Rafiq et al. demonstrate that the small G protein ARF1 and its activator, cytohesin 2 (ARNO), are required for podosome formation in macrophage-like cells and fibroblasts. Inhibition of ARNO-ARF1 signaling results in increased RhoA activity and disassembly of podosomes in a myosin-IIA–dependent fashion. In fibroblasts that normally do not form podosomes, constitutively active ARF1 induces actin-rich puncta associated with sites of matrix degradation, putative precursors of podosomes.
Abstract
Podosomes represent a class of integrin-mediated cell-matrix adhesions formed by migrating and matrix-degrading cells. We demonstrate that in macrophage-like THP1 cells and fibroblasts stimulated to produce podosomes, down-regulation of the G-protein ARF1 or the ARF1 guanine nucleotide exchange factor, ARNO, by small, interfering RNA or pharmacological inhibitors led to striking podosome elimination. Concomitantly, treatments inducing podosome formation increased the level of guanosine triphosphate (GTP)–bound ARF1. ARNO was found to colocalize with the adhesive rings of podosomes, whereas ARF1 was localized to vesicular structures transiently contacting podosome rings. Inhibition of ARF1 led to an increase in RhoA-GTP levels and triggered assembly of myosin-IIA filaments in THP1 cells, whereas the suppression of myosin-IIA rescued podosome formation regardless of ARF1 inhibition. Finally, expression of constitutively active ARF1 in fibroblasts induced formation of putative podosome precursors: actin-rich puncta coinciding with matrix degradation sites and containing proteins of the podosome core but not of the adhesive ring. Thus, ARNO-ARF1 regulates formation of podosomes by inhibition of RhoA/myosin-II and promotion of actin core assembly.
Introduction
Podosomes are a distinctive form of integrin-mediated cell-matrix adhesion typical of monocyte-derived cells but under some circumstances produced by cells of other lineages. They usually appear as micrometer-sized radially symmetrical protrusions containing central actin cores (height ∼2 µm) rooted in the cytoplasm surrounded by matrix-associated “adhesive rings” (∼1-µm diameter) enriched in integrins and plaque proteins such as talin, paxillin, vinculin, and Tks5 (Calle et al., 2006; Wiesner et al., 2010; Murphy and Courtneidge, 2011; Cox and Jones, 2013; Labernadie et al., 2014; Meddens et al., 2014; Seano et al., 2014). In the majority of cell types, podosomes form arrays consisting of numerous individual podosomes connected to each other via a mesh of F-actin–containing links containing myosin-II (Cox et al., 2011; van den Dries et al., 2013; Panzer et al., 2016). Individual podosome-like structures formed by invasive cancer cells are more stable, protrusive, and larger in size than normal podosomes and are often termed invadopodia (Gimona et al., 2008; Murphy and Courtneidge, 2011). Podosomes participate in the processes of cell migration and invasion as well as degradation of ECM via secretion of matrix metalloproteinases (MMPs; Gawden-Bone et al., 2010; Wiesner et al., 2010; Linder and Wiesner, 2015; El Azzouzi et al., 2016).
Cells of monocytic origin (for example, cultured macrophage-like THP1 cells) form numerous podosomes upon stimulation with TGFβ or increasing PKC activity by phorbol esters (e.g., PMA). Moreover, upon appropriate stimulation, even podosome-lacking cells can be forced to form podosome-like structures. In particular, expression of constitutively active Src in fibroblasts triggers formation of high-order adhesion structures termed podosome rosettes, which are capable of degrading the ECM (Tarone et al., 1985). More recently, we have shown that nontransformed fibroblasts that typically do not form podosomes develop podosome-like adhesions under conditions in which a cell cannot apply strong traction force to nascent integrin clusters, such as spreading on fluid arginylglycylaspartic acid (RGD)–functionalized lipid bilayers, where stress fibers fail to assemble (Yu et al., 2013).
A key process in podosome formation is a local polymerization of actin cores primarily mediated by Arp2/3 complex activated by Wiskott–Aldrich syndrome protein (WASP; Machesky and Insall, 1998; Linder et al., 1999; Burns et al., 2001). In turn, WASP activation depends largely on the activity of the small G protein Cdc42 and can be regulated by WASP-interacting protein (WIP; Abdul-Manan et al., 1999; Calle et al., 2004; Monypenny et al., 2011; Schachtner et al., 2013; Vijayakumar et al., 2015). Indeed, microinjection of dominant-negative Cdc42 has been shown to significantly impair podosome formation in human dendritic cells (Burns et al., 2001). Similarly, podosome formation is impaired in cells microinjected with dominant-negative Rac1 (Burns et al., 2001), as well as in Rac1- and especially Rac2-depleted cells (Wheeler et al., 2006), although the downstream pathways are not yet elucidated. Conversely, active RhoA, which typically promotes assembly of stress fibers and focal adhesions, has been generally described to be low in podosome-forming cells (Pan et al., 2011; Yu et al., 2013), and microinjection of active RhoA impairs podosome formation (Burns et al., 2001).
Although the role of Rho family GTPases in podosome formation is relatively well documented, the function of the ARF family of G proteins is essentially unknown. Even though these proteins are considered mainly as regulators of membrane traffic, some evidence exists that they also participate in a variety of processes related to regulation of the actin cytoskeleton and involved in cross talk with the G proteins of the Rho family. In particular, ARF1, the most abundant ARF family member, known to recruit the coatomer complexes for vesicle budding in the Golgi (Donaldson and Jackson, 2011), was shown to be required for clathrin-independent endocytosis (Kumari and Mayor, 2008), as well as for formation of “ventral actin structures” in some cell types (Caviston et al., 2014). Thus ARF1 is a potentially interesting candidate for function as a podosome regulator because it could control fundamental systems involved in podosome formation, actin cytoskeleton, and the plasma membrane.
In this study, we demonstrate that regardless of particular stimuli, ARF1 is required for inducing podosome formation in different cell types. Moreover, these stimuli, via an ARF exchange factor, ARNO, increase the fraction of GTP-bound ARF1 in cells. ARNO localizes to the adhesive ring of podosomes, and its inhibition interferes with podosome assembly. We demonstrate that the ARNO-ARF1 pathway regulates podosomes by inhibition of RhoA- and ROCK-dependent formation of myosin-II filaments, which antagonizes podosome integrity. In addition, constitutively active ARF1 induces formation of actin-rich puncta colocalizing with matrix degradation sites and containing podosome core markers. Our data strongly suggest a direct role for ARF1 in podosome-type adhesions and further extend the increasing number of roles for ARF1 at the plasma membrane.
Results
Depletion of endogenous ARF1 interferes with podosome formation
Stimulation by either TGFβ1 or the PKC activator, PMA, has been previously used as a model system to study podosome formation and dynamics in several cell types (Tatin et al., 2006; Varon et al., 2006; Burger et al., 2011; Monypenny et al., 2011). Consistent with numerous previous studies, we define podosomes as F-actin–rich spots with a diameter of ∼0.5 µm surrounded by an approximately ring-shaped vinculin-rich zone. We consider a cell as “podosome-forming” if it has more than 10 morphologically identifiable podosomes.
In this study, 85 ± 4.7% (mean ± SD, n = 3 independent experiments) of cells of the human monocytic cell line, THP1, plated on fibronectin-coated substrata in the presence of TGFβ1, formed podosomes after 24 h, with 55 ± 3.2 (mean ± SEM, n = 212 cells) podosomes per cell. The corresponding numbers for PMA-stimulated cells were 88 ± 5.5% and 140 ± 19.5 (n = 80 cells), respectively. For cells plated on fibronectin in the absence of any additional stimuli, the percentage of podosome-forming cells was only 15 ± 4.7% (n = 3 independent experiments), and even among these cells, the mean number of podosomes did not exceed 22 ± 8.7 (n = 58 cells).
To investigate the role of ARF1 in podosome dynamics, we depleted ARF1 in TGFβ1-treated THP1 cells by siRNA. Immediately before plating, cells were electroporated with ARF1 or control siRNA and seeded onto fibronectin in the presence of TGFβ1. We observed that maximum silencing (>95%) was achieved by 48 h (Fig. 1 A).
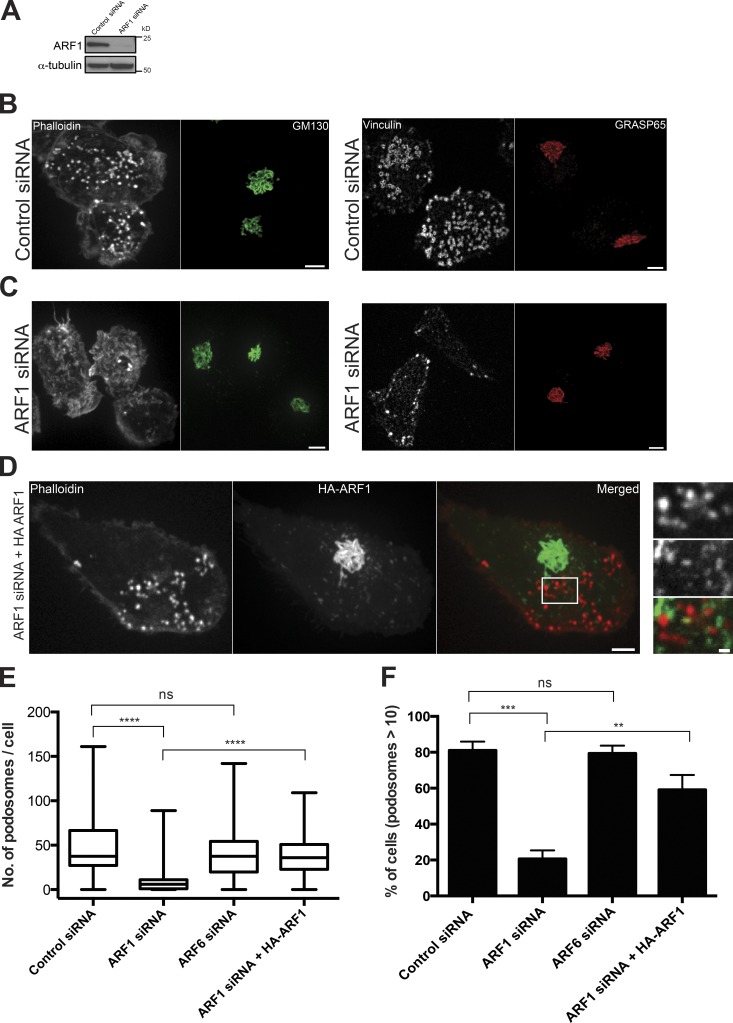
Figure 1.
Depletion of endogenous ARF1 disrupts podosomes. (A) Western blot showing ARF1 levels in cells treated with scrambled (control) or ARF1 siRNA; α-tubulin was used as a loading control. (B and C) ARF1 knockdown leads to disruption of podosomes but not the Golgi apparatus. Actin labeled with phalloidin (left) and vinculin visualized by antibody staining (right) in control (B) and ARF1 siRNA-transfected (C) THP1 cells 48 h after TGFβ1 stimulation. The Golgi apparatus in the same cells was visualized by staining with antibody against cis-Golgi proteins, GM130 (left, green) and GRASP65 (right, red). Bars, 5 µm. (D) Expression of HA-tagged bovine ARF1 in ARF1-depleted human THP1 cells rescues podosome formation. Podosomes are visualized by phalloidin staining (left) and HA-ARF1 by immunostaining with anti-HA antibody (middle); merged image is shown on the right. Bar, 5 µm. HA-ARF1 was localized to Golgi and to punctate structures shown with high magnification in the right panels representing the enlarged area boxed to the left. Bar, 1 µm. Labeling in the right panel of D shows actin (top), HA-ARF1 (middle), and merged image of both (bottom). Width of the images is 7 µm. (E and F) Quantification of the effect of ARF1 and ARF6 knockdown on podosome integrity. Both number of podosomes per cell (E) and percentage of cells having more than 10 podosomes (F) decreased upon ARF1 but not ARF6 knockdown. Mean ± SD is indicated.This effect was rescued by expression of exogenous HA-ARF1. The graphs represent results of three independent experiments with 100–200 cells used for each group. The numbers of podosomes per cell are presented as a box-and-whiskers plot. Values of median, lower and upper quartiles (box), minimum and maximum values (whiskers) are indicated. The percentages of cells with more than 10 podosomes are presented as mean ± SD. The significance of the difference between groups was estimated by two-tailed Student’s t test. ns, P > 0.05 (nonsignificant); **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Depletion of ARF1 led to a dramatic decrease of podosome number in TGFβ1-treated cells (Fig. 1, B and C). Both the number of podosomes per cell and the percentage of podosome-forming cells significantly dropped upon ARF1 depletion (Fig. 1, D–F). Although the mean number of podosomes per cell and the percentage of podosome-forming cells in cells transfected with control siRNA did not differ from aforementioned control numbers, the cells transfected with ARF1 siRNA had a mean of only 8 ± 1.7 (n = 96 cells) podosomes per cell. Total intensity of F-actin–containing nonpodosomal structures at the ventral surface of the cells became somewhat higher in ARF1-depleted cells (Fig. 1, B and C, left). At the same time, the vinculin-containing nonpodosomal structures at the cell periphery that can be classified as small focal adhesions were not apparently affected (Fig. 1, B and C, right). Depletion of ARF1 by siRNA did not affect the integrity of the Golgi apparatus as visualized by cis-Golgi markers GM130/GRASP65 (Fig. 1, B and C), in agreement with previous publications (Volpicelli-Daley et al., 2005; Szul et al., 2007; Nakai et al., 2013).
