Abstract Abstract
The Balkan Peninsula represents one of the hottest biodiversity spots in Europe. However, the invertebrate fauna of this region is still insufficiently investigated, even in respect of such well-studied organisms as Lepidoptera. Here we use a combination of chromosomal, molecular and morphological markers to rearrange the group of so-called anomalous blue butterflies (also known as ‘brown complex’ of the subgenus Agrodiaetus Hübner, [1822] and as the Polyommatus (Agrodiaetus) admetus (Esper, 1783) species group) and to reveal its cryptic taxonomic structure. We demonstrate that Polyommatus aroaniensis (Brown, 1976) is not as widespread in the Balkans as was previously thought. In fact, it has a dot-like distribution range restricted to the Peloponnese Peninsula in South Greece. Polyommatus orphicus Kolev, 2005 is not as closely related to the Turkish species Polyommatus dantchenkoi (Lukhtanov & Wiemers, 2003) as was supposed earlier. Instead, it is a Balkan endemic represented by two subspecies: Polyommatus orphicus orphicus (Bulgaria) and Polyommatus orphicus eleniae Coutsis & De Prins, 2005 (Northern Greece). Polyommatus ripartii (Freyer, 1830) is represented in the Balkans by an endemic subspecies Polyommatus ripartii pelopi. The traditionally recognized Polyommatus admetus (Esper, 1783) is shown to be a heterogeneous complex and is divided into Polyommatus admetus sensu stricto (the Balkans and west Turkey) and Polyommatus yeranyani (Dantchenko & Lukhtanov, 2005) (east Turkey, Armenia, Azerbaijan and Iran). Polyommatus nephohiptamenos (Brown & Coutsis, 1978) is confirmed to be a species with a dot-like distribution range in Northern Greece. Finally, from Central Greece (Timfristos and Parnassos mountains) we describe Polyommatus timfristos Lukhtanov, Vishnevskaya & Shapoval, sp. n. which differs by its haploid chromosome number (n=38) from the closely related and morphologically similar Polyommatus aroaniensis (n=47-48) and Polyommatus orphicus (n=41-42). We provide chromosomal evidence for three separate south Balkan Pleistocene refugia (Peloponnesse, Central Greece and Northern Greece/South Bulgaria) and stress the biogeographic importance of Central Greece as a place of diversification. Then we argue that the data obtained have direct implications for butterfly karyology, taxonomy, biogeography and conservation.
Keywords: Agrodiaetus, biodiversity, chromosome, COI, conservation, cryptic species, DNA barcode, ITS2, karyotype, mitochondrial marker, Polyommatus timfristos sp. n., protected species, red list
Introduction
The Balkan Peninsula is recognized as a European biodiversity hotspot, with high endemism found in animals and plants (Nicolić et al. 2014, Buj et al. 2015, Bregović and Zagmajster 2016). However, the invertebrate fauna of this region is still insufficiently investigated (Previšić et al. 2016), even in respect of such a well-studied group as Lepidoptera (butterflies and moths) (Sobezyk and Gligorović 2016).
Within Balkan Lepidoptera, the Agrodiaetus Hübner, [1822] blue butterflies are the most complicated group from the taxonomical point of view. The subgenus Agrodiaetus is a distinct monophyletic lineage within the species-rich genus Polyommatus Latreille, 1804 (Talavera et al. 2013a). Adult Agrodiaetus butterflies are small in size with wing span from 1.9 to 3.6 cm. Females are mostly warm brown on the upperside of the wings, whereas males can be either blue or brown. In the latter case, they resemble females. Thus, a species can be classified as either dimorphic or monomorphic depending on the wing color of the males. Most of the Agrodiaetus species have a white streak on the underside of hind wings, and this feature appears to be an apomorphic character of the subgenus Agrodiaetus. However, in a few species and populations this white streak is secondarily reduced or totally absent (Eckweiler and Bozano 2016).
The subgenus Agrodiaetus includes numerous species, subspecies and forms with uncertain taxonomic positions (Eckweiler and Häuser 1997). It was estimated to have originated only about 3 million years ago (Kandul et al. 2004) and radiated rapidly in the Western Palaearctic (Kandul et al. 2007). The last published review of the subgenus includes 120 valid species (Eckweiler and Bozano 2016). Many of them have extremely local ‘dot-like’ distributions that are restricted to particular mountain valleys in the Balkan Peninsula, Asia Minor, Transcaucasus, Iran and Central Asia (Vila et al. 2010, Eckweiler and Bozano 2016).
Although this group has attracted the attention of numerous researchers (e.g. de Lesse 1960a, b, Häuser and Eckweiler 1997, Oliver et al. 1999, Carbonell 2000, 2001, Dantchenko 2000, Przybyłowicz 2000, 2014, ten Hagen and Eckweiler 2001, Skala 2001, Lukhtanov and Dantchenko 2002a, Kandul et al. 2002, 2004, 2007, Wiemers 2003, Schurian and ten Hagen 2003, Vila et al. 2010, Talavera et al. 2013a), a large number of unresolved taxonomic problems still persist in Agrodiaetus.
In most cases, species identification in Agrodiaetus is extremely difficult. The morphology of male genitalia is uniform throughout most of the species and, with a few exceptions (see Coutsis 1985, 1986), at most it can help to separate groups of species, e.g. the Polyommatus dolus (Hübner, 1823) and Polyommatus admetus (Esper, 1783) species groups (see Kolev 2005), but not individual species. The differences in wing pattern and coloration between many Agrodiaetus species are very subtle or nearly lacking (Eckweiler and Bozano 2016).
Despite morphological similarity, the taxonomic and identification problems within the subgenus Agrodiaetus can be solved if chromosomal (de Lesse 1960a,b, Lukhtanov 1989) or molecular markers (Wiemers 2003, Kandul et al. 2004, 2007, Lukhtanov et al. 2005, Stradomsky and Fomina 2013), or their combination (Lukhtanov et al. 2006, 2014, 2015a, Vila et al. 2010, Lukhtanov and Tikhonov 2015, Shapoval and Lukhtanov 2015a,b) are applied. Although chromosome numbers are invariable in many groups of Lepidoptera (Robinson 1971, Lukhtanov 2014, Hernández-Roldán 2016), a few genera demonstrate chromosomal instability, a situation in which multiple closely related species differ drastically from each other by major chromosomal rearrangements, sometimes resulting in high variability in chromosome number (de Lesse 1960a,b, Talavera et al. 2013b). An unusual diversity of karyotypes is the most remarkable characteristic of the subgenus Agrodiaetus. Species of Agrodiaetus exhibit one of the highest ranges in chromosome numbers in the animal kingdom (Lukhtanov 2015). Haploid chromosome numbers in Agrodiaetus range from n=10 in Agrodiaetus caeruleus (Staudinger, 1871) to n=134 in Agrodiaetus shahrami (Skala, 2001) (Lukhtanov and Dantchenko 2002a, Lukhtanov et al. 2005). Additionally, this subgenus demonstrates a high level of karyotypic differentiation with respect to chromosome size (Lukhtanov and Dantchenko 2002b) and variation in number of chromosomes bearing ribosomal DNA clusters (Vershinina et al. 2015). The karyotype is generally stable within species although differences between closely related taxa are often high and provide reliable characters for species delimitation, description and identification (de Lesse 1960a,b, Lukhtanov and Dantchenko 2002a,b).
Molecular studies revealed that subgenus Agrodiaetus consists of 10 monophyletic clades: the Polyommatus transcaspicus (Heyne, 1895) group, the Polyommatus iphigenides (Staudinger, 1886) group, the Polyommatus ershoffii (Lederer, 1869) group, the Polyommatus poseidon (Herrich-Schäffer, 1844) group, the Polyommatus admetus group, the Polyommatus damone (Eversmann, 1841) group, the Polyommatus carmon (Herrich-Schäffer, 1851) group, the Plebejus damon (Denis & Schiffermüller, 1775) group, the Polyommatus dolus group and the Polyommatus actis (Herrich-Schäffer, 1851) group (Kandul et al. 2002, 2004, 2007, Wiemers 2003). They also demonstrated that many species are clearly differentiated with respect to mitochondrial and nuclear DNA sequences. However, this is not a general rule, as the standard mitochondrial DNA barcodes are often identical or nearly identical between closely related taxa and even between morphologically distinct species (Kandul et al. 2004, 2007, Wiemers and Fiedler 2007). Generally, chromosomal characters in Agrodiaetus evolve more quickly than standard DNA barcodes, and because they are usually present as fixed differences, provide better markers for recently evolved taxa than nucleotide substitutions (Lukhtanov et al. 2015a).
Species delimitation is especially difficult within a group of so-called anomalous blue species (known also as ‘brown complex’ of the subgenus Agrodiaetus and as the Polyommatus admetus species complex). This group is composed of multiple species in which both male and female butterflies have similar brown coloration on the upperside of the wings (Lukhtanov et al. 2003).
The group of anomalous blue species includes taxa belonging to two clearly monophyletic and most probably sister clades: the Polyommatus admetus clade (comprises only monomorphic species – Polyommatus admetus, Polyommatus demavendi, Polyommatus khorasanensis. Polyommatus nephohiptamenos, Polyommatus ripartii, Polyommatus pseudorjabovi) and the Polyommatus dolus clade (comprises both monomorphic – Polyommatus alcestis, Polyommatus karacetinae, Polyommatus eriwanensis, Polyommatus interjectus, Polyommatus dantchenkoi, Polyommatus humedasae, Polyommatus aroaniensis, Polyommatus orphicus, Polyommatus timfristos sp. n., Polyommatus fabressei, Polyommatus violetae, Polyommatus valiabadi, Polyommatus rjabovianus; and dimorphic species – Polyommatus dolus, Polyommatus fulgens, Polyommatus menalcas). The anomalous blue butterflies represent a real stumbling block in the Agrodiaetus taxonomy (Lukhtanov et al. 2003, Przybyłowicz et al. 2014). According to Eckweiler and Bozano (2016), the group is distributed in West Palearctic from Spain in the west to Mongolia in the east. The majority of the species have very localized distribution areas concentrated in (1) the Iberian Peninsula, (2) the Balkan Peninsula and (3) west Asia (mostly in the Middle East and Caucasus). Vila et al. (2010) studied in detail the European Agrodiaetus taxa distributed west of the 17th meridian, using a combination of molecular and chromosomal markers (Vila et al. 2010). Chromosomal and molecular markers were also applied to study the taxonomy of the Asian taxa (Lukhtanov et al. 2015a). It is paradoxical that systematic studies based on combined analysis of molecular and chromosomal markers have never been applied to Balkan taxa of the Polyommatus admetus species complex. However, some DNA data can be found in GenBank (Wiemers 2003, Wiemers et al. 2007, 2009, 2010, Lukhtanov et al. 2009, 2015a, Vila et al. 2010, Dincă et al. 2013, Przybyłowicz et al. 2014) and chromosome numbers are known for a few Balkan populations (Coutsis and De Prins 2005, 2007, Kolev 2005).
The goal of the present study is a simultaneous investigation of chromosomal, molecular and morphological diversity in the anomalous blue butterflies from the Balkan Peninsula and interpretation of this diversity in terms of taxonomy. To achieve this goal, the following tasks were set:
To collect specimens of all the taxa of the complex described from the territory of the Balkan Peninsula. To collect specimens from different populations of these taxa.
To study their karyotypes (chromosome number and structure) using standard protocols for staining.
To obtain data on the variability of molecular markers: mitochondrial DNA barcode (COI gene fragment) and nuclear internal transcribed spacer 2 (ITS2). These markers were selected because the usefulness of mitochondrial COI barcodes in taxonomic studies on species-level is generally recognized (Hebert et al. 2004, but see Wiemers and Fiedler 2007), and despite some limitations (Shapoval and Lukhtanov 2015c), internal transcribed spacer 2 was found to be a useful nuclear marker in butterfly taxonomy (Wiemers et al. 2009).
To study the variability of the wing pattern characters which can be potentially useful for delimitation of species and populations (presence/reduction/absence of the white streak on the underside of the hindwings, the development of the marginal marking on the underside of the wings, presence or absence of a white stroke on the underside of the forewings).
To interpret the discovered chromosomal, molecular and morphological diversity in terms of taxonomy using two original methodologies: (1) detecting and taxonomic interpretation of cryptic entities found in sympatry and allopatry using combined analysis of mitochondrial and chromosomal markers (Lukhtanov et al. 2015a), and (2) critical evaluation of pre-existing morphology-based taxonomic hypotheses using DNA barcodes (Lukhtanov et al. 2016).
Material and methods
Taxon sampling
Butterflies for this study were collected in 2008 in the Balkan Peninsula by V.A. Lukhtanov, N.A. Shapoval and L. Rieppel, in 2016 in Hvoyna village (Bulgaria) by E.A. Pazhenkova and in the Tigirekskiy Reservation (the Altai Mountains, Russia) by M.S.Vishnevskaya in 2007 (Fig. 1, Table 1). We paid special attention to collecting the taxa in their type localities: mount Chelmos (Greece: Peloponnese) (type locality of Agrodiaetus alcestis aroaniensis Brown, 1976), mount Falakró near Granítis (Greece, Makedonía, Dráma district) (type locality of Agrodiaetus eleniae Coutsis & De Prins, 2005) and Hvoyna (south Bulgaria, the Rhodopi mts) (type locality of Polyommatus dantchenkoi orphicus). Unfortunately, in our research we did not have an opportunity to study the holotypes of these taxa. Taking into account a possibility of multiple cryptic species within a local area even in well-studied European butterflies (Dincă et al. 2011, 2013b), in each place we managed to collect (and then to study) as many individuals as possible paying special attention to the specimens with unusual or intermediate morphology.
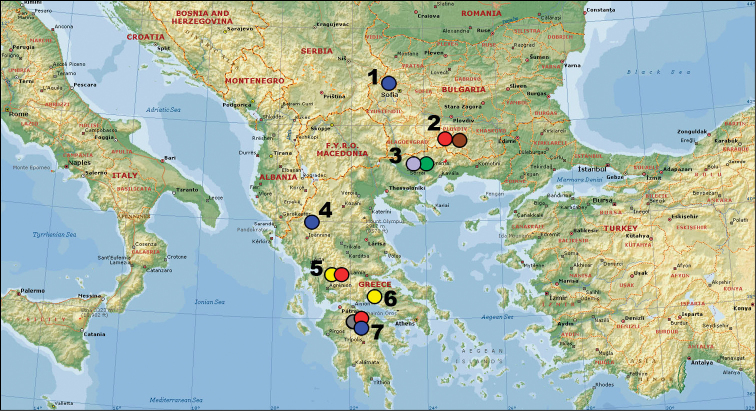
Figure 1.
Localities of the species collected for the study (the species list is presented in Table 1). 1 Bulgaria: Dragoman (Polyommatus admetus) 2 Bulgaria: Hvoyna (Polyommatus ripartii pelopi, Polyommatus orphicus orphicus) 3 Greece: Granitis (Polyommatus nephohiptamenos, Polyommatus orphicus eleniae) 4 Greece: Smolikas (Polyommatus admetus) 5 Greece: Timfristos Mt (Polyommatus ripartii pelopi, Polyommatus timfristos) 6 Greece: Parnassos Mt (Polyommatus timfristos) 7 Greece: Kalavrita (Polyommatus admetus, Polyommatus aroaniensis, Polyommatus ripartii pelopi). Colored circles match different taxa. Blue circle: Polyommatus admetus. Red circle: Polyommatus ripartii. Brown circle: Polyommatus orphicus orphicus. Lavender circle: Polyommatus orphicus eleniae. Yellow circle: Polyommatus timfristos sp. n. Grey circle: Polyommatus aroaniensis.
Table 1.
List of butterflies collected for the present study*
| Traditionally accepted name and combination | Proposed name and combination | Sample code | GenBank code COI | GenBank ITS2 | Locality and date |
|---|---|---|---|---|---|
| Polyommatus admetus | Polyommatus admetus | 08D109 | KY050594 | Greece, Kalavrita, 38°02.097'N; 22°07.085'E, 812 m, 17 July 2008 | |
| Polyommatus admetus | Polyommatus admetus | 08D211 | KY050595 | KY066732 | Greece, Kalavrita 38°02.097'N; 22°07.085'E, 1150 m, 19 July 2008 |
| Polyommatus admetus | Polyommatus admetus | 08D386 | KY050596 | KY066733 | Greece, Smolikas, 40°03.175'N; 20°53.941'E, 1497 m, 22 July 2008 |
| Polyommatus admetus | Polyommatus admetus | 08D655 | KY050597 | Bulgaria, Dragoman, 42°56.320'N; 22°56.038'E, 753 m, 29 July 2008 | |
| Polyommatus aroaniensis | Polyommatus aroaniensis | 08D102 | KY050598 | KY066734 | Greece, Kalavrita, 38°00.699'N; 22°11.554'E, 1640, 16 July 2008 |
| Polyommatus aroaniensis | Polyommatus timfristos | 08D205 | KY066724 | KY081278 | Greece, Parnassos, 38°33.311'N; 22°34.300'E, 1750m, 19 July 2008 |
| Polyommatus aroaniensis | Polyommatus timfristos | 08D247 | KY066725 | KY081279 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus aroaniensis | Polyommatus timfristos | 08D255 | KY066726 | KY081280 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus aroaniensis | Polyommatus timfristos | 08D258 | KY066727 | KY081281 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus aroaniensis | Polyommatus timfristos | 08D273 | KY066728 | KY081282 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus aroaniensis | Polyommatus timfristos | 08D274 | KY066729 | KY081283 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | 08D546 | KY066698 | KY081246 | Bulgaria, Hvoyna, Rodopi Mts, 41°52'14"N; 24°41'6"E, 800 m, 26 July 2008 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | 08D560 | KY066699 | KY081247 | Bulgaria, Hvoyna, Rodopi Mts, 41°52'14"N; 24°41'6"E, 800 m, 26 July 2008 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 002 | KY066700 | KY081266 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 003 | KY066701 | KY081267 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 006 | KY066702 | KY081268 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 010 | KY066705 | KY081271 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 011 | KY066706 | KY081272 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 012 | KY066707 | KY081273 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 013 | KY066708 | KY081274 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 014 | KY066709 | KY081275 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 015 | KY066710 | KY081276 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 016 | KY066711 | KY081277 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 007 | KY066703 | KY081269 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | PE 008 | KY066704 | KY081270 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 950 m, 3–7 July 2016 |
| Polyommatus dantchenkoi orphicus | Polyommatus orphicus orphicus | 08D545 | KY066697 | KY081245 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 800 m, 26 July 2008 |
| Polyommatus eleniae | Polyommatus orphicus eleniae | 08D431 | KY050599 | KY066735 | Greece, Granitis, 41°17.543'N; 23°56.265'E, 830 m, 23 July 2008 |
| Polyommatus eleniae | Polyommatus orphicus eleniae | 08D433 | KY050600 | KY066736 | Greece, Granitis, 41°17.543'N; 23°56.265'E, 830 m, 23 July 2008 |
| Polyommatus eleniae | Polyommatus orphicus eleniae | 08D434 | KY050601 | KY081243 | Greece, Granitis, 41°17.543'N; 23°56.265'E, 830 m, 23 July 2008 |
| Polyommatus eleniae | Polyommatus orphicus eleniae | 08D437 | KY050602 | KY081244 | Greece, Granitis, 41°17.543'N; 23°56.265'E, 830 m, 23 July 2008 |
| Polyommatus nephohiptamenos | Polyommatus nephohiptamenos | 08D471 | KY050603 | KY081248 | Greece, Granitis, 41°17.543'N; 23°56.265'E, 830 m, 23 July 2008 |
| Polyommatus nephohiptamenos | Polyommatus nephohiptamenos | 08D483 | KY050604 | KY081249 | Greece, Granitis, 41°13.485'N; 24°02.990'E, 1646 m, 23 July 2008 |
| Polyommatus nephohiptamenos | Polyommatus nephohiptamenos | 08D485 | Greece, Granitis, 41°13.485'N; 24°02.990'E 1646 m, 23 July 2008 | ||
| Polyommatus nephohiptamenos | Polyommatus nephohiptamenos | 08D494 | KY050605 | KY081250 | Greece, Granitis, 41°13.485'N; 24°02.990'E, 1450–1750 m, 24 July 2008 |
| Polyommatus nephohiptamenos | Polyommatus nephohiptamenos | 08D496 | KY050606 | KY081251 | Greece, Granitis, 41°13.485'N; 24°02.990'E, 1450–1750 m, 24 July 2008 |
| Polyommatus nephohiptamenos | Polyommatus nephohiptamenos | 08D498 | KY066694 | KY081252 | Greece, Granitis, 41°13.485'N; 24°02.990'E, 1450–1750 m, 24 July 2008 |
| Polyommatus nephohiptamenos | Polyommatus nephohiptamenos | 08D499 | KY066695 | KY081253 | Greece, Granitis, 41°13.485'N; 24°02.990'E, 1450–1750 m, 24 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D249 | KY066717 | KY081258 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D252 | KY066718 | KY081259 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D257 | KY066719 | KY081260 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D260 | KY066720 | KY081263 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D291 | KY066721 | KY081261 | Greece, Timfristos, 38°55.460'N; 21°47.605'E, 1267 m, 20 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D549 | KY066722 | KY081262 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 800 m |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D551 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N; 24°41.6'E, 800m, 26 July 2008 | ||
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D571 | KY066723 | KY081264 | Bulgaria, Hvoyna, Rodopi Mts yna, 41°52.14'N; 24°41.6'E, 800 m, 26 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D085 | KY066712 | KY081254 | Greece, Kalavrita, 38°02.097'N; 22°07.085'E, 812 m, 16 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D092 | KY066713 | KY081255 | Greece, Kalavrita, 38°02.097'N; 22°07.085'E, 812 m, 16 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D120 | KY066714 | KY081256 | Greece Kalavrita, 38°02.097'N; 22°07.085'E, 812 m, 17 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D144 | KY066715 | KY085933 | Greece Kalavrita, 38°01.617'N; 22°13.411'E, 1610–1700 m, 17 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | 08D145 | KY066716 | KY081257 | Greece, Kalavrita, 38°01.617'N; 22°13.411'E 1610–1700 m, 17 July 2008 |
| Polyommatus ripartii pelopi | Polyommatus ripartii pelopi | PE 009 | KY066696 | KY081265 | Bulgaria, Hvoyna, Rodopi Mts, 41°52.14'N 24°41.6'E, 950 m |
| Plebejus damon | Plebejus damon | VM237 | KY066730 | Russia, Altai Mts, Tigirek, 51°0'N; 82°55'E, 28 July 2007 | |
| Plebejus damon | Plebejus damon | VM196 | KY066731 | Russia, Altai Mts, Tigirek, 51°0'N; 82°55'E, 19 July 2007 |
The samples 08D485 and 08D551 were not used for molecular analysis since the sequences obtained were too short.
Before processing butterflies were put in glassine envelopes and kept alive for less than one hour. Testes were removed and put into a vial with a fresh fixative (3:1, 96% ethanol: glacial acetic acid). The wings were removed and put into a glassine envelope, and the body was placed into a vial with 96% ethanol for further molecular analysis. All chromosome preparations, butterfly bodies in ethanol and wings in glassine envelopes are stored in the Department of Karyosystematics (Zoological Institute of the Russian Academy of Sciences, St. Petersburg).
Analysis of karyotype
Testes were stored in the 3:1 fixative for several months at +4 °C and then stained with 2% acetic orcein for 30 days at 20 °C. We used a two-phase method of chromosome analysis following Lukhtanov and Dantchenko (2002b). In the first phase, stained testes were placed into a drop of 40% lactic acid on a slide where spermatocysts were dissected from testis membranes using entomological pins. Intact spermatocytes were transferred into a new drop of 40% lactic acid and covered with a coverslip. A Carl Zeiss Amplival light microscope was used for cytogenetic analysis. During the metaphase I stage, each spermatocyst was observed as a regular sphere consisting of 64 spermatocytes. In the second phase, different degrees of chromosome spreading were observed by gradually increasing pressure on the coverslip. The second phase was useful for studying the bivalent structure and counting the bivalent number. By scaling up the pressure on the coverslip, we were able to manipulate chromosomes, e.g. change their position and orientation on the slide, and consequently to resolve controversial cases of contacting or overlapping bivalents. Haploid chromosome numbers were counted in metaphase I (MI) and/or metaphase II (MII) of meiosis.
DNA extraction and sequencing
We used a 657-bp fragment within the mitochondrial COI gene and a 440-bp fragment within the ITS2 region. DNA was extracted using phenol-chloroform method according to the standard protocol (Sambrook and Russel 2006). The first two abdominal segments were homogenized in lysis buffer [25 mM EDTA, 75 mM NaCl, 10 mM Tris (pH 7.5)]. Then proteinase K (20 mg/ml) and 10% SDS were added and the samples were incubated for 2 h at 60 °C. DNA was extracted from lysate first with phenol/chloroform (1:1) and then with chloroform to remove any remaining phenol. DNA was precipitated with isopropyl alcohol in the presence of 0.1 M NaCl and pelleted by centrifugation. The pellets were washed with 70% ethanol, dried and dissolved in ddH2O. The extracted DNA was stored at -20 °C.
For COI amplification we used the self-designed primers COIF1 (5’-CCACAAATCATAAAGATATTGGAAC-3’) and COIR1 (5’-TGATGAGCTCATACAATAAATCCTA-3’). For ITS2 amplification we used the self-designed primers ITS2F (5’-CATATGCCACACTGTTCGTCTG-3’) and ITS2R (5’-GATATCCGTCAGCGCAACG-3’).
The polymerase chain reaction (PCR) was carried out with Taq-polymerase (Sileks) in 20 µl of PCR buffer containing MgCl2 [2.5 mM], dNTP [200 mM] and forward and reverse primers [20 pmol each]. Amplification of COI gene fragment was carried out with the following conditions: initial denaturation at 94 °C for 3 min, followed by 30 cycles of 30 sec at 94 °C, 30 sec at 51 °C (the annealing temperature) and 30 sec at 72 °C, and then final elongation 5 min for 72 °C. Amplification of ITS2 region fragment was carried out with the following conditions: initial denaturation at 94 °C for 2 min, followed by 30 cycles of 30 sec at 94 °C, 30 sec at 60 °C (the annealing temperature) and 30 sec at 72 °C, and then final elongation 5 min for 72 °C.
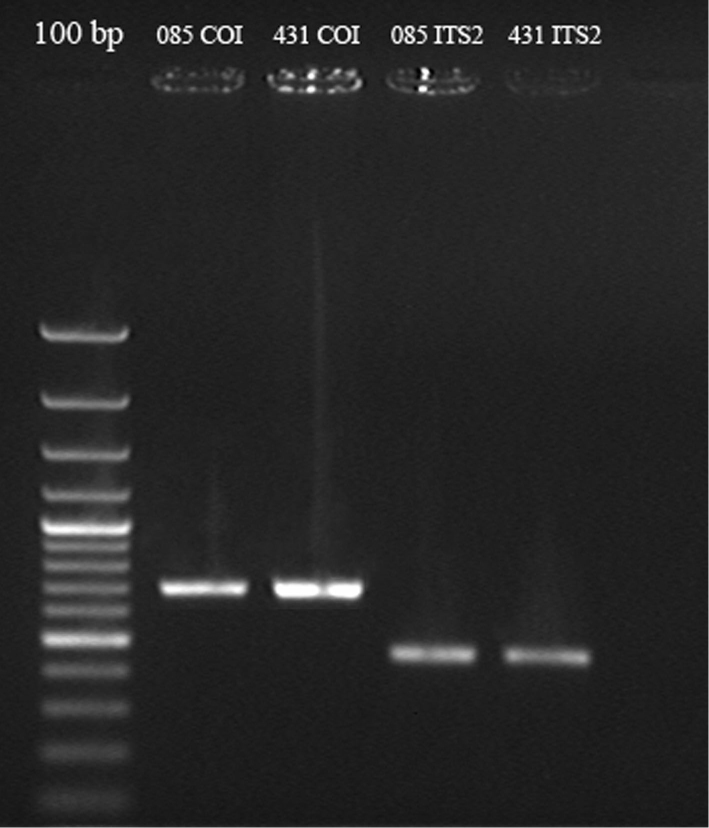
After amplification, PCR mix was loaded in 1% agarose gel and specific product was separated by gel electrophoresis (Fig. 2). Pieces of gel containing the DNA fragment of required length were cut out and then double-stranded DNA was purified using the method of ‘DNA purification from agarose gels with MP@SiO2 magnetic particles’ according to the manufacturer’s protocol (Sileks). Purified DNA fragments were extracted with ddH2O from magnetic particles pelleted with a magnetic rack and collected in a fresh tube. The concentration of purified DNA was estimated via gel electrophoresis (by comparing the brightness of the sample fragment to the brightness of the DNA marker (in our case 100 bp DNA Ladder, Thermo Fisher Scientific).
