Different forms of nuclear envelope–spanning LINC complexes have opposing roles in the transcription-independent control of the small GTPase RhoA. Competition between LINC complexes in the nuclear envelope may therefore dictate the outcome of signaling to cytoskeletal networks.
Abstract
Linker of Nucleoskeleton and Cytoskeleton (LINC) complexes span the nuclear envelope and transduce force from dynamic cytoskeletal networks to the nuclear lamina. Here we show that LINC complexes also signal from the nuclear envelope to critical regulators of the actin cytoskeleton. Specifically, we find that LINC complexes that contain the inner nuclear membrane protein Sun2 promote focal adhesion assembly by activating the small GTPase RhoA. A key effector in this process is the transcription factor/coactivator complex composed of SRF/Mkl1. A constitutively active form of SRF/Mkl1 was not sufficient to induce focal adhesion assembly in cells lacking Sun2, however, suggesting that LINC complexes support RhoA activity through a transcription-independent mechanism. Strikingly, we also find that the inner nuclear membrane protein Sun1 antagonizes Sun2 LINC complexes and inhibits RhoA activation and focal adhesion assembly. Thus different LINC complexes have opposing roles in the transcription-independent control of the actin cytoskeleton through the small GTPase RhoA.
INTRODUCTION
A hallmark of eukaryotic cells is the compartmentalization of the genome by a double membrane bilayer called the nuclear envelope. The presence of a membrane barrier requires that cells have specialized mechanisms to facilitate communication between the nuclear interior and surrounding cytoplasm. This is accomplished in part by nuclear pore complexes, which facilitate the specific transport of large (>∼40 kDa) macromolecules through the nuclear envelope (Capelson et al., 2011). In addition, Linker of Nucleoskeleton and Cytoskeleton (LINC) complexes transduce mechanical forces across the nuclear envelope (Isermann and Lammerding, 2013; Chang et al., 2015b). Composed in vertebrates of an outer nuclear membrane (ONM) nesprin protein and an inner nuclear membrane (INM) Sad1, Unc84 (SUN)–domain protein, LINC complexes span the nuclear envelope and directly connect all the major cytoskeletal networks to the nuclear interior (Padmakumar et al., 2005; Crisp et al., 2006; Haque et al., 2006; Stewart-Hutchinson et al., 2008). The primary force-bearing element in the nucleus is the nuclear lamina. Closely associated with the INM, the nuclear lamina is a polymeric network of lamin proteins that provide structural integrity to the nucleus (Shimi et al., 2011; Gruenbaum and Foisner, 2015). Together LINC complexes and the nuclear lamina harness forces generated by dynamic cytoskeletal networks to drive nuclear positioning and chromosome movement during meiosis.
Vertebrate genomes have six different nesprin genes, which are defined by the presence of a C-terminal Klarsicht, ANC-1, and Syne homology (KASH) sequence. SYNE-1 and SYNE-2, which encode nesprin 1 and nesprin 2, respectively, are widely expressed and alternatively spliced to produce several different protein isoforms (Zhang et al., 2005; Rajgor et al., 2012). The largest and most complex isoforms are nesprin1-giant (N1G) and nesprin2-giant (N2G). Both N1G and N2G contain >50 spectrin repeats and an N-terminal actin-binding domain that extend from the ONM into the cytoplasm. The KASH peptide resides in the nuclear envelope lumen, where it binds to a SUN domain–containing INM protein (Sosa et al., 2012; Wang et al., 2012). Five different SUN-domain proteins have been identified in vertebrates, of which only Sun1 and Sun2 are broadly expressed in somatic cells. No specificity has been demonstrated in the association of KASH peptides from different nesprins with Sun1 and Sun2, indicating that several distinct LINC complex types likely coexist within most vertebrate cell types (Stewart-Hutchinson et al., 2008; Sosa et al., 2012).
Genetic analysis in mice revealed that related SUN-domain and nesprins proteins are at least partially redundant (Ding et al., 2007; Zhang et al., 2007b, 2009b; Lei et al., 2009). At the same time, different LINC complexes play specific roles in nuclear positioning. For example, LINC complexes composed of N2G and Sun2 are required for actin-dependent rearward nuclear movement in polarizing fibroblasts (Luxton et al., 2010). In addition, nesprin 1 and nesprin 2 mediate nuclear clustering in skeletal muscle and nuclear migration in the retina and neocortex, respectively, and Sun2 is important to establish nuclear position in skin (Zhang et al., 2007b, 2009a; Yu et al., 2011; Stewart et al., 2015).
LINC complexes also play important and specific roles in controlling actin dynamics during cell migration and cell adhesion (Lammerding et al., 2004; Lee et al., 2007; Hale et al., 2008; Stewart-Hutchinson et al., 2008; Khatau et al., 2009; Luxton et al., 2010; Folker et al., 2011; Kim et al., 2012; Chang et al., 2013, 2015a). How signals are transmitted from the nuclear envelope to the actin cytoskeleton and the identity of the cytoplasmic targets of these signals are not well understood. One possibility is that nesprin proteins in the ONM directly regulate actin dynamics in the cytoplasm. Alternatively, gene expression programs regulated by LINC complexes could indirectly control the actin cytoskeleton. Supporting this possibility are recent data showing that LINC complexes promote the activity of the transcription factor/coactivator complex composed of Serum-Response Factor (SRF) and Megakaryoblastic Leukemia 1 (Mkl1; also known as MRTF-A/Mal; Ho et al., 2014; Plessner and Grosse, 2015; Plessner et al., 2015). SRF/Mkl1 target genes include actin isoforms, myosins, and numerous other actin-binding proteins that contribute to cell migration and contraction (Liu and Olson, 2006; Olson and Nordheim, 2010; Esnault et al., 2014; Rajakylä and Vartiainen, 2014).
Mutations in LINC complexes cause several human diseases, including Emery–Dreifuss muscular dystrophy (EDMD; Zhang et al., 2007a; Horn, 2014; Meinke et al., 2014). EDMD and the related dilated cardiomyopathy (DCM) are also caused by mutations in genes encoding lamin A and the INM protein emerin (Crisp et al., 2006; Haque et al., 2006, 2010; Worman, 2011; Ho et al., 2014). Cells isolated from EDMD and DCM patients and mouse models contain defects in nuclear morphology and nuclear position and in actin cytoskeleton organization (Lammerding et al., 2004; Lee et al., 2007; Hale et al., 2008; Stewart-Hutchinson et al., 2008; Folker et al., 2011). However, although nuclear defects are found in many cell types in vivo, the disease phenotypes manifest most severely in heart and skeletal muscle, which are critically dependent on the assembly of contractile actin structures called myofibrils for their development, function, and homeostasis. Thus, elucidating mechanisms of signaling to the actin cytoskeleton is critical to understanding the role of LINC complexes during development and in disease.
In tissue culture cells, the most prominent actin-based structures are contractile actomyosin bundles called stress fibers. Although rare in vivo, stress fibers resemble in structure and mechanism of assembly nascent myofibrils (Sanger et al., 2010). As a result, stress fibers have been extensively used to identify and interrogate conserved signaling networks that regulate the actin cytoskeleton during cardiovascular development (Jaffe and Hall, 2005; Burridge and Wittchen, 2013; Shimokawa et al., 2016). Here we take advantage of this fact to interrgate the mechanisms of actin cytoskeleton regulation by LINC complexes. We show that a critical target of LINC complex signaling is the small GTPase RhoA. Strikingly, however, we find that Sun1 and Sun2 have opposing roles in regulating RhoA activity. Moreover, our data suggest that whereas changes in transcription are important for LINC complex signaling to RhoA, SRF/Mkl1 is not the primary target in this pathway. On the basis of these data, we propose that competition between different LINC complexes in the nuclear envelope directly affects actin dynamics through regulation of RhoA activity.
