Abstract
Legumes are an important plant functional group since they can form a tripartite symbiosis with nitrogen-fixing Rhizobium bacteria and phosphorus-acquiring arbuscular mycorrhizal fungi (AMF). However, not much is known about AMF community composition in legumes and their root nodules. In this study, we analyzed the AMF community composition in the roots of three nonlegumes and in the roots and root nodules of three legumes growing in a natural dune grassland. We amplified a portion of the small-subunit ribosomal DNA and analyzed it by using restriction fragment length polymorphism and direct sequencing. We found differences in AMF communities between legumes and nonlegumes and between legume roots and root nodules. Different plant species also contained different AMF communities, with different AMF diversity. One AMF sequence type was much more abundant in legumes than in nonlegumes (39 and 13%, respectively). Root nodules contained characteristic AMF communities that were different from those in legume roots, even though the communities were similar in nodules from different legume species. One AMF sequence type was found almost exclusively in root nodules. Legumes and root nodules have relatively high nitrogen concentrations and high phosphorus demands. Accordingly, the presence of legume- and nodule-related AMF can be explained by the specific nutritional requirements of legumes or by host-specific interactions among legumes, root nodules, and AMF. In summary, we found that AMF communities vary between plant functional groups (legumes and nonlegumes), between plant species, and between parts of a root system (roots and root nodules).
Arbuscular mycorrhizal fungi (AMF) are among the most important plant symbionts. AMF, which are fungi belonging to the phylum Glomeromycota (34), form a symbiosis with about 60% of all terrestrial plant species. In exchange for carbohydrates they provide plants with phosphorus and other immobile nutrients. These mutualistic soil fungi also can protect plants against pathogens and drought (5, 9, 23, 36). As phosphorus is a major limiting nutrient in many ecosystems, AMF play a key role in ecosystem functioning. They enhance plant growth and can have a big influence on plant community structure due to the differential supply of nutrients (13, 44).
The identity and diversity of AMF also are important determinants of plant community structure. Plant species vary in their responses to AMF species, and AMF diversity promotes plant diversity, biomass acquisition, and nutrient capture (43, 44). As AMF have a great impact on plant community structure, it is important to investigate AMF community composition in different plant species. Different plant species are colonized by different AMF communities (4, 16, 41, 42), and some plant-AMF combinations are more likely to occur than others under field conditions. Whether these plant-AMF combinations have a functional relationship is unclear. Helgason et al. (16) showed that Glomus hoi, which was found almost exclusively in the roots of Acer pseudoplatanus in the field, was the only fungal species (of four species tested) to benefit this plant species in pot experiments.
Naturally occurring plant-AMF combinations may indicate functional relationships, and different plant types may host different AMF communities. Legumes form a special plant functional group due to their symbiosis with rhizobia, which can fix atmospheric nitrogen (37). Legumes contribute substantially to nitrogen input and the productivity of many terrestrial ecosystems (8, 45). Rhizobial nitrogen fixation provides legumes with an additional nitrogen source, but it requires large amounts of energy and phosphorus (2, 31). The amount of phosphorus delivered by different AMF species varies (20, 27, 36), and legumes might preferentially associate with specific AMF that are efficient in supplying phosphorus. AMF also enhance nodulation and nitrogen fixation, but the extent of these effects is dependent on the AMF species (19, 40).
There have been relatively few studies of AMF communities in legumes, and even less is known about AMF colonization of root nodules, the root organs in which nitrogen fixation takes place. AMF can colonize root nodules under laboratory conditions (3, 47), but it is not known whether root nodules are colonized by AMF under field conditions or which AMF species are responsible for the colonization. Nodules are different from roots in structure, function, and nutritional requirements and capabilities. Nodules result from dividing root cortical cells. They contain nitrogen-fixing bacteria and have higher energy and phosphorus demands than the roots (2, 31). Nodule formation also results in physiological changes, including responses related to plant defense reactions (33, 46). These differences suggest that the AMF communities in roots and nodules might differ as well.
Our objective in this study was to describe AMF communities in six co-occurring plant species that are common in dune grasslands, with an emphasis on the legumes and their root nodules. We hypothesized (i) that the AMF communities in legumes are different from those in nonlegumes, (ii) that AMF colonize root nodules under field conditions, (iii) that the AMF communities found in root nodules differ from those found in roots, and (iv) that AMF communities and AMF diversity are different in different plant species. Determining which AMF colonize legume roots and nodules and co-occurring plant species under natural conditions can contribute to our understanding of interactions between legumes and AMF within plant communities and may help identify specific nutritional interactions in the tripartite legume-AMF-Rhizobium symbiosis.
MATERIALS AND METHODS
Sampling.
