Abstract
Although genes contribute to colorectal cancer, the gut microbiota are an important player. Accumulating evidence suggests that chronic infection and the ensuing inflammation contributes to tumor initiation and tumor progression. A variety of bacterial species and tumor-promoting virulence mechanisms have been investigated. Significant advances have been made in understanding the composition and functional capabilities of the gut microbiota and its roles in cancer. In the current review, we discuss the novel roles of microbiota in the progression of colon cancer. Although microbiota technically include organisms other than bacteria e.g., viruses and fungi, this review will primarily focus on bacteria. We summarize epidemiological studies of human microbiome and colon cancer. We discuss the progress in the scientific understanding of the interplay between the gut microbiota, barrier function, and host responses in experimental models. Further, we discuss the potential application in prevention, diagnosis, and therapy of colon cancer by targeting microbiota. We discuss the challenges lie ahead and the future direction in studying gut microbiome in colon cancer to close the gap between the basic sciences and clinical application.
Keywords: Beta-catenin, Colon cancer, Cytokines, Dysbiosis, Epidemiologic, Gut barrier, Human microbiome, Inflammation
Introduction
Colorectal cancer is the 3rd most common cancer in both males and females in the US and the 2nd leading cause of cancer deaths with the estimated new cases of nearly 133,000 and deaths of 50,000 in 2015.1 Worldwide, 1,360,000 new cases and 694,000 deaths per year are estimated.2 Cancer incidence in the large intestine is also known to be approximately 12-fold higher than that of the small intestine, which has been attributed to several magnitude greater bacterial density in the large intestine (∼1012 cells per ml) compared with that in the small intestine (∼102 cells per ml).3 With advance in metagenomic technology, growing evidence now suggests that dysbiosis, i.e., imbalance in of normal intestinal microbiota, can promote chronic inflammatory conditions and the production of carcinogenic metabolites, leading to neoplasia.4, 5
Gut microbiota represents a complex ecosystem that develops in close parallel with hosts and depends on the physiological environment of hosts. Humans have coevolved with their microbes over thousands of years. The gut bacterial population stabilizes during the first years of life and then remains stable throughout our life in terms of the major populations. Human gut microbiota are dominated by four main phyla: Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria. The corporate number of microbial species in human gut is estimated to be 1000–1150, with each individual harboring at least 160 (Qin, Li et al 2010). The number of genes of gut microbiota exceeds the number of genes in the human genome by 150 times. A large portion (38%) of the total gene pool is commonly shared from individual to individual. The “core human microbiome” refers to the central part of microbial gene pool existing in all or most of humans. The “variable human microbiome” is the microbial genes in a specific cohort of people, which is determined by a combination of host factors (Turnbaugh, Ley et al 2007). In the modern society, the host-microbial relationship is now being dramatically affected by shifts in the collective human microbiome resulting from changes in the environment and societal norms (Sun and Chang 2014).
In this review, we will discuss the roles of gut microbiota in colorectal cancer, summarizing both epidemiologic observations and the data from experimental animals. Although microbiota technically include organisms other than bacteria e.g., viruses and fungi, this review will primarily focus on bacteria, of which significant recent progresses have been made in understanding their role in human health. Specifically, understanding of the interplay between the gut microbiota, barrier function, and inflammatory responses will uncover new therapeutic targets in colorectal cancer. We will discuss the potential application in prevention, diagnosis, and therapy of colorectal cancer by targeting gut microbiota. Moreover, we will also discuss challenges lie ahead and the future direction in studying gut microbiome in cancer to close the gap between the basic sciences and clinical application.
Epidemiological studies of microbiome and colorectal cancer
At least two approaches have been employed to study colorectal cancer-associated microbiome. One is the targeted, more hypothesis-testing studies to examine whether exposure to specific bacteria species of interest increases the risk of colorectal cancer. The second type is studies aiming to identify differences in overall microbial composition by disease status. The latter has gained more popularity recently with advances in genomic technology for high throughput sequencing and discussed here first.
Microbiome core structure, diversity, richness and colorectal cancer
Most common materials used in these types of investigation are fecal or mucosal biopsy/resection samples and have been analyzed primarily by pyrosequencing. But it is now clear that bacterial populations in feces and mucosa are distinct.6, 7 As summarized in Table 1, the majority of these studies have demonstrated beta diversity by principal coordinate or component analysis illustrating structural difference of gut microbiome, where samples belonging to different disease status (cancer, adenoma, or controls/normal adjacent tissue) cluster in different two dimensional spaces,7, 8, 9, 10, 11, 12 indicating the presence dysbiosis. Analysis of community diversity/richness indies based on 16SRNA gene sequencing has shown significantly reduced microbial diversity in feces of colorectal cancer patients than in controls13 and in cancer tissue compared with mucosa at least 10 cm apart from cancer.14 On the contrary a richness index was higher in rectal mucosa of colorectal cancer patients than in that of control subjects7 or in cancer tissues than paired normal tissue.11 Others did not find differences in these alpha diversity indices.9, 10, 15, 16 With or without using additional quantitative PCR (qPCR), these studies have also found that specific bacterial groups were more common or less common in colorectal cancer cases than control specimens.7, 8, 9, 10, 11, 12, 13, 14, 15, 16 Because each study has used different taxonomic levels/classifications for the comparison, there have little consistency in changes associated with colorectal cancer. However, there were multiple studies reporting overrepresentation of Fusobacterium and Porphyromonasand and underrepresentation of Faecalibacterium (Table 1). Yet, it should be noted that some of these studies were based on very small numbers of samples and control subjects were often not comparable with cases in terms of basic demographic factors (such as age). In summary, while these studies underscore marked differences in gut microbial membership between colorectal cancer patients and healthy controls, it is difficult to generalize characteristics of cancer associated gut microbiome.
Table 1.
Summary of 16rRNA pyrosequencing studies involving colorectal cancer (CRC) and control specimens addressing microbial community structure.
| Authors (year) | Study subjects (N) | Type of specimens | 16S rRNA region | Beta diversity | Alpha diversity | Overrepresentation | Underrepresentation |
|---|---|---|---|---|---|---|---|
| Sobhani et al (2011)8 | CRC (60), colonoscopy control (119) | Stool | V3–V4 | PCA | – | Bacteroides/Prevotella | |
| Ahn et al (2013)13 | CRC (47), surgical control (94) | Stool | V3–V4 | – | Shannon index down in CRC | Fusobacterium, Porphyromonas | Clostridia |
| Wang et al (2012)9 | CRC (46), healthy volunteers (56) | Stool | V3 | PCA | No difference in diversity and evenness | Porphyromonas, Escherichia/Shigella, Enterococcus, Streptococcus, Peptostreptococcus | Bacteroides, Roseburia, Alistipes, Eubacterium, Parasutterella |
| Wu et al (2013)10, 65 | CRC (19), healthy volunteers (20) | Stool | V3 | PCoA | No difference in diversity and richness | Bacteroides species Fusobacterium Campylobacter species | Faecalibacterium, Roseburia |
| Weir et al (2013)15 | CRC (11), healthy volunteers (10) | Stool | V4 | – | No difference in diversity and richness | Akkermansia muciniphila | Bacteroides, Prevotella, Ruminococcus |
| Chen et al (2012)14 | CRC (46), healthy volunteers (56) | Stool, rectal swab, cancer tissue, adjacent (2–5 cm and 10–20 cm apart) normal mucosa | V1–V3 | – | Shannon index down in CRC tissue vs paired mucosa 10–20 cm apart | Lactobacillales (tumor), Erysipelotrichaceae, Prevotellaceae, Coriobacteriaceae (stool) | Faecalibacterium (tumor) |
| Mira-Pascual et al (2015)7 | CRC (7), adenoma (11), healthy volunteer (10) | Tissue (tumor or rectal mucosa), stool | V1–V3 | PCoA (tissue) | Richness up in cancer tissue | Enterobacteriaceae (cancer tissue) | |
| Geng et al (2013)11 | CRC (8) | Paired tissue (cancer, normal) | V1–V2 | PCoA | Richness up in cancer | Roseburia | Microbacterium, Anoxybacillus |
| Geng et al (2014)12 | CRC (8), adenoma (10), healthy volunteer (10) | Normal and tumor tissue | V1–V2 | PLS-DA | – | Streptococcus, Porphyromonas, Veillonella (cancer vs control) | |
| Kostic et al (2012)16 | CRC (95) | Paired tissue (cancer, normal) | V3–V5 | – | No difference in richness | Fusobacterium | Bacteroides, Clostridia, Faecalibacterium |
PCA: Principal component analysis; PCoA: Principal coordinate analysis; PLS-DA; Partial least square discriminant analysis.
