Abstract
The spirochete Borrelia burgdorferi is the causative agent of Lyme disease, the leading vector-borne illness in the United States. Many of the genetic factors affecting spirochete morphology and physiology are unknown due to the limited genetic tools available and the large number of open reading frames with unknown functions. By adapting a mariner transposon to function in B. burgdorferi, we have developed a random mutagenesis system that tags the mutated locus for rapid identification. Transposition occurs at saturating levels in B. burgdorferi and appears to be random, targeting both linear and circular replicons. By combining the transposon system with a screen for factors affecting growth rate, mutations were readily identified in genes putatively involved in cell division and chemotaxis and a hypothetical open reading frame involved in outer membrane integrity. The successful adaptation of a mariner transposon to function in B. burgdorferi should aid in identifying virulence factors and novel gene products related to spirochete physiology.
Lyme disease is the leading vector-borne illness in the United States and is caused by the spirochete Borrelia burgdorferi. The class Spirochaetes contains many significant pathogens of humans and other animals, including Treponema pallidum (agent of syphilis), Treponema denticola (associated with periodontal disease), Brachyspira hyodysenteriae (associated with swine dysentery), Brachyspira pilosicoli (associated with porcine intestinal spirochetosis), Leptospira interrogans (associated with leptospirosis), and Borrelia spp. (associated with relapsing fever) (27). However, many of the genetic tools available for members of the Enterobacteriaceae have not yet been developed for the spirochetes, limiting the experimental identification of virulence factors and genes specific to spirochete physiology. Identifying new virulence factors and functions of hypothetical open reading frames (ORFs) in B. burgdorferi has been further hindered by the low efficiency of targeted allelic exchange (for a review, see reference 25).
An efficient and random mutagenesis system would facilitate functional identification of the large number of unknown ORFs identified in the genome sequence of B. burgdorferi (8, 12). Some transposon systems are capable of achieving saturation mutagenesis and are nearly random in their insertion sites, and the mutated locus is marked by the transposon insertion and therefore easily identified. Specifically, transposons of the mariner family have been used successfully for mutagenesis of a diverse range of organisms, including eukaryotes, archaea, and both gram-positive and gram-negative bacteria (1, 14, 16, 21, 29). The mariner elements do not require host cofactors for transposition, likely contributing to their wide host range (15). Further, mariner transposition is virtually random, requiring only a TA dinucleotide for target specificity. In addition, Lampe and colleagues derived hyperactive transposase mutants of Himar1 (a member of the mariner family), increasing the efficiency of transposition in heterologous hosts (16).
The advantages of the Himar1 element led us to adapt it for transposon mutagenesis in B. burgdorferi. Using this transposon mutagenesis system, we screened for factors affecting the growth rate of B. burgdorferi, as many aspects of spirochete physiology and morphology remain unknown. Analysis of mutants indicated that transposition was random and could achieve saturating levels from a single transformation. The high levels of transposon mutagenesis in B. burgdorferi, combined with a screen for mutants with slower growth rates, readily identified mutations in genes putatively involved in cell division and chemotaxis and a hypothetical ORF affecting outer membrane integrity. These results identify previously uncharacterized B. burgdorferi ORFs involved in spirochete morphology and physiology and establish the foundations for developing transposon mutagenesis in infectious strains of the Spirochaetes.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
B. burgdorferi strains were grown in liquid Barbour-Stoenner-Kelly (BSK)-II medium at 35°C or in solid BSK medium incubated at 35°C under 2.5% CO2 (20). Strain B31 (ATCC 35210) was originally isolated from a tick collected on Shelter Island, N.Y. (7). The genomic sequence of B. burgdorferi B31 MI culture has been determined (8, 12). Strain B31-AchbC72 is a culture-attenuated, noninfectious strain lacking lp25, lp56, and other plasmids and was derived from clone B31-A (26). The chbC locus of strain B31-AchbC72 has been disrupted by allelic exchange using the gyrB301 allele (conferring coumermycin resistance) (10). This strain was chosen because of its plasmid content, and the disruption of the chbC locus with the coumermycin resistance marker is not predicted to affect transposition. A3 and N40 are infectious strains; A3 is a clonal derivative of the type strain, B31, and contains all plasmids except cp9 (11), and N40 was isolated from a tick obtained in Westchester County, N.Y. (2, 3). The plasmid content of N40 has not been determined. Strain A3-89 is a low-passage-number, noninfectious derivative of A3 that lacks cp9, lp25, lp28-4, and lp56. TOP10 (Invitrogen, Carlsbad, Calif.) was the strain used in all Escherichia coli plasmid manipulations.
Construction of pMarGent.
David Lampe, Duquesne University, generously provided two hyperactive alleles of the Himar1 transposase, designated C9 and A7 (16). The genes encoding the transposase were fused to the B. burgdorferi flgB promoter by using PCR to incorporate unique restriction sites, as previously described by Bono et al. (6).
