Abstract
To address the poor prognosis of mixed lineage leukemia (MLL)-rearranged infant acute lymphoblastic leukemia (iALL), we generated a panel of cell lines from primary patient samples and investigated cytotoxic responses to contemporary and novel Food and Drug Administration-approved chemotherapeutics. To characterize representation of primary disease within cell lines, molecular features were compared using RNA-sequencing and cytogenetics. High-throughput screening revealed variable efficacy of currently used drugs, however identified consistent efficacy of three novel drug classes: proteasome inhibitors, histone deacetylase inhibitors and cyclin-dependent kinase inhibitors. Gene expression of drug targets was highly reproducible comparing iALL cell lines to matched primary specimens. Histone deacetylase inhibitors, including romidepsin (ROM), enhanced the activity of a key component of iALL therapy, cytarabine (ARAC) in vitro and combined administration of ROM and ARAC to xenografted mice further reduced leukemia burden. Molecular studies showed that ROM reduces expression of cytidine deaminase, an enzyme involved in ARAC deactivation, and enhances the DNA damage–response to ARAC. In conclusion, we present a valuable resource for drug discovery, including the first systematic analysis of transcriptome reproducibility in vitro, and have identified ROM as a promising therapeutic for MLL-rearranged iALL.
Introduction
Infants with acute lymphoblastic leukemia (iALL) comprise a high-risk group with inferior outcome compared with older children.1 Adverse risk factors include presence of a mixed lineage leukemia (MLL/KMT2A) gene rearrangement (MLL-r), hyperleukocytosis at presentation, age <90 days at diagnosis and poor response to initial prednisone therapy.2, 3 Contemporary treatment uses dose-intensive chemotherapy combinations. However, 5-year event-free survival remains <40% for MLL-r iALL,4, 5 highlighting the need to identify novel drugs to improve outcome.
Genomic profiling and high-throughput compound screening of patient-derived cell lines have identified personalized treatments in breast6 and lung7 cancer and catalogued predictive biomarkers of drug–responses across large collections of human cancer cell lines.8, 9, 10 However, in vitro cancer drug screening is limited by the absence of cell line characterization in relation to the primary disease. For example, over 40 leukemia cell lines have been reported as MLL-r, including monocytic (for example, MV4-11, MOLM-13, THP-1), immature T-ALL (for example, Karpas 45, SUP-T13) and B-cell precursor ALL (for example, SEM, RS4;11); but there are few reports verifying the molecular representation of cell lines derived from rare clinical sub-types, such as iALL.11
We previously demonstrated variable cytotoxic response between two iALL cell lines to contemporary chemotherapeutics12 highlighting the need to test multiple patient-derived lines. Thus, a panel of genetically characterized cell lines derived from iALL patients with defined clinical features is a crucial resource for drug discovery. To address these requirements, we established cell lines from infants with high-risk MLL-r iALL, performed a comprehensive molecular comparison with primary specimens and assessed drug sensitivity in vitro. We identified several novel drug classes with consistent efficacy, including histone deacetylase inhibitors (HDACi), proteasome inhibitors and cyclin-dependent kinase inhibitors. Pre-clinical assessment of romidepsin (ROM), a US Food and Drug Administration (FDA)-approved HDACi, revealed synergy with cytarabine (ARAC) in vitro and further reduction of leukemic burden in vivo. Thus, we describe a novel resource for drug discovery and identify promising new therapies for iALL.
Materials/subjects and methods
Patient specimens and cell lines
Cell lines were established as described12, 13 from bone marrow or peripheral blood leukocytes of five infants, diagnosed with iALL at Princess Margaret Hospital for Children, Perth, WA, Australia (Table 1). The Institutional Review Board approved all studies and parental informed consent was obtained. Pre-treatment patient specimens and cell lines were cultured, unstimulated, for 24 h for cytogenetic analysis. G-banded metaphases and clonal chromosomal abnormalities were reported according to the International System for Human Cytogenetic Nomenclature. Metaphase and interphase fluorescent in situ hybridization analysis was performed using the MLL break apart probe (Abbott Molecular, Des Plaines, IL, USA). Doubling times were determined by absolute cell counts measured by trypan blue exclusion over 10 days. DNA fingerprinting was performed by the Genetic Resources Core Facility at the Johns Hopkins School of Medicine, using the GenePrint 10 kit (Promega, Madison, WI, USA).
Table 1. Clinical characteristics of five infants with MLL-rearranged acute lymphoblastic leukemia and characterization of nine patient-derived cell lines.
| Patient ID | Sex | Age (days) at diagnosis | White blood count at presentation ( × 109/l) | CNS status at diagnosis | Upfront therapy | Relapse | Time from diagnosis to relapse (months) | Relapse therapy | Outcome | Cell line | Doubling time (hours) | Karyotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P272 | Female | 339 | 317 | CNS 1 | CCG 1883 | Bone marrow | 3 | CCG 1008 | Died of disease; overall survival 5 months | PER-485A | 48 | 47, XX,der(4)t(4;11)(q21;q23), add(4)(p16),+6, del(7)(p14),add(8)(q24.3),der(9)inv(9)(p11q12) del(9)(p24), der(11)t(4;11)(q21;q23) |
| P287 | Female | 9 | 4.2 | Unknown | CCG 1883 | Bone marrow | 3 | CCG 0922 | Died of disease; overall survival 4 months | PER-490A | 72 | 46, XX,t(4;11)(q21;q23)/46,XX,t(4;11),dup(1) (q12q44)/46,XX,t(4;11),der(2)t(1;2)(q12;q37); mosaic karyotype. |
| P337 | Female | 82 | 564 | CNS 2 | CCG 1901-HSCT | Bone marrow | 16 | CCG 1882-2nd HSCT | Alive without disease; 21 years of age at last follow up | PER-784A | ND | 46,XX,t(2;11)(q37;q23)[15].ish t(2;11)(3̀MLL+5̀MLL+)[10].nuc ish(MLLx2) (5̀MLLsep3̀MLLx1)[198/200] der(11), der(2) and der(13) in complex rearrangement (FISH). |
| PER-826A | 192 | Nuc ish(MLLx2)(5̀MLLsep3̀MLLx1)[100] der(11), der(2) and der(13) in complex rearrangement (FISH). | ||||||||||
| P399 | Female | 66 | 670 | Unknown | CCG 1953 | No | _ | _ | Alive without disease; 19 years of age at last follow up | PER-785A | 60 | 46,XX,t(4;11)(q21;q23),der(19)t(1;19)(q12;p13.3) [16]/46,idem,der(16)t(1;16)(q21;q12.1)[4].ish t(4;11)(3̀MLL+5̀MLL+)[5] mosaic karyotype. |
| PER-785S | ND | 46,XX,t(4;11)(q21;q23),der(19)t(1;19)(q12;p13.3) [16]/46,idem,der(16)t(1;16)(q21;q12.1)[2].ish t(4;11)(3̀MLL+5̀MLL+)[5] mosaic karyotype. | ||||||||||
| P810 | Female | 52 | 102 | CNS 2 | COG P9407-HSCT | No | _ | _ | Died from hepatic sinusoidal obstruction syndrome post HSCT; overall survival 4 months | PER-703A | 60 | 46~49,XX,+der(1)t(1;11)(p32;q23), t(1;11)(p32;q23),+3,+7,+8,-11[cp6] |
| PER-703S | ND | 48~51,XX,+der(1)t(1;11)(p32;q23),t(1;11) (p32;q23),+3,+7, +8[cp6] | ||||||||||
| PER-733S | ND | 47,XX,+der(1)t(1;11)(p32;q23),t(1;11)(p32;q23), add(22)(q13)[7] |
Abbreviations: CCG, Children's Cancer Group; CNS, central nervous system; COG, Children's Oncology Group; FISH, fluorescence in situ hybridization; HSCT, hematopoietic stem cell transplantation; MLL, mixed lineage leukemia; ND, not determined.