Individual bacterial species and colorectal cancer risk
Streptococcus bovis
Streptococcus bovis (SB) is a gram-positive bacterium and lower-grade opportunistic pathogen that can cause systemic infections (endocarditis or bacteremia) in humans. It is a group D streptococcus with the specific ability to grow in 40 percent bile.17 Intestinal mucosal lesions have been deemed to serve as a portal for these bacteria to the systemic circulation. Based on biochemical traits, DNA homology and divergence in 16S rRNA sequences, SB can be grouped into Streptococcus gallolyticus (SB biotype I and II/2) and Streptococcus infantarius (biotype II/1). Earlier studies suggest stronger association of S. gallolyticus with colorectal tumors18 in contract to stronger link of S. infantarius to non-colonic cancers, primarily in the pancreas and biliary tract.19
Although SB is a member of normal gastrointestinal flora in ruminants, e.g., cattle, sheep, horses, pigs, camels and deers, it is also found in human feces as well as gastric biopsy materials.20, 21 Approximately 10% of healthy individuals have been estimated to carry this bacterium asymptomatically in their digestive tract.20 While fecal–oral or oral–oral is a possible transmission route between humans, it may be acquired through dietary intake of ruminant-derived foods, such as unpasteurized dairy products,22 red meat and animal organs.20 In fact SB is a frequently detected contaminant in commercially available meat.23, 24 The correlation between SB and colonic disease has long been recognized. Besides case-reports for the patients who were diagnosed with asymptomatic colorectal neoplasia simultaneously with SB endocarditis or bacteremia,25, 26, 27, 28, 29, 30 investigators have reported increased prevalence of colorectal tumors (cancer and polyps) among patients diagnosed with SB endocarditis or bacteremia. The prevalence of colorectal tumors ranges from 10 to 60%,18, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45 although these are based on diverse study populations in terms of patient demographics and colorectal surveillance methods. These variations may also be due to the heterogeneous definition of the cases, as adenomas have been defined as diseased in some studies but not in the others.46 A more recent study found that 52% of SB bacteremia patients had advanced adenoma/cancer, which was approximately 2.5 fold more frequent than colonoscopy controls.47 Similar prevalence (60%) of advanced adenoma/cancer was reported in SB endocarditis patients by Sharara et al.48
The second set of evidence is derived from studies comparing SB prevalence among various patient groups with or without colonic diseases.49, 50, 51, 52, 53, 54, 55, 56 While 3 small studies including 13–46 controls and corresponding 11 colorectal cancer, 47 pediatric inflammatory bowel disease (IBD) and 56 polyp patients failed to show any association,52, 53, 54 five other studies found that SB carriage (either in stool or antibodies) rates were significantly higher in cancer patients than in controls. Interestingly, 3 studies also showed that patients with premalignant lesions (IBD or polyps) had intermediate SB carriage rate between cancer cases and controls. In addition, stronger associations observed in studies by Darjee & Gibb, Tjalsma et al and Abdulamir et al51, 55, 56 suggest that antibody assays may be a more powerful tool than fecal culture in assessing the associations between this bacterial infection and colorectal disease. Subsequent enzyme-linked immunosorbent assay (ELISA) based studies have demonstrated that seropositivity or higher antibody titer to specific SB antigens or their combinations was associated with early stage of colorectal cancer57, 58 or colorectal cancer diagnosed at younger age (<65 years),59 yielding odds ratios of 1.5–8.0. In summary, despite these observations it remains elusive whether colorectal neoplastic sites provide a specific niche for SB resulting in sustained colonization and survival or whether SB infection itself promotes colorectal carcinogenesis, or a combination of both.
Helicobacter pylori
H. pylori was designated as a group 1 human carcinogen by International Agency for Research on Cancer (IARC) in 1994 because an expert panel concluded that there was sufficient evidence in humans for the carcinogenicity of this bacterial infection and that its chronic infection causes non-cardia gastric adenocarcinoma and low-grade B-cell mucosa-associated lymphoid tissue lymphoma.60 H. pylori is a gastric pathogen that infect more than a half of the adult population in the world.61 Gastric carcinogenic pathway caused by H. pylori has been well documented, arising from stages of premalignant lesions, i.e., chronic gastritis, atrophic gastritis, intestinal metaplasia and dysplasia, and then progressing to adenocarcinoma.62, 63, 64 While the gold standard for diagnosis of active Helicobacter infection is histological detection in gastric biopsies, stool antigen tests have been clinically accepted as a non-invasive alternative, indicating H. pylori also resides in the large intestine. Although no Helicobacter induced intestinal pathologies have been established, a number of epidemiologic studies have been conducted to examine if HP infection increases the risk of colorectal cancer. A recent meta-analysis by Wu et al65 summarized the data for 3488 colorectal cancer cases, 3792 colorectal adenoma and 10,598 and 4348 corresponding controls from 27 studies. They reported significantly increased summary odds ratios for both, 1.39 (1.18–1.64) for cancer and 1.66 (1.39–1.97) for adenoma. However, two prospective studies66, 67 with the nested case–control design did not find any indication of increased risk, while all others were either cross-sectional or retrospective case–control studies. It is noteworthy that except one study by Jones et al,68 there was no histological confirmation of presence of H. pylori in colorectal mucosa as others used gastric histology, serology or breath test to assess H. pylori infection. Nevertheless, significantly increased risks of cancer and polyps were observed by Jones et al,68 as well as in an additional study among children with hamartomous (juvenile) colorectal polyps,69 respectively. Despite relatively consistent epidemiologic observations to date, there seems insufficient evidence to support causality of the events. Certain biases may be involved, such as publication bias as reported65 as well as surveillance bias particularly for adenoma. In addition, there may be indirect consequences from gastric pathology, such as hypergastrinemia, which is common in patients with Helicobacter infection and has been hypothesized to stimulate colorectal tumor growth.70
Escherichia coli
Escherichia (E) coli strains are aero-anaerobic Gram-negative bacteria in the normal intestinal flora. As a commensal, E. coli coexist harmoniously with their mammalian host, promote normal intestinal homeostasis and rarely cause disease. However, some virulent E. coli that have acquired pathogenicity islands can colonize the human gastrointestinal tract and induce disease.71 Mucosa-associated E. coli have been identified more frequently in colon tissue from patients with adenocarcinomas than in controls.72, 73, 74 Some E. coli strains harbor a ∼54-kb polyketide synthases (pks) pathogenicity island that encodes multi-enzymatic machinery for synthesizing a peptide-polyketide hybrid genotoxin named Colibactin.75 Carriage of E. coli positive to the pks island or genes in the island has been recently found more common in the mucosa of colorectal cancer and IBD patients than that of control subjects.71, 75, 76 Epithelial proliferation and E. coli colonization density were significantly correlated in the mucosa distant from cancer76 and psk positive cancer specimens showed higher levels of DNA damage than its negative counterparts,77 supporting potential causal link.
Bacteroides fragilis
The anaerobe B. fragilis is a colonic symbiote with an affinity for mucosal colonization that comprises a relatively small proportion of fecal microbiota (approximately 0.5%–1%). There are 2 molecular subtypes, nontoxigenic B. fragilis (NTBF) and enterotoxigenic B. fragilis (ETBF) and ETBF is now established as a cause of diarrheal disease.78 ETBF pathogenicity is due to the B. fragilis toxin (BFT), a 20 kDa zinc-dependent metalloprotease toxin with 3 isotypes (BFT-1, BFT-2, and BFT-3) and the bft gene is unique, only identified in B. fragilis.78 BFT binds to a specific colonic epithelial receptor activating Wnt and NF-κB signaling pathways with increased cell proliferation, epithelial release of proinflammatory mediators, and induction of DNA damage78, 79 and ETBF promotes tumor formation in experimental animals.79, 80 Despite these laboratory data, to date only limited data in humans support an association of ETBF with colorectal cancer. Ulger Toprak et al81 reported that 38% of fecal samples from cancer patients were positive to btf gene while only 12% of those from control patients were positive (P = 0.009). Boleij et al82 recently revealed more frequent detection (∼75%) of btf genes in colonoscopic biopsies, particularly among patients with no antibiotic pretreatment and the prevalence was significantly higher in cancer than controls.
Fusobacterium (F) nucleatum
F. nucleatum is a Gram negative, non-spore forming, obligate anaerobic of the Fusobacteriaceae family, which consists of 9 genera, including Fusobacterium and Leptotrichia. Fusobacterium genus includes at least 14 species, several of which (including F. nucleatum) are known pathogens.83 F. nucleatum is perhaps best appreciated for its role as a component of oral plaque, where by virtue of its adhesive abilities it serves as a bridge organism between early and late colonizers of this biofilm and consequently is implicated in various forms of periodontal diseases.84
Until recently, F. nucleatum was thought to primarily be a component of the oral microbiota of humans and only an occasional resident of the gut. However, this premise was built on culture-based examination of stool, which usually does not contain high numbers of live, epithelium-associated bacteria. Using metagenomic approaches recently, growing number of studies have reported an over-representation of sequences from F. nucleatum16, 85, 86 or genus F5, 87, 88 in tumors relative to control specimens. Two of these by Castellarin86 and by Warren87 were based on RNA, representing transcribing bacteria. These observations were further confirmed by quantitative (q) PCR and in situ hybridization in tumor tissue.16, 86, 89 Using qPCR, McCoy et al studied F counts in normal rectal mucosa of the cases with or without colorectal adenoma, revealing a 3 fold increase in risk of adenoma among subjects with highest tertile of F counts.90 Ito et al91 also demonstrated that F. nucleatum detection in formalin-fixed paraffin-embedded tissue by qPCR progressively increased with malignant grades of the lesions from hyperplastic polyps to colorectal cancer. Several others found higher fecal carriage of genus F13, 85, 92 or Fusobacteriaceae family10 in colorectal cancer patients than in control subjects, pointing to a potential tool for colorectal cancer screening.
All F. nucleatum strains may not equal in their virulence potential. F. nucleatum is naturally co-aggregative and would likely exist in the human gut microbiota as a feature of a larger microbial grouping. The ability to adhere to other bacterial species could also enable gene transfer and thus some F. nucleatum strains may acquire genes through horizontal transfer leading to increased virulence,93 which suggests that the involvement of F. nucleatum in disease may not be just a function of a direct result of its own virulence. Despite these accumulated evidences, however, whether this association is indeed involved in colorectal carcinogenesis, or simply the result of F. nucleatum exploitation of an ecological niche created as a result of the cancer/tumor microenvironment, remains to be tested in further studies.
