Abstract
ChIA-PET2 is a versatile and flexible pipeline for analyzing different types of ChIA-PET data from raw sequencing reads to chromatin loops. ChIA-PET2 integrates all steps required for ChIA-PET data analysis, including linker trimming, read alignment, duplicate removal, peak calling and chromatin loop calling. It supports different kinds of ChIA-PET data generated from different ChIA-PET protocols and also provides quality controls for different steps of ChIA-PET analysis. In addition, ChIA-PET2 can use phased genotype data to call allele-specific chromatin interactions. We applied ChIA-PET2 to different ChIA-PET datasets, demonstrating its significantly improved performance as well as its ability to easily process ChIA-PET raw data. ChIA-PET2 is available at https://github.com/GuipengLi/ChIA-PET2.
INTRODUCTION
A number of high-throughput methods based on nuclear proximity ligation have been developed to detect genome-wide chromatin interactions, including high-throughput chromosome conformation capture (Hi-C) and chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) (1,2). While Hi-C was developed to capture all chromatin interactions and is effective for mapping large-scale structures such as chromatin compartments and topologically associated domains (1,3), ChIA-PET is emerging as an important experimental method for detecting specific protein-mediated chromatin loops genome-wide at high resolution. In principle, ChIA-PET requires several main steps such as cross-linking the chromatin, proximity ligating the interacting fragments with linkers and sequencing the paired-ends of DNA fragments to estimate the frequency of chromatin interactions (2,4). The use of ChIA-PET has helped deepen our view of 3D genome organization and chromatin impact on gene regulation. For instance, ER-α-binding sites are anchored at gene promoters through long-range chromatin interactions to regulate target genes (2). CTCF, cohesin and ZNF143 are the key architectural factors of 3D chromatin structure (5,6). Enhancer-promoter interactions are highly cell-type specific (6). The CFTR gene promoter interacts with different distal cell-type specific regulatory elements that are all located within the same TAD (7). Cell identity controlling genes are located within CTCF–CTCF loops in mammalian chromosomes (8). Pivotal genes in the reprogramming process are transcribed within physical proximity to each other in embryonic stem cells (9). Chromatin loop disruption may alter gene expression (10), and more importantly, oncogene activation could be caused by genetic variants that disrupt chromatin loops (11). Allele-specific chromatin loops were observed at some imprinted loci, such as the H19/IGF2 locus (5). All these findings require the accurate detection of chromatin loops, in which robust ChIA-PET assays and efficient data analysis play an important role.
However, similar to other genome-wide high-throughput sequencing experiments, ChIA-PET usually requires hundreds of millions of paired-end sequencing reads. The unique protocol of ChIA-PET requires a specific bioinformatics workflow to process and analyze data, which makes the study of ChIA-PET data challenging. A robust, versatile and easy-to-use analysis pipeline for ChIA-PET data is in great need, especially for experimental biologists. A workflow for processing ChIA-PET raw data often requires these common steps, linker trimming, read alignment, paired-end tag (PET) filtering, PCR duplicate removal, peak calling and chromatin interaction calling. There exist several published tools, e.g., ChIA-PET Tool (CPT) (12), ChiaSig (13), MICC (14), Mango (15) and 3CPET (16), to process and analyze ChIA-PET data. As summarized in Table 1 for the comparison of these tools with our ChIA-PET2, CPT fails to correct the major sources of bias (14,15) while ChiaSig and MICC focus on significant loop calling which is only a part of the data analysis workflow. Though Mango was designed for correcting major sources of bias from genomic proximity and provides a complete pipeline, it was designed only for half-linker ChIA-PET data processing. More importantly, as shown below, we found that Mango is too conservative at the significant loop calling step and suffers a relatively low reproducibility between replicates. The models of these methods have been reviewed in detail (17). The newly improved (bridge linker) ChIA-PET protocol for long-reads has several critical differences from the original protocol (4), it has crippled those published ChIA-PET analysis pipelines as they are not capable of analyzing the datasets generated by the new ChIA-PET protocol. Additionally, none of those tools support allele-specific ChIA-PET analysis or comprehensive data quality control measures.
Table 1. Comparing solutions for ChIA-PET data analysis.
| Linker trimming | Fuzzy search | Read alignment | PETs detection | PCR removing | Peak calling | Loop detection | Significant loop calling | Quality control | Allele-specific | |
|---|---|---|---|---|---|---|---|---|---|---|
| CPT | Half-linker | batman | Y | Y | Y | Y | Y | |||
| ChiaSig | Y | |||||||||
| MICC | Y | |||||||||
| Mango | Half-linker | bowtie2 | Y | Y | Y | Y | Y | |||
| ChIA-PET2 | Half-linker, Bridge linker | Y | bwa | Y | Y | Y | Y | Y | Y | Y |
The blank means the function is not supported by the tool. CPT and Mango is only capable for half-linker ChIA-PET data. ChiaSig and MICC only perform the significant loop calling step. When searching linkers, CPT and Mango use exact search while ChIA-PET2 can apply fuzzy search. Only ChIA-PET2 offers allele-specific analysis and quality control of ChIA-PET data.
To fill these gaps, we developed ChIA-PET2, an easy-to-use and complete analysis pipeline to process both bridge-linker and half-linker ChIA-PET data from raw sequencing reads to significant chromatin loop calls. ChIA-PET2 can detect chromatin loops with a significantly higher sensitivity and reproducibility than the existing pipeline at the same false discovery rate. ChIA-PET2 integrates all steps required for processing ChIA-PET datasets. Mismatches are allowed at the linker trimming step, which rescues a large portion of pair-end tags (PETs). Multi-threading is supported to speed up the processing time. Quality control measures are supported at different steps of the ChIA-PET analysis. When phased genotype data are available, ChIA-PET2 is also able to detect allele-specific chromatin loops.