RNA-sequence analysis
RNA-seq (100 bp paired end) was performed using the Illumina TruSeq RNA Sample Preparation kit on a HiSeq 2000 (Illumina, Inc., San Diego, CA, USA) at the Australian Genome Research Facility, Melbourne. Raw (fastQ) files were filtered using Trimmomatic14 and aligned using TopHat215 with a reference annotation GTF from Gencode version(v)19. Gene-counts were generated with HTSeq16 and data were normalized using RUVseq (v1.1.1),17 implementing factor analysis of control genes. 'In-silico empirical' negative control genes were identified by fitting a linear model with grouping of primary and derived cell line data as a covariate. voom18 (limma v3.20.9) was used to normalize for library size. Count data from paired primary and derived cell lines was compared using the Irreproducible Discovery Rate (IDR) package (v1.2) with default settings.19 Analyses were performed using R (v3.1.2). Gene ontology enrichment analysis was performed using DAVID.20 MLL-fusion split-reads were identified with FusionFinder21 using default settings. Split-reads containing identical strings of MLL- and candidate fusion gene sequences were extracted using ‘grep', searching for chimeric sequences with 15 nucleotides flanking the fusion.
In vitro drug sensitivity
In vitro cell viability assays were performed using a modified alamarBlue assay with cells in logarithmic growth. After 72 h drug exposure, alamarBlue reagent was added and cell viability determined by fluorescence intensity (excitation 555 nm, emission 585 nm). Synergy experiments focused on drugs that form a key component of iALL therapy, ARAC and dexamethasone, combined with novel drugs identified from our screen, bortezomib and ROM, with biological replicates (n=2). Experimental conditions are provided in the Supplementary Information.
Antibodies, immunoblotting and fluorescence staining
The antibodies used were as follows: α-CD19-FITC (Cat 555412, BD Biosciences, Franklin Lakes, NJ, USA), goat α-rabbit–APC (Cat 3846 Santa Cruz Biotechnology, Santa Cruz, CA, USA), rabbit polyclonal α-cytidine deaminase (CDA) (SAB 1300717; Sigma, St Louis, MO, USA), rabbit polyclonal α-deoxycytidine kinase (DCK) (ab151966; Abcam, Cambridge, MA) and α-Actin (A1978-200UL, Sigma). Flow cytometry was performed with the BD LSRII (BD Biosciences). Leukemia burden was quantified by harvesting mouse organs and subjecting cell suspensions to flow cytometry with α-human CD19-FITC antibody or no stain. Data analysis was performed using FlowJo software v10.0.5 (Tree Star, Ashland, OR, USA). For dual PI-γ-H2AX staining, cells were fixed and permeabilised with 1% formaldehyde and 0.1% triton-X. Cells were incubated with 2 µl of primary α-Phospo-Histone γ-H2A.X rabbit mAb (Cat 9718, Cell Signaling Technology, Danvers, MA, USA) for 20 min; and with 1 ul of secondary goat α-rabbit–APC for 20 min. PI was added (final concentration 100 µg/m) before flow cytometric analysis. Cell lysates were prepared for immunoblot using Evans buffer and proteins subject to SDS-PAGE using the NuPage 4–12% Bis-Tris Gel (NP0321, Life Technologies, Carlsbad, CA, USA). Membranes were scanned using the ChemiDoc MP Imaging System (Bio-Rad, Philadelphia, PA, USA).
Drug–response in vivo
Eight-week-old female NOD/SCID mice were inoculated with 1 × 106 PER-785A or PER-826A cells by tail vein injection. Leukemia levels were assessed following inoculation to determine time of reproducible engraftment in bone marrow. Drugs were administered by intraperitoneal injection beginning on day 11 for PER-785A or day 18 for PER-826A xenografts. The potential toxicity of combined administration of ROM and ARAC was assessed with dose-finding experiments by treating NOD/SCID mice with 1 mg/kg ROM twice/week together with graded doses of ARAC (100 mg/kg, 75 mg/kg, 50 mg/kg) three times/week. This revealed that mice receiving the highest ARAC dose showed no overt signs of toxicity assessed by comprehensive monitoring of pain, distress, discomfort, weight gain or weight loss. To determine therapeutic outcome mice were randomized and treated for 3 weeks with either 100 mg/kg ARAC three times/week or 1 mg/kg ROM twice/week or ARAC and ROM combined. Combined ARAC–ROM drug administrations were performed by injecting ROM either 6 or 18 h before ARAC.22 Four days post completion of therapy leukemia burden was assessed by the proportion of human CD19+ positive live cells.
Statistical analysis
Dose–response curves were used to calculate IC50. The area above the dose–response curve (activity area) was determined as described.8 Bar graphs, dot-plots and statistics were generated using GraphPad Prism (v6.0c) with two-sample t-tests or one-way analysis of variance and Tukey's multiple comparison tests. Synergy was scored using Chalice software (Horizon CombinatoRx Inc., Cambridge, MA, USA), applying the Bliss-independence model.23 Bliss-excess synergy scores were summed across matrices to rank combination effects. Scatterplots and heatmaps were drawn with gplots and ggplot2 and hierarchical clustering and correlation analysis were performed in R (v3.1.2).
Results
Establishment and characterization of iALL cell lines
Cell lines were generated from four infant ALL patients diagnosed at <90 days of age and one relapse patient, who was initially diagnosed at 339 days (Table 1). Fluorescence in situ hybridization (FISH) identified the MLL-rearrangement in each patient specimen and matched cell line. G-banded chromosomal analysis revealed sideline clones in cell lines, with structural and numerical changes undetected in patient specimens.12, 13, 24 DNA fingerprinting revealed identical short tandem repeat profiles for each patient cell line pair except for cell line PER-703A. It lacked a single marker at the TH01 locus on chromosome 11 (Supplementary Table S1), which corresponded with loss-of-heterozygosity of chromosome 11 in this cell line. These results confirmed 100% concordance of DNA markers in cell lines and patient specimens. Immunophenotypic analysis of cell lines revealed a phenotype expressing B-lymphoid (CD19 or CD24) and myeloid (CD33) markers (Table 2). Cell lines PER-784A and PER-826A were also positive for CD7.
Table 2. Immunophenotypes of infant acute lymphoblastic leukemia cell lines.
| Cell line | CD7 | CD19 | CD24 | CD33 | CD34 | CD38 | HLA-DR | CD45 |
|---|---|---|---|---|---|---|---|---|
| PER-485A | −ve | −ve | 17% | 99% | −ve | 24% | −ve | 98% |
| PER-490A | −ve | 98% | −ve | 52% | −ve | 100% | 94% | 66% |
| PER-784A | 90% | 95% | 97% | 65% | −ve | 95% | 85% | 99% |
| PER-826A | 68% | 97% | 98% | 80% | −ve | 99% | 94% | 99% |
| PER-785A | −ve | 99% | 99% | 30% | 80% | 98% | 91% | 99% |
| PER-785S | −ve | 96% | 95% | 49% | 75% | 99% | ND | 99% |
| PER-703A | −ve | −ve | 98% | 99% | −ve | 90% | −ve | 98% |
| PER-703S | −ve | −ve | 99% | 99% | −ve | 95% | −ve | 97% |
| PER-733S | −ve | −ve | 70% | 94% | −ve | 30% | 70% | 92% |
Detection of MLL-fusion transcripts
Concordant MLL-fusion partners (Supplementary Table S2) were found for each patient cell line pair. Expression of the reciprocal fusion transcript was detected in two patient samples (P810 (EPS15-MLL) and P399 (AFF1-MLL)) and in each of their matched cell lines. Alternative splice junctions fusing variant MLL exons with partner genes were observed in three patients (P287, P377 and P399), with the same splice variants identified in matched cell lines indicating concordance of MLL-fusion transcripts in iALL cell lines.