Figure 2.
Gel electrophoresis with COI and ITS2 PCR products showing the length of the fragments.
All the preparations for sequencing were held in “The Laboratory of Animal Genetics” of Saint-Petersburg State University and “Chromas” Core Facility, Saint-Petersburg State University Research Park. Sequencing was carried out in the Research Resource Center for Molecular and Cell Technologies. GenBank codes of the studied samples are provided in Tables 1 and 2.
Table 2.
List of samples and haplogroups used for the present study.
| Taxon and field code | COI GenBank code | ITS2 GenBank code | COI haplogroup |
|---|---|---|---|
| Polyommatus admetus 08D109 | KY050594 | ad_1 | |
| Polyommatus admetus 08D211 | KY050595 | KY066732 | ad_2 |
| Polyommatus admetus 08D386 | KY050596 | KY066733 | ad_3 |
| Polyommatus admetus | AY556867 | AY556733 | ad_4 |
| Polyommatus admetus | AY556986 | ad_5 | |
| Polyommatus admetus | KC581753 | ad_6 | |
| Polyommatus admetus | KC581754 | ad_7 | |
| Polyommatus alcestis alcestis | AY557008 | AY556641 | alc_3 |
| Polyommatus aroaniensis 08D102 | KY050598 | KY066734 | ar_1 |
| Polyommatus aroaniensis | AY556856 | AY556725 | ar_2 |
| Polyommatus dantchenkoi | AY557072 | AY556678 | dan_1 |
| Polyommatus dantchenkoi | AY557081 | AY556685 | |
| Polyommatus dantchenkoi | AY557073 | AY556679 | |
| Polyommatus demavendi belovi | KR265493 | dem_1 | |
| Polyommatus demavendi belovi | KR265494 | dem_2 | |
| Polyommatus demavendi belovi | EF104630 | dem_3 | |
| Polyommatus demavendi lorestanus | AY557142 | AY556743 | dem_4 |
| Polyommatus dolus virgilius | HM210162 | HM210180 | dol_1 |
| Polyommatus dolus vittatus | AY496740 | dol_2 | |
| Polyommatus fabressei | AY496744 | fab_1 | |
| Polyommatus fabressei | AY556952 | AY556608 | |
| Polyommatus fabressei | AY556869 | AY556734 | fab_1 |
| Polyommatus fulgens | AY556941 | AY556601 | |
| Polyommatus fabressei | EF104605 | HM210186 | fab_4 |
| Polyommatus fulgens | AY556963 | AY556615 | ful_1 |
| Polyommatus fulgens | AY496746 | ||
| Polyommatus fulgens | AY496712 | ||
| Polyommatus fulgens | AY556954 | AY556610 | ful_2 |
| Polyommatus fulgens | AY556958 | ful_4 | |
| Polyommatus humedasae | AY557127 | AY556710 | hum_1 |
| Polyommatus humedasae | AY557128 | AY556711 | hum_2 |
| Polyommatus humedasae | HM210169 | HM210192 | |
| Polyommatus humedasae | HM210170 | HM210193 | hum_4 |
| Polyommatus karacetinae | AY556906 | alc_1 | |
| Polyommatus karacetinae | AY556907 | AY556574 | alc_1 |
| Polyommatus karacetinae | AY557090 | alc_4 | |
| Polyommatus karacetinae urmiaensis | EF104631 | urm | |
| Polyommatus khorasanensis | AY557138 | AY556737 | khor |
| Polyommatus menalcas | AY556982 | men_1 | |
| Polyommatus menalcas | AY557111 | men_2 | |
| Polyommatus menalcas | AY557001 | AY556635 | men_3 |
| Polyommatus nephohiptamenos 08D471 | KY050603 | KY081248 | ne_1 |
| Polyommatus nephohiptamenos 08D483 | KY050604 | KY081249 | |
| Polyommatus nephohiptamenos 08D499 | KY066695 | KY081253 | |
| Polyommatus nephohiptamenos 08D496 | KY050606 | KY081251 | |
| Polyommatus nephohiptamenos 08D494 | KY050605 | KY081250 | ne_3 |
| Polyommatus nephohiptamenos 08D498 | KY066694 | KY081252 | ne_5 |
| Polyommatus nephohiptamenos | KC581745 | ne_7 | |
| Polyommatus nephohiptamenos | AY556860 | ||
| Polyommatus nephohiptamenos | AY556859 | AY556728 | |
| Polyommatus orphicus eleniae 08D431 | KY050599 | KY066735 | orph_1 |
| Polyommatus orphicus eleniae 08D433 | KY050600 | KY066736 | |
| Polyommatus orphicus eleniae 08D437 | KY050602 | KY081244 | |
| Polyommatus orphicus eleniae 08D434 | KY050601 | KY081243 | orph_3 |
| Polyommatus orphicus orphicus 08D545 | KY066697 | KY081245 | orph_5 |
| Polyommatus orphicus orphicus 08D560 | KY066699 | KY081247 | |
| Polyommatus orphicus orphicus PE 003 | KY066701 | KY081267 | orph_5 |
| Polyommatus orphicus orphicus PE 011 | KY066706 | KY081272 | |
| Polyommatus orphicus orphicus PE 013 | KY066708 | KY081274 | |
| Polyommatus orphicus orphicus PE 014 | KY066709 | KY081275 | |
| Polyommatus orphicus orphicus PE 015 | KY066710 | KY081276 | |
| Polyommatus orphicus orphicus PE 007 | KY066703 | KY081269 | |
| Polyommatus orphicus orphicus PE 006 | KY066702 | KY081268 | |
| Polyommatus orphicus orphicus PE 012 | KY066707 | KY081273 | orph_6 |
| Polyommatus orphicus orphicus PE 008 | KY066704 | KY081270 | |
| Polyommatus orphicus orphicus 08D546 | KY066698 | KY081246 | |
| Polyommatus orphicus orphicus PE 002 | KY066700 | KY081266 | orph_8 |
| Polyommatus orphicus orphicus PE 010 | KY066705 | KY081271 | orph_11 |
| Polyommatus orphicus orphicus PE 016 | KY066711 | KY081277 | |
| Polyommatus pseudorjabovi | KR265487 | pse_1 | |
| Polyommatus pseudorjabovi | KR265489 | ||
| Polyommatus pseudorjabovi | KR265490 | ||
| Polyommatus pseudorjabovi | KR265491 | pse_1 | |
| Polyommatus pseudorjabovi | KR265484 | ||
| Polyommatus pseudorjabovi | KR265480 | ||
| Polyommatus pseudorjabovi | KR265496 | pse_2 | |
| Polyommatus pseudorjabovi | KR265483 | ||
| Polyommatus pseudorjabovi | KR265481 | ||
| Polyommatus pseudorjabovi | KR265488 | pse_3 | |
| Polyommatus pseudorjabovi | KR265482 | pse_9 | |
| Polyommatus pseudorjabovi | KR265500 | pse_12 | |
| Polyommatus ripartii pelopi 08D249 | KY066717 | KY081258 | rip_1 |
| Polyommatus ripartii pelopi 08D252 | KY066718 | KY081259 | |
| Polyommatus ripartii pelopi 08D257 | KY066719 | KY081260 | rip_3 |
| Polyommatus ripartii pelopi 08D260 | KY066720 | KY081263 | rip_4 |
| Polyommatus ripartii pelopi 08D291 | KY066721 | KY081261 | |
| Polyommatus ripartii pelopi 08D549 | KY066722 | KY081262 | |
| Polyommatus ripartii pelopi 08D085 | KY066712 | KY081254 | |
| Polyommatus ripartii pelopi 08D145 | KY066716 | KY081257 | |
| Polyommatus ripartii ripartii | AY556858 | AY556727 | |
| Polyommatus ripartii ripartii | KC581746 | ||
| Polyommatus ripartii ripartii | KC581747 | ||
| Polyommatus ripartii ripartii | KC581748 | ||
| Polyommatus ripartii ripartii | KC581749 | ||
| Polyommatus ripartii ripartii | KC581750 | ||
| Polyommatus ripartii ripartii | KC581751 | ||
| Polyommatus ripartii ripartii | KC581752 | ||
| Polyommatus ripartii pelopi 08D571 | KY066723 | KY081264 | |
| Polyommatus ripartii paralcestis | KC581715 | rip_8 | |
| Polyommatus ripartii paralcestis | KC581716 | rip_9 | |
| Polyommatus ripartii pelopi | AY557042 | rip_10 | |
| Polyommatus ripartii pelopi 08D092 | KY066713 | KY081255 | rip_12 |
| Polyommatus ripartii pelopi 08D120 | KY066714 | KY081256 | rip_13 |
| Polyommatus ripartii pelopi 08D144 | KY066715 | KY085933 | rip_14 |
| Polyommatus ripartii pelopi PE 009 | KY066696 | KY081265 | rip_82 |
| Polyommatus ripartii ripartii | HM210164 | rip_16 | |
| Polyommatus ripartii ripartii | HM210172 | ||
| Polyommatus ripartii riparii | HM210163 | HM210197 | |
| Polyommatus ripartii ripartii | AY556944 | AY556603 | rip_18 |
| Polyommatus ripartii ripartii | KC581717 | rip_19 | |
| Polyommatus ripartii ripartii | KC581718 | ||
| Polyommatus ripartii ripartii | AY556957 | ||
| Polyommatus ripartii ripartii | AY556962 | rip_20 | |
| Polyommatus ripartii ripartii | EF104603 | rip_21 | |
| Polyommatus ripartii ripartii | FJ663243 | rip_22 | |
| Polyommatus ripartii ripartii | FJ663244 | rip_23 | |
| Polyommatus ripartii ripartii | FJ663245 | ||
| Polyommatus ripartii ripartii | FJ663246 | ||
| Polyommatus ripartii ripartii | JN276883 | rip_26 | |
| Polyommatus ripartii ripartii | GU675760 | ||
| Polyommatus ripartii ripartii | GU676039 | rip_27 | |
| Polyommatus ripartii ripartii | GU676152 | ||
| Polyommatus ripartii ripartii | GU677012 | ||
| Polyommatus ripartii ripartii | GU677029 | ||
| Polyommatus ripartii ripartii | HM901559 | ||
| Polyommatus ripartii ripartii | HM901664 | ||
| Polyommatus ripartii ripartii | KC581736 | ||
| Polyommatus ripartii ripartii | KC581737 | ||
| Polyommatus ripartii ripartii | KC581738 | ||
| Polyommatus ripartii ripartii | KC581739 | ||
| Polyommatus ripartii ripartii | KC581740 | ||
| Polyommatus ripartii ripartii | GU676158 | ||
| Polyommatus ripartii ripartii | GU676213 | rip_30 | |
| Polyommatus ripartii ripartii | KC617793 | rip_31 | |
| Polyommatus ripartii ripartii | KC617794 | ||
| Polyommatus ripartii ripartii | GU676220 | rip_31 | |
| Polyommatus ripartii ripartii | HM210167 | rip_35 | |
| Polyommatus ripartii ripartii | KC581741 | rip_36 | |
| Polyommatus ripartii ripartii | KC581742 | ||
| Polyommatus ripartii ripartii | KC581743 | ||
| Polyommatus ripartii ripartii | HM210168 | ||
| Polyommatus ripartii ripartii | KC581723 | rip_37 | |
| Polyommatus ripartii ripartii | KC581724 | ||
| Polyommatus ripartii ripartii | KC581725 | ||
| Polyommatus ripartii ripartii | HM210171 | ||
| Polyommatus ripartii ripartii | KC567885 | rip_42 | |
| Polyommatus ripartii ripartii | KC581719 | ||
| Polyommatus ripartii ripartii | KC567883 | ||
| Polyommatus ripartii ripartii | KC567884 | rip_43 | |
| Polyommatus ripartii ripartii | KC581720 | rip_48 | |
| Polyommatus ripartii ripartii | KC581721 | rip_49 | |
| Polyommatus ripartii ripartii | KC581722 | rip_50 | |
| Polyommatus ripartii ripartii | KC581726 | rip_54 | |
| Polyommatus ripartii ripartii | KC581727 | rip_55 | |
| Polyommatus ripartii ripartii | KC581728 | ||
| Polyommatus ripartii ripartii | KC581729 | rip_57 | |
| Polyommatus ripartii ripartii | KC581730 | ||
| Polyommatus ripartii ripartii | KC581731 | ||
| Polyommatus ripartii ripartii | KC581732 | ||
| Polyommatus ripartii ripartii | KC581733 | ||
| Polyommatus ripartii ripartii | KC581734 | rip_62 | |
| Polyommatus ripartii ripartii | KC581735 | ||
| Polyommatus ripartii ripartii | KC581744 | rip_72 | |
| Polyommatus rjabovianus rjabovianus | KR265475 | rja_1 | |
| Polyommatus rjabovianus rjabovianus | KR265476 | ||
| Polyommatus rjabovianus rjabovianus 2014A10 | |||
| Polyommatus rjabovianus rjabovianus | KR265477 | ||
| Polyommatus rjabovianus masul | KR265497 | rja_4 | |
| Polyommatus rjabovianus masul | KR265485 | ||
| Polyommatus rjabovianus masul | KR265498 | ||
| Polyommatus rjabovianus masul | AY954006 | ||
| Polyommatus rjabovianus masul | KR265499 | ||
| Polyommatus rjabovianus rjabovianus | KR265478 | rja_5 | |
| Polyommatus rjabovianus rjabovianus | AY954019 | ||
| Polyommatus timfristos 08D205 | KY066724 | KY081278 | tim_1 |
| Polyommatus timfristos 08D247 Holotype | KY066725 | KY081279 | tim_2 |
| Polyommatus timfristos 08D273 | KY066728 | KY081282 | |
| Polyommatus timfristos 08D274 | KY066729 | KY081283 | |
| Polyommatus timfristos 08D255 | KY066726 | KY081280 | |
| Polyommatus timfristos 08D258 | KY066727 | KY081281 | tim_4 |
| Polyommatus valiabadi | KR265495 | val_1 | |
| Polyommatus valiabadi | KR265486 | ||
| Polyommatus valiabadi | AY556934 | AY556594 | |
| Polyommatus valiabadi | AY556882 | AY556557 | |
| Polyommatus violetae subbaeticus | EF104604 | HM210188 | viol_1 |
| Polyommatus violetae subbaeticus | HM210166 | HM210187 | viol_2 |
| Polyommatus violetae violetae | HM210173 | HM210200 | viol_3 |
| Polyommatus violetae violetae | HM210174 | HM210201 | |
| Polyommatus violetae violetae | HM210175 | HM210202 | viol_5 |
| Polyommatus yeranyani malyevi | KJ906515 | ad_8 | |
| Polyommatus yeranyani yeranyani | KR265492 | ad_9 |
Phylogenetic analysis
The analysis involved 221 COI sequences (169 GenBank sequences and 52 own material) and 117 ITS2 sequences (66 GenBank and 51 own data).
Sequences of different length (from 415 to 657 bp in case of COI and from 415 to 440 bp in case of ITS2) were included into the final dataset alignment. We used BioEdit 7.2.5 software (Hall 1999) to align the sequences and then edited them manually. The final COI alignment included 657 sites, with 137 variable sites and 112 parsimony-informative sites. The final ITS2 alignment included 440 sites, with 52 variable sites and 22 parsimony-informative sites.
Previously, no significant conflict was detected between the mitochondrial COI and nuclear ITS2 Agrodiaetus data sets (Vila et al. 2010). Thus, we combined mitochondrial and nuclear sequences to improve phylogenetic signal. This resulted in a concatenated alignment with a total of 1039 bp.
Phylogenetic relationships were inferred using Bayesian Inference (BI), maximum likelihood (ML) and maximum parsimony (MP) analyses. jModelTest was used to determine optimal substitution models for ML inference (Posada 2008).
Bayesian analyses were conducted using MrBayes, version 3.2 (Ronquist et al. 2012). Datasets were partitioned by codon position. Substitution models used for each partition were chosen according to jModelTest (Posada 2008): nst=2 and rates=invgamma for the first position, nst=2 and rates=gamma for the second position, and nst=6 and rates=gamma for the third position of COI barcodes. Substitution model nst=6 and rates=invgamm was chosen for ITS2. In evolution of ITS2 sequences, the mono, bi- and mullti-nucleotide insertions/deletions are frequent and contain phylogenetically important information. To account for this, each indel event was coded as a binary character (1/0, presence/absence of the gap independently of its length) and then used in the Bayesian analyses of ITS2 and concatenated data sets. Two runs of 10 000 000 generations with four chains (one cold and three heated) were performed. Chains were sampled every 10 000 generations, and burn-in was determined based on inspection of log likelihood over time plots using TRACER, version 1.4 (available from http://beast.bio.ed.ac.uk/Tracer).
The ML trees were inferred using MEGA6 under the GTR+G+I model. MP analysis was performed using a heuristic search as implemented in MEGA6 (Tamura et al. 2013). A heuristic search was carried out using the close-neighbor-interchange algorithm with search level 3 (Nei and Kumar 2000) in which the initial trees were obtained with the random addition of sequences (100 replicates). We used nonparametric bootstrap values (Felsenstein 1985) to estimate branch support for ML and MP trees. The bootstrap consensus tree was inferred from 500 replicates.
Haplotype network
Median network was constructed using the program Network 4.6.1.3. (Fluxus Technology, fluxus-engineering.com), with the Median Joining algorithm (Bandelt 1999). The algorithm picks close haplotype groups and finds hypothetical ancestors, to join the haplotypes in a common parsimony network. The program shows each haplotype with a colored circle. When the haplotypes are identical, they are united in one bigger circle under one name. Similar haplotypes then are combined in haplogroups (Table 2). The network was constructed on the base of COI alignment, with 191 sequences. The length of the sequences was 612 bp with 116 parsimony-informative sites. The final alignment included only sequences of equal length. Short and ambiguous sequences were excluded.
Karyotypes of the studied samples
Table 3
Table 3.
Chromosome numbers of the studied samples.
| Code | Species | Chomosome number | Country | Locality | Elevation | Date |
|---|---|---|---|---|---|---|
| LR08D109 | Polyommatus admetus | n=80 | Greece (South) | Mt. Chelmos (Aroania), Kalavrita, 38°02.097'N; 22°07.085'E | 812m | 2008.VII.17 |
| LR08D211 | Polyommatus admetus | n=80 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D386 | Polyommatus admetus | n=caca80 | Greece | Smolikas Mt, Pades, 40°03.175'N; 20°53.941'E | 1497m | 2008.VII.22 |
| LR08D655 | Polyommatus admetus | n=ca80 | Bulgaria | Dragoman, 42°56.320'N; 22°56.038'E | 753m | 2008.VII.29 |
| LR08D085 | Polyommatus ripartii pelopi | 2n=ca180 | Greece (South) | Mt. Chelmos (Aroania), Kalavrita, 38°02.097'N; 22°07.085'E | 812m | 2008.VII.16 |
| LR08D092 | Polyommatus ripartii pelopi | n=90 | Greece (South) | Mt. Chelmos (Aroania), Kalavrita, 38°02.097'N; 22°07.085'E | 812m | 2008.VII.16 |
| LR08D120 | Polyommatus ripartii pelopi | 2n=ca180 | Greece (South) | Mt. Chelmos (Aroania), Kalavrita, 38°02.097'N; 22°07.085'E | 812m | 2008.VII.17 |
| LR08D144 | Polyommatus ripartii pelopi | n=90 | Greece (South) | Mt. Chelmos (Aroania), Kalavrita, 38°01.617'N; 22°13.411'E | 1610–1700m | 2008.VII.17 |
| LR08D145 | Polyommatus ripartii pelopi | n=90 | Greece (South) | Mt. Chelmos (Aroania), Kalavrita, 38°01.617'N; 22°13.411'E | 1610–1700m | 2008.VII.17 |
| LR08D249 | Polyommatus ripartii pelopi | n=90 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D252 | Polyommatus ripartii pelopi | n=ca90 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D257 | Polyommatus ripartii pelopi | n=90 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D260 | Polyommatus ripartii pelopi | 2n=ca180 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D291 | Polyommatus ripartii pelopi | n=ca90 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D549 | Polyommatus ripartii pelopi | n=ca90 | Bulgaria | Rodopi Mts, Hvoyna, 41°15'N; 24°32'E | 800m | 2008.VII.26 |
| LR08D571 | Polyommatus ripartii pelopi | n=90 | Bulgaria | Rodopi Mts, Hvoyna, 41°15'N; 24°32'E | 800m | 2008.VII.26 |
| LR08D562 | Polyommatus ripartii pelopi | n=90 | Bulgaria | Rodopi Mts, Hvoyna, 41°15'N; 24°32'E | 800m | 2008.VII.26 |
| LR08D471 | Polyommatus nephohiptamenos | n=90 | Greece (North) | Granitis, 41°17.543'N; 23°56.265'E | 830m | 2008.VII.23 |
| LR08D483 | Polyommatus nephohiptamenos | n=ca90 | Greece (Northern) | Falakro Mt, 41°13.485'N; 24°02.990'E | 1646m | 2008.VII.23 |
| LR08D485 | Polyommatus nephohiptamenos | n=ca90 | Greece (North) | Falakro Mt, 41°13.485'N; 24°02.990'E | 1646m | 2008.VII.23 |
| LR08D494 | Polyommatus nephohiptamenos | n=90 | Greece (North) | Falakro Mt, 41°13.485'N; 24°02.990'E | 1450–1750m | 2008.VII.24 |
| LR08D496 | Polyommatus nephohiptamenos | n=ca90 | Greece (North) | Falakro Mt, 41°13.485'N; 24°02.990'E | 1450–1750m | 2008.VII.24 |
| LR08D498 | Polyommatus nephohiptamenos | n=ca90 | Greece (North) | Falakro Mt, 41°13.485'N; 24°02.990'E | 1450–1750m | 2008.VII.24 |
| LR08D102 | Polyommatus aroaniensis | n=47 | Greece (South) | Mt. Chelmos (Aroania), Kalavrita, 38°00.699'N; 22°11.554'E | 1640m | 2008.VII.16 |
| LR08D247 Holotype | Polyommatus timfristos | n=38 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D255 | Polyommatus timfristos | n=38 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D258 | Polyommatus timfristos | n=38 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D273 | Polyommatus timfristos | n=38 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D274 | Polyommatus timfristos | n=38 | Greece (Central) | Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605'E | 1267m | 2008.VII.20 |
| LR08D205 | Polyommatus timfristos | n=38 | Greece (Central) | Parnassos Mt, 38°33.311'N; 22°34.300'E | 1750m | 2008.VII.19 |
| LR08D545 | Polyommatus orphicus orphicus | n=ca41–42 | Bulgaria | Rodopi Mts, Hvoyna, 41°15'N; 24°32'E | 800m | 2008.VII.26 |
| LR08D546 | Polyommatus orphicus orphicus | n=ca41–42 | Bulgaria | Rodopi Mts, Hvoyna, 41°15'N; 24°32'E | 800m | 2008.VII.26 |
| LR08D560 | Polyommatus orphicus orphicus | n=41, n=42 | Bulgaria | Rodopi Mts, Hvoyna, 41°15'N; 24°32'E | 800m | 2008.VII.26 |
| LR08D561 | Polyommatus orphicus orphicus | n=41, n=42 | Bulgaria | Rodopi Mts, Hvoyna, 41°15'N; 24°32'E | 800m | 2008.VII.26 |
| LR08D431 | Polyommatus orphicus eleniae | n=42 | Greece (North) | Granitis, 41°17.543'N; 23°56.265'E | 830m | 2008.VII.23 |
| LR08D433 | Polyommatus orphicus eleniae | n=41, n=42 | Greece (North) | Granitis, 41°17.543'N; 23°56.265'E | 830m | 2008.VII.23 |
| LR08D434 | Polyommatus orphicus eleniae | n=ca42 | Greece (North) | Granitis, 41°17.543'N; 23°56.265'E | 830m | 2008.VII.23 |
| LR08D437 | Polyommatus orphicus eleniae | n=ca42 | Greece (North) | Granitis, 41°17.543'N; 23°56.265'E | 830m | 2008.VII.23 |
Polyommatus admetus
Fig. 3a–c
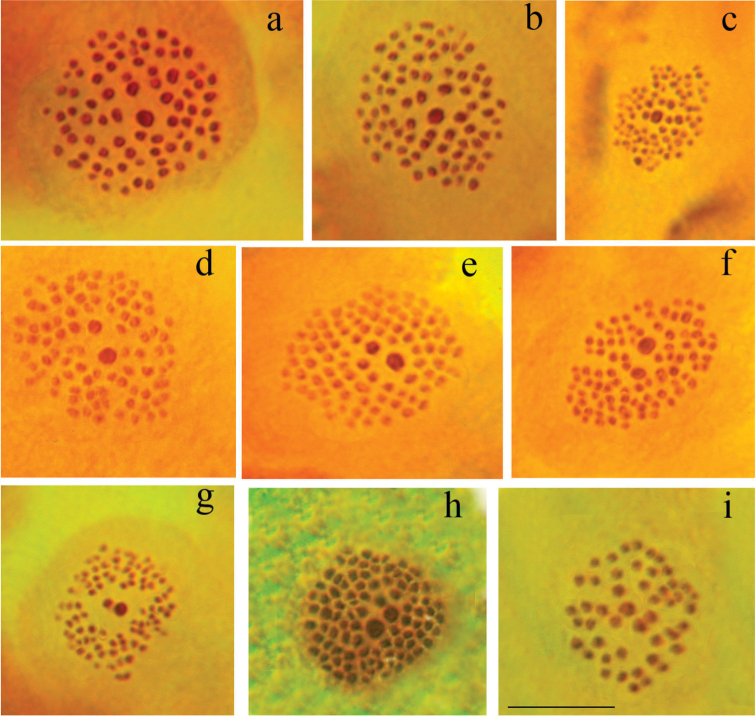
Figure 3.
