Abstract
Successful RNAi applications depend on strategies allowing robust and persistent expression of minimal gene silencing triggers without perturbing endogenous gene expression. Here, we propose a novel avenue which is integration of a promoterless shmiRNA, i.e. a shRNA embedded in a micro-RNA (miRNA) scaffold, into an engineered genomic miRNA locus. For proof-of-concept, we used TALE or CRISPR/Cas9 nucleases to site-specifically integrate an anti-hepatitis C virus (HCV) shmiRNA into the liver-specific miR-122/hcr locus in hepatoma cells, with the aim to obtain cellular clones that are genetically protected against HCV infection. Using reporter assays, Northern blotting and qRT-PCR, we confirmed anti-HCV shmiRNA expression as well as miR-122 integrity and functionality in selected cellular progeny. Moreover, we employed a comprehensive battery of PCR, cDNA/miRNA profiling and whole genome sequencing analyses to validate targeted integration of a single shmiRNA molecule at the expected position, and to rule out deleterious effects on the genomes or transcriptomes of the engineered cells. Importantly, a subgenomic HCV replicon and a full-length reporter virus, but not a Dengue virus control, were significantly impaired in the modified cells. Our original combination of DNA engineering and RNAi expression technologies benefits numerous applications, from miRNA, genome and transgenesis research, to human gene therapy.
INTRODUCTION
Originally discovered in plants and worms, RNA interference (RNAi) has become a powerful and versatile technology for gene regulation in biology and medicine (1,2). A major part of its popularity is due to the ease with which it is triggered in mammalian cells, as it merely requires the delivery of double-stranded RNA mimics of endogenous microRNAs (miRNAs), such as short hairpin RNAs, which engage the intrinsic cellular RNAi machinery for processing and target mRNA inhibition. Particularly appealing is that artificial RNAi triggers can be expressed from various promoters and encoded by non-viral or viral gene delivery vectors, yielding a comprehensive toolbox for applications ranging from fundamental gene annotation in cultured cells, to therapeutic (m)RNA suppression in humans. Very attractive clinical targets are viral pathogens such as the hepatitis B or C viruses (HBV or HCV, respectively) since they go through an obligatory RNA phase that is vulnerable to RNAi (3,4), and since many established treatments have limited efficacy and/or adverse side effects. Indeed, we and others have previously exemplified the tremendous potential of RNAi for robust and stable in vivo suppression of hepatitis viruses, including in HBV-transgenic mice that were infused with liver-specific Adeno-associated viral vectors of type 8 (AAV8) encoding anti-HBV shRNAs (5–9).
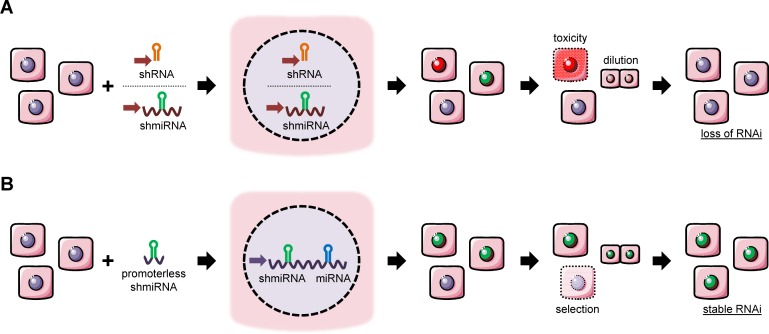
Still, RNAi applications and their clinical translation remain severely hampered by concerns over specificity, safety and longevity (Figure 1A). Particularly alarming are findings in different animal species, from mice to primates, that shRNA over-expression from strong RNA polymerase III promoters can cause cytotoxicity, organ failure and lethality (8,10–13). Possible reasons include adverse off-targeting and dose-dependent saturation of the cellular RNAi machinery which is needed for processing and functionality of miRNAs and other endogenous RNAi triggers. This has tempted us and others to improve RNAi expression strategies, for instance by pre-selecting inherently safe and potent shRNAs (8,14), or by placing shRNAs under weak and/or tissue-specific promoters (7). Another strategy to diminish in vivo RNAi toxicity is to embed an ectopic RNAi sequence within a cellular miRNA, by replacing one of the two arms of the double-stranded miRNA with the shRNA antisense arm, resulting in a so-called shmiRNA (15–19).
Figure 1.
Comparison of (A) conventional and (B) novel RNAi expression strategies. (A) Traditional delivery of exogenous RNAi sequences in the form of shRNAs (orange) or miRNA-embedded RNAi hairpins (shmiRNAs, green). Both strategies require an exogenous promoter (red arrow) for expression of the ectopic RNAi molecule. Depending on the strength of this promoter and on inherent features of the shRNA, this can result in adverse accumulation of RNAi triggers in the cell and ensuing cytotoxicity. Alternatively, cells will lose episomal shRNA or shmiRNA cassettes due to dilution during cell division. One solution (not depicted) can be stable integration using, e.g. retro- or lentiviruses, but this may cause insertional mutagenesis and oncogenesis. Moreover, insufficient control over the insertion events diminishes the options to govern RNAi expression levels. (B) In the new strategy introduced here, a minimal promoterless shmiRNA hairpin (green) is integrated into a cellular miRNA gene (blue), to be expressed from the cognate endogenous promoter at non-toxic levels. Co-integration of a selection marker (not shown) permits removal of cells that have failed to incorporate the RNAi hairpin, in turn resulting in a population of cells that stably and robustly express the shmiRNA.
While all these avenues can mitigate adverse RNAi effects to some extent, a major remaining challenge is to combine these advances with equally improved strategies for safe and specific ex or in vivo RNAi delivery. A promising option are vectors derived from AAV as they are non-pathogenic, easy-to-engineer and retargetable to tissues and cells of choice (20–24). However, because their genomes remain predominantly episomal and are rapidly lost in dividing cells (25) (Figure 1A), the usefulness of AAV/RNAi vectors is highest in quiescent tissues, such as the liver, or in applications requiring short-term RNAi expression. Conversely, integrating vectors derived from retro- or lentiviruses permit long-term RNAi expression, but due to their promiscuity bear a risk of insertional mutagenesis and progression to clonal expansion or oncogenesis (26–29). Mechanisms include disruption of tumor suppressor genes, or activation of neighboring oncogenes by vector-borne promoter/enhancer elements. The latter was observed in retro- or lentiviral gene therapy trials, and it was also noted in mice that developed hepatocellular carcinoma due to AAV vector integration into a certain locus (Rian) and activation of proximal small RNAs and genes (30–32). Additional concerns about imprecise vector integration include insufficient control over the number of insertion events and over local effects from the integration site, and consequently limited options to govern RNAi expression levels. Finally, ectopic promoters can vary in their strength between different cell types or become silenced over time, further complicating the fine-tuning of sh(mi)RNA expression levels and the establishment of lifelong RNAi phenotypes, even in non-dividing cells (33–35).
Here, we developed and validated a novel RNAi expression strategy that alleviates these vector-associated concerns and uncertainties, and that concurrently exploits the benefit of higher safety of miRNA-embedded shmiRNAs. To this end, we devised the original concept to stably integrate a minimal promoterless shmiRNA hairpin into an endogenous miRNA locus, in order to hijack the cellular miRNA promoter for long-term shmiRNA expression at robust yet non-toxic levels (Figure 1B). For proof-of-principle and convenient read-out, we harnessed TALE and CRISPR/Cas9 nucleases to insert a shmiRNA against HCV, a human pathogen that is susceptible to RNAi (36,37), into the genomic miR-122 locus (hcr) .The latter is abundantly and specifically expressed in liver cells, where HCV infection occurs (38). Using a comprehensive battery of assays including whole genome sequencing, we show that it is indeed possible to obtain engineered cellular clones that still largely resemble the parental cells, but that are stably protected against HCV replication and infection. As the identical strategy can likely be customized for many further miRNA loci and for any RNAi target, we expect the new concept introduced here to hold significant potential for a wide scope of basic or therapeutic RNAi applications.
MATERIALS AND METHODS
Details on plasmids, vectors, cell culture, transfections, PCRs, Northern blotting, luciferase assays, FISH and statistical analysis are found in Supplementary Methods.
Generation of stable shmiRNA cell lines
Huh7 cells were transfected as described in Supplementary Methods with TALEN or CRISPR/Cas9 expression plasmids together with the homologous recombination template pSSV9-hcr-donor-shmiRHCV318. Fourty-eight to 72 h post-transfection, cells were trypsinized and transferred to 15 cm dishes. They were then cultured for at least 15 days in Huh7 media supplemented with 500 μg/ml G418, before single colonies were picked and transferred to 96-well plates. Genomic DNA was extracted from these colonies using DirectPCR Lysis Reagent Cell (PeqLab, Erlangen, Germany) and analyzed by polymerase chain reaction (PCR) to assess the occurrence of proper homologous recombination. Positive colonies were expanded and eventually re-seeded in 24-well plates. Two wells of each cell line were transduced with an AAV vector expressing Cre recombinase, while a third well was treated with an AAV vector expressing YFP as a transduction control (see next chapter for details). Two days after transduction, genomic DNA was extracted from the cells of one of the Cre-treated wells and analyzed by PCR, to monitor the formation of minicircle DNA derived from Cre-mediated excision of the floxed NeoR cassette. The cells from the other well were trypsinized and transferred to 15 cm dishes to allow formation of single colonies for subcloning. The cells were then cultured for approximately seven days in Huh7 media without G418 (and diluted again, in case the initial cell density turned out to be too high for colony formation from a single cell). Afterward, colonies were again picked and analyzed by PCR to validate that the floxed NeoR cassette had been excised properly. Finally, the PCR product was sub-cloned and sequenced, and positive clones were characterized further.
To generate the stable HEK293 cells used in Supplementary Figure S9, no colonies were picked after treatment with the Cre-expressing AAV vector. Instead, based on the 100% efficiency of the AAV/Cre vector, the entire cell pool was used for further analysis.
AAV transductions
To eliminate the floxed NeoR cassette from the shmiRNA-engineered clones, they were transduced with AAV vectors expressing Cre recombinase. To this end, the different Huh7 or HEK293 clones were seeded at 5 × 104 cells per well in 24-well plates and 24 h later transduced with AAV vectors, expressing either Cre recombinase or a YFP control, at a multiplicity of infection (MOI) of 4 × 104 viral genomes per cell. Cells were re-seeded for colony picking (Huh7) or for luciferase assays (HEK293), or harvested for PCR analysis 48 h post-transduction.