MATERIALS AND METHODS
Public datasets used
Two public ChIA-PET datasets were processed: CTCF ChIA-PET data from human GM12878 cells (4) and POL2 ChIA-PET data from human K562 cells (18). The first is currently the largest dataset available, generated by the bridge-linker (long-reads) ChIA-PET protocol while the latter was generated by the half-linker (short-reads) ChIA-PET protocol and has two replicates. The ChIP-Seq data were retrieved from the ENCODE data repository site (19). Phased genotype data for the GM12878 genome were extracted from the Illumina Platinum Genomes Project (http://www.illumina.com/platinumgenomes/). The genomic view of different ChIP-Seq, DNaseq-Seq and RNA-Seq data was obtained using IGV (20,21).
ChIA-PET2 workflow
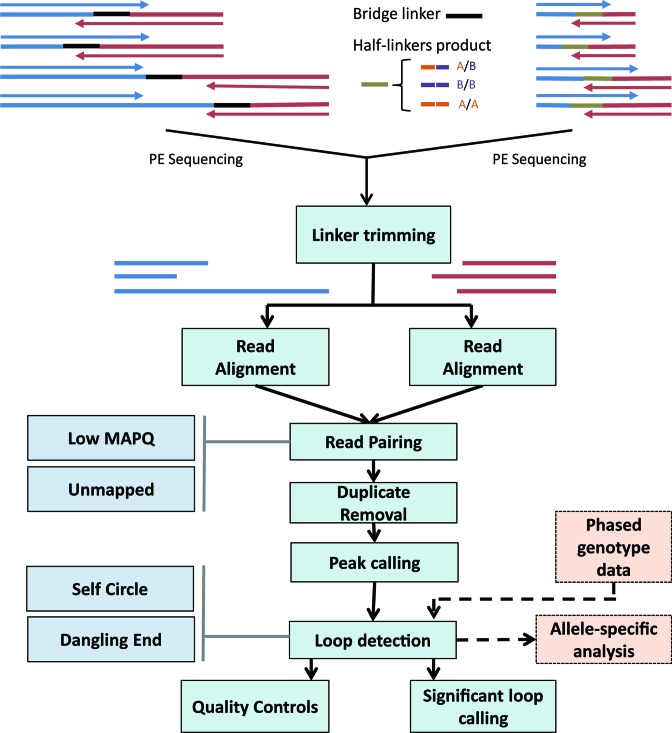
ChIA-PET2 is organized into six main steps (Figure 1): (i) linker trimming, (ii) read alignment, (iii) detection and filtering of valid PETs, (iv) peak calling, (v) chromatin loop calling and (vi) quality control. After the chromatin loops are detected, MICC is applied to reduce the random ligation and random collision noise and to estimate the statistical confidence of the chromatin loops. When phased genotype data are available, ChIA-PET2 allows users to call the allele-specific PETs and loops. Additionally, ChIA-PET2 can build a Hi-C style contact matrix, which is helpful for understanding the higher topological configuration of chromatin loops and 3D genome organization.
Figure 1.
ChIA-PET2 workflow. Linkers are first trimmed for each pair of reads in different ways according to different ChIA-PET protocols. Only read pairs that satisfy certain user-defined conditions are kept. Then, each end of the read pairs is aligned to the reference genome independently using bwa. PETs with high mapping quality scores are kept to call ChIP-Seq-like peaks by MACS2. The pairing information is not used in the peak calling step. Once peaks are called, PETs that link these peaks are clustered as loops in the loop detection step. If phased genotype data are available, ChIA-PET2 will assign the PETs to parental alleles. Statistical metrics are plotted as quality controls. After the loops are detected, a significant loop calling step is applied using MICC.
Linker trimming
We assume that the adapter sequences have been removed by users, such that the input paired-end reads are adapter-free. In this context, linker trimming is the first step in the ChIA-PET data analysis. As DNA fragment sizes vary, the sequenced reads may read through the linker or not. The linker sequence may occur within the sequenced reads entirely or partly; it may also not occur in the sequenced reads. ChIA-PET2 allows users to set the configuration of the types of reads to be kept. Trimmed reads that exceed the length threshold and satisfy the configuration requirements are kept and used for sequential steps. Users can choose the bridge-linker mode or half-linker mode according to their experiment protocols. Mismatches are allowed in the linker-searching step by implementation of the bitap fuzzy search algorithm (22). We checked the results of fuzzy searching of bridge linkers in the sequencing reads (Supplementary Table S1). By allowing one mismatch in the linker sequence, >16 million PETs were rescued, which resulted in a 12.85% improvement of the valid PET ratio. Since all the sequential steps are based on the valid PETs, a significant improvement of the valid PET ratio can result in a higher sensitivity of loop calling using the same dataset.
Read alignment and read pairing
Read pairs are independently aligned to the reference genome using bwa. Users can choose either bwa-aln or bwa-mem aligner. Once the reads are aligned, the reads are paired again to build the PETs. Only PETs uniquely mapped with a high mapping quality (MAPQ threshold) are retained. PETs with only one read aligned can also be retained optionally because these reads may be informative in the ChIP-Seq-like peak calling step.
Peak calling and Loop calling
Duplicated PETs are retained with only one copy before peak calling to reduce the PCR duplicate bias. MACS2 is applied to call peaks using the PETs, but the pair information is not used in the peak calling step. Once the peaks are called, they are used as loop anchors. A PET that links two different peaks is counted as one interaction between the two peaks. The PET counts of two different peaks measure the frequency of the chromatin interaction between these two peaks. Overall, there are four types of PETs in a ChIA-PET dataset: (i) PET linking two different peaks, (ii) PET in the same peak, (iii) PET with one and only one end inside a peak and (iv) PET with no ends inside peaks. The most informative PETs are the type-1 PETs. A PET with a small genomic span, e.g. <8 kb, if linked to two different peaks, will contribute to loop calling. In this way, we avoid to use an empirical distance cut-off of the PET. The size of the smallest loop one can detect depends on the nearest peaks one can distinguish and the supportive PET count. The PET counts and peak depths are calculated for the input of MICC. MICC systematically removes random ligation and random collision noise to call the significant chromatin loops. Details of the Bayesian mixture model in MICC can be found in our previous work (14). Additional hard filters, such as PET count threshold, can also be applied.