The effect of ARF1 siRNA on podosome number was specific because it could be fully reversed by expression of exogenous bovine HA-ARF1 insensitive to human ARF1 siRNA (Fig. 1, D–F). Interestingly, exogenous HA-ARF1 was often localized in the spots adjacent to the podosomes of transfected cells (Fig. 1 D, right). See Fig. 2 for a detailed analysis of localization dynamics. Significantly, we found that loss of podosome induction caused by ARF1 depletion was not exclusive to TGFβ1 stimulation, because after PMA stimulation (Fig. S1, A–I), ARF1-depleted cells demonstrated a significant decrease in both the number of podosomes and the percentage of podosome-forming cells compared with control cells (Fig. S1, A, D, F, and G).
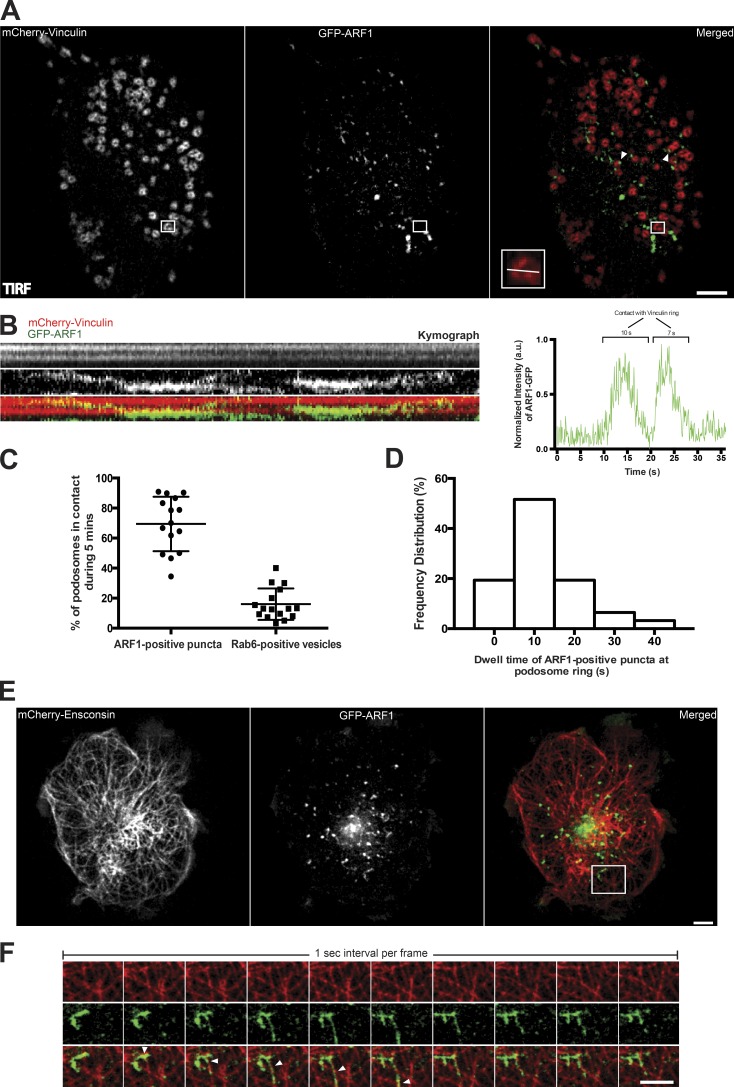
Figure 2.
Localization and dynamics of ARF1 puncta in TGFβ1-stimulated THP1 cells. (A) TIRF image of the ventral surface of cell with podosomes labeled by mCherry-vinculin (left) and ARF1 puncta labeled by GFP-ARF1 (middle). Merged image (right) shows nonrandom distribution of ARF1 puncta with a tendency to colocalize to podosome periphery (see white arrowheads). Bar, 5 µm. Boxed area (2.5 × 2.5 µm2) contains a podosome whose colocalization dynamics with ARF1 puncta is presented in B. (B, left) Kymograph representing fluorescent intensities in a line scan through the podosome boxed in A. Although mCherry-vinculin is stably labeled in the podosome ring (top), GFP-ARF1 was transiently concentrated at one side of the ring (middle, merged image at bottom). See also Video 1. (B, right) The time course of fluorescence intensity of GFP-ARF1 at the podosome ring. (C) Each dot corresponds to a single cell and represents percentage of podosome rings (labeled by vinculin) contacted by either ARF1-containing puncta or Rab6-containing vesicles within 5 min of image acquisition. Mean ± SD is indicated. (D) Frequency distribution of the durations of podosome contacts (in seconds) with ARF1-containing puncta (35 podosomes from 10 cells were filmed as shown in kymograph B). (E and F) GFP-ARF1 puncta moving along microtubules. (E) Left, microtubule labeling with 125-kD microtubule-associated protein, ensconsin (mCherry-ensconsin); middle, GFP-ARF1 puncta in the same cell; right, merged image. The dynamics of microtubules and ARF1 puncta in the boxed area (8 × 7.5 µm2) of C is shown in F. Bars, 5 µm. Movement of puncta along the microtubule is indicated by the arrowhead. See also Video 2.
In contrast to ARF1, depletion of ARF6 by siRNA with a silencing efficiency of >95% (Fig. S1 J) did not affect podosome induction in TGFβ1-treated THP1 cells (Fig. S2 K). Neither the mean number of podosomes per cell nor the percentage of cells forming more than 10 podosomes was significantly different from control siRNA-treated cells (Fig. 1, E and F). Collectively, these data indicate a specific role for ARF1 in podosome induction in stimulated THP1 cells.
We also examined the effect of expression of dominant-negative and constitutively active mutants of ARF1 on adhesion of THP1 cells in the presence of TGFβ1. The dominant-negative mutant, CFP-ARF1 (T31N), led to a significant decrease of cell adhesion to fibronectin under these conditions (control: 71 ± 5.5%; ARF1 T31N: 4 ± 0.3%), and the few adherent cells observed did not form podosomes (unpublished data). This behavior resembles a “non-adhesive phenotype,” described previously in the culture of normal human dendritic cells (Burns et al., 2004). Unexpectedly, the expression of a constitutively active mutant of ARF1, CFP-ARF1 (Q71L), also interfered with cell adhesion and completely prevented podosome formation. Thus, sustained high activity of ARF1 is also damaging for cell adhesion and podosome formation in THP1 cells. Overexpression of wild-type ARF1 or constitutively active ARF1 in unstimulated THP1 cells did not induce any apparent phenotypic changes. These cells remain poorly attached to the fibronectin and do not form podosomes.
ARF1-containing vesicles transiently contact podosomes
We used fluorescently tagged ARF1 to further elucidate the localization and dynamics of ARF1 in TGFβ1-stimulated THP-1 cells. Expression of GFP-ARF1 showed a predominant Golgi localization (Fig. 1 D) in agreement with previous publications (Lippincott-Schwartz et al., 1989; Sciaky et al., 1997). However, in addition to the Golgi localization, we found numerous irregular puncta throughout the cell, some of which were apparently associated with podosomes (Figs. 1 D [right] and 2 A).
We used total internal reflection fluorescence (TIRF) microscopy to explore the spatiotemporal dynamics of ARF1 puncta at the plasma membrane. GFP-ARF1 puncta displayed temporal localization at regions of podosome assemblies and made transient periodic contacts with the adhesive rings of podosomes, as marked by mCherry-vinculin in THP1 cells (Fig. 2 B and Video 1). Up to 80% of podosomes appeared to be in contact with ARF1-containing puncta during 5-min periods of observation (Fig. 2 C), with a mean dwell time of 10 ± 1.6 s (mean ± SEM; Fig. 2 D). In contrast, CFP-ARF6 does not form puncta-like structures in THP1 cells, and no preferential localization of CFP-ARF6 to regions of podosome assembly was found (Fig. S1 L). We further characterized the GFP-ARF1–associated puncta by determining whether their mobility was dependent on a cytoskeletal network. We found that GFP-ARF1 patches appeared to be traveling on microtubule tracks identified by labeling with mCherry-ensconsin (Fig. 2, E and F; and Video 2). To elucidate the nature of the ARF1 puncta, we coexpressed GFP-ARF1 with several markers of vesicular traffic carriers, Rab6, Rab7, Rab8, and Rab11 (Fig. S2, A–D). Among those, Rab11 (Welz et al., 2014) demonstrated significant colocalization with ARF1 (Fig. S2 E), suggesting that ARF1-containing puncta have a vesicular nature.
Inhibition of ARF1 activity interferes with formation of podosomes induced by diverse stimuli
To study the immediate effect of ARF1 inhibition on podosome formation, we used two inhibitors known to suppress ARF1 activity. Brefeldin A (BFA) promotes formation of complexes between GDP-bound ARF1 and Sec7 domains of ARF1 nucleotide exchange factors GBF1, BIG1, and BIG2 and prevents completion of the nucleotide exchange reaction (D’Souza-Schorey and Chavrier, 2006). SecinH3 inhibits activity of another group of ARF exchange factors, cytohesins 1–4, by binding to their Sec7 domain, without formation of a complex with ARF1 (Casanova, 2007).
Using a G-LISA assay for the measurement of ARF1-GTP levels, we demonstrate that TGFβ1 or PMA treatment of THP1 cells enhances the fraction of active, GTP-bound ARF1, whereas both SecinH3 and BFA significantly reduces it (Figs. 3 A and S1 C). Both SecinH3 and BFA treatment induced rapid disassembly of all podosomes in ∼30–40 min (Fig. 3, B–F, and Video 3). In the case of SecinH3, this process was accompanied by a burst of lamellipodial activity (Fig. 3 B); the integrity of the Golgi apparatus was not affected, nor was localization of ARF1 to Golgi and to vesicular structures in the cytoplasm (Fig. 3, B and C). Unlike SecinH3 treatment, disruption of podosomes with BFA was accompanied by loss of ARF1 localization at the Golgi and at cytoplasmic vesicular structures (Fig. 3 D), as well as structural disintegration of the Golgi apparatus, in agreement with numerous previous studies (Lippincott-Schwartz et al., 1989; Donaldson et al., 2005). Thus, comparison between the SecinH3 and BFA effects confirmed that active ARF1 is required for podosome integrity, and this function of ARF1 does not depend on its role in Golgi stabilization. A second confirmation of independence of podosomes from Golgi traffic can be inferred from experiments with knockdown of COPB1, a subunit of the COPI coatomer protein complex required for retrograde transport from trans-Golgi to cis-Golgi and ER (Beck et al., 2009). We found that COPB1 knockdown or its inhibition generated only minor effects on podosome integrity (Fig. S3, A–E). Finally, disruption of podosomes with SecinH3 or BFA still proceeded (albeit at a slower rate) in TGFβ1-stimulated THP1 cells expressing constitutively active Cdc42 (GFP–Cdc42 Q61L), a potent podosome-inducing signaling protein (Fig. S3 H). Moreover, treatment of cells with BFA or SecinH3, as well as knockdown of ARNO or ARF1, did not change the level of Cdc42-GTP in TGFβ-stimulated THP1 cells (Fig. S3, I and J).
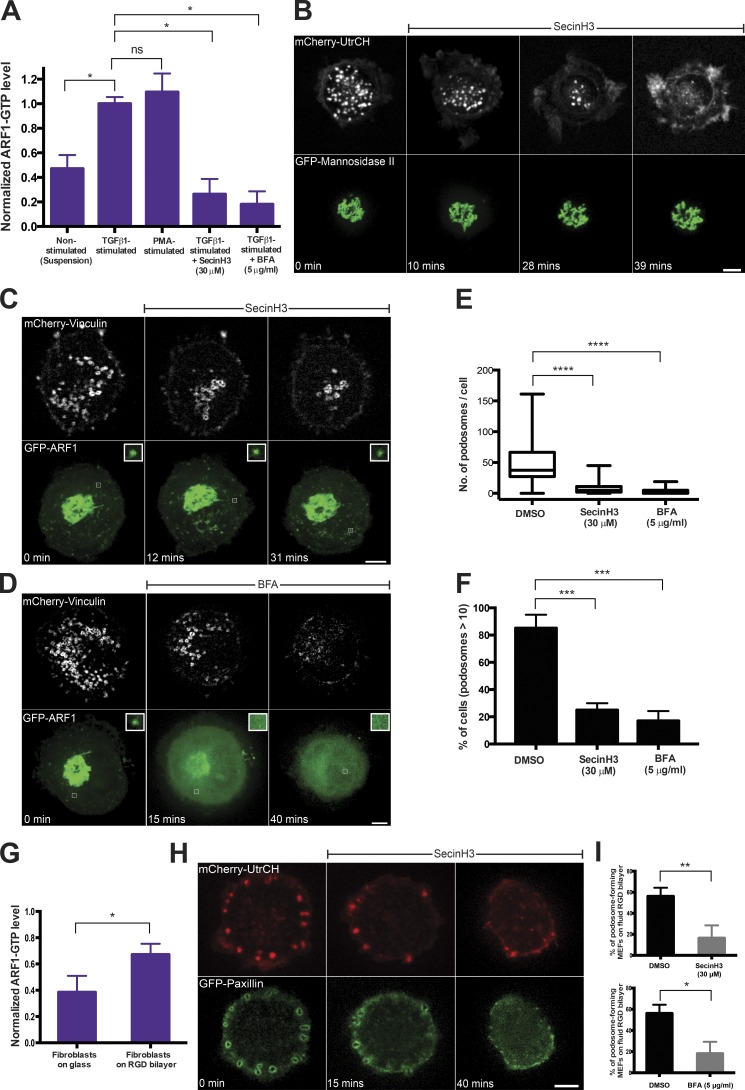
Figure 3.
