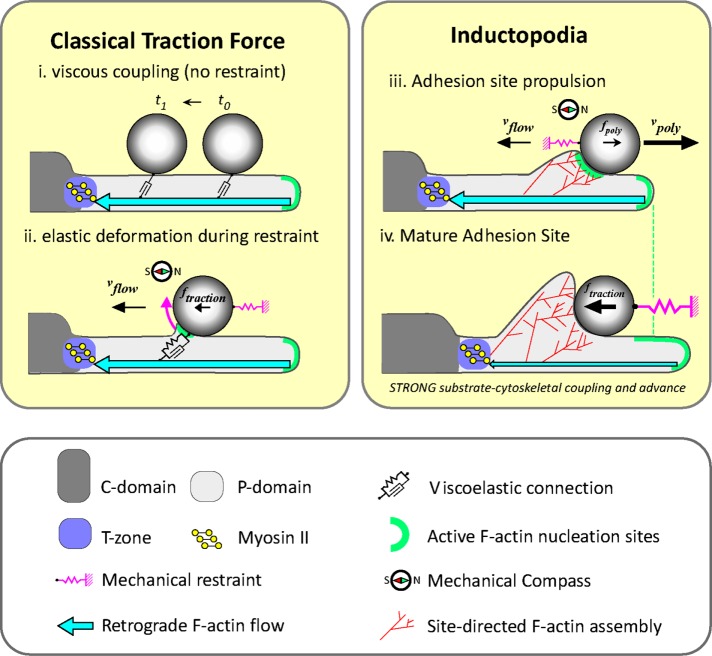
In growth cones, local Arp 2/3-dependent actin assembly mechanically buffers apCAM adhesions from retrograde flow–associated traction forces. The resulting propulsive forces drive the exploratory motility of inductopodia. Increasing the stiffness of apCAM targets induces an extensive 3D actin cup to form at the adhesion during evoked growth responses.
Abstract
Homophilic binding of immunoglobulin superfamily molecules such as the Aplysia cell adhesion molecule (apCAM) leads to actin filament assembly near nascent adhesion sites. Such actin assembly can generate significant localized forces that have not been characterized in the larger context of axon growth and guidance. We used apCAM-coated bead substrates applied to the surface of neuronal growth cones to characterize the development of forces evoked by varying stiffness of mechanical restraint. Unrestrained bead propulsion matched or exceeded rates of retrograde network flow and was dependent on Arp2/3 complex activity. Analysis of growth cone forces applied to beads at low stiffness of restraint revealed switching between two states: frictional coupling to retrograde flow and Arp2/3-dependent propulsion. Stiff mechanical restraint led to formation of an extensive actin cup matching the geometric profile of the bead target and forward growth cone translocation; pharmacological inhibition of the Arp2/3 complex or Rac attenuated F-actin assembly near bead binding sites, decreased the efficacy of growth responses, and blocked accumulation of signaling molecules associated with nascent adhesions. These studies introduce a new model for regulation of traction force in which local actin assembly forces buffer nascent adhesion sites from the mechanical effects of retrograde flow.
INTRODUCTION
Developing neurons extend neuritic processes tipped with motile growth cones into the embryonic environment to establish connectivity. As growth cones migrate, extracellular spatial information is relayed via transmembrane receptors and cell adhesion molecules to intracellular signaling pathways that modulate the cytoskeletal dynamics underlying growth cone motility and generation of traction forces (Lowery and Van Vactor, 2009). In growth cones, actin filaments (F-actin) are organized into filopodial bundles and a network of branched filaments, referred to as lamellipodia or actin veils, in the peripheral (P) domain (Goldberg and Burmeister, 1986; Lewis and Bridgman, 1992) while the central (C) domain contains organelles, microtubules, and nonmuscle myosin II-dependent contractile F-actin arcs. These actin arcs form at the base of recycling filopodia in the transition zone (T zone) between the P and C domains (Zhang et al., 2003). Other F-actin–containing structures termed intrapodia form on the “dorsal” plasma membrane and can dynamically explore the proximal P domain and T zone via spreading waves of motility driven by actin polymerization (Forscher et al., 1992; Rochlin et al., 1999). While filopodia and lamellipodia increase surface area and enable the growth cone to explore its environment, the physiological role intrapodia play in growth cone motility is not well understood.
The life cycle of actin filaments is defined by stereotyped events in space and time within the growth cone. Actin polymerization preferentially occurs in a band at the leading edge, with some polymerization distributed throughout the P domain, while depolymerization is distributed throughout the growth cone (Yang et al., 2012). Formation of new actin filaments from monomers is unfavorable unless actin-nucleating proteins such as the Arp2/3 complex (hereafter referred to as “Arp2/3”) and nucleation-promoting factors (NPFs) are present (Pollard and Cooper, 1986; Campellone and Welch, 2010). Pharmacological inhibition of Arp2/3 nucleation activity in growth cones decreases filament density at the leading edge, the integrity of actin veils, and activity of intrapodia (Yang et al., 2012). The combination of leading-edge polymerization, nonmuscle myosin II contractility in the T zone and ADF/cofilin-dependent actin turnover leads to the steady-state treadmilling of actin polymer in the P domain, a process commonly referred to as “retrograde actin flow” (Lin et al., 1997; Medeiros et al., 2006; Ohashi et al., 2011; Flynn et al., 2012; Van Goor et al., 2012). Arp 2/3 nucleation is spatially regulated by the NPF Scar/WASP-family verprolin-homologous protein (WAVE) regulatory complex (WRC), the small GTPase Rac, and acidic phospholipids (Lebensohn and Kirschner, 2009); Rac signaling is known to be important for guided neurite outgrowth (Yuan et al., 2003; Woo and Gomez, 2006; Tahirovic et al., 2010).
The relationship between cytoskeletal dynamics, adhesion, and cell migration has been assessed in various contexts. Growth cones and crawling cells such as keratocytes exert distributed traction forces through lamellipodial adhesions when migrating on two-dimensional surfaces (Lee et al., 1994; Bard et al., 2008; Fournier et al., 2010; Hyland et al., 2014; Nichol et al., 2016). Vertebrate cell adhesion molecules (CAMs) involved in neuronal adhesion, outgrowth and fasciculation include the N-cadherins and the immunoglobulin superfamily of CAMs (IgCAMs) (Maness and Schachner, 2007). Among the IgCAMs expressed in the nervous system are N-CAM; L-1, which interacts with the cytoskeletal adapter ezrin and Src-family kinases (Walsh and Doherty, 1997; Bard et al., 2008; Sakurai et al., 2008); and the Aplysia homologue apCAM. We recently characterized mechanical properties of nascent apCAM adhesions forming at optically trapped beads applied to the P domain of Aplysia bag cell growth cones (Mejean et al., 2013). Release of beads from traps reveals stochastic switching between states dominated by: 1) simple viscous drag or 2) elastic coupling to flow, characterized by an elastic length scale of up to 1 μM and elastic modulus in the range of 45–350 Pa, consistent with an actin polymer network bearing traction forces. In some cases, a third state is observed, in which restrained or unrestrained beads are associated with short bursts or sustained episodes of site-directed actin assembly. Sustained actin polymerization results in formation of actin tails that propel beads on the growth cone surface (Forscher et al., 1992; Thompson et al., 1996). The comet-like shape and motility of these “inductopodia” are reminiscent of the rocketing motility of intracellular pathogens such as Listeria (Welch et al., 1997) or beads coated with actin nucleation-promoting factors in cell-free assays (Loisel et al., 1999; van Oudenaarden and Theriot, 1999; Yarar et al., 1999), both of which use branched actin network assembly for forward propulsion. Inductopodial movement in growth cones is driven by local actin assembly, since acute wash-in of the barbed-end capping agent cytochalasin results in immediate arrest of actin tail/inductopodial lengthening and resumption of flow-coupling (Forscher et al., 1992). In some cases, when assembly matches the rate of retrograde flow and is in the opposite direction, there is little net bead movement; if assembly stops, the bead resumes flow coupling. The physiological significance of inductopodial/intrapodial activity for axon outgrowth remains poorly understood.
Growth cones of Aplysia bag cells are normally slow growing (1–2 μm/h) when plated on polylysine substrates that do not specifically engage adhesions. However, they exhibit dramatic motile responses after contacting attractive physiological targets such as neurites of other bag cells (Lin and Forscher, 1993) or mechanically restrained apCAM beads (Suter et al., 1998). Growth responses to apCAM beads have two well-characterized phases: 1) an initial “latency period” that involves clustering of endogenous apCAM, accumulation of active Src family kinase, and assembly of new actin filaments into a cuplike structure around the target site (Lin and Forscher, 1993; Thompson et al., 1996; Suter and Forscher, 2001); and 2) a later advance period characterized by attenuated retrograde actin flow and generation of traction force applied to the apCAM bead that depends on myosin II–dependent contractility of actin arc structures (Suter et al., 1998, 2004; Schaefer et al., 2008). Vertebrate hippocampal growth cones exhibit similar F-actin accumulation in response to N-cadherin beads, (Lambert et al., 2002; Bard et al., 2008). Furthermore, retrograde flow continues at similar rates near the target site during the latency period and slows in the axis of interaction between the bead and C domain during the advance period (Lin and Forscher, 1995; Suter et al., 1998; Schaefer et al., 2008). These findings led to the proposal of a “molecular clutch” model wherein the rate of advance is inversely proportional to the rate of retrograde flow (Suter and Forscher, 2000; Giannone et al., 2009).
In this study, we apply apCAM beads to growth cones at variable stiffnesses of restraint and show that branched actin assembly at apCAM adhesions drives formation and motility of inductopodia and reorientation of force from tractive to propulsive mode. Arp2/3 and Rac activity are required for building an extensive F-actin structure during the latency period and for growth responses to apCAM targets. Site-directed actin assembly adds a new force component to the traditional molecular clutch model that can buffer nascent adhesions from the normal mechanical drag of retrograde flow during adhesion site maturation.