Comparative transcriptome analysis
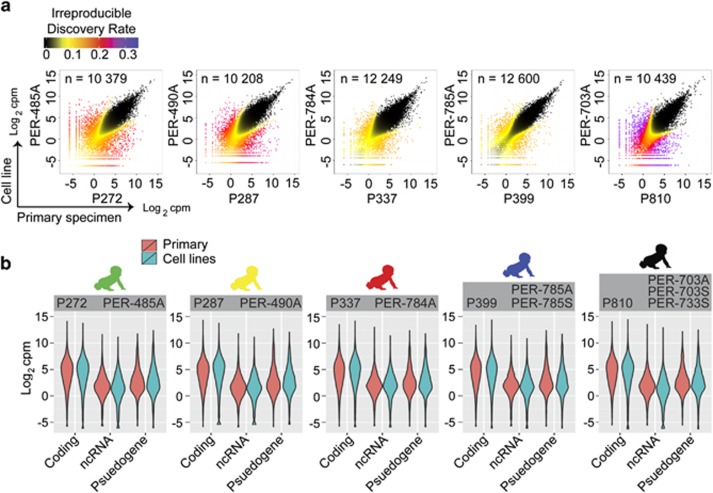
The RNA-seq data set comprised 614 million paired end reads, with a median of 47 million read pairs per sample. There were 15 612 expressed genes, defined with a detection cutoff of 75+ reads in two or more of the 13 samples analyzed (five primary and eight cell lines). Using IDR<0.05, concordant expression was observed in up to 81% of genes (range: 65–81% total transcripts; n=10208–12655; Figure 1a, Supplementary Figure S1A). By intersecting concordantly expressed genes in each cell line and primary sample pairing, we found 8677 genes (56% of all detectable transcripts), representing the iALL transcriptome with reproducible expression across primary samples and multiple cell cultures. Analysis of gene-sets characteristic of hematopoietic progenitors25 revealed similar expression distributions in primary specimens and corresponding cell lines (Supplementary Table S3; Supplementary Figure S1B).
Figure 1.
Comparative analysis of iALL patient and matched cell line transcriptomes. (a) Scatterplots of normalized RNA-seq count data (log2 counts per million) displaying correspondence between patient sample (x axes) and a representative matched cell line (y axes) defined using the IDR algorithm. Data points are coloured according to IDR value. The number of genes showing corresponding expression (below a cutoff IDR<0.05) for paired samples are shown within each plot. (b) Violinplots displaying RNA-seq count data from patient and matched cell lines partitioned by gene categories defined using Ensembl annotations, including protein-coding genes (n=12130), non-coding RNAs (n=1855) and pseudogenes (n=1261).
Discordant expression was defined as genes detected in paired samples at levels less than 0.0625 and greater than two counts per million (cpm), revealing 203 genes (1.3% total transcripts) expressed at higher levels in cell lines relative to the matched patient specimen, including 41 genes identified in two or more unrelated cell lines (Supplementary Figure S1C; Supplementary Table S4). There were 729 genes (4.7% total transcripts) expressed at lower levels in cell lines relative to patient specimens (Supplementary Figure S1D; Supplementary Table S5), including 233 genes in two or more unrelated cell lines. Ontology analysis of genes at higher levels in cell lines (152 genes mapped to Gene Ontology terms; Supplementary Table S4) suggests an over-representation of inflammatory response (15-fold enrichment; Benjamini–Hochberg (BH) adjusted P=0.37; for example, IL2RA, CD276) and cell signalling (3.1-fold enrichment; BH adjusted P=0.18; for example, SIGLEC6, PMP22) genes. In contrast, genes at lower levels in cell lines (569 genes mapped to Gene Ontology terms; Supplementary Table S5) are over-represented in immune response (3.0-fold enrichment; BH adjusted P=1.18 × 10−10; for example, CR1, CR2), antigen-binding (7.9-fold enrichment; BH adjusted P=3.49 × 10−5; for example, Fc-receptors, immunoglobulins and T-cell receptors) and hematopoietic cell lineage (3.9-fold enrichment; BH adjusted P=0.04; for example, KIT, CD14, IL1R2) genes.
Gene expression stratified by Ensembl gene-type annotations (n=15 250) identified 12 130 protein-coding genes (80% of annotated transcripts). Among non-coding (nc) RNA bio-types, processed pseudogenes (n=922) were the most numerous followed by anti-sense-RNA (n=915) and long intergenic RNA (n=556). The set of ncRNAs were consolidated to compare expression of the major RNA bio-types including protein-coding genes, pseudogenes (n=1261) and other ncRNA genes (n=1855) (Supplementary Table S6). We found lower expression of ncRNA genes compared with protein-coding genes in primary and matched cell lines, with a similar distribution of the ncRNA bio-types (Figure 1b), consistent with data from normal human tissues.26 When cataloguing conserved in vitro expression across all of the eight cell lines (8677 annotated genes with IDR<0.05; Supplementary Table S6), we identified 7666 protein-coding transcripts, 535 pseudogenes (including 419 processed pseudogenes) and 353 ncRNA (including 190 anti-sense-RNA and 86 long intergenic RNA bio-types). These results reveal similarities in expression of RNA bio-types in primary samples and iALL cell lines.
Effective drug classes comprise inhibitors of proteasome, cyclin-dependent kinase and histone deacetylase
Our screening strategy focused on identifying novel FDA-approved agents that may also enhance chemotherapeutics. A summary of the initial and secondary screens in Supplementary Figure S2A includes drug information and experimental conditions. The initial screen of 101 FDA-approved cancer drugs (Supplementary Table S7) was performed at three doses, revealing heterogeneity in responses among cell lines (Supplementary Figure S2B). However, we identified proteasome inhibitors and HDACi drug classes that were consistently effective at nanomolar concentrations. We next assessed IC50 concentrations on a panel of drugs, including proteasome inhibitors, HDACis, conventional chemotherapeutics and their analogues and rationally chosen targeted inhibitors (Supplementary Figure S2C). These data confirmed variable responses to chemotherapeutics, including dexamethasone, prednisone, ARAC, methotrexate, 6-mercaptopurine, l-asparaginase, thioguanine, etoposide, daunorubicin and 4-hydroxycyclophosphamide. Clofarabine and gemcitabine demonstrated higher efficacy compared with conventional agents within the same drug class (that is, antimetabolites and pyrimidines) used to treat iALL. In accordance with large-scale drug screens,8 we found consistent efficacy at nanomolar range of vincristine (Figure 2).
Figure 2.
Characterization of drug–responses of iALL cell lines. Response of iALL cell lines to 12 currently used drugs, novel FDA-approved drugs and rationally chosen targeted drugs. Cell viability following 72-h drug exposure was measured by alamarBlue assay. Dose resulting in 50% cell death (IC50) was determined by testing eight drug concentrations (from 3 nM up to 10 μM doses in threefold dilutions). The heatmap shows IC50 values (log10-scale) according to the colour key; cell lines (columns) are ordered by unsupervised clustering. Drugs are ordered according to mechanism of cytotoxicity or chemical structure, with conventional clinical iALL drugs displayed in the top section.
Consistent low IC50 concentration within nanomolar range, was confirmed for several drugs belonging to proteasome inhibitor and HDACi drug classes. These included three of four proteasome inhibitors, bortezomib (IC50 range: 4–10 nM), carfilzomib (3–20 nM) and delanzomib (8–10 nM), with higher IC50 doses for MG-132 (0.67–1.25 μM). Two HDACis had consistent nanomolar IC50 doses, panobinostat (4–50 nM) and ROM (3–10 nM), with higher IC50 doses for vorinostat (0.7–10 μM) and belinostat (0.14–1.1 μM). We also observed nanomolar IC50 doses for three of five cyclin-dependent kinase inhibitors. Dinaciclib was the most potent (8–12 nM), followed by flavopiridol, (200–350 nM) and SN-032, (213–295 nM). In contrast, PD0332991 was cytotoxic in one cell line, whereas roscovitine was not cytotoxic.
We also identified drugs showing efficacy within micromolar dose range. These included a BCL2-inhibitor, obatoclax (IC50 range: 0.2–3.8 μM), and bromodomain and extra-terminal domain family inhibitors, GSK1210151 (0.16–2.4 μM), GSK525762 (0.19–2.1 μM) and JQ1 (0.15–1.35 μM). The JMJD3/UTX H3K27-demethylase inhibitor, GSK-J4, was cytotoxic in four cell lines (1.6–2.3 μM). The EZH2-inhibitor, GSK-343, was not cytotoxic. Drugs targeting receptor tyrosine kinases were generally not cytotoxic, except for lestaurtinib (0.02–1.9 μM) and crizotinib (1.5–3.3 μM).
