Abstract
Angiogenesis, or new blood vessel formation, is critical for the growth and spread of tumors. Multiple phases of this process, namely, migration, proliferation, morphogenesis, and vascular stabilization, are needed for optimal tumor growth beyond a diffusion-limited size. The sphingosine 1–phosphate (S1P) receptor-1 (S1P1) is required for stabilization of nascent blood vessels during embryonic development. Here we show that S1P1 expression is strongly induced in tumor vessels. We developed a multiplex RNA interference technique to downregulate S1P1 in mice. The small interfering RNA (siRNA) for S1P1 specifically silenced the cognate transcript in endothelial cells and inhibited endothelial cell migration in vitro and the growth of neovessels into subcutaneous implants of Matrigel in vivo. Local injection of S1P1 siRNA, but not a negative control siRNA, into established tumors inhibited the expression of S1P1 polypeptide on neovessels while concomitantly suppressing vascular stabilization and angiogenesis, which resulted in dramatic suppression of tumor growth in vivo. These data suggest that S1P1 is a critical component of the tumor angiogenic response and argue for the utility of siRNA technology in antiangiogenic therapeutics.
Introduction
Sphingosine 1–phosphate (S1P), a potent lipid mediator produced from the metabolism of sphingomyelin, acts on a family of S1P G protein–coupled receptors (S1Pn) and transduces intracellular signals involved in numerous cellular processes (1, 2). In vascular endothelial cells, S1P binds to SIP receptors 1–3 (S1P1–3) to induce migration, proliferation, cell survival, and morphogenesis into capillary-like structures (3, 4). Such responses require the function of S1P1, which was originally isolated as an inducible gene from endothelial cells (4, 5). In addition to the well-characterized effects of S1P on endothelial cell migration and survival, it also induces the formation of cell-cell adherens junctions, thus inhibiting paracellular permeability of solutes and macromolecules (4, 6, 7). In vivo studies showed that S1P synergized with polypeptide angiogenic factors such as FGF-2 and VEGF to induce angiogenesis and vascular maturation in mouse models in which the extracellular matrix Matrigel was implanted subcutaneously (4). Moreover, S1P1-KO mice died in utero between embryonic days 13.5 and 14.5 due to a defect in vascular stabilization — entailing the reinforcement of nascent endothelial tubes with pericytes and vascular smooth muscle cells — which suggests that this receptor is required for vascular development (8). These studies form a basis for the emerging concept that S1P is a potent regulator of vascular growth and development, at least during embryogenesis.
Many regulators of embryonic vascular development also play important regulatory roles in pathologic angiogenesis in the adult. For example, the VEGF gene is not only critical for the various phases of embryonic vascular development but is also important in tumor angiogenesis (9). Indeed, a neutralizing antibody against VEGF has recently shown efficacy in colorectal cancer treatment in a combination regimen with conventional chemotherapeutic agents (10, 11). Given that S1P acts via G protein–coupled receptors, a class of receptors that are amenable to pharmacologic inhibition, it is of interest to ascertain whether this bioactive mediator plays a role in angiogenesis in the adult mouse. Indeed, little is known about the role of S1P in the adult vasculature, particularly with respect to angiogenesis. S1P1 is expressed in selected vascular beds; for example, in the cardiac ventricular microvessels of adult mice (12). Whether it is expressed in angiogenic vessels in vivo is not known. Here, we document that S1P1 is induced in angiogenic vessels in vivo.
Loss-of-function genetic and/or pharmacologic approaches are required to critically determine the function of S1P receptors in vivo. Lack of specific pharmacologic tools, coupled with the fact that the S1p1–/– mice die during mid-gestational stages of embryogenesis, have hampered efforts to decipher the postnatal functions of this receptor. We therefore utilized RNA interference (RNAi) technology, which was recently discovered to be a naturally occurring mechanism of posttranscriptional gene silencing (13). It was originally shown that long double-stranded RNA (dsRNA) species are cleaved into duplex RNAs of 21–23 nt with 2-bp overhangs, termed small interfering RNAs (siRNAs), which potently and specifically suppress gene expression by the induction of the RNA-induced silencing complex (14). RNAi occurs widely in nature and is implicated in embryonic development and normal adult physiology (15, 16). Administration of siRNAs into eukaryotic cells induces specific gene silencing in vitro and in vivo (17, 18). Thus, the potential of RNAi technology in therapeutic gene regulation has received significant interest (19).
The limitations of the synthetic siRNA technology include the empirical nature of target site selection and the expense associated with chemical synthesis of siRNA in the quantities required for in vivo administration (15). Recent work has shown that the use of multiplex siRNA species can achieve synergistic suppression of genes (20, 21). Such multiplex species of siRNAs can be produced by the eukaryotic enzyme Dicer or the E. coli RNaseIII enzyme. To overcome the problem of off-target suppression of related gene sequences in the protein-coding regions, we developed a method to suppress genes based on the 3′–untranslated region (3′-UTR) sequences, which are not under the same degree of evolutionary conservation as the coding regions (22). In this study, we used the E. coli RNaseIII enzyme to generate 3′-UTR–directed multiplex siRNA species to effectively downregulate gene expression. We show the utility of this method in the suppression of S1P1 expression in vitro and in vivo.
Results
Induction of S1P1 expression during tumor angiogenesis.