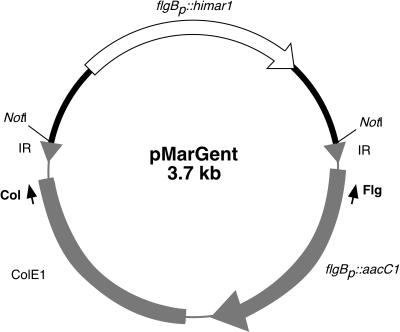
Vector pMarGent (Fig. 1) was constructed for delivery of the transposon on a suicide vector in B. burgdorferi. The inverted terminal repeats (binding sites for the transposase, shown in bold below) were incorporated into primers ITR.Flg(5′-GCGGCCGCTAACAGGTTGGCTGATAAGTCCCCGGTCTGCCCTAGGTAATACCCGAGC) and ITR.ColE1.RC (5′-GCGGCCGCTAACAGGTTGGCTGATAAGTCCCCGGTCTCCTAAGGATGAACTTGCCG), which were used for amplifying the ColE1 origin of replication together with the gentamicin-resistance cassette from pBSV2G (10). Primers ITR.Flg and ITR.ColE1.RC also included NotI restriction enzyme sites (underlined) used to produce compatible ends and circularize the PCR fragment. Finally, the C9 or A7 alleles encoding the hyperactive Himar1 transposase, fused to the borrelial flgB promoter, were cloned into the NotI restriction site, producing pMarGent (Fig. 1). The transposon portion of pMarGent (Fig. 1) confers the ability to replicate in E. coli only, but the gentamicin-selectable marker confers resistance in both E. coli and B. burgdorferi.
FIG. 1.
Plasmid map of pMarGent. The transposase, Himar1, is under the control of the borrelial promoter, flgBp. The region in grey represents the transposable element and is bounded by inverted repeats (IR), denoted by triangles. The arrows (Flg and Col) indicate oligonucleotides used for subsequent sequencing after rescue in E. coli. ColE1, E. coli origin of replication; flgBp::aacC1, the gentamicin resistance marker fused to the flgB promoter.
DNA sequencing and analysis.
Nucleotide sequences were determined with the ABI Big Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems, Foster City, Calif.) using an ABI 3700 DNA sequencer. Nucleotide sequences were analyzed with DNAstar (Madison, Wisc.) software.
Electroporation of B. burgdorferi and DNA isolation from transformants.
B. burgdorferi strains were transformed by electroporation with 20 μg of pMarGent (23, 24). Transformants were plated in the presence of 40 μg of gentamicin/ml. B. burgdorferi colonies grown on selective media were screened for the gentamicin resistance cassette by PCR, as previously described (24). Colonies positive by PCR analysis were aspirated from the agarose plate with sterile Pasteur pipettes, transferred to liquid BSK, and incubated at 35°C until the culture density reached >108 cells/ml.
Total genomic DNA was isolated from 5- to 10-ml cultures by using the Wizard genomic DNA purification kit (Promega, Madison, Wisc.) or by repeated phenol-chloroform extractions followed by ethanol precipitation. Genomic DNA was separated on a 0.3% 1× Tris-acetate-EDTA agarose gel to resolve intact DNA replicons (restriction-digested DNA was separated on a 0.7% 0.5× Tris-borate-EDTA agarose gel) and visualized by ethidium bromide staining. Gel electrophoresis and Southern hybridization analysis were carried out as previously described (22, 24). The plasmid contents of transposon mutants were determined by PCR (11).
Screen for growth rate phenotypes.
B. burgdorferi transformants were visually scored for altered colony morphology (i.e., compact versus diffuse colony shape) or delayed colony formation (i.e., transformants that arose at least 4 days after other colonies became visible). Selected transformants were aspirated with sterile Pasteur pipettes and transferred to liquid BSK medium containing 40 μg of gentamicin/ml. Those transformants that grew slower in liquid than the wild type were selected for further characterization (see below).
Microscopic characterization of transformants.
Mutants were visually examined by dark-field and scanning electron microscopy. Cells for scanning electron microscopy studies were allowed to settle on poly-l-lysine-coated Thermanox coverslips (Nunc, Naperville, Ill.) for 30 min and then fixed with 2.5% glutaraldehyde-4% paraformaldehyde in 0.1 M sodium cacodylate-0.1 M sucrose buffer for 2 h. Samples were postfixed for 1 h with 1% osmium tetroxide and dehydrated in a graded ethanol series. The samples were critical point dried under CO2 in a Bal-Tec (Balzers, Liechtenstein) model CPD 030 dryer, mounted on aluminum studs, and sputter coated with 100 Å of iridium in a model IBS/TM200S ion beam sputterer (South Bay Technologies, San Clemente, Calif.). Samples were viewed at 5 kV in an S-4500 field emission scanning electron microscope (Hitachi, Tokyo, Japan). The images were captured with Orion version 6.01 software (Focused Resolutions, Inc., Methuen, Mass.) and processed using Photoshop version 7 (Adobe Systems, Mountain View, Calif.).
