Abstract
Clubroot disease caused by Plasmodiophora brassicae is one of the most important diseases of cultivated brassicas. P. brassicae occurs in pathotypes which differ in the aggressiveness towards their Brassica host plants. To date no DNA based method to distinguish these pathotypes has been described. In 2011 polymorphism within the 28S rDNA of P. brassicae was reported which potentially could allow to distinguish pathotypes without the need of time-consuming bioassays. However, isolates of P. brassicae from around the world analysed in this study do not show polymorphism in their LSU rDNA sequences. The previously described polymorphism most likely derived from soil inhabiting Cercozoa more specifically Neoheteromita-like glissomonads. Here we correct the LSU rDNA sequence of P. brassicae. By using FISH we demonstrate that our newly generated sequence belongs to the causal agent of clubroot disease.
Keywords: Clubroot, FISH (fluorescence in-situ hybridisation), LSU rDNA, pathotyping, phytomyxea, plasmodiophorids
Introduction
Plasmodiophora brassicae is a soil-borne plant pathogen that causes clubroot disease in crucifers. Clubroot disease is an economically important disease of oilseed rape and Brassica spp. food and feed crops worldwide (Dixon 2009; Hwang et al. 2012). Phylogenetically, P. brassicae belongs to the Plasmodiophorida, a plant pathogenic group of protists in the Phytomyxea within the eukaryotic supergroup Rhizaria (Bass et al. 2009; Burki et al. 2010; Neuhauser et al. 2014). P. brassicae isolates differ in preferences and aggressiveness towards different Brassica hosts (Buczacki et al. 1975). Because of the huge yield losses caused by P. brassicae, tools for a rapid identification and characterisation of the different pathotypes are important for phytopathologists, plant breeders and plant growers alike. But despite various efforts, to date pathotypes of P. brassicae can still only be classified using time-consuming, labour intensive bioassays (Buczacki et al. 1975; Hatakeyama et al. 2004; Some et al. 1996; Williams 1966). Currently no conclusive DNA marker suitable for a rapid pathotype identification could be identified. A number of different DNA methods such as random amplified polymorphic DNA (RAPD) and amplified fragment length polymorphism (AFLP) have been used to characterise P. brassicae isolates (Möller and Harling 1996; Strehlow et al. 2010). Although these studies revealed high polymorphisms among as well as within P. brassicae populations, so far no correlations to their pathogenicity was found (Manzanares-Dauleux et al. 2001; Osaki et al. 2008; Strehlow et al. 2014). A report of polymorphism in the large subunit (LSU) of the rDNA in different Japanese isolates suggested considerable variation across hosts and geographic areas (Niwa et al. 2011). This result held great potential for the molecular distinction of P. brassicae isolates and pathotypes.
While analysing LSU rDNA sequences from P. brassicae isolates of cosmopolitan origin, we did not retrieve the intraspecific molecular diversity pattern reported by Niwa et al. (2011). After further analyses involving the LSU of other phytomyxids and related Rhizaria species, it became clear that the second half of the sequence published by Niwa and colleagues (2011) did not correspond to the LSU sequences we had obtained. The aim of this study was to identify the correct LSU sequence of P. brassicae. By using specific fluorescence in-situ hybridisation (FISH) we tested which LSU sequence belongs to the clubroot causing species. In addition, we analysed the LSU sequences in a wider context of rhizarian LSU sequences to verify the taxonomic position of these sequences.
Results
Phylogenetic Analyses
We obtained the nearly complete sequence of the full rDNA operon (18S + ITS1 + 5.8S + ITS2 + 28S) from P. brassicae (isolate AT from Kematen, Austria – the same site where the sample for the FISH analyses was collected), W. pythii (isolate PD0236), and M. ectocarpii (strain CCAP 1538/1). Because of the variable nature of the 3′-end of the 28S region we did not generate sequence data for approximately the last 230 bp out of a total length of ca. 3600 bp. A second nearly complete sequence of the rDNA operon of P. brassicae (isolate e3) was derived from the genomic data. In W. pythii and M. ectocarpii the length of the 18S and 28S regions of the rDNA operon corresponded to what is typically observed in most eukaryotes (about 1800 bp for the complete 18S and about 3200 bp for the nearly complete 28S). In P. brassicae, the rDNA operon proved to be significantly longer (3094 bp for the complete 18S, and 4354 bp for the nearly complete 28S). In 18S, this is due to the presence of three introns (of 374, 393, and 479 bp., Supplementary Material Fig. S1) which are also present in the sequence from the Japanese P. brassicae isolate NGY (AB526243). In 28S, this was due to introns (of 501 and 485 bp, respectively) present in the last third of the gene, downstream from where introns can be found in the sequences obtained by Niwa et al. (2011).
