Abstract
Presenilins (PSs) are required for Notch signaling in the development of vertebrates and invertebrates. Mutations in human PS1 and PS2 homologs are a cause of familial Alzheimer's disease (AD). The function of the recently identified ancient family of IMPAS proteins (IMP/SPP/PSH) homologous to PSs is not yet known. We show here that, unlike PSs, IMPs (orthologous C. elegans Ce-imp-2 and human hIMP1/SPP) do not promote Notch (C. elegans lin-12,glp-1) proteolysis or signaling. The knock-down of Ce-imp-2 leads to embryonic death and an abnormal molting phenotype in Caenorhabditis elegans. The molting defect induced by Ce-imp-2 deficiency was mimicked by depleting cholesterol or disrupting Ce-lrp-1 and suppressed, in part, by expression of the Ce-lrp-1 derivate. C. elegans lrp-1 is a homolog of mammalian megalin, lipoprotein receptor-related protein (LRP) receptors essential for cholesterol and lipoprotein endocytosis and signaling. These data suggest that IMPs are functionally distinct from related PSs and implicate IMPs as critical regulators of development that may potentially interact with the lipid-lipoprotein receptor-mediated pathway.
Presenilin 1 and presenilin 2 (PS1/PSEN1 and PS2/PSEN2) were initially identified by positional cloning of the genes for Alzheimer's disease (AD) (1, 2). Mutations in human PS1 are a frequent cause of early-onset familial AD, and mutations in human PS2 underlie more rare early- or late-onset AD cases (1-3). Many tested missense- or deletion/insertion-AD mutations in PSs increase production of fibrillogenic Aβ42, presumably by means of γ-secretase cleavage of amyloid precursor protein (APP) membrane-tethered derivates. The data support the role of amyloid in the etiology of AD (4). Many lines of evidence show that human PS1 and PS2 and their homologs in other organisms (SEL-12 and HOP-1 in Caenorhabditis elegans) are unusual polytopic endoproteases capable of cleaving different type I transmembrane proteins within their membrane-spanning domains (5-8). The primary evolutionary conserved function of PSs in vertebrates and invertebrates is regulation of the Notch (lin-12,glp-1 in C. elegans) signaling pathway during embryogenesis. Knock-out PS1 mice display a lethal phenotype similar to mice with Notch receptor and ligand knock-outs. Inactivation of both PS homologs hop-1 and sel-12 in C. elegans induces defects described for lin-12 and glp-1 loss-of-function mutants (9, 10). Moreover, the loss-of-function sel-12 mutant suppresses multivulva phenotype in “hyperfunctional” lin-12 mutants (11). The results demonstrate that PSs/sel-12,hop-1 facilitate signaling by means of Notch/lin-12,glp-1 receptors. Recently, genes encoding proteins homologous to PSs and termed IMPAS (or IMP/PSH/SPP,SPPL, hereafter referred to as IMPs) have been found (12-14, ††). The abundant ancient family of orthologous and paralogous IMP proteins is conserved from yeast to humans and share structural similarities with Archaea proteins and prokaryotic type 4 prepilin peptidases (TFPP) (15). The intramembrane proteolytic activity of human Impas 1 (hIMP1/SPP), an evolutionary conserved member of this protein family, has been shown. Direct examination in cultured cells or in vitro assays demonstrated that hIMP1/SPP is capable of cleaving hydrophobic signal peptide domains (14) or multipasss-transmembrane protein substrates (15). The presumable intramembrane cleavage may release potentially bioactive short peptides or may be involved in maturation or trafficking of the proteins (16, 17). However, the in vivo functions of IMP proteins are as yet unknown. Here, we describe the analysis of imp-genes in C. elegans (Ce-imp). We show that Ce-imp-2, an ortholog of human hIMP1/SPP, is required for embryonic development and larval molting in C. elegans but is not required for Notch/(lin-12,glp-1) signaling and, therefore, has a function distinct from homologous presenilins. Instead, our data suggest that Ce-imp-2 is involved in the cholesterol-lipoprotein receptor (Ch-LR)-dependent development pathway and raise the possibility that the mammalian ortholog, IMP1/SPP, may have similar function.
Materials and Methods
C. elegans Strains and Clones. The following strains were used: N2 Bristol as a wild-type strain; sel-12(ar131), sel-12(ar131)unc-1, sel-12(ar171)unc-1 (e538) (provided by I. Greenwald, Columbia University, New York), glp-1(e2142ts) (provided by C. Goutte, Amherst College, Amherst, MA), lin-12(n137)/unc-32(e189)III;him-5(e1467)V, unc-32(e189)lin-12(n676n930)III, lrp-1(ku156)/gld-1(q266) I (provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources).
cDNA clones yk671a5, yk336d12, yk1191b02, and yk260f8 were obtained from Y. Kohara (Genome Biology Laboratory, National Institute of Genetics, Mishima, Japan). We used standard methods of C. elegans handling and culture (18) with OP50 Escherichia coli strain as a food source. Worms were observed by using a dissecting microscope with a maximum magnification of ×40-60 or a fluorescent dissecting stereo-microscope with a Nomarski optic and magnification of up to ×100. Filipin staining was performed as described (19).
Double-Stranded RNA (dsRNA) Synthesis and Injections. To generate dsRNA for injection, a T7 sequence was added at both ends of the cDNA template by PCR reaction with corresponding primers. PCR product was then purified on Qiagen (Valencia, CA) columns and used for RNA synthesis and purification by using Ambion (Austin, TX) MEGAscript T7 Transcription Kit. RNA was then checked on standard agarose gel, was quantified (≈ 1-5 μg/μl), and was used for pseudocoelomic injections [in head and tail regions beyond the positions of the two gonad arms (20, 21)] with an average of 0.5-1 × 106 RNA molecules per animal. Four to six hours after injection, worms were transferred individually to fresh plates, and the phenotype of F1 progeny was analyzed within 2-3 days.
