diluted 1:1,000, washed as above, and finally mounted “ganglion cells up” in Vectashield medium. Retinas used for ChAT and RBPMS staining were also incubated in ethidium homodimer 1 diluted 1:1,000, for 2 hours at room temperature. For TH and GFAP ICCH, 1:1,000 ethidium or YOYO were used for nuclear counterstaining, depending on the fluorescence of the secondary antibody used.
TABLE 1.
Antibody Characterization
| Antibody name | Immunogen structure | Manufacturer | Species | Cat. no, lot no., RRID | Dilution | Cell type labeled |
|---|---|---|---|---|---|---|
| AP | Placental alkaline phosphatase | Ghent | Mouse monoclonal anti-AP | VIB E6 clone | 1:100 | GCs |
| Brn3a | Amino acids 186–224 of Brn3a fused to the T7 gene 10 protein |
Merck Millipore/Merck KGaA, Darmstadt, Germany |
Mouse monoclonal anti-Brn3a |
MAB1585, 2089417, RRID:AB_94166 |
1:10 | GCs |
| Brn3b | Human Brn3b amino acids 184–252 protein 10 fusion (pGEMEX) |
(Xiang et al., 1993, 1995) | Rabbit polyclonal anti-Brn3b | 1:20 | GCs | |
| Cabp5 | Calcium binding protein 5 bacterially expressed in New Zeeland rats (Haeseleer et al., 2000, Liu et al., 2009) |
Gift of F. Haeseleer and K. Palczewski |
Rabbit polyclonal anti-calcium binding protein 5 (CaBP5) |
RRID:AB_2314052 | 1:200 | CBCs + a few RBCs |
| Calbindin | Recombinant rat calbindin D-28k (CB) | SWANT, Bellinzona, Switzerland |
Rabbit polyclonal anti-calbindin D-28k |
CB38a, 9.03 RRID:AB_10000340 |
1:500 | HCs, ACs |
| ChAT | Human placental enzyme (choline acetyltransferase) |
Merck Millipore/Merck KGaA |
Goat polyclonal anti-ChAT | AB144P, 2280814, RRID:AB_2079751 |
1:800; 1:200 (WM) |
Synaptic molecules |
| Ctbp2/Ribeye | Sequence of amino acids 361–445 from mouse Ctbp2 (ID: 13017) |
BD Transduction Laboratories, Milan, Italy |
Mouse monoclonal anti-Ctbp2 |
612044, 3087931, RRID:AB_399431 |
1:500 | Ribbon synapses |
| Connexin36 | Peptide corresponding to a sequence located in the cytoplasmic loop between the second and third transmembrane domains of rat and mouse Connexin 36. (ID: 57369) |
Invitrogen, Carlsbad, CA | Rabbit polyclonal anti-Cx36 | 51–6200, 983780A, RRID:AB_2533912 |
1:500 | Gap junctions |
| GAD 67 | Recombinant glutamic acid decarboxylase (GAD)67 protein (ID: 2571) |
Merck Millipore/Merck KGaA |
Mouse monoclonal anti-GAD67 |
MAB5406, 2581753, RRID:AB_2278725 |
1:500 | ACs |
| GFAP | Glial fibrillary acidic protein from human brain |
Sigma-Aldrich St. Louis, MO |
Rabbit polyclonal anti-GFAP |
G9269, 129H4869, RRID:AB_477035 |
1:200 (WM) | Astrocytes |
| PKCα | Synthetic peptide corresponding to amino acids 659–672 from the C- terminal variable (V5) region of rat protein kinase C (PKC) α conjugated to KLH |
Sigma-Aldrich | Rabbit polyclonal anti-PKCα |
P4334, 033K4852, RRID:AB_477345 |
1:800 | RBCs |
| RBPMS | Synthetic peptide corresponding to amino acid residues from the N- terminal region of the rat RNA binding protein with multiple splicing (RBPMS) sequence conjugated to KLH (ID: 19663) |
PhosphoSolutions, Aurora, CO |
Guinea pig polyclonal antibody anti-RBPMS |
1832, NB6, RRID:AB_2492226 |
1:500 | ACs |
| TH | Denatured tyrosine hydroxylase from rat pheochromocytoma |
Merck Millipore/Merck KGaA |
Rabbit polyclonal anti-TH |
AB152, RRID:AB_390204 | 1:500; 1:200 (WM) |
ACs |
| ZNP1 | Synaptotagmin 2 from zebrafish homogenates |
Zebrafish International Resource Center, Eugene, OR |
Mouse monoclonal anti-synaptotagmin 2 (ZNP-1) |
ZNP-1, RRID:AB_10013783 | 1:1,000 | CBCs |
Abbreviations: AC, amacrine cell; CBC, cone bipolar cell; GC, ganglion cell; HC, horizontal cell; RBC, rod bipolar cell.
Retinal profiles of whole-mount preparations were imaged in brightfield with a Zeiss Axiocam camera attached to a Zeiss Axioplan microscope, using a 1.25 × objective. Retinal areas were then measured on image TIFF files using a Metamorph edge detector tool. These measures were obtained before and after confocal analysis of each retinal sample; the two measures were then averaged to compensate for possible tissue compression during prolonged microscope examination.
Electron microscopy
For electron microscopy (EM) studies, the animals were anesthetized and perfused through the heart with a solution of 2.5% glutaraldehyde and 2% PFA in 0.1 M PB; their eyes were enucleated and fixed for an additional hour in the same fixative. Subsequently, the retinas were separated from the choroid and dissected into eight triangular fragments, two for each quadrant, which were washed extensively in PB. The retinal blocks were postfixed for 2 hours at 4 °C in cold 2% osmium tetroxide (OsO4), then washed for 4 × 15 minutes in cold sodium maleate buffer 0.05 M, pH 5.15, and bloc stained with 2% uranyl acetate in the same buffer, for 1 hour at 4 °C. After an additional wash in maleate buffer, the blocks were dehydrated in an ascending series of ethanol for a total of 2 hours. Subsequently, the blocks were immersed in propylene oxide (2 × 10 minutes), infiltrated overnight in a mixture of 50:50 propylene oxide:Embed 812/Araldite plastics, and finally embedded in Embed 812/Araldite (Electron Microscopy Sciences, Hatfield, PA). Ultrathin sections were obtained with a Leica Ultracut ultramicrotome equipped with a diamond knife. Two separate series of sections were cut from central, nasal retinal areas of each of the Pax6α:Cre; Brn3bCKOAP/KO mice and controls. The sections were collected on 400-mesh grids, stained with uranyl acetate and lead citrate, and examined with a Jeol 1200EX II electron microscope (Jeol, Milan, Italy). Images were obtained with a charge-coupled device (CCD) Olympus Veleta Megaview (Olympus, Münster, Germany) camera at a magnification of 40,000×, covering systematically the entire width of the inner plexiform layer (IPL) at two adjacent positions per each sample. Image files were saved as TIFF, transferred to an image analysis workstation, and examined offline.
Antibody characterization
Table 1 lists the antibodies used in the present study.
Mouse monoclonal anti-AP (Flanders Institute for Biotechnologies, Ghent, Belgium), E6 clone, is an affinity-purified mouse monoclonal antibody recognizing only the human placental AP isoform.
Mouse monoclonal anti-Brn3a (Chemicon Millipore, Temecula, CA, MAB1585) was raised against human Brn3a amino acids 186–224/protein 10 fusion (pGEMEX). It recognizes specifically the Brn3a protein on western blots, but does not cross-react with Brn3b or Brn3c. It stains specifically retinas from WT but not Brn3a KO mice.
Rabbit polyclonal anti-Brn3b antiserum was previously described (Xiang et al., 1993, 1995). It was raised against a polypeptide from human Brn3b fused to T7 bacteriophage protein 10 (via the pGEMEX system) and affinity-purified using the same respective peptide fused with maltose binding protein (New England Biolabs, Ipswich, MA). The antiserum does not cross-react on western blots for the three proteins, and does not stain retinas from the Brn3b KO mice.
Rabbit polyclonal anti-calcium-binding protein 5 (Cabp5) was raised against bacterially expressed Cabp5 in New Zealand rats (Haeseleer et al., 2000). It detects a band of ~16–18 kDa on western blots. In the mouse retina, the Cabp5 antibody labels the rod bipolar cells and type 3 OFF and type 5 ON of cone bipolar cells (Haeseleer et al., 2000; Haverkamp et al., 2003).
Rabbit polyclonal anti-calbindin D-28K antibody (CB38a; Swant, Bellinzona, Switzerland) detects a band of approximately 27–28 kDa in western blots on brain extracts from different species, including mouse and rat, as well as a higher band, corresponding to calretinin (~29–30 kDa) when the concentration of the preparation reaches 10-fold higher protein concentrations then necessary to detect calbindin (Bardet et al., 2006). The specificity of the antibody in the rodent retina, including mouse, was proved by several studies to be in the horizontal and amacrine cells (Wässle et al., 1998; Haverkamp and Wässle, 2000; Strettoi et al., 2002).
Goat polyclonal anti-ChAT (Chen et al., 2007) shows bands of ~68 kDa on western blots on brain extracts from several species (rodents, birds, primates). In the retina of the rodents, the antibody detects the cholinergic/starburst amacrine cells (Strettoi et al., 2002; Haverkamp et al., 2009) in an identical pattern as described by initial studies of cholinergic amacrine cells in the rodent retina (Voigt, 1986; Brandon, 1987).
Rabbit anti-connexin 36 (Cx36) recognizes a peptide corresponding to a sequence located in the cytoplasmic loop between the second and third transmembrane domains of rat and mouse (Cx36) (Xu et al., 2014). In the mouse retina, Cx36 is located in the gap junctions in both the outer plexiform layer (OPL) and the inner nuclear layer (INL) (Kihara et al., 2009; Pan et al., 2010; Meyer et al., 2014).
Mouse monoclonal anti-glutamic acid decarboxylase (GAD; E.C. 4.1.1.15) is the enzyme responsible for the conversion of glutamic acid to γ-aminobutyric acid (GABA), the major brain inhibitory transmitter. Two molecular forms of GAD (65 kDa and 67 kDa, 64% amino acid identity) are highly conserved and expressed in the central nervous system (CNS), and are distributed cytoplasmically in the brains of rats and mice (Sheikh, 1999). GAD65 is an amphiphilic, membrane-anchored protein (585 amino acids), encoded on human chromosome 10, and is responsible for vesicular GABA production. GAD67 is cytoplasmic (594 amino acids), encoded on chromosome 2, and seems to be responsible for significant cytoplasmic GABA production.
Specificity of the rabbit anti-GFAP antibody was proved by western blots on lysates from human brain extracts (Encinas et al., 2011). The staining pattern of astrocyte networks was consistent with previous studies in the mouse and rat retina (Gargini et al., 2007; Xue et al., 2011).
The rabbit anti-protein kinase Cα (PKCα) antibody detects an ~80-kDa band on western blots from rat brain extracts or NIH 3T3 mouse fibroblasts (Dhingra et al., 2008). PKCα is a widely used marker for rod bipolar cells in the rat retina (Greferath et al., 1990) as well as the mouse retina (Strettoi et al., 2002; Haverkamp et al., 2003; Gargini et al., 2007).
Mouse monoclonal anti-C-terminal binding protein 2 (Ctbp2)/Ribeye recognizes an ~48-kDa band on western blots on cellular lysate of Ntera2 as well as on brain membrane fractions (Dieck et al., 1998). The specificity of the antibody was proved in several studies in the retina of a variety of species to be in the synaptic ribbons in the photoreceptors and bipolar cells (Schmitz et al., 2001; Jusuf et al., 2006; Puller et al., 2007; Hirano et al., 2007; Jakobs et al., 2008; Koizumi et al., 2011).
The guinea pig polyclonal anti-RBPMS, also known as HERMES, contains one RNA recognition motif (RRM) domain and belongs to the RRM family of RNA binding proteins. RNA binding proteins specific to GCs have been previously identified (Kwong et al., 2009). Recently antibodies against RBPMS have been demonstrated to exclusively label GCs in multiple mammalian species (Rodriguez et al., 2014).
The rabbit polyclonal anti-TH antibody detects an ~62-kDa band on lysate of PC12 cells when western blot is performed. The staining is consistent with previous studies in the retina (Strettoi et al., 2004; Noorian et al., 2011).
The mouse monoclonal anti ZNP-1/synaptotagmin 2 recognizes a band of ~60 kDa in western blots from zebrafish homogenate and on lysates from mouse cerebellum and synaptosomes (Fox and Sanes, 2007). In the rodents retina it has been previously shown to detect both type 2 (Fox and Sanes, 2007; Wässle et al., 2009) and type 6 (Wässle et al., 2009) cone bipolar cells, as well as the horizontal cells in the mouse retina (Fox and Sanes, 2007).
Statistical analysis
Statistical comparisons were run for data obtained from the cell population studies, counts of contacts in the IPL (ribbons, detected with anti-Ctbp2 antibody, and Cnx36-positive gap junctions), and the regularity indexes for the mosaic analyses. Whenever data passed a normality distribution test, comparisons were made using a double-tailed t-test analysis, with a confidence interval (CI) of 95% run on Microsoft Excel software. Number of replicates (n), means, and CIs are detailed below for each analysis.
Comparisons of conventional and ribbon synapses distributions in the two strains studied, based on EM analysis, was obtained by applying the two-sample Kolmogorov–Smirnov test, with a confidence interval of 95%, using the statistics toolbox of Sigmastat.
Cell population studies and morphometric analysis
Retinal whole mounts stained with the antibodies anti-ChAT and anti-TH were used to estimate the number of cholinergic and dopaminergic amacrine cells in KO and control mice, whereas the GABAergic population was analyzed in vertical sections.
ChAT-positive amacrine cells
For ChAT-positive cell counting, four images (covering an area of 375 × 375 µm) were acquired in each of the four retinal quadrants (dorsal, ventral, nasal, and temporal), at decreasing eccentricities from the periphery toward the head of the optic nerve, using a Leica TCS SL confocal microscope with a 40×/1.25 HCX PL APO objective and a resolution of 1,024 × 1,024 pixels. Two separate sets of images were acquired, respectively encompassing ChAT amacrine cells displaced in the ganglion cell layer (GCL) and ChAT cells in the INL. First, serial optical sections, 1 µm apart, covering the thickness of the GCL and of the innermost part of the INL were acquired. Then a projection image of the corresponding retinal width was used for counting ChAT cells. The surface of the retina sampled represents 15–20% of the average mouse retinal area. Images were saved as TIFF files and analyzed offline with the MetaMorph 5.0 r1 routine dedicated to automatic counting. Manual counting on the z stacks was used whenever overlapping cells were encountered. The mean number of cells/mm2 was calculated for each retina, and the total number was obtained by multiplying the mean value by the measured retinal area.
Statistical analysis was performed comparing Pax6α:-Cre; Brn3aCKOAP/KO (n = 3) and Pax6α:Cre; Brn3aCKOAP/WT (n = 4); as well as Pax6α:Cre; Brn3bCKOAP/KO (n = 6) and Pax6α:Cre; Brn3bCKOAP/WT (n = 3).