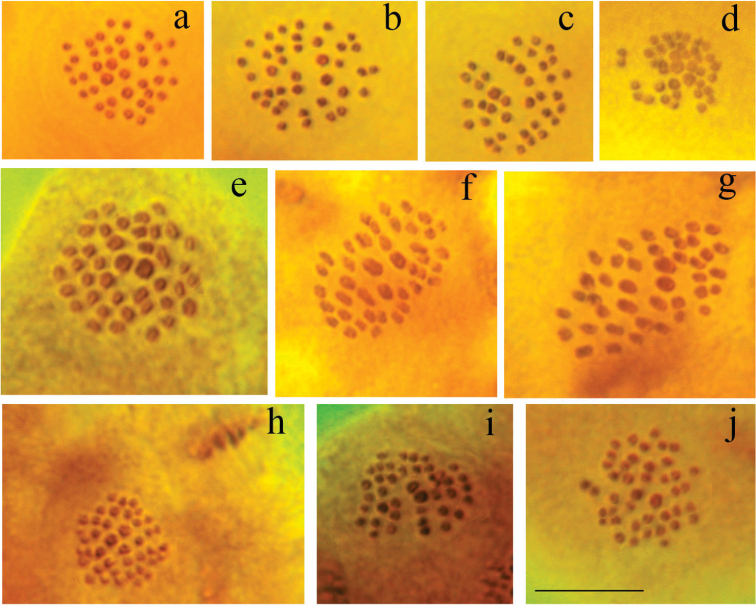
Polyommatus (Agrodiaetus) karyotypes. Bar =10 µ. a–b Polyommatus admetus, sample LR08D109, Greece, MI, n=80. One large bivalent in the centre of the plate can be seen c Polyommatus admetus, sample LR08D109, Greece, MII, n=80. One large chromosome in the centre of the plate can be seen d Polyommatus ripartii pelopi, sample LR08D249, Greece, MI, n=90. Two large bivalents in the centre of the plate can be seen e Polyommatus ripartii pelopi, sample LR08D144, Greece, MI, n=90. Two large bivalents in the centre of the plate can be seen f Polyommatus ripartii pelopi, sample LR08D145, Greece, MI, n=90. Two large bivalents in the centre of the plate can be seen g Polyommatus ripartii pelopi, sample LR08D92, Greece, MII, n=90. Two large chromosomes in the centre of the plate can be seen h Polyommatus nephohiptamenos, sample LR08D494, Northern Greece, MI, n=90. All the bivalents are situated in a plane with the largest elements in the centre of the circular metaphase plate. Bivalents are clearly separated from each other by gaps. Two bivalents are larger than the rest. i Polyommatus aroaniensis, sample LR08D102, Greece, MI, n=47.
The haploid chromosome number n=80 was found in MI and MII cells of two studied individuals from South and Central Greece. In two specimens (Greece, Smolikas Mt and Bulgaria) we counted approximately n=ca80 at MI. The last count was performed with an approximation due to the overlapping of some bivalents. The karyotype displayed one larger bivalent in the centre of the MI plate and one larger univalent in the centre of the MII plate.
Polyommatus ripartii pelopi
Fig. 3d–g
The haploid chromosome number was determined to be n=90 in MI and MII cells of seven studied individuals from different localities (Greece, Bulgaria). At MI, two bivalents were especially large and were situated in the centre of the metaphase plates. Bivalent 1 was 1.4–1.6 times larger than bivalent 2. The sizes of the remaining 88 bivalents decreased more or less linearly. At MII, two univalents were especially large and were situated in the centre of the metaphase plates. Chromosome 1 was 1.4–1.6 times larger than chromosome 2. The sizes of the remaining 88 chromosomes decreased more or less linearly. In three specimens we counted approximately n=ca 90 at MI. The last count was an approximation due to the overlapping of some bivalents. In three specimens, the diploid chromosome number was estimated as 2n=ca180 in male asynaptic meiosis.
Polyommatus nephohiptamenos
Fig. 3h
The haploid chromosome number was determined to be n=90 in MI and MII cells of two studied individuals. At MI, two bivalents (one big and one medium-sized) were larger than the others. At MII, two univalents (one big and one medium-sized) were larger than the rest. The sizes of the remaining 88 bivalents and univalents decreased more or less linearly. In four specimens we counted approximately n=ca90 at MI. The last count was an approximation due to the overlapping of some bivalents.
Polyommatus aroaniensis
Fig. 3i
In the single studied specimen collected in the type locality (Greece, Mt. Chelmos) haploid chromosome number n=47 was found in MI cells. Bivalents were fairly well differentiated with respect to their size. However, it was difficult to subdivide them objectively into size groups because the sizes of the 47 bivalents decrease more or less linearly.
Polyommatus timfristos Lukhtanov, Vishnevskaya & Shapoval, sp. n.
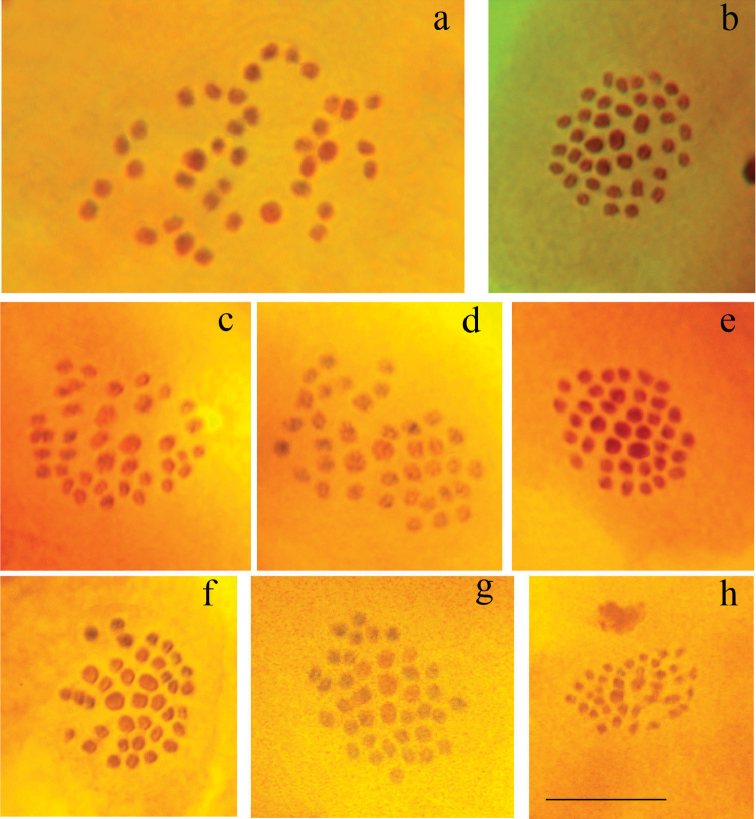
Figure 4.
Polyommatus (Agrodiaetus) timfristos karyotypes. Bar = 10 µ. a Polyommatus timfristos, sample LR08D205, Central Greece, Parnassos, first prometaphase of meiosis, n=38 b Polyommatus timfristos, sample LR08D205, Central Greece, Parnassos, MI, n=38 c Polyommatus timfristos, holotype, sample LR08D247, Central Greece, Timfristos, MI, n=38 d Polyommatus timfristos, sample LR08D255, Central Greece, Timfristos, MI, n=38 e Polyommatus timfristos, sample LR08D258, Central Greece, Timfristos, MI, n=38 f Polyommatus timfristos, sample LR08D258, Central Greece, Timfristos, MI, n=38 g Polyommatus timfristos, sample LR08D273, Central Greece, Timfristos, MI, n=38 h Polyommatus timfristos, sample LR08D274, Central Greece, Timfristos, MII, n=38.
Figure 5.
Polyommatus (Agrodiaetus) karyotypes. Bar = 10 µ. a Polyommatus timfristos, sample LR08D205, Central Greece, Parnassos, MII, n=38 b Polyommatus timfristos, sample LR08D205, Central Greece, Parnassos, MII, n=38 c Polyommatus timfristos, sample LR08D205, Central Greece, Parnassos, MII, n=38 d Polyommatus timfristos, sample LR08D258, Central Greece, Timfristos, MII, n=38 e Polyommatus orphicus eleniae, sample LR08D433, Northern Greece, MI, n=41 f Polyommatus orphicus eleniae, sample LR08D431, Northern Greece, MI, n=42 g Polyommatus orphicus eleniae, sample LR08D431, Northern Greece, MI, n=ca42 h Polyommatus orphicus eleniae, sample LR08D437, Northern Greece, MII, n=41 i Polyommatus orphicus eleniae, sample LR08D437, Northern Greece, MII, n=41 j Polyommatus orphicus eleniae, sample LR08D431, Northern Greece, MII, n=42.
The haploid chromosome number was determined to be n=38 in prometaphase, MI and MII cells of the holotype and six studied paratypes. Bivalents at MI and prometaphase and univalents at MII were fairly well differentiated with respect to their size; however, it was difficult to subdivide them objectively into size groups because the sizes of the 47 elements decrease more or less linearly.
Polyommatus orphicus orphicus
Two different haploid chromosome numbers (n=41 and n=42) were observed in MI and MII cells of the four specimens studied. This variation was most likely caused by polymorphism for one chromosome fussion/fission. This polymorphism resulted in three types of MI karyotype: n=41 (homozygous for chromosomal fusion/fission, one pair of fused chromosomes), n=42 (homozygous for chromosomal fusion/fission, two pairs of unfused chromosomes) and n=41 (heterozygous for chromosomal fusion/fission, 40 bivalents and one trivalent). Bivalents at MI and univalents at MII were fairly well differentiated with respect to their size; however, it was difficult to subdivide them objectively into size groups because the sizes of the elements decrease more or less linearly.
Polyommatus orphicus eleniae
Fig. 5e–j
Chromosome numbers (n=41 and n=42) were observed in MI and MII cells of the four specimens studied. This variation was most likely caused by polymorphism for one chromosome fussion/fission. This polymorphism resulted in three types of MI karyotype: n=41 (homozygous for chromosomal fusion/fission, one pair of fused chromosomes), n=42 (homozygous for chromosomal fusion/fission, two pairs of unfused chromosomes) and n=41 (heterozygous for chromosomal fusion/fission, 40 bivalents and one trivalent). Bivalents and univalents were fairly well differentiated with respect to their size; however, it was difficult to subdivide them objectively into size groups because the sizes of the elements decrease more or less linearly.
Phylogenetic reconstruction
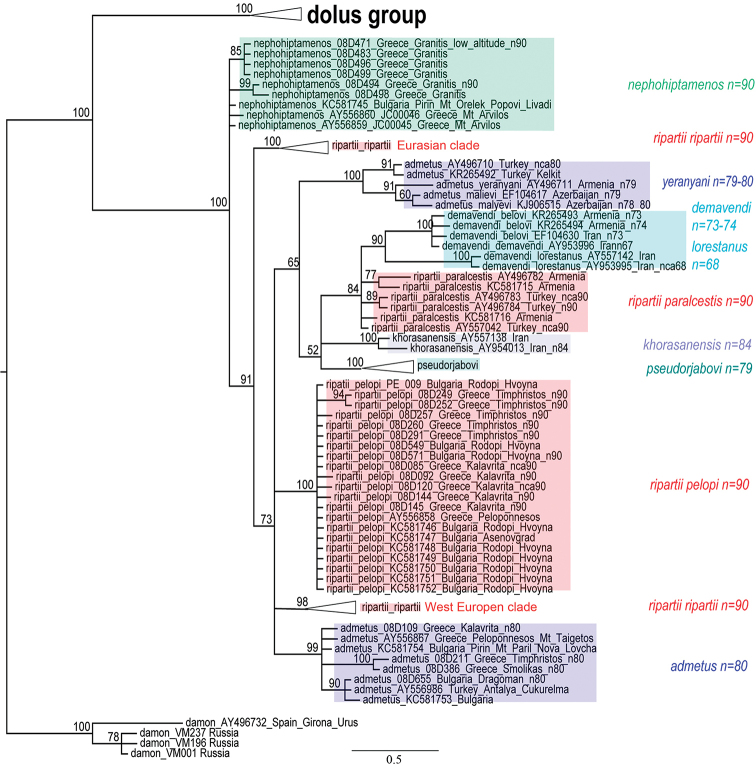
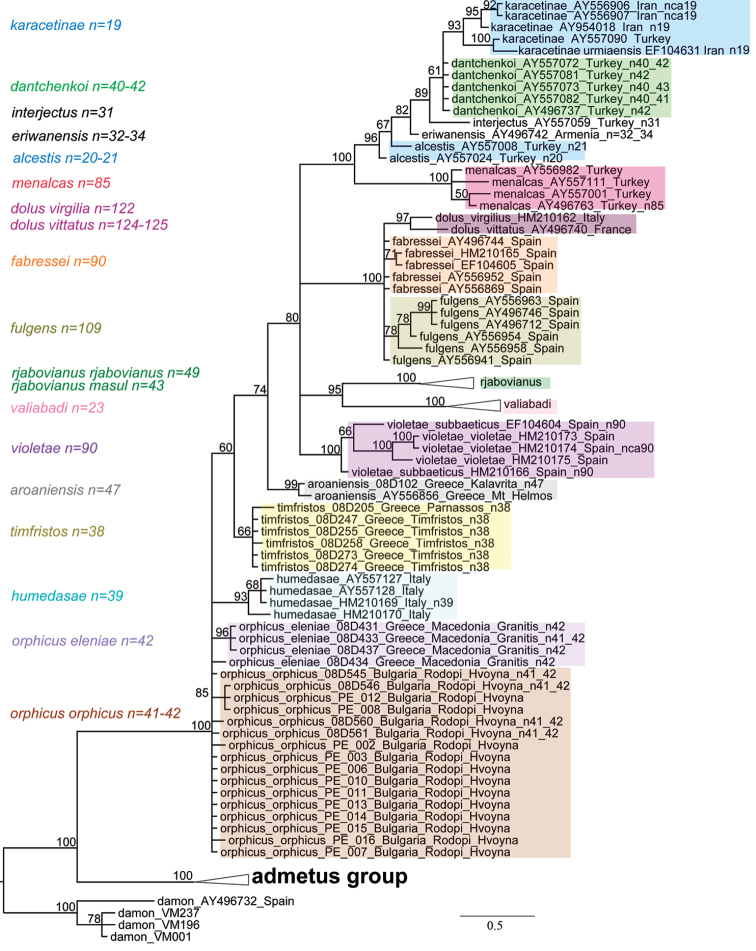
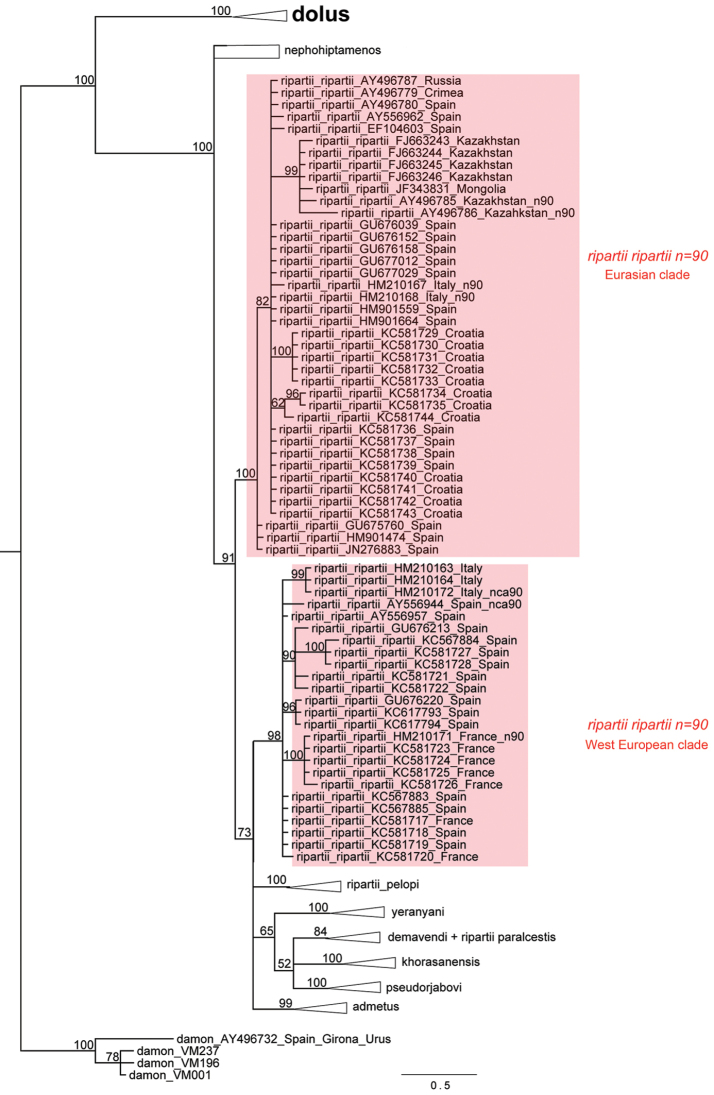
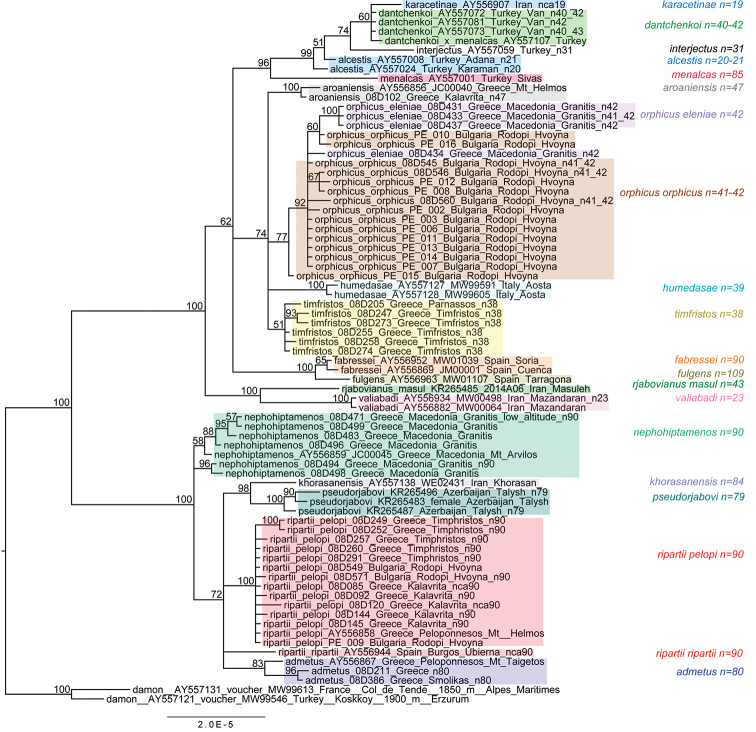
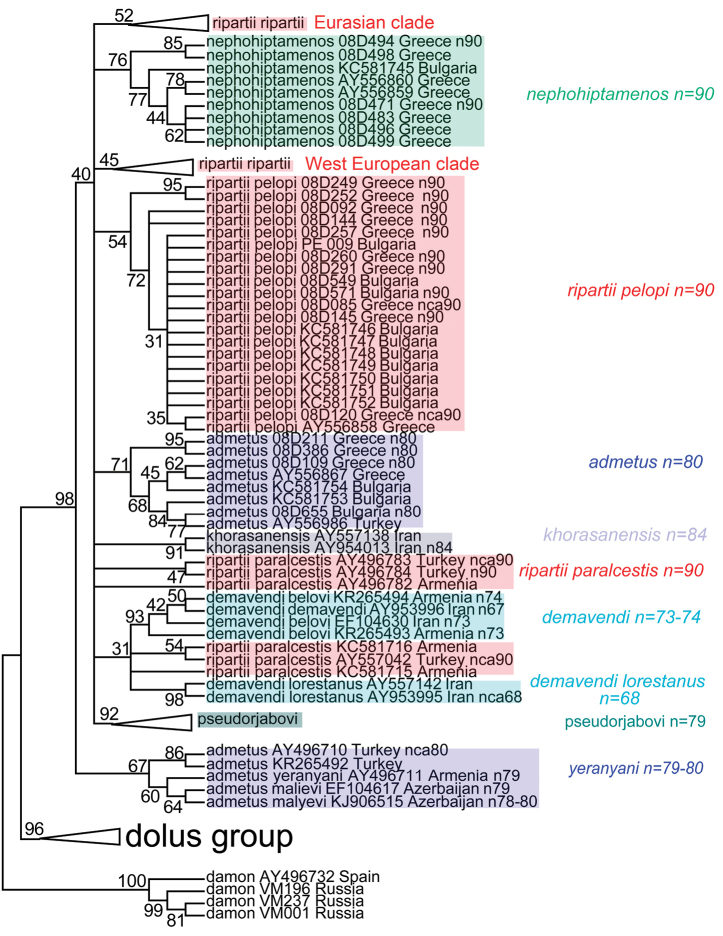
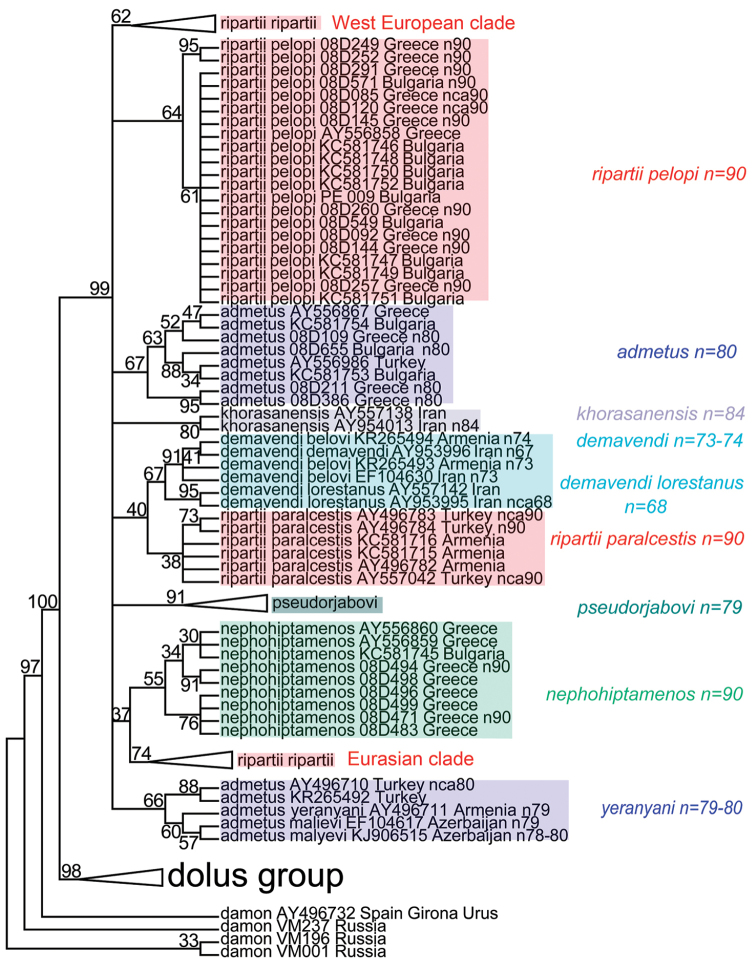
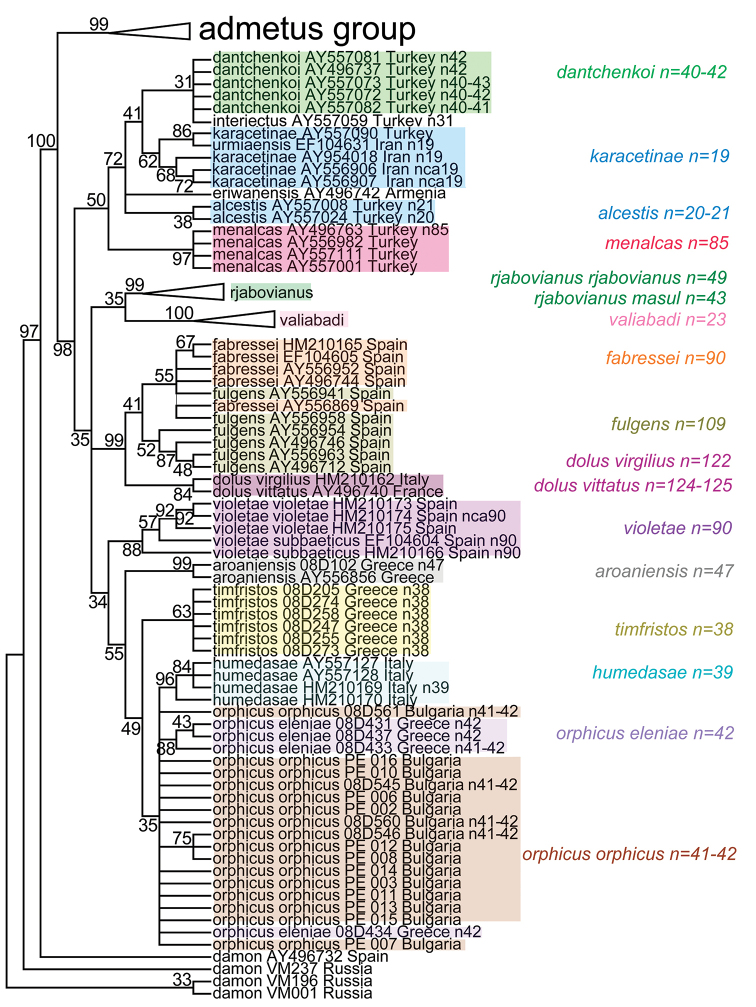
Bayesian analysis of the 657-bp region of COI gene resulted in a phylogram, showing a high level of posterior probability for the majority of the revealed clades. Analysis of the 221-specimen dataset recovered the Polyommatus admetus and Polyommatus dolus species groups as distinct monophyletic lineages. This is consistent with the previous conclusions (Wiemers 2003, Kandul et al. 2004, 2007, Lukhtanov et al. 2005, 2015, Vila et al. 2010, Dincă et al. 2013a). The tree divided into two parts (Polyommatus admetus and Polyommatus dolus groups) is shown in Figures 6–8.
Figure 6.
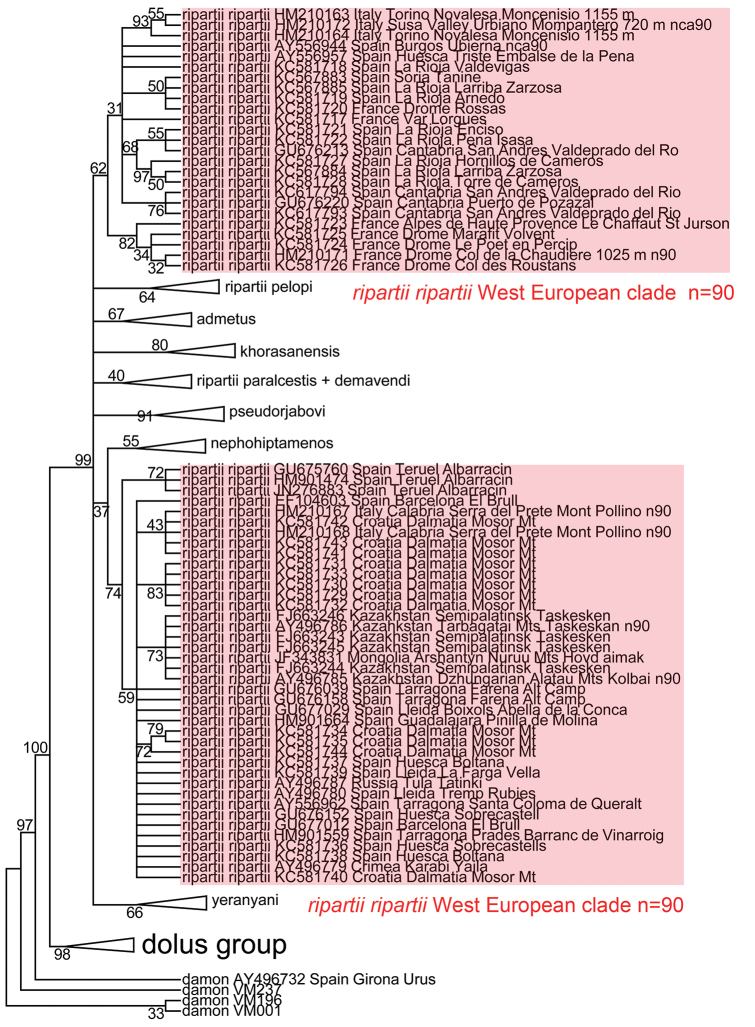
Fragment of the Bayesian tree of Polyommatus admetus and Polyommatus dolus complexes based on analysis of COI barcodes and focused on Polyommatus nephohiptamenos, Polyommatus admetus and Polyommatus ripartii pelopi. Polyommatus pseudorjabovi clade is not shown in details, for its composition see Lukhtanov et al. (2015a). The West-European and the “mixed” (Eurasian) clades of Polyommatus ripartii are shown in Fig. 7. Polyommatus dolus group is shown in Fig. 8. Numbers at nodes indicate Bayesian posterior probability.
Figure 8.