RESULTS
Sun1 and Sun2 have opposing roles in regulating the activity of RhoA
Focal adhesions are massive and dynamic protein assemblies that link cytoplasmic actin stress fibers to the extracellular matrix (Gardel et al., 2010). Contractile forces applied by actin stress fibers alter the protein composition of focal adhesions (Pasapera et al., 2010; Kuo et al., 2011). Thus we used changes in focal adhesion composition, which are easily assayed by immunofluorescence, as a simple and quantitative means to determine whether LINC complexes regulate the properties of actin stress fibers. Focal adhesion area and the relative levels of paxillin protein at focal adhesions were unchanged after small interfering RNA (siRNA)–mediated depletion of Sun1 or Sun2 in HeLa cells (Supplemental Figure S1, A and B). In contrast, vinculin recruitment to focal adhesions, which is highly force sensitive (Pasapera et al., 2010; Kuo et al., 2011), was significantly increased after depletion of Sun1 (Figure 1, A and B). The levels of vinculin at focal adhesions were not significantly changed in cells lacking Sun2 alone, but depletion of Sun2 suppressed the increase in vinculin recruitment to focal adhesions that was caused by depletion of Sun1. Expression of mouse Sun1 (mSun1) also suppressed the increase in vinculin recruitment to focal adhesions caused by depletion of endogenous Sun1, demonstrating the specificity of the siRNAs used in this experiment (Supplemental Figure S1, C–E). Thus Sun1 and Sun2 have opposing roles in focal adhesion assembly, which likely reflect a direct role for LINC complexes in establishing the contractile properties of actin stress fibers.
FIGURE 1:
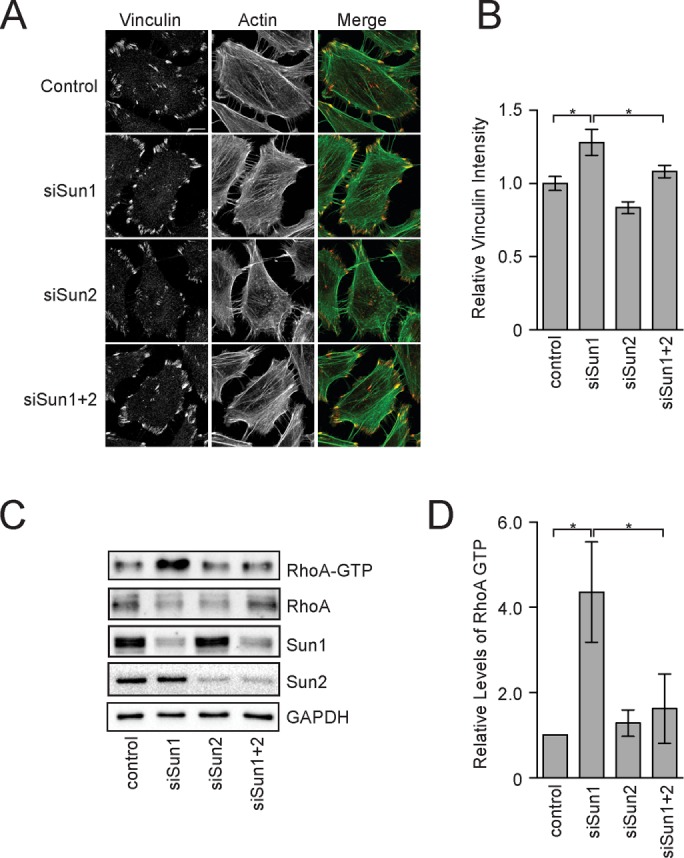
SUN-domain proteins have opposing roles in regulating RhoA. (A) F-actin and vinculin visualized in HeLa cells treated with the indicated siRNAs. (B) Quantitation of vinculin levels at individual focal adhesions in HeLa cells. Data represent mean ± SEM from four independent experiments. (C) Western blot of total and active RhoA in extracts from HeLa cells treated with the indicated siRNAs. (D) Quantitation of Western blots in C. Data represent mean ± SEM from four independent experiments. Statistical significance was determined using one-way analysis of variance (ANOVA). *p < 0.05. Scale bar, 5 μm.
A large family of RhoA GTPases cycle between an active, GTP-bound conformation and an inactive, GDP-bound conformation to control actin dynamics (Cherfils and Zeghouf, 2013; Goicoechea et al., 2015). RhoA activity is specifically rate limiting for stress fiber and focal adhesion assembly in tissue culture cells (Schwartz, 2004; Jaffe and Hall, 2005). We therefore used recombinant glutathione S-transferase–RhoA–binding domain (GST-RBD), which binds to only the GTP-bound form of RhoA, to measure RhoA activity in cell extracts lacking SUN-domain proteins (Ren et al., 1999). Depletion of Sun1 caused a 4.3-fold increase in RhoA activity relative to control cells (Figure 1, C and D). Depletion of Sun2 alone had no affect on RhoA activity, but Sun2 was required to sustain the increase in RhoA activity in cells also lacking Sun1. Together these data suggest that the opposing roles of Sun1 and Sun2 in regulating focal adhesion assembly are transduced through RhoA. Moreover, our data suggest that Sun1 dominates signaling to the actin cytoskeleton in HeLa cells in their basal state. The function of Sun2 in this process, in contrast, only emerges after Sun1 has been depleted with siRNAs.
Sun2 LINC complexes are required for RhoA activation
SUN-domain proteins typically perform their functions in nuclear envelope–spanning LINC complexes. We therefore asked whether nesprins also regulate focal adhesion assembly in HeLa cells. We generated a stable HeLa cell line that expressed from a doxycycline-inducible promoter a dominant-negative form of N2G called SR-KASH, which lacks most of the cytoplasmic domains (Luxton et al., 2010). An additional cell line was also created that expressed the mini-nesprin-2G (mN2G) protein, which contains the N-terminal actin-binding domain from N2G stitched onto the SR-KASH scaffold (Luxton et al., 2010). Neither the SR-KASH nor mN2G proteins were detected in HeLa cells before induction of transgene expression. After doxycycline addition, both nesprin 2 truncations were easily detected and displaced endogenous nesprin 2 from the nuclear envelope (Supplemental Figure S2, A and B). Expression of SR-KASH or mN2G in control HeLa cells did not affect focal adhesion composition as determined by vinculin immunofluorescence (Figure 2, A and B). However, since focal adhesions in cells lacking Sun2 alone or both Sun1 and Sun2 resemble those found in normal HeLa cells (Figure 1, A and B), a change in vinculin levels at focal adhesions would only be expected to occur in this case if the SR-KASH or mN2G protein specifically disrupted Sun1 LINC complex function. We therefore focused our analysis on cells lacking Sun1. Expression of the SR-KASH or mN2G proteins suppressed the increase in vinculin recruitment to focal adhesions that was observed after depletion of Sun1 (Figure 2, A and B). Moreover, the increase in GTP-bound RhoA in cells lacking Sun1 was eliminated by the expression of either the SR-KASH or mN2G protein (Figure 2, C and D). Thus the SR-KASH and mN2G proteins effectively inhibited the ability of Sun2 to sustain RhoA activation and support increased vinculin recruitment to focal adhesions, indicating that Sun2 performs these functions in the context of LINC complexes. Of interest, mN2G was shown to be under tension and sufficient for Sun2 LINC complexes to mediate actin-dependent rearward nuclear movement in polarizing fibroblasts (Luxton et al., 2010; Arsenovic et al., 2016). Thus force transduction across the nuclear envelope and signaling to RhoA by Sun2 LINC complexes have distinct nesprin requirements.
