Abstract
The enzyme 11β-hydroxysteroid dehydrogenase type 2 (11βHSD2) is selectively expressed in aldosterone target tissues, where it confers aldosterone selectivity for the mineralocorticoid receptor by inactivating 11β-hydroxyglucocorticoids. Variable activity of 11βHSD2 is relevant for blood pressure control and hypertension. The present investigation aimed to elucidate whether an epigenetic mechanism, DNA methylation, accounts for the rigorous control of expression of the gene encoding 11βHSD2, HSD11B2. CpG islands covering the promoter and exon 1 of HSD11B2 were found to be densely methylated in tissues and cell lines with low expression but not those with high expression of HSD11B2. Demethylation induced by 5-aza-2′-deoxycytidine and procainamide enhanced the transcription and activity of the 11βHSD2 enzyme in human cells in vitro and in rats in vivo. Methylation of HSD11B2 promoter–luciferase constructs decreased transcriptional activity. Methylation of recognition sequences of transcription factors, including those for Sp1/Sp3, Arnt, and nuclear factor 1 (NF1) diminished their DNA-binding activity. Herein NF1 was identified as a strong HSD11B2 stimulatory factor. The effect of NF1 was dependent on the position of CpGs and the combination of CpGs methylated. A methylated-CpG–binding protein complex 1 transcriptional repression interacted directly with the methylated HSD11B2 promoter. These results indicate a role for DNA methylation in HSD11B2 gene repression and suggest an epigenetic mechanism affecting this gene causally linked with hypertension.
Introduction
The intracellular access of glucocorticoids to their receptors is modulated by the 11β-hydroxysteroid dehydrogenase (11βHSD) enzymes, which interconvert biologically active 11β-hydroxyglucocorticoids and inactive 11-ketosteroids (1–3). The enzyme 11βHSD2 catalyzes the dehydrogenation of 11β-hydroxyglucocorticoids, has a nanomolar Km for glucocorticoids, utilizes NAD+ as a cofactor, and is localized in the endoplasmic reticulum membrane with a cytoplasmic orientation of its catalytic domain (4, 5). The enzyme exhibits cell-specific expression in mineralocorticoid target tissues, such as epithelial cells from colon or renal cortical collecting tubules, where its main function is to protect the nonselective mineralocorticoid receptor (MR) from activation by 11β-hydroxyglucocorticoids (1, 4). Reduced activity of 11βHSD2 leads to overactivation of the MR by cortisol with renal sodium retention, hypokalemia, and a salt-sensitive increase in blood pressure (6, 7). Compromised 11βHSD2 activity can be caused by two mechanisms: first, by loss-of-function mutations of the gene encoding 11βHSD2 (HSD11B2), and second, by xeno- or endobiotics or shear stress (4). Slightly reduced activity of the 11βHSD2 enzyme is associated with a salt-sensitive increase in blood pressure and polymorphic markers in the gene in normal volunteers or in patients with essential hypertension (8, 9). Furthermore, associations between microsatellites or variants of the HSD11B2 gene and the occurrence of end-stage renal failure have been observed (10, 11). All these findings suggest that 11βHSD2 is relevant for blood pressure control. However, the frequency of mutated exons in the HSD11B2 gene is extremely low (11). Thus, other mechanisms accounting for the inter-individual variable 11βHSD2 enzyme activities have to be considered.
The mechanism of the cell-specific constitutive expression of HSD11B2 in mineralocorticoid target tissues is poorly understood. Others and we have reported that the transcription factors Sp1 and/or Sp3 (Sp1/Sp3) interact with the HSD11B2 gene promoter (12, 13). Sp1/Sp3 are ubiquitously expressed and therefore cannot sufficiently account for the distinct cell type–specific expression of HSD11B2. The HSD11B2 promoter comprises a highly G + C–rich (or GC-rich) core, contains more than 80% GC, lacks a TATA-like element, and has two typical CpG islands. This raises the possibility that CpG dinucleotide methylation may play a role in the cell type–specific and possibly in the epigenetically determined inter-individual variable expression of HSD11B2. Here we present evidence that CpG methylation regulates HSD11B2 expression in vitro and in vivo.
Results
The HSD11B2 promoter comprises CpG islands.
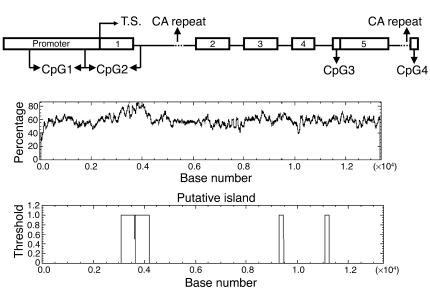
Analysis of a 13,400-bp DNA fragment ranging from nucleotide –3,731 to nucleotide +9,669 in the human HSD11B2 gene showed two CpG islands located in the promoter and exon 1 (nucleotides –633 to –97 and –77 to +460) and two islands in exon 5 and the downstream region (nucleotides +5,569 to +5,721 and +7,357 to +7,515) (Figure 1). Analysis of rat HSD11B2 revealed one CpG Island in the promoter and exon 1 (nucleotides –358 to +763) (Figure 7C).
Figure 1.
CpG plot of the human HSD11B2 gene. The top represents the putative CpG islands along the gene and the middle graph indicates the relative abundance of CpGs (Percentage) as a function of the base number. CpG islands were defined as a region in which the calculated percentage of CpGs over an average range of ten windows was over 50% and the calculated versus the expected CpG distribution higher than 0.6. White boxes in the bottom graph indicate CpG islands identified and investigated. T.S., transcription start site.
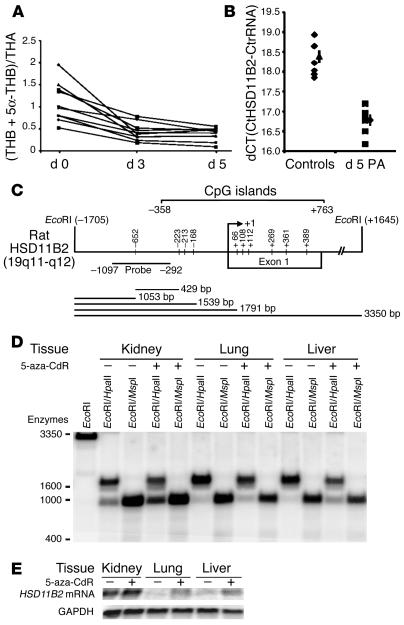
Figure 7.
Effect of procainamide and 5-aza-CdR on HSD11B2 in rats. (A and B) After treatment with procainamide (PA), the urinary (THB + 5α-THB) / THA ratios declined in all animals (A), indicating increased activity of 11βHSD2. This observation is in line with the elevated mRNA levels in kidney tissue of rats given procainamide (B). The y axis indicates the difference between the cycle threshold values (dCT) of HSD11B2 mRNA and ribosomal RNA as internal control. A low value indicates high content of HSD11B2 mRNA. (C) Schematic representation of rat HSD11B2 CpG islands spanning from nucleotide –358 to nucleotide +763 with respect to the transcription initiation site (arrow at +1). A fragment spanning nucleotides –1,097 to –292 was used for Southern blot analysis. Horizontal bars with numbers (below diagram) indicate the expected size of the hybridized fragments. (D) Methylation-sensitive restriction enzyme analyses. Genomic DNA from different tissues of control rats (–) and 5-aza-CdR–treated rats (+) was double-digested with EcoRI and either HpaII or MspI and was fractionated by agarose gel electrophoresis followed by Southern blotting. For interpretation, see the Results section. (E) Northern blot analyses without and with 5-aza-CdR treatment. An XhoI fragment of HSD11B2 was used as a probe. Expression of HSD11B2 increased after administration of 5-aza-CdR in the three tissues analyzed.
The HSD11B2 gene is upregulated by 5-aza-2′-deoxycytidine and procainamide.
