Abstract
Proper temporal epigenetic regulation of gene expression is essential for cell fate determination and tissue development. The Bromodomain-containing Protein-4 (BRD4) was previously shown to control the transcription of defined subsets of genes in various cell systems. In this study we examined the role of BRD4 in promoting lineage-specific gene expression and show that BRD4 is essential for osteoblast differentiation. Genome-wide analyses demonstrate that BRD4 is recruited to the transcriptional start site of differentiation-induced genes. Unexpectedly, while promoter-proximal BRD4 occupancy correlated with gene expression, genes which displayed moderate expression and promoter-proximal BRD4 occupancy were most highly regulated and sensitive to BRD4 inhibition. Therefore, we examined distal BRD4 occupancy and uncovered a specific co-localization of BRD4 with the transcription factors C/EBPb, TEAD1, FOSL2 and JUND at putative osteoblast-specific enhancers. These findings reveal the intricacies of lineage specification and provide new insight into the context-dependent functions of BRD4.
INTRODUCTION
The bromodomain containing protein-4 (BRD4) is the best characterized member of the bromo- and extra-terminal (BET) domain family of proteins. The BET proteins harbor two amino-terminal bromodomains (BD1 and BD2) that preferentially bind to diacetylated histone tails followed by one extraterminal (ET) domain (1). Additionally, the long isoform of BRD4 has a unique Positive Transcription Elongation Factor b (P-TEFb) Interaction Domain (PID) at its carboxy-terminus which associates with the P-TEFb complex components Cyclin T1 and CDK9 and serves to recruit them to chromatin (2,3). In turn, RNA Polymerase II (RNAPII) can be phosphorylated at Ser-2 residues within the carboxy-terminal heptapeptide repeat domain (CTD), which serves as a hallmark for transcriptional elongation to recruit various proteins involved in transcription-associated processes. In addition to binding to acetylated histone proteins, BRD4 has recently also been shown to directly interact with several transcription factors (TFs) including TWIST, p53, C/EBPα and C/EBPβ, ERG and NFκB in both acetylation-dependent and -independent manners (4–8). Moreover, proteomic analyses revealed the ability of the ET domain of BRD4 to interact with other chromatin-associated proteins like NSD3 and JMJD6 (9,10).
Gene expression is tightly regulated by the cooperative action of TFs. Generally, TFs bind to the promoter and/or regulatory DNA elements called enhancers. Enhancers determine the specificity of gene expression patterns in a spatiotemporal- and context-dependent manner (11). The mechanisms by which enhancers stimulate target gene expression still remain poorly understood. Application of chromosomal conformation capture analyses suggests that long-range chromosomal interactions facilitate the expression of target genes, for example by bridging distal enhancer regions with the transcriptional start site (TSS) via specific TFs and cofactors (12,13). In addition, recent findings uncovered non-coding transcripts produced from enhancer regions (eRNA) that might also promote enhancer-mediated long-range chromosomal interactions and/or P-TEFb activation (14–18). Notably, BRD4 also occupies intergenic regions containing putative enhancers and facilitates eRNA transcription (10,19–22).
Since the discovery of BET inhibitors, which prevent BET domain binding to acetyl-lysine containing sequences, the function of BRD4 has been widely studied in tumor-associated transcriptional programs (4,5,20,23–27). Interestingly, while BET inhibition (BETi) has been shown to work in multiple tumor types, the effects elicited are highly dependent upon the underlying transcriptional program defining the tumor cell phenotype (e.g. MYC in the context of hematological malignancies (26,28), ERα in the context of breast cancer (20), etc.). This supports a central role of BRD4 in controlling specific subsets of genes defined by tumor- or tissue-specific TFs. Consistently, CDK9 is also required for lineage-specific transcription and promotes the differentiation of human mesenchymal stem cells (hMSC) (29). However, apart from studies in embryonic stem (ES) cells (21,30,31), few reports have examined the significance and mechanisms of action of BRD4 in lineage maintenance and determination.
