Abstract
Auxin modulates diverse plant developmental pathways through direct transcriptional regulation and cooperative signaling with other plant hormones. Genetic and biochemical approaches have clarified several aspects of the auxin-regulated networks; however, the mechanisms of perception and subsequent signaling events remain largely uncharacterized. To elucidate unidentified intermediates, we have developed a high-throughput screen for identifying small molecule inhibitors of auxin signaling in Arabidopsis. Analysis of 10,000 compounds revealed several potent lead structures that abrogate transcription of an auxin-inducible reporter gene. Three compounds were found to interfere with auxin-regulated proteolysis of an auxin/indole-3-acetic acid transcription factor, and two impart phenotypes indicative of an altered auxin response, including impaired root development. Microarray analysis was used to demonstrate the mechanistic similarities of the two most potent molecules. This strategy promises to yield powerful tools for the discovery of unidentified components of the auxin-signaling networks and the study of auxin's participation in various stages of plant development.
The plant hormone auxin arbitrates plant development at multiple levels, from differentiation and division of individual cells to vascular patterning and flowering of the larger organism. Although auxin perception is a central biochemical event, it and the subsequent signaling events directing gene regulation are poorly understood. Several rapid responses to auxin have been identified; among these, the most enlightening studies of auxin activity have focused on immediate modulations in gene expression. Critical to auxin-coordinated genetic reprogramming are the auxin/indole-3-acetic acid (Aux/IAA) and auxin response factor (ARF) gene families (1, 2).
The Aux/IAA family consists of 29 homologous gene products in Arabidopsis thaliana that homo- and heterodimerize with other Aux/IAA proteins as well as members of the ARF family of transcriptional regulators (3-5). Although the Aux/IAA proteins have not been shown to bind DNA directly, members of the ARF family do interact with auxin-response elements in the promoter region of auxin-induced genes (6, 7). Little is known about the specificity of the Aux/IAA gene products for particular ARF proteins or whether additional proteins are involved in gene induction or modulating the Aux/IAA-ARF interaction.
The most well characterized components of the auxin-signaling network are those involved in the degradation of the Aux/IAA proteins (8). Ubiquitination by means of the coordinated action of the COP9 signalosome/E3 ubiquitin ligase SCFTIR1 complex is crucial for proper Aux/IAA proteolysis (9-11). An up-regulation of mitogen-activated protein kinase activity accompanies auxin treatment, and mitogen-activated protein kinase cascades also may modulate auxin activity (12). In addition, both a G protein (13) and GTPases (14) have been linked to the molecular activity of auxin. Most recently, the action of peptidyl-prolyl isomerases has been implicated in early auxin signaling and hypothesized to direct the Aux/IAA proteins to the proteolytic machinery (15, 16). The participation of other regulatory proteins and the mechanism that guides specificity of the SCFTIR1 complex for the Aux/IAA proteins are issues that remain to be addressed. The culmination of current evidence points to a model by which the Aux/IAA proteins coordinate the tissue-specific response to auxin by functioning as negative regulators of the ARF protein family; undefined signaling components trigger Aux/IAA proteolysis, thus altering ARF transcriptional activity and eliciting diverse developmental and regulatory consequences.
Traditional genetic approaches for studying auxin signaling have relied on mutant plant lines with aberrant auxin responses. Mutant characterization has led to the identification of several important regulatory proteins, including the auxin influx carrier AUX1 (17) and components of the ubiquitination machinery such as the E1-like RUB1 ligase AXR1 (18) and the F-box protein TIR1 (10). Several gain-of-function mutations in the regulatory domain of the Aux/IAA genes have illuminated the participation of the transcription factors in downstream pathways (19-23). The development of auxin-responsive reporter lines has facilitated targeted mutant screening. The Arabidopsis BA3 line containing the β-glucuronidase (GUS) reporter under the regulatory control of an auxin-responsive synthetic promoter derived from the Pisum sativum PS-IAA4/5 gene provided a necessary tool for such a screening strategy. This system was previously used to identify the auxin-hypersensitive mutant lines age1 and age2 (24). The power of transcriptional profiling has been harnessed to dissect the early modulations of gene expression induced by auxin treatment (25, 26). These studies have defined the gene set whose rapid, dramatic changes in expression levels trigger the downstream auxin-regulated developmental pathways.
Forward genetics has proven to be a powerful approach for studying signaling mechanisms in a variety of organisms, but it suffers from an inability to identify genes that are essential for embryogenesis and early development. Recently developed technologies, such as RNA interference methods, lack temporal control over the abrogation of gene product function. Auxin's role in tissue differentiation and organ development indicates that many components of the auxin-signaling network are essential; therefore, their participation in the auxin response might not be identified through traditional strategies. An alternative approach, forward chemical genetics, utilizes small molecules to perturb a signaling pathway, permitting the identification of relevant gene products at any stage of development (27-30). Cell-permeant inhibitors of auxin-regulated transcription would enable the study of disrupted signaling under a variety of environmental stimuli, facilitating the examination of hormone cross-talk and the interplay of auxin and stress responses. The use of synthetically tractable small-molecule libraries simplifies subsequent structure-activity studies and the production of appropriate derivatives for target identification. Indeed, recent work has demonstrated the feasibility of this approach with the identification of a small-molecule trigger of auxin signaling, sirtinol (16), and the isolation of yokonolide A and B, macrolide inhibitors of auxin signaling from Streptomyces diastatochromogenes B59 (31, 32).
