Abstract
Reverse genetics in Plasmodium, the genus of parasites that cause malaria, still faces major limitations. Only red blood cell stages of this haploid parasite can be transfected. Consequently, the function of many essential genes in these and subsequent stages, including those encoding vaccine candidates, cannot be addressed genetically. Here, we establish conditional mutagenesis in Plasmodium by using site-specific recombination and the Flp/FRT system of yeast. Site-specific recombination is induced after cross-fertilization in the mosquito vector of two clones containing either the target sequence flanked by two FRT sites or the Flp recombinase. Parasites that have undergone recombination are recognized in the cross progeny through the expression of a fluorescence marker. This approach should permit to dissect the function of any essential gene of Plasmodium during the haploid phase of its life, i.e., during infection of salivary glands in the mosquito and infection of both the liver and red blood cells in the mammal.
All symptoms and complications of malaria are caused by the multiplication of Plasmodium parasites inside the red blood cells (RBC) of a host. The parasite is transmitted between two mammalian hosts through mosquitoes, typically Anopheles, during blood feeding (Fig. 1). It is ingested as sexual forms (gametocytes), and fertilization occurs rapidly in the lumen of the mosquito midgut. It is then inoculated to a new mammalian host as haploid sporozoites, which transform inside hepatocytes into forms that invade RBC.
Fig. 1.
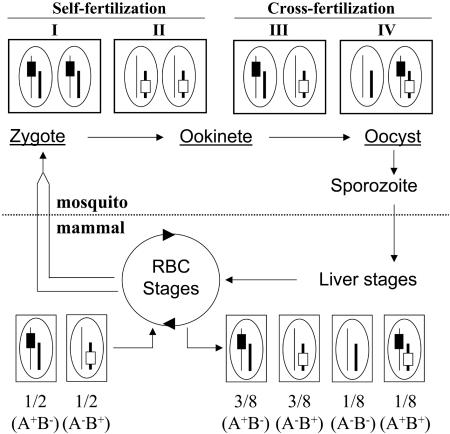
Strategy for conditional mutagenesis in Plasmodium. The life cycle of Plasmodium takes place in a mammalian and a mosquito host. The haploid, RBC stages of the parasite generate gametocytes (parallel lines). In the lumen of the mosquito midgut, released gametocytes transform into gametes, which fertilize to create a zygote. Zygotes transform into ookinetes, which differentiate into oocysts. These three parasite stages (underlined) contain the two parental genomes; oocysts contain thousands of copies of the genomes generated by meiotic reduction in the diploid zygote. Sporozoites bud off from the multinucleate oocysts, traverse mosquito salivary glands, and invade mammalian hepatocytes, where they generate RBC-infecting stages. Sporozoites, liver stages, and RBC stages are uninucleate, haploid stages of the parasite. Shown around the life cycle is a parasite cross for conditional mutagenesis; boxes and ellipses indicate parasitic cells and nuclei, respectively. Two parasite clones are mixed, each carrying one marker (black or open rectangles, symbolizing a flirted sequence and the Flp-encoding locus, respectively) on distinct chromosomes (thin or thick lines, respectively). Self-fertilization propagates the parental genotypes, A+/B- and A-/B+, in type I and II cells, respectively. Cross-fertilization either regenerates the parental genotypes within type III cells or creates new, hybrid genotypes (A-/B- and A+/B+) in type IV cells. Assuming similar frequencies of self- and cross-fertilizations, as well as random chromosome segregation during meiosis in the zygotes, then 1/8 of the sporozoites, and subsequent liver and RBC stages, should contain both markers in their haploid genome.
Genome manipulation is an essential tool for understanding key events in the Plasmodium life cycle in molecular terms. Stable transfection of the parasite (1, 2) and modification of its genome by homologous recombination (3-5) are now common procedures. The genome, however, can be manipulated only in RBC stages of the parasite, which are the only stages that can be produced in large amounts and subjected to selection. An important limitation of the current technology is that loss-of-function mutations cannot be selected in genes that play a role in parasite invasion of, or multiplication inside RBC, which are required for selection. Although evidence can be gained that a gene is important for the RBC cycle when its disruption cannot be selected, as has been reported for the leading vaccine candidates merozoite surface protein 1 (MSP-1) (6) and apical membrane antigen 1 (AMA-1) (7) and many other proteins (8), the actual function of the protein cannot be studied in the RBC or subsequent stages. Likewise, an increasing proportion of Plasmodium proteins are recognized as being produced at more than just one parasite stage, and gene inactivation in RBC stages can only reveal their earlier role in the cycle. Therefore, conditional procedures are needed for inactivating or activating genes at will during the Plasmodium life cycle.