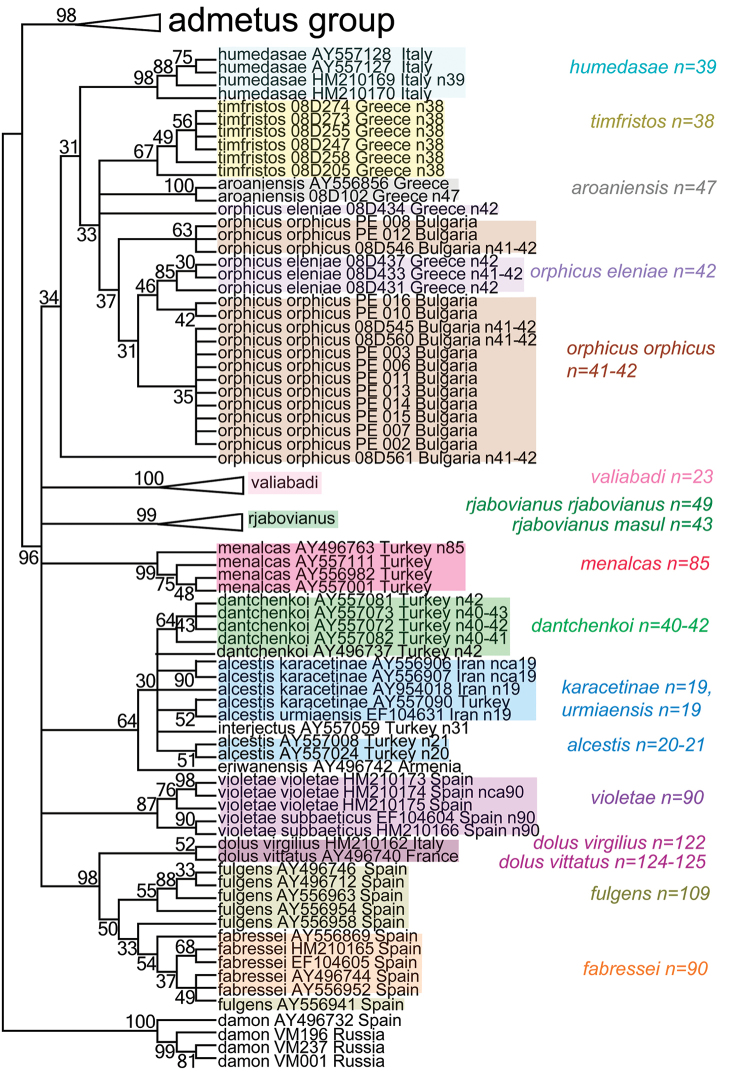
Fragment of the Bayesian tree based on analysis of COI barcodes and focused on details of the Polyommatus dolus group. Polyommatus rjabovianus and Polyommatus valiabadi clades are not shown in details, for their composition see Lukhtanov et al. (2015a). Numbers at nodes indicate Bayesian posterior probability.
Figure 7.
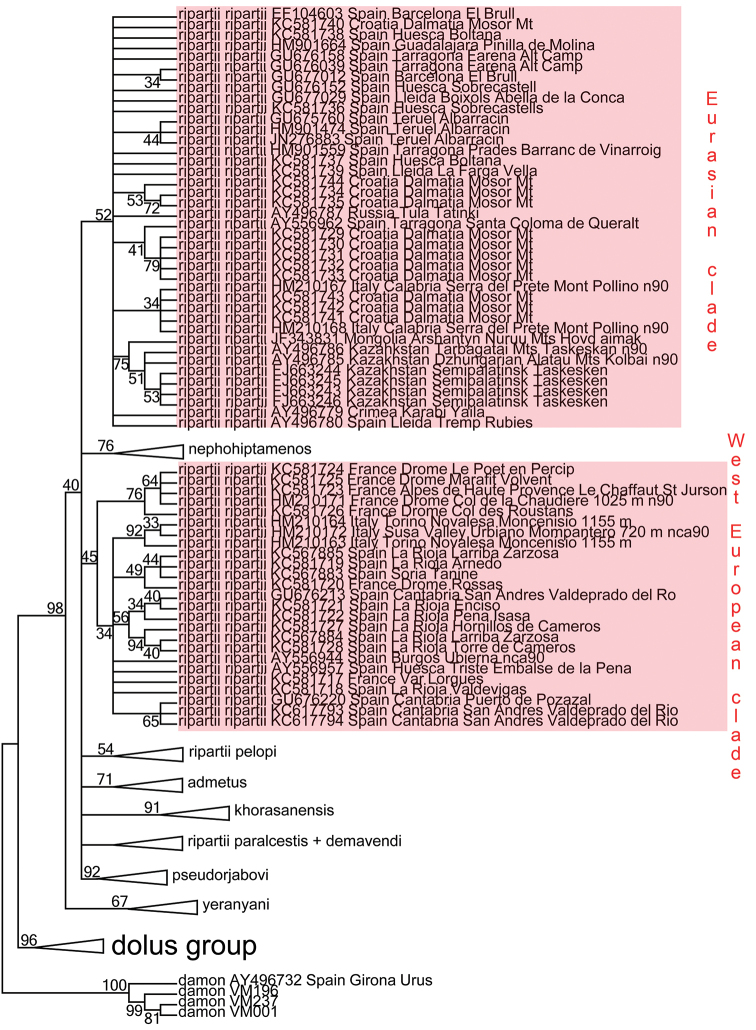
Fragment of the Bayesian tree of Polyommatus admetus and Polyommatus dolus complexes based on analysis of COI barcodes and focused on details of the West-European and the “mixed” (Eurasian) clades of Polyommatus ripartii. Numbers at nodes indicate Bayesian posterior probability.
Within the Polyommatus admetus group, the species Polyommatus ripartii appeared as a polyphyletic assemblage consisting of four monophyletic lineages: the “Balkan” clade, including specimens from Greece and Bulgaria, “West-European” clade, including butterflies from France, Italy and Spain, “mixed” (or Eurasian) clade, including butterflies distributed from Spain to Mongolia, and Turkish-Transcaucasian clade, including butterflies from Turkey and Armenia. The last clade formed an independent lineage, sister to the species Polyommatus demavendi (Pfeiffer, 1938) from east Turkey, Transcaususus and Iran.
Polyommatus admetus sensu auctorum formed two independent clades: one consisting of European and west Turkish specimens and another consisting of specimens from east Turkey, Armenia and Azerbaijan. Polyommatus nephohiptamenos appeared on the Bayesian tree as a paraphyletic group consisting of nine weakly differentiated individuals. On the MP and ML trees (Figs 18 and 21 in Appendix 2), Polyommatus nephohiptamenos tended to form a monophyletic clade, but the bootstrap support of this clade was very low.
The Polyommatus dolus group is interesting for its Balkan species position. Polyommatus aroaniensis formed an independent clade separate from Polyommatus timfristos sp. n., which formed a monophyletic clade as well. Specimens of Polyommatus orphicus orphicus and Polyommatus orphicus eleniae were closely related and formed together a paraphyletic cluster.
Because of low variability, it was difficult to use ITS2 as a single marker to construct the phylogeny of Agrodiaetus. Therefore, we decided to combine the sequence data on COI and ITS2 and constructed a tree on the base of these two markers (Fig. 9). We used 75 specimens for which we had data on both markers. Total length of the combined sequence was 1039 bp. The Bayesian tree constructed on the base of the concatenated alignment revealed generally the same topology as in the case of COI tree, however with a higher support for few clades, and Polyommatus orphicus orphicus + Polyommatus orphicus elenia formed a monophyletic clade with a posterior probability value 77.
Figure 9.
Bayesian tree of Polyommatus admetus and Polyommatus dolus complexes based on analysis of concatenated alignment (COI+ITS2). Numbers at nodes indicate Bayesian posterior probability
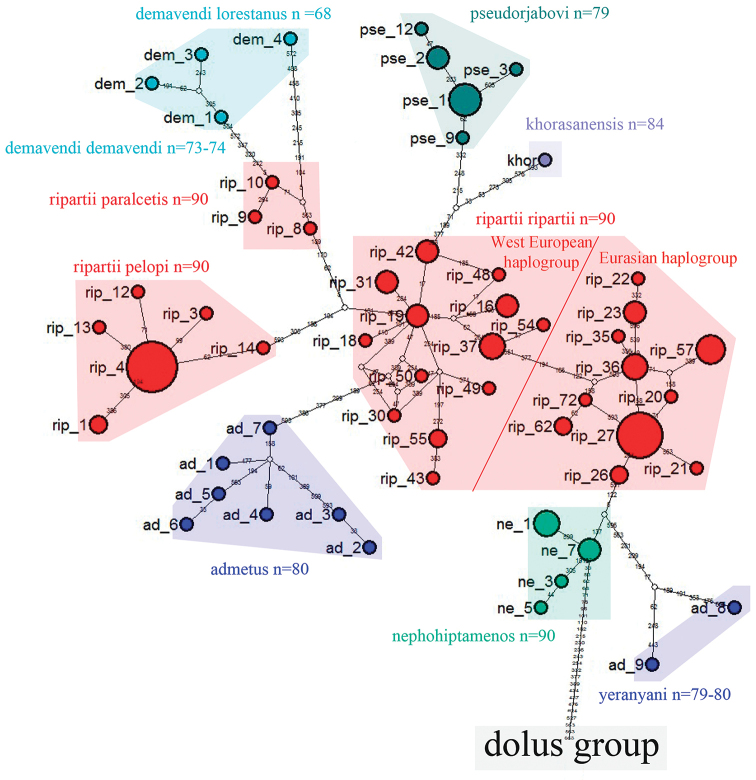
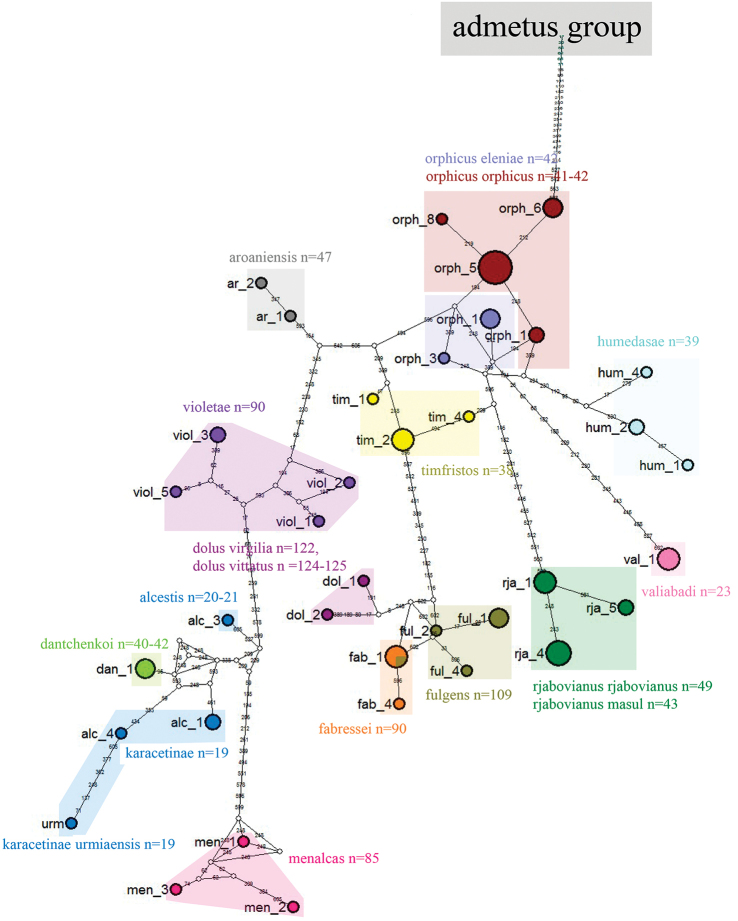
Haplotype network analysis
The complicated relationships between species of Polyommatus admetus and Polyommatus dolus groups were also reflected by a haplotype network (Figs 10 and 11) constructed on the base of COI. To construct the network we used 191 specimens that were collapsed in 96 haplotypes representing 26 haplogroups (Table 2): 10 haplogroups for Polyommatus admetus group and 16 haplogroups for Polyommatus dolus group.
Figure 10.
Haplotype network of Polyommatus admetus species group. Colored circles represent different taxa. Each line segment represents a mutation step, and white small circles represent “missing” haplotypes.
Figure 11.
Haplotype network of Polyommatus dolus species group. Colored circles represent different taxa. Each line segment represents a mutation step, and white small circles represent “missing” haplotypes.
Polyommatus ripartii was represented by 82 specimens divided in 38 haplotypes and four haplogroups which corresponded completely with the four clades revealed on the Bayesian tree (Fig. 10). Polyommatus admetus sensu auctorum was found to include two haplogroups. One haplogroup was represented by specimens from the Balkan and west Turkey (Polyommatus admetus admetus), and the other haplogroup was represented by specimens from Armenia and Azerbaijan (Polyommatus admetus yeranyani + Polyommatus admetus malievi). These two haplogoups were clearly distinct from one another as can be seen in the number of nucleotide substitutions between them. Polyommatus nephohiptamenos was represented by a distinct haplogroup most close to Polyommatus ripartii ripartii haplogroup.
As a by-product of our study, we also discovered that within our samples Polyommatus demavendi comprised two haplogroups. One haplogroup was represented by specimens of Polyommatus demavendi belovi, whilst the other was represented by Polyommatus demavendi lorestanus. Polyommatus pseudorjabovi was represented by a single differentiated haplogroup. A distinct haplogroup represented by a single haplotype was found within Polyommatus khorasanensis.
Concerning Polyommatus dolus group (Fig. 11) we would like to mention that all recognized species, except for Polyommatus fulgens and Polyommatus fabressei, were represented by clearly distinct COI haplogroups. Polyommatus fulgens and Polyommatus fabressei were closely related and even shared one haplotype, despite clear differences in butterfly wing color and karyotypes.
Haplotypes of our target taxa (Polyommatus aroaniensis, Polyommatus timfristos sp. n., Polyommatus orphicus and Polyommatus humedasae) formed together a single cluster. However, all these taxa were distinct, and they did not share any common haplotypes. Therefore, this cluster could be subdivided into four haplogroups: ar (Polyommatus aroaniensis), tim (Polyommatus timfristos), orph (Polyommatus orphicus) and hum (Polyommatus humedasae) (Table 2, Fig. 11).
Despite presumed conspecifity (Kolev 2005), Polyommatus orphicus and Polyommatus dantchenkoi were found to be in the opposite parts of the recovered net, being separated by a number of other species (Polyommatus alcestis, Polyommatus violetae, Polyommatus aroaniensis, Polyommatus timfristos). The chromosomally distinct taxa Polyommatus alcestis and Polyommatus karacetinae were found to be also distinct with respect to their COI haplotypes. These two taxa were already treated as different species by Wiemers et al. (2009).
Butterfly morphology
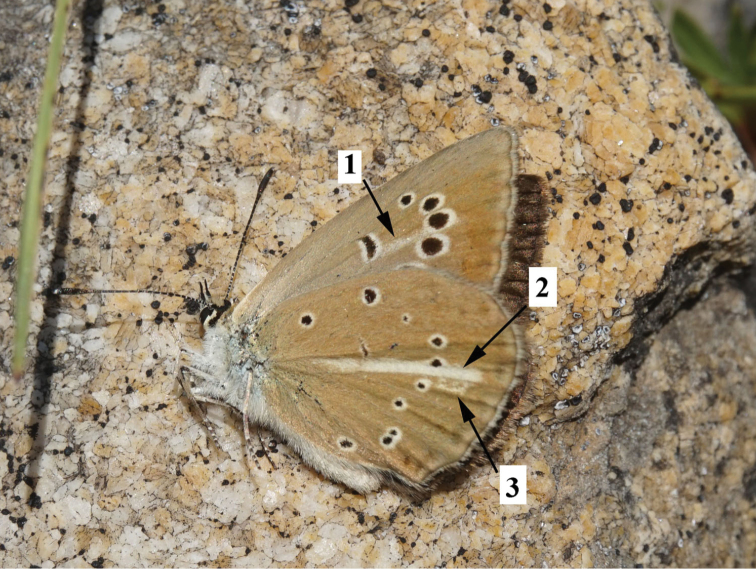
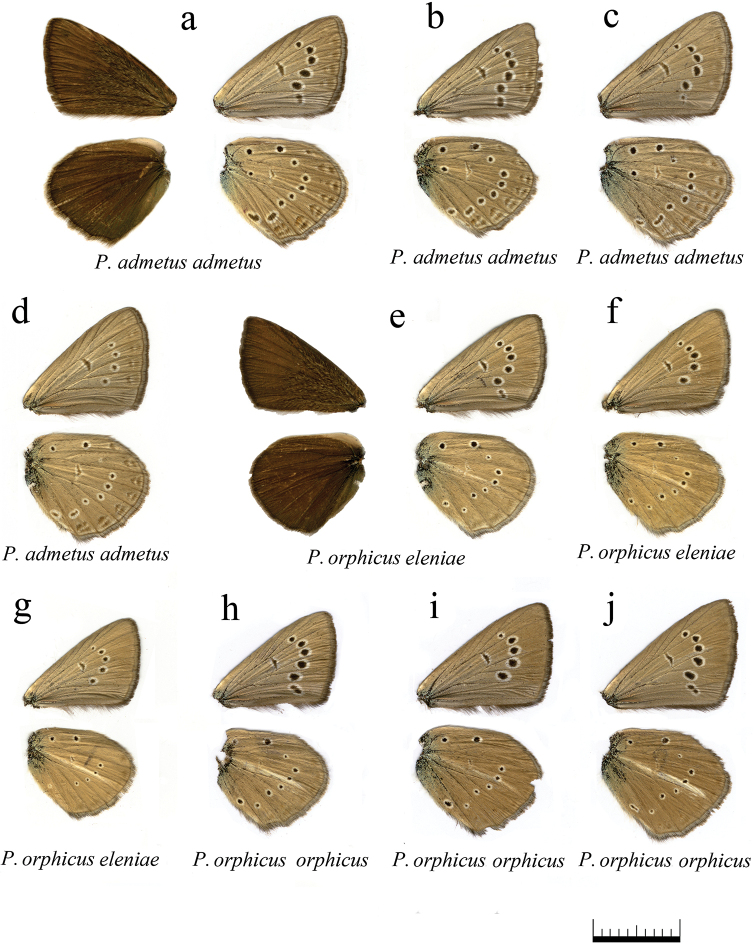
One of the main characteristic features of the anomalous blue butterflies is the upperside wing color. All males and females have brown upper side of the wings, and therefore the group is also called “brown” complex. As for the underside (Fig. 12), there are some differentiated characters of the wing pattern that allow the defining of seven morphological types.
Figure 12.
Polyommatus orphicus orphicus collected in the type locality (Bulgaria, Hvoyna, 3 July 2016). Photo by E. Pazhenkova. White postdiscal streak between discal spot and submarginal marking on the forewing underside (character 1), prominent white streak on the hindwing underside (character 2) and additional white short streak between postdiscal and submarginal areas of the hind wing underside (character 3) are shown.
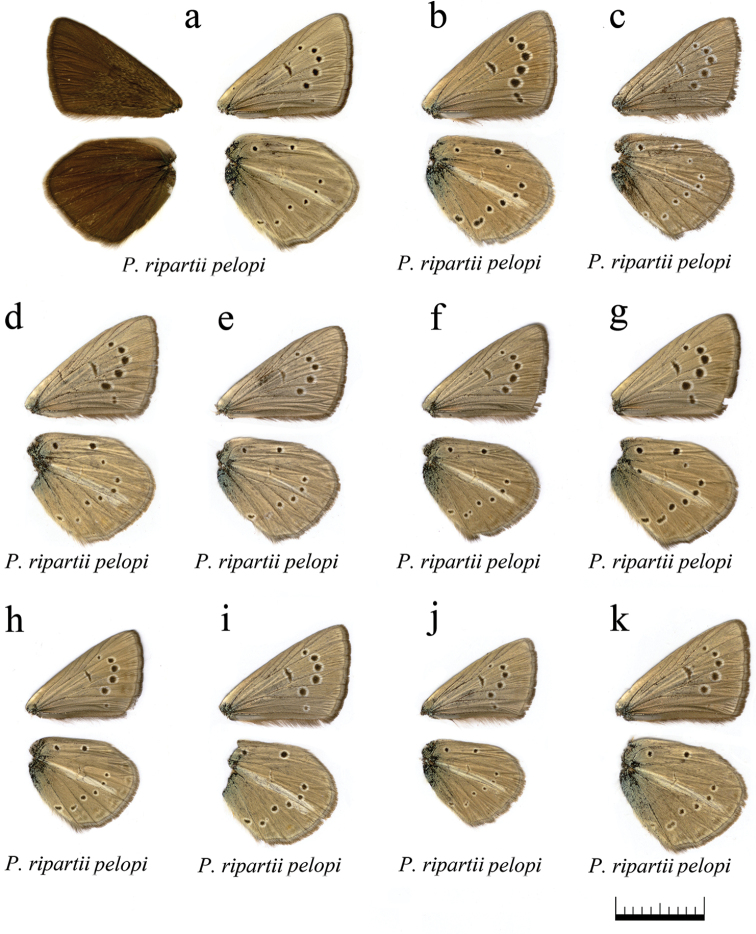
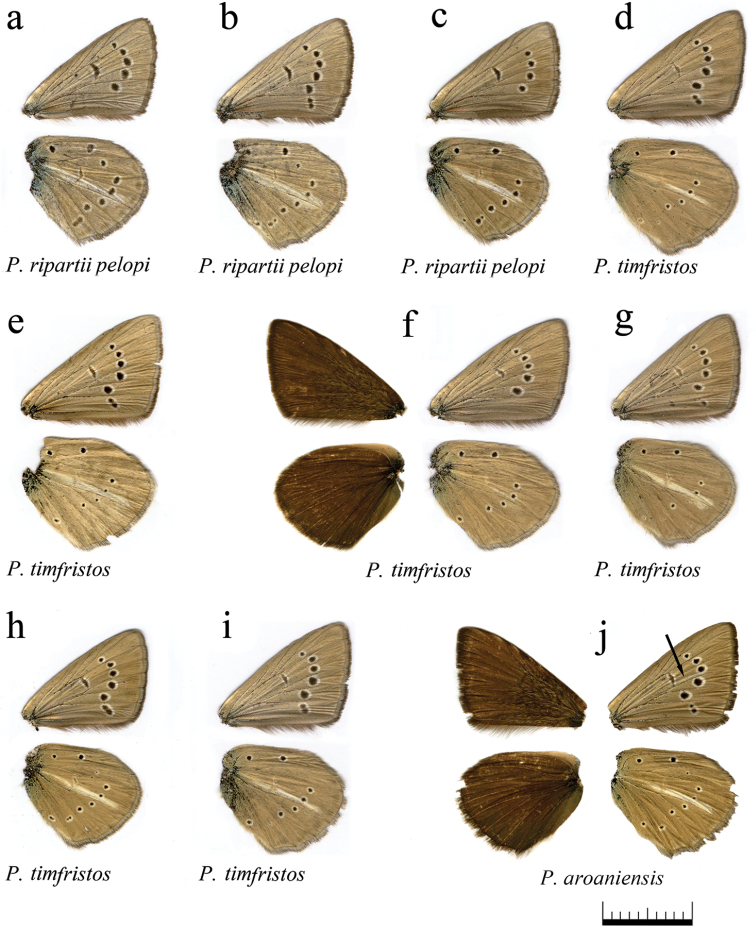
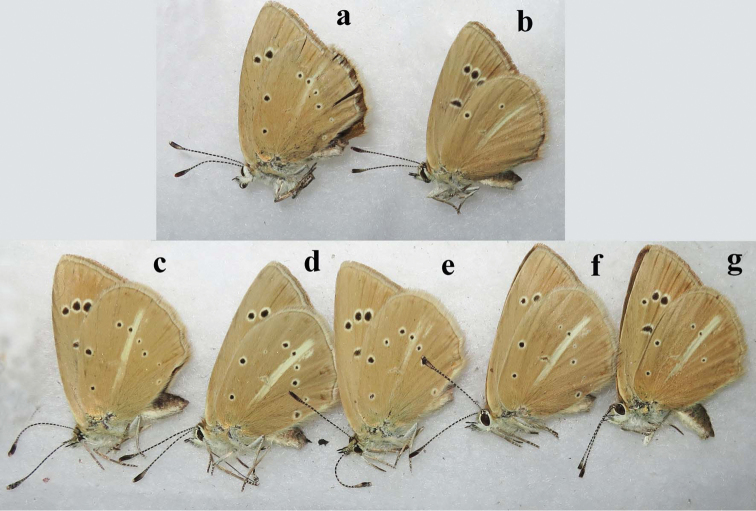
Polyommatus ripartii type: hindwing underside with well-developed white streak (character 2 in Fig. 12), spots are small or medium-sized, marginal marking is reduced. This type is found in different species of both Polyommatus admetus and Polyommatus dolus complexes, e.g. in Polyommatus orphicus orphicus (Fig. 13g), Polyommatus orphicus eleniae (Fig. 13j), Polyommatus nephohiptamenos (Fig. 14e), Polyommatus ripartii pelopi (Figs 15a, f, g, h, j, k, 16a, b, c) and Polyommatus timfristos (Fig. 16e, h).
Polyommatus valiabadi type: the wing underside with exaggerated spots, white streak on the hindwing underside is clearly visible and sharp. This type is found in Polyommatus valiabadi, Polyommatus rjabovianus and Polyommatus pseudorjabovi from Iran and Azerbaijan (Lukhtanov et al. 2015). This type is not found in European Agrodiaetus species.
Polyommatus admetus type: the hindwing has no white streak, marginal marking is very well pronounced. This type is found in Polyommatus admetus (Fig. 13a, b, c, d).
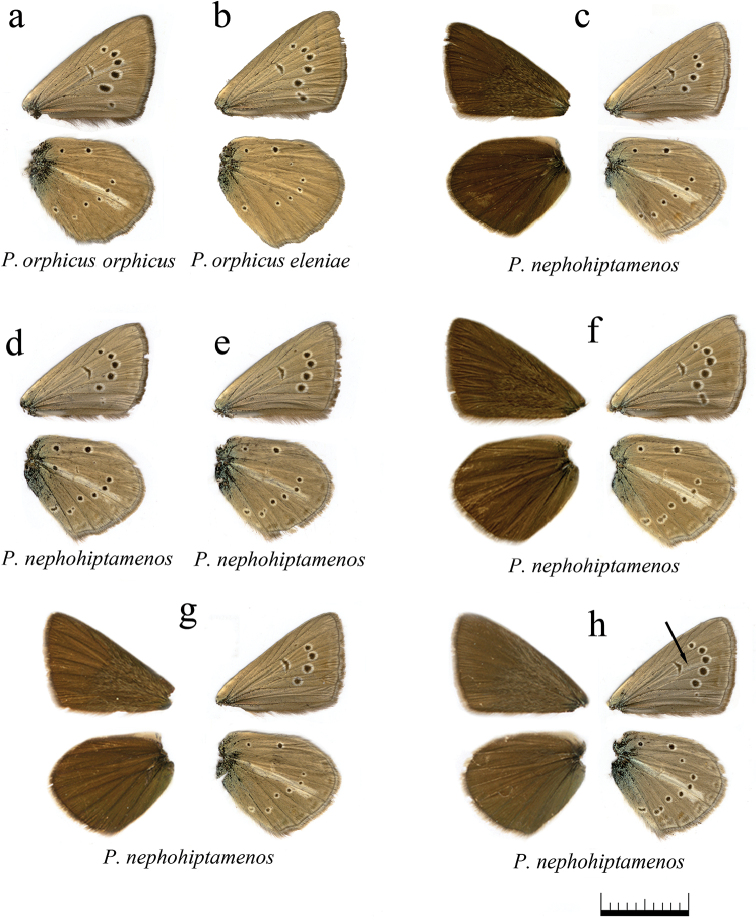
Polyommatus nephohiptamenos type: White streak is well pronounced and very broad on the hindwing, consisting of the main streak and an additional short streak between postdiscal and submarginal areas, just under the main streak. This type is common in Polyommatus nephohiptamenos (Fig. 14c, d, f, g, h), not rare in Polyommatus ripartii (Fig. 15b,d,i) and also found in Polyommatus orphicus orphicus (Fig. 14a) and Polyommatus timfristos (Fig. 16g).