Whole genome sequencing analysis
Whole genome sequencing was performed on Huh7 T2 31.3 and Huh7 wildtype (WT) cells to validate the specificity of shmiRHCV318 integration and to identify possible off-target integration sites. After library preparation, sequencing was performed using the Illumina HiSeq platform. Sequencing reads were aligned against the human reference genome hs37d5 (http://www.1000genomes.org/home) and the shmiRHCV318 as separate contig using the BWA-MEM (v0.7.8) software (39). Resulting BAM files were coordinate-sorted, merged and PCR duplicates marked by biobambam (v0.0.148) software (40). The resulting BAM files have an average coverage of 19.82x and 21.69x for WT and T2 31.3 cells, respectively (0x and 12.69x on the shmiRHCV318 contig). The integration site at chr18 and the shmiRHCV318 contig were visualized by using IGV (41).
Gene and miRNA expression profiling
Prior to using total RNA samples for gene and miRNA expression profiling, their quality was assessed via two methods. First, RNA concentration as well as A260/280 and A260/230 ratios were measured using a Nanovue Plus Spectrophotometer (GE Healthcare, Pasching, Austria). Second, the RNA integrity number (RIN) was determined using the Agilent 2100 Bioanalyzer and the Agilent RNA 6000 Nano Kit (both Agilent Technologies, Waldbronn, Germany) following the manufacturer's instructions. Samples with a high RNA quality (RIN near or equal to 10) were then subjected to gene and miRNA expression profiling at the German Cancer Research Center (DKFZ, Heidelberg, Germany), Genomics and Proteomics Core Facility. Three biological replicates (different cell passages) were provided for each cell clone.
Gene expression profiling was performed using Affymetrix Human Genome U133 Plus 2.0 arrays. Raw expression values were quantile-normalized and log2-transformed. A principal component analysis showed a cumulative R2 of 0.99 within the first two principal components. For hypothesis testing, log2 fold-changes of each sample (T2 31.3, TS 30.20 and U6 20.16) as compared to the WT samples and pairwise between the samples (after correction for the WT) were computed, and a fixed-effects linear model was fit for each individual gene to estimate expression differences. An empirical Bayes approach was used to moderate the standard errors of the normalized log2 fold-changes. Finally, we used a two-sided moderated t-test to compute P-values controlled by Benjamini–Hochberg for multiple testing. Genes with an absolute log2 fold-change larger than 1 and a corrected P-value < alpha = 0.05 were assigned to be differentially expressed with high significance.
MiRNA expression profiling was performed using the Agilent-046064 Unrestricted_Human_miRNA_V19.0_Microarray. Raw miRNA microarray data were pre-processed with background subtraction using the ‘normexp’ convolution standard method with subsequent quantile normalization as implemented in the Bioconductor limma package (v3.22.1) (42) for R statistical software (v3.1.2). Expression values were log2-transformed, and probe duplicates were averaged and quantile normalized to ensure comparability between samples. Potential batch effects between Agilent chips conducted at different dates were reduced by the ComBat-function in the Bioconductor sva-package (v3.12.0) on the basis of an empirical Bayesian framework (43).
We calculated differential gene expression on quantile-normalized Affymetrix mRNA expression profiles with the R statistical software (44) (v3.1.0). Data were filtered for low expression and variance with standard parameters, retaining 12 680 (of 54 675) probe IDs. Significance Analysis of Microarrays ((45), R-package ‘siggenes’ v1.40.0 (46)) was performed and differentially expressed probe IDs were called at a false discovery rate threshold of <0.05. The same procedure was applied to the Agilent microarray data to identify differentially expressed miRNAs.
HCV inhibition experiments
In vitro transcription of plasmids pFKi341PiLucNS3-3′-JFH1-dg, pFKi341PiLucNS3-3′-JFH1-dg-dGDD, pFKi389-JCR2a-dg-JC1 and pFKi389-JCR2a-dGDD-dg-JC1 (all 10 μg, MluI-linearized) was used to generate subgenomic or full-length HCV RNA, respectively. Likewise, in vitro transcription of 10 μg of XbaI-linearized plasmid pFK-sgDVs-R2A (47) resulted in subgenomic Dengue virus replicon RNA. DNA was purified by using the NucleoSpin Gel and PCR Clean up Kit (Macherey Nagel, Düren, Germany) according to the manufacturer's instructions. In vitro transcription reaction mixtures (total volume 100 μl) for use with T7 polymerase contained 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 3.125 mM of each nucleoside triphosphate, 1 U/μl RNasin (Promega), 0.1 μg/μl of plasmid DNA and 0.6 U/μl of T7 RNA polymerase (Promega). For HCV transcripts and use with SP6 polymerase, the mix contained 80 mM HEPES (pH 7.5), 16 mM MgCl2, 2 mM spermidine, 40 mM DTT, 3.125 mM each of rATP, rCTP and rUTP, 1.5625 mM of rGTP, 1 mM m7G(5′)ppp(5′)G RNA cap structure analog, 1 U/μl of RNasin, 0.1 μg/μl of plasmid DNA and 0.6 U/μl of SP6 RNA polymerase (Promega). After a 2 h incubation at 37°C (HCV) or 40°C (Dengue virus), 0.3 U of T7 RNA polymerase or 0.4 U of SP6 RNA polymerase, respectively, were added per μl of reaction mixture, and the reaction mixture was incubated overnight. Transcription was terminated by adding 1.2 U of RNase-free DNase (Promega) per μg plasmid DNA and 30 min incubation at 37°C. RNA was extracted with acidic phenol and chloroform, precipitated with isopropanol at room temperature and dissolved in RNase-free water.
For electroporations, single-cell suspensions of WT or engineered Huh7 clones were prepared by trypsinization, washing with 1x phosphate buffered saline (PBS) and resuspension at a concentration of 1 × 107 cells per ml in Cytomix (48) supplemented with 2 mM adenosine triphosphate (ATP) and 5 mM glutathione. Next, 2.5 μg in vitro transcripts were mixed with 100 μl cell suspension and transfected by electroporation using a GenePulser system (Bio-Rad, Hercules, CA, USA) and a 0.2 cm gap cuvette (Bio-Rad) at 500 μF and 166 V. Cells were diluted immediately in complete Dulbecco's modified Eagle's medium and seeded.
JCR2a virus was produced in Huh7.5 cells by electroporation of HCV RNA. Supernatants were harvested every 24 h between 48 and 96 h after electroporation. Infectious titer (median tissue culture infective dose, TCID50) was determined by limited dilution. For infection, Huh7 cells were seeded at 5 × 104 cells per well in a 24-well plate 1 day prior to infection with a MOI of 0.5.
For luciferase assays, cells were lysed in luciferase lysis buffer (1% [v/v] Triton X-100, 10% [v/v] glycerol, 25 mM glycyl-glycine pH 7.8, 15 mM μl MgSO4, 4 mM EGTA, kept at 4°C and freshly supplied with 1 mM DTT immediately before use). For luciferase measurements, cells were washed once with PBS, lysed directly in the 96-well plate with 30 μl lysis buffer per well and frozen at −80°C. Shortly before measurement, lysates were allowed to thaw at room temperature for 30 to 60 min. Firefly luciferase activity was measured for 3 s in a Mithras LB940 multimode microplate reader (Berthold Technologies, Bad Wildbad, Germany). Renilla luciferase activity was measured for 10 s. Firefly assay buffer (25 mM glycyl-glycine pH 7.8, 15 mM K2PO4 pH 7.8, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP) supplemented with 70 μM D-luciferin (PJK GmbH, Kleinblittersdorf, Germany), or Renilla assay buffer (25 mM glycyl-glycine pH 7.8, 15 mM K2PO4 pH 7.8, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT) supplemented with 1.5 μM Coelenterazine (PJK GmbH), was added to each well automatically prior to each measurement.
Data access
All microarray data are available in the NCBI Gene Expression Omnibus database (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE68638. The whole genome sequencing data have been deposited in the European Genome-phenome Archive (EGA, https://www.ebi.ac.uk/ega/home) under accession number EGAS00001001252.
RESULTS
Design and validation of an anti-HCV shmiRNA for integration into the hcr locus
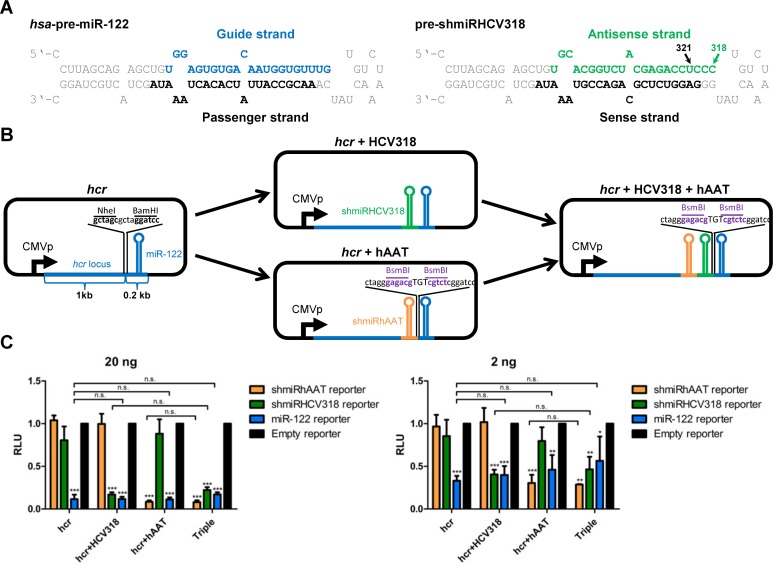
To create an effective anti-HCV shmiRNA, we harnessed the antisense sequence of the published HCV-specific shRNA HCV-321 (36) that binds a highly conserved target in the HCV 5′NTR (non-translated region) starting at nucleotide 321. For adaptation to the precursor of human miR-122 (used as scaffold for our shmiRNA design), we added three nucleotides to the 3′ end of the shRNA, yielding an extended 22 nucleotides long antisense sequence starting at HCV position 318 (accordingly named shmiRHCV318, Figure 2A). Importantly, we have recently validated anti-HCV efficacy of an analogous shRNA (shHCV318), proving that this part of the HCV genome is well accessible (49).
Figure 2.