Transposon recovery in E. coli.
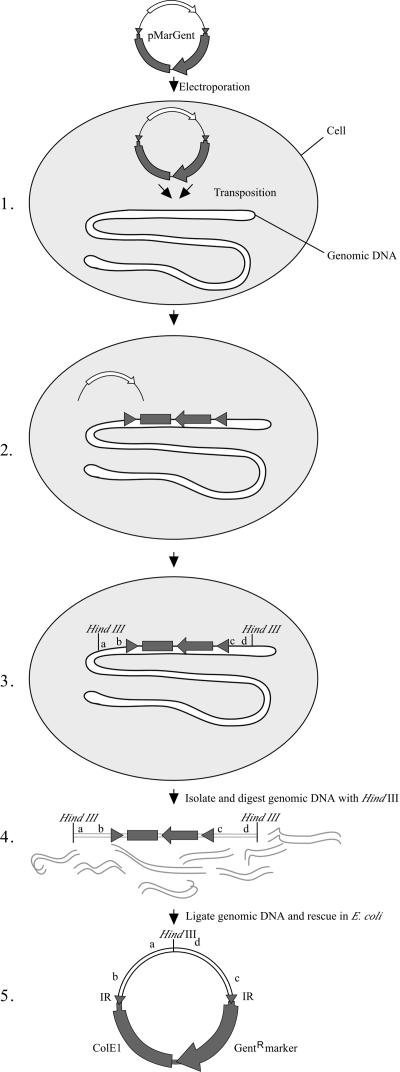
Vector pMarGent was constructed such that the transposon and B. burgdorferi DNA flanking the insertion site could be rescued easily in E. coli (Fig. 1 and 2). The recovery strategy entailed digestion of total genomic DNA with HindIII, a restriction enzyme with cleavage sites that were frequent throughout B. burgdorferi genomic DNA but absent from the transposon. Digested DNA was self ligated, transformed into E. coli, and plated in the presence of 5 μg of gentamicin/ml. Only circularized DNA containing the ColE1 origin of replication and the gentamicin resistance marker (carried on the transposon) would successfully transform E. coli. Specifically, 500 ng of total genomic DNA from B. burgdorferi transformants was digested with HindIII and ligated in a 10- to 15-μl volume at room temperature for >4 h (Fig. 2). Transposon insertions along with B. burgdorferi flanking DNA were recovered by transforming 5 μl of the ligation reaction mixture into chemically competent E. coli TOP10 cells (Invitrogen). Individual E. coli transformants were inoculated into 5-ml cultures, and plasmid DNA was isolated using the Qiaprep Spin Miniprep kit (QIAGEN, Valencia, Calif.). Primers Col (5′-CAGCAACGCGGCCTTTTTACG) and Flg (5′-TTTTTTGTTTGTTTTAAAAT) were used to sequence out from the transposon into the flanking B. burgdorferi DNA (Fig. 1). Sequences were identified using the BLAST nucleotide algorithm from the National Center for Biotechnology Information (27).
FIG. 2.
Transposon mutagenesis system for B. burgdorferi. Suicide vector pMarGent was electroporated into competent B. burgdorferi cells, allowing transient expression of the Himar1 transposase (1). After transposition, the Himar1 gene remains on a DNA fragment that is presumably degraded by intracellular nucleases (2). Mutants were selected in the presence of gentamicin, and those with desired phenotypes were transferred to liquid culture for DNA isolation. The B. burgdorferi DNA flanking the transposon insertion site was recovered by digestion with HindIII, an enzyme that does not cut within the transposon (3 and 4); self-ligation; and transformation into E. coli (5). Purified plasmid DNA was then isolated from E. coli clones and sequenced. IR, inverted repeat.
RESULTS
Construction of a transposon system for B. burgdorferi.
We searched the B. burgdorferi genome sequence for transposons from which an endogenous mutagenesis system could be derived. However, the type strain apparently lacks any complete transposable elements (8, 12). Therefore, we focused on the mariner transposons that have been used in a variety of heterologous hosts (1, 14, 16, 21, 29). Because of the low efficiency of transformation and allelic exchange in B. burgdorferi, we chose to avoid in vitro transposon mutagenesis (in which purified DNA is mutagenized by transposition followed by transformation and allelic exchange to introduce the mutation). Instead, we developed an in vivo expression system for the mariner element Himar1, thus avoiding the constraints imposed by the low frequency and efficiency of allelic exchange in B. burgdorferi.
