Abstract
We have used comparative genomic hybridization (CGH) on a full-genome Campylobacter jejuni microarray to examine genome-wide gene conservation patterns among 51 strains isolated from food and clinical sources. These data have been integrated with data from three previous C. jejuni CGH studies to perform a meta-analysis that included 97 strains from the four separate data sets. Although many genes were found to be divergent across multiple strains (n = 350), many genes (n = 249) were uniquely variable in single strains. Thus, the strains in each data set comprise strains with a unique genetic diversity not found in the strains in the other data sets. Despite the large increase in the collective number of variable C. jejuni genes (n = 599) found in the meta-analysis data set, nearly half of these (n = 276) mapped to previously defined variable loci, and it therefore appears that large regions of the C. jejuni genome are genetically stable. A detailed analysis of the microarray data revealed that divergent genes could be differentiated on the basis of the amplitudes of their differential microarray signals. Of 599 variable genes, 122 could be classified as highly divergent on the basis of CGH data. Nearly all highly divergent genes (117 of 122) had divergent neighbors and showed high levels of intraspecies variability. The approach outlined here has enabled us to distinguish global trends of gene conservation in C. jejuni and has enabled us to define this group of genes as a robust set of variable markers that can become the cornerstone of a new generation of genotyping methods that use genome-wide C. jejuni gene variability data.
Campylobacter jejuni is a human pathogen, a commensal inhabitant of many domestic animals, and globally, the most common cause of acute bacterial enteritis (for a review, see reference 31). Two well-established serotyping methods, namely, Penner typing based on heat-stable antigens and Lior typing based on heat-labile antigens, have been in use for more than two decades to study species diversity, to track epidemiological trends, and to determine important epidemiological correlations (15, 23). Technical limitations on the production of high-quality typing sera have limited the availability of these reagents. Culturing conditions can affect the expression of serotyping determinants, which affects serotyping results, and several strains are nontypeable (32). Additionally, serotype relatedness is not always indicative of genetic relatedness since members of different serotypes of C. jejuni are genetically related, despite differences in heat-stable antigen expression (16).
The need for alternative subtyping schemes has been recognized, leading to the development of a number of different methods based on differences at the DNA level (i.e., genotyping). The techniques used at present range from analysis of polymorphisms in groups of housekeeping genes (multilocus sequence typing [5, 26]), amplified fragment length polymorphism analysis (28), restriction fragment length polymorphism analysis of flaA or rRNA genes (for a review, see reference 32), and pulsed-field gel electrophoresis (PFGE) analysis of macrorestriction patterns (34). Despite the large number of competing approaches, PFGE (8) and, more recently, multilocus sequence typing (26) have emerged as the present “gold standard” genotyping methods, and considerable efforts have been made to standardize protocols in order to facilitate interlaboratory comparisons (32).
One potential weakness shared by these genotyping approaches is that strain relatedness is inferred on the basis of limited subsampling of the entire genome (14). Whole-genome sequencing provides the most complete data set for comparative genomics studies; and genome sequence data are available for more than one strain of an increasing number of bacterial species, which includes Helicobacter pylori (1) and Escherichia coli (24, 33), among others. The genomic sequence of C. jejuni NCTC 11168 was completed by Parkhill et al. (21); and preliminary genome sequence data for a second C. jejuni strain, RM1221 (18), has recently been made available by The Institute for Genomic Research (TIGR; http://www.tigr.org). Despite these efforts, species with multiple strain coverage number less than 20, and in most cases genome sequence data sets are restricted to two strains. Although sequencing of bacterial genomes has become technically straightforward, it remains expensive and logistically demanding. It is unlikely that whole-genome sequencing can be used for genotyping or large-scale comparative genomics.
Whole-genome sequence data have been used to construct full-genome DNA microarrays that include every open reading frame (ORF) in a genome strain. Microarray-based comparative genomic hybridization (CGH), in which labeled DNAs from two strains are competitively hybridized to a full-genome microarray, has been described in numerous reports (2-4, 6, 9, 11, 20, 27). Microarray-based CGH provides both rich data sets for whole-genome genotyping and an indirect approach for comparative genomics in the absence of whole-genome sequence data. Studies on the genetic diversity and the feasibility of using DNA microarrays as a tool for the genotyping of C. jejuni (7, 14, 22) have demonstrated the value of microarray-based CGH in comparative genomics.
Initial observations of C. jejuni genetic variability by Dorrell et al. (7) were based on a survey of 11 strains. Leonard et al. (14) and Pearson et al. (22) have recently analyzed an additional 16 and 18 strains, respectively. These small-scale studies have revealed extensive genetic variability in C. jejuni, underscoring the need to further characterize intraspecies variability through large-scale surveys involving data sets comprising greater epidemiological, phenotypic, and geographical strain diversities. With the 51 strains analyzed in the present study, the cumulative data on C. jejuni represent the largest and most diverse microarray-based comparative genomics data set to date. We describe here a detailed meta-analysis of all available C. jejuni CGH data. The data have provided us with a comprehensive picture of global C. jejuni gene conservation patterns that suggest low levels of genome plasticity. This analysis has also enabled us to define a highly robust set of variable genes for genotyping of C. jejuni. An increasing body of C. jejuni CGH data will enable us to begin formulating hypotheses about C. jejuni genome evolution and the development of the wide variation in virulence, pathogenicity, and host specificity observed in this economically and medically important human pathogen.
MATERIALS AND METHODS
Bacterial strains.
The backgrounds for the 51 strains that we analyzed by microarray-based CGH are presented in Table 1. C strains and F strains, obtained from the Bureau of Microbial Hazards (Health Canada, Ottawa, Ontario, Canada), represent clinical and food isolates, respectively, collected in Ottawa and the surrounding area over a period spanning from January 1998 to January 2001. For additional strain information, see http://ibs-isb.nrc-cnrc.gc.ca/ibs/immunochemistry/suppInfo_Taboada_2004a_e.html. Strains were selected to cover a wide range of different PFGE profiles (17). Cells were grown on Mueller-Hinton agar plates (Difco, Oakville, Ontario, Canada) for 36 h at 42°C under microaerophilic conditions prior to DNA isolation.
TABLE 1.
