Abstract
In flowering plants, many respiration-related proteins are encoded by the mitochondrial genome and the splicing of mitochondrion-encoded messenger RNA (mRNA) involves a complex collaboration with nuclear-encoded proteins. Pentatricopeptide repeat (PPR) proteins have been implicated in these RNA–protein interactions. Maize defective kernel 2 (dek2) is a classic mutant with small kernels and delayed development. Through positional cloning and allelic confirmation, we found Dek2 encodes a novel P-type PPR protein that targets mitochondria. Mitochondrial transcript analysis indicated that dek2 mutation causes reduced splicing efficiency of mitochondrial nad1 intron 1. Mitochondrial complex analysis in dek2 immature kernels showed severe deficiency of complex I assembly. Dramatically up-regulated expression of alternative oxidases (AOXs), transcriptome data, and TEM analysis results revealed that proper splicing of nad1 is critical for mitochondrial functions and inner cristaes morphology. This study indicated that Dek2 is a new PPR protein that affects the splicing of mitochondrial nad1 intron 1 and is required for mitochondrial function and kernel development.
Keywords: Zea mays, dek2, pentatricopeptide repeat protein, mitochondrion, RNA splicing
MITOCHONDRIAL respiration is the core for cell free energy releasing and ATP production (Siedow and Day 2000). The electron transport chain (ETC) is composed of five respiratory complexes in mitochondrion (Dudkina et al. 2006). The mitochondrial genome possesses genes encoding proteins involved in ETC. The mitochondrial genome-expressed pre-RNAs can be post-transcriptionally processed, including RNA editing, RNA cis- and trans-splicing, RNA cleavage, and RNA maturation processes (Knoop 2013; Hammani and Giege 2014). Nuclear genome expressed pentatricopeptide repeat (PPR) proteins are reported to play a critical role in these processes (Barkan and Small 2014). The PPR proteins are defined by the tandem repeats of a degenerate 35-amino-acid motif and are classified into two major subgroups: the P-type PPR members only possess tandem repeats of the 35-amino-acid PPR motif, and the PLS-type PPR members composed of sequential repeats of P, short (S), and long (L) PPR motifs and often carry an E or E–DYW domain extension in the C terminus (Lurin et al. 2004; Cheng et al. 2016). The PPR family comprises >450 members in plants, and a number of severe growth and development defects associated with loss-of-function PPR mutants were described before (Fujii and Small 2011; Sosso et al. 2012; Liu et al. 2013; Colas des Francs-Small and Small 2014; Hammani and Giege 2014; Li et al. 2014; Sun et al. 2015; Chen et al. 2016; Haili et al. 2016; Xiu et al. 2016). However, the regulatory functions are still unknown for great numbers of unidentified PPR proteins.
P-type PPR proteins are dedicated to RNA stabilization, cleavage, translational activation, and splicing (Barkan and Small 2014; Xiu et al. 2016). Groups I and II introns are two kinds of introns with distinct splicing patterns. Group II introns are found in ribosomal (rRNA), transfer RNA (tRNA), bacterial messenger RNA (mRNA) and mRNA of organelles in fungi, plants, and protests. They are self-splicing ribozymes (Eickbush 1999). In plants, >20 group II introns exist in the mitochondrial genome, and most of them belong to genes encoding subunits in complex I, including 15 cis-spliced introns in nad1, nad2, nad4, nad5, nad7, rps3, cox2, ccmFC and 7 trans-spliced introns in nad1, nad2, and nad5 (Burger et al. 2003; Berrisford and Sazanov 2009). Five P-type PPR proteins are reported to be necessary for group II intron splicing of mitochondrial transcripts in Arabidopsis, functioning in nad1 (de Longevialle et al. 2007), nad2 (Liu et al. 2010), nad5 (Colas des Francs-Small et al. 2014), and nad7 (Hsieh et al. 2015). In maize, EMP16 was reported to be involved in nad2 intron 4 cis-splicing (Xiu et al. 2016), and DEK35 is required for nad4 intron 1 cis-splicing (Chen et al. 2016). The important roles of more P-type PPR proteins in mitochondrial RNA cis- and trans-splicing processes can be revealed through studies on maize kernel mutants.
Maize (Zea mays) is suitable material for genetics research, partly because of its numerous easily observable phenotypes (Neuffer and Sheridan 1980). Defective kernel (dek) mutants are a major class of maize kernel mutants that are a good resource for investigating kernel development (Neuffer and Sheridan 1980). To date, many maize dek mutants have been identified: dek1 causes severe growth and development defects (Lid et al. 2002); reas1(dek*) causes only mild developmental delay (Qi et al. 2016); and the mutants of PPRs, including ppr2263, smk1, emp5, emp7, emp16, and dek35, always have an obvious small kernel phenotype, with arrested development of embryo and endosperm (Liu et al. 2013; Li et al. 2014; Sun et al. 2015; Chen et al. 2016). Dek mutants offer opportunities to investigate many basic biological processes during kernel development.
In this study, we characterized maize classic mutant dek2, a defective kernel mutant with small kernels and delayed development. We report the map-based cloning of Dek2 and demonstrate it encodes a P-type PPR in maize. We present evidence that Dek2 is specifically involved in the splicing of nad1 intron 1. Lack of these splicing processes affects complex I accumulation. Consequently, it arrests mitochondrial oxidative phosphorylation and kernel development.
Materials and Methods
Plant materials
The maize dek2-N1315A stock was obtained from the Maize Genetics Cooperation Stock Center (http://www.maizegdb.org/data_center/stock?id=13997) as an EMS-induced mutant first described by Neuffer and Sheridan (1980). The mutant was crossed into a W22 genetic background to produce the F2 populations. Kernels of the F2 ears exhibited a 3:1 segregation of wild-type (WT) kernels (dek2/+ or +/+) and homozygous mutant kernels (dek2/dek2) were used for analysis. All the plants were cultivated in the field on the Shanghai University campus.
Measurement of protein and starch
For the protein measurements, the endosperm of dek2 and WT mature kernels was separated from the embryo and pericarp by dissection after soaking the kernels in water. The samples were dried to constant weights, pulverized with a mortar and pestle in liquid N2, and then measured according to a previously described protocol (Wang et al. 2011). All the measurements were replicated at least three times.
For the starch measurements, five mature kernels of the WT and dek2 were ground in liquid N2. The resulting powders were dried to a constant weight. Finally, the total starch was measured by using an amyloglucosidase/α-amylase starch assay kit (Megazyme). The protocol referenced the method by Wang et al. (2014). All the measurements were replicated at least three times.