Design and validation of shmiRHCV318. (A) Sequence and secondary structure of human (hsa-) pre-miR-122 and pre-shmiRHCV318. The nucleotide positions in the 5′ HCV region where the original HCV-321 shRNA or shmiRHCV318 start binding are indicated (321 or 318, respectively). (B) Schematic representation of expression plasmids created and used to verify that the integration site chosen within the hcr locus allows for multiplexed shmiRNA co-expression together with miR-122. The parental plasmid (left) contains 1.2 kb of the hcr locus comprising the miR-122 hairpin driven by a CMV promoter (CMVp). NheI and BamHI restriction sites were inserted 76 nt upstream of the miR-122 5p arm, to facilitate shmiRNA cloning. These sites were used to insert the shmiRHCV318 hairpin into the hcr plasmid (center, top). The plasmids containing shmiRhAAT (center, bottom) or both, shmiRhAAT and shmiRHCV318 hairpins (right) upstream of miR-122 carry two BsmBI restriction sites (purple) for insertion of additional shmiRNA hairpins. (C) Luciferase reporter assays (n = 3) from HEK293T cells transfected with the shmiRNA expression plasmids from panel B at high (20 ng) or low (2 ng) doses, verifying the possibility to multiplex shmiRNAs within the hcr locus without compromising functionality of miR-122 or co-expressed shmiRNAs (HCV318 and/or hAAT). psiCheck-2 plasmids with perfect binding sites for miR-122, shmiRHCV318 or shmiRhAAT in the 3′ UTR of Renilla luciferase were used as reporters. Data are normalized to the corresponding Firefly luciferase signal from each plasmid and to a psiCheck-2 vector without miRNA/shmiRNA binding sites (empty, set to 1.0). Statistical analysis was a one-way ANOVA with Bonferroni's multiple comparison post-test in which each reporter was compared to the empty reporter (stars), or in which the expression plasmids were compared amongst each other (indicated with the capped lines). RLU, relative light units; ***, P < 0.001; n.s., not significant. Error bars are S.D.
While considering a suitable integration site within the miR-122 locus (hcr), we realized that the precursor (pre-)miR-122 hairpin is located close to the putative hcr polyadenylation signal (Supplementary Figure S1A). It thus appeared reasonable to integrate the shmiRNA upstream of the miR-122 hairpin, to ensure its proper inclusion into the full transcript. We moreover argued that targeting a site inherently lacking a local RNA structure would increase the likelihood of preserving folding and processing of the miR-122 stem-loop. We therefore computationally predicted the RNA secondary structure of the hcr 3′ end including the precursor (pre-)miR-122 hairpin, and then selected one exposed site 63 nt upstream of pre-miR-122 (or 76 nt upstream of the miR-122 5p arm, respectively) for shmiRNA integration (Supplementary Figures S1B and S2).
To verify that this position permits miR-122 and shmiRNA co-expression, we generated plasmids expressing 1.2 kb of the hcr gene (hg38: chr18: 58450012–58451248) comprising miR-122 alone or co-encoding shmiRHCV318 (manually cloned into position hg38: chr18: 58451012). Two further constructs contained a shmiRNA against a different target (human alpha-1-antitrypsin, hAAT), either in place of shmiRHCV318 or in combination, the latter resulting in a triple hairpin (Figure 2B). These constructs served as control for the anti-HCV shmiRNA, and to assess whether our strategy would enable multiplexing of three hairpins. Co-transfections of HEK293T cells (human embryonic kidney cells lacking miR-122 expression) with these expression plasmids and luciferase reporters carrying perfect binding sites for the different hairpins confirmed that each shmiRNA potently and specifically inhibited its cognate target (Figure 2C). Notably, this was observed for both configurations, double (either shmiRHCV318 or shmiRhAAT plus miR-122) and triple hairpins (shmiRHCV318, shmiRhAAT and miR-122). Activity of the co-encoded miR-122 hairpin was comparably high in all four plasmids, and only marginally and non-significantly reduced in the triple versus the single miR-122 expression plasmid (Figure 2C, blue bars; best visible in the right graph). Collectively, this demonstrates the feasibility to co-express at least two exogenous shmiRNAs together with miR-122 without significantly compromising its function.
Efficient and specific anti-HCV shmiRNA integration into the miR-122 locus via homologous recombination
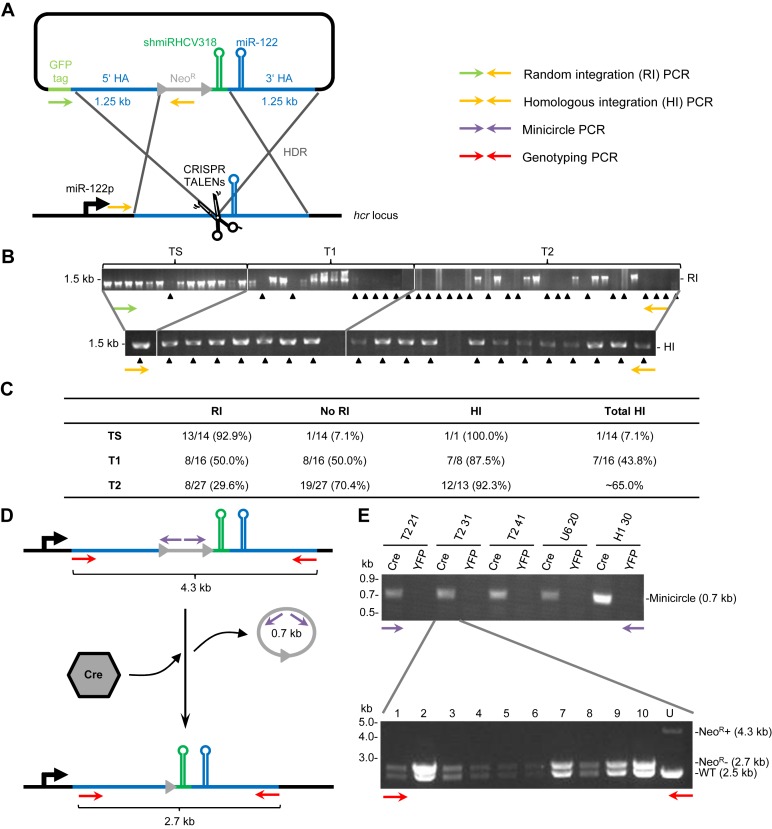
To site-specifically and efficiently integrate shmiRHCV318 into the hcr locus of liver cells, we devised a homologous recombination strategy based on potent TALEN and CRISPR constructs against human miR-122 from our lab (50) (Supplementary Figure S3 and Figure 3A). We chose the human hepatocarcinoma cell line Huh7 as target since it is susceptible to HCV infection and abundantly expresses miR-122 (38). Notably, despite being a tumor-derived cell line, Huh7 carries only two copies of the long arm of chromosome 18 where the hcr/miR-122 locus resides, as demonstrated by multiplex fluorescence-in-situ-hybridization (M-FISH) (Supplementary Figure S4). The recombination template contained the shmiRHCV318 hairpin plus a floxed neomycin resistance (NeoR) cassette for selection, flanked on each side by 1.25 kb of DNA homologous to the hcr locus (the 3′ homology arm comprised the miR-122 hairpin). Following nuclease and template co-transfection and G418 selection, single colonies were picked and analyzed by PCR, to detect and distinguish random versus directed shmiRHCV318 integration. Results from the TALEN-based approaches are shown in Figure 3B (for primer sequences see Supplementary Table S1), illustrating that roughly 90% of all clones that were negative for random integration showed proper insertion of the exogenous hairpin into the hcr locus. If we factor in that we pre-selected for, and excluded, negative clones (random integration), we still calculate around 65% positive clones (correct insertion) for the best TALEN pair T2 (Figure 3C).
Figure 3.
Integration of shmiRHCV318 into the hcr locus in Huh7 liver cells. (A) Schematic depiction of the strategy pursued to achieve this integration, including primer pairs used to detect integration or excision events, respectively. HA, homology arm; NeoR, neomycin resistance cassette; HDR, homology-directed repair; miR-122p, miR-122 promoter. (B) PCR to detect random integration (RI, top) or homologous integration (HI, bottom) after TALEN-mediated miR-122 editing. In the top panel, black arrow heads indicate clones that were negative for RI and hence further assessed for potential HI. A subset of results is shown in the bottom gel, where black arrow heads denote positive clones (no RI, but HI). Green or yellow arrows symbolize primers used (see panel A). TS, TALEN Seed; T1, TALEN 1; T2, TALEN 2. (C) Summary of observed integration events with the three TALEN pairs. Column ‘RI’ lists clones that were positive for RI and thus excluded from further analysis. Vice versa, column ‘No RI’ shows clones that are negative for RI (marked with arrow heads in the top gel in panel B) and thus potentially positive for HI. Out of these ‘No RI’ clones, a subset was further analyzed (panel B bottom); resulting numbers of clones with HI are summarized in column ‘HI’. Column ‘Total HI’ depicts numbers and percentages of clones positive for HI out of the total number of clones analyzed for each TALEN pair. For the T2 samples, this was estimated to be about 65% since we re-tested 13 of the 19 clones without RI from the original 27 (70.4%) and found 12 to be positive for HI (92.3%). (D) Schematic depiction of the strategy used to eliminate the NeoR cassette after selection, by treating the engineered cells with a Cre recombinase-expressing AAV vector. Also shown is the resulting 0.7 kb minicircle, whose successful detection by PCR is exemplified in the gel at the top of panel E. (E) Lanes YFP are negative controls where the cells were treated with an AAV vector expressing YFP instead of Cre recombinase. The bottom gel shows results from PCR reactions to genotype single clones picked from the Cre-treated Huh7 T2 31 pool (representative example). Expected bands were: wildtype amplicon (WT) 2.5 kb; integration amplicon without NeoR cassette (NeoR-) 2.7 kb, and integration amplicon with NeoR cassette (NeoR+) 4.3 kb. Note the 100% efficiency as all 10 clones show the pattern expected for a heterozygous clone (loss of the 4.3 kb and gain of the 2.7 kb band). The arrows in panel E indicate specific PCR primers as shown in panels A and D. U, untransduced Huh7 T2 31 cells (control).
Curiously, in our first experiments, the efficiencies of homologous integration were substantially lower with the TS TALEN pair as well as the CRISPR strategy, reaching only up to 7.1% or 3.3%, respectively (Figure 3B and C, and data not shown). We then realized that both, the TS TALEN pair and the anti-miR-122 gRNA, bind and inadvertently cleave a sequence that partially overlaps with the miR-122 hairpin and is thus fully conserved in the shmiRHCV318-containing repair template (see also Supplementary Figure S3), providing a likely explanation for their inferior performance. In line with this, the much more effective TALEN pairs T1 and T2 exclusively recognize and cleave the endogenous hcr locus, but not the repair DNA. For the gRNA-based strategy, a possible solution was to mutate the protospacer-adjacent motif (PAM, a critical three nucleotide sequence directly downstream of the actual target sequence) in the repair template. This should abolish its adverse Cas9-mediated cutting before, during or after recombination, and instead restrict Cas9 cleavage to the endogenous locus. Accordingly, we generated an improved repair template in which we point-mutated the gRNA PAM from the canonical TGG to a TGC that is not recognized by Streptococcus pyogenes Cas9 (Supplementary Figure S5A). Importantly, because the point mutation occurred in a small bulge of the predicted pre-miR-122 structure, it did not affect miR-122 functionality (Supplementary Figure S5B). In addition, we replaced the Cas9 cDNA in our original construct (50) with a more potently expressed variant (see Supplementary Methods). These two modifications boosted the efficiency by 2-fold (new repair template) or 5-fold (new Cas9 cDNA), respectively, or by roughly 10-fold altogether (Supplementary Figure S5C). Notably, the resulting 33% clones that were positive for homologous integration using the improved CRISPR strategy now approached the results obtained with our second-best TALEN pair T1 (43.8%, Figure 3B and C).