The sequences of the rDNA operons of our two isolates – AT obtained by direct sequencing of PCR amplicons, and e3 obtained from the genomic assembly (Schwelm et al. 2015) – differ only in three positions: (1) at position 58 of the first 18S intron, there is a 3 bp insertion (’ATT’) in isolate e3; (2) position 342 of the third 18S intron consists of an ‘A’ in isolate AT and a ‘G’ in isolate e3; (3) at the end of the ITS2, we found a 2 bp. difference in a region consisting of an ‘AC’ motif repeated five (isolate AT) or six (isolate e3) times (Supplementary Material Fig. S1). The 18S, ITS1, 5.8S, and 28S regions are 100% identical, and so are the second 18S intron and the two 28S introns. By contrast, the sequence from the Japanese P. brassicae isolate NGY (AB526243) only matches our sequences up to position 4422 of sequence KX01115, about one fourth (850 bp) within 28S. Upstream of that position, there are only a few differences in the three 18S introns, while the sequences are identical in 18S, ITS1, 5.8S, ITS2, and beginning of 28S. Downstream from position 4422, sequence AB526243 becomes significantly distinct from ours; it differs in many positions in conserved regions, exhibits numerous size and primary sequence differences in variable regions, and lacks both of the introns we found in the last third of the 28S in our isolates. When blasting this part of sequence AB526243 against GenBank, we found the closest related hit to be sequence FJ973380 from the glissomonad Neoheteromita globosa, and not that of a previously obtained sequence from an environmental Spongospora-like plasmodiophorid (KJ150290) from a soil sample collected in Warwick, UK (Hartikainen et al. 2014).
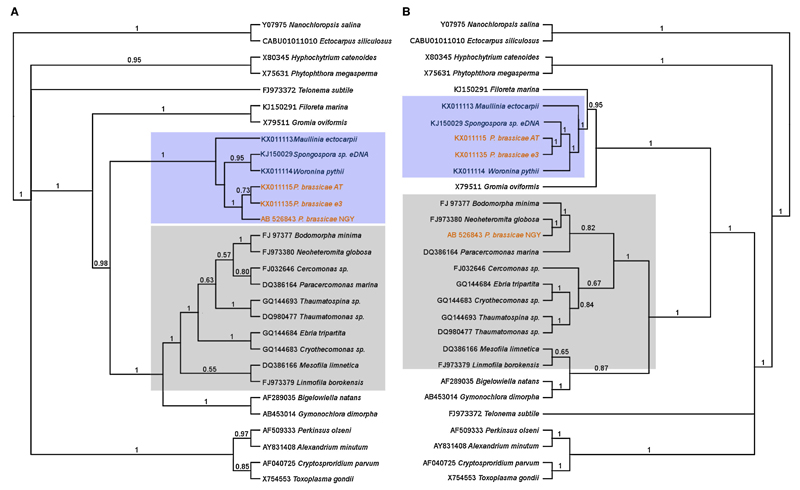
We then investigated the possible chimeric origin of sequence AB526243 by phylogenetic analysis. The four full-length sequences of phytomyxids obtained in this study were aligned with the sequence from the Japanese P. brassicae isolate NGY (AB526243), the sequence of the environmental Spongospora-like plasmodiophorid (KJ150290), and a selection of Stramenopiles, Alveolates and Rhizaria from GenBank. We focused on the regions of the rDNA operon encoding the large subunit of ribosomal RNA (5.8S and 28S), and performed phylogenetic analyses on two separate datasets consisting of the regions upstream and downstream from position 4422 of sequence AB526843, respectively (Supplementary Material Fig. S2). The overall topology of the trees we obtained from these two datasets was stable, highly similar and well supported. The phytomyxid sequences generated for this study formed a well-supported clade in both phylogenies, and the branching pattern within the Rhizaria corresponded to what was expected from earlier studies based on 18S rDNA (Fig. 1A). However, comparing the two tree topologies it became clear that whereas the first half of sequence AB526843 is identical to that of our isolates, its second half does not group with the other Phytomyxea but next to Neoheteromita globosa, within the glissomonad Cercozoa (Fig. 1B).
Figure 1.
Cladogram of trees generated from the posterior output tree from Bayesian analyses of the 5′ end (Fig. 1A left) and the 3′ end (Fig. 1B right) of the LSU rDNA. This shows that sequence P. brassicae NGY (AB526843) is a chimeric sequence of P. brassicae and a glissmonad sequence similar to Neoheteromita globosa. Fig. 1A was generated using an alignment of LSU sequences up to position 4400 of sequence (AB526843), the right tree was generated starting from position 4500. Cercozoan sequences are shaded in grey, phytomyxid sequences are shaded in light blue. Trees were generated using phytomyxid sequences plus a selection of Rhizaria, alveolates and stramenopiles from GenBank using .29 sequences, A: 5999 positions, B: 4358 positions. Trees showing distances are given in the Supplementary Material Figures S3 and S4.