Light microscopy
Immature WT and dek2 kernels at 12 days after pollination (DAP) and 18 DAP were collected from F2 ears and were cut along the longitudinal axis for paraffin and resin section preparation. The sections were fixed in formalin–acetic acid–alcohol mixture and were dehydrated in an ethanol gradient series of 50, 60, 70, 85, 95, and 100% ethanol. After replacement of acetone and infiltration with paraffin or resin, the sections were embedded and cut using Leica RM2265. The paraffin section was stained by fuchsin basic, and the resin section was stained by fuchsin basic (horizontal cut) and toluidine blue (longitudinal cut). The sections were observed using Leica DFC500.
Scanning electron microscopy and transmission electron microscopy
For scanning electron microscopy, dek2 and WT kernels were prepared according to Lending and Larkins (1992): mature maize kernels were rifted with a razor at the peripheral region and placed in 2.5% glutaraldehyde. Samples were critically dried and spray coated with gold. Gold-coated samples were then observed with a scanning electron microscope (S3400N; Hitachi).
For transmission electron microscopy (TEM), immature kernels of dek2 and WT were prepared according to Lending and Larkins (1992), with some modifications: 18 DAP kernels of dek2 and WT were fixed in paraformaldehyde and postfixed in osmium tetraoxide. After dehydrated in an ethanol gradient, samples were then transferred to a propylene oxide solution and gradually embedded in acrylic resin (London Resin Company). Sections (70 nm) of samples were made with a diamond knife microtome (Reichert Ultracut E). Sample sections were stained with uranyl acetate and poststained with lead citrate. Sample sections were observed with a Hitachi H7600 transmission electron microscope.
Map-based cloning
A population of 3358 homozygous mutant kernels from F2 ears was used for gene mapping. The chromosome arm location of dek2 was reported in Neuffer and Sheridan (1980) and this information enabled us to focus on using molecular markers distributed throughout maize (Zea mays) chromosome arm 1L for preliminary mapping. Molecular markers for fine mapping (Supplemental Material, Table S2) were developed to localize the dek2 locus to a 146-kb region. The corresponding DNA fragments were amplified from dek2 allele and WT plants using KOD Plus DNA polymerase (Toyobo) and sequenced using a MegaBACE 4500 DNA analysis system (Amersham Biosciences).
Subcellular localization of Dek2
The full-length Dek2 ORF without the stop codon was cloned into pB7YWG. The recombinant plasmid was extracted with ∼1 μg total amount and introduced into tobacco leaf epidermal cells through transient transformation using Bio-Rad PDS-1000/HeTM biolistic particle delivery system. The fluorescence signals were detected using LSM710 (Occult International).
RNA extraction and RT-PCR analysis
Total RNA was extracted with TRIzol reagent (Tiangen) and DNA was removed by a treatment with RNase free DNase I (Takara). Using ReverTra Ace reverse transcriptase (Toyobo) RNA was reverse transcribed to complementary DNA (cDNA). Quantitative real-time PCR (qRT-PCR) was performed with SYBR Green Real-Time PCR Master Mix (Toyobo) using a Mastercycler ep realplex 2 (Eppendorf) according to the standard protocol. Specific primers were designed (Table S2) and the experiments were performed to two independent RNA samples sets. A final volume of 20 ml contained 1 ml reverse transcribed cDNA (1–100 ng), 10 ml 23 SYBR Green PCR buffer, and 1.8 ml 10 mM/liter forward and reverse primers for each sample. Relative quantifiable differences in gene expression were analyzed as described previously (Livak and Schmittgen 2001).
Isolation of mitochondria
For blue native (BN)-PAGE, about 15 g of immature seeds at 18 DAP were harvested and were ground with a mortar and pestle in liquid nitrogen, adding 20 ml of extraction buffer (100 mM tricine, 300 mM sucrose, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA-K, 0.1% BSA, 5 mM DTT, pH 7.4) and 60 μl of plant protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). The samples were twice centrifuged at 2600 × g for 15 min, after filtration through a Miracloth membrane (Calbiochem, San Diego, CA), the supernatant were then centrifuged at 12,000 × g for 25 min to pellet crude mitochondria. The pellet was resuspended in wash buffer (100 mM tricine, 300 mM sucrose, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA-K, 0.1% BSA, pH 7.4) and loaded on sucrose density gradients of 1.5, 2.5, 2.5, 2, and 2 ml containing, respectively, 1.8, 1.45, 1.2, 0.9, and 0.6 M of sucrose diluted in wash buffer. After 90 min of centrifugation at 24,000 rpm at 4°, mitochondria were collected from the 1.2/1.45 M interface and diluted four times in wash buffer. The enriched mitochondria were collected after 20 min of centrifugation at 12,000 rpm at 4°.
BN-PAGE
The enriched mitochondria were resuspended in 50 μl of B25G20 solution (25 mM Bis-Tris, 20% glycerin, pH 7.0), adding 20% n-dodecyl-β-D-maltoside (DDM) to the final concentration of 1% DDM, and gently mixed on ice for 1 hr. After 15 min of centrifugation at 12,000 rpm at 4°, the supernatant was collected and added to the loading buffer before BN-PAGE. The concentration of separation gel was from 4 to 13%. At first electrophoresis was running at 50 V, adding 25 V every 20 min to the final 150 V until the loading dye migrated to the edge of the gel. The gel was stained by Coomassie brilliant blue (Zhang et al. 2015).
RNA-sequencing analysis
Total RNA (10 μg) was extracted from endosperm of dek2 and WT kernels were harvested at 18 DAP, and three dek2 or WT biological samples were pooled together. The poly-A selected RNA-sequencing (RNA-seq) library was prepared according to Illumina standard instruction (TruSeq Stranded RNA LT Guide). Library DNA was checked for concentration and size distribution in an Agilent2100 bioanalyzer before sequencing with an Illumina HiSequation 2500 system according to the manufacturer’s instructions (HiSequation 2500 User Guide). Paired-end reads were aligned to the maize B73 genome build Zea mays AGPv2.15 using TopHat 2.0.6 (Langmead et al. 2009). Data were normalized as fragments per kilobase of exon per million fragments mapped (FPKM), because the sensitivity of RNA-seq depends on the transcript length. Significant differentially expressed genes (DEGs) were identified as those with a fold change and P-value of differential expression above the threshold (fold change >2.0, P < 0.05).
Data availability
RNA-seq data are available from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the series entry GSE87067.