ARF1-GTP levels and podosome formation. (A) Quantification of ARF1-GTP levels by G-LISA assay in control, stimulated, and inhibitor-treated THP1 cells. Both TGFβ1 and PMA increased the fraction of GTP-bound ARF1 compared with control, whereas treatment with SecinH3 or BFA dramatically reduced it. Pooled results of three independent experiments are shown. Mean ± SD is indicated. (B) Disruption of podosomes labeled with mCherry-Utrophin (UtrCH) upon treatment with SecinH3 (top). Note that integrity of the Golgi apparatus labeled with GFP-mannosidase II was preserved in the same SecinH3-treated cell (bottom). See also Video 3. (C and D) Disruption of podosomes labeled with mCherry-vinculin by SecinH3 (C, top) and BFA (D, top). Although the effect of SecinH3 in these cells was not accompanied by changes in localization of ARF1 to the Golgi and cytoplasmic puncta (C, bottom), BFA disrupted both Golgi and ARF1 puncta (D, bottom); Bars, 5 µm. Insets (1 × 1 µm2) show evolution of individual ARF1 puncta in each case. (E and F) Quantification of the effect of SecinH3 and BFA on number of podosomes per cell (E) and percentage of cells with more than 10 podosomes (F). (G) ARF1-GTP level increase in fibroblasts plated on a RGD-functionalized fluid lipid bilayer compared with fibroblasts plated on glass coverslip. Mean ± SD is indicated. (H) Effect of SecinH3 on the integrity of podosome-like structures formed by fibroblasts plated on fluid lipid bilayer. (I) Quantification of the disruptive effect of SecinH3 and BFA on podosome-like structures formed by fibroblasts on lipid bilayer. The percentage of podosome-forming cells significantly decreased upon treatment by each of the inhibitors. The data in E, F, and I are presented as indicated in the legend to Fig. 1. Pooled data of three independent experiments are presented for each group. The significance of the difference between groups was estimated by two-tailed Student’s t test. ns, P > 0.05 (nonsignificant); *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Visualization of podosomes using structured-illumination microscopy (SIM) revealed a central F-actin core surrounded by patches enriched in adhesion proteins (talin and vinculin) as well as thin F-actin–rich links connecting neighboring podosomes (Fig. S4 A and Video 4) in agreement with previous studies (Cox et al., 2011; van den Dries et al., 2013). Treatment with SecinH3 led to the rapid disappearance of the connecting links and gradual concurrent disassembly of both the actin cores and surrounding adhesive rings (Fig. S4, B and C; and Video 5).
In view of the high podosome turnover rate (van den Dries et al., 2013), gradual disruption of podosomes upon addition of SecinH3 suggests that inactivation of ARF1 changes the balance between podosome assembly and disassembly rather than completely blocking the assembly processes. The process of disassembly often proceeds through podosome fission and is accompanied by an apparent increase of podosome mobility in the plane of the plasma membrane (Fig. S4, D–F).
We next studied the effect of inhibition of ARF1 on podosome-like structures formed by fibroblast-type cells. Under standard culture conditions, mouse embryonic fibroblasts (MEFs) generally form focal adhesions, which appeared to be resistant to treatment with either BFA (Bershadsky and Futerman, 1994) or SecinH3 (Fig. S5 A). It was recently shown that fibroblasts plated on a fluid substratum (supported RGD-functionalized lipid bilayer), under conditions where they cannot exert traction forces, by default formed podosome-like adhesion structures (Yu et al., 2013). We showed that the level of GTP-ARF1 increased in MEFs plated on supported lipid bilayers (Fig. 3 G). Similarly to classic podosomes, podosome-like structures formed by MEFs plated on the lipid bilayer underwent rapid disassembly upon treatment with either BFA or SecinH3 (Fig. 3, H and I).
A well-known method of induction of podosome-like structures in fibroblast-like cells is ectopic expression of constitutively active Src. In agreement with published results (Tarone et al., 1985), expression of Src Y527F in MEFs led to the formation of prominent rosettes formed as a result of fusion of numerous podosome-like structures (Fig. S5, B and C). Treatment of such cells with either BFA or SecinH3 resulted in the gradual disassembly of these rosettes and a decrease in the number of rosette-positive cells (Fig. S5, B and C).
Altogether, these data demonstrate that ARF1 activity is required for formation/maintenance of podosome-like structure irrespective of upstream stimuli (TGFβ1, PMA, active Src, or fluid substratum). We conclude that a role for ARF1 in podosome dynamics is independent of early signaling pathways that lead to initiation of cellular differentiation to a podosome-generating phenotype.
ARNO guanine nucleotide exchange factor activates ARF1 to drive podosome formation
The mammalian ARF GTPases are activated by 15 different guanine nucleotide exchange factors (GEFs) categorized in five classes. Among these, only seven GEFs can activate ARF1: three BFA-sensitive (GBF1, BIG1, and BIG2) and four SecinH3-sensitive (cytohesins 1–4; D’Souza-Schorey and Chavrier, 2006; Donaldson and Jackson, 2011). Both BFA- and SecinH3-sensitive GEFs share a common conserved SEC7 domain that promotes GDP release and subsequent GTP binding to ARF1. SecinH3-sensitive GEFs have in addition a pleckstrin homology domain that enables them to interact with phosphoinositides at the plasma membrane (Santy et al., 1999; DiNitto et al., 2007).
We examined the effect of inhibition of several ARF1 GEFs on the process of podosome formation (Fig. S3, B and D–G). We found that expression of dominant-negative mutants of two BFA-sensitive GEFs, HA–BIG1 (E793K) and HA–BIG2 (E738K), did not prevent formation of podosomes in THP1 cells treated with TGFβ1 (Fig. S3, F and G, bottom). Conversely podosome formation was not affected by overexpression of wild-type HA-BIG1 or HA-BIG2 (Fig. S3, F and G, top). Furthermore, both the wild-type HA-BIG1 and HA-BIG2 showed predominant Golgi localization (Fig. S3, F and G) and no podosome localization, consistent with previous studies (Citterio et al., 2006; Ishizaki et al., 2008). To inhibit the activity of the third BFA-sensitive GEF, GBF1, we used a small-molecule inhibitor, Golgicide A (Sáenz et al., 2009). We found only partial dissolution of podosomes in THP1 cells treated with TGFβ1 after application of Golgicide A in a concentration that induced visible fragmentation of the Golgi complex (Fig. S3, B, D, and E). Thus, in spite of profound inhibitory effect of BFA on podosome formation, selective inhibition of the BFA-sensitive GEFs produced only minor effect on podosomes.
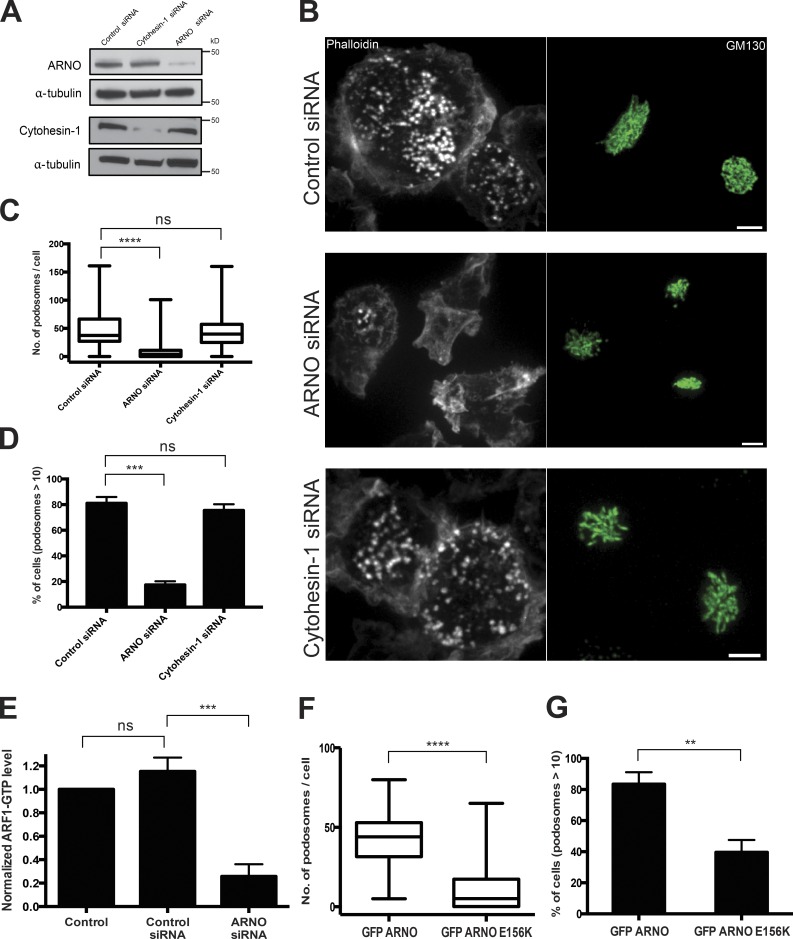
In contrast, knockdown of one of the SecinH3-sensitive GEFs, ARNO (cytohesin 2), significantly affected podosome formation in TGFβ1-activated (Fig. 4, A–D) or PMA-activated THP1 cells (Fig. S1, B, E, H, and I). Knockdown of ARNO led to a significant decrease in the number of podosomes per cell as well as the percentage of cells having more than 10 podosomes (Fig. 4, C and D). Additionally, we found that ARNO knockdown reduced ARF1 activity in THP1 cells stimulated by TGFβ1 (Fig. 4 E). Moreover, a dominant-negative mutant of ARNO (E156K) also suppressed podosome formation/maintenance (Fig. 4, F and G). The effect of ARNO knockdown on podosomes was specific, because knockdown of another SecinH3-sensitive GEF, cytohesin 1, did not inhibit podosome formation in stimulated THP1 cells (Fig. 4 B). Neither ARNO nor cytohesin 1 knockdown produced any significant effect on Golgi integrity (Fig. 4 B).
Figure 4.
Knockdown of ARF1 exchange factor ARNO (cytohesin 2), but not cytohesin 1, leads to podosome disruption. (A) Western blot showing ARNO and cytohesin 1 levels in cells treated with control (scrambled) siRNA, ARNO, or cytohesin 1 siRNAs; α-tubulin was used as a loading control. (B, top) TGFβ1-stimulated THP1 cells transfected with scrambled siRNAs; podosomes labeled with phalloidin and Golgi apparatus labeled by antibody against GM130. (middle) siRNA-mediated knockdown of ARNO disrupted podosomes, leaving the Golgi undisturbed. (bottom) Cytohesin 1 knockdown disrupted neither podosomes nor Golgi. Bars, 5 µm. (C and D) Quantification of the effect of ARNO and cytohesin 1 knockdown on number of podosomes per cell (C) and percentage of cells with more than 10 podosomes (D). (E) G-LISA quantification of ARF1-GTP level in nontransfected TGFβ1-stimulated THP1 cells (control), scrambled siRNA–transfected cells, and cells transfected with ARNO siRNA. Mean ± SD is indicated. (F and G) Quantification of the effect of expression of wild-type GFP-ARNO and dominant-negative ARNO mutant (GFP–ARNO E156K) on mean number of podosomes per cell (F) and percentage of cells with more than 10 podosomes (G). The data in C, D, F, and G are presented as indicated in the legend to Fig. 1. Pooled data of three independent experiments are presented for each group. The significance of the difference between groups was estimated by two-tailed Student’s t test. ns, P > 0.05 (nonsignificant); **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
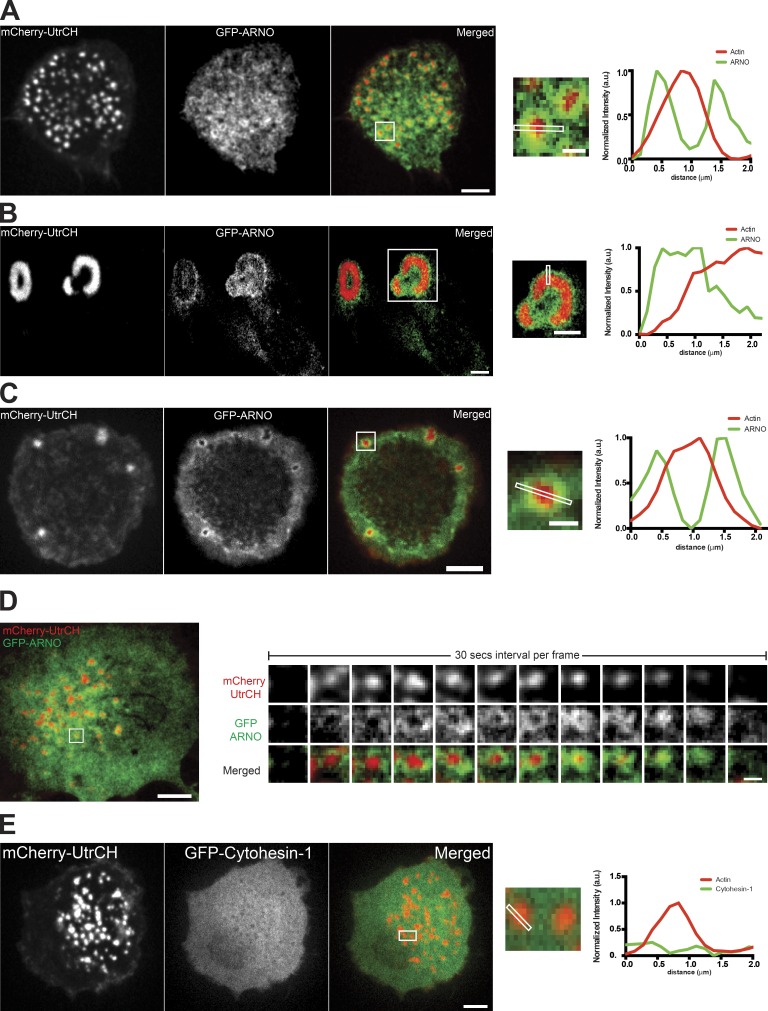
Next, we investigated the localization of ARNO and cytohesin 1 in TGFβ1-treated THP1 cells by expressing GFP-fusion construct of these GEFs. ARNO was found to localize to the rings surrounding the actin cores of podosomes (Fig. 5 A). Similarly, GFP-ARNO localized to the rim around the actin core of podosome rosettes (Fig. 5 B). Moreover, podosome-like structures formed by normal fibroblasts plated on RGD-functionalized lipid bilayers also contained ARNO in the rings surrounding the actin cores (Fig. 5 C). Live imaging of GFP-ARNO in all these situations revealed that ARNO localization at the podosome ring was stable and spanned the entire lifetime of a podosome (Fig. 5 D and Video 6). Unlike ARNO, cytohesin 1 showed diffuse localization over the plasma membrane and was not enriched at podosomes (Fig. 5 E).