Salmonella enterica
S. enterica is a Gram-negative, facultative anaerobe and an intracellular pathogen to both humans and animals, posing a major public health concern worldwide. Non-typhoidal Salmonella is a major foodborne pathogen, with an estimated 93.8 million cases and about 155,000 deaths globally per year.94a Common sources of infection include contaminated food, such as meat, eggs and produce.94b Outcomes of this bacterial infection vary widely, ranging from mild self-limiting gastroenteritis to the severe systemic infection that can be fatal. Some of these acute infections result in a chronic carrier state excreting the bacteria in stool and urine without symptoms, which represents another transmission mechanism of this bacterium to other humans. Salmonellosis has also been implicated in the development of various chronic sequelae, including reactive arthritis, irritable bowel syndrome, IBD95 and even cancer.96
Two studies from Scandinavian countries have found that the probability of new IBD diagnosis significantly (2–3 fold) increases compared with general population following an episode of non-typhoid salmonella infection, particularly within the first 10 years.97, 98 Although data directly linking to colorectal cancer are still limited, Salmonella typhi carries status is well recognized to increase the risk of gallbladder cancer. A meta-analysis by Nagaraja et al demonstrated the summary odds ratio of 3–496 regardless of salmonella detection methods. Furthermore, Kato et al99 recently reported that antibody against Salmonella flagellin was higher in colorectal cancer and pre-cancer cases than controls in two distinct populations in US and the Netherlands and that dietary intake is the one of the mediating factors, supporting a possible link of Salmonella to colorectal cancer.
Other miscellaneous
Several other species of bacteria have received research interest because their bacterial metabolites have potential detrimental effects against colorectal mucosa or may exert potentially beneficial or protective effect towards epithelial cells. These include Desulfovibrio, Enterococcus faecalis due to hydrogen sulfide and superoxide respectively,100 Faecalibacterium prausnitzii and Bifidobacteria due to butyrate and lactate, respectively.101, 102 The presence or density/quantity of these bacteria in feces or mucosa has been primarily studied by qPCR. However, there have been only sporadic studies reporting a significant association with colorectal cancer itself,101, 102 while others found higher prevalence or density of these bacteria in IBD than in controls, which was further correlated with disease activity.4, 103, 104 In addition to F. nucleatum, Porphyromonas gingivalis, another oral pathogen more tightly associated with periodontal disease has been linked to digestive tract cancer in a seroepidemiologic study. However, the study was too small to separate colorectal cancer from other cancers.105 The potential association of this bacterium with colorectal cancer may be further corroborated by several other metagenomic studies that observed the overrepresentation of genus Porphyromonas or Porphyromonadaceae family in colorectal cancer specimens than control specimens.10, 13, 92 Overall, the information available thus far for these bacteria is insufficient to address their etiological involvement in colorectal cancer.
Interactions between colorectal cancer risk factors and gut microbiome
As discussed above, growing evidence now point to differential gut microbial compositions or differential prevalence of specific bacteria in colorectal cancer patients in comparison with control subjects. However, there are also abundant data supporting the associations between gut microbiota and several established risk factors for colorectal cancer. Thus, one should consider a possibility that observed difference in microbiota mirror at least in part changes associated with such risk factors. The best example is obesity. Obese and lean individuals are known to harbor different types of gut microbiota.106 While low energy diet induces change in microbial compositions increasing gene richness,107 microbiome itself also contributes energy harvest to the host, as demonstrated in mice models where transfer of obese microbiome to lean animals led to an increase in body adiposity in a diet dependent manner.108, 109 Other dietary risk factors for colorectal cancer include low fiber and high red meat intake.110 Dietary fiber and resistant starch are well known to stimulate gut bacterial fermentation to generate short chain fatty acids (SCFA) as well as lactate and to increase relative abundance of bacterial groups with the relevant metabolic activities.111 Although meat intake itself has been rarely studied, removal of animal products (vegan diet) was recently tested in a few clinical trials, showing changes in the Firmicutes/Bacteroidetes ratio and abundance of bacteria capable of triggering inflammation.112, 113 Moreover, as discussed above, meats are one of the suspected sources of acquisition of specific pathogens, e.g., S. Bovis ad Salmonella enterica. There has been relatively sparse information as to the associations between other risk factors, physical activity, smoking and alcohol, and gut microbiome. A study from Ireland found that athletes hard significantly higher microbial diversity than controls.114 Alcoholics have been reported to carry greater abundance of Proteobacteria or its family Enterobacteriaceae115, 116 than control subjects. Smoking cessation led to changes in gut microbial composition, increasing some Firmicutes and decreasing some Bacteroidetes and Proteobacteria,117, 118 while Kato et al demonstrated a positive association between smoking and Desulfovibrio abundance.119 Since these risks factors are postulated to be involved in multiple mechanistic pathways, contribution of microbial changes to colorectal carcinogenesis remains to be determined.
Cautions in the interpretation of epidemiologic data
Despite the presence of certain biological mechanisms possibly contributing to colorectal carcinogenesis (discussed in later sections), the causal association cannot be inferred only from the data from retrospective or cross-sectional human studies. Except a few for H. pylori66, 67 and P. gingivalis,105 all other studies identified the exposure, i.e., the presence bacteria or their antibodies to bacteria, was assessed at or after diagnosis of the disease. This makes it difficult to establish the temporal sequence of the events, which came first, bacteria or cancer. Moreover, the presence of the organism may no longer necessary once carcinogenic pathways are activated by infection as seen in the case of HP and gastric cancer. Serum antibody assays can capture past and current infection and have played a vital role in establishing infectious etiology of several types of cancer, including H. pylori and hepatitis viruses,120 especially with use of prediagnostic blood samples from prospective cohorts. Thus, development of reliable serological assays is likely to greatly advance epidemiologic studies. However, due to the limitation of serology as well as fecal analyses, i.e., an inability to identify the location of colonization for the bacteria that can colonize at diverse anatomical sites, histological detection of bacteria in cancer and surrounding tissues would also be required to reinforce their causal involvement.
Mechanisms for microbially induced/promoted colorectal cancer
A systemic review summarizes the original articles studying microbiota and colorectal cancer until November 2014. It showed that some bacteria are consistently augmented (such as Fusobacteria, Alistipes, Porphyromonadaceae, Coriobacteridae, Staphylococcaceae, Akkermansia spp. and Methanobacteriales), while other are constantly diminished in colorectal cancer (such as Bifidobacterium, Lactobacillus, Ruminococcus, Faecalibacterium spp., Roseburia, and Treponema). It is clear that bacteria metabolites amino acids are increased and butyrate is decreased throughout colonic carcinogenesis.121
Identification of components of the microbiota and elucidation of the molecular mechanisms of their action in inducing pathological changes or exerting beneficial activities could aid in our ability to influence the composition of the microbiota and to find bacterial strains and components (e.g., probiotics and prebiotics) whose administration may aid in disease prevention and treatment.122
Experimental animal models to study microbiome in colon cancer
To study the microbiome in colon cancer, researchers have developed various Experimental animal models: gnotobiological model, antibiotic treatment, inflammatory model with increased risk of colon cancer, inoculation of specific bacteria or products in genetic engineering mice.
Gnotobiological model is an indispensable tool for studying the consequences of bacterial colonization. Animals (such as zebrafish, mouse, rat, pig) can be maintained in sterile conditions and colonized with defined microbes. The effects of the germ-free state or the effects of colonization on disease initiation and maintenance can be observed in these experimental models for disease initiation and progression. Using this approach, researchers have demonstrated direct involvement of components of the microbiota (including non-cultivable commensal bacteria) in chronic intestinal inflammation, development of colonic neoplasia, and other diseases.
A variety of bacterial species and tumor-promoting virulence mechanisms have been investigated, using mouse models. There involve bacterial metabolic products, Pathogenic bacterial toxins/virulence factors, and Immune reaction/modulation.
Bacterial metabolic products
Firmicutes and Bacteroidetes predominate the gut microbiota, followed by Proteobacteria and Actinobacteria, with minor contributors including Verrucomicrobia and Fusobacteria.123 Bacteroides and Ruminococcus are consistent with enriched intake of animal sources, while a plant-based diet favors Prevotella.124 Prevotella to Bacteroides ratio constitutes an important index for clinical diagnosis. Butyrate-producing bacteria, including Clostridium groups IV (Faecalibacterium prausnitzii) and XIVa, Roseburia spp., Butyricicoccus, and lactic acid bacteria (LAB), mainly Lactobacillus and Bifidobacterium, are believed to benefit the host through anti-inflammation, anti-tumorigenesis, and pathogen exclusion.125, 126, 127 There is also a metabolic interplay between LAB and butyrate-producing bacteria due to the ability of the latter to feed on lactate.128
It is known that gut microbiota could produce an enormous quantity of molecules interacting with the host. The beneficial effects of gut microbiota on the host are mainly mediated by its metabolites. Short-chain fatty acid (SCFA), including acetate, propionate, and butyrate, are the major end-products of gut bacteria fermentation of dietary fiber. SCFAs, particularly butyrate, are the preferred source of energy for colonic epithelial cells. SCFA promotes and maintains colonic epithelial health through maintaining barrier function,129 suppressing colonic cancer,130, 131, 132 inhibiting intestinal inflammation (Wu et al 2014), modulating immune response,133 regulating DNA methylation for proliferation,132 and diminishing oxidative DNA damage.134
The balance between two phyla (Firmicutes and Bacteroidetes) appears to be critical to regulating disease progression. Some bacterial species have been implicated in the development of colorectal carcinoma. Using culture methods, Moore and Moore observed that the abundance of Bacteroides and bifidobacteria was associated with increased risk of colon polyps, whereas Lactobacillus and Eubacterium aerofaciens were protective.135 An association between the abundance of Fusobacterium, E. coli, hydrogen sulfide (H2S)-, and bile salt-producing bacteria was associated with increased risk of colon cancer.5, 136 Cancer is associated with reduced abundances of Clostridium, Roseburia, Eubacteria spp., and other butyrate-producing bacteria in fecal samples of adenoma subjects compared with healthy controls. Zeller et al85 reported that a relative abundances of 22 gut microbial species, such as Fusobacterium collectively associated with CRC. This is the first paper based on the whole sequence of bacterial genes, not 16S. It also compared the bacterial markers with the results of the standard Hemoccult FOBT routinely applied for CRC screening and an experimental CRC screening test based on methylation of the wif-1 gene, a Wnt pathway member. The authors believe that there is a potential to use fecal microbiota markers for early-stage detection of colorectal cancer.