Initial attempts to constitutively express the Himar1 transposase by introduction on shuttle vector pBSV2 failed (24; data not shown), possibly because overexpression of the transposase was lethal to B. burgdorferi. Therefore, we constructed a suicide vector in which the transposase would be transiently expressed from a strong borrelial promoter (flgBp). Use of this vector should allow sufficient expression of the transposase for transposition to occur but presumably avoid problems related to constitutive expression. For this purpose, vector pMarGent (Fig. 1) was constructed for transient expression of the transposase. The transposon (mobile element) portion of pMarGent is shown in Fig. 2 and consists of the inverted terminal repeats flanking a gentamicin-selectable marker and a ColE1 origin of replication. Once transposition had occurred, the transposase gene would be left on a fragment of foreign DNA and presumably degraded by cellular nucleases, enhancing the likelihood of a single transposition event per cell. Placement of ColE1 between the inverted repeats (i.e., within the transposon) allowed facile rescue in E. coli for subsequent characterization of the insertion site (Fig. 2; see Materials and Methods).
Transposition frequencies in B. burgdorferi strains.
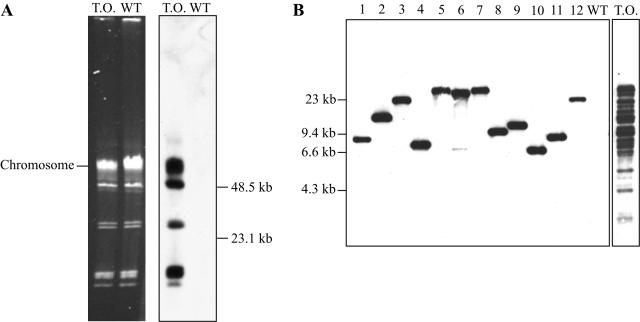
Derivatives of pMarGent constructed with either the A7 or C9 hyperactive transposase alleles were used in parallel for all transformations. Transpositions of the A7 and C9 alleles occurred at approximately the same frequency in B. burgdorferi (data not shown). Mutagenesis of strain B31-AchbC72 resulted in a transformation frequency of ∼5.5 × 10−5 (Table 1). Between 5,000 and 40,000 independent mutants were obtained per transformation, depending on the number of spirochetes surviving electroporation. Assuming a lower limit of ∼5,000 mutants per transformation and an approximate B. burgdorferi genome size of ∼1.52 Mbp (8), transposon insertions would occur, on average, every 300 bp. The ability of pMarGent to attain saturating levels of mutagenesis in B. burgdorferi is further supported by the Southern blot shown in Fig. 3B. Total genomic DNA was isolated from a transformation outgrowth (a mixed population of all mutants), digested with the restriction enzyme SpeI, blotted, and hybridized with the gentamicin resistance gene (aacC1). All visible restriction fragments hybridized with the probe, supporting a random, saturating mutagenesis (Fig. 3B, lane marked T.O.).
TABLE 1.
Transformation frequencies of B. burgdorferi strains electroporated with pMarGent
| Strain | Plasmids missingc | Transformation frequencya |
|---|---|---|
| Noninfectious strains | ||
| B31-AchbC72 | lp25, lp56, 1p28-4, 1p36, cp32-6, cp9 | 5.5 × 10−5 |
| A3-89 | lp25, lp56, 1p28-4, cp9 | 3.0 × 10−5 |
| Infectious strains | ||
| A3 | cp9 | No mutants detectedb |
| N40 | Plasmid content not determined | No mutants detectedb |
Transformation frequency is defined as the number of confirmed transposon mutants divided by the number of cells surviving electroporation, as determined by the number of CFU arising on plates without antibiotic selection.
Transformation frequency is ≤1 × 10−8, representing the limit of detection in these transformations.
Boldface type indicates plasmids encoding putative restriction-modification systems (17).
FIG. 3.
Southern blot analysis of genomic DNA from transposon mutants. Genomic DNA was purified from a pMarGent transformation outgrowth selected in the presence of gentamicin (T.O. [panel A]), wild-type B. burgdorferi (WT [panel A]), and 12 randomly chosen transposon mutants (panel B). All linear and circular replicons contained transposon insertions, as indicated in panel A. Genomic DNA was separated on 0.3% agarose gels, ethidium bromide stained, and blotted. Blots were hybridized with the labeled aacC1 gene, which encodes gentamicin resistance. As shown in panel B, genomic DNA from 12 mutants was digested with SpeI, which does not cleave within the transposon; separated on a 0.7% agarose gel; blotted; and hybridized as described above. A single major DNA fragment in each clone hybridized to the probe, indicating that transposition occurred only once per cell. The pMarGent system apparently reaches saturating levels of mutagenesis, as shown in the T.O. lane of panel B. SpeI-digested genomic DNA was prepared from the transformation outgrowth (shown in panel A), and virtually all restriction fragments hybridized to the probe.