Figure 5.
ARNO but not cytohesin 1 is localized to podosomes and podosome-like structures in different cell types. (A–C) Localization of F-actin marker, mCherry-UtrCH, and GFP-ARNO in TGFβ1-stimulated THP1 cell (A), active Src-transformed fibroblast (B), and fibroblast on a RGD-functionalized fluid lipid bilayer (C). Left, F-actin cores of podosomes (A), podosome rosettes (B), and podosome-like structures formed on fluid bilayer (C). (middle) GFP-ARNO localized to periphery of F-actin cores (A–C). (right) Merged images. The boxed areas (A and C: 2.5 × 2.5 µm2, bar, 1 µm; B: 14 × 14 µm2, bar, 5 µm) of merged images are enlarged and line scanned as shown in inset. The graphs on the right demonstrate intensity profiles of F-actin and ARNO in individual podosome (A), podosome rosette (B), and podosome-like structure on bilayer (C). (D) Time course of ARNO localization to the podosome periphery. Dynamics of F-actin (labeled by mCherry-UtrCH) and GFP-ARNO fluorescence intensities in the podosome shown in the boxed area (3 × 3 µm2) in the left panel are presented in the sequences in the right panels. Time interval between frames is 30 s. See also Video 6. (E) Cytohesin-1 is not localized to podosomes. (left) F-actin cores of podosomes in TGFβ1-stimulated THP1 cell. Middle, GFP–cytohesin 1 localization in the same cell. (right) Merged image. Line scanning through the individual podosome in the boxed area (4 × 1.5 µm2) of the merged image shown in inset is quantified in the graph on the right. No enrichment of GFP–cytohesin 1 at podosome core or periphery was detected. Bars, 5 µm.
Inhibition of ARF1 triggers podosome disassembly via activation of Rho and myosin-IIA
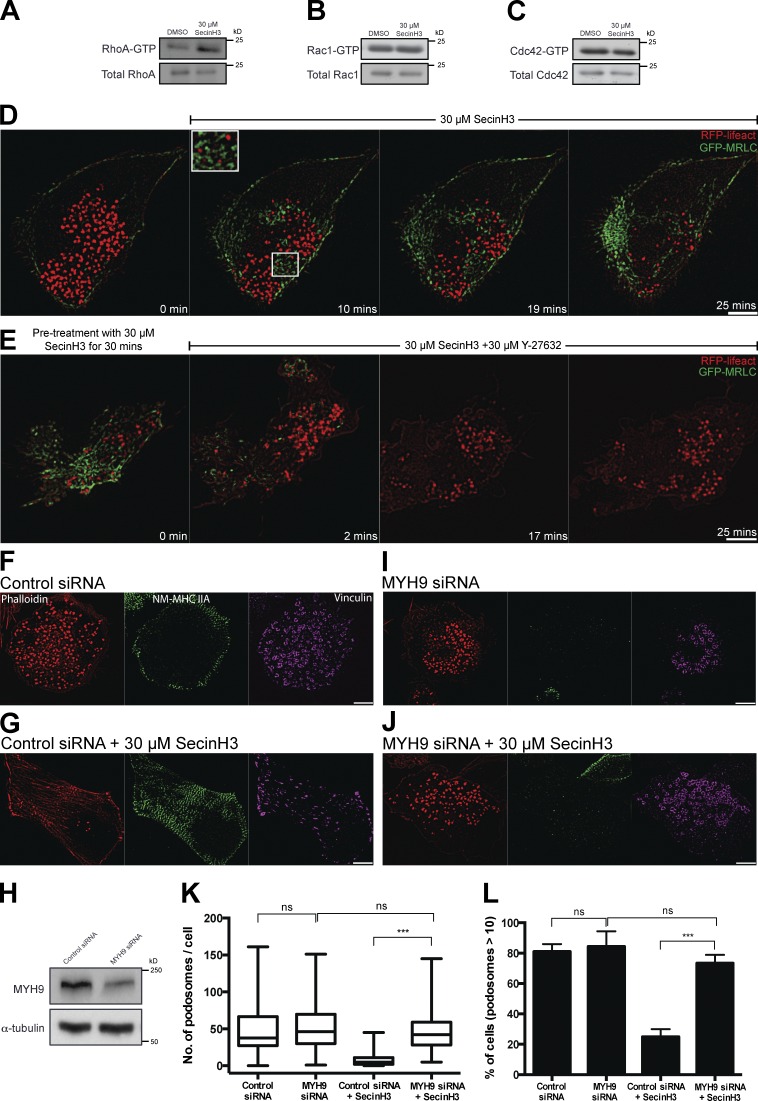
In search of downstream factors that mediate podosome disruption upon ARF1 inhibition, we checked the activity of three major Rho family G proteins in TGFβ1-treated THP1 cells. We found that the fraction of RhoA-GTP significantly increased upon inhibition of ARF1 by SecinH3 (Fig. 6 A), whereas activities of both Rac1 and Cdc42 did not change (Fig. 6, B and C). RhoA and Rho-associated kinase (ROCK) are master regulators of myosin-IIA–driven cell contractility, because ROCK-mediated activation of myosin regulatory light chain (MRLC) phosphorylation promotes assembly of myosin-II filaments as well as myosin-II motor activity (Vicente-Manzanares et al., 2009). Indeed, we have demonstrated that inhibition of ARF1 by SecinH3 promoted assembly of the myosin-II filaments as visualized by live imaging of GFP-MRLC using SIM (Fig. 6 D and Video 7). Simultaneous visualization of podosomes and myosin-II filaments revealed that podosome disappearance occurred in those cell regions enriched in myosin-II filaments (Fig. 6 D), suggesting that podosome disassembly is triggered by local activation of myosin-II–driven contractility. Indeed, treatment of ARF1-inhibited cells lacking a majority of podosomes with an inhibitor of ROCK, Y-27632, led to a burst of podosome formation concurrent with the disappearance of myosin-II filaments (Fig. 6 E and Video 8). To confirm that inhibition of ARF1 led to podosome disruption via activation of myosin-II filament assembly, we performed siRNA-mediated myosin-IIA heavy chain (MYH9) knockdown, which completely blocked formation of myosin-IIA filaments as visualized by antibody to nonmuscle (NM) myosin heavy chain IIA (Fig. 6, F–H). Myosin-II knockdown by itself did not affect podosome integrity (Fig. 6, F, I, K, and L). Although treatment of THP1 cells transfected with control siRNA by SecinH3 led to pronounced disassembly of podosomes (Fig. 6, G, K, and L), the same treatment on myosin-IIA knockdown cells did not disrupt podosomes (Fig. 6, J, K, and L).
Figure 6.
Inhibition of ARF1 activity induces RhoA activation. (A–C) 1-h incubation of TGFβ1-stimulated THP1 cells with 30 µM SecinH3 led to increase in RhoA-GTP (A) but not Rac1-GTP (B) or Cdc42-GTP (C) fractions as indicated by Western blots after pull-down assay. (D–J) SIM visualization of podosome dynamics in TGFβ1-stimulated THP1. (D and E) Live imaging of cell stably transfected with GFP-MRLC to visualize myosin-II filaments and RFP-Lifeact to visualize podosome cores. (D) Cell treated with 30 µM SecinH3 shows an increase in myosin-IIA filament assembly (green) and disruption of podosomes (red). Enlarged image of white-boxed area (5 × 4.5 µm2) of D shows colocalization between appearance of myosin-IIA filaments and podosome disruption (see Video 7). (E) Time course of podosome reappearance after addition of 30 µM ROCK inhibitor Y-27632 to cell incubated in SecinH3-containing medium. Note that podosomes (red) appeared after disassembly of myosin-II filaments (green). (F–J) TGFβ1-stimulated THP1 cells were transfected with either control scrambled siRNA (F and G) or with siRNA to NM-myosin-IIA heavy chain, MYH9 (I and J) and, after 48 h, treated with either 0.1% DMSO (F and I) or 30 µM SecinH3 (G and J) for 1 h. (F, G, I, and J) After fixation, the cells were stained with phalloidin (left) and antibodies to NM-myosin-IIA heavy chain (middle) and vinculin (right). Bars, 5 µm. (H) Western blot showing protein levels of NM-myosin-IIA heavy chain in control cells (transfected with scrambled siRNA) or in NM-myosin-IIA knockdown cells (transfected with MYH9 siRNA); α-tubulin was used as a loading control. (K and L) Effect of SecinH3 treatment of control and NM-myosin-IIA knockdown cells on number of podosomes per cell (K) and percentage of cells with more than 10 podosomes (L). The data in K and L are presented as indicated in the legend to Fig. 1. Pooled data of at least two independent experiments are presented for each group. The significance of the difference between groups was estimated by two-tailed Student’s t test. nonsignificant (ns), P > 0.05; ***, P ≤ 0.001.
Constitutively active ARF1 induces actin-rich puncta in fibroblasts
To test whether constitutively active ARF1 could induce formation of podosome-like adhesions in a more general context, we expressed constitutively active ARF1, CFP–ARF1 (Q71L), in cells that normally do not form podosomes, such as MEFs. Overexpression of constitutively active but not wild-type ARF1 induced formation of numerous actin-rich puncta localized to the ventral surface of these cells, in the same focal plane as focal adhesions (Fig. 7, A and B). Similar to mature podosomes, the actin-rich puncta induced by constitutively active ARF1 (CFP–ARF1 Q71L) were transiently associated with CFP–ARF1 (Q71L)–containing vesicles (Fig. 7 B, right). Formation of these puncta was accompanied by some reduction in the number of stress fibers and focal adhesions, but even total disassembly of these structures upon expression of dominant-negative RhoA (GFP–RhoA T19N) was not sufficient to induce actin-containing puncta (Fig. 7 C). At the same time, constitutively active ARF1 efficiently triggered formation of such puncta in cells also expressing dominant-negative RhoA (Fig. 7 D). Similarly, inhibition of Rho activity by cell-permeable C3 transferase (2 µg/ml) did not by itself induce formation of the actin puncta and did not interfere with the induction of these puncta by constitutively active ARF1 (Fig. S5, D and E).
Figure 7.