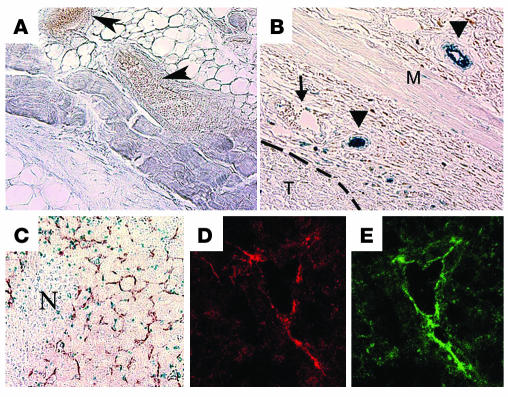
S1P1 mRNA is strongly induced in endothelial cells during embryonic angiogenesis (8). In the adult mouse, S1P1 is expressed in both vascular and nonvascular cells under basal conditions in selected tissues (12). However, the expression of S1P1 in angiogenic vessels in the adult has not been examined. We thus implanted Lewis lung carcinoma cells — which do not endogenously express the S1P1 transcript (Supplemental Figure 1; supplemental material available at http://www.jci.org/cgi/content/full/114/8/1082/DC1) and are capable of establishing tumors in immunocompetent mice — subcutaneously into S1p1+/– mice. We analyzed tissue sections for the expression of S1P1 by examining the activity of β-gal, which is directed from the endogenous S1p1 gene promoter (8, 12). In normal skin microvessels, S1P1 expression was not detected (Figure 1, A and B). However, S1P1 expression was induced in the vasculature upon the implantation of tumor cells. Endothelial cells from both small and large vessels surrounding the tumor expressed S1P1. Immunohistochemical analysis of X-gal–stained tumor sections with the endothelial cell marker CD31 and smooth muscle cell marker α-SMA showed that highest expression of S1P1 is in the endothelial cell compartment (data not shown). However, the intensity of X-gal staining was generally much lower in vascular smooth muscle cells. S1P1 expression was also noted in angiogenic vessels that had infiltrated the tumor. As shown in Figure 1C, many microvessel-like structures stained blue, which suggests that intratumoral vessels express S1P1. This was further confirmed by immunofluorescence staining with an antibody specific for the murine S1P1 polypeptide (Figure 1, D and E) (23). It should also be noted that some CD31-negative cells, which have the morphology of infiltrating monocytic and/or immune cells, also are positive for X-gal staining in both growing and necrotic areas of the tumor (Figure 1C). This is consistent with the knowledge that many cells derived from the hematopoietic lineage (such as monocytes, T cells, and mast cells) also express S1P1 (23, 24). These data suggest that S1P1 is induced in intratumoral capillaries and juxtatumoral large vessels during tumor angiogenesis.
Figure 1.
Induction of S1P1 expression in tumor xenografts. Normal skin tissue (A) and subcutaneous tumor nodules (B) from the back of S1p1+/–LacZ mice were excised and stained with X-gal to detect the expression of β-gal marker as described. Sections were cut and imaged under a bright-field microscope using a ×20 objective lens. β-gal expression (blue) was detected in blood vessels only in mice with the growing tumor (arrowheads in B). However, blood vessels in normal skin tissues (arrowheads in A) and few blood vessels with veinlike morphology in tumor-bearing tissue (arrow in B) were S1P1-negative. T, tumor; M, skeletal muscle layer. Dashed line indicates margin of the tumor. (C) X-gal–stained, fixed tissues were counterstained with anti–CD31 antibody to detect blood vessels, and the intratumor region is shown. At least 5 animals were used in this study. N, necrotic center. (D and E) Frozen sections of the subcutaneous implants of the tumor were analyzed in an immunofluorescence assay with the anti–S1P1 antibody (red) and anti–CD31 antibody (green) and imaged by a confocal microscope as described in Methods. Note that intratumoral blood vessels express S1P1, whereas tumor cells do not express this receptor.
Gene silencing by E. coli RNaseIII–generated siRNA.
To determine the function of S1P1 during tumor angiogenesis, we developed a novel RNAi method to downregulate its expression. We hypothesized that utilization of the 3′-UTR sequences as a template to derive multiplex siRNA species might overcome some of the problems associated with siRNA technology. Indeed, sequence homology analysis using the BLAST algorithm did not reveal significant similarity between the 3′-UTR of S1P1 and the 3′-UTR of the other four S1P receptors. In addition, we hypothesized that utilization of E. coli RNaseIII, a double stand–specific endoribonuclease, would produce duplex RNA species that can silence genes by an siRNA-like mechanism. In fact, recent studies published after the initiation of our work show that small duplex RNA produced by E. coli RNaseIII enzyme does indeed induce RNA interference in cultured cells (20).
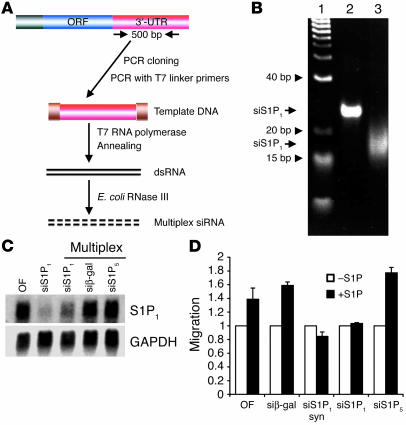
Our procedure is shown schematically in Figure 2A. We amplified a 500-bp fragment of the 3′-UTR from mouse S1P1 cDNA (25). Double-stranded RNA was produced by PCR with T7 RNA polymerase binding site–containing primers and in vitro transcription, followed by annealing of the sense and antisense RNA products. Extensive digestion with the E. coli RNaseIII resulted in the formation of multiplex RNA of approximately 15–20 bp (Figure 2B). RNaseIII did not digest single-stranded substrates of S1P1 3′-UTR. In addition, formation of smaller mol wt species was double strand–, RNaseIII-, and time-dependent. Complete digestion (500 nM enzyme, 4 hours treatment at 37°C) resulted in the formation of 15- to 20-bp double-stranded RNA species. This fraction had the RNA silencing activity, whereas we did not observe appreciable RNA silencing in larger or smaller regions of the gel (with much lower amounts of ethidium bromide–stained material). Similarly, we produced siRNAs for S1P5 and β-gal, both of which are not expressed in endothelial cells.
Figure 2.