In this study, we investigated the role of BRD4 in lineage specification during osteoblast differentiation. Perturbation of BRD4 function affects osteoblast differentiation both at early stages as well as later during mineralization by regulating skeleton- or extracellular matrix-specific gene expression. Moreover, we computationally identified putative tissue-specific BRD4-occupied enhancers and TFs associated with these regions. Subsequently, genome-wide occupancy studies confirmed preferential and differential binding patterns of activator protein-1 (AP1) TF family members, a Hippo/YAP signaling mediator (TEAD1) and CEBP/β with osteoblast-specific BRD4-occupied putative enhancers. Furthermore, depletion of these TFs impaired the recruitment of BRD4 to potential enhancer sites of BRD4-target genes. Taken together, our data suggest that BRD4 plays a major role in maintaining tissue specificity by serving as a hub to integrate the effects of multiple TFs at lineage-specific enhancers.
MATERIALS AND METHODS
Cell culture, differentiation, inhibitor treatments and siRNA transfections
Human fetal osteoblasts (hFOB) were maintained as previously described (32). In short, cells were maintained at 34°C in 5% CO2 in phenol red-free high-glucose Dulbecco's modified Eagle's medium (DMEM/F12, Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Sigma-Aldrich). For induction of differentiation, confluent cells were treated with differentiation cocktail as described previously (29) and shifted to 39°C. Cells were pretreated with JQ1 (250 nM) 1 h before the induction of differentiation and added again with every media change (every second day). hMSC were maintained in α-MEM (Invitrogen) and differentiated to the osteoblast lineage as described before (29). The JQ1 treatment was performed as described above for hFOBs. IDG-SW3 osteocyte cells were cultured and differentiated as previously described (33). Namely, cells were maintained at 34°C in collagen coated plates. For the experiment, cells were plated onto 12-well plates at 50% density and grown for 3–4 days until confluency before inducing differentiation and transferring to 37°C with a concomitant treatment with JQ1 or vehicle (DMSO). For day 0 harvest, the cells were maintained in differentiation media and treated with DMSO for 24 h at 37°C. For long term treatment, differentiation media was replaced every third day with fresh treatments of DMSO and JQ1 (250 nM). The cells were harvested following 14 and 26 days of treatment for harvesting RNA or performing alizarin red staining.
siRNA transfections were performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Control siRNAs (ON-TARGETplus, Non-targeting siRNA#2, target sequence: UGGUUUACAUGUUGUGUGA, used in hFOBs; Luciferase GL2 duplex, target sequence: CGUACGCGGAAUACUUCGA, used in hMSCs) and BRD4 siRNAs (siGENOME, D-004937-02 – D-004937-05, target sequences: #2 –GAACCUCCCUGAUUACUAU,#3 – UAAAUGAGCUACCCACAGA,#4 – UGAGAAAUCUGCCAGUAAU, #5 – AGCUGAACCUCCCUGAUUA) were purchased from Dharmacon, ThermoScientific.
Western blot, RNA isolations, qRT-PCR and stainings
Protein and RNA isolations, reverse transcription, western blot and qRT-PCR analyses were performed as previously described (20,29). The list of antibodies used in this study is provided in Supplementary Table S1. The primers are listed in Supplementary Table S2.
Alkaline phosphatase staining was performed following the manufacturer's instructions with the use of Leukocyte Alkaline Phosphatase Kit (Sigma). Pictures of stained wells were taken using an AxioScope.A1 microscope equipped with an AxioCam MRc under A-Plan 2.5× magnification. For alizarin red staining cells were fixed with neutral formalin for 24 h and stained with alizarin red as previously described (33). The stained 12-well plates were scanned and quantification of the stained areas was evaluated with the ImageJ software using the Threshold Color Plugin.
For statistical analysis of staining and gene expression values a two-tailed two-sample with equal variance Student's t-test and one-way ANOVA followed by Dunnett's or Turkey's test (as indicated in figure legends) were used.