Constitutively active ARF1 induces F-actin–rich puncta (labeled by mCherry-UtrCH) in mouse fibroblasts. (A) Transfection with wild-type CFP-ARF1 did not change actin cytoskeleton of fibroblast. (B) Numerous F-actin–rich puncta in the fibroblast transfected with constitutively active ARF1 mutant, CFP–ARF1 Q71L. (B, right) Images of the boxed area (6 × 4.5 µm2) in the left panel showing F-actin (red, top row), CFP–ARF1 Q71L (green, middle row), and their superimposition (bottom row) at three time points taken with a 3-s time interval. Transient contacts (yellow) of CFP–ARF1 Q71L–containing puncta with the F-actin–rich puncta are seen. (C) Cell transfected with dominant-negative RhoA (GFP–RhoA T19N) did not contain stress fibers. (D) Cell cotransfected with dominant-negative RhoA (GFP–RhoA T19N) and constitutively active ARF1 (CFP–ARF1 Q71L) formed numerous F-actin–rich puncta. (E) Control fibroblast forming stress fibers (left) did not demonstrate localization of mCherry-WIP (right). (F) F-actin–rich puncta in fibroblast expressing CFP–ARF1 Q71L are enriched with mCherry-WIP. (G and H) Fibroblasts transfected with constitutively active ARF1 (CFP–ARF1 Q71L) were plated on fluorescent gelatin-coated coverslips in control medium containing 0.2% DMSO (G) or in medium containing 25 µM MMP inhibitor GM6001 (H) and incubated for 4 h. The matrix degradation sites in the boxed areas are seen at high magnification on the left in the bottom panels of G but not H. Actin puncta were visualized by phalloidin staining. High magnifications of boxed areas in G and H as well as merged images of actin and fluorescent gelatin are shown at left, middle, and right in the bottom panels of G and H, respectively. Note that actin puncta colocalize with dark areas corresponding to degraded fluorescent gelatin in control fibroblasts (G, bottom, white arrowheads), whereas gelatin degradation is completely prevented in cells treated with GM6001 (H, bottom). Bars: (A and C–H) 5 µm; (B) 1 µm.
Proteins typically associated with podosome cores in different cell types (WIP, N-WASP, cortactin, Arp3, and dynamin-2) were found in the actin-rich puncta (Fig. S5, F–J). At the same time, protein components of the podosome ring, such as vinculin (unpublished data) and paxillin (Fig. S5 K), were not found to be associated with active ARF1-induced actin puncta, suggesting incomplete podosome formation. ARF1-induced actin puncta were not related to clathrin-depended endocytic activity because they did not colocalize with clathrin-coated pits (Fig. S5 L).
Unlike native podosomes, the puncta induced by constitutively active ARF1 were motile. Although podosomes of THP1 cells (Fig. S4 D, right insets), as well as podosome-like structures in fibroblasts plated on fluid substrate (Yu et al., 2013), are essentially stationary with respect to the substratum, the positions of puncta induced by active ARF1 are oscillating, with a mean velocity of 0.88 ± 0.28 µm/s (± SD). Thus, constitutively active ARF1 induced formation of actin-rich puncta in proximity to the ventral cell membrane that can be considered as incompletely anchored podosome-like structures and possibly podosome precursors.
In spite of the difference between authentic podosomes and the actin-rich puncta induced by constitutively active ARF1, the puncta mimic one important podosome function, namely MMP-dependent ability to degrade the matrix. Indeed, the positions of actin puncta induced in the fibroblasts by constitutively active ARF1 (Q71L) coincided with the sites of matrix degradation: dark areas on the substratum covered with fluorescently labeled gelatin (Fig. 7 G). Formation of such dark areas could be prevented by treatment with 25 µM of MMP inhibitor GM6001 (Fig. 7 H) and therefore depended on the exocytosis of MMPs by cells. Thus, our experiments show that actin puncta induced in fibroblasts by constitutively active ARF1 trigger local matrix degradation by facilitating exocytosis of MMPs, independently of podosome ring assembly.
Discussion
In this article, we demonstrate that an ARNO-ARF1 signaling axis is required for the maintenance of podosome integrity (see Fig. 8 for flow diagram). First, knockdown of ARF1 but not ARF6 prevents podosome formation by TGFβ1- or PMA-treated THP1 cells. In addition to these classic podosomes, we explored podosome-like structures induced in fibroblasts by either expression of constitutively active Src (Tarone et al., 1985) or plating cells on a fluid substratum (Yu et al., 2013). We checked that specific drugs inhibiting ARF1-activating GEFs, BFA (Yamaji et al., 2000; Niu et al., 2005) and SecinH3 (Hafner et al., 2006), led to rapid dissolution of podosomes in THP1 cells and the podosome-like structures in fibroblasts. In addition, we have shown that in both THP1 cells and fibroblasts, treatments inducing podosome formation augmented the fraction of active, GTP-bound ARF1.
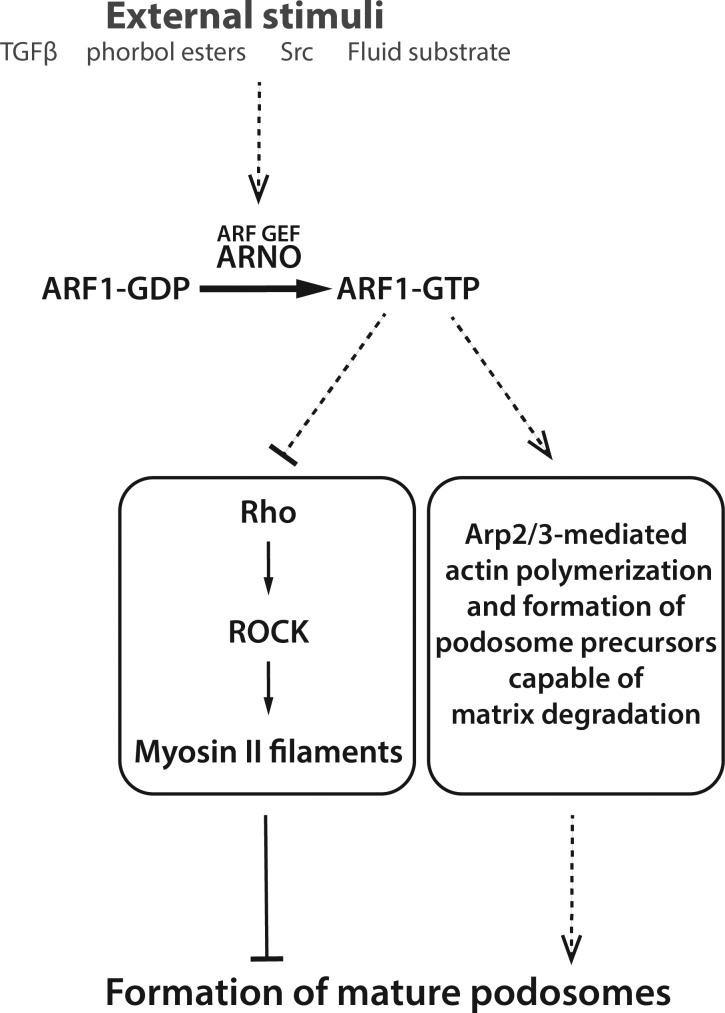
Figure 8.
Flow diagram illustrating the role of ARNO-ARF1 signaling axis in podosome formation. A variety of external factors known to switch cells toward podosome formation activate the ARF GEF, ARNO. ARNO activates ARF1, which in turn induces two major pathways regulating the podosomes. First, ARF1 inhibits Rho, which negatively regulates podosomes via ROCK-dependent formation of myosin-II filaments. Second, ARF1 promotes formation of Arp2/3- and actin-enriched podosome core-like structures associated with matrix degradation sites. The solid arrows represent the direct pathways, and the dashed arrows indicate the existence of unknown intermediate steps in the depicted pathways.
BFA and SecinH3 inhibit different classes of ARF1-activating GEFs (Donaldson and Jackson, 2011). In our experiments, the BFA-sensitive GEFs (GBF1, BIG1, and BIG2) appeared to be functionally unrelated to podosome regulation. The inhibitory effect of BFA could thus be explained by sequestration of ARF1 within the BFA-induced ternary complexes consisting of inhibited GEFs, ARF1-GDP and BFA (Peyroche et al., 1999; Mossessova et al., 2003; Zeghouf et al., 2005). At the same time, we found that one of the SecinH3-sensitive GEFs, ARNO (cytohesin 2), but not cytohesin 1, is indispensable for podosome integrity in THP1 cells. Possible functions of several other ubiquitous ARF family proteins (ARF3, ARF4, and ARF5) as well as other SecinH3-sensitive GEFs, cytohesin 3 and 4, remain to be studied in the context of podosome formation.
Localization studies revealed that ARNO (but not cytohesin 1) is stably colocalized with adhesion proteins in the ring domain of podosomes in THP1 cells as well as with podosome-like structures in fibroblasts. This is consistent with biochemical data showing direct association of ARNO with paxillin (Torii et al., 2010). Furthermore, live-cell imaging showed transient contacts of vesicles containing ARF1 with the periphery and ring domain of podosomes and podosome-like structures in THP1 cells. We demonstrated that ARF1-containing vesicles are moving along microtubules, so one of the functions of microtubules important for podosome maintenance (Linder et al., 2000) could be delivery of ARF1. It is worth noting that a negative regulator of ARF1 activity, ARF1 GTPase-activating protein ASAP1, was also shown to localize to podosomes (Shiba and Randazzo, 2011; Curtis et al., 2015).
How could active ARF1 affect podosome assembly and stability? The first possibility is based on well-documented functions of ARF1 in the Golgi complex and vesicular traffic (Donaldson et al., 2005). It can be conjectured that some of the ARF1-dependent functions of the Golgi are required for podosome integrity. This possibility cannot be entirely excluded; however, it is worth noting that in our experiments integrity of podosomes can be dissected from the integrity of the Golgi. In particular, inhibition of the ARF1 exchange factor GBF1, responsible for ARF1-dependent COPI recruitment to the Golgi, as well as knockdown of the COPI subunit, βCOP, only marginally affected podosome integrity. ARF1 in principle could be involved in integrin turnover and affect podosome formation via regulation of available integrin adhesion receptors. However, in our experiment, experimental manipulations with ARF1 did not affect the integrity or dynamics of another class of integrin-dependent adhesions, focal adhesions. This suggests that other mechanisms should be considered to explain the specific effect of ARF1 depletion/inhibition on podosome integrity.
Podosomes are part of the actin cytoskeleton and as such likely to be regulated by small G proteins of Rho family. We have shown that inhibition of ARF1 triggered significant activation of RhoA but not Rac or Cdc42. Activation of RhoA in turn triggers the assembly of numerous myosin-IIA filaments, which as we have demonstrated led to considerable disruption of podosomes. We have shown that suppression of myosin-IIA filament formation by either inhibition of ROCK or knockdown of myosin-IIA prevented the disruptive effect of ARF1 inhibition on podosome formation. Thus, our experiments suggest that ARF1 functions in podosome formation as an inhibitor of RhoA activity and subsequent myosin-IIA filament formation. This conclusion is consistent with our observation that ARF1-dependent activation of podosome formation by plating of cells on fluid bilayer led to inhibition of RhoA (Yu et al., 2013).
Interestingly, apparently the same mechanism based on suppression of Rho and myosin-II by the cytohesin family exchange factor, Steppke, and a Drosophila melanogaster ARF was found in a completely different system, during cellularization of Drosophila embryos (Lee and Harris, 2013). However, the pathway connecting ARF1 and RhoA remains unknown. It is perhaps worth noting that ARF1 can bind the RhoGAP ARHGAP10/21 and deliver it to the Golgi or plasma membrane (Dubois et al., 2005; Ménétrey et al., 2007; Kumari and Mayor, 2008). ARHGAP10/21 is known to inhibit Cdc42 but also shows some RhoA inhibitory activity in vitro (Dubois et al., 2005).
It is also not clear why an excess of myosin-II filaments antagonizes podosome integrity. Myosin-II has been shown to localize to actin links radiating from the podosomes (van den Dries et al., 2013), but its functional role in podosomes is yet to be established.
In addition to its function as a myosin-II regulator, ARF1 could affect podosomes via regulation of actin polymerization. There are several lines of evidence suggesting involvement of ARF1 in the regulation of Arp2/3 complex-driven actin polymerization: via recruitment of actin nucleation promoting WAVE complex (Humphreys et al., 2012a,b); via sequestration and inactivation of Arp2/3 inhibitor, PICK1 (Rocca et al., 2013); and via activation of Cdc42 (Dubois et al., 2005; Heuvingh et al., 2007). We were not able to find evidence of any of these mechanisms in the context of podosome regulation. Neither data from the literature nor our own observations indicate that WAVE or PICK1 is localized to podosomes. Moreover, ARF1 inhibition did not induce any changes in GTP-bound Cdc42 levels, and constitutively active Cdc42 did not prevent or overcome the disruption of podosomes seen upon ARF1 inhibition.
Nevertheless, the effect of ARF1 on the polymerization of actin in the context of podosome formation is seen in our experiments with expression of constitutively active ARF1 in fibroblast-type cells, which normally do not produce podosomes. Active ARF1 not only suppresses stress fiber formation, but also induces formation of numerous actin and Arp3-containing patches in such cells. The induction of actin polymerization at the plasma membrane by active ARF1 and ARF6 was previously demonstrated (Caviston et al., 2014). Many actin-associated proteins typical of podosomes (N-WASP, WIP, cortactin, and dynamin-II) were also found in these puncta. Moreover, a hallmark of podosome function, the local gelatin matrix degradation by MMPs, appeared to be associated with these puncta. At the same time, the puncta were not surrounded by any podosome adhesive ring components. The recruitment of certain adhesion components such as paxillin was shown to require dynamic GTP/GDP turnover of ARF1 (Liu et al., 2002, 2005). This may explain the lack of adhesive ring surrounding podosome core-like structures induced by constitutively active ARF1. Of note, our data show that matrix degrading and adhesion functions could be dissected under conditions of induction of podosome precursors by constitutively active ARF1.