Site-specific recombination (SSR) offers an effective way to inactivate a gene in a temporally defined manner. Two SSR systems have been widely used in eukaryotes, Cre/loxP of bacteriophage P1 and Flp/FRT of yeast. The recombinases, CRE and Flp, catalyze a reciprocal conservative recombination between two of their respective target sites, loxP and FRT, without the need for cofactors (9, 10). Depending on the position and relative orientation of the target sites, recombination can delete, invert, or insert DNA. This paper demonstrates that SSR can be used in Plasmodium berghei, a plasmodial species that infects rodents and can be cycled routinely through Anopheles stephensi mosquitoes. We developed procedures for inducing SSR in mosquito stages of the parasite by using the Flp/FRT system. The strategy is based on a cross between a parasite target clone bearing a “flirted” sequence of interest (i.e., flanked by two FRT sequences) and a deleter clone bearing the recombinase under the control of a stage-specific promoter. The progeny that has undergone the SSR event is fluorescent, allowing for both recognition of the SSR mutants and characterization of their phenotype in vitro and in vivo.
Materials and Methods
Construction of Plasmids. All targeting plasmids used in this study contain the pUC plasmid backbone and the human dihydrofolate reductase (hDHFR) selectable marker (11). Both the Flp gene and the FRT sequences originating from the yeast Saccharomyces cerevisiae have the WT sequence. Plasmids were transformed into XL10-Gold ultracompetent bacteria (Stratagene), and bacteria were grown overnight at 30°C in yeast extract tryptone medium (2YT, Becton Dickinson). For details of the strategies and sequences that are required for constructing pTARGET and pDELETER plasmids, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Parasite Transfection and Genotype Analysis. Transfection of WT P. berghei NK65 parasites, genomic DNA extraction, and analysis of the transgenic locus were performed as described in ref. 13 (see Supporting Materials and Methods).
Mosquito Infection. Equal amounts of the DELETER and TARGET parasites were mixed in a mouse to feed starved 4-day-old A. stephensi females. Midguts and salivary glands were dissected (at days 11, 13, and 15 or 15 and 18 postfeeding, respectively), and the percentage of fluorescent oocysts and sporozoites was determined, respectively. Natural transmission was performed at day 18 postfeeding. Infected mice were detected by blood smear analysis 5-6 days after transmission. The blood of positive animals was recovered and analyzed as described above (see Supporting Materials and Methods).
Three independent cross experiments were performed with the DELETER and TARGET parasites. At day 15 postfeeding, 25% (n = 252), 24% (n = 381), and 8.6% (n = 175) of fluorescent sporozoites were observed in the salivary glands. At day 18 postfeeding in the same experiments, 25% (n = 234), 29% (n = 296), and 24.5% (n = 208) of fluorescent sporozoites were detected in the glands. Averages of 19.2% and 26.2% of fluorescent sporozoites were found at days 15 and 18 postinfection, respectively.
Four independent cross experiments were performed with the TARGET and DELETER-EPI parasites. At day 15 postfeeding, an average of 29% (34%, 36%, 25%, and 22.3%; total n = 1,215) of fluorescent sporozoites were found in the salivary glands. At day 18 postfeeding, an average of 28% (27%, 27%, 25%, and 34%; total n = 8,569) of fluorescent sporozoites were observed in the salivary glands.
Results
Rationale of the Mutagenesis Procedure. Our initial goal was to develop procedures for inducing SSR in sporozoites, the parasite stage that is formed inside midgut oocysts, invades salivary glands, and is transmitted to the mammalian host. Because SSR was desired when the parasite is in the mosquito, we chose the Flp recombinase, which has an optimum activity near 30°C and remains active at the lower temperatures of parasite development in Anopheles mosquitoes (21-23°C) (14). Flp also is known to reach maximum excision levels of ≈100%, whereas Cre-mediated excision hardly exceeds 75% (15). To obtain both a flirted target and the Flp gene in the same parasite genome, we relied on fertilization between two parasite clones containing either the target or the recombinase. Fig. 1 schematizes the product of a cross between two clones having two different markers, e.g., the flirted target (A) or the Flp gene (B), on two distinct chromosomes. We assumed that (i) half of the fertilization events would be cross-fertilizations between the two parental genotypes, A+/B- and A-/B+, and that (ii) random chromosome segregation would occur during meiotic reduction in zygotes emerging from cross-fertilizations, creating the new A+/B+ and A-/B- genotypes (type IV cells in Fig. 1). After numerous mitotic divisions in the single-cell oocyst, one out of eight of the uninucleate and haploid sporozoites emerging from an A+/B- × A-/B+ cross, as well as subsequent liver and RBC stages, was expected to have the A+/B+ genotype.