Herein we report the development of a high-throughput screen for auxin-mediated signaling in whole Arabidopsis seedlings and the identification of potent inhibitors of auxin signaling. Our strategy used the BA3 line, in which exogenously applied auxin stimulates the tissue-specific expression of GUS in the root elongation zone. Our screen used a commercial library of 10,000 structurally diverse small molecules and identified 30 compounds with strong inhibition. We narrowed our focus to four structurally distinct compounds with activity in the low micromolar range. We have characterized the effects of these compounds on auxin-mediated transcriptional activation and Aux/IAA proteolysis in addition to exploring the effects in other phytohormone-signaling pathways. Two compounds impart similar growth phenotypes, and microarray studies were used to compare the transcriptional changes induced by these inhibitors. Our approach has provided tools for dissecting auxin biology and should prove instrumental in identifying new components in the auxin-signaling network.
Materials and Methods
Plant Material and Growth Conditions. We used A. thaliana ecotype Columbia-0 (Col-0) for the phenotype and microarray experiments. The BA3, DR5::GUS, ARR5::GUS, and HS::AXR3NT-GUS lines are described in refs. 1-4, respectively. All seedlings were grown under a 16-h light cycle at 25°C.
Chemical Genetic Screen and GUS Assays. The small-molecule library (Diverset) was purchased from ChemBridge (San Diego). Surface-sterilized seeds were distributed into GM liquid medium [0.5× Murashige and Skoog salts, Gamborg vitamins, and 2.5 mM 2-(4-morpholino)ethanesulfonic acid (Mes), pH 5.6] at 5-10 seeds per well. After stratification (48 h at 4°C), the plates were transferred to an orbital shaker with a 16-h light cycle at 25°C. After 5 days of growth, the medium was removed on a vacuum manifold and replaced with medium containing DMSO, 5 μM 1-naphthaleneacetic acid (NAA), or NAA and library component (≈20 μM). The plates were allowed to incubate for 8 h before the medium was removed, and the seedlings were washed with water and stained for GUS expression. The GUS staining protocol is described in Supporting Text, which is published as supporting information on the PNAS web site.
RT-PCR. Total RNA was extracted from treated tissues by using the RNAqueous purification kit (Ambion, Austin, TX) according to the manufacturer's instructions. cDNAs were synthesized, and gene fragments were amplified by PCR using gene-specific primers. The list of primers used is in Supporting Text.
Transcriptional Profiling and Data Analysis. Approximately 2.5 mg of dry Col-0 seedlings were surface-sterilized and stratified for 2 days at 4°C in liquid medium containing 1.5% (wt/vol) sucrose before being transferred to light with constant shaking at 100 rpm on an orbital shaker. After 7 days, the seedling clusters were subjected to the treatments for 1 h, followed by total RNA isolation using the RNAqueous kit (Ambion). Each treatment was performed in triplicate or quadruplicate. All labeling (Enzo Biochem) and hybridization (Affymetrix, Santa Clara, CA) procedures were performed as directed by the manufacturers. The analysis of the microarray data is described in Supporting Text.
Results
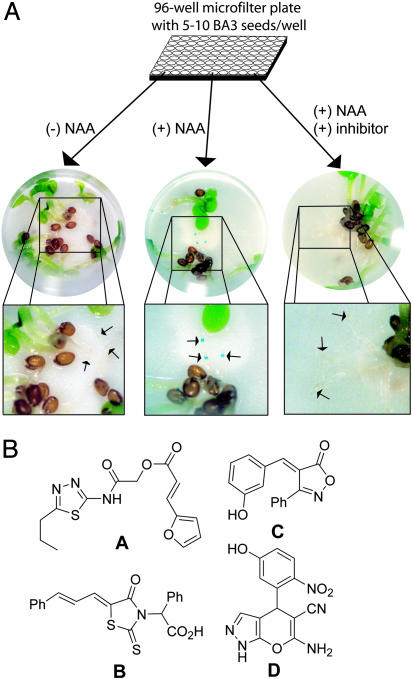
Targeted Chemical Genetics Screen Reveals Auxin-Signaling Inhibitors. A high-throughput whole-seedling assay was developed by using the BA3 reporter system in Arabidopsis. The seedlings (5-10 per well) were germinated, grown in liquid culture in 96-well microfilter plates, and assayed after 5 days of growth (Fig. 1A). Cell-permeant NAA was used instead of the endogenous auxin, IAA, to avoid the identification of transport inhibitors that prevent auxin uptake but not transcriptional activation (33). Inhibitors of auxin signaling obstruct the tissue-specific expression of GUS in the root elongation zone, easily detected by rapid examination of each well under a dissecting microscope. Control wells containing a gradient of NAA were included in each plate to qualitatively assess the relative potency of the compounds.