Pathogenic bacterial toxins/virulence factors
Salmonella infection in humans can become chronic which leads to low grade persistent inflammation.137 These chronic infections increase the risk of several gastrointestinal138 diseases, including chronic cholecystitis and gallbladder cancer.139, 140 Recently, Kato et al reported that antibody against Salmonella flagellin was higher in colorectal cancer and pre-cancer cases than controls in two distinct populations in US and the Netherlands and that dietary intake is the one of the mediating factors, suggesting a potential link of Salmonella to colorectal cancer.99 In animal models, Salmonella and its derivatives have been observed invading transformed tissue more efficiently than normal tissue.141, 142 Salmonella AvrA is a multifunctional protein that influences eukaryotic cell pathways by altering ubiquitination and acetylation of target proteins.143, 144, 145, 146, 147, 148, 149 We reported that AvrA acts as a deubiquitinase to stabilize β-catenin. By suppressing β-catenin degradation, AvrA enhances intestinal epithelial proliferation, thus promoting tumorigenesis.150 We reported that AvrA-enhanced tumor multiplicity and tumor progression. Our studies could suggest biomarkers (such as AvrA level in gut) to assess cancer risk in susceptible individuals and infection-related dysregulation of β-catenin signaling in colon cancer. Another novel finding in our study was that the pathogenicity factor altered tumor distribution. Uninfected mice treated with AOM/DSS developed tumors in the distal colon.150 In contrast, in mice infected with AvrA-expressing bacteria, tumors were found more in the proximal colon. AvrA alters the colonic milieu so as to enhance tumorigenesis in the right colon. Compared with the left colon, the cecum has a greater bacterial load and increased bacterial fermentation that we speculate contributes to this rightward shift in tumors. Increasing incidence in right-sided tumors has also been reported in the Western world. While increased endoscopic screening that probably clears distal colonic lesions more effectively than proximal colonic lesions, based on our studies, this shift might also reflect changes in the microbiome. However, it remains unclear how many human CRC cases can be attributed to bacterial agents, how these exactly interact with the human host or the microbial community in the gut.
Gut microbiota metabolism could be linked with polyp formation, using mice genetic model.151 A diet reduced in carbohydrates resulted in reduced polyp formation in APCMin/+ MSH2−/− mice. Butyrate, a bacterial product, induced aberrant proliferation and transformation of colon epithelial cells. Treatment with either antibiotics or a low-carbohydrate diet reduced cell proliferation as well as the number of tumors in the small intestines and colons. However as mice microbial ecology is different, compared to human, authors did not found Fusobacterium, which was shown to be link to CRC in humans.
A paper from Journal of Experimental Medicine152 reported that antibiotics prevented polyp formation. Most of the tumor-dwelling bacteria belonged to the Clostridiales family and an upregulation of inflammatory molecules near the polyps. FMT from the untreated mice to the once germ-free mice, the previous germ-free mice developed polyps. If transplanted early embryos of the transgenic mice into females of another, cancer-free mouse strain. Inoculated at birth with the bacteria of their surrogate mothers, these transplanted mice did not develop tumors until 25 weeks, whereas the genetically identical controls had tumors by 12 weeks. This showed that small changes in the gut microbiota could have a large influence on tumor growth. This study indicates that the same genetic mutation in different individuals may have a different outcome.152
One environmental factor – a diet low in fiber – may impact the intestinal microbiota in a way that affects host cell physiology, cellular homeostasis, energy regulation, and/or metabolism of xenobiotics. This in turn may lead to chronic inflammation and CRC. Cancer is associated with reduced abundances of some butyrate-producing species. Transplanting feces from mice with CRC into germ-free mice leads to increased tumorigenesis.153
While emerging evidence suggests a link between the gut microbiota and colon cancer, it is hard to say that certain bacteria strain(s) play a causal role in CRC. Evidence is still needed to determine whether those bacteria enhance the development of the disease or might even play a causal role.
Cancer is fueled by deregulation of signaling pathways in control of cellular growth and proliferation. These pathways are also targeted by infectious pathogens en route to establishing infection. It is established that a single infectious agent, namely H. pylori, hepatitis B virus, plays a causal role in human gastric and hepatic cancers, respectively. The exact roles and mechanisms of microbes on the development of colon cancer in are still unknown and of great interests.
Immune reaction/modulation
Although genes contribute to colorectal cancer (CRC), the gut microbiota are an important player. Accumulating evidence suggests that chronic infection and the ensuing inflammation contributes to tumor initiation and tumor progression.137, 154 A variety of bacterial species and tumor-promoting virulence mechanisms have been investigated, using mouse models. A recent study in mice showed that adenomas cause barrier defects in the colonic epithelium allowing microbial products to drive IL-23/IL-17-mediated tumor growth.155 Another study demonstrated that a human colonic commensal bacterium promoted tumorigenesis via activation of T helper type 17 T cell responses.80
Colitis was shown to promote tumorigenesis by altering microbial composition and inducing the expansion of microorganisms with genotoxic capabilities.75 Arthur et al reported the intestinal microbiota as a target of inflammation that affects the progression of CRC. Monocolonization with the commensal E. coli NC101 promoted invasive carcinoma in azoxymethane (AOM)–treated Il10−/− mice. Specifically, deletion of the polypetide synthase genotoxin from E. coli NC101 decreased tumor load and tumor invasion in AOM treated IL10 knockout mice. E. coli NC101 mutant without the polyketide synthase (pks) genotoxic island decreased tumor multiplicity and invasion in AOM/Il10−/− mice. Mucosa-associated pks+ E. coli were found in a significantly high percentage of inflammatory bowel disease and CRC patients. These studies have highlighted the essential roles of bacteria and/or their products in colonic tumorigenesis.
SCFA is known to modulate immune responses in intestine.133 Another bacterial product Peptidoglycan (PTGN) modulates peripheral immune function via a pattern-recognition receptor, oligomerization domain-containing protein-1 (NOD1) and depletion of the microbiota in mice.133 Lower systemic PTGN concentration leads to less ability to kill certain bacterial pathogens. Polysaccharide A, produced by a commensal bacteria, increases local interleukin 10 by inducing Foxp3+ regulatory T-cell and this effect is mediated by Toll like receptor 2 signaling.156, 157 Although recent studies provide insights into the roles of the bacterial products, the molecular mechanisms of the beneficial effects are not fully elucidated yet.
Analysis of the functions that significantly differed between healthy participants and cancer patients revealed a global metabolic shift from predominant utilization of dietary fiber in the tumor-free colon to more host-derived energy sources in CRC.85 They hypothesize that an increased degradation of host glycans might be related to the etiology of CRC. In healthy gut metagenomes, exclusively some fiber-degrading enzymes and fiber-binding domains are enriched, whereas in CRC metagenomes, the microbiota appeares to exploit growth substrates derived from host cells to a much larger extent.85
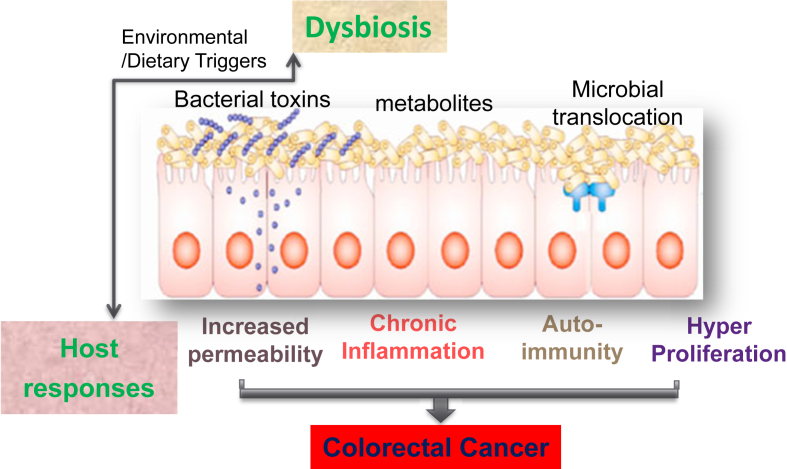
In summary, the general mechanisms for bacteria – associated (or induced) GI tumorigenesis are through enhancing toxic bacterial products, decreasing beneficial bacterial metabolites, disrupted tissue barriers. Abnormal immunity, chronic inflammation, and hyperpreliferation also contribute to the progression of cancer (Fig. 1). Microbial pathogens and intestinal inflammation can compromise intestinal barrier function and result in increased gut permeability, translocation of various microbial substances, and immune activation.158a Dysbiosis further enhances barrier failure and inflammation. The host factor, such as genetic defect, could enhance the dysbiosis along with the environment trigger and change of dietary (Fig.1). One unanswered question is how microbes affect the intestinal epithelium: Do the bacteria make it more permeable or just capitalize on its pre-existing weak spots?
Fig. 1.
Working models of general mechanisms for bacteria – associated (or induced) colon cancer. Through enhancing toxic bacterial products, decreasing beneficial bacterial metabolites, disrupted tissue barriers, translocation of microbes, dysbiosis leads to abnormal immune activation, chronic inflammation, and hyperpreliferation that contribute to the colorectal cancer. The host factor, such as genetic defect, could enhance the dysbiosis along with the environment trigger and change of dietary.