Gene silencing by siRNA. (A) Scheme for the generation of multiplex siRNA. ORF, open reading frame. (B) Gel electrophoresis of synthetic and multiplex siRNA. Synthetic siRNA or multiplex siRNA for S1P1 were analyzed by a non-denaturating gel electrophoresis and detected by ethidium bromide staining. Lane 1: DNA mol wt markers; lane 2: 21-bp synthetic siRNA for S1P1 (siS1P1); lane 3: RNaseIII-digested siRNA for S1P1. (C) Mouse endothelial cells were transfected with various siRNAs and total RNA was isolated and analyzed by Northern blot analysis using S1P1 or GAPDH probes as described. Data show the results of a representative experiment that was repeated at least 3 times. siβ-gal, siRNA for β-gal. (D) Mouse endothelial cells were transfected with various siRNAs and analyzed in a chemotaxis assay using S1P (100 nM) as a chemoattractant. Data represent mean ± SE from a representative experiment that was repeated twice. syn, synthetic; OF, oligofectamine alone.
The efficacy and specificity of the siRNA for S1P1 in the suppression of gene expression was tested next. We treated mouse embryonic ECs (MEECs) with either synthetic siRNA from the open reading frame region of the mouse S1P1 mRNA or the RNaseIII-derived siRNA for S1P1 at a concentration of 200 nM. The RNA was delivered into endothelial cells by lipid-mediated transfection as described (26). The effect of siRNA was assessed by measuring the steady-state levels of S1P1 mRNA by Northern blot analysis 48 hours after transfection. As shown in Figure 2C, both multiplex and synthetic siRNA for S1P1 strongly suppressed the expression of the endogenous S1P1 mRNA. In separate experiments, siRNA for β-gal potently suppressed β-gal activity and protein levels in murine endothelial cells from heterozygous S1P1 KO mice that had the β-gal gene knocked into the S1P1 locus (S1p1+/–LacZ mice), whereas treatment with transfection reagent was without effect (Supplemental Figure 2). S1P5 expression was not detected in these cells and therefore was used as an irrelevant siRNA. However, siRNA for S1P5 or β-gal siRNA did not suppress the S1P1 transcripts, which suggests that the siRNA species achieved sequence-specific suppression of gene expression.
To determine whether the siRNA for S1P1 is functional in endothelial cells, we tested its effect on endothelial cell migration in a Boyden chamber assay (3). As shown in Figure 2D, both synthetic siRNA and multiplex siRNA for S1P1 but not the multiplex siRNA for S1P5 or β-gal potently suppressed the S1P-induced migration of murine endothelial cells. These data suggest that multiplex siRNA for S1P1 is likely to influence angiogenic response, of which endothelial cell migration is an early critical step.
Inhibition of angiogenesis in vivo by S1P1 siRNA.
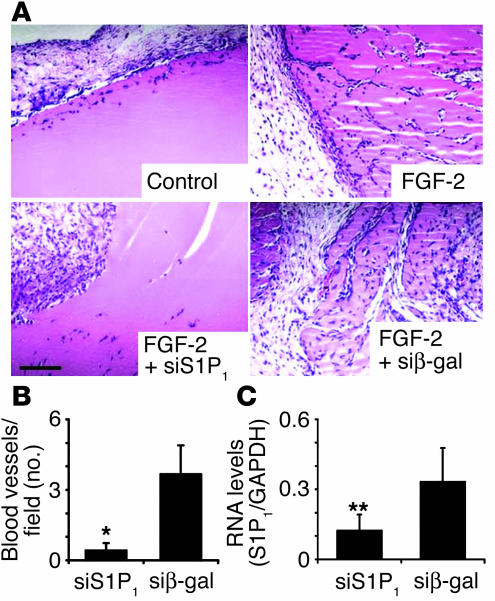
In previous studies, we have shown that antisense oligonucleotides against S1P1 suppressed FGF-2–induced angiogenesis in vivo (4). To test whether S1P1 siRNA regulates angiogenesis in the adult mouse, we performed subcutaneous angiogenesis assay using Matrigel. FGF-2 strongly induced cell infiltration and blood vessel formation (Figure 3). siRNA for S1P1 significantly inhibited FGF-2–induced angiogenesis and cell infiltration, whereas irrelevant siRNA (β-gal) did not have significant effect on angiogenesis (Figure 3A). Microvessel counting after immunohistochemical staining for CD31 showed that siRNA for S1P1 potently suppressed angiogenesis (Figure 3B). RNA was isolated from Matrigel plugs and analyzed for the expression of S1P1 transcript by Northern blot analysis. As shown in Figure 3B, siRNA for S1P1 significantly suppressed the expression of the cognate transcript in a specific manner.
Figure 3.
Inhibition of angiogenesis in vivo by S1P1 siRNA. (A) Matrigel mixture containing multiplex siRNA with or without FGF-2 was injected subcutaneously into athymic nude mice as described. The Matrigel plugs were harvested 7 days after implantation, fixed, sectioned, and H&E-stained as described. Images are obtained from a representative field of an experiment (n = 3) that was repeated twice. Scale bar: 100 μm. (B) The number of CD31-positive blood vessels in the Matrigel plugs was determined by immunohistochemistry and plotted (n = 4). Results represent mean ± SE. *P < 0.02. (C) Total RNA was isolated from the Matrigel plugs with siRNA for S1P1 or β-gal in the presence of FGF-2. S1P1 mRNA expression in the cells within Matrigel plugs was quantified by Northern blot analysis and normalized with respect to GAPDH expression (n = 3). **P < 0.1. In B and C, multiplex siS1P1 and siβ-gal indicate respective siRNAs in the presence of FGF-2 as described in A.
Inhibition of tumor growth and angiogenesis by S1P1 siRNA.