Polyommatus humedasae type: no white streak on the hindwing, marginal marking is pale. This type is quite common in Polyommatus aroaniensis, Polyommatus timfristos (Fig. 16f) and Polyommatus orphicus (Fig. 14b). It is typical for some populations of Polyommatus ripartii from West Europe (Vila et al. 2010) (but not from the Balkan Peninsula).
Polyommatus aroaniensis type: the white streak on the hindwing underside demonstrates different level of reduction. This type is found in Polyommatus aroaniensis (Fig. 16j), Polyommatus timfristos (Fig. 16d, e, i), Polyommatus orphicus orphicus (Fig. 14h, i) and Polyommatus orphicus eleniae (Fig. 13e, g). It is also found in the population of Polyommatus ripartii from the Crimea (Vila et al. 2010) (but not from the Balkan Peninsula).
Polyommatus orphicus type: forewing underside with clear white postdiscal streak between discal spot and submarginal marking, white streak on hindwing underside is prominent, often with additional small white streak (Fig. 12). This type is common in Polyommatus orphicus orphicus (Fig. 14a); nevertheless, the most characteristic feature (the white postdiscal streak between discal spot and submarginal marking on the forewing underside) can be found in other species, e.g. Polyommatus aroaniensis (Fig. 16j) and Polyommatus nephohiptamenos (Fig. 14h).
Figure 13.
The coloration and wing pattern of Polyommatus admetus, Polyommatus orphicus eleniae and Polyommatus orphicus orphicus. The letters correspond to the following sample numbers: a LR-08-D109 upperside and underside b LR-08-386 c LR-08-655 d LR-08-211 e LR-08-433 upperside and underside f LR-08-434 g LR-08-437 h LR-08-545 i LR-08-546 j LR-08-560. Scale bar corresponds to 10 mm in all figures.
Figure 14.
The coloration and wing pattern of Polyommatus orphicus orphicus, Polyommatus orphicus eleniae and Polyommatus nephohiptamenos. The letters correspond to the following sample numbers: a LR-08-D561 b LR-08-431 c LR-08-483 upperside and underside d LR-08-496 e LR-08-498 f LR-08-499 upperside and underside g LR-08-485 upperside and underside h LR-08-494 upperside and underside, white postdiscal streak between discal spot and submarginal marking on the forewing underside is shown by arrow. Bar = 10 mm.
Figure 15.
The coloration and wing pattern of Polyommatus ripartii pelopi. The letters correspond to the following sample numbers: a LR-08-D257 upperside and underside b LR-08-471 c LR-08-085 d LR-08-092 e LR-08-120 f LR-08-144 g LR-08-145 h LR-08-249 i LR-08-252 j LR-08-260 k LR-08-291. Bar = 10 mm.
Figure 16.
The coloration and wing pattern of Polyommatus ripartii pelopi, Polyommatus timfristos sp. n. and Polyommatus aroaniensis. The letters correspond to the following sample numbers: a LR-08-D549 b LR-08-551 c LR-08-571 d LR-08-273 e LR-08-205 f LR-08-274 upperside and underside g LR-08-258 h LR-08-247 (Holotype) i LR-08-255 j LR-08-102 upperside and underside. White postdiscal streak between discal spot and submarginal marking on the forewing underside is shown by arrow. Bar = 10 mm.
Species level monophyly, paraphyly and polyphyly
The studied taxa were found to demonstrate a relatively low level of COI and ITS2 differentiation in terms of genetic distances between species and numbers of evolutionary steps between the taxa on haplotype network (Figs 10 and 11). This result is not unexpected in light of our previous knowledge of this group (Wiemers and Fiedler 2007).
The low genetic differentiantion results in relatively low support for some recovered clades (e.g. for Polyommatus timfristos, Figs 8 and 9) and in non-monophyly of some taxa (Polyommatus nephohiptamenos, Polyommatus orphicus) with respect to COI gene or to combination of COI and ITS2. Species-level non-monophyly in DNA barcode gene trees can have multiple explanations (Mutanen et al. 2016). In our case, combination of low interspecific differentiation with low level of intraspecific variation indicates that preservation of ancestral polymorphism and incomplete lineage sorting (rather than interspecific hybridization) is the most likely mechanism explaining the pattern observed. This finding is also in agreement with the previous conclusion that the subgenus Agrodiaetus itself and its species represent young evolutionary entities (Kandul et al. 2004). We should also stress that despite the obvious paraphyly, the taxa Polyommatus nephohiptamenos and Polyommatus orphicus are distinct with respect to the COI barcodes, and this can be seen on both Bayesian tree (Figs 6–8) and haplotype network (Figs 10 and 11).
An entirely different situation was found in Polyommatus ripartii and Polyommatus admetus sensu auctorum. In these taxa polyphyly in COI trees arises as a result of deep intraspecific divergence. There are two theoretically possible explanations for this kind of non-monophyly. First, each taxon can be a mix of unrecognized multiple species (Dincă et al. 2011, 2013b). Second, a profound irregularity in barcodes can be caused by reasons other than speciation resulting in extraordinary intra-specific barcode variability (Pazhenkova and Lukhtanov 2016). Among these reasons, interspecific mitochondrial introgression (Lukhtanov et al. 2015b) and blending of deeply diverged mitochondrial lineages which evolved in allopatry in different Pleistocene refugia (Pazhenkova and Lukhtanov 2016) are most likely ones. The first explanation could be applied to Polyommatus admetus sensu auctorum which most probably comprises two allopatric species, Polyommatus admetus sensu stricto and Polyommatus yeranyani (see the section Taxonomy below). The situation with Polyommatus ripartii sensu lato seems to be much more complicated. A combination of the first and the second explanations could be applied to Polyommatus ripartii sensu lato, and West-European and Eurasian clades could represent sympatric (parapatric?) intraspecific lineages (Dinca et al. 2013) whereas Turkish-Transcaucasian clade could represent an allopatric species. Additioanl studies are required to solve this problem.
Chromosomal diversity
Polyommatus admetus
The chromosome number of Polyommatus admetus was first established by H. de Lesse who discovered n=80 in populations from Bulgaria (Kalotina) and W Turkey, and n=78-80 (with predominance of n=79) in populations from the eastern part of Turkey (de Lesse 1960a,b). The last count (n=78-80 with predominance of n=79) was later confirmed for populations from Armenia (Lukhtanov and Dantchenko 2002a), Turkey and Azerbaijan (Dantchenko and Lukhtanov 2005, Lukhtanov et al. 2015a). Here we confirm the haploid chromosome number n=80 for Dragoman near Kalotina (Bulgaria) and demonstrate that this karyotype occurs in other localities in Greece. The karyotype of the European samples (with predominance of n=80) seems to be similar, but not completely identical to the karyotype of samples from east Turkey, Armenia and Azerbaijan (with predominance of n=79).
Polyommatus ripartii
This transpalearctic species has been known to have a stable karyotype (n=90, including one large, one medium and 88 small elements) throughout its whole distribution range from Spain in the west to the Altai in the east (de Lesse 1960a,b, Kandul 1997, Lukhtanov and Datchenko 2002, Vila et al. 2010, Vershinina and Lukhtanov 2010, Przybyłowicz et al. 2014). The number n=90 was also found in Polyommatus ripartii pelopi (Coutsis et al. 1999), and we confirmed this count for samples from South and Central Greece and from Bulgaria.
Polyommatus nephohiptamenos
The haploid chromosome number was erroneously given for this taxon as n=8-11 by Brown and Coutsis (1978), and later corrected by Coutsis and De Prins (2007) who established the chromosome number with an approximation due bivalents overlaps as n=ca84-88. Here we were able to make a precise count of chromosome elements in this taxon and to demonstrate that n=90, exactly as in Polyommatus ripartii. We do not confirm the proposed difference between Polyommatus nephohiptamenos and Polyommatus ripartii in number of large chromosomes (Coutsis and De Prins 2007). In our squash preparations, both species demonstrate one big and one medium-sized element in the haploid chromosome set.
Polyommatus aroaniensis
The haploid chromosome number for this taxon was erroneously given as n=15-16 by Brown (1976a), and later corrected to be n=48 in few studied metaphase plates by Coutsis et al. (1999). In the single studied sample we were able to make a precise count of chromosome elements and found the haploid chromosome number to be n=47. Both counts (previous n=48 and n=47 in this study), are essentially different from those found in closely related Polyommatus timfristos and Polyommatus orphicus (Kolev 2005, this work) and Polyommatus humedasae (Troiano et al. 1979, Vila et al. 2010).
Polyommatus orphicus and Polyommatus eleniae
The chromosome number of Polyommatus orphicus was first established by Kolev (2005) who discovered n=41-42 in population from Hvoyna (Bulgaria), thus, similar to the karyotype found in Polyommatus dantchenkoi from remote east Turkey (Lukhtanov et al. 2003).
The chromosome number of Polyommatus eleniae was established first by Coutsis and De Prins (2005) who discovered n=41 in population from Falakro Mt near Granitis (Greece). Coutsis and De Prins reported that despite identical chromosome number, karyotypes of Polyommatus orphicus and Polyommatus eleniae were different in respect to their structure. Karyotype of Polyommatus eleniae was reported to be more asymmetrical than karyotype of Polyommatus orphicus (that is, the chromosomes were more differentiated with respect to their size).
Here we reinvestigated the karyotypes of Polyommatus orphicus and Polyommatus eleniae originating directly from their type-localities. Our data confirm previous chromosome number counts, but do not confirm the differences in karyotype structures. In our opinion, the presumed differences could appear because of differences in staining techniques used by Kolev (2005) for Polyommatus orphicus and Coutsis and De Prins (2005) for Polyommatus eleniae (see Wiemers and De Prins 2004). In our study, we used the same technique for both taxa, and we did not find any differences in the karyotype structure.
Polyommatus timfristos
The haploid chromosome number of this taxon is established first here as n=38 and thus differs by at least three fixed chromosome fussions/fixions from Polyommatus orphicus orphicus and Polyommatus orphicus eleniae (n=41-42). This number is similar (but not identical) to that found in Polyommatus humedasae (n=39, Vila et al. 2010). We are not sure that the karyotypes of Polyommatus timfristos and Polyommatus humedasae are related in their origin because they are not found in proximity and separated by an area where Polyommatus orphicus with n=41–42 is distributed.
Taxonomy
Polyommatus admetus
The Balkan and west Turkish populations of Polyommatus admetus have a unique hindwing underside pattern (Polyommatus admetus type, Fig. 13a, b, c, d) and can be easily separated on the basis of morphology from other species. However, some taxonomic and identification problems appear if oriental populations of Polyommatus admetus sensu lato are considered. In 2004, Polyommatus admetus yeranyani from Armenia and Polyommatus admetus malievi from Azerbaijan were described (Dantchenko and Lukhtanov 2005). The two last taxa differ from the nominative subspecies morphologically. They usually have a distinct white streak on the underside of the hindwing, and the marginal pattern of the wing underside is not as prominent as in Polyommatus admetus admetus. In fact, Polyommatus admetus yeranyani and Polyommatus admetus malievi are phenotypically similar to Polyommatus ripartii and Polyommatus demavendi, and their identification is not always easy. Karyological analysis revealed a minor difference between the western and oriental forms (see above), and molecular analysis demonstrated that they were differentiated with respect to COI barcodes and did not constitute together a monophyletic entity. This barcode distinctness is especially clearly expressed in the haplotype network (Fig. 10). Therefore, in accordance with the criterion of avoiding non-monophyletic groups in taxonomy (Vila et al. 2013), they should be treated as distinct species Polyommatus admetus and Polyommatus yeranyani.
Polyommatus ripartii
The distribution of COI haplotypes in Polyommatus ripartii demonstrates a very complex picture. This taxon is represented by several clades on the phylogenetic reconstructions. The West-European clade includes butterflies from France, Italy and Spain. Another clade (a “mixed”, or Eurasian clade) includes butterflies from the whole Western Palaearctic region from Spain to Mongolia. Eastern Turkish-Caucasian clade (Polyommatus ripartii paralcestis) is strongly differentiated and appears as a group close to Polyommatus demavendi. Complicated taxonomy and phylogeography of Polyommatus ripartii have recently been topics of several specific studies and publications (Vila et al. 2010, Vodolazhsky et al. 2011, Dincă et al. 2013a, Przybyłowicz et al. 2014) and are out of the focus of the present paper. The sequences obtained in our study confirm that Balkan samples represent one of the major clades within Polyommatus ripartii populations, thus Polyommatus ripartii pelopi is confirmed as a valid subspecies.
Polyommatus nephohiptamenos
Taxonomic interpretation of this local Balkan endemic is difficult since it is morphologically very similar and chromosomally seems to be identical to the close species Polyommatus ripartii. However, distinct COI barcodes in combination with ecological differentiation (Polyommatus nephohiptamenos is a high altitude species, whereas Polyommatus ripartii pelopi can be found usually at middle and low elevations) do not allow us to reject the pre-existing taxonomic hypotheis that Polyommatus nephohiptamenos represents a distinct taxonomic entity. The fact that Polyommatus nephohiptamenos retains its homogeneity with respect to COI being surrounded by closely related Polyommatus ripartii is additional indirect evidence for a presence of genetic boundaries between them. Further molecular and genetic studies are required to understand the real taxonomic status of Polyommatus nephohiptamenos.
Polyommatus orphicus
Polyommatus dantchenkoi orphicus was described (Kolev 2005) and later considered (Tshikolovets 2011, Eckweiler and Bozano 2016) as a subspecies of Polyommatus dantchenkoi, a species known from east Turkey, because Polyommatus dantchenkoi dantchenkoi and Polyommatus dantchenkoi orphicus shared a similar phenotype and number of chromosomes (Lukhtanov et al. 2003, Kolev 2005). At times, Polyommatus orphicus has been considered as a distinct species (e.g. Van Swaay et al. 2010); however, its species level status was not justified.
Our molecular data demonstrate that, despite similarity in number of chromosomes, Polyommatus dantchenkoi dantchenkoi and Polyommatus dantchenkoi orphicus are not closely related as was previously thought. In the haplotype network, these taxa were found to be placed in the opposite parts of the recovered net, being separated by a number of other species (Fig. 11). Their merging would result in a polyphyletic assemblage (Fig. 8). Avoiding non-monophyletic groups is a preferable option in practical taxonomy (Talavera et al. 2013a). Therefore, Polyommatus dantchenkoi and Polyommatus orphicus should be considered as two distinct species. We should also note that the COI barcodes alone (as in our study) can provide weak evidence for monophyly or non-monophyly of taxa since trees inferred from single markers sometimes display relationships that reflect the evolutionary histories of individual genes rather than of the species being studied. In case of Agrodiaetus, COI barcodes showing such a discrepancy between species and gene trees may be a result of interspecific mitochondrial introgression (Lukhtanov et al. 2008, 2015b). Despite this limitation, we argue that monophyletic clusters resulting from the DNA barcode analysis are better primary taxonomic hypotheses than para- or polyphyletic ones (Lukhtanov et al. 2016).
Polyommatus eleniae was described from a place located 80 km south-west from the type locality of Polyommatus orphicus. Polyommatus orphicus and Polyommatus eleniae have the same number of chromosomes, but it was supposed that they were different in karyotype structure (Coutsis and De Prins 2005). Additionally, it was supposed that Polyommatus eleniae differed from Polyommatus orphicus by the constant lack of a white postdiscal streak on the forewing underside (character 1 on Fig. 12) and by strong reduction or total lacking of a white streak on the hindwing underside (character 2 on Fig. 12) (Coutsis and De Prins 2005). In Polyommatus orphicus these streaks are supposed to be always sharply defined (Kolev 2005). Our study does not support the difference in karyotypes (see above). Our analyses showed that the supposed differences in morphology disappeared if individual variations were taken into account. Although the “typical” phenotype of Polyommatus orphicus (Figs 12 and 14a) often present in in Hvoyna, the individuals with different level of reduction of white streak on both fore- and hindwing underside are very common (Fig. 13h, i, j). These individuals with confidence can be identified as Polyommatus orphicus as they have the same karyotype and do not differ in mitochondrial haplotypes. Thus, the morphological difference between individuals from Hvoyna (Polyommatus orphicus) and Falakro Mt (Polyommatus eleniae) is not clear and is not based on fixed characters. The difference in karyotypes was also not confirmed in our analysis (see the section Chromosomal diversity above). Therefore, we conclude that the population from Falakro Mt is most probably conspecific with Polyommatus orphicus and can be treated as a subspecies Polyommatus orphicus eleniae.
Polyommatus aroaniensis
This taxon was first described by Brown (1976a) as a subspecies of Polyommatus alcestis and two years later was raised to species rank (Brown and Coutsis 1978). Despite its similarity to other taxa of the brown complex, especially with Polyommatus humedasae, Polyommatus orphicus orphicus and Polyommatus orphicus eleniae, it differs by its karyotype and COI barcodes. Its species distinctness confirmed by chromosomal analysis (Coutsis et al. 1999) has never been questioned. Thus, there has been no problem with treatment of Polyommatus aroanisnsis as a separate species. However, there are numerous identification problems associated with Polyommatus aroaniensis because several populations from Central and Northern Greece, as well as from other countries of the Balkan Peninsula were identified as Polyommatus aroaniensis (see the section Distribution areas below), but their karyotypes were not studied. In our work, we discovered that two of these populations (from Timfristos Mt and Parnassos Mt) represented a previously unrecognized species. Below we name it and provide its formal description.
Polyommatus (Agrodiaetus) timfristos
Lukhtanov, Vishnevskaya & Shapoval sp. n.
http://zoobank.org/58B77480-1FD0-423B-8FF9-BF39E79F177C
Holotype
(Fig. 16h). male, field code LR-08-247, GenBank accession number KY066725 for COI and KY081279 for ITS2; Greece, Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605 E, 1270 m, 20 July 2008, V.A. Lukhtanov and N.A. Shapoval leg., deposited in Zoological Institute of the Russian Academy of Science (St. Petersburg).
COI barcode sequence of the holotype, 657 base pairs.
ACATTATATTTTATTTTTGGAATTTGAGCAGGAATAGTAGGAACATCTCTAAGAATTTTAATTCGTATGGAATTAAGAACTCCTGGATCCTTAATTGGAAATGATCAAATTTATAATACTATTGTTACAGCCCATGCATTTATTATAATTTTTTTTATGGTTATACCTATTATAATTGGAGGATTTGGTAACTGATTAGTTCCCTTAATATTAGGAGCACCTGATATAGCTTTTCCACGATTAAATAATATGAGATTTTGATTATTACCGCCATCATTAATACTACTAATTTCTAGAAGAATTGTAGAAAATGGAGCAGGAACAGGATGAACAGTTTACCCCCCACTTTCATCAAATATTGCACATGGAGGATCATCTGTAGATTTAGCAATTTTCTCTCTTCATTTAGCGGGAATTTCTTCAATTTTAGGAGCAATTAATTTTATTACAACTATCATTAATATACGAGTAAATAATTTATCTTTTGATCAAATATCATTATTTATTTGAGCAGTGGGAATTACAGCATTATTATTACTTTTATCATTGCCTGTATTAGCTGGGGCAATTACCATATTATTAACAGATCGAAATCTTAATACCTCATTCTTTGACCCAGCTGGTGGAGGAGATCCAATTTTATATCAACATTTATTT
Haploid chromosome number of the holotype n=38 (Fig. 4c).
Paratypes.
Four males, field codes LR-08-255, LR-08-258, LR-08-273, LR-08-274, forewing length 17–18 mm, the same data as holotype. Male: field code LR-08-205, Greece, Parnassos, 38°33.311'N; 22°34.300 E, 1750 m, 19 July 2008, V.A. Lukhtanov and N.A.Shapoval leg. Five females: forewing length 15–16 mm; Greece, Timfristos Mt, Karpenisi, 38°55.554'N; 21°48.460 E, 1490 m, 21 July 2008, V.A. Lukhtanov and N.A. Shapoval leg. Two females: forewing length 14.5–15.5 mm; Greece, Parnassos, 38°33.311'N; 22°34.300 E, 1750 m, 19 July 2008, V.A. Lukhtanov and N.A. Shapoval leg.. All paratypes are deposited in Zoological Institute of the Russian Academy of Science (St. Petersburg).
Males
(Fig. 16d–i). Forewing length 16.2–18.2 mm. Upperside: ground color completely brown. Discoidal, submarginal and antemarginal marking absent on both fore- and hindwings. Forewings with a developed sex brand and scaletuft. Fringe brown as ground color.
Underside: ground color light brown with yellowish coffee-milk tint. Greenish blue basal suffusion very slight, nearly lacking. One basal black spot is present only on hindwings. Discoidal black spot is present on the forewings, but can be slightly seen on the hindwings (absent or vestigial). Postdiscal black ocelli are encircled by a whitish border. They are prominent on the forewings, forming a strongly curved row. Postdiscal black ocelli on the hindwing small. Submarginal and antemarginal marking is absent on the forewings, and absent or vestigial on the hindwings. White streak on hindwings clearly visible. In one specimen the white streak is vestigial, in one the white streak is almost absent (can be slightly distinguished), and in one specimen there is an additional short streak between postdiscal and submarginal areas of the wing, straight under the main white streak. Fringe brown, slightly darker than the underside ground color.
Genitalia: the male genitalia have a structure typical for other species of the subgenus Agrodiaetus (Coutsis, 1986).
Females
(Fig. 17a–g). Forewing length 15.8–17.5 mm.Upperside: ground color as in males, but lighter dark brown and without sex brand and scaletuft. Fringe greyish brown. Underside: ground color and general design as in males but fringes lighter-colored. Greenish blue basal suffusion almost invisible. White streak on hindwing underside is present in all paratypes and demonstrates a variable level of reduction.
Figure 17.
Paratypes of Polyommatus timfristos sp. n. (females). a, b samples from Parnassos Mt c–g samples from Timfristos Mt.
Diagnosis.
Polyommatus timfristos (n=38) differs by at least three fixed chromosome fusions/fissions from the most closely related and allopatric Polyommatus orphicus orphicus and Polyommatus orphicus eleniae (n=41-42). Polyommatus timfristos (n=38) differs by at least nine fixed chromosome fusions/fissions from allopatric Polyommatus aroaniensis (n=47). From the closely related Polyommatus orphicus and Polyommatus aroaniensis, Polyommatus timfristos differs also by a number of nucleotide substitutions within the studied 657-bp fragment of the mitochondrial COI gene.
The chromosome number in Polyommatus timfristos (n=38) is similar (but not identical) to that found in Polyommatus humedasae (n=39, Vila et al. 2010). However, we are not sure that these karyotypes are related in their origin because they are not found in proximity and separated by an area where Polyommatus orphicus with n=41-42 is distributed. With respect to COI barcodes, the pair Polyommatus timfristos/Polyommatus humedasae is more differentiated than pairs Polyommatus timfristos/Polyommatus aroaniensis and Polyommatus timfristos/Polyommatus orphicus.
From sympatric and syntopic Polyommatus ripartii pelopi the new species can usually be distinguished by the absence of submarginal marking and strong reduction of greenish blue basal suffusion. These characteristics are usually (but not always) better expressed in Polyommatus ripartii pelopi specimens. In doubtful cases, the separation is only possible on the base of chromosomal and molecular markers since these species are different: the chromosome number of Polyommatus ripartii pelopi is n=90; they also have fixed differences in 33 positions within the studied 657-bp fragment of COI gene.
Ecology.
Polyommatus timfristos sp. n. inhabits xerothermic and xeromontane localities and dry meadows from 1200 to 1800 m altitude (Figs 32–35). It was found in complete syntopy with Polyommatus ripartii pelopi and Polyommatus admetus.
Etymology.
Timfristos is a mountain in the eastern part of Evrytania and the western part of Phthiotis in Central Greece. The name is a noun.
Distribution areas
Polyommatus admetus
This species is widespread in the Balkan Peninsula. It is local in the northern part of Hungary (Ilonczai and Bálint 2001, Bálint et al. 2006) and recorded in Slovakia (Kulfan and Kulfan 1992, Eckweiler and Bozano 2016). It has been shown to be widely distributed in the western part of Romania, but no exact localities were provided (Eckweiler and Bozano 2016). It is common in Greece and found in Croatia, Bosnia and Herzegovina, Montenegro, Serbia, Bulgaria, The Republic of Macedonia, Albania and European Turkey (Sijaric and Mihljevic 1972, Hesselbarth et al. 1995, Abadjiev 2001, Tolman and Lewington 2008, Eckweiler and Bozano 2016).
Polyommatus ripartii
Fig. 27
Polyommatus ripartii is widespread in the southern part of the Balkan Peninsula (Greece and Bulgaria); however, it is more local in the north. It is known from Albania, the Republic of Macedonia, south Serbia (Kudrna et al. 2011, Eckweiler and Bozano 2016), Bosnia and Herzegovina (Koren 2010). It was mentioned for European Turkey (Hesselbarth et al. 1995, Tolman and Lewington 2008) and recently found in Croatia (Koren 2010, Dincă et al. 2013a).
Polyommatus nephohiptamenos
Polyommatus nephohiptamenos has a dot-like distribution area and is known from the high altitudes of north-east Greece (Mt Pangeon, Mt Phalakro and Mt Orvilos) and south-west Bulgaria (Mt Orvilos, also known as Mt Slavyanka, Mt Alibotush and Kitka Planina) (Kolev 1994, Tolman and Lewington 2008, Eckweiler and Bozano 2016).
Polyommatus aroaniensis
Fig. 31
Polyommatus aroaniensis has been considered as a relatively widespread species (Kolev and van der Poorten 1997). Apart from its type-locality (South Greece, Peloponnese), it has been recorded in different parts of Central and Northern Greece (Brown 1976a, Wakeham-Dawson and Spurdens 1994, Wakeham-Dawson 1998, Pamperis 2009), from a few areas in south Macedonia (Kolev and van der Poorten 1997, Melovski and Bozhinovska 2014) and from some localities in south-west Bulgaria and one isolated place in the central part of the country (Abadjiev 2001, Kolev 1994, Kolev and van der Poorten 1997). Tshikolovets (2011) and Eckweiler and Bozano (2016) show its distribution extending into Albania, although the species has not been recorded from this country in recent surveys (Verovnik and Popović 2013). Verovnik et al. (2015) recorded it in Bosnia and Herzegovina. Koren and Laus (2015) recorded it in Croatia; however, this record was not confirmed by molecular data (Lovrenčić et al. 2016).
Our chromosomal data confirm Polyommatus aroaniensis in South Greece (Peloponnese), but cannot confirm it in Central and Northern Greece and in Bulgaria where it is replaced by the closely related allopatric species Polyommatus timfritos and Polyommatus orphicus. In the light of the data obtained, the occurrence of Polyommatus aroaniensis in Bulgaria, Albania, Macedonia and Bosnia and Herzegovina seems to be doubtful and requires a confirmation based on chromosomal analysis. We cannot exclude that the populations from Albania, Macedonia and Bosnia and Herzegovina could represent Polyommatus orphicus or even undescribed taxa of the subgenus Agrodiaetus.
Polyommatus timfristos
This species is known from Timfristos and Parnassos Mts in Central Greece only.
Polyommatus orphicus
This species is known from South Bulgaria and Northern Greece only. However, its occurrence in other countries in the northern Balkan is theoretically possible (see above).