RNA Interference (RNAi) by dsRNA Feeding. RNAi by dsRNA feeding was performed as described (22-24). cDNAs for C. elegans Ce-imp-1, Ce-imp-2, and Ce-imp-3 were cloned directly from yk clones or as PCR subfragments into an L4440 vector containing a double T7 promoter sequence (Fig. 5, which is published as supporting information on the PNAS web site).
Germ-Line Injections and Extrachromosomal Array. For Ce-lrp-1 rescue experiments, germ-line injections of plasmid containing Ce-lrp-1 L-ICD were performed as described (25). Ce-lrp-1L-ICD fragment was cloned into BamHI-PstI L4440 vector. Ce-lrp-1L-ICD contains a 996-bp Ce-lrp-1 promoter genomic region upstream of the ATG and a 1,026-bp 3′ genomic region downstream of stop codon TAA. Encoding sequence contains ICD fragment of Ce-lrp-1 CDS with introduced ATG (5′-ATGGGACTTATTGGATTC) and ends on the last coding triplet of Ce-lrp-1 CDS fused to myc-epitope sequence (Fig. 6, which is published as supporting information on the PNAS web site). DNA was injected in concentrations of 10-50 ng/μl into gonadal distal cells of young adult animals. The coinjection marker sur-5-GFP for easy detection of transgenic progeny was used in concentration at 10 ng/μl. Four to six hours after injection, the worms were transferred individually to fresh plates. To establish individual lines with extrachromosomal maintenance of the transgene, marker-positive F1 animals were picked and placed separately.
Results
Imp-genes in C. elegans. Multiple IMP/SPP-related genes were identified in a variety of species from yeast to human. At least five IMP genes in human and three diverged homologous genes similar to presenilins in C. elegans were identified. Comparison of the most conserved domains suggests links to a family of proteins in
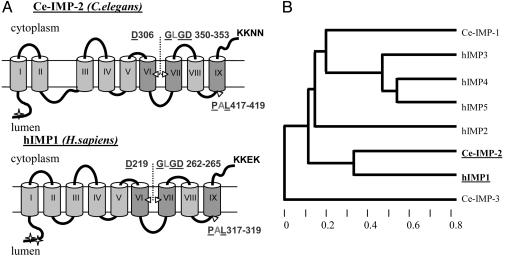
Archaea and to bacterial peptidases (12, 15), indicating that C. elegans Ce-IMP-1, Ce-IMP-2, and Ce-IMP-3, like their human orthologs, may be polytopic proteases. The C. elegans Ce-IMP-1, Ce-IMP-2, and Ce-IMP-3 have divergent N termini but have substantial amino acid conservation in three hydrophobic regions predicted to be transmembrane (Tm) domains (Fig. 1; Fig. 7, which is published as supporting information on the PNAS web site). The amino acid signature 5DxxxV-LGxGD-PxL (where 5-F,Y,W; “x” is any amino acid, and “-” connects the three most conserved domains of the signature) is invariant in C. elegans Ce-IMP-1 and Ce-IMP-2 and members of IMPAS or PS families from distant taxons. Ce-IMP-3 is a more diverged family member, lacking this consensus signature but still harboring conserved aspartate and proline residues. The predicted Tm protein structure and membrane topology also resemble those described for human/C. elegans PS/SEL-12,HOP-1 proteins (Fig. 1 A) (26, 27). Consistent with data for human IMP1/SPP, the sequences for endoplasmic reticulum retention signals were identified in the C-terminal part of Ce-IMP-2 and Ce-IMP-3, but not in Ce-IMP-1 (PSORT II prediction). The cluster algorithm and phylogenetic tree, limited to analysis of evolutionary conserved domains, demonstrate that Ce-IMP-2 has a more stringent homology to hIMP1/SPP than Ce-IMP-1 and Ce-IMP-3 (genebee analysis, Fig. 1B). The percentage of identity/similarity for the entire amino acid sequence [hIMP1/Ce-IMP-1 (14/27); hIMP1/Ce-IMP-2 (30/46); hIMP1/Ce-IMP-3 (12/29)] also supports the idea that Ce-IMP-2 is an ortholog of hIMP1/SPP.
Fig. 1.
Similarity of Ce-IMP-2 to other members of IMPAS and presenilin families of proteins. (A) Predicted transmembrane structure of Ce-IMP-2, C. elegans, and hIMP1,Homo sapiens. The arrowheads denote the positions of conserved aspartate and PAL-motif residues. The letters show invariant amino acid residues in eukaryotic PSs and IMPASes; the underlined letters are identical also in related proteins from Archaea and bacterial polytopic type 4 prepilin peptidases (TFPP). Most conserved domains in PSs and IMPAS families are shown in intense gray. The predicted sites of N-glycosylation (marked by asterisks) and endoplasmic reticulum-membrane retention signals (KKXX motif) are shown (netnglyc 1.0 and psort ii predictions). (B) Phylogenic tree (Cluster algorithm, genebee) for conserved domains (amino acid 269-444) of C. elegans Ce-IMP-2 and corresponding domains of C. elegans Ce-IMP-1, Ce-IMP-3, and human IMP (hIMP1-5) proteins.
