Abstract
In a previous bioinformatics-based search for novel small-RNA genes encoded by the Escherichia coli genome, we identified a region, IS063, located between the ompN and ydbK genes, that encodes an ∼100-nucleotide small-RNA transcript. Here we show that the expression of this small RNA is increased at a low temperature and in minimal medium. Twenty-two nucleotides at the 5′ end of this transcript have the potential to form base pairs with the leader sequence of the mRNA encoding the outer membrane protein OmpC. The deletion of IS063 increased the expression of an ompC-luc translational fusion 1.5- to 2-fold, and a 10-fold overexpression of the small RNA led to a 2- to 3-fold repression of the fusion. Deletion and overexpression of the IS063 RNA also resulted in increases and decreases, respectively, in OmpC protein levels. Taken together, these results suggest that IS063 is a regulator of OmpC expression; thus, the small RNA has been renamed MicC. The antisense regulation was further demonstrated by the finding that micC mutations were suppressed by compensatory mutations in the ompC mRNA. MicC was also shown to inhibit ribosome binding to the ompC mRNA leader in vitro and to require the Hfq RNA chaperone for its function. We suggest that the MicF and MicC RNAs act in conjunction with the EnvZ-OmpR two-component system to control the OmpF/OmpC protein ratio in response to a variety of environmental stimuli.
A burgeoning number of small RNAs have been identified in Escherichia coli over the past few years (4, 7, 21, 29, 33, 36). Some of these noncoding RNAs have been shown to act by forming base pairs with target sequences, while others bind specific proteins or have structural roles. However, the challenge remains to elucidate the functions of the vast majority of these RNAs.
Among the E. coli small RNAs, the most information is known about those that act as antisense riboregulators (reviewed in references 14, 31, 32, and 34). These RNAs exert their functions by forming base pairs with mRNAs that are generally encoded at separate loci. The net effect can be the up-regulation or down-regulation of target genes. Examples of small RNAs that block ribosome accessibility to target mRNAs and thereby decrease gene expression are MicF, which forms base pairs with the ompF mRNA; DicF, which forms base pairs with the ftsZ mRNA; OxyS, which forms base pairs with the fhlA mRNA; and Spot42, which forms base pairs with the galETKM mRNA. The DsrA and RprA small RNAs form base pairs with the rpoS mRNA and increase translation by preventing the formation of an inhibitory mRNA secondary structure. The formation of base pairs between the newly identified RyhB small RNA and its targets leads to the degradation of these mRNAs.
The MicF small RNA is encoded divergently from the gene encoding the OmpC porin and represses the expression of OmpF, another porin (1, 3, 15, 16). While their outer membranes prevent the passage of most molecules, E. coli cells possess three trimeric outer membrane porins, OmpC, OmpF, and PhoE, that allow fairly nonspecific passage of low-molecular-weight soluble molecules (reviewed in references 11 and 18). The ompN gene of E. coli encodes a homolog of the trimeric porins, but the OmpN protein has not yet been detected (20). The OmpC and OmpF porins are among the most abundant outer membrane proteins, and their expression is extensively regulated (reviewed in reference 19). OmpC, which has the smaller pore diameter of the two, is thought to be important in environments where nutrient and toxin concentrations are high, such as in the intestine, and it is the predominant porin at high temperatures and high osmolarities. OmpF, which has a larger pore diameter, is thought to be important in environments where nutrient and toxin concentrations are low, such as in fresh water, and it is more abundant at low temperatures and low osmolarities. Part of the differential response to environmental insults is regulated at the transcriptional level. The EnvZ sensor protein monitors external signals and modulates the activity of the OmpR response regulator by phosphorylation and dephosphorylation. The OmpR regulator binds to both the ompC and ompF promoters, but at low concentrations of OmpR-P, ompF is activated, while at high concentrations of OmpR-P, ompF is repressed and ompC is activated. Additional transcriptional control in response to nutrient availability is exerted by the leucine-responsive regulatory protein LRP, a global regulator that negatively regulates ompC transcription.
As mentioned above, ompF expression is also regulated at the posttranscriptional level via the MicF RNA, and many different environmental stresses impact the OmpC/OmpF ratio via changes in MicF levels (reviewed in reference 9). The expression of micF is increased along with that of ompC at high OmpR-P concentrations. The expression of micF is also induced by toxic agents, such as paraquat and weak acids, by the SoxS, MarA, and Rob transcription factors, which bind to the same sequence element in the micF promoter. MicF RNA levels increase at high temperatures, although the mechanism by which this occurs is not known. As a consequence of MicF induction, high osmolarities, exposures to toxic agents, and high temperatures all indirectly lead to decreased OmpF levels. Since the micF gene is repressed by LRP along with ompC, LRP repression indirectly leads to increased OmpF levels. Finally, the H-NS-like StpA protein appears to impact OmpF levels through the destabilization of micF RNA. Thus, multiple regulatory pathways converge at MicF, leading to changes in the OmpF/OmpC ratio.
In a previous genome-wide search for novel small RNAs, Chen et al. identified a DNA region denoted IS063 that is located between the ompN porin gene and a gene of unknown function (ydbK) and that is transcribed into a short RNA transcript that lacks the capacity to be translated (7). Using BLAST searches, we found that the small RNA shows complementarity to the leader sequence of the ompC mRNA. We postulated that the small RNA may regulate ompC expression in the same fashion that MicF regulates ompF, and thus we named the small RNA the MicC RNA. Here we present evidence that MicC, which is conserved in Shigella, Salmonella, and Klebsiella, inhibits ompC expression at the posttranscriptional level.
MATERIALS AND METHODS
Plasmids and bacterial strains.
Standard molecular biology procedures were used for the isolation of genomic DNAs and plasmids, for restriction digests, for molecular cloning, and for transformation by electroporation or heat shock. Taq polymerase was used to amplify DNA fragments. The ompR101 mutant allele linked to zhf37::Tn10 (22) was moved into strain JM109 by P1 transduction (to create GSO105). The hfq-1::Ω mutant allele (26) was moved into strain BW25113 by P1 transduction (GSO107). All other strains with gene deletions or mutations on the chromosome were constructed by a one-step method for gene inactivation in E. coli by using PCR products and BW25113 transformed with pKD46 and pCP20 (8). After mutants were generated, PCR was routinely used to confirm that the mutated regions had the expected sizes. Some of the PCR products also were sequenced to confirm the presence of mutations and fusions. All of the plasmids and bacterial strains used for this study are listed in Table 1. The sequences of all oligonucleotide primers used for this study are given at the following web site: http://dir2.nichd.nih.gov/nichd/cbmb/segr/segrPublications.html.