Intraspecific Biodiversity of Plasmodiophora brassicae
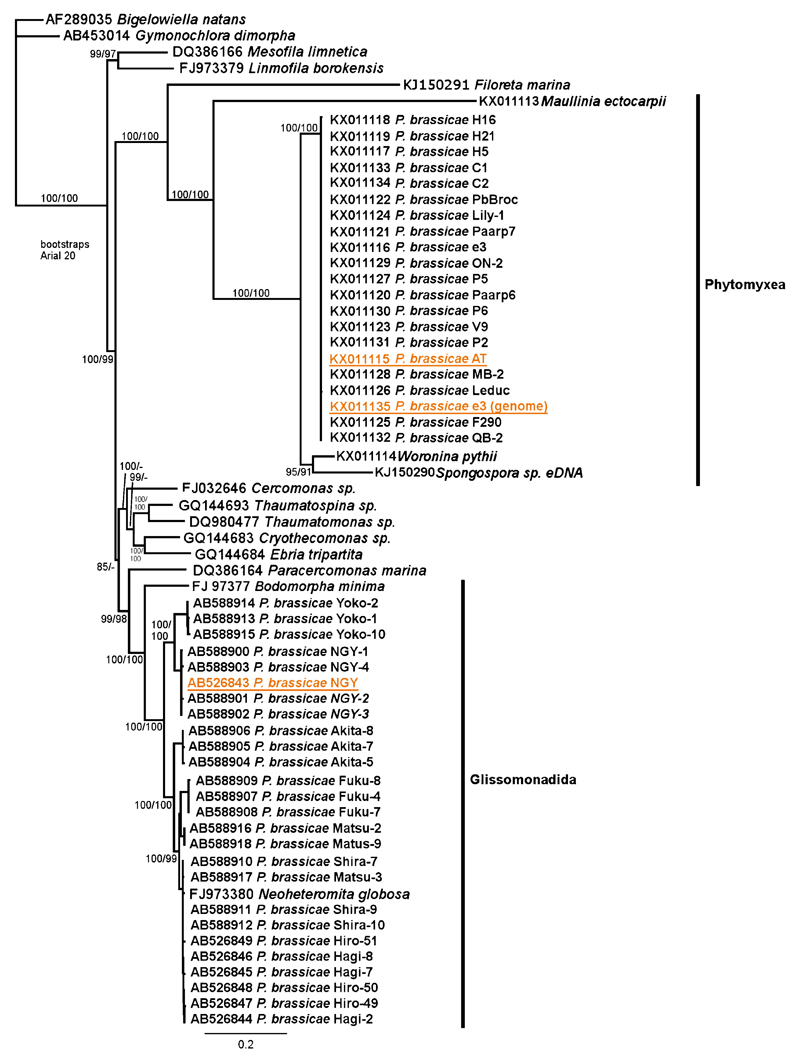
We generated 21 new sequences from P. brassicae isolates from different continents using primers LPR4, NDL22f, NDL22, 28s3r and 28s4r. These primers bind at the region of the LSU rDNA of P. brassicae which is located in the 3′ part of the alignment (Supplementary Material Fig. S2). From this position the “P. brassicae” NGY-type LSU sequences (AB588900–AB588918; AB526843–AB526848) group within Glissomonadida and with Neoheteromita globosa (sequence similarities between 99% and 92%). In contrast, the P. brassicae LSU sequences obtained in this study do form a well-supported clade with the sequences of other phytomyxids (Fig. 2). No intraspecific variation between the P. brassicae sequences generated in this study could be detected, all sequences of P. brassicae were 100% identical in this region. The diversity pattern of the previously assigned non-phytomyxid “P. brassicae” sequences is similar as described in the original study (Niwa et al. 2011). This diversity pattern is high enough in hyper-variable regions to be compatible with inter-specific differences.
Figure 2.
Intraspecific diversity of P. brassicae isolates of worldwide origin. Sequences generated in this study (KX011115-KX011135) form a well-supported clade with sequences of other Phytomyxea from this study (KX011113, KX011114) and genbank (KJ150290) while the NGY-type sequences (AB526844–AB526849; AB588900–AB588918) form a clade with Glissomonadida sequences from genbank (FJ97377, FJ973380). 63 Sequences, PhyML tree, support values are given Chi2 support values (%)/Posterior probabilities (%).