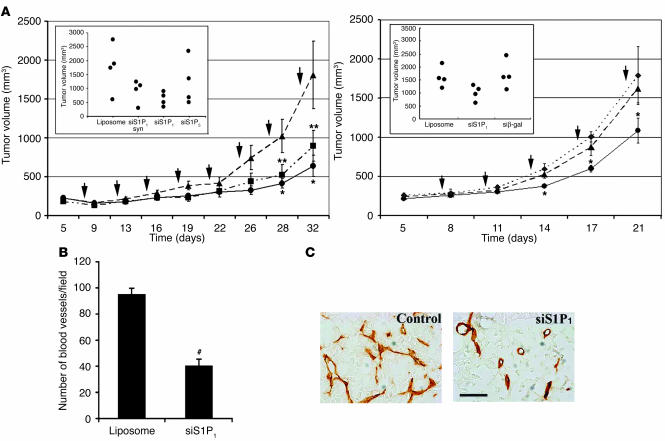
To determine the functional consequences of S1P1 function in tumor growth and angiogenesis, we injected multiplex S1P1 siRNA–liposome complexes locally into xenografts of Lewis lung carcinoma cells in nude mice. Synthetic siRNA–liposome complexes (positive control), liposome alone, and S1P5 siRNA–liposome complexes (negative controls) were also injected. As shown in Figure 4A, both synthetic and multiplex siRNA for S1P1 potently suppressed tumor growth. Liposome alone was not inhibitory, and siRNA for S1P5 had a minor but statistically insignificant effect on tumor growth rate. Furthermore, siRNA for β-gal did not suppress tumor growth. These data suggest that the expression of S1P1 is important for optimal tumor growth.
Figure 4.
Inhibition of tumor growth and angiogenesis by S1P1 siRNA. (A) Lewis lung carcinoma cells were implanted subcutaneously and allowed to establish as growing tumors, and various siRNA-liposome complexes were injected into the tumor every 3 days (where indicated by arrows) as described. Tumor volume was measured and plotted at various times following treatment with synthetic or multiplex siRNA for S1P1 (synthetic siS1P1, squares; multiplex siS1P1, circles) or liposome alone (triangles). The inset shows the tumor volume at 32 days for additional controls. Data represent mean ± SE of an experiment that was repeated twice. The right panel shows an independent experiment repeated with the inclusion of another control siRNA (multiplex β-gal siRNA, diamonds). n = 4; *P < 0.05; **P < 0.1. (B) Intratumoral microvessel density was quantified from multiple fields as described after CD31 staining. #P < 0.0003. (C) Morphology of tumor vessels is shown from a representative field. Note that vessel morphology and density are altered by S1P1, but not control, siRNA. Scale bar: 20 μm.
S1P1 was not detected in Lewis lung carcinoma cells by Northern blot analysis (Supplemental Figure 1). In addition, immunofluorescence microscopy of tumor sections with an anti–S1P1 antibody did not detect the expression of this receptor in implanted tumor cells in vivo (Figure 1 and below). Moreover, treatment of Lewis lung cancer cells in vitro with siRNA for S1P1 did not suppress growth of these cells, whereas it suppressed the growth of MEECs (Supplemental Figure 3). Thus, the effect of siRNA for S1P1 is likely to be on the host, namely, angiogenic endothelial cells, which express this receptor.
To determine the potential antiangiogenic action of the S1P1 siRNA, we quantified the microvessels in the tumor tissue. As shown in Figure 4B, microvessel density was significantly reduced by the local administration of siRNA for S1P1. Morphological analysis showed that branching and morphogenesis of tumor vasculature are profoundly affected (Figure 4C). These data suggest that S1P1 function in the angiogenic vessels is critical for optimal tumor angiogenesis, an essential process for tumor growth.
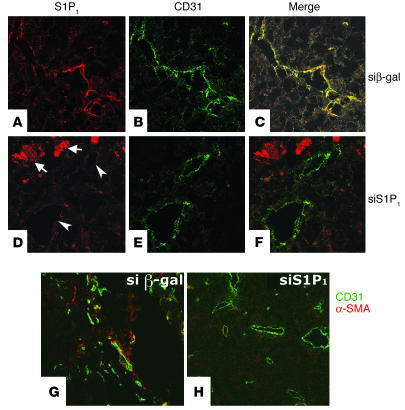
The suppression of S1P1 by intratumoral injection of siRNA was confirmed by immunostaining of the S1P1 polypeptide using the murine S1P1 antibody (23). Strong immunostaining of S1P1 was detected in endothelial cells within the tumor (Figure 5A). The expression of S1P1 in endothelial cells was significantly reduced by siRNA for S1P1, but not by the irrelevant siRNA, which suggests that downregulation of S1P1 contributed to the suppression of angiogenesis and tumor growth. (Figure 5D). In the necrotic areas of the tumor, diffuse S1P1 immunostaining was observed. Treatment with siRNA for S1P1 did not suppress this signal. The identity of these structures is not known but is likely to be cell debris.
Figure 5.
Inhibition of S1P1 expression and vascular stabilization in siRNA-injected tumor xenografts. Tumor xenografts were injected with multiplex siRNAs for β-gal (A–C and G) or S1P1 (D–F and H) as described. Tissue sections were harvested at 15 days and stained for S1P1 (red, A and D) and CD31 (green, B and E). Overlays are shown in C and F. siRNA for S1P1 potently suppressed the expression of S1P1 in endothelial cells (arrowheads). Arrows indicate S1P1 positivity in necrotic areas of the tumor. (G and H) Some sections were costained with anti–α-SMA and anti–CD31, and then α-SMA and CD31 were detected with secondary antibodies conjugated with alexa 568 (red, α-SMA) and alexa 488 (green, CD31) as described. Images were acquired using a confocal microscope. Note that α-SMA–positive cell association with angiogenic vessels were reduced after treatment with siRNA for S1P1.
Since vascular stabilization contributes to tumor angiogenesis (27, 28) and since S1P1 regulates vascular stabilization during development (8), we studied whether injection of siRNA for S1P1 would influence vascular stabilization of tumor vessels. Stabilization of tumor vessels was assessed by immunofluorescence microscopy of tumor tissue sections with the α-SMA antibody (Figure 5, G and H). Injection of siRNA for S1P1 strongly reduced mural cell coverage of tumor vessels (Figure 5H). These data suggest that siRNA for S1P1 inhibits the stabilization of nascent blood vessels and that this might be a mechanism via which vascular growth is inhibited.