TH-positive amacrine cells
For dopaminergic (TH-positive), amacrine cell counting, the entire surface of each retina was imaged using a Zeiss Axioscope fluorescence microscope with a 10 × objective and a Zeiss Axiocam CCD camera. Thirty to 40 overlapping images were combined with the Photomerge tool of Adobe Photoshop (RRID:SCR_014199) Cells were counted directly with a MetaMorph manual counting tool. Statistical analysis was performed as above comparing Brn3aKO (n = 3) with Brn3aWT (n = 3); and Brn3bKO (n = 3) with Brn3bWT (n = 3).
GAD67-positive amacrine cells
GABAergic cells were labeled on retinal vertical sections of Brn3KO and WT mice by means of a well-characterized GAD67 antibody. Sections from KO and WT mice with matching orientation and eccentricity were collected on the same slide, immunostained, and imaged at the confocal microscope using a 40×/1.5 oil objective. Five fields per sections (four sections per retina) were imaged, each field covering an extension of 148 µm linear). GAD67-positive cells with somas in the INL were counted in each image displayed on a high-definition monitor; the number of cells/mm was assessed for each retina, and the average number/strain was estimated and compared.
Astrocytes
To obtain quantitative information on astrocyte branching, the entire surface of retinas stained with GFAP antibodies was imaged with a Zeiss Apotome fluorescence microscope using a 10 × objective and obtaining extended-focus images encompassing the width of the astrocyte network in the innermost retinal layer. Images were then assembled with Photomerge as above. One field (1 mm2) was extracted from each retinal quadrant and divided into four subfields. A square grid with 9 horizontal and vertical lines (spaced 0.5 µm apart) was superimposed on each subfield. Intersections of astrocytic processes with the grid lines were counted.
For comparison of Pax6α:Cre; Brn3bCKOAP/KO (n = 3) with Pax6α:Cre; Brn3bCKOAP/WT (n = 3) retinas, because only one retina was available from one Pax6α:Cre; Brn3aCKOAP/KO mouse, this was sampled as above and compared with a Pax6α:Cre; Brn3aCKOAP/WT using individual retinal subfields.
Ganglion cells and displaced amacrine cells
For GC counting, whole-mount retinas from Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3aCKOAP/WT mice stained with anti-RBPMS antibodies and counterstained with ethidium homodimer were imaged with a Zeiss Apotome fluorescence microscope using a 40×/1.5× Plan Neofluar oil objective. As for ChAT-positive cells, four focal series of images (each covering a retinal area of 224 × 168 µm) were acquired in each of the four retinal quadrants at decreasing eccentricities from the periphery toward the ON. Serial optical sections, 1 µm apart, covering the thickness of the GCL were acquired. Projection images of the corresponding retinal width were used for counting RBPMS-positive cells (labeled green) and the nuclei of remaining cells (labeled red), belonging to displaced amacrine cells. Nuclei of blood vessels were excluded based on typical elongated shape and small diameter. Serial optical sections (rather than projection images) were used for counting in central retina areas characterized by high cell density.
Synaptic ribbons and gap junctions
Densities of ribbons and gap junction in the IPL were estimated on confocal images (1 µm thick) of vertical sections from Pax6α:Cre; Brn3bCKOAP/KO (n = 3) and Pax6α:Cre; Brn3bCKOAP/WT (n = 3) retinas stained with Ctbp2/Ribeye and Cx36 antibodies, respectively. Regions of interest of 2,295 µm2 (90 × 25.5-µm rectangles) were drawn on the IPL, encompassing the full thickness of this layer, and using images thresholded for a Metamorph tool based on contrast detection. Identical threshold parameters were applied in parallel on images from KO and WT samples. The density of the punctuated staining was then assessed, using three to five sections (separated by at least 50 µm) per retina. The density of ribbons in the OFF and ON sublaminae of the IPL was also determined. This was done by counting separately Ribeye-positive puncta respectively located in the portion of each image located within the outermost two-thirds and the innermost third of the IPL. Data from Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT samples were compared.
Synapse counting in EM pictures
Ribbon and conventional synapses in the IPL were counted on non-overlapping EM images of retinal sections obtained from Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT mice at similar eccentricities and at two different locations (dorsal and nasal quadrants), for each strain. Images were from retinal locations inside the area of full recombination. Ribbon synapses were identified from a dense (variable length) ribbon in close proximity to the presynaptic membrane and from the unequivocal presence of at least one postsynaptic process. The following criteria were used for the identification of conventional, chemical synapses: the presence of cluster(s) of vesicles abutting the presynaptic membrane; an enlargement of the extracellular space; the presence of synaptic densities; and the clear identification of a postsynaptic process. Counts were done on ~200 electron micrographs (56 µm2 each) per sample, per strain.
Mosaic analysis
The same retinal whole mounts stained with ChAT and TH antibodies were used for mosaic analysis. For ChAT-positive amacrine cells, each retina (n = 3 for all four genotypes analyzed) was sampled at four locations, along the dorsoventral and nasotemporal axis. Sampling fields were 0.035 mm2 (187.5 × 187.5 µm), located in the central and in the peripheral retina. Mosaics of both populations (i.e., in the INL and in the GCL, respectively) of cholinergic amacrine cells were analyzed.
Mosaic analysis of TH-positive amacrine cells was done for each retina at four locations (one for each quadrant), in fields of 1 mm2. Retinas for Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3aCKOAP/WT were n = 3 each; retinas for Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT were n = 3 each.
To each cell encountered in the sampling field, a centroid was assigned, and its X and Y coordinates were generated using the ImageJ image analyzer (Schneider et al., 2012). Two measures of cell tiling, the nearest neighbor index and the Voronoi domains, were computed. The regularity index (RI) was calculated as the ratio between the mean of the nearest neighbor distance and the standard deviation of the cellular array (NNRI = nearest neighbor regularity index and the ratio of the mean Voronoi domain areas for the cellular array and their standard deviation (VDRI = Voronoi domain regularity index) (Raven et al., 2003; Rossi et al., 2003, Whitney et al., 2008; Keeley et al., 2007). RIs from WT and KO in each case were plotted and the confidence intervals shown as error bars. Statistical significance was assessed by the double-tailed t-test.
RESULTS
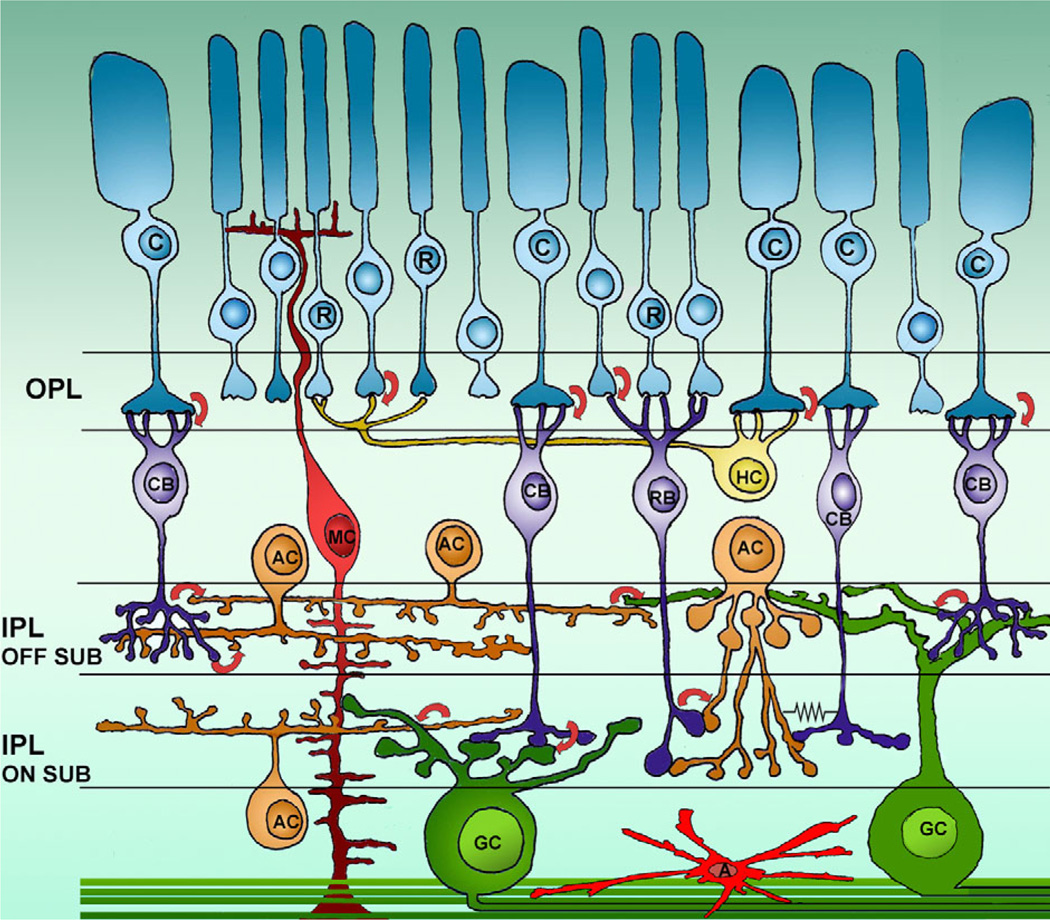
Figure 1 shows a schematic representation of retinal cell types and main synaptic networks. Here, we will first compare GCs in Brn3 KO and WT mice and proceed systematically to describe cells of all the other classes, moving from the outer to the inner retina.
Figure 1.
Retinal cells and wiring diagram. Schematic diagram of retinal cells with their main connections. R and C, rods and cones; RBC and CBC, rod and cone bipolar cells; AC, amacrine cells; GC, ganglion cells; MC, Muller cells; A, astrocytes; OPL and IPL, outer and inner plexiform layers. Arrows indicate synaptic contacts and the direction of communication. The symbol of an electric resistance shows the gap junction between an amacrine cell and a cone bipolar. The two sublaminae (on and off) of the IPL are indicated. Both wide-field and small-field amacrines are represented, as well as examples of cone bipolar cells branching at different depths in the IPL.
Morphology of ganglion cells
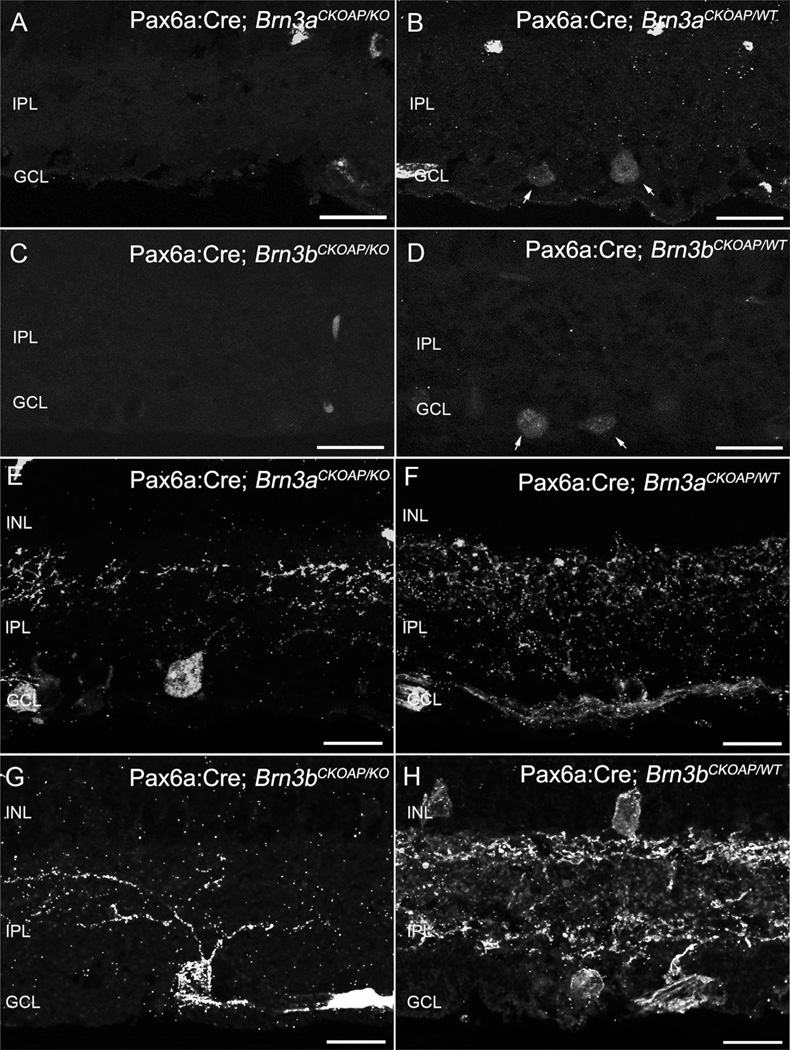
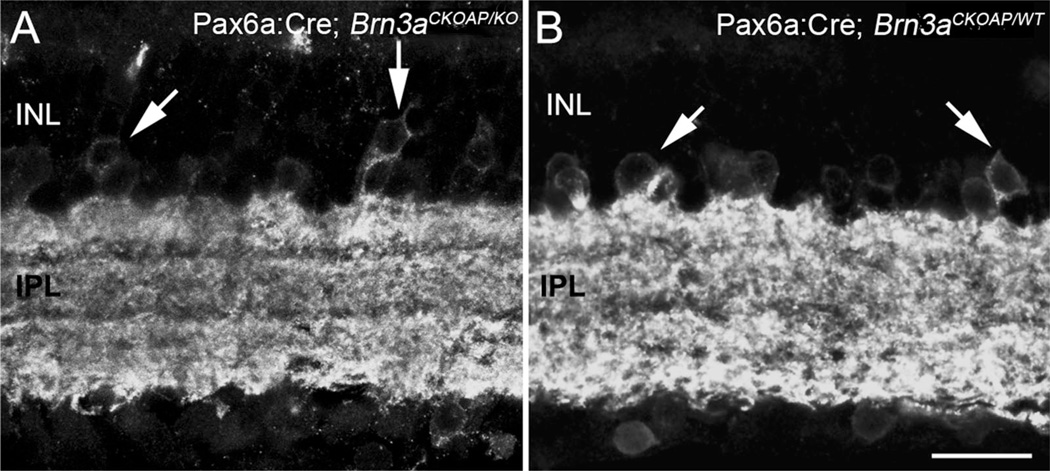
We used immunocytochemistry for AP, Brn3a, and Brn3b, and neurofilament 200 antibody staining (not shown), to confirm decrement in the numbers and altered dendritic morphology of GCs from fully recombined areas of Brn3 KO retinas used for the study. Immunocytochemistry with Brn3a (Fig. 2A,B) and Brn3b (Fig. 2C,D) antibodies shows the lack of staining in retinal areas with the full KO phenotype. AP antibody staining of the reporter gene shows impoverishment of Pax6α:Cre; Brn3aCKOAP/KO GC dendritic arbors (Fig. 2A,E), and visibly reduced numbers of GCs and a net decrement of dendritic staining in the IPL of Pax6α:Cre; Brn3bCKOAP/KO (Fig. 2C,D).
Figure 2.
