Abstract
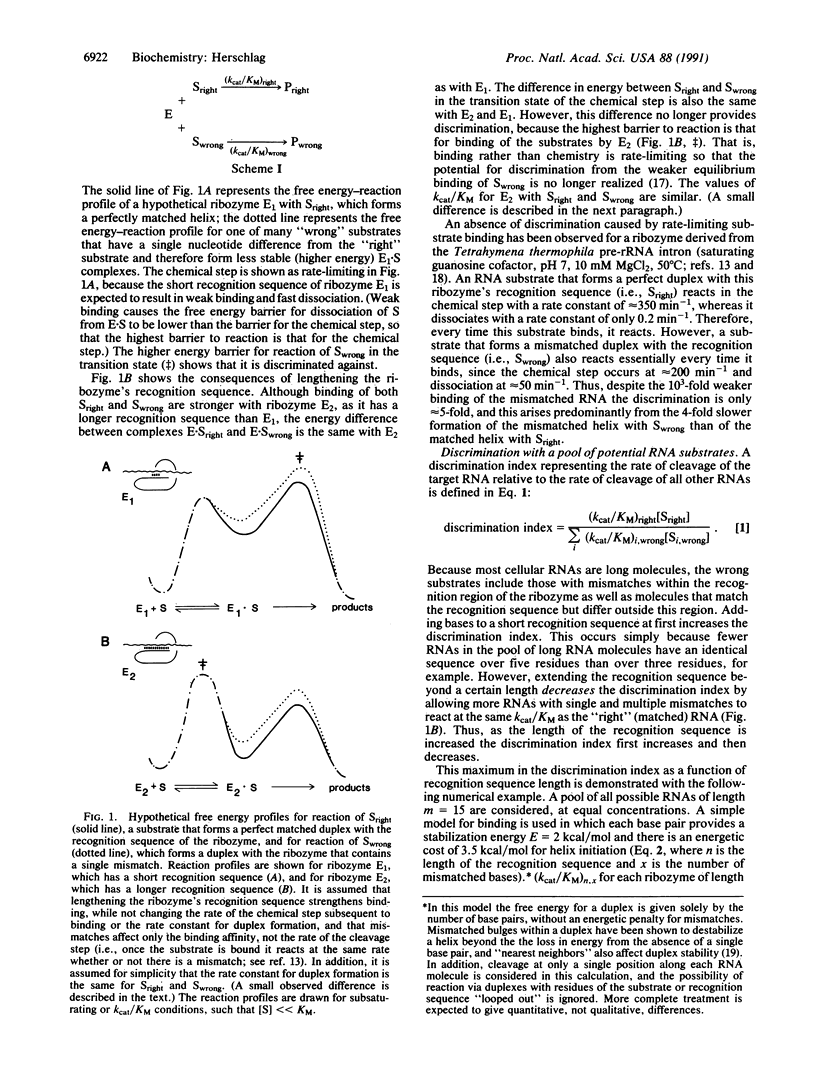
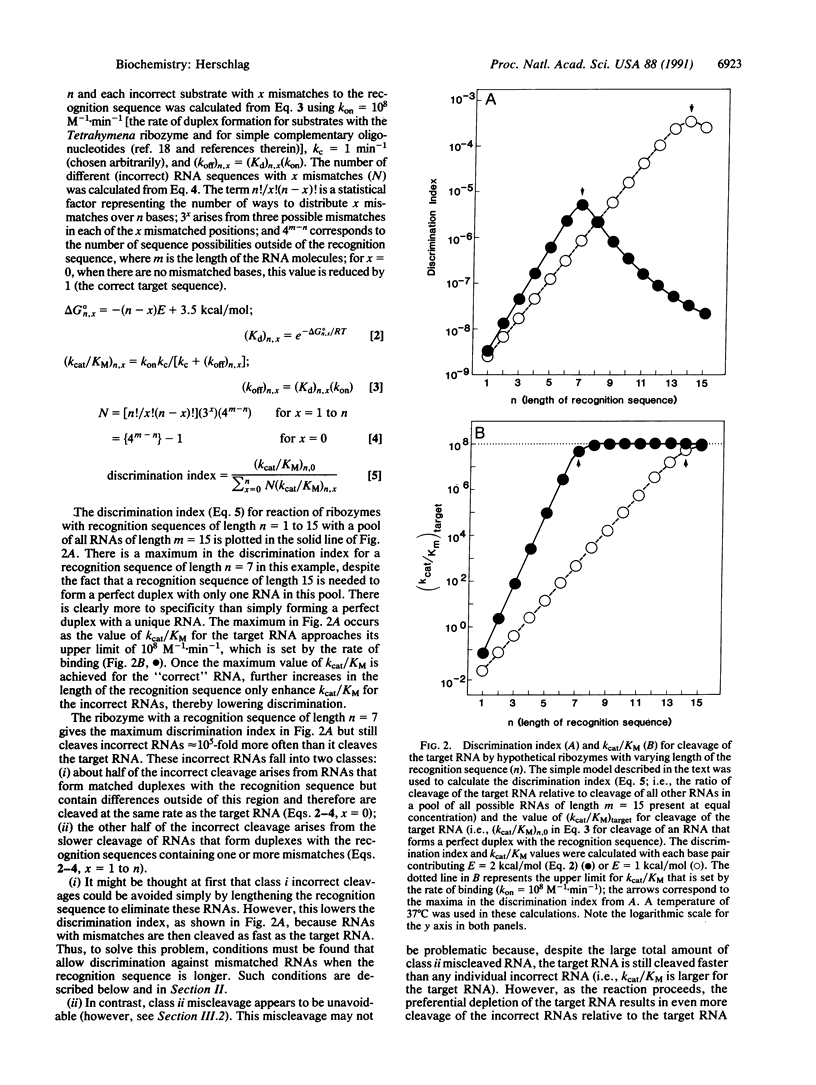
Kinetic and thermodynamic factors that determine specificity of RNA cleavage by ribozymes are illustrated with examples from recent work with a ribozyme derived from the group I intron of Tetrahymena thermophila pre-rRNA. The conclusions also apply to other ribozymes, to antisense oligonucleotide experiments, and to RNA and DNA cleavage agents that can recognize a single-stranded or double-stranded region of variable length. At first, adding bases to a ribozyme's recognition sequence is expected to increase cleavage of the target RNA relative to cleavage of other RNAs. However, adding more bases ultimately reduces this discrimination, as cleavage occurs essentially every time the target RNA or a mismatched RNA binds the ribozyme. This occurs despite the weaker binding of the mismatched RNA because dissociation becomes too slow (binding is too strong) to allow the ribozyme to "choose" between cleavage of the target RNA and a mismatched RNA. In summary, more (base pairing) isn't always better, because maximal discrimination requires equilibrium binding prior to cleavage. The maximum discrimination that can be obtained is expected to be greater with an A + U-rich recognition sequence than with a G + C-rich recognition sequence. This is because the weaker A.U base pairs (relative to G-C base pairs) allow recognition to be spread over a larger number of bases while preventing binding that is too strong. Finally, creating an A-rich ribozyme rather than a U-rich ribozyme avoids the loss in discrimination expected with U-rich ribozymes from the formation of U.G wobble pairs in addition to the "targeted" Watson-Crick U.A pair.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cech T. R. Ribozymes and their medical implications. JAMA. 1988 Nov 25;260(20):3030–3034. [PubMed] [Google Scholar]
- Colman A. Antisense strategies in cell and developmental biology. J Cell Sci. 1990 Nov;97(Pt 3):399–409. doi: 10.1242/jcs.97.3.399. [DOI] [PubMed] [Google Scholar]
