Abstract
The hypothalamus and neocortex are subdivisions of the mammalian forebrain, and yet, they have vastly different evolutionary histories, cytoarchitecture, and biological functions. In an attempt to define these attributes in terms of their genetic activity, we have compared their genetic repertoires by using the Serial Analysis of Gene Expression database. From a comparison of 78,784 hypothalamus tags with 125,296 neocortical tags, we demonstrate that each structure possesses a different transcriptional profile in terms of gene ontological characteristics and expression levels. Despite its more recent evolutionary history, the neocortex has a more complex pattern of gene activity. Gene identities and levels of gene expression were mapped to their chromosomal positions by using in silico definition of GC-rich and GC-poor genome bands. This analysis shows contrasting views of gene activity on a genome scale that is unique to each brain substructure. We show that genes that are more highly expressed in one tissue tend to be clustered together on a chromosomal scale, further defining the genetic identity of either the hypothalamus or neocortex. We propose that physical proximity of coregulated genes may facilitate transcriptional access to the genetic substrates of evolutionary selection that ultimately shape the functional subdivisions of the mammalian brain.
Of the various subdivisions of the mammalian forebrain, the neocortex and hypothalamus sit on opposite ends of the evolutionary scale. The six-layered neocortex is unique to mammals and not present in fish, amphibians, birds or reptiles (1). Although functionally divided into multiple areas, the neocortex is cytologically homogeneous across its entire breadth with characteristic distribution of glutamatergic and γ-aminobutyric acid neurons throughout its six layers (2-4). Whereas it is a relative newcomer in evolution, the neocortex has expanded dramatically in size throughout class Mammalia, and in primates is responsible for higher cognitive functions and traits associated with complex behavior (5). In contrast, the hypothalamus is evolutionarily ancient and complex in structure and function (6). Anatomically, it is a cluster of disparate areas comprising a dozen well defined nuclei and other less defined regions containing neurons, which release neuropeptides and monoamines (7-9). Functionally, the hypothalamus is an important integrator of homeostatic mechanisms, which regulate basic needs such as food and water intake, maintenance of body temperature and blood pressure, reproduction and parental care, and control of the autonomic nervous system (10-12).
In a nutshell, neurons in these two brain divisions appear to subserve either executive (in neocortex) or vegetative (in hypothalamus) functions, and although their structural and physiological differences are beginning to be understood, their genetic variances are largely unknown. Of particular interest is whether the hypothalamus has a molecular anatomy that is instantly recognizable as being distinctive and readily distinguishable from other brain regions such as the neocortex. Such a molecular portrait would correspond to its operating “transcriptome,” representing all of the genes that are expressed (qualitative and quantitative) in the hypothalamus and neocortex under standard conditions.
In the current study, we describe the use of the Serial Analysis of Gene Expression (SAGE) database, which allows simultaneous detection of the expression levels of the entire genome without a priori knowledge of gene sequences (13). SAGE takes advantage of the fact that a small sequence tag taken from a defined position within the transcript is sufficient to identify the gene (from known cDNA or EST sequences), and up to 40 tags can be concatenated and sampled in a single sequencing reaction. SAGE tag frequency is directly proportional to the originating mRNA copy number and is therefore a reliable measure of transcript abundance (13). We present data after comparative analyses of 78,784 tags from the hypothalamus with 125,296 tags from the neocortex. We show that each structure has a unique repertoire of expressed genes and their genetic correlates of cellular function are reflective of their separate neural identities. After mapping of gene identities and expression levels to their chromosomal positions, each structure can be further defined by a virtual map of expressed genes on all chromosomes except for the Y chromosome. Interestingly, genes that are differentially expressed for each structure are clustered together in small groups along the chromosomes, suggesting coregulation of genes in close proximity and such clusters may facilitate evolutionary selection of coexpressed genes.
Materials and Methods
RNA Isolation, SAGE Libraries, and SAGE Tag Extraction/Analysis. Total RNA was isolated from pooled adult hypothalamus tissue (8-week-old C57BL/6 male mice, n = 20) by using TRIzol reagent (Invitrogen). SAGE libraries were made according to SAGE protocol, Version 1.0d (14) with modifications (refs. 14 and 15, and see Supporting Text and Supporting Appendix, which are published as supporting information on the PNAS web site, for details of the methods that were used). For the neocortex, mRNA from female mice (5-6 mo, C57BL/6, n = 34) was obtained as above and SAGE libraries were made according to the LongSAGE protocol (16). Fisher's exact test was used to identify differentially expressed genes between the hypothalamic and neocortical libraries (see Supporting Text for more details).