Target gut microbiota in prevention diagnosis, and therapy of GI cancers
Based on current understandings of the roles of microbiota in GI cancer, targeting the gut microbiota is a promising avenue in order to prevent cancer or at least stop the increase of cancerous cells. O'Keefe et al158b investigated the role of fat and fiber in this association by conducting 2-week-long food changes in volunteers from both populations: African-Americans received an African-style diet high in fiber and low in fat, while rural Africans received a high-fat, low-fiber ‘Western’ diet. They found the food changes led to remarkable reciprocal changes in mucosal biomarkers of cancer risk. The dietary switch also changed the microbiota and metabolism in ways known to affect cancer risk.158b This study suggests the potential of dietary intervention or use of prebiotics in colorectal cancer prevention.
Insights into microbiome and cancer risk also provide the opportunities to use of fecal microbial detection for mass screening and diagnosis. By comparing the fecal CRC data to those of IBD patients the researchers could confirm that the microbial characteristics found in the feces were really specific to CRC and not just indicative of inflammatory intestinal conditions in general. The use of fecal microbial CRC detection for mass screening will depend on the development of procedures that are more cost-effective than the ones we used for research purposes.85
The idea of using bacteria as a potent cancer fighting therapy traces its roots back to the early nineteenth century, when French researchers first noticed that bacterial infections in people with cancer often led to shrinkage of their tumors. Increasing evidence has demonstrated that targeting microbiome can improve therapy effects of anti-cancer drugs. Wallace et al reported that inhibiting an enzyme beta-glucuronidase produced by gut microbiota can improve cancer therapy by preventing the intestinal metabolism of the anticancer drug irinotecan.159 More studies have also shown that gut microbes make three anticancer therapies most effective.160a Melanoma growth in mice harboring distinct commensal microbiota and observed differences in spontaneous antitumor immunity, which were eliminated upon cohousing or following fecal transfer. Bifidobacterium is identified to be associated with the antitumor effects. Oral administration of Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy (checkpoint blockade).160b This study also indicates the importance of gut microbiota in other cancers beyond the GI cancer. Although different bacterial groups are implicated in enhancing cancer therapy, the same endpoint through different drugs and different bugs further indicate the novel role of gut microbiome in health and diseases.
Cachexia is a multifactorial condition characterized by systemic inflammation and severe wasting of skeletal muscle, with or without wasting of adipose tissue that causes considerable morbidity and mortality in cancer patients. Infections and inflammation can lead to cachexia and wasting of skeletal muscle and fat tissue by as yet poorly understood mechanisms. Gut colonization by a strain of E. coli prevents wasting triggered by infections or physical damage to the intestine.161 During intestinal infection with Salmonella Typhimurium or pneumonic infection with Burkholderia thailandensis, the presence of this E. coli did not alter changes in host metabolism, caloric uptake, or inflammation but instead sustained signaling of the insulin-like growth factor 1/phosphatidylinositol 3-kinase/AKT pathway in skeletal muscle, which is required for prevention of muscle wasting. This effect was dependent on engagement of the NLRC4 inflammasome.161 Therefore, commensal bacteria in gut promote tolerance to diverse diseases.
Compromised gut barrier function because of dysbiosis or intestinal inflammation can lead to translocation of microbial substances and the development of systemic inflammation with potential consequences for patients prone to cachexia. A recent study showed that non digestible oligosaccharides modulate the gut microbiota may constitute a new nutritional strategy to modulate gut microbiota with positive consequences on cancer progression and associated cachexia.162 Research is needed to clarify the role of gut microbiota and systemic inflammation in the cause of cancer cachexia. Efforts to preserve the integrity of the gut epithelial barrier and/or limit intestinal inflammation in cancer patients may help avoid the serious metabolic alterations associated with cachexia. Multimodal treatment strategies that include interventions aimed at maintaining gut barrier function and correcting dysbiosis may be used to in controlling cachexia.
Microbiota-based cancer prevention, diagnosis, and therapy are beginning to emerge as researchers learn to ‘decode’ the meaning of human microbiota composition at different stages in cancer.
Conclusion and future direction
Growing evidence suggests that human microbiota play novel roles in the progression of colon cancer. The advance of current experimental models and methods allow us to obtain the scientific understanding of the interplay between the gut microbiota, barrier function, and host responses. These insights will leads to uncover new therapeutic targets in cancer. Despite these gains, many challenges lie ahead that make it difficult to close the gap between the basic sciences and clinical application.
We believe the following steps are needed in order to move the current microbiota research into clinical practice. First, we need focus on gaining mechanistic insights. Microbiota functions will be important to be considered. We already generated huge information from microbiota analyses. Based on the genomic analyses, we need analyze the microbiota of individuals. Second, we need simple and low-cost tools to identify key bacteria in patients with colon cancer. For GI patients who will undergo therapy – surgery, chemotherapy – we should follow-up of these bacteria and try to understand why some of those will have very good response to therapy and some others will not. Last, identification of components of the microbiota and elucidation of the mechanisms of their action in inducing pathological changes or exerting beneficial, disease-protective activities could aid in our ability to influence the composition of the microbiota. Understanding gut microbiota in cancer will open a door for the prevention, diagnosis and therapy.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
We would like to acknowledge the NIDDK grant R01 DK105118 (JS) and the UIC Cancer Center.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.gendis.2016.03.004.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Howlader N., Noone A.M., Krapcho M. 2015. SEER Cancer Statistics Review, 1975–2012. 2014; based on November 2014 SEER Data Submission, Posted to the SEER Web Site:http://seer.cancer.gov/csr/1975_2011/ Accessed 1.07.15. [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Tjalsma H., Boleij A., Marchesi J.R., Dutilh B.E. A bacterial driver–passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 4.Joossens M., Huys G., Cnockaert M. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 5.Marchesi J.R., Dutilh B.E., Hall N. Towards the human colorectal cancer microbiome. PLoS One. 2011;6:e20447. doi: 10.1371/journal.pone.0020447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albenberg L., Esipova T.V., Judge C.P. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147 doi: 10.1053/j.gastro.2014.07.020. 1055–1063.e1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mira-Pascual L., Cabrera-Rubio R., Ocon S. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2015;50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 8.Sobhani I., Tap J., Roudot-Thoraval F. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T., Cai G., Qiu Y. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu N., Yang X., Zhang R. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 11.Geng J., Fan H., Tang X., Zhai H., Zhang Z. Diversified pattern of the human colorectal cancer microbiome. Gut Pathog. 2013;5:1–5. doi: 10.1186/1757-4749-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geng J., Song Q., Tang X. Co-occurrence of driver and passenger bacteria in human colorectal cancer. Gut Pathog. 2014;6:1–5. doi: 10.1186/1757-4749-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn J., Sinha R., Pei Z. Human gut microbiome and risk for colorectal cancer. J Nat Cancer Inst. 2013;105:1907–1911. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W., Liu F., Ling Z., Tong X., Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weir T.L., Manter D.K., Sheflin A.M., Barnett B.A., Heuberger A.L., Ryan E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8:e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kostic A.D., Gevers D., Pedamallu C.S. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moellering R.C., Jr., Watson B.K., Kunz L.J. Endocarditis due to group D streptococci. Comparison of disease caused by Streptococcus bovis with that produced by the enterococci. Am J Med. 1974;57:239–250. doi: 10.1016/0002-9343(74)90448-3. [DOI] [PubMed] [Google Scholar]
- 18.Corredoira J., Alonso M.P., Coira A. Characteristics of Streptococcus bovis endocarditis and its differences with Streptococcus viridans endocarditis. Eur J Clin Microbiol Infect Dis. 2008;27:285–291. doi: 10.1007/s10096-007-0441-y. [DOI] [PubMed] [Google Scholar]
- 19.Corredoira J., Alonso M.P., Coira A., Varela J. Association between Streptococcus infantarius (formerly S. bovis II/1) bacteremia and noncolonic cancer. J Clin Microbiol. 2008;46:1570. doi: 10.1128/JCM.00129-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlegel L., Grimont F., Collins M.D., Régnault B., Grimont P.A., Bouvet A. Streptococcus infantarius sp. nov., Streptococcus infantarius subsp. infantarius subsp. nov. and Streptococcus infantarius subsp. coli subsp. nov., isolated from humans and food. Int J Syst Evol Microbiol. 2000;50:1425–1434. doi: 10.1099/00207713-50-4-1425. [DOI] [PubMed] [Google Scholar]