Discussion
S1P is beginning to be recognized as a critical mediator in the growth and maintenance of the vascular system. In vitro studies clearly show that it is one of the most potent inducers of endothelial cell migration (3, 29, 30). In addition, endothelial cell survival, morphogenesis, NO synthesis, and proliferation are induced by S1P in cultured endothelial cells (1). These effects are attributed to signal transduction pathways regulated by S1P receptors, particularly S1P1. The relevance of S1P1 in the vascular system is underscored by the fact that it is abundantly expressed in cultured endothelial cells and its expression is induced by tumor promoters, growth factors, and biomechanical shear stress (31, 32). Extension of these studies to in vivo models of angiogenesis also show the requirement for S1P1 in adult angiogenesis; for example, downregulation of S1P1 expression with antisense phosphothioate oligonucleotides resulted in the suppression of angiogenesis in the Matrigel model of angiogenesis in the mouse (4). However, during embryonic development, the S1p1 gene is not required for early developmental angiogenesis. Instead, this receptor is absolutely required for proper vascular stabilization, i.e., enforcement of nascent endothelial vessels with the mural cells, for example vascular smooth muscle cells (8, 27). Therefore it is important to assess the role of S1P1 in adult angiogenesis, a process that occurs in many pathological conditions such as solid tumor growth (33).
We show here that S1P1 is induced in endothelial cells during tumor angiogenesis. Using the Lewis lung carcinoma model of tumor growth, we show that microvessels within the tumor as well as juxtatumoral large vessels express S1P1, as indicated by the induction of β-gal activity, which is under the control of the endogenous S1P1 promoter. Immunofluorescence analysis with the S1P1 antibody also confirmed that S1P1 expression is induced in tumor vessels. Angiogenic vessels in this model also undergo vascular stabilization, since many endothelial cells have a mural cell coat, as detected by α-SMA staining. Tumor infiltrating cells with a leukocytic morphology also express S1P1. These data suggest that S1P/S1P1 signaling may be involved in the regulation of tumor angiogenesis.
To address the specific role of S1P1 in tumor angiogenesis, we developed a novel method to silence S1p1 gene expression using the siRNA method. This was essential because the null mutation of the S1p1 gene induced embryonic lethality. Endothelial cell–specific deletion of the S1p1 gene was also lethal and exhibited a very similar phenotype (34). Thus, epigenetic approaches are needed, especially in the absence of pharmacologic antagonists for S1P1.
RNAi has emerged as a potentially promising technology to analyze the function of specific genes by loss-of-function approaches. At present, RNAi technology is effective in downregulating specific gene products in tissue culture and in model organisms such as Caenorhabditis elegans (16). The use of RNAi to downregulate cellular and viral genes in vivo in mammals is limited at present (19). A recent report showed that local injection of synthetic siRNA against VEGF into established tumors suppressed angiogenesis and tumor growth (35).
We developed the multiplex siRNA method, which offers potential improvements over existing methodology. First, targeting the 3′-UTR of transcripts for gene silencing offers several advantages; for example, this strategy is anticipated to minimize the problem of off-target silencing, as the 3′-UTR sequences are not generally conserved among related transcripts in multigene families, including in the S1P receptors. Second, the use of the E. coli RNAseIII, a highly stable enzyme (36), facilitates the efficient production of siRNA species in amounts required for in vivo administration (20). Third, multiplex siRNA species may achieve a higher opportunity for efficient gene silencing in an unbiased manner (20).
Our data show that the siRNA for S1P1 suppressed the S1P1 transcript in endothelial cells. Irrelevant siRNAs, such as those for S1P5 and β-gal, were ineffective, which suggests the specificity and efficacy of our procedure. Downregulation of S1P1 in mouse endothelial cells resulted in the inhibition of S1P-induced chemotaxis, an important step in angiogenesis. These in vitro studies suggested that the siRNA for S1P1 is likely to suppress angiogenesis in vivo.
To test the efficacy of the siRNA for S1P1 in angiogenesis in vivo, we delivered it to the site of angiogenesis by mixing with the extracellular matrix Matrigel. It is known that extracellular matrix proteins such as Matrigel and atelocollagen facilitate the uptake and stability of plasmid DNA, antisense oligonucleotides, and siRNA in vivo (35, 37). We show that incorporation of siRNA into Matrigel followed by subcutaneous implantation results in the efficient suppression of S1P1 transcript in infiltrating cells. This resulted in the potent suppression of neovessel formation in the Matrigel plug, which suggests that siRNA is capable of silencing the S1P1 gene in vivo.
To evaluate the role of S1P1 in tumor angiogenesis, we injected siRNA in the established tumor xenografts implanted subcutaneously in mice. Local delivery of S1P1 siRNA inhibited tumor growth and angiogenesis concomitantly with the significant suppression of S1P1 in the intratumoral endothelial cells. Control siRNAs for S1P5 and β-gal did not significantly suppress tumor growth and angiogenesis. This tumor-suppressive effect of S1P1 siRNA is unlikely to be due to direct effects on tumor cells, as Lewis lung carcinoma cells do not express the S1P1, as determined by Northern blot analysis. In addition, proliferation of Lewis lung carcinoma cells was not altered by in vitro treatment with siRNA. Rather, S1P1 siRNA inhibited angiogenesis, as indicated by the suppression of microvessel density. In addition to the microvessel density within the tumor, vessel morphology is also different. Tumor vasculatu re treated with S1P1 siRNA has fewer vascular branch points, which might be due to impaired migration of endothelial cells or its precursor cells. S1P is a potent chemoattractant for endothelial cells, and S1P1 is a major S1P receptor regulating endothelial cell migration (3). It is therefore likely that S1P1 contributes to tumor angiogenesis, in part, by regulating the migration of endothelial cells. It is also possible that S1P1 regulates the homing/migration of monocytic cells into the sites of angiogenesis and thereby modulates endothelial cell functions. In addition, S1P1 in various cells regulates cell survival (4), and, therefore, downregulation of this receptor with specific siRNA could induce apoptosis within the tumor. Indeed, siRNA for S1P1-injected tumors contained numerous necrotic regions. Further, endothelial cell morphogenesis could be regulated by S1P1, which is consistent with the finding that the vessels in the siRNA for S1P1-treated tumors contain altered morphology with reduced branch points.