- Fedor M. J., Uhlenbeck O. C. Substrate sequence effects on "hammerhead" RNA catalytic efficiency. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1668–1672. doi: 10.1073/pnas.87.5.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Ellington A. D., Szostak J. W. In vitro genetic analysis of the Tetrahymena self-splicing intron. Nature. 1990 Sep 27;347(6291):406–408. doi: 10.1038/347406a0. [DOI] [PubMed] [Google Scholar]
- Griffin L. C., Dervan P. B. Recognition of thymine adenine.base pairs by guanine in a pyrimidine triple helix motif. Science. 1989 Sep 1;245(4921):967–971. doi: 10.1126/science.2549639. [DOI] [PubMed] [Google Scholar]
- Hampel A., Tritz R., Hicks M., Cruz P. 'Hairpin' catalytic RNA model: evidence for helices and sequence requirement for substrate RNA. Nucleic Acids Res. 1990 Jan 25;18(2):299–304. doi: 10.1093/nar/18.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel A., Tritz R. RNA catalytic properties of the minimum (-)sTRSV sequence. Biochemistry. 1989 Jun 13;28(12):4929–4933. doi: 10.1021/bi00438a002. [DOI] [PubMed] [Google Scholar]
- Haseloff J., Gerlach W. L. Simple RNA enzymes with new and highly specific endoribonuclease activities. Nature. 1988 Aug 18;334(6183):585–591. doi: 10.1038/334585a0. [DOI] [PubMed] [Google Scholar]
- Hearst J. E. A photochemical investigation of the dynamics of oligonucleotide hybridization. Annu Rev Phys Chem. 1988;39:291–315. doi: 10.1146/annurev.pc.39.100188.001451. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990 Nov 6;29(44):10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- Herschlag D., Cech T. R. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 2. Kinetic description of the reaction of an RNA substrate that forms a mismatch at the active site. Biochemistry. 1990 Nov 6;29(44):10172–10180. doi: 10.1021/bi00496a004. [DOI] [PubMed] [Google Scholar]
- Hélène C., Toulmé J. J. Specific regulation of gene expression by antisense, sense and antigene nucleic acids. Biochim Biophys Acta. 1990 Jun 21;1049(2):99–125. doi: 10.1016/0167-4781(90)90031-v. [DOI] [PubMed] [Google Scholar]