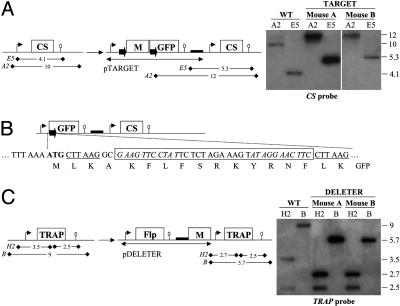
Construction and Characterization of the TARGET and DELETER Clones. To test the cross strategy, we constructed two P. berghei clones that contained either a flirted sequence or the Flp recombinase, called TARGET and DELETER, respectively. The TARGET clone was obtained after homologous integration of the plasmid pTARGET at the CS (circumsporozoite protein) locus on chromosome 4 of WT P. berghei NK65 (Fig. 2A). Plasmid pTARGET contained CS upstream region, the hDHFR selectable marker (M) flanked by FRT sequences, and the GFP gene followed by CS downstream region. In the recombinant locus called TARGET, the two 34-bp FRT sequences placed in direct orientation (i.e., leading to excision of intervening DNA upon SSR) separate the CS promoter from the GFP gene. The Flp-mediated excision of the cassette, creating the TARGET/Exc locus (Fig. 2B), was thus expected to direct fluorescence in sporozoites, where the CS promoter is active. After SSR, the residual FRT site and adjacent restriction sites would encode a 16-residue-long N-terminal extension to GFP (Fig. 2B).
Fig. 2.
Construction of the P. berghei TARGET and DELETER clones. (A)(Left) Schematic representation of the WT CS locus and the TARGET recombinant locus generated by homologous integration of plasmid pTARGET at the CS locus of WT P. berghei NK65 (not drawn to scale). Plasmid pTARGET contained 1.3 kb of CS upstream region (thin arrow), the hDHFR selectable marker (M, 1.7 kb, including its own expression sequences) flanked on either side by a FRT sequence (solid arrows), the GFP gene (0.7 kb) followed by 0.3 kb of CS downstream region (ellipse), and a pUC plasmid backbone (thick line). Plasmid pTARGET integrated via the CS promoter region, which is thus duplicated in the TARGET locus. The predicted size (in kilobases) of restriction fragments generated by digestion with EcoRV (E5) or AflII (A2) in the WT CS or the TARGET locus is shown. (Right) Southern hybridization of genomic DNA of the WT and TARGET P. berghei by using a CS internal probe. Mouse A and B correspond to parasite RBC stages collected before and after cycling through mosquitoes, respectively. (B) Schematic representation of the TARGET/Exc locus created by Flp-mediated SSR at the TARGET locus. In the TARGET/Exc locus, the 5′ promoter region of the CS gene is WT to the ATG start codon. The start codon is immediately followed by the CTTAAGGC sequence (AflII restriction site underlined), the FRT site (boxed, two inverted repeats in italics flanking the central spacer region), a CTTAAG sequence (AflII restriction site), and the full-length GFP sequence. The TARGET/Exc locus therefore encodes a GFP protein possessing the N-terminal extension MLKAKFLFSRKYRNFLK. (C) (Left) Schematic representation of the WT TRAP locus and the DELETER recombinant locus generated by homologous integration of the plasmid pDELETER at the TRAP locus of WT P. berghei NK65 (not drawn to scale). The plasmid pDELETER contained 1.5 kb of TRAP promoter region (thin arrow), the Flp gene (1.3 kb), 0.6 kb of TRAP downstream region (ellipse), the pUC backbone (thick line), and the hDHFR selectable marker (M, 1.7 kb). The plasmid pDELETER integrated via the TRAP promoter region, so that in the DELETER locus, expression of both Flp and TRAP is controlled by TRAP expression sequences. The predicted size (in kilobases) of restriction fragments generated by digestion with BamHI (B) or HincII (H2) in the WT TRAP or the DELETER locus is shown. (Right) Southern hybridization of genomic DNA of the WT and DELETER P. berghei by using a TRAP internal probe.
A DELETER clone was obtained after homologous integration of the plasmid pDELETER at the TRAP (thrombospondin-related adhesion protein) locus on chromosome 13 of WT P. berghei NK65 (Fig. 2C). The plasmid pDELETER contained both the Flp gene flanked by TRAP upstream and downstream regions and the hDHFR selectable marker. In the recombinant locus (DELETER), both the Flp and the TRAP genes were flanked by the 5′ and 3′ regulatory sequences of the TRAP gene, which are mainly active at the sporozoite stage.