RESULTS
Inductopodial movement is driven by Arp2/3-dependent actin assembly
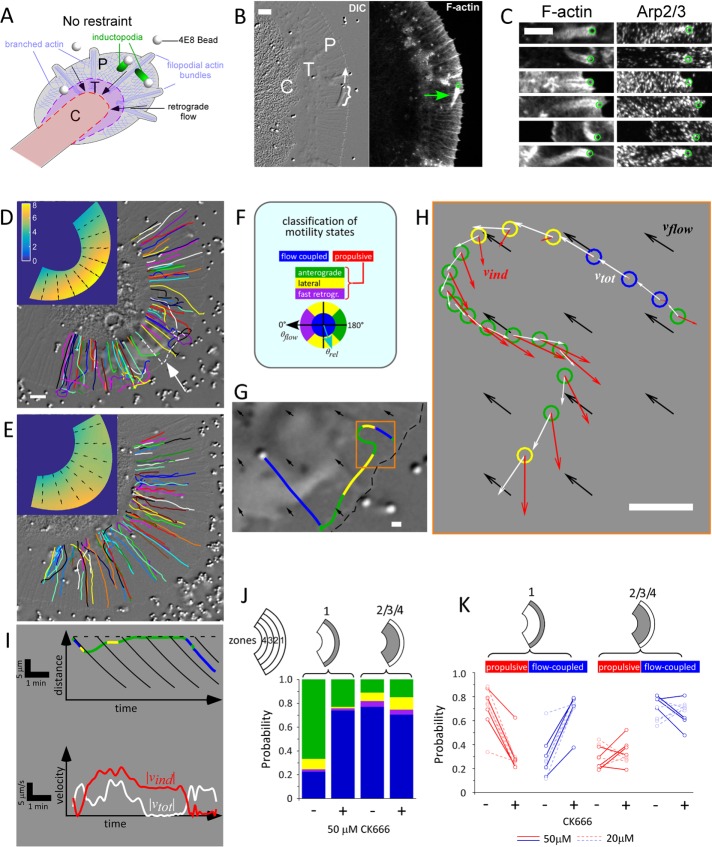
To study site-directed actin assembly evoked by apCAM, we used 800 nm silica beads functionalized with an antibody (4E8) that specifically binds the extracellular domain of apCAM. A suspension of 4E8 beads was applied to growth cones; 4E8 beads landed randomly on the P domain and their unrestrained motion was recorded by differential interference contrast microscopy (DIC) time-lapse imaging (Figure 1). Most beads that landed on the P domain coupled to retrograde flow and were carried centripetally toward the C domain; however, some beads formed inductopodia and moved in other directions (Figure 1B and Supplemental Movie 1). In agreement with our previous findings, (Forscher et al., 1992), inductopodial tails contained F-actin (Figure 1, B–C). Costaining with an antibody for the Arp2/3 complex (anti-p34Arc/ARPC2; Machesky et al., 1997) showed punctate labeling of actin tails (Figure 1C), prompting further investigation into the role of the Arp2/3 complex in inductopodial motility.
FIGURE 1:
Site-directed actin assembly drives inductopodial motility and requires Arp2/3 complex activity. (A) Inductopodia are induced by unrestrained beads derivatized with anti-apCAM (4E8) antibody landing on the growth cone. Growth cone domains: P, P domain; T, T zone; C, C domain. (B) DIC image (left, cf. Supplemental Movie 1) taken immediately before fixation and confocal phalloidin-stained fluorescent image (right) showing the actin tail of an inductopodium (bracket and green arrow; bead positions in B and C: green circles). Direction of inductopodial movement is indicated by white arrow. (C) Multiple inductopodia with F-actin visualized with phalloidin (left) and Arp2/3 complex with p34Arc antibody (right). (D) 4E8 beads were applied to a growth cone in control conditions (cf. Supplemental Movie 2). Arbitrarily colored bead tracks are shown superimposed over DIC image of the growth cone. Scale bar: 5 μm. Inset, Retrograde flow shown as a color map of flow speed |vflow| calculated from flow-coupled beads only, with arrows indicating flow direction. (E) Bead tracks for the same growth cone in the presence of 50 μM CK666 (cf. Supplemental Movie 2). Inset, Map of |vflow| in 50 μM CK666. (F) Bead-velocity classification scheme. Beads are classified as being flow coupled when the flow-subtracted bead/inductopodial speed |vind| is below a threshold of 2 μm/min (blue), or when |vind| is above threshold, as retrograde, lateral, or anterograde (purple, yellow, or green, respectively) according to flow-subtracted bead-velocity angles relative to flow, θrel (light blue arrow). The latter three classifications can be grouped into a nonflow or “propulsive” velocity classification category (red). (G) Track corresponding to the dotted white track indicated by arrow in D with color coding for classified motility states. Local flow direction is indicated by black arrows. Scale bar: 1 μm. (H) Plot of observed positions, bead velocities (white arrows), and flow-corrected velocities (red arrows) of the initial portion of the track (orange box in G) shown at 5 s intervals with color-coded motility classification (circles). Velocities are also color coded: flow velocity vflow in black, uncorrected bead velocity vtot in white, and flow-corrected bead/inductopodial velocity vind in red. Scale bar: 1 μm. (I) Kinematic analysis of bead track shown in G and H. Kymograph of bead position relative to leading edge (dotted line) vs. time (top panel) showing color-coded motility states, with retrograde flow (solid black lines); velocity vs. time graph (bottom panel) with uncorrected (white) and flow-corrected (red) speed traces. (J) The P domain of each growth cone was subdivided into four annular zones for analysis of motility (cf. Supplemental Figure S1). Probabilities of the four velocity states in the front one-fourth (zone 1) or rear three-fourths (zones 2/3/4) of the P domain of the growth cone in D and E are shown as stacked histograms in control vs. 50 μM CK666 conditions. (K) Changes in flow-coupled (blue) vs. propulsive (red) state probabilities resulting from Arp2/3 complex inhibition are shown for multiple growth cones. Each line represents paired probabilities for control vs. CK666; solid lines, experiments in which [CK666] = 50 μM; dashed lines, [CK666] = 20 μM. ngc = 9 paired growth cones; total number of beads nb = 592 (control), 247 (20 mM CK-666), and 342 (50 mM CK 666).
Site-directed actin assembly can propel beads associated with inductopodia: 1) against retrograde flow toward the leading edge, 2) laterally across the P domain, or 3) toward the C domain at velocities that can exceed retrograde flow (Forscher et al., 1992). To directly test a role for Arp2/3 activity in propulsive motility, we recorded 4E8 bead movements under control conditions or after inhibition of Arp2/3 activity and tracked their trajectories using a custom Matlab program. Tracks of 4E8 beads over a 25 min period under control conditions are shown in Figure 1D and the left panel of Supplemental Movie 2. While most of the bead tracks aligned with retrograde actin flow, some beads exhibited bouts of inductopodial motility, resulting in deviations from simple centripetal flow trajectories (e.g., arrow and dotted white line in Figure 1D). To test whether the nucleating activity of the Arp2/3 complex is required for this actin assembly–driven motility, we used the small molecule inhibitor CK-666, a specific small-molecule inhibitor of Arp2/3 (Nolen et al., 2009) employed previously in Aplysia growth cones (Yang et al., 2012). CK666 (50 μM) was applied to the same growth cone as in Figure 1D for 15 min before 4E8 beads + CK666 were washed in and motility recorded. Unlike control tracks, which exhibited a mixture of trajectories, all track trajectories in CK666 aligned with centripetal flow (Figure 1E and right side of Supplemental Movie 2).
To quantify motility, it was necessary to distinguish between beads in flow-coupled versus inductopodium-driven states. To this end, we used thresholds for bead-velocity angle and speed to extract flow-coupled velocities, then generated retrograde flow–velocity maps for each growth cone (Figure 1, D and E, insets; also Supplemental Figure 1A and Supplemental Methods). This flow extraction approach was validated by comparing flow maps of 4E8 beads, which exhibit both flow-coupled and actin assembly–driven movement, with those of ConA beads, which exhibit only flow-coupled movement and do not form inductopodia, despite the continued presence of intrapodia (Supplemental Figure 1, B and C). Inductopodial velocities (vind) over time were calculated by subtracting retrograde flow velocities (vflow) from total bead velocities (vtot); then motility states were classified and color coded (Figure 1F). Analysis of the bead track indicated by the dashed white line in Figure 1D (arrow) is shown in Figure 1, G–I, and Supplemental Movie 3. Under control conditions, beads often switched between flow-coupled states (blue) and propulsive states, exhibiting sustained bouts of anterograde (green) or lateral (yellow) movement (Supplemental Movie 3). Initially, the bead passively couples to flow (blue). Total bead velocity (Figure 1H, white vectors) was similar to the local flow velocity (black vectors) and vind was low (red vectors). Subsequently the bead transitioned to an active motile state, moving first laterally (yellow) and then toward the leading edge (green). As the bead moved laterally and deviated from the local flow trajectory, vtot slowed and vind continued to increase, eventually matching and exceeding local retrograde flow and resulting in net movement toward the leading edge. Note the continued presence of an actin tail in Supplemental Movies 2 and 3 associated with propulsive behavior. Finally, the bead became coupled to retrograde flow again and moved toward the transition zone (Figure 1, G and I, blue). Motility classifications for all tracked beads under control conditions are depicted in the left panel of Supplemental Movie 4. After Arp2/3 inhibition with CK666, beads exhibited predominantly flow-coupled motility, and sustained bouts of anterograde movement were absent in the P domain (Supplemental Movie 4, right panel).
We analyzed the probability of each classified motile state in the growth cone from Figure 2, A and B, as a function of its location (zones in Figure 1F) and Arp2/3 activity. The probability of observing non–flow coupled anterograde or lateral motility, that is, inductopodial movement, was highest in the most distal region (zone 1) under control conditions. Arp2/3 complex inhibition strongly decreased the probability of observing inductopodia (Figure 1J). Comparing inductopodial motility in control versus Arp2/3 inhibition with 20 or 50 μM CK666 in multiple growth cones revealed significantly reduced probability of active motility in zone 1, with a corresponding increase in flow-coupling probability (Figure 1K). Motility in the more proximal zones 2/3/4 did not exhibit a clear trend. Thus inductopodial formation and speed (vind) are dependent on branched actin network polymerization. These results suggest Arp2/3-dependent F-actin assembly is generating forces that drive inductopodium-based bead motility, particularly toward the leading edge in opposition to retrograde flow.