Fig. 1.
A high-throughput screen for auxin signaling inhibitors. (A) BA3 seeds expressing GUS from an auxin-sensitive promoter were arrayed into 96-well microfilter plates (5-10 seeds per well) and grown in liquid culture for 5 days. Incubation of the seedlings with 5 μM NAA results in the tissue-specific expression of GUS in the root elongation zone, easily visualized after incubation with 5-bromo-4-chloro-3-indolyl β-d-glucuronide (X-gluc). The inclusion of an inhibitor of auxin signaling prevents GUS expression. (B) Structures of the four inhibitors, compounds A-D, chosen for detailed analysis.
A total of 10,000 compounds were screened for their ability to impede GUS transcription from the auxin-regulated BA promoter. Follow-up screening used more stringent conditions, including higher concentrations of NAA or IAA, which induces more robust expression from the BA promoter. We narrowed our focus to four structurally unique compounds (A-D in Fig. 1B) demonstrating consistently strong inhibition under a variety of assay conditions. Compound A consists of a furyl acrylate ester of a thiadiazole heterocycle, and B is a 4-thiazolidinone appended with a derivatized acetic acid. Compound C contains an isoxazolone core, and D consists of a functionalized pyranopyrazole. Each compound contains a synthetically accessible scaffold for future derivative preparation and structure-activity studies.
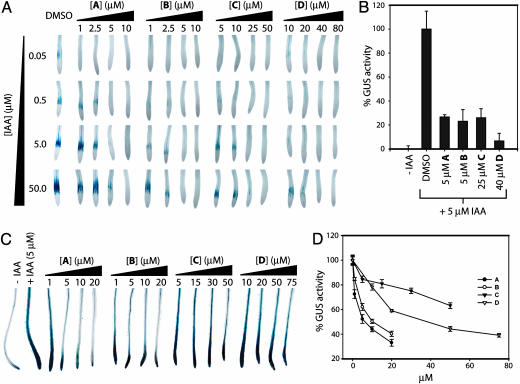
The Inhibitors Display Varied Potencies. We quantified the relative potency of our confirmed hits in silencing auxin-activated transcription. We analyzed the level of GUS activity in both the BA3 line and the DR5::GUS reporter line that harbors a more highly active synthetic auxin-responsive promoter element (5). Competition studies were performed using the BA3 line and various concentrations of both IAA and each inhibitor. These assays demonstrate that the inhibitory activity of all compounds can be negated with high concentrations of auxin (Fig. 2A). We next quantified the inhibitory activity of each compound in the BA3 line by using 5 μM IAA and the concentration of each compound that abrogated GUS staining in the qualitative assays. Fluorometric GUS assays revealed that under these conditions the compounds inhibit between 75% and 95% of GUS activity (Fig. 2B).
Fig. 2.
The compounds inhibit auxin transcriptional activation in the BA3 and DR5::GUS reporter lines. Five-day-old BA3 (A and B) or DR5::GUS (C and D) seedlings were subjected to the indicated concentrations of IAA and compound for 4 h, followed by staining with X-gluc (A and C) or fluorimetric determination of GUS activity (B and D). Error = SE of the mean (n = 6).
We next turned to the DR5::GUS line and characterized the inhibitory activity by using 5 μM IAA and various concentrations of compounds A-D (Fig. 2C). Dose-response curves were obtained for each of the four inhibitors (Fig. 2D). The relative inhibitory potency of the four compounds was similar in the BA3 and DR5::GUS reporter lines, with compounds A and B showing the most dramatic activity, reducing GUS expression to nearly background levels at 20 μM in the DR5::GUS line.
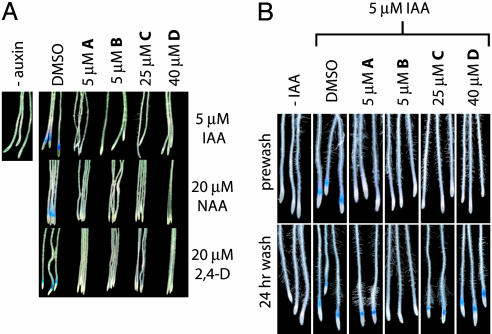
Additional characterization confirmed the inhibitory activity to be independent of auxin structure. The compounds were equally capable of inhibiting auxin signaling when IAA, NAA, or 2,4-dichlorophenoxyacetic acid was used to stimulate GUS expression (Fig. 3A). We also tested the reversibility of the inhibition. BA3 seedlings were treated with the compounds in liquid culture for 4 h followed by 24-h incubation in fresh medium. A subsequent 4-h treatment with IAA was sufficient to induce strong GUS expression in all seedlings except those incubated with B (Fig. 3B). This result could reflect slower detoxification for compound B or an irreversible inhibition mechanism. However, we were able to grow seedlings in the presence of all four compounds for >2 weeks before transplanting them to soil and obtaining healthy and fertile mature plants (data not shown).