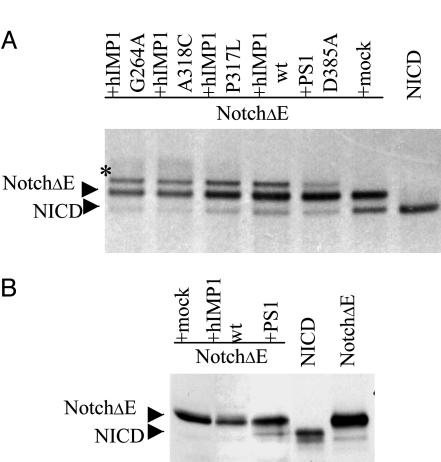
hIMP1 Does Not Promote S3-Cleavage of Notch1 in Mammalian Cells. Because PSs are major components of the protein complex regulating intramembrane Notch protein cleavage, we tested whether hIMP1/SPP, homologous to PSs, may also facilitate cleavage of the Notch1 receptor. Constructs with human IMP1 were generated that efficiently express hIMP1 in transiently and stably transfected mammalian cells. hIMP1 in cell lysates is detected by Western blots in predicted 42- to 48-kDa and 95- to 98-kDa protein fractions. The high molecular mass 95- to 98-kDa fraction of exogenous or endogenous hIMP1 is likely homodimeric hIMP1 complex resistant to protein complex disassociation during protein extract preparation and SDS/PAGE procedures. However, upon expression of Ce-IMP-2 in HEK293 cells, only the predicted protein of molecular mass 50-55 kDa was dectected (Fig. 8A, which is published as supporting information on the PNAS web site, and refs. 12 and 28). PS1 was shown to be required for intramembranous S3 cleavage of NH2-terminally truncated Notch derivates (NotchΔE) (6, 29). Mutations in the putative catalytic PS1 D257 and D385 residues inhibit proteolysis at the S3 site (30). We cotransfected HEK293 with NotchΔE (a substrate for intramembrane S3 cleavage) and hIMP1 or with the PS1 D385A construct, which inhibits the NotchΔE cleavage. The metabolic labeling and pulse-chase procedures demonstrated that hIMP1 does not induce or facilitate the NotchΔE S3 cleavage and generation of NOTCH1 intracellular domain (NICD) derivatives. Moreover, in cells overexpressing hIMP1, some inhibition of the NotchΔE S3 cleavage was detected comparable with the dominant negative effect of the PS1 D385A construct (Fig. 2A and Fig. 8 B, C, and D). Interestingly, a modified NotchΔE isoform was observed in cells overexpressing hIMP1. The wild-type and mutant IMP1 proteins had similar effects on modification of NotchΔE. The data demonstrate that hIMP1 does not facilitate NotchΔE S3 cleavage. In contrast to PS1wt, ectopic overexpression of hIMP1wt did not restore the S3 cleavage in PS1-/- PS2-/- mouse fibroblasts (Fig. 2B). Together, the data indicate that the substrate-dependent enzymatic function of hIMP1 is not redundant with PSs.
Fig. 2.
Expression of human IMP1 in mammalian cells and S3 cleavage of Notch1 receptor. (A) Overexpression of hIMP1wt or mutant forms does not facilitate cleavage of Notch1 as detected by pulse-chase analysis. HEK293 cells were cotransfected with the constructs (i) NotchΔE and (ii) wild or mutant hIMP1 or PS1 D385A or mock (pcLacZ). NotchΔE and NOTCH1 intracellular domain (NICD) (predicted derivate of NotchΔ S3-cleavage) are fused to c-myc epitope. Cells were pulse-labeled and chased for 60 min. Protein extracts were immunoprecipitated with anti-c-myc antibodies and subjected to electrophoresis and autoradiography. As described, the NICD product of S3 cleavage is clearly detected in 60 min of pulse-chase in HEK293 cells expressing membrane-tethering NotchΔE. The coexpression of NotchΔE with loss-of-function PS1 D385A mutant has dominant-negative effect suppressing S3 activity of endogenous PS. The hIMP1wt or the hIMP1 mutant isoforms (G264A, A318C, and P317L) do not increase, but reduce efficiency of S3-cleavage. The modified Notch1 fragment observed in cells overexpressing IMP1 constructs is designated by asterisk. (B) Expression of hIMP1 in PS1-/-,PS2-/- fibroblasts does not induce NotchΔE cleavage, unlike expression of exogenous PS1. Notch1 C-terminal fragments are detected by Western blot analysis with c-myc AB (Supporting information).
Ce-imp-2 RNAi Does Not Affect lin-12,glp-1 (Notch)-Signaling in Development in C. elegans. To gain further insights into the possible functions of IMPAS genes in vivo, we used the C. elegans model. In our preliminary experiments, Ce-imp-1 and Ce-imp-3 dsRNAi produced no obvious development abnormalities. Therefore, we focused further upon study of Ce-imp-2, the ortholog of mammalian IMP1/SPP. Wild-type worms were injected with dsRNA or fed with E. coli-expressing dsRNA corresponding to the Ce-imp-2 gene (Fig. 5). The search for the sel-12, lin-12, and glp-1 characteristic phenotypes was performed over the course of multiple replicate experiments. There are several major defects caused by single or concomitant reduction of lin-12 and glp-1 (Notch homologs) and their upstream and downstream regulators. The first of these defects involves cell fate changes between the 4-cell and 12-cell stages that result in embryos lacking a portion of the pharynx and failing to enclose and elongate the body (31). Although the earliest defect caused by Ce-imp-2 (RNAi) is embryonic arrest, <3% of Ce-imp-2(RNAi) embryos exhibited a failure to undergo body morphogenesis (Fig. 9A, which is published as supporting information on the PNAS web site). Instead, the majority of Ce-imp-2 RNAi animals died during late embryonic stages or after hatching (Fig. 9 B and C). The lack of an anterior pharynx was previously described as a characteristic specific to diminished maternal glp-1 activity, a decrease in the activity of factors required for glp-1,lin-12 signaling (e.g., sel-8, aph-1, and aph-2) or the simultaneous reduction of sel-12 and hop-1 (9, 32, 33). We found that the development of anterior pharynx is normal in arrested larvae of Ce-imp-2-deficient N2 worms. Both Notch homologs glp-1 and lin-12 function together during late embryogenesis in the specification of the rectum (32). However, development of the rectum was unaffected in Ce-imp-2-deficient worms. Reduction of Ce-imp-2 activity by RNAi did not induce defects in vulva formation similar to those described for mutants in lin-12/Notch or the Ce-imp-2 homolog sel-12 (10, 11, 32). Finally, we used sensitized genetic backgrounds to look for interactions between Ce-imp-2 and Notch-pathway components. None of the hypo- or hypermorphic glp-1, lin-12, or sel-12 mutant strains analyzed exhibited enhancement or suppression of their corresponding phenotypes upon induction Ce-imp-2 (RNAi) (Fig. 3A and Table 1, which is published as supporting information on the PNAS web site). The data suggest that, unlike its distant homologs sel-12 or hop-1, Ce-imp-2 is not a positive regulator of lin-12,glp-1 (Notch) signaling in cell-cell interaction and cell fate specification during development in C. elegans.
