Abstract
Objective/methods
DNA methylation plays an important role in obesity and related metabolic complications. We examined genome-wide DNA promoter methylation along with mRNA profiles in paired samples of human subcutaneous adipose tissue (SAT) and omental visceral adipose tissue (OVAT) from non-obese vs. obese individuals.
Results
We identified negatively correlated methylation and expression of several obesity-associated genes in our discovery dataset and in silico replicated ETV6 in two independent cohorts. Further, we identified six adipose tissue depot-specific genes (HAND2, HOXC6, PPARG, SORBS2, CD36, and CLDN1). The effects were further supported in additional independent cohorts. Our top hits might play a role in adipogenesis and differentiation, obesity, lipid metabolism, and adipose tissue expandability. Finally, we show that in vitro methylation of SORBS2 directly represses gene expression.
Conclusions
Taken together, our data show distinct tissue specific epigenetic alterations which associate with obesity.
Keywords: DNA methylation, Epigenetic mechanisms, Human adipose tissue depots, mRNA expression, Obesity-related co-morbidities
Highlights
-
•
Obesity-associated differences in DNA promoter methylation and transcriptome in human adipose tissue (ETV6).
-
•
Depot-specific analyses revealed novel/known genes (HAND2, HOXC6, PPARG, SORBS2, CD36, CLDN1).
-
•
EWAS revealed SSPN and CCDC125 associated to BMI in SAT or OVAT, respectively.
-
•
Differentially methylated genes overlap in part with GWAS hits for obesity and fat distribution.
1. Background
Abdominal omental visceral adipose tissue (OVAT) storage is more strongly associated with increased risk of obesity-related co-morbidities than subcutaneous (SAT) [1], [2]. Studying adipose tissue depot specific DNA methylation and concomitant alterations in mRNA expression patterns can help to better understand the intrinsic differences between SAT and OVAT [3], [4]. In addition to genetic factors ([5], [6]; reviewed in [7]), epigenetic mechanisms contribute to the unexplained heritability of obesity and fat distribution. Despite recent progress [8], [9], our current knowledge of adipose tissue depot specific methylation, especially in OVAT, and its impact on the development of co-morbidities is still limited. Recently, we reported significant differences in global DNA methylation levels between SAT and OVAT [10] and at candidates genes, such as TMEM18 [11]. Others demonstrated strong evidence for epigenetic mechanisms involved in obesity such as for HIF3A [12] and reported differences in DNA methylation and gene expression between human subcutaneous abdominal and gluteal adipose tissue [13] as well as between SAT and OVAT before and after gastric bypass and weight loss [14]. Furthermore, several studies reported methylation changes in skeletal muscle after bariatric surgery as well as after acute or long-term physical exercise in skeletal muscle or adipose tissue [15], [16], [17], [18]. Recently, Barrès and colleagues demonstrated dynamic remodeling of DNA methylation in the spermatozoal epigenome after bariatric surgery [19], while others reported indications for epigenetic inheritance in mice [20].
In order to acquire deep insights into adipose tissue specific biological principles of epigenetic gene regulation and to elucidate how these provoke the well-known physiological differences between SAT and OVAT, we tested the hypotheses that DNA promoter methylation levels in SAT and OVAT associate with BMI and obesity and that these profiles are adipose tissue depot specific.
2. Materials and methods
2.1. The Leipzig cohort
One hundred five Caucasian men (N = 39) and women (N = 66) were included in the study and paired samples of OVAT and SAT were obtained from patients who underwent open abdominal surgery for e.g. cholecystectomy or weight reduction surgery. Characteristics of the study population are summarized in Table 1. Seventy seven individuals out of 105 were included into the genome wide DNA-promoter methylation as well as in technical validation analyses. Initially, 82 individuals out of 105 were involved in mRNA expression profiling. Nineteen samples were excluded from the analysis due to insufficient RNA integrity leaving RNA expression values available from 63 individuals. Sixty three individuals out of 105 were included into mRNA expression profiling (SAT or OVAT). For a total number of 42 individuals, we were able to detect genome wide DNA promoter methylation in both tissue depots as well as genome wide expression profile in SAT or OVAT (overlap of methylation and expression in both tissues for 31 subjects, Table 1, Figure 1). Among the 105 subjects, 51 were non-obese; 44 of these were lean (mean age 65 ± 11 years, mean BMI 22.6 ± 2.2 kg/m2), 7 were overweight (mean age 61 ± 13 years, mean BMI 25.8 ± 0.8 kg/m2) and 54 were obese (mean age 50 ± 15 years, mean BMI 43.3 ± 10.9 kg/m2). Among the 77 individuals who were involved in methylation analyses, 54 did not have T2D, and 23 subjects had T2D. Eighty two individuals out of 105 were involved in mRNA expression profiling (mean age 55 ± 16 years, mean BMI 35.2 ± 13.7, non-obese = 36, obese = 46), including 61 non-diabetics and 21 subjects with T2D. Sixty three individuals out of 105 which were included into mRNA expression profiling (SAT or OVAT), included 49 non-diabetics and 14 subjects with T2D. Mature adipocytes and cells of the stromal vascular fraction were isolated from adipose tissue samples of 47 additional individuals (20 men, 27 women). Thirty three of these were obese (mean age 47 ± 11; mean BMI 55.4 ± 10.8) and 14 were lean (mean age 67 ± 9; mean BMI 23.4 ± 1.4). Paired samples of isolated adipocytes and SVF were available from 34 subjects (30 obese, mean age 47 ± 11; mean BMI 55.0 ± 11.1; 4 lean, mean age 70 ± 14, mean BMI 23.2 ± 0.4). Phenotyping was performed as previously described [21] and included anthropometric measurements, (weight, height, waist-to-hip-ratio (WHR)), body fat analysis using dual-energy X-ray absorptiometry, and laboratory parameters such as fasting plasma glucose and insulin, a 75 g oral glucose tolerance test (OGTT), and HbA1c. Insulin sensitivity was assessed with hyperinsulinemic-euglycemic clamps. Based on computed tomography scans measurement (L4–L5) of abdominal visceral and subcutaneous fat areas, obese subjects were further categorized as predominantly viscerally or subcutaneously obese as defined by a ratio of visceral/subcutaneously fat area of > or <0.5. Importantly, we included only individuals with a (in part self-reported) stable body weight at least 3 months prior to surgery (<2% fluctuations of body weight). All study protocols have been approved by the ethics committee of the University of Leipzig. All participants gave written informed consent before taking part in the study.
Table 1.
Main characteristics of the Leipzig and Italian cohorts.
| Subgroup | Leipzig |
Italian |
Italian vs. Leipzig |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-obese | Obese | P-value non-obese vs. obese | Total | Methylation total | Expression total | Overlap of methylation and expressiona total | Lean | Obese | P-value lean vs. obese | Lean vs. non-obese | Obese vs. obese | |
| N | 51 | 54 | – | 105 | 77 | 63 | 42 | 13 | 17 | – | – | – |
| Male/female (N) | 21/30 | 18/36 | 0.411 | 39/66 | 33/44 | 16/47 | 11/31 | 7/6 | 10/7 | 0.227 | – | – |
| Lean/overweight/obese (N) | 44/7/0 | 0/0/54 | – | 44/7/54 | 36/5/36 | 23/3/37 | 17/1/24 | 13/0/0 | 0/0/17 | – | – | – |
| Age (years) | 64 ± 11 | 50 ± 15 | <0.0001 | 57 ± 15 | 60 ± 10 | 53 ± 16 | 57 ± 10 | 42 ± 9 | 40 ± 10 | 0.576 | <0.0001 | 0.012 |
| BMI (kg/m2) | 23.05 ± 2.35 | 43.29 ± 10.90 | <0.0001 | 33.46 ± 12.9 | 31.14 ± 11.5 | 36.07 ± 13.9 | 34.31 ± 13.5 | 24.41 ± 1.7 | 39.54 ± 8.4 | <0.0001 | 0.021 | 0.198 |
| Body fat (%) | 22.3 ± 6.4 | 41.6 ± 10.4 | <0.0001 | 32.4 ± 13.0 | 31.5 ± 12.7 | 35.44 ± 13.9 | 35.74 ± 14.2 | – | – | – | – | – |
| CT-ratio (OVAT/SAT area) | 1.02 ± 0.89 | 0.32 ± 0.21 | <0.0001 | 0.69 ± 0.75 | 0.78 ± 0.83 | 0.62 ± 0.51 | 0.69 ± 0.55 | – | – | – | – | – |
| Waist (cm) | 81.2 ± 18.4 | 124.9 ± 20.1 | <0.0001 | 101.9 ± 29.1 | 96.9 ± 24.5 | 104.5 ± 30.1 | 99.0 ± 27.2 | – | – | – | – | – |
| Hip (cm) | 89.5 ± 11.8 | 124.9 ± 21.4 | <0.0001 | 106.3 ± 24.6 | 100.5 ± 17.3 | 108.7 ± 23.9 | 103.1 ± 18.3 | – | – | – | – | – |
| Waist-to-Hip ratio | 0.90 ± 0.12 | 1.01 ± 0.16 | <0.0001 | 0.95 ± 0.15 | 0.96 ± 0.15 | 0.95 ± 0.15 | 0.95 ± 0.14 | – | – | – | – | – |
| SAT area (cm) | 107.1 ± 197.2 | 1016.2 ± 585.3 | <0.0001 | 533.2 ± 622.2 | 364.5 ± 411.1 | 582.6 ± 635.3 | 393.5 ± 400.5 | – | – | – | – | – |
| OVAT area (cm) | 69.2 ± 73.1 | 254.7 ± 124 | <0.0001 | 156.2 ± 136.3 | 141.7 ± 125.7 | 164.0 ± 128.3 | 160.8 ± 136.0 | – | – | – | – | – |
| Fasting plasma glucose (mmol/l) | 5.45 ± 0.88 | 6.40 ± 1.82 | 0.001 | 5.94 ± 1.52 | 5.98 ± 1.48 | 5.87 ± 1.31 | 6.00 ± 1.39 | 4.90 ± 0.57 | 5.03 ± 0.63 | 0.564 | 0.036 | 0.003 |
| Fasting plasma insulin (pmol/l) | 17.94 ± 47.03 | 134.84 ± 146.72 | <0.0001 | 74.46 ± 121.92 | 75.02 ± 135.60 | 94.70 ± 141.10 | 107.59 ± 164.22 | 55.28 ± 35.84 | 111.12 ± 92.16 | 0.049 | 0.010 | 0.538 |
| HbA1c (%) | 5.42 ± 0.50 | 6.09 ± 0.76 | <0.0001 | 5.76 ± 0.73 | 5.79 ± 0.80 | 5.71 ± 0.66 | 5.81 ± 0.74 | – | – | – | – | – |
| HDL-C (mmol/l) | 1.49 ± 0.48 | 1.15 ± 0.26 | <0.0001 | 1.33 ± 0.43 | 1.34 ± 0.42 | 1.33 ± 0.39 | 1.37 ± 0.42 | 1.35 ± 0.29 | 1.14 ± 0.25 | 0.042 | 0.326 | 0.895 |
| LDL-C (mmol/l) | 3.06 ± 1.05 | 3.51 ± 1.38 | 0.107 | 3.27 ± 1.23 | 3.31 ± 1.23 | 3.28 ± 1.29 | 3.36 ± 1.38 | 2.98 ± 1.08 | 3.55 ± 1.07 | 0.161 | 0.812 | 0.916 |
| Triglycerides (mmol/l) | 1.28 ± 0.68 | 1.59 ± 0.94 | 0.150 | 1.41 ± 0.81 | 1.44 ± 0.79 | 1.20 ± 0.50 | 1.23 ± 0.51 | 1.11 ± 0.35 | 1.86 ± 0.65 | 0.001 | 0.398 | 0.313 |
| Free fatty acids (mmol/l) | 0.24 ± 0.25 | 0.48 ± 0.4 | 0.004 | 0.34 ± 0.34 | 0.29 ± 0.27 | 0.33 ± 0.36 | 0.23 ± 0.21 | – | – | – | – | – |
| Urates (mg/dl) | – | – | – | – | – | – | – | 5.02 ± 1.07 | 6.15 ± 1.07 | 0.008 | – | – |
All data are presented as mean ± SD.