Results
Kernel development is arrested in dek2
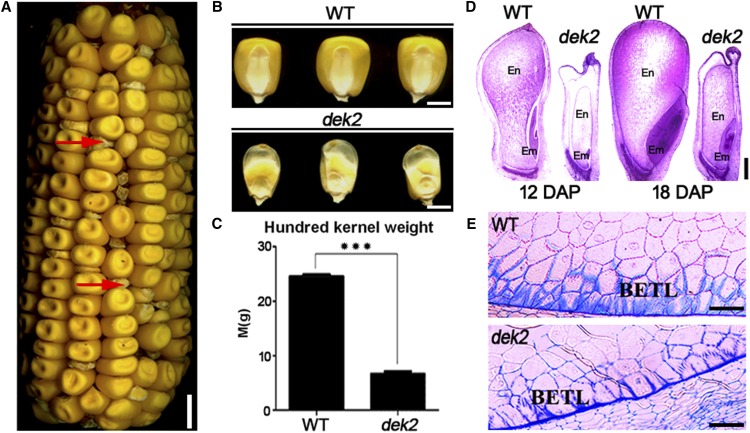
The dek2-ref (dek2-N1315A) mutant was obtained from the Maize Genetics Cooperation Stock Center. It was crossed to the W22 inbred line to produce an F2 population that displayed a 1:3 segregation of dek (dek2/dek2) and WT (+/+ and dek2/+) phenotypes (Figure 1A). Mature homozygous dek2 kernels were small and flat, along with an empty pericarp at the top (Figure 1B). The 100-kernel weight of dek2 was only ∼32% of WT (Figure 1C).
Figure 1.
Phenotypic features of maize dek2 mutant. (A) Mature F2 ear of dek2 × W22 population, red arrow identifies the dek2 kernel. Bar, 10 mm. (B) Randomly selected mature dek2 and WT kernels from segregated F2 population. Bar, 5 mm. (C) Comparison of 100-grain weight of randomly selected mature dek2 and WT kernels in segregated F2 population. Values are the mean values with SE, n = 3 individuals (*** P < 0.001, Student’s t-test). (D) Paraffin sections of 12 DAP and18 DAP dek2 and WT kernels. Bars, 500 μm. (E) Microstructure of developing endosperm BETL of dek2 and WT kernels (18 DAP). Bars, 100 μm.
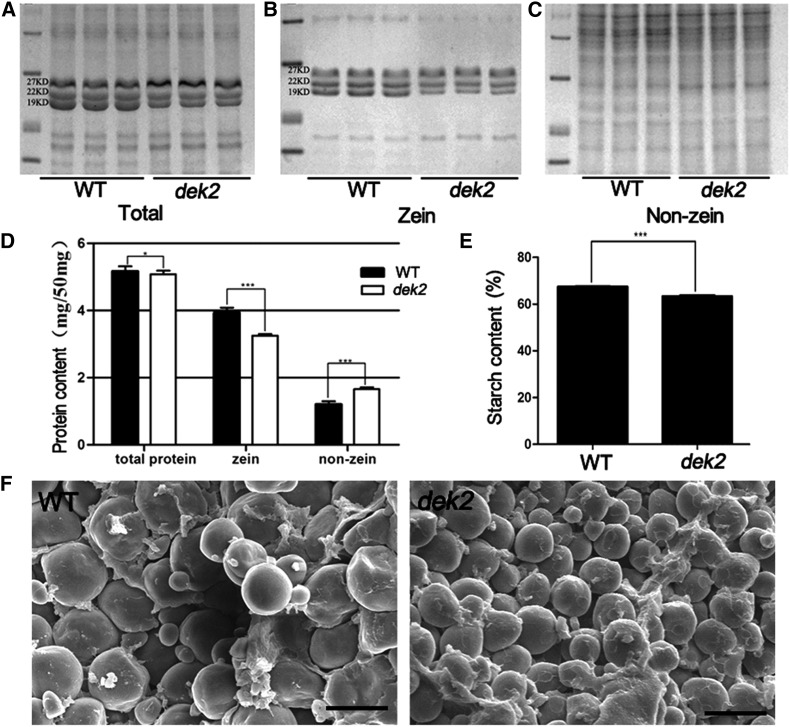
There was a slight decrease in the amount of total protein (4%), a significant decrease in the amount of zein (21%), and a significant increase in the amount of nonzein (38%) in mature dek2 endosperm (Figure 2, A–D). Among zein proteins, the 22 kDa α-zein and 19 kDa α-zein proteins were relatively less abundant in dek2 (Figure 2, A and B). We also found obvious difference in total starch content, and starch granule size was smaller in dek2 endosperm compared to WT (Figure 2, E and F).
Figure 2.
Biochemical analysis and scanning electron microscopy analysis of WT and dek2 endosperm. (A) SDS-PAGE analysis of total proteins from WT and dek2 mature endosperm. (B) SDS-PAGE analysis of zein proteins from WT and dek2 mature endosperm. (C) SDS-PAGE analysis of nonzein proteins from WT and dek2 mature endosperm. (D) Comparison of total, zein, and nonzein proteins from dek2 and WT mature endosperm. The measurements were done per milligram of dried endosperm. Values are the mean values with SE, n = 3 individuals (* P < 0.05, ** P < 0.01, *** P < 0.001, Student’s t-test). (E) Comparison of total starch content in WT and dek2 mature endosperm. The measurements were done per milligram of dried endosperm. Values are the mean values with SE, n = 3 individuals (*** P < 0.001, Student’s t-test). (F) Scanning electron microscopy analysis of WT and dek2 endosperm. Bars, 20 μm.
WT and dek2 kernels of 12 DAP and 18 DAP were analyzed by light microscopy to compare their development. Longitudinal sections of the whole kernel indicated a >6-day developmental delay in dek2 compared to WT (Figure 1D). Furthermore, paraffin sections of 18 DAP WT and dek2 endosperm showed the development of basal endosperm transfer layer (BETL) was dramatically arrested in dek2 kernels (Figure 1E). These results demonstrated that the growth and development of dek2 kernels are affected.