Allele-specific analysis
ChIA-PET2 is able to detect allele-specific chromatin loops when phased haplotype information is available. In this context, the sequencing reads are first aligned and PETs are built using the standard procedure. Then, ChIA-PET2 examines all PETs spanning polymorphic sites, checks the nucleotides at the appropriate positions and assigns the reads to either the paternal (p) or maternal (m) allele. PETs with conflicting SNPs are filtered out. Reads without SNPs are labelled as unassigned (u). The combination of assigned labels of PETs could be p-p, m-m, p-m, m-p, p-u, m-u, u-p and u-m. p-p, m-m, p-m and m-p together compose the phased PETs. p-u, m-u, u-p and u-m compose the extended phased PETs.
Quality control
To assess the quality of a ChIA-PET experiment, quality control at different steps of the ChIA-PET2 pipeline are performed. The linker trimming statistics quantify the efficiency of linker ligation and the occurrence of the linker in the sequencing reads. As the library fragment sizes vary, the linker may occur in either end of the paired reads or not. Users can choose whether to keep the empty reads (reads without linker) in the sequential analysis based on this metric. The second metric is the read alignment statistics. We use histograms to show the percentages of unmapped reads, reads with low MAPQ, and reads with high MAPQ at each end of the paired-end reads. The statistics also shows the number of PETs built from the paired-end reads and the duplicate rate. A high duplicate rate value indicates a potential PCR bias. The third metric shows the composition of different types of PETs. Intra-chromosomal PETs that link different peaks (Intra_BetweenPeak) are the most valuable type of PETs for chromatin loop calling. The fourth metric is the fraction of intra- and inter-chromosomal PETs, which is also important, since a high fraction of inter-chromosomal PETs usually indicates a low quality experiment. Finally, the PET count distributions of intra- and inter-chromosomal interactions are shown and compared. These quality control metrics provided by ChIA-PET2 help users to assess the quality of a ChIA-PET experiment. All these metrics are summarized in the QCplot figure and can be drawn automatically by ChIA-PET2.
Build contact matrix
Optionally ChIA-PET2 can build Hi-C style 2D contact matrix. Given a user-defined bin size, such as 10kb, ChIA-PET2 cuts the genome into bins and assigns the PETs to corresponding entries of the contact matrix. Because this matrix built from ChIA-PET data is usually very sparse, we store the matrix in the same tripe tuple format as HiC-Pro (23) does. Hi-C style contact map can be drawn by HiCPlotter (24) seamlessly. Comparisons between two examples of 2D contact matrixes generated by ChIA-PET2 and Hi-C contact maps are shown (Supplementary Figures S1 and S2).
RESULTS
Quality control in ChIA-PET data analysis
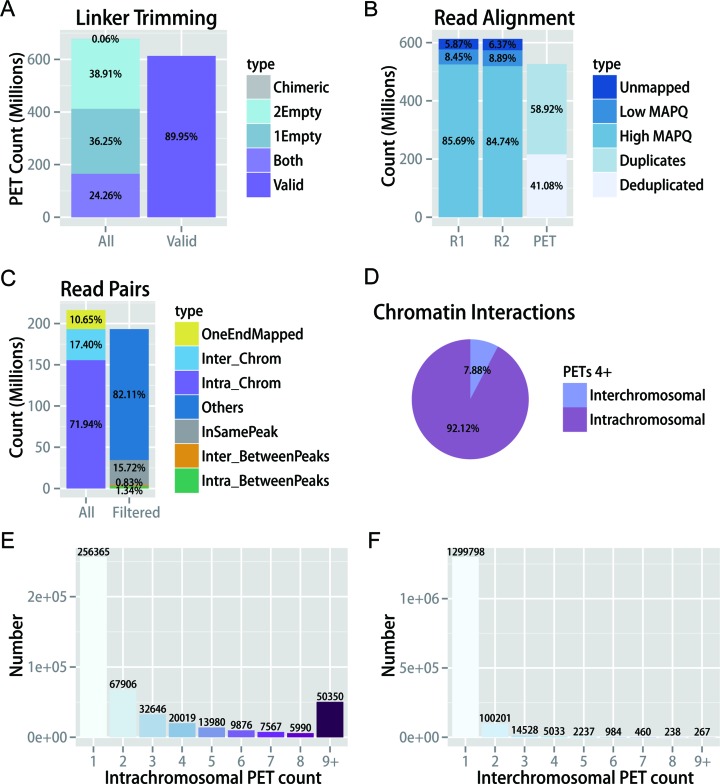
We first applied ChIA-PET2 to the GM12878 dataset (4). The quality controls are shown in Figure 2. Linker sequences were trimmed from raw PETs. Overall, 24.26% of the raw PETs have linkers in both ends. If we kept all the PETs regardless of whether they have linkers or not, 89.95% of the total raw PETs were valid as input for the sequential alignment step (Figure 2A). Both ends of the PETs were aligned to the hg19 reference genome independently. After filtering out the unmapped reads and low MAPQ reads, 85% of the input PETs were retained. After duplicate removal, 41.08% of PETs were retained and were ready to be used as input of MACS2 (Figure 2B). After peaks were called by MACS2, PETs were classified into different categories. There were 15.72% of PETs with both ends in the same peak, 0.83% of PETs linking different peaks from inter-chromosomes and 1.34% of PETs linking different peaks from intra-chromosomes (Figure 2C). In total, 92.12% of chromatin interactions that had at least four supportive PETs were intra-chromosomal, and the remaining interactions were inter-chromosomal (Figure 2D). The PET count distributions of intra and inter-chromosomal interactions show a large difference (Figure 2E and F). After the PET count and peak depth information were gathered, MICC was applied to estimate the statistical significance of each chromatin interaction. Another example of QCplot for the half-linker ChIA-PET dataset is shown in Supplementary Figure S3.