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| JM109 | Used as wild type | Stratagene |
| GSO105 | JM109 ompR101 zhf37::Tn10 | This study |
| BW25113 | Used as wild type | 8 |
| SC200 | BW25113 ΔmicC::kan (Kanr) | This study |
| SC201 | BW25113 ompC-luc (Cmr) | This study |
| SC204 | BW25113 ompC-lucΔmicC::kan (Kanr Cmr) | This study |
| SC209 | BW25113 ompCmutant-luc (Cmr) | This study |
| SC210 | BW25113 ompCmutant-luc ΔmicC::kan (Kanr Cmr) | This study |
| SC216 | BW25113 ompCmutant-luc ΔmicC (Cmr) | This study |
| SC218 | BW25113 ompC-luc ΔmicC (Cmr) | This study |
| GSO107 | BW25113 hfq-1::Ω (Cmr) | This study |
| Plasmids | ||
| pUC119 | Cloning vector (Ampr) | New England Biolabs |
| pUC-micC | pUC119 carrying ≈410-bp micC region (Ampr) | This study |
| pAlter-Ex2 | p15A origin cloning vector (Tetr) | Promega |
| pAE-micC | pAlter-Ex2 carrying ∼410-bp micC region (Tetr) | This study |
| pAE-micCmutant | pAlter-Ex2 carrying ∼410-bp mutated micC region (Tetr) | This study |
| pSP-luc+ | Cloning vector with luciferase reporter gene (Ampr) | Promega |
| pSP-ompC | ompC mRNA fused to luc in pSP-luc+ (Ampr) | This study |
| pSP-ompCmutant | Mutated ompC mRNA fused to luc in pSP-luc+ (Ampr) | This study |
| pSP-ompC (cat) | ompC mRNA fused to luc in pSP-luc+(Cmr) | This study |
| pSP-ompCmutant (cat) | Mutated ompC mRNA fused to luc in pSP-luc+ (Cmr) | This study |
| pKD3 | Plasmid carrying cat gene cassette and oriRγ origin (Cmr) | 8 |
| pKD4 | Plasmid carrying kan gene cassette and oriRγ origin (Kanr) | 8 |
| pKD46 | Red recombinase expression plasmid (Ampr) | 8 |
| pCP20 | FLP recombinase expression plasmid (Ampr) | 8 |
| pGEM-2 | Cloning vector (Ampr) | Promega |
| pGEM-micC | pGEM-2 carrying micC gene (Ampr) | This study |
| pGEM-micCmutant | pGEM-2 carrying micC mutant gene (Ampr) | This study |
| pGEM-ompC | pGEM-2 carrying ompC gene (Ampr) | This study |
| pGEM-micF | pGEM-2 carrying micF gene (Ampr) | 36 |
| pSP64 | Cloning vector (Ampr) | Promega |
| pSP64-dsrA | pSP64 carrying dsrA gene (Ampr) | This study |
For overexpression of the MicC RNA, an ∼410-bp DNA fragment (from 180 bp upstream to 110 bp downstream) of the micC gene was amplified from JM109 genomic DNA by the use of primers IS063-Bm and IS063-Ec and was then cloned into pUC119 (pUC-micC) and into pAlter-Ex2 (pAE-micC) after digestion with BamHI and EcoRI. For the generation of a micC mutant plasmid, a 200-bp fragment was amplified with primers IS063-Bm and MicC-PacRev and a 210-bp fragment was amplified with primers IS063-Ec and MicC-PacI. These two fragments were coligated into pAlter-Ex2 after digestion with PacI, BamHI, and EcoRI (pAE-micCmutant).
For construction of a micC deletion mutant (SC200), primers IS063-P1 and IS063-P2 were used to amplify the kanamycin resistance gene (Kanr) from pKD4 together with sequences flanking the micC gene. This fragment was then used for the linear transformation of strain BW25113. For unknown reasons, we found this insertion to be recalcitrant to transduction by P1.
The ompC-luc fusion strains were constructed by first creating the fusions in pSP-luc+ and then using amplified portions of the plasmids for linear transformation. For the wild-type ompC-luc fusion, a 250-bp fragment containing the transcriptional and translational elements of ompC was amplified from the genomic DNA with primers OmpC-Nde2 and OmpC-Nco and was then cloned into pSP-luc+ after digestion with NdeI and NcoI (pSP-ompC). For the mutant ompC-luc fusion, a 160-bp fragment was amplified with primers OmpC-Nde2 and OmpC-PacI and a second fragment was made by annealing two oligonucleotides, OmpC-Mut3 and OmpC-Mut4. The two fragments were coligated into pSP-luc+ after digestion with PacI, NdeI, and NcoI (pSP-ompCmutant). A 900-bp cat cassette obtained by digesting pKD3 with XbaI was cloned into pSP-ompC and pSP-ompCmutant to generate derivatives carrying chloramphenicol resistance (Cmr). The cat genes in both plasmids had the same orientations relative to the reporter genes. Fragments of the Cmr plasmids were generated by the use of primers Ybak-OmpC and Ybak-luc+ and were used to transform BW25113 to give strains SC201 and SC209. The micC deletion derivatives of SC201 (SC204) and SC209 (SC210) were generated with the PCR product used to make SC200. The Kanr markers in SC204 and SC210 were eliminated with pCP20 to create SC218 and SC216, respectively.
For in vitro synthesis of the ompC RNA, the 5′ end of the ompC gene (from −79 to +126) was amplified from MC4100 chromosomal DNA by a PCR with primers OmpC-Ec and OmpC-Hd. The PCR fragment was digested with EcoRI and HindIII and then cloned into the corresponding sites of pGEM-2 (pGEM-ompC). For in vitro synthesis of the wild-type MicC RNA, the entire micC coding sequence was amplified from MC4100 genomic DNA with primers MicC-Ec and MicC-Hd. The PCR fragment was digested with EcoRI and HindIII and then cloned into the corresponding sites of pGEM-2 (pGEM-micC). For the in vitro synthesis of mutant MicC RNA, site-directed mutagenesis (Stratagene, La Jolla, Calif.) was performed by using pGEM-micC as a DNA template and primers MicCmut2A and MicCmut2B (pGEM-micCmutant).
To generate a probe for DsrA, we amplified the dsrA gene from JM109 genomic DNA by using primers DsrA-Ec and DsrA-Hd and then cloned it into the pSP64 vector (Promega) after digestion with EcoRI and HindIII (pSP64-dsrA).
Growth conditions.
Unless otherwise specified, all strains were grown in Luria-Bertani (LB) broth or on LB agar containing appropriate antibiotics (23). M9-glycerol containing 1× M9 salts (23), 0.2% glycerol (vol/vol), 1 mM MgSO4, 0.1 mM CaCl2, and 2 μg of vitamin B1/ml was used as minimal medium.