We then assessed the stability of the TARGET and DELETER recombinant loci during a full parasite life cycle completed in the absence of drug pressure. The corresponding clones were separately transmitted from mouse A to A. stephensi mosquitoes by natural feeding (throughout this paper, “mouse A” and “mouse B” refer to mice infected with parasites before and after cycling through mosquitoes, respectively). The two clones produced similar numbers of sporozoites in salivary glands of mosquitoes as WT at days 15 and 18 after infective blood meal (not shown), indicating that both the CS and TRAP genes that are essential for salivary gland infection (16, 17) were normally expressed in the two clones. No fluorescent sporozoite was detected when the TARGET clone was cycled alone, indicating that SSR did not occur in the absence of exogenous Flp. Sporozoites from each of the TARGET and DELETER clones were then transmitted by natural mosquito feeding to mouse B at day 18 postinfection. In both cases, RBC stages in mouse B emerged 5-6 days postinoculation, as with WT, indicating that Flp expression throughout the sporozoite stage was not impairing parasite viability. The stability of the recombinant loci was confirmed by Southern blot analysis. In mouse B, reversion to a WT locus by plasmid excision was not detected at the CS locus of the TARGET clone (Fig. 2 A) or at the TRAP locus of the DELETER clone (Fig. 2C).
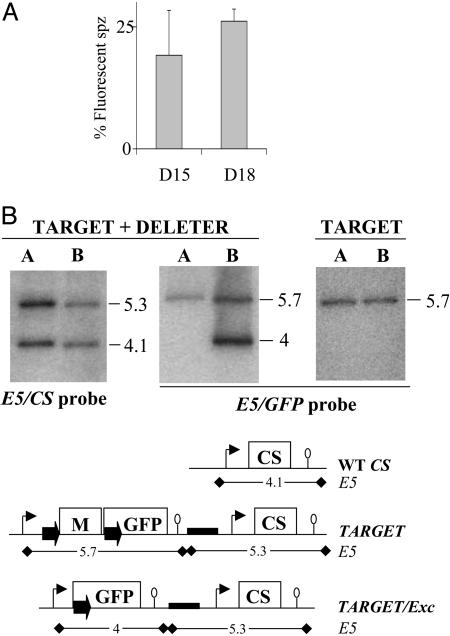
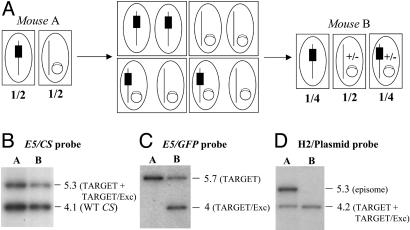
Characterization of the Progeny of TARGET × DELETER Crosses. The TARGET clone was then mixed 1:1 with the DELETER clone in mouse A, and the mixture was transmitted to mosquitoes. In three independent experiments, an average of 19% and 26% of the salivary gland sporozoites originating from the cross were brightly fluorescent at days 15 and 18 postinfection, respectively (Fig. 3A). Sporozoites from the cross, including the fluorescent ones, displayed normal gliding motility and infectivity (not shown). These sporozoites were transmitted to mouse B by natural feeding, and RBC stages in mouse A and B were analyzed by Southern blotting (Fig. 3B). When a CS probe (Left) was used, the fragments diagnostic of the TARGET loci (5.3-kb band) and of the WT CS (4.1-kb band) were detected with similar intensities in RBC stages from mouse A, in agreement with the equal proportions of the TARGET and DELETER clones fed to mosquitoes. In RBC stages from mouse B, these two fragments were still detected with similar intensities, confirming that expression of the recombinase and/or fluorescence was not impairing parasite cycling in vivo. The GFP probe (Fig. 3B Center and Right), which distinguishes the TARGET (5.7-kb band) and the TARGET/Exc (4-kb band) loci, showed the presence of the latter only in mouse B after the cross, as expected. Approximately 1/4 of all RBC stages from mouse B after the cross appeared to have a TARGET/Exc locus, i.e., the CS and GFP probes detected similar amounts of WT CS and TARGET loci, and among the latter similar amounts of nonexcised (TARGET) and excised (TARGET/Exc) alleles. Similar results were obtained in the three independent TARGET × DELETER crosses.
Fig. 3.
Progeny of a TARGET × DELETER cross. (A) The TARGET and DELETER clones were mixed in equal proportions in the same mouse and transmitted to 100 A. stephensi female mosquitoes. The percentage of fluorescent sporozoites (spz) observed in the salivary glands of infected mosquitoes at days 15 and 18 postinfection is ≈25% on average (mean of three experiments). Bars represent standard deviation values. (B) Southern hybridization of genomic DNA of the TARGET + DELETER mixture or the TARGET clone alone collected from mouse A (before mosquito infection) and mouse B (after mosquito infection). The predicted size (in kilobases) of restriction fragments generated by digestion with EcoRV (E5) at the CS, TARGET, and TARGET/Exc loci are shown. The CS probe shows a similar intensity of the EcoRV fragments corresponding to the WT CS (4.1 kb) and the TARGET + TARGET/Exc loci (5.3 kb) in both mouse A and mouse B. The GFP probe shows a similar intensity of the EcoRV fragments corresponding to the TARGET (5.7 kb) and the TARGET/Exc (4 kb) loci in mouse B. Therefore, ≈25% of all RBC stages from mouse B after the cross have a TARGET/Exc locus. The 4-kb band is not detected in mouse B when the TARGET clone is cycled alone.
