Summary
Systematic genetic access to GABAergic cell types will facilitate studying the function and development of inhibitory circuitry. However, single gene-driven recombinase lines mark relatively broad and heterogeneous cell populations. Although intersectional approaches improve precision, it remains unclear whether they can capture cell types defined by multiple features. Here we demonstrate that combinatorial genetic and viral approaches target restricted GABAergic subpopulations and cell types characterized by distinct laminar location, morphology, axonal projection, and electrophysiological properties. Intersectional embryonic transcription factor drivers allow finer fate mapping of progenitor pools that give rise to distinct GABAergic populations, including laminar cohorts. Conversion of progenitor fate restriction signals to constitutive recombinase expression enables viral targeting of cell types based on their lineage and birth time. Properly designed intersection, subtraction, conversion, and multi-color reporters enhance the precision and versatility of drivers and viral vectors. These strategies and tools will facilitate studying GABAergic neurons throughout the mouse brain.
eTOC Blurb
He et al. present strategies and tools for the combinatorial targeting of GABAergic neurons in the mouse brain. Specific cell type targeting can be achieved by combining 2 to 3 driver-reporter alleles with viral vectors that engage multiple cell-defining features.
Introduction
Understanding the functional organization of neural circuits requires comprehensive knowledge of the basic cellular components. Whereas glutamatergic pyramidal neurons make up ~80% of neurons in rodent neocortex and constitute the major cortical processing streams and output channels, GABAergic neurons regulate the delicate balance and dynamic operations of local circuitry and route information flow in cortical networks (Harris and Shepherd, 2015; Huang, 2014; Kepecs and Fishell, 2014). Although a minority, GABAergic neurons consist of diverse cell types that reflect an elaborate division of labor of an intricate inhibitory control system (Jiang et al., 2015; Klausberger and Somogyi, 2008; Markram et al., 2015; Markram et al., 2004; Roux and Buzsaki, 2015). However, comprehensive identification and characterization of GABAergic cell types have remained an unfulfilled goal, largely due to the lack of experimental access to specific cell types in a systematic way.
Although GABAergic neurons have been classified according to their morphology, connectivity, physiological properties, gene expression and developmental origin (Petilla Interneuron Nomenclature et al., 2008), a combination of multiple correlated features is likely more informative in cell type definition (Kepecs and Fishell, 2014). The generation of mouse recombinases driver lines begins to provide reliable access to multiple GABAergic cell populations and lineages (Taniguchi et al., 2011), and has facilitated progress in studying the development, connectivity, and function of cortical inhibitory interneurons (e.g. Cardin et al., 2009; Fu et al., 2014; Lee et al., 2013; Pfeffer et al., 2013; Pi et al., 2013; Taniguchi et al., 2013). However, as there is no simple correlation between single genes and specific cell types, most individual driver lines often include multiple cell types (Jiang et al., 2015; Munoz et al., 2014; Pronneke et al., 2015; Xu et al., 2013). Thus a key challenge of broad significance is to increase the specificity of cell type targeting. Enhanced specificity will facilitate multi-faceted studies toward elucidating the biological basis for defining GABAergic cell types, a fundamental issue in understanding cortical circuitry, and will provide a ground truth starting point for comprehensive identification of cortical cell types.
Intersectional methods based on 2–3 genes or other cell features improve the precision of cell targeting (Dymecki et al., 2010; Fenno et al., 2014; Madisen et al., 2015). However, strategically designed combinatorial driver and reporter lines are currently very limited. More importantly, few strategies have been demonstrated to incorporate multiple cell-defining phenotypes toward more specific cell targeting. Furthermore, there are practical limits to the number of driver/reporter allele combinations suitable for mouse engineering and breeding. Thus it remains unclear to what extent genetic methods will achieve sufficient specificity in targeting GABAergic subpopulations and cell types.
Here we have designed multiple combinatorial strategies, implemented a suite of genetic and viral tools, and demonstrated more precise targeting of GABAergic neurons and progenitors. We designed intersectional transcription factor drivers that allow more precise fate mapping of progenitor pools giving rise to more restricted GABAergic populations, including laminar cohorts. We showed that intersection of Cre and Flp lines driven by two marker genes targeted more restricted GABAergic subpopulations, and that incorporation of viral methods allowed capturing specific cell types. We further implemented a method that converted a transient fate restriction signal in progenitors to constitutive recombinase expression, which enabled viral targeting of cell types in the mature brain based on cell lineage and birth time. Therefore, highly specific targeting is feasible by combining 2 to 3 driver-reporter alleles with viral vectors to engage a spectrum of cell-defining features that include lineage, birth time, marker genes, and anatomy. Together these strategies and tools will accelerate progress in studying GABAergic circuits in the cerebral cortex and in other brain areas.
Results
Our strategy is to combine a small set of driver-reporter alleles and viral vectors that incorporate multiple cell features to increase targeting specificity (Figure 1, Table 1, Table S1–2). Our newly generated drivers include: 1) Nkx2.1-ires-FlpO (Nkx2.1-Flp) for intersectional fate mapping of medial ganglionic eminence and preoptic area progenitors, 2) vasointestinal peptide-ires-FlpO (VIP-Flp) and somatostatin-ires-FlpO (SST-Flp) for dissecting these two broad cell populations, 3) a highly efficient inducible PV-2A-CreER driver which is far superior to the previous version (Taniguchi et al., 2011). The new reporter lines include: 1) an intersectional reporter expressing nuclear-targeted GFP (HG), 2) a high performance intersection-subtraction reporter (IS), 3) a conversion reporter that converts Cre expression to Flp expression, 4) a RGBow reporter that expresses one of three membrane-tethered fluorescent proteins in a random manner upon Cre-mediated recombination. We have also designed several viral vectors tailored for combinatorial targeting with driver and reporter lines (Table 1, Table S1–2). All mouse lines are deposited to the Jackson Laboratory for distribution.
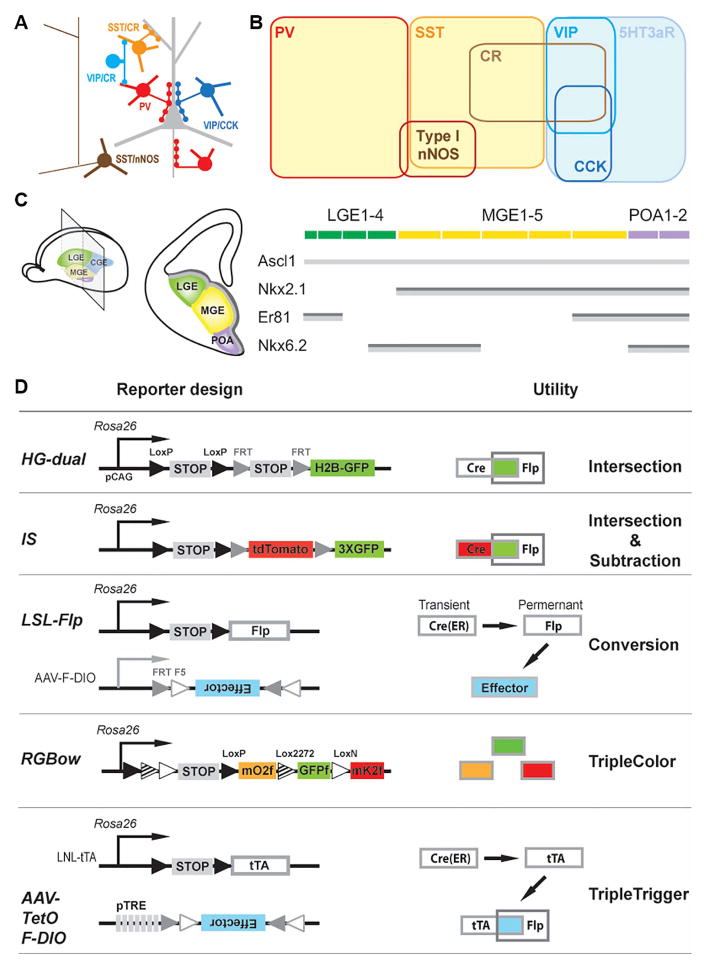
Figure 1. Combinatorial strategies and tools for genetic targeting of cortical GABAergic neurons.
(A) A schematic of several major GABAergic neuron types in the neocortex. (B) Major neurochemical markers expressed in cortical GABAergic neurons. (C) Left: Schematic of subpallium progenitor domains in the embryonic telencephalon and its coronal view at the indicated plane (middle). Right: Schematic representation of expression patterns for four transcriptional factors, Ascl1, Nkx2.1, Nkx6.2 and Er81, along the subpallium progenitor domains. Dark grey bar: ventricular zone consisting of mostly radial glia cells. Light grey bar: subventricular zone consisting of mostly intermediate progenitors. (D) Diagram of reporter and viral strategies. Newly generated tools are in larger font. PV, parvalbumin; SST, somatostatin; CR, calretinin; VIP, vasoactive intestinal polypeptide; CCK, cholecystokinin; nNOS, neuronal nitric oxide synthase; MGE, medial ganglionic eminence; CGE, caudal ganglionic eminence; LGE, lateral ganglionic eminence; POA, pre-optic area. (See also Table 1, Figure S1–2, Table S1–3.)
Table 1.