As shown in Table 1, the transformation frequency of the low-passage-number but noninfectious strain A3-89 was similar to that of the high-passage-number strain B31-AchbC72. In contrast, no transposon mutants were detected in infectious strains A3 and N40. Both B31-AchbC72 and A3-89 lack lp25 and lp56, but infectious strain A3 contains both linear plasmids (the plasmid content of N40 has not been determined). Lawrenz and colleagues demonstrated a barrier to shuttle vector transformation in strains that retain lp25 and lp56, presumably due to putative restriction-modification systems encoded on these plasmids (17). Potentially, these restriction-modification systems also act upon pMarGent, reducing the transformation frequency to undetectable limits.
However, it was theoretically possible that additional plasmids, other than lp25 and lp56, impeded efficient transposition in B. burgdorferi. A3-89 cells in which transposition had occurred may have lost additional plasmids, most likely during growth and competence preparation, and these other plasmids may potentially encode barriers to transposition. Therefore, several A3-89 transposition mutants were randomly chosen and analyzed for their plasmid content; all retained the same plasmid composition as the parental A3-89 strain (data not shown). This result further supports the possibility that a barrier to efficient transposition is imposed by the restriction-modification systems encoded on lp25 and lp56 but not by other plasmids.
Characterization of transposon mutants.
Transposon mutants were characterized to determine the target site preferences. Genomic DNA was isolated from 12 randomly chosen transposon mutants. The transposon, along with flanking B. burgdorferi DNA, was recovered from E. coli and sequenced to identify the insertion site (Fig. 2). No additional consensus sequence was observed beyond the canonical TA dinucleotide target site (Table 2 and data not shown), indicating that transposition is random in B. burgdorferi. When the gene conferring gentamicin resistance (aacC1) was used as a probe, it hybridized to all B. burgdorferi DNA replicons isolated from a pMarGent transformation outgrowth (Fig. 3A, lane marked T.O.). Together, the results indicate that Himar1-mediated transposition targets both linear and circular DNA forms without preference (Fig. 3 and Table 2).
TABLE 2.
Transposon insertion sites of B. burgdorferi mutants
| Clone | Sequence at insertion sitea | Insertion site | Function or description | Replicon |
|---|---|---|---|---|
| 1 | ATGTAGTGGTAAAAGCCTCA | BB0608 | Aminoacyl-histidine dipeptidase (putative) | Chromosome |
| 2 | GGGTATAAATAGAGTTTGTT | BBB18 | GuaA-GMP synthase | cp26 |
| 3 | GCCGATCTTTACTACACTCA | BB0347 | Fibronectin and fibrinogen binding protein (putative) | Chromosome |
| 4 | AATAGAAACTATTAATGACT | BBR41 | Probable pseudogene | cp32-4 |
| 5 | TAAATCCCTTAAAGGTTATT | BB0830 | Exonuclease SbcC homolog (putative) | Chromosome |
| 6 | TTTTTACAATATCTTTGATA | BB0562 | Unknown, hypothetical | Chromosome |
| 7 | AATAACATTTATGTAATAAA | BB0827 | ATP-dependent helicase homolog (putative) | Chromosome |
| 8 | AATTGGCTGTAATTACAAGT | BBN16 | Hypothetical pseudogene | cp32-9 |
| 9 | GTTCATATATATATATGCAT | BBD14/15 | Insertion in intergenic region | lp17 |
| 10 | ACATATATTTAGAGTACATT | BBN26 | Outer surface protein (putative) | cp32-9 |
| 11 | AGGAATACTTAATGGCACAT | BBO36 | Unknown, hypothetical | cp32-7 |
| 12 | ATCTAGGAATAATAATTAAA | BB0102 | Unknown, hypothetical | Chromosome |
The TA dinucleotide insertion site for each mutant is shown in bold letters.
Further, the Southern blot of genomic DNA from 12 randomly chosen transposon clones hybridized with the labeled gentamicin resistance gene, confirming a single transposon insertion per cell (Fig. 3B). If transposition occurred more than once per cell, B. burgdorferi colonies would presumably be mixed populations. However, the transposon and B. burgdorferi flanking DNA were rescued in E. coli from over 20 B. burgdorferi mutants; sequencing of multiple E. coli clones per rescue confirmed a single insertion site per mutant (data not shown).
Genomic DNA from a subset of mutants analyzed by Southern blotting as shown in Fig. 3 was also probed with the labeled transposase gene. The probe hybridized only to the pMarGent vector DNA included as a positive control, indicating that once transposition had occurred, the remaining portion of the vector (including the transposase gene) was lost from the cell, as anticipated (data not shown).
Screen for growth rate mutants.