Chromatin immunoprecipitation
ChIP analyses were performed as previously reported (20,29) with modifications as described. Briefly, cells were fixed in 1% formaldehyde in phosphate buffered saline for 20 min and quenched with 125 Glycine for 5 min at room temperature. Cells were scraped, lysed and washed twice with Nuclear Preparation buffer (150 mM NaCl, 50 mM Tris–HCl (pH 7.5), 5 mM ethylenediaminetetraacetic acid (EDTA), NP-40 (0.5% vol/vol), Triton X-100 (1.0% vol/vol)). The nuclear pellet was resuspended in Lysis Buffer (150 mM NaCl, 1% (v/v) NP-40, 0.5% w/v sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS), 50 mM Tris–HCl (pH 8), 20 mM EDTA, 20 mM sodium fluoride and protease inhibitor cocktail) and subjected to 20 cycles of sonication (30 s ON/OFF) using a Bioruptor Pico (Diagenode). Following sonication, the chromatin extract was precleared with 100 μl of 50% Sepharose 4B (GE Healthcare) in Lysis Buffer and incubated overnight with 1–2 μg of the corresponding antibody (Supplementary Table S2). The next day, 30 μl of bovine serum albumin-blocked 50% Protein-A Sepharose slurry (GE Healthcare) was added to capture the immunocomplexes. After additional 2 h of incubation, the ChIP immunocomplexes were washed once with Lysis Buffer, twice with Wash Buffer (100 mM Tris (pH 8.5), 500 mM LiCl, 1% (v/v) NP-40, 1% w/v sodium deoxycholate, 20 mM EDTA, 20 mM NaF), again twice with Lysis Buffer followed by two washes with TE Buffer. After the last step of washes, samples were subjected to RNAse A treatment (Qiagen) in 10 mM TRIS (pH 8) for 30 min at 37°C. Samples were subsequently diluted once with a buffer containing 20 mM EDTA, 100 mM Tris–HCl (pH 8), 2% SDS and 20 μg Proteinase K and incubated for at least 4 h at 65°C for reversing the crosslinking. DNA was precipitated with 0.4 M LiCl and linear polyacrylamide and isolated using phenol-–hloroform–isoamyl alcohol extraction.
The obtained DNA was utilized to confirm the efficiency of immunoprecipitation by qPCR on positive and negative sites using the primers listed in Supplementary Table S3. The PCR signals of the IP samples were normalized to input DNA (isolated from the same ChIP sample after the preclearing step) and displayed as enrichment relative to input in percent. The background signal was estimated using DNA from IgG immunoprecipitated DNA.
Library preparation and next-generation sequencing
After confirming the efficiency of ChIP with qPCR, DNA was subjected to an additional 40 cycles of sonication in 10 μl volume using a Bioruptor Pico (Diagenode) to achieve a size range of ≤200 bp. ChIP-Seq libraries were generated using the MicroPlex Library Preparation Kit (Diagenode) following the manufacturer's instructions. For RNA-sequencing, libraries were prepared using The NEXTflex Rapid Directional RNA-Seq Kit (Bioo Scientific) after verifying the RNA integrity on an agarose gel.
The library size was estimated using a Bioanalyzer 2100. Pooled libraries were used for cluster generation on cBot followed by 51 bp single-end sequencing on HiSeq 2000 from Illumina performed by the Transcriptome Analysis Laboratory in Göttingen, Germany. The sequencing BaseCaller bcl output files were further demultiplexed to fastq files using CASAVA 1.8.2.
Raw and processed ChIP-Seq data generated by the Encyclopedia of DNA Elements (ENCODE) Consortium (34) from primary normal human osteoblast cells (NHOST) for H3K4me1, H3K4me3, H3K27ac, H3K27me3, DNAse-Hypersensitivity as well as published RUNX2 ChIP-Seq data (35) were downloaded from Gene Expression Omnibus and the European Nucleotide Archive, respectively. The accession numbers for each dataset are listed in Supplementary Table S3.