Deletion of the S1p1gene in the mouse resulted in embryonic vascular stabilization defect, leading to vascular hemorrhage and lethality (8). Whether S1P1 is needed for vascular stabilization in the adult is not known. In fact, vascular stabilization is thought to be needed for optimal angiogenesis in tumors, since angiogenic vessels are ensheathed by mural cells (albeit abnormally) (27, 38). Secondly, inhibition of mural cell coverage of intratumoral vessels with PDGF receptor tyrosine kinase inhibitors either alone or together with VEGF inhibitors significantly attenuated tumor growth in vivo (28). Our data show that inhibition of S1P1 expression with siRNA inhibited mural cell coverage of neovessels, which suggests that this may constitute an additional mechanism involved in the inhibition of tumor vascular growth.
In conclusion, we show that S1P1 is induced in the tumor vasculature in vivo. We also show that downregulation of S1P1 expression with siRNA is effective in inhibiting angiogenesis and tumor growth in vivo. Inhibition of S1P1 expression in the tumor vasculature may be a novel therapeutic approach in the control of pathologic angiogenesis. Further, the siRNA approach may find general utility in the suppression of genes that are upregulated in pathologic conditions.
Methods
Preparation of multiplex siRNA.
E. coli RNaseIII plasmid was obtained from Allan Nicholson (Wayne State University, Detroit, Michigan, USA). The RNaseIII polypeptide was overexpressed in E. coli strain DH5α and purified using Ni2+ beads as previously described (36). Purified RNaseIII (>95% purity and comparable activity as described in ref. 23) was dialyzed and stored at –20°C in 50% glycerol, 0.5M NaCl, 30 mM Tris-HCl (pH 7.9), 0.5 mM EDTA, and 0.5 mM DTT. Template DNAs with the length of 500 nt of 3′-UTR were prepared by PCR using hybrid primers containing the T7 promoter sequence. Sense and antisense transcripts produced by in vitro transcription with T7 RNA polymerase were heated in boiling water bath for 5 minutes and cooled to room temperature for annealing in 30 mM HEPES-KOH (pH 7.4), 100 mM potassium acetate, and 2 mM magnesium acetate. Short dsRNA fragments were obtained by complete digestion with E. coli RNaseIII (500 nM) to the substrate dsRNA in 30 mM Tris-HCl (pH 8.0), 160 mM NaCl, 0.1 mM DTT, 0.1 mM EDTA, and 10 mM MgCl2 at 37°C for 4 hours. siRNA was extracted with phenol/chloroform/isopropanol, followed by ethanol precipitation. Products were analyzed by non-denaturation polyacrylamide gel electrophoresis and stained with ethidium bromide. Concentration of siRNA was determined using a standard extinction coefficient of dsRNA of approximately 20 bp.
PCR primer pairs were as follows: sense S1P1 3′-UTR: forward 5′-TCCTAATACGACTCACTATAGGGCTGTTGATACTGAGGGAAGC-3′, reverse 5′-TCCACAACCTCCTTCTGATGA-3′; antisense S1P1 3′-UTR: forward 5′-CTGTTGATACTGAGGGAAGC-3′, reverse 5′-TCCTAATACGACTCACTATAGGGTCCACAACCTCCTTCTGATGA-3′; sense S1P5 3′-UTR: forward 5′-TCCTAATACGACTCACTATAGGGCCCTTTGCAAGAACAGACTGA-3′, reverse 5′-CCAGCCCTTAGCACACTCTTA-3′; antisense S1P5 3′-UTR: forward 5′-CCCTTTGCAAGAACAGACTGA-3′, reverse 5′-TCCTAATACGACTCACTATAGGGCCAGCCCTTAGCACACTCTTA-3′; sense β-gal: forward 5′-TCCTAATACGACTCACTATAGGGACTGGCAGATGCACGGTTAC-3′, reverse 5′-CGTAGGTAGTCACGCAACTC-3′; antisense β-gal: forward 5′-ACTGGCAGATGCACGGTTAC-3′, reverse 5′-TCCTAATACGACTCACTATAGGGCGTAGGTAGTCACGCAACTC-3′.
Endothelial cell chemotaxis assay.
MEECs were isolated from S1p1+/+ and S1p1–/– mice as described (39). MEECs were transfected with 200 nM of siRNAs using oligofectamine (Invitrogen) as described (26). After 48 hours, cell migration assay was performed as previously described (3).
Preparation of cationic liposomes.
Cationic liposomes were prepared with an 11:9 molar ratio of 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP; Avanti Polar Lipids Inc.) and cholesterol (Sigma-Aldrich) in chloroform (10 mg/ml). Chloroform was evaporated using nitrogen stream, and residual solvent was removed by placing lipid mixtures under vacuum pump for 1 hour. Dried lipids were resuspended in 5% sucrose and sonicated for 20 minutes at 37°C. To generate unilamellar liposome vesicles, we passed the hydrated mixture through the Mini-Extruder (Avanti Polar Lipids Inc.).
Northern blot analysis.
Total RNA from tissues or MEECs was isolated using RNA-STAT (Tel-Test Inc.) as described by the manufacturer. Total RNA (5 μg) was subjected to Northern blot analysis using cDNA probes specific to mouse S1P1 or GAPDH as described (5).
Tumor angiogenesis and growth.