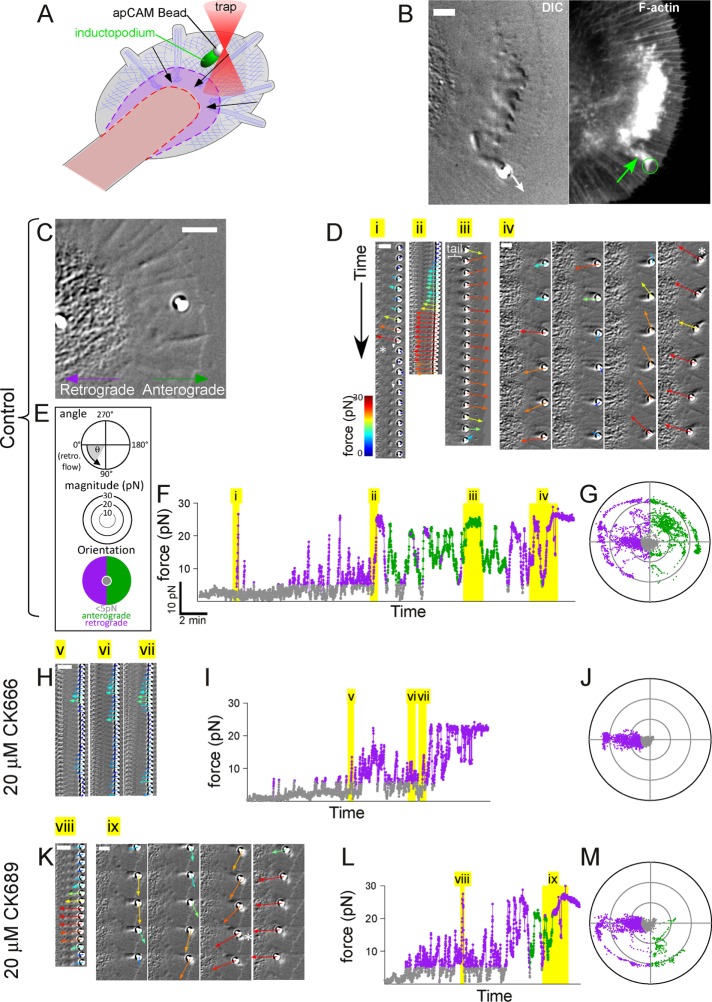
FIGURE 2:
Analysis of Arp2/3-dependent forces during apCAM adhesion formation using optical trapping. (A) Schematic of optical trapping approach to apply apCAM-coated beads. (B) DIC image (left) taken immediately before fixation and confocal phalloidin-stained fluorescent image (right) showing the actin tail of an inductopodium (green arrow) at an optically restrained bead. The direction of inductopodial movement is indicated by white arrow. Scale bar: 5 μm. (C) DIC image of growth cone at beginning of experiment (cf. Supplemental Movie 5). Retrograde and anterograde directions are noted and apply to subsequent panels. (D) Time montages of force vectors overlaid on DIC images from Supplemental Movie 5, control sequence. Each sequence corresponds to time periods highlighted in yellow in F denoted by roman numerals. The montage frames are displayed at 1 s intervals progressing downward in Di and Dii, and at 5 s intervals in Diii and Div. Asterisk in panel Di denotes breakage when the bead snapped to the center of the trap; white arrowheads, actin structure detaching from bead with flow. Bracket in Diii indicates inductopodium/tail. Asterisk in Div denotes protrusive advance of intrapodium in front of the bead as it becomes stably coupled to flow. (E) Scheme showing classification of forces (low, anterograde, retrograde) and color coding for subsequent panels. Force vector angles (θ) are relative to retrograde flow, defined as 0°; force vectors with magnitudes below a threshold of 5 pN (of all vector angles) are classified as low (gray). Force vectors with magnitudes > 5 pN are classified as anterograde (90° < θ < 270°, green) or retrograde (270° < θ < 90°; purple). (F) Force magnitude vs. time plot in control, color coded according to classification in E. (G) Polar plot distribution of force vector angles (θ) and magnitude (in pN) from the entire control recording. (H) Time montages of force vectors/DIC images in Arp2/3-inhibited (20 μM CK666) conditions (cf. Supplemental Movie 6). Each sequence (v, vi, vii, shown at 1 s intervals) corresponds to time periods highlighted in yellow. (I) Force magnitude vs. time plot in 20 μM CK666. (J) Polar plot distribution of force vectors in 20 μM CK666. (K) Time montage of force vectors/DIC images in the inactive analogue CK689 (20 μM; cf. Supplemental Movie 7). Time montage of 1 s (vii) and 5 s (ix) intervals corresponding to yellow highlighted areas of L. Asterisk, continued inductopodial protrusion during coupling. (L) Force vs. time plot of 20 μM CK869 recording. (M) Polar plot distribution of force vectors in 20 μM CK689. Scale bars (B, C, D, H, and K): 5 μm.
Forces associated with actin assembly near nascent apCAM adhesion sites depend on Arp2/3 complex activity
To measure forces generated by inductopodia, we used optical trapping, also referred to as optical tweezers, to apply and restrain apCAM beads and induce actin tail formation (Figure 2A). This technique provides low stiffness of restraint and force readout in the 10–70 pN/μm range (Block et al., 1989). After initial small fluctuations in the trap, likely reflecting transient coupling to flow, an inductopodium formed, and eventually the bead moved past the trap (Figure 2B). At this point, the cell was fixed and stained with phalloidin. As expected, the inductopodium evoked by optical restraint contained F-actin (Figure 2B, green arrow). To analyze the mechanical effects of site-directed actin assembly, we measured force development over time using closed-loop feedback that iteratively adjusts trap position to provide continuous force readout up to a maximum force threshold (Mejean et al., 2013). Cell-generated forces are plotted as color-coded vectors in the time-lapse image montages in Figure 2 and Supplemental Movies 5–7. Upon apCAM bead application, forces applied by the growth cone were initially negligible. After ∼2.5 min of optical restraint, traction force on the bead increased (Figure 2, D and F, cf. Supplemental Movie 5) and then returned to near zero (Figure 2D, asterisk). Interestingly, small DIC-refractive structures consistent with bursts of actin assembly were observed near the bead coincident with force reduction (white arrowheads). Later, retrograde forces again increased (Figure 2Dii); this was followed by forces reorienting to an anterograde direction and formation of a DIC-refractive inductopodium/tail behind the bead (Figure 2Diii, bracket). Outward forces associated with actin tail formation matched the maximal trapping force threshold of ∼30 pN, propelling the bead toward the leading edge for a period of ∼1.5 min (Figure 2Diii). For the rest of the recording, force magnitude and direction continued to fluctuate (Figure 2Div), until the bead became coupled to retrograde flow and was pulled consistently toward the C domain (Figure 2Div and Supplemental Movie 5, asterisk). To further characterize these events, we classified apCAM bead forces >5 pN as anterograde or retrograde according to their orientation (Figure 2, E and F) and plotted all force measurements using polar coordinates (Figure 2G). Under control conditions, a broad distribution of angular trajectories was observed as a result of the generally random nature of inductopodium-based bead propulsion (Forscher et al., 1992; Thompson et al., 1996; Figure 2, G and M). These results show that the net cellular force applied to apCAM beads fluctuates in magnitude and direction as a function of local actin polymerization dynamics.
Inductopodial motility was dependent on Arp2/3 activity (Figure 1); thus we investigated whether propulsive forces were Arp2/3 dependent as well. Following the control experiment above, the same growth cone was treated with 20 μM CK666 and a second optically restrained apCAM bead was applied (Figure 2, H–J, and Supplemental Movie 6). Traction forces were now consistently oriented in the direction of retrograde flow, and force reorientations to anterograde were absent. Under these conditions, no actin tail structures were observed (compare Figure 2H with Figure 2B). Washout of CK-666 into a solution containing the inactive drug analogue (CK869) resulted in reappearance of actin tail structures near apCAM beads and resumption of dynamic fluctuations in force amplitude and direction (Figure 2, K–M, and Supplemental Movie 7). Similar Arp2/3-dependent propulsive force measurements are shown for another growth cone in Supplemental Figure 2. Together these data strongly suggest that local Arp2/3-dependent actin assembly at apCAM bead binding sites leads to production of propulsive forces associated with inductopodial activity. Subsequently propulsive forces reorient and convert to traction as the bead again becomes coupled to flow.
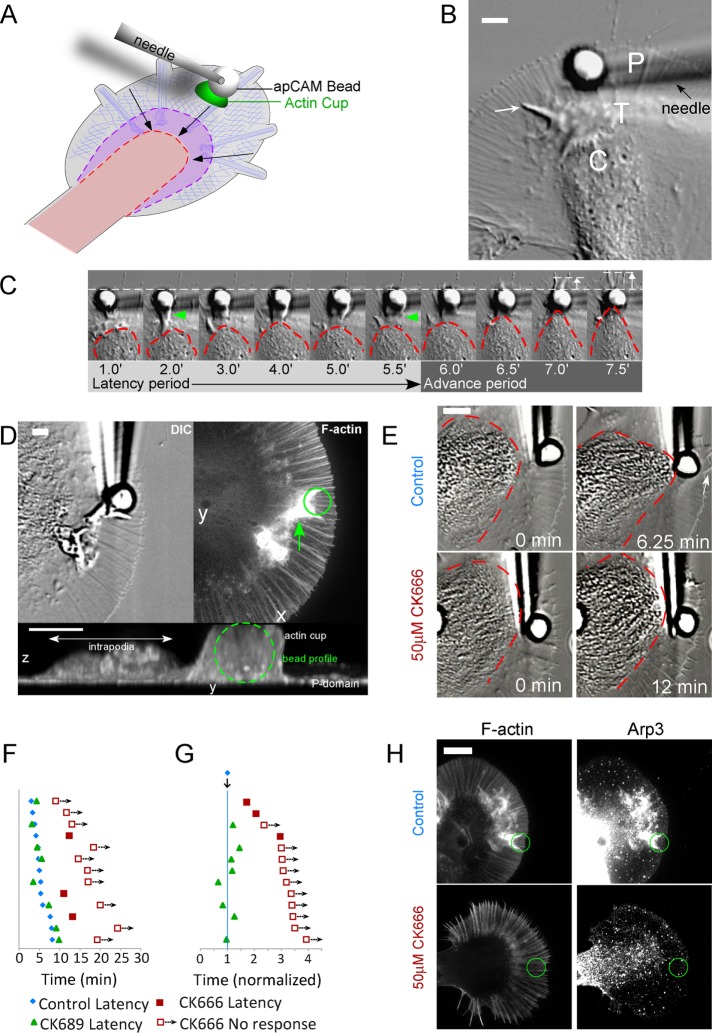
An extensive F-actin cup forms in response to stiffly restrained apCAM beads
The limited stiffness regime of optical trapping is not sufficient to elicit full-blown growth responses; stiffer restraint is required to resist the strong (>1 nN) forces generated during neurite elongation (Dennerll et al., 1989) and generate larger actin structures at the apCAM bead (Suter et al., 1998). To this end, we employed larger beads (5 μm diameter) restrained with a glass needle (Figure 3A), as previously described (Suter et al., 1998). We have referred to this approach as the “RBI” (restrained bead interaction) assay; here we will refer to these responses simply as “evoked growth.” We applied known external loads to measure the spring constant κ of glass needles, which was approximately 18 μN/μm. A typical apCAM bead–evoked growth response is shown in Figure 3, B and C, and Supplemental Movie 8. The growth cone C domain (dotted line in Figure 3C) and leading edge remained largely unchanged in shape during the initial latency period, which lasted 6 min. During this time, intrapodia (Rochlin et al., 1999) could be observed to propagate from the transition zone outward into the P domain (Figure 3B, arrow). In some cases, obvious actin tail-like structures formed on the proximal side of restrained beads (Figure 3C, green arrowheads). Actin tails were quite dynamic and often transient during the latency. Transition from the latency to the advance period was characterized by rapid advance of the central organelle-rich domain toward the restrained bead target (Figure 3C, right, and Supplemental Movie 8). Note that, after the 6.5 min time point, the leading edge of the P domain started to advance in front of the bead target (Figure 3C, white arrows).