Newly generated mouse lines and virus.
| Category | Name | Description | |
|---|---|---|---|
| Driver | Progenitor | Nkx2.1-ires-Flpo | Nkx2.1 locus; ires-Flpo targeted after STOP codon; JAX 028577 |
| Mature marker | VIP-ires-Flpo | VIP locus; ires-Flpo targeted after STOP codon; JAX 028578 | |
| SST-ires-Flpo | Somatostatin locus; ires-Flpo targeted after STOP codon; JAX 028579 | ||
| PV-2A-CreER | PV locus; 2A-CreER targeted before STOP codon; endogenous 3′UTR preserved; JAX 028580 | ||
| Reporter (Rosa26 locus; CAG promoter) | Intersection | HG-Dual | Cre dependent;express H2B-GFP; JAX 028581 |
| Intersection and subtraction | IS | Express tdTomato for “Cre NOT Flp” subtraction and GFP for “Cre AND Flp” intersection; JAX 028582 | |
| Cre to Flp “Conversion” | LSL-Flpo | Cre dependent; express Flpo; JAX 028584 | |
| Cre dependent triple color | RGBow | Cre dependent; express farnesylated derivatives of mOrange2, EGFP and mKate2; JAX 028583 | |
| AAV virus | tetO-F-DIO- GFP-2A-RabiesG | Respond to tTA activation; Flp dependent; Starter virus for monosynaptic retrograde rabies tracing. | |
| tetO-F-DIO-mCherry-2A-TVA | |||
Nkx2.1-Flp enables intersectional targeting of MGE subdomains and progenitor types
The embryonic subpallium contains a developmental plan embedded in progenitors along the lateral-, medial- and caudal- ganglionic eminence (LGE, MGE, CGE) and the preoptic area (POA) that orchestrates the reliable generation of all GABAergic neurons in rodent neocortex (Gelman and Marin, 2010; Rudy et al., 2011) (Figure 1C). Beyond these major domains, 18 subdomains have been identified according to combinatorial TF expression patterns (Flames et al., 2007). Furthermore, the subpallium contains different types of progenitors, including radial glial progenitors (RGs), which reside in the ventricular zone, (VZ) and intermediate progenitors (IPs), which reside in the subventricular zone (SVZ) (Brown et al., 2011; Hansen et al., 2013; Harwell et al., 2015). Whether these progenitor subdomains and cell types contribute to different aspects of GABAergic neuron production and diversity remains unclear.
The homeodomain transcription factor Nkx2.1 is expressed in MGE and POA throughout embryonic neurogenesis in both RGs and IPs (Marin and Rubenstein, 2001; Sussel et al., 1999). We tested our Nkx2.1-Flp driver with multiple reporters and demonstrated that it captured all major groups of MGE and POA derived cortical GABAergic neurons (Figure S1).
The expression patterns of Nkx6.2 and Er81 at E12.5-E13.5 mark the dorsal and ventral MGE, respectively (among other LGE, CGE and POA domains, Figure 1C) (Flames et al., 2007; Sousa et al., 2009). By combining the Nkx6.2-CreER or Er81-CreER driver with Nkx2.1-Flp and the Ai65 intersectional reporter, we fate mapped dorsal vs. ventral MGE progenitors at E13.5. Herein we refer to Nkx2.1-Flp;Nkx6.2-CreER;Ai65 allele combination as Nkx2.1/Nkx6.2 and Nkx2.1-Flp;Er81-CreER;Ai65 as Nkx2.1/Er81 (Figure 2A–B); the same naming convention will be followed for other allele combinations throughout the text. In adult progeny, typically several hundred neurons were sparsely labeled in each cortex across multiple cortical areas and layers (Figure 2C–D, Figure S3). While neurons derived from Nkx2.1/Nkx6.2 vs. Nkx2.1/Er81 progenitors show laminar bias in their cortical settlement, the most notable difference between them is the morphological characteristics of their cell composition. Significantly more putative layer2/3 (L2/3) Martinotti cells were labeled in Nkx2.1/Nkx6.2 (~20%, Figure 2Ci, Figure S3A) compared to Nkx2.1/Er81(<6%, Figure 2Di). Neurons residing in L1/2 with dense horizontally extending axons (Figure 2Cii) and a rarer type of L6 neurons that extended axons along the corpus callosum (Figure 2Ciii) were only observed in Nkx2.1/Nkx6.2 but not found in Nkx2.1/Er81. Most of the labeled cells in Nkx2.1/Er81 were multipolar cells that resided in deep layers (especially L5) with radiating axons (Figure 2Dii–iii, Figure S3C, D). While Nkx2.1/Nkx6.2 progenitors produced only a small number of chandelier cells (ChCs) in both superficial and deep layers (Figure 2Civ, S3F-H), Nkx2.1/Er81progenitors produced more that were almost exclusively superficial layer ChCs (Figure 2Div, Figure S3E). Together, these results suggested that progenitors in dorsal vs. ventral MGE are fated to generate distinct sets of GABAergic neurons. Nkx2.1-Flp thus allows intersectional fate mapping of finer grain progenitor pools in MGE and POA defined by additional transcription factors.
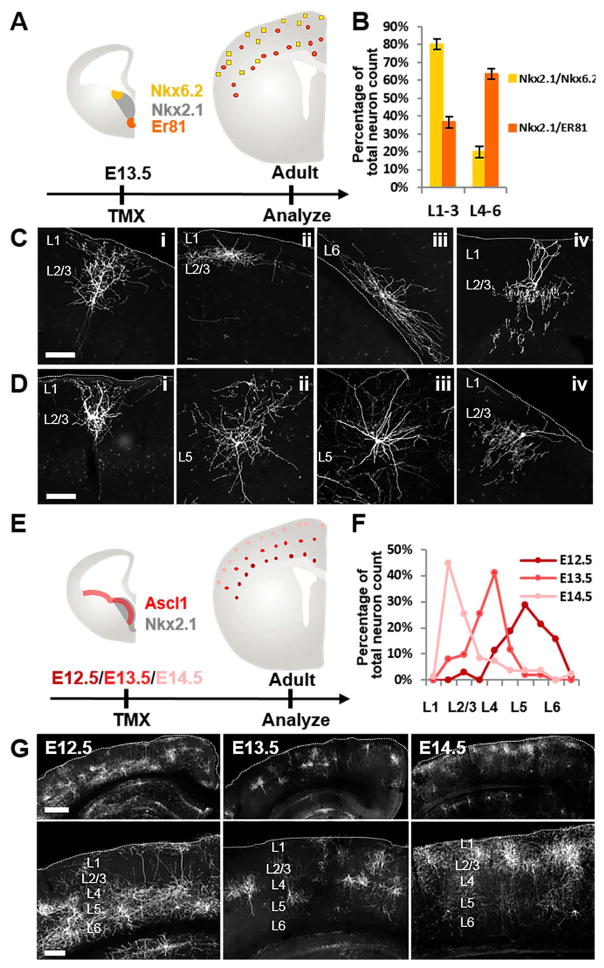
Figure 2. Intersectional targeting of spatial and temporal MGE progenitor pools.
(A) Diagram for intersectional fate mapping from dorsal vs. ventral MGE progenitor pools defined by Nkx2.1/Nkx6.2 and Nkx2.1/Er81, respectively. (B) Quantification of laminar distribution of neurons derived from Nkx2.1/Nkx6.2 and Nkx2.1/Er81 progenitors. (C) Representative neurons derived from Nkx2.1/Nkx6.2 progenitors. (D) Representative neurons derived from Nkx2.1/Er81 progenitors. (E) Diagram for intersectional fate mapping from MGE intermediate progenitors in Nkx2.1/Ascl1. (F) Quantification of laminar distribution of cortical GABAergic neurons labeled at three embryonic time points. (G) Representative images for each induction time point. Dashed line: pial surface or border of white matter. Scale bar in (C), (D): 100μm. Scale bar in (G), upper panel: 500μm. Scale bar in G, lower panel: 250μm. (See also Figure S3)
The bHLH transcription factor Ascl1 is primarily expressed by IPs throughout the embryonic subpallium. Inducible fate mapping from IPs should allow more precise labeling of temporal cohorts of neurons born at the time of CreER induction than from RGs that undergo multiple rounds of neurogenesis. By combining the Ascl1-CreER knockin driver (Kim et al., 2011) with Nkx2.1-Flp, we demonstrated the feasibility of fate mapping MGE IPs at multiple embryonic time points. Tamoxifen induction at E12.5, E13.5 and E14.5 labeled GABAergic neurons that occupied cortical layers in a clear “inside-out” pattern: neurons born at E12.5 mainly occupy L5 to L6, at E13.5 mainly L4, and at E14.5 L2/3 (Figure 2 E–H). These laminar patterns of cell distribution were significantly sharper than those fate mapped from the Olig2-CreER driver, which included both RGs and IPs (Miyoshi et al., 2007). Therefore, the Nkx2.1-Flp driver allows differential fate mapping of IPs in MGE and POA defined by appropriate TF drivers. This provides an improved tool for analyzing the relationship between neuronal birthdate and mature phenotypes that contribute to cell identity.
VIP-Flp-mediated intersectional targeting reveals multiple subpopulations
Among GABAergic neurons derived from CGE, the VIP expressing cells make up to 15% of all cortical interneurons (Miyoshi et al., 2010; Rudy et al., 2011) and display multiple morphological and physiological features (Cauli et al., 2004; Pronneke et al., 2015). The generation of a VIP-ires-Cre driver (Taniguchi et al., 2011) allowed a direct demonstration that certain VIP interneurons preferential inhibit other GABAergic neurons (Lee et al., 2013; Pfeffer et al., 2013; Pi et al., 2013), and enabled studies that revealed the function of this dis-inhibitory circuit in the context of behavior (Fu et al., 2014; Pi et al., 2013).