Ce-imp-2 Inactivation Induces Embryonic Lethality and Molting Defects During Development. Two major abnormalities induced by Ce-imp-2 RNAi were embryonic/early L1 lethality and L1-L4 larvae molting defects leading to larval arrest (Figs. 3B and 9 A-G). Studies of the progeny of adult worms injected with Ce-imp-2 dsRNA showed 80-100% lethality of their offspring at late embryonic or early larval stages [number of experiments n = 7; total number of analyzed progeny n = 138 (100%); total number of dead worms on early stages 120 (87%)]. In an effort to avoid maternal germ-line effects and to delineate potential Ce-imp-2 deficiency phenotypes in later development, we used RNAi by feeding.
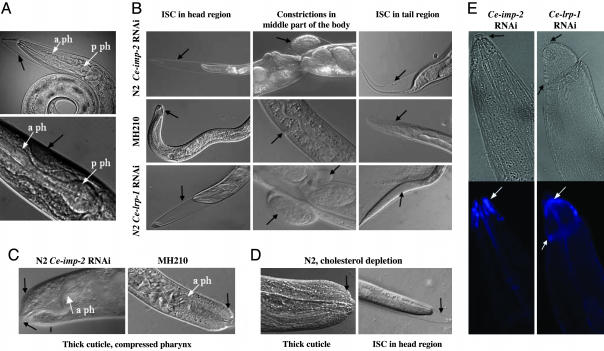
Fig. 3.
Phenotypes induced by Ce-imp-2 RNAi in C. elegans and cholesterol regulation-related pathway. Shown are Nomarski optic longitudinal views of C. elegans phenotypes. (A Upper) Ce-imp-2 RNAi and sel-12 reduced activities have no synergetic effects on lin-12,glp-1 signaling. ISC defect induced by Ce-imp-2 RNAi is observed in sel-12 (ar131) larvae, but pharyngeal anterior bulb is developed normally. (A Lower) Muscle detachment occasionally detected in N2 Ce-imp-2 RNAi worms and normal pharyngeal anterior bulb are shown. (B) Similar ISC phenotypes are detected in Ce-imp-2 RNAi N2 worms and in Ce-lrp-1-deficient worms (Ce-lrp-1 RNAi N2 worms or Ce-lrp-1 deletion strain MH210). (C) Thick cuticle and compressed pharynx in Ce-imp-2 RNAi N2 and MH210 worms. (D) ISC phenotype and thick cuticle caused by cholesterol depletion in N2 worms. (E) Filipin complexes are localized in anterior head region in P0 worms, which were fed with Ce-imp-2 or Ce-lrp-1 dsRNA (Upper, regular light, black arrows indicate ISC; Lower, UV-light picture, white arrows show filipin localization).
Staged populations of L1 worms were transferred to Ceimp-2 dsRNA plates and were analyzed for phenotypes in both initial generation (P0 population) and their progeny (the F1 generation). Defects in P0 development were observed at the L3-L4 and L4-adult molt stages. At least 20-30% of N2 worms at these stages (n = 9; n > 2 × 103) cannot properly shed the cuticle (Fig. 3B). In these cases, shedding of the cuticle was initiated, but the old cuticle remained attached to the body. Henceforth, we shall refer to this phenotype as the incomplete shedding of cuticle (ISC) phenotype. Analysis of individuals exhibiting this phenotype showed that ≈80% of these individuals failed to shed their cuticle and died, whereas the remaining 20% escape from the cuticle, which often remained attached to the tail region. These escapers matured to form sterile adults or Egl animals that could produce only a few eggs. The F1 generation of the dsRNA Ce-IMP-2-fed animals exhibited phenotypes similar to those observed after micro-injection of dsRNA. Approximately 3% of these embryos died at early stages before morphogenesis, whereas the majority died at late embryonic stages. Among the hatching F1 progeny, the ISC phenotype was observed at all molt stages (Fig. 9 D-G). In experiments using F1 embryos, we found that >60% of eggs died during late embryogenesis with elongated 3-fold embryos inside the egg shell or as early L1 larvae, shortly after hatching (n = 6, n > 103). The remaining animals reached later stages, and most of them arrested with the ISC phenotype. Other postembryonic pathologies in Ce-imp-2 RNAi worms included body constrictions (Fig. 3B), dumpy (Dpy), thick cuticle (Fig. 3C), exploded body (Rup, Fig. 9J), small body size (Sma), slow growth (Gro), slow pumping, uncoordinated movement (Unc), and muscle detachment (Fig. 3A).