Ganglion cells. Immunostaining for Brn3a and Brn3b transcription factors shows nuclei of cells located in the GCL layer (A–D). Immunoreactive nuclei are absent from the retinas of KO mice (A,C). Inner plexiform layer (IPL) of Brn3K_O mice (E,G) and of WT control (F,H) immunostained for AP revealing global Brn3a- or Brn3b–positive GCs dendritic arbors. For both KOs (D), loss of dendrites and decreased staining in the IPL are evident. A larger decrement in the number of GCs can be appreciated in the case of the Pax6α:Cre; Brn3bCKOAP/KO (G). Scale bar = 20 µm in A–H.
Morphology of ON and OFF cone bipolar cells
Several bipolar cell markers were used to investigate the effects of GC loss in Brn3 KO retinas on the morphology of cone bipolar cells, as they provide synaptic input to GC dendrites at various depths in the IPL and might remodel as an effect of major GC loss. Synaptotagmin (ZNP1- or Syt2)-positive type 2 and type 6 cone BCs (Pignatelli and Strettoi, 2004; Ghosh et al., 2004) were visualized using ZNP1 mouse monoclonal antibodies (Fox and Sanes, 2007) which highlight their cell bodies at different levels in the INL, their dendritic arborization in the outer retina, and two bands of axonal endings in the IPL. We found that neither cell body position nor the dendritic distribution in the OPL nor the lamination of the axonal arbors of these cone bipolar cells in the OFF and ON sublaminae of the IPL showed alterations or evident abnormalities in the retinas of Brn3 KO mice (green staining in Fig. 3E–G). These cells were indistinguishable from their WT counterparts.
Figure 3.
Bipolar cells. Retinal sections from Brn3KO mice (A,C) and WT controls (B,D) were immunostained for ZNP1 (green) and CaBP5 (red), resulting in labeling of cell bodies, dendritic arbors, and axonal arborizations of several types of cone bipolar cells. Axons of type 2 OFF bipolar and type 6 ON bipolar cells are labeled by ZNP1/Syt2, while CaBP5 marks the axonal processes of rod bipolar, type 3 OFF cone bipolar, and type 5 ON cone bipolar cells. No conspicuous differences in lamination were observed for the five labeled bipolar cell types. ONL, INL, and GCL nuclei were stained with Hoechst nuclear markers. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layers; Scale bar = 20 µm in A–D.
Two additional types of cone bipolar cells, type 3 (OFF) and type 5 (ON), were stained using an anti-Cabp5 antibody (Haverkamp et al., 2003), which labels cell bodies, axons, and axon terminals of these cone bipolar types. Cabp5 also labels brightly some large-size axonal endings of rod bipolar cells (Haverkamp et al., 2003; Haeseleer et al., 2000), resulting in a three-layered pattern of labeling in the IPL. Both the overall pattern of staining produced by Cabp5 antibodies and the detailed morphological features of the labeled cells appeared unaltered in the Pax6α:Cre; Brn3aCKOAP/KO ; and Pax6α:Cre; Brn3bCKOAP/KO; mice (Fig. 3A,C) compared with their WT counterparts (Fig. 3B,D).
Thus, bipolar cells belonging to both the ON and OFF morphological and functional types, known to be presynaptic to GCs in different portions of the IPL, have evidently not remodeled or rearranged in the retina of Brn3 KO mice.
Rod bipolar and horizontal cells
The morphology of horizontal cells and that of rod bipolar cells was used as an additional indication of possible rearrangements occurring in the outer and inner retina of Brn3 KO mice.
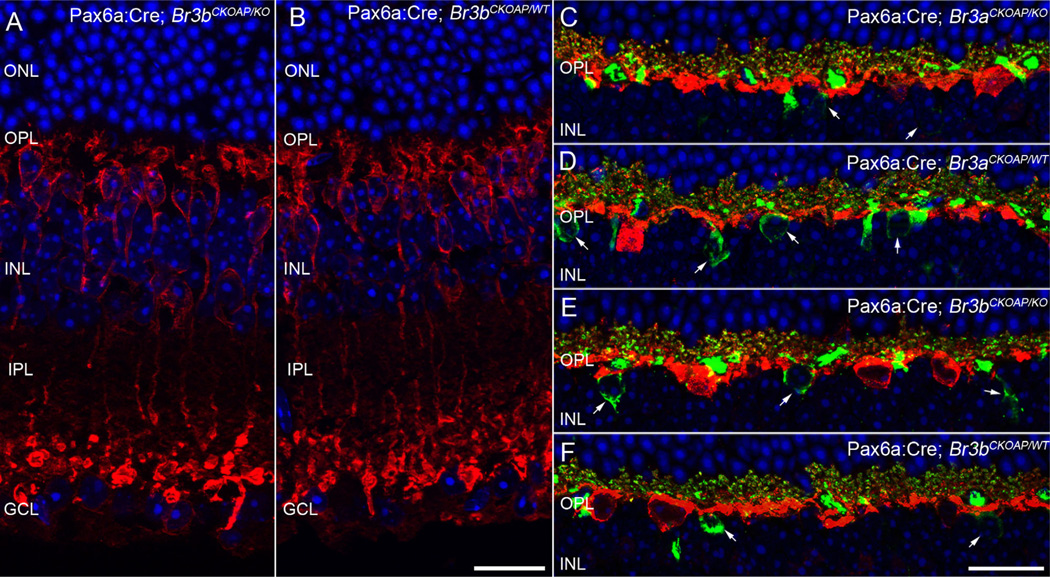
Rod bipolar cells were specifically studied using PKCα antibodies (Strettoi et al., 2002). These neurons are characterized by cell bodies located in the outer half of the INL, profuse dendritic arbors in the OPL, and axons terminating with large, bulbous endings in the innermost part of the IPL, where they intermingle with the apical aspect of the cell bodies of GCs, without establishing synaptic contacts with them. Rod bipolar cells were totally unaffected in their shape, level of stratification, and axonal ending position in the Brn3 KO retinas studied here (Fig. 4A,B).
Figure 4.
Rod bipolar cells and horizontal cells. PKC staining of rod bipolar cells in vertical sections shows the expected array of cell bodies lying in the outer aspect of the INL, sending chandelier-like dendritic arborizations to the OPL. The axons of RBCs run a vertical course to terminate in large, bulbous endings, in the innermost part of the IPL, partially intermingling with cell bodies of GCs. All the main morphological features of RBCs are clearly unvaried in the retina of the Pax6α:Cre; Brn3bCKOAP/KO mouse shown here (A vs B). The dendrites of horizontal cells labeled by calbindin antibodies (red signal) interdigitate with dendrites of cone bipolar cells (arrows) stained by ZNP1 antibodies (green signal). Some horizontal cell processes in the OPL are colabeled and appear yellow. This fine dendritic architecture can be observed in both Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/KO (C,E) as well as control mice (D,F) without visible differences among the strains. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer; OPL, outer plexiform layer. Scale bar = 20 µm in B (also applies to A) and F (applies to C–F).
Calbindin staining, labeling horizontal cells in their entirety, confirms the expected organization of widely spaced horizontal cell bodies from which thick, primary dendrites emerge with a tangential course in the OPL and is undistinguishable between KO mice and WT controls (red signal in Fig. 4). A combination of calbindin and ZNP1 labeling to reveal the details of cone bipolar and horizontal cell dendritic arbors shows processes of the two cell types forming a fine mesh in the OPL, and is identical in KO mice and WT controls (see ZNP1 in green and calbindin labeling in red in Fig. 4).
Selected amacrine cell numbers and mosaic regularity
Cholinergic (starburst), dopaminergic, and GABAergic amacrine cells were analyzed quantitatively as single populations, using anti-ChAT, anti-TH, and anti-GAD67 antibodies.
ChAT cells represent about 5% of the total number of amacrine cells in a C57B6 mouse retina (Jeon et al., 1998; Whitney et al., 2008) and are comprised of two mirror-symmetric populations whose cell bodies are located in the INL or displaced in the GCL, respectively. Their dendritic arbors form two plexa correspondingly positioned at one-third and two-thirds of the IPL, in the OFF and ON sublaminae, respectively.
Dopaminergic (interplexiform) amacrine cells that can be stained with TH antibodies in the mouse retina constitute a single type (type 1). These are wide-field neurons with large, sparse cell bodies in the INL and processes forming a thin plexus rigorously stratified in the outermost tier (S1) of the IPL, in sublamina OFF. Additional loose ramifications with an irregular course can be visualized at various depths in the IPL, while a rich plexus of thin processes is clearly visible in the OPL (Contini and Raviola, 2003). These amacrine cells are numerically few (~600 cells in the mouse retina), but their dendrites cover the retina profusely and their spatial arrangement has been studied in detail (Badea et al., 2009b; Keeley and Reese, 2010b).
GABAergic amacrine cells represent 25.4% among the amacrine cells displaced in the GCL, and their percentage in the INL is estimated to be 40% in the human (Crooks and Kolb, 1992), 33–55% in the monkey (Nishimura et al., 1985; Grünert and Wässle, 1990; Koontz and Hendrickson, 1990), 25–40% in the cat (Pourcho, 1980; Wässle and Chun, 1989), 38% in the rabbit (Strettoi and Masland, 1996), and ~21.3% in the mouse (May et al., 2008).
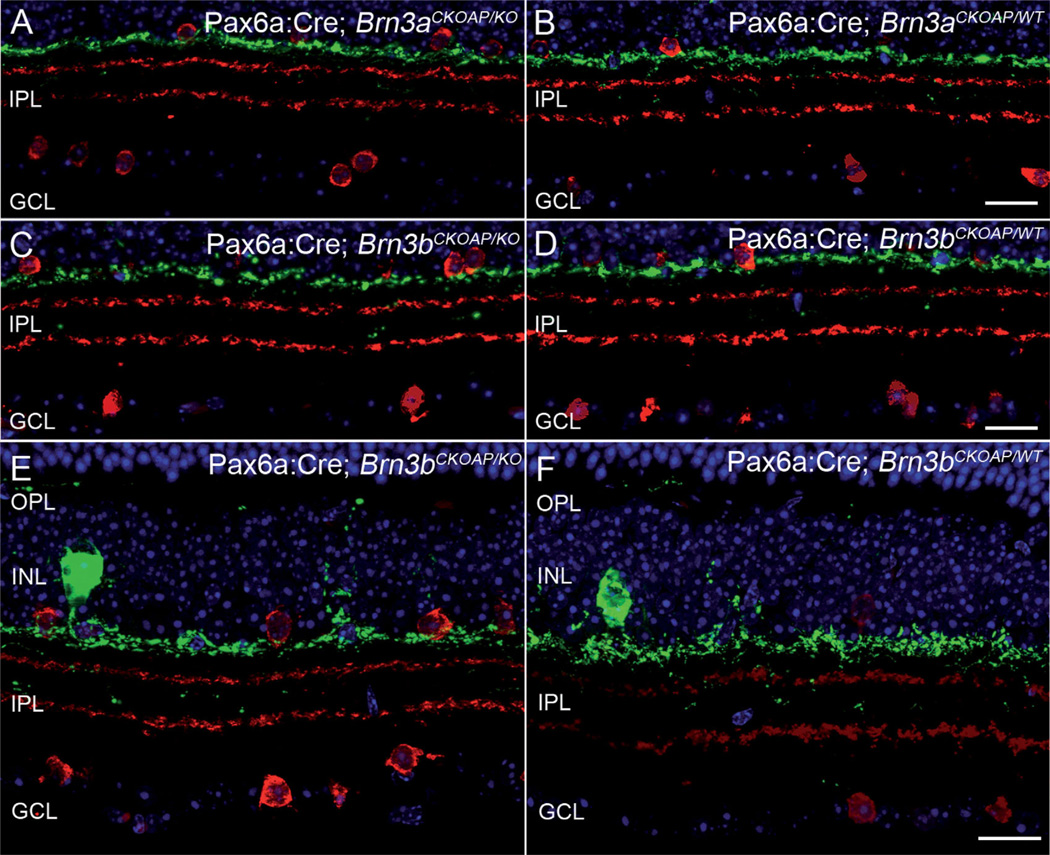
ChAT- and TH-positive amacrine cells were first analyzed in retinal vertical sections. No lamination differences between the retinas of KO and WT mice were found (Fig. 5).
Figure 5.
ChAT and TH amacrines. Stratification of amacrine cells, labeled by ChAT (red) and TH (green). The green-labeled plexus in S1 corresponds to the dendrites of TH cells in the IPL, while the two red plexa are the well-known ChAT bands, located at 1/3 and 2/3 of the IPL depth. Both Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/KO mice (A,C) show identical amacrine cell patterning indistinguishable from that of controls (B,D). Same preparations as above, at slightly higher magnification, showing the thin plexus of TH processes in the outer retina, as well as rare dendritic profiles in the deeper part of the IPL. Staining of amacrine cells in the Pax6α:Cre; Brn3bCKOAP/KO mouse (E) matches that of the control (F) completely. GCL, ganglion cell layer; INL, inner nuclear layer; IPL, inner plexiform layer; OPL, outer plexiform layer. Scale bar = 20 µm in D (applies to A–D) and F (applies to E,F).
Considering the recombination pattern of the Pax6α: Cre; Br3aCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/KO retinas, the density of both ChAT cell populations were analyzed initially in the nasal and temporal quadrants of the retinas only (data not shown). As the analyses yielded similar results among the strains compared, we extended the analysis of ChAT cells over the entire retinal surface. However, the examples chosen (Fig. 6A,B,D,E,G,J) are from the nasal quadrant of the retina.
Figure 6.