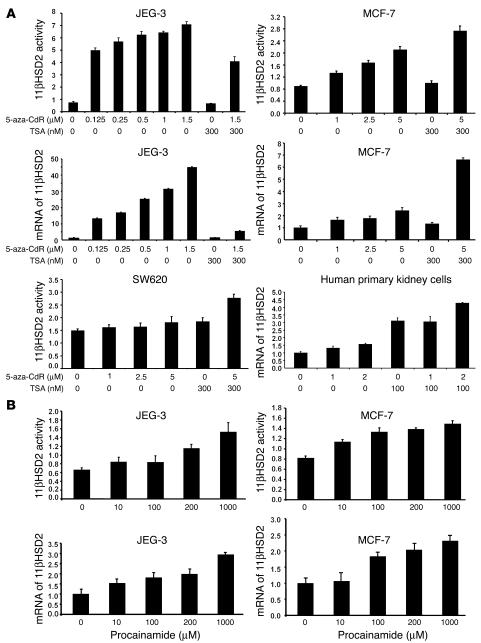
To examine the role of DNA methylation in HSD11B2 expression, we grew cell lines expressing various levels of the HSD11B2 with and without 5-aza-2′-deoxycytidine (5-aza-CdR), a methyltransferase inhibitor. Dose-dependent activation of the gene was observed (Figure 2A), suggesting that demethylation enhances the expression of HSD11B2. Promoters with methylated CpG islands are hypoacetylated and repressed (14). To test whether histone hyperacetylation enhances HSD11B2 expression, we treated cells with the histone deacetylase (HDAC) inhibitor trichostatin A (TSA). TSA alone did not induce HSD11B2 gene expression in JEG-3, MCF-7, and SW620 cells (Figure 2A), but the addition of TSA for 12–24 hours after 48 hours of treatment with 5-aza-CdR increased the expression in MCF-7 and SW620 cells, an effect not observed in JEG-3 cells (Figure 2A). When carcinoma cell lines are established, certain genes are methylated de novo. Therefore, we examined the effect of 5-aza-CdR on HSD11B2 expression in primary human kidney cells and again observed a dose-dependent activation of the gene (Figure 2A). Like 5-aza-CdR, procainamide, a less effective DNA methyltransferase inhibitor (15), increased the activity and mRNA of 11βHSD2 in a dose-dependent way (Figure 2B).
Figure 2.
The effect of 5-aza-CdR, TSA, or procainamide on the activity and mRNA of HSD11B2. (A and B) The activity of 11βHSD2 is expressed as pmol/mg/h and the abundance of mRNA is normalized to S18 rRNA. The activity and/or mRNA of HSD11B2 increased after the addition of 5-aza-CdR (A) or procainamide (B) in all cells analyzed. TSA increased the activity and/or mRNA when the MCF-7, SW620, and human primary cells were pretreated with 5-aza-CdR (A). The abundance of mRNA paralleled the changes in activity (mRNA in SW620 cells and activity in human primary cells were not measured). The data are given as the mean ± SD of triplicate samples of a representative experiment repeated at least three times.
Reduced glucocorticoid receptor transactivation by cortisol after 5-aza-CdR treatment.
The intracellular access of glucocorticoids to the receptor is modulated by 11βHSD2. In humans it converts cortisol into biologically inactive cortisone. To demonstrate that demethylation of the HSD11B2 gene causes a relevant enhanced enzyme activity in intact cells, we used a transactivation assay (3). For that purpose, the mouse mammary tumor virus (MMTV) promoter, which contains essential glucocorticoid response elements, was linked to a luciferase gene (MMTV-Luc) and was cotransfected with a glucocorticoid receptor (GR) expression vector and a thymidine kinase–Renilla luciferase (TK–Renilla luciferase) internal control plasmid in JEG-3 cells. Transfected cells express luciferase only when an intracellular 11β-hyroxyglucocorticoid such as cortisol, but not when an 11-ketosteroid such as cortisone, is present (3). Increased 11βHSD2 activity enhances the conversion of cortisol to cortisone, thus causing a diminished luciferase expression. Diminished transactivation of MMTV-Luc was consistently observed after 5-aza-CdR treatment (Figure 3D), indicating an enhanced 11βHSD2 activity. That the reduced transactivation upon 5-aza-CdR treatment was caused by decreased cortisol concentrations due to increased 11βHSD2 activity rather than by modulation of the GR itself was demonstrated by the fact that dexamethasone-mediated transactivation by GR was not altered by 5-aza-CdR treatment (Figure 3D). Both dexamethasone and 11-keto-dexamethasone, the product of 11βHSD2, are potent GR agonists (16).
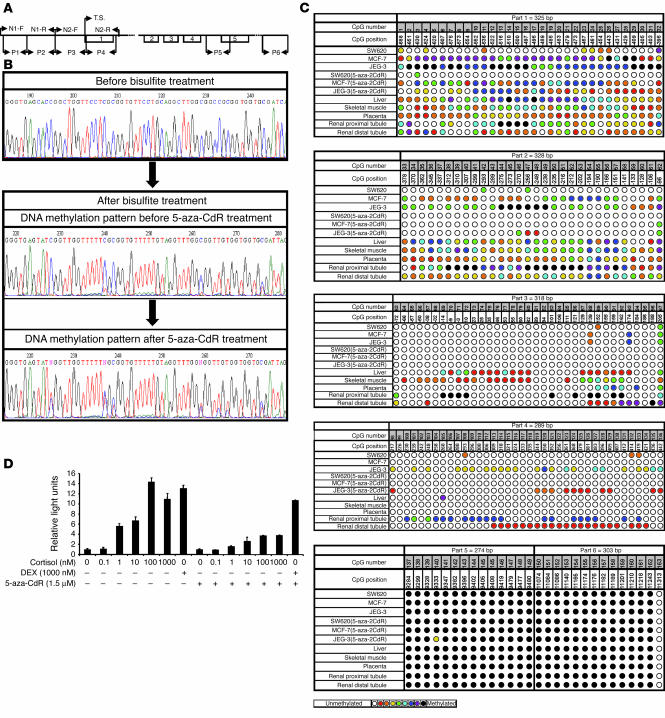
Figure 3.
The effect of 5-aza-CdR on the DNA methylation pattern of HSD11B2 and glucocorticoid-mediated transactivation of MMTV-Luc. (A) Schematic representation of HSD11B2 gene. P1–P6 indicate the stretches of DNA amplified for methylation analyses and refer to parts 1–6 in C. N1-F, N1-R, N2-F, and N2-R are nested primers. (B) Example of bisulfite sequencing (nucleotides –456 to –391). The sequences before (top) and after bisulfite treatment either without (middle) or with (bottom) 5-aza-CdR treatment are presented. After bisulfite treatment, C-to-T conversion did not occur at several CG sites, indicating methylation of those CpG dinucleotides. Treatment with 5-aza-CdR increased unmethylated forms of alleles, as shown by increased T versus C signals (compare middle and bottom). (C) Methylation pattern of the HSD11B2 promoter and exonic and downstream CpG islands in various cells and tissues. The CpG number indicates the number of the CpG dinucleotides along the promoter and CpG position denotes the position of this CpG in relation to the transcription start site. The color of the circle reflects the degree of methylation (key). (D) Effect of 5-aza-CdR on glucocorticoid-mediated MMTV-Luc transactivation in JEG-3 cells. The dose-response curve of cortisol was blunted by 5-aza-CdR. Transfections were performed in triplicate; all results were confirmed by at least two different independent experiments.
Hypermethylation of the HSD11B2 promoter correlates with an absence of HSD11B2 expression.
Hypermethylation of normally unmethylated CpG islands correlates with transcriptional repression (17, 18). To define the methylation status of HSD11B2 CpG islands, we performed bisulfite sequencing (19). Examples of sequencing are shown in Figure 3B. The cytosines in the CpG dinucleotides remained unchanged at several positions after bisulfite treatment and therefore were methylated. In contrast, all non-CpG dinucleotide cytosines appeared after PCR as thymidines, indicating complete bisulfite modification. After 5-aza-CdR treatment, the thymidine signals (unmethylated CpGs) increased compared with the cytosine signals (methylated CpGs), indicating reduced DNA methylation (Figure 3B). Figure 3C shows the methylation pattern of the HSD11B2 CpG islands in normal human tissues and in cell lines before and after 5-aza-CdR treatment. The methylation levels were low in SW620 cells, placenta, and distal renal tubules when part 1 (nucleotides –704 to –380) and part 2 (nucleotides –403 to –76) were considered (Figure 3C). In contrast, the same regions were methylated in proximal tubules, liver, and skeletal muscle as well as in JEG-3 and MCF-7 cells. Methylation of CpGs was reduced by 5-aza-CdR in JEG-3 and MCF-7 cells (Figure 3C, parts 1 and 2), an effect that correlated with higher expression of the HSD11B2 gene (Figure 2A). In part 3 (nucleotides –102 to +215), normal tissues but not cell lines were slightly methylated, whereas in part 4 (nucleotides +187 to +476), only JEG-3 cells presented evidence of 5-aza-CdR reversible methylation (Figure 3C). Two downstream CpG islands (part 5, nucleotides +5,510 to +5,784 and part 6, nucleotides +7,295 to +7,597) were fully methylated in all samples before and after 5-aza-CdR treatment. Liver, muscle, and proximal kidney tubules, with strong CpG methylation, are well known to express almost no HSD11B2, whereas placenta and distal kidney tubules, with a lower degree of CpG methylation, express HSD11B2 (20). Thus, these data reveal a strong negative correlation between the methylation status and gene expression of the HSD11B2 promoter in tissues, especially in parts 1 and 2, an observation in line with the lower degree of methylation and higher 11βHSD2 activity in SW620 than in JEG-3 or MCF-7 cells (Figure 2).