On the other hand, the VIP population consists of several subpopulations that show distinct morphology (e.g. bipolar type, small basket type (Bayraktar et al., 2000)), intrinsic electrophysiological properties (Lee et al., 2013; Pronneke et al., 2015) and differential marker expression (e.g. calretinin, cholecystokinin, corticotropin-releasing hormone (Kubota, 2014; Kubota et al., 2011)). To demonstrate the morphological heterogeneity within the VIP population, we used the VIP-ires-Cre driver to activate a triple-color reporter RGBow (Figure 1D) that we have generated to achieve multi-color labeling of VIP cells in the same animal (Figure 3B).
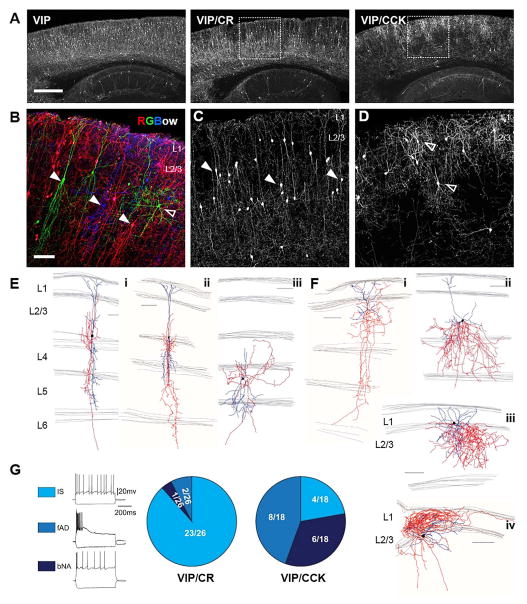
Figure 3. Intersectional targeting of VIP subpopulations.
(A–C) Representative cortical sagittal sections from VIP-Flp;tdTomato-FRT (VIP), VIP-Flp;CR-Cre;Ai65 (VIP/CR) and VIP-Flp;CCK-Cre;Ai65 (VIP/CCK) mice. (A) Lower magnification view of reporter expression in sagittal sections. (B) Triple color labeling by RGBow reporter via VIP-Cre activation revealed several morphologically distinct neuron types intermingled in superficial layers. (C–D) Higher magnification images of boxed area in (A). Arrow heads: neurons with vertically restricted axons and dendrites. Open arrow heads: neurons with multipolar, small basket cell like morphology. (E–F) Neurolucida reconstruction of biocytin filled neurons recorded in VIP/CR neocortex (E) and VIP/CCK neocortex (F). Axons were traced in red and dendrites in blue. (G) Current clamp recordings in somatosensory cortex of VIP/CR and VIP/CCK mice show different proportions of cells with various intrinsic firing patterns. Representative recording traces are shown on the left. Irregular spiking cells (IS) are characterized by action potentials elicited with irregular intervals at near threshold. Fast adapting cells (fAD) are characterized by their failure to fire throughout the 600 ms step during suprathreshold depolarization. Burst non-adapting cells (bNA) exhibit bursts of two or three spikes, followed by regular spiking. Proportions of each firing pattern in the two intersection lines are shown as pie charts on the right. Scale bar in A: 500μm. Scale bar in B–F: 100μm. (See also Figure S4, Table S2–5.).
To further define the VIP cell subpopulations, we generated a VIP -Flp line (Figure 3A) which showed a near identical recombination specificity and efficiency as VIP-ires-Cre (Figure S4 A, B). We combined this line with calretinin(CR) - and cholecystokinin(CCK)-ires-Cre (Taniguchi et al., 2011), to carry out intersectional targeting (Figure 3, Figure S4, Table S2–4). The VIP/CR intersection resulted in a significant enrichment of cells located in lower L2/3, with smaller fractions in deep layers (Figure 3A, C). These cells showed prominent vertically oriented bipolar morphology (18 out of 24 biocytin filled single cells recovered from patch clamp recording in brain slice), consistent with previous studies (Cauli et al., 2014; Jiang et al., 2015); they extended vertically oriented dendrites that span much of the cortical thickness and projected vertically restricted axons (Figure 3Ei–ii). In rare cases, neurons with multipolar morphology can also be observed (Figure 3Eiii). Previous studies reported that VIP interneurons as a group show a range of different firing patterns including irregular spiking (IS) which has also been frequently reported for CR expressing interneurons (Cauli et al., 1997; Cauli et al., 2000), bursting non-adapting (b-NA), and fast adapting (fAD) (Figure 3G) (Jiang et al., 2015; Lee et al., 2010; Miyoshi et al., 2010; Pronneke et al., 2015). The VIP/CR intersection significantly enriched for cells with IS firing pattern, with a subpopulation also showing b-NA or fAD (Figure 3G). Both the prominently bipolar axon morphology and the irregular-spiking intrinsic features are characteristic of interneuron-selective cells (Lee et al., 2013), suggesting that the VIP/CR intersection significantly enriched for this subpopulation.
In contrast, most of the VIP/CCK cell somata were enriched in upper L2/3 near the L1 and L2 boundary, with a smaller fraction in deep layers (Figure 3A, D). 15 out of 17 VIP/CCK neurons recovered from cell filling show multipolar morphology. Many of these cells extend highly exuberant local axon arbors within L2/3, but their multi-polar dendrites extended to L1 (Figure 3Fi–iii). We also recovered rare cells that projected axons to L1 (Figure 3Fiv). The intrinsic properties of a substantial proportion of VIP/CCK cells were either bursting or fast-adapting, which were less frequently observed in VIP/CR cells (Figure 3G, Table S3). These morphological and physiological features suggest that VIP/CCK cells likely correspond to a set of VIP− and CCK-expressing small basket cells (Kawaguchi and Kubota, 1998; Kubota, 2014; Ma et al., 2011). It is notable that this population appears to be found in a large scale recording in juvenile rat (Markram et al., 2015) but was undetected by similar studies in the mouse even using the VIP-ires-Cre driver (Jiang et al., 2015; Pronneke et al., 2015). Together, these results demonstrate the presence of multiple distinct subpopulations within the VIP population and the feasibility of intersectional targeting of these subpopulations.
SST-Flp and CR-Cre intersection enriches L1-targeting Martinotti cells
SST-expressing neurons constitute approximately half of MGE-POA derived GABAergic neurons and up to ~30% of all GABAergic neurons in many cortical areas (Rudy et al., 2011). SST cells are present throughout L2 to L6, with notable enrichment in deep layers (Pfeffer et al., 2013), and consist of multiple heterogeneous subpopulations (Ma et al., 2006; Munoz et al., 2014; Tomioka et al., 2005; Xu et al., 2013). Although the SST-ires-Cre driver confers experimental access to SST cells, the diverse cell types within this broad population are not well understood.
There is evidence that some SST and CR double positive cells may correspond to dendrite-targeting Martinotti cells (Wang et al., 2004; Xu et al., 2006) and SST and nNOS double positive cells to long projection cells (Tomioka et al., 2005). To examine these correlations, we generated a SST-Flp driver that allows intersectional dissection of the SST population. The SST-Flp driver gave rise to a nearly identical recombination pattern to SST-ires-Cre and labeled SST cells across L2 to L6 (Figure 4A). In sharp contrast, SST/CR intersection revealed a subpopulation with striking laminar distribution patterns: SST/CR cells were highly enriched in L2/3 and L5a, with less abundance in L5b and L6 but were virtually absent in L4 (Figure 4A, B, Figure 5B). These cells often had prominent laminar-targeted axon projection and arborization, with L2/3 and L5a cells projecting axons to L1 (Figure 4C), and L6 cells likely projecting axons to L4/5a (Markram et al., 2015). Single cell reconstruction confirmed that L2/3 and L5 SST/CR cells have Martinotti cell morphology with an ascending axon that arborizes in L1 with additional branching in L2/3 (Figure 4C), consistent with the population pattern.
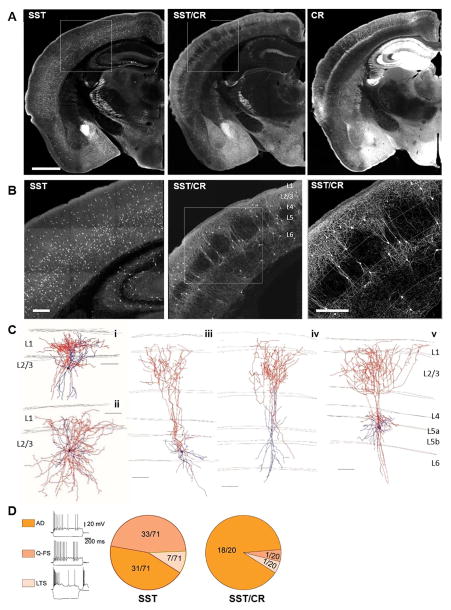
Figure 4. Intersectional targeting of the SST/CR subpopulation.
(A) SST-Flp;tdTomato-FRT, (SST) SST-Flp;CR-Cre;Ai65 (SST/CR) and CR-Cre;Ai14(CR) mice show distinct cell distributions in numerous brain areas. (B) Higher magnification images of boxed area in (A) highlight laminar difference between SST and SST/CR cells. (C) Neurolucida reconstruction of biocytin filled L2/3 (i, ii) and L5 (iii-v) neurons with Martinotti type morphology. Axons were traced in red and dendrites in blue. (D) More homogenous firing patterns were seen in SST/CR than in SST cells. AD: adapting firing pattern, characterized by a strong firing frequency adaptation, and triphasic afterhyperpolarization following single spikes. Q-FS: “quasi” fast spiking, characterized by low input resistance (≤200 MΩ), stuttering firing pattern, ability to sustain high firing frequencies, and deep and short duration afterhyperpolarizations. LTS: low-threshold spiking characterized by high input resistance (> 400 MΩ), rebound bursting in response to hyperpolarizing current steps, and low-threshold bursting in response to depolarizing current steps from hyperpolarized potentials. Scale bar in (A): 1mm. Scale bar in (B): 250μm. Scale bar in (C), (D): 100μm. (See also Figure S5, Table S2–5.)