Northern analysis.
For Fig. 1, total cellular RNAs were isolated by the use of Trizol reagent (Invitrogen). For Fig. 7, total and immunoprecipitated RNAs were isolated by phenol-chloroform extraction and ethanol precipitation. The RNAs were fractionated in an 8% polyacrylamide-8 M urea gel and transferred to Zeta Probe blotting membranes for MicC blots (Bio-Rad Laboratories, Hercules, Calif.) or to Hybond N membranes for MicF and DsrA blots (Amersham Biosciences, Piscataway, N.J.). A MicC Northern oligonucleotide capable of hybridizing to MicC was labeled at the 5′ end with 32P by the use of T4 polynucleotide kinase. The uniformly 32P-labeled MicF RNA probe was generated by using T7 RNA polymerase to transcribe plasmid pGEM-micF (36) linearized with EcoRI. The uniformly 32P-labeled DsrA RNA probe was generated by using SP6 RNA polymerase to transcribe plasmid pSP64-dsrA linearized with EcoRI. Zeta Probe blotting membranes were prehybridized and hybridized in Ultrahyb Oligo buffer (Ambion, Austin, Tex.) at 45°C. Hybond N membranes were prehybridized and hybridized in buffer containing 50% formamide, 1.5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 1% sodium dodecyl sulfate (SDS), and 0.5% dry milk at 55°C. All membranes were washed twice with 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at room temperature followed by two 25-min washes with 0.1× SSC-0.1% SDS at 45°C for Zeta Probe blotting membranes and at 55°C for Hybond N membranes.
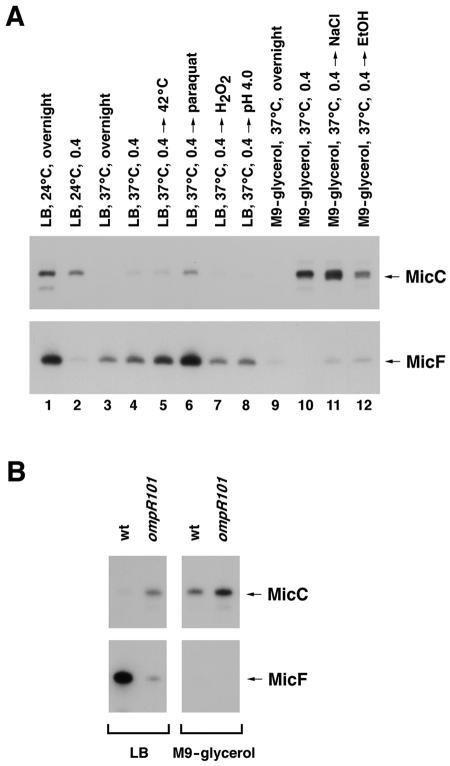
FIG. 1.
MicC and MicF RNA levels under various growth conditions. (A) Levels of MicC and MicF RNAs in JM109 cells grown under different growth conditions as follows: stationary phase (overnight; lane 1) or exponential phase (OD600 = 0.4; lane 2) in LB medium at 24°C; stationary phase (lane 3) or exponential phase (lane 4) in LB medium at 37°C; growth in LB medium at 37°C to exponential phase and then a switch to 42°C for 20 min (lane 5), a treatment with 0.5 mM paraquat for 20 min (lane 6), a treatment with 0.2 mM H2O2 for 5 min (lane 7), or a switch to pH 4.5 for 20 min (lane 8); stationary phase (lane 9) or exponential phase (lane 10) in M9-glycerol medium at 37°C; growth in M9-glycerol medium at 37°C to exponential phase and then a treatment with 0.3 mM NaCl (lane 11) or 10% ethanol (lane 12) for 20 min. The bands corresponding to the MicC and MicF RNAs are denoted by arrows. (B) Levels of MicC and MicF RNAs in wild-type and ompR mutant strains grown to exponential phase (OD600 = 0.4) in LB medium or M9-glycerol medium at 37°C. For both panels, 10-μg samples of total RNA were fractionated in 8% polyacrylamide-urea gels and analyzed by Northern hybridization with a labeled oligonucleotide complementary to MicC or a labeled RNA complementary to MicF.
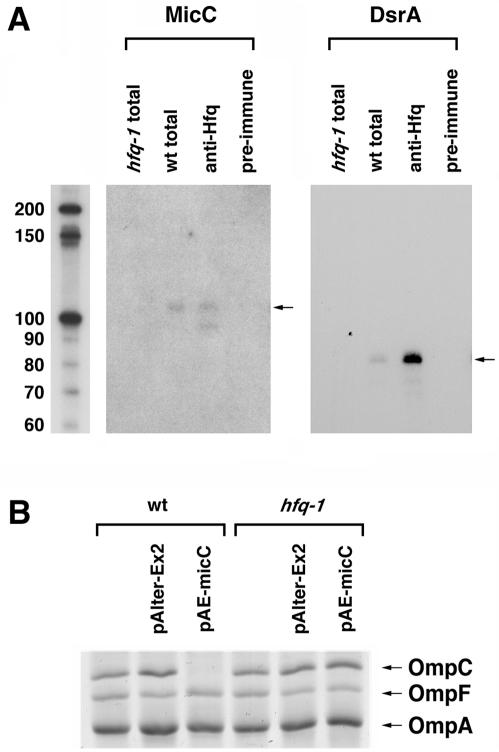
FIG. 7.
Requirement of Hfq for MicC repression of ompC. (A) Hfq binding to MicC RNA. Cell extracts were prepared from wild-type (BW25113) or hfq-1 mutant (GSO107) cells grown to an OD600 of 0.6 in LB medium at 24°C. Immunoprecipitations were carried out with the wild-type extracts and an Hfq antiserum or a preimmune serum and were compared to total RNAs from 1/10 extract equivalents of the wild-type and hfq-1 mutant RNAs. The levels of MicC and the DsrA positive control were determined by Northern hybridization. (B) Levels of OmpC, OmpF, and OmpA proteins in wild-type and hfq-1 mutant strains without and with pAlter-Ex2 or pAE-micC grown to an OD600 of 0.6 in LB medium at 24°C.
Primer extension analysis.
Primer extension assays were performed with primer OmpC-N1 labeled at the 5′ end with P33, RetroScript reverse transcriptase (Ambion), and total RNA isolated from JM109 cells grown overnight in LB medium (to an optical density at 600 nm [OD600] of 1.2) at room temperature. The total RNA (∼10 μg) was isolated from JM109 by use of an RNeasy kit (Qiagen, Valencia, Calif.) and was then incubated with ∼10 ng of the primer, 100 U of enzyme, a 0.12 μM concentration of each deoxynucleoside triphosphate, and 0.5 U of RNase inhibitor in 50 mM Tris buffer, pH 8.3, containing 75 mM KCl, 5 mM dithiothreitol, and 3 mM MgCl2 (20-μl reaction volume). The reaction was terminated after 1 h and loaded onto a QuickPoint mini sequencing gel (Invitrogen) together with a sequencing ladder generated from plasmid pIS063 by using the same primer and the DNA sequencing kit of the fmol DNA cycle sequencing system (Promega, Madison, Wis.).