Figure 2.
ChIA-PET2 quality controls. (A) Statistics metrics of linker trimming. (B) Statistics metrics of read alignment. (C) Different types of PETs and their percentages. (D) Intra/Inter-chromosomal interactions percentage with at least four supported PETs. (E) PET count distribution of all candidate intra-chromosomal loops before statistical confidence estimation. (F) PET count distribution of all candidate inter-chromosomal loops before statistical confidence estimation.
Higher sensitivity and reproducibility in chromatin loop calling
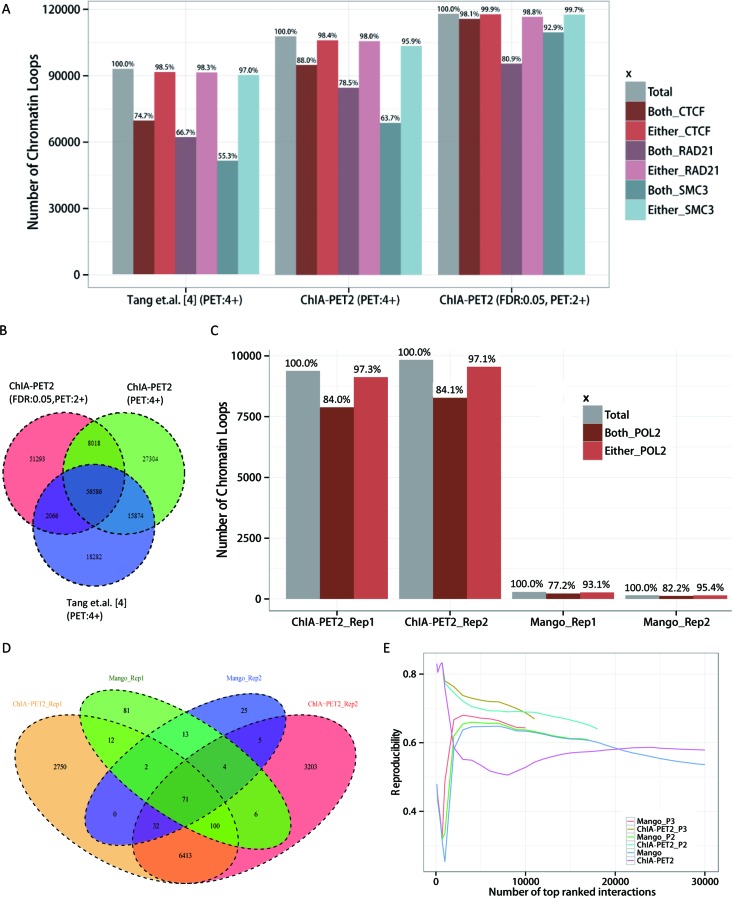
By using the ChIA-PET2 pipeline we were able to detect an additional 25,155 high-confidence intra-chromosome interactions at an FDR of 0.05 followed by an additional filter, i.e., a PET count cut-off of 2. We noticed that by allowing mismatches in the linker searching step, the rescued PETs may contain some noise. But the noise is random and submerges in the peak calling and loop calling step (Figure 3A). To check the specificity of loop calling, we used independent ChIP-Seq datasets. Because the cohesin protein complex is closely associated with CTCF in chromatin looping (6,25), we examined the CTCF-anchor sites for occupancy by CTCF and co-occupancy by cohesin using independent ChIP-seq data of CTCF and cohesin subunits RAD21 and SMC3. Approximately 99.9% of the CTCF loops have CTCF occupancy in either one (n = 2,091) or both anchors (n = 115,726). The vast majority (98.8% and 99.7%) of the loops have RAD21 or SMC3 co-occupancy in either one or both anchors, respectively (Figure 3A). The ratios of CTCF or cohesin occupancy in loop anchors are all higher than those calculated from the in-house pipeline in the Tang et al. paper (4), as shown in Figure 3A. This is also true in the case in which we use the same PET count cut-off of 4 without statistical confidence estimation, as performed by Tang et al. A large percentage of the chromatin interactions detected by the two pipelines are also consistent with each other (Figure 3B).
Figure 3.
Results of the ChIA-PET2 pipeline on real ChIA-PET datasets. (A) Number of CTCF-mediated chromatin loops detected by different methods from bridge-linker ChIA-PET data. CTCF, RAD21 and SMC3 occupancy in the anchors of loops are shown above the histogram. (B) Overlap of CTCF-mediated chromatin loops detected by ChIA-PET2 and the pipeline by Tang et al. (4). (C) Number of RNA POL2-mediated chromatin loops detected by different methods. POL2 occupancy in the anchors of loops is shown above the histogram. (D) Overlap of POL2-mediated chromatin loops detected by ChIA-PET2 and Mango. (E) Reproducibility of ChIA-PET2 and Mango.
We also compared the power of ChIA-PET2 in analyzing half-linker ChIA-PET datasets. Since Mango has been shown to perform better than CPT and is a published pipeline for half-linker (short-reads) ChIA-PET data, we compared our pipeline with Mango. The K562 POL2 half-linker (short-read) ChIA-PET dataset (18) was used in this comparison analysis. This dataset comprises two replicates. We detected high confidence chromatin loops at an FDR of 0.05 in both pipelines. While our pipeline detected >9,000 of chromatin loops, Mango only detected less than 3 hundred chromatin loops (Figure 3C). The percentages of loops occupied by POL2 in either one anchor or both anchors are all higher than those detected by Mango (Figure 3C). This finding shows that ChIA-PET2 could detect chromatin loops with a significantly higher sensitivity than Mango at the same false discovery rate. Our pipeline also recovered a large portion of loops detected by Mango (64% in replicate 1 and 74% in replicate 2), as shown in Figure 3D. The Jaccard index of loops detected from these two replicates for ChIA-PET2 is 52.5%, which is much higher than 25.6% for Mango. The Jaccard index is defined as the size of the intersection divided by the size of the union of the sample sets and shows that ChIA-PET2 detects loops with a higher reproducibility than Mango.