FIGURE 2:
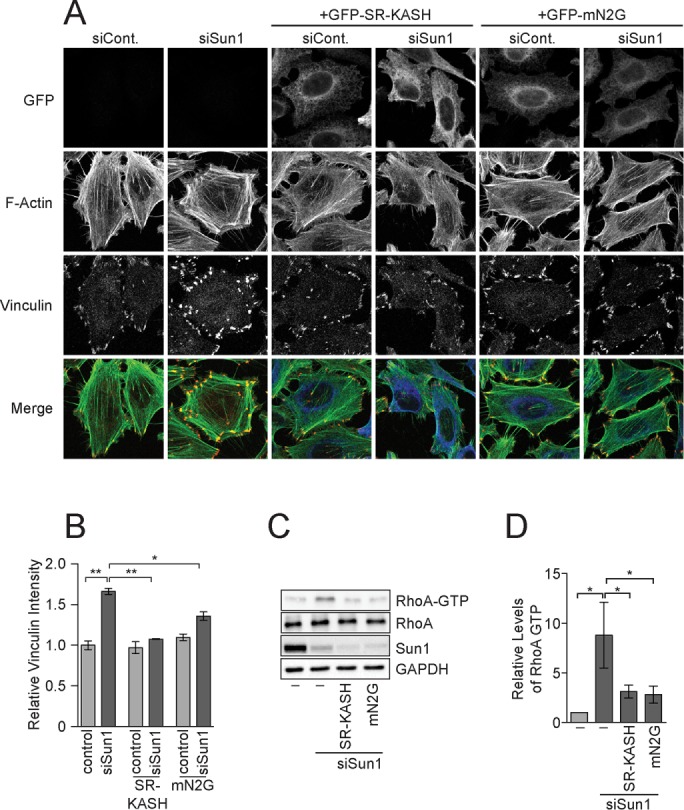
Sun2 LINC complexes promote RhoA activity. (A) F-actin and vinculin visualized in HeLa cells treated with the control or Sun1 siRNAs and expressing the indicated nesprin transgene. (B) Quantitation of vinculin levels at individual focal adhesions in HeLa cells. Data represent mean ± SEM from four independent experiments. (C) Western blot of total and active RhoA in extracts from HeLa cells treated with control or Sun1 siRNAs expressing the indicated nesprin transgene. (D) Quantitation of Western blots in C. Data represent mean ± SEM from four independent experiments. Statistical significance was determined using one-way ANOVA. *p < 0.05; **p < 0.01. Scale bar, 5 μm.
SRF/Mkl1-dependent gene expression is required for RhoA activation by Sun2
We considered two potential mechanisms for how LINC complexes could regulate focal adhesion assembly. First, they could signal directly from the nuclear envelope through the cytoplasm to RhoA. Alternatively, LINC complexes could regulate the transcription of genes that feed back to control RhoA activity. The best-described gene expression program important for focal adhesion assembly is driven by SRF/MKl1 (Small, 2012). We therefore asked whether cells lacking Sun1 have increased levels of SRF/Mkl1 target gene expression. The mRNA levels of several SRF/Mkl1 target genes, including VCL (which encodes vinculin) and SM22, were increased after depletion of Sun1 as determined by quantitative PCR (qPCR; Figure 3A). Sun2 depletion alone did not cause significant changes in VCL or SM22 expression. Depletion of Sun2 did, however, suppress the increase in mRNA levels that was observed in cells lacking Sun1.
FIGURE 3:
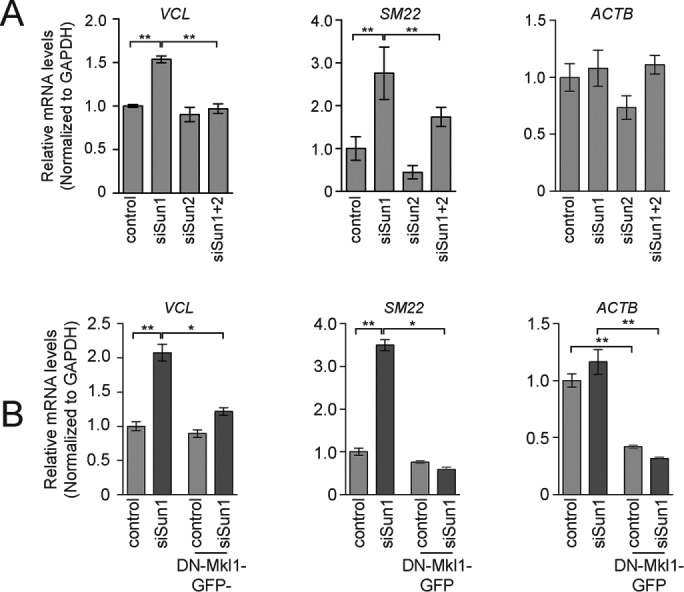
SUN-domain proteins regulate SRF/Mkl1-dependent gene expression. (A) qPCR of Mkl1/SRF target genes using mRNA isolated from HeLa cells treated with the indicated siRNA. Data represent Mean ± SEM from four independent experiments. (B) qPCR of Mkl1/SRF target genes using mRNA isolated from HeLa cells treated with control or Sun1 siRNA in the presence or absence of DN-Mkl1. Data represent mean ± SEM from four independent experiments. Statistical significance was determined using one-way ANOVA. *p < 0.05; **p < 0.01.
The mRNA levels of ACTB, which is also a direct transcriptional target of SRF/Mkl1, were not affected by depletion of Sun1 (Figure 3A), raising the possibility that SRF/Mkl1-independent mechanisms led to the changes in SM22 and VCL expression in cells lacking Sun1. To test this possibility directly, we introduced into HeLa cells under control of an inducible promoter a dominant-negative form of Mkl1 (DN-Mkl1; Cen et al., 2003; Miralles et al., 2003). When expressed, DN-Mkl1 did not significantly reduce VCL or SM22 mRNA levels in control cells (Figure 3B). DN-Mkl1 did, however, reduce the expression of ACTB by 2.2-fold. Thus, established target genes in HeLa cells have varying thresholds of sensitivity to SRF/Mkl1 activity, which likely accounts for the differential response of these genes to Sun1 depletion. Importantly, DN-Mkl1 eliminated the increase in VCL and SM22 expression that was induced after depletion of Sun1 (Figure 3B). Sun1 depletion also led to an increase in vinculin and Sm22 protein levels, which was reversed by expression of DN-Mkl1 (Supplemental Figure S3, A and B). Thus Sun1 inhibits SRF/Mkl1 activity in HeLa cells.
We next determined whether the increased expression of SRF/Mkl1 target genes was essential to stimulate focal adhesion assembly in cells lacking Sun1. As described earlier, vinculin recruitment to focal adhesions was significantly higher in cells lacking Sun1 than it was in control cells. No increase in vinculin at focal adhesions occurred, however, in cells lacking Sun1 after the expression of DN-Mkl1 (Figure 4, A and B). Sun1 therefore limits focal adhesion assembly in part by repressing the expression of SRF/Mkl1 target genes.