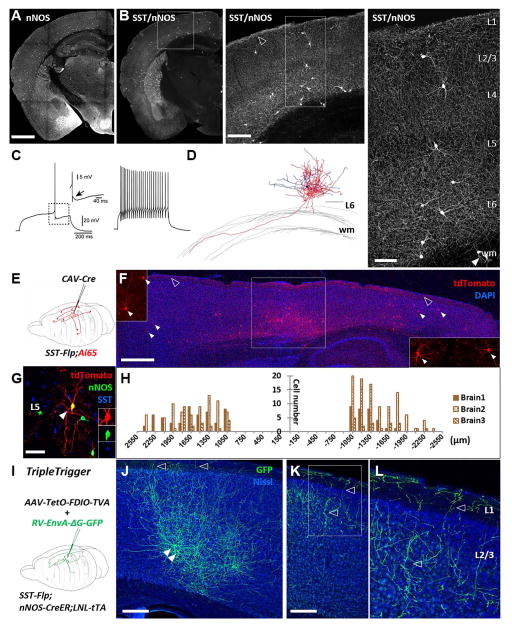
Figure 5. Combinatorial genetic-viral targeting of long projection GABAergic neurons.
(A) nNOS neurons distribute throughout the cortical thickness as labeled in nNOS-CreER;Ai14 mice. (B) SST/nNOS intersection labels long projection GABAergic neurons which mainly reside in deep layers and extend long range axons that cover the entire neocortex. The dense axons in L1 are visible as bright signal at the pial surface (open arrowhead in middle panel). Note neurons in the white matter in the right panel (arrowheads), consistent with previous reports. (C) Representative recording traces. All recorded neurons showed very homogenous firing patterns, characterized by irregular accommodating spiking profile with delayed first spike (left panel) and an afterdepolarization (ADP) following the fast AHP (black arrow). (D) Neurolucida reconstruction of biocytin filled deep layer neuron in SST/nNOS reveals the presence of long projection axons extending into the white matter. Axons were traced in red and dendrites in blue. (E) Scheme for retrograde labeling of long projection GABAergic neurons via CAV2-Cre intersection with SST-Flp;Ai65. Retrograde transport of the CAV2-Cre virus allowed activation of RFP expression in SST neurons, including long projection neurons that were millimeters away from the injection site. (F) Representative sagittal section. Box in the middle: injection site in M1. Insets: higher magnification view of retrogradely labeled neurons (arrow heads) rostral (left) and caudal (right) to the injection site. Open arrowheads: L1 axons. (G) A retrogradely labeled SST neuron that also expressed nNOS. (H)Quantification of the distribution of retrogradely labeled neurons away from injection site. Injection site was defined as a 900μm radius region from the section with the highest number of infected neurons. (I) Experiment scheme of “Triple-Trigger”. (J) GFP labeled SST/nNOS neurons (arrow heads) and their axons (open arrow heads) at injection site. (K) Long range projecting axons several hundred microns away from the cell bodies labeled at the injection site. Open arrow heads: descending branches from L1 axons. (L) Higher magnification image of the boxed area (K). Scale bars in (A) (from left to right): 1mm, 250μm, 100μm. Scale bars in (D), (G): 100μm. Scale bar in (F):1mm. Scale bar in (J), (K): 200μm. wm, white matter. (See also Figure S6, Table S2–3.)
We compared the intrinsic properties of cells sampled in SST/CR with those sampled from the whole SST population in SST-Cre;Ai14 mice. In the SST population (n= 71) we observed: 1) cells with a strongly adapting firing pattern (~44%), 2) cells with narrower spikes that can sustain much higher firing frequencies resembling the L4-targeting SST cells in the X-94 mouse line (~47%) (Ma et al., 2006; Xu et al., 2013), and 3) cells with very high input resistance that produced rebound spikes following a hyperpolarization, resembling the SST cells reported in the X-98 mouse line (~9%) (Ma et al., 2006) (Figure 4D). In the SST/CR population, intrinsic electrophysiological properties are much more homogeneous, with 90% strongly characterized as adapting firing pattern (AD) (Figure 4D). Therefore, the SST/CR intersection significantly enriched, although did not exclusively purify, Martinotti cells in L2/3 and L5, and it essentially excluded L4-targeting and long range projection SST neurons.
Combinatorial SST, nNOS and viral targeting captures cortical GABAergic projection neurons
Nitric oxide (NO) is a versatile signaling molecule in the brain synthesized by the neuronal isoform of nitric oxide synthase (nNOS) (Tricoire et al., 2013). In mature cerebral cortex, all nNOS cells are GABAergic and consist of two major populations (Kubota et al., 2011; Perrenoud et al., 2012). Type II neurons weakly stain for nNOS and have small somata distributed in multiple layers; they include several subpopulations (e.g. neurogliaform cells) (Magno et al., 2012; Olah et al., 2009; Perrenoud et al., 2012). In contrast, type I neurons have heavy nNOS stain and large somata distributed predominantly in L5 and L6 and, to a lesser extent, L2 (Kubota et al., 1994; Tricoire et al., 2013). These highly unique GABAergic neurons are not interneurons but extend axons to distant cortical and subcortical regions(Tomioka et al., 2005). They are selectively activated during slow wave sleep (Kilduff et al., 2011). The distribution, connectivity and function of type I nNOS neurons are poorly understood due to the lack of specific experimental access.
In SST/nNOS mice, a single dose tamoxifen induction at P16 labeled ~85% of type I nNOS neurons characterized by strong nNOS immunostaining and large cell bodies (Figure 5B, Figure S5B–C). SST/nNOS cells were prominently localized to L5 and L6, and to a much lesser extent at the L1/2 border, with a small number of cells also scattered in L3 and the white matter (Figure 5B, Figure S6A). The abundance and laminar distribution of type I cells varied according to cortical areas (Figure 5B, Table S5). At the population level, the axon arbors of type I cells were dense and extensive, forming a matrix that appeared to cover the entire cortical volume (Figure 5B, Figure S6 A–F). These axons were dense in L1 and also extended into the white matter (Figure S6B–E). This particularly rare cell type has not been described by previous physiological studies (Jiang et al., 2015; Markram et al., 2015). We found that the intrinsic properties of type I cells are quite homogeneous, characterized by an irregular accommodating spiking profile with a delayed first spike; their action potentials have relatively narrow spike widths (0.51 ms) and always showed a biphasic after potential consisting of a fast afterdepolarization(ADP) followed by an afterhyperpolarization (AHP) (Figure 5C). Single cell reconstruction of L6 SST/nNOS cells revealed that while their multipolar dendrites were largely restricted to the same layer, their axons arborized extensively with branches entering the white matter that could not be fully traced in brain slice (Figure 5D).
We next designed a genetic and viral intersection strategy to examine the distribution pattern and axon trajectory of SST/nNOS cells that make distant projections to a specific cortical area. In a SST-Flp;Ai65 mouse, we injected a retrograde-transported canine virus CAV2-Cre (Hnasko et al., 2006; Kremer, 2005) in the primary motor cortex (M1) (Figure 5E–H). Only Flp expressing SST cells that were infected with CAV2-Cre would activate the Ai65 reporter allele and express tdTomato. At the injection site, many SST cells expressed tdTomato as their local axons were infected by CAV2-Cre (Figure 5E square boxed area). In more distant cortical areas from M1, only SST cells that projected across a long distance to the injection site could acquire CAV2-Cre and activate tdTomato expression. We found that M1-projecting SST neurons were distributed in widespread cortical areas ranging from the rostral orbital frontal cortex to the primary and secondary visual cortex in the caudal end (Figure 5E–H, Movie S1). Their rostro-caudal axon projection distance exceeded 3 millimeters across several cortical areas, and cells were labeled more than 2.5 mm away laterally in the somatosensory area. We detected only 1 cell in the contralateral hemisphere in 3 mice. Approximately half of the SST projection neurons were strongly nNOS positive (Figure 5G), while the other half were either negative or very weakly stained, consistent with an earlier report (Tamamaki and Tomioka, 2010). As expected, these M1 long projection cells were highly enriched in L5 and L6, with another minor group of cells in L2/3 (Movie S1). In areas distant from the injection site, sparse cell labeling revealed their extensive axon arbors, which often projected branches into L1 (Figure S6F, G). Thus SST/nNOS neurons likely project to distant cortical areas through L1 as well as the white matter. The SST-Flp and CAV2-Cre intersection strategy thus provides a reliable and efficient method to systematically map the areal organization and projection pattern of this unique and intriguing class of cortical GABAergic projection neurons.