An alternative classification and conservation
Theoretically, the main groupings in the Polyommatus humedasae – Polyommatus orphicus – Polyommatus timfristos – Polyommatus aroaniesnsis subcomplex can be interpreted as subspecies-level taxa, if the polytypic species concept is applied. None of them appears to be sympatric in distribution, and taken together they form a moderately supported monophyletic lineage on the COI+ITS2 tree (Fig. 9). Under this scenario, this subcomplex would be considered a diverse array of allopatric populations, each of which possesses unique genetic attributes (karyotypes and molecular markers) and is distributed in a particular area within the Alp-Balkan region. As possible theoretical support for this alternative classification, one can argue that differences in chromosome numbers in Agrodiaetus do not necessarily result in complete reproductive isolation, and, at least in some particular cases, do not prevent interspecific hybridization and genetic introgression (Lukhtanov et al. 2015b).
However, even if the last statement is true, it does not mean that chromosomal rearrangements are irrelevant to formation of genetic barriers between populations. Chromosome changes have been shown to be important in speciation in the blue butterflies (Lukhtanov et al. 2005, 2015b, Kandul et al. 2007, Talavera et al. 2013b). Even a weak decrease in fertility in heterozygotes for multiple chromosomal rearrangenments can result in selection against them and in formation of a boundary between chromosomally diverged homozygous populations. Additional studies are required to shed light on this topic. Recent studies have treated Polyommatus humedasae, Polyommatus aroaniensis and Polyommatus orphicus as species-level taxa (Eckweiler and Bozano 2016), which our study suggests is a reasonable interpretation although distribution areas of Polyommatus aroaniensis and Polyommatus orphicus should be corrected. Based on our current knowledge, if Polyommatus humedasae, Polyommatus aroaniensis and Polyommatus orphicus are considered species-level taxa, Polyommatus timfristos should be treated as a species-level taxon as well.
Regardless of its taxonomic status as a species or subspecies, Polyommatus timfristos represents a unique entity within the genus Polyommatus that deserves additional study. A better understanding of its evolutionary history may be helpful in understanding mechanisms of chromosomal diversification within the genus, and may further elucidate the biogegraphy of the south Balkan and Aegean regions. As a distinct taxonomic entity occupying a very restricted area in Central Greece it should be considered a candidate on the list of protected species in Greece and the whole of Europe.
Biogeography
Analysis of distribution areas and phylogeny of the Polyommatus dolus lineage shows that the phylogeograpic history of this complex involved a combination of dispersal and vicariance events with a clear general trend of dispersal from the East (Iran), where the group most likely arose, to the West: to the Mediterranean area and to the Iberian Peninsula (Vila et al. 2010). The Europe was estimated to be colonized approximately 1.24 Mya (range 0.88–1.64 Mya). Approximately 1.15 Mya (range 0.80–1.51 Mya), the European lineage was divided into three subclades located (1) in the Balkan Mountains and Alps (Polyommatus aroaniensis sensu auctorum: the Balkans; Polyommatus humedasae: the Alps), (2) southern Spain (Polyommatus violetae), and (3) the Iberian-Italian region (Polyommatus fabressei + Polyommatus dolus), respectively (Vila et al. 2010).
Three chromosomal sublineages discovered in our study (Polyommatus aroaniensis sensu stricto + Polyommatus orphicus +Polyommatus timfristos) represent late Pleistocene splits of the Balkan subclade that evolved in allopatry within the Balkan refugium. Given the deep level of chromosomal divergence between these sublineages, we assume that there was a long period of allopatric differentiation when they were separated by geographic or/and ecological barriers. In our opinion, this is evidence for presence of three separate Balkan subrefugia in the past (Pelonnese, Central Greece and Northern Grecee/South Bulgaria).
Greece, as a part of the Balkan Peninsula, has been already reported to harbor genetically differentiated lineages from the rest of the Balkans for a number of animal species as a result of evolution in multiple separate refugia (Kasapidis et al. 2005, Alexandri et al. 2012, Karaiskou et al. 2014). Thus, our data provide a chromosomal evidence for this refugia-within-refugia concept (Gòmez and Lunt 2007, Karaiskou et al. 2014), and the discovery of a new, chromosomally diverged species Polyommatus timfristos stresses the biogeographic importance of Central Greece as a separate Pleistocene refugium within the Balkans.
Taxonomic conclusion
We propose the following taxonomic arrangement of the Polyommatus dolus and Polyommatus admetus lineages (chromosome numbers are in parentheses when known, the Balkan taxa are in bold):
Polyommatus dolus lineage
Polyommatus dolus (Hübner, [1823])
Polyommatus dolus dolus (Hübner, [1823]) (n=123-125)
Polyommatus dolus vittatus (Oberthür, 1892) (n=124-125)
Polyommatus dolus virgilia (Oberthür, 1910) (n=122)
Polyommatus dolus gargano (Wimmers, 1931) (n=122)
Polyommatus dolus paravirgilia Verity, 1943 (n unknown)
Polyommatus fulgens (Sagarra, 1925)
Polyommatus fulgens fulgens (Sagarra, 1925) (n=109)
Polyommatus fulgens ainsae (Forster, 1961) (n=108-110)
Polyommatus fulgens pseudovirgilia (de Lesse, 1962) (n=108)
Polyommatus fulgens leonensis (Verhulst, 2004) (n unknown)
Polyommatus menalcas (Freyer, [1837]) (n=85)
Polyommatus fabressei (Oberthür, 1910) (n=90)
Polyommatus violetae (Gómez-Bustillo, Expósito & Martínez, 1979)
Polyommatus violetae violetae (Gómez-Bustillo, Expósito & Martínez, 1979) (n=90)
Polyommatus violetae subbaeticus (Gil-T. & Gil-Uceda, 2005) (n=90)
Polyommatus humedasae (Toso & Balletto, 1976) (n=39)
Polyommatus orphicus Kolev, 2005
Polyommatus orphicus orphicus Kolev, 2005 (n=41-42)
Polyommatus orphicus eleniae Coutsis & De Prins, 2005 (n=41-42)
Polyommatus timfristos Lukhtanov, Vishnevskaya & Shapoval, sp. n. (n=38)
Polyommatus aroaniensis (Brown, 1976) (n=47)
Polyommatus alcestis (Zerny, 1932) (n=20-21)
Polyommatus karacetinae (Lukhtanov & Dantchenko, 2002)
Polyommatus karacetinae karacetinae (Lukhtanov & Dantchenko, 2002) (n=19)
Polyommatus karacetinae urmiaensis Schurian & Ten Hagen, 2003, comb. n. (n=19)
Polyommatus dantchenkoi (Lukhtanov & Wiemers, 2003) (n=40-42)
Polyommatus eriwanensis (Forster, 1960) (n=32-34)
Polyommatus interjectus (de Lesse, 1960) (n=29-32)
Polyommatus rjabovianus (Koҫak, 1980) (= rjabovi (Forster, 1960)
Polyommatus rjabovianus rjabovianus (Koҫak, 1980) (n=49)
Polyommatus rjabovianus masul Lukhtanov, Dantchenko, Vishnevskaya & Saifitdinova, 2015 (n=43)
Polyommatus valiabadi (Rose & Schurian, 1977) (n=24)
Polyommatus admetus lineage
Polyommatus ripartii (Freyer, 1830)
Polyommatus ripartii ripartii (Freyer, 1830) (= agenjoi Forster, 1965; = budashkini Kolev & de Prins, 1995; = exuberans Verity, 1926; = montanesa Gómez-Bustillo, 1971; = mozuelica Agenjo, 1973; = ovchinnikovi Lukhtanov & Dantchenko, 2002; = ramonagenjo Koçak & Kemal, 2001; = rippertii Boisduval, 1832; = sarkani Lukhtanov & Dantchenko, 2002; = susae Bertaccini, 2003) (n=90)
Polyommatus ripartii pelopi (Brown, 1976) (n=90)
Polyommatus ripartii paralcestis (Forster, 1960) (n=90)
Polyommatus ripartii colemani (Lukhtanov & Dantchenko, 2002) (n=90)
Polyommatus ripartii tengritaghicus Koҫak & Kemal, 2001 (n unknown)
Polyommatus nephohiptamenos (Brown & Coutsis, 1978) (n=90)
Polyommatus demavendi (Pfeiffer, 1938) (n=67-74)
Polyommatus demavendi demavendi (Pfeiffer, 1938)
Polyommatus demavendi amasyensis (de Lesse, 1961)
Polyommatus demavendi belovi (Dantchenko & Lukhtanov, 2005)
Polyommatus demavendi ahmadi (Carbonell, 2001)
Polyommatus demavendi lorestanus Eckweiler, 1997
Polyommatus khorasanensis (Carbonell, 2001) (n=74)
Polyommatus pseudorjabovi Lukhtanov, Dantchenko, Vishnevskaya & Saifitdinova, 2015 (n=79)
Polyommatus admetus (Esper, [1783]) (= anatoliensis Forster, 1960) (n=80)
Polyommatus yeranyani (Dantchenko & Lukhtanov, 2005), stat. n.
Polyommatus yeranyani yeranyani (Dantchenko & Lukhtanov, 2005) (n=78-80)
Polyommatus yeranyani malievi (Dantchenko & Lukhtanov, 2005), comb. n. (n=78-80)
Supplementary Material
Acknowledgements
The complete financial support for this study was provided by the grant from the Russian Science Foundation N 14-14-00541 to The Zoological Institute of the Russian Academy of Sciences. The work was held in the laboratory of “Chromas” Core facility and Centre for Molecular and Cell Technologies of St. Petersburg State University, and in “The Laboratory of animal genetics” of Biological Institute. We thank Nazar Shapoval, Elena Pazhenkova and Lukas Rieppel for their help in material collecting in the Balkan Peninsula, and Svetlana Nedoshivina and Andrei Barabanov for their help in material collecting in Tigireksky Reservation, Russia. We thank Lukasz Przybyłowicz and Nazar Shapoval for critical comments. We also would like to mention, that the expedition to the Balkan Peninsula in 2008 would have been impossible without the help of Naomi Pierce.
Appendix 1
Alphabetical list of the nominal taxa described within the Polyommatus (Agrodiaetus) admetus and Polyommatus (Agrodiaetus) dolus complexes.
admetus (Esper, [1783])
Original combination: P.[apilio] Pl.[ebeius] Rur.[alis] Argus Admetus
In: Esper EJC (1777-1807) Die Schmetterlinge in Abbildungen nach der Natur mit Beschreibungen (I)2: 148; Tab. LXXXII, figs 2, 3.
Type locality: “Ungarn ... bis nach Semlin an die Gränze von Sclavonien” [Hungary].
Syntypes in Naturwissenschaftliche Sammlung, Wiesbaden, Germany, and in Zoologische Staatssammlung, Munich, Germany (after Häuser and Eckweiler 1997).
Current status: species.
ahmadi (Carbonell, 2001)
Original combination: Agrodiaetus ahmadi
In: Carbonell F (2001) Contribution à la connaissance du genre Agrodiaetus Hübner (1822), Agrodiaetus ahmadi et Agrodiaetus khorasanensis nouvelles espèces dans le Nord de l‘Iran (Lepidoptera, Lycaenidae). Linneana Belgica 18: 105–110.
Type locality: environs d’Alulak, 1600 m., E. Prov. De Zanjan, N. Iran [N Iran, Alilak, Qazvin].
Holotype in Muséum National d’Histoire Naturelle, Paris.
Current status: subspecies of Polyommatus (Agrodiaetus) demavendi (after Eckweiler and Bozano 2016).
agenjoi (Forster, 1965)
Original combination: Agrodiaetus admetus agenjoi
In: Forster W (1965) Agrodiaetus admetus agenjoi ssp. n. Entomologische Zeitschrift 75(18): 198.
Type locality: “Barcelona, Taradell” [Spain: Barcelona].
Holotype in Museo Nacional de Ciencias Naturales, Madrid, Spain (after Häuser and Eckweiler 1997).
Current status: preoccupied name (see ramonagenjoi).
ainsae (Forster, 1961)
Original combination: Agrodiaetus dolus ainsae
In: Forster W (1961) Bausteine zur Kenntnis der Gattung Agrodiaetus Scudd. (Lep. Lycaen.) II. Zeitschrift der Wiener Entomologischen Gesellschaft 46: 76. Taf. 14, 15, figs 5, 6.
Type locality: “Spanien, Pyrenäen, Ainsa” [Spain: Huesca].
Holotype in Naturhistorisches Museum, Wien, Austria.
Current status: subspecies of Polyommatus (Agrodiaetus) fulgens (after Eckweiler and Bozano 2016).
alcestis (Zerny, 1932)
Original combination: Lycaena (Hirsutina) ripperti alcestis
In: Zerny H (1932) Lepidopteren aus dem nördlichen Libanon. Deutsche Entomologische Zeitschrift Iris 46(4): 186.
Type locality: Becharré [Lebanon].
Lectotype in The Natural History Museum, London (after Bálint 1999: 9).
Current status: species.
amasina (Neuburger, 1900)
Original combination: Lycaena menalcas ab. amasina
In: Neuburger W (1900) Lycaena menalcas Frr. ♂ aberr. (Lep.). Illustrierte Zeitschrift für Entomologie 5: 370.
Type locality: “Aus der Gegend von Amasia” [Turkey: Amasya].
Holotype in coll. W. Neuburger [?] (after Häuser and Eckweiler 1997).
Current status: unavailable (infrasubspecific name).
amasyensis (de Lesse, 1961)
Original combination: Agrodiaetus demavendi amasyensis
In: de Lesse H (1961) Variations géographiques des caractères externes chez les espèces autrefois réunies sous le nom d’Agrodiaetus ripartii Freyer (Lep. Lycaenidae). Revue Française d’Entomologie 28(2): 96.
Type locality: “Amasya” [Turkey: Amasya].
Holotype in Museum National d’Histoire Naturelle, Paris, France (after Häuser and Eckweiler 1997).
Current status: subspecies of Polyommatus (Agrodiaetus) demavendi (after Eckweiler and Bozano 2016).
anatoliensis (Forster, 1960)
Original combination: Agrodiaetus admetus anatoliensis
In: Forster W (1960) Einige neue Formen der Gattung Agrodiaetus Scudd. (Lep. Lycaen.). Entomologische Zeitschrift 70(3): 20.
Type locality: “Akshehir” [Turkey: Akşehir].
Holotype in Zoologische Staatssammlung, München, Germany (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) admetus admetus (after Eckweiler and Bozano 2016).
anticodiscoelongata (Verity, 1943)
Original combination: Agrodiaetus dolus anticodiscoelongata
In: Verity R (1943) Farfalle diurne d’Italia 2: 323.
Type locality: “Monti Sibillini” [Central Italy].
Current status: synonym of Polyommatus (Agrodiaetus) dolus virgilius (after Eckweiler and Bozano 2016).
aroaniensis (Brown, 1976)
Original combination: Agrodiaetus alcestis aroaniensis
In: Brown J (1976) Notes regarding previously undescribed European taxa of the genera Agrodiaetus Hübner, 1822 and Polyommatus Klug, 1801 (Lep., Lycaenidae). Entomologist’s Gazette 27(2): 78.
Type locality: “Kamena Allonia, Mt. Chelmos” [Greece: Peloponnese].
Holotype in coll. J. Brown, Sutton (after Häuser and Eckweiler 1997).
Current status: species.
belovi (Dantchenko & Lukhtanov, 2005)
Original combination: Agrodiaetus belovi
In: Dantchenko A, Lukhtanov V (2005) New taxa of the brown species-complex of the genus Agrodiaetus Hübner, (1822) from Transcaucasia (Lepidoptera, Lycaenidae). Atalanta 35: 327–334, 472–475.
Type locality: Armenia, Gegamsky mts.
Holotype in Museum of Comparative Zoology (Harvard University, Cambridge, MA, USA).
Current status: subspecies of Polyommatus (Agrodiaetus) demavendi (after Eckweiler and Bozano 2016).
budashkini (Kolev & De Prins, 1995)
Original combination: Polyommatus (Agrodiaetus) budashkini
In: Kolev Z, De Prins W (1995) A new species of the “brown Agrodiaetus” complex from the Crimea (Lepidoptera: Lycaenidae). Phegea 23(2): 121.
Type locality: “Crimea, Sudak” [Ukraine: Crimea].
Holotype in Vlaamse Lepidoptera Collectie, Antwerpen.
Current status: synonym of Polyommatus (Agrodiaetus) ripartii ripartii.
colemani (Lukhtanov & Dantchenko, 2002)
Original combination: Agrodiaetus ripartii colemani
In: Lukhtanov V, Dantchenko A (2002) Descriptions of new taxa of the genus Agrodiaetus Hübner, [1822] based on the karyotype investigation (Lepidoptera, Lycaenidae). Atalanta 33(1/2): 81–107.
Type locality: Kazakhstan, Shymkentskaya oblast’, Ugamskiy Khrebet, Saryaigyr, 1600 m.
Holotype in Museum of Comparative Zoology, Harvard University, Cambridge, MA, USA.
Current status: subspecies of Polyommatus (Agrodiaetus) ripartii.
crassipuncta (Stauder, 1921)
Original combination: Lycaena dolus virgilia Obth. f. n. crassipuncta
In: Stauder (1921) Deutsche Entomologische Zeitschrift [Iris] 35: 31.
Type locality: “Unteritalien” [South Italy].
Current status: synonym of Polyommatus (Agrodiaetus) dolus virgilius (after Eckweiler and Bozano 2016).
dantchenkoi (Lukhtanov & Wiemers, 2003)
Original combination: Agrodiaetus dantchenkoi
In: Lukhtanov V, Wiemers M, Meusman K (2003) Description of a new species of the “brown” Agrodiaetus complex from South-East Turkey (Lycaenidae). Nota lepidopterologica 26(1/2): 65–71.
Type locality: Turkey, Prov. Van, 34 km N Catak [SE Turkey (Van)].
Holotype in Museum of Comparative Zoology, Harvard University, Cambridge, MA, USA.
Current status: species.
demavendi (Pfeiffer, 1938)
Original combination: Lycaena ripertii Frr. ssp. n. demavendi m.
In: Pfeiffer E (1938) Notizen über persische Lycaenidae (Lepid.). Mitteilungen der Münchner Entomologischen Gesellschaft 28(2): 194.
Type locality: Ort Demavend (Tar-Tal) 2200-2500 m [Iran: Elburs mts.].
Holotype in Zoologische Staatssammlung, München, Germany (after Häuser and Eckweiler 1997).
Current status: species.
discoelongata (Courvoisier, 1914)
Original combination: Lycaens dolus disco-elongata
In: Courvoisier (1914) Deutsche Entomologische Zeitschrift [Iris] 28: 186.
Type locality: not stated.
Current status: synonym of Polyommatus (Agrodiaetus) dolus (after Eckweiler and Bozano 2016).
dolus (Hübner, [1823])
Original combination: Papilio dolus
In: Hübner J (1796-[1838]) Sammlung europäischer Schmetterlinge I. Taf. 159, figs 793–796.
Type locality: [S France].
Current status: species.
elachista (Dannehl, 1927)
Original combination: Lycaena dolus Hb. elachista
In: Dannehl F (1927) Neue Formen und geografische Rassen aus meinem Rhopaloceren-Ausbeuten der letzten Jaren. Mitteilungen der Münchner Entomologischen Gesellschaft 17: 7-8.
Type locality: “Mt. Sabini (Gennaro), Simbruini, Velino, Sirente,… bei Aquila, Gran-Sasso, Morrone, Agatone, Majella” [Central and South Italy].
Current status: synonym of Polyommatus (Agrodiaetus) dolus virgilius (after Eckweiler and Bozano 2016).
eleniae Coutsis & De Prins, 2005
Original combination: Polyommatus (Agrodiaetus) eleniae
In: Coutsis J, De Prins J (2005) A new brown Polyommatus (Agrodiaetus) from northern Greece (Lepidoptera: Lycaenidae). Phegea 33(4): 129–137.
Type locality: Greece, Makedonía, Dráma district, Mt. Falakró, eastern foothills, near Granítis, 900 m.
Holotype in Zöologisch Museum, Universiteit van Amsterdam, Netherlands.
Current status: subspecies of Polyommatus (Agrodiaetus) orphicus.
epidolus (Boisduval, 1840)
Original combination: Lycaena epidolus
In: Boisduval [JBA] (1840): Genera et Index Methodictis Europaeorum Lepidopterorurn, p. 13.
Type locality: “Turcia” [Turkey].
Lectotype in The Natural History Museum, London (after Bálint 1999: 28)
Current status: synonym of Polyommatus (Agrodiaetus) menalcas (after: Bálint 1999: 28; Eckweiler and Bozano 2016).
eriwanensis (Forster, 1960)
Original combination: Agrodiaetus ripartii eriwanensis
In: Forster W (1960) Einige neue Formen der Gattung Agrodiaetus Scudd. (Lep. Lycaen.). Entomologische Zeitschrift 70(3): 19.
Type locality: “Eriwan” [Armenia, Yerevan].
Holotype in Zoologische Staatssammlung, München, Germany (after Häuser and Eckweiler 1997).
Current status: species.
exuberans (Verity, 1926)
Original combination: Hirsutina admetus race exuberans
In: Verity R (1926) Zygaenae, Grypocera and Rhopalocera of the Cottian Alps compared with other races. The Entomologist’s Record and Journal of Variation 38(9): 121.
Type locality: “Oulx” [Italy: Piemonte: Torino].
Syntypes in Museo Zoologico de la Specola, Firenze, Italy (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) ripartii ripartii (after Vila et al. 2010).
fabressei (Oberthür, 1910)
Original combination: Lycaena rippertii fabressei
In: Oberthür C (1910) Notes pour servir à établir la Faune Franςaise et Algérienne des Lepidopteres (suite). Etudes de Lépidoptérologié Comparee 4(1): 260.
Type locality: “à Albarracin” [Spain: Teruel].
Lectotype in The Natural History Museum, London (after Bálint 1999: 30).
Current status: species.
fulgens (De Sagarra, 1925)
Original combination: Hirsutina dolus rassa fulgens
In: De Sagarra I (1925) Anotacions a la lepidopterologia ibèrica III (1). Formes noves dignes d’esment. Butlleti de la Institución Catalana de Historia Natural 5(9): 271.
Type locality: “Santa Coloma de Queralt” [Spain: Cataluňa].
Holotype in Museu de Catalunya, Barcelona, Spain (after Häuser and Eckweiler 1997).
Current status: species.
galloi (Balletto & Toso, 1979)
Original combination: Agrodiaetus galloi
In: Balletto E, Toso G (1979) On a new species of Agrodiaetus (Lycaenidae) from Southern Italy. Nota Lepidopterologica 2(1/2): 13–25.
Type locality: Pollino, Lucania, Southern Italy, loc. Piano di Ruggio, 1550 m.
Holotype in Museo Civico di Storia Naturale Giacomo Doria, Genova, Italy.
Current status: synonym of Polyommatus (Agrodiaetus) ripartii ripartii (after Vila et al. 2010).
gargano (Wimmers, 1931)
Original combination: Lycaena dolus var. gargano
In: Wimmers C (1931) Aus der Lepidopteren-Fauna Italiens (Apulien). Entomologische Zeitschrift 45(7): 96.
Type locality: “Mte. Gargano” [Italy: Puglia: Foggia].
Current status: subspecies of Polyommatus (Agrodiaetus) dolus (after Eckweiler and Bozano 2016).
humedasae (Toso & Balletto, 1976)
Original combination: Agrodiaetus humedasae
In: Toso G, Balletto E (1976) Una nuova specie del genere Agrodiaetus Hübn. (Lepidoptera Lycaenidae). Annali del Museo Civico di Storia Naturale Giacomo Doria 81: 125.
Type locality: dintorni di Cogne, Val d'Aosta [Italy: Valle d'Aosta].
Holotype in Museo Civico di Storia Naturale “Giacomo Doria”, Genova, Italy (after Häuser and Eckweiler 1997).
Current status: species.
infralunulata (Verity, 1943)
Original combination: Agrodiaetus dolus infralunulata
In: Verity R (1943) Farfalle diurne d’Italia 2: 323.
Type locality: not stated.
Syntypes possibly in The Natural History Museum, London (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) dolus virgilius (after Eckweiler and Bozano 2016).
interjectus (de Lesse, 1960)
Original combination: Agrodiaetus interjectus
In: de Lesse H (1960) Les nombres de chromosomes dans la classification du groupe d'Agrodiaetus ripartii Freyer (Lepidoptera Lycaenidae). Revue Franςaise d'Entomologie 27(3): 253.
Type locality: “Erzincan ... 1200 m” [Turkey: Erzincan].
Holotype in Museum National d’Histoire Naturelle, Paris, France (after Häuser and Eckweiler 1997).
Current status: species.
iris (Agenjo, [1973])
Original combination: Plebejus (Agrodiaetus) damon iris
In: Agenjo R (1971) Nuevas subespecies de ropalóceros ibéricos. Graellsia 26: 30.
Type locality: “Puerto de Pozazal, a 1 001 m., en Valdeprado del Río, provincia de Santander” [Spain: Santander].
Holotype in coll. G. Pardo, Torrelavega (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) fulgens pseudovirgilia (after Eckweiler and Bozano 2016).
karacetinae (Lukhtanov & Dantchenko, 2002)
Original combination: Agrodiaetus alcestis karacetinae
In: Lukhtanov V, Dantchenko A (2002) Description of new taxa of the genus Agrodiaetus (Hübner, 1822) based on karyotype investigation. Atalanta 33(1/2): 81–107.
Type locality: Turkey, Hakkari, Dez Valley, 1500 m.
Holotype in Institute of Systematic and Population Biology (Zoological Museum), Amsterdam, Netherlands.
Current status: species.
khorasanensis (Carbonell, 2001)
Original combination: Agrodiaetus khorasanensis
In: Carbonell F (2001) Contribution à la connaissance du genre Agrodiaetus Hübner (1822), Agrodiaetus ahmadi et Agrodiaetus khorasanensis nouvelles espèces dans le Nord de l’Iran (Lepidoptera, Lycaenidae). Linneana Belgica 18: 105–110.
Type locality: North-East Iran, Khorasan.
Holotype in Muséum National d’Histoire Naturelle in Paris.
Current status: species.
lefebvrii (Godart, [1824])
Original combination: Polyommatus lefebvrii
In: Latreille [PA], Godart [J B] (1819[-1824]) Encyclopédie Métho-dique. Histoire Naturelle. 9. Entomologie, ou Histoire Naturelle des Crustaces, des Arachnides et des Insectes, p. 696.
Type locality: “aux environs de Toulon ... et dans les Bouches-du-Rhône” [France: Var/Bouches-du-Rhône].
Syntypes in [Toulon] and [Bouches-du-Rhône] [?] (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) dolus dolus (after Eckweiler and Bozano 2016).
leonensis (Verhulst, 2004)
Original combination: Agrodiaetus ainsae leonensis
In: Verhulst J (2004) Description d’une nouvelle sous-espèce d’Agrodiaetus ainsae Forster, 1961 provenant de la province de Leon (Lep. Lycaenidae). Linneana Belgica 19(5): 209–212.
Type locality: the vicity of Cistierna, Leon province, northern Spain.
Holotype in Institut des Sciences Naturalles de Bruxelles (after Verhulst 2004).
Current status: subspecies of Polyommatus (Agrodiaetus) fulgens (after Eckweiler and Bozano 2016).
lorestanus Eckweiler, 1997
Original combination: Polyommatus (Agrodiaetus) demavendi lorestanus
In: Eckweiler W (1997) Neue Taxa von Polyommatus (Agrodiaetus) (Lepidoptera: Lycaenidae). Nachrichten des Entomologischen Vereins Apollo Supplementum 16: 8.
Type locality: “Iran, Lorestan, Dorud, Saravand, 2000-2300 m” [Iran: Zagros mts.].
Holotype in coll. W. Eckweiler in Naturkundemuseum, Karlsruhe, Germany.
Current status: subspecies of Polyommatus (Agrodiaetus) demavendi (after Eckweiler and Bozano 2016).
magnabrillata (Gomez-Bustillo, 1971)
Original combination: Plebejus dolus magnabrillata
In: Gomez-Bustillo M (1971) For un mejor conosimiento de los ropaloceros españoles. Sociedad de Ciencias Naturales Aranzadi, Publicacion N° 19: 8
Type locality: “Puerto de Pozazal (1000 m), T. M. de Valdeprado, Prov. de Santander” [Spain: Santander].