Fig. 3.
The inhibitory activity is independent of auxin structure and reversible for most compounds. (A) Five-day-old BA3 seedlings were incubated with the indicated concentration of inhibitor and auxin for 4 h, followed by staining with X-gluc. 2,4-D, 2,4-dichlorophenoxyacetic acid. (B) Five-day-old BA3 seedlings were incubated with the indicated concentration of inhibitor (24-h wash) or inhibitor/IAA (prewash) for 4 h. The seedlings were then stained with X-gluc (prewash) or incubated in fresh medium before an additional 4-h treatment with IAA and subsequent staining with X-gluc (24-h wash).
Inhibitors Affect Cytokinin but Not Abscisic Acid (ABA) Signaling. We ascertained the effects of the compounds in two alternative hormone-signaling cascades. First, we verified that the compounds were not inhibiting reporter gene activity in vivo. We used a transgenic line expressing GUS under the regulatory control of the constitutive cauliflower mosaic virus 35S promoter (35S::GUS). After an 8-h incubation, total protein extracts were prepared and GUS activity was measured. No inhibition of GUS activity was detected (Fig. 6A, which is published as supporting information on the PNAS web site).
We then explored the effects of the compounds on cytokinin signaling by using the ARR5::GUS line, which displays a cytokinin-dependent, ubiquitous up-regulation of GUS expression. Seedlings were incubated for 6 h in the presence of the cytokinin N6-benzyladenine and one of the compounds A-D and subsequently were examined for GUS activity. Although histochemical staining revealed little effect on the GUS expression in the hypocotyl or cotyledons (data not shown), the compounds did have inhibitory effects on the expression of GUS in the root (Fig. 6B). The compounds may interfere with a common component of auxin and cytokinin signaling, or the inhibition may represent the downstream cross-talk between the hormone pathways (34), as was suggested for the inhibition of cytokinin signaling observed with yokonolide B (32).
Next, we examined the effects of the compounds on ABA-induced RAB18 expression by using semiquantitative RT-PCR (Fig. 6C). RAB18 is induced by cold, water stress, or exogenous ABA treatment (35). None of the compounds influenced RAB18 expression when applied alone or in conjunction with ABA. This result indicates that the compounds neither act as general inhibitors of phytohormone signaling nor nonspecifically inhibit gene transcription.
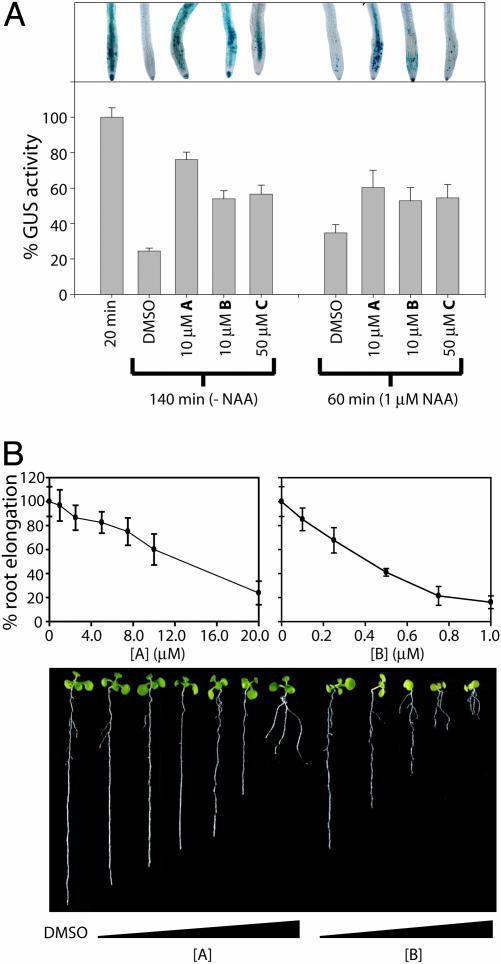
Inhibition of Aux/IAA Proteolysis. Auxin has been shown to regulate the proteolytic degradation of representative members of the Aux/IAA protein family (36). We used a reporter line containing a heat shock-inducible translational fusion of the N terminus of AXR3 (Aux/IAA17) and GUS (HS::AXR3NT-GUS) to examine the effects of compounds A-D on auxin-mediated proteolysis (10) (Fig. 4A). Only compound D failed to significantly inhibit proteolysis. Compounds A, B, and C diminished the proteolysis both in the presence and absence of exogenously applied NAA. Compound A demonstrated the greatest inhibitory activity in this assay. These results suggest that the inhibition of auxin signaling occurs upstream in the signaling pathway, either by means of inhibition of the proteolytic machinery or components acting before the SCFTIR1 complex.
Fig. 4.