To remove the floxed NeoR cassette from positive clones, selected candidates were treated with an AAV vector expressing Cre recombinase under the strong CAG (chicken beta-actin) promoter (Figure 3D). Successful excision was then confirmed by PCR-based detection of resulting 0.7 kb minicircles (Figure 3E top). From positive pools of cells, individual cellular clones were expanded further and re-analyzed using PCR primers that spanned the entire integration site (red arrows in Figure 3D). As exemplified for clone T2 31 in Figure 3E (bottom), this procedure was highly efficient, resulting in elimination of the NeoR cassette in all of the tested AAV/Cre-treated progeny clones (10 out of 10, 100%). As expected, this restored G418 sensitivity of these cells, further evidencing complete removal of the selection marker (Supplementary Figure S6).
Sequencing analysis of integration events and nuclease-induced modifications in the miR-122 locus
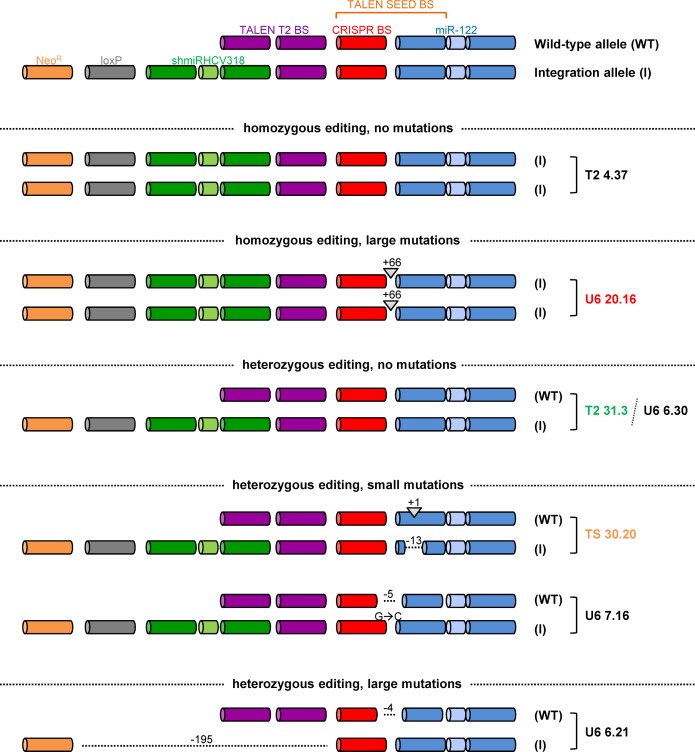
To verify proper integration of the shmiRHCV318 hairpin, PCR products spanning the targeted region were amplified from genomic DNA from some of the engineered cellular clones and then sequenced. Surprisingly, we observed a broad spectrum of nuclease-induced modifications within the edited miR-122 hairpin, as illustrated for seven clones in Figure 4 (complete sequences are shown in Supplementary Figure S7). Importantly, successful integration of the exogenous shmiRHCV318 hairpin into at least one miR-122 allele was detected in all clones. The only exception was the Cas9/gRNA-derived clone U6 6.21 which carried a deletion of 4 nucleotides in the WT allele, and a 195 nucleotide deletion in the edited allele that spanned the shmiRHCV318 hairpin (but not the NeoR cassette, explaining why this clone was G418-resistant).
Figure 4.
Schematic depiction of the results from sequencing of seven selected Huh7 clones after shmiRHCV318 integration. Shown on top are the different elements that are found in WT miR-122 alleles or expected after successful integration of the shmiRHCV318 hairpin plus floxed NeoR cassette, respectively. BS, binding site. Illustrated underneath are the different cellular clones, grouped by homo- versus heterozygous editing, and by no, small or large mutations. The G > C conversion in clone U6 7.16 was expected from the use of a PAM-mutated repair template. Blue: miR-122; red: CRISPR BS; purple: TALEN T2 BS; green: shmiRHCV318; grey: loxP site; orange: neomycin resistance cassette. See Supplementary Figure S7 for more details on all sequences.
Amongst the six other clones, two (T2 4.37 and U6 20.16) were homozygous for shmiRHCV318 integration, while the remaining four were heterozygous. This was concluded from detection of either one (homozygous) or two (heterozygous) PCR products after AAV/Cre treatment (see Figure 3E above for an example for a heterozygous clone), and from sequencing of at least five independent bacterial colonies after subcloning of each product. The miR-122 hairpin had remained fully intact in three of these six clones, including one of the clones with a homozygous shmiRHCV318 integration (T2 4.37). The other homozygous clone, U6 20.16, carried an identical 66 nucleotide insertion right upstream of the PAM sequence in both alleles. Two of the four heterozygous clones (T2 31.3 and U6 6.30) had intact miR-122 alleles, while the other two (TS 30.20 and U6 7.16) showed modifications ranging from a 1 nucleotide insertion, to a 13 nucleotide deletion. Possible explanations for this spectrum of on-target mutations are provided in Supplementary Discussion part A.
Originally, we had intended to derive and characterize a clone with minimal modification of the hcr locus, i.e. heterozygous shmiRHCV318 integration and preserved miR-122 sequence, as is indeed the case with clones T2 31.3 and U6 6.30. However, as miR-122 is a host factor for HCV (38), we reasoned that partial miR-122 disruption might in fact inhibit viral replication synergistically, and thus decided to also study clones with miR-122 modifications. Overall, we had to restrict our selection to three different clones in order to maintain feasibility of the comprehensive downstream analysis, including HCV infection experiments, miRNA profiling and whole genome sequencing (see below). We therefore selected the following three representative clones for all further studies: (i) T2 31.3 (heterozygous for shmiRHCV318, both miR-122 alleles intact), (ii) TS 30.20 (heterozygous for shmiRHCV318, minor modifications in both miR-122 alleles) and (iii) U6 20.16 (homozygous for shmiRHCV318, also homozygous for a 66 nucleotide insertion upstream of miR-122).
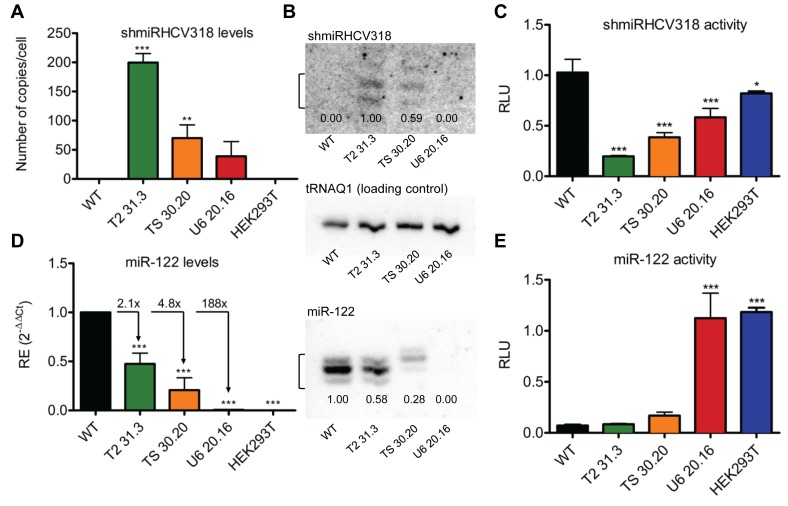
Characterization of anti-HCV shmiRNA levels and of the different miR-122 phenotypes
Steady-state levels and functionality of the two hairpins (shmiRHCV318 and miR-122) were first assessed by quantitative real-time PCR (qRT-PCR), Northern blotting and luciferase reporter assays (Figure 5). All three clones expressed shmiRHCV318 at different levels as detected by qRT-PCR that correlated with the degree of miR-122 mutation (compare Figure 4 and Supplementary Figure S7), from about 200 copies per cell for the fully intact clone T2 31.3, to about 25 for the largely miR-122-mutated clone U6 20.16 (Figure 5A). These numbers were mirrored by different relative levels of mature shmiRHCV318 as measured by Northern blotting (Figure 5B top), and they moreover corresponded well to the extent of inhibition of a shmiRHCV318-sensitive luciferase reporter, reaching around 80% for the best clone T2 31.3 (Figure 5C). A similar pattern was noted for endogenous miR-122, which was most prominently detected in clone T2 31.3 but almost absent in clone U6 20.16 (Figure 5B bottom). Mature miR-122 steady-state levels were reduced by 2.1- or 4.8-fold (by qRT-PCR) in clones T2 31.3 and TS 30.20, respectively (Figure 5D), congruent with 2-fold lower levels of precursor miR-122 in clone T2 31.3 (Supplementary Figure S8). See Supplementary Discussion part B for possible reasons for the mild reduction in clone T2 31.3 despite the absence of mutations in both miR-122 alleles. Notably, this barely affected knockdown of a miR-122-sensitive luciferase reporter (Figure 5E). In contrast, clone U6 20.16 which showed 188-fold lower miR-122 expression in qRT-PCR (Figure 5D) and no detectable expression by Northern blotting (Figure 5B bottom) also exhibited no miR-122 reporter inhibition (Figure 5E), characterizing this clone as a complete miR-122 knock-out.
Figure 5.
Characterization of selected shmiRHCV318-engineered Huh7 clones. (A and D) qRT-PCR-based quantification of (A) shmiRHCV318 or (D) miR-122 expression in the three clones or in HEK293T cells (negative control). Total RNA harvested at three different cell passages (n = 3 each) was used. Data were normalized (ΔΔCt method) to the small nucleolar RNA SNORD25 and to Huh7 WT RNA. Error bars are S.D. To calculate absolute numbers of shmiRHCV318 copies per cell (c/c) in panel A, a standard curve was prepared using a shmiRHCV318 mimic. One outlier was discarded for clone TS 30.20 with a 90% confidence level applying Dixon's Q-Test. (B) Northern blot to determine shmiRHCV318 and miR-122 expression in the three selected clones. tRNAQ1 served as loading control. Numbers underneath the bands represent expression of shmiRHCV318 and miR-122 relative to T2 31.3 and WT cells, respectively. The miR-122 shift in clone TS 30.20 was expected from the one bp insertion (see Figure 4 and Supplementary Figure S7). (C and E) Luciferase reporter assays (n = 3 each) to determine functionality of shmiRHCV318 and miR-122 in the engineered selected clones at three different passages. psiCheck-2 reporters with appropriate binding sites were used (see Figure 2C for more details). Error bars are S.D. Statistical analysis was a one-way ANOVA with a Bonferroni's multiple comparison post-test. *P < 0.05; **P < 0.01; ***P < 0.001.