Expression either in SAT or OVAT.
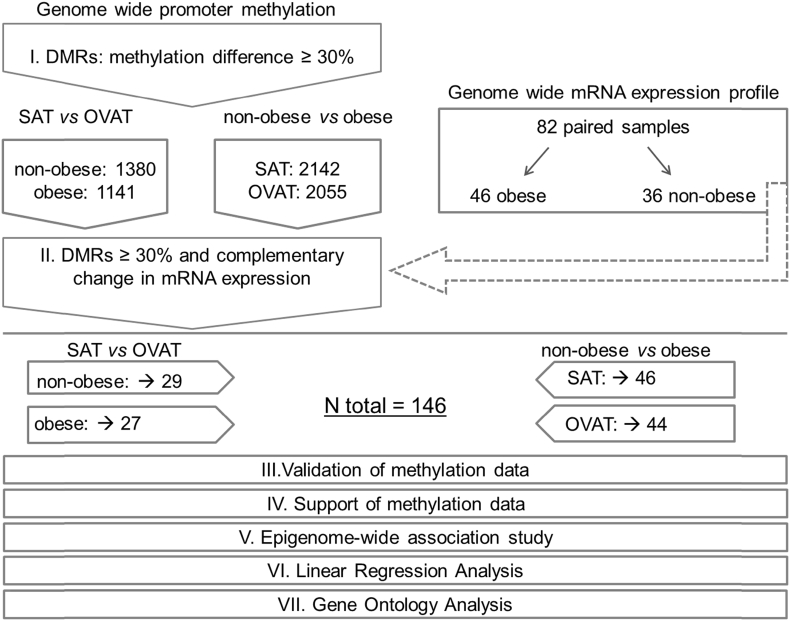
Figure 1.
Experimental workflow and study design. The figure shows the experimental workflow and study design. I. Genome wide promoter methylation was generated and an arbitrarily chosen cut off of 30% methylation differences (=DMR; differential methylated region) in the comparisons. Identified genes are given as numbers in non-obese and obese subgroups (SAT vs. OVAT) and in SAT and OVAT subgroups (non-obese vs. obese). II. The identified transcripts were tested for overlapping changes in mRNA expression. The final number of genes with negatively correlation between methylation and expression is underlined. Additionally, methylation data was validated using pyrosequencing for two genes (step III.) and supported in 3 independent data sets (step IV.). Furthermore, we conducted an epigenome-wide association study (EWAS) (step V.). Finally most promising candidates (N = 24) were tested for association with anthropometric and metabolic variables using linear regression analysis (step VI.) and analyzed using gene ontology analyses (step VII.).
2.2. Independent Italian cohort to support methylation effects
A total of 30 Caucasian individuals with paired samples of SAT and OVAT from an Italian cohort were included in the replication analysis. The cohort comprised 17 non-diabetic obese subjects and 13 lean (Table 1). SAT and OVAT biopsies where obtained either during bariatric surgery interventions (in obese individuals) or from abdominal surgery for non-inflammatory diseases (lean individuals). The study was approved by the Ethics Committee of Istituto Auxologico Italiano, Milan, Italy, and all patients gave written informed consent for sampling during surgical procedures. Compared to lean individuals, obese subjects were similar in terms of age and sex distribution, glucose, and total cholesterol and LDL levels. Obese subjects had a respectively significant higher mean value for BMI, triglycerides, urates levels, and white blood cells count (Table 1). HOMA-IR was ≥2 in 11 out of 17 obese enrolled patients and 3 had a diagnosis for hypertension. One patient was on fluoxetin treatment.
2.3. Sample preparation
Adipose tissue samples from the Leipzig cohort were taken from abdominal regions (SAT in the 5 cm periumbilical area, OVAT from the upper left part of omentum), directly frozen in liquid nitrogen after explantation, and stored at −80 °C. DNA and RNA were extracted using standard approaches (SIGMA ALDRICH, Saint Louis, USA and Qiagen, Hilden, Germany). DNA from the confirmatory cohort (Italy) was extracted usingDNeasy Blood and Tissue Kit (Qiagen, Milano, Italy). DNA was bisulfite converted using the EZ DNA Methylation kit (Zymo Research, Irvine, CA).
2.4. Genome wide promoter methylation
2.4.1. The Leipzig cohort
DNA promoter methylation was measured by using methylated DNA immunoprecipitation (MeDIP) combined with subsequent hybridization on tiling arrays. In brief, DNA shearing was performed by using a Bioruptor Plus, and MeDIP was performed using the MagMeDIP kit (both Diagenode, Seraing, Belgium). PCR primers for known methylated (TSH2B) and unmethylated regions (GAPDH) (MagMeDIP kit, Diagenode) were used to test MeDIP specificity by qPCR. Enriched methylated DNA was amplified using WGAII, purified using GenElute Clean-UP kit, and re-amplified using a modified protocol [22] of WGAIII (all SIGMA-ALDRICH, Saint Louis, USA). The amount of dUTP was increased to 1.8 μl to ensure length (about 66 bp) and uniformity of the double stranded DNA fragments. DNA was fragmented, labeled, and hybridized on GeneChip Human Promoter 1.0R Arrays (Affymetrix Inc., Santa Clara, USA). Arrays comprised ∼4.6 million probes covering ∼25.500 human promoter regions.
2.4.2. Italian cohort
2.4.2.1. Differential methylation (DM) analysis using Infinium HumanMethylation450 BeadChips
Infinium HumanMethylation450K BeadChips (Illumina) were used to detect methylation levels in paired SAT and OVAT samples. At gene level 450K, microarray covers 99% of RefSeq genes with multiple sites in the annotated promoter (1500 bp or 200 bp upstream of transcription start site), 5′-UTR, first exon, gene body and 3′-UTR. From the CpG context, it covers 96% of CpG islands with multiple sites in the annotated CpG islands, shores (regions flanking island) and shelves (regions flanking shores).
2.5. Genome wide expression – Leipzig cohort
Genome wide expression profiling was performed using Illumina human HT-12 expression chips. RNA integrity and concentration was examined using Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). RNA samples with RNA integrity values (RIN) of less than five were discarded from further analysis [23].
2.6. Pyrosequencing
Pyrosequencing was performed for HOXD3 and HOXD4 as described elsewhere [10], [11]. PyroMark assays were designed for: NM_006898; probe number: 176736323; NM_014621, probe numbers: 176723238, 176724282. Pyrosequencing was conducted in duplicates and sequencing results were checked for consistence with MeDIP methylation array results (Supplementary Table 9).
2.7. Luciferase assays
2000 bp sized DNA fragments of the human SORBS2 and EMX2 promoter region (hg38-SORBS2: chr4:186969043-186971042; EMX2: chr10:119289946-119291945) were inserted into a CpG-free firefly luciferase reporter vector (pCpGL-basic) [24]. We performed luciferase assays as described elsewhere [18]. Briefly, DNA sequences were inserted into the vector and constructs were methylated either using SssI (methylation of double stranded dinucleotide CG sequence; results in complete promoter methylation) or HpaII (methylation of the internal cytosine residue in the CCGG sequence) methyltransferases and S-adenosylmethionine (SAM) as methyldonor (New England Biolabs, Frankfurt am Main, Germany). Unmethylated controls were treated exactly as methylated constructs including application of SAM but without using methylation enzymes. Constructs were transfected into human MCF7 cells (human breast adenocarcinoma cell line). Additionally, transfection of MCF7 cells with an empty-reporter vector serves as background control of firefly activity. The unmethylated promoter construct generates significantly more luciferase signal than the empty reporter vector (data not shown). All cells were co-transfected with pRL renilla luciferase control-reporter vector (pRL-CMV vector, Promega, Madison, USA) to control transfection efficiency. FuGene® HD transfection reagent (Promega, Madison, USA) was used for transfection of cells. Furthermore, untransfected cells were used for background correction of renilla luciferase activity. Finally, luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega, Madison, USA). P-values were calculated using two sided, unpaired t-tests.
2.8. qPCR experiments in adipocytes vs. stromal vascular fractions
Adipocytes were isolated by collagenase (1 mg/ml) digestion. For expression analysis of SORBS2, HAND2, HOXC6, EMX2, PPARG, CLDN1, CD36, and ETV6 in adipocytes and stromal vascular fractions, total RNA was isolated from adipocytes and stromal vascular fractions extracted from subcutaneous and visceral adipose tissue. Relative quantification was performed with the Quantstudio 6 System (Thermo Fisher Scientific), using commercially available TaqMan Probes (Assay-on-Demand; Thermo Fisher Scientific). Relative quantities of target transcripts were calculated from samples after normalization of the data against the endogenous control, HPRT1 rRNA (Thermo Fisher Scientific).
2.9. Data analysis and statistics
2.9.1. The Leipzig cohort
Normalized probe intensities were used. Differential methylation (two-class comparisons) was calculated using Model-based analysis of tiling-arrays for ChIP–chip (MAT) [25] as implemented in the R package ‘rMAT’ (http://www.bioconductor.org/packages/release/bioc/manuals/rMAT/man/rMAT.pdf). We included all probes interrogating methylation of cytosines 2000 bp upstream to 200 bp downstream the transcription start site. rMAT provides regions of differential methylation for each promoter. They were transformed into differential methylation percentage-values, ranging from 0% (no differential methylation) to 100% (all interrogated regions are affected; Table 2, Table 3). Only promoters with a minimum differential methylation of larger than 30% were considered in further analysis (Supplementary Tables 1–4). Significance was estimated by permutation tests providing P-values, which were multitest adjusted in terms of false discovery rates (FDR) using the R package fdr-tool [26]. We applied a cut-off of FDR < 0.3.
Table 2.