FIGURE 3:
(A) Schematic of evoked growth assay elicited using stiff restraint of apCAM bead with glass needle. (B) DIC image taken from the beginning of evoked growth assay (cf. Supplemental Movie 8) showing 5 μm apCAM-functionalized bead being restrained by a glass needle. White arrow, intrapodium extending in the P domain. (C) Time montage of apCAM-evoked growth response of growth cone in B. Red dashed line outlines the shape of the C-domain; arrowheads indicate site-directed actin cup; dotted white line indicates original extent of P domain. Forward protrusion of P domain toward end of advance period is indicated by white arrow in front of bead. (D) F-actin accumulation at restrained apCAM target site. A growth cone was fixed at the end of the latency period (DIC, left panel) and stained with phalloidin (average projection of confocal z-stack, right panel; cf. Supplemental Movie 9). Bottom, y-z view of 3D confocal reconstruction taken from Supplemental Movie 9 showing height of F-actin cup in relation to 5 μm bead (outlined by dashed green circle). (E) CK-666 blocks apCAM-evoked growth. Top row, DIC images of the same growth cone in control conditions at the beginning (0 min) and end (6.25 min) of the evoked growth response. Bottom row, the same growth cone in the presence of 50 mM CK666 Arp2/3 inhibitor, at the beginning (0 min) and end (12 min) of the recording. Red dashed line indicates outline of the C domain. Scale bar: 5 μm. (F) Pictogram of evoked growth response times. Responses of individual growth cones in different conditions are shown in horizontally aligned groups showing the control latency (blue diamonds) and latency in 50 μM CK666 (filled red squares); for experiments in which no C-domain targeting or peripheral advance was observed, the length of the recording is noted (empty squares with arrows). A subset of growth cones were also challenged with restrained apCAM beads in the presence of the inactive analogue CK689 (green triangles). (G) The data from F are shown normalized to control latency times. (H) F-actin and Arp2/3 complex localization in control (top row) vs. 50 μM CK666 (bottom row). Growth cones were fixed at 0.5× latency and stained for F-actin with phalloidin (left panels) or Arp2/3 complex (right panels, anti Arp-3).
For characterization of the structure of the nascent apCAM adhesion site, growth cones were fixed during the latency period, and actin filament structure was visualized by phalloidin labeling (Figure 3D, right panel). A remarkably dense actin filament network was associated with the side of the bead, and a tail-like actin projection extended proximally (green arrow). The bottom panel of Figure 3D is a y-z projection of the same field; note that the actin filament structure associated with the bead has a vertical dimension similar to the 5 μm bead and is an order of magnitude thicker than the nearby P domain. Three-dimensional (3D) reconstruction revealed the actin filament structure associated with the bead is cup shaped, conforming to the spherical shape of the apCAM coated bead (Supplemental Movie 9) and that the proximal actin filament “tail” extends from the bead into the transition zone (Figure 3D, arrow). Actin tail formation during the latency and associated bead propulsion has been correlated with longer latency periods during RBI experiments (Suter et al., 1998). We confirmed the presence of Arp2/3 complex in the actin cup by immunostaining (Figure 3H, top right panel); the Arp 2/3 complex displayed much higher intensity near the bead than anywhere else, including the leading edge. These results suggest that Arp2/3 nucleation of branched actin filaments is involved in actin cup formation during the latency period.
Arp2/3 activity is required for actin cup generation during apCAM-evoked growth responses
To investigate a role for Arp2/3 complex activity in evoked growth, we challenged growth cones with needle-restrained apCAM beads in various Arp2/3 backgrounds. The top row of Figure 3E shows the beginning (0 min) and end (6.25 min) of an apCAM-evoked growth response under control conditions; the latency period was 5 min, and the advance phase was 1.25 min, during which time the P domain advanced distal to the bead (Figure 3E, arrow). After a recovery period, cells were pretreated with 50 μM CK666 for 20 min, and then a second restrained bead was applied to the same growth cone (Figure 3E, bottom row). There was no evidence of C- or P-domain advance observed during the 12 min test period—more than double the duration of the latency period observed in the control. Supplemental Figure 3A shows C-domain boundaries for multiple growth cones under control conditions (left), in the presence of an inactive CK666 analogue (CK689; center), and during Arp2/3 inhibition by CK666 (right). Note the absence of C-domain narrowing and advance toward the apCAM bead in CK666. Figure 3F is a latency period plot for multiple individual growth cones under control conditions (blue diamonds) or CK689 (green triangles). After Arp2/3 inhibition with CK666, no targeting response/advance was observed by the times indicated (red open squares) for the majority of trials; in the other cases observed, the latency period was longer (filled squares). Normalizing responses to the control latency period (Figure 3G) shows that while latencies in inactive CK869 were similar to control, most growth cones failed to mount growth responses in CK-666, even when bead restraint times were extended to more than twice the control latency period. Beads released from restraint in CK-666 conditions immediately coupled to retrograde flow (unpublished data), indicating drug treatment did not interfere with apCAM binding. The probability of growth responses at various latency times under the various treatment conditions used are summarized in Table 1.
TABLE 1:
Inhibition of apCAM-evoked growth responses in the presence of Arp2/3 or Rac inhibition.
| Normalized time (t) | ||||||
|---|---|---|---|---|---|---|
| Perturbation | 1.0× | 1.5× | 2.0× | 2.5× | 3.0× | 3.5× |
| 50 μM CK666 | 0% (n = 13) | 0% (n = 13) | 8% (n = 13) | 17% (n = 12) | 25% (n = 12) | |
| 100 μM CK689 | 38% (n = 8) | 100% (n = 8) | ||||
| 100 μM NSC23766 | 0% (n = 25) | 13% (n = 24) | 16% (n = 19) | 21% (n = 14) | 30% (n = 10) | 44% (n = 9) |
| 5 μM EHT 1864 | 12% (n = 17) | 25% (n = 17) | 53% (n = 17) | 59% (n = 17) | 59% (n = 17) | |
Each column indicates normalized time (t). Responses are noted as probability at normalized time, P(t) (see Materials and Methods).
In parallel experiments, we investigated the effects of Arp2/3 inhibition on Arp2/3 complex distribution and actin structure near restrained bead binding sites by fixing growth cones at the end of the latency period and using retrospective immunocytochemistry. Normal accumulation of F-actin near the bead binding site (Figure 3H, top left panel) was strongly suppressed in the presence of 50 mM CK666 (bottom left panels). Similarly, CK666 blocked accumulation of Arp2/3 complex (Figure 3H, compare top and bottom right panels).
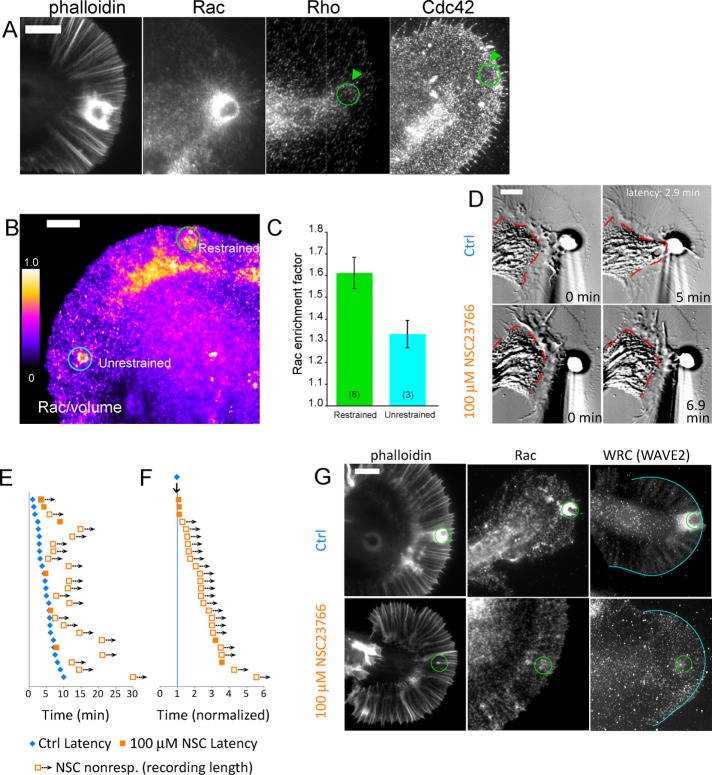
Rac is enriched at apCAM target sites
Given the presence of Arp2/3 and actin networks at apCAM beads and the well-known roles of Rho GTPase family proteins in subcellular control of F-actin structures, we investigated whether Rho, Rac, and Cdc42 accumulate near bead target sites during the latency period. High levels of Rac immunofluorescence were observed near restrained beads, consistent with a possible role for Rac in actin cup formation (Figure 4A). In contrast, Rho protein was observed predominantly in the C domain but did not appear to be enriched at bead target sites. Similarly, Cdc42 protein did not exhibit accumulation at target beads during the latency (Figure 4A).