3′ RACE analysis.
We performed 3′ rapid amplification of cDNA ends (3′ RACE) as described previously (4), using total RNA isolated from JM109 cells grown in LB medium to stationary phase (OD600 = 2.0) at 24°C. The total RNA (20 μg) was dephosphorylated with bacterial alkaline phosphatase (Invitrogen) and ligated with a 3′ RNA adapter (E1). cDNA was synthesized by reverse transcription with a primer complementary to the E1 RNA adapter (E4) by use of the Thermoscript RT-PCR system (Invitrogen) according to the manufacturer's instructions. After an RNase H treatment, the reverse transcription products were amplified by PCR with a micC gene-specific primer (MicC-RACE) and the adapter-specific primer (E4) and were cloned into the pCR2.1 TOPO TA vector (Invitrogen). The 3′ end of the MicC RNA was identified by DNA sequencing.
Luciferase assays.
For assays of the strains carrying ompC-luc fusions, overnight cultures were diluted 1:100 in fresh LB medium and incubated at 37°C for 6 h before being harvested. Activities were measured by use of a luciferase assay system (Promega) for bacteria, with slight modifications. The cell culture was diluted four times immediately before being mixed with carrier cells (BW25113), flash frozen, and lysed. The luminescence obtained after mixing 20 μl of the cell lysate and 100 μl of the assay reagent was measured in a microplate TopCount NXT scintillation counter (Perkin-Elmer LAS, Shelton, Conn.). The protein concentration of each cell lysate was measured by use of the noninterfering BCA protein assay reagent (Pierce, Rockford, Ill.). The luciferase activity is given in counting units normalized to the protein concentration for each protein extract. For strains expressing MicC and its mutant from plasmids, the host strains were freshly transformed. Cultures that were started from single colonies picked within 18 h after transformation and grown in LB broth with antibiotics for 12 h were used for the dilutions into fresh LB broth.
SDS-polyacrylamide gel electrophoresis.
To determine the levels of the OmpC protein, we incubated cells in LB medium to exponential phase (OD600 = 0.6) at 24°C and then prepared the cell envelopes as described previously (6, 17). The proteins were separated by SDS-polyacrylamide gel electrophoresis in 12% polyacrylamide gels containing 4 M urea and were stained with GelCode Blue stain (Pierce).
Toeprinting assays.
Toeprinting assays were performed by a modification of a previously described method (2). For the generation of OmpC, MicC, and MicCmutant RNAs, pGEM plasmids were linearized with HindIII, and the RNAs were synthesized by in vitro transcription with SP6 RNA polymerase. Annealing mixtures contained 0.2 pmol of ompC RNA, 0.6 pmol of 5′-end-labeled oligonucleotide OmpC-RT complementary to ompC RNA, and 1.2 pmol of wild-type MicC oligo, mutant MicC oligo #1, mutant MicC oligo #2, or in vitro-synthesized wild-type MicC or mutant MicC RNA. The mixtures were heated for 3 min at 65°C and then chilled in ice water for 15 min. The extension reactions contained the annealing mixtures, a 0.5 mM concentration of each deoxynucleoside triphosphate, 3 pmol of preactivated (30 min at 37°C) 30S ribosomal subunits (kindly provided by Steven Ringquist), 20 mM Tris-HCl, 10 mM magnesium acetate, 0.1 M NH4Cl, 0.5 mM EDTA, and 2.5 mM β-mercaptoethanol. After preincubation for 5 min at 37°C, 12 pmol of uncharged fMet-tRNA was added, and the reactions were incubated for an additional 10 min. Reverse transcriptase (1 U; Invitrogen) was added, and cDNA synthesis was allowed to proceed for 15 min. The cDNA products were analyzed in 8% polyacrylamide gels containing 8 M urea.
Immunoprecipitation.
Cell extracts were prepared from cultures of BW25113 or GS107 grown in LB medium at 24°C to exponential phase. Immunoprecipitations were performed as described previously (35), using 20 μl of an Hfq antiserum or preimmune serum, 24 mg of protein A-Sepharose (Amersham Biosciences), and 200 μl of cell extract.
RESULTS
A 109-nucleotide RNA encoded between ompN and ydbK.
In a previous computational search for novel small RNA genes in the E. coli genome, Chen et al. predicted that a small RNA might be expressed from a σ70 promoter with a −10 sequence at position 1435142 and with a putative terminator at position 1435259, and indeed they detected a small RNA of ∼100 nucleotides on Northern blots probed with a PCR-generated fragment (7). For this study, we set out to characterize this small RNA by examining its expression, determining its 5′ and 3′ ends, searching for homologs and complementary sequences by comparative genomic analysis, and assaying the phenotypes associated with decreased and increased expression of the RNA. Although the RNA was initially designated IS063, we have renamed it MicC for the reasons described below.
To examine the expression profile of the MicC RNA, we prepared total cellular RNAs from JM109 cells grown under a series of growth conditions and analyzed them on Northern blots probed with a labeled antisense oligonucleotide specific for the predicted transcript. As shown in Fig. 1A, expression of the MicC RNA was induced by growth at 24°C in both stationary- and exponential-phase cells (lanes 1 and 2). The level of the RNA was also elevated in cells grown to exponential phase in minimal medium with glycerol as the carbon source (lane 10) and was elevated further when these cells were exposed to osmotic shock (lanes 11). To a lesser degree, the expression of the RNA was induced in cells grown in LB broth and exposed to paraquat (lane 6). The MicC RNA was undetectable in stationary-phase cells grown in either LB or minimal medium at 37°C (lanes 3 and 9) and was only present at very low levels during exponential growth in LB medium at 37°C (lane 4), with no further induction by heat shock (lane 5) or exposure to H2O2 (lane 7) or a low pH (lane 8). Interestingly, under most of the conditions tested, the expression of the MicC RNA was opposite to the expression of the MicF RNA. Since OmpR has been reported to modulate MicF expression (reviewed in reference 9), we also examined MicC expression in an ompR mutant strain (Fig. 1B). As expected, MicF RNA levels were lower in the ompR mutant strain. On the other hand, MicC RNA levels were increased, indicating that OmpR, directly or indirectly, represses MicC expression.