Top genes differentially methylated and expressed between non-obese and obese individuals.
| Depot | Gene name | Percentage of hyper/hypomethylation |
P-value |
logFC | ||
|---|---|---|---|---|---|---|
| Obese | Non-obese | Obese | Non-obese | |||
| SAT | SETMAR | 53.5 | 0.0 | 2.00E-04 | 1 | −0.032 |
| HSD17B8 | 45.0 | 0.0 | 2.00E-04 | 1 | −0.103 | |
| GOLGA6L4 | 42.9 | 0.0 | 2.00E-04 | 1 | −0.040 | |
| NEDD4L | 35.6 | 0.0 | 2.00E-04 | 1 | −0.036 | |
| THNSL2 | 34.7 | 0.0 | 4.00E-04 | 0.9996 | −0.050 | |
| RTN1 | 34.0 | 0.0 | 2.00E-04 | 1 | −0.120 | |
| NET1 | 33.4 | 0.0 | 2.00E-04 | 1 | −0.158 | |
| C8orf46 | 33.0 | 0.0 | 2.00E-04 | 1 | −0.063 | |
| TTLL7 | 32.1 | 0.0 | 2.00E-04 | 1 | −0.046 | |
| BCKDHB | 31.4 | 0.0 | 2.00E-04 | 1 | −0.085 | |
| EPDR1 | 0.0 | 72.8 | 1 | 2.00E-04 | 0.169 | |
| RUNX1 | 0.0 | 64.9 | 1 | 2.00E-04 | 0.051 | |
| ATP6V1B2 | 0.0 | 61.7 | 1 | 2.00E-04 | 0.028 | |
| GPR137B | 0.0 | 50.8 | 1 | 2.00E-04 | 0.064 | |
| SLC2A5 | 0.0 | 49.7 | 1 | 2.00E-04 | 0.141 | |
| HCK | 0.0 | 49.0 | 1 | 2.00E-04 | 0.106 | |
| CD99L2 | 0.0 | 46.0 | 1 | 2.00E-04 | 0.011 | |
| FAM46A | 0.0 | 44.8 | 1 | 2.00E-04 | 0.104 | |
| EXOSC7 | 0.0 | 44.1 | 1 | 2.00E-04 | 0.175 | |
| CD9 | 0.0 | 42.8 | 1 | 2.00E-04 | 0.116 | |
| OVAT | SORBS2 | 81.9 | 0.0 | 2.00E-04 | 1 | −0.021 |
| CASQ2 | 74.6 | 0.0 | 2.00E-04 | 1 | −0.043 | |
| DUSP22 | 59.7 | 12.6 | 2.00E-04 | 1 | −0.059 | |
| KCNT2 | 58.9 | 0.0 | 2.00E-04 | 1 | −0.007 | |
| SETMAR | 57.1 | 0.0 | 2.00E-04 | 1 | −0.056 | |
| GOLGA6L4 | 50.4 | 0.0 | 2.00E-04 | 1 | −0.026 | |
| MMRN1 | 46.7 | 0.0 | 2.00E-04 | 1 | −0.053 | |
| CPE | 42.2 | 0.0 | 2.00E-04 | 1 | −0.024 | |
| KIAA1217 | 40.7 | 0.0 | 2.00E-04 | 1 | −0.098 | |
| APMAP | 40.1 | 0.0 | 2.00E-04 | 1 | −0.107 | |
| RGS1 | 0.0 | 64.2 | 1 | 2.00E-04 | 0.110 | |
| TNFRSF21 | 0.0 | 57.0 | 1 | 2.00E-04 | 0.039 | |
| DSP | 0.0 | 47.2 | 1 | 2.00E-04 | 0.134 | |
| AQP9 | 0.0 | 44.8 | 1 | 2.00E-04 | 0.133 | |
| NKX3-2 | 0.0 | 43.3 | 1 | 2.00E-04 | 0.087 | |
| TLE1 | 0.0 | 43.0 | 1 | 2.00E-04 | 0.004 | |
| KCTD8 | 2.2 | 44.8 | 1 | 2.00E-04 | 0.034 | |
| CYFIP2 | 14.2 | 56.1 | 1 | 2.00E-04 | 0.087 | |
| SERPINE2 | 0.0 | 41.7 | 1 | 2.00E-04 | 0.091 | |
| VAV3 | 0.0 | 39.7 | 1 | 2.00E-04 | 0.014 | |
Table 2 presents the top 10 candidate genes in SAT and OVAT comparing promoter methylation levels between non-obese and obese individuals. Differences in mRNA expression are given as logarithmic fold change (logFC), consistently in comparison to non-obese subjects.
Table 3.
Top genes differentially methylated and expressed between SAT and OVAT.
| Gene name | Percentage of hyper/hypomethylation |
P-value |
logFC | |||
|---|---|---|---|---|---|---|
| SAT | OVAT | SAT | OVAT | |||
| Obese | FAM25C | 54.9 | 0.0 | 2.00E-04 | 1 | 0.509 |
| CKMT1B | 53.2 | 0.0 | 2.00E-04 | 1 | 0.468 | |
| SORBS2 | 51.8 | 0.0 | 2.00E-04 | 1 | 0.192 | |
| TCF21 | 45.7 | 0.0 | 2.00E-04 | 1 | 0.521 | |
| DFNA5 | 36.3 | 0.0 | 2.00E-04 | 1 | 0.270 | |
| OLFML1 | 35.2 | 0.0 | 2.00E-04 | 1 | 0.142 | |
| ANGPTL7 | 33.7 | 0.0 | 2.00E-04 | 1 | 0.278 | |
| HAND2 | 33.5 | 0.0 | 2.00E-04 | 1 | 0.226 | |
| PAPPA | 31.0 | 0.0 | 2.00E-04 | 1 | 0.204 | |
| CKMT1A | 30.5 | 0.0 | 2.00E-04 | 1 | 0.357 | |
| HOXC6 | 0.0 | 66.4 | 1 | 2.00E-04 | −0.479 | |
| AFF3 | 0.0 | 58.2 | 1 | 2.00E-04 | −0.166 | |
| PHLDB1 | 0.0 | 53.2 | 1 | 2.00E-04 | −0.100 | |
| EMX2 | 0.0 | 45.8 | 1 | 2.00E-04 | −0.362 | |
| COL12A12 | 0.0 | 44.5 | 1 | 2.00E-04 | −0.210 | |
| SORT1 | 0.0 | 43.5 | 1 | 2.00E-04 | −0.191 | |
| AOC3 | 0.0 | 42.5 | 1 | 2.00E-04 | −0.117 | |
| DEFB1 | 0.0 | 41.2 | 1 | 2.00E-04 | −0.289 | |
| PPARG | 0.0 | 41.7 | 1 | 2.00E-04 | −0.132 | |
| ANXA1 | 0.0 | 39.3 | 1 | 2.00E-04 | −0.138 | |
| Non-obese | TCF21 | 81.5 | 0.0 | 2.00E-04 | 1 | 0.399 |
| CKMT1A | 61.0 | 0.0 | 2.00E-04 | 1 | 0.147 | |
| SORBS2 | 56.8 | 0.0 | 2.00E-04 | 1 | 0.142 | |
| CPE | 53.8 | 0.0 | 2.00E-04 | 1 | 0.106 | |
| GREM1 | 49.0 | 0.0 | 2.00E-04 | 1 | 0.338 | |
| CLDN1 | 48.5 | 0.0 | 2.00E-04 | 1 | 0.543 | |
| PFKM | 42.8 | 0.0 | 2.00E-04 | 1 | 0.175 | |
| PAMR1 | 42.4 | 0.0 | 2.00E-04 | 1 | 0.301 | |
| PNMA2 | 40.2 | 0.0 | 2.00E-04 | 1 | 0.190 | |
| DFNA5 | 38.3 | 0.0 | 2.00E-04 | 1 | 0.173 | |
| HOXC6 | 0.00 | 72.2 | 1 | 2.00E-04 | −0.416 | |
| MEOX1 | 0.00 | 48.3 | 1 | 2.00E-04 | −0.186 | |
| GOS2 | 0.00 | 47.5 | 1 | 2.00E-04 | −0.121 | |
| TMEM139 | 0.00 | 46.9 | 1 | 2.00E-04 | −0.063 | |
| BHMT | 0.00 | 46.3 | 1 | 2.00E-04 | −0.085 | |
| PPARG | 0.00 | 46.2 | 1 | 2.00E-04 | −0.035 | |
| AOC3 | 0.00 | 44.2 | 1 | 2.00E-04 | −0.097 | |
| COL12A1 | 0.00 | 43.1 | 1 | 2.00E-04 | −0.067 | |
| ANGPT1 | 0.00 | 42.8 | 1 | 2.00E-04 | −0.055 | |
| EMX2 | 0.00 | 41.3 | 1 | 2.00E-04 | −0.260 | |
Table 3 presents the top 10 candidate genes in non-obese and obese individuals comparing promoter methylation levels between SAT and OVAT. Corresponding negatively correlated mRNA expression values are shown. Changes in mRNA expression are given as logFC, consistently standardized in relation to OVAT. Genes which are hypermethylated in SAT show increased mRNA expression in OVAT and are represented by a positive logFC value.
IlluminaBeadChipsHT-12, expression data were background-corrected, log-transformed, and quantile-normalized before downstream analysis. Expression data were then matched to methylation data and ranked by using the Mann–Whitney U test. Differential expression analysis was performed using the R package oposSOM [27]. Results were listed as log fold change (logFC; Table 2, Table 3). Differential expression and methylation were then analyzed together to identify genes being either up- or down-regulated and either hyper- or hypo-methylated. For further analyses, we focused exclusively on genes showing negatively correlated DNA methylation and expression values (Figure 2). For visualization by means of circle plots, we used circos software package (version circos-0.65-pre5) [28].
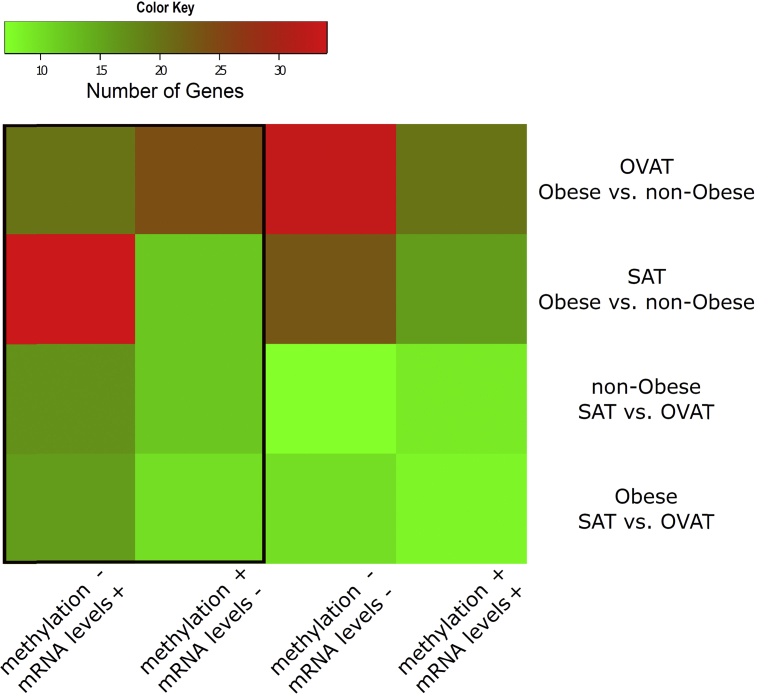
Figure 2.