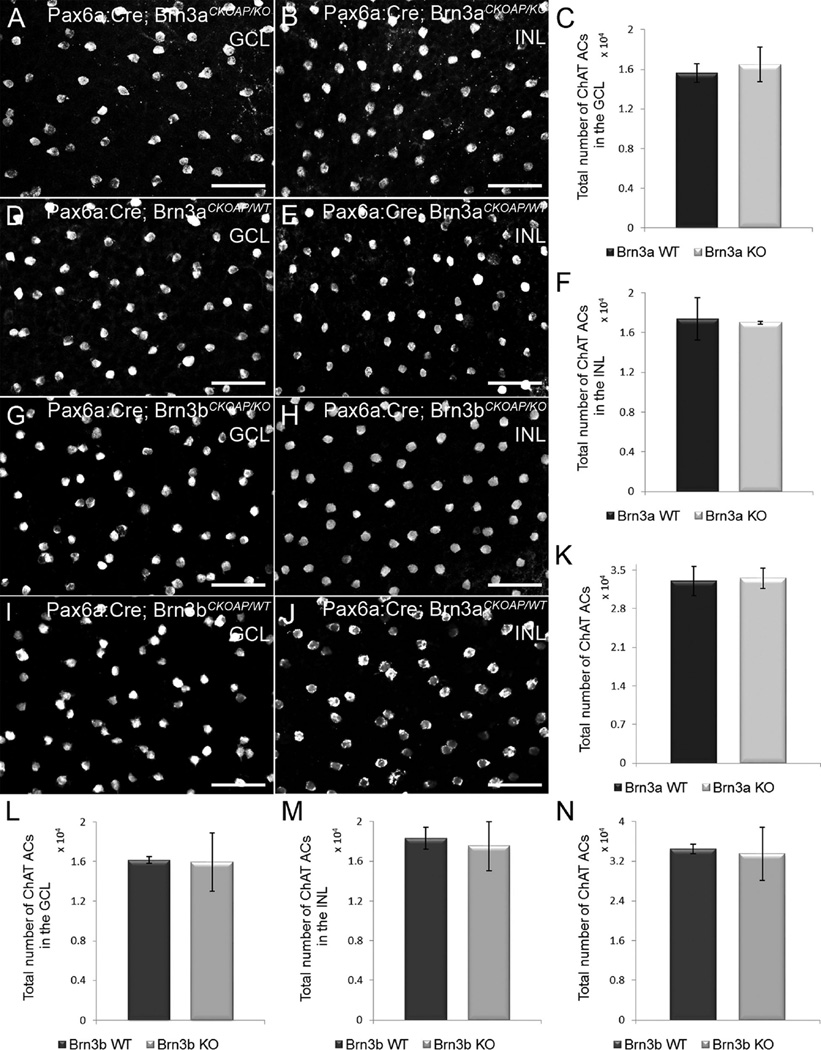
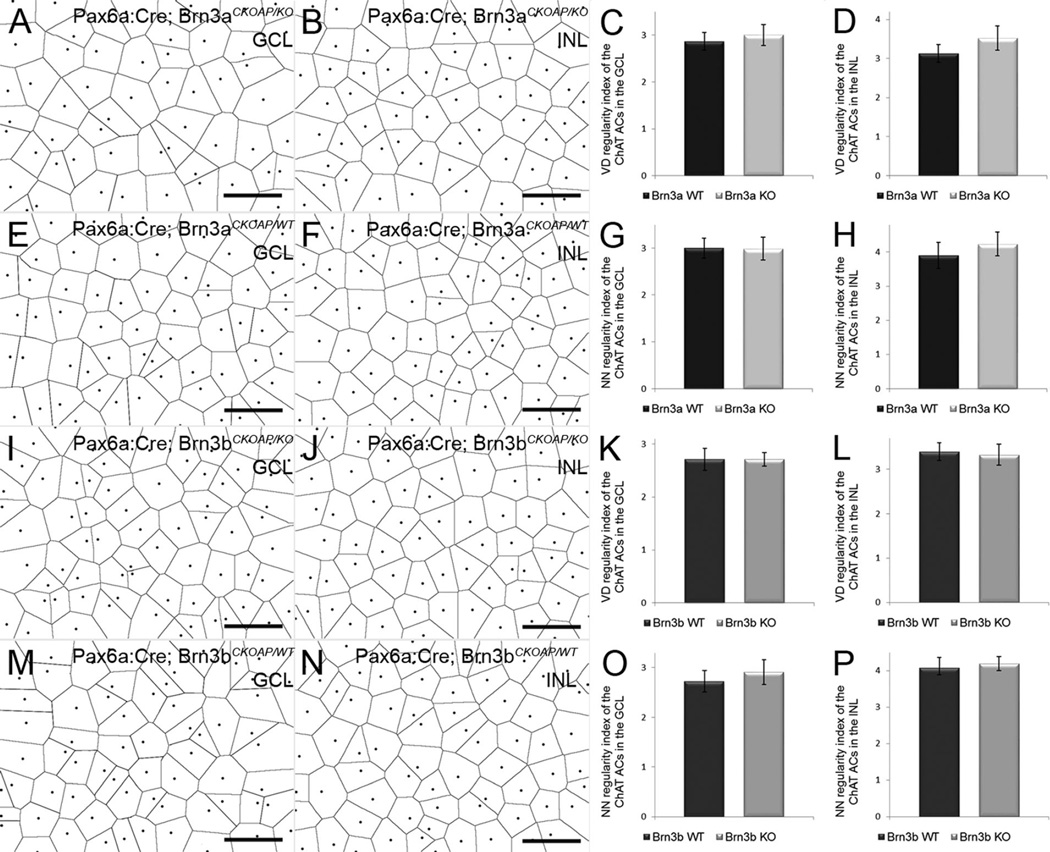
Population analysis of ChAT amacrine cells. Whole-mount immunostaining for ChAT was used to assess their numbers in the GCL (A,D,G,I) and in the INL (B,E,H,J). There are no differences in the number of ChAT-positive cells in Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/KO mice (A,B,G,H) compared with their controls (D,E,I,J) in both the GCL and INL. The total number of ChAT cells/retina shows no differences among the strains as well (K,N). In the bar plots, the nomenclature of the mouse strains has been abbreviated in Brn3a and Brn3b KO and WT, respectively. The comparison of ChAT cells in the INL of the Pax6α:Cre; Brn3aCKOAP/KO and (n = 3) and Pax6α:Cre; Brn3bCKOAP/WT control mice (n = 4) yielded a P = 0.75 (F); P = 0.44 for displaced Amacrine cells (C); and P = 0.77 for the total number of ACs in the retinas of the Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3aCKOAP/WT mice (K). The comparison of ACs in the INL of the Pax6α:Cre; Brn3bCKOAP/KO (n = 3) and Pax6α:Cre; Brn3bCKOAP/WT (n = 6), gave a P = 0.61; for displaced ACs, P = 0.90 (L); for the total number of ACs in the retinas of the Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT mice, P = 0.75 (N). The 95% confidence intervals are presented as error bars. GCL, ganglion cell layer; INL, inner nuclear layer. Scale bar = 50 µm in A,B,D,E,G,H.
ChAT cell counting using retinal whole mounts showed that the average number of displaced ChAT amacrine cells in Pax6α:Cre; Brn3aCKOAP/KO mice (1,648 × 10e4 cells, CI ± 0.172 × 10e4) was not significantly different from that of Pax6α:Cre; Brn3aCKOAP/KO mice (1,562 × 10e4, CI ± 0.094 × 10e4) (P = 0.44; Fig. 6A,C,D). Similarly, ChAT cell numbers in the INL were comparable for Pax6α:Cre; Brn3aCKOAP/KO (1,701 × 10e4, CI ± 0.0138 × 10e4) and Pax6α:Cre; Brn3aCKOAP/KO mice (1,7384 × 10e4, CI ± 0.2129 × 10e4) (P = 0.75; Fig. 6B,E,F). The average cumulative number of ChAT-positive amacrine cells was 3,349 × 10e4 (CI ± 0.182 × 10e4) per retina for Pax6α:Cre; Brn3aCKOAP/KO and 3,300 × 10e4 (CI ± 0.266 × 10e4) for Brn3a WT mice. No statistically significant difference was observed when the two groups were compared (P = 0.77; Fig. 6K).
Analysis of ChAT amacrine cells in Brn3b mutants yielded a similar result, with no statistically significant differences for both populations analyzed. In Pax6α:Cre; Brn3bCKOAP/KO animals, displaced ChAT cells were on average 1,596 × 10e4 (CI ± 0.201 × 10e4), while Pax6α:Cre; Brn3bCKOAP/KO mice had 1,616 × 10e4 (CI ± 0.0345 × 10e4) cells (P = 0.90; Fig. 4G,I,L). Similarly, ChAT cells in the INL were 1,752 × 10e4 (CI ± 0.248 × 10e4) for Pax6α:Cre; Brn3bCKOAP/KO mice and 1,831 × 10e4 (CI ± 0.112 × 10e4) for Brn3b WT mice (P = 0.61; Fig. 6H,J,M). Cumulative counts confirmed that there was no significant difference between the number of ChAT-positive amacrine cells from Pax6α: Cre; Brn3bCKOAP/KO mice, at 3,348 × 10e4 (CI ± 0.540 × 10e4) and Pax6α:Cre; Brn3bCKOAP/KO mice, at 3,447 × 10e4 (CI ± 0.098 × 10e4) (P = 0.75; Fig. 6N).
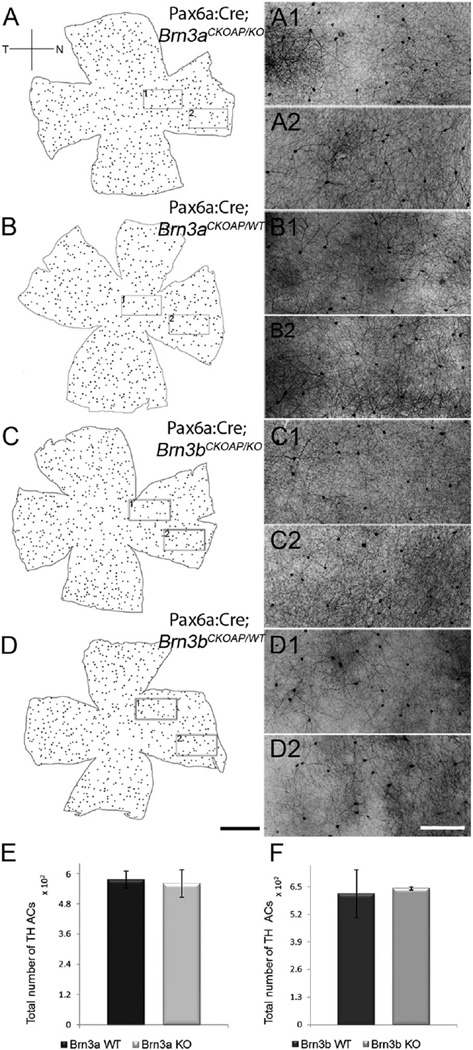
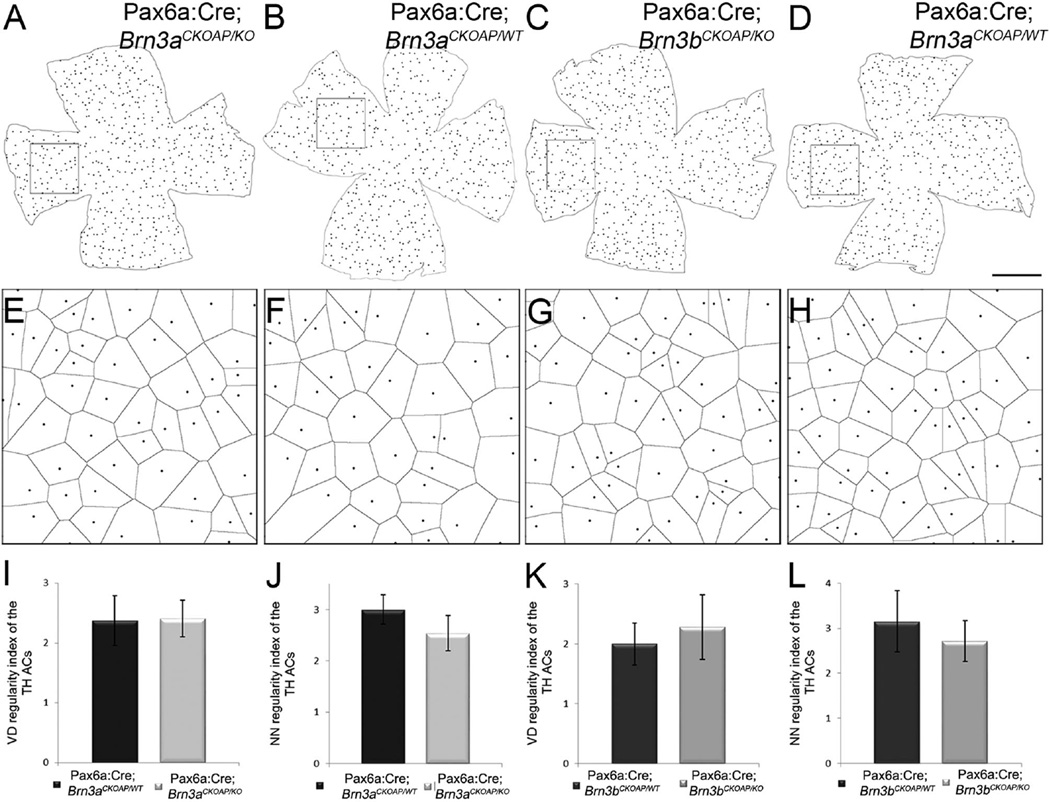
Type 1 dopaminergic amacrine cells were counted in retinal whole mounts stained for TH. We found no significant differences between Pax6α:Cre; Brn3aCKOAP/KO mice, in which the average number was 561.1 cells/retina (CI ± 55.28) and Pax6α:Cre; Brn3aCKOAP/WT mice, at 576 (CI ± 33.67) (P = 0.68; Fig. 7A,B,E). Similar findings were obtained for the Brn3b comparison, which gave the following average numbers of cells per retina: Pax6α:Cre; Brn3bCKOAP/KO, 642.16 (CI ± 6.85) and Pax6α:Cre; Brn3bCKOAP/WT, 617 (CI ± 113.02) (P = 0.70; Fig. 7C,D,F).
Figure 7.
TH cells. Maps of the dopaminergic amacrine cells labeled with antibodies against TH in the four genotypes analyzed: Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3aCKOAP/WT (A–B2); Pax6α:Cre; Brn3bCKOAP/KO andPax6α:Cre; Brn3bCKOAP/WT (C–D2). For each map on the left, two images (with the same letter) are shown on the right, obtained at central (1) and peripheral (2) locations in the nasal quadrant, as shown by the rectangular frames. TH cells show a similar morphology in all the examples illustrated. Total number of TH amacrine cells in the retina of the Pax6α:Cre; Brn3aCKOAP/KO (n = 3) and Pax6α: Cre; Brn3aCKOAP/WT (n = 3), (E) and Pax6α:Cre; Brn3bCKOAP/KO (n = 3) and Pax6α:Cre; Brn3bCKOAP/WT (n = 3) (F). No significant differences were observed in cell numbers between Brn3a_KOs and controls (P = 0.68) and for Brn3b_KOs and controls (P = 0.73), respectively. The 95% confidence intervals are presented as error bars. Scale bar = 1 mm in D (applies to A–D) and D2 (applies to A1–D2).
Similarly, both the pattern of staining of GABAergic amacrine cells labeled on retinal sections by means of GAD67 antibodies and their density estimated by cell counting in the INL demonstrated a close correspondence of Brn3a KO and WT retinas (Fig. 8). As previously described, GAD67 immunostaining revealed a large population of cell bodies in the amacrine cell layer, plus additional cell bodies in the GCL (Vardi and Auerbach, 1995). A dense plexus of processes spanning the whole IPL was strongly labeled. Occasional profiles of putative horizontal cells were observed in the outer retina. The prevalence of GAD67-positive profiles in the INL of Pax6α:Cre; Brn3aCK-OAP/KO mice was 136.621 (CI ± 27.724) cells/mm, and 149.465 (CI ± 14.658) cells/mm for the retina of corresponding Pax6α:Cre; Brn3aCKOAP/WT animals (P = 0.40). In the case of Pax6α:Cre; Brn3bCKOAP/KO mice, the number of GAD67 cells was 126.742 (CI ± 18.812) compared with their Pax6α:Cre; Brn3aCKOAP/WT controls, at 110.401 (CI ± 16.657) (P = 0.22).
Figure 8.
GAD67-positive amacrine cells. GAD67-positive, GABAergic amacrine cells labeled on vertical sections from Pax6α:Cre; Brn3aCKOAP/KO and Brn3aCKOAP/WT retinas have similar morphologies: cells with large size somata line the innermost border of the INL and are also found in the GCL. The IPL is densely stained. Their density is similar in the Brn3 KOs and in their controls (see text). INL, inner nuclear layer; IPL, inner plexiform layer. Scale bar = 20 µm in B (applies to A,B).