FIGURE 4:
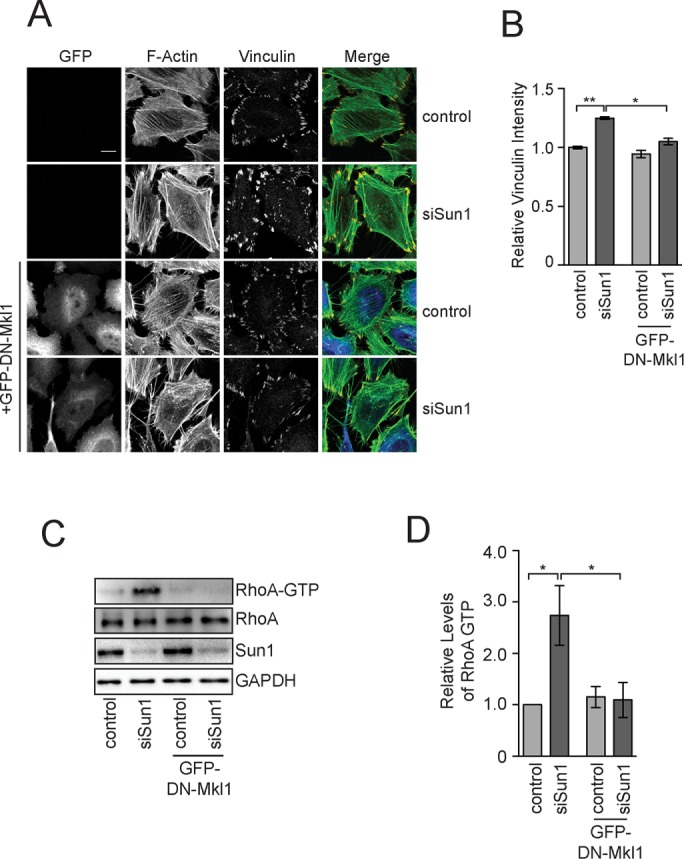
Mkl1 is required for focal adhesion assembly after Sun1 knockdown. (A) F-actin and vinculin visualized in HeLa cells treated with the control or Sun1 siRNAs without or with DN-MKl1. (B) Quantitation of vinculin levels at individual focal adhesions in HeLa cells treated as in A. Data represent mean ± SEM from four independent experiments. (C) Western blot of total and active RhoA in extracts from HeLa cells treated with control or Sun1 siRNAs without or with DN-MKl1. (D) Quantitation of Western blots in C. Data represent mean ± SEM from four independent experiments. Statistical significance was determined using one-way ANOVA. *p < 0.05; **p < 0.01. Scale bar, 5 μm.
RhoA is critical for Mkl1 localization to the nucleus and SRF/Mkl1 activity, and an increase in GTP-bound RhoA is believed to occur before the initiation of SRF/Mkl1-dependent gene expression in most contexts (Hill et al., 1995; Miralles et al., 2003; Vartiainen et al., 2007). Nevertheless, we found that DN-Mkl1 expression also eliminated the increase in GTP-bound RhoA that occurred in cells lacking Sun1 (Figure 4, C and D). One interpretation of these data is that Sun1 in HeLa cells directly imhibits SRF/Mkl1 activity to suppress RhoA activation and focal adhesion assembly. Alternatively, a positive-feedback loop between RhoA in the cytoplasm and SRF/Mkl1 activity in the nucleus could be required for increased vinculin recruitment to focal adhesions. In this case, LINC complexes could target RhoA, SRF/MKl1, or both to regulate actin organization.
Transcription-independent control of stress fiber assembly by LINC complexes
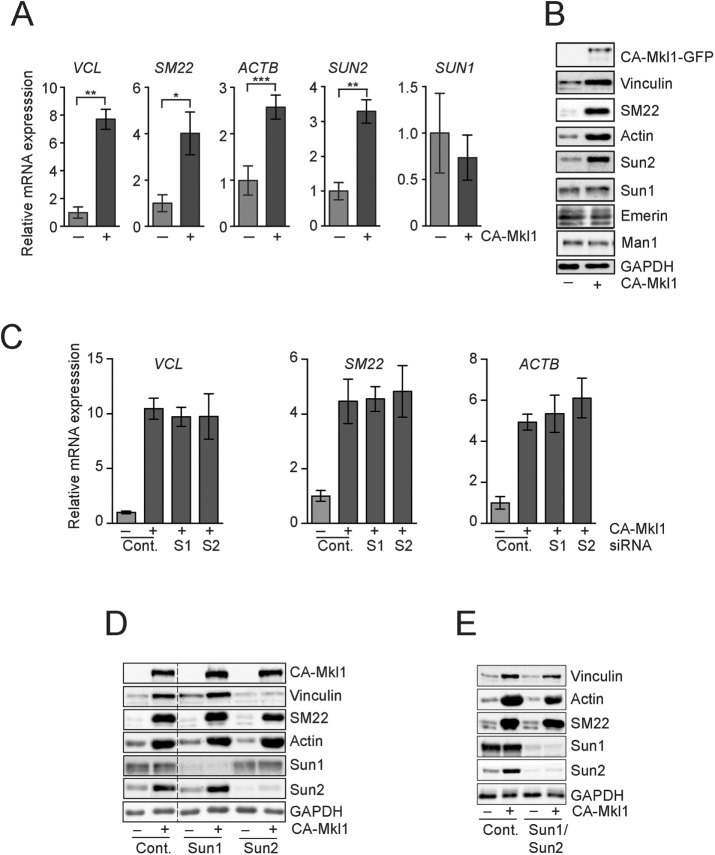
To directly determine whether SRF/Mkl1 is the primary target of LINC complex signaling, we generated a HeLa cell line that expressed a constitutively active Mkl1 (CA-Mkl1) transgene under control of a doxycycline-inducible promoter (Cen et al., 2003; Miralles et al., 2003). Our goal was to create a system in which SRF/Mkl1-dependent gene expression was no longer sensitive to LINC complex function. We began by first characterizing the response of HeLa cells to CA-MKl1 expression. Induction of CA-Mkl1 led to an increase in the mRNA and protein levels of numerous SRF/Mkl1 target genes, including VCL, SM22, and ACTB (Figure 5, A and B). Of interest, qPCR revealed that SUN2, but not SUN1, mRNA levels were also significantly higher after CA-Mkl1 expression, suggesting that SUN2 is a transcriptional target of SRF/Mkl1 (Figure 5A). Moreover, we found that the levels of Sun2 protein were substantially higher in cells expressing CA-Mkl1 than they were in control cells (Figure 5B). Sun1 protein levels, on the other hand, were not affected by CA-Mkl1 expression.
FIGURE 5:
CA-Mkl1 is insensitive to LINC complex function. (A) qPCR of Mkl1/SRF target genes using mRNA isolated from control and CA-Mkl1–expressing HeLa cells. Data represent Mean ± SEM from four independent experiments. (B) Western blot of indicated proteins in control and CA-Mkl1–expressing cells. (C) qPCR of Mkl1/SRF target genes using mRNA isolated from HeLa cells expressing CA-Mkl1 and treated with the indicated siRNA. Data represent Mean ± SEM from four independent experiments. (D, E) Western blot in control and CA-Mkl1–expressing cells treated with the indicated siRNAs. The dashed line in D represents where two regions from the same Western exposure were stitched together to omit two lanes that are not relevant to this study. Statistical significance was determined using one-way ANOVA. *p < 0.05; **p < 0.01; ***p < 0.005.
We next determined whether CA-Mkl1 activity was refractory to regulation by LINC complexes. Depletion of Sun1 in control HeLa cells led to an increase in the expression of several SRF/Mkl1 target genes (Figure 3, A and B). The mRNA levels of VCL, SM22, and ACTB were not different, however, between control and Sun1-depleted cells expressing CA-Mkl1 (Figure 5C). Sun2 depletion also did not affect the expression of SRF/Mkl1 target genes, suggesting that CA-Mkl1 is not sensitive to LINC complexes that regulate endogenous SRF/Mkl1 activity. Of interest, Western blotting revealed that, whereas the levels of actin and SM22 were unaffected by depletion of SUN-domain proteins, vinculin abundance was dramatically reduced in cells expressing CA-Mkl1 after depletion of Sun2 (Figure 5D). The failure to accumulate vinculin protein, but not mRNA, in CA-Mkl1–expressing cells lacking Sun2 suggested that LINC complexes have essential transcription-independent functions in focal adhesion assembly. To test this possibility directly, we visualized focal adhesions by immunofluorescence. CA-Mkl1 expression in HeLa cells induced the assembly of focal adhesions that were larger and had significantly higher levels of vinculin protein than in control cells (Figure 6, A and B, and Supplemental Figure S4). The ability of CA-Mkl1 to drive these changes in focal adhesions was entirely dependent upon Sun2 (Figure 6, C and D). Depletion of Sun1 alone with siRNAs did not affect focal adhesions in cells expressing CA-Mkl1. Sun1 depletion did, however, suppress the defects in focal adhesion assembly that were caused by the depletion of Sun2. Moreover, we found that total vinculin protein levels were restored to nearly normal levels in cells lacking both Sun1 and Sun2 (Figure 5E).