Quantitative Real-Time PCR (qRT-PCR) Validation. qRT-PCR using Sybr green chemistry (Applied Biosystems) was performed on five biological replicates on a subset (21 of 82) of National Center for Biotechnology Information Reference Sequence (RefSeq) database-matched differentially expressed genes (17). The 18s gene was used as a reference. Control reactions with no reverse transcriptase were performed to detect genomic contamination and comparative cycle threshold calculations were performed as described (18).
Gene Ontology (GO) Database Annotations. To link tag identity with putative gene function, all identified genes were annotated by using the Mouse Genome Informatics (which can be accessed at www.informatics.jax.org) and GO (which can be accessed at www.geneontology.org) databases. See Supporting Text for detailed descriptions.
SAGE Tag Mapping to Genes and Chromosomal Positions. A comparative chromosomal view of expression levels in hypothalamus and neocortex was generated by assigning chromosomal positions for tags corresponding to known genes after matching to the National Center for Biotechnology Information Refseq database (February 2003 release; Mm3, which can be accessed at http://genome.ucsc.edu). See Supporting Text for detailed descriptions.
Results
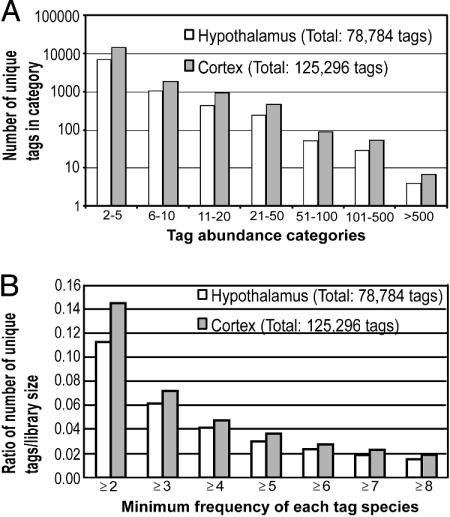
Gene Expression and Distribution. A total of 78,784 hypothalamus and 125,296 neocortex SAGE tags (14 nt) were analyzed. A large proportion (65%) of expressed tags occurred at lower frequencies (f ≤10), whereas tags of intermediate abundance levels (11 ≥ f ≥100) contributed to 22% of the total tag count (Fig. 1A). Tags with the highest frequency (f ≥101) contributed to 13% of the transcript pool. In total, the entire pool represents 33,149 unique tags (including tags observed once), but there are 8,850 unique tags after single tags are removed. The corresponding percentages for the neocortex dataset are 60%, 27%, and 13%, respectively. There are 18,149 unique tags (f ≥2) in the pooled neocortical library of 125,296 tags. This distribution of tag abundance categories in hypothalamus and neocortex closely parallels the distribution patterns reported for other transcriptomes (e.g., T cells, retina, and whole brain) (19-21).
Fig. 1.
The neocortex is genetically more complex compared with the hypothalamus. (A) Distribution of SAGE tags in abundance categories. Shown is the number of unique transcripts (tags) for each abundance category. (B) Genetic complexity of the mouse hypothalamus in comparison with the mouse neocortex.
The genetic complexity of the hypothalamus and neocortex can be viewed as a range of expressed tag frequencies, ranging from two or more tags. This analysis reveals that the neocortex is more complex than the hypothalamus for these higher tag abundance categories. The chart is presented as a ratio to account for the size difference between the two libraries (Fig. 1B). This analysis reveals that the neocortex is more complex at all tag cutoff frequencies. To correlate this data with the published database, we queried 8,850 unique hypothalamic tags (f >1) against the RefSeq database. This procedure revealed that only a minor proportion of the hypothalamus transcriptome has a match to peer-reviewed, provisional, or predicted genes in the database (see Fig. 5, which is published as supporting information on the PNAS web site). Overall, 39% of unique hypothalamic tags produced a match. A similar analysis by using the neocortex dataset (18,149 unique tags) provided a similar picture, albeit at a lower tag-to-gene match at 25%. This analysis would suggest that the vast majority of expressed genes uncovered by SAGE in the hypothalamus (and neocortex) remain uncharacterized.
Identification of Specific and Enriched Genes. Table 1 lists 21 genes that were extracted from the RefSeq database and were independently verified for differential expression by using qRT-PCR. (P < 0.10). A full list of 82 RefSeq-matched tags are presented in Table 2, which is published as supporting information on the PNAS web site. The level of enrichment varies between 2- to 24-fold (in neocortex) and 2- to 20-fold (in hypothalamus). Beyond this finding, another 117 unique tag identities were found differentially expressed but because these tags did not have a RefSeq record, they were excluded from further study. Hypothalamus-enriched genes include those with nonspecific functions such as chaperones, structural genes, enzyme regulators, growth suppressors, and transport molecules. The genes enriched in the neocortex are associated with cell growth, energy generation, vesicle-associated neurotransmission, cytoskeletal structure, and signal transduction.