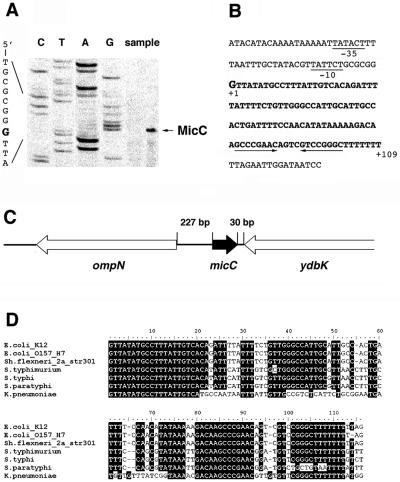
We also determined the 5′ and 3′ ends of the small RNA. To identify the 5′ end, we performed a primer extension analysis with total RNAs isolated from JM109 cells (Fig. 2A). A strong signal was detected corresponding to transcription starting at a G at position 1435145 (shown in bold and in a larger font in Fig. 2B), in agreement with our promoter prediction. The 3′ end was mapped to position 1435253 by 3′ RACE. Thus, the 3′ stem-loop terminator structure was retained even though some of the 3′ U residues were removed, which is a common feature among the mature small RNAs of E. coli (12). The 5′ and 3′ ends corresponded to a 109-nucleotide RNA, in agreement with our Northern analysis.
FIG. 2.
Sequence of MicC RNA. (A) Primer extension analysis of MicC RNA. Reverse transcriptase reactions were carried out as described in Materials and Methods, using total RNA isolated from JM109 cells grown overnight in LB medium at room temperature. The transcription initiation site corresponding to a G is indicated with an arrow. (B) Sequence of micC gene in E. coli K-12. The −10 and −35 promoter sequences are underlined, and bold letters denote the micC coding sequence. The stem-loop of the predicted terminator is indicated by arrows. (C) Chromosomal position of micC. micC is transcribed clockwise on the chromosome on the strand opposite the adjacent ompN and ydbK genes. (D) Alignment of micC homologs by the CLUSTALW program (http://molbio.info.nih.gov/molbio/gcglite/clustalw18.html).
The small RNA is encoded by the intergenic region between two open reading frames whose gene products are poorly characterized (Fig. 2C). The ompN gene is transcribed divergently upstream of micC. This gene encodes an outer membrane protein that has the highest similarity to OmpC (65% identity) (20). The ydbK gene, which is located downstream of micC and transcribed in the same direction as ompN, encodes a probable pyruvate-flavodoxin oxidoreductase. The ompN-micC-ydbK region on the K-12 chromosome is well conserved in E. coli O157:H7, with over 98% identity. MicC homologs were also found in all of the sequenced Shigella, Salmonella, and Klebsiella genomes, with the highest level of conservation encompassing the first 20 nucleotides and the terminator structure (Fig. 2D). The −10 and −35 sequences of the micC promoter are also conserved in the alignment of the entire intergenic sequence (http://dir2.nichd.nih.gov/nichd/cbmb/segr/segrPublications.html).
Complementarity between MicC and 5′ leader of ompC mRNA.
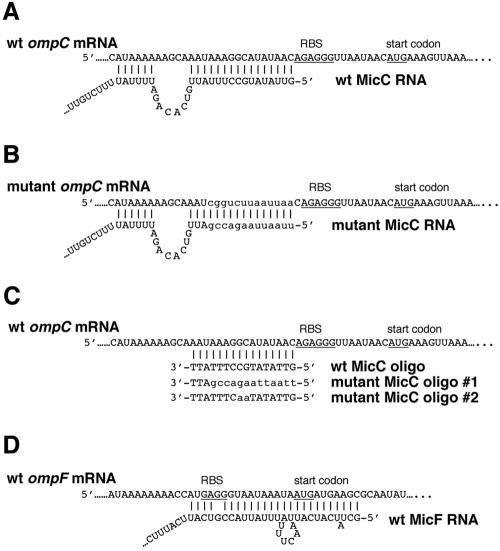
We next looked for sequences that are complementary to MicC in the E. coli genome. A BLASTN search revealed that 16 continuous nucleotides starting from the 5′ end of the small RNA are complementary to the sequence adjacent to the presumed ribosomal binding site of the ompC gene, which encodes major outer membrane protein C (Fig. 3A). Thus, MicC could potentially form base pairs with the ompC mRNA, suggesting that it may act as an antisense regulator. Upon allowing a gap, the potential antisense region can be extended another six nucleotides upstream. Since the micC and ompC genes are located at different genetic loci and are produced from two separate transcriptional units, the situation strongly mimics the MicF regulation of ompF (Fig. 3D). We also noted that the micC-ompN gene organization resembles the micF-ompC gene organization. These similarities encouraged us to test the hypothesis that MicC regulates ompC expression.
FIG. 3.
Proposed formation of MicC-ompC and MicF-ompF duplexes. (A) Base pairing between wild-type ompC mRNA leader and wild-type MicC RNA (pAE-micC) identified by a BLASTN search (http://www.ncbi.nlm.nih.gov/BLAST/) of the E. coli genome. (B) Base pairing between a mutant ompC mRNA leader and a mutant MicC RNA (pAE-micCmutant). The sequence of the PacI restriction site is UUAAUUAA. (C) Base pairing between wild-type ompC mRNA leader and wild-type and mutant oligonucleotides. (D) Base pairing between wild-type ompF mRNA leader and wild-type MicF RNA (24). Ribosome-binding sites (RBS) and start codons for ompC and ompF are underlined, and the mutant sequences are indicated with lowercase letters.
Effects of micC overexpression and deletion.
To assay the effects of eliminating MicC RNA expression, we created a mutant with the entire micC sequence deleted (positions 1435155 to 1435241) in the background of E. coli K-12 strain BW25113 (SC200). Since the stem-loop sequence comprising the micC terminator is likely to also serve as a terminator for the adjacent ydbK gene, the complete micC deletion may have affected expression of the unknown YdbK protein. Thus, we constructed a second deletion strain in which only the 5′ end of micC was deleted (positions 1435155 to 1435201), but we found that the two strains had similar phenotypes in all of the assays that we performed (data not shown). The micC region (∼410 bp) was also cloned into a plasmid, pAlter-Ex2 (pAE-micC), in strain BW25113 to create an overexpression strain. Northern analysis confirmed that MicC was absent from SC200 and that the micC RNA was overexpressed about 10-fold by pAE-micC (data not shown). Neither the deletion nor the overexpression strain showed significant variations in growth compared to the parent strain under various growth conditions.