Positional cloning of Dek2
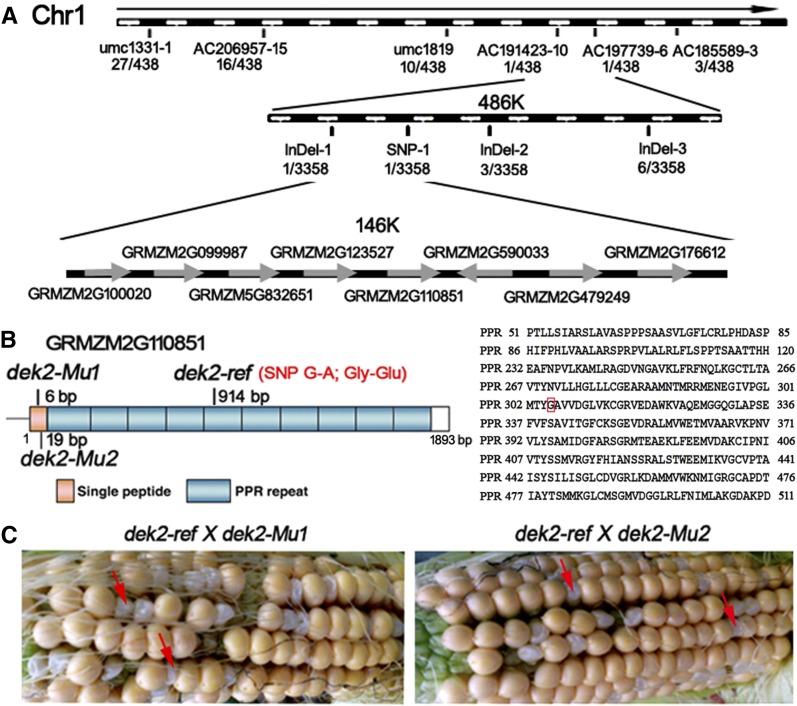
We performed a map-based approach to clone Dek2. After characterizing a population of 438 mutant kernels from the F2 population, the dek2 gene was placed between the molecular markers AC191423-10 and AC197739-6, which encompasses a physical region of 486 kb. We further characterized a population of 3358 mutant kernels and placed the dek2 gene between newly developed molecular markers InDel-1 and SNP-1, which encompasses a physical region of 146 kb. There are eight candidate genes within the interval (gene1: GRMZM2G100020, gene2: GRMZM2G099987, gene3: GRMZM5G832651, gene4: GRMZM2G123527, gene5: GRMZM2G110851, gene6: GRMZM2G590033, gene7: GRMZM2G479249 and gene8: GRMZM2G176612) (Figure 3A). Sequence comparison of the eight candidate genes between WT and mutant alleles revealed a single nucleotide polymorphism (SNP) in gene5 (GRMZM2G110851), resulting in an amino acid replacement between the alleles of dek2-ref and WT (Figure 3B). There is no sequence difference in other candidate genes. Therefore, GRMZM2G110851 appeared to be the candidate for the Dek2.
Figure 3.
Map-based cloning and identification of Dek2. (A) The Dek2 locus was mapped to a 146-kb region between molecular markers InDel-1 and SNP-1 on chromosome 1, which contained eight candidate genes. See Table S2 online for primer information. (B) Structure and mutation sites of the Dek2 gene. (C) Heterozygous dek2-ref and dek2-Mu1 and heterozygous dek2-ref and dek2-Mu2 were used in allelism test. Red arrow identifies the mutant kernel.
Dek2 encodes a P-type PPR protein
The genomic DNA sequence of GRMZM2G110851 produces a transcript containing a 1893-bp coding sequence and encodes a protein of 630 amino acids (Figure 3B). BLASTP searches of GenBank indicated that GRMZM2G110851 encodes a P-type PPR protein with 10 PPR repeats (Figure 3B). SNP mutation (G–A) at 914 bp results in an amino acid replacement (Gly–Glu) in the fifth PPR domain in dek2-ref.
Two UniformMu insertion lines (UFMu08366, UFMu00444) were identified to carry a Mu insertion at 6 bp (dek2-Mu1) and 19 bp (dek2-Mu2) downstream of the start codon in GRMZM2G110851, respectively (Figure 3B). To confirm that GRMZM2G110851 is the Dek2 gene, UFMu08366 (dek2-Mu1) and UFMu00444 (dek2-Mu2) were used for a subsequent allelic test. The allelic test crosses between heterozygous +/dek2-ref and +/dek2-Mu generated ears exhibiting 3:1 segregation that confirmed that dek2-Mu1 and dek2-Mu2 are allelic to dek2-ref (Figure 3C). The results demonstrated that GRMZM2G110851 is the causative gene for dek2.
Dek2 is a conserved mitochondria localized protein
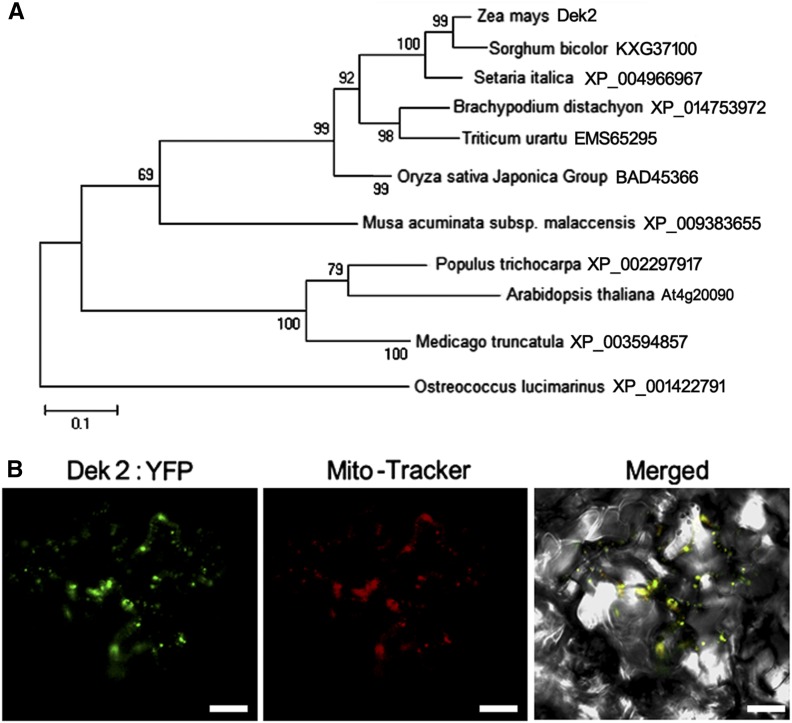
The PPR family is prevalently expanded in plants (Fujii and Small 2011). We constructed a phylogenetic tree on the basis of the maize Dek2 full-length protein sequence and homolog protein sequences from other organisms (Figure 4A). The results suggest that Dek2 homologues are highly conserved in angiosperms. And there is no paralog of Dek2 in maize.
Figure 4.
Phylogenetic analysis and subcellular localization of Dek2. (A) Phylogenetic relationships of Dek2 and its homologs. Maize Dek2 and identified homologous proteins in Sorghum bicolor, Setaria italica, Brachypodium distachyon, Triticum urartu, Oryza sativa, Musa acuminate, Populus trichocarpa, Arabidopsis, Medicago truncatula, and Ostreococcus were aligned by MUSCLE method in MEGA 5.2 software package. The phylogenetic tree was constructed using MEGA 5.2. The numbers at the nodes represent the percentage of 1000 bootstraps. (B) Subcellular localization of Dek2. The Dek2 fusion protein with YFP at the C terminus (green) and Mito-Tracker pBIN20-MT-RK (red) were transiently expressed in tobacco leaf epidermal cells. Bar, 5 µm.