Table 1. Genes differentially expressed between adult mouse hypothalamus and neocortex and validated by using qRT-PCR.
| RefSeq ID code | Tag-to-gene match (Mus musculus) | SAGE tag | Hypothalamus/100,000 tags (SAGE) | Neocortex/100,000 tags (SAGE) | Neocortex fold-enrichment (SAGE) | P value (SAGE) | Neocortex fold-enrichment (qRT-PCR), corrected | P value (qRT-PCR) |
|---|---|---|---|---|---|---|---|---|
| NM_009366 | Transforming growth factor β-1-induced transcript 4 (TGF-βi4) | TCCCCCACAC | 0 | 34 | Neocortex-specific | 0.00 | 1.41 | <0.001 |
| NM_144828 | Protein phosphatase 1, regulatory (inhibitor) subunit 1B (Ppplrlb) | TCCCTCCCTT | 0 | 24 | Neocortex-specific | 0.00 | −1.41 | 0.300 NS |
| NM_008451 | Kinesin light-chain 2 (Klc2) | GCCTGACCCC | 0 | 16 | Neocortex-specific | 0.04 | 1.31 | 0.013 |
| NM_016801 | Syntaxin 1A (brain) (Stx1a) | CAGCGGGAGC | 1 | 30 | 30.00 | 0.00 | 5.01 | <0.001 |
| NM_144900 | ATPase, Na+/K+ transporting, α-1 polypeptide (Atp1α1) | TAGCTGTAAC | 10 | 68 | 6.80 | 0.00 | 2.99 | <0.001 |
| NM_174998 | Hippocalcin-like 4 (Hpcal4) | CTGCTTCTAA | 10 | 42 | 4.20 | 0.01 | 1.88 | <0.001 |
| NM_177407 | Calcium/calmodulin-dependent protein kinase II α (Camk2α) | GCTTCCCCAC | 15 | 49 | 3.27 | 0.03 | 2.32 | <0.001 |
| NM_031161 | Cholecystokinin (Cck) | GGCTGGATGG | 17 | 49 | 2.88 | 0.05 | 2.88 | <0.001 |
| NM_031158 | Ankyrin 1, erythroid (Ank1) | GGCTGGATGG | 17 | 49 | 2.88 | 0.05 | 2.88 | <0.001 |
| NM_011428 | Synaptosomal-associated protein 25 (Snap25) | TATATTAAAT | 79 | 226 | 2.86 | 0.00 | 2.15 | <0.001 |
| NM_022029 | Neurogranin (Nrgn) | TTACCATACT | 62 | 26 | −2.38 opposite | 0.03 | 3.78 opposite | <0.001 |
| NM_011986 | Neurochondrin (Ncdn-pending) | TGGACACTCA | 121 | 37 | −3.27 | 0.00 | −1.64 | 0.004 |
| NM_009976 | Cystatin C (Cst3) | CCTTGCTCAA | 352 | 106 | −3.32 | 0.00 | −1.12 | 0.129 NS |
| NM_016755 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F (Atp5j) | AATTAGTTGT | 46 | 14 | −3.29 | 0.01 | −1.15 | 0.012 |
| NM_008791 | Purkinje cell protein 4 (Pcp4) | AAGAGAAACC | 47 | 14 | −3.36 | 0.01 | −2.11 | <0.001 |
| NM_008722 | Nucleophosmin 1 (Npm1) | TGAAATAAAC | 29 | 7 | −4.14 | 0.07 | −2.14 | <0.001 |
| NM_015744 | Ectonucleotide pyrophosphatase/phosphodiesterase 2 (Enpp2) | GTGCTGCCAG | 39 | 9 | −4.33 | 0.00 | −3.07 | <0.001 |
| NM_010758 | Myelin-associated glycoprotein (Mag) | AAATAAATGT | 29 | 6 | −4.83 | 0.03 | −2.30 | <0.001 |
| NM_010882 | Necdin (Ndn) | TATGCAACCC | 53 | 10 | −5.30 | 0.00 | −2.38 | <0.001 |
| NM_013746 | Pleckstrin homology domain containing, family B (evectins) member 1 (Plekhb1) | ATTGGCCCCA | 62 | 10 | −6.20 | 0.00 | −1.77 | <0.001 |
| NM_007705 | Cold-inducible RNA-binding protein (Cirbp) | CATACTCCAT | 11 | 0 | Hypothalamus-specific | 0.07 | −1.29 | 0.009 |
Note that multiple SAGE tags can be mapped to the same gene identity and vice versa. A negative sign preceding fold-enrichment value indicates enrichment in the hypothalamus. NS, not significant.