Cholinergic and dopaminergic amacrine cells were analyzed in retinal whole mounts to study their mosaic regularity, calculating the area of the Voronoi domain of each cell and the NNRI.
Table 2 shows the results of comparisons of parameters of ChAT amacrine cells (both INL and displaced) and of TH amacrine cells from retinas of the four genotypes (see also Figs. 9A–P, 10A–L). No statistically significant difference was detected in any of the mosaic indicators used.
TABLE 2.
ChAT and TH Cell Voronoi Domain Regularity Index (VDRI) and Nearest Neighbor Regularity Index (NNRI)
| GCL | P | INL | P | |
|---|---|---|---|---|
| ChAT ACs | ||||
| VDRI | ||||
| Pax6a:Cre; Brn3aCKOAP/KO | 3.000 ± 0.22 | 0.365 | 3.520 ± 0.311 | 0.054 |
| Pax6a:Cre; Brn3aCKOAP/WT | 2.863 ± 0.19 | 3.126 ± 0.229 | ||
| Pax6a:Cre; Brn3bCKOAP/KO | 2.574 ± 0.28 | 0.284 | 3.325 ± 0.237 | 0.657 |
| Pax6a:Cre; Brn3bCKOAP/WT | 2.711 ± 0.13 | 3.395 ± 0.196 | ||
| NNRI | ||||
| Pax6a:Cre; Brn3aCKOAP/KO | 2.987 ± 0.245 | 0.927 | 4.233 ± 0.348 | 0.219 |
| Pax6a:Cre; Brn3aCKOAP/WT | 3.002 ± 0.214 | 3.901 ± 0.286 | ||
| Pax6a:Cre; Brn3bCKOAP/KO | 2.910 ± 0.252 | 0.290 | 4.192 ± 0.316 | 0.583 |
| Pax6a:Cre; Brn3bCKOAP/WT | 2.729 ± 0.214 | 4.077 ± 0.281 | ||
| TH ACs | ||||
| VDRI | ||||
| Pax6a:Cre; Brn3aCKOAP/KO | 2.407 ± 0.300 | 0.899 | ||
| Pax6a:Cre; Brn3aCKOAP/WT | 2.374 ± 0.416 | |||
| Pax6a:Cre; Brn3bCKOAP/KO | 2.279 ± 0.541 | 0.425 | ||
| Pax6a:Cre; Brn3bCKOAP/WT | 1.996 ± 0.397 | |||
| NNRI | ||||
| Pax6a:Cre; Brn3aCKOAP/KO | 2.541 ± 0.343 | 0.064 | ||
| Pax6a:Cre; Brn3aCKOAP/WT | 3.001 ± 0.282 | |||
| Pax6a:Cre; Brn3bCKOAP/KO | 2.719 ± 0.453 | 0.334 | ||
| Pax6a:Cre; Brn3bCKOAP/WT | 3.153 ± 0.680 |
Abbreviations: AC, amacrine cell; ChAT, choline acetyltransferase; GCL, ganglion cell layer; INL, inner nuclear layer; TH, tyrosine hydroxylase.
Figure 9.
ChAT amacrines mosaics. Voronoi domains (VD) of ChAT cells in the GCL (A,E,I,M) and in the INL (B,F,J,N). Pax6α:Cre; Brn3aCKOAP/KO, Pax6α:Cre; Brn3bCKOAP/KO and control samples compared individually show no statistically significant differences. In the bar plots on the right, the nomenclature of the mouse strains has been abbreviated in Brn3a and Brn3b KO and WT, respectively. For the VD regularity index, P = 0.365 when GCL ChATs of the Pax6α:Cre; Brn3aCKOAP/KO are compared with those of Pax6α:Cre; Brn3aCKOAP/WT controls (C), and P = 0.284 for the comparison of the Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT (K). For the ChAT-positive ACs in the INL, P = 0.054 for comparison of the Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3aCKOAP/WT (D) and P = 0.658 for comparison of Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT (L).The nearest neighbor (NN) regularity index comparison gives the following results: P = 0.927 when comparing the displaced ACs population in the Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3aCKOAP/WT with the WT (G) and P = 0.291 in the case of the Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT (O). Comparison of the INL AC population shows a P = 0.219 in the case of the Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3aCKOAP/WT (H) and a P = 0.583 for INL ChAT cells of the Pax6α:Cre; Brn3bCKOAP/KO versus Pax6α:Cre; Brn3bCKOAP/WT (P). For each sample n = 3, and 95% confidence intervals are shown as error bars. GCL, ganglion cell layer; INL, inner nuclear layer. Scale bar = 50 µm in A,B,E,F,I,J,M,N.
Figure 10.
TH mosaics. Voronoi domains (VD) of TH cells. Comparison of Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/KO (A,E,C,G) with corresponding controls Pax6α:Cre; Brn3aCKOAP/WTand Pax6α:Cre; Brn3bCKOAP/WT (C,F,D,H) showed no statistically significant differences. For the VD regularity index, P = 0.899 when comparing TH cells of Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/KO with Pax6α:Cre; Brn3aCKOAP/WT controls (A), and P = 0.425 for comparison of Pax6α:Cre; Brn3bCKOAP/KO with Pax6α:Cre; Brn3bCKOAP/WT(K). The nearest neighbor (NN) regularity index comparison gives the following results: P = 0.343 when comparing the TH mosaics in Pax6α:Cre; Brn3aCKOAP/KO and Pax6α:Cre; Brn3aCKOAP/WT (J) and P = 0.334 in the case of Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT (L). For each sample, n = 3; 95% confidence intervals are presented as error bars. Scale bar = 1 mm in D (applies to A–D).
Distribution of contacts in the IPL
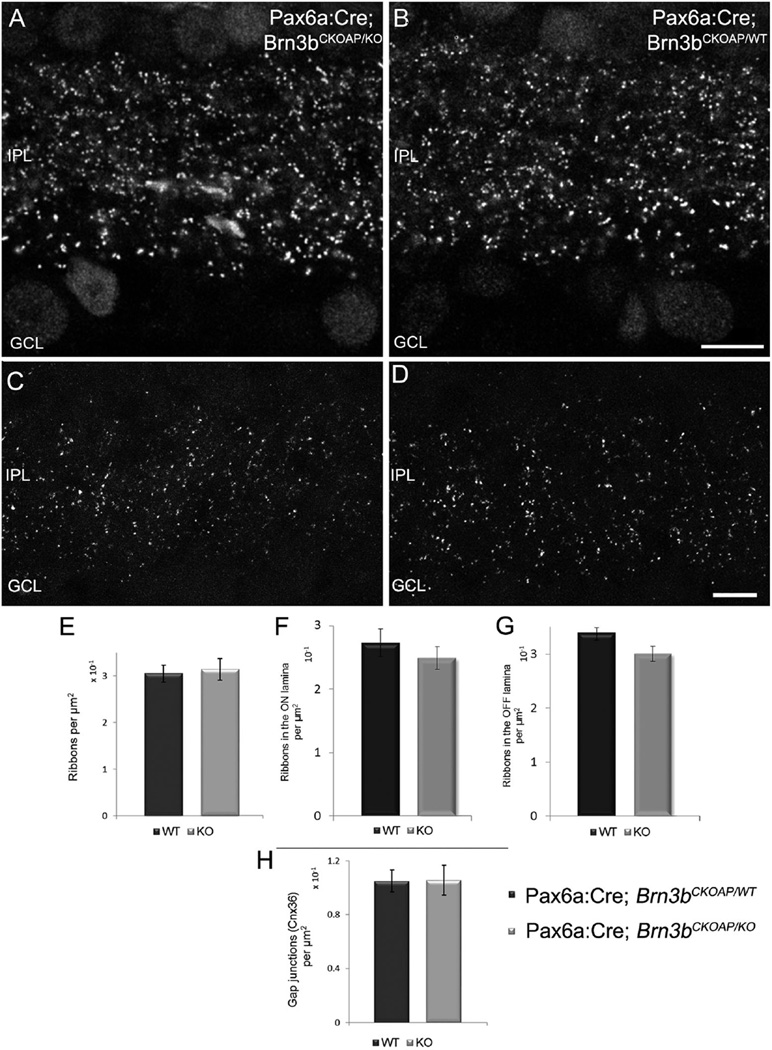
Glutamatergic synapses in the retina are established by photoreceptors in the OPL and by bipolar cells (both rod and cone bipolar cells) in the IPL and are characterized by the presence of typical ribbons that can be labeled specifically with antibodies. We used anti-Ctbp2 (Ribeye) antibodies, high-definition confocal microscopy, and densitometry analysis on vertical sections to estimate the density of Ctbp2-positive puncta in the IPL of KO and WT retinas (Fig. 11A,B). The analysis yielded an average total density of 0.314 (CI ± 0.022) puncta/µm2 in Pax6α:Cre; Brn3bCKOAP/KO retinas and 0.304 (CI ± 0.018) puncta/µm2 in Pax6α:Cre; Brn3bCKOAP/WT animals (P = 0.44; Fig. 9B). To reveal possible differences in ribbon distributions between OFF and ON sublaminae of the IPL in the two strains, we counted ribeye-positive puncta in the outermost two-thirds and the innermost third of the IPL (representing fiducial limits of the “a”, or OFF sublamina and “b” or ON sublamina of the IPL, respectively), in the Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT retinas. The comparison of ON sublaminae resulted in 0.249 (CI ± 0.01) puncta/µm2 in mutants and 0.273 (CI ± 0.22) puncta/µm2 in controls (P = 0.179). Similar results were observed in the comparison of the OFF sublaminae: 0.301 (CI ± 0.014) puncta/µm2 for Pax6α:Cre; Brn3bCKOAP/KO and 0.340 (CI ± 0.09) puncta/µm2 for Pax6α:Cre; Brn3bCKOAP/WT mice (P = 0.097). Hence, no significant differences were found (Fig. 11E,F).
Figure 11.
Ribbons and gap junctions in the IPL. Confocal micrographs showing the distribution of ribbon synapses in the IPL of Pax6α: Cre; Brn3bCKOAP/KO (A) and Pax6α:Cre; Brn3bCKOAP/WT (B) retinas labeled with an antibody against CtBP2. Quantitative analysis showed no significant differences between the density of ribbons in the IPL of the two strains, P = 0.44 (C), when the entire thickness of the IPL was measured. No differences were observed also when the ON sublamina of the KO was compared with the one of the control, P = 0.179 (E); or in the case of the OFF sublaminae, P = 0.09 (F). Analogous data were obtained for estimates of the density of IPL gap junctions expressing Connexin36. Comparison of Pax6α:Cre; Brn3bCKOAP/KO and Pax6α:Cre; Brn3bCKOAP/WT animals returned a value of P = 0.99 (D). Scale bar = 10µm in B (applies to A,B); 5 µm in D (applies to C,D).
A similar strategy was applied to estimate the density of gap junctions containing Cx36; this component is largely expressed in the IPL and particularly by GC den-drites (Pan et al., 2010). Cx36 antibody staining results in a highly punctate pattern, adequate for densitometric analysis. Again, no differences were detected between the Cx36 distribution in Pax6α:Cre; Brn3bCKOAP/KO retinas, where puncta had an average density of 0.106 (CI ± 0.011)/µm2 and that in Pax6α:Cre; Brn3bCKOAP/WT retinas, at 0.105 (CI ± 0.008) puncta/µm2 (P = 0.99; Fig. 11D).
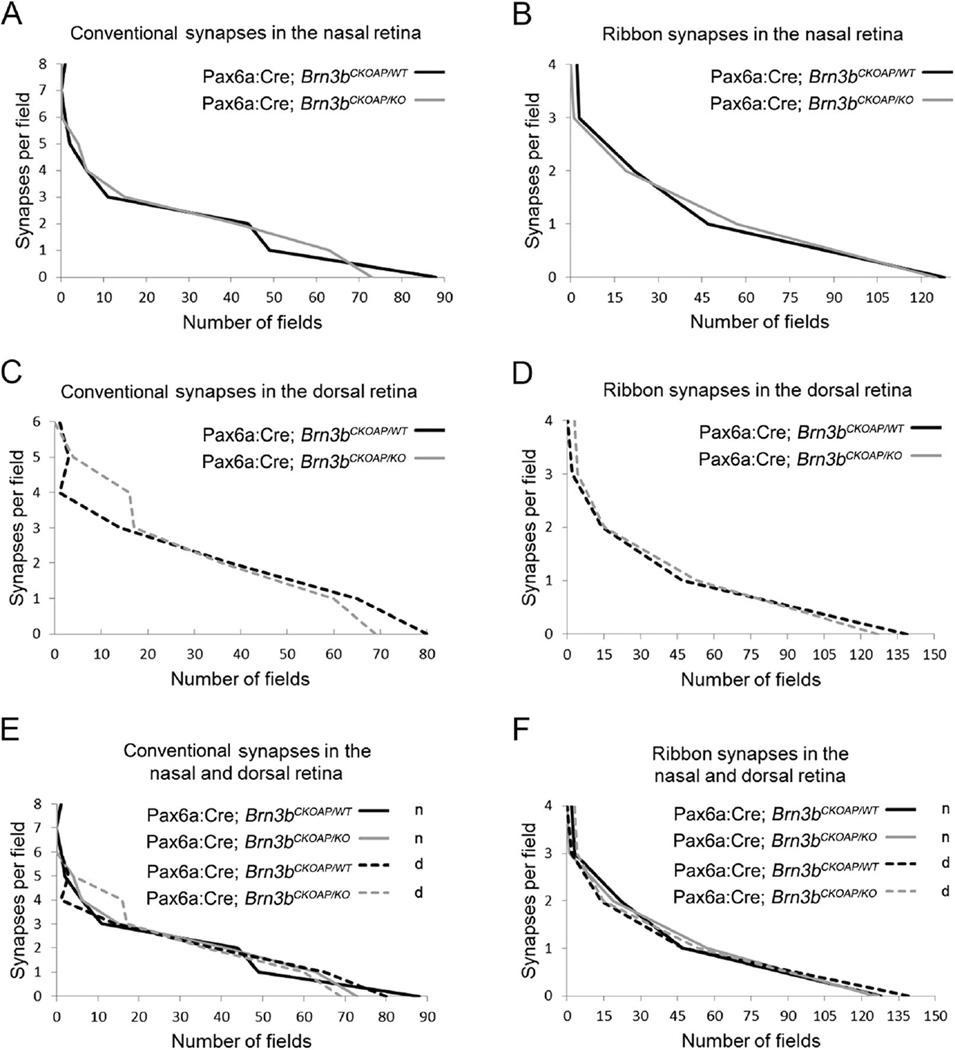
In addition, we used transmission EM to give an account of the type and distribution of synapses in the IPL of Brn3b KO mice (Fig. 12A–E). We counted conventional chemical synapses (known to originate from amacrine cells, and thus to be inhibitory), and ribbon synapses (from excitatory, glutamatergic bipolar cells) at two matching retinal locations and in IPL samples of similar areas. Statistical comparison was done with the Kolmogorov–Smirnov test (Fig. 13), which showed no differences for conventional synapses in both the dorsal retina (P = 0.882; Fig. 13C) and the nasal retina (P = 0.957; Fig. 13A). Similarly, nonstatistically significant differences were detected when the distribution of ribbon synapses was compared, in both the nasal retina (P = 0.69; Fig. 13B) and the dorsal retina (P = 0.69; Fig. 13D).