Compounds A-C inhibit auxin-mediated proteolysis, and compounds A and B inhibit primary root elongation. (A) Seven-day-old HS::AXR3NT-GUS seedlings were heat shocked at 37°C for 2 h, followed by a 20-min incubation at 23°C. The indicated concentration of compound and/or NAA was added to the medium, and the seedlings were incubated for an additional 2 h (-NAA) or 40 min (+NAA) before quantification of GUS activity (Lower). Error = SE of the mean (n = 6). (Upper) Root tips from a representative seedling of each treatment stained with X-gluc. (B) Seedlings (Col-0) were germinated and grown vertically on medium containing increasing concentrations of compound A or B. (Upper) Primary root length was measured after 8 days of growth. Error = sample SD (n > 12). (Lower) Representative seedlings from each inhibitor treatment.
Phenotype Analysis. Compounds A and B impaired root growth in developing seedlings, with compound B imparting effects at significantly lower concentrations (Fig. 4B). When Arabidopsis seedlings were germinated on medium containing either compound, a dose-dependent inhibition of primary root length was observed. In addition, adventitious root growth was induced at a concentration of 20 μM A and 0.5 μM B. Compound A had little effect on cotyledon expansion or hypocotyl length; cotyledons of seedlings grown in the higher concentrations of B appeared slightly chlorotic. The difference in effective concentrations may reflect the efficiency of detoxification pathways for each compound, given the similar potencies in our transient reporter assay.
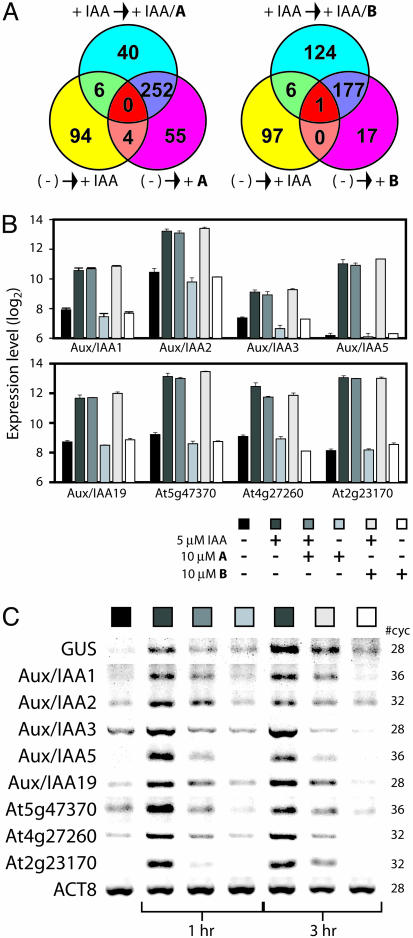
Transcriptional Profiling. The similar phenotypes induced by compounds A and B prompted us to perform detailed transcriptional profiling experiments on both compounds by using Affymetrix whole-genome ATH1 microarrays. Seven-day-old seedlings grown in liquid culture were treated for 1 h with the carrier solvent DMSO, 5 μM IAA, or a combination of IAA and 10 μM compound A or B. Each treatment was carried out in triplicate, and total RNA from each sample was independently isolated and used to generate labeled cRNA for subsequent hybridization. We generated differentially regulated gene lists by using the log2 expression values from the Robust Multichip Analysis output file. We used strict statistical criteria, namely a differential regulation of at least 2-fold and a corresponding P value of < 0.001. Auxin differentially regulated a total of 104 genes (0.5% of probes) under these conditions (Fig. 5A). The two compounds induced or repressed a similar number of genes, irrespective of whether they were applied independently or in concert with auxin. Compound A affected the expression of 347 genes (1.5% of probes) when applied alone or with auxin, and compound B similarly affected 318 genes (1.4% of probes). A comparison of the differentially expressed gene lists revealed 132 genes modulated by both compounds A and B. Similarly, a comparison of the IAA/A- and IAA/B-treated samples revealed an overlap of 179 genes differentially expressed under both conditions. Based on these sets of genes, Fisher's exact test was applied to the null hypothesis that the effects of the two compounds were independent. The null hypothesis was rejected, both in the absence (P = 8.6 × 10-13) and in the presence (P < 1 × 10-13) of IAA. These data imply that the compounds have comparable effects on whole seedling tissue, although the global effects most likely reflect both auxin-specific and xenobiotic responses to the treatment. An examination of the gene classes [Gene Ontology (GO) slim annotation, www.geneontology.org] affected by compounds A and B reveals a similar pattern, one that mimics the relative distribution of the gene classes across the genome (Fig. 7, which is published as supporting information on the PNAS web site). In addition, the compounds seem to impart their transcriptional effects independently of auxin because the effect of adding compound A or B on the transcriptional profiles is similar regardless of whether IAA was present in the medium. A more detailed analysis of the gene ontology gene classifications revealed many highly overrepresented functional classes relative to the genome (see Table 1, which is published as supporting information on the PNAS web site). In particular, several gene classes involved in transport, heat response, and stress response were significantly overrepresented in the differentially regulated compound-treated gene lists. Similar genes were induced by yokonolide B treatment (32) and may be part of xenobiotic response or could be specific to the inhibitory activity of the compounds.