FISH Analyses
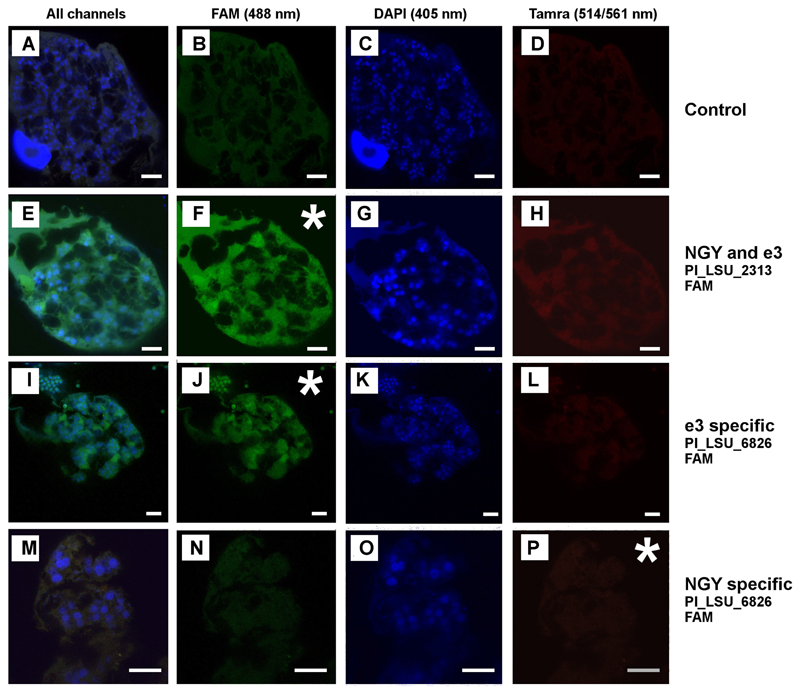
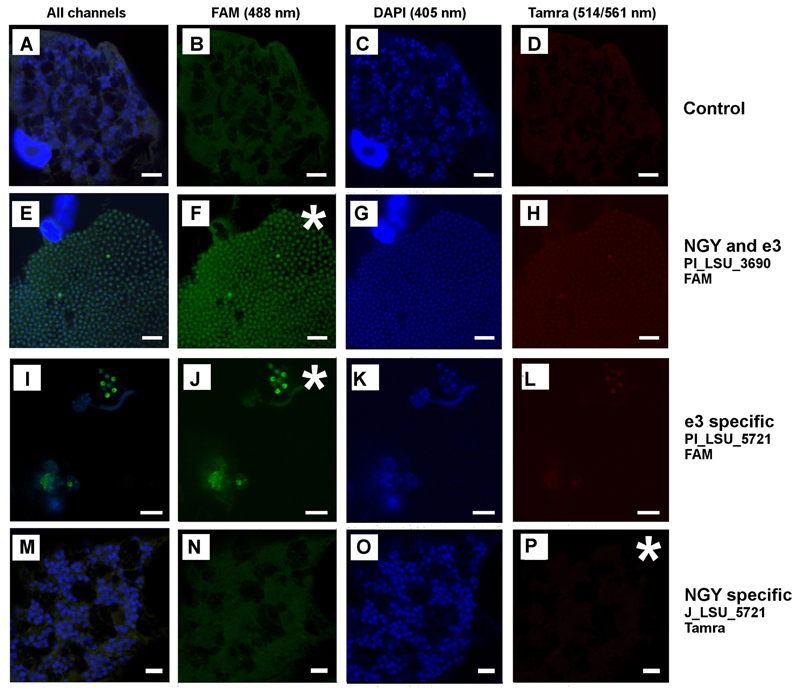
Plasmodiophora brassicae shows a weak autofluorescence at 488 nm (Fig. 3B) and a very weak autofluorescence at 514/561 nm (Fig. 3D). But this autofluorescence is considerably weaker than a true signal obtained from using FISH probes (e.g. Fig. 3F). The FISH probes binding the region in which the sequence of the Japanese isolate NGY (AB526843) and our sequences are identical (Pl_LSU_2313, Pl_LSU_3690) did work well, returning a clear positive signal at 488 nm on both the lobose plasmodia (Fig. 3F) and the resting spores (Fig. 4F) of P. brassicae. In the region where sequence AB526843 differs from ours, the two FISH probes matching the sequence from isolate e3 but not isolate NGY did both return a clear positive signal (Figs 3J, 4J). However, the two probes designed to match only isolate NGY did not yield any positive results in FISH (Figs 3P, 4P). These results strongly support the molecular phylogenetic results that the sequence of the Japanese isolate NGY is chimeric from position 4400 onwards.
Figure 3.
FISH images of P. brassicae, an asterisk is used to highlight the channel where the respective probe should be visible. All images which were created using the same excitation wavelength are arranged vertically, while horizontally all samples were treated with the identical probe. A–H and M–P show sporogenic plasmodia of P. brassicae. Images I–L show both, plasmodia and resting spores. The nuclei of actively growing plasmodia resemble doughnuts, which is caused by the characteristic cruciform nuclear division (e.g. Fig. 3C, G, K). Bars = 10 µm.
Figure 4.