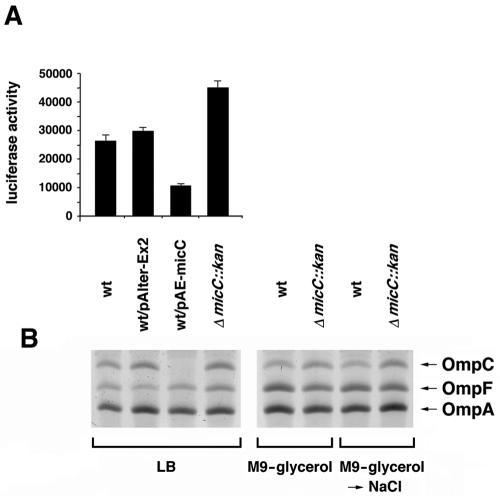
To test whether MicC had effects on the expression of the proposed target ompC mRNA, we constructed an ompC-luc translational fusion in which luc was fused in frame to the start codon of ompC (Fig. 4A). In this construct, the expression of luciferase was under the control of the transcriptional and translational elements of ompC, including three OmpR binding sites (27). The fusion was integrated into the intergenic region between genes ybaK and ybaP (positions 506382 to 506473) to avoid compensatory changes in the expression of other porin genes associated with the disruption of ompC. A previous deletion of the ybaK-ybaP region on the chromosome did not result in any observable phenotypes (data not shown). The ompC-luc fusion strain was transformed with the MicC overexpression plasmid (pAE-micC) and the corresponding control plasmid. The micC deletion was also moved into the fusion strain. The different ompC-luc strains were grown for 6 h in LB medium, and luciferase activities were assayed and normalized to the total protein concentrations. These assays showed that there was a 60% reduction in the expression of the ompC-luc fusion when MicC was overexpressed, while there was a 1.5- to 2.0-fold increase in ompC expression when micC was deleted (Fig. 4A). Similar increases and decreases in activity were observed for cells grown in minimal medium, although the overall levels of activity were lower, possibly due to some unknown expression problem associated with the luc gene in minimal medium (data not shown).
FIG. 4.
Effects of increased and decreased expression of MicC on ompC expression. (A) Luciferase activities (luminescence counts per microgram of protein) for the wild-type strain (SC201), the wild-type strain carrying the control pAlter-Ex2 vector, the wild-type strain carrying pAE-micC, and the corresponding micC deletion strain (SC204), with each grown for 6 h in LB medium at 37°C. The experiment was repeated three times, and averages and standard deviations are presented. (B) Levels of OmpC, OmpF, and OmpA proteins in the wild-type strain (BW25113), the wild-type strain carrying the control pAlter-Ex2 vector, the wild-type strain carrying pAE-micC, and the corresponding micC deletion strain (SC200), with each grown to an OD600 of 0.6 in LB medium at 24°C or an OD600 of 0.4 in M9-glycerol medium at 37°C and then treated with 0.3 mM NaCl for 20 min. The strains assayed in panel B were the same as the strains assayed in panel A, except they did not carry the ompC-luc fusion.
The effects of MicC overexpression and the micC deletion on ompC expression were also examined by monitoring OmpC protein levels directly in urea gels by using outer membrane protein preparations from cells grown to exponential phase. As shown in Fig. 4B, the overexpression of MicC clearly resulted in reduced OmpC protein levels, while the deletion of micC led to a twofold increase in OmpC levels, again suggesting that the MicC RNA represses ompC expression. Overexpression of the small RNA and deletion of the micC gene did not impact the expression of the OmpA and OmpF porins, which were also detected under these conditions.
Duplex formation between MicC and ompC-luc.
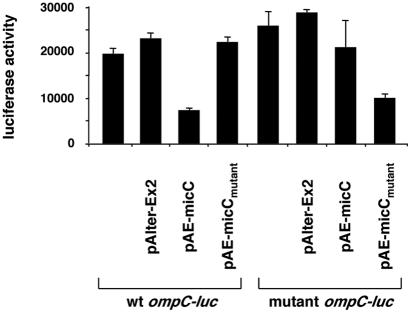
To further test for base pairing between MicC and the ompC mRNA, we separately mutated the regions of base pairing and investigated the effects of the mutations on ompC-luc gene expression. We scrambled 10 nucleotides in the base pairing region of the ompC mRNA leader while retaining the GC composition (25%) to create a mutated ompC-luc fusion in the plasmid. The mutated region had a PacI restriction site as its signature (Fig. 3B). The wild-type and mutant ompC-luc constructs were integrated into the ybaK-ybaP intergenic region on the chromosome by a double crossover. The micC deletion mutation subsequently was also recombined into these strains. In addition, we mutated the complementary nucleotides of the micC gene encoded on pAE-micC (generating pAE-micCmutant) to allow the mutant MicC RNA to form base pairs with the mutant, but not the wild-type, ompC-luc fusion (Fig. 3B). The mutant micC gene also carried a signature PacI site. The pAlter-Ex2 vector, pAE-micC, and pAE-micCmutant were separately transformed into the wild-type and mutant ompC-luc fusion strains carrying the micC deletion. The transformants and the nontransformed parental strains were then grown for 6 h, and luciferase activities were measured as described above.
As shown in Fig. 5, we again observed that wild-type MicC repressed wild-type ompC-luc expression about twofold (wt ompC-luc/pAE-micC). The overexpression of mutant MicC repressed the mutated ompC-luc fusion to a similar degree (mutant ompC-luc/pAE-micCmutant). This repression was not seen when wild-type MicC was expressed in the mutant ompC-luc strain (mutant ompC-luc/pAE-micC) or when mutant MicC was expressed in the wild-type ompC-luc strain (wt ompC-luc/pAE-micCmutant). The luciferase activities in the last two strains were almost identical to the activity levels seen for the untransformed strains as well as for the strains carrying the control vector. We thus concluded that the wild-type ompC-luc mRNA forms base pairs with and is repressed by wild-type MicC and that the antisense regulation of this reporter gene expression was only restored in a strain carrying the corresponding compensatory mutations.
FIG. 5.
Effects of compensatory mutations. Luciferase activities (luminescence counts per microgram of protein) for the micC deletion strain carrying the wild-type ompC-luc fusion (SC218) or the mutant ompC-luc fusion (SC216) and transformed with pAlter-Ex2, pAE-micC, or pAE-micCmutant are given. The experiment was repeated three times, and averages and standard deviations are presented.
MicC inhibits 30S ribosome binding to ompC mRNA.