We further developed a combinatorial genetic-AAV strategy, termed “Triple-Trigger”, to establish viral-based experimental access that allows integration of multiple tools for studying the connectivity and function of SST/nNOS neurons (Figure 1D, Figure 5I). SST-Flp;nNOS-CreER mice were first bred with a Rosa26-loxPNeoSTOPloxP-tTA (LNL-tTA) (Wang et al., 2008) line to generate SST-Flp;nNOS-CreER;LNL-tTA mice, in which tamoxifen induction can convert CreER into constitutive tTA expression in nNOS neurons. We further generated a new AAV vector in which TVA-mcherry expression is dependent on both tTA and Flp activities (Figure 1D, Table 1, Figure 5I). We injected this AAV-tetO-fDIO-TVA-mcherry into the motor cortex of SST-Flp;nNOS-CreER;LNL-tTA mice, followed by tamoxifen induction of Cre recombination to activate tTA expression. Three weeks later, a pseudo-typed glycoprotein deficient rabies virus expressing GFP (RV-EnvA-ΔG-GFP) was injected in the same area to infect TVA expressing neurons. This method yielded intense GFP labeling of a small number (~10) of SST/nNOS neurons in L5/6 of motor cortex in a restricted region (<1mm diameter) (Figure 5J), which extended dense and widespread axon arbors towards L1. Descending axon branches from L1 were observed in sections far away from their parent cell bodies. The longest projecting axons were detected 2.4 mm away from the center of the injection site (Figure 5K, L).
In summary, nNOS-CreER and SST-Flp intersection captures a specific and unique type of GABAergic projection neurons that likely contributes to the regulation of neuronal populations in widespread cortical regions. Further incorporation of intersectional viral vectors allows versatile experimental access enabling investigators to map the projection and connectivity patterns, monitor the activity and manipulate the function of this enigmatic and likely important cell type.
Laminar distribution pattern of GABAergic subpopulations
In contrast to glutamatergic pyramidal neurons, which are organized with laminar patterns characteristic to the neocortex, GABAergic neurons appear more evenly distributed across the cortical thickness. However, it is unclear to what extent more specific GABAergic subpopulations or cell types are deployed to more restricted cortical layers.
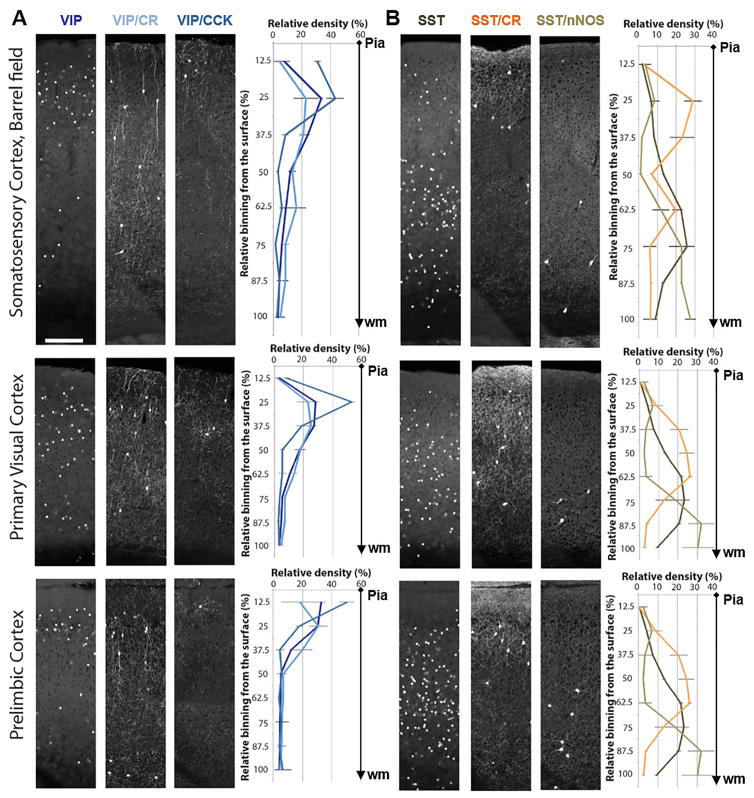
Using serial two-photon tomography (Ragan et al., 2012), we quantified the laminar distribution of six GABAergic populations in somatosensory, motor and medial prefrontal cortices (Figure 6). We found that, while the broad VIP and SST populations showed laminar biases, each of the four subpopulations revealed significantly more restricted laminar distributions, with some variations in different cortical areas. Whereas VIP/CCK cells were highly enriched in upper L2/3, most VIP/CR cells were located in deeper L2/3 (Figure 3, 6). SST/CR showed striking enrichment in L2/3, L5a, L6, and were nearly absent in L4 in barrel cortex (Figure 6, note the dip at 50% of cortical depth in barrel cortex, which was not present in the other two cortical regions; Table S5). Lastly, SST/nNOS neurons were concentrated in deep L5/6 (~74% combined in bin 6–8, Table S5), with another much smaller cohort in upper L2/3 (~8% in bin 2, Table S5). Together, these results indicate that GABAergic subpopulations and cell types are in fact deployed to restricted cortical layers with characteristic distribution patterns. It is possible that further purification of cell types might reveal even more specific laminar distribution patterns (e.g. ChCs (Taniguchi et al., 2013)). As the location of cell somata anchors the spatial distribution of their mostly local dendritic and axonal arbors that mediate connectivity (with a notable exception for the long projection axons of type I nNOS neurons), the laminar restriction of GABAergic neurons suggests that they are likely embedded in spatially segregated subnetworks.
Figure 6. Laminar distribution pattern of GABAergic subpopulations in barrel, primary visual and prelimbic cortices.
(A) Laminar distribution of VIP neurons and their two subpopulations VIP/CR and VIP/CCK. (B) Laminar distribution of SST neurons and their two subpopulations SST/CR and SST/nNOS. VIP and SST neurons are labeled by a Cre activated H2B-GFP reporter (LSL-HG), while intersectional lines are labeled by a Cre and Flp activated tdTomato reporter (Ai65). Data points: mean ± standard deviation (see Table S5 for actual numbers). N = 4 in VIP, VIP/CR, SST, SST/CR. N=3 in VIP/CCK and SST/nNOS. Scale bar: 200μm. (See also Table S5.)
Conversion of an embryonic fate specification signal to constitutive recombinase expression enables viral targeting of chandelier cells
ChCs are one of the most distinctive types of cortical GABAergic neurons that specifically innervate pyramidal neurons at their axon initial segment (Somogyi et al., 1982), the site of action potential generation, and might exert powerful control over the firing of a pyramidal neuron ensemble (Woodruff et al., 2010). Genetic fate mapping using the Nkx2.1-CreER driver revealed the developmental origin of ChCs and allowed reliable morphological labeling (Taniguchi et al., 2013). Since Nkx2.1 is mainly expressed in progenitors and no specific markers have been identified for postmitotic ChCs, it remains difficult to monitor and manipulate ChCs in mature cortex.
We have designed a strategy that converts the developmentally transient Nkx2.1-CreER induction to a constitutive Flp expression, which allows specific and reliable viral targeting of ChCs in mature animals. We have generated a mouse line, Rosa26-loxPSTOPloxP-Flpo (LSL-Flp), in which Flp expression is conditional upon Cre-mediated excision of a transcription STOP cassette (Figure 1C). In Nkx2.1-CreER;LSL-Flp mice, late embryonic tamoxifen induction results in constitutive Flp expression in postmitotic ChCs throughout the mouse’s life span (Figure 7A). Flp-dependent AAV-fDIO-GFP injection in the mature primary somatosensory cortex of E17.5 tamoxifen-induced Nkx2.1-CreER; LSL-Flp mice resulted in specific and intense labeling of ChCs (Figure 7B). Morphological reconstruction revealed that, interestingly, L2 ChCs consisted of at least two subsets defined by their axon morphology. Most L2 ChCs extended dense axon arbors within L2/3, characterized by highly distinct vertically oriented cartridges of synaptic boutons (Figure 7C). Another more rare subset projected additional axonal branches to L5 and L6, with elaborate local arbors studded with cartridges of boutons (Figure 7D), consistent with a previous report using in utero electroporation labeling of ChCs (Tai et al., 2014). Such dual laminar-projecting ChCs likely coordinate the firing of different sets of pyramidal neuron ensembles spatially segregated in different cortical layers. Combined with other viral tools that express functional effectors such as ChR2, DREADDs or pseudo-typed rabies virus receptor TVA, the conversion method allows manipulation of ChCs’ activity in vivo and tracing their local and long range inputs (J.T and J.L et al, in preparation).
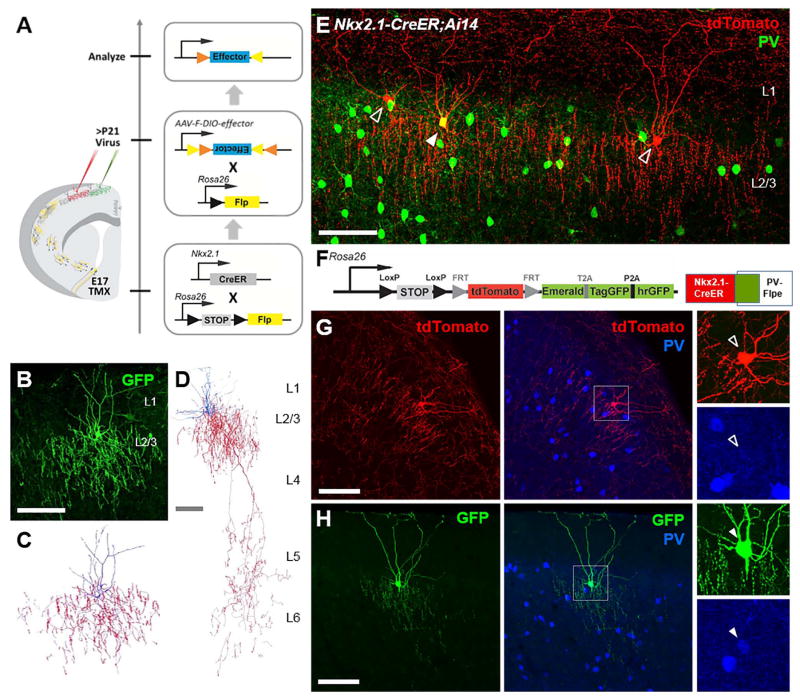
Figure 7. Combinatorial targeting of chandelier cells.