PPR proteins are predominantly targeted to plastids or mitochondria (Colcombet et al. 2013). To determine the subcellular localization, full-length Dek2 was fused to YFP in a binary vector pB7YWG. The fusion was transiently expressed in tobacco leaf epidermal cells by bombarding, and the fluorescent signal was detected by confocal laser microscopy. The YFP signals were detected in small dots that were identified as mitochondria by red fluorescence of Mito-Tracker pBIN20-MT-RK (Nelson et al. 2007; Figure 4B). This result indicated that Dek2 is mitochondrion-targeted protein.
dek2 affects splicing of mitochondrial nad1 intron 1
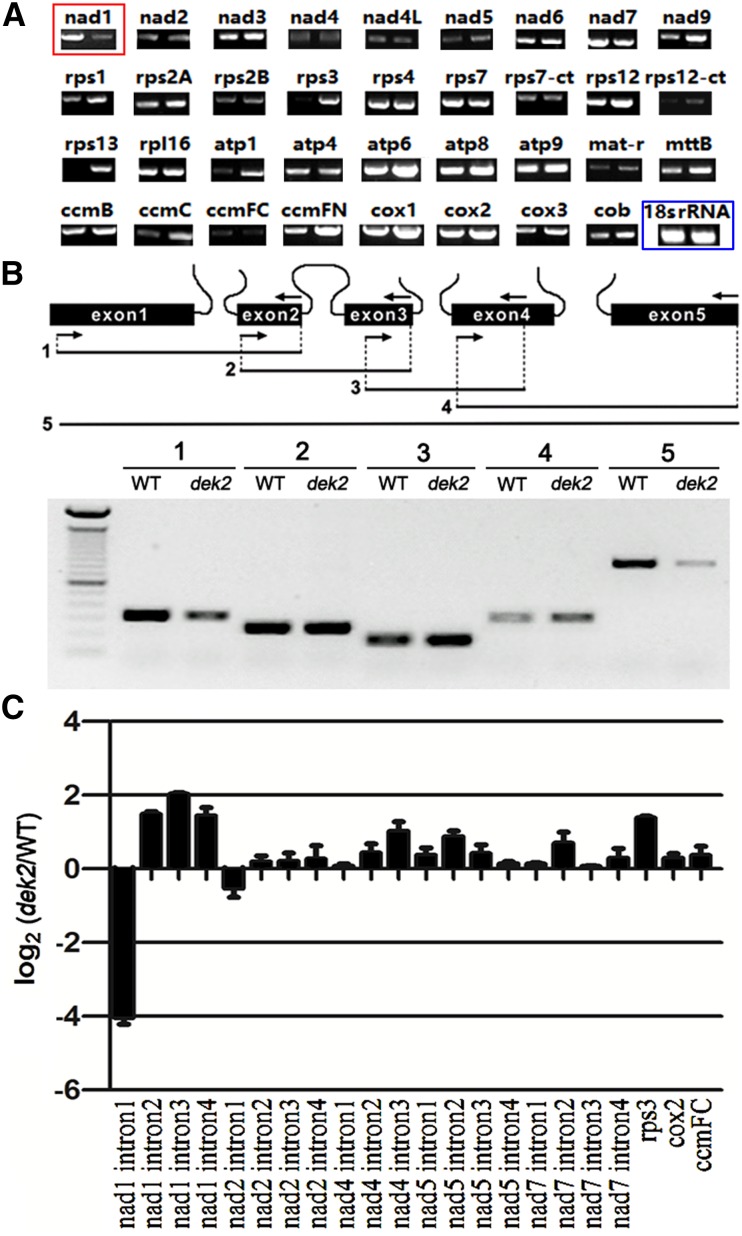
Previous genetic studies have implicated that a PPR protein may interact with a corresponding mitochondrial or chloroplast RNA (Chen et al. 2016; Xiu et al. 2016). Because Dek2 is a mitochondrion-targeted P-type PPR protein, we examined the mitochondrial transcripts of 18 DAP dek2 and WT endosperm. We used specific primers to amplify transcripts with cDNA template for all mitochondrial protein-encoding genes (Table S2). Among 35 genes, only the nad1 mature transcript was dramatically decreased in dek2 (Figure 5A).
Figure 5.
The decrease of nad1 mature transcript and the splicing deficiency of nad1 intron 1 in dek2. (A) RT-PCR analysis of 35 mitochondria-encoded transcripts in 18 DAP WT (left) and dek2 (right) endosperm. The RNA was isolated from the same ear segregating for WT and dek2 mutants.The 18s rRNA served as internal control. Red square identifies nad1 with decreased transcript abundance; blue square identifies internal control. (B) RT-PCR analysis of nad1 intron-splicing efficiency in the WT and dek2 mutant. Structure of the maize mitochondrial nad1 gene. Intron 1 is a trans-spliced intron. The expected amplification products using different primer pairs are indicated. The PCR products were confirmed by sequencing. (C) qRT-PCR analysis of all 22 group II introns in maize mitochondrial genes. Primers spanned adjacent exons, and differences in each spliced fragment were measured. Values are the mean values with SE, n = 3 individuals.
Then we designed specific primers across adjacent exons of the four introns in nad1 transcript, respectively (Figure 5B). The result revealed the splicing efficiency of nad1 intron 1 was decreased in dek2 compared to WT, while the splicing efficiency of nad1 intron 2 was not affected and the splicing efficiency of nad1 intron 3 and intron 4 were even increased. The mature full-length transcript was also reduced (Figure 5B).
P-type PPR proteins were shown to be necessary for splicing of group II introns (Chen et al. 2016; Xiu et al. 2016). Maize mitochondria have 22 group II introns, including 4 introns of the nad1 transcript. To investigate the splicing alterations in dek2 mutants, specific primers were designed for qRT-PCR to inspect the 22 mitochondrial group II introns in dek2 (Table S2). The quantitative differences in spliced exons between dek2 and WT were compared with amplifying primers across adjacent exons. The results showed the common and single reduction of the nad1 spliced exon 1–2 fragment in dek2 (Figure 5C). This result indicated the splicing efficiency of nad1 intron 1 was decreased in dek2 and suggested that Dek2 is required for the splicing of mitochondrial nad1 intron 1 as well as the formation of the nad1 mature transcript.