As a control experiment to verify the liver lineage specificity of our strategy, we also integrated the shmiRHCV318 hairpin into human HEK293 kidney cells in which the miR-122 locus is inherently silent. With up to 81% (Supplementary Figure S9A and B), the efficiencies of homologous integration even surpassed those observed in Huh7 cells (Figure 3C), perhaps related to the higher transfectability of HEK293 cells. Interestingly, we noted a moderate read-through effect from the Rous Sarcoma Virus (RSV) promoter driving the NeoR cassette that resulted in shmiRHCV318 and miR-122 expression, as detected by luciferase reporter assays (Supplementary Figure S9C and D). Most importantly, however, this effect was eliminated once the NeoR cassette and hence the ectopic promoter were floxed out (compare dark and light blue bars in Supplementary Figure S9D). Accordingly, the fully engineered HEK293 cells no longer exhibited suppression of the shmiRHCV318 or miR-122 luciferase reporters, identical to the parental HEK293 cells and in contrast to the Huh7 clone T2 31.3 that was included as positive control. We thus conclude that expression of integrated shmiRHCV318 is strictly dependent on the activity of the endogenous miR-122 promoter (silent in HEK293, but active in Huh7 cells), and that, vice versa, integration of the exogenous hairpin does not trans-activate the silent hcr locus in non-hepatic cells, both adding to the stringency of our approach.
Comprehensive gene and miRNA expression analysis to verify the similarity of lead clone T2 31.3 to WT Huh7 cells
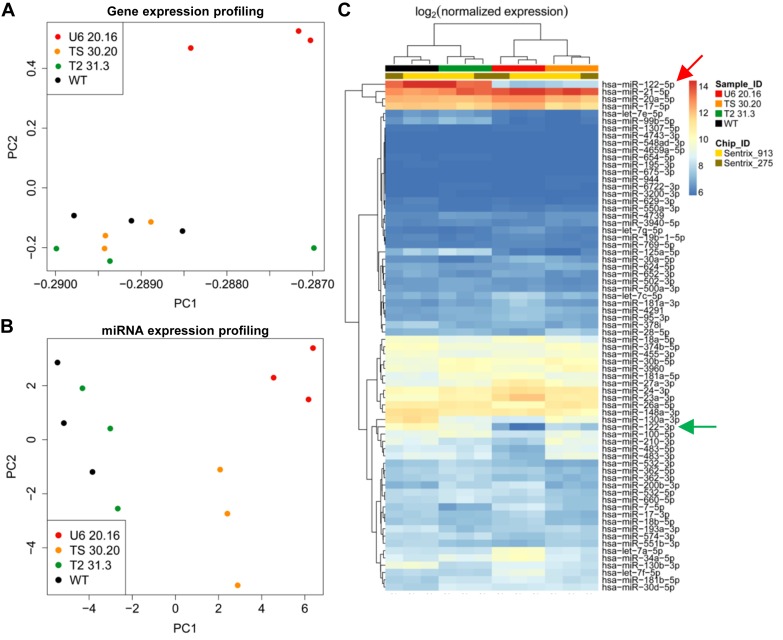
To further assess the similarity of the three selected cellular clones and WT Huh7 cells, gene expression profiling of all cells was performed at three different passages. Analyses of significantly differentially expressed genes (DEG) revealed a minor number of DEG in clones T2 31.3 (24 DEG) and TS 30.20 (16 DEG) (all genes are listed in the Supplementary Expression profiles). Conversely, a much larger number of genes were affected in clone U6 20.16 (1797 DEG). This is evident from Supplementary Figure S10 that visualizes the quantiles of the observed versus the expected statistics. The higher similarity of the two TALEN-derived clones to WT Huh7 cells, especially of clone T2 31.3, is moreover illustrated by principal component analysis (PCA) which showed that clones T2 31.3 and TS 30.20 cluster together and resemble WT Huh7 cells, but are distinct from clone U6 20.16 (Figure 6A).
Figure 6.
Transcriptome profiles of shmiRHCV318-engineered Huh7 clones. (A and B) Principal component analysis (PCA) performed after (A) gene expression profiling and (B) miRNA expression profiling of the shmiRHCV318-engineered Huh7 clones. (C) Heat map depicting the most differentially expressed miRNAs in the three selected shmiRHCV318-engineered Huh7 clones and WT Huh7. Clustering of differentially expressed miRNAs was based on empirical Bayes analysis (n = 70, false discovery rate < 0.05). The greatest difference is seen for miR-122-5p (red arrow), which is highly expressed in Huh7 and T2 31.3, but not in TS 30.20 and U6 20.16 cells (the latter showed the lowest expression). Similar differences can be observed for miR-122-3p (green arrow).
Congruent with this, global miRNA expression profiling confirmed a notable similarity between clone T2 31.3 and WT Huh7, as opposed to clone U6 20.16 in which numerous miRNAs were expressed aberrantly (Figure 6B; see Supplementary Discussion part C for possible reasons). The largest discrepancies in miRNA expression between this clone and the parental cells were noted for the two strands of miR-122 (Figure 6C, arrows), concordant with the qRT-PCR, Northern blotting, sequencing and functional data (see above). In clone U6 20.16, miR-122 is strongly down-regulated (fold-change FC = 0.06), while the least difference was found in T2 31.3 (FC = 0.48); clone TS 30.20 was in between (FC = 0.18).
Altogether, these data consistently indicate that shmiRHCV318 expression per se (even the high levels in clone T2 31.3), and also the additional mutations of miR-122 in clone TS 30.20, only mildly affected global gene and miRNA expression. This is in contrast to the largely miR-122-mutated clone U6 20.16 in which we noted 100-fold more dysregulated genes (Supplementary Expression profiles) and poorer clustering (Figure 6) with the parental cells.
Next, we performed whole genome sequencing of T2 31.3, which had emerged as our lead clone at this point based on its superior similarity to WT Huh7 cells. We mapped the whole genome sequencing reads of T2 31.3 and WT Huh7 cells to a concatenated reference genome consisting of GRCh37 version hs37d5 and the shmiRNA contig. This identified 37 read pairs which mapped to shmiRHCV318 and the expected integration locus on chromosome 18 (Supplementary Figure S11). In addition, all reads overlapping the shmiRNA contig borders support the intended integration site. Importantly, we detected no read pair or read overhang mapping to a different genomic position or being unmappable, which would have indicated off-target integration in the Huh7 genome.
Furthermore, we identified single nucleotide variants and short indels via Platypus (51), and structural variants via Manta (52), again using GRCh37 version hs37d5 as reference genome. Only the variants that passed all internal filters within Platypus and Manta were then used in comparisons of WT Huh7 cells and our lead clone T2 31.3. Supplementary Table S2 shows that in all categories, parental Huh7 cells had even more private genetic variants than clone T2 31.3. This suggests that the private variants in T2 31.3 accumulated during Huh7 clonal derivation rather than representing specific nuclease-induced variations.
As a whole, these complementary analyses suggested the presence of a single shmiRHCV318 integration within an intact miR-122 locus at the expected position in clone T2 31.3. This independently verifies our sequencing data in Figure 4 and Supplementary Figure S7, and illustrates the possibility to juxtapose high precision with low genotoxicity through our nuclease-mediated miRNA engineering strategy.
Validation of efficient and specific impairment of HCV replication in engineered Huh7 cells
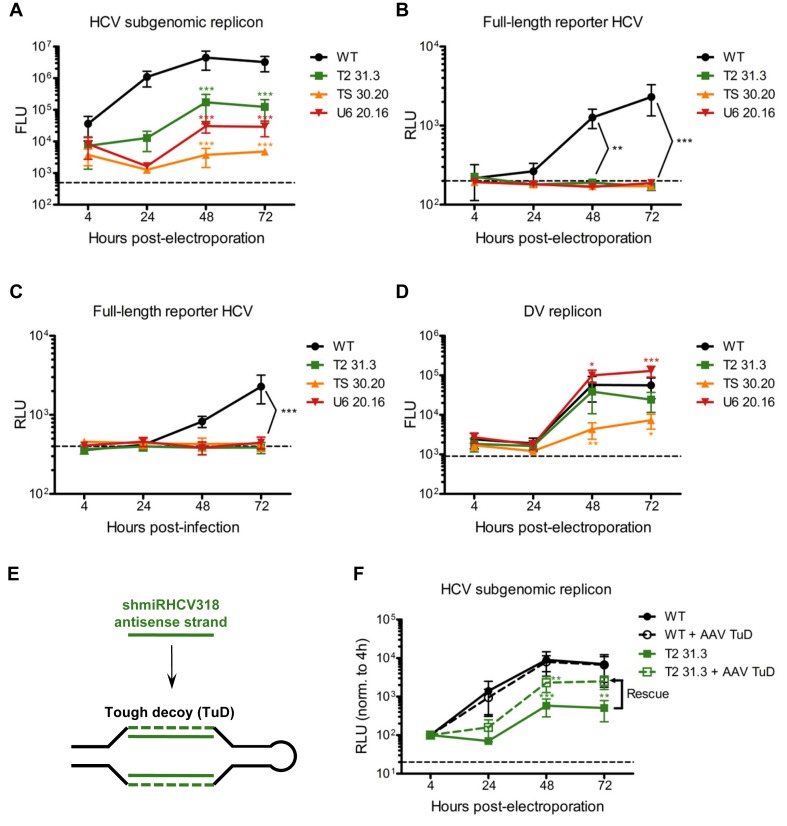
Finally, we challenged all three cell clones with HCV to study whether their genetic engineering had induced the expected resistance against HCV. Fourty-eight and 72 h after electroporation of a subgenomic HCV replicon encoding a luciferase reporter (surrogate marker for HCV replication (53)), we noted about 10- (T2 31.3) to 100-fold (TS 30.20) reduced viral replication in all clones as compared to WT Huh7 (Figure 7A and Supplementary Figure S12A). HCV inhibition was likewise pronounced when we electroporated a full-length reporter virus genome encoding luciferase and able to form infectious HCV particles (54) (Figure 7B). In fact, remaining luciferase levels were indistinguishable from mock-electroporated cells (dashed line in Figure 7B) or a replication-deficient full-length HCV control (Supplementary Figure S12B). Identical findings were made after infection with purified full-length reporter HCV, which was also completely blocked in our engineered cells (Figure 7C). Curiously, a Dengue virus (DV) subgenomic replicon used as control also amplified inefficiently in the TS 30.20 cells, albeit it still expressed above the background levels observed in the mock control (Figure 7D) or expected with fully replication-deficient DV (55). This indicates that this clone, which also showed the highest inhibition of the HCV subgenomic replicon (Figure 7A), has a defect that broadly impairs replication of flaviviruses. Importantly, the same DV control behaved normally in clone T2 31.3 and replicated even slightly better in clone U6 20.16, verifying that the HCV inhibition observed in these two clones is truly specific and mediated by the integrated anti-HCV hairpin. This was further validated through sequestration of the anti-HCV shmiRHCV318 in clone T2 31.3 with a ‘tough decoy’ inhibitor (56), which rescued the inhibition of HCV replication (Figure 7E and F and Supplementary Figure S13).