C. jejuni strains analyzed by CGH in this study
| Strain | Source | Isolate origin |
|---|---|---|
| C strainsab | Bureau of Microbial Hazards, Health Canada | Human clinical isolate |
| F strainscd | Bureau of Microbial Hazards, Health Canada | Raw chicken |
| ATCC 43429 | American Type Culture Collection | Human feces |
| ATCC 43430 | American Type Culture Collection | Calf feces |
| ATCC 43431 | American Type Culture Collection | Human feces |
| ATCC 43432 | American Type Culture Collection | Human feces |
| ATCC 43438 | American Type Culture Collection | Human feces |
| ATCC 43446 | American Type Culture Collection | Human feces |
| ATCC 43449 | American Type Culture Collection | Human feces |
| ATCC 43455 | American Type Culture Collection | Marmoset feces |
| ATCC 43456 | American Type Culture Collection | Human feces |
| ATCC 43460 | American Type Culture Collection | Gazelle feces |
| ATCC 49302 | American Type Culture Collection | Human clinical isolate |
| RM1221d | Food Safety and Health Research Unit, U.S. Department of Agriculture | Raw chicken |
C strains are C015, C028, C064, C078, C089, C092, C102, C105, C115, C132, C133, C150, C164, C170, C193, C207, C228, C230, C235, C236, C245, C254, C256, C279, C288, C293, C310, C314, C321, and F002.
Data are from reference 17.
F strains are F006, F008, F009, F012, F013, F036, F037, F042, and F043.
Data are from reference 18.
Construction of a C. jejuni NCTC 11168 ORF DNA microarray.
PCR primers were designed for each of the 1,634 ORFs described in the annotated sequence of C. jejuni NCTC 11168 (21). Primers were selected by using the Primer3 program (25). Primer selection parameters were standardized and included a similar predicted melting temperature (58 to 62°C), uniform length (25 nucleotides), and a minimum amplicon size of 100 bp. A computer program designed in-house (the make_primers program) was used to administer Primer3 submissions, collect acceptable primer pairs, and resubmit requests for ORFs that failed to yield acceptable primer pairs (http://ibs-isb.nrc-cnrc.gc.ca/ibs/immunochemistry/group_software_e.html). Primer synthesis was performed in a high-throughput 96-well LCDR/MerMade oligonucleotide synthesizer (BioAutomation Corporation, Court Plano, Tex.). ORF-specific amplicons were produced from 30 ng of a genomic DNA template in a 96-well plate format by standard PCR amplification methods. Amplicon quality and sample tracking were performed with BRIDNA, an in-house database developed by the Biotechnology Research Institute, National Research Council of Canada, Montreal, Quebec, Canada (http://www.bri.nrc-cnrc.gc.ca/business/microarraylab/bridna_e.html). Amplicons were purified in Multiscreen-FB plates (Millipore), lyophilized, and resuspended to an average concentration of 0.1 to 0.2 μg/μl in spotting buffer (50% dimethyl sulfoxide, 1 mM EDTA [pH 8.0]). Microarrays were printed onto CMT-GAPS II slides (Corning) by using a Chipwriter spotting robot (Bio-Rad, Mississauga, Ontario, Canada). Details on the construction and content of this microarray (CampyChip1.2) are available at http://ibs-isb.nrc-cnrc.gc.ca/ibs/immunochemistry/campychips_e.html.
Isolation of genomic DNA.
C. jejuni strains were harvested from the growth on plates that had been incubated for 24 h, resuspended in 10 mM Tris-10 mM EDTA (pH 8.0), and treated with lysozyme (Roche, Laval, Quebec, Canada) and RNase A (Qiagen, Mississauga, Ontario, Canada) for 10 min at room temperature. The cell suspensions were then digested with proteinase K (MBI Fermentas, Burlington, Ontario, Canada) for 1 h at 37°C, and complete lysis was obtained by addition of sodium dodecyl sulfate to a final concentration of 0.1% (wt/vol). Genomic DNA was extracted from the cell lysates by three extractions with phenol-chloroform-isoamyl alcohol (25:24:1) and was precipitated in isopropanol.
Genomic DNA labeling.
Genomic DNA was restricted to an average size of 2 to 5 kb by double digestion with EcoRI and HindIII. A total of 5 μg of DNA was fluorescently labeled by direct chemical coupling with the Label-IT (Mirus Corp., Madison, Wis.) dyes cyanine 3 (Cy3) and Cy5, as recommended by the manufacturer. Probes were purified from the incorporated dyes by sequentially passing samples through SigmaSpin (Sigma, Oakville, Ontario, Canada) and Qiaquick (Qiagen) columns. Labeled DNA sample yields and dye incorporation efficiencies were calculated by using an ND-1000 spectrophotometer (Nanodrop, Rockland, Del.).
Microarray hybridizations.
The hybridization profile for each strain was obtained by cohybridizing labeled DNA from the test strain and from strain NCTC 11168 (control) to our microarray. By convention, the DNA from test strains was labeled with Cy5 and that from the control strain was labeled with Cy3, although reciprocal labeling was performed with selected strains to test for potential dye incorporation bias. Labeled samples were normalized by selecting test and control sample pairs with similar dye incorporation efficiencies. Equivalent amounts (1 to 2 μg) of labeled test and control samples were pooled, lyophilized, and then resuspended in 35 μl of hybridization buffer (1× DIGEasy hybridization solution [Roche, Laval, Quebec, Canada], 0.5 μg of torulla yeast tRNA per μl, 0.5 μg of denatured salmon sperm genomic DNA per μl). The probes were denatured at 65°C for 5 min, cooled to room temperature, and applied to the microarray. Hybridizations were performed overnight at 37°C under glass coverslips (24 by 42 mm) in a high-humidity chamber. Microarrays were washed two times for 10 min each time at 50°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate, two times for 5 min each time at 50°C in 0.5× SSC, and once for 5 min at 50°C in 0.1× SSC. The slides were spun dry (500 × g, 5 min) and stored in light-tight containers until they were scanned.
Data acquisition and analysis.