- Latham J. A., Zaug A. J., Cech T. R. Self-splicing and enzymatic cleavage of RNA by a group I intervening sequence. Methods Enzymol. 1990;181:558–569. doi: 10.1016/0076-6879(90)81151-j. [DOI] [PubMed] [Google Scholar]
- Martin F. H., Tinoco I., Jr DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980 May 24;8(10):2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierhaus K. H. The allosteric three-site model for the ribosomal elongation cycle: features and future. Biochemistry. 1990 May 29;29(21):4997–5008. doi: 10.1021/bi00473a001. [DOI] [PubMed] [Google Scholar]
- Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57(5):587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Pontius B. W., Berg P. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8403–8407. doi: 10.1073/pnas.87.21.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson D. L., Joyce G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990 Mar 29;344(6265):467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- Rossi J. J., Sarver N. RNA enzymes (ribozymes) as antiviral therapeutic agents. Trends Biotechnol. 1990 Jul;8(7):179–183. doi: 10.1016/0167-7799(90)90169-x. [DOI] [PubMed] [Google Scholar]
- Ruffner D. E., Stormo G. D., Uhlenbeck O. C. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990 Nov 27;29(47):10695–10702. doi: 10.1021/bi00499a018. [DOI] [PubMed] [Google Scholar]
- Takayama K. M., Inouye M. Antisense RNA. Crit Rev Biochem Mol Biol. 1990;25(3):155–184. doi: 10.3109/10409239009090608. [DOI] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990 Aug 3;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Walker G. T. Deoxycytidine methylation does not affect DNA.RNA hybrid formation or B-A transitions of (dG)n.(dC)n sequences. Nucleic Acids Res. 1988 Apr 11;16(7):3091–3099. doi: 10.1093/nar/16.7.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A. J., Been M. D., Cech T. R. The Tetrahymena ribozyme acts like an RNA restriction endonuclease. Nature. 1986 Dec 4;324(6096):429–433. doi: 10.1038/324429a0. [DOI] [PubMed] [Google Scholar]