Numbers of identified genes showing co-regulated changes in methylation and gene expression. The heatmap [57] shows the number of genes showing significant differences in methylation and mRNA expression. The framed columns on the left side represent genes conferring negatively correlated methylation and expression levels, which were taken forward to replication analyses.
Linear regression analyses were performed using R [58] adjusted for sex, age, lnBMI (except for BMI), and type 2 diabetes. Methylation data were used as normalized probe intensities, and non-normally distributed phenotypes were log transformed to approximate normal distribution.
To analyze differences in expression levels between visceral and subcutaneous adipocytes vs. stromal vascular fractions as well as for group differences (non-obese vs. obese), unpaired two-tailed t-tests were applied.
2.9.2. Independent Italian cohort to support methylation effects
In order to identify differentially methylated cytosines (DMCs), we used a method based on F-test. We first focused on DMCs between SAT and VAT and then on those between lean and obese groups for each adipose depot. While DMCs analysis among sample group (SAT/VAT) resulted in about 100,000 DMC positions (multiple testing corrected q-value < 0.05), only about 1800 DMC positions (multiple testing corrected q-value < 0.05) could be identified between lean and obese individuals.
2.9.2.1. Data filtering
The R package minfi was used to read differential methylation values (describing methylation level between 0 and 100%) from the .idat files. A detection P-value was determined for every cytosine probed in every sample. Then, the cytosine positions with P-value > 0.05 in more than 20% of total samples (60) were removed from the further analysis. In total, 3532 cytosines were removed out of 485512. The data were then subjected to within array normalization method SWAN which reduces technical variation within and between arrays.
3. Results
3.1. General methylome and transcriptome differences
Differential methylation was estimated in the promoter range of each gene. By comparing non-obese vs. obese individuals in the same adipose tissue depot, we found 2142 genes which were differentially methylated in SAT, while 2055 genes were identified in OVAT (Figure 1).
In non-obese subjects, we identified 1381 differentially methylated genes when comparing SAT and OVAT. The same comparison in obese individuals yielded 1141 genes (Figure 1). All these identified genes passed through a correction for multiple testing using FDR (Supplementary Tables 1–4).
To further substantiate the results from this discovery approach, we focused on genes showing negative correlations in mRNA expression profiles along with the described methylation differences. We finally identified 29 genes differentially regulated in SAT vs. OVAT in the non-obese subgroup and 27 in obese individuals. Similarly, in our obesity-specific analysis, we focused only on genes fulfilling these stringent filter criteria, and, by comparing non-obese vs. obese subjects, we found 46 differentially regulated genes in SAT and 44 genes in OVAT (all Figure 1, Figure 2).
3.2. Obesity associated differences in DNA methylation and mRNA expression
The top ten candidate genes showing the biggest differences in the ratio of hyper/hypomethylation between non-obese and obese individuals in SAT and OVAT are presented in Table 2. Effect directions of methylation and expression of differentially regulated genes and their distribution over the genome are visualized in circle plots (Figure 3C,D). We observed several novel candidates such as the empty spiracles homeobox 2 (EMX2), EPDR1 (ependymin related 1), RUNX1 (runt-related transcription factor 1), DUSP22 (dual specificity phosphatase 22), and RGS1 (regulator of G-protein signaling 1). Complete lists of differentially methylated and expressed transcripts are presented in Supplementary Tables 4 and 5
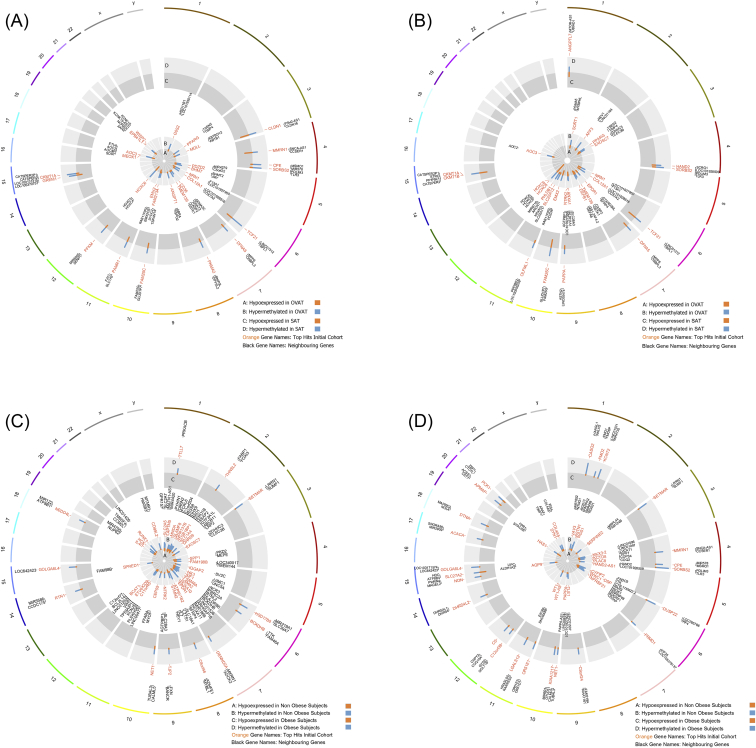
Figure 3.
Circle plots showing the genome wide distribution of differentially regulated genes. The figure shows all genes conferring significant changes in DNA methylation and negatively correlated gene expression levels. Blue bars represent methylation intensities; orange bars represent expression levels (logFC values). Outside circles represent chromosomes, the light grey circles represent methylation levels, and the grey circles represent expression values. A) non-obese subjects hypermethylated/lower expressed in SAT (middle circle) and hypermethylated/lower expressed in OVAT (inner circle); B) obese subjects hypermethylated/lower expressed in SAT (middle circle) and hypermethylated/lower expressed in OVAT (inner circle); C) SAT: genes hypermethylated/lower expressed in obese (middle circle) and hypermethylated/lower expressed in non-obese individuals (inner circle); D) OVAT: genes hypermethylated/lower expressed in obese (middle circle) and hypermethylated/lower expressed in non-obese individuals (inner circle).
3.3. Adipose tissue depot specific differences
The top ten candidate genes showing the largest differences in the ratio of hyper/hypomethylation between SAT and OVAT in both non-obese and obese subjects along with corresponding changes in gene expression are presented in Table 3. Further, effect directions and their distribution over the genome are visualized in circle plots (Figure 3A,B). We identified novel candidates that may play a role in adipose tissue development and differentiation such as the transcription factor 21 (TCF21), also known as an epigenetically regulated white adipocyte marker [29], which was strongly hypermethylated and less expressed in SAT among non-obese subjects. Another identified candidate was claudin 1 (CLDN1) encoding an integral membrane protein and a component of tight junction strands. We further observed the heart and neural crest derivatives expressed 2 (HAND2) which may play a role in adipogenic differentiation via NOTCH signaling [30], [31] and the cluster of differentiation 36 (CD36), which is involved in lipid metabolism [32], [33]. Further, we also confirmed known candidate genes such as the homeobox C6 (HOXC6) and the peroxisome proliferator-activated receptor gamma (PPARG). Complete lists of differentially methylated and expressed genes are presented in Supplementary Tables 7 and 8
3.4. Gene expression analysis in isolated adipocytes and SVF
To further substantiate our findings and to shed more light on potential functional effects of these methylation events, we analyzed gene expression of the top genes (SORBS2, HAND2, HOXC6, EMX2, PPARG, CLDN1, CD36, and ETV6) separately in mature adipocytes and stromal vascular fractions (Supplementary Figure 1). Our results largely confirm the initially observed effects performed in adipose tissue biopsies. However, although non-significant we discovered different effects in adipocytes compared to SVF for two genes (PPARG and CD36), which might have influenced our initial analyses.
3.5. Technical validation of methylation data
Pyrosequencing was used to validate methylation data. Two genes demonstrating high differences of DNA methylation between SAT and OVAT (Supplementary Tables 1 and 2) were selected for validation. The results revealed similar directions in DNA methylation changes of the analyzed transcripts of homeobox D3 (HOXD3); and homeobox D4 (HOXD4) compared to the genome wide array derived data (Supplementary Table 9).
3.6. Support of methylation effects in independent cohorts
We further sought to support our methylation data in three independent data sets from analyses of DNA methylation pattern in SAT vs. OVAT and/or in non-obese vs. obese subjects. We used a data set comprising six subjects with data available for SAT and OVAT (obtained post mortem) [34] publicly available via MARMAL-Aid [35]. We observed nine genes showing similar effect directions of differential methylation in SAT vs. OVAT. Further, we used another independent cohort from Italy comprising 30 individuals, which supported methylation effects of 44 genes compared to our initial methylation data. Among these three cohorts, we finally observed 6 genes, which were consistently supported: HAND2, HOXC6, PPARG, sorbin and SH3 domain containing 2 (SORBS2), CD36, and CLDN1 (all Table 4).
Table 4.
In-silico replication of methylation data.