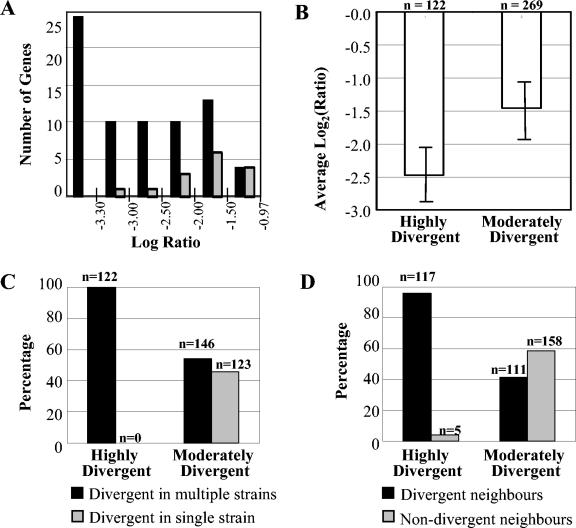
The microarrays were scanned with a Chipreader laser scanner (Bio-Rad), according to the recommendations of the manufacturer. Spot quantification, signal normalization, and data visualization were performed with the program ArrayPro Analyzer (version 4.5; Media Cybernetics, Silver Spring, Md.). Net signal intensities were obtained by performing local-ring background subtraction, and spots with a signal less than five times greater than the background signal were excluded from the analysis. Signal intensities for triplicate spots were averaged, and the data from each channel were adjusted by subarray normalization by using cross-channel Loess regression. The ratio of the signal for the test strain to that for the control strain for each gene was transformed to its base 2 logarithm (29), log2(tester signal/C. jejuni NCTC 11168 signal), hereafter referred to as the “log ratio,” and genes with log ratios less than −0.97 were considered divergent. Technical variations in our methodology were tested for by selecting a subset of strains for replicate hybridizations and treating the data from replicates separately throughout the various analyses. Consistency in the data was assessed by direct comparison of the lists of variable genes obtained from each replicate. In order to examine mapping of variable genes to genomic regions, we organized all CGH data by assuming conservation of gene order (synteny) with C. jejuni NCTC 11168. Genes were assigned to the highly variable (HV) group if they were divergent in more than one strain. On the basis of our unpublished observations of CGH with C. jejuni RM1221, log ratios less than −3.3 are likely to represent genes that are highly divergent (HD) or absent in the tester strain (see Fig. 4A). Genes were assigned to the HD group if the lowest observed log ratio for the gene was less than −3.3 for any of the strains in the data set. All non-HD genes were assigned to the moderately divergent (MD) group.
FIG. 4.
Association between high levels of divergence, the occurrence of codivergent clusters, and divergence in multiple strains. (A) Genes which were divergent in experiments with strain RM1221 and NCTC 11168 CGH (log ratio < −0.97) were binned by log ratio value. The BLAST server at TIGR was used to examine the homology of the corresponding NCTC 11168 genes to the RM1221 genome sequence. Gray bars, the number of genes in each bin for which BLAST hits indicated detectable sequence identities; black bars, genes without BLAST hits in the RM1221 genome. On the basis of these data, a log ratio <−3.3 was used as a cutoff to define HD or absent genes from CGH data. (B) Average log ratios [log2(test signal/control signal)] for HD and MD genes. A statistically significant difference in the average log ratios for the two groups can be observed. (C) Percentage of divergent genes that were variable in multiple strains. HD genes are exclusively found to be divergent in multiple strains (100%; 122 of 122). In contrast, MD genes have a similar likelihood of being divergent in a single strain or in multiple strains (45.7 and 54.3%, respectively). (D) Percentage of divergent genes that were adjacent in the C. jejuni NCTC 11168 genome. The majority of HD genes have codivergent neighbors (95.9%; 117 of 122), whereas this value is only 41.3% among MD genes. The figure is based on raw microarray CGH data for data sets II and III.
Analysis of additional C. jejuni data sets.
The results analyzed for data set I (7), in which divergent genes had previously been determined by using a log ratio threshold of −1.0, were obtained from http://www.sghms.ac.uk/depts/medmicro/bugs/GR-1858/index.htm. Raw log ratio data from data set II (14) were obtained from the Stanford Microarray Database website (http://genome-www5.stanford.edu/); the data were reanalyzed by using a log ratio threshold of −0.97 to define divergent genes. Highly divergent genes (log ratio < −3.3) were determined by using data sets for which raw microarray CGH data were available (data sets II and III). The results are summarized at http://ibs-isb.nrc-cnrc.gc.ca/ibs/immunochemistry/suppInfo_Taboada_2004a_e.html. The results for data set IV (22), in which the authors applied a novel algorithm to determine divergent genes from CGH data, were obtained from http://www.sciencedirect.com/science/MiamiMultiMediaURL/B6T36-49SW7D4-9/B6T36-49SW7D4-9-4/4938/Table.xls. A list of genes which are highly variable and highly divergent, as determined from our meta-analysis of the four data sets, is provided at http://ibs-isb.nrc-cnrc.gc.ca/ibs/immunochemistry/suppInfo_Taboada_2004a_e.html Only data for 1,597 genes common to all data sets were considered for analysis. The BLAST server at TIGR (http://tigrblast.tigr.org/ufmg/index.cgi?database=c_jejuni|seq) was used to determine the levels of homology between NCTC 11168 genes and the draft RM1221 genome sequence. Preliminary sequence data were obtained from the Institute for Genomic Research through the website at http://www.tigr.org. Our clusters of orthologous genes (COGs) analysis was performed by using COG assignments of the C. jejuni NCTC 11168 genome available from http://www.ncbi.nlm.nih.gov/sutils/coxik.cgi?gi=152&target=a. COG X was created by the authors to denote genes that do not fall under any other COG groups.
RESULTS
Determination of divergent genes in C. jejuni genome.