RNA-sequencing bioinformatic analysis
For RNA-Seq in hFOBs, seven conditions were used: undifferentiated hFOBs treated with DMSO and non-targeting siRNA (siCNTR); differentiated hFOBs treated with DMSO or JQ1; differentiated hFOBs transfected with a non-targeting siRNA (siCNTR) or two independent siRNAs (#3 and #4) against BRD4. The FASTQ files were mapped to the human genome (assembly hg19) using TopHat Gapped-read mapper (36) for RNA-seq data with very sensitive Bowtie2 settings on Galaxy Platform (Version 0.9) (37). The read counting was performed via HTSeq (38) (version 0.6.0) with the following parameters: -f bam -r pos -s reverse -a 10 -t exon -m union. The count files were subsequently subjected for differential analysis using the DESeq2 Package (39) on R (Bioconductor version 3.2.2). The Wald Test was utilized for a comparison of one-factor conditions (in case of DIF_JQ1 or DIF_siBRD4 #3+#4 versus DIF_DMSO or DIF_siCNTR, respectively), whereas for differential analysis of conditions differing by two factors (in case of DIF_DMSO + DIF_siCNTR versus UND_DMSO + UND_siCNTR) Likelihood Ratio Test reduced by ‘treatment’ was used. The same model was implemented in case of comparison across all conditions and samples for estimation of sample-to-sample distances depicted in principal component analysis (PCA) plot. The threshold values to determine the differentially expressed genes during differentiation (DIF_siCNTR+DIF_DMSO versus UND_siCNTR+UND_DMSO) were as follows: DESeq2 ‘average baseMean’ >20, abs(log2fold change) ≥ 1 and false discovery rate (FDR) < 0.05. For differentiation-unregulated genes the thresholds were set to abs(log2 fold change value) ≤ 0.2 and FDR > 0.8. For JQ1-regulated genes (DIF_JQ1 versus DIF_DMSO) the thresholds were set to ‘average baseMean’ >20, abs(log2 fold change) ≥ 0.8, FDR < 0.05 and abs(log2 fold change) ≤ 0.2 and FDR > 0.85 in case of unregulated genes. For siBRD4 regulated genes (DIF_siBRD4 #3 + #4 versus DIF_siCNTR), following thresholds were used: ‘average baseMean’ > 20, abs(log2 fold change) ≥ 0.7, FDR < 0.05 and in case of siBRD4 unregulated genes the same thresholds as for JQ1 unregulated genes were used. The gene ontology (GO) analysis was performed using the ‘goseq’ (40) Bioconductor package version 3.2.2. The significantly enriched GO categories were identified based on the FDR values < 0.05 using the Benjamini and Hochberg test. The resulting list of GO categories were further synthesized using REVIGO (41) which clusters the categories based on semantics.
For the gene set enrichment analysis (GSEA) (42) the variance stabilization transformed read counts obtained through DESeq2 analysis were used. GSEA was performed with default settings (1000 permutations of gene sets, Signal2Noise ranking metric). For enrichment analysis GO sets (C5.all) were used.
The RNA-Seq heatmaps were performed with heatmap.2 from gplots R package (43).
ChIP-sequencing bioinformatic analysis
The FASTQ files were mapped to the human reference genome assembly hg19 using Bowtie2 (44) with very-sensitive presets in end-to-end mode. For visualization the corresponding BAM files of the replicates were merged within each condition. For visualization in the genome browser and to generate the heatmaps, all the BAM files were filtered on MAPQ ≥ 10 (MAPQ ≥ 2 for TF ChIPs) with removal of duplicates and normalized to fragments (reads) per kilobase per million (RPKM) using the bamcoverage tool on deepTools (45) (Galaxy Version 1.1.4 and later for TF BAMs 1.5.9.1.0). The visualization was performed using the Integrative Genomics Viewer (46). For the differential binding analysis and chromatin segmentation, only the reads with MAPQ ≥ 2 (Samtools 0.1.19) were retained for each BAM file and the BAM files were further deduplicated with MarkDuplicates Picard Tools (version 1.140). Heatmaps were generated using deepTools, the profile and correlation plots were made with deepTools and Cistrome Galaxy. The annotation file containing the genomic coordinates for all the human genes (assembly hg19) were downloaded from the UCSC Table Browser (47). For correlations of ChIP-seq signal with gene expression, duplicate TSS (from different transcripts) were removed. Based on gene expression values from hFOB RNA-Seq data, genes with DESeq ‘average baseMean’ values < 20 and gene length < 1000 bp were further removed from the analysis. Moreover, for genes bearing more than one TSS, the TSS bearing the highest BRD4 signal was considered.
The peak calling step was performed with Model-based Analysis of ChIP-Seq 2 (MACS2) (48) on Galaxy Cistrome (version 2.1.0.20140616.0) using default settings with q-cutoff < 0.05. For BRD4, H3K27ac, H3K27me3, H2Bub1 and RNAPII –broad option was used, whereas for TF ChIP the narrow peak option was utilized. For each condition the corresponding input samples were used as control.