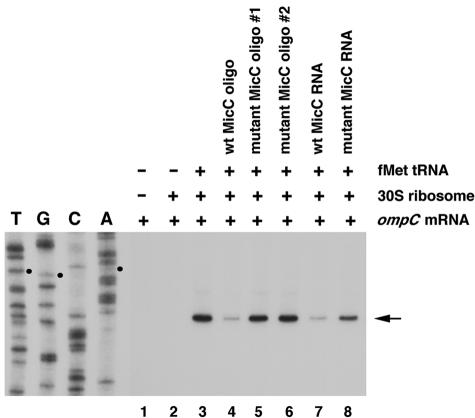
Unlike the region of MicF-ompF base pairing (Fig. 3D), the region of MicC-ompC base pairing does not overlap the ribosome binding site and the ompC start codon (Fig. 3A). Interactions involving nucleotides in the mRNA leader other than those of the ribosome binding site and the start codon have been shown to inhibit translation in other cases, such as that of the Trap-regulated genes in Bacillus and CsrA-regulated genes in E. coli (5, 13). However, we wanted to directly test whether MicC could block ribosome binding. Thus, we performed a primer extension inhibition assay, also called a toeprinting assay, to examine whether MicC-ompC RNA base pairing would inhibit the formation of the translational initiation complex. In vitro-synthesized ompC mRNA was annealed to an end-labeled primer complementary to a region 97 to 121 nucleotides downstream of the ompC translation start site. This complex was then incubated with 30S ribosome subunits in the presence or absence of uncharged fMet-tRNA. An analysis of the extension products revealed one ribosome-induced, fMet-tRNA-dependent termination site at the C residue 15 nucleotides downstream of the AUG start codon (Fig. 6, lane 3). This toeprint signal was decreased when a wild-type MicC oligonucleotide or RNA was added prior to incubation with the 30S subunits and the uncharged fMet-tRNA (lanes 4 and 7). In contrast, the addition of MicC mutant oligonucleotides (lanes 5 and 6) or a mutant RNA (lane 8) did not lead to a decrease in the toeprint, indicating that the mutant oligonucleotides and RNA were unable to repress 30S binding. The mutant RNA and mutant oligonucleotide 1 carried the same mutations as pAE-micCmutant, while mutant oligonucleotide 2 contained a GC→AA change (Fig. 3B and C). Interestingly, we did not detect an obvious difference between the two mutant oligonucleotides (Fig. 6, lanes 5 and 6), despite that fact that oligonucleotide 1 had a 10-nucleotide mismatch while oligonucleotide 2 only had a 2-nucleotide mismatch. This suggests that the GC dinucleotide contributes substantial energy to maintaining the small RNA-mRNA duplex. Together, the results demonstrate that MicC inhibits the formation of an ompC mRNA-30S ribosome initiation complex.
FIG. 6.
Toeprinting analysis of 30S ribosomal subunit binding to ompC mRNA. The arrow indicates the toeprint signal at the C residue, and the three small dots indicate the AUG start codon. The DNA sequencing reactions were carried out with the same end-labeled oligonucleotide used in the toeprinting assay.
Hfq requirement for MicC function.
All of the E. coli small RNAs that act by base pairing have been found to bind the Sm-like Hfq protein, and many have been shown to require Hfq for their function (reviewed in references 25 and 28). Given that MicC represses ompC by base pairing, the paradigms suggest that MicC also should bind to Hfq. To directly test whether the MicC RNA is bound by Hfq, we performed a Northern analysis with RNAs isolated from wild-type and hfq mutant cells as well as with RNAs immunoprecipitated with control and Hfq-specific antisera. MicC was indeed detected in the samples from Hfq immunoprecipitation, and the levels of the RNA were decreased in the hfq mutant strain, similar to what has been found for other Hfq binding RNAs such as the control DsrA RNA (Fig. 7A). We did note that the MicC RNA was extremely unstable in the cell extracts used for the immunoprecipitation assays. This may explain why MicC was not identified in a global screen for Hfq binding RNAs (36).
To test whether Hfq was required for MicC function, we also examined the effect of MicC overexpression on OmpC protein levels in wild-type and hfq mutant cells. The repression of OmpC levels observed for the wild-type strain was diminished in the hfq mutant, indicating that Hfq is needed for MicC base pairing with the ompC mRNA (Fig. 7B).
DISCUSSION
After the discovery of tens of potential small RNA genes in E. coli by our bioinformatics-based strategies and various approaches employed by many other groups (4, 7, 21, 29, 33, 36), work is starting to focus on the characterization of these novel small RNAs. In this paper, we show that a 109-nucleotide RNA identified in a computational screen contributes to the regulation of synthesis of the major outer membrane protein OmpC. MicC inhibits ompC expression at the posttranscriptional level by an antisense mechanism that involves the formation of base pairs between 22 nucleotides at the 5′ end of MicC and nucleotides just before the ribosome binding site of the ompC mRNA (Fig. 3).
Many parallels can be drawn between the 109-nucleotide MicC RNA and the 93-nucleotide MicF RNA. Both repress the expression of abundant porins by base pairing near the ribosome binding site, thereby blocking translation. Both are also encoded opposite other porin genes in E. coli. In addition, the MicF RNA and the corresponding ompF target sequence have been detected in Salmonella enterica serovar Typhi, S. enterica serovar Typhimurium, and Klebsiella pneumoniae (10), the same organisms in which MicC homologs are evident. It should be noted, however, that while an outer membrane protein-like open reading frame is present immediately upstream of micC in Klebsiella, a similar gene is absent from Salmonella sp.
Interestingly, the MicC and MicF RNAs were generally expressed under different environmental conditions. As shown in Fig. 1, MicF levels were elevated under most conditions in which MicC levels were low, while MicC levels were highest under those conditions in which MicF levels were low. These observations are in agreement with the previous observation that OmpC and OmpF show reciprocal expression under many environmental conditions (reviewed in reference 19). It should be noted, however, that the regulation of porin expression is complex and that there may be conditions in which similar levels of OmpC and OmpF are needed for growth and survival.
Given the analogies between the MicF regulation of ompF and the MicC regulation of ompC, it is tempting to speculate that there is a dual transcriptional regulation of micC and the adjacent ompN gene similar to what is observed for micF and the adjacent ompC gene. OmpN was characterized as a quiescent porin, with the amounts present in cells grown in rich media under normal laboratory conditions being too small to be detected (20). It was noted that OmpN resembles OmpC in its primary amino acid sequence and channel conductance, while OmpN is more similar to OmpF with respect to the differential uptake of mono- and disaccharides (20). There is no further information concerning the regulation of ompN and the functional relevance of OmpN to OmpC. The intergenic space between the ompN and micC genes is 229 bp, which is slightly shorter than the ∼256-bp spacer region between ompC and micF, and we could not find any sequence elements in the micC and ompN promoter region that were similar to the OmpR binding sites in the micF and ompC promoter region (27). However, given the coinduction of micF and ompC under conditions of high osmolarity, it might be interesting to test whether OmpN expression is increased under conditions in which MicC is maximally induced.