The pathways downstream of ARF1 underlying formation of these podosome precursors are not the same as Rho and myosin-II inhibitory activity of ARNO-ARF1 characterized earlier, because expression of dominant-negative RhoA did not by itself induce formation of the actin-rich puncta in fibroblasts. We cannot exclude that local changes in Cdc42 activity may still play a role in this process (Heuvingh et al., 2007), even though ARF1 did not affect the total level of Cdc42 activity in our experiments.
In summary, we demonstrate that the signaling axis ARNO-ARF1 plays a critical role in the control of podosome integrity and find that this pathway in macrophage-like cells operates via inhibition of RhoA and myosin-II activity. Other pathways found in fibroblasts downstream of active ARF1 induce formation of F-actin–rich puncta resembling podosome actin cores that are not associated with the matrix adhesion components but are involved in matrix degradation. These findings open new features of the processes of podosome formation and matrix degradation. Investigation of ARNO-ARF1 upstream and downstream pathways provides a rich source of future studies.
Materials and methods
Cell culture and transfection procedures
THP1 human monocytic leukemia cell line was obtained from Health Protection Agency Culture Collections and cultured using RPMI-1640 supplemented with 10% heat-inactivated FBS and 50 µg/ml 2-mercaptoethanol (Sigma-Aldrich) at 37°C and 5% CO2.
The suspended THP-1 cells were differentiated into adherent macrophage-like cells with either 1 ng/ml human recombinant cytokine TGFβ1 (R&D Systems) or 50 nM PMA (Sigma-Aldrich) for 24 or 48 h on fibronectin-coated glass substrates. No apparent difference between the phenotypes of cells stimulated for 24 or 48 h was detected. 35-mm glass-bottomed dishes (81158; Ibidi) were coated with 1 µg/ml fibronectin (EMD Millipore) in PBS for 1–2 h at 37°C, washed with PBS twice, and incubated in complete medium before seeding of cells.
Cells were transiently transfected before stimulation with DNA plasmids using electroporation (Neon Transfection System; Thermo Fisher Scientific) in accordance with manufacturer’s instructions. Specifically, two pulses of 1,400 V for 20 ms were used.
For siRNA transfection, THP1 cells were treated with 100 nM ARF1 siRNA (L-011580-00-0005; GE Healthcare), 150 nM ARF6 siRNA (L-004008-00-0005; GE Healthcare), 100 nM MYH9 siRNA (L-007668-00-000; GE Healthcare), or 100 nM COPB1 siRNA (L-017940-00-0005; GE Healthcare). For control experiments, cells were transfected with 100–150 nM non-targeting control siRNA (D-001810-10; GE Healthcare) at a concentration similar to individual gene-targeted siRNAs.
For knockdown of ARF GEFs in THP1 cells, siRNA duplex 5′-GCAAUGGGCAGGAAGAAGU-3′ (Oh and Santy, 2010) against human ARNO sequence and 5′-AUGGAGGAGGACGACAGCUAC-3′ (Sendide et al., 2005) against human cytohesin 1 with dT-dT overhangs were purchased from Sigma-Aldrich. For rescue experiments in Fig. 1 D, ARF1 siRNA–transfected THP1 cells were cotransfected with HA-ARF1 (bovine origin, nonsensitive to aforementioned ARF1 siRNA) and fixed 48 h after plating on fibronectin.
Immortalized rptp-α+/+ MEFs (Su et al., 1999) were obtained from the Sheetz laboratory (Mechanobiology Institute, Singapore, Singapore). MEFs were cultured in high-glucose DMEM supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 1% l-glutamine, and 100 IU/mg penicillin-streptomycin (Invitrogen) at 37°C and 5% CO2. MEFs were transiently electroporated with a single pulse of 1,400 V for 20 ms. MEFs were seeded on either fibronectin-coated 35-mm Ibidi or 27-mm Iwaki glass-bottomed dishes for 24 h posttransfection. For plating on supported lipid bilayer membrane, transfected MEFs were seeded on six-well Nunc plastic dishes (Thermo Fisher Scientific) for 24 h posttransfection. These MEFs were then treated with trypsin solution, TrypLE (Thermo Fisher Scientific), for 5 min and kept in suspension for 15 min in complete medium to recover from trypsinization before seeding on supported lipid bilayer membrane.
Plasmids
mCherry-WIP and GFP-WASP were described in Vijayakumar et al. (2015); GFP–β-actin and mCherry-talin, in Cox et al. (2011). The following plasmids described in corresponding references were provided by the listed researchers. EGFP-ARNO (Santy et al., 1999) and EGFP-ARNO E156K (Hernández-Deviez et al., 2004), J. Casanova (University of Virginia, Charlottesville, VA); EGFP–cytohesin 1 (Bourgoin et al., 2002), S. Bourgoin (University of Laval, Quebec, Canada); ARF1-RFP (Hsu et al., 2010), N. Altan-Bonnet (National Institutes of Health, Bethesda, MD); GFP-vinculin and mCherry-vinculin, M. Davidson (Florida State University, Tallahassee, FL); GFP-Paxillin and mApple-Paxillin (Kanchanawong et al., 2010), P. Kanchanawong (Mechanobiology Institute, Singapore, Singapore); constitutively active Src Y527F, K. Kawauchi (Mechanobiology Institute, Singapore, Singapore); EGFP-Rab6A (Miserey-Lenkei et al., 2010), S. Miserey-Lenkei (Institut Curie, Paris, France); GFP–Mannosidase II (van Galen et al., 2014), V. Malhotra (Center of Genomic Regulation, Barcelona, Spain); mApple-Rab11A (verified in-house), V. Allan (University of Manchester, Manchester, England, UK); GFP–Dynamin II (Ochoa et al., 2000), P. De Camilli (Yale University, New Haven, CT); and Myosin regulatory light chain–GFP, M. Dodding (King’s College London, London, England, UK). All BIG constructs (HA-BIG1, HA-BIG2, HA–BIG1 E793K, and HA–BIG2 E738K; Ishizaki et al., 2008) were obtained from H. Shin (Kyoto University, Kyoto, Japan).
The following plasmids described were purchased from Addgene: ARF1-GFP (39554), ARF1-ECFP (11381), ARF1(T31N)-ECFP (11384), ARF1(Q71L)-ECFP (11385), HA-ARF1 (10830), GFP-Rab11 (12674), mCherry UtrCH (26740), EMTB-mCherry (26742), GFP-RhoA (T19N; 12967), ARF6-CFP (11382), mCherry-Arp3 (27682), mCherry-Cortactin (27676), mCherry–clathrin light chain (27680), dsRed-Rab7 (12661), GFP-Rab8A (31803), and EMTB (ensconsin)-mCherry (26742).
Supported lipid bilayer membrane
Methodologies of supported lipid bilayer preparation and membrane functionalization have been described in Yu et al. (2011) and Yu et al. (2013). In brief, 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) 16:0 (biotinyl-Cap-PE) were purchased from Avanti Polar Lipids, Inc. The lipids (0.2 mol% biotinyl-Cap-PE and 99.8 mol% DOPC) were mixed with an equal volume of PBS and pipetted onto cleaned glass substratum with a 25-mm coverslip placed on top for self-assembly of lipid vesicles. The lipid-coated coverslips were immersed into a deionized water bath and then placed and sealed in an Attofluor cell chamber (Thermo Fisher Scientific). The supported lipid bilayer membrane ensemble was kept under aqueous environment at all times. For membrane functionalization, the supported lipid membrane was blocked with 50 µg/ml Casein. A total of 0.1 µg/ml Cascade blue neutravidin (Thermo Fisher Scientific) was added onto supported lipid membranes, followed by 1 µg/ml biotinylated RGD, cyclo (Arg-Gly-Asp-d-Phe-Lys [Biotin-PEG-PEG]) (Peptides International). Cells were then added onto the RGD-functionalized lipid bilayer membrane and imaged or fixed within 2–3 h of preparation.
Drug treatment
For drug inhibition studies, cells were treated with 30 µM SecinH3 (Tocris Bioscience), 10 µM Golgicide A (Santa Cruz Biotechnology, Inc.), 5 µg/ml BFA (Sigma-Aldrich), 30 µM Y-27632 (Sigma-Aldrich), 25 µM GM6001 (Enzo Life Sciences), and 2 µg/ml C3 transferase (Cytoskeleton, Inc.) in complete medium for 1–2 h or 4 h for GM6001 at 37°C with 5% CO2 and subsequently fixed with 4% PFA. For live-cell imaging, cells were imaged immediately after addition of appropriate inhibitors, which remained in the medium during the entire period of image acquisition. To study the effect of inhibitors on podosomes formed by MEFs plated on RGD lipid bilayer, the cells were treated with appropriate inhibitors 30–45 min after cell seeding on the bilayer.
Immunoblotting
For verification of knockdown experiments, cells were lysed in RIPA buffer 48 h after transfection. Extracted proteins were separated by 4–20% SDS-PAGE (Thermo Fisher Scientific), transferred to PVDF membranes (Bio-Rad Laboratories), incubated at 75 V for 2 h, and blocked for 1 h with 5% nonfat milk (Bio-Rad Laboratories) or BSA (Sigma-Aldrich). The PVDF membranes were incubated overnight at 4°C with appropriate antibodies: anti-ARF1 (dilution 1:1,000; ab108347; Abcam); anti-ARF6 (dilution 1:1,000; ab77581; Abcam); anti-ARNO (dilution 1:1,000; ab56510; Abcam); anti–cytohesin 1 (dilution 1:500; MABT14; EMD Millipore); anti–α-tubulin (dilution 1:3,000; T6199; Sigma-Aldrich); anti-βCOP (dilution 1:1,000; ab2899; Abcam); anti-HA (dilution 1:1,000; 2367; Cell Signaling Technology); anti-Cdc42 (dilution 1:1,000; 2462; Cell Signaling Technology); anti-RhoA (dilution 1:1,000; sc-418; Santa Cruz Biotechnology, Inc.); anti-Rac1 (dilution 1:1,000; 610650; BD); and anti–NM myosin-IIA (dilution 1:1,000; M8064; Sigma-Aldrich).
After three washes (10 min each), appropriate secondary antibodies conjugated with HRP (Bio-Rad Laboratories) were incubated with the membrane for 1 h, washed three times (15 min at RT), and detected by ECL Western blotting substratum (Thermo Fisher Scientific) using CL-Xposure film (Thermo Fisher Scientific).
Small G protein activity assay
Total cell lysates were collected and immediately quantified by the G-LISA ARF1 or Cdc42 Activation Assay Biochem kit (colorimetric-based) and performed as per manufacturer’s protocol (Cytoskeleton, Inc.). Samples were run in duplicates per sample, means were calculated, and values were normalized to the total ARF1 or Cdc42 levels detected by immunoblotting. For each set of experiments, data were normalized to TGFβ1-treated THP1 cells giving a fold-change value from 0 (minimum) to 1 (maximum). A pull-down assay using GST-tagged RhoA-binding domain of Rhotekin was used to precipitate GTP-bound RhoA, and GST-tagged Rac1/Cdc42-binding domain of PAK1 (PBD) beads were used to precipitate GTP-bound Rac1 or Cdc42 in THP1 cells. Pulled-down RhoA, Rac1, and Cdc42 were immunoblotted using respective antibodies as described in the previous section.
Immunofluorescence
Cells were fixed with 3.7% PFA in PBS, washed twice, permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) in PBS for 10 min, and washed twice. Fixed cells were blocked with 5% BSA or 5% FBS for 1 h at RT or overnight at 4°C before incubation with appropriate primary antibodies: anti-GM130 (dilution 1:400; 610822; BD); anti-GRASP65 (dilution 1:500; ab30315; Abcam); anti-HA (dilution 1:400; 2367; Cell Signaling Technology), anti-vinculin (dilution 1:400; V9131; Sigma-Aldrich); anti-ARF6 (dilution 1:200; ab77581; Abcam); anti-βCOP (dilution 1:200; ab2899; Abcam); and anti–NM myosin-IIA (dilution 1:800; M8064; Sigma-Aldrich). Cells were washed three times with PBS and incubated with Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific) for 1 h at RT followed by three washes in PBS. Actin staining was performed using Alexa Fluor 488 Phalloidin (Thermo Fisher Scientific), Phalloidin-TRITC (Sigma-Aldrich), or Alexa Fluor 647 Phalloidin (Thermo Fisher Scientific).
Matrix degradation assay
50% sulfuric acid–washed coverslips were coated with 50 mg/ml poly-d-lysine for 30 min at RT and fixed with 0.5% glutaraldehyde for 15 min. 0.2% gelatin warmed at 37°C was mixed with Oregon green 488–conjugated pig gelatin at a 6:1 ratio. Coverslips were coated with gelatin mix for 10 min, washed with 1× PBS, and quenched with 5 mg/ml sodium borohydride for 15 min followed by numerous washes. For the matrix degradation assay, MEFs were seeded on these coated coverslips for 4 h and fixed for immunofluorescence imaging as described earlier. Dark spots corresponding to areas of cells indicate degradation of the matrix.