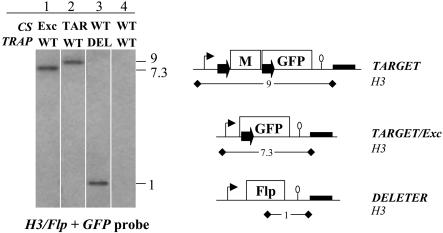
The proportion of parasites having undergone SSR after the cross at both the sporozoite and RBC stages was thus approximately twice the 1/8 ratio of double Flp/FRT segregants. In all cross experiments, fluorescence was first detected in late oocysts (see below), the multinucleate cell that gives rise to the uninucleate sporozoites. This observation indicated that the TRAP promoter controlling Flp expression was active in oocysts and suggested that SSR targets may originate not only from type IV oocysts (Fig. 1), where the flirted locus and the Flp gene are located in the same nucleus, but also from type III oocysts, where the flirted locus and the Flp gene are located in distinct nuclei in the same oocyst. We thus sought the predicted uninucleate parasites having a TARGET/Exc locus but no Flp in their genome, i.e., originating from type III oocysts. For this, RBC stages from mouse B were cloned by limiting dilution and analyzed by Southern blotting using simultaneously a GFP probe for TARGET locus analysis and an Flp probe (Fig. 4). Clones were indeed found that displayed only the 7.3-kb band diagnostic of the TARGET/Exc locus with no Flp at the TRAP locus recognized as a 1-kb band (Fig. 4, lane 1). This finding demonstrated that SSR occurred in nuclei lacking the Flp gene. We conclude that SSR occurred with ≈100% efficiency in the TARGET nuclei of cross-fertilization oocysts, by means of Flp specified by the same (type IV cells) or a distinct (type III cells) nucleus, yielding ≈25% sporozoites after the cross with a TARGET/Exc locus.
Fig. 4.
Analysis of clones from of a TARGET × DELETER cross. The RBC stages of mouse B after the TARGET × DELETER cross were cloned by limiting dilution, and their CS and TRAP loci were analyzed by Southern hybridization after HindIII (H3) digestion by using a mix of Flp and GFP internal probes. The predicted sizes (in kilobases) of restriction fragments generated by digestion with HindIII at the TARGET, TARGET/Exc, and DELETER loci are shown on the right. A clone carrying a TARGET/Exc locus without the Flp gene at the TRAP locus is shown in lane 1. Clones carrying a TARGET locus and a WT TRAP, a WT CS and DELETER locus, and a WT CS and a WT TRAP are shown in lanes 2, 3, and 4, respectively.
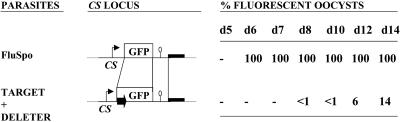
Stage Specificity of Recombinase Expression. We then assessed stage specificity of Flp expression by means of the TRAP regulatory sequences. For this assessment, we compared the onset of fluorescence in parasitic oocysts of the P. berghei clone FluSpo (12) with that of the TARGET × DELETER cross progeny (Fig. 5). In the FluSpo clone, the GFP gene is preceded by natural CS upstream sequences and followed by 300 bp of CS downstream sequences. In the TARGET/Exc parasites that emerge from the TARGET × DELETER cross, the GFP gene is flanked by identical 5′ and 3′ regulatory sequences, as in FluSpo parasites. However, GFP expression also is controlled by the TRAP regulatory sequences, which direct the formation of the TARGET/Exc locus upon Flp expression. All FluSpo oocysts were fluorescent from day 6 onward, in agreement with the known CS (circumsporozoite protein) production in young oocysts (18). In contrast, among the TARGET × DELETER progeny, significant numbers of fluorescent oocysts were not detected before day 12, and the proportion of fluorescent oocysts increased after day 12. The difference in the timing of fluorescence emergence demonstrates that Flp expression and SSR were indeed controlled by stage-specific regulatory sequences.
Fig. 5.
Stage specificity of recombinase expression. The percentage of fluorescent oocysts of the FluSpo clone and of the progeny of a TARGET × DELETER parasite cross was estimated at various days postinfection. GFP expression controlled by the CS regulatory sequences, as in the FluSpo recombinant locus, results in 100% fluorescent oocysts from day 6 onward. When a similar locus is created upon SSR controlled by the TRAP regulatory sequences (note that the residual FRT site in the TARGET/Exc locus is located inside the GFP coding sequence, not in the 5′ CS upstream region), the first fluorescent oocysts are detected in significant numbers only from day 12 onward.