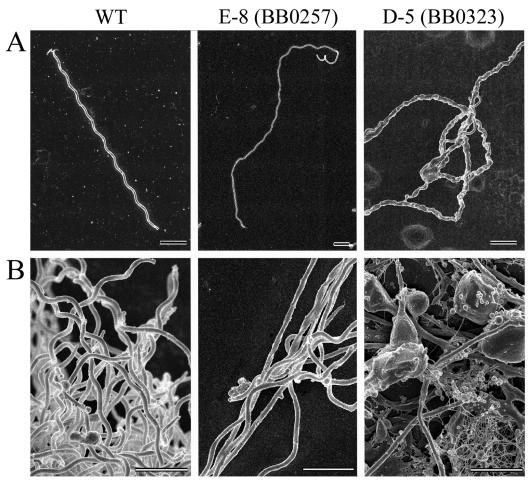
The utility of the pMarGent transposon system was demonstrated by designing a simple screen to identify mutations affecting growth, since many aspects of spirochete physiology and morphology remain unknown. A3-89 colonies that arose late or had compact colony morphologies (compared to the parental diffuse colony morphology) were selected for further characterization. Although many slow-growing mutants such as E-2 appeared normal, visual examination of mutants E-8 and D-5 by dark-field and electron microscopy revealed significant alterations in spirochete morphology. Rescue of the transposon in E. coli followed by sequencing identified the genetic loci disrupted. Mutant E-2 contained a transposon insertion into BB0414 (cheR-2), which encodes a putative methyltransferase protein. In other organisms, methyltransferases are involved in regulating the chemotactic response (9), but B. burgdorferi homologs have not been characterized. Elongated cells of mutant E-8 resulted from insertion of the transposon into BB0257, a putative cell division ORF (Fig. 4). The protein encoded by BB0257 has not been experimentally characterized, but by sequence similarity it was assigned to the FtsK/SpoIIIE family of proteins (12). Mutations in the FtsK/SpoIIIE family affect cell division and intercellular DNA transfer (5, 28). The third mutant, D-5, was significantly impaired in outer membrane integrity (Fig. 4). The disrupted ORF, BB0323, is designated as a hypothetical open reading frame lacking database homologs but containing a LysM domain at the carboxy terminus. The LysM domain is a peptidoglycan-binding module present in a variety of different bacterial proteins (4). These three mutants, and potentially others identified in this screen, represent new inroads into spirochete physiology and warrant further investigation to complete our understanding of their functions.
FIG. 4.
Scanning electron micrographs of B. burgdorferi cells. Images of wild-type (WT) B. burgdorferi, cell division mutant E-8 (BB0257), and hypothetical ORF mutant D-5 (BB0323) are shown at low (A) and high (B) magnifications. The scale bar represents 2 μm in all micrographs. Note that the cell division mutant E-8 is shown at approximately one-half the magnification of the WT.
DISCUSSION
Spirochetes compose a diverse and ancient class of bacteria, including both free-living and parasitic species. Although there are many spirochetes that are pathogenic for humans, relatively little is known concerning the physiology and morphology of these distinctive bacteria. Compounding this lack of fundamental knowledge is a limited repertoire of genetic tools available to elucidate gene function in this group of organisms. Here, we report the development of a new genetic tool, a random mutagenesis system based on a mariner transposon, that we used to address a basic physiological question about spirochetes.
The pMarGent system allows for random-tagged, saturating mutagenesis in B. burgdorferi. Transposition in high- and low-passage-number strains occurs at near saturating levels, with an insertion occurring, on average, every 300 bp. Construction of pMarGent was designed for easy recovery of the transposon and flanking Borrelia DNA sequences in E. coli, allowing rapid identification of the insertion site. Characterization of mutants, by Southern blot analysis and sequencing of recovered transposons, supports a single transposition event per cell (Fig. 3, Table 2, and data not shown). The utility of this mutagenesis system was demonstrated by applying a straightforward screen for mutants with altered growth rates. The mutants isolated by using this screen included those impaired in cell division, chemotaxis, and morphology.
The morphology and mechanism(s) of motility of spirochetes are unusual. Comparison of the three sequenced spirochete genomes (those of B. burgdorferi, T. pallidum, and L. interrogans) reveals that chemotaxis and motility genes represent a significant portion of the total genome of these organisms (over 5% for B. burgdorferi and T. pallidum) (8, 12, 13, 19). One reason for this substantial dedication of genetic and cellular machinery to directed motion might relate to survival and growth within the organisms' eukaryotic hosts. B. burgdorferi, however, must survive in both the mammalian host and the tick vector. The presence, then, of an additional chemotaxis operon in B. burgdorferi, not found in either of the other two spirochetes, may relate to motility in two very different eukaryotic environments (9). Likewise, a duplicated set of sensory transducing homologs in B. burgdorferi, including a cheR-2 homolog, may also pertain to nutrient acquisition in different hosts (9). The transposon mutagenesis system presented here, combined with a screen for slower growth rates, identified an insertion in cheR-2 (BB0414), which encodes a methyltransferase involved in regulating the chemotactic response. The cheR-2 mutant should be useful for studying the function of this additional copy of the methyltransferase, as T. pallidum, L. interrogans, and most other bacteria contain only a single copy.