Holotype in coll. M. Gomez-Bustillo, Madrid (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) dolus pseudovirgilia (after Eckweiler and Bozano 2016).
malievi (Dantchenko & Lukhtanov, 2005)
Original combination: Agrodiaetis admetus malievi
In: Dantchenko A, Lukhtanov V (2005) New taxa of the brown species-complex of the genus Agrodiaetus Hübner, (1822) from Transcaucasia (Lepidoptera, Lycaenidae). Atalanta 35: 327–334, 472–475.
Type locality: Azerbaijan, Talysh mts.
Holotype in Museum of Comparative Zoology, Harvard University, Cambridge, MA, USA.
Current status: subspecies of Polyommatus (Agrodiaetus) yeranyani.
masul Lukhtanov, Dantchenko, Vishnevskaya & Saifitdinova, 2015
Original combination: Polyommatus (Agrodiaetus) rjabovianus masul
In: Lukhtanov V, Dantchenko A, Vishnevskaya M, Saifitdinova A (2015) Detecting cryptic species in sympatry and allopatry: analysis of hidden diversity in Polyommatus (Agrodiaetus) butterflies (Lepidoptera: Lycaenidae). Biological Journal of the Linnean Society 116: 468–485.
Type locality: North Iran, Gilan, vicinity of Masuleh, 2200 m.
Holotype in Zoological Institute of the Russian Academy of Science, St. Petersburg, Russia.
Current status: subspecies of Polyommatus (Agrodiaetus) rjabovianus.
menalcas (Freyer, [1837])
Original combination: Lycaena Menalcas
In: Freyer C ([1836]-1839) Neuere Beiträge zur Schmetterlingskunde. 3. Band, 38. Heft, p. 46; tab. 223, figs 2–3.
Type locality: “bei Konstantinopel” [Turkey: Istanbul].
Lectotype in The Natural History Museum, London (after Bálint 1999: 43).
Current status: species.
montanesa (Gomez-Bustillo, 1971)
Original combination: Plebejus rippartii montanesa
In: Gomez-Bustillo M (1971) Por un mejor conosimiento de los ropaloceros espanoles. Sociedad de Ciencias Naturales Aranzadi, Publicacion N° 19: 8.
Type locality: “T. M. de Valdeprado, Prov. de Santander” [Spain: Santander].
Holotype in coll. M. Gomez-Bustillo,Madrid [?] (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) ripartii (after Eckweiler and Bozano 2016).
mozuelica (Agenjo, [1973])
Original combination: Plebejus (Agrodiaetus) ripartii mozuelica
In: Agenjo R (1971 [1973]) Nuevas subespecies de ropalóceros ibéricos. Graellsia 26: 31.
Type locality: “Mozuelos, at 840 m., provincia de Burgos” [Spain: Burgos].
Holotype in National Institute of Entomology (Institute Español de Entomologia), Madrid, Spain (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) ripartii ripartii (after Eckweiler and Bozano 2016).
nephohiptamenos (Brown & Coutsis, 1978)
Original combination: Agrodiaetus nephohiptamenos
In: Brown J, Coutsis JG (1978) Two newly discovered lycaenid butterflies (Lepidoptera: Lycaenidae) from Greece, with notes on allied species. Entomologist's Gazette 29(4): 207.
Type locality: “mountains of NE. Greece, 1800 m” [Greece].
Holotype in coll. J. Brown, Sutton (after Häuser and Eckweiler 1997).
Current status: species.
obsoleta (Stauder, 1921)
Original combination: Lycaena dolus virgilia f.n. obsoleta
In: Stauder (1921) Deutsche Entomologische Zeitschrift [Iris] 35: 31
Type locality: “Unteritalien” [South Italy].
Syntypes [possibly in BMNH, London] (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) dolus virgilius (after Eckweiler and Bozano 2016).
orphicus Kolev, 2005
Original combination Polyommatus dantchenkoi orphicus
In: Kolev Z (2005) Polyommatus dantchenkoi (Lukhtanov et Wiemers, 2003) tentatively identified as new to Europe, with a description of a new taxon from the Balkan Peninsula (Lycaenidae). Nota lepidopterologica 28(1): 25–34.
Type locality: South Bulgaria, Rhodopi mts, Hvoyna.
Holotype in National Museum of Natural History, Sofia, Bulgaria.
Current status: species.
ovchinnikovi (Lukhtanov & Dantchenko, 2002)
Original combination: Agrodiaetus ripartii ovchinnikovi
In: Lukhtanov V, Dantchenko A (2002) Descriptions of new taxa of the genus Agrodiaetus Hübner , [1822] based on the karyotype investigation (Lepidoptera, Lycaenidae). Atalanta 33(1/2): 81–107.
Type locality: Kazakhstan, Vostochno-Kazakhstanskaya oblast’, Zyryanovskij raion, Kremnyukha, 450 m.
Holotype in Zoological Institute of the Russian Academy of Science, St. Petersburg, Russia.
Current status: synonym of Polyommatus (Agrodiaetus) ripartii ripartii.
paralcestis (Forster, 1960)
Original combination: Agrodiaetus ripartii paralcestis
In: Forster W (1960) Einige neue Formen der Gattung Agrodiaetus Scudd. (Lep. Lycaen.). Entomologische Zeitschrift 70(3): 17.
Type locality: “Akschehir, Sultan Dagh 1700-2200 m” [Turkey: Konya].
Holotype in Zoologische Staatssammlung, München, Germany (after Häuser and Eckweiler 1997).
Current status: subspecies of Polyommatus (Agrodiaetus) ripartii.
paravirgilius (Verity, 1943)
Original combination: Agrodiaetus dolus sattorazza paravirgilia
In: Verity R (1943) Farfalle diurne d’Italia 2: 325.
Type locality: “Monte Faito della penisola Sorrentina” [S. Italy, Sorrento peninsula].
Current status: subspecies of Polyommatus (Agrodiaetus) dolus (after Eckweiler and Bozano 2016).
paucipuncta Lhomme, 1927
Original combination: Polyommatus dolus paucipuncta
In: Lhomme (1927) Amateur De Papillons 3: 192.
Type locality: not stated.
Current status: synonym of Polyommatus (Agrodiaetus) dolus (after Eckweiler and Bozano 2016).
pelopi (Brown, 1976)
Original combination: Agrodiaetus ripartii pelopi
In: Brown J (1976) On two previously undescribed subspecies of Lycaenidae (Lepidoptera) from Greece. Entomologische Berichten 36(3): 47.
Type locality: Troupa, Mt. Chelmos, Greece, 1300 m [Greece: Peloponnese].
Holotype in coll. J. Brown, Sutton (after Häuser and Eckweiler 1997).
Current status: subspecies of Polyommatus (Agrodiaetus) ripartii.
pseudorjabovi Lukhtanov, Dantchenko, Vishnevskaya & Saifitdinova, 2015
Original combination: Polyommatus (Agrodiaetus) pseudorjabovi
In: Lukhtanov V, Dantchenko A, Vishnevskaya M, Saifitdinova A (2015) Detecting cryptic species in sympatry and allopatry: analysis of hidden diversity in Polyommatus (Agrodiaetus) butterflies (Lepidoptera: Lycaenidae). Biological Journal of the Linnean Society 116: 468–485.
Type locality: Azerbaijan, Talysh mt., Zuvand plateau, vicinity of Mistan village.
Holotype in Zoological Institute of the Russian Academy of Science, St. Petersburg.
Current status: species.
pseudovirgilia (de Lesse, 1962)
Original combination: Agrodiaetus dolus pseudovirgilia
In: de Lesse H (1962) Variation chromosomique chez Agrodiaetus dolus HB. (Lep. Lycaenidae). Alexanor 2(7): 285.
Type locality: “25 km W. Burgos, près Villanueva de Aragon” [Spain: Burgos].
Holotype in Museum National d’Histoire Naturelle, Paris, France (after Häuser and Eckweiler 1997).
Current status: subspecies of Polyommatus (Agrodiaetus) fulgens.
punctigera (Dannehl, 1927)
Original combination: Lycaena dolus Hb. punctigera
In: Dannehl F (1927) Neue Formen und geografische Rassen aus meinem Rhopaloceren-Ausbeuten der letzten Jaren. Mitteilungen der Münchner Entomologischen Gesellschaft 17: 7–8.
Type locality: “Mt. Sabini (Gennaro), Simbruini, Velino, Sirente,… bei Aquila, Gran-Sasso, Morrone, Agatone, Majella” [Central and South Italy].
Current status: synonym of Polyommatus (Agrodiaetus) dolus virgilius (after Eckweiler and Bozano 2016).
ramonagenjo Koçak & Kemal, 2001
Original combination: Polyommatus (s. str. (Agrodiaetus (Admetusia)) ripartii ssp. ramonagenjo
In: Koçak A, Kemal M (2001) Polyommatus Latr. Cinsideki Agrodiaetus Seksiyonunun Biyolojik Çestiligi Zoocografyasi ve Taksonomisi Üzerine bir Arastirma (Lycaenidae, Lepidoptera). CESA Miscellaneous Papers 78/79: 1–11.
Current status: a replacement name for Agrodiaetus admetus agenjoi nec Polyommatus escheri race/form agenjoi Higgins, 1948. Synonym of Polyommatus ripartii ripartii (Vila et al. 2010) or separate subspecies of Polyommatus ripartii (Eckweiler and Bozano 2016).
ripartii (Freyer, 1830)
Original combination: Lycaena ripartii
In: Freyer C (1830) Beiträge zur Geschichte europäischer Schmetterlinge mit Abbildungen nach der Natur, 3. Band, 23. Heft: 128; tab. 133, fig. 3.
Type locality: “Spanien” [Spain].
Lectotype in The Natural History Museum, London (after Bálint 1999: 54).
Current status: species.
rippertii (Boisduval, 1832)
Original combination: Polyommatus rippertii
In: Boisduval [JBA] (1832-1843) Icones historique des Lépidoptères nouveaux ou peu connus 1 : 68. Pl. 16, figs 4–6.
Type locality: “aux environs de Digne” [France: Alpes de Haute Provence].
Lectotype in The Natural History Museum, London (after Bálint 1999: 54).
Current status: synonym of Polyommatus (Agrodiaetus) ripartii (Bálint 1999: 54, Eckweiler and Bozano 2016).
rjabovi (Forster, 1960)
Original combination: Agrodiaetus rjabovi
In: Forster W (1960) Agrodiaetus rjabovi sp. n. Entomologische Zeitschrift 70(14): 157.
Type locality: “Talysh, distr. Lenkoran, Ljulakeran” [Azerbaijan].
Holotype in Zoologische Staatssammlung, München, Germany (after Häuser and Eckweiler 1997).
Current status: invalid name, junior secondary homomym of Polyommatus thersites rjabovi Obraztsov, 1936, replaced by Polyommatus (Agrodiaetus) rjabovianus.
rjabovianus (Koҫak, 1980)
Original combination: Agrodiaetus valiabadi nom. n. rjabovianus
In: Koҫak A (1980) On the nomenclature of some genus- and species-group names of Lepidoptera. Nota lepidopterologica 2(4): 142.
Type locality: see rjabovi Forster, 1960.
Type material: see rjabovi Forster, 1960.
Current status: Replacement name for Agrodiaetus rjabovi Forster, 1960.
rufomaculata (Dannehl, 1927)
Original combination: Lycaena dolus Hb. ♀ rufomaculata
In: Dannehl F (1927) Neue Formen und geografische Rassen aus meinem Rhopaloceren-Ausbeuten der letzten Jahren. Mitteilungen der Münchner Entomologischen Gesellschaft 17: 7–8.
Type locality: “Mt. Sabini (Gennaro), Simbruini, Velino, Sirente,… bei Aquila, Gran-Sasso, Morrone, Agatone, Majella” [Central and South Italy].
Current status: synonym of Polyommatus (Agrodiaetus) dolus virgilius (after Eckweiler and Bozano 2016).
sarkani (Lukhtanov & Dantchenko, 2002)
Original combination: Agrodiaetus ripartii sarkani
In: Lukhtanov V, Dantchenko A (2002) Descriptions of new taxa of the genus Agrodiaetus Hübner, [1822] based on the karyotype investigation (Lepidoptera, Lycaenidae). Atalanta 33 (1/2): 81–107.
Type locality: Kazakhstan, Taldy-Kurganskaya oblast’, Dzhungarian Alatau, Andreevskij rayon (Kabanbai), Kolbai vic., 800 m.
Holotype in Museum of Comparative Zoology, Harvard University, Cambridge, MA, USA.
Current status: synonym of Polyommatus (Agrodiaetus) ripartii ripartii (after Vila et al. 2010)
splendens (Gomez-Bustillo, 1971)
Original combination: Plebejus damon subsp. splendens
In: Gomez-Bustillo M (1971) For un mejor conosimiento de los ropaloceros españoles. Sociedad de Ciencias Naturales Aranzadi, Publicacion N° 19: 6.
Type locality: “Puerto de Pozazal (1000 m), T. M. de Valdeprado, Prov. de Santander” [SPAIN: Santander].
Holotype in coll. M. Gomez-Bustillo, Madrid [?] (after Häuser and Eckweiler 1997).
Current status: synonym of Polyommatus (Agrodiaetus) fulgens pseudovirgilia (after Eckweiler and Bozano 2016).
splendida (Dannehl, 1927)
Original combination: Lycaena dolus Hb. splendida
In: Dannehl F (1927) Neue Formen und geografische Rassen aus meinem Rhopaloceren-Ausbeuten der letzten Jaren. Mitteilungen der Münchner Entomologischen Gesellschaft 17: 7–8. Type locality: “Mt. Sabini (Gennaro), Simbruini, Velino, Sirente,… bei Aquila, Gran-Sasso, Morrone, Agatone, Majella” [Central and South Italy].
Syntypes [possibly in BMNH, London] (after Häuser and Eckweiler 1997).
Current status: synomym of Polyommatus (Agrodiaetus) dolus virgilius (after Eckweiler and Bozano 2016).
subbaeticus (Felipe Gil-T. & Talia Gil-Uceda, 2005)
Original combination: Agrodiaetus fabressei subbaeticus
In: Felipe Gil-T, Talia Gil-Uceda (2005) Agrodiaetus violetae (Gómez-Bustillo, Expósito et Martínez, 1979): morfología comparada y descripción de Agrodiaetus fabressei subbaeticus ssp. n. del sureste de la península Ibérica (Lepidoptera, Lycaenidae). Boletín Sociedad Entomológica Aragonesa 36(4): 361.
Type locality: Spain, Iberica peninsula, N. Sierra de la Sagra.
Holotype in coll. of Dr. U. Eitschberger, McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, USA.
Current status: subspecies of Polyommatus (Agrodiaetus) violetae.
subtusradiata (Oberthür, 1910)
Original combination: Lycaena Ripertii Subtus radiata
In: Oberthür (1910) Études de Lépidopterologie Comparee 4: 261.
Type locality: “Fort-Naryne” [Kyrgyzstan].
Current status: unavailable name and synonym of Polyommatus (Agrodiaetus) ripartii colemani (after Eckweiler and Bozano 2016).
susae (Bertaccini, 2003)
Original combination: Agrodiaetus ripartii susae
In: Bertaccini E (2003) Prima segnalazione in piemonte di Agrodiaetus ripartii (Freyer, 1831) e descrizione di Agrodiaetus ripartii susae ssp. nova (Insecta Lepidoptera Lycaenidae). Quoderno di Studi e Notizie di Storio Noturole dello Romogno 17: 127–138.
Type locality: “valle di susa (To) loc. Mompantero”, Italy.
Current status: synonym of Polyommatus (Agrodiaetus) ripartii ripartii.
tengritaghicus Koҫak & Kemal, 2001
Original combination: Polyommatus (Admetusia) tengritaghicus
In: Koҫak A, Kemal M (2001) Lepidoptera Cograpiyesi llstide tetqiqatlar. 2. Qazaqistan Kepinekliring Zoocograpiyesi ve Taksonomiyesi Llstide Tetqiqatlar. (Lepidoptera, Papilionoidea, Hesperioidea). Priamus 10: 111–163.
Type locality: Kazakhstan.
Current status: likely subspecies of Polyommatus (Agrodiaetus) ripartii.
timfristos Lukhtanov, Vishnevskaya & Shapoval, sp. n.
Original combination: Polyommatus timfristos
In the present paper.
Type locality: Greece, Timfristos Mt, Karpenisi, 38°55.460'N; 21°47.605 E, 1270 m.
Holotype in Zoological Institute of the Russian Academy of Science, St. Petersburg.
Current status: species.
urmiaensis Schurian & Ten Hagen, 2003
Original combination: Polyommatus (Agrodiaetus) urmiaensis
In: Schurian K, Ten Hagen W (2003) Polyommatus (Agrodiaetus) urmiaensis sp. n. aus Nordwestiran (Lepidoptera: Lycaenidae). Nachrichten des Entomologischen Vereins Apollo 24(1/2): 1–5.
Type locality: Iran, Azarbaygan-e Garbi, vic. Salamas, 60 km N of Orumieh, 1700–1800 m.
Holotype in Senckenberg Museum, Frankfurt am Main, Germany.
Current status: subspecies of Polyommatus (Agrodiaetus) karacetinae.
valiabadi (Rose & Schurian, 1977)
Original combination: Agrodiaetus rjabovi ssp. valiabadi
In: Rose K., Schurian K (1977) Beitraege zur Kenntnis der Rhopaloceren Irans. 7. Beitrag: Eine neue Unterart von Agrodiaetus rjabovi Forster. Journal of Entomological Society of Iran 4(1/2): 63.
Type locality: “Elburs-Nordseite (Chalus-Tal), Umgebung Vali-Abad, 1900- 2100 m NN, 25 km nördlich Kandevantunnel” [Iran: Elburs mts.].
Holotype in coll. K. Schurian, Schwalbach [Kelkheim] (after Häuser and Eckweiler 1997).
Current status: species.
violetae (Gómez-Bustillo, Expósito & Martínez, 1979)
Original combination: Agrodiaetus violetae
In: Gómez-Bustillo M, Expósito HA, Martínez BP (1979): Una nueva especie para la Ciencia: Agrodiaetus violetae (Lep. Lycaenidae). Shilap Revista de Lepidopterología 7(1): 51.
Type locality: “Sierra de Almijara (a 1150 m.), Prov. de Málaga” [Spain: Malaga].
Holotype in coll. M. Gómez-Bustillo, Madrid [?] (after Häuser and Eckweiler 1997)
Current status: species.
violetapunctata (Gómez-Bustillo, Exposìto & Martinéz, 1979)
Original combination: Agrodiaetus violetae f. violetapunctata
In: Gómez-Bustillo M, Expósito HA, Martínez BP (1979): Una nueva especie para la Ciencia: Agrodiaetus violetae (Lep. Lycaenidae). Shilap Revista de Lepidopterología 7(1): 53.
Type locality: “Sierra de Almijara 9°1150 m), Prov. de Málaga” [Spain].
Current status: synonym of Polyommatus (Agrodiaetus) violetae.
virgilius (Oberthür, 1910)
Original combination: Lycaena dolus race virgilia
In: Oberthür C (1910) Notes pour servir à établir la Faune Franҫaise et Algérienne des Lépidoptères (suite). Études de Lépidoptérologie Comparée 4(l): 263.
Type locality: “Italic méridionale . . . notamment à Sulmona” [Italy: Abruzzi e Molise: L'Aquila].
Lectotype in The Natural History Museum, London (after Bálint 1999: 63).
Current status: subspecies of Polyommatus (Agrodiaetus) dolus.
vittatus (Oberthür, 1892)
Original combination: Lycaena dolus forma geographica vittata
In: Oberthür C (1892) Bulletin de la Société Entomologique de France, 1892: X.
Type locality: “Lozère” [France: Lozère].
Syntypes possibly in The Natural History Museum, London (after Häuser and Eckweiler 1997).
Current status: subspecies of Polyommatus (Agrodiaetus) dolus.
yeranyani (Dantchenko & Lukhtanov, 2005)
Original combination: Agrodiaetus admetus yeranyani
In: Dantchenko A, Lukhtanov V (2005) New taxa of the brown species-complex of the genus Agrodiaetus Hübner, (1822) from Transcaucasia (Lepidoptera, Lycaenidae). Atalanta 35: 327–334, 472–475.
Type locality: Armenia, Zangezur mts, Kajaran distr., right bank Vokhtchi River, Pkhrut vic. 1900 m.
Type material: Museum of Comparative Zoology (Harvard University, Cambridge, MA, USA).
Current status: species.
Appendix 2
Additional phylogenetic trees
Figure 18.
Fragment of the ML tree of Polyommatus admetus and Polyommatus dolus complexes based on analysis of COI barcodes and focused on Polyommatus nephohiptamenos, Polyommatus admetus and Polyommatus ripartii pelopi. Polyommatus pseudorjabovi clade is not shown in details, for its composition see Lukhtanov et al. (2015a). Details of the West-European and the “mixed” (Eurasian) clades of Polyommatus ripartii are shown in Fig. 19. Details of the Polyommatus dolus group are shown in Fig. 20. Numbers at nodes indicate bootstrap support.
Figure 19.
Fragment of the ML tree of Polyommatus admetus and Polyommatus dolus complexes based on analysis of COI barcodes and focused on details in the structure of the West-European and the “mixed” (Eurasian) clades of Polyommatus ripartii. Numbers at nodes indicate bootstrap support.
Figure 20.
Fragment of the ML tree based on analysis of COI barcodes and focused on details of the Polyommatus dolus group. Polyommatus rjabovianus and Polyommatus valiabadi clades are not shown in details, for their composition see Lukhtanov et al. (2015a). Numbers at nodes indicate bootstrap support.
Figure 21.
Fragment of the MP tree of Polyommatus admetus and Polyommatus dolus complexes based on analysis of COI barcodes and focused on Polyommatus nephohiptamenos, Polyommatus admetus and Polyommatus ripartii pelopi. Polyommatus pseudorjabovi clade is not shown in details, for its composition see Lukhtanov et al. (2015a). The West-European and the “mixed” (Eurasian) clades of Polyommatus ripartii are shown in Fig. 22. Polyommatus dolus group is shown in Fig. 23. Numbers at nodes indicate bootstrap support.
Figure 22.
Fragment of the MP tree of Polyommatus admetus and Polyommatus dolus complexes based on analysis of COI barcodes and focused on details of the West-European and the “mixed” (Eurasian) clades of Polyommatus ripartii. Numbers at nodes indicate bootstrap support.
Figure 23.
Fragment of the MP tree based on analysis of COI barcodes and focused on details of the Polyommatus dolus group. Polyommatus rjabovianus and Polyommatus valiabadi clades are not shown in details, for their composition see Lukhtanov et al. (2015a). Numbers at nodes indicate bootstrap support.
Appendix 3
Habitats of the studied species
Figure 24.
Habitat of Polyommatus admetus. Greece, Peloponnesse Peninsula, Mt. Chelmos, near Kalavrita, 800 m, 16 july 2008. Photo by V.A. Lukhtanov.
Figure 25.
Habitat of Polyommatus admetus. Greece, Smolikas Mt, Konitsa, 950 m, 22 July 2008. Photo by V.A. Lukhtanov.
Figure 26.
Habitat of Polyommatus admetus. W Bulgaria, near Dragoman, 700 m, 29 July 2008. Photo by V.A. Lukhtanov.
Figure 27.
Habitat of Polyommatus ripartii pelopi. Greece, Peloponnesse Peninsula, Mt. Chelmos, near Kalavrita, 800 m, 17 July 2008. Photo by V.A. Lukhtanov.
Figure 28.
Polyommatus nephohiptamenos. Northern Greece, Falakro Mt, near Granitis, 1700 m, 23 July 2008. Photo by V.A. Lukhtanov.
Figure 29.
Habitat of Polyommatus nephohiptamenos. Northern Greece, Falakro Mt, near Granitis, 1700 m, 24 July 2008. Photo by V.A. Lukhtanov.
Figure 30.
Habitat of Polyommatus nephohiptamenos. Northern Greece, Falakro Mt, near Granitis, 1700 m, 24 July 2008. Photo by V.A. Lukhtanov.
Figure 31.
Habitat of Polyommatus aroaniensis in its type locality. Greece, Peloponnesse Peninsula, Mt. Chelmos, near Kalavrita, 1600 m, 16 July 2008. Photo by V.A. Lukhtanov.
Figure 32.
Habitat of Polyommatus timfristos. Central Greece, Mt. Timfristos, near Karpenisi, 1200 m, 20 July 2008. Photo by V.A. Lukhtanov.
Figure 33.
Habitat of Polyommatus timfristos. Central Greece, Mt. Timfristos, near Karpenisi, 1200 m, 20 July 2008. Photo by V.A. Lukhtanov.
Figure 34.
Habitat of Polyommatus timfristos. Central Greece, Mt. Parnassos, 19 July 2008. Photo by V.A. Lukhtanov.
Figure 35.
Habitat of Polyommatus timfristos. Central Greece, Mt. Parnassos, 19 July 2008. Photo by V.A. Lukhtanov.
Figure 36.
Hvoyna, Bulgaria, type locality of Polyommatus orphicus orphicus, 2 July 2016. Photo by E. Pazhenkova.
Figure 37.
Habitat of Polyommatus orphicus orphicus in its type locality. Bulgaria, Hvoyna, 3 July 2016. Photo by E. Pazhenkova.
Figure 38.
Habitat of Polyommatus orphicus orphicus. Bulgaria, Hvoyna, 3 July 2016. Photo by I. Zubkova.
Figure 39.
Habitat of Polyommatus orphicus eleniae in its type locality. Northern Greece, Makedonía, Dráma district, near Granítis, 900 m, 23 July 2008. Photo by V.A. Lukhtanov.