Comparison of Phenotypes Caused by Depletion of Ce-imp-2, Ce-lrp-1, and Cholesterol. A similar ISC phenotype has been described for strains deprived of exogenous dietary cholesterol (34). However, this phenotype was not observed in another study in which cholesterol was completely removed from media (19). Recessive mutations in C. elegans Ce-lrp-1 also result in defects in shedding the old cuticle during larval development (34). The C. elegans Ce-LRP-1 encoded by Ce-lrp-1 is an epidermal protein homologous to mammalian low density lipoprotein receptors (LDLR), LRP1 (lipoprotein receptor related protein), and, in particular, megalin (termed also as LRP2/gp330). The LDLRs endocytose lipoproteins and regulate lipid homeostasis in mammals. We compared phenotypes caused by removal of supplementary cholesterol and by Ce-imp-2 and Ce-lrp-1 RNAi in parallel experiments. Under similar conditions, the Ce-lrp-1 mutant MH210 [lrp-1(ku156)/gld-1(q266) strain] was also tested. Worms growing on plates with no supplementary cholesterol (as compared with standard plates with 5 μg/ml supplementary cholesterol) developed the ISC phenotype patterns similar to those found for Ce-imp-2 RNAi animals (Fig. 3 B-D). As described in the original report (34), the ISC phenotype was not evident in F1 but was displayed in successive generations after extended cultivation in cholesterol-free medium. Only a small amount of sterol is required for growth of insect cells and free-living nematodes (19, 35-37). Traces of cholesterol in agar, E. coli, or maternal supply in the eggs may be sufficient for normal development of F1 (34), but not the following generations. We tested this prediction preparing ether-extracted peptone (19) for cholesterol-free agarose plates. As a result, we were able to detect 3.7% (75/2020) of animals with ISC in the P0 generation in a synchronized population of worms. Many worms growing under these conditions remained small and weak. One possible model from our studies is that the Ce-imp-2 is required for the production of a cholesterol-derived hormone important for molting. For example, the steroid hormone, 20-hydroxyecdysone, regulates molting in Drosophila (reviewed in ref. 38). However, these hormones have not yet been found in C. elegans.
We next compared phenotypes in Ce-lrp-1 RNAi and Ce-imp-2 RNAi worms. The effects of Ce-lrp-1 RNAi were similar to Ce-imp-2 RNAi, producing the ISC phenotypes during larval development. The partially detached cuticle found on all L1-L4 molt stages was seen frequently in the head and less frequently in the tail. Constrictions in the middle part of body were also found in both Ce-lrp-1 and Ce-imp-2 RNAi worms (Fig. 3B and Table 2, which is published as supporting information on the PNAS web site). The most noticeable difference was observed during the late embryonic/early L1 stages; death was much less frequent or not detected in F1 progeny of Ce-lrp-1 RNAi-treated animals. Animals homozygous for lrp-1(ku156) showed the same ISC phenotypes observed in Ce-lrp-1 (RNAi) (Fig. 3B). Arrested growth (with and without visual ISC), Unc, Sma, and occasional Dpy,Rup phenotypes were also identified in both Ce-lrp-1(RNAi) and Ce-imp-2(RNAi) animals. These effects may be related to the overly thick cuticle found in cholesterol-, Ce-imp-2-, and Ce-lrp-1-deficient worms (Fig. 3 C and D) and the inability to shed old cuticle compressing the growing body. The reduced brood size has been described in progeny of worms maintained on a reduced cholesterol diet (19, 39). We examined the brood size of individual worms on Ce-imp-2 RNAi food. Worms that developed ISC at the L4-adult stage died, some of them having a low number of progeny hatched inside dead mothers. The surviving worms were able to lay eggs over the next 2 or 3 days, but with a mean brood size at least 3-fold less than control RNAi worms (Supporting Text, which is published as supporting information on the PNAS web site). In summary, our data showed that Ce-imp-2 deficiency phenotype resembles abnormalities induced by cholesterol depletion and closely mimics the Ce-lrp-1 deficiency phenotype. Because Ce-lrp-1 regulates endocytosis of cholesterol in mammals, it is conceivable that both Ce-imp-2 and Ce-lrp-1 may contribute to cholesterol uptake in C. elegans. Filipin is a fluorescent polyene antibiotic capable of forming complexes with cholesterol and other sterols in accumulation sites (19). Consistent with the idea that Ce-imp-2 and Ce-lrp-1 functions may contribute to cholesterol uptake or homeostasis pathway, filipin complexes accumulated in the body locations of Ce-imp-2 RNAi animals where displacement of old cuticle normally is initiated (Fig. 3E).