Methylated CpG sites in the HSD11B2 promoter are targets for protein binding.
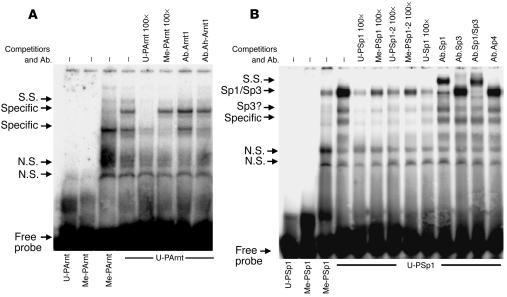
Sites with high scores for potential binding for transcription factor complexes including nuclear factor 1 (NF1), AP2, SP1, Ah-Arnt, Arnt1, AP4, GC-box, and Ik2 in methylated CpG sites were identified. To prove that methylation-dependent protein-DNA interactions occur, we performed electrophoretic mobility-shift assays (EMSAs) with methylated (Me-) and unmethylated (U-) probes (Ps) containing binding sites for Arnt and Ah-Arnt (nucleotides –518 to –489, including six CpGs), Sp1/Sp3 (nucleotides –287 to –268, including three CpGs), and NF1 (nucleotides –419 to –397, including three CpGs) (Table 1).
Table 1.
Oligonucleotides for bisulfite sequencing, EMSA, or cloning
EMSA with MCF-7 cell nuclear protein extracts revealed two specific complexes with the unmethylated probe (U-PArnt) and one with the methylated probe (Me-PArnt) (Figure 4A). Unmethylated excess competitor (U-PArnt 100×) displaced both complexes, but methylated competitor (Me-PArnt 100×) displaced only the lower one, indicating methylation sensitivity of the upper complex. The lower complex comprises Ah-Arnt, because antibody against Ah-Arnt (Ab.Ah-Arnt) but not against Arnt1 (Ab.Arnt1) inhibited formation of the lower complex. EMSA with MCF-7 nuclear protein extracts using an Sp1/Sp3 probe (nucleotides –287 to –268) revealed three abundant protein-DNA complexes when the probe was unmethylated (U-PSp1), while the complexes were reduced when the methylated counterpart (Me-PSp1) was used (Figure 4B). Unmethylated probe in excess (U-PSp1 100×) competed more efficiently with U-PSp1 than did methylated probe (Me-PSp1 100×), an observation confirmed by another set of methylated (Me-PSp1-2 100×) and unmethylated (U-PSp1-2 100×) competitors and an unmethylated probe containing the Sp1/Sp3 recognition binding site (U-Sp1 100×) (Figure 4B). Supershift studies showed antibodies against Sp1, Sp3, or both together (Figure 4B, Ab.Sp1, Ab.Sp3, and Ab.Sp1/Sp3), but not the unrelated antibody (Ab.Ap4), shifted the complexes. Thus, methylation decreases Sp1/Sp3 binding.
Figure 4.
Binding of nuclear proteins on methylated or unmethylated putative Arnt- or Sp1/Sp3-binding sites in the HSD11B2 gene. (A and B) The positions of specific complexes (Specific, Sp1/Sp3, and Sp3) or nonspecific bands (N.S.) are indicated along the left margins. Supershifts (S.S.) were performed with specific antibodies (Ab.Arnt, Ab.Ah-Arnt, Ab.Sp1, or Ab.Sp3), combinations (Ab.Sp1/Sp3), or anti-Ap4 as a nonspecific control (Ab.Ap4). For competition studies, a 100-fold (100×) molar excess of cold methylated or unmethylated probe was used.
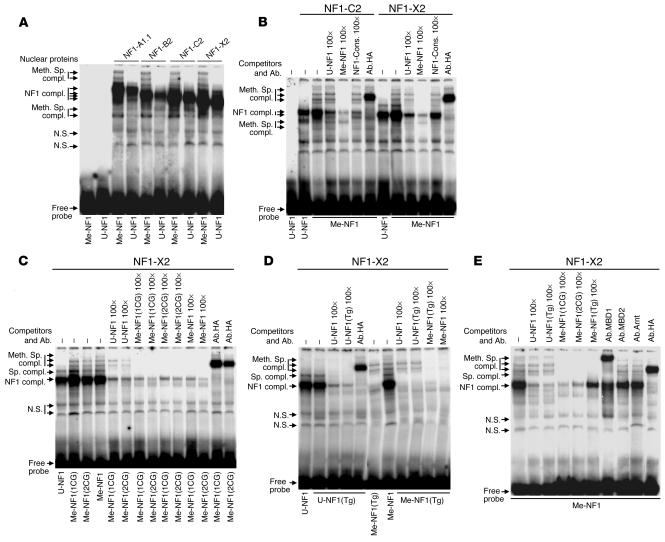
NF1 isoforms interact with methylated-CpG binding proteins at NF1 binding sites.
EMSA performed on SW620 cell nuclear protein extracts after overexpression of different murine HA-tagged NF1 isoforms with an NF1 recognition site (nucleotides –419 to –397) revealed the formation of a main complex with all four NF1 isoforms, whether fully methylated (Me-NF1) or unmethylated (U-NF1) NF1 probes were considered (Figure 5, A and B). Above and below the main complex, several additional bands restricted to the methylated NF1 probe (Me-NF1) appeared, suggesting presence of methylated-CpG binding proteins (Figure 5, A and B). Me-NF1 was completely displaced by excess of unlabeled Me-NF1 (Me-NF1 100×) (Figure 5B), while unmethylated competitor (U-NF1 100×) caused only a partial displacement, an effect confirmed by the use of an excess of unmethylated probe containing an NF1 binding site (U-NF1-Cons. 100×) (Figure 5B). The main complex and methylation-specific complexes were shifted by antibody against HA (Figure 5B). Similar DNA-protein interactions were observed with two additional isoforms of NF1 (A1.1 and B2) (data not shown). Taken together, these results indicate that methylated-CpG binding proteins interact with the NF1 at methylated NF1 binding sites.
Figure 5.
EMSA with differentially methylated or unmethylated NF1 probes. (A and B) EMSA with fully methylated (Me-NF1) or unmethylated (U-NF1) NF1 probes of the human HSD11B2 promoter was performed on SW620 cell nuclear protein extracts after overexpression of HA-tagged murine NF1 isoforms NF1-A1.1, NF1-B2, NF1-C2, or NF1-X2 (A) and NF1-C2 or NF1-X2 (B). The position of methylation-specific complexes (Meth. Sp. compl.) and NF1-specific complexes (NF1 compl.) or nonspecific bands (N.S.) are indicated along the left margins. Supershifts were performed with HA-specific antibodies (Ab.HA). For competition studies, a 100-fold (×100) molar excess of a cold methylated (Me-), unmethylated (U-) or a consensus unmethylated (NF1-Cons. 100×) NF1 probe was used. The sequences and methylation sites of the probes are given in Table 1. (C–E) EMSA with site-specific methylated probes. EMSA was performed on SW620 cell nuclear protein extracts after overexpression of the HA-tagged murine NF1-X2 isoform. Different DNA probes [Me-NF1(1CG), Me-NF1(2CG), Me-NF1, and U-NF1], including mutated probes [U-NF1(Tg) or Me-NF1(Tg)] and their corresponding cold competitors (×100), were used (see Table 1). Supershifts were performed with specific (Ab.HA, Ab.MBD1) or unrelated (Ab.MBD2 and Ab.Arnt) antibodies. Whereas C demonstrates that the position of CpG methylation and D, that the composition of core neighboring nucleotides affect the binding of NF1, E suggests that MBD1 interacts with NF1 at the NF1 binding site. Sp. compl.; specific complexes.