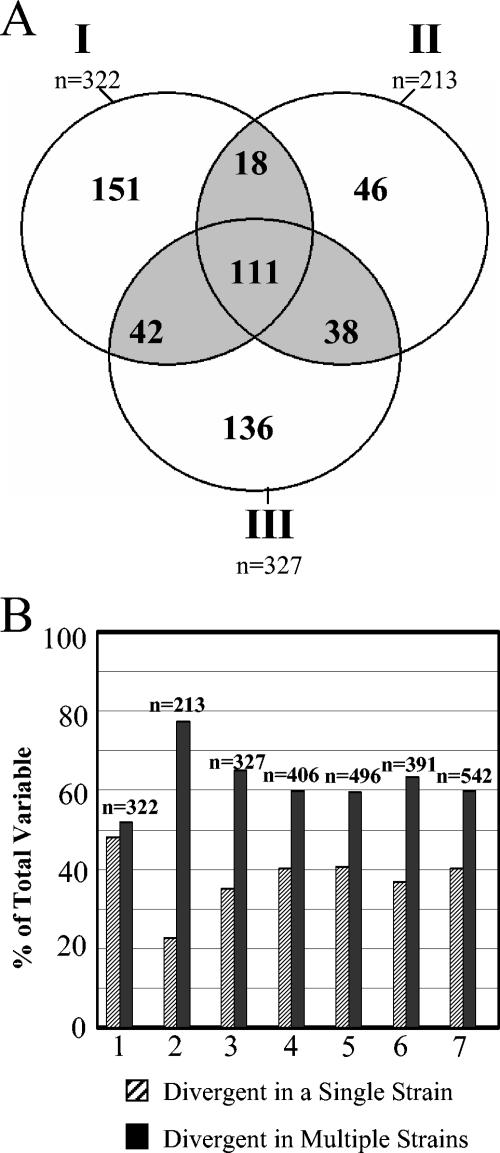
The hybridization profile for each of 51 strains analyzed in this study was obtained by hybridizing labeled genomic DNA to a C. jejuni microarray based on sequence data from the genome strain NCTC 11168. Divergent and absent genes are expected to show decreased hybridization signals with respect to those obtained with the genome strain, and we designated genes as divergent using a threshold log ratio of −0.97. By using this threshold, approximately 20% of the genes (327 of 1,634) in C. jejuni NCTC 11168 were divergent in at least 1 of the 51 strains that we examined. Although we obtained a number of variable genes similar to that in a previous study of C. jejuni (7), 327 compared to 322, only 153 genes were found to be variable in both data sets. Because of the uniqueness encountered in each data set, we combined our CGH data (data set III) with those from two previous studies (data set I, results from reference 7; data set II, results from reference 14) to produce a data set of 79 strains for the meta-analysis in order to reduce the effect of sampling biases. We determined that 542 genes were divergent in at least one of the strains in the meta-analysis data set (Fig. 1A), a value that is considerably higher than the number of variable genes within each data set (n = 322, n = 213, and n = 327 for data sets I, II, and III, respectively). The increase in the number of variable genes in the collective data is due to the large number of genes (333 of 542) that were variable in only one of the three data sets (Fig. 1A, sum of unshaded values).
FIG. 1.
Cumulative data from microarray-based CGH surveys of C. jejuni. (A) Divergent genes observed in data sets I, II, and III analyzed in this survey (see text). Of the 542 divergent genes observed in the collective data set, 209 are variable in two or more data sets (gray shading). The remaining 333 genes were variable in only one of the data sets. (B) Prevalence of genes divergent in a single strain (singletons) in three different C. jejuni CGH data sets. Whether analyzed separately or in various combinations (1, data set I; 2, data set II; 3, data set III; 4, data sets I and II; 5, data sets I and III; 6, data sets II and III; 7, data sets I, II, and III), a significant number of divergent singletons (striped bars) were obtained in the various data sets.
An additional factor contributing to the large number of variable genes observed in the meta-analysis data set is the number of genes, 217 of 542 (40%), which were divergent in a single strain (termed “singletons”). A high prevalence of singletons was found in each of the three strain sets analyzed here (data set I, 48.1% of 322 genes; data set II, 22.6% of 213 genes; data set III, 35.2% of 327 genes; Fig. 1B). Although the biological significance of the large numbers of divergent singletons in each data set is unknown, a lower number of singletons was observed in data set II, which is composed of epidemiologically related strains (14). The incidence of singletons is likely a reflection of the degree of genetic relatedness of the members of a data set. Although many divergent singletons in one study mapped onto genes that are variable in another data set, use of a combination of data from any two studies led to a high but stable proportion (35 to 39%) of cumulative singletons (Fig. 1B). It would therefore appear that any group of genetically diverse strains is likely to collectively harbor a significant repertoire of genes found to be uniquely divergent in individual strains.
Although differences in the methodology used to define divergent genes prevented us from directly incorporating data set IV (22) into the meta-analysis data set, we were able to compare the gene conservation trends obtained in each data set (Fig. 2). Of 266 genes that were variable in data set IV, 78% (209 of 266) had variable counterparts in the meta-analysis data set. Despite the overlap between data sets, of 542 genes that were variable in the meta-analysis data set, 61% (333 of 542) had no variable counterparts in data set IV. Similarly, 57 genes that were variable in data set IV had no variable counterparts in the meta-analysis data set. Of note, more than half of the variable genes unique to data set IV (33 of 57) were divergent singletons. The remaining singletons in data set IV (n = 31) mapped to variable genes previously identified in the meta-analysis data set.
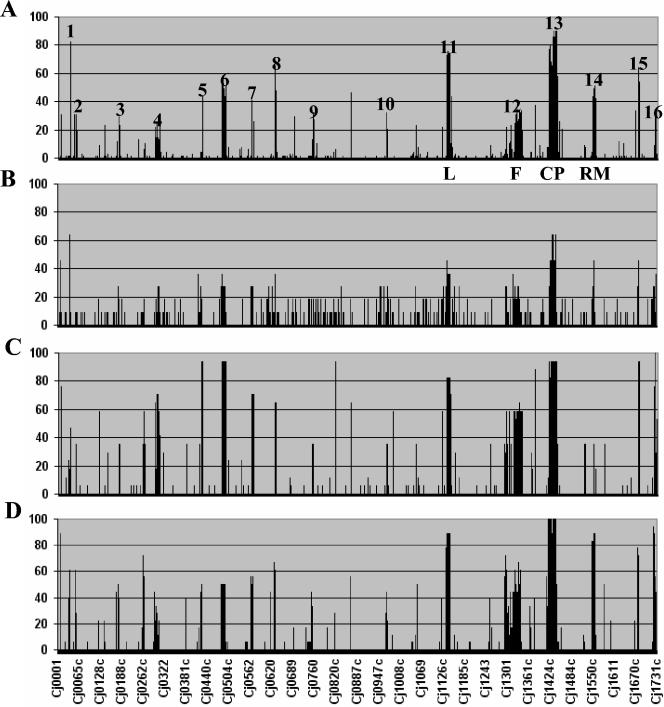
FIG. 2.
Survey of genetic variability in C. jejuni. Divergent genes were determined for each strain, and the percentage of strains showing divergence at each gene position was calculated and plotted as a histogram according to their position on the genome strain NCTC 11168 (18). The results for data set III (A), data set I (7) (B), data set II (14) (C), and data set IV (22) (D) are shown. Most variable genes map to 16 hypervariable regions in the C. jejuni NCTC 11168 genome (Table 2). These include the lipooligosaccharide biosynthesis locus (L), the flagellar biosynthesis locus (F), the capsular polysaccharide biosynthesis locus (CP), and the restriction-modification locus (RM). Although similar trends were observed for all four data sets, data set III (A) shows better resolution because of the larger sample size.
Analysis of gene conservation patterns in C. jejuni genome.