Experiments using animals were reviewed and approved by the Institutional Animal Care and Use Committee of the University of Connecticut Health Center. Lewis lung carcinoma cells (106; American Type Culture Collection) were mixed with 100 μl of Matrigel solution (BD Biosciences — Discovery Labware) at 4°C and injected subcutaneously into the dorsal skin of nude mice and allowed to grow. After 5 days, siRNA-liposome complexes were locally administered into growing tumors by intratumoral injection on every 3rd day. siRNA-liposome complexes (60 μl) were obtained by incubating 15 μg of siRNA and 25 μl of liposomes (10 mg/ml) in 5% glucose for 20 minutes at room temperature. The complexes were injected into 4–6 sites on tumor. Tumor growth was determined using a caliper to measure tumor diameter. Tumor volume was calculated by the equation, V = (L × W2) × 0.5 (V, volume; L, length; and W, width). Angiogenesis in tumor sections was evaluated by counting the number of CD31-positive blood vessels in the 5 most densely vascularized areas within tumor as described previously (26). All CD31 positive vessels (regardless of shape, branch points, size, lumens) were considered as a single vascular unit and were counted. Tumor vascular stabilization was assessed by counting all positive cells that expressed the α-SMA antigen in the immunofluorescence assay.
β-gal (LacZ) staining of tissues. Tissues were dissected in cold PBS and fixed in 0.2% glutaraldehyde and 1.5% paraformaldehyde in PBS, pH 7.4, at room temperature for 90 minutes, then washed in PBS. Staining was performed at 37°C or room temperature in 0.02% X-gal, 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, and 2 mM MgCl2 in PBS overnight. The stained tissues were washed, postfixed in 4% paraformaldehyde, and embedded in paraffin; 5 μm-sections were then cut and imaged as described (40).
In vivo Matrigel angiogenesis assay.
In vivo angiogenesis assay was performed as previously described (4). Briefly, female athymic nude mice were injected subcutaneously with 0.2 ml of Matrigel mixture containing 2 μM of siRNA with or without FGF-2 (4 μg/ml). After 7 days, Matrigel plugs were harvested, fixed with 10% formalin in PBS, and embedded in paraffin. Sections were stained with H&E or anti–CD31 antibody as described below.
Immunohistochemistry.
Tissues sections were postfixed in 4% paraformaldehyde for 20 minutes, treated in 3% hydrogen peroxide/methanol for 30 minutes, blocked in 10% normal goat serum for 20 minutes, incubated with anti–CD31 antibody (BD Biosciences — Pharmingen) for 1 hour at room temperature, biotinylated goat-anti mouse IgG (DakoCytomation) for 30 minutes, and VECTASTAIN Elite ABC biotin-avidin-peroxidase complex (Vector Laboratories) for 30 minutes. Sections were then developed with diaminobenzidine and diaminobenzidine enhancer (Vector Laboratories). Smooth muscle cells were stained with anti–α-SMA antibody (Sigma-Aldrich). Rabbit polyclonal antibodies against murine S1P1 were obtained from S. Mandala at Merck Research Laboratories. The S1P1 staining was performed as previously described (23).
Statistical analysis.
All results are expressed as means ± SE, and n represents the number of individual experiments. Statistical analyses were performed with Student’s t test.
Supplementary Material
Acknowledgments
We thank Richard L. Proia for the gift of S1p1+/LacZ mice, Allan Nicholson for the RNaseIII construct, and Suzanne Mandala for the anti–S1P1 antibody. This work is supported by NIH grants HL70694 and HL67330. Timothy Hla dedicates this work to the memory of Thomas Maciag.
Footnotes
Nonstandard abbreviations used: dsRNA, double-stranded RNA; MEEC, mouse embryonic EC; RNAi, RNA interference; S1P, sphingosine 1–phosphate; S1P1, S1P receptor-1; siRNA, small interfering RNA; 3′-UTR, 3′–untranslated region.Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Saba JD, Hla T. Point-counterpoint of sphingosine 1-phosphate metabolism. Circ. Res. 2004;94:724–734. doi: 10.1161/01.RES.0000122383.60368.24. [DOI] [PubMed] [Google Scholar]
- 2.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 3.Paik JH, Chae S, Lee MJ, Thangada S, Hla T. Sphingosine 1-phosphate-induced endothelial cell migration requires the expression of EDG-1 and EDG-3 receptors and Rho-dependent activation of alpha vbeta3- and beta1-containing integrins. J. Biol. Chem. 2001;276:11830–11837. doi: 10.1074/jbc.M009422200. [DOI] [PubMed] [Google Scholar]
- 4.Lee MJ, et al. Vascular endothelial cell adherens junction assembly and morphogenesis induced by sphingosine-1-phosphate. Cell. 1999;99:301–312. doi: 10.1016/s0092-8674(00)81661-x. [DOI] [PubMed] [Google Scholar]
- 5.Hla T, Maciag T. An abundant transcript induced in differentiating human endothelial cells encodes a polypeptide with structural similarities to G-protein- coupled receptors. J. Biol. Chem. 1990;265:9308–9313. [PubMed] [Google Scholar]
- 6.Sanchez T, et al. Phosphorylation and action of the immunomodulator FTY720 inhibits vascular endothelial cell growth factor-induced vascular permeability. J. Biol. Chem. 2003;278:47281–47290. doi: 10.1074/jbc.M306896200. [DOI] [PubMed] [Google Scholar]