Plants were collected in June 2002. The sampling area was a species-rich dry dune grassland in the north of Holland (Provinciale Waterleidingduinen; coordinates, 52°40′N, 4°39′W). The plant community was dominated by Festuca ovina and Anthoxanthum odoratum and contained a large number of subordinate species, including Plantago lanceolata, Hieracium pilosella, and Lotus corniculatus. Three samples (27 by 27 cm) of turf with vegetation (samples A, B, and C) were collected from locations that were approximately 2 m apart. Fresh-looking roots of six plant species, one grass (F. ovina), two herbs (P. lanceolata and H. pilosella), and three legumes (L. corniculatus, Trifolium repens, and Ononis repens), were collected from each turf sample. O. repens was analyzed in only two turf samples, because this plant species was not present in turf sample C. Consequently, we used two replicates for O. repens and three replicates for the other plant species. The roots were washed, and the legume roots separated into roots and root nodules. If sufficient root material could be obtained from the turf samples, then 15 to 30 cm of roots was analyzed per sample. For O. repens, it was not possible to evaluate more than 10 cm of roots. The number of root nodules obtained from the turf samples was different for different plant samples (Table 1). Roots and nodules were dried at room temperature (18 to 22°C) in petri dishes containing silica gel that were closed with Parafilm, and they were stored for about 2 weeks until DNA was extracted. A subset of each sample was used to determine the degree of root colonization by AMF. Shoots of the plants also were stored on silica gel for nitrogen analyses.
TABLE 1.
Number of nodules analyzed in each sample
| Plant species | No. of nodules in turf sample:
|
||
|---|---|---|---|
| A | B | C | |
| L. corniculatus | 10 | 3 | 2 |
| T. repens | 15 | 8 | 13 |
| O. repens | 4 | 1 | |
Molecular analysis.
DNA was extracted from roots and nodules by a cetyltrimethylammonium bromide extraction method (12), followed by an additional purification step with a Strataprep PCR purification kit (Stratagene, La Jolla, Calif.). Partial AMF 18S ribosomal DNA was amplified from total DNA extracts with the universal eukaryotic primer NS31 (35) and primer AM1, designed to amplify AMF 18S ribosomal DNA sequences but not plant sequences (14). PCRs were performed by using mixtures (final volume, 50 μl) containing 1.2 U of Pfu proofreading enzyme (Promega, Madison, Wis.), each deoxynucleoside triphosphate at a concentration of 0.2 mM, 10 pmol of each primer, 0.05% nonfat dry milk powder, and the reaction buffer supplied. The cycling regimen was one cycle of 94°C for 3 min, 58°C for 1 min, and 72°C for 1.5 min, followed by 29 cycles of 94°C for 30 s, 60°C for 1 min, and 72°C for 1.5 min. PCR products were cloned into pPCR-Script Amp SK(+) and transformed into Escherichia coli XL10-Gold according to the manufacturer's instructions (PCR-Script Amp cloning kit; Stratagene). Positive clones were reamplified with the NS31-AM1 primer pair. Restriction fragment length polymorphism (RFLP) patterns were determined for 20 to 24 clones per sample by digesting the PCR products with HinfI or Hsp92II. A total of 562 clones were examined, and depending on the relative abundance, one to eight clones per RFLP pattern were sequenced. Forty-seven clones were sequenced (BaseClear, Leiden, The Netherlands) by using the T3 and T7 plasmid primers after PCR amplification with the T3-T7 primer pair.
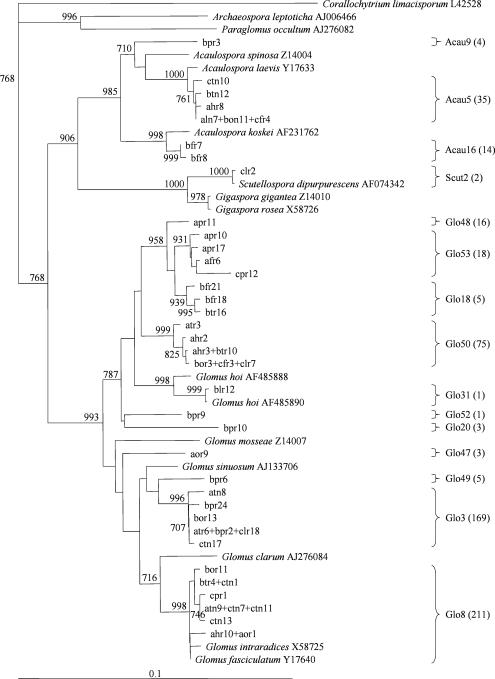
The CLUSTALX program (39) was used for multiple alignment and neighbor-joining phylogenetic analysis (32); Corallochytrium limacisporum, a putative choanozoan (6), was used as the outgroup. The phylogenetic tree (Fig. 1) includes all of the sequences found in this study and sequences of cultured AMF species from different taxa. For each of our sequences, the AMF species from the GenBank database that is most closely related was included in the tree.
FIG. 1.
Neighbor-joining phylogenetic tree showing the relationship among the AMF sequence types obtained in this study and sequences from cultured AMF species. Bootstrap values of >70% are indicated at the nodes (1,000 replicates). The cultured AMF species that are most closely related to each sequence are included. Individual clones are identified by turf sample (a, turf sample A; b, turf sample B; c, turf sample C), plant species (f, F. ovina; p, P. lanceolata; h, H. pilosella; l, L. corniculatus; t, T. repens; o, O. repens), and plant organ (r, roots; n, nodules). The total number of clones attributed to each sequence type on the basis of their RFLP patterns is indicated in parentheses.
We used a phylogenetic approach to classify the new AMF sequence types (7, 41). AMF communities are characterized by the percentages of clones with the different AMF sequence types. The percentage of clones with a certain sequence type is assumed to represent the relative abundance of this sequence type in the root. This analysis assumes that all DNA is equally likely to be extracted, amplified, ligated, and transformed (15).
AMF infection percentage.
Dried roots were rehydrated, cleared in 10% KOH, and stained with trypan blue (25). The modified line intersection method (22) was used to determine the percentage of root length colonized by AMF. For each sample, 50 intersections were examined. The amount of root material of O. repens examined was not sufficient to determine AMF infection.