Position, number of methylated CpGs, and type of neighboring nucleotides determine NF1 binding.
The following NF1 probes were incubated with SW620 nuclear protein overexpressing NF1-X2: Me-NF1(1CG), with the CpG next to the core methylated; Me-NF1(2CG), with two adjacent CpGs methylated; Me-NF1, with all three CpGs methylated; and U-NF1, with no CpG methylated (Table 1). These experiments revealed that, similar to the fully methylated NF1 probe (Me-NF1), the “positionally methylated” probes Me-NF1(1CG) and Me-NF1(2CG) were able to bind NF1 and methylated-CpG binding proteins (Figure 5C). Compared with Me-NF1, Me-NF1(1CG) increased but Me-NF1(2CG) slightly decreased affinity for the complexes. Competition experiments revealed that Me-NF1(1CG) and Me-NF1(2CG) were highly displaced from the main but less so from the methylation-specific complexes by an excess of U-NF1 100×, while Me-NF1(1CG) 100×, Me-NF1(2CG) 100×, and Me-NF1 100× completely displaced the Me-NF1(1CG) and Me-NF1(2CG) probes (Figure 5C). Unmethylated and methylated cold probes competed better with the Me-NF1(2CG) probe than with the Me-NF1(1CG) probe, confirming that Me-NF1(1CG) has a higher affinity for the NF1 main complex.
All NF1 proteins bind as a dimer to DNA in the major groove and recognize a dyad symmetric consensus sequence (Cons.) TTGGC(N)5GCCAA and with less affinity to individual “half-sites” (TTGGC or GCCAA) (21). The core binding site of NF1 (TTGGC) contains a cytosine at the 3′ end, with the potential to be the cytosine of another CpG. In the HSD11B2 gene promoter we identified both configurations: the C followed by a G or the C followed by another nucleotide. To investigate the relevance of these two possible configurations with respect to the methylation status by EMSA, we used two mutated NF1 probes, U-NF1(Tg) and Me-NF1(Tg), and SW620 nuclear extracts overexpressing NF1-X2 (Table 1). The U-NF1(Tg) and U-NF1 probes produced an identical pattern of shifted bands (Figure 5D). NF1-specific binding was confirmed by competition assays using cold U-NF1 100× and U-NF1(Tg) 100× probes and by supershift assays with an antibody against HA (Figure 5D). Interestingly, when methylation occurred at CpGs outside of the core NF1 binding site of the mutated probe Me-NF1(Tg), the NF1 main complex and the methylation-specific complexes decreased when compared with the complexes obtained when unmethylated [U-NF1 and U-NF1(Tg)] and methylated (Me-NF1) probes were used (Figure 5D). The specific binding of the complexes was confirmed using an excess of U-NF1 100×, U-NF1(Tg) 100×, Me-NF1(Tg) 100×, or Me-NF1 100× (Figure 5D). These data demonstrate the pivotal role of the core neighboring nucleotide for DNA-NF1 interactions. This conclusion was confirmed by the use of end-labeled Me-NF1 probe (Figure 5E).
To study the type of methylated-CpG binding protein involved (22), we demonstrated by supershift studies that methylation-specific complexes and parts of the main complex shifted with antibodies against methylated-CpG–binding domain protein 1 (MBD1) but not against MBD2 and Arnt1 (Figure 5E). Antibody against HA again showed a supershifted band. These data indicate that the MBD1 interacts with NF1 at the NF1 binding site.
In vitro methylation represses the HSD11B2 promoter activity.
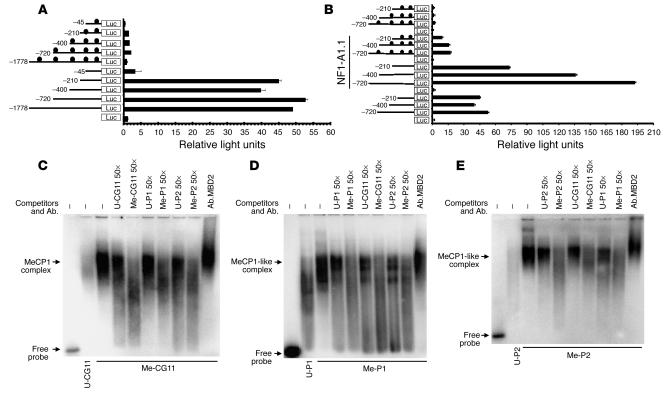
In order to determine whether the HSD11B2 promoter activity is regulated by CpG methylation and/or NF1, we performed reporter gene assays. Promoter-luciferase constructs containing various lengths of upstream regulatory sequences between nucleotides –1,778 and +117 (Figure 6, A and B) were fully methylated by SssI methylase and were subsequently transfected with and without an NF1-A1.1 expression vector into SW620 cells. The construct containing nucleotides –210 to +117 produced strong luciferase activity (Figure 6, A and B), whereas the construct containing nucleotides –400 to +117 showed consistently less activity, suggesting presence of a repressor/silencer between nucleotides –400 and –210 (12). This repression was overcome by the construct containing nucleotides –720 to +117, indicating the presence of an activator/enhancer between nucleotides –720 and –400. Methylation of the constructs completely suppressed their activity (Figure 6A). The major activity of the HSD11B2 promoter appears to be within sequences between nucleotides –720 and +117 (Figure 6, A and B). To assess the effect of methylation on the promoter alone, we separately methylated the promoter fragment (nucleotides –720 to +117), religated this into the unmethylated plasmid, and transfected cells with this construct. The methylation decreased the relative luciferase activity from 50 to less than 3, a decrease comparable to the values in Figure 6, A and B, in which whole plasmids were methylated. In cotransfection studies (see Methods), NF1 upregulated mock-methylated and, less so, methylated promoter-luciferase constructs (Figure 6B), indicating that the activity of HSD11B2 promoter is regulated by NF1 and/or methylation status.
Figure 6.
Relevance of NF1 and/or DNA methylation on HSD11B2 promoter activity and binding of MeCP1-like complex to methylated HSD11B2 promoter probes. (A and B) Repression of promoter-driven HSD11B2 transcription by CpG methylation with and without NF1 overexpression. Different luciferase (Luc) constructs were methylated (filled circles) or mock-methylated (no circles) in vitro and were transfected into SW620 cells. Results are given as luciferase activity normalized to cotransfected pCMV-LacZ activity. The reporter plasmids were cotransfected with pCHNFI A1.1 for NF1 overexpression. Results are the mean ± SE of at least three experiments. (C–E) Binding of MeCP1-like complex to methylated HSD11B2 promoter probes. 32P-labeled mock-methylated (C, U-CG11; D, U-P1; E, U-P2) or methylated (C, Me-CG11; D, Me-P1; E, Me-P2) probes were incubated with MCF-7 nuclear extracts in absence (–) or presence of a 50-fold molar excess (×50) of the corresponding cold unmethylated (U-) or methylated (Me-) probes. Supershifts were performed with N-terminal anti-MBD2 (Ab.MBD2).
Densely methylated regions of HSD11B2 promoter binds to methylated-CpG–binding protein complex 1.
The complexes known to bind preferentially to methylated CpGs include methylated-CpG–binding protein complex 1 (MeCP1) and MeCP2 (22). MeCP1 binds to DNA containing at least 12 symmetrically methylated CpGs (23), whereas MeCP2 binds to a single methylated CpG (22). Because the HSD11B2 promoter has multiple densely methylated CpGs where the gene is repressed, we focused on MeCP1. An end-labeled methylated CG11 probe (Me-CG11), which is known to bind to MeCP1 (23), and a mock-methylated CG11 probe (U-CG11), and two methylated and mock-methylated fragments of the HSD11B2 promoter (Me-P1 and U-P1, nucleotides –573 to –380, and Me-P2 and U-P2, nucleotides –403 to –76) were used for EMSA with MCF-7 nuclear extracts. A MeCP1 complex was observed with Me-CG11 (Figure 6C), indicating that MCF-7 contains MeCP1. Formation of the MeCP1 complex was abrogated more effectively by competition with Me-CG11-50×, Me-P1-50×, and Me-P2-50× than by competition with U-CG11-50×, U-P1-50×, and U-P2-50× (Figure 6C), indicating a higher affinity of the complex for methylated probes. MeCP1 has been shown to contain MBD2 (24). Therefore, the shift induced with an anti-MBD2 (Figure 6C) confirmed that the complex formed by Me-CG11 is MeCP1 (23, 24). Formation of a MeCP1-like complex on densely methylated HSD11B2 promoter fragments was confirmed using end-labeled methylated and mock-methylated P1 and P2 (Me-P1, U-P1, Me-P2, and U-P2) probes (Figure 6, D and E).