- 7.Garcia JG, et al. Sphingosine 1-phosphate promotes endothelial cell barrier integrity by Edg-dependent cytoskeletal rearrangement. J. Clin. Invest. 2001;108:689–701. doi:10.1172/JCI200112450. doi: 10.1172/JCI12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y, et al. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Invest. 2000;106:951–961. doi: 10.1172/JCI10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat. Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 10.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 11.Hurwitz H, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 12.Chae SS, Proia RL, Hla T. Constitutive expression of S1P1 receptor in adult tissues. Prostaglandins Other Lipid Mediat. 2004;73:141–150. doi: 10.1016/j.prostaglandins.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 13.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milhavet O, Gary DS, Mattson MP. RNA interference in biology and medicine. Pharmacol. Rev. 2003;55:629–648. doi: 10.1124/pr.55.4.1. [DOI] [PubMed] [Google Scholar]
- 16.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 17.McCaffrey AP, et al. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen DR, Leirdal M, Sioud M. Gene silencing by systemic delivery of synthetic siRNAs in adult mice. J. Mol. Biol. 2003;327:761–766. doi: 10.1016/s0022-2836(03)00181-5. [DOI] [PubMed] [Google Scholar]
- 19.Dave RS, Pomerantz RJ. RNA interference: on the road to an alternate therapeutic strategy! Rev. Med. Virol. 2003;13:373–385. doi: 10.1002/rmv.407. [DOI] [PubMed] [Google Scholar]
- 20.Yang D, et al. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuersten S, Goodwin EB. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet. 2003;4:626–637. doi: 10.1038/nrg1125. [DOI] [PubMed] [Google Scholar]
- 23.Forrest M, et al. Immune cell regulation and cardiovascular effects of sphingosine 1-phosphate receptor agonists in rodents are mediated via distinct receptor subtypes. J. Pharmacol. Exp. Ther. 2004;309:758–768. doi: 10.1124/jpet.103.062828. [DOI] [PubMed] [Google Scholar]
- 24.Allende ML, Dreier JL, Mandala S, Proia RL. Expression of the sphingosine-1-phosphate receptor, S1P1, on T-cells controls thymic emigration. J. Biol. Chem. 2004;279:15396–15401. doi: 10.1074/jbc.M314291200. [DOI] [PubMed] [Google Scholar]
- 25.Liu CH, Hla T. The mouse gene for the inducible G-protein-coupled receptor edg-1. Genomics. 1997;43:15–24. doi: 10.1006/geno.1997.4759. [DOI] [PubMed] [Google Scholar]
- 26.Ancellin N, et al. Extracellular export of sphingosine kinase-1 enzyme. Sphingosine 1-phosphate generation and the induction of angiogenic vascular maturation. J. Biol. Chem. 2002;277:6667–6675. doi: 10.1074/jbc.M102841200. [DOI] [PubMed] [Google Scholar]
- 27.Jain RK. Molecular regulation of vessel maturation. Nat. Med. 2003;9:685–693. doi: 10.1038/nm0603-685. [DOI] [PubMed] [Google Scholar]
- 28.Bergers G, Song S, Meyer-Morse N, Bergsland E, Hanahan D. Benefits of targeting both pericytes and endothelial cells in the tumor vasculature with kinase inhibitors. J. Clin. Invest. 2003;111:1287–1295. doi:10.1172/JCI200317929. doi: 10.1172/JCI17929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.English D, Garcia JG, Brindley DN. Platelet-released phospholipids link haemostasis and angiogenesis. Cardiovasc. Res. 2001;49:588–599. doi: 10.1016/s0008-6363(00)00230-3. [DOI] [PubMed] [Google Scholar]
- 30.Wang F, et al. Sphingosine 1-phosphate stimulates cell migration through a G(i)-coupled cell surface receptor. Potential involvement in angiogenesis. J. Biol. Chem. 1999;274:35343–35350. doi: 10.1074/jbc.274.50.35343. [DOI] [PubMed] [Google Scholar]
- 31.Hla T. Sphingosine 1-phosphate receptors. Prostaglandins. 2001;64:135–142. doi: 10.1016/s0090-6980(01)00109-5. [DOI] [PubMed] [Google Scholar]
- 32.Igarashi J, Erwin PA, Dantas AP, Chen H, Michel T. VEGF induces S1P1 receptors in endothelial cells: Implications for cross-talk between sphingolipid and growth factor receptors. Proc. Natl. Acad. Sci. U. S. A. 2003;100:10664–10669. doi: 10.1073/pnas.1934494100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 34.Allende ML, Yamashita T, Proia RL. G-protein-coupled receptor S1P1 acts within endothelial cells to regulate vascular maturation. Blood. 2003;102:3665–3667. doi: 10.1182/blood-2003-02-0460. [DOI] [PubMed] [Google Scholar]
- 35.Takei Y, Kadomatsu K, Yuzawa Y, Matsuo S, Muramatsu T. A small interfering RNA targeting vascular endothelial growth factor as cancer therapeutics. Cancer Res. 2004;64:3365–3370. doi: 10.1158/0008-5472.CAN-03-2682. [DOI] [PubMed] [Google Scholar]
- 36.Amarasinghe AK, Calin-Jageman I, Harmouch A, Sun W, Nicholson AW. Escherichia coli ribonuclease III: affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol. 2001;342:143–158. doi: 10.1016/s0076-6879(01)42542-0. [DOI] [PubMed] [Google Scholar]
- 37.Ochiya T, et al. New delivery system for plasmid DNA in vivo using atelocollagen as a carrier material: the Minipellet. Nat. Med. 1999;5:707–710. doi: 10.1038/9560. [DOI] [PubMed] [Google Scholar]
- 38.Morikawa S, et al. Abnormalities in pericytes on blood vessels and endothelial sprouts in tumors. Am. J. Pathol. 2002;160:985–1000. doi: 10.1016/S0002-9440(10)64920-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bussolino F, et al. Murine endothelioma cell lines transformed by polyoma middle T oncogene as target for and producers of cytokines. J. Immunol. 1991;147:2122–2129. [PubMed] [Google Scholar]
- 40.Chae SS, Paik JH, Allende ML, Proia RL, Hla T. Regulation of limb development by the sphingosine 1-phosphate receptor S1p1/EDG-1 occurs via the hypoxia/VEGF axis. Dev. Biol. 2004;268:441–447. doi: 10.1016/j.ydbio.2004.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.