Nitrogen analysis.
Dried shoots of plants used for AMF community analysis were ground for 5 min at 30 Hz in a mixer mill (MM200; Retsch, Haan, Germany). Nitrogen concentrations were determined with a continuous-flow isotope ratio mass spectrometer (Delta Plus; ThermoQuest Finnigan, Bremen, Germany) coupled with an elemental analyzer (NC2500; ThermoQuest Italia, Rodana, Italy).
Microscopic analysis.
The numbers of nodules obtained from the turf samples were very small, so we also collected nodules from pots containing L. corniculatus. These pots were inoculated with root nodules from the same field site. The nodules were cleared in 10% KOH and stained with trypan blue (25). Nodules were examined with an Axioplan 2 imaging microscope (Carl Zeiss, Göttingen, Germany), and pictures were taken with an AxioCam MRc5 camera coupled to the AxioVision software (Carl Zeiss).
Statistical analysis.
We tested whether the abundance of AMF sequence types varied between legumes and nonlegumes and between legume roots and root nodules. For this analysis, data for the three nonlegumes, for the three legumes, and for the nodules of the three legumes were pooled, which resulted in nine, eight, and eight replicates, respectively. Our data had neither a normal distribution of error terms nor constant error variance among treatments, so a nonparametric test was required. We used the Mann-Whitney U test (SPSS, version 10.1; exact significance). However, multiple testing (every AMF sequence type was tested separately) increases the chance of finding a significant result, so a sequential Bonferroni analysis was performed as a post hoc test (17, 30). Rare AMF sequence types may have shown nonsignificant differences because of the small numbers involved, but including them would have severely reduced the statistical power of the sequential Bonferroni analysis. Thus, we excluded sequence types with five clones or less, which resulted in two tests with each of six sequence types (Glo3, Glo8, Glo50, Glo53, Glo48, and Acau16 when legumes were compared with nonlegumes; and Glo3, Glo8, Glo50, Glo48, Acau5, and Acau16 when legume roots and root nodules were compared). Linear regression analysis (SPSS, version 10.1) was used to test whether there was a correlation between the nitrogen concentration in the plants (shoots) and the percentage of AMF sequence type Glo8 in the roots.
Nucleotide sequence accession numbers.
Nucleotide sequences determined in this study have been deposited in the GenBank database under accession numbers AY512345 to AY512380. One representative was deposited for each sequence.
RESULTS
Phylogeny.
Twenty-one RFLP patterns were identified for the 562 AMF 18S ribosomal DNA fragments. This number was reduced to 15 sequence types after representatives of the 21 RFLP patterns were sequenced (Fig. 1). A number of sequence types were related to cultured AMF species. These were Glo8 for Glomus intraradices and Glomus fascicultum, Glo31 for G. hoi, Acau5 for Acaulospora laevis, and Scut2 for Scutellospora dipurpurescens. The other 11 sequence types represented AMF species not included in the databases. Our sequences represent three different genera, Glomus (507 clones), Acaulospora (53 clones), and Scutellospora (2 clones). No sequences of the Archaeosporaceae or the Paraglomaceae were detected, which was expected since the AM1 primer does not amplify the 18S ribosomal DNA sequences of these AMF taxa (28).
AMF communities in legumes.
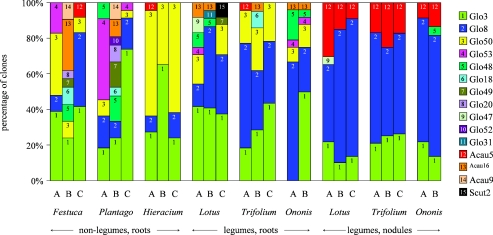
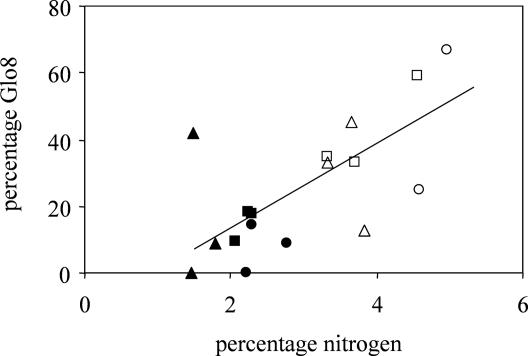
The compositions of AMF communities from roots and nodules of six different plant species in three turf samples were determined based on the percentages of clones of the 15 AMF sequence types (Fig. 2). The most common sequence type, Glo3, was present in 23 of the 24 samples, and 30% of all clones had this sequence type. Glo8 was common in legume roots (39%) and nodules (63%) but was significantly less common in nonlegumes (13%) (Table 2). The relative abundance of Glo8 in roots was positively correlated with the plant nitrogen concentration (Fig. 3), but this relationship was confounded since Glo8 was more common in legumes, which contain more nitrogen than nonlegumes. Glo8 was even more common in nodules, which have higher nitrogen concentrations than the rest of the root (2, 31).
FIG. 2.
AMF communities in roots and nodules of different plant species. AMF communities are represented by the percentages of clones of AMF sequence types. Each color corresponds to a sequence type. Festuca, F. ovina; Plantago, P. lanceolata; Hieracium, H. pilosella; Lotus, L. corniculatus; Trifolium, T. repens; Ononis, O. repens; A, turf sample A; B, turf sample B; C, turf sample C.