To obtain independent verification of differential gene expression, we interrogated a sample subset (21 genes in Table 1) of differentially expressed genes by using qRT-PCR. To ensure robustness of the data, each gene was queried by using five biological replicates of unpooled hypothalamus or neocortex RNA. The results show that in the vast majority (18 of 21), SAGE tag enrichment in a particular tissue is confirmed by qRT-PCR, although the fold-enrichment detected by qRT-PCR is generally lower. These results indicate that at mid to high levels of penetration, SAGE libraries provide reliable estimates of differential gene expression.
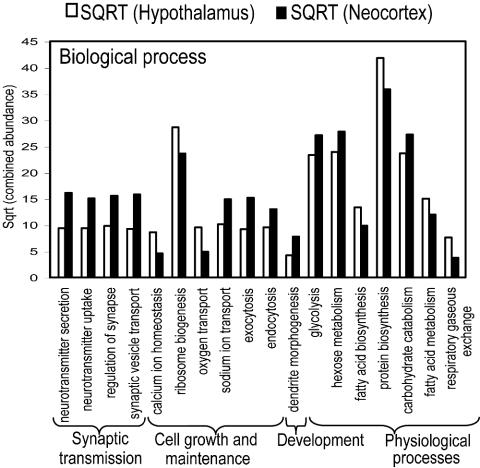
Gene Ontology. By using functional descriptors provided by the GO Consortium, GO classifications for the hypothalamus and neocortex were analyzed in three different groups: biological process, cellular classification, and molecular function (Fig. 2, and see Fig. 6, which is published as supporting information on the PNAS web site). The GO analysis displays differential enrichment of genes within particular functional groups (GO terms), where pairwise comparisons at the single-gene level may not illustrate the broader trend. The analysis shows that gene abundance levels of all ontology classes are consistently higher in the hypothalamus, with the exception of the sodium/potassium-exchanging ATPase complex, suggesting the neocortex to be more highly involved in the directed cellular movement of potassium and sodium ions. The levels of genes annotated as ribosomal and Golgi-related genes are higher in the hypothalamus, suggesting that although the neocortex is a more complex organ, its functionality in protein biosynthesis and processing is somewhat lower.
Fig. 2.
GO database classifications for biological processes. These categories include only the highly significantly differentially expressed gene ontologies (P < 0.05). The combined abundance is calculated as the sum of the normalized abundances of all tags associated with its GO database term. Gene ontologies are listed on the horizontal axis. SQRT, square root.
All genes classified under the “biological processes” framework of the GO database (Fig. 2) show domination of synaptic transmission and cell signaling-related (e.g., neurotransmitter secretion and exocytosis) genes by the cortex whereas homeostasis-, ribosome-, and gas-transport-associated genes are highly expressed in the hypothalamus. Many of the secretory and ion transport genes are also highly expressed in the cortex. Genes associated with energy generation (e.g., carbohydrate catabolism/metabolism) and neural developmental controls are expressed at higher levels in the cortex. Conversely, fat, protein, and organic acid biosynthesis and metabolism are expressed at higher levels in the hypothalamus. These differences mirror the contrasting functional characteristics of neocortex and hypothalamus, by relegating cell communication and energy use genes to the neocortex and vegetative level genes to the hypothalamus. The functions dominated by the neocortex are wide ranging, covering cation transport/exchange, DNA binding, receptor binding, and various enzymatic activities. Exceptions include RNA binding, oxygen transport, pyrophosphatase, apoptotic, ribosomal, and protease activities, with the latter being significantly higher in the hypothalamus.