Figure 7.
Demonstration of the HCV resistance of shmiRHCV318-engineered Huh7 cells. (A–D, F) Luciferase reporter assays (n = 3 each) to measure inhibition of HCV replication and anti-viral specificity. Huh7 cells were (A, B, D, F) electroporated or (C) infected with a (A, F) subgenomic HCV replicon, (B, C) a full-length reporter HCV, or (D) a Dengue virus (DV) replicon. All co-encoded luciferase reporters. Dashed lines indicate background levels of the assays obtained with mock electroporations/infections. (E) Scheme illustrating the mode of action of the shmiRHCV318 antisense strand-specific tough decoy (TuD), i.e. stable sequestration of this strand by two complementary sequences within the TuD hairpin. (F) Huh7 WT or T2 31.3 cells were transduced with AAV-DJ encoding a shmiRHCV318 antisense strand-specific TuD at 4 × 104 particles per cell and 1.5 days later electroporated with a subgenomic HCV replicon. At 72 h post-electroporation, TuD expression had partially rescued the shmiRHCV318-mediated inhibition of HCV replication, resulting in HCV replication levels that are not significantly different from those in Huh7 WT cells. Samples were harvested at the indicated time points post-electroporation/-infection, and all data were normalized to the 4 h time point. Error bars are S.D. FLU, Firefly luciferase units; RLU, Renilla luciferase units. Statistical analysis was a two-way ANOVA with a Bonferroni's multiple comparison post-test in which every engineered Huh7 clone was compared to WT Huh7 cells. *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
Here, we demonstrated the feasibility to usurp an endogenous miRNA locus for stable, precise and potent expression of an exogenous promoterless RNAi hairpin, using nucleases for its site-specific integration. Our lead cellular clone, T2 31.3, (i) carries a single targeted shmiRHCV318 integration, (ii) shows neither mutations in the miR-122 alleles nor substantial changes in gene or miRNA expression (including miR-122), (iii) stably expresses shmiRHCV318 for at least 20 passages (data not shown) and (iv) specifically and robustly inhibits HCV, together illustrating the great potential of our new RNAi expression strategy.
Compared to conventional stable transfection or transduction of RNAi cassettes, our unique approach essentially differs in that it obviates the need for ectopic regulatory elements for RNAi hairpin transcription, especially vector-borne RNA polymerase II or III promoters. This is highly advantageous considering that exogenous promoters can dysregulate cellular genes via cis- or trans-activation, compete for transcription factors, get silenced over time and/or vary in strength between different cell types (35). A recent example strikingly illustrating these concerns are findings in mice that AAV vector integration into the Rian locus can cause hepatocellular carcinoma (HCC), depending on vector dose and promoter/enhancer elements (31). Accordingly, HCC incidences reached over 70% when the transgene was expressed from a chicken beta-actin or a thyroxine-binding globuline promoter, but 0% with a human alpha-1-antitrypsin promoter (the same used in clinical AAV trials (57)). For reasons unknown, the first two promoters but not the third had induced an upregulation of various small non-coding RNAs including miR-543 as well as of the Rtl1 gene (associated with HCC (58)). Importantly, these adverse localized transcriptional perturbations were not due to the integration per se since all three vector variants were found in the Rian locus. Similarly, another study provided evidence that integration of the WT AAV2 3′ inverted terminal repeat (ITR), believed to have intrinsic enhancer activity, might be associated with HCC in humans (59). It must be noted that to date, over 100 clinical trials with AAV vectors yielded no evidence that this putative tumorigenic property is conserved in recombinant ITRs (30,60). Nonetheless, these latest studies highlight dangers and malignancy risks that can be associated with ectopic DNA cassettes comprising strong promoters/enhancers and capable of chromosomal integration. Our new strategy, where the shmiRNA lacks its own promoter and is placed under an endogenous miRNA promoter, helps to alleviate such concerns about cis or trans perturbation of cellular gene expression, and concomitantly promises persistent and cell-specific expression at physiological levels (depending on the selected miRNA).
With these features, our work complements and expands on recent efforts to integrate exogenous promoterless cDNAs into tissue-specific genes that likewise used homologous recombination and followed the same rationale. In one example, the Porteus lab integrated fluorescent reporters under the β- or γ-globin promoters in human K562 cells (61), to screen small compounds for effects on transcription of the endogenous β-/γ-globin genes. A second example comes from the Kay lab who integrated the human factor IX cDNA into the abundantly expressed albumin locus in mouse livers, using AAV8 vectors for cDNA delivery (33). Another study also used AAV vectors and harnessed the same locus, but integrated other (partial) therapeutic cDNAs (62). Moreover, in this work, the efficiency of cDNA integration was boosted by co-expressing Zinc-finger nucleases (ZFNs) against the albumin locus from AAV vectors. Finally noteworthy are two studies that generated transgenic animals via CRISPR-mediated integration of promoterless ectopic sequences into cellular genes. In one, the human albumin cDNA was inserted into the albumin locus in swine zygotes, creating transgenic pigs secreting human albumin in their blood (63). In the other study, transgenic mice were created by inserting an artificial miRNA into an intron of the eEF-2 gene, via microinjection of mouse embryos and subsequent PCR-based selection of positive offspring (34).
Our own work adds a unique and versatile concept to this rapidly growing list of strategies that harness endogenous elements to control exogenous sequences, by exemplifying the possibility to engineer and repurpose a human miRNA locus for shmiRNA expression. Like some of the aforementioned studies, we employed TALEN and CRISPR nucleases to induce double-stranded DNA breaks and thus boost the efficiency of homology-directed repair. This benefit of including nucleases was also highlighted by Sharma et al. who failed to obtain detectable factor IX levels in mice when trying to integrate the cDNA in the absence of co-delivered ZFNs (62). Further informative is that Barzel et al. achieved only 0.5% gene editing in their nuclease-free in vivo targeting approach (33). Concurrently, the Zhang lab reported over 40% gene editing with AAV/CRISPR vectors in mouse livers (64), the same organ targeted by Sharma or Barzel et al. (33,62). Importantly, this high number only reflects gene cleavage and faulty repair, leaving open what the frequencies of targeted integration of foreign DNA will be in an in vivo setting.
Generally, one may conclude that a common asset of promoterless cDNA or RNAi hairpin integration strategies is a gain in specificity and safety, compared to promiscuous chromosomal insertion of vectors. Concomitantly, we note that our use of current nuclease generations resulted in sporadic insertions or deletions within the targeted region. Moreover, we found a slight, 2-fold reduction of mature miR-122 in our lead clone T2 31.3 despite the absence of mutations in both miR-122 alleles (see Supplementary Discussion parts A and B for likely explanations). Together, this illustrates the spectrum of possible on-target modifications that can be found and that should be considered in forthcoming iterations of our new approach. Whether or not such extra alterations can be tolerated will depend on the exact target and application. In the case of miR-122 and HCV, they might in fact be beneficial in view of this miRNA's critical role as viral dependency factor and of in vivo data that miR-122 inhibition suppresses viremia in HCV-infected chimpanzees and patients (65–67). Congruent with this, clone U6 20.16 with a large mutation in both miR-122 alleles even outperformed our lead clone T2 31.3 at inhibiting subgenomic HCV replicons. However, U6 20.16 also differed in global gene and miRNA expression profiles (see Supplementary Discussion part C for possible reasons), hampering direct comparison. One should moreover consider that miR-122 plays various essential roles in the liver and acts as tumor suppressor (68,69), suggesting the importance of keeping at least one allele intact when engineering this locus. As noted, this was indeed the case in clone T2 31.3 and in three others: T2 4.37 and U6 6.30 (both miR-122 alleles intact), and U6 7.16 (one allele intact); i.e. 4 out of 7 (57%).
Next to on-target effects, one must consider possible adverse off-target modifications, although they seem rare in our lead clone T2 31.3 within the detection limits of our assays. Fortunately, nuclease specificity is an area of highly active inquiry, and there are already numerous encouraging options to minimize off-targeting. Most notable are the latest generations of CRISPR/Cas9 systems with improved specificity and safety, including truncated gRNAs, double-nicking approaches, Cas9 orthologs, high-fidelity Cas9 mutants, the Cpf1 nuclease or split Cas9 variants (70–80). Looming efforts to enhance CRISPR specificity will concomitantly benefit from the expanding repertoire of experimental options to detect these adverse events, such as ChiP-Seq, Digenome-Seq or GUIDE-Seq (81–86). While our present focus was to provide proof-of-concept for our novel strategy including a first characterization of possible outcomes, we readily anticipate that future iterations will greatly profit from these tools and hence see an increase in specificity and thus safety.
These improved CRISPR systems and their compatibility with gene delivery vehicles (87,88) also imply a broad applicability of our new RNAi expression strategy beyond our feasibility study in cultured liver cells. One intriguing use could be ex vivo engineering of induced pluripotent stem cells before differentiation into liver cells, or direct modification of primary human hepatocytes. In both cases, integration of RNAi hairpins could genetically ‘immunize’ these cells against infection with hepatitis viruses prior to transplantation into a patient with hepatitis. Likewise, engineering an anti-HIV RNAi hairpin into a miRNA locus in hematopoietic progenitor cells could yield HIV-resistant cells for autologous transplantation. This would be similar to, but probably genetically safer than, clinical trials in which such hairpins were stably delivered with lentiviral vectors under exogenous promoters (89). Interestingly, miR-122 is induced during HIV infection (90), implying that our miR-122-specific tools and strategies could be directly applied. Moreover, notable is our evidence that the miR-122 locus (and likely others as well) tolerates at least two different additional hairpins, which could be useful to inhibit highly mutagenic viruses such as HCV or HIV with combinatorial RNAi (91,92). Such attempts will profit from the vast experience in multiplexing of RNA hairpins gained in other contexts, including long hairpin RNA precursors, engineered miRNA polycistrons, extended short hairpin RNAs or clusters of TuD inhibitors, to name a few (92–96). A third attractive example for clinical applications are human cancers which are frequently characterized by dysregulation of cellular miRNAs (97), suggesting the possibility to exploit viral CRISPR vectors (50,64,87,88) to integrate cytotoxic RNAi hairpins into miRNA genes that are aberrantly up-regulated.