FISH images of P. brassicae, an asterisk is used to highlight the channel where the respective probe should be visible. All images which were created using the same excitation wavelength are arranged vertically, while horizontally all samples were treated with the identical probe. Fig. 4M–P show sporogenic plasmodia of P. brassicae. Images E–H show resting spores. In Fig. 4E–H a germinating fungal spore can be seen in between the resting spores and the plasmodium. The difference in the appearance of the nuclei is linked to the different life cycle stages: Nuclei are very small and condensed in the resting spores (Fig. 4G, K) while the nuclei of actively growing plasmodia resemble doughnuts, which is caused by the characteristic cruciform nuclear division (Fig. 4C, O, see also Fig. 3G, K). Differences in the intensity of the FISH signal such as in in Fig. 4F or J, but also like in Fig. 3F or J can be explained by the aging of P. brassicae, with the strongest signal in the most active cells, the dividing plasmodia and a fading signal in the resting spores. The control images in Fig. 4A–D are identical with Fig. 3A–D. Bars = 10 µm
Discussion
Our results demonstrate that there is no genetic diversity in the LSU sequence of P. brassicae in isolates from around the world. Therefore, the LSU is not suitable to distinguish between different isolates or pathotypes of P. brassicae. FISH analyses confirmed that the LSU sequences we are presenting here do belong to the causal agent of clubroot disease, P. brassicae. Our phylogenetic analyses performed on different parts of the LSU show that downstream of the binding site of primers NDL22/NDL22F, the sequences of the NGY-type (AB525843) describing differences in the “P. brassicae” LSU sequences Niwa et al. (2011) most likely originated from a glissomonad cercozoan. The lack of overlap with the sequence they had obtained upstream of that binding site prevented them from identifying the different biological origin of these sequences. The specific primers designed by Niwa et al. (2011) for their diversity study are situated in the downstream region of that point and as a result instead of amplifying the LSU of P. brassicae isolates, the LSU of glissomonads that were present in the rhizosphere of the clubroot-infected plants was amplified. Glissomonad Cercozoa are highly abundant and diverse organisms in soil (Howe et al. 2009), it is therefore not surprising that they could be found in most/all rhizosphere samples. Glissomonads are also – like phytomyxids – known to have introns in their ribosomal gene clusters (Howe et al. 2009). The presence of an intron in the downstream fragment obtained by Niwa et al. (2011) was therefore in concordance with a plasmodiophorid origin of the sequence. We did find one long intron in the downstream sequence of P. brassicae that made amplification and sequencing of that fragment difficult, but not in the same position as that present in the glissomonad sequence. At the time when the study of Niwa et al. (2011) was published only a small number of LSU sequences was available for Cercozoa. Given this limitation and the fact that P. brassicae and glissomonads both belong to Cercozoa, the chimeric nature of the P. brassicae sequence did result in a plausible phylogenetic tree and did not arouse suspicion. However, with an enlarged dataset as the one presented here, it became possible to highlight this. This emphasises the need to generate more sequence data for less studied eukaryotic groups to increase the accuracy of phylogenetic analyses. Our results also suggest that the described differences in the LSU sequences of the Japanese “P. brassicae” NGY-type isolates do actually represent genetic diversity in the LSU between different glissomonad species. By contrast, we could not identify any differences in the LSU sequences of P. brassicae from cosmopolitan isolates and therefore this gene is not suitable to distinguish between different isolates or pathotypes of P. brassicae.
Methods
Pathogen material and DNA extraction
DNA from P. brassicae (AT) and Woronina pythii was extracted and sequenced as described by Neuhauser et al. (2014). DNA from the single spore isolate e3 grown in the roots of Brassica napus was extracted as described by Schwelm et al. (2015). DNA from the Swedish isolates was obtained in the same way. DNA-extracts for other isolates were kindly provided by the sources listed in Table 1, where also the origin of P. brasssicae isolates used in this study is given.
Table 1.
P. brassicae isolates analysed in this study. Isolates marked with * are single spore isolates.
| Accession number | P. brassicae isolate | Origin | Source | Reference |
|---|---|---|---|---|
| Germany | ||||
| KX011116 (PCR) | e3* | Germany | J. Ludwig-Müller (TU Dresden, Germany) | Fahling et al. (2004) |
| KX011135 (genomic) | ||||
| Sweden | ||||
| KX011120 | Påarp6 | Påarp | A.C. Wallenhammar (HS Konsult AB, Sweden) | This study |
| KX011121 | Påarp7 | Påarp | ||
| KX011119 | H21 | Hallsberg | ||
| KX011117 | H5 | Hallsberg | ||
| KX011118 | H16 | Hallsberg | ||
| KX011123 | V9 | Veneberg | ||
| KX011122 | PbBro | Skara | ||
| New Zealand | ||||
| KX011124 | Lily-1 | NZ South Island | S. Bulman (PFR Lincoln, New Zealand) | |
| Canada | ||||
| KX011129 | ON-2 | Ontario | S. Strelkov (University of Alberta, Canada) | |
| KX011128 | MB-2 | Manitoba | Cao et al. (2009) | |
| KX011131 | P2 | Quebec | ||
| KX011132 | QB | Quebec | ||
| KX011127 | P5 | Alberta | ||
| KX011130 | P6 | Ontario | ||
| KX011125 | F290-07 | Alberta | Cao et al. (2009) | |
| KX011126 | Leduc-SS2* | Alberta | Xue et al. (2008) | |
| Austria | ||||
| KX011115 | AT | Kematen, Tyrol | M. Kirchmair, S. Neuhauser | This study |
| South Korea | ||||
| KX011133 | C1 | South Korea | P.Y. Lim (Chungnam National University, South Korea) | |
| KX011134 | C2 |
single spore isolate.