The original number of variable genes reported by Dorrell et al. (7), data set I, was ∼20% (n = 322) of the genes in strain NCTC 11168, with ∼80% of genes showing high degrees of conservation among all strains. The investigators acknowledged that this level of conservation was likely to be overestimated because of the small number of strains studied. When we combined our data for 51 strains with data for 46 strains from three previous studies (7, 14, 22), 599 genes, or 36.6% of the 1,634 genes in C. jejuni NCTC 11168, were detectably divergent in at least one strain. The almost 10-fold increase in the number of strains included in this meta-analysis uncovered an additional 277 divergent genes.
Even though the microarray data cannot provide evidence on the conservation of gene order, if we assume only localized regions of synteny between test strains and genome strain NCTC 11168, more than half of all divergent genes in our meta-analysis mapped to 16 well-defined genomic regions likely to represent functionally related groups of genes (Fig. 2; Table 2). Most of the variable genes in the meta-analysis data set converged onto variable loci previously defined by Dorrell et al. (7) and Pearson et al. (22); these include the lipooligosaccharide, capsular polysaccharide, and flagellar biosynthetic loci and the restriction-modification locus (Fig. 2). In several cases, variable loci were expanded by the additional data. For example, the variable locus between Cj0295 and Cj0309c (Fig. 2, region 4) was increased by an additional seven genes, on the basis of the CGH meta-analysis data set. Similarly, the restriction-modification locus (Fig. 2, region 14), which spanned from Cj1549c to Cj1556 in the original data set of Dorrell et al. (7), was increased by an additional 10 genes (from Cj1543 to Cj1563c) when the cumulative data were taken into account.
TABLE 2.
| Region | Start | End |
|---|---|---|
| 1 | Cj0032 | Cj0036 |
| 2 | Cj0055c | Cj0059c |
| 3 | Cj0177 | Cj0182 |
| 4 | Cj0294 | Cj0310c |
| 5 | Cj0421c | Cj0425 |
| 6 | Cj0480c | Cj0490 |
| 7 | Cj0561c | Cj0571 |
| 8 | Cj0625 | Cj0629 |
| 9 | Cj0727 | Cj0755 |
| 10 | Cj0967 | Cj0975 |
| 11 | Cj1135 | Cj1151c |
| 12 | Cj1293 | Cj1343 |
| 13 | Cj1414c | Cj1449c |
| 14 | Cj1543c | Cj1563c |
| 15 | Cj1677 | Cj1679 |
| 16 | Cj1717c | Cj1729c |
Endpoints for each hypervariable region were determined by extending each region to include the longest contiguous block of neighboring variable genes determined from the meta-analysis data set, assuming the NCTC 11168 gene order. Regions 4, 6, 9, 11, 12, 13, and 16 represent plasticity regions 1 to 7 (22), respectively.
Global trends in C. jejuni gene conservation.
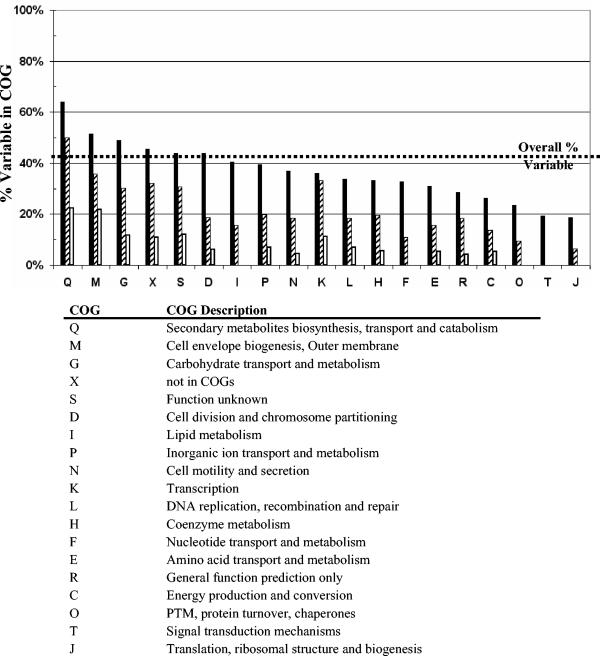
The results from our large-scale meta-analysis of C. jejuni CGH data (data sets I through IV) show that 36.6% of the genes in C. jejuni NCTC 11168 were variable in at least 1 of the 97 strains from the four data sets. Of these, 350 showed significant intraspecies variability in the form of detectable divergence across multiple strains. On the basis of their high degrees of intraspecies conservation, the remaining 1,284 genes (78.6%) are likely to form a core set of genes required by C. jejuni. We carried out an analysis of the variability levels observed within each group of COGs to illustrate variability across different functional groups of genes and also calculated the percentage of HV and HD genes within each COG (Fig. 3). Many of the genes involved in utilization of secondary metabolites, cell envelope biogenesis, and carbohydrate transport and metabolism (COGs Q, M, and G, respectively) showed both high degrees of divergence and high degrees of intraspecies variability. As variation in the complement of genes in the last two COGs can generate heterogeneity in cell surface structures (10, 12), a low level of conservation was expected. Genes involved in DNA replication, recombination, and repair (COG L) had lower than expected conservation levels due to the inclusion of the restriction-modification system in this COG group. Nearly all HV genes (eight of nine) and all HD genes (five of five) in this COG were restriction-modification enzymes. Whereas 65.5% (915 of 1,398) of the C. jejuni genes that have homologs in other species are conserved in all strains, this value decreases to only 49.3% (98 of 199) in the subset of genes that are unique to C. jejuni. This accounts for a lower-than-average conservation level in COGs S and X, which have high proportions of genes unique to C. jejuni.
FIG. 3.
Gene conservation levels across different COG groups. Conservation levels were calculated as the percentage of variable genes in each COG group on the basis of the cumulative data from data sets I, II, III, and IV (black bars). The percentage of variable genes belonging to the HV group (striped bars) and the HD group (white bars) was also calculated for each COG. The HV and HD genes are defined in the text. COG X was created by the authors to denote all genes that do not fall under all other defined COG groups. HD and HV genes are not mutually exclusive, as their sum can exceed the total number of variable genes. PTM, posttranslational modification.
Determination of a robust set of variable genes for C. jejuni genotyping.
To be useful in a molecular typing context, the genes to be used for genotyping should show high degrees of intraspecies variability and should provide unambiguous results. We carried out a detailed analysis of the raw microarray CGH data available (data sets II and III) and coupled the information for genes with high degrees of intraspecies variability with two additional criteria that should enhance the reliability of the CGH observations: highly negative log ratio values (i.e., the HD group) and the occurrence of adjacent genes that are divergent within the same strain (i.e., divergent neighbors).