| SAT vs. OVAT (combined non-obese and obese) |
Obese vs non-obese (combined SAT and OVAT) |
||||
|---|---|---|---|---|---|
| Own initial data | Replication Italy | Replication Slieker et al. (33) | Own initial data | Replication Italy | Replication Benton et al. (27) (SAT) |
| TMEM139 | TCF21 | DMRT2 | SETMAR | BCKDHB | AIF1 |
| EPDR1 | CKMT1B | DMRT3 | ETV6 | ETV6 | ETV6 |
| HAND2 | HAND2 | HAND2 | HSD17B8 | HSD17B8 | |
| HOXC6 | HOXC6 | HOXC6 | NEDD4L | NEDD4L | |
| PPARG | PPARG | PPARG | THNSL2 | NET1 | |
| SORBS2 | SORBS2 | SORBS2 | TTLL7 | SETMAR | |
| CD36 | CD36 | CD36 | C8orf46 | TJP2 | |
| CLDN1 | CLDN1 | CLDN1 | RTN1 | C11orf45 | |
| DEFB1 | TMEM139 | TBX15 | NET1 | DHRS9 | |
| AOC3 | ANGPTL7 | TJP2 | C8orf46 | ||
| COL12A1 | EPDR1 | DENND2A | FAM46A | ||
| EMX2 | NTRK2 | BCKDHB | GPR137B | ||
| AFF3 | EXOSC7 | SPP1 | HOXA10 | ||
| NTRK2 | ANXA1 | STMN2 | IL4I1 | ||
| CKMT1A | HOXC8 | LHCGR | IQGAP2 | ||
| PAPPA | AOC3 | FAM198B | LAPTM5 | ||
| HOXC8 | EMX2 | NPL | LHCGR | ||
| ANGPTL7 | OLFML1 | LAPTM5 | RUNX1 | ||
| OLFML1 | DEFB1 | SLAMF8 | SLAMF8 | ||
| DFNA5 | NRN1 | EXOSC7 | ACACA | ||
| TCF21 | COL12A1 | HK3 | C9orf24 | ||
| NRN1 | AFF3 | EPDR1 | CPE | ||
| CKMT1B | GREM1 | MNDA | DHRS4L2 | ||
| FAM25C | CKMT1A | C11orf45 | DTNA | ||
| TPGS2 | CPE | GOLGA6L4 | DUSP22 | ||
| CDH13 | MMRN1 | BCAT1 | FMO2 | ||
| EXOSC7 | PFKM | SLC2A5 | FRMD1 | ||
| PDZD2 | PAPPA | DHRS9 | KIAA1217 | ||
| EPB41L1 | PAMR1 | ITGB1BP1 | LGALS12 | ||
| WISP2 | FAM25C | HLA-DRA | NDN | ||
| PHLDB1 | PNMA2 | CD9 | OR51E1 | ||
| ANGPT1 | CDH13 | IL4I1 | SORBS2 | ||
| FAM213A | DFNA5 | IQGAP2 | AQP9 | ||
| LHX6 | PDZD2 | CEP55 | ASTN1 | ||
| G0S2 | EPB41L1 | AIF1 | CYFIP2 | ||
| BHMT | WISP2 | HCK | HAS1 | ||
| TMEM139 | PHLDB1 | LAMA2 | KCTD8 | ||
| MGLL | FAM213A | FAM46A | NKX3-2 | ||
| MEOX1 | G0S2 | SLC1A4 | SERPINE2 | ||
| ANXA1 | BHMT | NTRK2 | WT1 | ||
| GREM1 | ANGPT1 | HOXA10 | WT1-AS | ||
| PAMR1 | LHX6 | SPRED1 | WWC1 | ||
| CPE | MGLL | GPR137B | |||
| MMRN1 | MEOX1 | RUNX1 | |||
| PFKM | CD99L2 | ||||
| ATP6V1B2 | |||||
| SORBS2 | |||||
| CASQ2 | |||||
| KCNT2 | |||||
| DUSP22 | |||||
| CPE | |||||
| MMRN1 | |||||
| KIAA1217 | |||||
| CS | |||||
| APMAP | |||||
| NDN | |||||
| OR51E1 | |||||
| FMO2 | |||||
| C9orf24 | |||||
| LGALS12 | |||||
| FRMD1 | |||||
| DTNA | |||||
| SLC27A2 | |||||
| DHRS4L2 | |||||
| PCK1 | |||||
| C12orf39 | |||||
| ACACA | |||||
| HAS1 | |||||
| WWC1 | |||||
| DSP | |||||
| WT1-AS | |||||
| AQP9 | |||||
| WT1 | |||||
| SYN1 | |||||
| PLAC8 | |||||
| RGS1 | |||||
| CYFIP2 | |||||
| SERPINE2 | |||||
| COL4A5 | |||||
| NKX3-2 | |||||
| PAPPA | |||||
| ASTN1 | |||||
| KCTD8 | |||||
| HAND2-AS1 | |||||
| VAV3 | |||||
| TNFRSF21 | |||||
| TLE1 | |||||
Replication analyses are summarized. Input data are genes from the initial sample set (Leipzig cohort) conferring negatively correlated DNA methylation and expression levels. Genes marked in bold were successfully replicated in two independent cohorts.
Moreover, to add further weight to our finding of methylation differences between non-obese vs. obese subjects, we used publicly available data of 15 individuals as reported by Benton et al. (E-MTAB-3052) [14]. By considering individuals as obese before bariatric surgery while categorizing subjects after the operation as lean(er), we used this dataset to look for genes showing similar effect directions as compared to our initial methylation data in SAT and OVAT. Our analysis revealed two genes exhibiting the same effect direction. Again, we also used the Italian cohort and supported methylation directions at 42 genes. Among all three cohorts, ets variant 6 (ETV6) (Table 4) showed consistent effects.
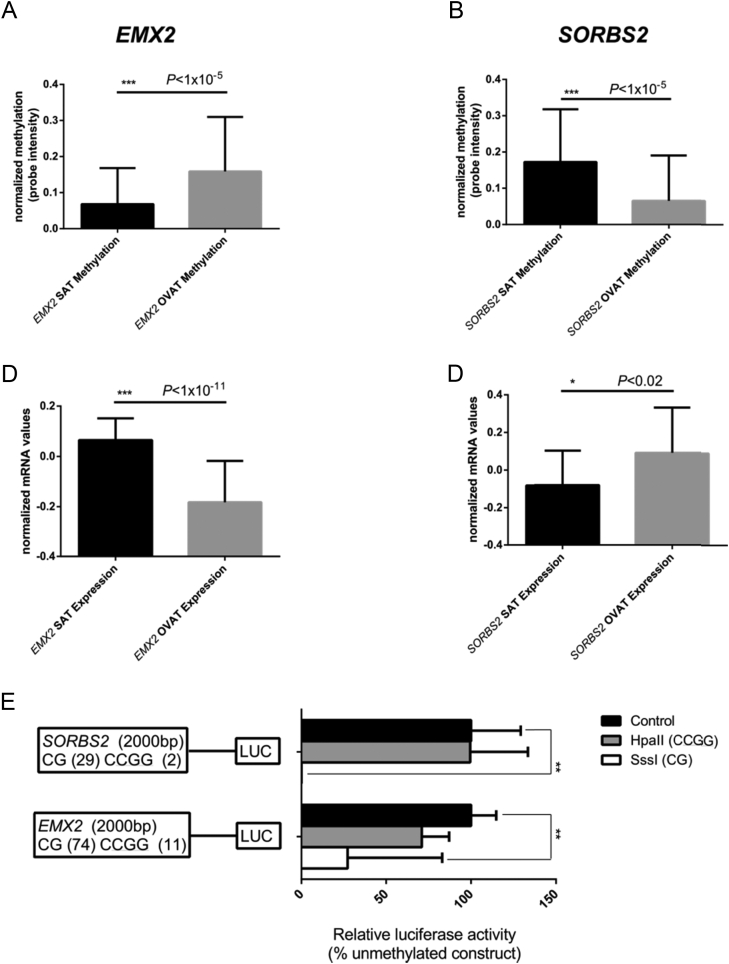
3.7. In-vitro methylation decreases gene expression
Next, we tested whether induced changes in promoter DNA methylation of two identified top genes SORBS2 and EMX2 truly affect the transcriptional activity in vitro using a firefly luciferase assay as described elsewhere [18]. The results clearly show that methylation by SssI methyltransferase significantly reduced the luciferase activity for both constructs as shown in Figure 4.
Figure 4.
Luciferase assays for SORBS2 and EMX2. The figure shows effect directions of methylation and expression in SAT and OVAT, respectively. Data shown represent results from the two luciferase reporter plasmids which were used to test effects of DNA methylation in SORBS2 and EMX2 promoter regions on transcriptional activity. Both plasmids comprised 2000 bp of either SORBS2 or EMX2 promoter regions inserted into a pCpGl-basic vector which was either methylated by HpaII (which methylates the internal Cs of the CCGG sequence) or SssI (which methylates all CpG sites) and then transfected into MCF7 cells. Data were normalized using a co-transfected renilla luciferase vector and presented as methylated promoter constructs (grey and white bars) relative to un-methylated constructs (black bars). Experiments were performed six times (SORBS) or eight times (EMX2) including four replicates in each experiment. Cells transfected with an empty pCpGL-basic vector and untransfected cells activities were used as background correction for firefly and renilla luciferase activity, respectively. *P < 0.05 **P < 0.005. Data are shown as mean ± SEM.
3.8. Epigenome wide analysis (EWAS) for BMI
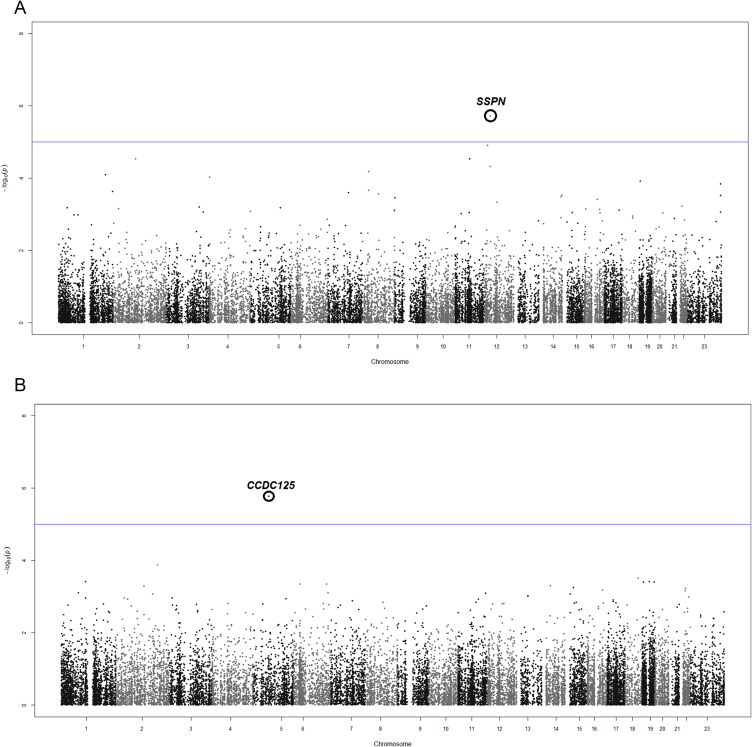
EWAS was conducted in the total cohort for which methylation data were available (N = 77). We tested for association of mean DNA methylation levels per promoter (for 22.625 transcripts) with BMI as a continuous variable in SAT and OVAT, separately. In SAT, we identified the sarcospan gene (SSPN) significantly and negatively associated to BMI (Figure 5, Table 5). In OVAT, we observed the coiled-coil domain containing protein 125 (CCDC125) showing the strongest positive effect on BMI (Figure 5, Table 5). Both top hits reached genome wide statistical significance according to multiple testing (Bonferroni corrected 0.05/22.625 transcripts = P < 2.20 × 10−6).
Figure 5.
Epigenome wide association study (EWAS) between DNA promoter methylation and BMI. EWAS was applied to test for a relationship (linear regression) of DNA promoter methylation per transcript with BMI. P-values for epigenome wide association analysis with BMI were calculated using an R (version 3.0.2) package called CpGassoc [58]. All analyses were adjusted for age, gender, and type 2 diabetes, separately in a) SAT and b) OVAT in the samples from the Leipzig cohort, for which methylation data were available (N = 77). Different transcripts per gene showing exactly the same association results are summarized in the same dot. Bonferroni correction was used to correct for multiple testing.
Table 5.