DNA methyltransferase inhibitors activate HSD11B2 and decrease methylation of the HSD11B2 promoter in vivo.
Procainamide decreased the ratio of urinary corticosterone (THB and 5α-THB) to 11-dehydrocorticosterone (THA) ([THB + 5α-THB] / THA) in all rats (Figure 7A), indicating enhanced 11βHSD2 activity. The 11βHSD2 mRNA was higher in kidneys from rats treated with procainamide than in those without procainamide treatment (Figure 7B). Similarly, in 10 rats treated with 5-aza-cytidine (10 mg/kg/day i.p.), the urinary (THB + 5α-THB) / THA ratio declined in all animals (mean ± SD: 2 ± 0.4 versus 1.2 ± 0.2, day 0 versus day 3). No further analyses beyond day 3 could be performed because the animals appeared sick after day 4 on 5-aza-cytidine. A third demethylating agent, 5-aza-CdR, given i.p. (1 mg/kg/day) for 3–7 days also caused a steady decline in the (THB + 5α-THB) / THA ratio of the same magnitude as that observed for procainamide, whereas no changes were observed in control rats given vehicle only (results not shown).
For methylation-sensitive restriction enzyme analyses, genomic DNA from different tissues of control and 5-aza-CdR–treated rats were digested with EcoRI and either HpaII, a methylation-sensitive enzyme, or MspI, a methylation-insensitive enzyme, and was probed in Southern blots with an 805-bp fragment derived from the rat HSD11B2 promoter (nucleotides –1,097 to –292). DNA from kidney digested with EcoRI alone produced the anticipated 3,350-bp fragment (Figure 7D, lane 1). In both control rats and rats treated for 7 days with 5-aza-CdR, digestion of genomic DNA with the methylation-insensitive enzyme MspI together with EcoRI yielded the predicted 1,053-bp band in all three tissues analyzed (Figure 7D). When instead of MspI the methylation-sensitive HpaII was used, a different pattern of bands was observed (about 1,791 bp, 1,539 bp, and 1,053 bp). In rats not given 5-aza-CdR, digestion of DNA from kidney with EcoRI and HpaII revealed a more intense band at 1,053 bp than did digestion of DNA from lung and liver (Figure 7D). This observation is in line with results derived from bisulfite sequencing (Figure 3C) and indicates that tissues not expressing HSD11B2 (lung and liver) exhibit a higher level of DNA methylation on the HSD11B2 promoter than does the HSD11B2-expressing kidney. Treatment with 5-aza-CdR and digestion with EcoRI and HpaII resulted in an increase in the intensity of the 1,053-bp band compared with the intensity of the band from DNA of the same organ from untreated rats (Figure 7D, kidney, lung, and liver) and a slight decrease in the corresponding 1,791-bp band (Figure 7D, lung and liver). These changes induced by 5-aza-CdR are compatible with a decline in HSD11B2 promoter DNA methylation and confirm the effect in cell lines assessed by bisulfite sequencing (Figure 3C). The decreased level of promoter methylation causing a higher expression of the HSD11B2 gene was further evidenced by Northern blotting (Figure 7E).
Discussion
The HSD11B2 gene promoter resembles the promoters of housekeeping genes, characterized by a GC-rich region, multiple GC boxes, and a lack of TATA or CCAAT sequences with CpG Islands along the gene (Figure 1). Thus, the cell type–specific expression of HSD11B2 is rather surprising, because housekeeping genes are usually ubiquitously expressed. The region between nucleotides –213 and +156, which contains several Sp1/Sp3 binding sites, is critical for basal promoter activity of 11βHSD2 (12, 13). Sp1/Sp3 are ubiquitously expressed. Therefore, the mechanism accounting for the cell type–specific expression of the HSD11B2 gene is not completely understood. The presence of CpG islands suggested that the HSD11B2 gene might be regulated at least in part through CpG methylation. In general, hypermethylation of normally unmethylated CpG islands correlates with transcriptional repression. Here we have provided evidence that HSD11B2 expression and activity are inversely correlated with the presence of methylation at the promoter. In support of the idea of methylation suppression, DNA methyltransferase inhibitors enhanced transcription and activity of 11βHSD2 in different cell types. Moreover, these inhibitors increased mRNA abundance in various tissues and decreased the urinary glucocorticoid metabolite ratios in rats, indicating higher 11βHSD2 activity. Furthermore, the relevance of DNA methylation for the tissue-specific expression and regulation of the HSD11B2 gene was demonstrated by methylation-sensitive restriction enzyme analyses followed by Southern blotting using genomic DNA from different tissues of control and 5-aza-CdR–treated rats.
The present investigation supports the notion that DNA methylation is dominant to histone acetylation, since treatment with an HDAC inhibitor increased the expression of HSD11B2 only after removal of the cytosine methylation in most of the cells analyzed (Figure 2) (14). In JEG-3 cells, TSA did not increase HSD11B2 expression, suggesting that chromatin modification in the HSD11B2 promoter may vary between cell types. The mechanisms discussed previously that lead to the differential effect of TSA include a cooperative inhibition of histone deacetylation and DNA methylation linked through the DNA methyl-binding proteins and/or an effect on other transcription factors (25, 26). An inverse correlation between promoter methylation and HSD11B2 expression is apparent when JEG-3, MCF-7, SW620, placenta, skeletal muscles, liver, lung, and renal proximal and distal tubules are consider as a group (20). DNA methylation in the upstream region was decreased by 5-aza-CdR, an effect associated with an enhanced expression of HSD11B2. In contrast, the exonic and downstream CpG islands were fully methylated in all normal tissues and cell lines before and after treatment with 5-aza-CdR (Figure 3C). The active expression of HSD11B2 despite the presence of fully methylated exonic and downstream CpG islands is in agreement with observations about other genes (17).
One mechanism by which promoter methylation affects expression of the corresponding gene is to modulate the binding of transcription factors, as shown, for instance, for Myc/Myn or AP2 (27, 28). Because the HSD11B2 promoter contains several Sp1/Sp3 binding sites that are methylated at core and neighboring CpGs, we investigated the effect of methylation on Sp1/Sp3 binding. In agreement with a study about the P21Cipl gene (29), the fully methylated Sp1/Sp3 probe with methylated-CpG dinucleotides at the core and outside of the consensus Sp1/Sp3 element reduced Sp1/Sp3 binding (Figure 4B). Similarly, methylation of the CpG dinucleotides at Ah-Arnt/Arnt binding sites in the HSD11B2 promoter appears to be relevant. In line with observations about the erythropoietin gene (30), the fully methylated Ah-Arnt/Arnt probe formed only one of two specific complexes (Figure 4A).
Because we found here that NF1 upregulates HSD11B2 (Figure 6B) and several methylated CpGs are present within or immediately around the NF1 binding sites, we explored the relevance of CpG methylation to NF1 binding. Previously the effect of methylation on NF1 binding was investigated by Ben-Hattar et al. (31), who showed that methylation-specific cytosine residues within or immediately around the CCAAT-box binding transcription factor (CTF) binding site of herpes simplex virus TK promoter had no effect on the affinity of CTF (28, 31), but the efficiency of transcription was reduced at least 50-fold compared with the efficiency when the unmethylated promoter was used. It is conceivable that methylated cytosines in a different configuration in the consensus NF1 binding site might modulate NF1 binding. All NF1 proteins bind as a dimer to DNA in the major groove and recognize a dyad symmetric consensus sequence [TTGGC(N)5GCCAA] on duplex DNA. NF1 binds specifically to individual half-sites (TTGGC or GCCAA) (21). Previous investigations focusing on the effect of the NF1 half-palindrome and mutations in the spacer region between the two palindromes revealed that using only an NF1 half-palindrome decreased the affinity of the NF1 and mutations of the first two nucleotides in the spacer modulated the NF1 binding (32, 33). The core binding site of NF1 (TTGGC) contains a cytosine at the 3′ end, which has the potential to be a part of a CpG dinucleotide. Our investigation demonstrates for the first time, to our knowledge, that probes with fully methylated or “positionally methylated” CpGs either at the core and/or adjacent to the core increase the main NF1 complex and recruit additional methylation-specific complexes compared with unmethylated probe (Figure 5), whereas methylation of adjacent CpGs considerably decreases binding of the NF1 main complex and methylation-specific complexes when the core CpG is mutated concomitantly (Figure 5D). These observations indicate that both the position of CpGs and the combination of CpGs methylated determine NF1 binding.