(i) HD group.
Our preliminary observations on the microarray data for C. jejuni RM1221 compared with those for NCTC 11168 CGH showed that all highly negative log ratios (log ratios < −3.3) corresponded to genes which are found in strain NCTC 11168 and which are absent from strain RM1221 (Fig. 4A). A detailed analysis of CGH data from data sets II and III revealed genes in which a minimum log ratio of less than −3.3 was observed in at least one strain. These genes were assigned to the HD group, and all other variable genes were assigned to the MD group. Of the variable genes in data sets II and III, 122 showed highly negative log ratios for at least one strain, providing unambiguous evidence for either high levels of sequence divergence or gene absence. For each variable gene, we calculated the average log ratio for all strains in which the gene was divergent and found that the average log ratio of the HD genes is approximately 1 log2 unit lower than that of the MD genes, and thus, HD genes have a tendency toward highly negative log ratios (Fig. 4B).
(ii) HV group.
More than two-thirds (268 of 391) of the variable genes found in data sets II and III were HV in multiple strains. Although 54.3% (146 of 268) of the MD genes were also HV, every HD gene was also HV (122 of 122) (Fig. 4C).
(iii) Divergent neighbors.
Although genomic rearrangements cannot be detected by CGH analysis, there is growing evidence that variable genes in C. jejuni are found in clusters (22). Thus, the presence of genes that have decreased hybridization signals within a single strain and that are adjacent in NCTC 11168 provides stronger evidence of gene divergence or gene absence than does the occurrence of genes without divergent neighbors. Fifty-eight percent of all variable genes that are adjacent to each other in the C. jejuni NCTC 11168 genomic sequence were found to be codivergent in at least one of the strains studied, and these divergent neighbors were often functionally related. Although only 41.3% of the MD genes had divergent neighbors, 95.9% of the genes in the HD group had divergent neighbors within the same strain (Fig. 4D).
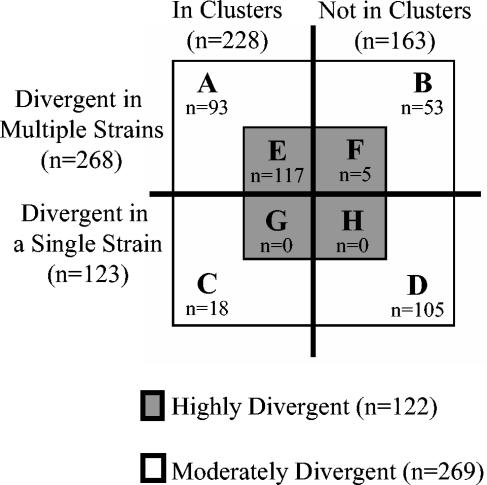
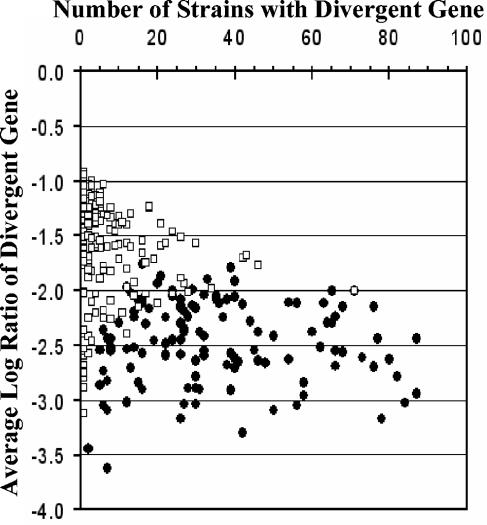
The results summarized in Fig. 5 show that of the 122 HD genes, 117 (95.9%) fulfilled the two additional criteria, i.e., HV and divergent neighbors (Fig. 5E), whereas this value was only 34.6% (93 of 269) among the MD genes (Fig. 5A). In addition, whereas the MD genes tended to vary in small numbers of strains (n = 269, mean = 4.0, standard deviation = 6.3), HD genes tended to be variable in larger numbers of strains (n = 122, mean = 26.5, standard deviation = 15.9) (Fig. 6). As genes with detectable divergence across various data sets are likely to represent a useful set of typing markers, it is significant that ∼57% (70 of 122) of the HD genes were also found to be HV in all four C. jejuni data sets included in the meta-analysis. Thus, it appears that our selection criteria independently converge on the HV and HD genes common to all four data sets.
FIG. 5.
Summary of HD genes obtained from microarray-based CGH of C. jejuni. Of 391 variable genes in data sets II and III, 122 genes were HD in one or more strains (E to H). A very high percentage (95.9%; 117 of 122) of all HD genes have variable neighbors within a single strain and are divergent in multiple strains (HV) (E). The results are based on raw microarray CGH data from data sets II and III.
FIG. 6.
Relationship between gene divergence and intraspecies variability. The distributions of MD and HD genes (white squares and black circles, respectively) show little overlap. MD genes tended to be variable in small numbers of strains (n = 269, mean = 4.0, standard deviation = 6.3), whereas HD genes displayed high degrees of intraspecies variability (n = 122, mean = 26.5, standard deviation = 15.9).
DISCUSSION
Although many of the early observations on C. jejuni gene conservation patterns (7) have been corroborated by additional data (14, 22; this study), the meta-analysis of cumulative CGH data reported here has enabled us to uncover global trends that would be difficult to obtain from the individual data sets. Wassenaar et al. (30) have shown that C. jejuni can undergo significant but rare genomic rearrangements, and it is very unlikely that microarray-based CGH would detect these. However, even if we assume that only small genomic regions are able to maintain synteny, the global gene divergence patterns observed in the expanded CGH data set show that gene divergence in C. jejuni is largely restricted to a small number of genomic regions and provides indirect evidence that large portions of the C. jejuni genome are stable. On the basis of observations for 18 strains, Pearson et al. (22) have shown that the locations of variable genes in C. jejuni are not random, and the results of our meta-analysis confirm these findings. The clustered nature of variable genes is also consistent with an analysis of genome sequences of multiple strains which has shown that closely related genomes are largely colinear and that most of the genomic differences between two strains are in the form of multigene insertions or deletions that are likely the result of homologous recombination events (1, 24, 33). Mira et al. (19) have shown that the synteny displayed by closely related genomes is higher for genomes with few or no repeated genetic elements. The genome stability predicted from the CGH data is consistent with the lack of mobile genetic elements or repeated sequences in the genome sequence of C. jejuni NCTC 11168 (21) and appears to be a feature of C. jejuni evolution. Direct evidence from comparative genomics with whole-genome sequence data will be required to confirm these indirect observations, and the forthcoming C. jejuni RM1221 genome sequence is likely to shed light on this issue.