To determine osteoblast-specific BRD4 binding regions differential binding analysis of all the BRD4 binding sites and regions among four different cell lines (differentiated hFOB, MCF7, MCF10A and L3.6) was performed by using ‘DiffBind’ (49) R package version 3.2.2. To determine the regions bound by BRD4 that are specific to differentiated hFOB the DiffBind thresholds were set to: Fold Concentration Change (hFOB versus the rest of the cell lines) > 1 and FDR < 0.05. These specific regions were further subdivided via Epigenome Count-based Segmentation (EpiCSeg) (50) software. The states were determined based on the combination of five histone modifications H3K27ac, H3K27me3, H3K79me2, H3K4me3, H3K4me1 from NHOST (34) as well as H3K27ac and H3K27me3 from differentiated hFOBs. Segments 3 and 4 were used as potential enhancer sites (marked by BRD4, H3K27ac and H3K4me1). Consecutive segments 3 and 4 were joined and considered as one region using Bedtools (51) merge (book end option). The resulting region file was analyzed using the Genomic Regions Enrichment of Annotations Tool (GREAT) (52) to identify genes associated with these putative enhancer regions and perform GO enrichment analysis (biological processes) on these genes. The same file was used as input for ReMap (Regulatory Map of TF Binding Sites) (53) to calculate the enrichment of TFs based on publicly available ChIP-seq data. Because of the larger size of some of the segments, we further intersected this file with DNAse-seq summit bed file from NHOST (ENCODE) to specifically focus on the accessible portions of the regions (based on DNAse summits). We then performed de novo motif discovery using peak-motifs from regulatory sequence analysis tool (RSAT) suite (54,55) on the regions flanking ±250 bp around the identified accessible regions of ‘Segment 3+4’. The differential mode was used between ‘Segment 3+4’ versus ‘Segment 1+2+5’ using oligo-analysis 6 and 7 nt outputting 5 motifs.
The BRD4 RPKM values, identified using the ‘reference-point’ mode of the computeMatrix tool on deepTools Galaxy (version 1.5.9.1.0), flanking ±500 bp around the TSS of genes in differentiated hFOBs were used to group the genes into ‘Very Low’ – log2BRD4 RPKM < 2.3; ‘Low’ – 2.3 < log2BRD4 RPKM < 2.8; ‘Medium’ – 2.8 < log2BRD4 RPKM < 3.3; ‘High’ – log2BRD4 RPKM > 3.3. The H3K27ac values within the groups were similarly identified on the same regions as BRD4. For RNAPII, regions ranging from TSS to +300 bp downstream were used. The H2Bub1 occupancy values were identified on the whole gene bodies (using the ‘scale-regions’ mode of computeMatrix tool on deepTools Galaxy).
RESULTS
BRD4 promotes osteoblast differentiation
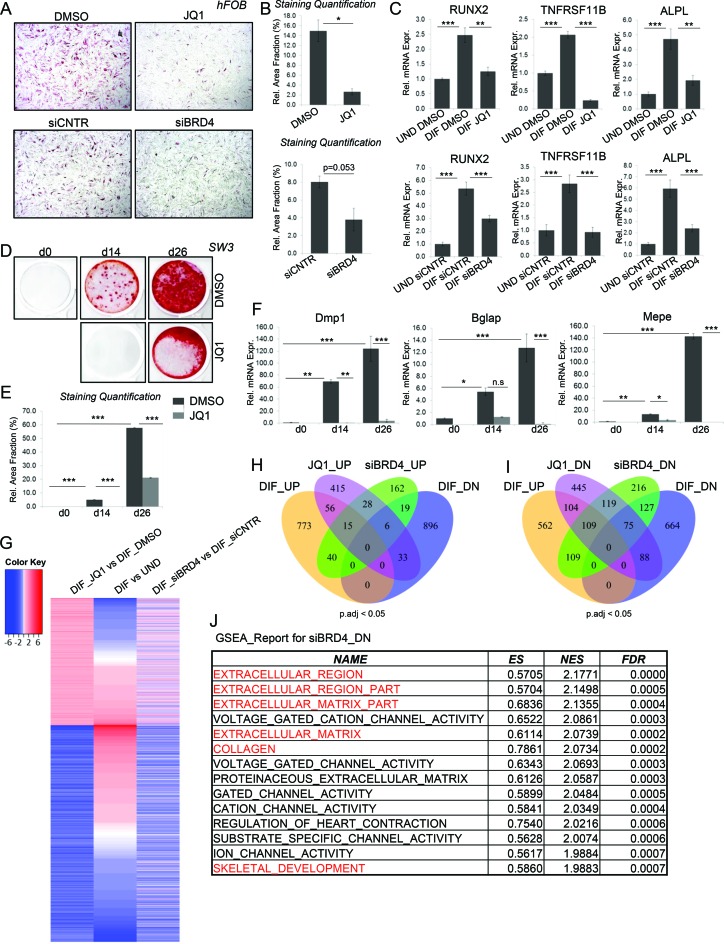