FIGURE 6:
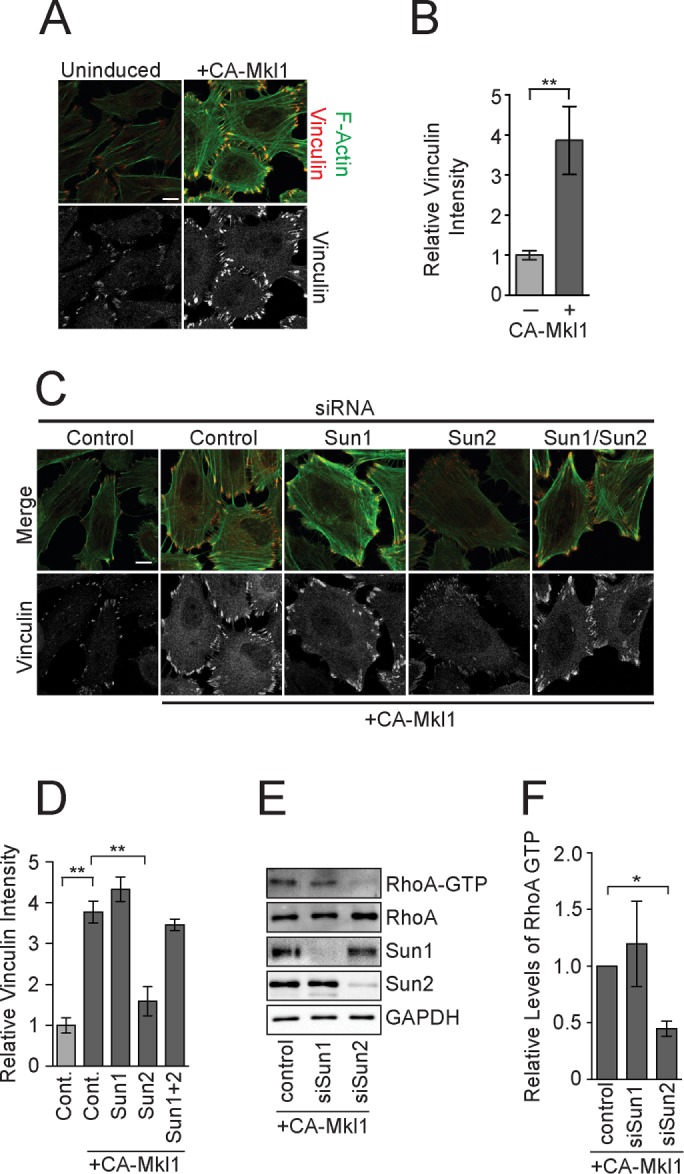
CA-Mkl1–induced focal adhesion assembly requires Sun2 LINC complexes. (A, B) Visualization and quantitation of vinculin levels at individual focal adhesions in control and CA-Mkl1–expressing cells. Data represent Mean ± SEM from four independent experiments. (C) F-actin and vinculin visualized in control and CA-Mkl1–expressing cells treated with the indicated siRNAs. (D) Quantitation of vinculin levels at individual focal adhesion after siRNA treatment. (E) Western blot of total and active RhoA in extracts from HeLa cells treated with the indicated siRNAs. (F) Quantitation of Western blots in E. Data represent mean ± SEM from four independent experiments. Statistical significance was determined using one-way ANOVA. *p < 0.05; **p < 0.01. Scale bar, 5 μm.
We also examined the dependence of RhoA activity on LINC complexes in cells expressing CA-Mkl1. In contrast to control cells, depletion of Sun1 did not lead to a significant increase in the levels of GTP-bound RhoA in HeLa cells that expressed CA-Mkl1 (Figure 6, E and F). Sun2, however, was required to maintain RhoA activity in CA-Mkl1 cells; depletion of Sun2 in cells expressing CA-Mkl1 led to a 2.3-fold reduction in RhoA-GTP levels relative to control cells.
Together these data show that LINC complexes have opposing roles in the transcription-independent control of focal adhesion assembly, suggesting that direct signaling from the nuclear envelope to modulators of RhoA activity is critical for proper actin dynamics. Moreover, whereas the function of different SUN-domain proteins is similar in control and CA-Mkl1–expressing cells, there is a difference in the dominant LINC complex form in these two conditions. Specifically, in control cells, Sun1 acts to limit RhoA activation and focal adhesion assembly, whereas the function of Sun2 LINC complexes in promoting focal adhesion assembly is only revealed in cells lacking Sun1. In contrast, in CA-Mkl1–expressing cells, Sun1 does not appreciably regulate focal adhesion assembly or RhoA activity, whereas Sun2 is essential to promote both processes. In this case, the inhibitory role of Sun1 is only revealed after depletion of Sun2. The switch from a Sun1 to a Sun2 LINC complex dominant state coincides with the transcriptional induction by SRF/Mkl1 of SUN2 expression. Thus it is likely the balance of different LINC complexes in the nuclear envelope that dictates the outcome of signaling to RhoA, and therefore, actin dynamics. To test this possibility directly, we overexpressed Sun2 in HeLa cells. Sun2 overexpression alone, however, was not sufficient to stimulate vinculin recruitment to focal adhesions or increase SRF/Mkl1-dependent gene expression, suggesting that additional proteins are also necessary to overcome the inhibitory function of Sun1 (Supplemental Figure S5, A and B).
DISCUSSION
We show here that different LINC complexes have opposing roles in the transcription-independent regulation of focal adhesions and RhoA activity in HeLa cells and propose, based on these data, a model for how LINC complexes regulate actin dynamics (Figure 7). A key feature of our model is that Sun2 LINC complexes signal from the nuclear envelope through the cytoplasm to increase the pool of active RhoA. RhoA, in turn, directly promotes actin stress fiber and focal adhesion assembly and initiates an SRF/Mkl1-dependent positive feedback loop that further amplifies RhoA activity. Sun1 antagonizes this network, perhaps by inhibiting Sun2 LINC complexes in the nuclear envelope. Of importance, a critical target of SRF/Mkl1 in the positive feedback loop is SUN2 itself, which is required to tilt the balance of LINC complex activity to favor RhoA activation.
FIGURE 7:
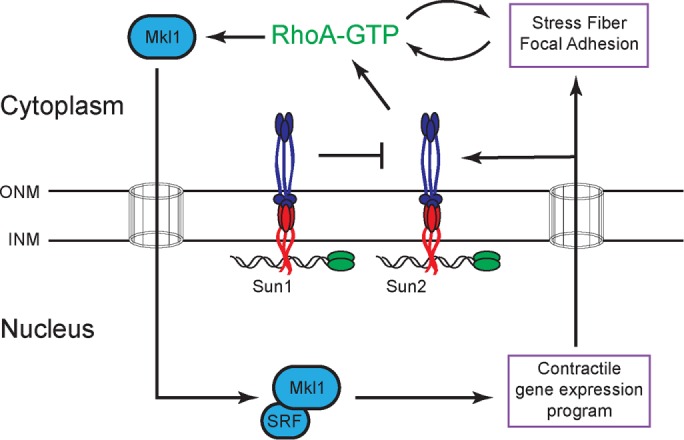
Working model for the regulation of actin organization by LINC complexes (see the text for details).
How do Sun2 LINC complexes promote RhoA activation?