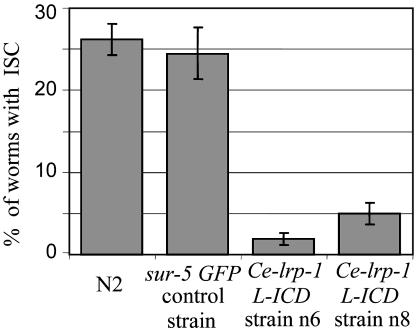
The Cytoplasmic Terminus of Ce-LRP-1 Can Suppress the Ce-imp-2 Defect Induced by RNAi. We tested further whether Ce-imp-2 may be an upstream regulator of Ce-lrp-1- or Ce-lrp-1-dependent pathway in development. We asked whether the Ce-imp-2 deficiency can be compensated by expression of Ce-LRP-1 protein derivate. The intracellular fragment of megalin contains FxN-PxY and PxxPxxP conserved motifs essential for endocytosis and association with adaptor proteins (e.g., Ce-DAB-1) (Fig. 6). A construct encoding a cytoplasmic fragment of Ce-LRP-1 truncated in the N-transmembrane domain (Ce-lrp-1 L-ICD) was designed (Fig. 6 and Supporting Materials and Methods). The transgenic worm strains with extrachromosomal maintenance of Ce-lrp-1 L-ICD under Ce-lrp-1 regulatory regions were generated. Sur-5 GFP plasmid was used as a coinjection marker. The wild-type N2 and strains with extrachromosomal maintenance of transgenic marker sur-5 GFP alone were used as controls. The synchronized population of worms at the L1 stage were transferred to Ce-imp-2 dsRNA plates, and, on day 3, worms were observed for the ISC phenotype as discussed above. The truncated Ce-lrp-1 fragment caused profound rescue of the ISC defect caused by Ce-imp-2 deficiency. This result was found and confirmed in two independent transgenic strains (Fig. 4). The rate of Ce-imp-2 RNAi-induced ISC molting defect in Ce-lrp-1 L-ICD strains was reduced 5- to 10-fold. These data suggest that Ce-imp-2 may be directly involved in the regulation of Ce-LRP-1 (by means of regulation of processing or trafficking) or may regulate other upstream elements interacting with the Ce-lrp-1 pathway (Fig. 10, which is published as supporting information on the PNAS web site).
Fig. 4.
Histogram showing compensatory effect of the NH2-terminally truncated Ce-lrp-1 fragment on the ISC defect caused by Ce-imp-2 deficiency. Two independent transgenic strains (n6 and n8) with extrachromosomal maintenance of Ce-lrp-1 L-ICD construction and control N2 or sur-5 GFP control strains were analyzed. Synchronized strains on L1 stage were placed on Ce-imp-2 dsRNA plates, and P0 worms with ISC phenotype were counted. There was significant reduction in ISC phenotype in Ce-lrp-1 L-ICD transgenic animals as compared with the controls (P < 10-6, n = 2-4 independent trials, n = 1,262 and 1,084 for n6 and n8; n = 1932 and 707 for N2 and sur-5 GFP strains, correspondingly. Bar graphs represent the fractions and 95% confident intervals (CI) expressed in percentage of animals with ISC.
Discussion
Presenilins and IMPAS: Two Related Families of Proteins Regulating Distinct Development Pathways. We reported here a primary in vivo analysis of the functional role of the IMPAS (IMP) family of proteins, which are distinctly homologous to PSs. Both families (IMPs and PSs) share conserved amino acid signatures and structure identities thought to be critical for their intramembrane protease activities. Although a role of PSs in protein trafficking is also postulated, there are many arguments supporting the view that PSs and IMPs are unusual bi-aspartyl polytopic proteases with many structural and functional similarities. Moreover, many amino acid residues in PSs, which are targets for multiple mutations in AD, also occur and are evolutionary conserved in IMPs (15). Nevertheless, we demonstrate here that the Notch protein regulated by PSs is not a major target for the IMP proteins. C. elegans Ce-imp-2 does not facilitate sel-12,hop-1-dependent lin-12,glp-1 signaling. Our data, instead, indicated that IMPs and PSs are functionally distinct families of proteins regulating two different pathways in development. Phylogenic analysis demonstrated that IMPs have a more ancient ancestry prototype (found in Archaea and yeast) than PSs (found in Protista) (12). Thus, beyond the role of IMPs in the development of multicellular organisms, the common cellular functions of IMPs (e.g., in regulation of cell maintenance, proliferation, or cycles) remain be elucidated.
Ce-imp-2-Regulated Development Pathway. Ce-imp-2 is the ortholog of human IMP1/SPP, which is capable of cleaving or, at least, promoting the cleavage of some proteins within hydrophobic domains (14, 15, 17). The proteolytic properties of IMP1/SPP were demonstrated by direct assays in vitro and in cultured cells. The signal peptide peptidase activity of IMP1/SPP is thought to produce short peptide fragments that serve as HLA-E epitopes in mammals (16). However, the major function of human IMP1/SPP or their orthologs or paralogs remained to be uncovered.
The knock-down of Ce-imp-2 performed herein revealed that Ce-imp-2 is required for proper embryonic development and completion of shedding of old cuticle in all four larval molts in C. elegans. A similar ISC phenotype was described for cholesterol-deficient worms. We noticed that loss-of-function in several other genes (daf-9,nhr-23 and nhr-25,Ce-lrp-1,let-512,dab-1) has been described to result in abnormalities that include molting defects with ISC. Intriguingly, all of these genes seem to be involved in lipid steroid homeostasis or signaling. For example, the C. elegans daf-9 gene regulates adult longevity and encodes a P450-cytochrome hydroxylase homologous to human CYP17 and CYP21 involved in biosynthesis of steroids (40). The dauer arrest found in daf-9 mutants was not evident in Ce-imp-2 RNAi worms. Thus, DAF-9 is unlikely to be the direct target regulated by Ce-imp-2. NHR-23 and NHR-25 are nuclear receptors, and NHR-25 is a homolog of mammalian FTZ-F1 receptor or FTZ-F1-like protein involved in regulation of cholesterol homeostasis (41-43). LET-512 is a member of the lipid kinase family that regulates localization and expression of Ce-LRP-1 in C. elegans (44). Ce-DAB-1 is related to a disabled family of cytoplasmic adaptor proteins interacting with LRPs and regulating the endocytosis of megalin and lipoprotein receptors in mammals. Recently, unshed cuticle molting defects induced by Ce-dab-1 RNAi have been reported (45). Finally, Ce-LRP-1 is an ortholog of LRP-mammalian receptors and is suggested to be a close counterpart of megalin (34). One possibility is that Ce-IMP-2 promotes trafficking of Ce-LRP-1 to the cell surface. It is intriguing that expression of the cytoplasmic Ce-LRP-1 domain partially suppressed the molting defect induced by Ce-IMP-2 RNAi (Fig. 4). This finding suggests that signal transduction may be impaired in Ce-imp-2 RNAi worms.