Top EWAS hits in SAT and OVAT.
| Depot | Gene ID | Transcript | T-statistic | P-value | Function | Literature |
|---|---|---|---|---|---|---|
| SAT | SSPN | NM_005086 | −5.19 | 1.9 × 10−06 | Sarcospan | Obesity, WHR PMID: 20935629 PMID: 26449484 |
| OVAT | CCDC125 | NM_176816 | 5.22 | 1.7 × 10−06 | Coiled-coil domain containing 125 | Isaac's syndrome PMID: 19787194 |
Top candidate transcripts from EWAS analyses in SAT and OVAT are shown approaching genome-wide significance (P < 2.2 × 10−6). T-statistic and P-values were generated by applying linear regression analysis.
3.9. Gene ontology analysis
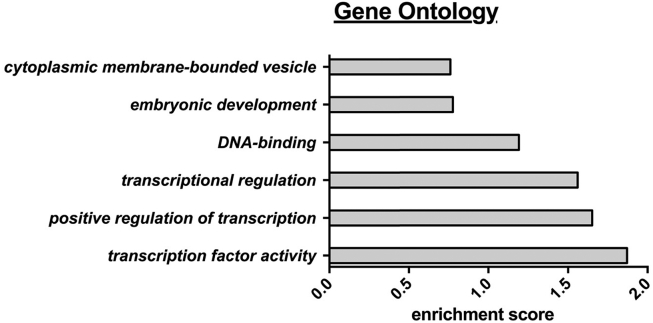
Next, we used the successfully replicated genes from i) non-obese vs. obese and ii) SAT vs. OVAT (Table 2, Table 3; total N = 7), the top three candidates selected from our top ten hits (N = 17) and the top candidates from the EWAS (N = 2) to generate a final list of the most promising candidate genes (N = 24), which were taken forward to gene ontology analyses and extended association studies with multiple variables of anthropometric and metabolic phenotypes. We performed gene ontology analyses (https://david.ncifcrf.gov/) and found that identified genes are most likely involved in transcription factor activity (enrichment score ∼ 1.87), regulation of transcription (enrichment score ∼ 1.65), transcriptional regulation (enrichment score ∼ 1.56), and DNA-binding (enrichment score ∼ 1.19) (Figure 6, Supplementary Table 10).
Figure 6.
Gene ontology analysis. Gene ontology analysis was performed using the DAVID program (http://david.abcc.ncifcrf.gov/summary.jsp) with default settings, high classification stringency and 0.5 as cut off for the enrichment score. Enriched pathways include the following genes: transcription factor activity: HAND2, PPARG, RUNX1, HSD17B8, HOXC6, TCF21; positive regulation of transcription: TCF21, MEOX1, HAND2, PPARG, RUNX1; transcriptional regulation: HOXC6, TCF21, MEOX1, HAND2, PPARG, ETV6, RUNX1, HSD17B8, AFF3; DNA-binding: HOXC6, TCF21, MEOX1, HAND2, PPARG, AFF3, ETV6, RUNX1.
3.10. Association with phenotypic traits
Linear regression analyses were performed in the total cohort for which methylation data were available (N = 77) (Supplementary Tables 11–14). We found several genes associated to BMI in OVAT (nine out of 24) while only four genes show similar effects in SAT. The strongest associations with BMI in OVAT were observed for SORBS2 and CASQ2 (Supplementary Table 12), although these genes did not show any additional associations to anthropometric traits. Recently, it was reported that DNA methylation at SORBS2 and CASQ2 is related to BMI in SAT [36]. We observed methylation of RUNX1 in OVAT associated significantly with parameters of fat distribution such as CT-ratio (OVAT/SAT area), waist, waist-to-hip ratio (WHR), and visceral fat area (Supplementary Table 12). Interestingly, RUNX1 methylation in SAT was also related to CT-ratio but showed an effect opposite in direction on subcutaneous fat area.
Several of the here reported associations were still significant after correction for multiple testing according to Bonferroni (0.05/16 traits = P < 3.1 × 10−3). Notably, we found SAT derived SSPN methylation levels significantly related to WHR, while methylation levels in OVAT correlated with serum levels of free fatty acids (FFA). Further, methylation at four genes (HAND2, SORBS2, CASQ2, and CCDC125) in OVAT was significantly associated with BMI (Supplementary Tables 11 and 12).
Next, we sought to investigate the association of methylation levels with variables related to glucose and lipid metabolism (Supplementary Tables 13 and 14). We observed the strongest relationship between RGS1 methylation and HbA1c levels in OVAT. Similar associations were found for HOXC6 and HAND2 with HbA1c in OVAT. However, the observed relationships do not withstand correction for multiple testing.
3.11. Differential methylation of GWAS loci, imprinted genes, and epigenetic regulators
We further compared our methylation data with known GWAS genes/loci for BMI [5] and fat distribution (WHR) [6]. In 16 BMI loci, we found differential methylation levels between SAT vs. OVAT and in 24 genes when comparing non-obese vs. obese in either depot (Supplementary Table 15). In 22 genes of the WHR loci, we found differential methylation in each comparison (Supplementary Table 16). Further, we mapped our differentially methylated genes against a catalogue of known imprinted loci (http://www.geneimprint.org/). As demonstrated in Supplementary Table 17, most of the imprinted genes were differentially methylated between non-obese vs. obese subjects (SAT N = 37, OVAT N = 37). To test whether genes encoding enzymes regulating epigenetic mechanisms or adapter proteins may be regulated by DNA methylation themselves, we extracted genes from the literature and searched for potential overlaps with our methylation data (Supplementary Table 18).
4. Discussion
4.1. Adipose tissue depot specific candidate genes
We discovered fat distribution candidate genes and, for 6 of these genes, methylation effects were further supported in additional cohorts; HAND2, HOXC6, PPARG, SORBS2, CD36, and CLDN1. Among them, the best studied candidate is PPARG, which is widely known as a key regulator of adipogenesis and differentiation (reviewed in [37]). Our data demonstrate that the PPARG promoter region is significantly hypermethylated in OVAT along with decreased gene expression in OVAT compared to SAT. This may indicate reduced adipogenesis in OVAT leading to impaired triglyceride uptake and insulin sensitivity as previously reported [38]. However, although non-significant, we observed different expression effects in isolated adipocytes. As we were unable to analyze adipocytes and SVF from the same individuals as in our discovery cohort, this may have contributed to the observed inconsistencies.
We also identified HOXC6 among the top ranked candidate genes with highest methylation levels in OVAT along with decreased mRNA expression, which was further supported in pure adipocytes. Homeobox genes are involved in developmental processes [39]. HOXC6 was also demonstrated by others to be differentially expressed in human SAT before and after bariatric surgery [40], between SAT and gluteofemoral adipose tissue [13] and skeletal muscle [17]. Our findings largely support these data.
Furthermore, HAND2 was higher methylated in SAT from obese subjects along with decreased expression levels in both adipose tissue biopsies and isolated adipocytes. Hand2 is involved in Notch signaling in heart development of mice [41]. Considering that NOTCH signaling also plays a major role in adipogenic differentiation [30], [31], mainly through inhibiting ASC (Adipose tissue-Derived-Stem Cells) differentiation to adipocytes and thereby affecting the adipose tissue expansion capacity [30], this might indicate reduced adipocyte differentiation in OVAT. In line with this, increased Notch signaling in mice blocked the expansion of white adipose tissue, ectopic fat accumulation and insulin resistance [42], which further supports the potential dysfunctional role of OVAT [1], [2].
Among the successfully replicated hits was SORBS2, which was more methylated and less expressed in SAT versus OVAT in both non-obese and obese subjects. We substantiated these results in isolated adipocytes. Linear regression analyses revealed a strong association of methylation with BMI for SORBS2 in OVAT. SORBS2 might be involved in insulin mediated translocation of GLUT4 [43] and t thereby might affect energy storage. Furthermore, we clearly show that in vitro promoter methylation of SORBS2 directly represses the transcriptional activity of the gene-reporter constructs. We observed similar effects for EMX2, a developmental gene which was already shown by others to be upregulated after weight loss in SAT [40]. Our data provide functional evidence that promoter methylation in these genes directly influences gene activity and thereby contributes to the well-known biological distinctions between SAT and OVAT.
The CD36 gene was more methylated and less expressed in OVAT among non-obese subjects and affects metabolism through several mechanisms. First, CD36 increases attraction to fatty foods in rodents while it is also expressed in human taste receptor cells, which, in turn, may also affect human eating behavior [44]. Secondly, it is involved in lipid metabolism by taking up long chain fatty acids and oxidized low-density lipoproteins [32], [33]. Finally, CD36 silencing prevents lipid accumulation and reduces proliferation in vascular smooth muscle cells induced by adipocyte-conditioned medium or oleic acid [45]. These data support our finding of reduced gene expression in OVAT among non-obese individuals, which, however, we could not confirm in isolated adipocytes.
We identified increased methylation of the tight junction protein CLDN1 together with lower expression levels in SAT compared to OVAT from non-obese individuals. CLDN1 is expressed in the intestinal membrane, and the expression can be reduced by fat emulsion, which, in turn, results in increased membrane permeability as demonstrated in rats [46]. Considering the fact that OVAT is located close to the intestine and recent reports about the leaky gut hypothesis [47], the increased expression of CLDN1 in OVAT compared to SAT suggest a functional role of CLDN1 in visceral fat. Similar effects were seen in isolated adipocytes, which adds further evidence to these findings.
4.2. Candidate genes differentially regulated between non-obese and obese subjects
We found several candidate genes and further supported ETV6 in two additional independent cohorts. ETV6 functions as a transcriptional repressor and is involved in acute lymphoblastic leukemia [48]. Moreover, ETV6 was reported in several GWAS influencing human height [49], [50]. In our dataset, ETV6 is hypermethylated in SAT and pure adipocytes from non-obese subjects compared to obese individuals. A potential role for ETV6 in obesity or related phenotypes has not been reported so far but seems plausible in light of its association with other anthropometric measures such as height. Consistently, RUNX1 is also significantly hypermethylated in SAT from non-obese subjects compared to obese individuals. Others have shown that RUNX1 DNA methylation is significantly decreased in response to exercise training in skeletal muscle [17]. Interestingly, in our data, RUNX1 showed a wide range of associations to parameters of fat distribution in SAT and OVAT, further strengthening its potential role in obesity. Although we confirmed several genes in additional cohorts, we consider data from our initial analysis as the most important original results. Therefore, we also consider genes as true signals that have not shown similar effect directions in other cohorts. Among these newly discovered obesity candidate genes is DUSP22, which is hypermethylated in OVAT from obese subjects and was previously shown to be differentially methylated between high and low responders to a weight loss intervention [51]. It was suggested, that reduced methylation along with increased expression of DUSP22 might be indirectly involved in obesity by inhibiting the IL6/LIF/STAT3 pathway [52]. However, we observed no correlation between methylation levels of DUSP22 and IL6-serum levels in our subjects.