Conserved in vitro expression of drug target genes
We used RNA-seq data to investigate the expression of drug targets in eight cell lines (Supplementary Table S8). Highly conserved expression (IDR<0.05; and expression>0.0625 cpm) of targets of chromatin inhibitors was observed in all cell lines, including genes encoding HDACs (HDAC-1 to −10; 74/80 reproducible observations), bromodomain and extra-terminal domain family proteins (BRD-2,-3,-4; 24/24), H3K27-demethylases (JMJD3 and UTX; 16/16) and H3K27-methylases (EZH1 and EZH2; 16/16; Supplementary Figure S3; Supplementary Table S8). We detected 22 transcripts encoding 26S proteasome subunits and five related pseudogenes.27 Proteasome subunit encoding genes showed broadly conserved expression in cell lines (208/216 reproducible observations; Supplementary Figure S4; Supplementary Table S8). The majority of cyclin-dependent kinase inhibitor targets were reproducibly expressed (CDK-1 to −9; 62/64 reproducible observations; Supplementary Figure S5; Supplementary Table S8), however receptor tyrosine kinase inhibitor targets showed variable expression with only 3/10 receptors (RAF1, BRAF and JAK2) expressed at reproducible levels across the cell line panel (43/75 reproducible observations; Supplementary Figure S6; Supplementary Table S8). Thus, we found broadly concordant in vitro RNA expression of molecules targeted by HDACis, proteasome inhibitors, cyclin-dependent kinase inhibitors and histone H3K27 methylase/demethylase inhibitors and variable expression of receptor tyrosine kinase-encoding genes.
Patient cell line-specific drug interactions
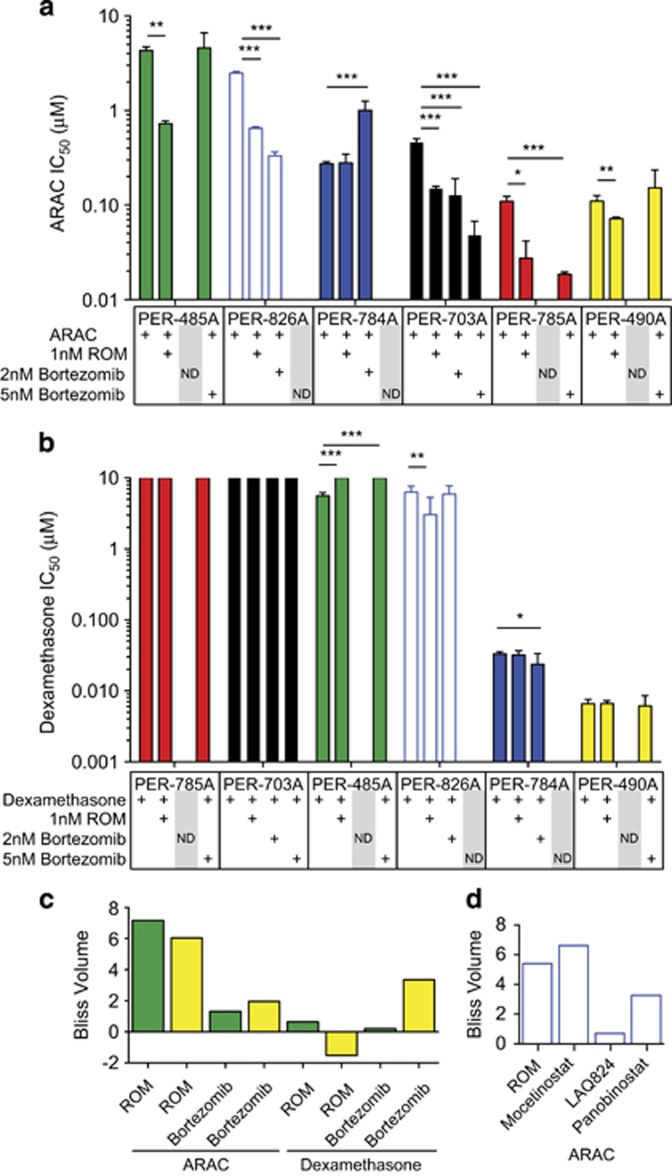
Bortezomib and ROM were the novel drugs chosen for further assessment, owing to their consistent effect in our screen, availability in a formulation suitable for infants and as they have translational potential given they are FDA-approved and undergoing clinical assessment for hematological malignancies. ROM consistently and significantly enhanced the effect of ARAC in five of six cell lines (Figure 3a), reducing the concentration of ARAC required for 50% growth inhibition by 5.9-fold (PER-485A), 4.0-fold (PER-785A), 3.8-fold (PER-826A) 3.1-fold (PER-703A) and 1.5-fold (PER-490A). ROM enhanced dexamethasone efficacy in PER-826A cell line by 1.1-fold and adversely affected dexamethasone cytotoxicity in PER-485A such that 50% growth inhibition was not achieved (Figure 3b). The effects of bortezomib on ARAC and dexamethasone efficacy were variable. Bortezomib enhanced ARAC efficacy in PER-785A (5 nM; 5.9-fold IC50 decrease), PER-703A (2 nM: 3.6-fold IC50 decrease; 5 nM: 9.7-fold IC50 decrease) and PER-826A (2 nM; 7.5-fold IC50 decrease). In contrast, bortezomib reduced ARAC efficacy in PER-784A (2 nM; 3.7-fold IC50 increase) and had no significant effect on ARAC efficacy in PER-490A or PER-485A (Figure 3a). Bortezomib enhanced the efficacy of dexamethasone in PER-784A (2 nM: 1.4-fold IC50 decrease) and in PER-785A (activity area shift from 1.5 (dexamethasone only) to 4.9 (dexamethasone with 5 nM bortezomib)) (Supplementary Figure S7); however, this difference was not captured by IC50 change because cell death did not reach 50%. Bortezomib adversely affected the efficacy of dexamethasone in PER-485A such that 50% growth inhibition was not achieved (Figure 3b). These results demonstrate consistent enhancement of ARAC efficacy by ROM with variable effects for other drug combinations. Our results confirm the inverse-correlation between IC50 and activity area across all cell lines, with r=−0.924 (Supplementary Figure S7A–C).
Figure 3.
ROM synergizes with ARAC in vitro. Cell lines (PER-485A, PER-490A, PER-703A, PER-784A, PER-826A and PER-785A) were treated for 72 h with increasing concentrations of (a) ARAC or (b) dexamethasone in the presence of ROM (1 nM) or bortezomib (2 or 5 nM). Cell viability was measured by alamarBlue and IC50 values were determined by normalizing to either DMSO controls (for dexamethasone or ARAC only treatment groups) or treatment with ROM or bortezomib only (for co-treated drug groups). Graphs display means of IC50, error bars represent 95% confidence intervals and P-values calculated by two-sample t-test (*P<0.05, **P<0.01, ***P<0.001). Drug combinations were evaluated in three replicate experiments; single-agent dexamethasone and ARAC IC50 were evaluated in six (PER-485A, PER-490A, PER-784A, PER-826A and PER-785A) or nine (PER-703A) replicates. Drug combination effects were not determined (ND) for bortezomib doses causing excessive cell death. (c) PER-485A or PER-490A cells were treated with increasing doses of ARAC or dexamethasone combined with ROM or bortezomib, at a fixed ratio with 1.2-fold incremental differences in drug concentrations. Cell viability was measured by alamarBlue at 72 h and synergy calculated by excess over Bliss. Bar graph displaying the sum of Bliss-excess scores (Bliss Volume) for each drug combination in cell lines PER-485A (green) and PER-490A (yellow). (d) PER-826A cells were treated with ARAC combined with HDACi drugs (ROM, panobinostat, mocetinostat and LAQ824) and synergy determined as described for (c).
HDAC-inhibitors synergize with ARAC in MLL-r iALL
Combination drug testing in matrix format confirmed that the ARAC–ROM combination provided the strongest synergy in both PER-485A and PER-490A cell lines (Figure 3c), when ranked by the sum of synergy scores. ARAC synergized with ROM and bortezomib in both cell lines at all doses, with ROM showing consistently larger effects (Supplementary Figure S8A–D). In contrast, antagonism was observed with ROM-dexamethasone at several doses in both cell lines (Supplementary Figure S8E and F). The bortezomib–dexamethasone combination, exceeded additive effects in PER-485A, but was antagonistic at higher bortezomib doses (Supplementary Figure S8G-H).