To examine the function of BRD4 during lineage specification and maintenance, we examined the effects of BETi on the differentiation of hFOBs (32). Perturbation of BRD4 function by BETi (JQ1) treatment or siRNA-mediated knockdown (Supplementary Figure S1A and B) resulted in a significant decrease of differentiation-induced alkaline phosphatase activity (Figure 1A and B; Supplementary Figure S1C and D). Gene expression analyses confirmed these findings and showed a reduced expression of the osteoblast-specific genes RUNX2, TNFRSF11B (Osteoprotegerin) and ALPL following JQ1 treatment or BRD4 knockdown (Figure 1C). To investigate whether this effect was limited to early stages of lineage-specification, we also tested the effect of BRD4 inhibition on IDG-SW3 mouse osteocytes. Despite their advanced stage of differentiation, mineralization was significantly impaired following JQ1 treatment (Figure 1D and E). Consistently, BETi treatment resulted in reduced expression of the osteoblast/osteocyte-specific genes Mepe, Bglap and Dmp1 (Figure 1F). The role of BET proteins in controlling osteoblast differentiation was differentiation-stage independent since similar effects were also observed in multipotent hMSC, where knockdown or inhibition of BRD4 also potently affected the expression of osteoblast lineage-specific genes (ALPL, COL1A1 and BGLAP; Supplementary Figure S1E and F).
Figure 1.
BRD4 is required for osteoblast differentiation. (A and B) Alkaline Phosphatase staining of hFOBs differentiated into the osteoblast lineage following DMSO or JQ1 (250nM) (upper panel) and siCNTR or siBRD4 (SmartPool) (lower panel) treatments (A). The stained areas were quantified and displayed as percentage of whole area. Mean ± SD, n = 2 (B). Student's t-test was performed for statistical analysis where ***P < 0.001, **P < 0.01, *P < 0.05 (C). The relative expression of osteoblast marker genes in hFOBs in undifferentiated (UND) and differentiated (DIF) state following siBRD4 or JQ1 treatment was evaluated via qRT-PCR and normalized to expression level of RPLP0 and the expression in the undifferentiated state. Mean ± SD, n = 3. One-way ANOVA followed by Turkey's test was performed for statistical analysis where ***P < 0.001, **P < 0.01, *P < 0.05. (D and E). Mouse IDG-SW3 osteocyte cells were differentiated for 14 and 26 days following DMSO or JQ1 treatment and stained with alizarin red for detection of mineralized nodules and the stained areas (D) were quantified as described in (B). Mean ± SD, n = 2. One-way ANOVA followed by Turkey's test was performed for statistical analysis where ***P < 0.001, **P < 0.01, *P < 0.05. (F). The relative expression of mouse osteocyte marker genes was evaluated via qRT-PCR and normalized to Gapdh and expression in the undifferentiated state. Mean ± SD, n = 2. One-way ANOVA followed by Turkey's test was performed for statistical analysis where ***P < 0.001, **P < 0.01, *P < 0.05. (G). Heatmap depicting log2fold change values of RNA-Seq data from hFOBs. The genes for the heatmap were selected based on JQ1 regulation, grouped into JQ1 up- and downregulated genes and sorted according to their regulation during differentiation. (H and I). Venn diagrams showing overlap of significantly (p.adj<0.05) up- (H) and downregulated (I) genes following JQ1 or siBRD4 treatment and regulation during differentiation (refer to the ‘Experimental Procedures’ section for description of thresholds used) (J). GSEA report for genes downregulated following BRD4 depletion. The table shows the enrichment score (ES), normalized enrichment score (NES) and FDR values for the top 15 hits.