One major challenge of microarray-based CGH remains data interpretation. Two biological processes, gene divergence and gene loss, are inferred solely on the basis of differential hybridization signals that are largely the result of a spectrum of degrees of gene divergence that can culminate in gene loss. Kim et al. (13) have argued that the use of arbitrary thresholds tends to underestimate the true number of outliers (divergent genes) in microarray-based CGH analysis and have devised a method that increases the sensitivity of outlier detection by dynamically computing a threshold based on the distribution of log ratio values for each hybridization experiment. For the purposes of exploratory comparative genomics, the increased sensitivity of this method is clearly superior to that from the use of a static threshold, but one drawback of this approach is that narrow log ratio distributions can produce thresholds with decreased stringencies that may overestimate the numbers of outliers. In addition, the use of the log ratio distribution ignores the effects of signal intensity and dynamic range on outlier detection, especially as low-intensity data are inherently less reliable because of low signal-to-noise ratios. For strain classification and genotyping, it is crucial that only unambiguous gene divergence data be used for analysis, and new methods which incorporate dynamic threshold determination but which apply intensity-dependent corrections to the threshold will need to be developed. In their absence, we have chosen to use a conservative linear threshold, a log ratio of −0.97, to assign gene divergence. Our decision to select this value was based on a high-resolution exploration of the range of thresholds from −0.75 to −3.0 (results not shown). Whereas increasing the stringency of the threshold to values below −0.97 led to a modest but steady drop in the number of genes assigned as divergent, even small decreases in the stringency of the threshold to values above −0.97 led to large increases in the number of potential false-positive results. It would therefore appear that thresholds at or near this value represent a good compromise between maximizing the sensitivity of divergent gene detection and minimizing the number of false-positive results.
Previous microarray-based CGH studies with C. jejuni and other species have grouped divergent and deleted genes into a single category because present analytical tools are unable to make the distinction between the two (7, 13). On the basis of empirical CGH work with strain RM1221 (Fig. 4A), we have determined that moderately negative log ratios are more likely to represent gene divergence events, whereas genes with highly negative log ratios are more likely to represent gene absence events. A detailed examination of the microarray CGH data allowed us to make the distinction between genes that exhibit high levels of divergence and genes that exhibit moderate levels of divergence on the basis of differences in the amplitudes of their respective log ratio values. Of the 599 genes observed to be divergent in the four data sets, 122 qualified as HD on the basis of the highly negative log ratios observed in at least one of the strains in data sets II and III.
In order to identify a set of divergent genes that would be most useful for genotyping, we coupled the data for genes with high intraspecies variability, as detected by CGH, with additional lines of evidence of potential biological importance: high degrees of divergence in one or more strains and the occurrence of adjacent divergent genes within a single strain. Although the distinction between highly and moderately divergent genes based on highly negative log ratios requires further refinement, it is worth noting that 96% (117 of 122) (Fig. 5E) of the HD genes that we identified have divergent neighbors and are divergent in multiple strains. More significantly, as these genes tend to provide unambiguous microarray results and tend to have high intraspecies variabilities, they represent an excellent set of polymorphic markers that could form the basis for a highly discriminatory genotyping method. Of the 84 genes that were HV in each of the four data sets analyzed, 70 were also HD (see http://ibs-isb.nrc-cnrc.gc.ca/ibs/immunochemistry/suppInfo_Taboada_2004a_e.html). Well-known polymorphic regions (e.g., lipooligosaccharide biosynthesis, flagellar biosynthesis, and capsular polysaccharide biosynthesis) are a significant source of HV and HD genes. These loci (Fig. 2, regions 11, 12, and 13, respectively) contain 7, 15, and 14 HV and HD genes, respectively. Another region contributing a large number of HV and HD genes (n = 10) is the region from Cj0480c to Cj490 (Fig. 2, region 6), which Pearson et al. (22) have termed plasticity region 2. This region contains truncated genes for altronate hydrolase and aldehyde dehydrogenase, a putative sugar transporter and a putative oxidoreductase, respectively, among others. Hypervariable region 3 (Fig. 2, region 3), which contains several iron uptake transporters and a putative membrane siderophore, contains three contiguous HV and HD genes (Cj0178-Cj0179-Cj0180). While many HV and HD genes have known function, a large number of HV and HD genes (16 of 70; 23%) represent putative or hypothetical genes of unknown function. While the preponderance of well-established polymorphic genes among the list of 70 HV and HD genes validates the results of our CGH meta-analysis, our results also show that a significant number of HV and HD genes represent novel polymorphic typing targets.
The main advantage of the meta-analysis approach described here is that increased sample sizes tend to comprise a greater degree of genetic diversity, reducing the effect of sampling biases. As the manuscript was being finalized, data set IV (22) became publicly available, which enabled us to perform a comparison of the global trends obtained from large-scale sampling using our original meta-analysis data set (data sets I, II, and III) and those obtained from the 18 strains evaluated in that study. There was significant overlap in the variable genes obtained from both data sets, as 78% of the variable genes (208 of 266) in data set IV had variable counterparts in the meta-analysis data set. However, despite similar gene conservation profiles, data set IV contained 58 variable genes that had shown no variability among the 79 strains in the original meta-analysis data set. Thus, despite the large sample size used in this study, we recognize that a more comprehensive study of C. jejuni comparative genomics will require targeted sampling of strains expected to be genetically diverse on the basis of a number of different epidemiological or phenotypic parameters.
Using a meta-analysis approach, we have been able to identify genes that have high degrees of intraspecies variability in C. jejuni and that can be targeted for genotyping purposes. Since most present molecular typing methodologies are based on DNA polymorphisms with poorly defined biological significance, a new generation of genotyping methods that incorporate data from microarray-based CGH will have the advantage of being founded on tracking of the conservation of genes of interest at the whole-genome level. This fundamental difference represents a major leap toward the rational design of molecular typing methods that couple biologically relevant information, namely, the gene conservation profiles that ultimately govern a strain's phenotype, to epidemiological surveillance.
Acknowledgments
We thank Laura Brown for helpful discussions and critical reading of the manuscript and Martin Young and Jim Richards for support.
Funding for this work has been provided to E.N.T., R.R.A., C.D.C., W.A.F., O.L.M., M.J.R., and J.H.E.N. through the National Research Council Genomics and Health Initiative and to D.T.M. and J.M.F. through Health Canada and the University of Ottawa. Preliminary sequence data were obtained from the Institute for Genomic Research at http://www.tigr.org.