NF1 and MBD1 coexist in the main and methylation-specific NF1 complexes, because either NF1 or MBD1 antibody shifted the complexes (Figure 5E). Therefore, we speculate that MBD1 interacts with NF1 to stabilize its binding to the methylated DNA sequence and mediates transcriptional repression. MBD1 (formerly PCM1) is the least characterized of the methylated-CpG binding proteins. MBD1 comprises an N-terminal MBD that binds to methylated DNA and a C-terminal transcriptional repression domain associated with HDAC (34). Earlier studies demonstrated that MBD1 partners with the p150 subunit of chromatin assembly factor 1 to form a multiprotein complex that also contains HP1a, suggesting a role for MBD1 in methylation-mediated transcriptional repression and inheritance of epigenetically determined chromatin states (34). A role for NF1 in chromatin remodeling, in addition to its directly enhancing transcriptional effect, has been reported by Hebbar et al. (35). NF1 interacts with a variety of coactivator or cosuppressor proteins in a cell-type– and/or promoter-specific manner (21). Our observations indicate for the first time, to our knowledge, that NF1 interacts with MBD1.
Direct binding of a specific transcriptional repressor complex is the second major mechanism of transcriptional repression of genes exhibiting methylated CpGs (22, 27). A conserved family of proteins that bind to methylated CpGs with little apparent specificity for flanking nucleotide sequences has been identified (22). MeCP1 was the first complex identified that discriminates between methylated and unmethylated DNA and represses transcription through preferential binding to, remodeling of, and deacetylation of methylated nucleosomes (23, 36). Here we have presented evidence that a similar complex binds to the methylated HSD11B2 promoter. Blocking MBD2 with anti-MBD2 shifted the MeCP1-like complex (Figures 6, C–E), supporting a previous report that MBD2 is a component of the MeCP1 and/or related complexes (24). A definite role for transcriptional repressors such as MeCP1 and MBD1 should be established by studies with the corresponding knockout animals in the future.
In vitro methylation of promoter-luciferase constructs considerably reduced the promoter activity after transfection. Reporter assays provided evidence for enhancer elements between nucleotides –720 and –400. The higher methylated CpGs in part 1 (nucleotides –704 to –380) and its effect on HSD11B2 expression is consistent with this notion. Procainamide increased 11βHSD2 activity and mRNA levels in cell lines and in vivo. Given the known effect of procainamide (15), it is likely that this effect is attributable to decreased DNA methylation. The increased mRNA abundance and activity of HSD11B2 observed within 5 days of procainamide dosing in vivo might be unexpected, as the best investigated inhibitor of DNA methyltransferase, 5-aza-CdR, acts mainly in the S phase of the cell cycle and the proliferation rate of the renal tubular cells is generally considered to be low. However, the mechanism of the demethylating effect of the procainamide is not well defined and is possibly not confined to the S phase. Furthermore, quantitative data of the proliferation rate of cortical collecting ducts without and with procainamide are not available. The enhancing effect of procainamide on the expression of HSD11B2 in vivo might also be the consequence of activation of the gene in other dividing cells. Of interest, evidence is growing that expression of HSD11B2 exhibits a “pro-proliferative” effect on its own (37).
Our investigation has demonstrated evidence of an epigenetic mechanism, providing a tentative explanation for the highly cell-specific expression of 11βHSD2. Potentially more importantly, changes in methylation patterns might explain the inter-individual differences in HSD11B2 expression in mineralocorticoid target tissues and by that mechanism modulate blood pressure. The HSD11B2 gene was induced by three structurally different DNA methyltransferase inhibitors. The magnitude of these changes appears to be clinically important, a contention supported by the observation that moderate changes in 11βHSD2 activity affected blood pressure significantly in normal volunteers (38). DNA methylation of many genes changes with age, disease states, and environmental signals, including diet (18). Thus, we hypothesize that epigenetic modifications of genes with pivotal relevance for blood pressure control, as shown here for HSD11B2, are linked to the development of hypertension. Future studies in humans and/or knockout mice with distinct deficiencies in compounds relevant to CpG methylation must be performed to define the ultimate relevance of this epigenetic mechanism to blood pressure.
Methods
Supplies.
Supplies were obtained from the following companies: chemicals and radiochemicals, from Sigma, Roche Diagnostics, Fluka, Merck, or Amersham Pharmacia Biotech; oligonucleotides, from Microsynth AG; Taq polymerase, from QIAGEN; enzymes and vectors, from Roche Diagnostics, Gibco BRL, New England Biolabs, or Promega; and TLC plates, from Macherey-Nagel.
Human cell line and tissues.
SW620, a colon carcinoma cell line, and MCF-7, a breast adenocarcinoma cell line, were grown in DMEM (13), and JEG-3, a placenta carcinoma cell line, in MEM (39). Human tissues were provided by the Department of Surgery (University Hospital of Berne, Switzerland). Informed consent was obtained from the patients. Renal proximal and distal tubular cells were isolated as described (40).
Animal studies.
The committee on animal research (University Hospital of Berne, Switzerland) approved the protocol. Wistar rats weighing 190–210 g were kept in metabolic cages under controlled conditions for 5 days for adaptation. Thereafter, animals were fed powdered normal chow diet with or without the addition of a watery solution of procainamide (250 mg/day/rat) or were fed powdered normal chow diet and injected daily with 5-aza-CdR (1 mg/kg in 0.4 ml of PBS) or PBS alone (0.4 ml) i.p. for 7 days. Urine was collected over 24 hours from 2 days before until 5–7 days after treatment. At the end animals were sacrificed and organs were removed and kept at –80°C.
Real-time PCR and Northern blotting.
RNA extraction and TaqMan assays were performed (13) using a forward primer of positions 802–821, a reverse primer of positions 850–869, and a probe of positions 823–847 for human HSD11B2 mRNA, and a forward primer of positions 1,269–1,288, a reverse primer of positions 1,336–1,313, and a probe of positions 1,290–1,311 for rat HSD11B2 mRNA. Total cellular RNA (20 μg) was separated by electrophoresis through a 1.2% formaldehyde agarose gel for Northern blotting and was transferred to a nylon membrane. A 529-bp XhoI fragment of 11βHSD2 cDNA was used for hybridization, with GAPDH cDNA as control.
Assay of 11 βHSD2 activity.
The conversion of cortisol to cortisone was assessed (39). The assay was repeated up to four times using different protein concentrations.
Nuclear transactivation assay.
To demonstrate that reduced CpG methylation enhances the 11βHSD2 enzyme activity in whole cells, thereby reducing the intracellular availability of cortisol to the receptor, we performed nuclear transactivation assays (3). JEG-3 cells were plated onto 24-well plates and 12–18 hours thereafter were transfected using Fugene-6. Each well contained MMTV-Luc (0.8 μg), TK–Renilla luciferase internal control (0.11 μg), and rat GR vector (0.11 μg). At 16 hours after transfection, cells were treated with 5-aza-CdR (1.5 μM) or PBS for 48 hours, were washed with PBS, and were incubated for 24 hours in charcoal-stripped media with cortisol or dexamethasone. Firefly and Renilla luciferase assays were performed (Dual-Luciferase Reporter Assay System; Promega). Transfections were performed in triplicate and results were confirmed with at least two independent experiments.
Treatment of cells with 5-aza-CdR, TSA, and procainamide.
At 12–24 hours before treatment with 5-aza-CdR or procainamide, cells were split at low density (15). Various concentrations of the drugs were added for 72 hours, and the medium was changed every 24 hours. TSA was added the last day for an additional 12–24 hours. On day 4, mRNA and enzyme activity were determined.