Progeny of a TARGET × DELETER-EPI Cross. We next tested whether the Flp gene could be brought into the cross by an episome. For this, we crossed the TARGET clone with a deleter clone called DELETER-EPI, in which the Flp gene under the control of the TRAP regulatory sequences was carried by a pUC plasmid. As depicted in Fig. 6A, assuming that after cross-fertilizations the episome would be transmitted to the TARGET nuclei that initially lacked the episome, then 1/4 of the emerging sporozoites were expected to be fluorescent.
Fig. 6.
Progeny of a TARGET × DELETER-EPI cross. (A) Schematics of the parasite cross. In mouse A, the two clones are mixed in equal proportions, one having the integrated TARGET locus (black box) and the other carrying the Flp gene on an episome (open box on a circle). After transmission of the parasite mixture to mosquitoes, nuclei fusion during cross-fertilization should transmit the episome to the nuclei containing the TARGET locus. Assuming similar frequencies of self- and cross-fertilizations and stable maintenance of the episome in parasites resulting from cross-fertilizations, then 1/4 of the emerging sporozoites, as well as RBC stages in mouse B, are expected to carry both the TARGET locus and the episome-borne Flp. Episomes can be lost (+/-) during parasite multiplication in the oocyst, liver, and RBC in mouse B. Boxes indicate parasitic cells, and ellipses indicate nuclei. (B) Southern hybridization of the parasite mixture TARGET + DELETER-EPI collected from mouse A and mouse B by using a CS probe. The probe shows a similar intensity in mice A and B of the EcoRV (E5) fragments corresponding to the WT CS (4.1 kb) and the TARGET + TARGET/Exc loci (5.3 kb) (Fig. 3B). (C) Southern hybridization of the parasite mixture TARGET + DELETER-EPI collected from mouse A and mouse B by using a GFP probe. The probe shows a similar intensity in mice A and B of the EcoRV (E5) fragments corresponding to the TARGET (5.7 kb) and the TARGET/Exc (4 kb) loci (Fig. 3B). (D) Southern hybridization of the parasite mixture TARGET + DELETER-EPI collected from mouse A and mouse B by using a plasmid pUC probe. The probe detects a HincII (H2) fragment of 4.2 kb, corresponding to the TARGET or TARGET/Exc locus, in mouse A and mouse B. In contrast, the probe detects a HincII fragment of 5.3 kb (corresponding to the episome, not shown) in mouse A that is absent in RBC stages from mouse B.
After mixing the TARGET and DELETER-EPI parasites in a 1:1 ratio in mouse A and transmission to mosquitoes, an average of 29% and 27% of salivary gland sporozoites were fluorescent at days 15 and 18 postinfection, respectively. Parasites were transmitted to mouse B, and RBC stages in mouse A and B were analyzed by Southern blotting. The CS probe (Fig. 6B) showed the 1:1 clone ratio before parasite transmission to mosquitoes (mouse A) and the similar cycling efficiencies of the two parental CS genotypes (mouse B). As expected, the GFP probe (Fig. 6C) indicated that the TARGET/Exc locus was present in mouse B. As with the previous cross, the relative intensities of the three CS loci in mouse B (WT, TARGET, and TARGET/Exc) detected by the CS and GFP probes indicated that ≈1/4 of the parasites had the desired TARGET/Exc locus.
Finally, the plasmid probe (Fig. 6D) showed that most parasites had lost the Flp-expressing episome during parasite cycling from mouse A to B. This loss could have occurred during parasite multiplication inside oocysts (with sufficient Flp being produced for SSR efficiency to be ≈100%), or in hepatocytes or RBC of mouse B. We conclude that SSR also occurs when the Flp gene is borne by an episome, at least if SSR is sought at the late oocyst-sporozoite stages. In addition, the episome-based approach generates a majority of parasites that have lost the Flp-containing episome at the subsequent RBC stages. It may therefore be particularly useful for analyzing mutants at the RBC stage after complete cycling of the parasite.
Discussion
We have developed approaches for conditional mutagenesis in P. berghei based on Flp-mediated SSR. We have shown that Flp acts with high fidelity and efficiency in the parasite without causing deleterious effects on its life cycle in vivo. The strategy based on crossing two clones having either a TARGET or a DELETER locus ensures that potential lethality/premature SSR in RBC stages of the parasite are bypassed. Depending on the timing of expression of the Flp recombinase, the strategy should allow for studying the in vivo function of any Plasmodium protein at the parasite haploid stages, i.e., from the sporozoite in the mosquito midgut to the RBC stages in the mammalian host. If the recombinase is first produced before or after sporozoite budding off from the oocyst, then 1/4 or 1/8 of the subsequent stages should have the desired gene modification, respectively. The other genotypes created serve as internal controls, monitoring the variable efficiency of mosquito infection and allowing for quantitative assessment of the mutant phenotype.