RGS1 methylation is higher in OVAT from non-obese subjects compared to obese individuals and negatively associated with HbA1c (Supplementary Table 14). Our results are in line with animal data showing an increased expression of Rgs1 in epididymal white adipose tissue due to high-fat diet (HFD)-induced obesity in mice [53].
4.3. EWAS
A EWAS for BMI in paired samples of SAT and OVAT has never been performed. We identified the two candidate genes SSPN and CCDC125 as significantly associated to BMI in SAT and OVAT, respectively. The SSPN gene has been previously described as one of 13 WHR loci [54]. SSPN encodes for a protein which is part of the dystrophin-glycoprotein complex and is predominantly expressed in muscle, adipose tissue, thyroid, and retina. In muscle dystrophy, reduced muscle mass is compensated for by increased fat tissue and connective tissue. Another sub-complex of the dystrophin-glycoprotein complex is the sarcoglycan-complex which is also present in adipose tissue. β/δ-sarcoglycan null mice show reduced sarcoglycan-complexes along with reduced protein levels of SSPN in white adipocytes [55]. While no established role for CCDC125 in obesity is known so far, it is involved in Isaac's syndrome [56], a movement disorder caused by increased sensitivity of peripheral motor nerves. CCDC125 is mainly expressed in immune associated tissues such as thymus, spleen and bone marrow [56].
No association was found for HIF3A, which was recently reported to be a significant correlate to BMI, most likely due to the fact that we did not measure the same CpG sites but measured mean DNA promoter methylation levels per transcript [12]. Therefore, we cannot rule out that HIF3A may also be related to BMI in our samples.
4.4. Limitations
Albeit greater compared to other studies, our sample size may still limit our ability to identify small effects and may have led to false positive and false negative results. Moreover, we included 77 paired samples of SAT and OVAT in the methylation profiling and 63 in the mRNA profiling. However, overlapping methylation and expression data (either SAT or OVAT) are only available for 42 subjects (overlap with expression from both SAT and OVAT, 31 subjects), which may have prevented us from identifying causal relationships. Further, in our discovery cohort, we used MeDIP to enrich methylated regions followed by array hybridization, which naturally has lower coverage than NGS based methods. Therefore, we may have missed important, physiologically meaningful, differentially methylated regions in our analysis. However, we further substantiated the results from our initial dataset by comparing those with effect directions derived from 450K arrays in several independent cohorts. The here presented results from functional analyses such as luciferase assays originate from MCF7 cells and need to be established in adipocytes in the future. We have only limited knowledge of the individual environmental factors, which may have theoretically influenced our results. Importantly, adipose tissue is a heterogeneous sample per se containing multiple cell types, and we cannot rule out that methylation levels originating from other cell types such as macrophages may have an impact on our results. However, for most of our top genes, we provide similar effect directions when analyzing gene expression in isolated adipocytes. As we were unable to perform these experiments in the same individuals included in our initial analyses, this may be one reason causing the observed differences.
5. Conclusions
Using a combined genome wide epigenetic and transcriptomic analysis, we confirmed obesity and fat distribution candidate genes and identified genes which have been previously unrecognized in the pathophysiology of obesity. Our data suggest that DNA promoter methylation of specific genes is directly associated with BMI and obesity while we clearly demonstrate adipose tissue depot specific differences. Confirming known candidate genes such as PPARG underlines the credibility of the here identified genes.
Author contributions
Y.B. and M.K. conceptualized, designed, and conceived the study. M.B. and Y.B. supervised the study. M.K. performed sample preparation for the arrays and gene ontology- and validation analyses, designed tables and figures, and wrote the first draft of the manuscript. L.H. performed bioinformatics analyses of methylation and expression array data. X.L. performed EWAS analyses, designed Manhattan plots and circle plots, and performed methylation in-silico replications by using publicly available data sets. T.W. performed linear regression analyses. R.C. performed methylation in-silico replications in the Italian cohort. K.R. performed luciferase experiments, edited the manuscript, and contributed to the discussion and data interpretation. M.Klös. assisted in sample preparation for expression analyses. F.E. assisted in validation analyses by using pyrosequencing. A.D., M.R.S., D.G., T.L., and M.D. contributed samples. M.S. and P.K. edited the manuscript. A.M.D. supervised in-silico replication analyses (PI of the Italian cohort). C. L. and K. B. supervised in-silico methylation analyses and luciferase assays, and contributed to data interpretation. M. Kern performed gene expression analyses in isolated adipocytes and SVF (supervised by M.B.) H.B. supervised bioinformatics analyses and contributed to discussion and data interpretation. M.B. edited the manuscript, contributed to the data analysis and discussion, and is PI of the Adipose Tissue Biobank (Leipzig Cohort). Y.B. conceived and designed experiments, edited the manuscript, and contributed to discussion and data interpretation. All authors contributed to the final version of the manuscript, guarantee for the data and analyses, and agreed to publish the data.
Financial disclosure
This work was supported by grants from the German Diabetes Association (to Y.B. and to P.K.) and from the DDS Foundation to Y.B. Further funds were provided by The Swedish Research Council, The Regional Research Council (ALF), Påhlsson Foundation and The Swedish Diabetes Foundation to C.L. Y.B was further supported by research grants from the IFB Adiposity Diseases, supported by German BMBF K6e-96 and K6e-97 and by a research fellowship from the EFSD (European Foundation for the Study of Diabetes). K.R. was supported by K6e-96. IFB Adiposity Diseases is supported by the Federal Ministry of Education and Research (BMBF), Germany, FKZ 01EO1501. Further funding for this study came from the Italian Ministry of Health to the Istituto Auxologico Italiano (to R.C and A.D.). This work was also supported by the Kompetenznetz Adipositas (Competence network for Obesity) funded by the Federal Ministry of Education and Research (German Obesity Biomaterial Bank; FKZ 01GI1128), and a grant from German Research Foundation, the SFB 1052/1: “Obesity mechanisms” (projects A01 to M.S, B01 to M.B. and B03 to P.K.). Y.B and M.B. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgements
We thank all those who participated in the studies. We thank PD. D Knut Kohn who is heading the Core Unit DNA technologies; IZKF, University of Leipzig, for excellent technical support. We further thank Karl Bacos (Department of Clinical Sciences, Lund University Diabetes Centre) for his excellent advice in luciferase experiments.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.11.003.
Contributor Information
Matthias Blüher, Email: matthias.blueher@medizin.uni-leipzig.de.
Yvonne Böttcher, Email: yvonne.boettcher@medizin.uni-leipzig.de, yvonne.bottcher@medisin.uio.no.
Conflict of interest
The authors have no conflict of interests.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.St-Pierre J., Lemieux I., Perron P., Brisson D., Santuré M., Vohl M.-C. Relation of the “Hypertriglyceridemic Waist” phenotype to earlier manifestations of coronary artery disease in patients with glucose intolerance and type 2 diabetes mellitus. The American Journal of Cardiology. 2007;99(3):369–373. doi: 10.1016/j.amjcard.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 2.Vega G.L., Adams-Huet B., Peshock R., Willett D., Shah B., Grundy S.M. Influence of body fat content and distribution on variation in metabolic risk. The Journal of Clinical Endocrinology & Metabolism. 2006;91(11):4459–4466. doi: 10.1210/jc.2006-0814. [DOI] [PubMed] [Google Scholar]
- 3.Gerhard G.S., Styer A.M., Strodel W.E., Roesch S.L., Yavorek A., Carey D.J. Gene expression profiling in subcutaneous, visceral and epigastric adipose tissues of patients with extreme obesity. International Journal of Obesity (Lond) 2014 Mar;38(3):371–378. doi: 10.1038/ijo.2013.152. [Epub 2013 Aug 16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trujillo M.E., Scherer P.E. Adipose tissue-derived factors: impact on health and disease. Endocrine Reviews. 2006;27(7):762–778. doi: 10.1210/er.2006-0033. [DOI] [PubMed] [Google Scholar]
- 5.Locke A.E., Kahali B., Berndt S.I., Justice A.E., Pers T.H., Day F.R. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schleinitz D., Böttcher Y., Blüher M., Kovacs P. The genetics of fat distribution. Diabetologia. 2014;57(7):1276–1286. doi: 10.1007/s00125-014-3214-z. [DOI] [PubMed] [Google Scholar]
- 8.Dahlman I., Sinha I., Gao H., Brodin D., Thorell A., Rydén M. The fat cell epigenetic signature in post-obese women is characterized by global hypomethylation and differential DNA methylation of adipogenesis genes. International Journal of Obesity (Lond) 2015 Jun;39(6):910–919. doi: 10.1038/ijo.2015.31. [Epub 2015 Mar 18] [DOI] [PubMed] [Google Scholar]
- 9.Nilsson E., Jansson P.A., Perfilyev A., Volkov P., Pedersen M., Svensson M.K. Altered DNA methylation and differential expression of genes influencing metabolism and inflammation in adipose tissue from subjects with type 2 diabetes. Diabetes. 2014;63(9):2962–2976. doi: 10.2337/db13-1459. [DOI] [PubMed] [Google Scholar]
- 10.Keller M., Kralisch S., Rohde K., Schleinitz D., Dietrich A., Schön M.R. Global DNA methylation levels in human adipose tissue are related to fat distribution and glucose homeostasis. Diabetologia. 2014;57(11):2374–2383. doi: 10.1007/s00125-014-3356-z. PubMed Central PMCID: PMC25145546. [DOI] [PubMed] [Google Scholar]
- 11.Rohde K., Keller M., Klös M., Schleinitz D., Dietrich A., Schön M.R. Adipose tissue depot specific promoter methylation of TMEM18. Journal of Molecular Medicine. 2014;92(8):881–888. doi: 10.1007/s00109-014-1154-1. [DOI] [PubMed] [Google Scholar]
- 12.Dick K.J., Nelson C.P., Tsaprouni L., Sandling J.K., Aïssi D., Wahl S. DNA methylation and body-mass index: a genome-wide analysis. The Lancet. 2014;383(9933):1990–1998. doi: 10.1016/S0140-6736(13)62674-4. [DOI] [PubMed] [Google Scholar]