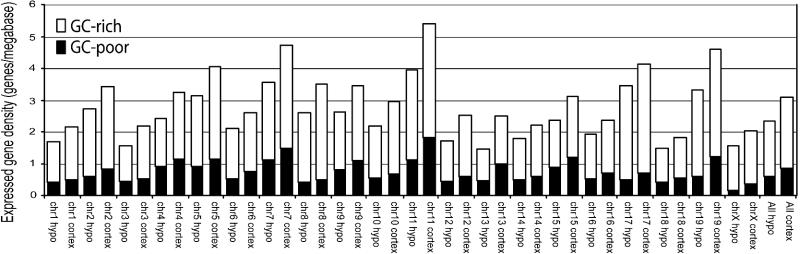
Mapping of Gene Identities and Expression Levels to Chromosomal Positions. Giemsa bands normally observed on metaphase chromosomes can be predicted in silico from the published genome sequence (22). Human studies indicate that Giemsa-dark (G) bands are relatively gene-poor, whereas Giemsa-light (R) bands are gene-rich and have higher GC content (22, 23). To map expression levels of hypothalamus relative to neocortex across the entire genome, and to detect correlations with G and R bands, gene positions and gene densities were computed for all tags after in silico generation of a mouse map of GC-rich and GC-poor bands according to the method of Niimura and Gojobori (22). Bands were assigned as GC-rich or GC-poor, based on the difference in GC content between a local window of 2.5 Mb and a regional window of 9.3 Mb (22). Several interesting observations emerged from this analysis. First, the gene density (expressed as genes per megabase) was consistently higher in GC-rich bands (0.97-3.50; compared with GC-poor bands, 0.15-1.80) for both hypothalamus and neocortex (Fig. 3). As expected, 82% of differentially expressed genes between the two brain structures are situated in the GC-rich bands, where most of the genes reside. Second, GC-poor or in silico G bands have been presumed to contain mainly tissue-specific genes (as opposed to housekeeping) (22, 24). We observed that our reconstructed GC-poor bands have greater gene densities for the neocortex compared with the hypothalamus (Student's t test in Fig. 3). This trend was also detected when comparing GC-rich bands between the neocortex and hypothalamus. Thus, the neocortex consistently displays greater gene densities across the chromosomes.
Fig. 3.
Gene densities of hypothalamus and neocortex genes localized in in silico-approximated GC-rich and GC-poor bands. Only genes with an expressed tag frequency of >1 are examined.
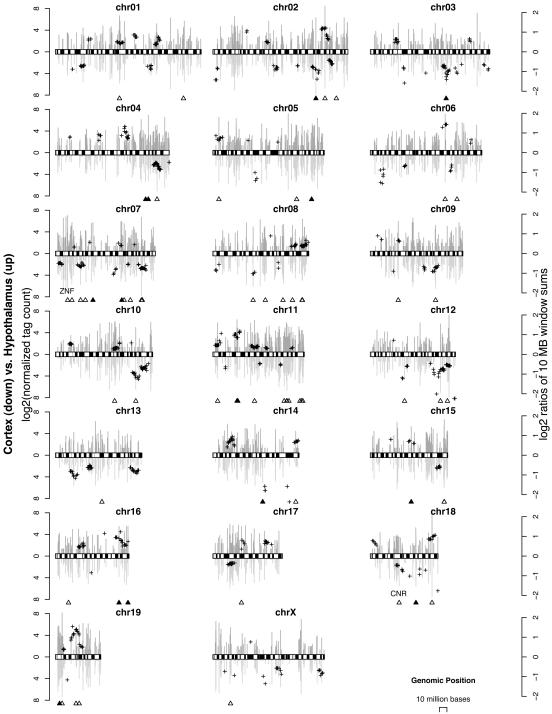
On a chromosomal scale, both tissues have roughly similar but not identical levels of gene activity across the 19 autosomes and the X chromosome (the Y chromosome was not evaluated because the cortex was obtained from female mice). Fig. 4 depicts the tag counts (vertical bars, log2 scale, left axis) for all known genes expressed in the hypothalamus (above) and neocortex (below), superimposed on top of GC-poor and GC-rich bands. In any given chromosome, gene expression levels are not uniform and many clusters signifying increased gene activity can be detected in either the neocortex or hypothalamus in every chromosome studied (Fig. 4). These clusters are indicated by a group of plus (+) symbols; each plus symbol records the ratio of gene expression levels (log2 scale, right axis) between two tissues in a given 10-Mb genome window. To compute the significance of these ratios, an identical assessment by using random gene order was carried out to detect random clustering. Gene order was randomized for the entire genome by random permutations (10,000 times) and the ratio of gene expression levels (hypothalamus versus necortex) was measured by using the same 10-Mb window. This permutation analysis indicates that the observed ratios are unlikely to reflect random differences in gene position (P < 0.1). The positions of individual genes that are significantly different in expression levels (P < 0.10, Fisher's exact test) are indicated by an open triangle, and positions of genes that have been independently confirmed by qRT-PCR are marked by a filled triangle. Taken together, these results indicate divergent gene expression signatures between the hypothalamus and neocortex. In addition, we detected clusters of genes that are differentially coexpressed; these genes are specific to either the hypothalamus or neocortex, suggesting coordinated regulation of transcriptional activity among genes in physical proximity.
Fig. 4.