LINC complexes have been shown to be required for proper actin organization in a number of cell types (Lammerding et al., 2004; Lee et al., 2007; Hale et al., 2008; Stewart-Hutchinson et al., 2008; Folker et al., 2011; Kim et al., 2012). The targets and mechanisms of LINC complex signaling, however, have not been defined. Our data suggest that Sun2 LINC complexes promote RhoA activation, which induces the increased assembly of focal adhesions in tissue culture cells. Moreover, by constructing cell lines that contained DN-Mkl1 or CA-Mkl1 transgenes, we showed that whereas SRF/Mkl1 activity is essential for focal adhesion assembly, other activities are likely the primary targets of Sun2 LINC complexes in this signaling network. We therefore propose that Sun2 LINC complexes signal through the cytoplasm to increase RhoA-GTP levels by stimulating the activity of one or more RhoA guanine-nucleotide exchange factors (GEFs). Sun2 LINC complexes could stimulate GEF activity in a manner that is analogous to integrins within focal adhesions, which both transduce actin-dependent forces across the plasma membrane and act as force-sensitive signaling platforms that feed back to amplify RhoA activity (Guilluy et al., 2011). Alternatively, Sun2 LINC complexes could indirectly control actin organization through the microtubule cytoskeleton. Extensive cross-talk between microtubule and actin networks has been demonstrated, including a requirement for microtubules to promote focal adhesion turnover (Ezratty et al., 2005). Microtubules are also known to limit the levels of active RhoA in HeLa cells by inhibiting GEF-H1 (Ren et al., 1998; Krendel et al., 2002; Chang et al., 2008). We did not find any evidence for altered microtubule organization in HeLa cells lacking SUN-domain proteins by immunofluorescence (unpublished data). A more detailed analysis, however, is likely required to address this possibility.
The specific nesprin protein that functions with Sun2 to promote stress fiber assembly in HeLa cells has not been identified, although several lines of evidence suggest that nesprin 2 is likely to be important in this process. N2G interacts with both actin filaments and microtubule motor proteins and is expressed in HeLa cells, whereas nesprin 1 is not (Randles et al., 2010). Loss-of-function approaches are unlikely to reveal a direct role for nesprin 2 in our system, however, because control cells and cells that lack both Sun1 and Sun2 appear similar using several critical readouts of signaling to the actin cytoskeleton, including RhoA-GTP levels, SRF/Mkl1-dependent gene expression, and focal adhesion composition (Figures 1 and 3). Thus it would be difficult to discern a role for N2G, or any other nesprin protein in regulating actin dynamics if it was important for the function of both Sun1 and Sun2 LINC complexes. The use of dominant proteins that contain specific cytoplasmic domains will likely be essential to dissect the contribution of individual nesprins to the transcription-independent control of actin stress fiber assembly. Our initial studies using this approach demonstrated that the actin-binding domain of nesprin 2 is not sufficient to support Sun2 LINC complex function. Other nesprin domains are therefore minimally required for RhoA activation by Sun2 LINC complexes in HeLa cells.
The identification of minimal nesprins capable of supporting Sun2 LINC complex functions will also be essential to clarifying our understanding of the relationship between Sun1 and Sun2 in this system. Our data indicate that a critical target of CA-Mkl1 in promoting focal adhesion assembly is SUN2, which, we suggest, tilts the balance of LINC complex activity in the nuclear envelope to a state that favors RhoA activation. Overexpression of Sun2 was unable, however, to overcome the inhibitory affect of Sun1. Thus other targets of SRF/MKl1 are necessary to stimulate signaling to the actin cytoskeleton. Of interest, SYNE2, which encodes nesprin 2, has been identified as a direct transcriptional target of SRF/Mkl1 (Esnault et al., 2014). Moreover, the quantitative analysis of global protein abundance in HeLa cells revealed that whereas Sun1 and Sun2 proteins are present at similar levels, nesprin 2 and nesprin 3 are ∼100-fold less abundant (Nagaraj et al., 2011). Thus simply overexpressing Sun2 is unlikely to significantly increase the concentration of Sun2-LINC complexes in the nuclear envelope. It will be interesting to determine whether cooverexpression of a functional nesprin minigene and Sun2 is sufficient to the overcome Sun1 inhibition.
How do Sun1 LINC complexes inhibit Sun2 LINC complexes?
Both overlapping and unique functions for Sun1 and Sun2 were previously defined. This is, however, to our knowledge, the first demonstration that SUN-domain proteins in the INM have opposing roles in a biological process. Establishing how Sun1 inhibits RhoA activation will therefore be critical to understand the mechanisms that control actin dynamics in HeLa cells. Our data suggest that Sun1 inhibits stress fiber assembly by limiting the activity of Sun2 LINC complexes in the INM. One possibility is that Sun1 acts as a nesprin sink, sequestering a limiting pool of KASH peptide–containing proteins away from Sun2. No direct comparison of the affinity of the SUN domains from Sun1 and Sun2 for KASH peptides has been described (Sosa et al., 2013). An autoinhibitory interaction between the coiled-coil and SUN domains of Sun2 has recently been demonstrated (Nie et al., 2016), however, which could bias the binding of KASH peptides toward Sun1 in cells that express both SUN-domain proteins. An alternative but related possibility is that Sun1 and Sun2 compete for binding to other critical interacting proteins, such as lamins or INM proteins.
It is important to note that the relationship between Sun1 and Sun2 in regulating RhoA activity outlined in our model, although wholly consistent with the data obtained using control HeLa cells, is not strictly supported by our results in cells expressing CA-Mkl1. Specifically, we show that depletion of Sun1 in cells expressing CA-Mkl1 suppressed the defects in focal adhesion assembly that were present in cells also lacking Sun2, which is not predicted by our model. Thus Sun1 may inhibit signaling to the actin cytoskeleton by multiple, independent mechanisms. In support of this possibility, we found that the modest overexpression of Sun1 did not affect focal adhesions but did reduce SRF/Mkl1 transcription (Supplemental Figure S5). Alternatively, these data could be a by-product of the fact that CA-Mkl1 expression leads to a substantial increase in Sun2 levels, resulting in sufficient levels of Sun2 even after siRNA-mediated knockdown to support RhoA activation.
An additional consideration relevant to our model is how Sun1 and Sun2 assume redundant, specific, or opposing functions, depending on the cellular and developmental context. Although many possibilities could account for these behaviors, including differences in the levels of expression of various LINC complex proteins or posttranslational modifications, it is noteworthy that numerous alternative spliced isoforms of Sun1 have been identified (Göb et al., 2011). Thus some isoforms of Sun1 could be redundant with Sun2, whereas others may have unique or inhibitory functions, and we are exploring this possibility.
Implications for LINC complex function in health and disease
Exploring the mechanisms of stress fiber and focal adhesion assembly in tissue culture cells was critical to discovering the molecular functions of RhoA and its effectors, including RhoA-activated protein kinase (ROCK; Jaffe and Hall, 2005). Those studies have been translated into an improved understanding of the role that RhoA and ROCK play in a broad range of biological contexts, including cardiovascular development (Jaffe and Hall, 2005; Shimokawa et al., 2016). Our observation that different LINC complex proteins have opposing roles in regulating RhoA in HeLa cells is therefore likely relevant to understanding the role of LINC complexes in health and disease. A central paradox of EDMD and CDM caused by mutations in nuclear envelope proteins is that defects in nuclear morphology and mechanics are found in virtually all cell types, whereas the disease phenotypes emerge specifically in the heart and skeletal muscle. Our data suggest that the muscle-specific pathologies arise in part due to the failure to properly assemble contractile actin-based structures, which are critical for the function of heart and skeletal muscle. Indeed, stress fibers and nascent muscle myofibrils are similar in composition and mechanism of assembly, including their mutual dependence on RhoA (Sanger et al., 2010; Burridge and Wittchen, 2013). It will be interesting to explore RhoA activity in experimental models for EDMD and DCM.