Figure 12.
Electron microscopy of the IPL. Electron micrographs showing similar examples of ribbon (R) and conventional synapses (arrows) in the IPL of retinal samples obtained at matching locations from Pax6α:Cre; Brn3bCKOAP/KO (A–C) and Pax6α:Cre; Brn3bCKOAP/WT mice (D–E). RBe, rod bipolar ending; CBe, cone bipolar endings. Arrows indicate the direction of synaptic transmission. Ribbons label the presynaptic endings of rod and cone bipolars, while conventional synapses are established by amacrine cells only. Scale bar = 1 µm in B (applies to A,B), C, and E (applies to D,E).
Figure 13.
Synaptic distribution in the IPL. Analysis of the distribution of conventional (A,C) and ribbon (B,D) synapses in the IPL of the nasal and dorsal retina of Pax6α:Cre; Brn3bCKOAP/KO (KO n, KO d) and Pax6α:Cre; Brn3bCKOAP/WT control (WT n, WT d) mice. Distributions of the data sets were compared using the Kolmogolov-Smirnov test, yielding no statistical significant differences between the samples (P = 0.697 for the ribbon synapses [B] and P = 0.957 for the conventional synapses [A] in the nasal retina; P = 0.697 for the ribbon synapses [D] and P= 0.882 for the conventional synapses [C]) in the dorsal retina. No statistical significant differences were observed among the four samples when compared individually (WT d vs KO d, WT n vs KO n and KO d vs KO n). Values were: P = 0.697 for KO d vs KO, and P = 0.99 for WT d vs WT for the ribbon synapses (F). For the conventional synapses of WT d vs WT n: P = 0.808; for KO d vs KO n: P =1 (E).
The ganglion cell layer in Brn3 KO retinas
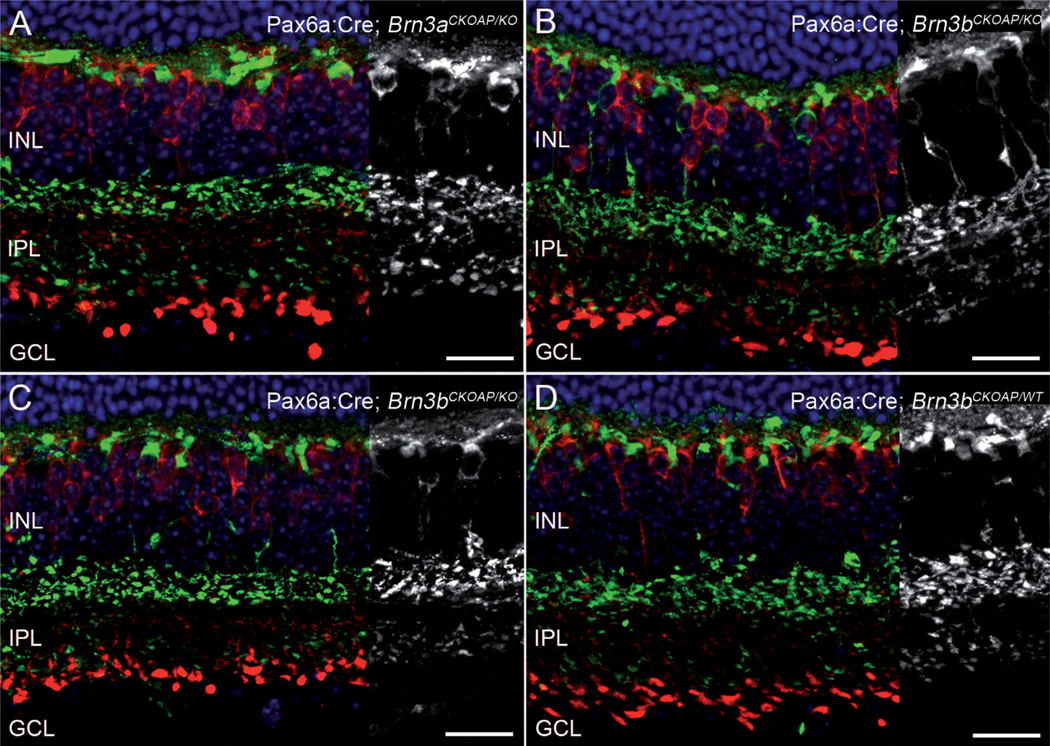
A 30% decrement in the number of GCs in Pax6α:Cre; Brn3aCKOAP/KO mice was confirmed by counting RBPMS-positive cells in retinal whole mounts (Fig. 14A–C). This finding is in agreement with previous studies documenting the phenotypes of Brn3a KO mice (Badea et al., 2009a; Shi et al, 2013).
Figure 14.
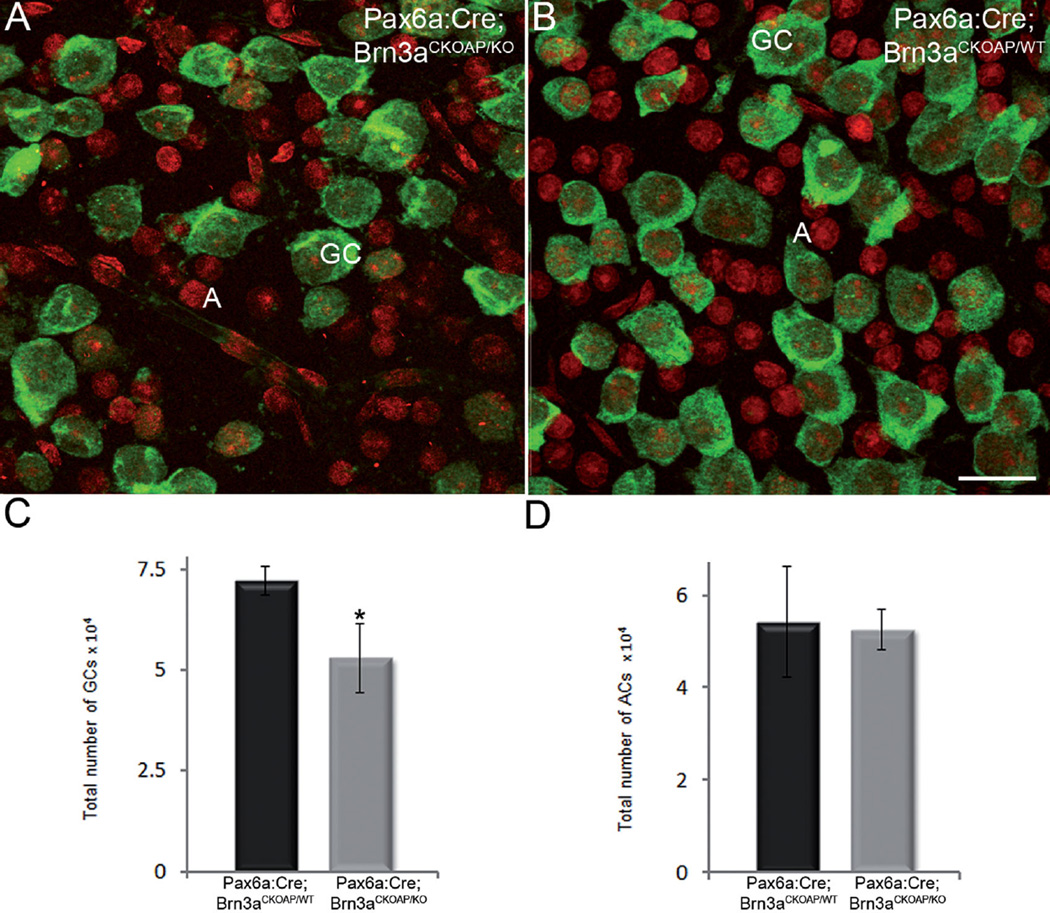
Ganglion cell layer. Immunostaining for RBPMS (green signal) identifies cell bodies of GCs in retinal whole mounts of Pax6α: Cre; Brn3aCKOAP/KO and (A) and Pax6α:Cre; Brn3aCKOAP/WT (B) retinas. Ethidium homodimer nuclear staining (red signal) shows round nuclei of RBPMS-negative cells, displaced amacrine cells. Counting of both GCs cells (C) and displaced amacrines (D) shows the expected decrement in GC number in the Pax6α:Cre; Brn3aCKOAP/KO retina, with 52,859 (CI ± 8,525.19) GCs in the Pax6α:Cre; Brn3aCKOAP/KO versus 72,055 (CI ± 3,493.44) GCs in the Pax6α:Cre; Brn3aCKOAP/WT retina (P = 0.03). Counts of the same samples show that the number of displaced amacrine cells is unvaried in the two strains (D), with 52,592 (CI ± 4,356.09) cells in the Pax6α:Cre; Brn3aCKOAP/KO versus 54,200 (CI ± 11,992.07) cells in the Pax6α:Cre; Brn3aCKOAP/WT retina, P = 0.08. GC, ganglion cell; A, amacrine cell. Scale bar = 20 µm in B (applies to A,B).
Cell counting on retinal whole mounts also showed that the number of displaced amacrines was virtually identical in Pax6α:Cre; Brn3aCKOAP/WT and Pax6α:Cre; Brn3aCKOAP/KO retinas and corresponded to over 40% of the total number of cells in the GCL (Fig. 14D). Thus, from population studies, we concluded that, apart from the expected change in GC morphology and number, no evident abnormalities could be detected in the overall number of two large cohorts of amacrine cells, namely, displaced amacrine cells (known to comprise multiple types) and GAD67-positive, GABAergic amacrine cells. The latter are known to include mostly large-field cells, such as cholinergic, starburst amacrine cells and indoleamine-accumulating amacrine cells (Strettoi and Masland, 1996; Masland, 2012b).
Finally, we analyzed Brn3a and Brn3b KO retinas for possible morphological changes in the array of astrocytes, as these glial cells, located at the boundary between the GC and the nerve fiber layers, organize GC axons in bundles, and they were expected to rearrange as a consequence of major GC structural modifications (Chan-Ling and Stone, 1991).
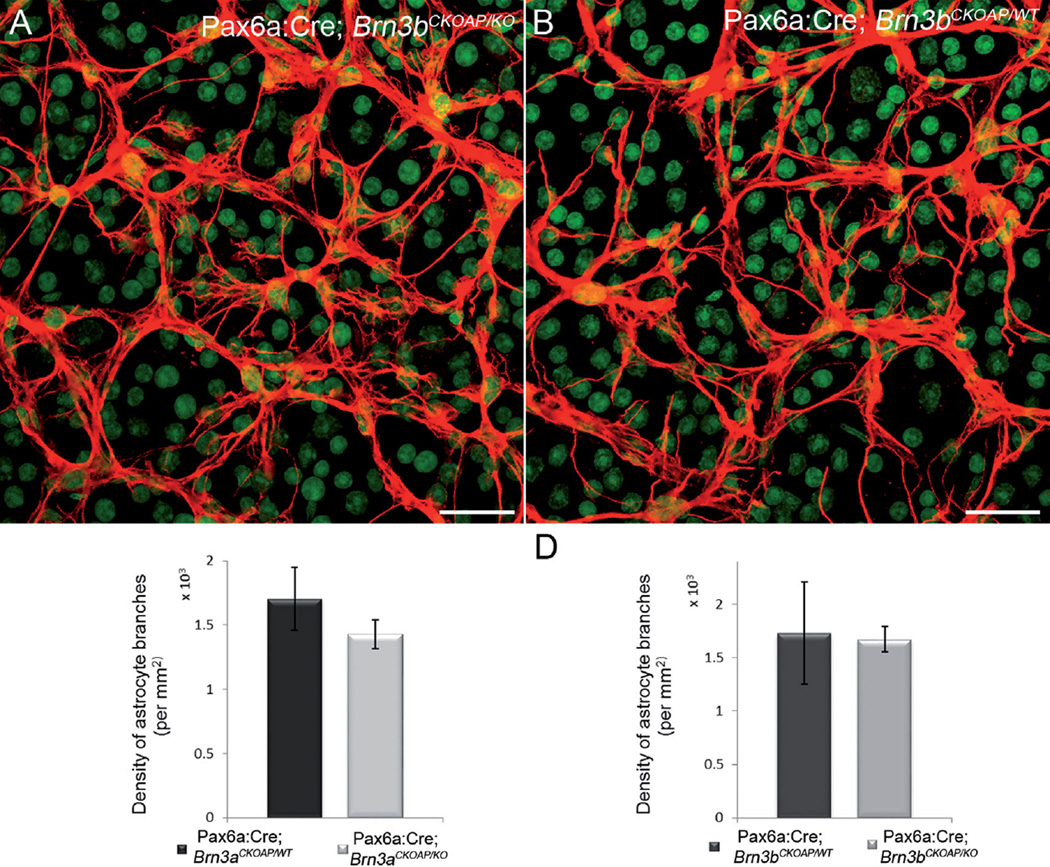
We analyzed the astrocytic branching pattern using a quantitative method that gives indications of the complexity of their network. We found that the average number of intersections of astrocytic processes with the standard sampling grid used was 1,672 (CI ± 122) for the Pax6α:Cre; Brn3bCKOAP/KO and 1,732 (CI ± 478.3) for the Pax6α:Cre; Brn3bCKOAP/WT group (P = 0.823; Fig. 15D). Similarly, branching of the astrocytic Pax6α:Cre; Brn3aCKOAP/WT network was not statistically different compared with controls, at 1,408.3 (CI ± 62.62) versus 1,592.5 (CI ± 170.46) (P = 0.0614; Fig. 15C).
Figure 15.
Astrocyte network. Confocal micrographs showing the arborization of the astrocytes labeled with GFAP in whole-mount preparations. There is no significant difference between the Pax6α:Cre; Brn3bCKOAP/KO and the Pax6α:Cre; Brn3bCKOAP/WT retinas in terms of pattern of arborization (A,B) and number of branches (C) quantified for these cells (P = 0.83). A similar result was obtained from analysis of the Pax6α:Cre; Brn3aCKOAP/KO and the Pax6α:Cre; Brn3aCKOAP/WT retinas (D) (P = 0.11). Green signal is YOYO nuclear staining. Scale bar = 30 µm in A,B.