FIGURE 4:
Rac accumulates at apCAM target sites, and its activity is required for evoked growth responses. (A) Growth cones were challenged with needle-restrained apCAM beads, fixed at the end of the latency period, and stained for F-actin (phalloidin) or with antibodies to Rac, Rho, or Cdc42 and fluorescent secondary antibodies. (B) Rac1 enrichment at apCAM target adhesions is enhanced by force restraint. Bag cells were injected with fixable Texas red–dextran before apCAM target assay, then fixed during latency and probed with monoclonal Rac1 antibody and fluorescent secondary antibodies for volume-corrected ratio imaging. Positions of restrained and unrestrained beads are noted. (C) Rac enrichment is enhanced at force-restrained beads. Rac enrichment factor is calculated by dividing the Rac/volume ratio of either a restrained bead or unrestrained bead by the Rac/volume ratio of nonbead P-domain region of interest. Numbers in parentheses denote number of experiments. Error bars are SEM. (D) Top row, DIC images of control-evoked growth assay at beginning (0 min) and end (5 min) of the evoked growth response. Bottom row, DIC images of the same growth cone subjected to restrained apCAM bead in media containing 100 μM NSC23766 Rac inhibitor. Red dashed line outlines the C domain. (E) Pictogram of evoked growth responses of individual growth cones in control and Rac-inhibited conditions, plotted as in Figure 3F, showing control latency period (blue diamonds), latency of growth cone responses in 100 μM NSC23766 (filled orange squares), or length of recordings in which no growth response was observed (empty squares). (F) Responses from E shown normalized to control latencies. (G) Rac activity is required for F-actin cup and localization of Rac and WRC protein. Growth cones were fixed at 0.5× latency and stained for F-actin (phalloidin), Rac, or WRC (WAVE2 antibody) in the absence (top row) or presence of 100 mM NSC23766 (bottom row). Scale bars: 10 μm (A and B); 5 μm (D and G).
Actin polymerization occurs near the target site early in the latency period. For testing whether Rac activity is important for this process, apCAM beads were applied to growth cones, and the neurons were fixed early in the latency period after 1–3 min of bead restraint. Unrestrained apCAM beads that underwent retrograde movement were used as force-free controls. For assessing local Rac protein enrichment and account for path length differences, which can vary by more than an order of magnitude (Grzywa et al., 2006), Rac intensities were divided by that of a fixable fluorescent dextran volume marker, and local enrichment was quantified relative to nonbead areas in the P domain. Rac enrichment was ∼30% higher at restrained bead sites when compared with unrestrained control beads (Figure 4C). These results suggest that application of restraining force during the latency period increases Rac protein recruitment to apCAM–bead interaction sites. It should be noted that unrestrained beads are in the P domain for only ∼3 min before being cleared by retrograde flow; thus they do not have as long to interact with the grown cone as restrained beads. On the basis of these findings, we further investigated the role of Rac activity in apCAM-evoked neurite growth.
Rac activity is required for apCAM-evoked growth
To address the role of Rac signaling, we assessed the kinetics of apCAM bead interactions under control conditions and in the presence of NSC23766, a small-molecule inhibitor that disrupts Rac interactions with GEFs Tiam1 and Trio (Gao et al., 2004). Figure 4D shows a growth cone at the beginning (0 min) and completion (5 min) of a control growth response with a latency phase of 2.9 min. After incubation with 100 μM NSC23766, apCAM beads became coupled to retrograde flow; however, no advance of the C (red dashed line) or P domains was observed during 6.9 min of bead restraint, which is more than twice the control latency time (Figure 4D, bottom row). Although growth cone retrograde flow and morphology were not perturbed by Rac inhibition, consistent with observations of growth cones injected with dominant negative Rac (Zhang et al., 2003), evoked growth responses were strongly suppressed (Figure 4, E and F, and Supplemental Figure 3B). In 22 out of 25 trials, the growth response was delayed or absent after Rac inhibition. In NSC23766, no growth cones responded faster than controls, and less than 30% of growth cones responded in two to three times the normalized latency period (Table 1). Similar results were obtained with EHT 1864, a biochemically distinct Rac inhibitor that displaces GTP and inhibits nucleotide exchange (Shutes et al., 2007; Table 1). These results demonstrate a requirement for Rac activity during growth responses to apCAM substrates.
To test whether Rac activity is required for formation of the F-actin cup around restrained apCAM beads, we compared F-actin levels in growth cones fixed during the latency period under control conditions or in the presence of NSC-23766 (Figure 4G). Site-directed actin assembly near the bead binding site (Figure 4G, top left) was strikingly absent after Rac inhibition (Figure 4G, bottom left). Similar results were observed using EHT 1864 (unpublished data). Active Rac targets to the plasma membrane, where it can activate Arp2/3 via binding WRC (Lebensohn and Kirschner, 2009). We found that Rac inhibition blocked accumulation of both Rac and WRC at restrained apCAM beads (Figure 4G, center and right columns). Thus Rac activity plays an instructive role at the apCAM adhesion to activate WAVE and Arp2/3 to build the actin cup during the latency period.
DISCUSSION
Mechanistic scenarios for apCAM bead targets interacting with retrograde actin flow are shown in Figure 5. When an unrestrained apCAM bead is initially placed in the growth cone P domain, viscous coupling to actin flow via apCAM-apCAM interactions results in retrograde bead transport (Figure 5i; Lin and Forscher, 1995). If the bead is mechanically restrained, the resulting traction force ftraction causes elastic deformation of the bead–actin linkage and bead rotation (curved arrow in Figure 5ii; Mejean et al., 2013), leading to increased bead–membrane contact area. Classical integrin adhesions have been observed to undergo similar rotational moment under contractile stress in fibroblasts (Legant et al., 2013). An interesting consequence of the increased contact area is that local actin assembly will also be spatially biased, later resulting in propulsion of beads in the opposite direction with respect to retrograde flow (green arrow, Figure 5iii) through a thermal ratchet mechanism (Peskin et al., 1993). This is consistent with previous observations of a directional bias in inductopodial formation during restrained apCAM bead–target interactions (Suter et al., 1998) and our observation that the highest probability of propulsive movement is in the anterograde direction (Figure 1J). Thus viscoelastic coupling to retrograde flow promotes symmetry breaking and a sense of directionality to the nascent adhesion. One emergent theme from studies of cell migration is the “chemical compass” (Weiner, 2002), in which locally regulated molecular activities/states provide “front” versus “back” cell polarization during migration in response to gradients of extracellular chemokines. We propose that, by biasing actin assembly toward the proximal bead surface (Figure 5, ii and iii), retrograde flow acts as a kind of “mechanical compass” that informs the nascent adhesion of directionality.
FIGURE 5:
Schematic depicting apCAM bead interactions with retrograde flow and site-directed actin assembly events. See Discussion for details.
The above direction sensing leads the anterograde bias of inductopodial propulsion we observed (Figures 1J and 2F) and a significant mechanical effect: local actin assembly forces will tend to cancel out traditional traction forces. Specifically, when the rate of actin polymerization (Figure 5iii, vpoly) is greater than retrograde flow speed (vflow), a net propulsive force (fpoly) results; moreover, the adhesion site is isolated from the normal effects of traction force. Measurements of protrusive forces powered by actin polymerization in advancing lamellipodia and filopodia have been reported (Cojoc et al., 2007; Shahapure et al., 2010); but, to our knowledge, this is the first study to assess how propulsive forces associated with site-directed actin assembly (inductopodia) could modulate traction force production at adhesion sites. Note that propulsive forces associated with in vitro actin assembly can be significant, extending into the nanonewton range (Marcy et al., 2004); thus it is not surprising that inductopodium-associated forces often exceeded the 30 pN optical trapping–force restraint limit used in this study.
How do the current findings affect interpretation of our previous results? We previously provided evidence that growth cone advance is inversely related to retrograde flow rates and involves engagement of a “molecular clutch” to generate traction forces (Lin and Forscher, 1995; Suter et al., 1998; Suter and Forscher, 2000). Frictional coupling between retrograde actin flow and cell adhesion complexes has been the mechanistic focus in studies that invoke the molecular clutch model of substrate–cytoskeletal coupling (Mitchison and Kirschner, 1988; Jay, 2000; Suter and Forscher, 2000; Nishimura et al., 2003; Bard et al., 2008; Chan and Odde, 2008; Shimada et al., 2008; Giannone et al., 2009; Myers and Gomez, 2011; Thievessen et al., 2013; Gomez and Letourneau, 2014; San Miguel-Ruiz and Letourneau, 2014; Yamashiro and Watanabe, 2014; Garcia et al., 2015; Nichol et al., 2016). Here we show that apCAM adhesions can fluctuate between states dominated by traditional traction forces and forces derived from site-directed actin assembly. We provide evidence that inductopodial motility depends on Arp2/3 complex activity localized to bead target sites (Figure 2, G–M) that can buffer the adhesion site from the mechanical effects of traction force. It follows that a possible mechanism for increasing traction force (ftraction) would be to attenuate local Arp2/3-dependent actin assembly (Figure 5iv) and/or move the locus of actin assembly away from the mature apCAM adhesion site (Figure 2, Div and Kix). Attenuation of local actin assembly may be a normal part of the apCAM maturation process and may be analogous to what occurs at the end of the latency phase in a restrained bead–target interaction. Attenuation of actin assembly would also result in cessation of mechanical buffering effects and traction force generation as adhesion sites mature (Figure 5iv). We speculate that growth cones moving on extended apCAM substrates, such as those found in vivo, use site-directed actin assembly to generate protrusive forces when vpoly > vflow.
In the presence of stiff mechanical restraint, the actin structure at the bead grows much more extensive as the adhesion site matures (Figures 1 and 5iv and Supplemental Movie 9). Eventually, strong traction forces develop, retrograde actin flow slows, and growth cones advance (Lin and Forscher, 1995; Schaefer et al., 2008). We speculate that the temporary mechanical isolation from traditional traction forces we describe here is a normal part of IgCAM adhesion site maturation that facilitates assembly of actin structures robust enough to handle the nanonewton-level forces typical of growth cone–axon towing (Dennerll et al., 1989). Site-directed actin assembly has also been observed at clustered ICAM-1 docking sites during transendothelial migration of neutrophils (van Rijssel et al., 2012) and in vertebrate neurons at restrained beads engaging N-cadherin (Bard et al., 2008), suggesting that this phenomenon may be more widespread. Potential mechanical effects of local actin assembly in these systems and the mechanical and/or signaling mechanism(s) involved in switching from mechanical buffering to molecular clutch engagement are interesting questions for future studies.