- 21.Ribeiro M.L., Godoy A.P.O., Benvengo Y.H.B., Ecclissato C.C., Mendonça S., Pedrazzoli J., Jr. The influence of endoscopic procedures upon the contamination of Helicobacter pylori cultures. Arq Gastroenterol. 2004;41:100–103. doi: 10.1590/s0004-28032004000200006. [DOI] [PubMed] [Google Scholar]
- 22.Randazzo C.L., Vaughan E.E., Caggia C. Artisanal and experimental Pecorino Siciliano cheese: microbial dynamics during manufacture assessed by culturing and PCR–DGGE analyses. Int J Food Microbiol. 2006;109:1–8. doi: 10.1016/j.ijfoodmicro.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Knudtson L.M., Hartman P.A. Comparison of fluorescent gentamicin-thallous-carbonate and KF streptococcal agars to enumerate enterococci and fecal streptococci in meats. Appl Environ Microbiol. 1993;59:936–938. doi: 10.1128/aem.59.3.936-938.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thian T.S., Hartman P.A. Gentamicin-thallous-carbonate medium for isolation of fecal streptococci from foods. Appl Environ Microbiol. 1981;41:724–728. doi: 10.1128/aem.41.3.724-728.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMahon A.J., Auld C.D., Dale B.A.S., Walls A.D.F., McCormick J.S. Streptococcus bovis septicaemia associated with uncomplicated colonic carcinoma. Br J Surg. 1991;78:883–885. doi: 10.1002/bjs.1800780734. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen S.D., Christensen J.J., Laerkeborg A., Haunso S., Knudsen J.D. Molecular-biological methods of diagnosing colon-related Streptococcus bovis endocarditis. Ugeskr Laeger. 2007;169:610–611. [PubMed] [Google Scholar]
- 27.Wentling G.K., Metzger P.P., Dozois E.J., Chua H.K., Krishna M. Unusual bacterial infections and colorectal carcinoma–Streptococcus bovis and Clostridium septicum: report of three cases. Dis Colon Rectum. 2006;49:1223–1227. doi: 10.1007/s10350-006-0576-4. [DOI] [PubMed] [Google Scholar]
- 28.Gupta A., Madani R., Mukhtar H. Streptococcus bovis endocarditis, a silent sign for colonic tumour. Colorectal Dis. 2010;12:164–171. doi: 10.1111/j.1463-1318.2009.01814.x. [DOI] [PubMed] [Google Scholar]
- 29.Kahveci A., Ari E., Arikan H., Koc M., Tuglular S., Ozener C. Streptococcus bovis bacteremia related to colon adenoma in a chronic hemodialysis patient. Hemodial Int. 2010;14:91–93. doi: 10.1111/j.1542-4758.2009.00400.x. [DOI] [PubMed] [Google Scholar]
- 30.Kim S.Y., Joo S.I., Yi J., Kim E.C. A case of Streptococcus gallolyticus subsp. gallolyticus infective endocarditis with colon cancer: identification by 16S ribosomal DNA sequencing. Korean J Lab Med. 2010;30:160–165. doi: 10.3343/kjlm.2010.30.2.160. [DOI] [PubMed] [Google Scholar]
- 31.Murray H.W., Roberts R.B. Streptococcus bovis bacteremia and underlying gastrointestinal disease. Arch Intern Med. 1978;138:1097–1099. [PubMed] [Google Scholar]
- 32.Klein R.S., Catalano M.T., Edberg S.C., Casey J.I., Steigbigel N.H. Streptococcus bovis septicemia and carcinoma of the colon. Ann Intern Med. 1979;91:560–562. doi: 10.7326/0003-4819-91-4-560. [DOI] [PubMed] [Google Scholar]
- 33.Reynolds J.G., Silva E., McCormack W.M. Association of Streptococcus bovis bacteremia with bowel disease. J Clin Microbiol. 1983;17:696–697. doi: 10.1128/jcm.17.4.696-697.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pigrau C., Lorente A., Pahissa A., Martinez-Vazquez J.M. Streptococcus bovis bacteremia and digestive system neoplasms. Scand J Infect Dis. 1988;20:459–460. doi: 10.3109/00365548809032490. [DOI] [PubMed] [Google Scholar]
- 35.Ruoff K.L., Miller S.I., Garner C.V., Ferraro M.J., Calderwood S.B. Bacteremia with Streptococcus bovis and Streptococcus salivarius: clinical correlates of more accurate identification of isolates. J Clin Microbiol. 1989;27:305–308. doi: 10.1128/jcm.27.2.305-308.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarridge J.E., Attorri S.M., Zhang Q., Bartell J. 16S Ribosomal DNA sequence analysis distinguishes biotypes of Streptococcus bovis: Streptococcus bovis biotype II/2 is a separate genospecies and the predominant clinical isolate in adult males. J Clin Microbiol. 2001;39:1549–1552. doi: 10.1128/JCM.39.4.1549-1552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzlez-Quintela A., Martínez-Rey C., Castroagudín J.F., Rajo-Iglesias M.C., Domínguez-Santalla M.J. Prevalence of liver disease in patients with Streptococcus bovis bacteraemia. J Infect. 2001;42:116–119. doi: 10.1053/jinf.2001.0799. [DOI] [PubMed] [Google Scholar]
- 38.Gold J.S., Bayar S., Salem R.R. Association of Streptococcus bovis bacteremia with colonic neoplasia and extracolonic malignancy. Arch Surg. 2004;139:760–765. doi: 10.1001/archsurg.139.7.760. [DOI] [PubMed] [Google Scholar]
- 39.Lee R.A., Woo P.C.Y., To A.P.C., Lau S.K.P., Wong S.S.Y., Yuen K.-Y. Geographical difference of disease association in Streptococcus bovis bacteraemia. J Med Microbiol. 2003;52:903–908. doi: 10.1099/jmm.0.05199-0. [DOI] [PubMed] [Google Scholar]
- 40.Zarkin B.A., Lillemoe K.D., Cameron J.L., Effron P.N., Magnuson T.H., Pitt H.A. The triad of Streptococcus bovis bacteremia, colonic pathology, and liver disease. Ann Surg. 1990;211:786–792. doi: 10.1097/00000658-199006000-00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jean S.S., Teng L.J., Hsueh P.R., Ho S.W., Luh K.T. Bacteremic Streptococcus bovis infections at a university hospital, 1992–2001. J Formos Med Assoc. 2004;103:118–123. [PubMed] [Google Scholar]
- 42.Corredoira J.C., Alonso M.P., Garcia J.F. Clinical characteristics and significance of Streptococcus salivarius bacteremia and Streptococcus bovis bacteremia: a prospective 16-year study. Eur J Clin Microbiol Infect Dis. 2005;24:250–255. doi: 10.1007/s10096-005-1314-x. [DOI] [PubMed] [Google Scholar]
- 43.Alazmi W., Bustamante M., O'Loughlin C., Gonzalez J., Raskin J.B. The association of Streptococcus bovis bacteremia and gastrointestinal diseases: a retrospective analysis. Dig Dis Sci. 2006;51:732–736. doi: 10.1007/s10620-006-3199-7. [DOI] [PubMed] [Google Scholar]
- 44.Giannitsioti E., Chirouze C., Bouvet A. Characteristics and regional variations of group D streptococcal endocarditis in France. Clin Microbiol Infect. 2007;13:770–776. doi: 10.1111/j.1469-0691.2007.01753.x. [DOI] [PubMed] [Google Scholar]
- 45.Beck M., Frodl R., Funke G. Comprehensive study of strains previously designated Streptococcus bovis consecutively isolated from human blood cultures and emended description of Streptococcus gallolyticus and Streptococcus infantarius subsp. coli. J Clin Microbiol. 2008;46:2966–2972. doi: 10.1128/JCM.00078-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boleij A., Schaeps R.M.J., Tjalsma H. Association between Streptococcus bovis and colon cancer. J Clin Microbiol. 2009;47:516. doi: 10.1128/JCM.01755-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corredoira-Sánchez J., García-Garrote F., Rabuñal R. Association between bacteremia due to Streptococcus gallolyticus subsp. gallolyticus (Streptococcus bovis I) and colorectal neoplasia: a case-control study. Clin Infect Dis. 2012;55:491–496. doi: 10.1093/cid/cis434. [DOI] [PubMed] [Google Scholar]
- 48.Sharara A.I., Abou Hamdan T., Malli A. Association of Streptococcus bovis endocarditis and advanced colorectal neoplasia: a case–control study. J Dig Dis. 2013;14:382–387. doi: 10.1111/1751-2980.12059. [DOI] [PubMed] [Google Scholar]
- 49.Burns C.A., McCaughey R., Lauter C.B. The association of Streptococcus bovis fecal carriage and colon neoplasia: possible relationship with polyps and their premalignant potential. Am J Gastroenterol. 1985;80:42–46. [PubMed] [Google Scholar]
- 50.Klein R.S., Recco R.A., Catalano M.T., Edberg S.C., Casey J.I., Steigbigel N.H. Association of Streptococcus bovis with carcinoma of the colon. N Engl J Med. 1977;297:800–802. doi: 10.1056/NEJM197710132971503. [DOI] [PubMed] [Google Scholar]
- 51.Darjee R., Gibb A.P. Serological investigation into the association between Streptococcus bovis and colonic cancer. J Clin Pathol. 1993;46:1116–1119. doi: 10.1136/jcp.46.12.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dubrow R., Edberg S., Wikfors E. Fecal carriage of Streptococcus bovis and colorectal adenomas. Gastroenterology. 1991;101:721–725. doi: 10.1016/0016-5085(91)90531-o. [DOI] [PubMed] [Google Scholar]
- 53.Potter M.A., Cunliffe N.A., Smith M., Miles R.S., Flapan A.D., Dunlop M.G. A prospective controlled study of the association of Streptococcus bovis with colorectal carcinoma. J Clin Pathol. 1998;51:473–474. doi: 10.1136/jcp.51.6.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teitelbaum J.E., Triantafyllopoulou M. Inflammatory bowel disease and Streptococcus bovis. Dig Dis Sci. 2006;51:1439–1442. doi: 10.1007/s10620-005-9053-5. [DOI] [PubMed] [Google Scholar]
- 55.Tjalsma H., Schöller-Guinard M., Lasonder E., Ruers T.J., Willems H.L., Swinkels D.W. Profiling the humoral immune response in colon cancer patients: diagnostic antigens from Streptococcus bovis. Int J Cancer. 2006;119:2127–2135. doi: 10.1002/ijc.22116. [DOI] [PubMed] [Google Scholar]
- 56.Abdulamir A.S., Hafidh R.R., Mahdi L.K., Al-jeboori T., Abubaker F. Investigation into the controversial association of Streptococcus gallolyticus with colorectal cancer and adenoma. BMC Cancer. 2009;9:1–12. doi: 10.1186/1471-2407-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boleij A., Roelofs R., Schaeps R.M. Increased exposure to bacterial antigen RpL7/L12 in early stage colorectal cancer patients. Cancer. 2010;116:4014–4022. doi: 10.1002/cncr.25212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boleij A., Roelofs R., Danne C. Selective antibody response to Streptococcus gallolyticus pilus proteins in colorectal cancer patients. Cancer Prev Res (Phila) 2012;5:260–265. doi: 10.1158/1940-6207.CAPR-11-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Butt J., Romero-Hernández B., Pérez-Gómez B. Association of Streptococcus gallolyticus subspecies gallolyticus with colorectal cancer: serological evidence. Int J Cancer. 2016;138:1670–1679. doi: 10.1002/ijc.29914. [DOI] [PubMed] [Google Scholar]