(A) Conversion strategy allows viral targeting of ChCs by engaging lineage and birth timing. The transient embryonic Nkx2.1-CreER activity induced by tamoxifen is converted into constitutive Flp recombinase expression, which enables viral targeting of ChCs in mature neocortex using a Flp-dependent (F-DIO) virus. (B) A superficial layer ChC labeled by AAV-F-DIO-GFP in the somatosensory cortex of an E17 induced Nkx2.1-CreER;LSL-Flp mouse. (C) Neurolucida reconstruction of the ChC in (F). (D) Neurolucida reconstruction of a bi-laminar targeting ChC which elaborates axonal arbors in both L2/3 and L5/6. (E) PV+ and PV− ChCs are found next to each other without obvious morphological differences. Cells were labeled in Nkx2.1-CreER;Ai14 mouse brain via tamoxifen induction at E18.5. Arrow head: PV+ ChCs. Open arrow head: PV− ChCs. Scale bar: 100μm. (F) Experiment scheme for labelling PV+ and PV− ChCs with the IS reporter in combination with Nkx2.1-CreER and PV-Flpe. (G) PV− ChCs expressing tdTomato. (H) PV+ ChCs expressing GFP. (F) and (G) were in the same brain. Scale bar: 100μm (See also Figure S7)
Our previous study revealed that ChCs consist of both PV immunopositive (PV+) and PV immunonegative (PV−) subsets (Taniguchi et al., 2013)(Figure 7E). As PV levels may reflect different functional subsets of hippocampal and neocortical basket cells (Dehorter et al., 2015; Donato et al., 2013), it is possible that PV+ and PV− ChCs have different physiological and functional properties. We designed a novel intersection and subtraction reporter (IS reporter): Rosa26-loxPSTOPloxP-Frt-Tdtomato-Frt-3XGFP. When combined with Nkx2.1-CreER; PV-Flp, the IS reporter differentially labeled PV+ ChCs with tdTomato and PV− ChCs with GFP (Figure 7F–H). This simultaneous and differential labeling will facilitate the physiological characterization of PV+ vs PV− ChCs. More generally, the high performance IS reporter will allow differential labeling of cell types using appropriate Cre and Flp driver lines. For example, using the Nkx2.1-CreER; SST –Flp;IS mouse, we were able to differentially label SST+ vs. SST− cells born at similar time from the Nkx2.1 progenitors (Figure S7).
Discussion
Efforts toward achieving a comprehensive catalogue of neuronal cell types in brain circuits face two fundamental challenges: 1) specificity – defining and working with pure cell types rather than mixtures of types, and 2) comprehensiveness – identifying many, if not all, cell types in the circuit such that their connectivity and interactions can be examined systematically toward understanding circuit operations (Seung and Sumbul, 2014). While researchers working in certain brain areas (e.g. the retina) have achieved consensus in solving these two issues and a cell catalogue appears within reach (Sanes and Masland, 2015; Seung and Sumbul, 2014), a cell catalogue of the neocortex is a longer term goal, and even the definition of cortical cell types remains contentious (DeFelipe et al., 2013). Framing these challenges in the context of understanding cortical GABAergic neurons, achieving sufficient specificity in cell targeting and cell type definition is a prerequisite for progress toward comprehensive cell type discovery. Given the scope of GABAergic neuron diversity, a major technical difficulty is simply the lack of reliable experimental access to a set of well-parsed cell populations such that different investigators can obtain and discuss results concerning the same cell population. As demonstrated by studies in the retina (Sanes and Masland, 2015) and invertebrate (Mauss et al., 2015) circuits, the more specific cell populations are studied, the more clear answers concerning cell types are likely to emerge.
Given the practical limitations in mouse engineering that severely constrain the number of gene combinations, it has remained unclear to what extent genetic methods will achieve sufficient specificity in targeting cell types, which are better defined by multiple features beyond 1 or 2 molecular markers. The key strength of our approach is not only to intersect gene expression patterns but, more importantly, to combine 2 or 3 strategically designed driver-reporter alleles with viral vectors to achieve multi-feature targeting of cell types. Further, in our scheme, gene expression patterns at different developmental stages of a cell’s life span, from transcription factors in progenitors to mature cell markers, are recruited for combinatorial targeting. These strategies and tools can be applied and modified for specific cell targeting in many brain circuits.
Intersectional genetic and viral targeting of GABAergic subpopulations
Cortical GABAergic neurons comprise three largely non-overlapping populations marked by PV, SST and 5HT3aR (~40% of the 5HT3aR population are marked by VIP) (Rudy et al., 2011). The generation of PV− (Madisen et al., 2015), SST− and VIP-Flp drivers allows intersectional targeting of more restricted subpopulations within these broad classes as well as simultaneous targeting of non-overlapping populations by combining with proper Cre lines. These knock-in Flp lines faithfully recapitulated the endogenous gene expression pattern and have identical recombination pattern as the corresponding knockin Cre lines (Taniguchi et al., 2011). This reliability of gene targeting-based approach is critical for the rational design of combinatorial cell targeting. Here we demonstrate that intersectional targeting with VIP− and SST-Flp substantially increases specificity. On the other hand, most subpopulations captured by two-gene intersections (e.g. VIP/CR, VIP/CCK, SST/CR) may still contain more than one cell type defined by axonal morphology and/or firing patterns (Figure 3, 4). SST/nNOS may represent an exception and appears to capture a rare type of categorically distinct GABAergic projection neurons (Figure 5). Recent large-scale recordings in the mouse brain slice have yielded impressive progress in characterizing neocortical GABAergic cell types (Jiang et al., 2015), yet appear to have missed rare types such as SST/nNOS and VIP/CCK. A coordinated effort that combines large scale recording and intersectional genetic targeting will likely facilitate further advances. An increased repertoire of conditional viral vectors that confer tool-gene expression (e.g.ChR2) upon Cre, Flp, and/or tTA activation (Fenno et al., 2014; Madisen et al., 2015) will further enhance the versatility and experimental power of cell targeting and manipulation.
Intersectional fate mapping of MGE progenitors
Previous studies using BrdU-based birth dating have provided evidence that cortical GABAergic neurons tend to populate the cortex in an inside-out sequence (Fairen et al., 1986; Miller, 1985; Valcanis and Tan, 2003). More recent experiments using the Olig2-CreER driver to fate map MGE progenitors (including both RGs and IPs) showed that early- and late-born interneurons do follow an inside-out layering trend and further display distinct, though diverse physiological properties (Miyoshi et al., 2007). It remains unclear to what extent cell birth date correlates and contributes to their laminar positioning and other cell phenotypes. Because IPs marked by Ascl1 are likely more fate-restricted than RGs and often only divide once to produce two neurons (Brown et al., 2011), Ascl1-CreER and Nkx2.1-Flp intersection allows more precise fate mapping of MGE IPs and birth dating of their progenies. Indeed our results revealed tight and striking laminar patterns and an inside-out sequence. This method thus enables studying the relationship among cell lineage, birth time/order, laminar location and cell phenotypes at a much higher resolution. By intersecting with other IP drivers (e.g. Dlx1-CreER), this will address whether IPs defined by different transcription factors generate distinct set of interneurons. This strategy further allows comprehensive discovery of MGE derived GABAergic neurons through sparse labeling and phenotyping.
Specific cell targeting by combining lineage, birth order, marker genes and anatomy
Genetic strategies that engage the molecular mechanisms of developmental programs such as cell lineage and birth order are particularly effective in fate mapping and targeting cell types (Harris et al., 2015; Taniguchi et al., 2013). However, transcription factors that act in progenitors often cease expression or change expression patterns in postmitotic neurons and thus are not directly suited for gaining experimental access to mature cell types. The essence of our conversion strategy is to convert a transient fate restriction signal in progenitors to a permanent “genetic handle” (e.g. Flp, tTA) in progeny neurons that allows viral access to engage anatomical features. In addition, we demonstrate that TF expression in progenitors (e.g. Nkx2.1) can be combined with phenotype-defining mature markers (e.g. PV) to target highly specific subtypes (e.g. PV+ vs. PV− ChCs) using appropriate reporters (e.g. IS). Together, these strategies combine 2 or 3 driver-reporter alleles with viral vectors to engage a spectrum of cell-defining features, thereby significantly expanding the versatility and enhancing the specificity of cell type targeting.
Toward a comprehensive discovery of MGE-derived cortical GABAergic neurons
The MGE gives rise to >60% of cortical GABAergic neurons that are categorically distinct from those generated in the CGE (Kessaris et al., 2014; Rudy et al., 2011). Because all MGE progenitors express Nkx2.1 and most MGE-derived interneurons can be parsed by PV or SST into largely non-overlapping populations (Kessaris et al., 2014; Rudy et al., 2011), these boundary-defining molecular markers provide the basis and scheme for a systematic genetic dissection. First, by combining PV− or SST-Flp drivers with proper Cre drivers, intersection/subtraction reporters, and viral vectors, it is possible to target increasingly restricted subpopulations and cell types based on mature cell markers. Second, by combining Nkx2.1-Flp driver with proper TF Cre drivers, it is possible to target restricted progenitor pools (e.g. spatial domains, RG vs. IP) and capture more specific cell populations, including laminar cohorts. Third, by combining a progenitor driver (e.g. Ascl1-CreER), a postmitotic cell driver (e.g. PV-Flp, SST-Flp), and proper reporters and viral vectors, it is possible to target cells jointly defined by lineage, birth order, marker expression, and anatomy. With increasing knowledge of cellular gene expression revealed by transcriptome analysis (Tasic et al., 2016; Zeisel et al., 2015), even a modest number of additional driver lines that expand combinatorial targeting will enable decisive progress.