These observations illuminate the intriguing possibility that similar patterns of interacting molecules and pathways are involved in sterol homeostasis and cholesterol-lipoprotein receptor (Ch-LR)-dependent development in C. elegans and mammals. The ISC phenotype may be caused by partial deficiency in Ch-LR signaling and may serve as an excellent phenotype marker in the search for other molecular components of the C. elegans Ch-LR pathway. Known and yet unknown proteins interacting with the Ce-lrp-1 pathway may be potential targets or downstream elements regulated by Ce-imp-2 (Figs. 6 and 10). The role of sel-12 in Notch-signaling is established (46). The Ce-imp-2 controls distinct pathway. In summary, the data described here imply that Ce-IMP-2 and, perhaps, the human ortholog IMP1/SPP are essential regulators of development. Given the ancient origin and diversity of genes of the IMPAS family, their pleotropic functions are anticipated in both vertebrates and invertebrates
Supplementary Material
Acknowledgments
We thank members of our laboratories for technical assistance, Dr. I. Greenwald and Dr. C. Goutte for providing C. elegans mutant strains, Dr. Y. Kohara for providing C. elegans cDNA clones, Dr. R. Kopan (Washington University School of Medicine, St. Louis) for providing Notch1 constructs, and Dr. B. De Strooper (Center for Human Genetics, Katholieke Universiteit Leuven, and Flanders Interuniversity Institute for Biotechnology, Leuven, Belgium) for providing PS1 and PS2 knockout mouse fibroblasts. This work was supported by Alzheimer's Association Grant TLL-03-5777, and, in part, by the Howard Hughes Medical Institute, the Russian Foundation for Basic Research, and the National Institute of Neurological Disorders and Stroke.
Author contributions: C.C.M. and E.I.R. designed research; A.P.G., Y.K.M., and M.C.S. performed research; A.P.G., Y.K.M., C.C.M., and E.I.R. analyzed data; and E.I.R. wrote the paper.
Abbreviations: AD, Alzheimer's disease; ISC, incomplete shedding of cuticle; PS, presenilin; dsRNA, double-stranded RNA; RNAi, RNA interference; LRP, lipoprotein receptor-related protein.
Footnotes
Rogaev, E. I., Grigorenko, A., Ryazanskaya, N., Sherbatich, T., Molyaka, Y., Korovaitseva, G. I. & Dvoryanchikov G. (2001) Sixth Human Genome Meeting, April 19-22, 2001, Edinburgh, N101, p. 28 (abstr.).
References
- 1.Sherrington, R., Rogaev, E. I., Liang, Y., Rogaeva, E. A., Levesque, G., Ikeda, M., Chi, H., Lin, C., Li, G., Holman, K., et al. (1995) Nature 375, 754-760. [DOI] [PubMed] [Google Scholar]
- 2.Rogaev, E. I., Sherrington, R., Rogaeva, E. A., Levesque, G., Ikeda, M., Liang, Y., Chi, H., Lin, C., Holman, K., Tsuda, T., et al. (1995) Nature 376, 775-778. [DOI] [PubMed] [Google Scholar]
- 3.Levy-Lahad, E., Wasco, W., Poorkaj, P., Romano, D. M., Oshima, J., Pettingell, W. H., Yu, C. E., Jondro, P. D., Schmidt, S. D., Wang, K., et al. (1995) Science 269, 973-977. [DOI] [PubMed] [Google Scholar]
- 4.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353-356. [DOI] [PubMed] [Google Scholar]
- 5.De Strooper, B., Saftig, P., Craessaerts, K., Vanderstichele, H., Guhde, G., Annaert, W., Von Figura, K. & Van Leuven, F. (1998) Nature 391, 387-390. [DOI] [PubMed] [Google Scholar]
- 6.De Strooper, B., Annaert, W., Cupers, P., Saftig, P., Craessaerts, K., Mumm, J. S., Schroeter, E. H., Schrijvers, V., Wolfe, M. S., Ray, W. J., et al. (1999) Nature 398, 518-522. [DOI] [PubMed] [Google Scholar]
- 7.Okochi, M., Eimer, S., Bottcher, A., Baumeister, R., Romig, H., Walter, J., Capell, A., Steiner, H. & Haass, C. (2000) J. Biol. Chem. 275, 40925-40932. [DOI] [PubMed] [Google Scholar]
- 8.Xia, W. & Wolfe, M. S. (2003) J. Cell Sci. 116, 2839-2844. [DOI] [PubMed] [Google Scholar]
- 9.Li, X. & Greenwald, I. (1997) Proc. Natl. Acad. Sci. USA 94, 12204-12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Westlund, B., Parry, D., Clover, R., Basson, M. & Johnson, C. D. (1999) Proc. Natl. Acad. Sci. USA 96, 2497-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levitan, D. & Greenwald, I. (1995) Nature 377, 351-354. [DOI] [PubMed] [Google Scholar]
- 12.Grigorenko, A. P., Moliaka, Y. K., Korovaitseva, G. I. & Rogaev, E. I. (2002) Biochemistry (Mosc.) 67, 826-835. [DOI] [PubMed] [Google Scholar]