REFERENCES
- 1.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 2.Anjum, M. F., S. Lucchini, A. Thompson, J. C. D. Hinton, and M. J. Woodward. 2003. Comparative genomic indexing reveals the phylogenomics of Escherichia coli pathogens. Infect. Immun. 71:4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Broekhuijsen, M., L. Par, A. Johansson, M. Byström, U. Eriksson, E. Larsson, R. G. Prior, A. Sjöstedt, R. W. Titball, and M. Forsman. 2003. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 41:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dingle, K. E., F. M. Colles, D. R. A. Wareing, R. Ure, A. J. Fox, F. E. Bolton, H. J. Bootsma, R. J. L. Willems, R. Urwin, and M. C. J. Maiden. 2001. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 39:14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobrindt, U., F. Agerer, K. Michaelis, A. Janka, C. Buchrieser, M. Samuelson, C. Svanborg, G. Gottschalk, H. Karch, and J. Hacker. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorrell, N., J. A. Mangan, K. G. Laing, J. Hinds, D. Linton, H. Al-Ghusein, B. G. Barrell, J. Parkhill, N. G. Stoker, A. V. Karlyshev, P. D. Butcher, and B. W. Wren. 2001. Whole genome comparison of Campylobacter jejuni human isolates using a low-cost microarray reveals extensive genetic diversity. Genome Res. 11:1706-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald, C., L. O. Helsel, M. A. Nicholson, S. J. Olsen, D. L. Swerdlow, R. Flahart, J. Sexton, and P. I. Fields. 2001. Evaluation of methods for subtyping Campylobacter jejuni during an outbreak involving a food handler. J. Clin. Microbiol. 39:2386-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert, M., M. F. Karwaski, S. Bernatchez, N. M. Young, E. Taboada, J. Michniewicz, A. M. Cunningham, and W. W. Wakarchuk. 2002. The genetic bases for the variation in the lipo-oligosaccharide of the mucosal pathogen, Campylobacter jejuni. Biosynthesis of sialylated ganglioside mimics in the core oligosaccharide. J. Biol. Chem. 277:327-337. [DOI] [PubMed] [Google Scholar]
- 11.Hakenbeck, R., N. Balmelle, B. Weber, C. Gardès, W. Keck, and A. de Saizieu. 2001. Mosaic genes and mosaic chromosomes: intra- and interspecies genomic variation of Streptococcus pneumoniae. Infect. Immun. 69:2477-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kharlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529-541. [DOI] [PubMed] [Google Scholar]
- 13.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:0065.1-0065.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard, E. E., II, T. Takata, M. J. Blaser, S. Falkow, L. S. Tompkins, and E. C. Gaynor. 2003. Use of an open-reading frame-specific Campylobacter jejuni DNA microarray as a new genotyping tool for studying epidemiologically related isolates. J. Infect. Dis. 187:691-694. [DOI] [PubMed] [Google Scholar]
- 15.Lior, H., D. I. Woodward, J. A. Edgar, I. J. Laroche, and P. Gill. 1982. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J. Clin. Microbiol. 15:761-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenz, E., A. Lastovica, and R. J. Owen. 1998. Subtyping of Campylobacter jejuni Penner serotypes 9, 38 and 63 from human infections, animals and water by pulsed field gel electrophoresis and flagellin gene analysis. Lett. Appl. Microbiol. 26:179-182. [DOI] [PubMed] [Google Scholar]
- 17.Medeiros, D. T. 2001. Epidemiological typing of Campylobacter clinical and food isolates using pulsed-field gel electrophoresis. M.Sc. thesis. University of Ottawa, Ottawa, Ontario, Canada.
- 18.Miller, W. G., A. H. Bates, S. T. Horn, M. T. Brandl, M. R. Wachtel, and R. E. Mandrell. 2000. Detection on surfaces and in Caco-2 cells of Campylobacter jejuni cells transformed with new gfp, yfp, and cfp marker plasmids. Appl. Environ. Microbiol. 66:5426-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mira, A., L. Klasson, and S. G. E. Andersson. 2002. Microbial genome evolution: sources of variability. Curr. Opin. Microbiol. 5:506-512. [DOI] [PubMed] [Google Scholar]
- 20.Murray, A. E., D. Lies, G. Li, K. Nealson, J. Zhou, and M. J. Tiedje. 2001. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc. Natl. Acad. Sci. USA 98:9853-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 22.Pearson, B. M., C. Pin, J. Wright, K. I'Anson, T. Humphreys, and J. M. Wells. 2003. Comparative genome analysis of Campylobacter jejuni using whole genome DNA microarrays. FEBS Lett. 554:224-230. [DOI] [PubMed] [Google Scholar]
- 23.Penner, J. L., and J. N. Hennessy. 1980. Passive hemagglutination technique for serotyping Campylobacter fetus subsp. jejuni on the basis of soluble stable heat-stable antigens. J. Clin. Microbiol. 12:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Pósfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 25.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 26.Sails, A. D., B. Swaminathan, and P. I. Fields. 2003. Utility of multilocus sequence typing as an epidemiological tool for investigation of outbreaks of gastroenteritis caused by Campylobacter jejuni. J. Clin. Microbiol. 41:4733-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schouls, L. M., S. Reulen, B. Duim, J. A. Wagenaar, R. J. L. Willems, K. E. Dingle, F. M. Colles, and J. D. A. van Embden. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth, G. K., Y. H. Yang, and T. Speed. 2003. Statistical issues in cDNA microarray data analysis. Methods Mol. Biol. 224:111-136. [DOI] [PubMed] [Google Scholar]
- 30.Wassenaar, T. M., B. Geihausen, and D. G. Newell. 1998. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl. Environ. Microbiol. 64:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wassenaar, T. M., and M. J. Blaser. 1999. Pathophysiology of Campylobacter jejuni infections in humans. Microbes Infect. 1:1023-1033. [DOI] [PubMed] [Google Scholar]
- 32.Wassenaar, T. M., and D. G. Newell. 2000. Genotyping of Campylobacter spp. Appl. Environ. Microbiol. 66:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S.-R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. T. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan, W., N. Chang, and D. E. Taylor. 1991. Pulsed-field gel electrophoresis of Campylobacter jejuni and Campylobacter coli genomic DNA and its epidemiologic application. J. Infect. Dis. 163:1068-1072. [DOI] [PubMed] [Google Scholar]