The base pairing between the MicC and MicF RNAs and their targets was more extensive than that observed for most of the E. coli small RNAs that act by base pairing. The ompF mRNA-MicF RNA duplex has been enzymatically and chemically characterized, and the region of base pairing was found to encompass 24 nucleotides (24). The ompC mRNA-MicC RNA interaction involves 16 contiguous nucleotides just before the ompC ribosome binding site in the upstream direction and an additional 6 nucleotides further upstream. The consequences of this extensive pairing are not known. As shown in Fig. 7, MicC binds to Hfq and requires the RNA chaperone to repress ompC. MicF has also been shown to bind to the RNA chaperone and is likely to require the protein for its function (36).
It is intriguing that small RNAs regulate the expression of two abundant outer membrane proteins, OmpC and OmpF, at a posttranscriptional level. The expression of yet another abundant outer membrane protein, OmpA, has also been shown to be posttranscriptionally regulated. In this case, Hfq binding has been found to stimulate ompA mRNA decay (30). Given the association of small RNAs with OmpC and OmpF expression, it is tempting to speculate that the regulation of OmpA expression also involves a trans-encoded RNA. A broader issue for future research is determining the advantage of antisense regulation for controlling the synthesis of outer membrane proteins.
Acknowledgments
We are grateful to M. Goulian for advice on separating the Omp proteins in urea gels, to S. Ringquist for 30S subunits, to B. Wanner and the E. coli Genetic Stock Center (EGSC) for providing the E. coli one-step gene inactivation system, and to T. A. Hall for providing software to analyze DNA sequences. We thank S. Gottesman and members of the laboratory for helpful discussions and comments on the manuscript.
REFERENCES
- 1.Aiba, H., S.-I. Matsuyama, T. Mizuno, and S. Mizushima. 1987. Function of micF as an antisense RNA in osmoregulatory expression of the ompF gene in Escherichia coli. J. Bacteriol. 169:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altuvia, S., D. Weinsterin-Fischer, A. Zhang, L. Postow, and G. Storz. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43-53. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, J., S. A. Forst, K. Zhao, M. Inouye, and N. Delihas. 1989. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264:17961-17970. [PubMed] [Google Scholar]
- 4.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. H. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 5.Babitzke, P., and P. Gollnick. 2001. Posttranscription initiation control of tryptophan metabolism in Bacillus subtilis by the Trp RNA-binding attenuation protein (TRAP), anti-TRAP, and RNA structure. J. Bacteriol. 183:5795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batchelor, E., and M. Goulian. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. USA 21:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S., E. A. Lesnik, A. T. Hall, R. Sampath, R. H. Griffey, D. J. Ecker, and L. B. Blyn. 2002. A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. BioSystems 65:157-177. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delihas, N., and S. Forst. 2001. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313:1-12. [DOI] [PubMed] [Google Scholar]
- 10.Esterling, L., and N. Delihas. 1994. The regulatory RNA gene micF is present in several species of gram-negative bacteria and is phylogenetically conserved. Mol. Microbiol. 12:639-646. [DOI] [PubMed] [Google Scholar]
- 11.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 12.Li, Z., S. Pandit, and M. P. Deutscher. 1998. 3′-Exoribonucleolytic trimming is a common feature of the maturation of small, stable RNAs in Escherichia coli. Proc. Natl. Acad. Sci. USA 95:2856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, M. Y., G. Gui, B. Wei, J. F. I. Preston, L. Oakford, U. Yuksel, D. P. Giedroc, and T. Romeo. 1997. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 272:17502-17510. [DOI] [PubMed] [Google Scholar]
- 14.Massé, E., N. Majdalani, and S. Gottesman. 2003. Regulatory roles for small RNAs in bacteria. Curr. Opin. Microbiol. 6:120-124. [DOI] [PubMed] [Google Scholar]
- 15.Matsuyama, S.-I., and S. Mizushima. 1985. Construction and characterization of a deletion mutant lacking micF, a proposed regulatory gene for OmpF synthesis in Escherichia coli. J. Bacteriol. 162:1196-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizuno, T., M.-Y. Chou, and M. Inouye. 1984. A unique mechanism regulating gene expression: translational inhibition of a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. USA 81:1966-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morona, R., and P. Reeves. 1982. The tolC locus of Escherichia coli affects the expression of three major outer membrane proteins. J. Bacteriol. 150:1016-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 20.Prilipov, A., P. S. Phale, R. Koebnik, C. Widmer, and J. P. Rosenbusch. 1998. Identification and characterization of two quiescent porin genes, nmpC and ompN, in Escherichia coli BE. J. Bacteriol. 180:3388-3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rivas, E., R. J. Klein, T. A. Jones, and S. R. Eddy. 2001. Computational identification of noncoding RNAs in E. coli by comparative genomics. Curr. Biol. 11:1369-1373. [DOI] [PubMed] [Google Scholar]
- 22.Russo, F. D., and T. J. Silhavy 1991. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 5:567-580. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Schmidt, M., P. Zheng, and N. Delihas. 1995. Secondary structures of Escherichia coli antisense micF RNA, the 5′-end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry 34:3621-3631. [DOI] [PubMed] [Google Scholar]
- 25.Storz, G., J. A. Opdyke, and A. Zhang. 2004. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 7:140-144. [DOI] [PubMed] [Google Scholar]
- 26.Tsui, H. C., G. Feng, and M. E. Winkler. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J. Bacteriol. 179:7476-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsung, K., R. E. Brissette, and M. Inouye. 1989. Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA foot printing. J. Biol. Chem. 264:10104-10109. [PubMed] [Google Scholar]
- 28.Valentin-Hansen, P., M. Eriksen, and C. Udesen. 2004. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 51:1525-1533. [DOI] [PubMed] [Google Scholar]
- 29.Vogel, J., V. Bartels, T. H. Tang, G. Churakov, J. G. Slagter-Jäger, A. Hüttenhofer, and E. G. H. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vytvytska, O., I. Moll, V. R. Kaberdin, A. von Gabain, and U. Blasi. 2000. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 14:1109-1118. [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, E. G. H., S. Altuvia, and P. Romby. 2002. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 46:361-398. [DOI] [PubMed] [Google Scholar]
- 32.Wassarman, K. M. 2002. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell 109:141-144. [DOI] [PubMed] [Google Scholar]
- 33.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wassarman, K. M., A. Zhang, and G. Storz. 1999. Small RNAs in Escherichia coli. Trends Microbiol. 7:37-45. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, A., K. M. Wassarman, J. Ortega, A. C. Steven, and G. Storz. 2002. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell 9:11-22. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, A., K. M. Wassarman, C. Rosenow, B. C. Tjaden, G. Storz, and S. Gottesman. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 50:1111-1124. [DOI] [PubMed] [Google Scholar]