Fig. 5.
The transcriptional effects of inhibitor treatment. (A) Venn diagrams specify the number of 2-fold differentially regulated genes (P < 0.001) (induced or repressed) when comparing the indicated data sets. (B) Log2 expression values for selected auxin-induced genes from the microarray analysis for compounds A and B. Seven-day-old liquid culture-grown seedling clusters (Col-0) were treated for 1 h with IAA and/or inhibitor as indicated. (C) Semiquantitative RT-PCR analysis of the selected genes by using RNA from excised root tissue of BA3 seedlings treated with IAA and/or inhibitor for the indicated times. ACT8, ACTIN8.
A total of six genes displayed differential expression patterns when comparing IAA- and IAA/compound A-treated samples. Similarly, seven genes were differentially expressed in the IAA- and IAA/compound B-treated samples. However, we were perplexed by the general lack of synergism between IAA and compound treatment. The clear inhibition of auxin signaling in the auxin-responsive reporter lines suggested that the compounds would impede the up-regulation of auxin-responsive genes, including the Aux/IAA family of transcription factors. The microarray analysis revealed little inhibition of the auxin-induced expression of these genes. We hypothesized that this result could be due to either a time lag in the inhibitory activity of the compounds or a tissue-specific effect for the compounds. To clarify this issue, we performed semiquantitative RT-PCR experiments and examined the effects of IAA and/or compound A/B application on the expression level of selected auxin-induced genes from excised root tissue. The results clarified the mechanism of action for both compounds. Although the microarray data revealed little effect on the expression levels of the selected genes when RNA from whole tissue was examined (Fig. 5B), RT-PCR using RNA from root tissue revealed clear inhibition of the auxin-induced gene expression at 1 h with compound A (Fig. 5C). The bulk of the RNA from whole seedlings is derived from the green tissues, masking these root-specific effects in the microarray studies.
The situation was slightly more complicated with compound B. RT-PCR studies from root tissue at a 1-h time point revealed little effect on the expression levels of the examined genes (data not shown); however, the expression levels at 3 h confirmed a clear inhibitory effect. The delay in inhibition may be due to slow uptake of the anionic compound.
Discussion
We have reported the identification of several potent inhibitors of auxin transcriptional activation by means of a high-throughput forward chemical genetic screen in Arabidopsis. We have focused on four structurally unique compounds that modulate auxin-regulated transcription and have characterized their effects on orthogonal signaling pathways and auxin-mediated proteolysis.
Our whole-seedling screening strategy provides the opportunity for high-throughput analysis of the auxin-signaling response. The screen is robust, requires little preparation time, and is amenable to the analysis of up to 400 compounds in duplicate per day. The screen provided reliable results, and the tissue-specific expression of the GUS gene in the root elongation zone facilitated the rapid identification of active compounds. By using a whole-seedling assay, we bypassed potential difficulties from identifying compounds with in vitro activity but low cell permeability. The use of the membrane-permeant auxin analog NAA (instead of the endogenous hormone IAA) biased the screen toward inhibitors of transcription, not carrier proteins associated with hormone uptake or transport. The platform should find broad applicability in similar screens targeting alternative hormone-signaling pathways. The 96-well microfilter plates are well suited for the growth of 5- to 7-day-old Arabidopsis seedlings in liquid culture and facilitate rapid medium exchange for the addition of hormones, compounds, or enzyme assay buffers.
We obtained a commercially available library of highly diverse small molecules from ChemBridge for our chemical genetic screen. Several confirmed hits with potent activity were identified in our assay. After several rounds of follow-up screening, we focused our efforts on the characterization of four inhibitors containing diverse structural features. The compounds display various levels of activity in our fluorimetric GUS assay, with two that nearly abolish auxin-mediated transcription of the GUS reporter gene at 20 μM. Although we have concentrated our efforts on only a few of the hits, future endeavors that explore the biological effects of the other active compounds could lead to the discovery of multiple unidentified components of the auxin-signaling network.
We used several reporter lines, RT-PCR, and whole-genome transcriptional profiling to probe the mechanism of action for the inhibitors. The compounds display a range of potencies in our auxin-responsive reporter lines with two compounds, A and B, completely abrogating reporter gene expression at 20 μM. Specificity studies revealed that the compounds had no effect on ABA signaling but did interfere with cytokinin signaling, an effect that could involve a target protein shared by both pathways. Compounds A-C impaired auxin-mediated proteolytic degradation of the translational fusion protein AXR3NT-GUS. This inhibition implies an inhibitory mechanism targeting an upstream component of the signaling pathway.
Compounds A and B induced similar developmental phenotypes on light-grown seedlings. Both inhibited primary root elongation and induced adventitious root formation in a dose-dependent manner. This phenotype closely matches that observed with yokonolide B, a bacterially produced macrolide inhibitor of auxin signaling, and may be due to the impaired degradation of particular Aux/IAA proteins involved in root growth and development.