Citation
Vishnevskaya MS, Saifitdinova AF, Lukhtanov VA (2016) Karyosystematics and molecular taxonomy of the anomalous blue butterflies (Lepidoptera, Lycaenidae) from the Balkan Peninsula. Comparative Cytogenetics 10(5): 1–85. https://doi.org/10.3897/CompCytogen.v10i5.10944
References
- Abadjiev SP. (2001) An atlas of the distribution of the butterflies in Bulgaria (Lepidoptera: Hesperioidea and Papilionoidea). Pensoft Publishers, Sofia, 344 pp. [Google Scholar]
- Alexandri P, Triantafyllidis A, Papakostas S, Chatzinikos E, Platis P, Papageorgiou N, Larson G, Abatzopoulos TJ, Triantaphyllidis C. (2012) The Balkans and the colonization of Europe: the post-glacial range expansion of the wild boar, Sus scrofa. Journal of Biogeograpjhy 39: 713–723. https://doi.org/10.1111/j.1365-2699.2011.02636.x [Google Scholar]
- Bálint Z. (1999) Annotated list of type specimens of Polyommatus sensu Eliot of the Natural History Museum, London (Lepidoptera, Lycaenidae). Neue Entomologische Nachrichten 46: 1–89. [Google Scholar]
- Bálint Z, Gubányi A, Pitter G. (2006) Magyarország Védett Pillangóalakú Lepkéinek. Katalógusa. Magyar Természettudományi Múzeum, Budapest, 136 pp. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. (1999) Median–joining networks for inferring intraspecific phylogenies. Molecular biology and evolution 16(1): 37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036 [DOI] [PubMed] [Google Scholar]
- Bregović P, Zagmajster M. (2016) Understanding hotspots within a global hotspot – identifying the drivers of regional species richness patterns in terrestrial subterranean habitats. Insect Conservation and Diversity 9(4): 268–281. https://doi.org/10.1111/icad.12164 [Google Scholar]
- Brown J. (1976a) Notes regarding previously undescribed European taxa of the genera Agrodiaetus Hübner, 1822 and Polyommatus Klus 1801 (Lep., Lycaenidae). Entomologist’s Gazette 27: 77–84. [Google Scholar]
- Brown J. (1976b) On two previously undescribed subspecies of Lycaenidae (Lepidoptera) from Greece. Entomologische Berichten 36(3): 46–47. [Google Scholar]
- Brown J, Coutsis J. (1978) Two newly discovered Lycaenid butterflies (Lepidoptera, Lycaenidae) from Greece, with notes on applied species. Entomologist’s Gazette 28: 201–213. [Google Scholar]
- Buj I, Caleta M, Marcic Z, Sanda R, Vukic J, Mrakovcic M. (2015) Different histories, different destinies–impact of evolutionary history and population genetic structure on extinction risk of the Adriatic spined loaches (Genus Cobitis; Cypriniformes, Actinopterygii). PLoS ONE 10(7): e0131580 https://doi.org/10.1371/journal.pone.0131580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell F. (2000) Contribution a la conaissance du genre Agrodiaetus Hübner (1822), A. barmifiruze n. sp. et A. musa esfahensis n. ssp, en Iran meridional. Linneana Belgica 17: 211–217. [Google Scholar]
- Carbonell F. (2001) Contribution à la connaissance du genre Agrodiaetus Hübner (1822), A. ahmadi et A. khorasanensis nouvelles espèces dans le Nord de l’Iran (Lepidoptera, Lycaenidae). Linneana Belgica 18: 105–110. [Google Scholar]
- Coutsis JG. (1985) Notes concerning the taxonomic status of Agrodiaetus tankeri de Lesse. Nota lepidopterologica 8: 8–14. [Google Scholar]
- Coutsis JG. (1986) The blue butterflies of the genus Agrodiaetus Hübner (Lepidoptera, Lycaenidae): symptoms of taxonomic confusion. Nota Lepidopterologica 9: 159–169. [Google Scholar]
- Coutsis JG, De Prins J. (2005) A new brown Polyommatus (Agrodiaetus) from northern Greece (Lepidoptera, Lycaenidae). Phegea 33(4): 129–137. [Google Scholar]
- Coutsis JG, De Prins J. (2007) The chromosome number and karyotype of Polyommatus (Agrodiaetus) nephohiptamenos (Lepidoptera, Lycaenidae). Phegea 35: 27–29. [Google Scholar]
- Coutsis JG, De Prins W, Puplesiene J. (1999) The chromosome number and karyotype of Polyommatus (Agrodiaetus) ripartii and Polyommatus (Agrodiaetus) aroaniensis from Greece (Lepidoptera, Lycaenidae). Phegea 27(1): 81–84. [Google Scholar]
- Dantchenko AV. (2000) Genus Agrodiaetus. In: Tuzov VK. (Ed.) Guide to the butterflies of Russia and adjacent territories, Vol. 2 Pensoft, Sofia, Moscow, 196–214. [Google Scholar]
- Dantchenko AV, Lukhtanov VA. (2005) New taxa of the “brown” species–complex of the genus Agrodiaetus Hübner, [1822] from Transcaucasia (Lepidoptera, Lycaenidae). Atalanta 35(3/4): 327–334. [Google Scholar]
- de Lesse H. (1960a) Les nombres de chromosomes dans la classification du groupe d’Agrodiaetus ripartii Freyer (Lepidoptera, Lycaenidae). Revue Francaise d’Entomologie 27: 240–264. [Google Scholar]
- de Lesse H. (1960b) Spéciation et variation chromosomique chez les Lépidoptères Rhopalocères. Annales des Sciences Naturelles Zoologie et Biologie Animale, 12e série 2: 1–223. [Google Scholar]
- Dincă V, Lukhtanov VA, Talavera G, Vila R. (2011) Unexpected layers of cryptic diversity in Wood White Leptidea butterflies. Nature Communications 2: 234 https://doi.org/10.1038/ncomms1329 [DOI] [PubMed] [Google Scholar]
- Dincă V, Runquist M, Nilsson M, Vila R. (2013a) Dispersal, fragmentation and isolation shape: the phylogeography of the European lineages of Polyommatus (Agrodiaetus) ripartii (Lepidoptera: Lycaenidae). Biological Journal of the Linnean Society 109: 817–829. https://doi.org/10.1111/bij.12096 [Google Scholar]
- Dincă V, Wiklund C, Lukhtanov VA, Kodandaramaiah U, Norén N, Dapporto L, Wahlberg N, Vila R, Friberg M. (2013b) Reproductive isolation and patterns of genetic differentiation in a cryptic butterfly species complex. Journal of Evolutionary Biology 26(10): 2095–2106. https://doi.org/10.1111/jeb.12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckweiler W, Bozano GC. (2016) Guide to the butterflies of the Palearctic region. Lycaenidae part IV. Omnes artes, Milano, 132 pp. [Google Scholar]
- Eckweiler W, Häuser CL. (1997) An illustrated checklist of Agrodiaetus Hübner, 1822, a subgenus of Polyommatus Latreille, 1804 (Lepidoptera: Lycaenidae). Nachrichten des Entomologischen Vereins Apollo, Suppl. 16: 113–168.
- Felsenstein J. (1985) Phylogenies and the comparative method. The American Naturalist 125: 1–15. https://doi.org/10.1086/284325 [DOI] [PubMed] [Google Scholar]
- Ganev J. (1984) Zwei neue Arten Lycaenidae fiir die bulgarische Fauna (Lepidoptera, Lycaenidae). Articulata 2(5): 125–126. [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Häuser CL, Eckweiler W. (1997) A catalogue of the species-group taxa in Agrodiaetus Hübner, 1822, a subgenus of Polyommatus Latreille, 1804 (Lepidoptera: Lycaenidae). Nachrichten des Entomologischen Vereins Apollo, Suppl. 16: 53–112.
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences, USA 101: 14812–14817. https://doi.org/10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Roldán JL, Dapporto L, Dincă V, Vicente JC, Hornett EA, Šíchová J, Lukhtanov VA, Talavera G, Vila R. (2016) Integrative analyses unveil speciation linked to host plant shift in Spialia butterflies. Molecular Ecology 25(17): 4267–4284. https://doi.org/10.1111/mec.13756 [DOI] [PubMed] [Google Scholar]
- Hesselbarth G, Oorschot H, Wagener S. (1995) Die Tagfalter der Türkei unter Berücksichtigung der angrenzenden Länder. Selbstverlag Siegbert Wagener, Bocholt, Vol. 1‒3, 1354 pp. [Google Scholar]
- Ilonczai Z, Balint Z. (2001) Ujabb adatok a Magyarorszagon vedett nappali lepkek ismeretehez (Lepidoptera: Lycaenidae, Nymphalidae) [New data to the knowledge of butterflies (Lepidoptera: Lycaenidae, Nymphalidae) protected in Hungary]. Termeszetvedelmi Kozlemenyek 9: 209–218. [Google Scholar]
- Kandul NP. (1997) The karyology and the taxonomy of the blue butterflies of the genus Agrodiaetus Hübner, [1822] from the Crimea (Lepidoptera: Lycaenidae). Atalanta 28(1–2): 111–119. [Google Scholar]
- Kandul NP, Lukhtanov VA, Pierce NE. (2007) Karyotypic diversity and speciation in Agrodiaetus butterflies. Evolution 61(3): 546–559. https://doi.org/10.1111/j.1558–5646.2007.00046.x [DOI] [PubMed] [Google Scholar]
- Kandul NP, Lukhtanov VA, Dantchenko AV, Coleman J, Haig D, Sekercioglu C, Pierce NE. (2002) The evolution of karyotype diversity: a molecular phylogeny of Agrodiaetus Hübner, 1822 (Lepidoptera: Lycaenidae) inferred from mtDNA sequences for COI and COII. 4th International Conference on the Biology of Butterflies Leeuwenhorst, the Netherlands, 23 March 2002, 33–34. [Google Scholar]
- Kandul NP, Lukhtanov VA, Dantchenko AV, Coleman JWS, Sekercioglu CH, Haig D, Pierce NE. (2004) Phylogeny of Agrodiaetus Hübner 1822 (Lepidoptera: Lycaenidae) inferred from mtDNA sequences of COI and COII and nuclear sequences of EF1–α: Karyotype diversification and species radiation. Systematic Biology 53(2): 278–298. https://doi.org/10.1080/10635150490423692 [DOI] [PubMed] [Google Scholar]
- Kasapidis P, Suchentrunk F, Magoulas A, Kotoulas G. (2005) The shaping of mitochondrial DNA phylogeographic patterns of the brown hare (Lepus europaeus) under the combined influence of Late Pleistocene climatic fluctuations and anthropogenic translocations. Molecular Phylogenetics and Evolution 34: 55–66. https://doi.org/10.1016/j.ympev.2004.09.007 [DOI] [PubMed] [Google Scholar]
- Kolev Z. (1994) Two Polyommatus (Agrodiaetus) species new to Bulgaria, with notes on the related Bulgarian taxa (Lepidoptera: Lycaenidae). Phegea 22(2): 61–71. [Google Scholar]
- Kolev Z. (2005) Polyommatus dantchenkoi (Lukhtanov et Wiemers, 2003) tentatively identified as new to Europe, with a description of a new taxon from the Balkan Peninsula (Lycaenidae). Nota lepidopterologica 28(1): 25–34. [Google Scholar]
- Kolev Z, De Prins W. (1995) A new species of the “brown Agrodiaetus” complex from the Crimea (Lepidoptera, Lycaenidae). Phegea 23(2): 119–132. [Google Scholar]
- Kolev Z, Van der Poorten D. (1997) Review of the distribution of the Balkan endemic Polyommatus (Agrodiaetus) aroaniensis (Lepidoptera: Lycaenidae), with notes on its sympatry with related species. Phegea 25(1): 35–40. [Google Scholar]
- Koren T. (2010a) New data about the distribution of anomalous blue Polyommatus admetus (Esper, 1783) (Lepidoptera: Lycaenidae) in Croatia. Acta Entomologica Serbica 15(2): 221–226. [Google Scholar]
- Koren T. (2010b) First finding of Ripart’s Anomalous Blue Polyommatus (Agrodiaetus) ripartii (Freyer, 1830) (Lepidoptera, Lycaenidae) in Croatia. Natura Croatica 19(2): 463–467. [Google Scholar]
- Koren T, Laus B. (2015) The Grecian anomalous blue Polyommatus (Agrodiaetus) aroaniensis (Brown, 1976) (Lepidoptera: Lycaenidae) discovered in Croatia, at the north–western edge of its distribution. Natura Sloveniae 17(2): 47–57. [Google Scholar]
- Kulfan M, Kulfan J. (1992) Changes of distribution of thermophilous butterflies in Slovakia. Journal of Research on the Lepidoptera 29(4): 254–266. [Google Scholar]
- Kudrna O, Harpke A, Lux K, Pennerstorfer J, Schweiger O, Settele J, Wiemers M. (2011) Distribution atlas of butterflies in Europe. Gesellschaft für Schmetterlingschutz, Halle, Germany, 576 pp. [Google Scholar]
- Lovrenčić L, Podnar M, Šašić M, Koren T, Tvrtković N. (2016) Molecular data do not confirm the Grecian anomalous blue Polyommatus (Agrodiaetus) aroaniensis (Brown, 1976) as a member of the Croatian fauna. Natura Croatica 25(1): 119–129. https://doi.org/10.20302/NC.2016.25.8 [Google Scholar]
- Lukhtanov VA. (1989) Karyotypes of some blue butterflies of the Agrodiaetus species groups (Lepidoptera, Lycaenidae). Annales Entomologici Fennici 55: 137–144. [Google Scholar]
- Lukhtanov VA. (2014) Chromosome number evolution in skippers (Lepidoptera, Hesperiidae). Comparative Cytogenetics 8(4): 275–291. https://doi.org/10.3897/CompCytogen.v8i4.8789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA. (2015) The blue butterfly Polyommatus (Plebicula) atlanticus (Lepidoptera, Lycaenidae) holds the record of the highest number of chromosomes in the non–polyploid eukaryotic organisms. Comparative Cytogenetics 9(4): 683–690. https://doi.org/10.3897/CompCytogen.v9i4.5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Dantchenko AV. (2002a) Descriptions of new taxa of the genus Agrodiaetus Hübner, [1822] based on karyotype investigation (Lepidoptera, Lycaenidae). Atalanta 33(1/2): 81–107, 224–225. [Google Scholar]
- Lukhtanov VA, Dantchenko AV. (2002b) Principles of highly ordered metaphase I bivalent arrangement in spermatocytes of Agrodiaetus (Lepidoptera). Chromosome Research 10: 5–20. https://doi.org/10.1023/A:1014249607796 [DOI] [PubMed] [Google Scholar]
- Lukhtanov VA, Tikhonov VV. (2015) Chromosomal and molecular evidence for presence of Polyommatus (Agrodiaetus) poseidon (Lepidoptera, Lycaenidae) in Caucasus region. Comparative Cytogenetics 9(2): 249–255. https://doi.org/10.3897/CompCytogen.v9i2.5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Shapoval NA, Dantchenko AV. (2008) Agrodiaetus shahkuhensis sp. n. (Lepidoptera, Lycaenidae), a cryptic species from Iran discovered by using molecular and chromosomal markers. Comparative Cytogenetics 2(2): 99–114. [Google Scholar]
- Lukhtanov VA, Shapoval NA, Dantchenko AV. (2014) Taxonomic position of several enigmatic Polyommatus (Agrodiaetus) species (Lepidoptera, Lycaenidae) from Central and Eastern Iran: insights from molecular and chromosomal data. Comparative Cytogenetics 8(4): 313–322. https://doi.org/10.3897/CompCytogen.v8i4.8939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Sourakov A, Zakharov EV. (2016) DNA barcodes as a tool in biodiversity research: testing pre-existing taxonomic hypotheses in Delphic Apollo butterflies (Lepidoptera, Papilionidae). Systematics and Biodiversity 14(6): 599–613. https://doi.org/10.1080/14772000.2016.1203371 [Google Scholar]
- Lukhtanov VA, Vila R, Kandul NP. (2006) Rearrangement of the Agrodiaetus dolus species group (Lepidoptera, Lycaenidae) using a new cytological approach and molecular data. Insect Systematics and Evolution 37(3): 325–334. https://doi.org/10.1163/187631206788838563 [Google Scholar]
- Lukhtanov VA, Wiemers M, Meusemann K. (2003) Description of a new species of the “brown” Agrodiaetus complex from South–East Turkey (Lycaenidae). Nota Lepidopterologica 26(1/2): 65–71. [Google Scholar]
- Lukhtanov VA, Dantchenko AV, Vishnevskaya MS, Saifitdinova AF. (2015a) Detecting cryptic species in sympatry and allopatry: analysis of hidden diversity in Polyommatus (Agrodiaetus) butterflies (Lepidoptera: Lycaenidae). Biological Journal of the Linnean Society 116: 468–485. https://doi.org/10.1111/bij.12596 [Google Scholar]
- Lukhtanov VA, Sourakov A, Zakharov EV, Hebert PD. (2009) DNA barcoding Central Asian butterflies: increasing geographical dimension does not significantly reduce the success of species identification. Molecular Ecology Resources 9: 1302–1310. https://doi.org/10.1111/j.1755–0998.2009.02577.x [DOI] [PubMed] [Google Scholar]
- Lukhtanov VA, Shapoval NA, Anokhin BA, Saifitdinova AF, Kuznetsova VG. (2015b) Homoploid hybrid speciation and genome evolution via chromosome sorting. Proceedings of the Royal Society B: Biological Sciences 282(1807): 20150157 https://doi.org/10.1098/rspb.2015.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA, Kandul NP, Plotkin JB, Dantchenko AV, Haig D, Pierce NE. (2005) Reinforcement of pre-zygotic isolation and karyotype evolution in Agrodiaetus butterflies. Nature 436: 385–389. https://doi.org/10.1038/nature03704 [DOI] [PubMed] [Google Scholar]
- Melovski D, Bozhinovska E. (2014) New records for four butterfly species (Lepidoptera: Papilionoidea and Hesperioidea) in the Republic of Macedonia. Journal of Natural Sciences Research 4(7): 40–44. [Google Scholar]
- Miljević M, Popović M. (Eds) (2014) Alciphron - database on insects of Serbia, Lepidoptera: Papilionoidea. http://www.habiprot.org.rs/Alciphron
- Mutanen M, Kivelä SM, Vos RA, Doorenweerd C, Ratnasingham S, Hausmann A, Huemer P, Dincă V, van Nieukerken EJ, Lopez-Vaamonde C, Vila R, Aarvik L, Decaëns T, Efetov KA, Hebert PD, Johnsen A, Karsholt O, Pentinsaari M, Rougerie R, Segerer A, Tarmann G, Zahiri R, Godfray HC. (2016) Species-level para- and polyphyly in DNA barcode gene trees: strong operational bias in European Lepidoptera. Systematic Biology 65(6): 1024–1040. https://doi.org/10.1093/sysbio/syw044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. (2000) Molecular Evolution and Phylogenetics. Oxford University Press, New York, 333 pp. [Google Scholar]
- Nikolić T, Mitić B, Ruščić M, Milašinović B. (2014) Diversity, knowledge and spatial distribution of the vascular flora of Croatia. Plant Biosystems 148(4): 591–601. https://doi.org/10.1080/11263504.2013.788091 [Google Scholar]
- Olivier A, Puplesiene J, van der Pooten D, De Prins W, Wiemers M. (1999) Revision of some taxa of Polyommatus (Agrodiaetus) transcaspicus group with description of a new species from Central Anatolia (Lepidoptera, Lycaenidae). Phegea 27(1): 1–24. [Google Scholar]
- Pazhenkova EA, Lukhtanov VA. (2016) Chromosomal and mitochondrial diversity in Melitaea didyma complex (Lepidoptera, Nymphalidae): eleven deeply diverged DNA barcode groups in one non-monophyletic species? Comparative Cytogenetics 10(4): 697–717. https://doi.org/10.3897/CompCytogen.v10i4.11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamperis LN. (2009) The butterflies of Greece. Edition Pamperis, Athens, 768 pp. [Google Scholar]
- Posada D. (2008) jModel Test: phylogenetic model averaging. Molecular Biology and Evolution 25: 1253–1256. https://doi.org/10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- Previšić A, Gelemanović A, Urbanič G, Ternjej I. (2016) Cryptic diversity in the Western Balkan endemic copepod: Four species in one? Molecular phylogenetics and evolution 100: 124–134. https://doi.org/10.1016/j.ympev.2016.04.010 [DOI] [PubMed] [Google Scholar]
- Przybyłowicz L. (2000) Polish butterflies of the subgenus Polyommatus (Agrodiaetus) (Lepidoptera: Lycaenidae). Polish Journal of Entomology 69: 329–334. [Google Scholar]
- Przybyłowicz L. (2014) Polyommatus ripartii: The biological basis for the conservation and the morphology of the developmental stages of a critically endangered, relict population in Central Europe. Journal of Insect Science 14: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybyłowicz Ł, Lukhtanov VA, Lachowska-Cierlik D. (2014) Towards the understanding of the origin of the Polish remote population of Polyommatus (Agrodiaetus) ripartii (Lepidoptera: Lycaenidae) based on karyology and molecular phylogeny. Journal of Zoological Systematics and Evolutionary Research 52: 44–51. https://doi.org/10.1111/jzs.12040 [Google Scholar]
- Robinson R. (1971) Lepidoptera genetics. Pergamon Press, Oxford, 698 pp. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. (2006) Purification of nucleic acids by extraction with phenol: chloroform. Cold Spring Harbour Protocols; https://doi.org/10.1101/pdb.prot4455 [DOI] [PubMed] [Google Scholar]
- Schurian K, ten Hagen W. (2003) Polyommatus (Agrodiaetus) urmiaensis sp. n. aus Nordwestiran (Lepidoptera: Lycaenidae). Nachrichten des Entomologischen Vereins Apollo 24(1/2): 1–5. [Google Scholar]
- Shapoval NA, Lukhtanov VA. (2015a) Taxonomic interpretation of chromosomal and mitochondrial DNA variability in the species complex close to Polyommatus (Agrodiaetus) dama (Lepidoptera, Lycaenidae). ZooKeys 538: 1–20. https://doi.org/10.3897/zookeys.538.6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapoval NA, Lukhtanov VA. (2015b) Taxonomic position and status of Polyommatus (Agrodiaetus) iphigenia (Lepidoptera, Lycaenidae) from the Peloponnese, Southern Greece. Folia Biologica-Krakow 63(4): 295–300. https://doi.org/10.3409/fb63_4.295 [DOI] [PubMed] [Google Scholar]
- Shapoval NA, Lukhtanov VA. (2015c) Intragenomic variations of multicopy ITS2 marker in Agrodiaetus blue butterflies (Lepidoptera, Lycaenidae). Comparative Cytogenetics 9(4): 483–497. https://doi.org/10.3897/CompCytogen.v9i4.5429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijarić R, Mihljević B. (1972) Prilog poznavanju faune Rhopalocera i Hesperioidea (Lepidoptera) primorskog podrucja Crne gore. Glasnik Republičkog zavoda za zaštitu prirode i Prirodnjačkog muzeja u Titogradu 5: 103–114. [Google Scholar]
- Skala P. (2001) New taxa of the subgenus Agrodiaetus Hübner, 1822 from Iran Polyommatus (Agrodiaetus) faramarzii sp. n., P. (A.) shahrami sp. n. and P. (A.) pfeifferi astyages ssp. n. (Lepidoptera, Lycaenidae). Nachrichten des Entomologischen Vereins Apollo 22(2): 101–108. [Google Scholar]
- Sobezyk R, Gligorović B. (2016) Diversity of Butterflies in the Zeta–Skadar Plain, Montenegro. Acta zoologica Bulgarica 68(2): 183–190. [Google Scholar]
- Stradomsky BV, Fomina ES. (2013) The comparison of taxa Polyommatus yurinekrutenko Kocak, 1996 and P. shamil (Dantchenko, 2000) (Lepidoptera: Lycaenidae). Caucasian Entomological Bulletin 9(1): 181–182. [Google Scholar]
- Talavera G, Lukhtanov VA, Pierce NE, Vila R. (2013a) Establishing criteria for higher–level classification using molecular data: the systematics of Polyommatus blue butterflies (Lepidoptera, Lycaenidae). Cladistics 29: 166–192. https://doi.org/10.1111/j.1096–0031.2012.00421.x [DOI] [PubMed] [Google Scholar]
- Talavera G, Lukhtanov VA, Rieppel L, Pierce NE, Vila R. (2013b) In the shadow of phylogenetic uncertainty: the recent diversification of Lysandra butterflies through chromosomal change. Molecular Phylogenetics and Evolution 69: 469–478. https://doi.org/10.1016/j.ympev.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution 30(12): 2725–2729. https://doi.org/10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hagen W, Eckweiler W. (2001) Eine neue Art von Polyommatus (Agrodiaetus) aus Zentraliran (Lepidoptera, Lycaenidae). Nachrichten des Entomologischen Vereins Apollo 22(2): 53–56. [Google Scholar]
- Tolman T, Lewington R. (2008) Collins butterfly guide: the most complete field guide to the butterflies of Britain and Europe. Collins guide. HarperCollins Publishers, London, 384 pp. [Google Scholar]
- Troiano G, Balletto E, Toso GG. (1979) The karyotype of Agrodiaetus humedasae Toso & Balletto, 1976. Bollettino della Societe Entomologica Italiana 111(7–10): 141–143. [Google Scholar]
- Tschikolovets VV. (2011) Butterflies of Europe and the Mediterranean area. Tschikolovets Publications, Pardubice, 544 pp. [Google Scholar]
- Van Swaay C, Cuttelod A, Collins S, Maes D, López Munguira ML, Šašić M, Settele J, Verovnik R, Verstrael T, Warren M, Wiemers M, Wynhoff I. (2010) European Red List of Butterflies. Publications Office of the European Union, Luxembourg, 47 pp. [Google Scholar]
- Verovnik R, Popović M. (2013) Annotated checklist of Albanian butterflies (Lepidoptera, Papilionoidea and Hesperioidea). ZooKeys 323: 75–89. https://doi.org/10.3897/zookeys.323.5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verovnik R, Franeta F, Popović M, Gascoigne–Pees M. (2015) The discovery of Polyommatus aroaniensis (Brown, 1976) in Bosnia and Herzegovina (Lepidoptera: Lycaenidae). Nachrichten des Entomologischen Vereins Apollo 36(4): 177–180. [Google Scholar]
- Vershinina AO, Lukhtanov VA. (2010) Geographical distribution of the cryptic species Agrodiaetus alcestis alcestis, A. alcestis karacetinae and A. demavendi (Lepidoptera, Lycaenidae) revealed by cytogenetic analysis. Comparative Cytogenetics 4(1): 1–11. https://doi.org/10.3897/compcytogen.v4i1.21 [Google Scholar]
- Vershinina AO, Anokhin BA, Lukhtanov VA. (2015) Ribosomal DNA clusters and telomeric (TTAGG)n repeats in blue butterflies (Lepidoptera, Lycaenidae) with low and high chromosome numbers. Comparative Cytogenetics 9(2): 161–171. https://doi.org/10.3897/CompCytogen.v9i2.4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila R, Lukhtanov VA, Talavera G, Gil-T F, Pierce NE. (2010) How common are dot–like distribution ranges? Taxonomical oversplitting in Western European Agrodiaetus (Lepidoptera, Lycaenidae) revealed by chromosomal and molecular markers. Biological Journal of the Linnean Society 101: 130–154. https://doi.org/10.1111/j.1095-8312.2010.01481.x [Google Scholar]
- Vodolazhsky DI, Yakovlev RV, Stradomsky BV. (2011) Study of taxonomic status of some specimens of subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus) from Western Mongolia based on mtDNA markers. Caucasian Entomological Bulletin 7(1): 81−82. [Google Scholar]
- Wakeham-Dawson A. (1998) Butterflies in southern Greece, June 1997, with notes on species of Agrodiaetus Hübner (Lepidoptera: Lycaenidae). Entomologist’s Gazette 49(4): 249–252. [Google Scholar]
- Wakeham-Dawson A, Benton T. (2002) Agrodiaetus nephohiptamenos Brown & Coutsis (Lep.: Lycaenidae) in North Greece. Entomologist’s Record and Journal of Variation 114(5): 219–220. [Google Scholar]
- Wakeham-Dawson A, Spurdens P. (1994) Anomalous blue butterflies of the genus Agrodiaetus Hübner (Lepidoptera: Lycaenidae) in southern Greece. Entomologist’s Gazette 45(1): 13–20. [Google Scholar]
- Wiemers M. (2003) Chromosome differentiation and the radiation of the butterfly subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus) a molecular phylogenetic approach. Ph.D. Dissertation, University of Bonn, Bonn, Germany, 203 pp http://hss.ulb.uni-bonn.de/2003/0278/0278.htm [Google Scholar]
- Wiemers M, De Prins J. (2004) Polyommatus (Agrodiaetus) paulae sp. n. (Lepidoptera: Lycaenidae) from Northwest Iran, discovered by means of molecular, karyological and morphological methods. Entomologische Zeitschrift 114(4): 155–162. [Google Scholar]
- Wiemers M, Fiedler K. (2007) Does the DNA barcoding gap exist? – a case study in blue butterflies (Lepidoptera: Lycaenidae). Frontiers in Zoology 4: 8 https://doi.org/10.1186/1742-9994-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers M, Keller A, Wolf M. (2009) ITS2 secondary structure improves phylogeny estimation in a radiation of blue butterflies of the subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus). BMC Evolutionary Biology 9: 300 https://doi.org/10.1186/1471-2148-9-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers M, Stradomsky BV, Vodolazhsky DI. (2010) A molecular phylogeny of Polyommatus s. str. and Plebicula based on mitochondrial COI and nuclear ITS2 sequences (Lepidoptera: Lycaenidae). European Journal of Entomology 107: 325–336. https://doi.org/10.14411/eje.2010.041 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.