In vitro methylation of reporter plasmids.
SssI methylase was used for the methylation of HSD11B2 promoter–luciferase constructs in pGl3-Basic. The constructs contained various lengths of the promoter and 117 bp of exon 1 (12). The reporter plasmid p–720/+117 was obtained by PCR cloning using the NR-F and NR-R primers (Table 1). All fragments had at their 3′ end an NcoI site corresponding to the translation initiation codon. The efficiency of methylation was determined with the methylation-sensitive restriction enzyme HpaII. Region-specific methylation was carried out on the promoter fragment of p–720/+117 after excision and isolation of the fragment (30, 41). In each case, half of the DNA was methylated with Sss1 methylase and the other half was incubated with methylase in the absence of S-adenosylmethionine as a mock methylation. Methylated and mock-methylated fragments were religated into the vector from which they had been excised.
Transfections and reporter gene assays.
These techniques have been described previously (13). The normalized values of β-galactosidase using the Dual-Light system (Tropix) in triplicate samples varied by less than 5%. Plasmid DNA was prepared using QIAfilter columns (QIAGEN).
Bisulfite sequencing.
Bisulfite genomic sequencing allows discrimination of 5-methyl-cytosine versus cytosine (19). Genomic DNA was isolated with the DNeasy tissue kit (QIAGEN) and bisulfite treatment carried out as described (19). Briefly, 2.5 mg of DNA cut with EcoRI was denaturated for 10 minutes at 37°C with 4.5 ml of 3 M NaOH. Then, 750 ml of sodium bisulfite (40.5%) and 42 ml of hydroxyquinone (10 mM) were added. The reaction was allowed to proceed at 55°C for 5 hours with the following program: 95°C for 1 minute for the first cycle, followed by 94°C for 30 seconds and 55°C for 59 minutes and 30 seconds (repeated five times). After the addition of 160 ml of 100% ethanol, DNA was purified with DNeasy tissue kit columns. Desulfonation was performed for 20 minutes after samples were washed on a column with 500 ml of 0.3 M NaOH/80% ethanol. After being washed, the modified DNA was eluted from the column with 50 ml of 10 mM Tris-HCl (pH 8.0). Modified DNA was amplified by nested PCR with the HotStarTaqTM PCR kit from QIAGEN using two sets of forward and reverse primers, which were designed for an area without or with a single CpG only (N1-F, N1-R, N2-F, and N2-R; Table 1). The sequences of strand-specific primers containing the modified cytosine bases are summarized in Table 1. The PCR fragments amplified were gel-purified and were directly sequenced or were cloned using the pCR4-TOPOTA Cloning kit (Invitrogen Corp.). Eight clones were sequenced to assess the level of methylation in each CpG site.
EMSA.
Nuclear protein extractions and assays for short synthetic probes (Table 1) were performed as described (13), except these used 0.7–1 μg of poly(dI-dC). Probes with larger sequences were CG11, a 135-bp fragment containing 20 GCGC sites and 7 CCGG sites (23, 41), and P1 (nucleotides –573 to –380; 193 bp) and P2 (nucleotides –403 to –76; 328 bp) from the HSD11B2 promoter. For long probes, EMSA was performed using 0.3 ng labeled probe, 2–3 mg nuclear protein, and sonicated genomic DNA of Escherichia coli as a nonspecific competitor on 1.5% agarose gels (23, 30, 41). Unlabeled probes were used to compete for specific binding. For supershift experiments, 1–2 μl of the following antibodies were added and kept on ice (for 20 minutes) before being mixed with labeled probes: N18 (anti-MBD2), anti-Sp1, anti-Sp3, anti-AP4, anti-Ah-Arnt, and anti-Arnt1 (Santa Cruz Biotechnology Inc.); anti-MBD1 (Abcam Ltd); and anti-HA (Roche Diagnostics). Gels were dried and the DNA-protein complexes were visualized and analyzed on a phosphorimager.
Methylation analysis with Southern blotting.
Genomic DNA (15 μg) was digested sequentially with excess of either HpaII or MspI together with EcoRI in optimal buffer (42). The digested DNA fragments were separated by 0.8% agarose gel electrophoresis and were blotted onto a nylon membrane. The rat HSD11B2 promoter probe (nucleotides –1,097 to –292) was prepared by PCR and labeled with [α32P]dCTP by random primer extension after gel purification.
Bioinformatics.
The human HSD11B2 gene (13,400 bp; nucleotides –3,731 to +9,669) was analyzed for CpG islands using software of the European Bioinformatics Institute (http://www.ebi.ac.uk/emboss/cpgplot/) and accepting their default criteria, but considering 100 bp CpG length, or using the CpG Island Explorer, CpGIE V1.6 (http://bioinfo.hku.hk/cpgieintro.html). Searches for transcription factor binding sites were performed with Match (http://www.gene-regulation.com/pub/programs.html#match). The rat HSD11B2 promoter sequence was extracted from the genome bank by Genome Blast (http://www.ncbi.nlm.nih.gov/BLAST/) and the region from nucleotide –3,000 to nucleotide +10,000 was analyzed for CpG islands.
Analysis of urinary steroid metabolites by gas chromatography-mass spectrometry.
The in vivo activity of 11βHSD2 was estimated by the urinary ratio of excreted metabolites THB and 5α-THB to THA ([THB + 5α-THB] / THA) in rats (43, 44).
Acknowledgments
We thank R. Gronostajski (State University of New York, Buffalo, New York, USA) for NF1 vectors; G. Ginders (Virginia Commonwealth University, Richmond, Virginia, USA) for the CG11 plasmid; and J. Benhattar (Institut de Pathologie, Lausanne, Switzerland) and B. Trueb, B. Steiner, B. Dick, A. Nawrocki, A. Rebuffat, and A. Odermatt (all of the University of Bern, Bern, Switzerland) for laboratory support and comments. Grant support was provided by the Swiss National Foundation for Scientific Research (3100-61505.00 and 3100A0-102153/1, to F.J. Frey and B.M. Frey).