Chromosomal localization of expressed genes in hypothalamus and neocortex. The chromosome display shows in silico chromosome banding (GC-rich, white; GC-poor, black). The bar chart refers to the left axis (dark gray bars above the chromosome represent the hypothalamus, and the light gray bars represent the neocortex), representing the gene expression levels of the two tissue types. The plus (+) symbols refer to the right axis, with each symbol representing the ratio of expressed gene levels contained within a 10-Mb window between the two tissues. The positions of qRT-PCR validated tags/genes (▴) and tags that are differentially expressed between the hypothalamus and neocortex (▵)(P < 0.10, corrected Fisher's exact test) are indicated. Note that the physical positions of cadherin-related neuronal receptor (CNR) and zinc finger-containing (ZNF) gene families are closely linked to predicted gene clusters indicated by the plus symbols.
Discussion
We have shown that, at medium to high levels of SAGE penetration, the hypothalamus and neocortex have contrasting patterns of gene expression. If tags occurring only once are removed from the hypothalamus library, there are at least 8,850 expressed tag identities in the hypothalamus compared with 18,149 expressed tag identities in the neocortex. Assuming a gene has an average of 1.6 tags (differential splicing and polyadenylation signal usage; ref. 24) this finding would imply that of the possible 30,000-40,000 genes in the mouse genome, the hypothalamus contains at least 14-18% of all genes in the mouse genome, whereas the neocortex has a higher number (28-38%). These assumptions are likely to be vast underestimates because tags occurring only once have been excluded, and because SAGE sequencing around the 100,000 mark is efficient at uncovering mainly medium to high abundance transcripts (>5 copies per cell, refs. 13 and 21). The greater number of unique tags encountered in the neocortex cannot be attributed simply to the larger size of the neocortical library because the probability of encountering unique tags would be biased in favor of the smaller hypothalamic library. This finding is because conversion of tag numbers to proportions overcompensates for library sizes and the proportion of unique tags in the larger library is expected to decrease. Because the larger neocortex library shows a higher proportion of unique tags, this is double evidence of greater complexity. The higher genetic complexity of the neocortex has also been reported in single-chromosome studies. Up to 41% of mouse genes syntenic to human chromosome 21 are expressed in the neocortex, with this figure being two to four times higher in neocortex than any other brain region surveyed (25). Thus, we find that only 25% of cortical tags match the largest genetic database (compared with 39% of hypothalamus tags), and of the tags that match, there are consistently more identified genes (whether high or low abundance) in the neocortex compared with the hypothalamus. This genetic complexity cannot be inferred from simply viewing their contrasting structural attributes; the neocortex is uniform in appearance with repetitive cytology across its six layers (26) compared with the more disparate hypothalamus comprising of separated and interconnected nuclei (e.g., arcuate, paraventricular, and supraoptic nuclei).
To ascertain whether gene composition provides a good indication of gene function, we performed gene ontology analyses for each structure (Fig. 2). The output shows a global scaling of gene activity that is unique to either hypothalamus or neocortex. This analysis also highlights both expected and unexpected differences in functional activity present in each of the two structures. Predictably, the hypothalamus shows a higher representation of genes involved in endocrine activities (proteases, peptidases, and enzyme inhibitors) that are the hallmarks of hypothalamic function such as blood pressure regulation, and water and salt balance. Similarly, the hypothalamus has a greater representation of genes associated with fatty acids (fatty acid metabolism and fatty acid biosynthesis), which correlates with what is known about lipid oxidation and the modulation of nutrients into the circulation. These representations are expected, based on what is currently known about the hypothalamus and regulation of the internal environment of the cell. Conversely, the neocortex appears to be more highly involved in the directed cellular movement of potassium and sodium ions, and although these functions are also present in the hypothalamus, they appear to be through indirect mechanisms involving regulatory molecules. The neocortex has more genes involved in glycolysis and glucose metabolism, possibly reflecting greater energy requirements by cortical metabolism. On the other hand, the hypothalamus exhibits higher representation of protein metabolism and ribosome biogenesis, indicating increased synthesis of ribosomes, although it is unclear why protein synthesis should be higher in the hypothalamus. A potential pitfall in comparing male hypothalamic with female neocortical tissues is that the perceived differences might reflect sexual dimorphism in brain structures. This possibility appears to be unlikely, given that in both human and mouse, the male and female hypothalamus has little, if any, sex-specific transcription (27).