Transcriptional profiling experiments revealed similar global effects on gene expression for both compounds A and B. A number of gene classes were overrepresented in the differentially regulated gene lists. These included heat shock proteins, transporter proteins, and transcriptional activators. These genes may be involved in a general xenobiotic response or may be relevant to the specific activity of the compounds. Additional comparative studies with root tissue-derived RNA by using semiquantitative RT-PCR revealed a root-specific effect for compound A and a delayed mechanism of inhibition for compound B. Additional studies will be necessary to deconvolute the tissue-specific effects of the compounds and more closely study the mechanism of inhibition.
Synthetic compound libraries represent a rich resource for the identification of new tools to probe biological signaling pathways. The identification of structurally unique small-molecule inhibitors of auxin transcriptional activation should facilitate the elucidation of novel components of the pathway. The compounds identified in this study are synthetically tractable, facilitating structure-activity studies to generate more potent and specific second-generation analogs and the preparation of matrix-tagged derivatives for protein affinity chromatography and target identification (37, 38). The optimized inhibitors can be used in forward genetic screens to generate new auxin-response mutants, furthering the use of genetics to study the auxin-signaling pathway.
The successful application of chemical genetics to plant biology will demand interdisciplinary approaches to overcome specificity issues and the difficulties associated with target identification. Future efforts will require an investment in synthetic chemistry to modulate the activity profile of the confirmed hits, increasing potency and eliminating undesired side effects resulting from interactions with multiple targets. Whole-genome expression microarrays offer a powerful method for examining the downstream consequences of compound treatment and should provide critical insight into the mechanism of these pharmacological tools. Recent advances in genomics methods, including yeast haploinsufficiency screens, may aid in the identification of target proteins common to Arabidopsis and Saccharomyces (39). Perhaps the most attractive future tool for target identification lies with the development of protein chips representing the entire Arabidopsis proteome. Protein chips would assist in the direct identification of desired and undesired targets in vitro. This technology hinges on the availability of complete cDNA collections such as that initiated by the ORFeome project (40).
Supplementary Material
Acknowledgments
We thank Joseph Kieber (University of North Carolina, Chapel Hill), Mark Estelle (University of Texas, Austin), and Tom Guilfoyle (University of Missouri, Columbia) for providing the ARR5-GUS, HS::AXR3NT-GUS, and DR5-GUS lines, respectively; and Yutaka Oono (Japan Atomic Energy Research Institute, Gunma, Japan) for helpful discussions. J.I.A. was supported by a National Science Foundation Postdoctoral Fellowship in Biological Informatics. This work was supported by National Institutes of Health Grant (GM0354A7) (to A.T.).
Abbreviations: ABA, abscisic acid; ARF, auxin-response factor; IAA, indole-3-acetic acid; NAA, 1-naphthaleneacetic acid; GUS, β-glucuronidase; X-gluc, 5-bromo-4-chloro-3-indolyl β-D-glucuronide.
Data deposition: The microarray data reported in this paper have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database (accession no. GSE1491).
References
- 1.Hagen, G. & Guilfoyle, T. (2002) Plant Mol. Biol. 49, 373-385. [PubMed] [Google Scholar]
- 2.Liscum, E. & Reed, J. W. (2002) Plant Mol. Biol. 49, 387-400. [PubMed] [Google Scholar]
- 3.Kim, J., Harter, K. & Theologis, A. (1997) Proc. Natl. Acad. Sci. USA 94, 11786-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilfoyle, T. J., Ulmasov, T. & Hagen, G. (1998) Cell. Mol. Life Sci. 54, 619-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulmasov, T., Murfett, J., Hagen, G. & Guilfoyle, T. J. (1997) Plant Cell 9, 1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulmasov, T., Hagen, G. & Guilfoyle, T. J. (1999) Plant J. 19, 309-319. [DOI] [PubMed] [Google Scholar]
- 7.Ulmasov, T., Hagen, G. & Guilfoyle, T. J. (1999) Proc. Natl. Acad. Sci. USA 96, 5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kepinski, S. & Leyser, O. (2002) Plant Cellxn 14, Suppl., S81-S95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray, W. M. & Estelle, M. (2000) Trends Biochem. Sci. 25, 133-138. [DOI] [PubMed] [Google Scholar]