This screen also identified a slow-growing mutant with a transposon insertion in a putative cell division ORF (BB0257). As shown in Fig. 4, the BB0257 mutant forms long chains of incompletely separated spirochetes, suggesting a defect in cell division. This result represents the first experimental support for the role of BB0257 in cell division. BB0257 has similarity to cell division genes in other organisms, including the E. coli ftsK gene. A mutation in ftsK blocks cell division but allows DNA replication (5). If the BB0257 mutant has a similar phenotype, then it will be a useful strain for studying DNA location and segregation in B. burgdorferi, an issue that remains largely unaddressed.
An insertion into the hypothetical ORF BB0323 resulted in a dramatic defect in outer membrane integrity. Electron micrographs of BB0323 mutant cells reveal spirochetes with ruptured outer membranes that often form enormous blebs (Fig. 4). Few individual spirochetes were observed, suggesting that there may be an aborted fission phenotype associated with this mutation. The Institute for Genomic Research annotation of this gene (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=gbb) recognizes a conserved lysin motif, designated LysM and present in a number of eukaryotic and prokaryotic proteins. This widespread motif apparently corresponds to a domain with general peptidoglycan-binding functions and includes proteins involved in cell wall degradation, binding and anchoring IgG to avoid the host immune system, and attachment to host cells (4). The function of the BB0323 ORF remains unknown, but the transposon insertion has a significant impact on the ability of B. burgdorferi cells to grow as individual spirochetes and form an intact outer membrane. Investigation into the function of the BB0323 product may elucidate a new protein function, perhaps necessary for critical outer membrane interactions with mammalian and tick hosts.
Although these transposon mutants were generated in A3-89, a noninfectious strain lacking lp25, infectivity can be assessed in this genetic background. As demonstrated by Purser and colleagues, reintroduction of the BBE22 product is sufficient to restore murine infectivity in lp25− strains (18). Therefore, the effect on infectivity of a specific transposon mutation can be tested by reintroduction of BBE22 and complementation with a wild-type copy of the mutated ORF. Complementation would ensure that the observed phenotypes of these mutants are directly linked to the disrupted locus, as transposon insertions may have polar effects on flanking genes.
Use of the pMarGent transposon system, in conjunction with a screen for slower growth rates, identified several previously uncharacterized genes that warrant further investigation and may enhance our understanding of spirochete cell division, chemotaxis, and spirochete membrane structure. This transposon system indicates the utility of the Himar1 element for mutagenesis in other spirochetes and provides the foundation for identifying B. burgdorferi loci relevant to pathogenesis, infectivity, and transmission in the tick vector and the mammalian host.
ADDENDUM IN PROOF
Y. Ostberg, J. A. Carroll, M. Pinne, J. G. Krum, P. Rosa, and S. Bergstrom (J. Bacteriol. 186:2074-2084, 2004) reported that the BB0323 protein may serve as a substrate for a carboxyl-terminal protease of B. burgdorferi.
Acknowledgments
We are grateful to Mark Fisher, Michael Otto, and Roberto Rebeil for manuscript review; Nyles Charon, Scott Samuels, and Kit Tilly for helpful discussions; and Dave Lampe for the generous gift of the Himar1 hyperactive mutants. Additionally, we are thankful to Pamela Ohler, Sandy Stewart, and Gail Sylva for expert technical assistance and Anita Mora and Gary Hettrick for graphic design.