Live-cell imaging and microscopy
Cells were imaged in complete medium (unless stated otherwise) at an acquisition rate from 5-s to 1-min intervals using a spinning-disc confocal microscope (Ultraview VoX; PerkinElmer) attached to an inverted microscope (IX81; Olympus), equipped with a 100× oil-immersion objective (1.40 NA, UPlanSApo), an EMCCD camera (C9100-13; Hamamatsu Photonics) for image acquisition, and Volocity software (PerkinElmer) to control the acquisition protocol. Fixed samples and live cells were also imaged with a Nikon confocal A1R system and Nikon SIM attached to a Ti-E inverted microscope (Nikon) with Perfect Focus System using a 100× oil immersion objective (1.40 NA, CFI Plan-ApochromatVC). The cameras (Neo sCMOS and DU-897; Andor Technology) were used to acquire images for confocal A1R and SIM systems, respectively, with NIS-Elements AR software (Nikon) to control the acquisition protocol. For z-stack images, cells were imaged at a step size of 0.2–0.5 µm with a total height of 15–20 µm.
Image processing and data analysis
Image processing and analysis were performed with ImageJ or Volocity software. The number of podosomes (marked by core or ring marker) was quantified automatically using an ImageJ-based plugin for counting nuclei, which was manually verified for the first 10 cells in the specimen to account for undetected podosomes (<10%). Line intensity measurements (arbitrary units) of GFP-ARNO, GFP–Cytohesin 1, and mCherry-UtrCH were quantified by measuring the mean intensity of GFP or mCherry fluorescence per area (square micrometers), background subtracted, and normalized with values ranging from 0 (lowest) to 1 (highest).
Statistical analyses
Prism version 6 (GraphPad Software) was used to plot, analyze, and represent the data. Significance of the differences was determined using two-tailed unpaired Student’s t test or one-way analysis of variance for more than two groups. The methods for statistical analysis and sizes of the samples (n) are specified in Results or the figure legends for all of the quantitative data. Differences were accepted as significant for P < 0.05.
Online supplemental material
Fig. S1 shows that knockdown of either ARF1 or ARNO prevents formation of podosomes in THP1 cells stimulated by PMA, whereas ARF6 is not involved in podosome formation. Fig. S2 shows that ARF1-containing puncta are positive for Rab11 but not Rab6, Rab7, or Rab8. Fig. S3 shows that inhibition of β-COP as well as ARF exchange factors GBF1, BIG1, and BIG2 does not lead to podosome disruption in TGFβ1-stimulated THP1 cells, whereas the ARF1-mediated pathway of podosome formation does not involve Cdc42. Fig. S4 is a visualization of podosome dynamics in TGFβ1-stimulated THP1 cells stably transfected with GFP–β-actin (A, B, D, and E) and mCherry-talin by structured illumination microscopy. Fig. S5 shows that drugs reducing the level of GTP-ARF1 disrupt podosome rosettes induced by constitutively active Src in fibroblasts, but do not disrupt focal adhesions. In addition, this figure shows formation of actin-rich puncta upon fibroblast transfection with constitutively active ARF1 and colocalization of podosome core proteins to these puncta. Video 1 shows transient association of GFP-ARF1–positive puncta and mCherry-vinculin–labeled podosomes as visualized by TIRF microscopy. Video 2 shows the movements of GFP-ARF1–positive puncta along microtubules labeled by mCherry as visualized by spinning disk confocal microscopy. Video 3 shows that the disruption of podosomes labeled with mCherry-UtrCH in cells treated with 30 µM SecinH3 (left) is not accompanied by changes in Golgi apparatus morphology and dynamics. Video 4 shows the dynamics of podosomes in control DMSO-treated cells labeled with GFP–β-actin as visualized by SIM. Video 5 shows the dynamics of podosomes labeled with GFP–β-actin in cells treated with 30 µM SecinH3 as visualized by SIM. Video 6 shows the dynamics of GFP-ARNO localization to podosome ring in the course of podosome formation and disassembly as visualized by spinning disk confocal microscopy. Video 7 shows the burst of myosin II filament formation that accompanies podosome disruption in cells treated with 30 µM SecinH3 as visualized by SIM. Video 8 shows that the disassembly of myosin II filaments by 30 µM Y-27632 results in recovery of podosomes in cells pretreated with 30 µM SecinH3 as visualized by SIM.
Supplementary Material
Acknowledgments
This research has been supported by the National Research Foundation Singapore, Ministry of Education - Singapore (grant R-714-006-006-271), and administrated by the National University of Singapore. A.D. Bershadsky also holds the Joseph Moss Professorial Chair in Biomedical Research at the Weizmann Institute of Science, Israel. N.B.M. Rafiq is funded by a joint National University of Singapore–King’s College London graduate studentship. G.E. Jones is supported by the Medical Research Council, UK (G1100041 and MR/K015664), and the generous provision of a visiting professorship from the Mechanobiology Institute, Singapore.
The authors declare no competing financial interests.
Author contributions: A.D. Bershadsky conceived and designed the project together with G.E. Jones. N.B.M. Rafiq designed and performed all experiments and prepared the manuscript; Z.Z. Lieu, T. Jiang, and C.H. Yu provided assistance in carrying out experiments and discussed results. A.D. Bershadsky, G.E. Jones, and P.T. Matsudaira discussed results and prepared the manuscript.
Footnotes
Abbreviations used:
- GEF
- guanine nucleotide exchange factor
- MEF
- mouse embryonic fibroblast
- MMP
- matrix metalloproteinase
- RGD
- arginylglycylaspartic acid
- ROCK
- Rho-associated kinase
- SIM
- structured-illumination microscopy
References
- Abdul-Manan N., Aghazadeh B., Liu G.A., Majumdar A., Ouerfelli O., Siminovitch K.A., and Rosen M.K.. 1999. Structure of Cdc42 in complex with the GTPase-binding domain of the ‘Wiskott-Aldrich syndrome’ protein. Nature. 399:379–383. 10.1038/20726 [DOI] [PubMed] [Google Scholar]
- Beck R., Rawet M., Wieland F.T., and Cassel D.. 2009. The COPI system: Molecular mechanisms and function. FEBS Lett. 583:2701–2709. 10.1016/j.febslet.2009.07.032 [DOI] [PubMed] [Google Scholar]
- Bershadsky A.D., and Futerman A.H.. 1994. Disruption of the Golgi apparatus by brefeldin A blocks cell polarization and inhibits directed cell migration. Proc. Natl. Acad. Sci. USA. 91:5686–5689. 10.1073/pnas.91.12.5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgoin S.G., Houle M.G., Singh I.N., Harbour D., Gagnon S., Morris A.J., and Brindley D.N.. 2002. ARNO but not cytohesin-1 translocation is phosphatidylinositol 3-kinase-dependent in HL-60 cells. J. Leukoc. Biol. 71:718–728. [PubMed] [Google Scholar]
- Burger K.L., Davis A.L., Isom S., Mishra N., and Seals D.F.. 2011. The podosome marker protein Tks5 regulates macrophage invasive behavior. Cytoskeleton (Hoboken). 68:694–711. 10.1002/cm.20545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns S., Thrasher A.J., Blundell M.P., Machesky L., and Jones G.E.. 2001. Configuration of human dendritic cell cytoskeleton by Rho GTPases, the WAS protein, and differentiation. Blood. 98:1142–1149. 10.1182/blood.V98.4.1142 [DOI] [PubMed] [Google Scholar]
- Burns S., Hardy S.J., Buddle J., Yong K.L., Jones G.E., and Thrasher A.J.. 2004. Maturation of DC is associated with changes in motile characteristics and adherence. Cell Motil. Cytoskeleton. 57:118–132. 10.1002/cm.10163 [DOI] [PubMed] [Google Scholar]
- Calle Y., Chou H.C., Thrasher A.J., and Jones G.E.. 2004. Wiskott-Aldrich syndrome protein and the cytoskeletal dynamics of dendritic cells. J. Pathol. 204:460–469. 10.1002/path.1651 [DOI] [PubMed] [Google Scholar]
- Calle Y., Burns S., Thrasher A.J., and Jones G.E.. 2006. The leukocyte podosome. Eur. J. Cell Biol. 85:151–157. 10.1016/j.ejcb.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Casanova J.E. 2007. Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic. 8:1476–1485. 10.1111/j.1600-0854.2007.00634.x [DOI] [PubMed] [Google Scholar]
- Caviston J.P., Cohen L.A., and Donaldson J.G.. 2014. Arf1 and Arf6 promote ventral actin structures formed by acute activation of protein kinase C and Src. Cytoskeleton (Hoboken). 71:380–394. 10.1002/cm.21181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citterio C., Jones H.D., Pacheco-Rodriguez G., Islam A., Moss J., and Vaughan M.. 2006. Effect of protein kinase A on accumulation of brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) in HepG2 cell nuclei. Proc. Natl. Acad. Sci. USA. 103:2683–2688. 10.1073/pnas.0510571103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox S., and Jones G.E.. 2013. Imaging cells at the nanoscale. Int. J. Biochem. Cell Biol. 45:1669–1678. 10.1016/j.biocel.2013.05.010 [DOI] [PubMed] [Google Scholar]
- Cox S., Rosten E., Monypenny J., Jovanovic-Talisman T., Burnette D.T., Lippincott-Schwartz J., Jones G.E., and Heintzmann R.. 2011. Bayesian localization microscopy reveals nanoscale podosome dynamics. Nat. Methods. 9:195–200. 10.1038/nmeth.1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis J., Luo Y., Zenner H.L., Cuchet-Lourenço D., Wu C., Lo K., Maes M., Alisaac A., Stebbings E., Liu J.Z., et al. 2015. Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nat. Genet. 47:523–527. 10.1038/ng.3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C., and Chavrier P.. 2006. ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7:347–358. 10.1038/nrm1910 [DOI] [PubMed] [Google Scholar]
- DiNitto J.P., Delprato A., Gabe Lee M.T., Cronin T.C., Huang S., Guilherme A., Czech M.P., and Lambright D.G.. 2007. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol. Cell. 28:569–583. 10.1016/j.molcel.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G., and Jackson C.L.. 2011. ARF family G proteins and their regulators: Roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12:362–375. 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G., Honda A., and Weigert R.. 2005. Multiple activities for Arf1 at the Golgi complex. Biochim. Biophys. Acta. 1744:364–373. 10.1016/j.bbamcr.2005.03.001 [DOI] [PubMed] [Google Scholar]
- Dubois T., Paléotti O., Mironov A.A., Fraisier V., Stradal T.E., De Matteis M.A., Franco M., and Chavrier P.. 2005. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat. Cell Biol. 7:353–364. 10.1038/ncb1244 [DOI] [PubMed] [Google Scholar]
- El Azzouzi K., Wiesner C., and Linder S.. 2016. Metalloproteinase MT1-MMP islets act as memory devices for podosome reemergence. J. Cell Biol. 213:109–125. 10.1083/jcb.201510043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawden-Bone C., Zhou Z., King E., Prescott A., Watts C., and Lucocq J.. 2010. Dendritic cell podosomes are protrusive and invade the extracellular matrix using metalloproteinase MMP-14. J. Cell Sci. 123:1427–1437. 10.1242/jcs.056515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimona M., Buccione R., Courtneidge S.A., and Linder S.. 2008. Assembly and biological role of podosomes and invadopodia. Curr. Opin. Cell Biol. 20:235–241. 10.1016/j.ceb.2008.01.005 [DOI] [PubMed] [Google Scholar]
- Hafner M., Schmitz A., Grüne I., Srivatsan S.G., Paul B., Kolanus W., Quast T., Kremmer E., Bauer I., and Famulok M.. 2006. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 444:941–944. 10.1038/nature05415 [DOI] [PubMed] [Google Scholar]
- Hernández-Deviez D.J., Roth M.G., Casanova J.E., and Wilson J.M.. 2004. ARNO and ARF6 regulate axonal elongation and branching through downstream activation of phosphatidylinositol 4-phosphate 5-kinase alpha. Mol. Biol. Cell. 15:111–120. 10.1091/mbc.E03-06-0410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuvingh J., Franco M., Chavrier P., and Sykes C.. 2007. ARF1-mediated actin polymerization produces movement of artificial vesicles. Proc. Natl. Acad. Sci. USA. 104:16928–16933. 10.1073/pnas.0704749104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu N.Y., Ilnytska O., Belov G., Santiana M., Chen Y.H., Takvorian P.M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T., et al. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 141:799–811. 10.1016/j.cell.2010.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D., Davidson A., Hume P.J., and Koronakis V.. 2012a Salmonella virulence effector SopE and Host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe. 11:129–139. 10.1016/j.chom.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D., Liu T., Davidson A.C., Hume P.J., and Koronakis V.. 2012b The Drosophila Arf1 homologue Arf79F is essential for lamellipodium formation. J. Cell Sci. 125:5630–5635. 10.1242/jcs.108092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizaki R., Shin H.W., Mitsuhashi H., and Nakayama K.. 2008. Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol. Biol. Cell. 19:2650–2660. 10.1091/mbc.E07-10-1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., and Waterman C.M.. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S., and Mayor S.. 2008. ARF1 is directly involved in dynamin-independent endocytosis. Nat. Cell Biol. 10:30–41. 10.1038/ncb1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labernadie A., Bouissou A., Delobelle P., Balor S., Voituriez R., Proag A., Fourquaux I., Thibault C., Vieu C., Poincloux R., et al. 2014. Protrusion force microscopy reveals oscillatory force generation and mechanosensing activity of human macrophage podosomes. Nat. Commun. 5:5343 10.1038/ncomms6343 [DOI] [PubMed] [Google Scholar]