- 10.Gray, W. M., Kepinski, S., Rouse, D., Leyser, O. & Estelle, M. (2001) Nature 414, 271-276. [DOI] [PubMed] [Google Scholar]
- 11.Schwechheimer, C., Serino, G., Callis, J., Crosby, W. L., Lyapina, S., Deshaies, R. J., Gray, W. M., Estelle, M. & Deng, X. W. (2001) Science 292, 1379-1382. [DOI] [PubMed] [Google Scholar]
- 12.Mockaitis, K. & Howell, S. H. (2000) Plant J. 24, 785-796. [DOI] [PubMed] [Google Scholar]
- 13.Ullah, H., Chen, J. G., Temple, B., Boyes, D. C., Alonso, J. M., Davis, K. R., Ecker, J. R. & Jones, A. M. (2003) Plant Cell 15, 393-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao, L. Z., Cheung, A. Y. & Wu, H. M. (2002) Plant Cell 14, 2745-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dharmasiri, N., Dharmasiri, S., Jones, A. M. & Estelle, M. (2003) Curr. Biol. 13, 1418-1422. [DOI] [PubMed] [Google Scholar]
- 16.Zhao, Y., Dai, X., Blackwell, H. E., Schreiber, S. L. & Chory, J. (2003) Science 301, 1107-1110. [DOI] [PubMed] [Google Scholar]
- 17.Marchant, A., Bhalerao, R., Casimiro, I., Eklof, J., Casero, P. J., Bennett, M. & Sandberg, G. (2002) Plant Cell 14, 589-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.del Pozo, J. C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W. M. & Estelle, M. (2002) Plant Cell 14, 421-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian, Q., Uhlir, N. J. & Reed, J. W. (2002) Plant Cell 14, 301-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ouellet, F., Overvoorde, P. J. & Theologis, A. (2001) Plant Cell 13, 829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagpal, P., Walker, L. M., Young, J. C., Sonawala, A., Timpte, C., Estelle, M. & Reed, J. W. (2000) Plant Physiol. 123, 563-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukaki, H., Tameda, S., Masuda, H. & Tasaka, M. (2002) Plant J. 29, 153-168. [DOI] [PubMed] [Google Scholar]
- 23.Tian, Q., Nagpal, P. & Reed, J. W. (2003) Plant J. 36, 643-651. [DOI] [PubMed] [Google Scholar]
- 24.Oono, Y., Chen, Q. G., Overvoorde, P. J., Köhler, C. & Theologis, A. (1998) Plant Cell 10, 1649-1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redman, J. C., Haas, B. J., Tanimoto, G. & Town, C. D. (2004) Plant J. 38, 545-561. [DOI] [PubMed] [Google Scholar]
- 26.Pufky, J., Qiu, Y., Rao, M. V., Hurban, P. & Jones, A. M. (2003) Funct. Integr. Genomics 3, 135-143. [DOI] [PubMed] [Google Scholar]
- 27.Haggarty, S. J., Mayer, T. U., Miyamoto, D. T., Fathi, R., King, R. W., Mitchison, T. J. & Schreiber, S. L. (2000) Chem. Biol. 7, 275-286. [DOI] [PubMed] [Google Scholar]
- 28.Peterson, R. T., Link, B. A., Dowling, J. E. & Schreiber, S. L. (2000) Proc. Natl. Acad. Sci. USA 97, 12965-12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Specht, K. M. & Shokat, K. M. (2002) Curr. Opin. Cell Biol. 14, 155-159. [DOI] [PubMed] [Google Scholar]
- 30.Zouhar, J., Hicks, G. R. & Raikhel, N. V. (2004) Proc. Natl. Acad. Sci. USA 101, 9497-9501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashi, K., Ogino, K., Oono, Y., Uchimiya, H. & Nozaki, H. (2001) J. Antibiot. 54, 573-581. [DOI] [PubMed] [Google Scholar]
- 32.Hayashi, K. I., Jones, A. M., Ogino, K., Yamazoe, A., Oono, Y., Inoguchi, M., Kondo, H. & Nozaki, H. (2003) J. Biol. Chem. 278, 23797-23806. [DOI] [PubMed] [Google Scholar]
- 33.Rahman, A., Hosokawa, S., Oono, Y., Amakawa, T., Goto, N. & Tsurumi, S. (2002) Plant Physiol. 130, 1908-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swarup, R., Parry, G., Graham, N., Allen, T. & Bennett, M. (2002) Plant Mol. Biol. 49, 411-426. [DOI] [PubMed] [Google Scholar]
- 35.Lang, V. & Palva, E. T. (1992) Plant Mol. Biol. 20, 951-962. [DOI] [PubMed] [Google Scholar]
- 36.Dharmasiri, S. & Estelle, M. (2002) Plant Mol. Biol. 49, 401-409. [PubMed] [Google Scholar]
- 37.Zheng, X. S., Chan, T. F. & Zhou, H. H. (2004) Chem. Biol. 11, 609-618. [DOI] [PubMed] [Google Scholar]
- 38.Wang, S., Sim, T. B., Kim, Y.-S. & Chang, Y.-T. (2004) Curr. Opin. Chem. Biol. 8, 1-7. [DOI] [PubMed] [Google Scholar]
- 39.Lum, P. Y., Armour, C. D., Stepaniants, S. B., Cavet, G., Wolf, M. K., Butler, J. S., Hinshaw, J. C., Garnier, P., Prestwich, G. D., Leonardson, A., et al. (2004) Cell 116, 121-137. [DOI] [PubMed] [Google Scholar]
- 40.Yamada, K., Lim, J., Dale, J. M., Chen, H., Shinn, P., Palm, C. J., Southwick, A. M., Wu, H. C., Kim, C., Nguyen, M., et al. (2003) Science 302, 842-846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.