- 13.Gehrke S., Brueckner B., Schepky A., Klein J., Iwen A., Bosch T.C.G. Epigenetic regulation of depot-specific gene expression in adipose tissue. PLoS One. 2013;8(12):e82516. doi: 10.1371/journal.pone.0082516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benton M.C., Johnstone A., Eccles D., Harmon B., Hayes M.T., Lea R.A. An analysis of DNA methylation in human adipose tissue reveals differential modification of obesity genes before and after gastric bypass and weight loss. Genome Biology. 2015;16:8. doi: 10.1186/s13059-014-0569-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barres R., Kirchner H., Rasmussen M., Yan J., Kantor F.R., Krook A. Weight loss after gastric bypass surgery in human obesity remodels promoter methylation. Cell Reports. 2013;3(4):1020–1027. doi: 10.1016/j.celrep.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 16.Barrès R., Yan J., Egan B., Treebak J.T., Rasmussen M., Fritz T. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metabolism. 2012;15(3):405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Nitert M.D., Dayeh T., Volkov P., Elgzyri T., Hall E., Nilsson E. Impact of an exercise intervention on DNA methylation in skeletal muscle from first-degree relatives of patients with type 2 diabetes. Diabetes. 2012;61(12):3322–3332. doi: 10.2337/db11-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rönn T., Volkov P., Davegårdh C., Dayeh T., Hall E., Olsson A.H. A six months exercise intervention influences the genome-wide DNA methylation pattern in human adipose tissue. PLoS Genetics. 2013;9(6):e1003572. doi: 10.1371/journal.pgen.1003572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donkin I., Versteyhe S., Ingerslev L.R., Qian K., Mechta M., Nordkap L. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metabolism. 2015;23(2):369–378. doi: 10.1016/j.cmet.2015.11.004. PubMed Central PMCID: PMC26669700. [DOI] [PubMed] [Google Scholar]
- 20.Huypens P., Sass S., Wu M., Dyckhoff D., Tschop M., Theis F. Epigenetic germline inheritance of diet-induced obesity and insulin resistance. Nature Genetics. 2016;48(5):497–499. doi: 10.1038/ng.3527. Epub 2016/03/15. PubMed PMID: 26974008. [DOI] [PubMed] [Google Scholar]
- 21.Kloting N., Fasshauer M., Dietrich A., Kovacs P., Schon M.R., Kern M. Insulin-sensitive obesity. AJP: Endocrinology and Metabolism. 2010;299(3):E506–E515. doi: 10.1152/ajpendo.00586.2009. [DOI] [PubMed] [Google Scholar]
- 22.O'Geen H., Nicolet C., Blahnik K., Green R., Farnham P. Comparison of sample preparation methods for ChIP-chip assays. Biotechniques. 2006;41(5):577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Molecular Biology. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klug M., Rehli M. Functional analysis of promoter CPG-methylation using a CpG-free luciferase reporter vector. Epigenetics. 2014;1(3):127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 25.Johnson W.E., Li W., Meyer C.A., Gottardo R., Carroll J.S., Brown M. Model-based analysis of tiling-arrays for ChIP-chip. Proceedings of National Academy of Sciences United States of America. 2006;103(33):12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strimmer K. A unified approach to false discovery rate estimation. BMC Bioinformatics. 2008;9:303. doi: 10.1186/1471-2105-9-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Löffler-Wirth H., Kalcher M., Binder H. oposSOM: R-package for high-dimensional portraying of genome-wide expression landscapes on bioconductor. Bioinformatics (Oxford, England) 2015;31(19):3225–3227. doi: 10.1093/bioinformatics/btv342. [DOI] [PubMed] [Google Scholar]
- 28.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D. Circos: an information aesthetic for comparative genomics. Genome Research. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elsen M., Raschke S., Tennagels N., Schwahn U., Jelenik T., Roden M. BMP4 and BMP7 induce the white-to-brown transition of primary human adipose stem cells. AJP: Cell Physiology. 2014;306(5):C431–C440. doi: 10.1152/ajpcell.00290.2013. [DOI] [PubMed] [Google Scholar]
- 30.Ferrer-Lorente R., Bejar M.T., Badimon L. Notch signaling pathway activation in normal and hyperglycemic rats differs in the stem cells of visceral and subcutaneous adipose tissue. Stem Cells and Development. 2014;23(24):3034–3048. doi: 10.1089/scd.2014.0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song B.-Q., Chi Y., Li X., Du W.-J., Han Z.-B., Tian J.-J. Inhibition of notch signaling promotes the adipogenic differentiation of mesenchymal stem cells through autophagy activation and PTEN-PI3K/AKT/mTOR pathway. Cellular Physiology and Biochemistry: International Journal of Experimental Cellular Physiology, Biochemistry, and Pharmacology. 2015;36(5):1991–2002. doi: 10.1159/000430167. [DOI] [PubMed] [Google Scholar]
- 32.Kennedy D.J., Kuchibhotla S., Westfall K.M., Silverstein R.L., Morton R.E., Febbraio M. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovascular Research. 2011;89(3):604–613. doi: 10.1093/cvr/cvq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love-Gregory L., Abumrad N.A. CD36 genetics and the metabolic complications of obesity. Current Opinion in Clinical Nutrition and Metabolic Care. 2011;14(6):527–534. doi: 10.1097/MCO.0b013e32834bbac9. PubMed Central PMCID: PMC21912245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slieker R.C., Bos S.D., Goeman J.J., Bovée J.V., Talens R.P., van der Breggen R. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics & Chromatin. 2013;6(1):26. doi: 10.1186/1756-8935-6-26. PubMed Central PMCID: PMC23919675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowe R., Rakyan V.K. Marmal-aid–a database for Infinium HumanMethylation450. BMC Bioinformatics. 2013;14:359. doi: 10.1186/1471-2105-14-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronn T., Volkov P., Gillberg L., Kokosar M., Perfilyev A., Jacobsen A.L. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Human Molecular Genetics. 2015;24(13):3792–3813. doi: 10.1093/hmg/ddv124. PubMed Central PMCID: PMC25861810. [DOI] [PubMed] [Google Scholar]
- 37.Kawai M., Rosen C.J. PPARγ: a circadian transcription factor in adipogenesis and osteogenesis. Nature Reviews Endocrinology. 2010;6(11):629–636. doi: 10.1038/nrendo.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosen E.D., Sarraf P., Troy A.E., Bradwin G., Moore K., Milstone D.S. PPARγ is required for the differentiation of adipose tissue in vivo and in vitro. Molecular Cell. 1999;4(4):611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto Y., Gesta S., Lee K.Y., Tran T.T., Saadatirad P., Ronald Kahn C. Adipose depots possess unique developmental gene signatures. Obesity. 2010;18(5):872–878. doi: 10.1038/oby.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dankel S.N., Fadnes D.J., Stavrum A.-K., Stansberg C., Holdhus R., Hoang T. Switch from stress response to homeobox transcription factors in adipose tissue after profound fat loss. PLoS One. 2010;5(6):e11033. doi: 10.1371/journal.pone.0011033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.VanDusen N.J., Casanovas J., Vincentz J.W., Firulli B.A., Osterwalder M., Lopez-Rios J. Hand2 is an essential regulator for two notch-dependent functions within the embryonic endocardium. Cell Reports. 2014;9(6):2071–2083. doi: 10.1016/j.celrep.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chartoumpekis D.V., Palliyaguru D.L., Wakabayashi N., Khoo N.K.H., Schoiswohl G., O'Doherty R.M. Notch intracellular domain overexpression in adipocytes confers lipodystrophy in mice. Molecular Metabolism. 2015;4(7):543–550. doi: 10.1016/j.molmet.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimura A., Baumann C.A., Chiang S.H., Saltiel A.R. The sorbin homology domain: a motif for the targeting of proteins to lipid rafts. Proceedings of National Academy of Sciences United States of America. 2001;98(16):9098–9103. doi: 10.1073/pnas.151252898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Degrace-Passilly P., Besnard P. CD36 and taste of fat. Current Opinion in Clinical Nutrition and Metabolic Care. 2012;15(2):107–111. doi: 10.1097/MCO.0b013e32834ff19c. [DOI] [PubMed] [Google Scholar]
- 45.Schlich R., Lamers D., Eckel J., Sell H. Adipokines enhance oleic acid-induced proliferation of vascular smooth muscle cells by inducing CD36 expression. Archives of Physiology and Biochemistry. 2015;121(3):81–87. doi: 10.3109/13813455.2015.1045520. [DOI] [PubMed] [Google Scholar]
- 46.Suzuki T., Hara H. Dietary fat and bile juice, but not obesity, are responsible for the increase in small intestinal permeability induced through the suppression of tight junction protein expression in LETO and OLETF rats. Nutrition & Metabolism. 2010;7:19. doi: 10.1186/1743-7075-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geurts L., Neyrinck A.M., Delzenne N.M., Knauf C., Cani P.D. Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: novel insights into molecular targets and interventions using prebiotics. Beneficial Microbes. 2014;5(1):3–17. doi: 10.3920/BM2012.0065. [DOI] [PubMed] [Google Scholar]
- 48.Ellinghaus E., Stanulla M., Richter G., Ellinghaus D., te Kronnie G., Cario G. Identification of germline susceptibility loci in ETV6-RUNX1-rearranged childhood acute lymphoblastic leukemia. Leukemia. 2012;26(5):902–909. doi: 10.1038/leu.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berndt S.I., Gustafsson S., Mägi R., Ganna A., Wheeler E., Feitosa M.F. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nature Genetics. 2013;45(5):501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467(7317):832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moleres A., Campión J., Milagro F.I., Marcos A., Campoy C., Garagorri J.M. Differential DNA methylation patterns between high and low responders to a weight loss intervention in overweight or obese adolescents: the EVASYON study. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2013;27(6):2504–2512. doi: 10.1096/fj.12-215566. [DOI] [PubMed] [Google Scholar]
- 52.Sekine Y., Tsuji S., Ikeda O., Sato N., Aoki N., Aoyama K. Regulation of STAT3-mediated signaling by LMW-DSP2. Oncogene. 2006;25(42):5801–5806. doi: 10.1038/sj.onc.1209578. PubMed Central PMCID: PMC16636663. [DOI] [PubMed] [Google Scholar]
- 53.Choi M.-S., Kim Y.-J., Kwon E.-Y., Ryoo J.Y., Kim S.R., Jung U.J. High-fat diet decreases energy expenditure and expression of genes controlling lipid metabolism, mitochondrial function and skeletal system development in the adipose tissue, along with increased expression of extracellular matrix remodelling- and inflammation-related genes. The British journal of nutrition. 2015;113(6):867–877. doi: 10.1017/S0007114515000100. PubMed Central PMCID: PMC25744306. [DOI] [PubMed] [Google Scholar]
- 54.Heid I.M., Jackson A.U., Randall J.C., Winkler T.W., Qi L., Steinthorsdottir V. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nature Genetics. 2010;42(11):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Groh S., Zong H., Goddeeris M.M., Lebakken C.S., Venzke D., Pessin J.E. Sarcoglycan complex: implications for metabolic defects in muscular dystrophies. The Journal of Biological Chemistry. 2009;284(29):19178–19182. doi: 10.1074/jbc.C109.010728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Araya N., Arimura H., Kawahara K.-I., Yagishita N., Ishida J., Fujii R. Role of Kenae/CCDC125 in cell motility through the deregulation of RhoGTPase. International Journal of Molecular Medicine. 2009;24(05):605–611. doi: 10.3892/ijmm_00000271. [DOI] [PubMed] [Google Scholar]
- 57.Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A. 2015. gplots: Various R programming tools for plotting data. R package version 2.17.0 ed: CRAN.R-project.org. [Google Scholar]
- 58.Barfield R.T., Kilaru V., Smith A.K., Conneely K.N. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics (Oxford, England) 2012;28(9):1280–1281. doi: 10.1093/bioinformatics/bts124. PubMed Central PMCID: PMC22451269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.