- 60.IARC . Vol 61. 1994. Schistosomes, Liver Flukes and Helicobacter pylori. (IARC monographs on the evaluation of carcinogenic risks to humans). Lyon. [PMC free article] [PubMed] [Google Scholar]
- 61.Parkin D.M. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118:3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 62.Suerbaum S., Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 63.Correa P., Haenszel W., Cuello C. Gastric precancerous process in a high risk population: cohort follow-up. Cancer Res. 1990;50:4737–4740. [PubMed] [Google Scholar]
- 64.Konturek P.C., Konturek S.J., Brzozowski T. Helicobacter pylori infection in gastric cancerogenesis. J Physiol Pharmacol. 2009;60:3–21. [PubMed] [Google Scholar]
- 65.Wu Q., Yang Z.P., Xu P., Gao L.C., Fan D.M. Association between Helicobacter pylori infection and the risk of colorectal neoplasia: a systematic review and meta-analysis. Colorectal Dis. 2013;15:e352–e364. doi: 10.1111/codi.12284. [DOI] [PubMed] [Google Scholar]
- 66.Limburg P.J., Stolzenberg-Solomon R.Z., Colbert L.H. Helicobacter pylori seropositivity and colorectal cancer risk: a prospective study of male smokers. Cancer Epidemiol Biomarkers Prev. 2002;11:1095–1099. [PubMed] [Google Scholar]
- 67.Thorburn C.M., Friedman G.D., Dickinson C.J., Vogelman J.H., Orentreich N., Parsonnet J. Gastrin and colorectal cancer: a prospective study. Gastroenterology. 1998;115:275–280. doi: 10.1016/s0016-5085(98)70193-3. [DOI] [PubMed] [Google Scholar]
- 68.Jones M., Helliwell P., Pritchard C., Tharakan J., Mathew J. Helicobacter pylori in colorectal neoplasms: is there an aetiological relationship? World J Surg Oncol. 2007;5 doi: 10.1186/1477-7819-5-51. 51–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng H., Zhang T., Gu W. The presence of Helicobacter pylori in colorectal polyps detected by immunohistochemical methods in children. Pediatr Infect Dis J. 2012;31:364–367. doi: 10.1097/INF.0b013e3182467538. [DOI] [PubMed] [Google Scholar]
- 70.Burnett-Hartman A.N., Newcomb P.A., Potter J.D. Infectious agents and colorectal cancer: a review of Helicobacter pylori, Streptococcus bovis, JC virus, and human papillomavirus. Cancer Epidemiol Biomarkers Prev. 2008;17:2970–2979. doi: 10.1158/1055-9965.EPI-08-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buc E., Dubois D., Sauvanet P. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS One. 2013;8:e56964. doi: 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin H.M., Campbell B.J., Hart C.A. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 73.Swidsinski A., Khilkin M., Kerjaschki D. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–286. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 74.Maddocks O.D.K., Short A.J., Donnenberg M.S., Bader S., Harrison D.J. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4:e5517. doi: 10.1371/journal.pone.0005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arthur J.C., Perez-Chanona E., Mühlbauer M. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonnet M., Buc E., Sauvanet P. Colonization of the human gut by E. coli and colorectal cancer risk. Clin Cancer Res. 2014;20:859–867. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 77.Cougnoux A., Dalmasso G., Martinez R. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932–1942. doi: 10.1136/gutjnl-2013-305257. [DOI] [PubMed] [Google Scholar]
- 78.Sears C.L. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22:349–369. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goodwin A.C., Shields C.E.D., Wu S. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu S., Rhee K.-J., Albesiano E. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ulger Toprak N., Yagci A., Gulluoglu B.M. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 82.Boleij A., Hechenbleikner E.M., Goodwin A.C. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allen-Vercoe E., Jobin C. Fusobacterium and enterobacteriaceae: important players for CRC? Immun Lett. 2014;162(2, Part A):54–61. doi: 10.1016/j.imlet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Han Y.W. Fusobacterium nucleatum: a commensal-turned pathogen. Curr Opin Microbiol. 2015;23:141–147. doi: 10.1016/j.mib.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zeller G., Tap J., Voigt A.Y. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10 doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Castellarin M., Warren R.L., Freeman J.D. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Warren R.L., Freeman D.J., Pleasance S. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:1–12. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dejea C.M., Wick E.C., Hechenbleikner E.M. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc Natl Acad Sci. 2014;111:18321–18326. doi: 10.1073/pnas.1406199111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tahara T., Yamamoto E., Suzuki H. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCoy A.N., Araújo-Pérez F., Azcárate-Peril A., Yeh J.J., Sandler R.S., Keku T.O. Fusobacterium is associated with colorectal adenomas. PLoS One. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ito M., Kanno S., Nosho K. Association of Fusobacterium nucleatum with clinical and molecular features in colorectal serrated pathway. Int J Cancer. 2015;137:1258–1268. doi: 10.1002/ijc.29488. [DOI] [PubMed] [Google Scholar]
- 92.Zackular J.P., Rogers M.A.M., Ruffin M.T., Schloss P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev Res (Phila) 2014;7:1112–1121. doi: 10.1158/1940-6207.CAPR-14-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Allen-Vercoe E., Strauss J., Chadee K. Fusobacterium nucleatum. Gut Microbes. 2011;2:294–298. doi: 10.4161/gmic.2.5.18603. [DOI] [PubMed] [Google Scholar]
- 94.(a) Majowicz S.E., Musto J., Scallan E. The global burden of nontyphoidal salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]; (b) Crum-Cianflone N.F. Salmonellosis and the gastrointestinal tract: more than just peanut butter. Curr Gastroenterol Rep. 2008;10:424–431. doi: 10.1007/s11894-008-0079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Keithlin J., Sargeant J.M., Thomas M.K., Fazil A. Systematic review and meta-analysis of the proportion of non-typhoidal Salmonella cases that develop chronic sequelae. Epidemiol Infect. 2015;143:1333–1351. doi: 10.1017/S0950268814002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagaraja V., Eslick G.D. Systematic review with meta-analysis: the relationship between chronic Salmonella typhi carrier status and gall-bladder cancer. Aliment Pharmacol Ther. 2014;39:745–750. doi: 10.1111/apt.12655. [DOI] [PubMed] [Google Scholar]
- 97.Ternhag A., Törner A., Svensson Å., Ekdahl K., Giesecke J. Short- and long-term effects of bacterial gastrointestinal infections. Emerg Infect Dis. 2008;14:143–148. doi: 10.3201/eid1401.070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jess T., Simonsen J., Nielsen N.M. Enteric salmonella or campylobacter infections and the risk of inflammatory bowel disease. Gut. 2011;60:318–324. doi: 10.1136/gut.2010.223396. [DOI] [PubMed] [Google Scholar]
- 99.Kato I., Boleij A., Kortman G.A. Partial associations of dietary iron, smoking and intestinal bacteria with colorectal cancer risk. Nutr Cancer. 2013;65:169–177. doi: 10.1080/01635581.2013.748922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huycke M.M., Gaskins H.R. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med. 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- 101.Balamurugan R., Rajendiran E., George S., Samuel G.V., Ramakrishna B.S. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23(8 pt 1):1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- 102.Gueimonde M., Ouwehand A., Huhtinen H., Salminen E., Salminen S. Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J Gastroenterol. 2007;13:3985–3989. doi: 10.3748/wjg.v13.i29.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fite A., Macfarlane G.T., Cummings J.H. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut. 2004;53:523–529. doi: 10.1136/gut.2003.031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fite A., Macfarlane S., Furrie E. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J Clin Microbiol. 2013;51:849–856. doi: 10.1128/JCM.02574-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahn J., Segers S., Hayes R.B. Periodontal disease, Porphyromonas gingivalis serum antibody levels and orodigestive cancer mortality. Carcinogenesis. 2012;33:1055–1058. doi: 10.1093/carcin/bgs112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Turnbaugh P.J., Hamady M., Yatsunenko T. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cotillard A., Kennedy S.P., Kong L.C. Dietary intervention impact on gut microbial gene richness. Nature. 2013;500:585–588. doi: 10.1038/nature12480. [DOI] [PubMed] [Google Scholar]
- 108.Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 109.Duca F.A., Sakar Y., Lepage P. Replication of obesity and associated signaling pathways through transfer of microbiota from obese-prone rats. Diabetes. 2014;63:1624–1636. doi: 10.2337/db13-1526. [DOI] [PubMed] [Google Scholar]
- 110.WCRF/AICR Continuous update project report: food, nutrition, physical activity, and the prevention of colorectal cancer. Colorectal Cancer 2011 Rep Lond WCRF/AICR. 2011:1–40. [Google Scholar]
- 111.Maukonen J., Saarela M. Human gut microbiota: does diet matter? Proc Nutr Soc. 2015;74:23–36. doi: 10.1017/S0029665114000688. [DOI] [PubMed] [Google Scholar]
- 112.David L.A., Maurice C.F., Carmody R.N. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim M.-S., Hwang S.-S., Park E.-J., Bae J.-W. Strict vegetarian diet improves the risk factors associated with metabolic diseases by modulating gut microbiota and reducing intestinal inflammation. Environ Microbiol Rep. 2013;5:765–775. doi: 10.1111/1758-2229.12079. [DOI] [PubMed] [Google Scholar]