We further evaluated the reproducibility by overlapping top-ranked interactions from two replicates for these two methods. Only intra-chromosomal loops were evaluated since Mango could not deal with inter-chromosomal loops. ChIA-PET2 shows a higher reproducibility than Mango when using a PET count cut-off of 2 or 3 (Figure 3E). The reproducibility would decrease substantially if we had kept all chromatin loops with only one supported PET. This is because it is very hard to distinguish true functional chromatin loops from random noise based upon only one supported PET.
Functional short-range chromatin loops can be detected
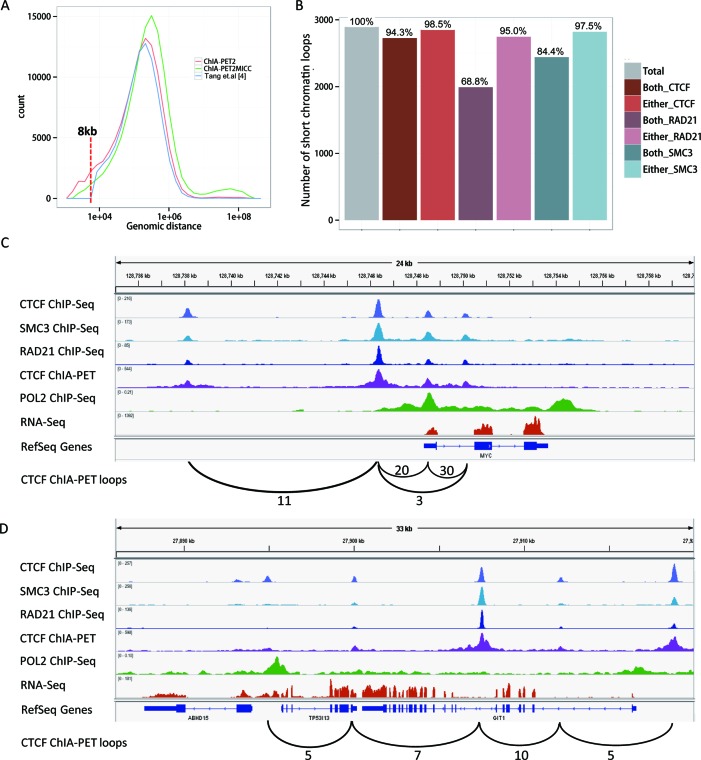
Since PETs with a genomic span less than a threshold, e.g., 8 kb, were classified as self-ligated PETs and were not used for the loop calling step in Tang et al. (4), it cannot detect chromatin loops with a genomic span smaller than 8 kb. We avoided this empirical cut-off in the ChIA-PET2 pipeline (see methods below), such that it is possible for ChIA-PET2 to detect smaller chromatin loops. The distribution of genomic distances between loop anchors detected by different pipelines is shown in Figure 4A.
Figure 4.
Short-range chromatin loops. (A) The distribution of genomic distance between loop anchors. POL2 occupancy in the anchors of RNA POL2 mediated chromatin loops detected by different methods. Red dashed line marks the 8kb distance. (B) CTCF, RAD21 and SMC3 occupancy in the anchors of CTCF mediated short (<8 kb) chromatin loops detected by ChIA-PET2. (C) Example of short chromatin loops near the Myc gene and ChIP-Seq/RNA-Seq signal along the window. PET count number is labelled at the loop. (D) Another example of short chromatin loops near gene TP53I13.
By using ChIA-PET2, we were able to detect 2,893 CTCF-mediated chromatin loops with genomic spans less than 8 kb (Figure 4B). Most of these short chromatin loops are also occupied with CTCF and cohesin in the loop anchors (Figure 4B). The percentages of short loops with anchors occupied by proteins are as similarly high as those for all significant loops (Figure 3A). This indicates that most of the short-range chromatin loops we detected may also be functional. For instance, the shortest one among these loops is approximately 1.6 kb, which is located near the MYC gene. This loop has 30 supported PETs and both anchors of this loop have CTCF, RAD21 and SMC3 occupancy (Figure 4C). The loop anchor is also associated with RNA POL2 binding at the promoter of the MYC gene. Adjacent loops are also shown in this region. Interestingly, CTCF was first identified as a transcriptional repressor of the chicken c-myc gene (26) and is also an exceptionally conserved transcriptional repressor of avian and mammalian c-myc oncogenes (27). Targeted deletion of human CTCF binding elements also revealed a requirement for CTCF in c-myc expression (28). Another example of short-range loop (Figure 4D) is located near TP53I13, also known as tumor protein P53 inducible protein 13. Again, adjacent loops are shown in the region. One of the loop anchors is located at the shared promoter of TP53I13 and ABHD15, which is occupied by POL2. These two bidirectional genes are both expressed. A third example of NFKB2 is shown (Supplementary Figure S4). Based on the multiple lines of evidence of ChIP-Seq, RNA-Seq and ChIA-PET signals, these loops indicate that at least some of, if not all, the extremely short-range chromatin loops are also likely to be important and functional.
Recently, chromatin was estimated to be bendable at the kilobase scale. The Kuhn length, the minimum fiber length that allows the beginning and the end of the fiber segment to point in the same direction, was measured to be about 1 kb based upon Hi-C data (29). Our result generated from ChIA-PET data shows that chromatin loops only slightly longer than the 1kb minimum theoretical estimate can be detected.