PCR amplification, cloning, and sequencing
Intraspecific variation within a range of P. brassicae isolates was analysed by PCR using primers LPR4 and primer 28s4r (Niwa et al. 2011). PCR amplification was performed in a 50 µl reaction mixture with the Expand High-Fidelity PLUS PCR System (New England Biolabs) containing 0.5 µM primers and 5–10 ng DNA template. All reactions were performed with an initial denaturation step of 98 °C for 2 min followed by 10 cycles of denaturation, annealing, and extension as follows: 98 °C for 15s, initial annealing temperature of 60 °C for 15 s with reduction of annealing temperature of 0.5 °C per cycle, and 72 °C for 90 s, followed by 20 cycles of 98 °C for 15 s, annealing temperature of 54 °C for 15 s, and 72 °C for 90 s and a final step of 5 min at 72 °C. The PCR products were analyzed on a 1.2% agarose gel in 0.5 × TBE buffer and gel or PCR purified. Products were cloned into pJET1.2 vector and sequenced by Macrogen sequencing service (Netherland) using pJET sequencing primer and primers LPR4, NDL22f, NDL22, 28s4r, 28s3r (Niwa et al. 2011). The sequence of the single spore isolate e3 was confirmed by the genome sequence (Schwelm et al. 2015).
Nearly complete sequences of the full rDNA operon were obtained in two or three overlapping PCR fragments from P. brassicae (isolate AT), Woronina pythii (isolate PD0236), and Maullinia ectocarpii (strain CCAP 1538/1), using a (semi-)nested PCR approach. The nearly complete 18S part of the operon was amplified using a first round of PCRs performed with universal primers sA1n (forward) and sB1n (reverse), followed by (semi-)nested PCRs with sA1n and semi-specific reverse primer sB2phy (P. brassicae); sA1n and Maullinia-specific reverse primer V9r-Mau (M. ectocarpii); and Phytomyxea-specific forward primer sA6-phy and reverse primer C9r-Phyt (W. pythii). The remaining part of the operon (from the 3’ end of 18S to the 3’ end of 28S) was obtained in two overlapping fragments in P. brassicae and a single fragment in W. pythii and M. ectocarpii. For P. brassicae, a first PCR was performed with plasmodiophorid-specific forward primer V7f-Plas and universal reverse primer 22R (about two-thirds inside 28S), followed by a nested PCR with plasmodiophorid-specific forward primer C9f-Plas and universal reverse primer 21R. Then the last third of 28S was obtained using a first PCR performed with Phytomyxea-specific forward primer D14f-phy1 and universal reverse primer nH4r (about 250 bp. before the 3′ end of 28S), followed by a semi-nested PCR with Phytomyxea-specific forward primer D14f-phy2 and nH4r. For W. pythii, a first PCR was performed with V7f-Plas and nH4r, followed by a nested PCR with C9f-Plas and universal reverse primer nH2r (about 330 bp. before the 3′ end of 28S). For M. ectocarpii, a first PCR was performed with V7f-Phag and nH4r, followed by a semi-nested PCR with phagomyxid-specific forward primer C9f-Phag and nH4r. Supplementary Table S1 lists all the PCR primers cited above.
PCR amplifications were done in a total volume of 30 µl in reaction mixtures containing a final concentration of 0.4 mM dNTPs, 0.5 µM of each primer, 1 × PCR-reaction buffer, 2 µg/ml Bovine Serum Albumin, 2.5 mM MgCl2, and 0.2 U Phusion-Polymerase (Finnzymes), using as template 1 µl of genomic DNA (first round PCRs) or 2 µl from the first round PCR product (nested PCRs). First round PCR conditions were: initial denaturation for 2 min at 95 °C, followed by 36 cycles with 30 sec at 95 °C, 30 sec at 58 °C, and 2 min at 72 °C, followed by a final elongation step of 10 min at 72 °C. Nested PCR conditions were the same, but with 32 cycles and an annealing temperature of 63 °C. Nested PCR products were run on 1.5% TAE agarose gels; bands of the appropriate lengths were excised and cleaned following the protocol of the QIAquick® Gel Extraction Kit (Qiagen). Direct Sanger sequencing of all cleaned PCR amplicons was done using the PCR primers and appropriate internal sequencing primers (details of which are available from the authors upon request). Sequencing was performed with the ABI-PRISM Big Dye Terminator Cycle Sequencing Kit, and analysed with an ABI-377 DNA sequencer (Perkin-Elmer, Rotkreuz, Switzerland).
Sequence analysis
A primary alignment of the full length phytomyxid LSU sequences was created using MAFFT (Katoh and Standley 2013) implemented in Geneious 9.0.5 (Drummond et al. 2012) and this alignment was improved manually using BioEdit (version 7.0.5.3; Hall 1999). This alignment was then split into two at position 4400 of sequence AB526843 (Isolate NGY, Japan) where the sequence AB526843 and the phytomyxid sequences generated in our study start to diverge. Bayesian consensus trees were constructed using MrBayes v 3.1.2 in parallel mode (Ronquist et al. 2012). Two separate MC3 runs with randomly generated starting trees were carried out for 3M generations each with one cold and three heated chains. The evolutionary model included a GTR substitution matrix, a four-category auto-correlated gamma correction and the covarion model. All parameters were estimated from the data. Trees were sampled every 1000 generations. 300000 generations were discarded as “burn-in” (trees sampled before the likelihood plots reached a plateau) and consensus trees constructed from the returning sample. The two returning trees were rearranged visually by rotating branches to mirror each other as much as possible. Sequences generated to analyse intraspecific variation were aligned, the alignment trimmed and analysed as described above.