We moreover see great potential of our new approach for generation of transgenic knockdown animals that stably express RNAi hairpins, to study gene function in normal physiology or disease. For these models to be stringent, it is essential to avoid local effects of integration sites, multiple single or tandem insertions, or transgene silencing. All these side effects are typical for conventional transgenesis technologies, but may be overcome by repurposing cellular promoters for expression of minimal RNAi hairpins. One study exploited a ubiquitously expressed mouse gene for this purpose (34), but our own strategy that uses a tissue-specific miRNA locus may be even more beneficial as it combines robust expression with high lineage specificity. Finally, our approach to link exogenous to endogenous RNAi triggers, or, generally, to precisely engineer and harness miRNA loci for expression of detectable foreign RNA sequences, should prove valuable for investigations into fundamental cellular RNAi and genome biology. This could include a direct replacement of a target miRNA with another artificial or natural miRNA, to study effects on physiology, differentiation or other basic cellular parameters.
We acknowledge that all these future applications, especially direct in vivo use and other advances toward clinical translation, require improvements in efficiency and accuracy of nuclease-mediated DNA modification. Ideally, these will alleviate the necessity for clonal selection, which results from the possibility of undesirable on- and off-target modifications that is inherent to all current nucleases. We are aware of these restrictions but reiterate that these are not intrinsic to our original strategy, and that some concerns about efficiency and specificity will be resolved with newer generations of nucleases and delivery systems. Particularly encouraging is the aforementioned report of greater than 40% targeted gene editing in adult mouse livers, using moderate doses of an AAV8 vector expressing Cas9 from S. aureus and delivered via peripheral infusion (64). Similar optimism is raised by the successful AAV/ZFN-mediated in vivo integration of promoterless cDNA in mouse livers (62). We further note an intriguing study showing cell-to-cell transfer of small RNAs in livers of mice (98), implying that therapeutic benefit may be achieved from editing only a fraction of cells in the liver (and potentially other organs). It may also be rewarding to replace Cre recombinase with alternatives posing a lower inherent risk of chromosomal translocation (albeit not observed in our study), e.g. hyperactive piggyBac transposase delivered as protein (99). Finally promising is that homology-directed repair can be promoted in mammalian cells and mice by suppressing key factors in the non-homologous end joining pathway, such as DNA ligase IV, which expands the options to enhance frequencies of targeted modification of miRNA loci (100–102). In light of these and other advances, and of the extreme pace with which the entire genome engineering field is currently progressing, we are optimistic that the way will soon be paved for a wealth of exciting ex or in vivo applications of our new concept for safe and accurate expression of exogenous RNAi hairpins.
Acknowledgments
The authors thank the microarray and sequencing units of the DKFZ Genomics and Proteomics Core Facility for providing gene expression, miRNA expression as well as whole genome sequencing analysis. The authors are grateful to Sarah Klinnert for help with cloning, as well as to members of the Grimm lab for critical reading.
Footnotes
Present address: Marc P. Windisch, Hepatitis Research Laboratory, Institut Pasteur Korea, Seongnam-si, Gyeonggi-do, Rep. of Korea.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Research Foundation [DFG, EXC81 (Cluster of Excellence CellNetworks) to E.S., S.G., S.M. and D.G., SFB1129 (Collaborative Research Center 1129) to D.G. (TP2), TRR179 (Transregional Collaborative Research Center 179) to R.B. (TP9) and D.G. (TP18), SPP1395 (InKoMBio, TH 900/6-1) to B.K.]; Helmholtz Initiative for Synthetic Biology [E.S and D.G.]; Heidelberg University Graduate Academy [PhD completion grant to E.S.]; HBIGS International Graduate School [MD/PhD program to D.R.]; European Research Council [Starting grant Latent Causes (259294) to F.J.T.]; Federal Ministry of Education and Research [BMBF, e:Med program for systems biology (PANC-STRAT consortium, 01ZX1305) to T.B.]. Funding for open access charge: Heidelberg University Hospital.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grimm D., Kay M.A. Therapeutic application of RNAi: Is mRNA targeting finally ready for prime time? J. Clin. Invest. 2007;117:3633–3641. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castanotto D., Rossi J.J. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimm D., Kay M.A. Therapeutic short hairpin RNA expression in the liver: viral targets and vectors. Gene Ther. 2006;13:563–575. doi: 10.1038/sj.gt.3302727. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg M.S., Arbuthnot P. Progress in the use of RNA interference as a therapy for chronic hepatitis B virus infection. Genome Med. 2010;2:28. doi: 10.1186/gm149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCaffrey A.P., Meuse L., Pham T.T., Conklin D.S., Hannon G.J., Kay M.A. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 6.Chen C.C., Sun C.P., Ma H.I., Fang C.C., Wu P.Y., Xiao X., Tao M.H. Comparative study of anti-hepatitis B virus RNA interference by double-stranded adeno-associated virus serotypes 7, 8, and 9. Mol. Ther. 2009;17:352–359. doi: 10.1038/mt.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giering J.C., Grimm D., Storm T.A., Kay M.A. Expression of shRNA from a tissue-specific pol II promoter is an effective and safe RNAi therapeutic. Mol. Ther. 2008;16:1630–1636. doi: 10.1038/mt.2008.144. [DOI] [PubMed] [Google Scholar]
- 8.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 9.Michler T., Grosse S., Mockenhaupt S., Roeder N., Stueckler F., Knapp B., Ko C., Heikenwalder M., Protzer U., Grimm D. Blocking sense-strand activity improves potency, safety and specificity of anti-hepatitis B virus short hairpin RNA. EMBO Mol. Med. 2016;8:1082–1098. doi: 10.15252/emmm.201506172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn M., Witting S.R., Ruiz R., Saxena R., Morral N. Constitutive expression of short hairpin RNA in vivo triggers buildup of mature hairpin molecules. Hum. Gene Ther. 2011;22:1483–1497. doi: 10.1089/hum.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimm D. The dose can make the poison: lessons learned from adverse in vivo toxicities caused by RNAi overexpression. Silence. 2011;2:8. doi: 10.1186/1758-907X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimm D., Wang L., Lee J.S., Schurmann N., Gu S., Borner K., Storm T.A., Kay M.A. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J. Clin. Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suhy D.A., Kao S.C., Mao T., Whiteley L., Denise H., Souberbielle B., Burdick A.D., Hayes K., Wright J.F., Lavender H., et al. Safe, long-term hepatic expression of anti-HCV shRNA in a nonhuman primate model. Mol. Ther. 2012;20:1737–1749. doi: 10.1038/mt.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schopman N.C., Liu Y.P., Konstantinova P., ter Brake O., Berkhout B. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral Res. 2010;86:204–211. doi: 10.1016/j.antiviral.2010.02.320. [DOI] [PubMed] [Google Scholar]
- 15.Boudreau R.L., Martins I., Davidson B.L. Artificial microRNAs as siRNA shuttles: Improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maczuga P., Lubelski J., van Logtenstein R., Borel F., Blits B., Fakkert E., Costessi A., Butler D., van Deventer S., Petry H., et al. Embedding siRNA sequences targeting apolipoprotein B100 in shRNA and miRNA scaffolds results in differential processing and in vivo efficacy. Mol. Ther. 2013;21:217–227. doi: 10.1038/mt.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B., et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. U.S.A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y., Wagner E.J., Cullen B.R. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 19.Maczuga P., Verheij J., van der Loos C., van Logtenstein R., Hooijer G., Martier R., Borel F., Lubelski J., Koornneef A., Blits B., et al. Therapeutic expression of hairpins targeting apolipoprotein B100 induces phenotypic and transcriptome changes in murine liver. Gene Ther. 2014;21:60–70. doi: 10.1038/gt.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asokan A., Schaffer D.V., Samulski R.J. The AAV vector toolkit: Poised at the clinical crossroads. Mol. Ther. 2012;20:699–708. doi: 10.1038/mt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimm D., Kay M.A. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 2003;3:281–304. doi: 10.2174/1566523034578285. [DOI] [PubMed] [Google Scholar]
- 22.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotterman M.A., Schaffer D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimm D., Zolotukhin S. E Pluribus Unum: 50 Years of Research, Millions of Viruses, and One Goal-Tailored Acceleration of AAV Evolution. Mol. Ther. 2015;23:1819–1831. doi: 10.1038/mt.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakai H., Yant S.R., Storm T.A., Fuess S., Meuse L., Kay M.A. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J. Virol. 2001;75:6969–6976. doi: 10.1128/JVI.75.15.6969-6976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aiuti A., Biasco L., Scaramuzza S., Ferrua F., Cicalese M.P., Baricordi C., Dionisio F., Calabria A., Giannelli S., Castiello M.C., et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deichmann A., Hacein-Bey-Abina S., Schmidt M., Garrigue A., Brugman M.H., Hu J., Glimm H., Gyapay G., Prum B., Fraser C.C., et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hacein-Bey-Abina S., Garrigue A., Wang G.P., Soulier J., Lim A., Morillon E., Clappier E., Caccavelli L., Delabesse E., Beldjord K., et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothe M., Schambach A., Biasco L. Safety of gene therapy: new insights to a puzzling case. Curr. Gene Ther. 2014;14:429–436. doi: 10.2174/1566523214666140918110905. [DOI] [PubMed] [Google Scholar]
- 30.Buning H., Schmidt M. Adeno-associated Vector Toxicity-To Be or Not to Be? Mol. Ther. 2015;23:1673–1675. doi: 10.1038/mt.2015.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandler R.J., LaFave M.C., Varshney G.K., Trivedi N.S., Carrillo-Carrasco N., Senac J.S., Wu W., Hoffmann V., Elkahloun A.G., Burgess S.M., et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J. Clin. Invest. 2015;125:870–880. doi: 10.1172/JCI79213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donsante A., Miller D.G., Li Y., Vogler C., Brunt E.M., Russell D.W., Sands M.S. AAV vector integration sites in mouse hepatocellular carcinoma. Science. 2007;317:477. doi: 10.1126/science.1142658. [DOI] [PubMed] [Google Scholar]
- 33.Barzel A., Paulk N.K., Shi Y., Huang Y., Chu K., Zhang F., Valdmanis P.N., Spector L.P., Porteus M.H., Gaensler K.M., et al. Promoterless gene targeting without nucleases ameliorates haemophilia B in mice. Nature. 2015;517:360–364. doi: 10.1038/nature13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura H., Gurumurthy C.B., Sato T., Sato M., Ohtsuka M. CRISPR/Cas9-based generation of knockdown mice by intronic insertion of artificial microRNA using longer single-stranded DNA. Sci. Rep. 2015;5:12799. doi: 10.1038/srep12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.Z., Lahn B.T. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kronke J., Kittler R., Buchholz F., Windisch M.P., Pietschmann T., Bartenschlager R., Frese M. Alternative approaches for efficient inhibition of hepatitis C virus RNA replication by small interfering RNAs. J. Virol. 2004;78:3436–3446. doi: 10.1128/JVI.78.7.3436-3446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lisowski L., Elazar M., Chu K., Glenn J.S., Kay M.A. The anti-genomic (negative) strand of Hepatitis C Virus is not targetable by shRNA. Nucleic Acids Res. 2013;41:3688–3698. doi: 10.1093/nar/gkt068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 39.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tischler G., Leonard S. biobambam: Tools for read pair collation based algorithms on BAM files. Source Code Biol. Med. 2014;9:13. [Google Scholar]
- 41.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wettenhall J.M., Smyth G.K. limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
- 43.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 44.Team R.C. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 45.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwender H. siggenes: Multiple testing using SAM and Efron's empirical Bayes approaches. 2012. R package version 1.36.0.