Synergy testing was extended using four different HDACi drugs in an independent MLL-r iALL cell line (PER-826A). We confirmed synergistic interactions at all ARAC–ROM doses tested and also for ARAC-panobinostat, ARAC-mocetinostat and ARAC-LAQ824 combinations with varying magnitudes (Figure 3d). In addition, we examined ARAC–ROM interaction in two pediatric ALL cell lines, of Pre-B and T-cell origin, lacking MLL-r and found additive effects in Pre-B-ALL cells (PER-278 with 1 nM ROM; 1.4-fold ARAC IC50 decrease) and antagonistic effects in T-ALL cells (PER-117 with 1 nM ROM; 2.6-fold ARAC IC50 increase). Thus, we found ROM enhances ARAC killing in five of six MLL-r iALL cell lines; three additional HDACi drugs capable of enhancing ARAC cytotoxicity; and ROM reduces ARAC efficacy in a germline-MLL T-ALL cell line and enhances ARAC efficacy in a germline-MLL Pre-B-ALL cell line.
ROM modulates cytidine-metabolizing enzymes and ARAC-induced DNA damage–response
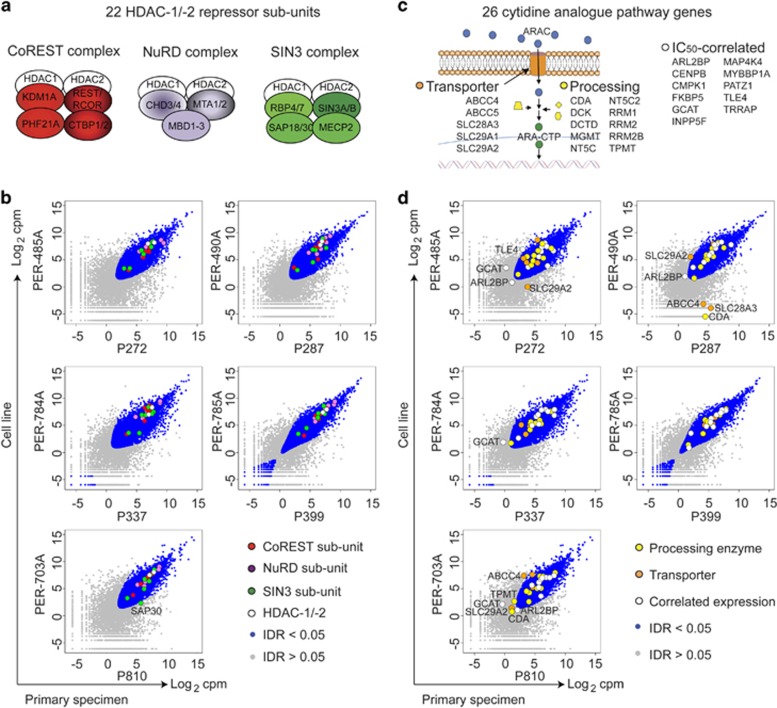
Next, we examined the expression of genes and pathways that could mediate the observed ARAC–HDACi combination drug effects (Supplementary Table S7) at steady state, and in perturbation experiments. HDAC-1/-2 repressor complex subunits (Figure 4a) were analyzed, as these are potential targets of ROM, panobinostat and mocetinostat.28 We detected 22 HDAC-repressor subunits,29 at a cutoff of 0.0625 cpm. These include seven NuRD subunits, six CoREST subunits, seven SIN3 subunits and two core enzymes (HDAC-1/-2) with highly conserved in vitro expression (175/176 reproducible observations; Figure 4b; Supplementary Table S7). In addition, we examined genes within the 'cytidine analogue pathway' involved in ARAC metabolism30 detecting expression of up to 26/32 genes in primary samples (Figure 4c); the majority with conserved expression in vitro (172/201 reproducible observations; Figure 4d; Supplementary Table S7). We subjected the data to cluster analysis focusing on genes within the 'cytidine analogue pathway', which revealed separation of cell lines according to ARAC sensitivity (Supplementary Figure S9). DCK was expressed highest in the cell lines most sensitive to ARAC, whereas CDA showed lower expression in the most sensitive cell lines, consistent with reports that ARAC cytotoxicity is limited by lower DCK activity and higher CDA activity.31, 32
Figure 4.
Conserved in vitro expression of HDAC repressors and modulators of ARAC cytotoxicity. (a) 22 subunits of HDAC-1/-2 repressor complexes were detected by RNA-seq. (b) Scatterplots of RNA-seq counts from primary samples (x axes) versus cell lines (y axes) with points coloured to show components of CoREST (red), NuRD (purple) and SIN (green) or IDR value (blue: IDR<0.05; grey: IDR>0.05). Genes of interest with IDR>0.05 are labelled within plot. (c) Twenty-six genes associated with the 'cytidine pathway'30 were detected including five membrane-bound transporters, 10 cytosolic processing enzymes and 11 genes whose expression levels correlate with ARAC sensitivity. (d) Scatterplots as for (b) with points coloured to show genes whose expression correlates with ARAC cytotoxicity (white), genes involved in ARAC processing (yellow) and transport (orange) or IDR value (blue/grey).
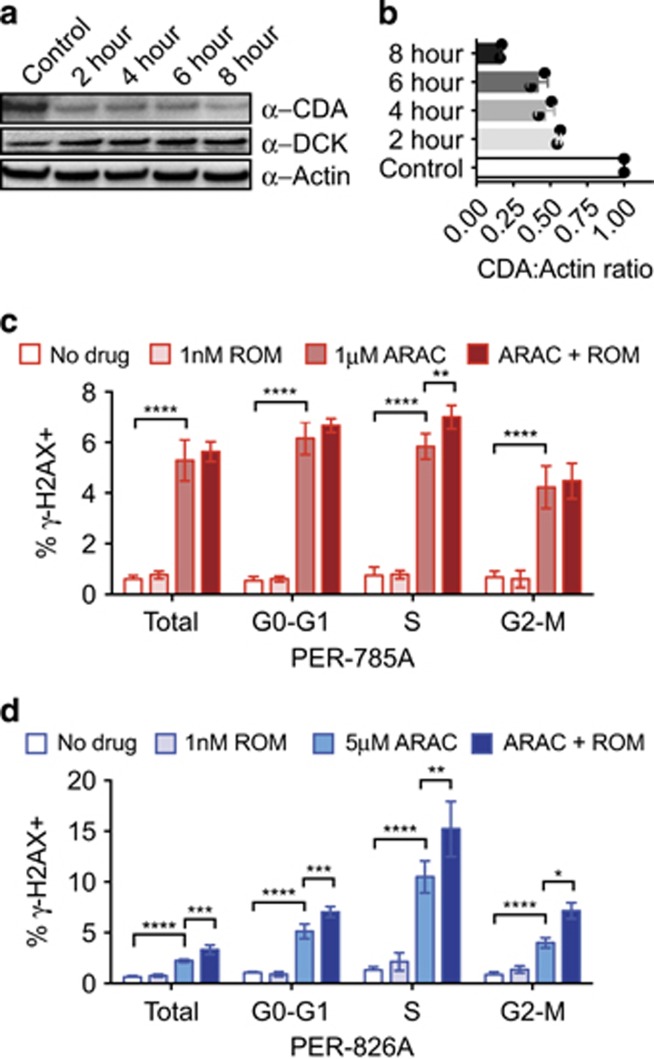
We hypothesized that ARAC–ROM synergy could be mediated by expression of cytidine-metabolizing enzymes and/or alterations to DNA damage–response pathways. We first monitored global histone changes (H3K27-acetylation and H3K79-dimethylation) with time-course experiments and found increases in H3K27-acetylation and H3K79-dimethylation detected at 6 h ROM exposure in iALL cells (data not shown). We next examined kinetics of DCK and CDA, key proteins in the cytidine metabolism pathway in response to ROM. CDA expression was reduced after 2 h incubation with ROM and decreased further at 4, 6 and 8 h (Figures 5a and b; n=2). In contrast, we did not detect reproducible changes in DCK expression (n=3). As ARAC induces DNA damage during S-phase33, 34 we tested whether ROM exposure modulates this response as measured by γ-H2AX and PI (DNA-content) staining in two cell lines with ~16-fold differing ARAC-sensitivities. PER-826A (ARAC IC50=1.79 μM) cells were treated with 5 μM ARAC, and PER-785A (IC50=0.11 μM) were treated with 1 μM ARAC to measure DNA damage after 24 h. Single-agent ROM showed no significant effect on γ-H2AX levels (Figure 5c; ROM vs saline P>0.05). In contrast, ARAC treatment significantly increased γ-H2AX in all phases of the cell cycle (ARAC vs saline P<0.0001). Co-dosing with ARAC–ROM induced significant γ-H2AX accumulation in S-phase compared with ARAC in both cell lines (PER-785A P<0.01, PER-826A P<0.01) and enhanced γ-H2AX across the cell cycle in PER-826A (G0-G1 P<0.001, G2-M P<0.05). Overall, these results reveal highly conserved in vitro expression of genes that could mediate ARAC–ROM interactions, and suggest ROM modulates the expression of cytidine-processing enzymes and enhances the DNA damage–response to ARAC.