Allele-specific analysis
We used ChIA-PET2 to call allele-specific chromatin loops for the human GM12878 cell line. Phased genotype data were extracted from the Illumina Platinum Genomes Project. Only heterozygous phased single nucleotide polymorphisms (SNPs) with good quality were retained. By comparing the nucleotides of mapped reads at the SNP location with the phased genotype, ChIA-PET2 is able to label the reads as maternal, paternal or unknown. Details of the allele-specific analysis are described in the Materials and Methods section. Supplementary Table S2 shows a summary of the allele-specific PETs in different parameter settings. Among the input of 681 million paired-end reads, about 193 million were classified as the valid PETs by ChIA-PET2 if we kept those PETs without linker sequences. More than 6 million (3.15%) of valid PETs were assigned to either maternal or paternal allele.
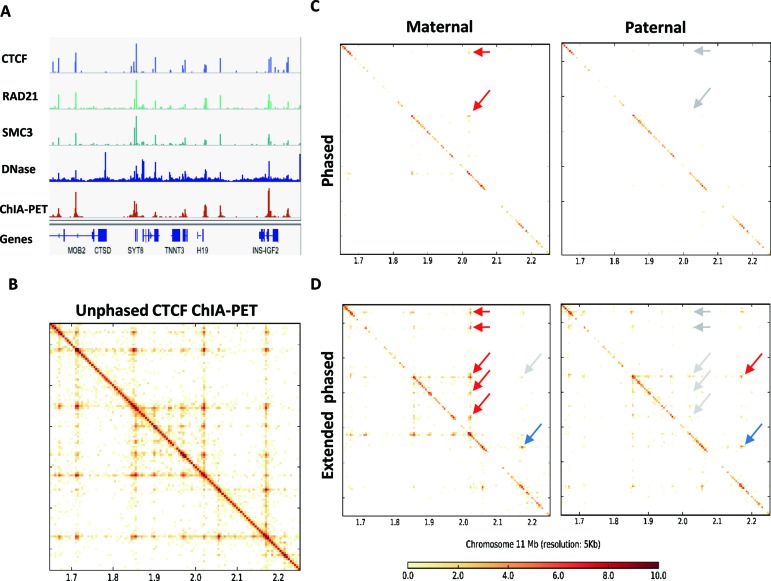
To illustrate the power of ChIA-PET2 in exploring the haplotype-specific chromatin loops, we plotted the CTCF-mediated chromatin interactions near the H19/IGF2 gene loci in chromosome 11 at a 5 kb resolution (Figure 5). The ChIP-Seq and DNase-Seq signals (Figure 5A) validate the corresponding grid-like peak pattern in the contact heatmap (Figure 5B). There is one loop that occurs in both alleles in the extended phased contact map. We also found that an obvious difference exists between maternal and paternal chromatin loops in this region. There are five maternal-specific chromatin loops whose anchors are close to the H19 gene while the loop to the IGF2 region is absent or greatly attenuated. The pattern is opposite on the paternal chromosome (Figure 5C and D). This result is consistent with the Hi-C data (5), although the contact matrix is at higher resolution and the pattern is much clearer. Notice that H19 is only transcribed from the maternally inherited allele and not expressed from the paternal allele while IGF2 is expressed only from the paternal allele (30). This example clearly demonstrated how ChIA-PET2 may be used to explore allele-specific chromatin loops and imprinted gene regulation.
Figure 5.
Allele-specific analysis. (A) Signal of CTCF,RAD21, SMC3 ChIP-Seq, DNase-Seq and ChIA-PET peaks near the H19/IGF2 region. (B) Contact matrix generated from all valid ChIA-PET PETs. (C) Maternal contact matrix generated from maternal phased PETs (left panel) and maternal extended phased PETs (right panel). Red arrow indicates the chromatin loop and gray arrow indicates absent or attenuated peak signal. (D) Paternal contact matrix generated from paternal phased PETs (left panel) and paternal extended phased PETs (right panel). The red arrow indicates the chromatin loop and the gray arrow indicates an absent or attenuated peak signal and the blue arrow indicates that both alleles have the peak signal.
Hi-C style contact map for ChIA-PET data to gain insight into 3D genome organization
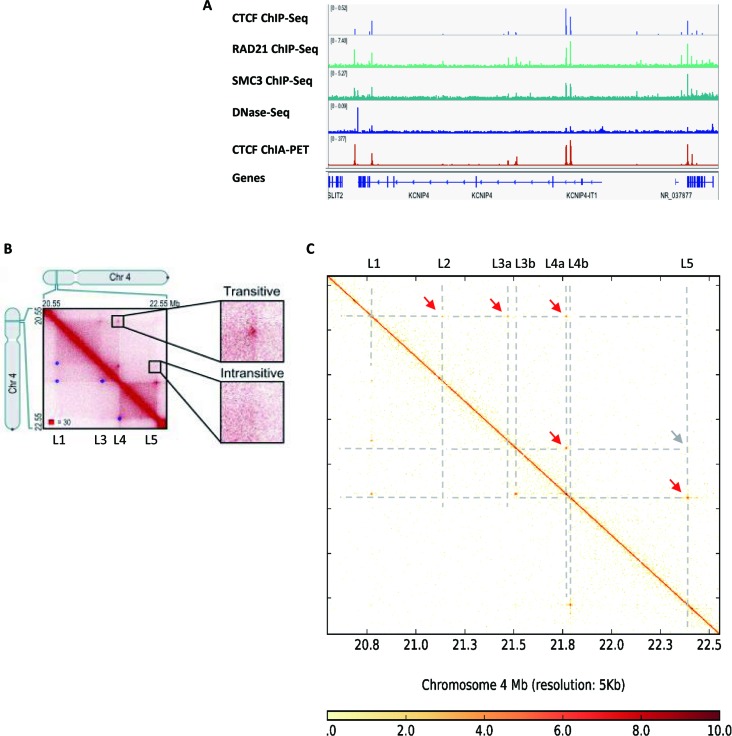
To further illustrate the ability of ChIA-PET2 to gain insight into 3D genome organization, we compared the Hi-C style contact matrix generated by ChIA-PET2 with those from the corresponding Hi-C data in the same cell type. Figure 6 shows ChIP-Seq signal in this region and contact matrix from the ChIA-PET and the Hi-C data. All four loops detected in Hi-C matrix were also captured by ChIA-PET2: L1-L3, L1-L4, L3-L4 and L4-L5. Using ChIA-PET2, we also detected one more loop in this region (L1-L2). All these loops are supported by CTCF and cohesin ChIP-Seq data, although the ChIP-Seq signal in L2 locus is relatively low.