Genomic studies of transcription from bacteria to humans have revealed higher orders of genetic organization with chromosomal clusters or genomic neighborhoods. These clusters may correspond to genes that are under common spatiotemporal control or have common biological functions or unique to specific lineages (28-30). Given sufficient data to distinguish between these options, coregulation may imply considerable information on unknown transcripts within the clusters. A comparative chromosomal view of expression levels between hypothalamus and neocortex emphasizes known facts about genome organization but also raises additional concepts. For example, the majority of the expressed genes in both tissues fall into GC-rich regions that are likely to correspond to transcriptionally active heterochromatin domains (31). However, our clustering analysis also revealed genes that are more highly expressed in one brain region over the other tend to be uniquely clustered together. Familial clustering of genes in mammalian brains has been demonstrated for genes encoding olfactory receptors, cadherin-related neuronal receptors, and Krüppel-type zinc finger-containing proteins (32-34). Testing of the last two categories indicate that both zinc finger-containing and cadherin-related neuronal receptor families are in close proximity to clusters of overexpressed genes in the cortex (Fig. 4; chromosomes 7 and 18, respectively).
It is likely that this coregulation is a result of epigenetic regulation and supports the notion that chromatin organization can be propagated along a chromosome and must require both initiation sites and boundary elements to properly delineate expression domains (35). This finding suggests that structural subdivisions of the mammalian brain are defined developmentally and may be further defined by a higher order of genetic organization. A complementary interpretation of these results is that these clustered domains may be regarded as evolutionary outcomes of genetic selection on functional subdivisions of the brain.
In conclusion, we have provided contrasting views of gene expression and regulation between an ancient brain structure with a more recent structure (neocortex). The hypothalamus and neocortex are derivatives of the embryonic prosencephalon, yet they diverge in terms of their organization, function, and evolution. The current study provides a global scaling of their respective genetic activity. We demonstrate many instances of genetic variation particularly in respect of gene ontology underlying different cellular activities. This variation is reflected at the chromosomal level, with clustering of genes that are differentially expressed in either structure.
Supplementary Material
Acknowledgments
We thank C. Augustine, V. Spirkoska, M. Brown, M. McBurnie, and L. Walker for technical assistance; Dr. M. McKinley for assistance with tissue dissections; and Nick Tan and the Walter and Eliza Hall Institute Information Technology Department for information technology support. This work was supported by a fellowship from the Deutsche Forschungsgemeinschaft (to T.B.); National Health and Medical Research Council Transitional Institute Grant 215499 (to T.B.); a National Health and Medical Research Council fellowship (to H.S.); National Health and Medical Research Council Grants 219176, 257501, 215201, and 257529 (to H.S., G.S., and S.-S.T.); and the Walter and Eliza Hall Institute Nossal Leadership Award (to H.S.). Most DNA sequencing of SAGE libraries was performed by the Australian Genome Research Facility, which was established through the Commonwealth-funded Major National Research Facilities program. Further assistance was provided by endowments from the Kenneth Myer Foundation, the G. Harold and Leila Y. Mathers Charitable Foundation, the Robert J., Jr., and Helen C. Kleberg Foundation, and the Search Foundation.
Author contributions: W.-M.B., T.B., D.A.D., H.S., and S.-S.T. designed research; W.-M.B. and T.B. performed research; W.-M.B., T.B., L.H., G.S., and H.S. analyzed data; W.-M.B., T.B., and S.-S.T. wrote the paper; J.G. provided technical expertise with the SAGE technique; D.A.D., H.S., and S.-S.T. funded research; and research was conducted in S.-S.T.'s laboratory.
Abbreviations: SAGE, serial analysis of gene expression; qRT-PCR, quantitative real-time PCR; RefSeq; Reference Sequence; GO, Gene Ontology.
References
- 1.Northcutt, R. G. & Kaas, J. H. (1995) Trends Neurosci. 18, 373-379. [DOI] [PubMed] [Google Scholar]
- 2.Jones, E. G. (1986) J. Neurosurg. 65, 135-153. [DOI] [PubMed] [Google Scholar]
- 3.Rakic, P. (1988) Science 241, 170-176. [DOI] [PubMed] [Google Scholar]
- 4.Dori, I., Petrou, M. & Parnavelas, J. G. (1989) J. Comp. Neurol. 290, 169-184. [DOI] [PubMed] [Google Scholar]
- 5.Finlay, B. L. & Darlington, R. B. (1995) Science 268, 1578-1584. [DOI] [PubMed] [Google Scholar]
- 6.Le Gros Clark, W. E. (1938) in The Hypothalamus: Morphological, Functional, Clinical, and Surgical Aspects (Oliver and Boyd, London).