Figure 5.
ROM modulates CDA expression and ARAC-induced γ-H2AX. (a) Protein extracted from PER-703A cells incubated with 1 nM ROM for 0, 2, 4, 6 or 8 h, were separated by SDS-PAGE and immunoblotted with antibodies against CDA, DCK and Actin. (b) Histogram of densitometric ratios for CDA:Actin immunoblots (n=2 independent experiments). (c) PER-785A and (d) PER-826A cells were treated with ROM and/or ARAC for 24 h, stained with PI and γ−H2AX and analyzed by flow cytometry. Total cells were defined excluding debris and dead cells; G0-G1, S- and G2-M phases of the cell cycle were gated on PI intensity; γ−H2AX-positive gates were defined by comparison to no drug controls. Graphs display the means of four independent experiments, error bars show s.d. and P-values calculated by one-way ANOVA and Tukey's multiple comparison tests (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
ROM and ARAC activity in iALL xenografts
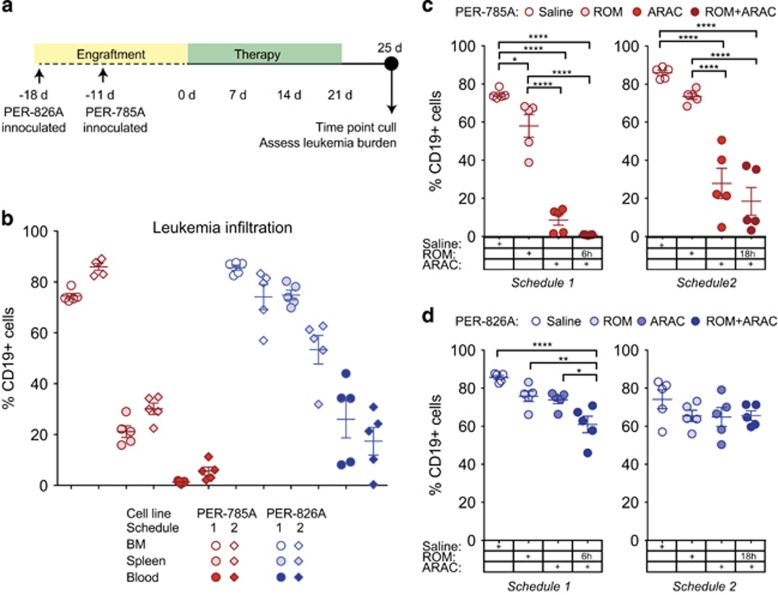
PER-785A and PER-826A were injected into NOD/SCID mice and consistent engraftment in bone marrow was observed for PER-785A on day 11 and for PER-826A on day 18. Single-agent activity was assessed for ROM and bortezomib, but did not achieve extension of event-free survival when administered as single drug 5 days after mice were injected with PER-785A or PER-826A (data not shown). Hence, further examination of in vivo activity of the ARAC–ROM combination was prioritized owing to consistent enhancement of ARAC by ROM in vitro (5/6 cell lines; Figure 3a) and the magnitude of synergy scores (Figure 3c). For combination drug testing, mice were randomized on day 11 for PER-785A xenografts and on day 18 for PER-826A xenografts and drug treatments initiated with ROM, ARAC, ROM and ARAC or saline for 3 weeks (Figure 6a; Material and Methods). Four days following the last drug injection leukemia burden was measured in bone marrow, spleen and blood. Leukemia infiltration in saline-treated animals shows the highest levels in bone marrow, followed by spleen and peripheral blood in both xenografts (Figure 6b), with comparison between the two models revealing distinct features.
Figure 6.
ARAC and ROM combination reduces leukemia burden in iALL xenografts. (a) Schematic depiction of the timing of injections and dosing schedules in xenografted mice inoculated with 1 × 106 cells. (b) Leukemia infiltration in bone marrow, spleen and peripheral blood in mice xenografted with PER-785A (red) or PER-826A (blue) from two independent experiments. Leukemia burden in (c) PER-785A and (d) PER-826A xenografted mice was measured after 3-week treatment with 1 mg/kg twice/week ROM; 100 mg/kg ARAC three times/week; ROM and ARAC combined; or saline (n=5 for each group). ROM was given either 6 h (left panels 'Schedule 1') or 18 h (right panels 'Schedule 2') before ARAC. Mice were culled 4 days post last injection of drug, and bone marrow analyzed by flow cytometry with anti-human CD19-FITC antibody. Percentage of human leukemia cells and s.d. are plotted with P-values calculated by one-way ANOVA and Tukey's multiple comparison tests (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Separate experiments were performed with animals given ROM 6 h ('Schedule 1') or 18 h ('Schedule 2') before ARAC (Figures 6c and d). Following treatment with ARAC alone, mice xenografted with PER-785A showed a mean reduction of leukemic cells in bone marrow of 66% (P<0.0001; n=5) with Schedule 1 and 58% (P<0.0001; n=5) with Schedule 2 (Figure 6c). Single-agent activity of ROM was observed with Schedule 1 in PER-785A xenografted mice with a mean reduction of leukemic cells of 16% (P<0.05; n=5). Addition of ROM prior to ARAC in PER-785A xenografts resulted in a mean reduction of leukemic cells of 73% (P<0.0001; n=5) with Schedule 1 and 67% (P<0.0001; n=5) with Schedule 2 (Figure 6c).
In PER-826A xenografts, single-agent ROM or ARAC did not reduce leukemia burden significantly in either experiment. However, when ROM was administered 6 h before ARAC (Schedule 1) the combined treatment reduced leukemia burden by 25% (95% C.I. 13–36% P<0.0001; n=5) compared with saline, by 15% (3.5–26% P<0.01) compared with ROM-only and by 13% (1.6–25% P <0.05) compared with ARAC only (Figure 6d). Treatment with ROM 18 h before ARAC (Schedule 2) did not show significant differences compared with saline, or single-agent treated groups in PER-826A xenografts (Figure 6d right panel). Therefore, PER-826A xenografts showed a poor response to ARAC, which could be enhanced by ROM.
Discussion
Novel cancer drugs frequently fail to demonstrate the expected benefits when administered to patients, hence the need to review and improve pre-clinical drug screening.35 Contributing factors include limitations of models used for cancer drug discovery, such as their representation of human disease.36 A major criticism of cancer cell lines is that they do not fully represent the primary tumor.37 Although the paradigm of molecular divergence in vitro is accepted, there are few reports comparing genomic profiles of primary specimens and derived cell lines. This is in contrast to patient-derived xenografts, for which several reports have investigated genomic changes following engraftment.38, 39
In this study we focused on MLL-r iALL because of the urgent need for novel therapeutic approaches to improve outcome. We document the genetic and transcriptional landscape of a panel of cell lines and defined over 8000 genes expressed at reproducible levels across eight pairs of cell line/primary samples. This gene set comprised almost 7500 coding and hundreds of non-coding genes (pseudogenes and long non-coding RNAs) and is likely to include essential genes for iALL viability. Importantly, virtually all drug targets from our chemical screen were reproducibly expressed in the iALL cell lines when compared with matched primary specimens. These include targets of HDACis, proteasome inhibitors and cyclin-dependent kinase inhibitors. We also found evidence for multiple MLL-fusion isoforms in three cell lines present in matched primary samples. The MLL-fusion isoforms were due to alternative 5' MLL exon junctions, fused to an invariant 3' partner gene exon suggesting splicing heterogeneity in primary samples and iALL cell lines. Furthermore, we observe conserved in vitro expression of the major RNA bio-types. Altogether, these data support the utility of this panel of iALL cell lines for modelling molecular responses to chemical hits from our screen.