dek2 exhibits deficiency of mitochondrial complex I assembly
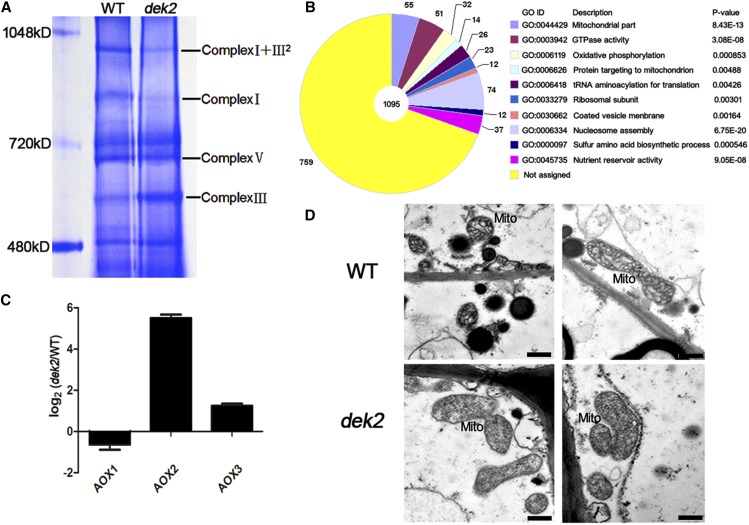
To further investigate the assembly of respiratory complexes, mitochondrial proteins were isolated from 18 DAP dek2 and WT endosperm and were analyzed by BN-PAGE. The bands of different complexes were recognized according to Zsigmond et al. (2008). The two profiles showed decrease of complex I band and complex I + III2 super band in dek2 mutant. And there was a slight increase of complex III band, which might be because of responsible up-regulation to the functional defect of complex I, as previously reported by Xiu et al. (2016) (Figure 6A). This result demonstrated that splicing defect in nad1 intron 1 results in functional reduction of complex I of the respiratory chain.
Figure 6.
Disrupted mitochondrial function in dek2 kernels. (A) BN-PAGE analysis of mitochondrial complexes. The positions of supercomplex I + III2, complex I, complex V, and complex III are indicated. (B) The most significantly related GO terms of the 1095 functional annotated DEGs. The significance and number of genes classified within each GO term are shown. (C) qRT-PCR analysis of genes associated with the alternative respiratory pathway, including Aox1, Aox2, and Aox3. Actin served as internal control. Values are the mean values with SE, n = 3 individuals. (D) Ultrastructure of developing endosperms of WT and dek2 (18 DAP) for mitochondria observation. Bars, 1 μm. Mito, mitochondrion.
dek2 exhibits impaired mitochondrial function
We compared the transcript profile of 18 DAP dek2 and WT endosperm using RNA-seq. Among the 53,520 gene transcripts detected by RNA-seq, significantly DEGs were identified as those with a threshold fold change >2 and P-value <0.05. Based on this criterion, 2065 genes showed significant altered expression between dek2 and the WT. Within the DEGs, 1095 genes could be functionally annotated (annotations were found using BLASTN and BLASTX analyses against the GenBank (http://www.ncbi.nlm.nih.gov/) database). Gene Ontology (GO; http://bioinfo.cau.edu.cn/agriGO/) analysis indicated that DEGs were mostly related to 10 GO terms (Figure 6B and Table S1). Among them, 4 GO terms were closely related to mitochondrial function: GO: 004429 (mitochondrial part, P-value = 8.43E-13); GO: 0003942 (GTPase activity, P-value = 3.08E-08); GO: 0006119 (oxidative phosphorylation, P-value = 0.000853); and GO: 0006626 (protein targeting to mitochondrion, P-value = 0.00488).
Fifty-five DEGs were classified to GO: 004429 (mitochondrial part), including Alternative oxidase 2 (Aox2, GRMZM2G125669). In plant, alternative oxidases (AOXs) can be activated to maintain the tricarboxylic acid cycle and electron transport when there is an electron transport defect in the cytochrome c pathway (Vanlerberghe and Ordog 2002). The expression of Aox2 was 1162-fold up-regulated in dek2 (Table S1). To further validate the expression differences of three Aox genes, we performed qRT-PCR and the result showed that both Aox2 and Aox3 were dramatically up-regulated (Figure 6C), indicating the alternative respiratory pathway was activated for the inefficient mitochondrial oxidative phosphorylation in dek2.
A mitochondrial function defect in the cytoplasm of 18 DAP dek2 endosperm was also observed by TEM analysis. Normal activation of the ETC was required for the proper formation of the inner envelope cristaes in mitochondria (Logan 2006). The mitochondria in the WT endosperm formed distinct inner envelope cristaes surrounded with dense matrix, while the internal structure of mitochondria in dek2 mutant was vague. There was no typical cristaes but only small dots (Figure 6D). The functional reduction of complex I of the respiratory chain causes abnormal morphology of mitochondria in the dek2 mutant.
Discussion
Dek2 is a newly identified P-type PPR protein required for nad1 intron 1 splicing
Here we define the function of a novel P-type PPR protein, Dek2, that is required for splicing of nad1 intron 1 in maize mitochondria. To date, at least 46 PPRs involved in pre-RNA editing in the plant mitochondria have been characterized (Hammani and Giege 2014; Li et al. 2014; Sun et al. 2015). These editing factors mostly belong to the E and DYW subgroups. Fewer splicing-involved PPRs have been studied. The P-type PPR proteins with splicing effect have mainly been studied in Arabidopsis mitochondria, including OTP43 (de Longevialle et al. 2007), ABO5 (Liu et al. 2010), OTP439 and TANG2 (Colas des Francs-Small et al. 2014), SLO3 (Hsieh et al. 2015), MTL1 (Haili et al. 2016), and three nuclear Mat (nMat) genes (Keren et al. 2009, 2012; Cohen et al. 2014). In maize, much chloroplast transcript splicing involving P-type PPR proteins has been studied. PPR4 was reported to encode a chloroplast-targeted PPR protein that associated with trans-splicing of rps12 intron 1 (Schmitz-Linneweber et al. 2006). PPR5 is another PPR protein that binds to the trnG-UCC precursor (Beick et al. 2008). THA8 is a short P-type PPR protein associated with splicing of introns in chloroplast ycf3-2 and trnA (Khrouchtchova et al. 2012). Recently, a few P-type PPR proteins were found to be involved in maize mitochondrial RNA splicing. EMP16 is specifically required for intron 4 cis-splicing of the nad2 transcript (Xiu et al. 2016). Dek35 is the first identified PPR protein responsible for nad4 transcript splicing (Chen et al. 2016).
Both cis- and trans-splicing of group II introns in the chloroplast and mitochondria require RNA splicing factors. RNA trans-splicing probably requires more protein factors than cis-splicing, and many more may remain to be identified (de Longevialle et al. 2007). The study on OTP43 showed it is specifically required for trans-splicing of the first intron in the mitochondrial nad1 transcript in Arabidopsis. Furthermore, nMAT1 and nMAT4 were also reported to be required in splicing of nad1 intron 1 in Arabidopsis (Keren et al. 2012; Cohen et al. 2014).
dek2-ref is a maize small kernel mutant with a SNP in the fifth PPR domain causing arrested endosperm and embryo development (Figures 1 and 3). The splicing efficiency of the nad1 intron 1 was affected in dek2, and this alteration of splicing efficiency was not observed in all other mitochondrial group II introns (Figure 5). Phylogenetic analysis showed Dek2 is not the homologous gene of previously reported OTP43 (At1g74900), nMAT1 (At1g30010), or nMAT4 (At1g74350), which is required for the trans-splicing process of nad1 intron 1 in Arabidopsis (Figure 4). So, Dek2 is a newly identified nad1 intron 1 trans-splicing factor in plant. Further analysis of mitochondrial complexes showed a significant decrease of complex I and supercomplex I + III2, suggesting a severe defect occurred in complex I assembling as a consequence of a deficient nad1 transcript (Figure 6). The functional characterization of Dek2 enlarges the understanding of PPR proteins acting in organelle RNA splicing in maize.