REFERENCES
- 1.Ashour, J., and M. K. Hondalus. 2003. Phenotypic mutants of the intracellular actinomycete Rhodococcus equi created by in vivo Himar1 transposon mutagenesis. J. Bacteriol. 185:2644-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthold, S. W., M. S. de Souza, J. L. Janotka, A. L. Smith, and D. H. Persing. 1993. Chronic Lyme borreliosis in the laboratory mouse. Am. J. Pathol. 143:959-972. [PMC free article] [PubMed] [Google Scholar]
- 3.Barthold, S. W., K. D. Moody, G. A. Terwilliger, P. H. Duray, R. O. Jacoby, and A. C. Steere. 1988. Experimental Lyme arthritis in rats infected with Borrelia burgdorferi. J. Infect. Dis. 157:842-846. [DOI] [PubMed] [Google Scholar]
- 4.Bateman, A., and M. Bycroft. 2000. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD). J. Mol. Biol. 299:1113-1119. [DOI] [PubMed] [Google Scholar]
- 5.Begg, K. J., S. J. Dewar, and W. D. Donachie. 1995. A new Escherichia coli cell division gene, ftsK. J. Bacteriol. 177:6211-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bono, J. L., A. F. Elias, J. J. Kupko III, B. Stevenson, K. Tilly, and P. Rosa. 2000. Efficient targeted mutagenesis in Borrelia burgdorferi. J. Bacteriol. 182:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burgdorfer, W., A. G. Barbour, S. F. Hayes, J. L. Benach, E. Grunwaldt, and J. P. Davis. 1982. Lyme disease—a tick-borne spirochetosis? Science 216:1317-1319. [DOI] [PubMed] [Google Scholar]
- 8.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 9.Charon, N. W., and S. F. Goldstein. 2002. Genetics of motility and chemotaxis of a fascinating group of bacteria: the spirochetes. Annu. Rev. Genet. 36:47-73. [DOI] [PubMed] [Google Scholar]
- 10.Elias, A. F., J. L. Bono, J. J. Kupko III, P. E. Stewart, J. G. Krum, and P. A. Rosa. 2003. New antibiotic resistance cassettes suitable for genetic studies in Borrelia burgdorferi. J. Mol. Microbiol. Biotechnol. 6:29-40. [DOI] [PubMed] [Google Scholar]
- 11.Elias, A. F., P. E. Stewart, D. Grimm, M. J. Caimano, C. H. Eggers, K. Tilly, J. L. Bono, D. R. Akins, J. D. Radolf, T. G. Schwan, and P. Rosa. 2002. Clonal polymorphism of Borrelia burgdorferi strain B31 MI: implications for mutagenesis in an infectious strain background. Infect. Immun. 70:2139-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidmann, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 13.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, and J. C. Venter. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 14.Gueiros-Filho, F. J., and S. M. Beverley. 1997. Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276:1716-1719. [DOI] [PubMed] [Google Scholar]
- 15.Lampe, D., M. Churchill, and H. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 16.Lampe, D. J., B. J. Akerley, E. J. Rubin, J. J. Mekalanos, and H. M. Robertson. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. USA 96:11428-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrenz, M. B., H. Kawabata, J. E. Purser, and S. J. Norris. 2002. Decreased electroporation efficiency in Borrelia burgdorferi containing linear plasmids lp25 and lp56: impact on transformation of infectious Borrelia. Infect. Immun. 70:4851-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 19.Ren, S. X., G. Fu, X. G. Jiang, R. Zeng, Y. G. Miao, H. Xu, Y. X. Zhang, H. Xiong, G. Lu, L. F. Lu, H. Q. Jiang, J. Jia, Y. F. Tu, J. X. Jiang, W. Y. Gu, Y. Q. Zhang, Z. Cai, H. H. Sheng, H. F. Yin, Y. Zhang, G. F. Zhu, M. Wan, H. L. Huang, Z. Qian, S. Y. Wang, W. Ma, Z. J. Yao, Y. Shen, B. Q. Qiang, Q. C. Xia, X. K. Guo, A. Danchin, I. Saint Girons, R. L. Somerville, Y. M. Wen, M. H. Shi, Z. Chen, J. G. Xu, and G. P. Zhao. 2003. Unique physiological and pathogenic features of Leptospira interrogans revealed by whole-genome sequencing. Nature 422:888-893. [DOI] [PubMed] [Google Scholar]
- 20.Rosa, P., D. S. Samuels, D. Hogan, B. Stevenson, S. Casjens, and K. Tilly. 1996. Directed insertion of a selectable marker into a circular plasmid of Borrelia burgdorferi. J. Bacteriol. 178:5946-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi, p. 253-259. In J. A. Nickoloff (ed.), Methods in molecular biology, vol. 47: electroporation protocols for microorganisms. Humana Press, Inc., Totowa, N.J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart, P. E., R. Thalken, J. L. Bono, and P. Rosa. 2001. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol. Microbiol. 39:714-721. [DOI] [PubMed] [Google Scholar]
- 25.Tilly, K., A. F. Elias, J. L. Bono, P. Stewart, and P. Rosa. 2000. DNA exchange and insertional inactivation in spirochetes. J. Mol. Microbiol. Biotechnol. 2:433-442. [PubMed] [Google Scholar]
- 26.Tilly, K., A. F. Elias, J. Errett, E. Fischer, R. Iyer, I. Schwartz, J. L. Bono, and P. Rosa. 2001. Genetics and regulation of chitobiose utilization in Borrelia burgdorferi. J. Bacteriol. 183:5544-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tramont, E. C., J. D. Chulay, W. E. Farrar, W. D. Johnson, Jr., A. C. Steere, and R. G. Washburn. 1990. Spirochetes, p. 1794-1827. In G. L. Mandell, R. G. Douglas, and J. E. Bennett (ed.), Principles and practice of infectious diseases, 3rd ed. Churchill Livingstone Inc., New York, N.Y.
- 28.Wu, L. J., and J. Errington. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, J. K., M. A. Pritchett, D. J. Lampe, H. M. Robertson, and W. W. Metcalf. 2000. In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc. Natl. Acad. Sci. USA 97:9665-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]