Cancer cell lines inevitably carry molecular alterations, and documenting them is essential to appropriately interpret the utility of these models. Thus, we also identified genes expressed at lower or higher levels in cell lines compared with primary samples. Genes at higher levels included signalling/inflammatory response genes, which may reflect changes associated with in vitro adaptation. Genes at lower levels were enriched for blood lineage and antigen binding, suggesting non-leukemic cells as a possible source. However, we also noted lower expression of drug-targetable receptor tyrosine kinases, including FLT1, FLT4, PDGFRB, ERBB2 and NTRK1 in two or more unrelated cell lines, suggesting the need to carefully select and validate models for growth factor-receptor tyrosine kinase perturbation studies. These results provide a catalogue of clinically relevant and conserved transcriptional features in vitro, and is the first report quantifying reproducibility of RNA-seq measurements in cell lines and matched primary material for any cancer type.
Our in vitro chemical screen identified consistent synergy between ROM and ARAC, the latter a key component of iALL therapy, where it is administered at high doses. In contrast, bortezomib had variable effects on ARAC and dexamethasone efficacy, although combinations of bortezomib and dexamethasone have been effective in other diseases such as multiple myeloma40 and light-chain amyloidosis.41 ROM also had variable effects on dexamethasone, suggesting that ROM or bortezomib may not be suitable for co-administration with dexamethasone in some iALL patients. Importantly, we found consistent synergistic enhancement of ARAC by three additional HDACis, supporting HDACs as therapeutic targets to potentiate ARAC killing, consistent with observations in pediatric acute myeloid leukemia cells.42 The potentiation of ARAC by three HDACis is highly relevant given the current consideration to include HDACis in front-line treatment for iALL patients. Further studies are necessary to determine whether HDACis enhance ARAC in other malignancies treated with these agents such as acute myeloid leukemia.43
Several studies indicate efficacy of drugs targeting chromatin-associated proteins in hematological malignancies.44 By performing an unbiased screen of FDA-approved compounds, we confirmed in vitro sensitivity to a subset of HDACis. Sensitivity to HDACis, including ROM, has previously been reported against t(4,11) MLL-r iALL,45 whereas sensitivity to HDAC-1/-2 genetic inhibition has also been shown in pediatric B-ALL.46 We also recorded cytotoxic activity of additional agents targeting chromatin features, including three bromodomain and extra-terminal domain family inhibitors within the upper range of doses reported previously in MLL-r AML47 and the histone H3K27-demethylase inhibitor, GSK-J4, in four cell lines, with an IC50 dose range consistent with responses of T-ALL cells.48 However, we did not detect growth inhibition activity of the EZH2 H3K27 methylase inhibitor GSK-343, consistent with findings that dual EZH1 and EZH2 inhibition is necessary for anti-leukemic effects in a murine model of MLL-r AML.49 Taken together, our findings implicate the chromatin landscape as a therapeutic target in MLL-r iALL.
We observed variable efficacy of drugs currently used to treat iALL, with the exception of vincristine. Clofarabine and gemcitabine were more potent than currently used nucleoside analogues of the same respective class. These results emphasize the limitation of contemporary MLL-r iALL chemotherapeutics and indicate the potential of structurally related compounds as noted previously.50 Importantly, not all of the novel drugs in the three identified classes (HDACis, proteasome inhibitors and cyclin-dependent kinase inhibitors) were efficacious, but those that were effective have demonstrable clinical activity against human hematological malignancies.40, 51, 52, 53, 54 For example, dinaciclib in relapsed/refractory chronic lymphocytic leukemia52 and relapsed multiple myeloma;53 ROM in cutaneous and peripheral T-cell lymphoma;54 and bortezomib for multiple myeloma40 and mantle cell lymphoma.51
The potential benefit of combining chromatin-modifying drugs with currently administered chemotherapeutic drugs is very attractive. Understanding their actions is fundamental to optimizing combination drug therapy. Our studies show that ROM potentiated the action of ARAC by affecting the cytidine-metabolizing pathway. Interestingly, the ARAC-deactivating enzyme, CDA was depleted after 2 h ROM treatment and fell below 20% within 8 h. We demonstrated that ROM also affects the DNA damage–response, and observed significantly increased γ-H2AX over and above the effect of ARAC alone. We noted distinct differences in response patterns in two iALL cell lines, which also differ by a factor of 16 in sensitivity to ARAC. Taken together, the drug effects are evident within hours under in vitro conditions and appear to be dependent on the features of each cell line. These observations are reflected by our xenograft studies, whereby the iALL cell line with a higher in vitro IC50 (PER-826A) showed a poorer response to ARAC treatment in vivo, indicating that in vivo efficacy recapitulates in vitro sensitivity. Furthermore, the two xenograft models showed distinct responses to combined ROM-ARAC treatment. We demonstrated that the enhancing effect of ROM was evident when the drugs were administered 6 h apart, but diminished when administered 18 h apart. We conclude from in vitro and in vivo studies that the timing of drug administration is of critical importance, and warrants further optimization. Further studies are required integrating analyses of chromatin, transcription and protein expression in order to characterize the sequence of changes induced by this drug combination to modulate the DNA damage–response and induce cytoxicity.
In conclusion, we have generated a novel panel of molecularly characterized MLL-r iALL cell lines. Our data using this panel of cell lines showed variable efficacy of several drugs currently used to treat iALL. We have identified several FDA-approved anti-cancer drugs showing cytotoxic effects in MLL-r iALL cells, which are not presently used to treat this aggressive cancer. Our results highlight the clinical potential of drugs targeting chromatin for iALL therapy, in particular the FDA-approved HDACis.
Acknowledgments
This work was supported by project grants from the Children's Leukaemia and Cancer Research Foundation, Western Australia and The Kids Cancer Project. Mark N Cruickshank is supported by the Woolworths Children's Leukaemia and Cancer Research Foundation Fellowship. We thank the patients, families and hospital staff for providing leukemic samples.