Loss of Dek2 resulted in defects in mitochondrial function and kernel development
Most of the characterized PPR mutants affect transcripts encoding proteins of complex I. The defect of complex I assembly always results in severe deficiency in mitochondrial function (Colas des Francs-Small and Small 2014; Hammani and Giege 2014). AOXs can reduce the reactive oxygen species (ROS) levels in situations when ETC complexes are unable to function properly for the maintenance of electron flux. There is rapid activation of AOXs in previously reported PPR mutants (Sun et al. 2015; Chen et al. 2016; Xiu et al. 2016). Respiratory metabolism was blocked in dek2 as Aox2, Aox3, and other mitochondrial function-related genes were dramatically up-regulated to rescue the functional tricarboxylic acid cycle (Figure 6 and Table S1). ETC biogenesis was reported to be required for the proper morphology of the cristaes in mitochondria (Logan 2006). In the maize ppr2263 mutant, the cristaes formed by the inner membrane are strongly reduced and the structurally altered mitochondria were likely less functional than mitochondria with a normal inner structure (Sosso et al. 2012). Abnormal morphology of mitochondria was also observed in the dek2 mutant (Figure 6). The loss of Nad1 (complex I) function results in a defect of ETC biogenesis, which is affecting not only respiratory metabolism but also inner structure of mitochondria.
Dek2 as a nuclear gene-encoded mitochondrial protein is having a profound effect on kernel development when the protein is defective and this has multiple effects, including the delayed formation of BETL and gross retardation of embryo morphogenesis.
BETL develops extensive cell wall ingrowths supporting an enlarged plasma membrane surface that promotes primary nutrient uptake of the endosperm (Pate and Gunning 1972; Thompson et al. 2001; Offler et al. 2003), which requires high metabolic rates. Therefore, transfer cells are typically rich in mitochondria. The development of BETL was dramatically arrested in dek2 endosperm (Figure 1). Mutations of the maize EMP4 gene, EMP16 gene, and DEK35 gene encoding PPR proteins also result in a defective transfer cell layer (Gutierrez et al. 2007; Chen et al. 2016; Xiu et al. 2016). The absence of a properly formed transfer cell layer is always correlated with reduced rates of grain filling and seed abortion (Brink and Cooper 1947; Charlton et al. 1995). These mutants with arrested BETL development exhibit small and hollow kernels.
The mitochondrial function defect in dek2 also brings about changes in the expression level of other important energy-consuming biological process-associated genes, including sulfur amino acid biosynthetic process, tRNA aminoacylation for protein translation, ribosomal subunit, nutrient reservoir activity, nucleosome assembly, and coated vesicle membrane (Figure 6 and Table S1). The large amount of transcriptionally regulated genes is not the consequence of developmental delay, according to the expression data for developing maize kernels (Chen et al. 2014). Sulfur amino acid biosynthetic process (GO:0000097), tRNA aminoacylation for protein translation (GO:0008418), and ribosomal subunit (GO:0033279) are important biological processes for cellular protein accumulation, and genes belong to these classifications were up-regulated might be the responsible regulation to the decreased total protein content in dek2 (Figure 2). Nutrient reservoir activity in the endosperm, the main storage tissue, largely determines the nutritional value of maize. The most abundant storage protein is zein, which accounts for 50–70% of the total protein, and α-zein is the largest class of zein protein (Holding and Larkins 2006). The genes encoding zein proteins were widely down-regulated in dek2, which might be the answer for the dramatically decreased zein protein content (Figure 2). Nucleosome assembly is essential for a variety of biological processes, such as cell cycle progression, development, and senescence (Gal et al. 2015), which are also energy-consuming processes. Hence, maize dek2 mutant affects kernel development, especially BETL formation and embryo morphogenesis, because of mitochondrial function defects and other secondary biological influences.
Acknowledgments
This work was supported by the National Natural Sciences Foundation of China (91335208 and 31425019). There is no conflict of interest.
Author contributions: R.S. and W.Q. designed the experiment. W.Q., Y.Y., X.F., and X.C. performed the experiments. W.Q., Y.Y., and R.S. analyzed the data. W.Q. and R.S. wrote the article.
Footnotes
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.116.196105/-/DC1.
These authors contributed equally to this work.