Figure 6.
Comparison with Hi-C contact map. (A) Signal of CTCF,RAD21, SMC3 ChIP-Seq, DNase-Seq and ChIA-PET peaks in a 2Mb region in chromosome 4. (B) Heatmap of contact matrix directly from Rao et al. (5) with additional annotation. (C) Heatmap of the contact matrix generated from ChIA-PET data by the ChIA-PET2 pipeline. The red arrow marks the chromatin loop and the gray arrow marks the absence of a peak signal.
Rao et al. proposed the concept of transitivity in the Hi-C matrix (5), i.e., three adjacent loci form three focal peaks between each corresponding pair. Figure 6B shows examples of transitive and intransitive looping behaviour in the contact matrix from Hi-C data in GM12878. The authors claimed that loci L1 and L3 form loop (L1–L3) while loci L3 and L4 form loop (L3–L4) such that the transitivity implies loci L1 and L4 form the transitive loop (L1–L4). The CTCF ChIA-PET contact matrix generated by our pipeline shows additional details in this region. Our analysis reveals that L3 and L4 loci comprise two different loop anchors respectively. This result is validated by independent CTCF, cohesion ChIP-Seq signal (Figure 6A and C). Interestingly, the transitivity (L1–L3a and L1–L4a implies L3a–L4a) does not exist because L3a and L4a do not form any loop. Instead, L3b and L4a form a loop (Figure 6C).
In addition to the transitivity, we found that there exists redundancy in some of the chromatin loops (Supplementary Figure S2). While some of the Hi-C loops are the same as the ChIA-PET CTCF loops (Supplementary Figure S2, left panel), some of the Hi-C loops may contain several ChIA-PET CTCF loops (Supplementary Figure S2, middle and right panel). Since the resolution of Hi-C may not be sufficiently high, multiple nearby peaks in the Hi-C heatmap could merge into one single wider peak. This redundancy phenomenon may indicate the robustness of the chromatin structure because if one of the CTCF loops collapses, the nearby loop still function and maintain the 3D structure.
Finally, we provide ChIA-PET CTCF contact matrix in the CTCF gene locus (Supplementary Figure S5) as an example of how ChIA-PET2 could help better understand gene regulation in the 3D genome context.
DISCUSSION
As the ChIA-PET technique is improving and maturing, it is of great importance to develop publicly available bioinformatics tools that can be used for a wide range of projects. ChIA-PET2 is a versatile and flexible pipeline for ChIA-PET data analysis. ChIA-PET2 can process different types of ChIA-PET data, from raw sequencing reads to significant chromatin-loop calls. Fuzzy search can be applied in the linker trimming step to obtain improved number of valid PETs. ChIA-PET2 could detect chromatin loops with a significantly higher sensitivity and reproducibility than existing pipelines at the same false discovery rate. Extremely short-range chromatin loops, e.g. <8 kb, can be detected too. These short-range chromatin loops can not be detected by previous ChIA-PET data analysis due to methodology limitation. ChIA-PET2 also provides quality control at different step. In addition, when phased genotype information is available, ChIA-PET2 allows the detection of allele-specific chromatin loops.
By applying ChIA-PET2 on real datasets, we accurately detected thousands of additional chromatin loops genome-wide, among which were extremely short-range chromatin loops which are likely to be functional. By comparison with Hi-C contact maps, we also show how the ChIA-PET2 pipeline can transform ChIA-PET data into insightful knowledge for a better understanding of gene regulation in the 3D genome context. Overall, we proposed a versatile and flexible pipeline for ChIA-PET2 data analysis to gain better insight into 3D genome organization.
Transitivity, redundancy and robustness of chromatin loops were also introduced and discussed in this article. By identifying chromatin loops more comprehensively, more details on the loop anchors and combinations of loops were observed. For example, chromatin loop previously identified by Hi-C were actually composed of multiple loops linking different nearby loop anchors. This may indicate a relationship between the redundancy and robustness of chromatin loops. However, since we focus on introducing the utility of the ChIA-PET pipeline, only a few examples of the transitivity, redundancy and robustness of chromatin loops were provided. It is interesting to systematically study these properties of chromatin loops to better understand the underlying mechanism of 3D genome organization.
In the end, we notice that there still may be potential limitation in the ChIA-PET2 method. ChIA-PET2 calls chromatin loops based on ChIP-Seq-like peaks. This strategy is based on the assumption and observation that the anchors of most chromatin loops are located at the ChIP-Seq-like peaks. The sensitivity of chromatin loop calling partly depends on the sensitivity of peak calling. However, in practice this will not be problematic since users can choose different thresholds in the ChIA-PET2 pipeline to balance the trade-off between sensitivity and specificity of chromatin loop calling.
AVAILABILITY
ChIA-PET2 was implemented by using C++, R and shell scripts. ChIA-PET2 was named not only because it is a tool for ChIA-PET data analysis but also because it supports at least 2 different ChIA-PET protocols (bridge linker protocol or half-linker protocol) data, 2 modes of read alignments (short or long read alignment) and 2D contact map output. The following additional software and libraries are required: bwa (31,32), samtools (33), bedtools (34), MACS2 (35) and g++ compiler. To generate the quality controls figure, R and ggplot2 are required. To use MICC, R package VGAM is also required. The ChIA-PET2 pipeline can be installed on a Linux/UNIX-like operating system. ChIA-PET2 is available at https://github.com/GuipengLi/ChIA-PET2.