TABLE 2.
Results of Mann-Whitney U tests comparing AMF sequence types for nonlegumes and legumes and for legume roots and legume nodules
| Sequence type | % of AMF community
|
P value
|
|||
|---|---|---|---|---|---|
| Nonlegumes | Legumes | Nodules | Nonlegumes-legumes | Legumes-Nodules | |
| Glo3 | 37 | 33 | 19 | 0.963 | 0.050 |
| Glo8 | 13 | 39 | 63 | 0.006a | 0.010a |
| Glo50 | 25 | 15 | 0.0 | 0.815 | 0.002a |
| Glo53 | 7.5 | 1.6 | 0.815 | ||
| Glo48 | 3.7 | 4.2 | 0.6 | 0.815 | 0.195 |
| Acau5 | 0.6 | 17 | 0.000a | ||
| Acau16 | 4.2 | 3.2 | 0.0 | 0.370 | 0.038 |
Values are significantly different as determined by a sequential Bonferroni post hoc test (P ≤ 0.05).
FIG. 3.
Relationship between plant nitrogen concentration and proportion of sequence type Glo8 in an AMF community. R2 = 0.46; P = 0.003; y = 12x − 10.5. ▴, F. ovina; ▪, P. lanceolata; •, H. pilosella; ▵, L. corniculatus; □, T. repens; ○, O. repens. Closed symbols represent nonlegumes, and open symbols represent legumes.
AMF communities in root nodules.
Legume root nodules had a characteristic AMF community that was clearly different from that of legume roots (Fig. 2). Significant differences were found in the relative frequencies of three sequence types, Glo8, Glo50, and Acau5 (Table 2). Sequence type Acau5 is of particular interest since it was found very frequently (31 of 35 clones) in root nodules. Sequence type Glo50 was never found in nodules, even though it was found in almost all of the root samples (15 of 17 samples). Nodules contained fewer sequence types (5 types) than legume roots (12 types). More than 50% of the AMF community both in root nodules and in legume roots consisted of Glo3 and Glo8. The AMF community composition was similar in all nodules and independent of the host species, turf sample, or the number of nodules (Table 1 and Fig. 2).
Microscopic analysis of root nodules from L. corniculatus pot cultures showed that AMF originating from the field site were able to colonize root nodules (Fig. 4). Hyphae were observed in the outer cortical layers and also outside the nodules (Fig. 4A). Spores were also present in some nodules (Fig. 4B), confirming the findings of Vidal-Dominguez et al. (47). Deeper layers of the nodules stained heavily blue, which made them difficult to examine.
FIG. 4.
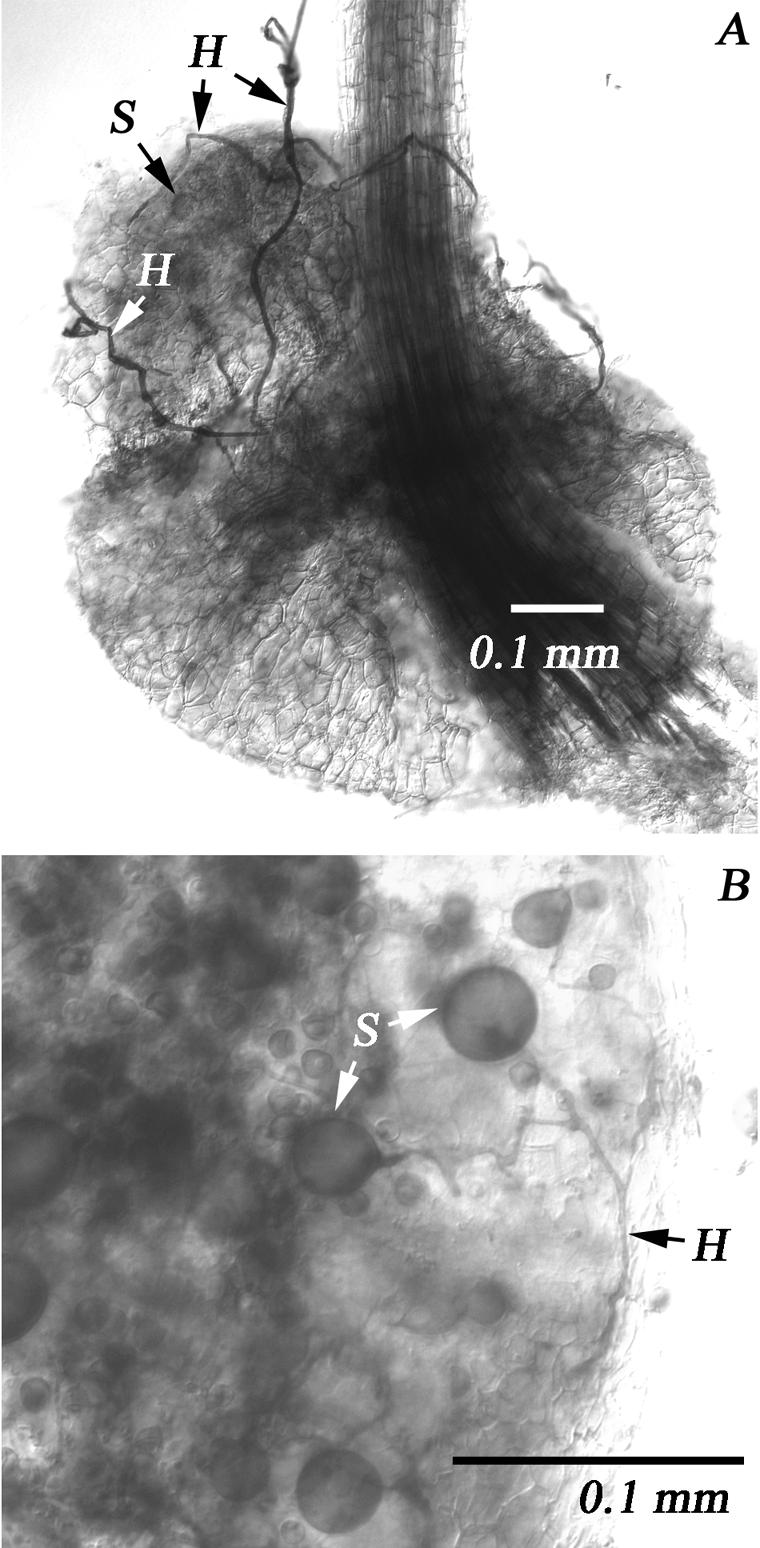
Mycorrhizal infection of root nodules. (A) Overview of a root nodule with attached hyphae (H) and a spore (S) inside the nodule. (B) Detail of a root nodule colonized by hyphae and spores.