Communicating editor: J. A. Birchler
Literature Cited
- Barkan A., Small I., 2014. Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A., 2008. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28: 5337–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrisford J. M., Sazanov L. A., 2009. Structural basis for the mechanism of respiratory complex I. J. Biol. Chem. 284: 29773–29783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink R. A., Cooper D. C., 1947. Effect of the de17 allele on development of the maize caryopsis. Genetics 32: 350–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G., Gray M. W., Lang B. F., 2003. Mitochondrial genomes: anything goes. Trends Genet. 19(12): 709–716. [DOI] [PubMed] [Google Scholar]
- Chen J., Zeng B., Zhang M., Xie S., Wang G., et al. , 2014. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 166: 252–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Feng F., Qi W., Xu L., Yao D., et al. , 2016. Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Mol. Plant. DOI: 10.1016/j.molp.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Cheng S., Gutmann B., Zhong X., Ye Y., Fisher M. F., et al. , 2016. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 85: 532–547. [DOI] [PubMed] [Google Scholar]
- Charlton W. L., Keen C. L., Merriman C., Lynch P., Greenland A. J., et al. , 1995. Endosperm development in Zea mays: implication of gametic imprinting and paternal excess in regulation of transfer layer development. Development 121: 3089–3097. [Google Scholar]
- Cohen, S., M. Zmudjak, C. Colas des Francs-Small, S. Malik, F. Shaya et al., 2014 nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 78: 253–268. [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C., Small I., 2014. Surrogate mutants for studying mitochondrially encoded functions. Biochimie 100: 234–242. [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C., Falcon de Longevialle A., Li Y., Lowe E., Tanz S. K., et al. , 2014. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiol. 165: 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J., Lopez-Obando M., Heurtevin L., Bernard C., Martin K., et al. , 2013. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol. 10: 1557–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Longevialle A. F., Meyer E. H., Andres C., Taylor N. L., Lurin C., et al. , 2007. The pentatricopeptide repeat gene OTP43 is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19: 3256–3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina N. V., Heinemeyer J., Sunderhaus S., Boekema E. J., Braun H. P., 2006. Respiratory chain supercomplexes in the plant mitochondrial membrane. Trends Plant Sci. 11: 232–240. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., 1999. Mobile introns: retrohoming by complete reverse splicing. Curr. Biol. 9: R11–R14. [DOI] [PubMed] [Google Scholar]
- Fujii S., Small I., 2011. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 191: 37–47. [DOI] [PubMed] [Google Scholar]
- Gal C., Moore K. M., Paszkiewicz K., Kent N. A., Whitehall S. K., 2015. The impact of the HIRA histone chaperone upon global nucleosome architecture. Cell Cycle 14: 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez L., VanWuytswinkel O., Castelain M., Bellini C., 2007. Combined networks regulating seed maturation. Trends Plant Sci. 12: 294–300. [DOI] [PubMed] [Google Scholar]
- Haili N., Planchard N., Arnal N., Quadrado M., Vrielynck N., et al. , 2016. The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrial NADH DEHYDROGENASE SUBUNIT7 mRNA in Arabidopsis. Plant Physiol. 170: 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K., Giege P., 2014. RNA metabolism in plant mitochondria. Trends Plant Sci. 19: 380–389. [DOI] [PubMed] [Google Scholar]
- Holding D. R., Larkins B. A., 2006. The development and importance of zein protein bodies in maize endosperm. Maydica 51: 243–254. [Google Scholar]
- Hsieh W. Y., Liao J. C., Chang C. Y., Harrison T., Boucher C., et al. , 2015. The SLOW GROWTH3 pentatricopeptide repeat protein is required for the splicing of mitochondrial NADH Dehydrogenase Subunit7 intron 2 in Arabidopsis. Plant Physiol. 168: 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren, I., A. Bezawork-Geleta, M. Kolton, I. Maayan, E. Belausov et al., 2009 AtnMat2, a nuclearencoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15: 2299–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren I., Tal L., Colas des Francs-Small C., Araujo W. L., Shevtsov S., et al. , 2012. nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 71: 413–426. [DOI] [PubMed] [Google Scholar]
- Khrouchtchova A., Monde R. A., Barkan A., 2012. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 18: 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V., 2013. Plant mitochondrial genome peculiarities evolving in the earliest vascular plant lineages. J. Syst. Evol. 51: 1–12. [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lending C. R., Larkins B. A., 1992. Effect of the floury-2 locus on protein body formation during maize endosperm development. Protoplasma 171: 123–133. [Google Scholar]
- Li, X. J., Y. F. Zhang, M. Hou, F. Sun, Y. Shen et al., 2014 Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 79: 797–809. [DOI] [PubMed] [Google Scholar]
- Lid S. E., Gruis D., Jung R., Lorentzen J. A., Ananiev E., et al. , 2002. The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc. Natl. Acad. Sci. USA 99: 5460–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., He J., Chen Z., Ren X., Hong X., et al. , 2010. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. Plant J. 63: 749–765. [DOI] [PubMed] [Google Scholar]
- Liu Y. J., Xiu Z. H., Meeley R., Tan B. C., 2013. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. Plant Cell 25: 868–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C (T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Logan D. C., 2006. The mitochondrial compartment. J. Exp. Bot. 57: 1225–1243. [DOI] [PubMed] [Google Scholar]
- Lurin C., Andres C., Aubourg S., Bellaoui M., Bitton F., et al. , 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organdie biogenesis. Plant Cell 16: 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B. K., Cai X., Nebenführ A., 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Neuffer M. G., Sheridan W. F., 1980. Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 95: 929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offler C. E., McCurdy D. W., Patrick J. W., Talbot M. J., 2003. Transfer cells: cells specialized for a special purpose. Annu. Rev. Plant Biol. 54: 431–454. [DOI] [PubMed] [Google Scholar]
- Pate J. S., Gunning B. E. S., 1972. Transfer cells. Annu. Rev. Plant Physiol. 23: 173–196. [Google Scholar]
- Qi W., Zhu J., Wu Q., Wang Q., Li X., et al. , 2016. Maize reas1 mutant stimulates ribosome use efficiency and triggers distinct transcriptional and translational responses. Plant Physiol. 170: 971–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R. E., Williams-Voelker P. M., Kroeger T. S., Vichas A., et al. , 2006. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow J. A., Day D. A., 2000. Respiration and photorespiration, pp. 676–728 in Biochemistry and Molecular Biology of Plants, edited by Buchanan B. B., Gruissem W., Jones R. L. American Society of Plant Physiologists, Rockville, MD. [Google Scholar]
- Sosso, D., S. Mbelo, V. Vernoud, G. Gendrot, A. Dedieu et al., 2012 PPR2263, a DYW-subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. Plant Cell 24: 676–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Wang X., Bonnard G., Shen Y., Xiu Z., et al. , 2015. Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J. 84: 283–295. [DOI] [PubMed] [Google Scholar]
- Thompson R. D., Hueros G., Becker H., Maitz M., 2001. Development and functions of seed transfer cells. Plant Sci. 160: 775–783. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe G. C., Ordog S. H., 2002. Alternative oxidase: integrating carbon metabolism and electron transport in plant respiration, pp. 173–191 in Advances in Photosynthesis and Respiration, Photosynthetic Nitrogen Assimilation and Associated Carbon and Respiratory Metabolism, Vol. 12, edited by Foyer G. H., Noctor G. Kluwer Academic Publishers, Dordrect, The Netherlands. [Google Scholar]
- Wang G., Sun X., Wang G., Wang F., Gao Q., et al. , 2011. Opaque7 encodes an acyl-activating enzyme-like protein that affects storage protein synthesis in maize endosperm. Genetics 189: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang G., Qi W., Wu Q., Yao D., Zhang J., et al. , 2014. Identification and characterization of maize floury4 as a novel semidominant opaque mutant that disrupts protein body assembly. Plant Physiol. 165: 582–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu, Z., F. Sun, Y. Shen, X. Zhang, R. Jiang et al., 2016 EMPTY PERICARP16 is required for mitochondrial nad2 Intron 4 cis‐splicing, complex I assembly and seed development in maize. Plant J. 85(4): 507–519. [DOI] [PubMed] [Google Scholar]
- Zhang H. D., Cui Y. L., Huang C., Yin Q. Q., Qin X. M., et al. , 2015. PPR protein PDM1/SEL1 is involved in RNA editing and splicing of plastid genes in Arabidopsis thaliana. Photosynth. Res. 126: 311–321. [DOI] [PubMed] [Google Scholar]
- Zsigmond L., Rigó G., Szarka A., Székely G., Otvös K., et al. , 2008. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiol. 146: 1721–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq data are available from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under the series entry GSE87067.