- 47.Fischl W., Bartenschlager R. High-throughput screening using dengue virus reporter genomes. Methods Mol. Biol. 2013;1030:205–219. doi: 10.1007/978-1-62703-484-5_17. [DOI] [PubMed] [Google Scholar]
- 48.van den Hoff M.J., Moorman A.F., Lamers W.H. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res. 1992;20:2902. doi: 10.1093/nar/20.11.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mockenhaupt S., Grosse S., Rupp D., Bartenschlager R., Grimm D. Alleviation of off-target effects from vector-encoded shRNAs via codelivered RNA decoys. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E4007–E4016. doi: 10.1073/pnas.1510476112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Senis E., Fatouros C., Grosse S., Wiedtke E., Niopek D., Mueller A.K., Borner K., Grimm D. CRISPR/Cas9-mediated genome engineering: an adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 2014;9:1402–1412. doi: 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- 51.Rimmer A., Phan H., Mathieson I., Iqbal Z., Twigg S.R., Wilkie A.O., McVean G., Lunter G. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., Schulz-Trieglaff O., Shaw R., Barnes B., Schlesinger F., Cox A.J., Kruglyak S., Saunders C.T. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32:1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 53.Lohmann V., Hoffmann S., Herian U., Penin F., Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poenisch M., Metz P., Blankenburg H., Ruggieri A., Lee J.Y., Rupp D., Rebhan I., Diederich K., Kaderali L., Domingues F.S., et al. Identification of HNRNPK as regulator of hepatitis C virus particle production. PLoS Pathog. 2015;11:e1004573. doi: 10.1371/journal.ppat.1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chatel-Chaix L., Fischl W., Scaturro P., Cortese M., Kallis S., Bartenschlager M., Fischer B., Bartenschlager R. A combined genetic-proteomic approach identifies residues within Dengue virus NS4B critical for interaction with NS3 and viral replication. J. Virol. 2015;89:7170–7186. doi: 10.1128/JVI.00867-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haraguchi T., Ozaki Y., Iba H. Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res. 2009;37:e43. doi: 10.1093/nar/gkp040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manno C.S., Pierce G.F., Arruda V.R., Glader B., Ragni M., Rasko J.J., Ozelo M.C., Hoots K., Blatt P., Konkle B., et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 2006;12:342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 58.Ranzani M., Cesana D., Bartholomae C.C., Sanvito F., Pala M., Benedicenti F., Gallina P., Sergi L.S., Merella S., Bulfone A., et al. Lentiviral vector-based insertional mutagenesis identifies genes associated with liver cancer. Nat. Methods. 2013;10:155–161. doi: 10.1038/nmeth.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nault J.C., Datta S., Imbeaud S., Franconi A., Mallet M., Couchy G., Letouze E., Pilati C., Verret B., Blanc J.F., et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 2015;47:1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 60.Berns K.I., Byrne B.J., Flotte T.R., Gao G., Hauswirth W.W., Herzog R.W., Muzyczka N., VandenDriessche T., Xiao X., Zolotukhin S., et al. Adeno-associated virus Type 2 and hepatocellular carcinoma? Hum. Gene Ther. 2015;26:779–781. doi: 10.1089/hum.2015.29014.kib. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voit R.A., Hendel A., Pruett-Miller S.M., Porteus M.H. Nuclease-mediated gene editing by homologous recombination of the human globin locus. Nucleic Acids Res. 2014;42:1365–1378. doi: 10.1093/nar/gkt947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sharma R., Anguela X.M., Doyon Y., Wechsler T., DeKelver R.C., Sproul S., Paschon D.E., Miller J.C., Davidson R.J., Shivak D., et al. In vivo genome editing of the albumin locus as a platform for protein replacement therapy. Blood. 2015;126:1777–1784. doi: 10.1182/blood-2014-12-615492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng J., Wang Y., Jiang J., Zhou X., Song L., Wang L., Ding C., Qin J., Liu L., Wang W., et al. Production of human albumin in pigs through CRISPR/Cas9-Mediated knockin of human cDNA into Swine Albumin Locus in the Zygotes. Sci. Rep. 2015;5:16705. doi: 10.1038/srep16705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S., et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janssen H.L., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., et al. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 66.Lanford R.E., Hildebrandt-Eriksen E.S., Petri A., Persson R., Lindow M., Munk M.E., Kauppinen S., Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van der Ree M.H., van der Meer A.J., de Bruijne J., Maan R., van Vliet A., Welzel T.M., Zeuzem S., Lawitz E.J., Rodriguez-Torres M., Kupcova V., et al. Long-term safety and efficacy of microRNA-targeted therapy in chronic hepatitis C patients. Antiviral Res. 2014;111:53–59. doi: 10.1016/j.antiviral.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 68.Hsu S.H., Wang B., Kota J., Yu J., Costinean S., Kutay H., Yu L., Bai S., La Perle K., Chivukula R.R., et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J. Clin. Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsai W.C., Hsu S.D., Hsu C.S., Lai T.C., Chen S.J., Shen R., Huang Y., Chen H.C., Lee C.H., Tsai T.F., et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J. Clin. Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 71.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A., et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zetsche B., Volz S.E., Zhang F. A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 2015;33:139–142. doi: 10.1038/nbt.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kleinstiver B.P., Pattanayak V., Prew M.S., Tsai S.Q., Nguyen N.T., Zheng Z., Joung J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kleinstiver B.P., Prew M.S., Tsai S.Q., Nguyen N.T., Topkar V.V., Zheng Z., Joung J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P., Li Z., Peterson R.T., Yeh J.R., et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wyvekens N., Topkar V.V., Khayter C., Joung J.K., Tsai S.Q. Dimeric CRISPR RNA-guided FokI-dCas9 nucleases directed by truncated gRNAs for highly specific genome editing. Hum. Gene Ther. 2015;26:425–431. doi: 10.1089/hum.2015.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cencic R., Miura H., Malina A., Robert F., Ethier S., Schmeing T.M., Dostie J., Pelletier J. Protospacer adjacent motif (PAM)-distal sequences engage CRISPR Cas9 DNA target cleavage. PLoS One. 2014;9:e109213. doi: 10.1371/journal.pone.0109213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fujita T., Fujii H. Isolation of specific genomic regions and identification of associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Methods Mol. Biol. 2015;1288:43–52. doi: 10.1007/978-1-4939-2474-5_4. [DOI] [PubMed] [Google Scholar]
- 83.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 84.Kuscu C., Arslan S., Singh R., Thorpe J., Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014;32:677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 85.O'Geen H., Henry I.M., Bhakta M.S., Meckler J.F., Segal D.J. A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res. 2015;43:3389–3404. doi: 10.1093/nar/gkv137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P., et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gaj T., Epstein B.E., Schaffer D.V. Genome engineering using adeno-associated virus: Basic and clinical research applications. Mol. Ther. 2015;24:458–464. doi: 10.1038/mt.2015.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schmidt F., Grimm D. CRISPR genome engineering and viral gene delivery: A case of mutual attraction. Biotechnol. J. 2015;10:258–272. doi: 10.1002/biot.201400529. [DOI] [PubMed] [Google Scholar]
- 89.DiGiusto D.L., Krishnan A., Li L., Li H., Li S., Rao A., Mi S., Yam P., Stinson S., Kalos M., et al. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci. Transl. Med. 2010;2:36ra43. doi: 10.1126/scitranslmed.3000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Triboulet R., Mari B., Lin Y.L., Chable-Bessia C., Bennasser Y., Lebrigand K., Cardinaud B., Maurin T., Barbry P., Baillat V., et al. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science. 2007;315:1579–1582. doi: 10.1126/science.1136319. [DOI] [PubMed] [Google Scholar]
- 91.Grimm D., Kay M.A. Combinatorial RNAi: A winning strategy for the race against evolving targets? Mol. Ther. 2007;15:878–888. doi: 10.1038/sj.mt.6300116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saayman S., Arbuthnot P., Weinberg M.S. Deriving four functional anti-HIV siRNAs from a single Pol III-generated transcript comprising two adjacent long hairpin RNA precursors. Nucleic Acids Res. 2010;38:6652–6663. doi: 10.1093/nar/gkq460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hollensen A.K., Bak R.O., Haslund D., Mikkelsen J.G. Suppression of microRNAs by dual-targeting and clustered Tough Decoy inhibitors. RNA Biol. 2013;10:406–414. doi: 10.4161/rna.23543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu Y.P., Haasnoot J., ter Brake O., Berkhout B., Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y.P., von Eije K.J., Schopman N.C., Westerink J.T., ter Brake O., Haasnoot J., Berkhout B. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol. Ther. 2009;17:1712–1723. doi: 10.1038/mt.2009.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang X., Marcucci K., Anguela X., Couto L.B. Preclinical evaluation of an anti-HCV miRNA cluster for treatment of HCV infection. Mol. Ther. 2013;21:588–601. doi: 10.1038/mt.2012.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kong Y.W., Ferland-McCollough D., Jackson T.J., Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13:e249–258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 98.Pan Q., Ramakrishnaiah V., Henry S., Fouraschen S., de Ruiter P.E., Kwekkeboom J., Tilanus H.W., Janssen H.L., van der Laan L.J. Hepatic cell-to-cell transmission of small silencing RNA can extend the therapeutic reach of RNA interference (RNAi) Gut. 2012;61:1330–1339. doi: 10.1136/gutjnl-2011-300449. [DOI] [PubMed] [Google Scholar]
- 99.Cai Y., Bak R.O., Krogh L.B., Staunstrup N.H., Moldt B., Corydon T.J., Schroder L.D., Mikkelsen J.G. DNA transposition by protein transduction of the piggyBac transposase from lentiviral Gag precursors. Nucleic Acids Res. 2014;42:e28. doi: 10.1093/nar/gkt1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chu V.T., Weber T., Wefers B., Wurst W., Sander S., Rajewsky K., Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat. Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 101.Ho T.T., Zhou N., Huang J., Koirala P., Xu M., Fung R., Wu F., Mo Y.Y. Targeting non-coding RNAs with the CRISPR/Cas9 system in human cell lines. Nucleic Acids Res. 2015;43:e17. doi: 10.1093/nar/gku1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Maruyama T., Dougan S.K., Truttmann M.C., Bilate A.M., Ingram J.R., Ploegh H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]