AMF diversity in different plant species.
AMF communities (Fig. 2) and their diversity (Table 3) varied by plant species. The AMF communities of H. pilosella had the lowest diversity with the lowest number of AMF sequence types. Sequence type Glo50 was common in H. pilosella roots, on average accounting for 52% of the AMF community in this host, but accounted for only 13% of the communities from the roots of other plant species. H. pilosella had similar AMF communities in all three turf samples. In contrast, F. ovina and P. lanceolata had diverse AMF communities and contained a large number of different AMF sequence types (Table 3). These AMF communities were different in the different turf samples, but they were similar in the same turf sample. In turf sample A, F. ovina and P. lanceolata each had five sequence types, four of which were the same. In turf sample B, they had eight and nine sequence types, respectively, seven of which were the same. In turf sample C, they shared three of four sequence types.
TABLE 3.
Shannon diversity indices and total number of AMF sequence types for AMF communities from roots of six plant species from a dune grassland
| Plant species | Shannon diversity index
|
No. of AMF sequence types | |
|---|---|---|---|
| Avg ± SE | Totala | ||
| F. ovina | 1.43 ± 0.23 | 1.92 | 11 |
| P. lanceolata | 1.43 ± 0.37 | 1.89 | 11 |
| H. pilosella | 0.86 ± 0.11 | 0.97 | 4 |
| L. corniculatus | 1.40 ± 0.15 | 1.62 | 10 |
| T. repens | 1.19 ± 0.12 | 1.29 | 6 |
| O. repens | 1.20 ± 0.15 | 1.44 | 6 |
| All plants | 1.25 ± 0.09 | 1.77 | 15 |
The total Shannon diversity index was calculated by pooling data from three turf samples.
The infection percentages for the different root samples ranged from 42 to 100%, showing that all plants were well colonized and indicating that the data in Fig. 2 are ecologically relevant. The infection levels (average ± standard error) were 63% ± 1.3%, 83% ± 4.4%, 47% ± 3.5%, 91% ± 5.9%, and 85% ± 2.4% for F. ovina, P. lanceolata, H. pilosella, L. corniculatus, and T. repens, respectively.
DISCUSSION
AMF communities in legumes.
We found that the AMF communities in legumes differ from those in nonlegumes, and one AMF sequence type, Glo8, is significantly more frequent in legumes than in nonlegumes (Fig. 2 and Table 2). The characteristic responsible for this association is not known. The positive correlation between Glo8 and plant nitrogen concentration (Fig. 3), although not significant when legumes and nonlegumes were analyzed separately, suggests that this AMF sequence type is specialized in plants with high nitrogen concentrations. This conclusion is consistent with previous observations that changes in AMF community composition occurred after N fertilization (11) and that Glomus intraradices, which corresponds to the Glo8 sequence type, was the only AMF species whose level increased after fertilization with N plus P (21). However, AMF associated with legumes also might be more efficient than other AMF in supplying phosphorus or other limiting nutrients (e.g., Cu and Zn) that are important for nodulation and nitrogen fixation. If this is true, then successful legumes must associate with nitrogen-fixing bacteria and with specific AMF. In a study of a single grass, Agrostis capillaris, and a legume, T. repens, in an upland grassland in Scotland, specific AMF were associated with each plant species (41). These AMF sequence types were not the same as those which we observed, however, perhaps reflecting the difference in the environments.
The importance of the tripartite legume-AMF-Rhizobium symbiosis for agriculture and ecology has been recognized, and several attempts have been made to find the most effective combinations of AMF and Rhizobium species (29, 40, 48). Because of its abundance in nonagricultural legumes, Glo8 is a good candidate for further studies of the relative effects of different AMF on legume growth.
AMF communities in root nodules.