Another approach for obtaining mutants by SSR would be to introduce both the flirted target and the recombinase in the same genome, sequentially or in a single construct. In this case, 100% of the parasites could be the desired mutants. However, the stage-specific promoter that controls expression of the recombinase would need to be effectively off before the stage of interest, particularly during the erythrocytic cycle, to avoid premature SSR.
The nature of the SSR event depends on the relative orientation of the asymmetric FRT sites (Fig. 2B). The intervening DNA is excised when the target sites are positioned as direct repeats, allowing for gene knock-out construction (see Fig. 7, which is published as supporting information on the PNAS web site, for an example of conditional gene deletion). The excision reaction is effectively irreversible because the circular reaction product is lost and reintegration of the excised circle, which is a bimolecular reaction, is kinetically less favorable than excision. Thus, excision products accumulate and the maximum recombination can approach 100% in conditions of excess recombinase (15). Conversely, the intervening DNA is inverted when the FRT sites are placed as inverted repeats. In this case, the recombination reaction product harbors two identical target sites in cis, which are themselves substrates for further recombination. However, mutant FRT sites have now been generated for engineering stable DNA inversions, as spacer variants (19-21) or inverted-repeat variants (22, 23). Such sites allow efficient SSR between homotypic, but not heterotypic, sites created by the first inversion event, making inversion reactions effectively irreversible. They should permit stage-specific expression of a modified version of a protein for structure-function analysis, such as by flipping the orientation of a promoter between two gene versions at a given time point in the parasite's life.
Linking gene deletion to expression of a fluorescent protein, as reported here, should be crucial for characterizing the phenotype of the mutant in vivo, and therefore understanding the function of the target gene product. GFP is expressed under control of the regulatory sequences of the target gene and is therefore expressed in targetless parasites only within the window of time that the target is normally expressed. With the development of techniques for in vivo imaging of various parasite stages (24, 25), fluorescence will allow the study of the behavior of SSR mutants in vivo.
The suitability of the technique will ultimately depend on construction of deleter clones for timely expression of the recombinase and induction of SSR. The TRAP promoter used here should prove useful to induce SSR in midgut sporozoites before infection of mosquito salivary glands, for example, to assess the role in salivary gland invasion of parasite molecules that are common to sporozoites and RBC stages. Another promoter that should prove useful is one active specifically in sporozoites located inside salivary glands, for example, the promoter of the spect gene (26) or of genes identified by differential expression screens between sporozoites and RBC stages (27). SSR induction in salivary gland sporozoites, the most highly motile and invasive stage of the parasite, should open the way to a functional analysis of the parasite surface motor, which cannot be studied by using conventional gene-targeting techniques. It also should permit addressing the function of proteins that are involved in sporozoite invasion of and differentiation inside hepatocytes, most of which also are involved in merozoite invasion of and differentiation inside RBC. To study RBC stages, a promoter that is specifically active in liver stages may be necessary, but any promoter that is active in the mosquito stages (such as TRAP) may be sufficient to inactivate genes that are specifically expressed in RBC stages.
As more stage-specific promoters are being identified by whole-genome expression profiling (28, 29), deleter clones expressing the recombinase at precise times of the parasite life cycle will become available for crossing with target clones of interest. Conditional mutagenesis by means of Flp-mediated SSR now allows us to reach beyond the first required function of a gene, providing a useful tool for further dissecting the molecular basis of key steps in the parasite life cycle.
Supplementary Material
Acknowledgments
We thank the Centre for Production and Infection of Anopheles for mosquito production; Sabrina Spitz for help in infecting mice; and Patricia Baldacci, Catherine Bourgouin, and Freddy Frischknecht for a critical review of the manuscript. T.G.C. was supported by fellowships from the Fondation Schlumberger and the Fondation pour la Recherche Médicale. R.M. is a Howard Hughes Medical Institute International Scholar.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: SSR, site-specific recombination.
References
- 1.Wu, Y., Sifri, C. D., Lei, H. H., Su, X. Z. & Wellems, T. E. (1995) Proc. Natl. Acad. Sci. USA 92, 973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Dijk, M. R., Waters, A. P. & Janse, C. J. (1995) Science 268, 1358-1362. [DOI] [PubMed] [Google Scholar]
- 3.Wu, Y., Kirkman, L. A. & Wellems, T. E. (1996) Proc. Natl. Acad. Sci. USA 93, 1130-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Dijk, M. R., Janse, C. J. & Waters, A. P. (1996) Science 271, 662-665. [DOI] [PubMed] [Google Scholar]
- 5.Crabb, B. S. & Cowman, A. F. (1996) Proc. Natl. Acad. Sci. USA 93, 7289-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Donnell, R. A., Saul, A., Cowman, A. F. & Crabb, B. S. (2000) Nat. Med. 6, 91-95. [DOI] [PubMed] [Google Scholar]
- 7.Triglia, T., Healer, J., Caruana, S. R., Hodder, A. N., Anders, R. F., Crabb, B. S. & Cowman, A. F. (2000) Mol. Microbiol. 38, 706-718. [DOI] [PubMed] [Google Scholar]
- 8.Cowman, A. F., Baldi, D. L., Healer, J., Mills, K. E., O'Donnell, R. A., Reed, M. B., Triglia, T., Wickham, M. E. & Crabb, B. S. (2000) FEBS Lett. 476, 84-88. [DOI] [PubMed] [Google Scholar]
- 9.Dymecki, S. M. (2000) in Gene Targeting: A Practical Approach, ed. Joyner, A. L. (Oxford Univ. Press, London), pp. 37-99.