MATERIALS AND METHODS
Cell culture and stable cell line production
HeLa cells that contain a single copy of a Flp-recombination target (FRT) site within the genome and express the tetracycline repressor protein were maintained in DMEM supplemented with 10% tetracycline-free fetal bovine serum (FBS; Clontech), 15 μg/ml blasticidin (InvivoGen, San Diego, CA), 200 μg/ml Zeocin (InvivoGen) and penicillin/streptomycin (ThermoFisher, Carlsbad, CA) at 37°C in a humidified incubator with 5% CO2. Stable cell lines containing doxycycline-inducible transgenes were generated using the Flp-In system (ThermoFisher). Briefly, the described cDNA was cloned into pcDNA/FRT/TO and transfected with pOG44 (which expressed the Flp recombinase) at a ratio of 1:10 using FuGENE (Promega, Madison, WI) into HeLa cells at ∼50% confluence. All transfections were performed in DMEM plus FBS in the absence of antibiotics. After 48 h, cells were split to low density in medium with the antibiotics described except that Zeocin was replaced by 200 μg/ml hygromycin B (Invitrogen, Carlsbad, CA). Multiple independent clones from each transfection were pooled after 2 wk of selection to create a polyclonal stable cell line. Doxycycline at 0.1 μg/ml was added to cell lines for 24–48 h to induce transgene expression.
RNA interference
HeLa cells were transfected with control, Sun1, or Sun2 small interfering RNA (siRNA) SMARTpools (GE/Dharmacon, Lafayette, CO) at a concentration of 50 nM using Lipofectamine RNAiMAX (Invitrogen) according to manufacturer’s instructions. siRNA treatments were allowed to proceed for 48–60 h before cells were prepared for analysis by immunofluorescence, Western blotting, or qPCR.
Antibodies and reagents
Rabbit polyclonal antibodies against Man1, Sun2, emerin, and GFP were made by the Carroll lab and are available upon request. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH; 2118; Cell Signaling, Danvers, MA), Sun1 (124770; Abcam, Cambridge, MA), lamin A/C (Santa Cruz Biotechnology, Santa Cruz, CA; 20681 for Western blots and 7292 for immunofluorescence), SM22 (14106; Abcam), vinculin (V4505; Sigma-Aldrich, St. Louis, MO), and β-actin (47778; Santa Cruz Biotechnology) were purchased and used as directed by the manufacturer. For immunofluorescence, goat anti-mouse or goat anti-rabbit Alexa 647, Alexa 555, Alexa 568, and Alexa 488 (Life Technologies, Carlsbad, CA) secondary antibodies were used at a dilution of 1:500. For immunoblotting, horseradish peroxidase–coupled goat anti-mouse or goat anti-rabbit (Cell Signaling) secondary antibodies were used at an appropriate dilution according to the manufacturer’s instructions. Actin filaments were visualized using Alexa 488– or Alexa 568–conjugated phalloidin (Life Technologies) according to the manufacturer’s instructions.
Preparation of protein extracts for Western blotting and RhoA-GTP isolation
Cells from one well of a six-well plate were washed twice with ice-cold phosphate-buffered saline (PBS), scraped from the well in 1 ml of cold PBS, and collected by centrifugation at 200 × g for 5 min at 4°C. Lysis was performed in 60 μl using 50 mM Tris, 8 M urea, pH 7.4, or RIPA (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100) in the presence of protease inhibitors. Protein concentrations were measured by Bradford’s assay. A 20-μg amount of total protein from each sample was used for SDS–PAGE and Western blotting. The level of RhoA GTP in cells extracts was measured as previously described (Ren et al., 1999).
Indirect immunofluorescence and confocal microscopy
Cells were grown on acid-washed glass coverslips coated with 50 μg/ml fibronectin (Sigma-Aldrich) before preparation for immunofluorescence and imaging. For immunofluorescence, cells were fixed in PBS plus 4% formaldehyde for 10 min, washed twice with PBS, and permeabilized for 5 min in PBS plus 0.2% Triton X-100. After permeabilization, blocking buffer (BB; 1× PBS plus 2% bovine serum albumin) was added to the cells for 30 min, followed by primary antibody in BB for 60 min at room temperature. After three 5-min washes with BB, cells were treated with the appropriate secondary antibodies in BB for 60 min. Cells were then washed three times for 5 min in BB, stained with Hoechst stain (2 μg/ml in BB) for 5 min, washed twice in BB and twice times in PBS, and then mounted on a slide using ProLong Gold antifade reagent (Molecular Probes) for imaging. Images were captured on a Zeiss LSM 710 Duo laser scanning confocal microscope using a 63× oil immersion objective. The analysis of focal adhesion size and intensity was performed using a custom macro developed in ImageJ. Briefly, after subtracting the background fluorescence, focal adhesions were segmented using an automated threshold function, and the size and intensity of individual focal adhesions were quantified.
RNA isolation, first-strand cDNA synthesis, and quantitative real-time PCR
mRNA was isolated from HeLa cells using the RNeasy Mini Kit (Qiagen, Hilden, Germany) with on-column DNase treatment according to manufacturer’s protocol. cDNA was synthesized with a SuperScript First-Strand synthesis system (Invitrogen) using 200 ng of mRNA and an oligo dT oligonucleotide according to manufacturer’s protocol. Quantitative real-time PCR was performed using SYBR green detection in a CFX96 Real-Time system (Bio-Rad, Hercules, CA). qPCR primers were designed using National Center for Biotechnology Information Primer-BLAST software as follows: vinculin, 5′ATTTGATGAGAGGGCAGCTA and 5′ATGCCTTCCACTGTTGATTT; SRF, 5′ACTGCCTTCAGTAGGAACAA and 5′TTCAAGCACACACACTCACT; lamin A, 5′ACCCATCTCCTCTGGCTCTT and 5′TTCTGGGGGCTCTGGGTT; Sun2, 5′TTAACCAGGCGCTTCTCGTC and 5′ACCAAAGCAGGGTGGAATGT; Sun1, 5′TCGACGGCCTCCTGTATTG and 5′TGGCAGCTTTATTTCCACCTTTA; SM22, 5′GCAGAGGAATTGATGGAAACCAC and 5′CTGGGGAAAGCTCCTTGGAAGT; and GAPDH, 5′TGCACCACCAACTGCTTAGC and 5′GGCATGGACTGTGGTCATGAG. mRNA levels were calculated using the dCT method after normalization to GAPDH mRNA.
Supplementary Material
Acknowledgments
We thank Megan King and Patrick Lusk for comments on the manuscript and the Carroll, King, Lusk, Yao, and Bahnmanyar labs (Yale University) for thoughtful suggestions during the course of this work. We also thank Gant Luxton (University of Minnesota) for providing the mN2G and SR-KASH constructs. This work was supported by an Innovator Award from the Progeria Research Foundation (C.C.).
Abbreviations used:
- EDMD
Emery–Dreifuss muscular dystrophy
- INM
inner nuclear membrane
- KASH
Klarsicht, ANC-1, and Syne homology
- LINC
Linker of Nucleoskeleton and Cytoskeleton
- ONM
outer nuclear membrane
- SUN
Sad1, UNC84.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0467) on November 9, 2016.