AMF can colonize legume root nodules under laboratory conditions (3, 47), but the AMF community composition of root nodules under field conditions had not been examined previously. Root nodules had a characteristic AMF community that clearly differed from the root community, but the communities were similar for nodules from three different legume species collected in three spatially discrete samples (Fig. 2 and Table 2). Sequence type Glo8 was relatively more abundant in root nodules, and sequence type Acau5 was found almost exclusively in root nodules. In contrast, sequence type Glo50 was common in roots and was never found in nodules. These data suggest that nodules inhibit or enhance colonization by certain AMF sequence types.
There are several possible explanations for the root nodule specificity. One hypothesis is that the ability to procure one or more nutrients (e.g., P, Cu, or Zn) might predispose an AMF to nodule colonization, because nodules have a relatively high nutrient demand. Alternatively, some AMF (e.g., Glo8 and Acau5 sequence types) might be attracted by the relatively high nitrogen concentrations in nodules. AMF found in nodules also could preferentially interact with the Rhizobium infection process, as the AMF and Rhizobium symbioses have many similarities and share some important steps (1, 26, 38). Legume mutants that cannot form symbioses with rhizobia also usually have difficulty establishing AMF symbioses (10), and rhizobial signals can influence AMF colonization (49). Finally, the physiology in nodules differs from that in the roots, and rhizobial infection alters root exudation and induces defense-like responses in plants (33, 46). These processes could affect the ability of some AMF to colonize the root nodules. Our microscopic studies confirmed that AMF from the field site used could colonize root nodules and that AMF were present as hyphae surrounding the nodules and as hyphae and spores inside the nodules (Fig. 4).
AMF diversity in different plant species.
We confirmed that the AMF community composition depends on the host plant species (4, 16, 41, 42). We also found differences in terms of both the diversity and the variability of the AMF communities (Fig. 2 and Table 3). H. pilosella hosted AMF communities with low diversity which were similar in all three turf samples, while F. ovina and P. lanceolata hosted diverse AMF communities that differed among the turf samples. Thus, plant species may have various degrees of selectivity for AMF species that range from selective specialists to nonselective generalists. P. lanceolata often is used for AMF isolation, a practice which we encourage due to our observation of its low AMF selectivity.
Although the AMF communities of F. ovina and P. lanceolata were highly variable among the three turf samples, these plant species had similar AMF communities in the same turf sample. This result suggests that the local availability of fungi, rather than a species-specific interaction, is the primary determinant of AMF community composition for these species. However, the sample sizes were small, and more extensive studies are needed to confirm these observations with statistical tests. Relatively large differences in AMF communities can be seen across distances as small as a few meters. Thus, generalists such as F. ovina and P. lanceolata probably are good indicators of which AMF are present at a particular location.
The most abundant fungal types in this study, Glo3 and Glo8, were present in almost all plant species and in roots and root nodules. These fungi appear to be generalist fungi, because they have been detected in a wide range of plant species and in almost all ecosystems investigated so far (24).
Data from several studies, in all of which the same PCR-RFLP technique was used to determine the AMF communities, are now available. The AMF diversity as measured with a Shannon diversity index ranges from 0.40 for an arable field to 2.3 for a tropical rain forest (14, 18). The Shannon diversity index in our study was moderately high, 1.8. Factors such as sample size, sample area, and number of clones analyzed influence the AMF diversity values. The number of plant species analyzed also is a factor, but we now think that the identity of the plant species examined could be even more important (Table 3). For example, a Shannon diversity index of 1.7 was reported in a grassland study conducted with Agrostis capillaris and T. repens (41). The Shannon diversity index based on our data for only F. ovina and T. repens was 1.7, but with H. pilosella and T. repens the Shannon diversity index was only 1.2, with F. ovina and P. lanceolata the Shannon diversity index was 2.0, and with all six plant species combined the Shannon diversity index was 1.8. Thus, AMF diversity is more dependent on the specific plant species than on the number of plant species, and plant species for AMF diversity studies should be carefully selected, especially if different ecosystems are compared.
In conclusion, AMF communities may vary between plant functional groups (e.g., legumes and nonlegumes), between plant species, and between parts of a root system (e.g., roots and root nodules). Further studies are needed to determine whether the differences in AMF communities are functionally important for plant growth, nodule performance, or the composition and structure of plant communities.
Acknowledgments
We thank Martin Braster and Henk van Verseveld of the microbial physiology group of Vrije Universiteit Amsterdam for allowing T.R.S. to work in their laboratory, Gerda Lamers of the Institute of Biology, Leiden University, for her help with the microscopic pictures, and NV PWN Waterleidingbedrijf Noord-Holland for providing access to the field site.
This research was supported by a grant from the Dutch Science Foundation (NWO-Vernieuwingsimpuls; grant 016.001.023). The collaboration between the University of York and Vrije Universiteit Amsterdam was supported by a LINKECOL grant provided by the European Science Foundation.