Author contributions
Conception and design: MNC, JF, JH, RSK, URK. Acquisition of data: MNC, JF, LCC, SS, JH, JW, TWF, GMA, RSK, URK. Analysis and interpretation of data: MNC, JF, JH, TWF, DA, AG, KWC, JO, TL, RSK, URK. Writing, review and/or revision of the manuscript: MNC, DA, TL, TWF, GMA, KWC, JO, TL, CHC, RSK, URK. Administrative, technical or material support: RKP, NS.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
RK Prinjha and N Smithers are employees and shareholders of GlaxoSmithKline, which is carrying out clinical development of epigenetic inhibitors. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Hunger SP, Lu X, Devidas M, Camitta BM, Gaynon PS, Winick NJ et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children's oncology group. J Clin Oncol 2012; 30: 1663–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters R, Schrappe M, De Lorenzo P, Hann I, De Rossi G, Felice M et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet 2007; 370: 240–250. [DOI] [PubMed] [Google Scholar]
- Dreyer ZE, Hilden JM, Jones TL, Devidas M, Winick NJ, Willman CL et al. Intensified chemotherapy without SCT in infant ALL: results from COG P9407 (Cohort 3). Pediatr Blood Cancer 2015; 62: 419–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotecha RS, Gottardo NG, Kees UR, Cole CH. The evolution of clinical trials for infant acute lymphoblastic leukemia. Blood Cancer J 2014; 4: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer ZE, Dinndorf PA, Camitta B, Sather H, La MK, Devidas M et al. Analysis of the role of hematopoietic stem-cell transplantation in infants with acute lymphoblastic leukemia in first remission and MLL gene rearrangements: a report from the Children's Oncology Group. J Clin Oncol 2011; 29: 214–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science 2014; 345: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science 2014; 346: 1480–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klijn C, Durinck S, Stawiski EW, Haverty PM, Jiang Z, Liu H et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol 2015; 33: 306–312. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012; 483: 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG, Quentmeier H, MacLeod RA. Malignant hematopoietic cell lines: in vitro models for the study of MLL gene alterations. Leukemia 2004; 18: 227–232. [DOI] [PubMed] [Google Scholar]
- Beesley AH, Palmer ML, Ford J, Weller RE, Cummings AJ, Freitas JR et al. Authenticity and drug resistance in a panel of acute lymphoblastic leukaemia cell lines. Br J Cancer 2006; 95: 1537–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kees UR, Ford J, Watson M, Murch A, Ringner M, Walker RL et al. Gene expression profiles in a panel of childhood leukemia cell lines mirror critical features of the disease. Mol Cancer Ther 2003; 2: 671–677. [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. HTSeq—-a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Ngai J, Speed TP, Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 2014; 32: 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15: R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. Ann Appl Stat 2011; 5: 1752–1779. [Google Scholar]
- Huang, da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57. [DOI] [PubMed] [Google Scholar]
- Francis RW, Thompson-Wicking K, Carter KW, Anderson D, Kees UR, Beesley AH. FusionFinder: a software tool to identify expressed gene fusion candidates from RNA-Seq data. PLoS One 2012; 7: e39987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panicker J, Li Z, McMahon C, Sizer C, Steadman K, Piekarz R et al. Romidepsin (FK228/depsipeptide) controls growth and induces apoptosis in neuroblastoma tumor cells. Cell Cycle 2010; 9: 1830–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald JB, Schoeberl B, Nielsen UB, Sorger PK. Systems biology and combination therapy in the quest for clinical efficacy. Nat Chem Biol 2006; 2: 458–466. [DOI] [PubMed] [Google Scholar]
- Henderson MJ, Choi S, Beesley AH, Baker DL, Wright D, Papa RA et al. A xenograft model of infant leukaemia reveals a complex MLL translocation. Br J Haematol 2008; 140: 716–719. [DOI] [PubMed] [Google Scholar]
- Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science 2002; 298: 601–604. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 2012; 22: 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Li J, Wang E. Signaling network analysis of ubiquitin-mediated proteins suggests correlations between the 26S proteasome and tumor progression. Mol Biosyst 2009; 5: 1809–1816. [DOI] [PubMed] [Google Scholar]
- Bradner JE, Mak R, Tanguturi SK, Mazitschek R, Haggarty SJ, Ross K et al. Chemical genetic strategy identifies histone deacetylase 1 (HDAC1) and HDAC2 as therapeutic targets in sickle cell disease. Proc Natl Acad Sci USA 2010; 107: 12617–12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans 2013; 41: 741–749. [DOI] [PubMed] [Google Scholar]
- Li L, Fridley B, Kalari K, Jenkins G, Batzler A, Safgren S et al. Gemcitabine and cytosine arabinoside cytotoxicity: association with lymphoblastoid cell expression. Cancer Res 2008; 68: 7050–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam RW, den Boer ML, Meijerink JP, Ebus ME, Peters GJ, Noordhuis P et al. Differential mRNA expression of Ara-C-metabolizing enzymes explains Ara-C sensitivity in MLL gene-rearranged infant acute lymphoblastic leukemia. Blood 2003; 101: 1270–1276. [DOI] [PubMed] [Google Scholar]
- Abraham A, Varatharajan S, Karathedath S, Philip C, Lakshmi KM, Jayavelu AK et al. RNA expression of genes involved in cytarabine metabolism and transport predicts cytarabine response in acute myeloid leukemia. Pharmacogenomics 2015; 16: 877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell JT, Edwards H, Buck SA, Ge Y, Taub JW. Targeting the wee1 kinase for treatment of pediatric Down syndrome acute myeloid leukemia. Pediatr Blood Cancer 2014; 61: 1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Boyson CA, Di Liberto M, Huang X, Hannah J, Dorn DC et al. CDK4/6 inhibitor PD 0332991 sensitizes acute myeloid leukemia to cytarabine-mediated cytotoxicity. Cancer Res 2015; 75: 1838–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Ferrara N. Modeling and predicting clinical efficacy for drugs targeting the tumor milieu. Nat Biotechnol 2012; 30: 648–657. [DOI] [PubMed] [Google Scholar]
- Wilding JL, Bodmer WF. Cancer cell lines for drug discovery and development. Cancer Res 2014; 74: 2377–2384. [DOI] [PubMed] [Google Scholar]
- Herter-Sprie GS, Kung AL, Wong KK. New cast for a new era: preclinical cancer drug development revisited. J Clin Invest 2013; 123: 3639–3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien S, Merino-Trigo A, Lacroix L, Pocard M, Goere D, Mariani P et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin Cancer Res 2012; 18: 5314–5328. [DOI] [PubMed] [Google Scholar]
- Klco JM, Spencer DH, Miller CA, Griffith M, Lamprecht TL, O'Laughlin M et al. Functional heterogeneity of genetically defined subclones in acute myeloid leukemia. Cancer Cell 2014; 25: 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painuly U, Kumar S. Efficacy of bortezomib as first-line treatment for patients with multiple myeloma. Clin Med Insights Oncol 2013; 7: 53–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood S, Palladini G, Sanchorawala V, Wechalekar A. Update on treatment of light chain amyloidosis. Haematologica 2014; 99: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Edwards H, Xu X, Zhou H, Buck SA, Stout ML et al. Mechanisms of synergistic antileukemic interactions between valproic acid and cytarabine in pediatric acute myeloid leukemia. Clin Cancer Res 2010; 16: 5499–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015; 373: 1136–1152. [DOI] [PubMed] [Google Scholar]
- Fong CY, Morison J, Dawson MA. Epigenetics in the hematologic malignancies. Haematologica 2014; 99: 1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpel DJ, Schneider P, Seslija L, Osaki H, Williams O, Pieters R et al. Connectivity mapping identifies HDAC inhibitors for the treatment of t(4;11)-positive infant acute lymphoblastic leukemia. Leukemia 2012; 26: 682–692. [DOI] [PubMed] [Google Scholar]
- Stubbs MC, Kim W, Bariteau M, Davis T, Vempati S, Minehart J et al. Selective Inhibition of HDAC1 and HDAC2 as a Potential Therapeutic Option for B-ALL. Clin Cancer Res 2015; 21: 2348–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011; 478: 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P, Tsirigos A, Welstead GG, Trimarchi T, Bakogianni S, Xu L et al. Contrasting roles of histone 3 lysine 27 demethylases in acute lymphoblastic leukaemia. Nature 2014; 514: 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, On DM, Ma A, Parton T, Konze KD, Pattenden SG et al. Selective inhibition of EZH2 and EZH1 enzymatic activity by a small molecule suppresses MLL-rearranged leukemia. Blood 2015; 125: 346–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpel DJ, Schneider P, Pieters R, Stam RW. The potential of clofarabine in MLL-rearranged infant acute lymphoblastic leukaemia. Eur J Cancer 2015; 51: 2008–2021. [DOI] [PubMed] [Google Scholar]
- Bose P, Batalo MS, Holkova B, Grant S. Bortezomib for the treatment of non-Hodgkin's lymphoma. Expert Opin Pharmacother 2014; 15: 2443–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J, Jones J, Johnson AJ, Andritsos L, Maddocks K, Jaglowski S et al. Dinaciclib is a novel cyclin-dependent kinase inhibitor with significant clinical activity in relapsed and refractory chronic lymphocytic leukemia. Leukemia 2015; 29: 1524–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SK, LaPlant B, Chng WJ, Zonder J, Callander N, Fonseca R et al. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood 2015; 125: 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekarz RL, Frye R, Prince HM, Kirschbaum MH, Zain J, Allen SL et al. Phase 2 trial of romidepsin in patients with peripheral T-cell lymphoma. Blood 2011; 117: 5827–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.