- 10.Branda, C. S. & Dymecki, S. M. (2004) Dev. Cell 6, 7-28. [DOI] [PubMed] [Google Scholar]
- 11.de Koning-Ward, T. F., Fidock, D. A., Thathy, V., Ménard, R., van Spaendonk, R. M., Waters, A. P. & Janse, C. J. (2000) Mol. Biochem. Parasitol. 106, 199-212. [DOI] [PubMed] [Google Scholar]
- 12.Natarajan, R., Thathy, V., Mota, M., Hafalla, J. C., Ménard, R. & Vernick, K. D. (2001) Cell. Microbiol. 3, 17-23. [DOI] [PubMed] [Google Scholar]
- 13.Thathy, V. & Ménard, R. (2002) Methods Mol. Med. 72, 317-331. [DOI] [PubMed] [Google Scholar]
- 14.Buchholz, F., Ringrose, L., Angrand, P. O., Rossi, F. & Stewart, A. F. (1996) Nucleic Acids Res. 24, 4256-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ringrose, L., Lounnas, V., Ehrlich, L., Buchholz, F., Wade, R. & Stewart, A. F. (1998) J. Mol. Biol. 284, 363-384. [DOI] [PubMed] [Google Scholar]
- 16.Ménard, R., Sultan, A., Cortes, C., Altszuler, R., van Dijk, M. R., Janse, C. J., Waters, A. P., Nussenzweig, R. S. & Nussenzweig, V. (1997) Nature 385, 336-340. [DOI] [PubMed] [Google Scholar]
- 17.Sultan, A., Thathy, V., Frevert, U., Robson, K., Crisanti, A., Nussenzweig, V., Nussenzweig, R. S. & Ménard, R. (1997) Cell 90, 511-522. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton, A. J., Davies, C. S. & Sinden, R. E. (1988) Parasitology 96, 273-280. [DOI] [PubMed] [Google Scholar]
- 19.Schlake, T. & Bode, J. (1994) Biochemistry 33, 12746-12751. [DOI] [PubMed] [Google Scholar]
- 20.Bode, J., Schlake, T., Iber, M., Schubeler, D., Seibler, J., Snezhkov, E. & Nikolaev, L. (2000) Biol. Chem. Hoppe-Seyler 381, 801-813. [DOI] [PubMed] [Google Scholar]
- 21.Baer, A. & Bode, J. (2001) Curr. Opin. Biotechnol. 12, 473-480. [DOI] [PubMed] [Google Scholar]
- 22.Senecoff, J. F. & Cox, M. M. (1986) J. Biol. Chem. 261, 7380-7386. [PubMed] [Google Scholar]
- 23.Senecoff, J. F., Rossmeissl, P. J. & Cox, M. M. (1988) J. Mol. Biol. 201, 405-421. [DOI] [PubMed] [Google Scholar]
- 24.Frischknecht, F., Baldacci, P., Martin, B., Zimmer, C., Thiberge, S., Olivo-Marin, J. C., Shorte, S. L. & Ménard, R. (2004) Cell. Microbiol. 6, 687-694. [DOI] [PubMed] [Google Scholar]
- 25.Vlachou, D., Zimmermann, T., Janse, C. J., Waters, A. P. & Kafatos, F. C. (2004) Cell. Microbiol. 6, 671-685. [DOI] [PubMed] [Google Scholar]
- 26.Ishino, T., Yano, K., Chinzei, Y. & Yuda, M. (2004) PLoS Biol. 2, 77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaiser, K., Matuschewski, K., Camargo, N., Ross, J. & Kappe, S. (2004) Mol. Microbiol. 51, 1221-1232. [DOI] [PubMed] [Google Scholar]
- 28.LeRoch, K. G., Zhou, Y., Blair, P. L., Grainger, M., Moch, J. K., Haynes, J. D., De la Vega, P., Holder, A., Batalov, S., Carucci, D. J. & Winzeler, E. A. (2003) Science 301, 1503-1507. [DOI] [PubMed] [Google Scholar]
- 29.Bozdech, Z., Llinas, M., Pulliam, B. L., Wong, E. D., Zhu, J. & DeRisi, J. L. (2003) PLoS Biol. 1, 85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.