- 13.Ponting, C. P., Hutton, M., Nyborg, A., Baker, M., Jansen, K. & Golde, T. E. (2002) Hum. Mol. Genet. 11, 1037-1044. [DOI] [PubMed] [Google Scholar]
- 14.Weihofen, A., Binns, K., Lemberg, M. K., Ashman, K. & Martoglio, B. (2002) Science 296, 2215-2218. [DOI] [PubMed] [Google Scholar]
- 15.Moliaka, Y. K., Grigorenko, A., Madera, D. & Rogaev, E. I. (2004) FEBS Lett. 557, 185-192. [DOI] [PubMed] [Google Scholar]
- 16.Lemberg, M. K., Bland, F. A., Weihofen, A., Braud, V. M. & Martoglio, B. (2001) J. Immunol. 167, 6441-6446. [DOI] [PubMed] [Google Scholar]
- 17.McLauchlan, J., Lemberg, M. K., Hope, G. & Martoglio, B. (2002) EMBO J. 21, 3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenner, S. (1974) Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merris, M., Wadsworth, W. G., Khamrai, U., Bittman, R., Chitwood, D. J. & Lenard, J. (2003) J. Lipid Res. 44, 172-181. [DOI] [PubMed] [Google Scholar]
- 20.Rocheleau, C. E., Downs, W. D., Lin, R., Wittmann, C., Bei, Y., Cha, Y. H., Ali, M., Priess, J. R. & Mello, C. C. (1997) Cell 90, 707-716. [DOI] [PubMed] [Google Scholar]
- 21.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 22.Timmons, L. & Fire, A. (1998) Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- 23.Tabara, H., Hill, R. J., Mello, C. C., Priess, J. R. & Kohara, Y. (1999) Development (Cambridge, U.K.) 126, 1-11. [DOI] [PubMed] [Google Scholar]
- 24.Timmons, L., Court D. L. & Fire, A. (2001) Gene 263, 103-112. [DOI] [PubMed] [Google Scholar]
- 25.Mello, C. C., Kramer, J. M., Stinchcomb, D. & Ambros, V. (1991) EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, X. & Greenwald, I. (1996) Neuron 17, 1015-1021. [DOI] [PubMed] [Google Scholar]
- 27.Dewji, N. N. & Singer, S. J. (1997) Proc. Natl. Acad. Sci. USA 94, 14025-14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyborg, A. C., Kornilova, A. Y., Jansen, K., Ladd, T. B., Wolfe, M. S. & Golde, T. E. (2004) J. Biol. Chem. 279, 15153-15160. [DOI] [PubMed] [Google Scholar]
- 29.Schroeter, E. H., Kisslinger, J. A. & Kopan, R. (1998) Nature 393, 382-386. [DOI] [PubMed] [Google Scholar]
- 30.Wolfe, M. S., Xia, W., Ostaszewski, B. L., Diehl, T. S., Kimberly, W. T. & Selkoe, D. J. (1999) Nature 398, 513-517. [DOI] [PubMed] [Google Scholar]
- 31.Priess, J. R., Schnabel, H. & Schnabel, R. (1987) Cell 51, 601-611. [DOI] [PubMed] [Google Scholar]
- 32.Doyle, T. G., Wen, C. & Greenwald, I. (2000) Proc. Natl. Acad. Sci. USA 97, 7877-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, G., Nishimura, M., Arawaka, S., Levitan, D., Zhang, L., Tandon, A., Song, Y. Q., Rogaeva, E., Chen, F., Kawarai, T., et al. (2000) Nature 407, 48-54. [DOI] [PubMed] [Google Scholar]
- 34.Yochem, J., Tuck, S., Greenwald, I. & Han, M. (1999) Development 126, 597-606. [DOI] [PubMed] [Google Scholar]
- 35.Silberkang, M., Havel, C. M., Friend, D. S., McCarthy, B. J. & Watson, J. A. (1983) J. Biol. Chem. 258, 8503-8511. [PubMed] [Google Scholar]
- 36.Bottjer, K. P., Weinstein, P. P. & Thompson, M. J. (1985) Comp. Biochem. Physiol. B 82, 99-106. [DOI] [PubMed] [Google Scholar]
- 37.Kurzchalia, T. V. & Ward, S. (2003) Nat. Cell Biol. 5, 684-688. [DOI] [PubMed] [Google Scholar]
- 38.Thummel, C. S. (1996) Trends Genet. 12, 306-310. [DOI] [PubMed] [Google Scholar]
- 39.Shim, Y. H., Chun, J. H., Lee, E. Y. & Paik, Y. K. (2002) Mol. Reprod. Dev. 61, 358-366. [DOI] [PubMed] [Google Scholar]
- 40.Jia, K., Albert, P. S. & Riddle, D. L. (2002) Development (Cambridge, U.K.) 129, 221-231. [DOI] [PubMed] [Google Scholar]
- 41.Kostrouchova, M., Krause, M., Kostrouch, Z. & Rall, J. E. (2001) Proc. Natl. Acad. Sci. USA 98, 7360-7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nitta, M., Ku, S., Brown, C., Okamoto, A. Y. & Shan, B. (1999) Proc. Natl. Acad. Sci. USA 96, 6660-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gissendanner, C. R. & Sluder, A. E. (2000) Dev. Biol. 221, 259-272. [DOI] [PubMed] [Google Scholar]
- 44.Roggo, L., Bernard, V., Kovacs, A. L., Rose, A. M., Savoy, F., Zetka, M., Wymann, M. P. & Muller, F. (2002) EMBO J. 21, 1673-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamikura, D. M. & Cooper, J. A. (2003) Genes Dev. 17, 2798-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selkoe D. & Kopan R. (2003) Annu. Rev. Neuroscience. 26, 565-597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.