FISH analyses
Fresh clubroots were collected at Kematen (Tyrol, Austria) in September 2013. The roots were rinsed and fixed in 1% PFA solution overnight and subsequently dehydrated in an ascending Ethanol series (50%, 80%, 96%). Clubroots were then cut by hand into thin sections suitable for light microscopy using a sterile blade. The sections were rinsed in sterile, distilled water and subsequently equilibrated for 10 minutes in hybridisation buffer (900 mM NaCl, 20 mM Tris HCl pH 7.5, 35% formamide, 0.01% SDS). Sections were transferred into fresh tubes containing hybridisation buffer; 50 ng of the respective probe was added (Table 2) and the samples were incubated overnight at 46 °C. Samples were then washed twice for 10 min in washing buffer (900 mM NaCl, 20 mM Tris HCl pH 7.5, 5 mM of NaEDTA pH8, 0.01% SDS) and mounted in Vectashield containing 0.05 mg mL−1 DAPI. Samples were analysed using a Leica TCS SP5 II confocal microscope using excitation wavelength of 405 nm (DAPI), 488 nm (FAM) and 514/561 nm (TAMRA). Controls containing only Hybridisation buffer and DAPI were included to ensure no false positives caused by autofluorescence of the samples.
Table 2.
FISH probes used. Probes were labelled on the 5′ end using either 6-FAM or Tamra. The table also indicates where on the LSU rDNA fragment these probes bind and for which sequence-types they are specific.
| Probe | Probe sequence | Dye | (KX011135) Isolate e3, Germany | AB526843 Isolate NGY, Japan |
|---|---|---|---|---|
| Pl_LSU_3690 | GCGGCAGTGATTTTCGGTTT | 6-FAM | bp 3675 | bp 3684 |
| Pl_LSU_2313 | CCAGGCCTTTCAGCCAAGTA | 6-FAM | bp 2313 | bp 2317 |
| Pl_LSU_5721 | CAGCGTCACCCAACCCTTAG | 6-FAM | bp 5711 | N |
| J_LSU_5721 | TCCATCCGCTCAACCCTTAG | Tamra | N | bp 5723 |
| Pl_LSU_6826 | ATTCCAGAAGTCTGCCGCTC | 6-FAM | bp 6819 | N |
| J_LSU_6826 | AGTCTAGAAACAAAGGCCTC | Tamra | N | bp 6336 |
Probes were designed using the ARB software package (Ludwig et al. 2004) to bind to the regions of 28S rDNA (i) where the sequence of the Japanese isolate NGY (AB526843) and our sequences (KX011115, KX011135) are identical (i.e. before position 4429of sequence AB526843) (ii) the region where the sequence AB526843 differs from our newly determined sequences (i.e. after position 4400), but avoiding the first 400 bp up- and downstream from this position. Downstream of position 4400, pairs of probes were designed in the same two regions, one matching sequence AB526843, and one matching the LSU sequences we determined (Supplementary Material Fig. S1).
Probes were synthesised and labelled with FAM (excitation 488 nm, green) or TAMRA (excitation 514/561 nm, red) at the 5′ end by Biomers (Ulm, Germany). All slides were initially screened using the same settings of the LSM, but the energy settings of the laser were adjusted for each sample to avoid excessive over- or underexposure of features. Microscopic images were processed using GIMP 2.8.16 (www.gimp.org). Images were trimmed and resized, overlay images of the three recorded channels were produced, and contrast and colour settings were adjusted to ensure a high quality of figures 1–28. All images of the same excitation wavelength were modified together to ensure that differences between the FISH-probes were not under- or overstated. The original, unprocessed images were deposited at Figshare (http://dx.doi.org/10.6084/m9.figshare.3184399.v1).
Appendix A. Supplementary Data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.protis.2016.08.008.
Acknowledgements
The authors would like to thank S. Strelkov (University of Alberta, Edmonton, Canada), S.Lim (Chungnam National University, South Korea), S. Bulman (Food and Plant Research, Lincoln, New Zealand) Ludwig-Müller (TU Dresden, Germany) and A.C. Wallenhammar (HS Konsult AB, Sweden) for kindly providing DNA samples and M. Kirchmair (University of Innsbruck) for collecting clubroots for FISH as well as A. Sandbichler (University of Innsbruck) for technical support using the confocal microscope. A. S. was financially supported by BioSoM. S. N. was funded by the Austrian Science Fund (FWF): grant J3175-B20 (Erwin Schrödinger Fellowship). We thank NERC for a Standard Research Grant (NE/H009426/1) supporting D. B. and C. B. and a New Investigator Grant (NE/H000887/1) supporting D. B.