Rac and Arp2/3 complex activity are necessary for formation of the robust actin cups characteristic of mature apCAM adhesion sites (Figures 3 and 4). Rac, WAVE, and Arp2/3 complex activity has also been extensively characterized in the context of lamellipodial extension (Weiner et al., 2007; Millius et al., 2012) and neurite outgrowth and pathfinding (Yuan et al., 2003; Woo and Gomez, 2006; San Miguel-Ruiz and Letourneau, 2014). When bag cell growth cones maintained on a flat surface are presented with an apCAM-coated bead, they initiate vertical actin-based protrusions that track the bead surface (Figure 3D and Supplemental Movie 9). The protrusive forces that drive formation of the resulting cuplike actin structure may be analogous to those used by growth cones for exploration of 3D space in situ. At the very least, our results show that the entire growth cone P domain is capable of generating robust 3D actin-based protrusive activity when presented with the appropriate signal.
Peripheral actin networks in growth cones exhibit dynamics based not only on the constant flux and turnover of polymer that constitutes retrograde flow but also, as we have shown here, on their ability to initiate a new structure, the inductopodium, whose dynamics are superimposed on underlying actin network flow. The presence of Arp2/3-dependent branched actin structures within inductopodia could endow them with useful biomechanical properties. For example, increasing branching and/or capping of Arp2/3 networks increases network stiffness without large changes in filament density (Pujol et al., 2012), and local curvature of actin filaments under compressive load has been shown to bias branch formation in the direction of compression, leading to local reinforcement and adaptation (Risca et al., 2012). In related studies, application of nanonewton-level pulling forces on polymerizing branched F-actin comets increased comet elongation rates, whereas compressive forces reduced but did not halt elongation, even at 4.3 nN force levels (Marcy et al., 2004). Taken together, these observations suggest that the presence of branched actin networks in IgCAM adhesion sites may endow them with unique biophysical attributes, including the ability to adapt to changes in internal force and substrate compliance.
MATERIALS AND METHODS
The following reagents with catalogue numbers were acquired from the indicated vendors: EHT1864 #E1657 (Sigma-Aldrich, St. Louis, MO); monoclonal mouse anti-Rac1 #ARC-03, polyclonal rabbit anti-Rac #ARC-01, polyclonal rabbit anti-Rho #ARH-02, monoclonal mouse anti-Cdc42 #ACD-03 (Cytoskeleton, Denver, CO); Alexa Fluor 594 Phalloidin #A12381, Alexa Fluor 488 Phalloidin #A12379 (ThermoFisher Scientific, Waltham, MA); monoclonal mouse anti-apCAM clone 4E8 (gift from E. Kandel, Columbia University); CK666 #182515, CK689 #182517, polyclonal rabbit anti-p34 Arc/ARPC2 #07-227, monoclonal mouse anti-cortactin (p80/85, clone 4F11) #05-180 (EMD Millipore, Billerica, MA), rabbit monoclonal anti-WAVE2 (clone D2C8) XP #3659 (Cell Signaling Technology, Danvers, MA).
Cell culture
Bag cell ganglia were dissected from adult Aplysia (obtained from National Resource for Aplysia at University of Miami, FL, or Marinus Scientific, Garden Grove, CA) as previously described (Forscher and Smith, 1988; Suter et al., 1998). Individual bag cells were plated on acid-washed, round 25 mm diameter #1 glass coverslips (VWR, Radnor, PA; for evoked growth assays) or 22 mm square #1 coverslips (ThermoFisher Scientific; for inductopodia and optical trapping assays) coated with 30–70 kDa molecular-weight polylysine (Sigma) and kept at 14°C overnight before experiments the next day. Laminin (Gemini Bio-Products, West Sacramento, CA) was added to cultures for trapping experiments (see section Force profiling with holographic optical tweezers (HOT) below). Bag cells were maintained in Leibowitz's L-15 medium supplemented with ASW, pH 7.9 (L15-ASW; Forscher and Smith, 1988), or a 50:50 mixture of L15-ASW:low ionic strength ASW (LIS-ASW, composed of 100 mM NaCl, 10 mM KCl, 5 mM MgCl2, 15 mM HEPES, 5 mM CaCl2, 628 mM Betaine, pH 7.9); both formulations were supplemented with 0.01% heat-inactivated fetal bovine serum (FBS; Gemini Bio-Products).
Bead preparation
Nickel-NTA derivatized beads (Micromod) of diameter 5 μm (for RBI/evoked growth assays) or 2 μm (for optical trapping assays) were functionalized with recombinant His-tagged apCAM protein as previously described (Suter et al., 2004). For inductopodia experiments, a 1% (wt/vol) stock of 800 nm silica beads coupled to protein A (Micromod) was functionalized in 0.5 mg/ml anti-apCAM antibody 4E8 as previously described (Thompson et al., 1996); control beads with concanavalin A (ConA) surfaces were made by incubating 0.54 μm streptavidin silica beads (Bangs Labs) with 2.5 mg/ml biotin–ConA (Sigma-Aldrich) in 1x ASW. Beads were diluted in imaging medium containing a low concentration of FBS/bovine serum albumin (BSA) to block nonspecific binding, and washed as described below.
Evoked growth assay
RBI assays were performed as previously described (Suter et al., 1998) using 5 μm diameter Ni-NTA/his-apCAM beads. Before the experiments, culture dishes were mounted into custom-machined chambers and washed with culture medium supplemented with 2 mg/ml BSA to block nonspecific binding. To avoid potential acute effects of serum addition/withdrawal, FBS was maintained at all times at a concentration of 0.01%. Nickel-NTA/apCAM beads were stored at 1% (wt/vol) in an excess of his-apCAM (300 μM, in PBS); immediately before evoked growth assay, beads were diluted 1:400 in 0.5x PBS and rinsed twice by spinning 3 min at ∼1000 x g in a tabletop centrifuge; this was followed by two to three rinses in culture medium before being added to culture chambers. For manipulating beads, a glass microneedle was made from a glass capillary microdispenser (Drummond Scientific) using a PB-10 two-stage puller (Narishige International, East Meadow, NY). The needle was secured to a hydraulic XYZ micromanipulator (Siskiyou Devices, Grants Pass, OR) that was mounted onto the microscope stage. For estimation of typical needle stiffness, a micromanipulator was used to depress the needle onto an electronic balance, and the force versus displacement relationship was fitted for seven needles and averaged.
Video microscopy was performed on a Zeiss Axiovert 10 with a 63×/1.40 NA objective as previously described (Forscher and Smith, 1988; Lin and Forscher, 1993, 1995). Video-enhanced DIC images were acquired at 5 s intervals, background subtracted and averaged on-the-fly by a 151-AT video processor (Image Technologies), and recorded on optical media. Video output was captured and digitized onto a PC equipped with a video-capture card (MiroVideo). Alternatively, background-subtracted, time-lapse DIC images were acquired from a CoolSnap charge-coupled device (CCD) camera (Photometrics) via a PC running Micromanager (micromanager.org; Edelstein et al., 2014) or a custom Matlab (MathWorks) function and stored as TIFF stacks.
Inductopodial motility assay
Before experiments, 22 mm square coverslips with bag cells were mounted in imaging chambers, and the medium was replaced with filtered imaging media, consisting of LIS-ASW supplemented with 2 mg/ml BSA, 2 mg/ml carnosine, and 0.01% FBS. The 800 nm 4E8 beads were diluted 1:10, and ConA beads were diluted 1:100 in imaging medium and washed by spinning at 4000 x g for 5–10 min and sonicated briefly with a probe sonicator for 5–10 min, resuspended and sonicated briefly with a probe sonicator (VirTis, Gardinier, NY) with 10–20 short pulses at low power (setting 2) prior to washing 150 µl (approximately one chamber volume) of resuspended beads into the chamber. Imaging was performed on a Nikon microscope equipped with 100× oil objective, a zoom lens set to 1.0 or 1.5×, and a CoolSnap CCD camera (Photometrics). After beads started landing on the growth cone, DIC images were acquired using Micromanager at 1 frame/s for ∼1500 frames/25 min (the recording length was adjusted depending on the landing rate of beads). Experiments were recorded first in control conditions (media containing 0.005% dimethyl sulfoxide [DMSO]); this was followed by wash-in and incubation of 20 or 50 μM CK-666 for 15–20 min before addition of beads and recording. Tracking and computational analysis of bead tracks are described in the Supplemental Methods.
Force profiling with holographic optical tweezers (HOT)
To increase the probability of inductopodial formation in optical trapping experiments with single beads, we added 1.25 μg/ml murine laminin (Sigma) to the medium after cell attachment overnight to coat the surface, and unbound laminin was washed out the following morning. Arp2/3-complex branched actin networks and intrapodial formation are enhanced when growth cones are cultured in the presence of laminin (our unpublished observations). Before experiments, 22 mm square coverslips were mounted in imaging chambers, and the medium was replaced with filtered imaging medium, consisting of LIS-ASW supplemented with 2 mg/ml BSA, 2 mg/ml carnosine, and 0.01% FBS. Two-micrometer Ni-NTA–derivatized polystyrene beads (Micromod) were functionalized with recombinant his-apCAM (as in Suter et al., 2004), washed, and suspended in imaging medium. We used a custom-built system combining holographic optical tweezers (HOT) with a Nikon Ti inverted microscope equipped with a confocal spinning disk (Yokogawa; described in Mejean et al., 2013). A 1064 nm laser was used to illuminate a spatial light modulator (SLM) to generate trap holograms; holographic control of the SLM was provided by a computer running gui_feedback, a custom Matlab graphical user interface (GUI). A second computer running iQ software (Andor) was used to acquire DIC images. A dichroic mirror was used to direct near-infrared brightfield images to a Marlin camera on the back port of the microscope (these images were used by gui_feedback to track bead positions), while visible light was directed to the confocal head and an iXon camera for DIC time-lapse images of beads and cell morphology.