REFERENCES
- Arsenovic PT, Ramachandran I, Bathula K, Zhu R, Narang JD, Noll NA, Lemmon CA, Gundersen GG, Conway DE. Nesprin-2G, a component of the nuclear LINC complex, is subject to myosin-dependent tension. Biophys J. 2016;110:34–43. doi: 10.1016/j.bpj.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge K, Wittchen ES. The tension mounts: stress fibers as force-generating mechanotransducers. J Cell Biol. 2013;200:9–19. doi: 10.1083/jcb.201210090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Doucet C, Hetzer MW. Nuclear pore complexes: guardians of the nuclear genome. Cold Spring Harb Symp Quant Biol. 2011;75:585–597. doi: 10.1101/sqb.2010.75.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen B, Selvaraj A, Burgess RC, Hitzler JK, Ma Z, Morris SW, Prywes R. Megakaryoblastic leukemia 1, a potent transcriptional coactivator for serum response factor (SRF), is required for serum induction of SRF target genes. Mol Cell Biol. 2003;23:6597–6608. doi: 10.1128/MCB.23.18.6597-6608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Antoku S, Östlund C, Worman HJ, Gundersen GG. Linker of nucleoskeleton and cytoskeleton (LINC) complex-mediated actin-dependent nuclear positioning orients centrosomes in migrating myoblasts. Nucleus. 2015a;6:77–88. doi: 10.1080/19491034.2015.1004947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Folker ES, Worman HJ, Gundersen GG. Emerin organizes actin flow for nuclear movement and centrosome orientation in migrating fibroblasts. Mol Biol Cell. 2013;24:3869–3880. doi: 10.1091/mbc.E13-06-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Worman HJ, Gundersen GG. Accessorizing and anchoring the LINC complex for multifunctionality. J Cell Biol. 2015b;208:11–22. doi: 10.1083/jcb.201409047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y-C, Nalbant P, Birkenfeld J, Chang Z-F, Bokoch GM. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol Biol Cell. 2008;19:2147–2153. doi: 10.1091/mbc.E07-12-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Xu R, Yu J, Xu T, Zhuang Y, Han M. SUN1 is required for telomere attachment to nuclear envelope and gametogenesis in mice. Dev Cell. 2007;12:863–872. doi: 10.1016/j.devcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014;28:943–958. doi: 10.1101/gad.239327.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezratty EJ, Partridge MA, Gundersen GG. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- Folker ES, Östlund C, Luxton G, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci USA. 2011;108:131. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Mechanical integration of actin and adhesion dynamics in cell migration. Annu Rev Cell Dev Biol. 2010;26:315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göb E, Meyer-Natus E, Benavente R, Alsheimer M. Expression of individual mammalian Sun1 isoforms depends on the cell type. Commun Integr Biol. 2011;4:440–442. doi: 10.4161/cib.4.4.15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea SM, Awadia S, Garcia-Mata R. I’m coming to GEF you: regulation of RhoGEFs during cell migration. Cell Adh Migr. 2015;8:535–549. doi: 10.4161/cam.28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu Rev Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- Guilluy C, Swaminathan V, Garcia-Mata R, O’Brien ET, Superfine R, Burridge K. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat Cell Biol. 2011;13:724–729. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, Hodzic D, Wirtz D. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J. 2008;95:5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Racl, CDC42Hsregulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1–SRF activity by modulating actin dynamics. Nature. 2014;497:507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF. LINC complex proteins in development and disease. Curr Top Dev Biol. 2014;109:287–321. doi: 10.1016/B978-0-12-397920-9.00004-4. [DOI] [PubMed] [Google Scholar]
- Isermann P, Lammerding J. Nuclear mechanics and mechanotransduction in health and disease. Curr Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Khatau SB, Feng Y, Walcott S, Sun SX, Longmore GD, Wirtz D. Actin cap associated focal adhesions and their distinct role in cellular mechanosensing. Sci Rep. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Zenke FT, Bokoch GM. Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat Cell Biol. 2002;4:294–301. doi: 10.1038/ncb773. [DOI] [PubMed] [Google Scholar]
- Kuo J-C, Han X, Hsiao C-T, Yates JR, Waterman CM. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol. 2011;13:383–393. doi: 10.1038/ncb2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–378. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JSH, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–2552. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K, Zhang X, Ding X, Guo X, Chen M, Zhu B, Xu T, Zhuang Y, Xu R, Han M. SUN1 and SUN2 play critical but partially redundant roles in anchoring nuclei in skeletal muscle cells in mice. Proc Natl Acad Sci USA. 2009;106:10207–10212. doi: 10.1073/pnas.0812037106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Olson EN. Coactivator control of cardiovascular growth and remodeling. Curr Opin Cell Biol. 2006;18:715–722. doi: 10.1016/j.ceb.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke P, Mattioli E, Haque F, Antoku S, Columbaro M, Straatman KR, Worman HJ, Gundersen GG, Lattanzi G, Wehnert M, et al. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014;10:e1004605–e1004618. doi: 10.1371/journal.pgen.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou A-I, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:1–8. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Ke H, Gao F, Ren J, Wang M, Huo L, Gong W, Feng W. Coiled-coil domains of sun proteins as intrinsic dynamic regulators. Structure. 2016;24:80–91. doi: 10.1016/j.str.2015.10.024. [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci. 2005;118:3419–3430. doi: 10.1242/jcs.02471. [DOI] [PubMed] [Google Scholar]
- Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plessner M, Grosse R. Extracellular signaling cues for nuclear actin polymerization. Eur J Cell Biol. 2015;94:359–362. doi: 10.1016/j.ejcb.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Plessner M, Melak M, Chinchilla P, Baarlink C, Grosse R. Nuclear F-actin formation and reorganization upon cell spreading. J Biol Chem. 2015;290:11209–11216. doi: 10.1074/jbc.M114.627166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakylä EK, Vartiainen MK. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases. 2014;5:e27539–00. doi: 10.4161/sgtp.27539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajgor D, Mellad JA, Autore F, Zhang Q, Shanahan CM. Multiple novel nesprin-1 and nesprin-2 variants act as versatile tissue-specific intracellular scaffolds. PLoS One. 2012;7:e40098. doi: 10.1371/journal.pone.0040098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles KN, Lam LT, Sewry CA, Puckelwartz M, Furling D, Wehnert M, McNally EM, Morris GE. Nesprins, but not sun proteins, switch isoforms at the nuclear envelope during muscle development. Dev Dyn. 2010;239:998–1009. doi: 10.1002/dvdy.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem. 1998;273:34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Wang J, Fan Y, White J, Sanger JM. Assembly and dynamics of myofibrils. J Biomed Biotechnol. 2010;2010:858606–00. doi: 10.1155/2010/858606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M. Rho signalling at a glance. J Cell Sci. 2004;117:5457–5458. doi: 10.1242/jcs.01582. [DOI] [PubMed] [Google Scholar]
- Shimi T, Butin-Israeli V, Adam SA, Goldman RD. Nuclear lamins in cell regulation and disease. Cold Spring Harb Symp Quant Biol. 2011;75:525–531. doi: 10.1101/sqb.2010.75.045. [DOI] [PubMed] [Google Scholar]
- Shimokawa H, Sunamura S, Satoh K. RhoA/Rho-kinase in the cardiovascular system. Circ Res. 2016;118:352–366. doi: 10.1161/CIRCRESAHA.115.306532. [DOI] [PubMed] [Google Scholar]
- Small EM. The Actin-MRTF-SRF gene regulatory axis and myofibroblast differentiation. J Cardiovasc Trans Res. 2012;5:794–804. doi: 10.1007/s12265-012-9397-0. [DOI] [PubMed] [Google Scholar]
- Sosa BA, Kutay U, Schwartz TU. Structural insights into LINC complexes. Curr Opin Struct Biol. 2013;23:285–291. doi: 10.1016/j.sbi.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RM, Zubek AE, Rosowski KA, Schreiner SM, Horsley V, King MC. Nuclear-cytoskeletal linkages facilitate cross talk between the nucleus and intercellular adhesions. J Cell Biol. 2015;209:403–418. doi: 10.1083/jcb.201502024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Wang W, Shi Z, Jiao S, Chen C, Wang H, Liu G, Wang Q, Zhao Y, Greene MI, Zhou Z. Structural insights into SUN-KASH complexes across the nuclear envelope. Cell Res. 2012;22:1440–1452. doi: 10.1038/cr.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2011;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft CM, Xu G, Xu T, Zhuang Y, Xu R, Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Felder A, Liu Y, Guo LT, Lange S, Dalton ND, Gu Y, Peterson KL, Mizisin AP, Shelton GD, et al. Nesprin 1 is critical for nuclear positioning and anchorage. Hum Mol Genet. 2009a;19:329–341. doi: 10.1093/hmg/ddp499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007a;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009b;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007b;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.