ACCESSION NUMBERS
The ChIP-Seq data were retrieved from the ENCODE data repository site (19). Phased genotype data for GM12878 genome were extracted from the Illumina Platinum Genomes Project (http://www.illumina.com/platinumgenomes/). ChIA-PET data used in this study are available in NCBI Gene Expression Omnibus under accession number GSE72816, GSM970213.
Acknowledgments
We thank Dr Gucci J. Gu Urban at Stanford University and other users for using and testing ChIA-PET2 and thank Prof. Minping Qian at Peking University for helpful discussion and suggestion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China [2012CB316503]; National High Technology Research and Development Program of China [2012AA020401]; National Natural Science Foundation of China [91519326, 31361163004 and 31301044]; Tsinghua National Laboratory for Information Science and Technology Cross-discipline Foundation; National Institutes of Health grants [HG001696, 5U01HG007919-02, 5U54HG00699604 and 5P50HG00773502]. Funding for open access charge: National Basic Research Program of China [2012CB316503].
Conflict of interest statement. Michael Snyder is a founder and consultant for Personalis, a member of the scientific advisory board of GenapSys and a consultant for Illumina.
REFERENCES
- 1.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O., et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fullwood M.J., Liu M.H., Pan Y.F., Liu J., Xu H., Mohamed Y.B., Orlov Y.L., Velkov S., Ho A., Mei P.H., et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B., et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heidari N., Phanstiel D.H., He C., Grubert F., Jahanbanian F., Kasowski M., Zhang M.Q., Snyder M.P. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith E.M., Lajoie B.R., Jain G., Dekker J. Invariant TAD boundaries constrain cell-type-specific looping interactions between promoters and distal elements around the CFTR locus. Am. J. Hum. Genet. 2016;98:185–201. doi: 10.1016/j.ajhg.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowen J.M., Fan Z.P., Hnisz D., Ren G., Abraham B.J., Zhang L.N., Weintraub A.S., Schuijers J., Lee T.I., Zhao K., et al. Control of cell identity genes occurs in insulated neighborhoods in Mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Wong C.H., Birnbaum R.Y., Li G., Favaro R., Ngan C.Y., Lim J., Tai E., Poh H.M., Wong E., et al. Chromatin connectivity maps reveal dynamic promoter-enhancer long-range associations. Nature. 2013;504:306–310. doi: 10.1038/nature12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wit E., Vos E.S., Holwerda S.J., Valdes-Quezada C., Verstegen M.J., Teunissen H., Splinter E., Wijchers P.J., Krijger P.H., de Laat W. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 11.Hnisz D., Weintraub A.S., Day D.S., Valton A.L., Bak R.O., Li C.H., Goldmann J., Lajoie B.R., Fan Z.P., Sigova A.A., et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G., Fullwood M.J., Xu H., Mulawadi F.H., Velkov S., Vega V., Ariyaratne P.N., Mohamed Y.B., Ooi H.S., Tennakoon C., et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paulsen J., Rodland E.A., Holden L., Holden M., Hovig E. A statistical model of ChIA-PET data for accurate detection of chromatin 3D interactions. Nucleic Acids Res. 2015;42:e143. doi: 10.1093/nar/gku738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He C., Zhang M.Q., Wang X. MICC: an R package for identifying chromatin interactions from ChIA-PET data. Bioinformatics. 2015;31:3832–3834. doi: 10.1093/bioinformatics/btv445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phanstiel D.H., Boyle A.P., Heidari N., Snyder M.P. Mango: a bias-correcting ChIA-PET analysis pipeline. Bioinformatics. 2015;31:3092–3098. doi: 10.1093/bioinformatics/btv336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djekidel M.N., Liang Z., Wang Q., Hu Z., Li G., Chen Y., Zhang M.Q. 3CPET: finding co-factor complexes from ChIA-PET data using a hierarchical Dirichlet process. Genome Biol. 2015;16:288. doi: 10.1186/s13059-015-0851-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He C., Li G., Djekidel M.N., Chen Y., Wang X., Zhang M.Q. Advances in computational ChIA-PET data analysis. Quant. Biol. 2016 doi:10.1007/s40484-016-0080-3. [Google Scholar]
- 18.Li G., Ruan X., Auerbach R.K., Sandhu K.S., Zheng M., Wang P., Poh H.M., Goh Y., Lim J., Zhang J., et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers G. A fast bit-vector algorithm for approximate string matching based on dynamic programming. J. ACM. 1999;46:395–415. [Google Scholar]
- 23.Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.J., Vert J.P., Heard E., Dekker J., Barillot E. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akdemir K.C., Chin L. HiCPlotter integrates genomic data with interaction matrices. Genome Biol. 2015;16:198. doi: 10.1186/s13059-015-0767-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ong C.T., Corces V.G. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filippova G.N., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gombert W.M., Krumm A. Targeted deletion of multiple CTCF-binding elements in the human C-MYC gene reveals a requirement for CTCF in C-MYC expression. PLoS One. 2009;4:e6109. doi: 10.1371/journal.pone.0006109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanborn A.L., Rao S.S., Huang S.C., Durand N.C., Huntley M.H., Jewett A.I., Bochkov I.D., Chinnappan D., Cutkosky A., Li J., et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E6456–E6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rachmilewitz J., Goshen R., Ariel I., Schneider T., de Groot N., Hochberg A. Parental imprinting of the human H19 gene. FEBS Lett. 1992;309:25–28. doi: 10.1016/0014-5793(92)80731-u. [DOI] [PubMed] [Google Scholar]
- 31.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing, S. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]