To monitor force development, we used a closed-loop feedback protocol (Mejean et al., 2009) in which the trapped beads were placed on the surface of growth cones by focusing the microscope stage. Once the program detected that the tracked bead had moved beyond a threshold displacement (set by the user to be equivalent to the maximum linear extent of the trap), the trap positions were iteratively adjusted to maintain the trap-versus-bead displacement, and hence the force, at the trap maximum. In the event the cytoskeletal linkage broke, the trap was moved back to its original position. By using a smaller region of interest for tracking and limiting the GUI display refresh rate, we were able to reduce loop cycles to ∼0.4–0.8 s. Recorded trap and bead positions were stored in a Matlab file; after completion of the experiment, force-versus-time profiles were calculated and plotted as previously described (Mejean et al., 2009). For displaying vectors on DIC time-lapse movies or montages, position and force data time points were selected that were closest to the DIC acquisition frame rate. A frame offset was manually adjusted during analysis to synchronize the Matlab data to the DIC frames. Vectors encoding force magnitude and direction were overlaid on DIC TIFF images using a custom Matlab script. Time montages and movie files (.avi or .mp4 format) were created in ImageJ; vectors were redrawn in Canvas X for visibility/clarity on the montages. Movie files were converted to quicktime .mov format and compressed using MOVAVI video editor (www.movavi.com).
Pharmacological inhibition and quantification of inhibitory effects
For HOT experiments, one or multiple trapped beads were placed on the surface of growth cones by focusing the stage and were allowed to contact and adhere to the surface while the force profile was measured over time. Following the control recording, approximately five chamber volumes of medium containing 20 or 50 μM CK666 was washed through the chamber and allowed to incubate 20 min before addition of new beads and subsequent force-profiling experiments. For some experiments, the active drug was washed out into control medium or into inactive analogue (CK689) for recovery force profiling.
For evoked growth assay, growth cones were first subjected to the restrained beads in control conditions (DMSO) to determine the control latency time, defined by the elapsed time between the beginning of bead restraint to the start of concerted C-domain forward targeting. Following control recordings, drug was washed through the chamber to an appropriate final concentration (100 μM NSC23766, 5 μM EHT 1864, 50 μM CK666, or 100 μM CK689) and allowed to incubate for 20 min before a second bead was restrained in the continued presence of drug and the experimental latency was measured. The experimental latency was normalized by dividing by the control latency; if no growth response was observed, the recording length was normalized in the same way. The probability of interaction in drug treatments was calculated at different normalized times (t) (e.g., 1.0× latency, 1.5× latency) by the formula
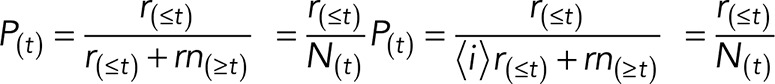 |
where P(t) is the probability of interaction at normalized time (t) expressed as a percentage, r(≤t) is number of growth cones exhibiting evoked growth responses with normalized latencies equal to or less than (t), and nr(≥t) is the number of quiescent nonresponding growth cones recorded for a normalized duration equal to or greater than (t). The denominator term, r(≤t) + nr(≥t), is equivalent to the total number of experiments N(t), performed at (t).
Immunofluorescence
Bag cells undergoing target interaction experiments were fixed by quick exchange with 3.7% formaldehyde in 1× ASW, 400 mM sucrose. Cells were fixed for 20 min, permeabilized with 0.1% saponin in fixative 15 min, and washed three times with 0.01% saponin in phosphate-buffered saline (PBS; wash solution). For experiments with labeling of F-actin, Alexa Fluor 594– or 488–conjugated phalloidin (Invitrogen) was applied at 1 U/ml in PBS for 15 min. Following three washes and blocking with 5% BSA in PBS/0.01% saponin for 30 min, primary antibodies were incubated at 1:100 for 1 h, washed three more times, and secondary antibodies conjugated to Alexa dyes were incubated an additional 1 h. After being rinsed, samples were mounted in 9% MOWIOL/90 mM Tris (pH 8.5) and imaged on a Nikon TE-200 microscope equipped with a 60×/1.3 NA oil-immersion objective with a 1.5× zoom lens, CoolSnapHQ CCD camera (Photometrics), and epifluorescence illumination with appropriate excitation and emission filters for fluorescein isothiocyanate (FITC) and tetramethylrhodamine isothiocyanate (TRITC) (Chroma Technology). Image acquisition, background subtraction, and analysis were performed with MetaMorph. For purposes of comparison between conditions (pairs of control vs. NSC23766-treated growth cones), the same acquisition parameters (magnification, exposure time, neutral density) and image display (background subtraction, image scaling) were applied to both images in a pair. For volumetric correction, bag cells were injected with fixable Texas red–conjugated dextran (10 kDa molecular weight; Invitrogen) before experiments. Rac1 staining and volume images were background subtracted, and a cell mask was created from the volume channel before dividing the Rac1 signal by the volume signal to yield the corrected image.
Supplementary Material
Acknowledgments
We thank Rostislav Boltyanskiy, Alla Shundrovsky, and Cecile Mejean for hardware and software development, technical assistance, and guidance with operation of the optical trapping system; Omer Mano for helpful discussions during development of flow-tracking/mapping code; and members of the Dufresne and Forscher groups for supportive discussion. This work was supported by a National Science Foundation Research Instrumentation grant (DBI-0619674) to P.F. and E.R.D., a National Institutes of Health (NIH) award (R01NS028695) to P.F., and an NIH fellowship (NS054487) to K.B.B.
Abbreviations used:
- apCAM
Aplysia cell adhesion molecule
- BSA
bovine serum albumin
- CAM
cell adhesion molecule
- C domain
central domain
- ConA
concanavalin A
- 3D
three-dimensional
- DIC
differential interference contrast microscopy
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- fflow
force associated with flow coupling
- find
force associated with inductopodial actin assembly
- ftraction
traction force
- HOT
holographic optical tweezers
- Ig
immunoglobin
- IgCAM
immunoglobin superfamily cell adhesion molecule
- NPF
actin nucleation promotion factor
- PBS
phosphate-buffered saline
- P domain
peripheral domain
- RBI
restrained bead interaction
- SLM
spatial light modulator
- T zone
transition zone
- vflow
rate of retrograde flow
- vind
rate of inductopodial site-directed actin assembly
- vtot
total bead velocities
- WAVE
WASP-family verprolin-homologous protein
- WRC
Scar/Wave regulatory complex.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-04-0228) on November 16, 2016.
REFERENCES
- Bard L, Boscher C, Lambert M, Mege RM, Choquet D, Thoumine O. A molecular clutch between the actin flow and N-cadherin adhesions drives growth cone migration. J Neurosci. 2008;28:5879–5890. doi: 10.1523/JNEUROSCI.5331-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM, Blair DF, Berg HC. Compliance of bacterial flagella measured with optical tweezers. Nature. 1989;338:514–518. doi: 10.1038/338514a0. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–1691. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- Cojoc D, Difato F, Ferrari E, Shahapure RB, Laishram J, Righi M, Di Fabrizio EM, Torre V. Properties of the force exerted by filopodia and lamellipodia and the involvement of cytoskeletal components. PLoS One. 2007;2:e1072. doi: 10.1371/journal.pone.0001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennerll TJ, Lamoureux P, Buxbaum RE, Heidemann SR. The cytomechanics of axonal elongation and retraction. J Cell Biol. 1989;109:3073–3083. doi: 10.1083/jcb.109.6.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using muManager software. J Biol Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn KC, Hellal F, Neukirchen D, Jacob S, Tahirovic S, Dupraz S, Stern S, Garvalov BK, Gurniak C, Shaw AE, et al. ADF/cofilin-mediated actin retrograde flow directs neurite formation in the developing brain. Neuron. 2012;76:1091–1107. doi: 10.1016/j.neuron.2012.09.038. [DOI] [PubMed] [Google Scholar]
- Forscher P, Lin CH, Thompson C. Novel form of growth cone motility involving site-directed actin filament assembly. Nature. 1992;357:515–518. doi: 10.1038/357515a0. [DOI] [PubMed] [Google Scholar]
- Forscher P, Smith SJ. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J Cell Biol. 1988;107:1505–1516. doi: 10.1083/jcb.107.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier MF, Sauser R, Ambrosi D, Meister JJ, Verkhovsky AB. Force transmission in migrating cells. J Cell Biol. 2010;188:287–297. doi: 10.1083/jcb.200906139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Leduc C, Lagardere M, Argento A, Sibarita JB, Thoumine O. Two-tiered coupling between flowing actin and immobilized N-cadherin/catenin complexes in neuronal growth cones. Proc Natl Acad Sci USA. 2015;112:6997–7002. doi: 10.1073/pnas.1423455112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Mege RM, Thoumine O. Multi-level molecular clutches in motile cell processes. Trends Cell Biol. 2009;19:475–486. doi: 10.1016/j.tcb.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Goldberg DJ, Burmeister DW. Stages in axon formation: observations of growth of Aplysia axons in culture using video-enhanced contrast-differential interference contrast microscopy. J Cell Biol. 1986;103:1921–1931. doi: 10.1083/jcb.103.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TM, Letourneau PC. Actin dynamics in growth cone motility and navigation. J Neurochem. 2014;129:221–234. doi: 10.1111/jnc.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzywa EL, Lee AC, Lee GU, Suter DM. High-resolution analysis of neuronal growth cone morphology by comparative atomic force and optical microscopy. J Neurobiol. 2006;66:1529–1543. doi: 10.1002/neu.20318. [DOI] [PubMed] [Google Scholar]