In summary, although the diversity of cortical GABAergic neurons has been difficult to untangle, this diversity likely results from the manifestation of developmental programs that reliably generate a large but not infinitely large set of neuronal cardinal types, which subsequently further differentiate into diverse and distinct cell types. With genetic and viral strategies that engage these developmental and molecular programs, as well as anatomic features, it is increasingly feasible to enhance the specificity of cell targeting and move toward a more comprehensive identification of MGE-derived cell types - a major fraction of cortical GABAergic neurons.
EXPERIMENTAL PROCEDURES
Generation of new mouse lines
New driver and reporter mouse lines were generated as described before (Taniguchi et al., 2011). All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of CSHL or NYU School of Medicine in accordance with NIH guidelines, or in accordance with institutional guidelines of Institutes of Brain Science, Fudan University, China.
Immunohistochemistry
Mice were anaesthetized (using avertin or chloral hydrate) and intracardially perfused with saline followed by 4% paraformaldehyde (PFA) in 0.1 M PB. Following 24hrs of post fixation at 4°C, brain slices at 50μm or 75μm thickness were sectioned with a Leica 1000s vibratome, stained and imaged as described before (Taniguchi et al., 2011).
Viral injection and analysis
Adult mice were anesthetized using a ketamine/xylazine mixture and stereotactic injections were performed via rodent stereotax as described (Huang et al., 2014).
Slice recording and cell morphology reconstruction
Neurons in coronal slices (300 μm) containing the barrel field of the somatosensory cortex from P21–30 mice were recorded, filled with biocytin and reconstructed as described before (Xu et al., 2013).
STP imaging
Perfused and post-fixed brain from adult mice were embedded in oxidized agarose and imaged with TissueCyte 1000 (Tissuevision) as described before (Kim et al., 2015; Ragan et al., 2012).
For more information, please refer to the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Designed strategies for combinatorial targeting of GABAergic neurons and progenitors
Generated new drivers, reporters, viral vectors that improve targeting specificity
Reported intersectional fate mapping of GABAergic progenitors and laminar cohorts
Combined genetic and viral method to target cells based on lineage and birth time
Acknowledgments
We are grateful to Hongkui Zeng for providing Ai3 and Ai65 vectors and the Ai65 mouse line, Dawen Cai, Jeff Lichtman and Joshua Sanes for sharing the Brainbow vector and antibodies, Sang Yong Kim for help with generation of knock in mice, Adam Kepecs and Hiroki Taniguchi, Dhananjay Huilgol for comments on the manuscript. This work was supported in part by NIH 5U01 MH078844-05, CSHL Robertson Neuroscience Fund to Z.J.H., National Natural Science Foundation of China 31471037, 91432106, 31421091 and Shanghai Yangfan 14YF1400500 to M.H., NARSAD Young Investigator Grant to Y.K., NIH U01MH105971 to P.O., and NIH R01NS30989 and P01NS074972 to B.R., J.T, S.M.K. and J.M.L. are supported by NRSA F30 Medical Scientist Predoctoral Fellowships.
Footnotes
AUTHOR CONTRIBUTIONS
Z.J.H conceived and organized the study. M.H. and Z.J.H. designed the experiments. M.H., P.W and Z.J.H designed and generated all new knockin mouse lines. J.T., M.H. and P.W. designed and generated all new AAVs. M.H., S.M.K, Y.C., Y.H., L.G., and J.M.L. conducted mouse breeding, tamoxifen induction, immunostaining, imaging and quantification. J.T., J.M.L., and L.G. conducted virus injection experiments. Y. K. P.O, M.H., J.M.L. performed STP experiments. S.L, M.N. I.K. and B.R performed electrophysiology recording and single cell reconstruction. M.H. and Z.J.H wrote the manuscript, with input from all other co-authors.
References
- Bayraktar T, Welker E, Freund TF, Zilles K, Staiger JF. Neurons immunoreactive for vasoactive intestinal polypeptide in the rat primary somatosensory cortex: morphology and spatial relationship to barrel-related columns. J Comp Neurol. 2000;420:291–304. [PubMed] [Google Scholar]
- Brown KN, Chen S, Han Z, Lu CH, Tan X, Zhang XJ, Ding L, Lopez-Cruz A, Saur D, Anderson SA, et al. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 2011;334:480–486. doi: 10.1126/science.1208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin JA, Carlen M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauli B, Audinat E, Lambolez B, Angulo MC, Ropert N, Tsuzuki K, Hestrin S, Rossier J. Molecular and physiological diversity of cortical nonpyramidal cells. J Neurosci. 1997;17:3894–3906. doi: 10.1523/JNEUROSCI.17-10-03894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauli B, Porter JT, Tsuzuki K, Lambolez B, Rossier J, Quenet B, Audinat E. Classification of fusiform neocortical interneurons based on unsupervised clustering. Proc Natl Acad Sci U S A. 2000;97:6144–6149. doi: 10.1073/pnas.97.11.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauli B, Tong XK, Rancillac A, Serluca N, Lambolez B, Rossier J, Hamel E. Cortical GABA interneurons in neurovascular coupling: relays for subcortical vasoactive pathways. J Neurosci. 2004;24:8940–8949. doi: 10.1523/JNEUROSCI.3065-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauli B, Zhou X, Tricoire L, Toussay X, Staiger JF. Revisiting enigmatic cortical calretinin-expressing interneurons. Front Neuroanat. 2014;8:52. doi: 10.3389/fnana.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelipe J, Lopez-Cruz PL, Benavides-Piccione R, Bielza C, Larranaga P, Anderson S, Burkhalter A, Cauli B, Fairen A, Feldmeyer D, et al. New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat Rev Neurosci. 2013;14:202–216. doi: 10.1038/nrn3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehorter N, Ciceri G, Bartolini G, Lim L, del Pino I, Marin O. Tuning of fast-spiking interneuron properties by an activity-dependent transcriptional switch. Science. 2015;349:1216–1220. doi: 10.1126/science.aab3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato F, Rompani SB, Caroni P. Parvalbumin-expressing basket-cell network plasticity induced by experience regulates adult learning. Nature. 2013;504:272–276. doi: 10.1038/nature12866. [DOI] [PubMed] [Google Scholar]
- Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods in enzymology. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- Fairen A, Cobas A, Fonseca M. Times of generation of glutamic acid decarboxylase immunoreactive neurons in mouse somatosensory cortex. J Comp Neurol. 1986;251:67–83. doi: 10.1002/cne.902510105. [DOI] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nature methods. 2014 doi: 10.1038/nmeth.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JL, Puelles L, Marin O. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Tucciarone JM, Espinosa JS, Sheng N, Darcy DP, Nicoll RA, Huang ZJ, Stryker MP. A cortical circuit for gain control by behavioral state. Cell. 2014;156:1139–1152. doi: 10.1016/j.cell.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman DM, Marin O. Generation of interneuron diversity in the mouse cerebral cortex. Eur J Neurosci. 2010;31:2136–2141. doi: 10.1111/j.1460-9568.2010.07267.x. [DOI] [PubMed] [Google Scholar]
- Hansen DV, Lui JH, Flandin P, Yoshikawa K, Rubenstein JL, Alvarez-Buylla A, Kriegstein AR. Non-epithelial stem cells and cortical interneuron production in the human ganglionic eminences. Nat Neurosci. 2013;16:1576–1587. doi: 10.1038/nn.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KD, Shepherd GM. The neocortical circuit: themes and variations. Nat Neurosci. 2015;18:170–181. doi: 10.1038/nn.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RM, Pfeiffer BD, Rubin GM, Truman JW. Neuron hemilineages provide the functional ground plan for the Drosophila ventral nervous system. Elife. 2015:4. doi: 10.7554/eLife.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwell CC, Fuentealba LC, Gonzalez-Cerrillo A, Parker PR, Gertz CC, Mazzola E, Garcia MT, Alvarez-Buylla A, Cepko CL, Kriegstein AR. Wide Dispersion and Diversity of Clonally Related Inhibitory Interneurons. Neuron. 2015;87:999–1007. doi: 10.1016/j.neuron.2015.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko TS, Perez FA, Scouras AD, Stoll EA, Gale SD, Luquet S, Phillips PEM, Kremer EJ, Palmiter RD. Cre recombinase-mediated restoration of nigrostriatal dopamine in dopamine-deficient mice reverses hypophagia and bradykinesia. P Natl Acad Sci USA. 2006;103:8858–8863. doi: 10.1073/pnas.0603081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ. Toward a genetic dissection of cortical circuits in the mouse. Neuron. 2014;83:1284–1302. doi: 10.1016/j.neuron.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang ZJ, Taniguchi H, He M, Kuhlman S. Cre-dependent adeno-associated virus preparation and delivery for labeling neurons in the mouse brain. Cold Spring Harb Protoc. 2014;2014:190–194. doi: 10.1101/pdb.prot080382. [DOI] [PubMed] [Google Scholar]
- Jiang X, Shen S, Cadwell CR, Berens P, Sinz F, Ecker AS, Patel S, Tolias AS. Principles of connectivity among morphologically defined cell types in adult neocortex. Science. 2015;350:aac9462. doi: 10.1126/science.aac9462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Kubota Y. Neurochemical features and synaptic connections of large physiologically-identified GABAergic cells in the rat frontal cortex. Neuroscience. 1998;85:677–701. doi: 10.1016/s0306-4522(97)00685-4. [DOI] [PubMed] [Google Scholar]
- Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, Magno L, Rubin AN, Oliveira MG. Genetic programs controlling cortical interneuron fate. Curr Opin Neurobiol. 2014;26:79–87. doi: 10.1016/j.conb.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilduff TS, Cauli B, Gerashchenko D. Activation of cortical interneurons during sleep: an anatomical link to homeostatic sleep regulation? Trends Neurosci. 2011;34:10–19. doi: 10.1016/j.tins.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Ables JL, Dickel LK, Eisch AJ, Johnson JE. Ascl1 (Mash1) defines cells with long-term neurogenic potential in subgranular and subventricular zones in adult mouse brain. PLoS One. 2011;6:e18472. doi: 10.1371/journal.pone.0018472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Venkataraju KU, Pradhan K, Mende C, Taranda J, Turaga SC, Arganda-Carreras I, Ng L, Hawrylycz MJ, Rockland KS, et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 2015:292–305. doi: 10.1016/j.celrep.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausberger T, Somogyi P. Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science. 2008;321:53–57. doi: 10.1126/science.1149381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer EJ. Gene transfer to the central nervous system: Current state of the art of the viral vectors. Curr Genomics. 2005;6:13–37. [Google Scholar]
- Kubota Y. Untangling GABAergic wiring in the cortical microcircuit. Curr Opin Neurobiol. 2014;26:7–14. doi: 10.1016/j.conb.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Hattori R, Yui Y. Three distinct subpopulations of GABAergic neurons in rat frontal agranular cortex. Brain Res. 1994;649:159–173. doi: 10.1016/0006-8993(94)91060-x. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Shigematsu N, Karube F, Sekigawa A, Kato S, Yamaguchi N, Hirai Y, Morishima M, Kawaguchi Y. Selective coexpression of multiple chemical markers defines discrete populations of neocortical GABAergic neurons. Cereb Cortex. 2011;21:1803–1817. doi: 10.1093/cercor/bhq252. [DOI] [PubMed] [Google Scholar]
- Lee S, Hjerling-Leffler J, Zagha E, Fishell G, Rudy B. The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J Neurosci. 2010;30:16796–16808. doi: 10.1523/JNEUROSCI.1869-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kruglikov I, Huang ZJ, Fishell G, Rudy B. A disinhibitory circuit mediates motor integration in the somatosensory cortex. Nat Neurosci. 2013;16:1662–1670. doi: 10.1038/nn.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Hioki H, Konno M, Pan S, Nakamura H, Nakamura KC, Furuta T, Li JL, Kaneko T. Expression of gap junction protein connexin36 in multiple subtypes of GABAergic neurons in adult rat somatosensory cortex. Cereb Cortex. 2011;21:2639–2649. doi: 10.1093/cercor/bhr051. [DOI] [PubMed] [Google Scholar]
- Ma Y, Hu H, Berrebi AS, Mathers PH, Agmon A. Distinct subtypes of somatostatin-containing neocortical interneurons revealed in transgenic mice. J Neurosci. 2006;26:5069–5082. doi: 10.1523/JNEUROSCI.0661-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magno L, Oliveira MG, Mucha M, Rubin AN, Kessaris N. Multiple embryonic origins of nitric oxide synthase-expressing GABAergic neurons of the neocortex. Front Neural Circuits. 2012;6:65. doi: 10.3389/fncir.2012.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- Markram H, Muller E, Ramaswamy S, Reimann MW, Abdellah M, Sanchez CA, Ailamaki A, Alonso-Nanclares L, Antille N, Arsever S, et al. Reconstruction and Simulation of Neocortical Microcircuitry. Cell. 2015;163:456–492. doi: 10.1016/j.cell.2015.09.029. [DOI] [PubMed] [Google Scholar]
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- Mauss AS, Pankova K, Arenz A, Nern A, Rubin GM, Borst A. Neural Circuit to Integrate Opposing Motions in the Visual Field. Cell. 2015;162:351–362. doi: 10.1016/j.cell.2015.06.035. [DOI] [PubMed] [Google Scholar]
- Miller MW. Cogeneration of retrogradely labeled corticocortical projection and GABA-immunoreactive local circuit neurons in cerebral cortex. Brain Res. 1985;355:187–192. doi: 10.1016/0165-3806(85)90040-9. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJ, Takebayashi H, Fishell G. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig2-expressing precursors. J Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, Johnson JE, Machold RP, Fishell G. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz W, Tremblay R, Rudy B. Channelrhodopsin-assisted patching: in vivo recording of genetically and morphologically identified neurons throughout the brain. Cell Rep. 2014;9:2304–2316. doi: 10.1016/j.celrep.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olah S, Fule M, Komlosi G, Varga C, Baldi R, Barzo P, Tamas G. Regulation of cortical microcircuits by unitary GABA-mediated volume transmission. Nature. 2009;461:1278–1281. doi: 10.1038/nature08503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrenoud Q, Geoffroy H, Gauthier B, Rancillac A, Alfonsi F, Kessaris N, Rossier J, Vitalis T, Gallopin T. Characterization of Type I and Type II nNOS-Expressing Interneurons in the Barrel Cortex of Mouse. Front Neural Circuits. 2012;6:36. doi: 10.3389/fncir.2012.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petilla Interneuron Nomenclature G. Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsaki G, Cauli B, Defelipe J, et al. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 2008;9:557–568. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat Neurosci. 2013;16:1068–1076. doi: 10.1038/nn.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi HJ, Hangya B, Kvitsiani D, Sanders JI, Huang ZJ, Kepecs A. Cortical interneurons that specialize in disinhibitory control. Nature. 2013;503:521–524. doi: 10.1038/nature12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronneke A, Scheuer B, Wagener RJ, Mock M, Witte M, Staiger JF. Characterizing VIP Neurons in the Barrel Cortex of VIPcre/tdTomato Mice Reveals Layer-Specific Differences. Cereb Cortex. 2015;25:4854–4868. doi: 10.1093/cercor/bhv202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan T, Kadiri LR, Venkataraju KU, Bahlmann K, Sutin J, Taranda J, Arganda-Carreras I, Kim Y, Seung HS, Osten P. Serial two-photon tomography for automated ex vivo mouse brain imaging. Nat Methods. 2012;9:255–258. doi: 10.1038/nmeth.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux L, Buzsaki G. Tasks for inhibitory interneurons in intact brain circuits. Neuropharmacology. 2015;88:10–23. doi: 10.1016/j.neuropharm.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Developmental neurobiology. 2011;71:45–61. doi: 10.1002/dneu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Seung HS, Sumbul U. Neuronal cell types and connectivity: lessons from the retina. Neuron. 2014;83:1262–1272. doi: 10.1016/j.neuron.2014.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somogyi P, Freund TF, Cowey A. The axo-axonic interneuron in the cerebral cortex of the rat, cat and monkey. Neuroscience. 1982;7:2577–2607. doi: 10.1016/0306-4522(82)90086-0. [DOI] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cereb Cortex. 2009;19(Suppl 1):i1–10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 1999;126:3359–3370. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- Tai Y, Janas JA, Wang CL, Van Aelst L. Regulation of chandelier cell cartridge and bouton development via DOCK7-mediated ErbB4 activation. Cell Rep. 2014;6:254–263. doi: 10.1016/j.celrep.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Tomioka R. Long-Range GABAergic Connections Distributed throughout the Neocortex and their Possible Function. Front Neurosci. 2010;4:202. doi: 10.3389/fnins.2010.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Lu J, Huang ZJ. The spatial and temporal origin of chandelier cells in mouse neocortex. Science. 2013;339:70–74. doi: 10.1126/science.1227622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka R, Okamoto K, Furuta T, Fujiyama F, Iwasato T, Yanagawa Y, Obata K, Kaneko T, Tamamaki N. Demonstration of long-range GABAergic connections distributed throughout the mouse neocortex. Eur J Neurosci. 2005;21:1587–1600. doi: 10.1111/j.1460-9568.2005.03989.x. [DOI] [PubMed] [Google Scholar]
- Tricoire L, Kubota Y, Cauli B. Cortical NO interneurons: from embryogenesis to functions. Front Neural Circuits. 2013;7:105. doi: 10.3389/fncir.2013.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcanis H, Tan SS. Layer specification of transplanted interneurons in developing mouse neocortex. J Neurosci. 2003;23:5113–5122. doi: 10.1523/JNEUROSCI.23-12-05113.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Sharma K, Deng HX, Siddique T, Grisotti G, Liu E, Roos RP. Restricted expression of mutant SOD1 in spinal motor neurons and interneurons induces motor neuron pathology. Neurobiol Dis. 2008;29:400–408. doi: 10.1016/j.nbd.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Wang Y, Toledo-Rodriguez M, Gupta A, Wu C, Silberberg G, Luo J, Markram H. Anatomical, physiological and molecular properties of Martinotti cells in the somatosensory cortex of the juvenile rat. J Physiol. 2004;561:65–90. doi: 10.1113/jphysiol.2004.073353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff AR, Anderson SA, Yuste R. The enigmatic function of chandelier cells. Front Neurosci. 2010;4:201. doi: 10.3389/fnins.2010.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Jeong HY, Tremblay R, Rudy B. Neocortical somatostatin-expressing GABAergic interneurons disinhibit the thalamorecipient layer 4. Neuron. 2013;77:155–167. doi: 10.1016/j.neuron.2012.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Roby KD, Callaway EM. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol. 2006;499:144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.