Footnotes
Nonstandard abbreviations used: × (50×, 100×), excess competitor; Ab., antibody against; 5-aza-CdR, 5-aza-2′-deoxycytidine; Cons., consensus sequence; EMSA, electrophoretic mobility-shift assay; GR, glucocorticoid receptor; HDAC, histone deacetylase; 11βHSD, 11β-hydroxysteroid dehydrogenase; HSD11B2, gene encoding 11βHSD2; MBD, methylated-CpG–binding domain protein; Me-, methylated; MeCP, methylated-CpG–binding protein complex; MMTV, mouse mammary tumor virus; MMTV-Luc, MMTV-luciferase; MR, mineralocorticoid receptor; NF1, nuclear factor 1; P, probe; THA, 11-dehydrocorticosterone; THB, 5α-THB, corticosterone; TK, thymidine kinase; TSA, trichostatin A; U-, unmethylated.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Albiston AL, Obeyesekere VR, Smith RE, Krozowski ZS. Cloning and tissue distribution of the human 11 beta-hydroxysteroid dehydrogenase type 2 enzyme. Mol. Cell. Endocrinol. 1994;105:R11–R17. doi: 10.1016/0303-7207(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 2.Tannin GM, Agarwal AK, Monder C, New MI, White PC. The human gene for 11 beta-hydroxysteroid dehydrogenase. Structure, tissue distribution, and chromosomal localization. J. Biol. Chem. 1991;266:16653–16658. [PubMed] [Google Scholar]
- 3.Escher G, Galli I, Vishwanath BS, Frey BM, Frey FJ. Tumor necrosis factor alpha and interleukin 1beta enhance the cortisone/cortisol shuttle. J. Exp. Med. 1997;186:189–198. doi: 10.1084/jem.186.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frey FJ, Odermatt A, Frey BM. Glucocorticoid-mediated mineralocorticoid receptor activation and hypertension. Curr. Opin. Nephrol. Hypertens. 2004;13:451–458. doi: 10.1097/01.mnh.0000133976.32559.b0. [DOI] [PubMed] [Google Scholar]
- 5.Odermatt A, Arnold P, Stauffer A, Frey BM, Frey FJ. The N-terminal anchor sequences of 11beta-hydroxysteroid dehydrogenases determine their orientation in the endoplasmic reticulum membrane. J. Biol. Chem. 1999;274:28762–28770. doi: 10.1074/jbc.274.40.28762. [DOI] [PubMed] [Google Scholar]
- 6.Kotelevtsev Y, et al. Hypertension in mice lacking 11beta-hydroxysteroid dehydrogenase type 2. J. Clin. Invest. 1999;103:683–689. doi: 10.1172/JCI4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart PM, Corrie JE, Shackleton CH, Edwards CR. Syndrome of apparent mineralocorticoid excess. A defect in the cortisol-cortisone shuttle. J. Clin. Invest. 1988;82:340–349. doi: 10.1172/JCI113592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lovati E, et al. Molecular basis of human salt sensitivity: the role of the 11beta-hydroxysteroid dehydrogenase type 2. J. Clin. Endocrinol. Metab. 1999;84:3745–3749. doi: 10.1210/jcem.84.10.6098. [DOI] [PubMed] [Google Scholar]
- 9.Agarwal AK, et al. CA-Repeat polymorphism in intron 1 of HSD11B2: effects on gene expression and salt sensitivity. Hypertension. 2000;36:187–194. doi: 10.1161/01.hyp.36.2.187. [DOI] [PubMed] [Google Scholar]
- 10.Watson B, Jr, et al. Genetic association of 11 beta-hydroxysteroid dehydrogenase type 2 (HSD11B2) flanking microsatellites with essential hypertension in blacks. Hypertension. 1996;28:478–482. doi: 10.1161/01.hyp.28.3.478. [DOI] [PubMed] [Google Scholar]
- 11.Zaehner T, Plueshke V, Frey BM, Frey FJ, Ferrari P. Structural analysis of the 11beta-hydroxysteroid dehydrogenase type 2 gene in end-stage renal disease. Kidney Int. 2000;58:1413–1419. doi: 10.1046/j.1523-1755.2000.00303.x. [DOI] [PubMed] [Google Scholar]
- 12.Agarwal AK, White PC. Analysis of the promoter of the NAD+ dependent 11 beta-hydroxysteroid dehydrogenase (HSD11K) gene in JEG-3 human choriocarcinoma cells. Mol. Cell. Endocrinol. 1996;121:93–99. doi: 10.1016/0303-7207(96)03855-5. [DOI] [PubMed] [Google Scholar]
- 13.Nawrocki AR, Goldring CE, Kostadinova RM, Frey FJ, Frey BM. In vivo footprinting of the human 11beta-hydroxysteroid dehydrogenase type 2 promoter: evidence for cell-specific regulation by Sp1 and Sp3. J. Biol. Chem. 2002;277:14647–14656. doi: 10.1074/jbc.M111549200. [DOI] [PubMed] [Google Scholar]
- 14.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 15.Lin X, et al. Reversal of GSTP1 CpG island hypermethylation and reactivation of pi-class glutathione S-transferase (GSTP1) expression in human prostate cancer cells by treatment with procainamide. Cancer Res. 2001;61:8611–8616. [PubMed] [Google Scholar]
- 16.Rebuffat AG, et al. The 11-ketosteroid 11-ketodexamethasone is a glucocorticoid receptor agonist. Mol. Cell. Endocrinol. 2004;214:27–37. doi: 10.1016/j.mce.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 17.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 18.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 19.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal AK, Rogerson FM, Mune T, White PC. Gene structure and chromosomal localization of the human HSD11K gene encoding the kidney (type 2) isozyme of 11 beta-hydroxysteroid dehydrogenase. Genomics. 1995;29:195–199. doi: 10.1006/geno.1995.1231. [DOI] [PubMed] [Google Scholar]
- 21.Gronostajski RM. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 22.Ballestar E, Wolffe AP. Methylated-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem. 2001;268:1–6. doi: 10.1046/j.1432-1327.2001.01869.x. [DOI] [PubMed] [Google Scholar]
- 23.Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 24.Ng HH, et al. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- 25.Mulholland NM, Soeth E, Smith CL. Inhibition of MMTV transcription by HDAC inhibitors occurs independent of changes in chromatin remodeling and increased histone acetylation. Oncogene. 2003;22:4807–4818. doi: 10.1038/sj.onc.1206722. [DOI] [PubMed] [Google Scholar]
- 26.Sowa Y, et al. Sp3, but not Sp1, mediates the transcriptional activation of the p21/WAF1/Cip1 gene promoter by histone deacetylase inhibitor. Cancer Res. 1999;59:4266–4270. [PubMed] [Google Scholar]
- 27.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3:226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 28.Constancia M, Pickard B, Kelsey G, Reik W. Imprinting mechanisms. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- 29.Zhu WG, et al. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol. Cell. Biol. 2003;23:4056–4065. doi: 10.1128/MCB.23.12.4056-4065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin H, Blanchard KL. DNA methylation represses the expression of the human erythropoietin gene by two different mechanisms. Blood. 2000;95:111–119. [PubMed] [Google Scholar]
- 31.Ben-Hattar J, Beard P, Jiricny J. Cytosine methylation in CTF and Sp1 recognition sites of an HSV tk promoter: effects on transcription in vivo and on factor binding in vitro. Nucleic Acids Res. 1989;17:10179–10190. doi: 10.1093/nar/17.24.10179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Osada S, Daimon S, Nishihara T, Imagawa M. Identification of DNA binding-site preferences for nuclear factor I-A. FEBS Lett. 1996;390:44–46. doi: 10.1016/0014-5793(96)00622-9. [DOI] [PubMed] [Google Scholar]
- 33.Gronostajski RM. Site-specific DNA binding of nuclear factor I: effect of the spacer region. Nucleic Acids Res. 1987;15:5545–5559. doi: 10.1093/nar/15.14.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reese BE, Bachman KE, Baylin SB, Rountree MR. The methylated-CpG binding protein MBD1 interacts with the p150 subunit of chromatin assembly factor 1. Mol. Cell. Biol. 2003;23:3226–3236. doi: 10.1128/MCB.23.9.3226-3236.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hebbar PB, Archer TK. Nuclear factor 1 is required for both hormone-dependent chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter. Mol. Cell. Biol. 2003;23:887–898. doi: 10.1128/MCB.23.3.887-898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rabbitt EH, et al. Prereceptor regulation of glucocorticoid action by 11beta-hydroxysteroid dehydrogenase: a novel determinant of cell proliferation. FASEB J. 2002;16:36–44. doi: 10.1096/fj.01-0582com. [DOI] [PubMed] [Google Scholar]
- 38.Ferrari P, Sansonnens A, Dick B, Frey FJ. In vivo 11beta-HSD-2 activity: variability, salt-sensitivity, and effect of licorice. Hypertension. 2001;38:1330–1336. doi: 10.1161/hy1101.096112. [DOI] [PubMed] [Google Scholar]
- 39.Lanz CB, et al. Fluid shear stress reduces 11ss-hydroxysteroid dehydrogenase type 2. Hypertension. 2001;37:160–169. doi: 10.1161/01.hyp.37.1.160. [DOI] [PubMed] [Google Scholar]
- 40.Baer PC, Nockher WA, Haase W, Scherberich JE. Isolation of proximal and distal tubule cells from human kidney by immunomagnetic separation. Technical note. Kidney Int. 1997;52:1321–1331. doi: 10.1038/ki.1997.457. [DOI] [PubMed] [Google Scholar]
- 41.Singal R, Ferris R, Little JA, Wang SZ, Ginder GD. Methylation of the minimal promoter of an embryonic globin gene silences transcription in primary erythroid cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13724–13729. doi: 10.1073/pnas.94.25.13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song SH, et al. Transcriptional silencing of Cyclooxygenase-2 by hyper-methylation of the 5′ CpG island in human gastric carcinoma cells. Cancer Res. 2001;61:4628–4635. [PubMed] [Google Scholar]
- 43.Schumacher M, et al. Salt-sensitivity of blood pressure and decreased 11beta-hydroxysteroid dehydrogenase type 2 activity after renal transplantation. Transplantation. 2002;74:66–72. doi: 10.1097/00007890-200207150-00012. [DOI] [PubMed] [Google Scholar]
- 44.Shackleton CH. Mass spectrometry in the diagnosis of steroid-related disorders and in hypertension research. J. Steroid Biochem. Mol. Biol. 1993;45:127–140. doi: 10.1016/0960-0760(93)90132-g. [DOI] [PubMed] [Google Scholar]