- Hyland C, Mertz AF, Forscher P, Dufresne E. Dynamic peripheral traction forces balance stable neurite tension in regenerating Aplysia bag cell neurons. Sci Rep. 2014;4:4961. doi: 10.1038/srep04961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay DG. The clutch hypothesis revisited: ascribing the roles of actin-associated proteins in filopodial protrusion in the nerve growth cone. J Neurobiol. 2000;44:114–125. doi: 10.1002/1097-4695(200008)44:2<114::aid-neu3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Lambert M, Choquet D, Mege RM. Dynamics of ligand-induced, Rac1-dependent anchoring of cadherins to the actin cytoskeleton. J Cell Biol. 2002;157:469–479. doi: 10.1083/jcb.200107104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Leonard M, Oliver T, Ishihara A, Jacobson K. Traction forces generated by locomoting keratocytes. J Cell Biol. 1994;127:1957–1964. doi: 10.1083/jcb.127.6.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, Chen CS. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc Natl Acad Sci USA. 2013;110:881–886. doi: 10.1073/pnas.1207997110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis AK, Bridgman PC. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J Cell Biol. 1992;119:1219–1243. doi: 10.1083/jcb.119.5.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Espreafico EM, Mooseker MS, Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Biol Bull. 1997;192:183–185. doi: 10.2307/1542600. [DOI] [PubMed] [Google Scholar]
- Lin CH, Forscher P. Cytoskeletal remodeling during growth cone-target interactions. J Cell Biol. 1993;121:1369–1383. doi: 10.1083/jcb.121.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Forscher P. Growth cone advance is inversely proportional to retrograde F-actin flow. Neuron. 1995;14:763–771. doi: 10.1016/0896-6273(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Van Vactor D. The trip of the tip: understanding the growth cone machinery. Nat Rev. 2009;10:332–343. doi: 10.1038/nrm2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Reeves E, Wientjes F, Mattheyse FJ, Grogan A, Totty NF, Burlingame AL, Hsuan JJ, Segal AW. Mammalian actin-related protein 2/3 complex localizes to regions of lamellipodial protrusion and is composed of evolutionarily conserved proteins. Biochem J. 1997;328:105–112. doi: 10.1042/bj3280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- Marcy Y, Prost J, Carlier MF, Sykes C. Forces generated during actin-based propulsion: a direct measurement by micromanipulation. Proc Natl Acad Sci USA. 2004;101:5992–5997. doi: 10.1073/pnas.0307704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros NA, Burnette DT, Forscher P. Myosin II functions in actin-bundle turnover in neuronal growth cones. Nat Cell Biol. 2006;8:215–226. doi: 10.1038/ncb1367. [DOI] [PubMed] [Google Scholar]
- Mejean CO, Schaefer AW, Buck KB, Kress H, Shundrovsky A, Merrill JW, Dufresne ER, Forscher P. Elastic coupling of nascent apCAM adhesions to flowing actin networks. PLoS One. 2013;8:e73389. doi: 10.1371/journal.pone.0073389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejean CO, Schaefer AW, Millman EA, Forscher P, Dufresne ER. Multiplexed force measurements on live cells with holographic optical tweezers. Opt Express. 2009;17:6209–6217. doi: 10.1364/oe.17.006209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millius A, Watanabe N, Weiner OD. Diffusion, capture and recycling of SCAR/WAVE and Arp2/3 complexes observed in cells by single-molecule imaging. J Cell Sci. 2012;125:1165–1176. doi: 10.1242/jcs.091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Cytoskeletal dynamics and nerve growth. Neuron. 1988;1:761–772. doi: 10.1016/0896-6273(88)90124-9. [DOI] [PubMed] [Google Scholar]
- Myers JP, Gomez TM. Focal adhesion kinase promotes integrin adhesion dynamics necessary for chemotropic turning of nerve growth cones. J Neurosci. 2011;31:13585–13595. doi: 10.1523/JNEUROSCI.2381-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichol RH IV, Hagen KM, Lumbard DC, Dent EW, Gomez TM. Guidance of axons by local coupling of retrograde flow to point contact adhesions. J Neurosci. 2016;36:2267–2282. doi: 10.1523/JNEUROSCI.2645-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Yoshihara F, Tojima T, Ooashi N, Yoon W, Mikoshiba K, Bennett V, Kamiguchi H. L1-dependent neuritogenesis involves ankyrinB that mediates L1-CAM coupling with retrograde actin flow. J Cell Biol. 2003;163:1077–1088. doi: 10.1083/jcb.200303060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Fujiwara S, Watanabe T, Kondo H, Kiuchi T, Sato M, Mizuno K. LIM kinase has a dual role in regulating lamellipodium extension by decelerating the rate of actin retrograde flow and the rate of actin polymerization. J Biol Chem. 2011;286:36340–36351. doi: 10.1074/jbc.M111.259135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskin CS, Odell GM, Oster GF. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- Pujol T, du Roure O, Fermigier M, Heuvingh J. Impact of branching on the elasticity of actin networks. Proc Natl Acad Sci USA. 2012;109:10364–10369. doi: 10.1073/pnas.1121238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risca VI, Wang EB, Chaudhuri O, Chia JJ, Geissler PL, Fletcher DA. Actin filament curvature biases branching direction. Proc Natl Acad Sci USA. 2012;109:2913–2918. doi: 10.1073/pnas.1114292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochlin MW, Dailey ME, Bridgman PC. Polymerizing microtubules activate site-directed F-actin assembly in nerve growth cones. Mol Biol Cell. 1999;10:2309–2327. doi: 10.1091/mbc.10.7.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Gil OD, Whittard JD, Gazdoiu M, Joseph T, Wu J, Waksman A, Benson DL, Salton SR, Felsenfeld DP. Interactions between the L1 cell adhesion molecule and ezrin support traction-force generation and can be regulated by tyrosine phosphorylation. J Neurosci Res. 2008;86:2602–2614. doi: 10.1002/jnr.21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Miguel-Ruiz JE, Letourneau PC. The role of Arp2/3 in growth cone actin dynamics and guidance is substrate dependent. J Neurosci. 2014;34:5895–5908. doi: 10.1523/JNEUROSCI.0672-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer AW, Schoonderwoert VT, Ji L, Mederios N, Danuser G, Forscher P. Coordination of actin filament and microtubule dynamics during neurite outgrowth. Dev Cell. 2008;15:146–162. doi: 10.1016/j.devcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahapure R, Difato F, Laio A, Bisson G, Ercolini E, Amin L, Ferrari E, Torre V. Force generation in lamellipodia is a probabilistic process with fast growth and retraction events. Biophys J. 2010;98:979–988. doi: 10.1016/j.bpj.2009.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Toriyama M, Uemura K, Kamiguchi H, Sugiura T, Watanabe N, Inagaki N. Shootin1 interacts with actin retrograde flow and L1-CAM to promote axon outgrowth. J Cell Biol. 2008;181:817–829. doi: 10.1083/jcb.200712138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shutes A, Onesto C, Picard V, Leblond B, Schweighoffer F, Der CJ. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. [DOI] [PubMed] [Google Scholar]
- Suter DM, Errante LD, Belotserkovsky V, Forscher P. The Ig superfamily cell adhesion molecule, apCAM, mediates growth cone steering by substrate-cytoskeletal coupling. J Cell Biol. 1998;141:227–240. doi: 10.1083/jcb.141.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Forscher P. Substrate-cytoskeletal coupling as a mechanism for the regulation of growth cone motility and guidance. J Neurobiol. 2000;44:97–113. [PubMed] [Google Scholar]
- Suter DM, Forscher P. Transmission of growth cone traction force through apCAM-cytoskeletal linkages is regulated by Src family tyrosine kinase activity. J Cell Biol. 2001;155:427–438. doi: 10.1083/jcb.200107063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter DM, Schaefer AW, Forscher P. Microtubule dynamics are necessary for SRC family kinase-dependent growth cone steering. Curr Biol. 2004;14:1194–1199. doi: 10.1016/j.cub.2004.06.049. [DOI] [PubMed] [Google Scholar]
- Tahirovic S, Hellal F, Neukirchen D, Hindges R, Garvalov BK, Flynn KC, Stradal TE, Chrostek-Grashoff A, Brakebusch C, Bradke F. Rac1 regulates neuronal polarization through the WAVE complex. J Neurosci. 2010;30:6930–6943. doi: 10.1523/JNEUROSCI.5395-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thievessen I, Thompson PM, Berlemont S, Plevock KM, Plotnikov SV, Zemljic-Harpf A, Ross RS, Davidson MW, Danuser G, Campbell SL, Waterman CM. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J Cell Biol. 2013;202:163–177. doi: 10.1083/jcb.201303129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C, Lin CH, Forscher P. An Aplysia cell adhesion molecule associated with site-directed actin filament assembly in neuronal growth cones. J Cell Sci. 1996;109:2843–2854. doi: 10.1242/jcs.109.12.2843. [DOI] [PubMed] [Google Scholar]
- Van Goor D, Hyland C, Schaefer AW, Forscher P. The role of actin turnover in retrograde actin network flow in neuronal growth cones. PLoS One. 2012;7:e30959. doi: 10.1371/journal.pone.0030959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oudenaarden A, Theriot JA. Cooperative symmetry-breaking by actin polymerization in a model for cell motility. Nat Cell Biol. 1999;1:493–499. doi: 10.1038/70281. [DOI] [PubMed] [Google Scholar]
- van Rijssel J, Kroon J, Hoogenboezem M, van Alphen FP, de Jong RJ, Kostadinova E, Geerts D, Hordijk PL, van Buul JD. The Rho-guanine nucleotide exchange factor Trio controls leukocyte transendothelial migration by promoting docking structure formation. Mol Biol Cell. 2012;23:2831–2844. doi: 10.1091/mbc.E11-11-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh FS, Doherty P. Neural cell adhesion molecules of the Ig superfamily: role in axon growth and guidance. Annu Rev Cell Dev Biol. 1997;13:425–456. doi: 10.1146/annurev.cellbio.13.1.425. [DOI] [PubMed] [Google Scholar]
- Weiner OD. Regulation of cell polarity during eukaryotic chemotaxis: the chemotactic compass. Curr Opin Cell Biol. 2002;14:196–202. doi: 10.1016/s0955-0674(02)00310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An actin-based wave generator organizes cell motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Woo S, Gomez TM. Rac1 and RhoA promote neurite outgrowth through formation and stabilization of growth cone point contacts. J Neurosci. 2006;26:1418–1428. doi: 10.1523/JNEUROSCI.4209-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashiro S, Watanabe N. A new link between the retrograde actin flow and focal adhesions. J Biochem (Tokyo) 2014;156:239–248. doi: 10.1093/jb/mvu053. [DOI] [PubMed] [Google Scholar]
- Yang Q, Zhang XF, Pollard TD, Forscher P. Arp2/3 complex-dependent actin networks constrain myosin II function in driving retrograde actin flow. J Cell Biol. 2012;197:939–956. doi: 10.1083/jcb.201111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarar D, To W, Abo A, Welch MD. The Wiskott-Aldrich syndrome protein directs actin-based motility by stimulating actin nucleation with the Arp2/3 complex. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- Yuan XB, Jin M, Xu X, Song YQ, Wu CP, Poo MM, Duan S. Signalling and crosstalk of Rho GTPases in mediating axon guidance. Nat Cell Biol. 2003;5:38–45. doi: 10.1038/ncb895. [DOI] [PubMed] [Google Scholar]
- Zhang X-F, Schaefer AW, Burnette DT, Schoonderwoert VT, Forscher P. Rho-dependent contractile responses in the neuronal growth cone are independent of classical peripheral retrograde actin flow. Neuron. 2003;40:931–944. doi: 10.1016/s0896-6273(03)00754-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.