- 114.Clarke S.F., Murphy E.F., O'Sullivan O. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- 115.Mutlu E.A., Gillevet P.M., Rangwala H. Colonic microbiome is altered in alcoholism. Am J Physiol – Gastrointest Liver Physiol. 2012;302:G966–G978. doi: 10.1152/ajpgi.00380.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tuomisto S., Pessi T., Collin P., Vuento R., Aittoniemi J., Karhunen P.J. Changes in gut bacterial populations and their translocation into liver and ascites in alcoholic liver cirrhotics. BMC Gastroenterol. 2014;14:1–8. doi: 10.1186/1471-230X-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Biedermann L., Zeitz J., Mwinyi J. Smoking cessation induces profound changes in the composition of the intestinal microbiota in humans. PLoS One. 2013;8:e59260. doi: 10.1371/journal.pone.0059260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Biedermann L., Brulisauer K., Zeitz J. Smoking cessation alters intestinal microbiota: insights from quantitative investigations on human fecal samples using FISH. Inflamm Bowel Dis. 2014;20:1496–1501. doi: 10.1097/MIB.0000000000000129. [DOI] [PubMed] [Google Scholar]
- 119.Kato I., Nechvatal J.M., Dzinic S., Basson M.D., Majumdar A.P., Ram J.L. Smoking and other personal characteristics as potential predictors for fecal bacteria populations in humans. Med Sci Monit. 2010;16:CR1-7. [PMC free article] [PubMed] [Google Scholar]
- 120.de Martel C., Ferlay J., Franceschi S. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 121.Borges-Canha M., Portela-Cidade J.P., Dinis-Ribeiro M., Leite-Moreira A.F., Pimentel-Nunes P. Role of colonic microbiota in colorectal carcinogenesis: a systematic review. Rev Esp Enferm Dig. 2015;107 doi: 10.17235/reed.2015.3830/2015. [DOI] [PubMed] [Google Scholar]
- 122.Tlaskalova-Hogenova H., Stepankova R., Kozakova H. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8:110–120. doi: 10.1038/cmi.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eckburg P.B., Bik E.M., Bernstein C.N. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Damman C.J., Miller S.I., Surawicz C.M., Zisman T.L. The microbiome and inflammatory bowel disease: is there a therapeutic role for fecal microbiota transplantation. Am J Gastroenterol. 2012;107:1452–1459. doi: 10.1038/ajg.2012.93. [DOI] [PubMed] [Google Scholar]
- 125.Sokol H., Pigneur B., Watterlot L. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Veerappan G.R., Betteridge J., Young P.E. Probiotics for the treatment of inflammatory bowel disease. Curr Gastroenterol Rep. 2012;14:324–333. doi: 10.1007/s11894-012-0265-5. [DOI] [PubMed] [Google Scholar]
- 127.Tirandaz H., Mohammadi E. Efficient tumor targeting by anaerobic butyrate-producing bacteria. Med Hypotheses. 2013;80:675–678. doi: 10.1016/j.mehy.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 128.Belenguer A., Holtrop G., Duncan S.H. Rates of production and utilization of lactate by microbial communities from the human colon. FEMS Microbiol Ecol. 2011;77:107–119. doi: 10.1111/j.1574-6941.2011.01086.x. [DOI] [PubMed] [Google Scholar]
- 129.Suzuki T., Yoshida S., Hara H. Physiological concentrations of short-chain fatty acids immediately suppress colonic epithelial permeability. Br J Nutr. 2008;100:297–305. doi: 10.1017/S0007114508888733. [DOI] [PubMed] [Google Scholar]
- 130.Hu S., Dong T.S., Dalal S.R. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS One. 2011;6:e16221. doi: 10.1371/journal.pone.0016221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Winter J., Nyskohus L., Young G.P. Inhibition by resistant starch of red meat-induced promutagenic adducts in mouse colon. Cancer Prev Res (Phila) 2011;4:1920–1928. doi: 10.1158/1940-6207.CAPR-11-0176. [DOI] [PubMed] [Google Scholar]
- 132.Worthley D.L., Whitehall V.L., Le Leu R.K. DNA methylation in the rectal mucosa is associated with crypt proliferation and fecal short-chain fatty acids. Dig Dis Sci. 2011;56:387–396. doi: 10.1007/s10620-010-1312-4. [DOI] [PubMed] [Google Scholar]
- 133.Maslowski K.M., Vieira A.T., Ng A. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rosignoli P., Fabiani R., De Bartolomeo A. Protective activity of butyrate on hydrogen peroxide-induced DNA damage in isolated human colonocytes and HT29 tumour cells. Carcinogenesis. 2001;22:1675–1680. doi: 10.1093/carcin/22.10.1675. [DOI] [PubMed] [Google Scholar]
- 135.Moore W.E., Moore L.H. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Scanlan P.D., Shanahan F., Marchesi J.R. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol. 2009;69:213–221. doi: 10.1111/j.1574-6941.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 137.Mantovani A. Cancer: inflaming metastasis. Nature. 2009;457:36–37. doi: 10.1038/457036b. [DOI] [PubMed] [Google Scholar]
- 138.Song M., Wu K., Ogino S., Fuchs C.S., Giovannucci E.L., Chan A.T. A prospective study of plasma inflammatory markers and risk of colorectal cancer in men. Br J Cancer. 2013;108:1891–1898. doi: 10.1038/bjc.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gradel K.O., Nielsen H.L., Schonheyder H.C., Ejlertsen T., Kristensen B., Nielsen H. Increased short- and long-term risk of inflammatory bowel disease after salmonella or campylobacter gastroenteritis. Gastroenterology. 2009;137:495–501. doi: 10.1053/j.gastro.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 140.Kumar S. Infection as a risk factor for gallbladder cancer. J Surg Oncol. 2006;93:633–639. doi: 10.1002/jso.20530. [DOI] [PubMed] [Google Scholar]
- 141.Arrach N., Zhao M., Porwollik S., Hoffman R.M., McClelland M. Salmonella promoters preferentially activated inside tumors. Cancer Res. 2008;68:4827–4832. doi: 10.1158/0008-5472.CAN-08-0552. [DOI] [PubMed] [Google Scholar]
- 142.Zhao M., Yang M., Li X.M. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci U S A. 2005;102:755–760. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Collier-Hyams L.S., Zeng H., Sun J. Cutting edge: Salmonella AvrA effector inhibits the key proinflammatory, anti-apoptotic NF-kappa B pathway. J Immunol. 2002;169:2846–2850. doi: 10.4049/jimmunol.169.6.2846. [DOI] [PubMed] [Google Scholar]
- 144.Du F., Galan J.E. Selective inhibition of type III secretion activated signaling by the Salmonella effector AvrA. PLoS Pathog. 2009;5:e1000595. doi: 10.1371/journal.ppat.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hardt W.D., Galan J.E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci U S A. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Liao A.P., Petrof E.O., Kuppireddi S. Salmonella type III effector AvrA stabilizes cell tight junctions to inhibit inflammation in intestinal epithelial cells. PLoS One. 2008;3:e2369. doi: 10.1371/journal.pone.0002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun J., Hobert M.E., Rao A.S., Neish A.S., Madara J.L. Bacterial activation of beta-catenin signaling in human epithelia. Am J Physiol Gastrointest Liver Physiol. 2004;287:G220–G227. doi: 10.1152/ajpgi.00498.2003. [DOI] [PubMed] [Google Scholar]
- 148.Ye Z., Petrof E.O., Boone D., Claud E.C., Sun J. Salmonella effector AvrA regulation of colonic epithelial cell inflammation by deubiquitination. Am J Pathol. 2007;171:882–892. doi: 10.2353/ajpath.2007.070220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu X., Lu R., Wu S., Sun J. Salmonella regulation of intestinal stem cells through the Wnt/beta-catenin pathway. FEBS Lett. 2010;584:911–916. doi: 10.1016/j.febslet.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lu R., Wu S., Zhang Y. Enteric bacterial protein AvrA promotes colonic tumorigenesis and activates colonic beta-catenin signaling pathway. Oncogenesis. 2014 doi: 10.1038/oncsis.2014.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Belcheva A., Irrazabal T., Robertson S.J. Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell. 2014;158:288–299. doi: 10.1016/j.cell.2014.04.051. [DOI] [PubMed] [Google Scholar]
- 152.Bongers G., Pacer M.E., Geraldino T.H. Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. J Exp Med. 2014;211:457–472. doi: 10.1084/jem.20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Keku T.O., Dulal S., Deveaux A., Jovov B., Han X. The gastrointestinal microbiota and colorectal cancer. Am J Physiol Gastrointest Liver Physiol. 2015;308:G351–G363. doi: 10.1152/ajpgi.00360.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Grivennikov S.I., Wang K., Mucida D. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491:254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Round J.L., Mazmanian S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Nat Acad Sci U S A. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Round J.L., O'Connell R.M., Mazmanian S.K. Coordination of tolerogenic immune responses by the commensal microbiota. J Autoimmun. 2010;34:J220–J225. doi: 10.1016/j.jaut.2009.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.(a) Schwabe R.F., Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) O'Keefe S.J., Li J.V., Lahti L. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wallace B.D., Wang H., Lane K.T. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.(a) Pennisi E. Biomedicine. Cancer therapies use a little help from microbial friends. Science. 2013;342:921. doi: 10.1126/science.342.6161.921. [DOI] [PubMed] [Google Scholar]; (b) Sivan A., Corrales L., Hubert N. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schieber A.M., Lee Y.M., Chang M.W. Disease tolerance mediated by microbiome E. coli involves inflammasome and IGF-1 signaling. Science. 2015;350:558–563. doi: 10.1126/science.aac6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bindels L.B., Neyrinck A.M., Salazar N. Non digestible oligosaccharides modulate the gut microbiota to control the development of leukemia and associated cachexia in mice. PLoS One. 2015;10:e0131009. doi: 10.1371/journal.pone.0131009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.