- Lee D.M., and Harris T.J.. 2013. An Arf-GEF regulates antagonism between endocytosis and the cytoskeleton for Drosophila blastoderm development. Curr. Biol. 23:2110–2120. 10.1016/j.cub.2013.08.058 [DOI] [PubMed] [Google Scholar]
- Linder S., and Wiesner C.. 2015. Tools of the trade: Podosomes as multipurpose organelles of monocytic cells. Cell. Mol. Life Sci. 72:121–135. 10.1007/s00018-014-1731-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S., Nelson D., Weiss M., and Aepfelbacher M.. 1999. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc. Natl. Acad. Sci. USA. 96:9648–9653. 10.1073/pnas.96.17.9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S., Hüfner K., Wintergerst U., and Aepfelbacher M.. 2000. Microtubule-dependent formation of podosomal adhesion structures in primary human macrophages. J. Cell Sci. 113:4165–4176. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Yuan L.C., Bonifacino J.S., and Klausner R.D.. 1989. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: Evidence for membrane cycling from Golgi to ER. Cell. 56:801–813. 10.1016/0092-8674(89)90685-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Loijens J.C., Martin K.H., Karginov A.V., and Parsons J.T.. 2002. The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol. Biol. Cell. 13:2147–2156. 10.1091/mbc.E02-01-0018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Yerushalmi G.M., Grigera P.R., and Parsons J.T.. 2005. Mislocalization or reduced expression of Arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling. J. Biol. Chem. 280:8884–8892. 10.1074/jbc.M412200200 [DOI] [PubMed] [Google Scholar]
- Machesky L.M., and Insall R.H.. 1998. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol. 8:1347–1356. 10.1016/S0960-9822(98)00015-3 [DOI] [PubMed] [Google Scholar]
- Meddens M.B., van den Dries K., and Cambi A.. 2014. Podosomes revealed by advanced bioimaging: what did we learn? Eur. J. Cell Biol. 93:380–387. 10.1016/j.ejcb.2014.09.002 [DOI] [PubMed] [Google Scholar]
- Ménétrey J., Perderiset M., Cicolari J., Dubois T., Elkhatib N., El Khadali F., Franco M., Chavrier P., and Houdusse A.. 2007. Structural basis for ARF1-mediated recruitment of ARHGAP21 to Golgi membranes. EMBO J. 26:1953–1962. 10.1038/sj.emboj.7601634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miserey-Lenkei S., Chalancon G., Bardin S., Formstecher E., Goud B., and Echard A.. 2010. Rab and actomyosin-dependent fission of transport vesicles at the Golgi complex. Nat. Cell Biol. 12:645–654. 10.1038/ncb2067 [DOI] [PubMed] [Google Scholar]
- Monypenny J., Chou H.C., Bañón-Rodríguez I., Thrasher A.J., Antón I.M., Jones G.E., and Calle Y.. 2011. Role of WASP in cell polarity and podosome dynamics of myeloid cells. Eur. J. Cell Biol. 90:198–204. 10.1016/j.ejcb.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E., Corpina R.A., and Goldberg J.. 2003. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol. Cell. 12:1403–1411. 10.1016/S1097-2765(03)00475-1 [DOI] [PubMed] [Google Scholar]
- Murphy D.A., and Courtneidge S.A.. 2011. The ‘ins’ and ‘outs’ of podosomes and invadopodia: Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai W., Kondo Y., Saitoh A., Naito T., Nakayama K., and Shin H.W.. 2013. ARF1 and ARF4 regulate recycling endosomal morphology and retrograde transport from endosomes to the Golgi apparatus. Mol. Biol. Cell. 24:2570–2581. 10.1091/mbc.E13-04-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu T.K., Pfeifer A.C., Lippincott-Schwartz J., and Jackson C.L.. 2005. Dynamics of GBF1, a Brefeldin A-sensitive Arf1 exchange factor at the Golgi. Mol. Biol. Cell. 16:1213–1222. 10.1091/mbc.E04-07-0599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa G.C., Slepnev V.I., Neff L., Ringstad N., Takei K., Daniell L., Kim W., Cao H., McNiven M., Baron R., and De Camilli P.. 2000. A functional link between dynamin and the actin cytoskeleton at podosomes. J. Cell Biol. 150:377–389. 10.1083/jcb.150.2.377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.J., and Santy L.C.. 2010. Differential effects of cytohesins 2 and 3 on beta1 integrin recycling. J. Biol. Chem. 285:14610–14616. 10.1074/jbc.M109.043935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y.-R., Chen C.-L., and Chen H.-C.. 2011. FAK is required for the assembly of podosome rosettes. J. Cell Biol. 195:113–129. 10.1083/jcb.201103016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer L., Trübe L., Klose M., Joosten B., Slotman J., Cambi A., and Linder S.. 2016. The formins FHOD1 and INF2 regulate inter- and intra-structural contractility of podosomes. J. Cell Sci. 129:298–313. 10.1242/jcs.177691 [DOI] [PubMed] [Google Scholar]
- Peyroche A., Antonny B., Robineau S., Acker J., Cherfils J., and Jackson C.L.. 1999. Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: Involvement of specific residues of the Sec7 domain. Mol. Cell. 3:275–285. 10.1016/S1097-2765(00)80455-4 [DOI] [PubMed] [Google Scholar]
- Rocca D.L., Amici M., Antoniou A., Blanco Suarez E., Halemani N., Murk K., McGarvey J., Jaafari N., Mellor J.R., Collingridge G.L., and Hanley J.G.. 2013. The small GTPase Arf1 modulates Arp2/3-mediated actin polymerization via PICK1 to regulate synaptic plasticity. Neuron. 79:293–307. 10.1016/j.neuron.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáenz J.B., Sun W.J., Chang J.W., Li J., Bursulaya B., Gray N.S., and Haslam D.B.. 2009. Golgicide A reveals essential roles for GBF1 in Golgi assembly and function. Nat. Chem. Biol. 5:157–165. 10.1038/nchembio.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santy L.C., Frank S.R., Hatfield J.C., and Casanova J.E.. 1999. Regulation of ARNO nucleotide exchange by a PH domain electrostatic switch. Curr. Biol. 9:1173–1176. 10.1016/S0960-9822(00)80019-6 [DOI] [PubMed] [Google Scholar]
- Schachtner H., Calaminus S.D., Thomas S.G., and Machesky L.M.. 2013. Podosomes in adhesion, migration, mechanosensing and matrix remodeling. Cytoskeleton (Hoboken). 70:572–589. 10.1002/cm.21119 [DOI] [PubMed] [Google Scholar]
- Sciaky N., Presley J., Smith C., Zaal K.J., Cole N., Moreira J.E., Terasaki M., Siggia E., and Lippincott-Schwartz J.. 1997. Golgi tubule traffic and the effects of brefeldin A visualized in living cells. J. Cell Biol. 139:1137–1155. 10.1083/jcb.139.5.1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seano G., Chiaverina G., Gagliardi P.A., di Blasio L., Puliafito A., Bouvard C., Sessa R., Tarone G., Sorokin L., Helley D., et al. 2014. Endothelial podosome rosettes regulate vascular branching in tumour angiogenesis. Nat. Cell Biol. 16:931–941: 1–8. 10.1038/ncb3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendide K., Reiner N.E., Lee J.S.I., Bourgoin S., Talal A., and Hmama Z.. 2005. Cross-talk between CD14 and complement receptor 3 promotes phagocytosis of mycobacteria: Regulation by phosphatidylinositol 3-kinase and cytohesin-1. J. Immunol. 174:4210–4219. 10.4049/jimmunol.174.7.4210 [DOI] [PubMed] [Google Scholar]
- Shiba Y., and Randazzo P.A.. 2011. GEFH1 binds ASAP1 and regulates podosome formation. Biochem. Biophys. Res. Commun. 406:574–579. 10.1016/j.bbrc.2011.02.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su J., Muranjan M., and Sap J.. 1999. Receptor protein tyrosine phosphatase alpha activates Src-family kinases and controls integrin-mediated responses in fibroblasts. Curr. Biol. 9:505–511. 10.1016/S0960-9822(99)80234-6 [DOI] [PubMed] [Google Scholar]
- Szul T., Grabski R., Lyons S., Morohashi Y., Shestopal S., Lowe M., and Sztul E.. 2007. Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J. Cell Sci. 120:3929–3940. 10.1242/jcs.010769 [DOI] [PubMed] [Google Scholar]
- Tarone G., Cirillo D., Giancotti F.G., Comoglio P.M., and Marchisio P.C.. 1985. Rous sarcoma virus-transformed fibroblasts adhere primarily at discrete protrusions of the ventral membrane called podosomes. Exp. Cell Res. 159:141–157. 10.1016/S0014-4827(85)80044-6 [DOI] [PubMed] [Google Scholar]
- Tatin F., Varon C., Génot E., and Moreau V.. 2006. A signalling cascade involving PKC, Src and Cdc42 regulates podosome assembly in cultured endothelial cells in response to phorbol ester. J. Cell Sci. 119:769–781. 10.1242/jcs.02787 [DOI] [PubMed] [Google Scholar]
- Torii T., Miyamoto Y., Sanbe A., Nishimura K., Yamauchi J., and Tanoue A.. 2010. Cytohesin-2/ARNO, through its interaction with focal adhesion adaptor protein paxillin, regulates preadipocyte migration via the downstream activation of Arf6. J. Biol. Chem. 285:24270–24281. 10.1074/jbc.M110.125658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Dries K., Meddens M.B., de Keijzer S., Shekhar S., Subramaniam V., Figdor C.G., and Cambi A.. 2013. Interplay between myosin IIA-mediated contractility and actin network integrity orchestrates podosome composition and oscillations. Nat. Commun. 4:1412 10.1038/ncomms2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Galen J., Campelo F., Martínez-Alonso E., Scarpa M., Martínez-Menárguez J.A., and Malhotra V.. 2014. Sphingomyelin homeostasis is required to form functional enzymatic domains at the trans-Golgi network. J. Cell Biol. 206:609–618. 10.1083/jcb.201405009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varon C., Tatin F., Moreau V., Van Obberghen-Schilling E., Fernandez-Sauze S., Reuzeau E., Kramer I., and Génot E.. 2006. Transforming growth factor beta induces rosettes of podosomes in primary aortic endothelial cells. Mol. Cell. Biol. 26:3582–3594. 10.1128/MCB.26.9.3582-3594.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S., and Horwitz A.R.. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790. 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayakumar V., Monypenny J., Chen X.J., Machesky L.M., Lilla S., Thrasher A.J., Antón I.M., Calle Y., and Jones G.E.. 2015. Tyrosine phosphorylation of WIP releases bound WASP and impairs podosome assembly in macrophages. J. Cell Sci. 128:251–265. 10.1242/jcs.154880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli-Daley L.A., Li Y., Zhang C.J., and Kahn R.A.. 2005. Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol. Biol. Cell. 16:4495–4508. 10.1091/mbc.E04-12-1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz T., Wellbourne-Wood J., and Kerkhoff E.. 2014. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 24:407–415. 10.1016/j.tcb.2014.02.004 [DOI] [PubMed] [Google Scholar]
- Wheeler A.P., Wells C.M., Smith S.D., Vega F.M., Henderson R.B., Tybulewicz V.L., and Ridley A.J.. 2006. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J. Cell Sci. 119:2749–2757. 10.1242/jcs.03024 [DOI] [PubMed] [Google Scholar]
- Wiesner C., Faix J., Himmel M., Bentzien F., and Linder S.. 2010. KIF5B and KIF3A/KIF3B kinesins drive MT1-MMP surface exposure, CD44 shedding, and extracellular matrix degradation in primary macrophages. Blood. 116:1559–1569. 10.1182/blood-2009-12-257089 [DOI] [PubMed] [Google Scholar]
- Yamaji R., Adamik R., Takeda K., Togawa A., Pacheco-Rodriguez G., Ferrans V.J., Moss J., and Vaughan M.. 2000. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc. Natl. Acad. Sci. USA. 97:2567–2572. 10.1073/pnas.97.6.2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.H., Law J.B., Suryana M., Low H.Y., and Sheetz M.P.. 2011. Early integrin binding to Arg-Gly-Asp peptide activates actin polymerization and contractile movement that stimulates outward translocation. Proc. Natl. Acad. Sci. USA. 108:20585–20590. 10.1073/pnas.1109485108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.H., Rafiq N.B., Krishnasamy A., Hartman K.L., Jones G.E., Bershadsky A.D., and Sheetz M.P.. 2013. Integrin-matrix clusters form podosome-like adhesions in the absence of traction forces. Cell Reports. 5:1456–1468. 10.1016/j.celrep.2013.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeghouf M., Guibert B., Zeeh J.C., and Cherfils J.. 2005. Arf, Sec7 and Brefeldin A: A model towards the therapeutic inhibition of guanine nucleotide-exchange factors. Biochem. Soc. Trans. 33:1265–1268. 10.1042/BST0331265 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.