REFERENCES
- 1.Albrecht, C., R. Geurts, and T. Bisseling. 1999. Legume nodulation and mycorrhizae formation; two extremes in host specificity meet. EMBO J. 18:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida, J. P. F., U. A. Hartwig, M. Frehner, J. Nosberger, and A. Lüscher. 2000. Evidence that P deficiency induces N feedback regulation of symbiotic N2 fixation in white clover (Trifolium repens L.). J. Exp. Bot. 51:1289-1297. [PubMed] [Google Scholar]
- 3.Baird, L. M., and K. J. Caruso. 1994. Development of root nodules in Phaseolus vulgaris inoculated with Rhizobium and mycorrhizal fungi. Int. J. Plant Sci. 155:633-639. [Google Scholar]
- 4.Bidartondo, M. I., D. Redecker, I. Hijri, A. Wiemken, T. D. Bruns, L. Domínguez, A. Sérsic, J. R. Leake, and D. J. Read. 2002. Epiparasitic plants specialized on arbuscular mycorrhizal fungi. Nature 419:389-392. [DOI] [PubMed] [Google Scholar]
- 5.Borowicz, V. A. 2001. Do arbuscular mycorrhizal fungi alter plant-pathogen relations? Ecology 82:3057-3068. [Google Scholar]
- 6.Cavalier-Smith, T., and M. T. E. P. Allsopp. 1996. Corallochytrium, an enigmatic non-flagellate protozoan related to choanoflagellates. Eur. J. Protistol. 32:306-310. [Google Scholar]
- 7.Clapp, J. P., T. Helgason, T. J. Daniell, and J. P. W. Young. 2002. Genetic studies of the structure and diversity of arbuscular mycorrhizal fungal communities, p. 201-224. In M. G. A. van der Heijden and I. R. Sanders (ed.), Mycorrhizal ecology. Springer-Verlag, Berlin, Germany.
- 8.Cleveland, C. C., A. R. Townsend, D. S. Schimel, H. Fisher, R. W. Howarth, L. O. Hedin, S. S. Perakis, E. F. Latty, J. C. Von Fischer, A. Elseroad, and M. F. Wasson. 1999. Global patterns of terrestrial biological nitrogen (N2) fixation in natural ecosystems. Global Biogeochem. Cycles 13:623-645. [Google Scholar]
- 9.Davies, F. T., J. R. Potter, and R. G. Linderman. 1993. Drought resistance of mycorrhizal pepper plants independent of leaf P concentration—response in gas exchange and water relations. Physiol. Plant. 87:45-53. [Google Scholar]
- 10.Duc, G., A. Trouvelot, V. Gianinazzi-Pearson, and S. Gianinazzi. 1989. First report of non-mycorrhizal plant mutants (myc-) obtained in pea (Pisum sativum L.) and faba bean (Vicia faba L.). Plant Sci. 60:215-222. [Google Scholar]
- 11.Eom, A. H., D. C. Hartnett, G. W. T. Wilson, and D. A. H. Figge. 1999. The effect of fire, mowing and fertilizer amendment on arbuscular mycorrhizas in tallgrass prairie. Am. Midl. Nat. 142:55-70. [Google Scholar]
- 12.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for Basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 13.Grime, J. P., J. M. L. Mackey, S. H. Hillier, and D. J. Read. 1987. Floristic diversity in a model system using experimental microcosms. Nature 328:420-422. [Google Scholar]
- 14.Helgason, T., T. J. Daniell, R. Husband, A. H. Fitter, and J. P. W. Young. 1998. Ploughing up the wood-wide web? Nature 394:431. [DOI] [PubMed] [Google Scholar]
- 15.Helgason, T., A. H. Fitter, and J. P. W. Young. 1999. Molecular diversity of arbuscular mycorrhizal fungi colonising Hyacinthoides non-scripta (bluebell) in a seminatural woodland. Mol. Ecol. 8:659-666. [Google Scholar]
- 16.Helgason, T., J. W. Merryweather, J. Denison, P. Wilson, J. P. W. Young, and A. H. Fitter. 2002. Selectivity and functional diversity in arbuscular mycorrhizas of co-occurring fungi and plants from a temperate deciduous woodland. J. Ecol. 90:371-384. [Google Scholar]
- 17.Holm, S. 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat. 6:65-70. [Google Scholar]
- 18.Husband, R., E. A. Herre, S. L. Turner, R. Gallery, and J. P. W. Young. 2002. Molecular diversity of arbuscular mycorrhizal fungi and patterns of host association over time and space in a tropical forest. Mol. Ecol. 11:2669-2678. [DOI] [PubMed] [Google Scholar]
- 19.Ianson, D. C., and R. G. Linderman. 1993. Variation in the response of nodulating pigeonpea (Cajanus cajan) to different isolates of mycorrhizal fungi. Symbiosis 15:105-119. [Google Scholar]
- 20.Jakobsen, I., L. K. Abbott, and A. D. Robson. 1992. External hyphae of vesicular-arbuscular mycorrhizal fungi associated with Trifolium subterraneum L. I. Spread of hyphae and phosphorus inflow into roots. New Phytol. 120:371-380. [Google Scholar]
- 21.Johnson, N. C. 1993. Can fertilization of soil select less mutualistic mycorrhizae? Ecol. Appl. 3:749-757. [DOI] [PubMed] [Google Scholar]
- 22.McGonigle, T. P., M. H. Miller, D. G. Evans, G. L. Fairchild, and J. A. Swan. 1990. A new method which gives an objective measure of colonisation of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol. 115:495-501. [DOI] [PubMed] [Google Scholar]
- 23.Newsham, K. K., A. H. Fitter, and A. R. Watkinson. 1995. Multi-functionality and biodiversity in arbuscular mycorrhizas. Tree 10:407-411. [DOI] [PubMed] [Google Scholar]