References
- Bass D, Chao EEY, Nikolaev S, Yabuki A, Ishida K-i, Berney C, Pakzad U, Wylezich C, Cavalier-Smith T. Phylogeny of novel naked filose and reticulose Cercozoa: Granofilosea cl. n. and Proteomyxidea revised. Protist. 2009;160:75–109. doi: 10.1016/j.protis.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Buczacki ST, Toxopeus H, Mattusch P, Johnston TD, Dixon GR, Hobolth LA. Study of physiologic specialization in Plasmodiophora brassicae—Proposals for attempted rationalization through an international approach. Trans Br Mycol Soc. 1975;65:295–303. [Google Scholar]
- Burki F, Kudryavtsev A, Matz MV, Aglyamova GV, Bulman S, Fiers M, Keeling PJ, Pawlowski J. Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol Biol. 2010;10:377. doi: 10.1186/1471-2148-10-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao T, Manolii VP, Hwang S-F, Howard RJ, Strelkov SE. Virulence and spread of Plasmodiophora brassicae clubroot in Alberta, Canada. Can J Plant Pathol-Rev Canad De Phytopathol. 2009;31:321–329. [Google Scholar]
- Dixon GR. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J Plant Growth Regul. 2009;28:194–202. [Google Scholar]
- Drummond A, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, et al. Geneious, v5. 6 ed. 2012. [Google Scholar]
- Fahling M, Graf H, Siemens J. Characterization of a single-spore isolate population of Plasmodiophora brassicae resulting from a single club. J Phytopathol. 2004;152:438–444. [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hartikainen H, Ashford OS, Berney C, Okamura B, Feist SW, Baker-Austin C, Stentiford GD, Bass D. Lineage-specific molecular probing reveals novel diversity and ecological partitioning of haplosporidians. ISME J. 2014;8:177–186. doi: 10.1038/ismej.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K, Fujimura M, Ishida M, Suzuki T. New classification method for Plasmodiophora brassicae field isolates in Japan based on resistance of F-1 cultivars of Chinese cabbage (Brassica rapa L. ) to clubroot. Breeding Sci. 2004;54:197–201. [Google Scholar]
- Howe AT, Bass D, Vickerman K, Chao EE, Cavalier-Smith T. Phylogeny, taxonomy, and astounding genetic diversity of Glissomonadida ord. nov., The dominant gliding zooflagellates in soil (Protozoa: Cercozoa) Protist. 2009;160:159–189. doi: 10.1016/j.protis.2008.11.007. [DOI] [PubMed] [Google Scholar]
- Hwang S-F, Strelkov SE, Feng J, Gossen BD, Howard RJ. Plasmodiophora brassicae: a review of an emerging pathogen of the Canadian canola (Brassica napus) crop. Mol Plant Pathol. 2012;13:105–113. doi: 10.1111/j.1364-3703.2011.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzanares-Dauleux M, Divaret I, Baron F, Thomas G. Assessment of biological and molecular variability between and within field isolates of Plasmodiophora brassicae. Plant Pathol. 2001;50:165–173. [Google Scholar]
- Möller M, Harling R. Randomly amplified polymorphic DNA (RAPD) profiling of Plasmodiophora brassicae. Lett Appl Microbiol. 1996;22:70–75. [Google Scholar]
- Neuhauser S, Kirchmair M, Bulman S, Bass D. Cross-kingdom host shifts of phytomyxid parasites. BMC Evol Biol. 2014;14:33. doi: 10.1186/1471-2148-14-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R, Kawahara A, Murakami H, Tanaka S, Ezawa T. Complete structure of nuclear rDNA of the obligate plant parasite Plasmodiophora brassicae: Intraspecific polymorphisms in the exon and Group I intron of the large subunit rDNA. Protist. 2011;162:423–434. doi: 10.1016/j.protis.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Osaki K, Fujiyama S, Nakayama A, Shimizu Y, Ito S-i, Tanaka S. Relation between pathogenicity and genetic variation within Plasmodiophora brassicae. J Gen Plant Pathol. 2008;74:281–288. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3. 2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwelm A, Fogelqvist J, Knaust A, Juelke S, Lilja T, Bonilla-Rosso G, Karlsson M, Shevchenko A, Dhandapani V, Choi SR, Kim HG, et al. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci Rep. 2015;5:11153. doi: 10.1038/srep11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Some A, Manzanares MJ, Laurens F, Baron F, Thomas G, Rouxel F. Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol. 1996;45:432–439. [Google Scholar]
- Strehlow B, de Mol F, Struck C. History of oilseed rape cropping and geographic origin affect the genetic structure of Plasmodiophora brassicae populations. Phytopathology. 2014;104:532–538. doi: 10.1094/PHYTO-07-13-0210-R. [DOI] [PubMed] [Google Scholar]
- Strehlow B, Preiss U, Horn R, Struck C. Genetic variability of the causal agent of clubroot Plasmodiophora brassicae in different regions of Germany. Julius-Kühn-Archiv. 2010;428:396–397. [Google Scholar]
- Williams PH. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology. 1966;56:624–626. [Google Scholar]
- Xue S, Cao T, Howard RJ, Hwang SF, Strelkov SE. Isolation and variation in virulence of single-spore isolates of Plasmodiophora brassicae from Canada. Plant Dis. 2008;92:456–462. doi: 10.1094/PDIS-92-3-0456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.