DISCUSSION
Retinas of Brn3 KO mice have been characterized before with regard to the effects of genetic ablation of these transcription factors on the distribution, number, dendritic morphology, and central projections of GCs. Decreases in GC number and defects in dendritic trees were previously reported (Erkman et al., 1996; Gan et al., 1996; Xiang 1996, 1997; Badea et al., 2009a; Badea and Nathans, 2011; Shi et al., 2013). Here, we exploit two of these mouse models to investigate whether the abnormalities of GCs translate into alterations of cellular architecture and number and types of synaptic contacts established by retinal neurons that are presynaptic partners to GCs, as well as by cells located several synapses away from them in the retina. We find that Brn3 KO mice are essentially indistinguishable from their controls. Presynaptic cell types (cone bipolar cells and amacrine cells), and also neurons positioned upstream along the retinofugal pathway (horizontal cells and rod bipolar cells) maintain constancy in general morphology, number, spacing, type, and density of connections established in the inner retina. Data obtained here for both cell numbers and mosaic parameters for the two populations of ChAT amacrine cells, as well as for TH cells, fully agree with quantitative data available for these cell types from previous studies (Jeon et al., 1998; Galli-Resta and Novelli, 2000; Masland, 2001; Whitney et al., 2008). Our results are also in line with previous literature addressing the role of specific transcription factors (in both mammalian and other vertebrates, including zebrafish and chicken), demonstrating that retinal cells of various types and belonging to different classes retain appropriate laminar positions even in the absence of major fractions of GCs (Nguyen-Ba-Charvet and Chédotal 2014). Years ago, a completely different type of manipulation, i.e., removal of GCs by optic nerve transection, demonstrated that these neurons do not provide signals that are critical for IPL stratification (Williams et al., 2001).
Quantitative data on the effects of abnormal numbers of GCs on the numbers of afferent neurons are available from a few studies. A study using a Bcl-2–overexpressing mouse under a neuron-specific enolase promoter shows that, when developmental cell death is largely inhibited, different retinal cell types are represented in the adult retina in completely different fractions (Strettoi and Volpini, 2002). Neurons normally engaged in synaptic contacts do not covary, and their final number appears to be irrespective of the large increase in the absolute number of GCs typical of this transgenic. Similarly, studies on Bax KO mice demonstrate the lack of covariation in INL cells despite a much higher number of GCs (Lee et al., 2011; Keeley et al., 2014a).
Specific studies on the determinants of cell number for various retinal cell types (Keeley et al., 2014b) show that this parameter is mostly controlled by dedicated genes. Indeed, retinal cells are not generated and do not migrate to their final destination in a sequential order that reflects their final wiring diagram. For instance, in the mouse retina, bipolar cells are born in a time window (between E16 and P10) partially overlapping but largely exceeding the time frame of genesis of GCs, to which they send synaptic connections. Obviously the program establishing the number of these cell types is not directly bound to their relative frequencies.
Indeed, it is known that neuronal numbers are the outcome of the combination of various processes acting during development, which include proliferation, fate determination, and rate of naturally occurring cell death. The number of cells for a given retinal type is specified with high precision within a mouse strain but is variable among different strains (Keeley et al., 2014b). Covariation between even developmentally, or synaptically, connected cells types is virtually absent. One noticeable exception is represented by OFF and ON ChAT cells, which covary within a given strain of mice, so that numbers of cholinergic amacrine cells in the INL and in the GCL are highly correlated (Whitney et al., 2014). This correlation is maintained in Brn3 KO mice, which show identical ChAT cell distribution as WT controls. Genetic factors modulate the size of any cell population: for neurons connected in a network, the extent of covariations of pre- and postsynaptic partners is obviously going to affect the architecture of the network and thus the physiological properties of cells.
Irregardless of the absence of covariation between perspective synaptic partners, refinement of connections is obtained by adjustments in the fine morphology of dendritic and axonal arborizations. For instance, the fundamental organization of type 7 cone bipolar cells is maintained in coneless and conefull mutant retinas despite large modulation of afferent density. However, the fine structure of dendritic terminals is afferent dependent, and terminations of these bipolar cells in the OPL are abnormal (Keeley and Reese, 2010a).
In the present study, we find that types and density of synapses in the IPL of Brn3 KO mice are undistinguishable from their WT counterparts, both in the overall distribution and when OFF and ON sublaminae of the IPL are studied separately. This could be explained by the occurrence of a physiologically low rate of synapses onto GC dendrites, thus weighing on total counts only marginally. Quantitative data on the relative occurrence of ribbon and conventional synaptic contacts onto dendrites of GCs only are not available for the mouse retina. This measurement would require labeling of the whole GC population with an EM dense dye, followed by identification and counting of all the synaptic contacts received by GC dendrites selectively. This laborious process goes beyond the purpose of the present study. However, we can safely conclude that, in Brn3 KO retinas, processes of other retinal neurons, including residual GCs, have replaced dendrites of the missing ones in synaptic contacts. Indeed, ribbons in the IPL are usually associated with dyads of postsynaptic processes, one of which can be a GC dendrite. Monads were equally rare in the IPL of Brn3 KO and WT mice, indicating that processes of other cells had “filled in” the available postsynaptic space at ribbon synapses.
Previous work has shown that Brn3b KO mice exhibit deficits in pupillary light responses and optokinetic responses but not in circadian photoentrainment, most likely due to the massive loss of retinal GC projections to the optic nerve and accessory optic tract (Badea et al., 2009a). However, a significant number of retinal GCs remain in the Brn3bKO/KO retinas (Badea et al., 2009a; Shi et al., 2011), and their function has not yet been thoroughly investigated. It will be interesting to understand whether the presumable synapse redistribution resulting from significant missing numbers of retinal GC dendrites will result in functional alterations in bipolar–amacrine circuits, and whether the remaining retinal GC dendrites receive increased numbers of contacts of the correct or incorrect type.
Similarly, gap junctions containing Cx36, the predominant subunit in GCs, were largely unaffected. We did not find any change in the density of Cx36-immunoreactive puncta, indicating a compensatory replacement with the dendrites of other cells (presumably the remaining GCs).
The present observations confirm previous studies in which the fate of photoreceptor precursors was altered genetically, resulting in retinas with abnormally high numbers of rods or cones. Bipolar and horizontal cells were shown to rewire with the new partners, maintaining a fairly normal morphology and lamination (Strettoi et al., 2004). However, their fine dendritic organization was altered, unmasking a role that afferents play in instructing morphologies of partners to match connectivity constrains (Raven et al., 2007).
Our data suggest that the concept of lack of covariance in cell numbers of retinal synaptic partners extends to the number of synaptic contacts in the IPL. In the Brn3 KO mice studied here, we find that a physiologically very relevant parameter, namely, the relative frequency of excitatory (i.e., ribbon synapses) and inhibitory contacts (chemical synapses established by amacrine cells), is unvaried. Possibly, a developmental program setting synaptic density exists, for which numerical constancy of contacts prevails over numerical matching.
Invariance extends also to astrocytes, a population of glial cells “dedicated” to GCs and residing in the nerve fiber layer. The astrocyte network was expected to be influenced by the density of cell bodies in the GCL and of their axons in the fiber layer, similarly to what is known for oligodendrocytes in the optic nerve. The number and organization of these cells depend, among other factors, on the number of GC axons, which can directly influence oligodendrocyte survival and the proliferation of their precursor cells (Barres and Raff, 1994).
We find no differences in astrocyte organization of Brn3 KO and WT mice, suggesting that, during development, and similarly to retinal neurons, astrocytes follow an intrinsic, stereotyped program, not affected by nearby cells. This is obviously very different from the well-documented Lye-Barthel et al., 2013) astrocytic reaction and rearrangement following GC loss in glaucoma. Indeed, no linkage has ever been established between Brn3 genes and a retinal pathology. In glaucoma, major GC loss is followed by functional and morphological alterations of the inner and outer retinal layers (Vidal-Sanz et al., 2012), suggesting that, in addition to a primary insult to the optic nerve head, secondary mechanisms (including ischemia) damage the remaining retinal layers. Learning from the Brn3 mice, in which a similar loss of GCs is observed, without concomitant inner retinal changes, one would argue that abnormalities found in glaucoma in cells other than GCs have their own pathogenic process, independent of GC loss.
Overall, our observations show that presynaptic partners of GCs (i.e., bipolar and amacrine cells) are not affected by the absence of their output neurons, as they maintain their morphology and pattern of connectivity. The general lamination of the retina appears to be unaffected as well. These data strongly suggest that the genetic, intrinsic program of each cell type is a fundamental feature of retinal neurons and that even major loss of one cell population can leave neighbors and main connectivity unaffected. This confirms and extends the results from zebrafish studies demonstrating that no single retinal cell type is essential for the formation of a synaptic neuropil in the IPL, which forms without apparent influence from the presynaptic or postsynaptic processes (Randlett et al., 2013).
The intrinsic stability of the inner retina might be considered when attempting vision restoration in retinal degenerative diseases, with rescue approaches based on neurons located downstream to lost photoreceptors and relying on the ability of inner retinal cells to adapt to unconventional (bionic or cellular) sources of input (Francis et al., 2013).
Acknowledgments
Grant sponsor: Sectorial Operational Program for Human Resources Development 2007–2013 fellowship (to M.G.G.); Grant sponsor: the European Social Fund (project POSDRU/107/1.5/S/76841—Modern Doctoral Studies: Internationalization and Interdisciplinarity (to M.G.G.); Grant sponsor: Italian National Research Council (to E.S.); Grant sponsor: Macula Vision Research Foundation (to E.S.); Grant sponsor: Fondazione Roma (to E.S.); Grant sponsor: intramural funds from the National Eye Institute (to T.B. and S.S.).
We thank Lucia Galli-Resta at the CNR Neuroscience Institute, Pisa and Anand Swaroop at N-NRL, NEI-NIH, Bethesda, for helpful suggestions throughout the course of the study, and Nick Brecha for guidance in the use of the RBPMS antibody. We thank Martina Biagioni at the CNR Neuroscience Institute for valuable help with ganglion cell counting and Cosmin Tegla for critically reading the manuscript.
ROLE OF AUTHORS
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: ES, MGG, TCB. Acquisition of data: MGG, EN. Mouse work and tissue collection: SS. Analysis and interpretation of data: MGG, ES, TCB. Drafting of the manuscript: MGG, ES. Critical revision of the manuscript for important intellectual content: ES, TCB. Statistical analysis: MGG, EN. Obtained funding: ES, TCB. Technical support: EN. Study supervision: ES, TCB.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare they have no conflicts of interest.
LITERATURE CITED
- Badea TC, Nathans J. Morphologies of mouse retinal ganglion cells expressing transcription factors Brn3a, Brn3b, and Brn3c: Analysis of wild type and mutant cells using genetically-directed sparse labeling. Vision Res. 2011;51:269–279. doi: 10.1016/j.visres.2010.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Ecker J, Hattar S, Nathans J. Distinct roles of transcription factors Brn3a and Brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron. 2009a;61:852–864. doi: 10.1016/j.neuron.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea T, Hua Z, Smallwood P, Williams J, Rotolo T, Ye X, Nathans J. New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. Plos One. 2009b;4:e7859. doi: 10.1371/journal.pone.0007859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Williams J, Smallwood P, Shi M, Motajo O, Nathans J. Combinatorial expression of Brn3 transcription factors in somatosensory neurons: genetic and morphologic analysis. J Neurosci. 2012;32:995–1007. doi: 10.1523/JNEUROSCI.4755-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardet SM, Cobos I, Puelles E, Martinez-De-La-Torre M, Puelles L. Chicken lateral septal organ and other circumventricular organs form in a striatal subdominal abutting the molecular striatopallidal border. J Comp Neurol. 2006;499:745–767. doi: 10.1002/cne.21121. [DOI] [PubMed] [Google Scholar]
- Barres B, Raff M. Control of oligodendrocyte number in the developing rat optic nerve. Neuron. 1994;12:935942. doi: 10.1016/0896-6273(94)90305-0. [DOI] [PubMed] [Google Scholar]
- Brandon C. Cholinergic neurons in the rabbit retina: dendritic branching and ultrastructural connectivity. Brain Res. 1987;426:119130. doi: 10.1016/0006-8993(87)90431-8. [DOI] [PubMed] [Google Scholar]
- Brown N, Kanekar S, Vetter M, Tucker P, Gemza D, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Chan-Ling T, Stone J. Factors determining the morphology and distribution of astrocytes in the cat retina: a “contact-spacing” model of astrocyte interaction. J Comp Neurol. 1991;303:387–399. doi: 10.1002/cne.903030305. [DOI] [PubMed] [Google Scholar]
- Chen H, Liu B, Neufeld A. Epidermal growth factor receptor in adult retinal neurons of rat, mouse, and human. J Comp Neurol. 2007;500:299–310. doi: 10.1002/cne.21161. [DOI] [PubMed] [Google Scholar]
- Contini M, Raviola E. GABAergic synapses made by a retinal dopaminergic neuron. Proc Natl Acad Sci U S A. 2003;100:1358–1363. doi: 10.1073/pnas.0337681100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks J, Kolb H. Localization of GABA, glycine, glutamate and tyrosine hydroxylase in the human retina. J Comp Neurol. 1992;315:287–302. doi: 10.1002/cne.903150305. [DOI] [PubMed] [Google Scholar]
- Dhingra A, Sulaiman P, Xu Y, Fina M, Veh R, Vardi N. Probing neurochemical structure and function of retinal ON bipolar cells with a transgenic mouse. J Comp Neurol. 2008;510:484–496. doi: 10.1002/cne.21807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieck S, Sanmartí-Vila L, Langnaese K, Richter K, Kindler S, Soyke A, Wex H, Smalla K-H, Kämpf U, Fränzer J-T, Stumm M, Garner C, Gundelfinger E. Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J Cell Biol. 1998;142:499–509. doi: 10.1083/jcb.142.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas J, Hamani C, Lozano A, Enikolopov G. Neurogenic hippocampal targets of deep brain stimulation. J Comp Neurol. 2011;519:6–20. doi: 10.1002/cne.22503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkman L, McEvilly R, Luo L, Ryan A, Hooshmand F, O’Connell S, Keithley E, Rapaport D, Ryan A, Rosenfeld M. Role of transcription factors a Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature. 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- Fox M, Sanes J. Synaptotagmin I and II are present in distinct subsets of central synapses. J Comp Neurol. 2007;503:280–296. doi: 10.1002/cne.21381. [DOI] [PubMed] [Google Scholar]