- 7.Cechetto, D. F. & Saper, C. B. (1988) J. Comp. Neurol. 272, 579-604. [DOI] [PubMed] [Google Scholar]
- 8.Moga, M. M. & Saper, C. B. (1994) J. Comp. Neurol. 346, 137-150. [DOI] [PubMed] [Google Scholar]
- 9.Elias, C. F., Saper, C. B., Maratos-Flier, E., Tritos, N. A., Lee, C., Kelly, J., Tatro, J. B., Hoffman, G. E., Ollmann, M. M., Barsh, G. S., et al. (1998) J. Comp. Neurol. 402, 442-459. [PubMed] [Google Scholar]
- 10.Saper, C. B. & Breder, C. D. (1992) Prog. Brain Res. 93, 419-428. [DOI] [PubMed] [Google Scholar]
- 11.Denton, D. A., McKinley, M. J. & Weisinger, R. S. (1996) Proc. Natl. Acad. Sci. USA 93, 7397-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saper, C. B., Loewy, A. D., Swanson, L. W. & Cowan, W. M. (1976) Brain Res. 117, 305-312. [DOI] [PubMed] [Google Scholar]
- 13.Velculescu, V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484-487. [DOI] [PubMed] [Google Scholar]
- 14.Kenzelmann, M. & Mühlemann, K. (1999) Nucleic Acids Res. 27, 917-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margulies, E. H., Kardia, S. L. R. & Innis, J. W. (2001) Nucleic Acids Res. 29, 1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha, S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508-512. [DOI] [PubMed] [Google Scholar]
- 17.Benjamini, Y. & Hochberg, Y. (1995) J. R. Stat. Soc. B 57, 289-300. [Google Scholar]
- 18.Johnston, H., Koukoulas, I., Jeyaseelan, K., Armugam, A., Earnest, L., Baird, R., Dawson, N., Ferraro, T. & Wintour, E. M. (2000) Placenta 21, 88-99. [DOI] [PubMed] [Google Scholar]
- 19.Margulies, E. H., Kardia, S. L. R. & Innis, J. W. (2001) Genome Res. 11, 1686-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackshaw, S., Fraioli, R. E., Furukawa, T. & Cepko, C. L. (2001) Cell 107, 579-589. [DOI] [PubMed] [Google Scholar]
- 21.Chrast, R., Scott, H. S., Papasavvas, M. P., Rossier, C., Antonarakis, E. S., Barras, C., Davisson, M. T., Schmidt, C., Estivill, X., Dierssen, M., et al. (2000) Genome Res. 10, 2006-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niimura, Y. & Gojobori, T. (2002) Proc. Natl. Acad. Sci. USA 99, 797-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craig, J. M. & Bickmore, W. A. (1993) BioEssays 15, 349-354. [DOI] [PubMed] [Google Scholar]
- 24.Velculescu, V. E., Madden, S. L., Zhang, L., Lash, A. E., Yu, J., Rago, C., Lal, A., Wang, C. J., Beaudry, G. A., Ciriello, K. M., et al. (1999) Nat. Genet. 23, 387-388. [DOI] [PubMed] [Google Scholar]
- 25.Gitton, Y., Dahmane, N., Baik, S., Ruiz, I., Altaba, A., Neidhardt, L., Scholze, M., Herrmann, B. G., Kahlem, P., Benkahla, A., et al. (2002) Nature 420, 586-590. [DOI] [PubMed] [Google Scholar]
- 26.Jones, E. G. (2000) Proc. Natl. Acad. Sci. USA 97, 5019-5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinn, J. L., Rozowsky, J. S., Laurenzi, I. J., Petersen, P. H., Zou, K., Zhong, W., Gerstein, M. & Snyder, M. (2004) Dev. Cell 6, 791-800. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J. M. & Sonnhammer, E. L. (2003) Genome Res. 13, 875-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spellman, P. T. & Rubin, G. M. (June 18, 2002) J. Biol., doi 10.1186/1475-4924-1-5.
- 30.Roy, P. J., Stuart, J. M., Lund, J. & Kim, S.K. (2002) Nature 418, 975-979. [DOI] [PubMed] [Google Scholar]
- 31.Caron, H., van Schaik, B., van der Mee, M., Baas, F., Riggins, G., van Sluis, P., Hermus, M. C., van Asperen, R., Boon, K., Voute, P. A., et al. (2001) Science 291, 1289-1292. [DOI] [PubMed] [Google Scholar]
- 32.Shannon, M., Hamilton, A. T., Gordon, L., Branscomb, E. & Stubbs, L. (2003) Genome Res. 13, 1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamada, S. & Yagi, T. (2001) Neurosci. Res. 41, 207-215. [DOI] [PubMed] [Google Scholar]
- 34.Mombaerts, P. (1999) Science 286, 707-711. [DOI] [PubMed] [Google Scholar]
- 35.Alekseyenko, A. A. & Kuroda, M. I. (2004) Science 303, 1148-1149. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.