- 24.Öpik, M., M. Moora, J. Liira, U. Kõljalg, M. Zobel, and R. Sen. 2003. Divergent arbuscular mycorrhizal fungal communities colonize roots of Pulsatilla spp. in boreal Scots pine forest and grassland soils. New Phytol. 160:581-593. [DOI] [PubMed] [Google Scholar]
- 25.Phillips, J. M., and D. S. Hayman. 1970. Improved procedure for clearing roots and staining parasitic and vesicular-arbuscular fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55:158-161. [Google Scholar]
- 26.Provorov, N. A., A. Y. Borisov, and I. A. Tikhonovich. 2002. Developmental genetics and evolution of symbiotic structures in nitrogen-fixing nodules and arbuscular mycorrhiza. J. Theor. Biol. 214:215-232. [DOI] [PubMed] [Google Scholar]
- 27.Ravnskov, S., and I. Jakobsen. 1995. Functional compatibility in arbuscular mycorrhizas measured as hyphal P transport to the plant. New Phytol. 129:611-618. [Google Scholar]
- 28.Redecker, D., J. B. Morton, and T. D. Bruns. 2000. Ancestral lineages of arbuscular mycorrhizal fungi (Glomales). Mol. Phylogenet. Evol. 14:276-284. [DOI] [PubMed] [Google Scholar]
- 29.Requena, N., E. Perez-Solis, C. Azcón-Aguilar, P. Jeffries, and J. M. Barea. 2001. Management of indigenous plant-microbe symbioses aids restoration of desertified ecosystems. Appl. Environ. Microbiol. 67:495-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice, W. R. 1989. Analyzing tables of statistical tests. Evolution 43:223-225. [DOI] [PubMed] [Google Scholar]
- 31.Sa, T. M., and D. W. Israel. 1991. Energy status and functioning of phosphorus-deficient soybean nodules. Plant Physiol. 97:928-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 33.Schultze, M., and A. Kondorosi. 1998. Regulation of symbiotic root nodule development. Annu. Rev. Genet. 32:33-57. [DOI] [PubMed] [Google Scholar]
- 34.Schüssler, A., D. Schwarzott, and C. Walker. 2001. A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105:1413-1421. [Google Scholar]
- 35.Simon, L., M. Lalonde, and T. D. Bruns. 1992. Specific amplification of 18S fungal ribosomal genes from vesicular-arbuscular endomycorrhizal fungi colonizing roots. Appl. Environ. Microbiol. 58:291-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, S. E., and D. J. Read. 1997. Mycorrhizal symbiosis, 2nd ed. Academic Press, London, United Kingdom.
- 37.Sprent, J. I. 2001. Nodulation in legumes. Royal Botanical Gardens, Kew, United Kingdom.
- 38.Staehelin, C., C. Charon, T. Boller, M. Crespi, and A. Kondorosi. 2001. Medicago truncatula plants overexpressing the early nodulin gene enod40 exhibit accelerated mycorrhizal colonization and enhanced formation of arbuscules. Proc. Natl. Acad. Sci. USA 98:15366-15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valdenegro, M., J. M. Barea, and R. Azcón. 2001. Influence of arbuscular-mycorrhizal fungi, Rhizobium meliloti strains and PGPR inoculation on the growth of Medicago arborea used as model legume for re-vegetation and biological reactivation in a semi-arid Mediterranean area. Plant Growth Regul. 34:233-240. [Google Scholar]
- 41.Vandenkoornhuyse, P., R. Husband, T. J. Daniell, I. J. Watson, J. M. Duck, A. H. Fitter, and J. P. W. Young. 2002. Arbuscular mycorrhizal community composition associated with two plant species in a grassland ecosystem. Mol. Ecol. 11:1555-1564. [DOI] [PubMed] [Google Scholar]
- 42.Vandenkoornhuyse, P., K. P. Ridgway, I. J. Watson, A. H. Fitter, and J. P. W. Young. 2003. Co-existing grass species have distinctive arbuscular mycorrhizal communities. Mol. Ecol. 12:3085-3095. [DOI] [PubMed] [Google Scholar]
- 43.van der Heijden, M. G. A., T. Boller, A. Wiemken, and I. R. Sanders. 1998. Different arbuscular mycorrhizal fungal species are potential determinants of plant community structure. Ecology 79:2082-2091. [Google Scholar]
- 44.van der Heijden, M. G. A., J. N. Klironomos, M. Ursic, P. Moutoglis, R. Streitwolf-Engel, T. Boller, A. Wiemken, and I. R. Sanders. 1998. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396:69-72. [Google Scholar]
- 45.Vandermeer, J. H. 1989. The ecology of intercropping. Cambridge University Press, New York, N.Y.
- 46.Vasse, J., F. de Billy, and G. Truchet. 1993. Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J. 4:555-566. [Google Scholar]
- 47.Vidal-Dominguez, M. T., C. Azcón-Aguilar, and J. M. Barea. 1994. Preferential sporulation of Glomus fasciculatum in the root nodules of herbaceous legumes. Symbiosis 16:65-73. [Google Scholar]
- 48.Xavier, L. J. C., and J. J. Germida. 2002. Response of lentil under controlled conditions to co-inoculation with arbuscular mycorrhizal fungi and rhizobia varying in efficacy. Soil Biol. Biochem. 34:181-188. [Google Scholar]
- 49.Xie, Z., C. Staehelin, H. Vierheilig, A. Wiemken, S. Jabbouri, W. J. Broughton, R. Vögeli-Lange, and T. Boller. 1995. Rhizobial nodulation factors stimulate mycorrhizal colonization of nodulating and nonnodulating soybeans. Plant Physiol. 108:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]