- Francis P, Mansfield B, Rose S. Proceedings of the First International Optogenetic Therapies for Vision Symposium. Trans Vis Sci Technol. 2013;2:4. doi: 10.1167/tvst.2.7.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli-Resta L, Novelli E. The effects of natural cell loss on the regularity of the retinal cholinergic arrays. J Neurosci. 2000;20:RC60. doi: 10.1523/JNEUROSCI.20-03-j0005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner D, Klein W, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargini C, Terzibasi E, Mazzoni F, Strettoi E. Retinal organization in the retinal degeneration 10 (rd10) mutant mouse: a morphological and ERG study. J Comp Neurol. 2007;500:222–238. doi: 10.1002/cne.21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh K, Bujan S, Haverkamp S, Feigenspan A, Wässle H. Types of bipolar cells in the mouse retina. J Comp Neurol. 2004;469:70–82. doi: 10.1002/cne.10985. [DOI] [PubMed] [Google Scholar]
- Greferath U, Grünert U, Wässle H. Rod bipolar cells in the mammalian retina show protein kinase C-like immunoreactivity. J Comp Neurol. 1990;301:433–442. doi: 10.1002/cne.903010308. [DOI] [PubMed] [Google Scholar]
- Grünert U, Wässle H. GABA-like immunoreactivity in the macaque monkey retina: a light and electron microscopic study. J Comp Neurol. 1990;297:509–524. doi: 10.1002/cne.902970405. [DOI] [PubMed] [Google Scholar]
- Haeseleer F, Sokal I, Verlinde CLMJ, Erdjument-Bromage H, Tempst P, Pronin AN, Benovic JN, Fariss RN, Palczewski K. Five members of a novel Ca2+-binding protein (CABP) subfamily with similarity to calmodulin. J Biol Chem. 2000;275:1247–1260. doi: 10.1074/jbc.275.2.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Wässle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- Haverkamp S, Ghosh K, Hirano A, Wässle H. Immunocytochemical description of five bipolar cell types of the mouse retina. J Comp Neurol. 2003;455:463–476. doi: 10.1002/cne.10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haverkamp S, Inta D, Monyer H, Wässle H. Expression analysis of green fluorescent protein in retinal neurons of four transgenic mouse lines. Neuroscience. 2009;160:126–139. doi: 10.1016/j.neuroscience.2009.01.081. [DOI] [PubMed] [Google Scholar]
- Hirano A, Brandstatter J, Vila A, Brecha N. Robust syntaxin-4 immunoreactivity in mammalian horizontal cell processes. Vis Neurosci. 2007;24:489–502. doi: 10.1017/S0952523807070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobs T, Koizumi A, Masland R. The spatial distribution of glutamatergic inputs to dendrites of retinal ganglion cells. J Comp Neurol. 2008;510:221–236. doi: 10.1002/cne.21795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci. 1998;18:8936–8946. doi: 10.1523/JNEUROSCI.18-21-08936.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jusuf P, Martin P, Grünert U. Synaptic connectivity in the midget-parvocellular pathway of primate central retina. J Comp Neurol. 2006;494:260–274. doi: 10.1002/cne.20804. [DOI] [PubMed] [Google Scholar]
- Keeley P, Reese B. Role of afferents in the differentiation of bipolar cells in the mouse retina. J Neurosci. 2010a;30:1677–1685. doi: 10.1523/JNEUROSCI.5153-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley P, Reese B. Morphology of dopaminergic amacrine cells in the mouse retina: independence from homotypic interactions. J Comp Neurol. 2010b;518:1220–1231. doi: 10.1002/cne.22270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley P, Whitney I, Raven M, Reese B. Dendritic spread and functional coverage of starburst amacrine cells. J Comp Neurol. 2007;505:539–546. doi: 10.1002/cne.21518. [DOI] [PubMed] [Google Scholar]
- Keeley P, Madsen N, St John A, Reese B. Programmed cell death of retinal cone bipolar cells is independent of afferent or target control. Dev Biol. 2014a;394:191196. doi: 10.1016/j.ydbio.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeley P, Whitney I, Madsen N, St John A, Borhanian S, Leong S, Williams R, Reese B. Independent genomic control of neuronal number across retinal cell types. Dev Cell. 2014b;30:103–109. doi: 10.1016/j.devcel.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara AH, Paschon V, Cardoso CM, Higa GSV, Castro LM, Hamassaki DE, Britto LRG. Connexin36, an essential element in the rod pathway, is highly expressed in the essentially rodless retina of Gallus gallus. J Comp Neurol. 2009;512:651–663. doi: 10.1002/cne.21920. [DOI] [PubMed] [Google Scholar]
- Koizumi A, Jakobs T, Masland R. Regular mosaic of synaptic contacts among three retinal neurons. J Comp Neurol. 2011;519:341–357. doi: 10.1002/cne.22522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz MA, Hendrickson AE. Distribution of GABA-immunoreactive amacrine cell synapses in the inner plexiform layer of macaque monkey retina. Vis Neurosci. 1990;5:17–28. doi: 10.1017/s0952523800000043. [DOI] [PubMed] [Google Scholar]
- Kwong J, Caprioli J, Piri N. RNA binding protein with multiple splicing: a new marker for retinal ganglion cells. Invest Ophthalmol Vis Sci. 2009;51:1052–1058. doi: 10.1167/iovs.09-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cowgill E, Al-Nabulsi A, Quinn E, Evans S, Reese B. Homotypic regulation of neuronal morphology and connectivity in the mouse retina. J Neurosci. 2011;31:14126–14133. doi: 10.1523/JNEUROSCI.2844-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Khare S, Liang X, Peters M, Liu X, Cepko C, Xiang M. All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development. 2000;127:3237–3247. doi: 10.1242/dev.127.15.3237. [DOI] [PubMed] [Google Scholar]
- Liu X, Robinson M, Schreiber A, Wu V, Lavail M, Cang J, Copenhagen D. Regulation of neonatal development of retinal ganglion cell dendrites by neurotrophin-3 overexpression. J Comp Neurol. 2009;514:449–458. doi: 10.1002/cne.22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye-Barthel M, Sun D, Jakobs T. Morphology of astrocytes in a glaucomatous optic nerve. Invest Ophthalmol Vis Sci. 2013;54:909917. doi: 10.1167/iovs.12-10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Masland RH. The fundamental plan of the retina. Nat Neurosci. 2001;4:877–886. doi: 10.1038/nn0901-877. [DOI] [PubMed] [Google Scholar]
- Masland RH. The neuronal organization of the retina. Neuron. 2012a;76:266–280. doi: 10.1016/j.neuron.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. The tasks of amacrine cells. Vis Neurosci. 2012b;29:3–9. doi: 10.1017/s0952523811000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May C, Nakamura K, Fujiyama F, Yanagawa Y. Quantification and characterization of GABA-ergic amacrine cells in the retina of GAD67-GFP knock-in mice. Acta Ophthalmol. 2008;86:395–400. doi: 10.1111/j.1600-0420.2007.01054.x. [DOI] [PubMed] [Google Scholar]
- Meyer A, Hilgen G, Dorgau B, Sammler E, Weiler R, Monyer H, Dedek K, Hormuzdi S. All amacrine cells discriminate between heterocellular and homocellular locations when assembling connexin36-containing gap junctions. J Cell Sci. 2014;127:1190–1202. doi: 10.1242/jcs.133066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ba-Charvet K, Chédotal A. Development of retinal layers. C R Biol. 2014;337:153–159. doi: 10.1016/j.crvi.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Schwartz M, Rakic P. Localization of γ-aminobutyric acid and glutamic acid decarboxylase in rhesus monkey retina. Brain Res. 1985;359:351–355. doi: 10.1016/0006-8993(85)91449-0. [DOI] [PubMed] [Google Scholar]
- Noorian A, Taylor G, Annerino D, Greene J. Neurochemical phenotypes of myenteric neurons in the rhesus monkey. J Comp Neurol. 2011;519:3387–3401. doi: 10.1002/cne.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Paul D, Bloomfield S, Völgyi B. Connexin36 is required for gap junctional coupling of most ganglion cell subtypes in the mouse retina. J Comp Neurol. 2010;518:911–927. doi: 10.1002/cne.22254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli V, Strettoi E. Bipolar cells of the mouse retina: a gene gun, morphological study. J Comp Neurol. 2004;476:254–266. doi: 10.1002/cne.20207. [DOI] [PubMed] [Google Scholar]
- Pourcho RG. Uptake of [3H]glycine and [3H]GABA by amacrine cells in the cat retina. Brain Res. 1980;198:33–46. doi: 10.1016/0006-8993(80)90748-9. [DOI] [PubMed] [Google Scholar]
- Puller C, Haverkamp S, Grünert U. OFF midget bipolar cells in the retina of the marmoset, Callithrix jacchus, express AMPA receptors. J Comp Neurol. 2007;502:442–454. doi: 10.1002/cne.21315. [DOI] [PubMed] [Google Scholar]
- Randlett O, MacDonald R, Yoshimatsu T, Almeida A, Suzuki S, Wong R, Harris W. Cellular requirements for building a retinal neuropil. Cell Rep. 2013;3:282–290. doi: 10.1016/j.celrep.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven M, Eglen S, Ohab J, Reese B. Determinants of the exclusion zone in dopaminergic amacrine cell mosaics. J Comp Neurol. 2003;461:123–136. doi: 10.1002/cne.10693. [DOI] [PubMed] [Google Scholar]
- Raven M, Oh E, Swaroop A, Reese B. Afferent control of horizontal cell morphology revealed by genetic respecification of rods and cones. J Neurosci. 2007;27:3540–3547. doi: 10.1523/JNEUROSCI.0372-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockhill R, Daly F, MacNeil M, Brown SP, Masland RH. The diversity of ganglion cells in a mammalian retina. J Neurosci. 2002;22:3831–3843. doi: 10.1523/JNEUROSCI.22-09-03831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, de Müller L, Brecha N. The RNA binding protein RBPMS is a selective marker of ganglion cells in the mammalian retina. J Comp Neurol. 2014;522:1411–1443. doi: 10.1002/cne.23521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roska B, Werblin F. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature. 2001;410:583–587. doi: 10.1038/35069068. [DOI] [PubMed] [Google Scholar]
- Rossi C, Strettoi E, Galli-Resta L. The spatial order of horizontal cells is not affected by massive alterations in the organization of other retinal cells. J Neurosci. 2003;23:9924–9928. doi: 10.1523/JNEUROSCI.23-30-09924.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz F, Königstorfer A, Südhof T. RIBEYE, a component of synaptic ribbons: a protein’s journey through evolution provides insight into synaptic ribbon function. Neuron. 2001;28:857–872. doi: 10.1016/s0896-6273(00)00159-8. [DOI] [PubMed] [Google Scholar]
- Schneider C, Rasband W, Eliceiri K. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert S, Scherf B, Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nat Neurosci. 2009;12:1197–1204. doi: 10.1038/nn.2370. [DOI] [PubMed] [Google Scholar]
- Sheikh S, Martin S, Martin D. Regional distribution and relative amounts of glutamate decarboxylase isoforms in rat and mouse brain. Neurochem Int. 1999;35:73–80. doi: 10.1016/s0197-0186(99)00063-7. [DOI] [PubMed] [Google Scholar]
- Shi M, Kumar SR, Motajo O, Kretschmer F, Mu X, Badea TC. Genetic interactions between Brn3 transcription factors in retinal ganglion cell type specification. PLoS One. 2013;8:e76347. doi: 10.1371/journal.pone.0076347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strettoi E, Masland R. The number of unidentified amacrine cells in the mammalian retina. Proc Natl Acad Sci USA. 1996;93:14906–14911. doi: 10.1073/pnas.93.25.14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strettoi E, Volpini M. Retinal organization in the bcl-2-overexpressing transgenic mouse. J Comp Neurol. 2002;446:1–10. doi: 10.1002/cne.10177. [DOI] [PubMed] [Google Scholar]
- Strettoi E, Porciatti V, Falsini B, Pignatelli V, Rossi C. Morphological and functional abnormalities in the inner retina of the rd/rd mouse. J Neurosci. 2002;22:5492–5504. doi: 10.1523/JNEUROSCI.22-13-05492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strettoi E, Mears A, Swaroop A. Recruitment of the rod pathway by cones in the absence of rods. J Neurosci. 2004;24:7576–7582. doi: 10.1523/JNEUROSCI.2245-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardi N, Auerbach P. Specific cell types in cat retina express different forms of glutamic acid decarboxylase. J Comp Neurol. 1995;351:374–384. doi: 10.1002/cne.903510305. [DOI] [PubMed] [Google Scholar]
- Vidal-Sanz M, Salinas-Navarro M, Nadal-Nicolás F, Alzrcón-Martínez L, Valiente-Soriano F, Imperial J, Avilés-Trigueros M, Agudo-Barriuso M, Villegas-Pérez M. Understanding glaucomatous damage: anatomical and functional data from ocular hypertensive rodent retinas. Prog Retin Eye Res. 2012;31:1–27. doi: 10.1016/j.preteyeres.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Voigt T. Cholinergic amacrine cells in the rat retina. J Comp Neurol. 1986;248:19–35. doi: 10.1002/cne.902480103. [DOI] [PubMed] [Google Scholar]
- Wässle H. Parallel processing in the mammalian retina. Nat Rev Neurosci. 2004;5:747–757. doi: 10.1038/nrn1497. [DOI] [PubMed] [Google Scholar]
- Wässle H, Chun M. GABA-like immunoreactivity in the cat retina: light microscopy. J Comp Neurol. 1989;279:43–54. doi: 10.1002/cne.902790105. [DOI] [PubMed] [Google Scholar]
- Wässle H, Peichl L, Airaksinen M, Meyer M. Calcium-binding proteins in the retina of a calbindin-null mutant mouse. Cell Tissue Res. 1998;292:211–218. doi: 10.1007/s004410051052. [DOI] [PubMed] [Google Scholar]
- Wässle H, Heinze L, Ivanova E, Majumdar S, Weiss J, Harvey R, Haverkamp S. Glycinergic transmission in the mammalian retina. Front Mol Neurosci. 2009;2:6. doi: 10.3389/neuro.02.006.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney I, Keeley P, Raven M, Reese B. Spatial patterning of cholinergic amacrine cells in the mouse retina. J Comp Neurol. 2008;508:1–12. doi: 10.1002/cne.21630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney I, Keeley P, St John A, Kautzman A, Kay J, Reese B. Sox2 regulates cholinergic amacrine cell positioning and dendritic stratification in the retina. J Neurosci. 2014;34:10109–10121. doi: 10.1523/JNEUROSCI.0415-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R, Cusato K, Raven M, Reese B. Organization of the inner retina following early elimination of the retinal ganglion cell population: effects on cell numbers and stratification patterns. Visual Neurosci. 2001;18:233–244. doi: 10.1017/s0952523801182088. [DOI] [PubMed] [Google Scholar]
- Xiang M. Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev Biol. 1998;197:155–169. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Peng Y, Eddy R, Shows T, Nathans J. Brn-3b: a POU domain gene expressed in a subset of retinal ganglion cells. Neuron. 1993;11:689701. doi: 10.1016/0896-6273(93)90079-7. [DOI] [PubMed] [Google Scholar]
- Xiang M, Zhoui L, Macke JP, Yoshioka T, Hendry SHC, Roger LE, Shows TB, Nathans J. The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci. 1995;15:4762–4785. doi: 10.1523/JNEUROSCI.15-07-04762.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Gan L, Zhou L, Klein W, Nathans J. Targeted deletion of the mouse POU domain gene Brn-3a causes selective loss of neurons in the brainstem and trigeminal ganglion, uncoordinated limb movement, and impaired suckling. Proc Natl Acad Sci U S A. 1996;93:11950–11955. doi: 10.1073/pnas.93.21.11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Gan L, Li D, Chen Z-Y, Zhou L, O’Malley B, Klein W, Nathans J. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc Natl Acad Sci U S A. 1997;94:9445–9450. doi: 10.1073/pnas.94.17.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Cheong Y-K, He S-Q, Tiwari V, Liu J, Wang Y, Raja S, Li J, Guan Y, Li W. Suppression of spinal connexin 43 expression attenuates mechanical hypersensitivity in rats after an L5 spinal nerve injury. Neurosci Lett. 2014;566:194–199. doi: 10.1016/j.neulet.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Ding P, Xiao L, Hu M, Hu Z. Nestin is induced by hypoxia and is attenuated by hyperoxia in Müller glial cells in the adult rat retina. Int J Exp Pathol. 2011;92:377–381. doi: 10.1111/j.1365-2613.2011.00786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]