In order to gain a more complete overview of the effects of BRD4 on the regulation of lineage-specific gene expression we performed transcriptome-wide mRNA-seq analyses following BRD4 depletion or JQ1 treatment in hFOB cells (Supplementary Figure S1G). Inhibition of BRD4 by JQ1 resulted in the differential regulation of a selected subset of genes. Consistent with the positive role of BET proteins in gene transcription, we observed more downregulated genes compared to upregulated genes following JQ1 treatment (Figure 1G). Interestingly, a large fraction of BETi-regulated genes were also differentially regulated during osteoblast differentiation, further supporting a lineage-specific function of BRD4 (Figure 1G–I). Consistent with the fact that JQ1 targets all BET proteins including BRD2, - 3 and -4 (56) and the potential differences induced by inhibition of bromodomain-mediated protein-protein interactions versus effects observed following a complete loss of BRD4 protein expression, the effects of JQ1 and BRD4 knockdown were not identical (Figure 1G–I). However, both treatments showed a similar effect of preferential downregulation of differentiation-induced genes (Figure 1I). Moreover, GSEA on siBRD4-downregulated genes indicated a significant enrichment for pathways associated with extracellular matrix, collagen and skeletal development (Figure 1J). JQ1-downregulated genes were similarly enriched for developmental pathways (Supplementary Figure S1H). Taken together, these results suggest a role of BRD4 in maintaining and directing osteoblast-specific lineage gene expression.
BRD4 is recruited to the TSS region of differentiation-induced genes and correlates with gene expression
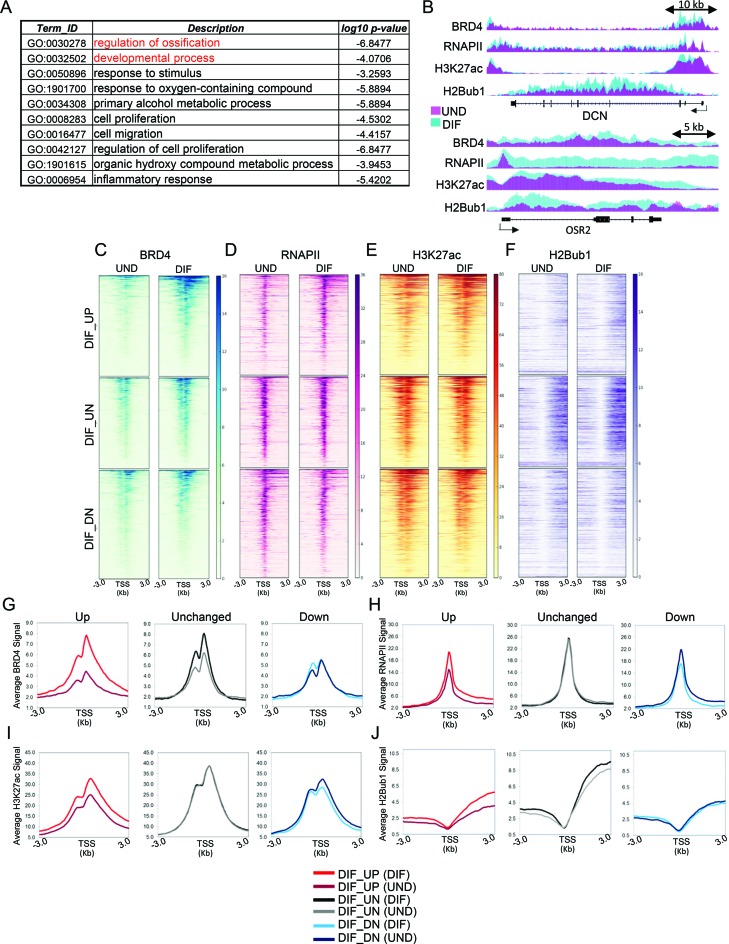
In order to understand the mechanisms by which BRD4 functions in osteoblast differentiation, we performed genome-wide occupancy studies using chromatin immunoprecipitation for BRD4, RNA Polymerase II (RNAPII) and histone modifications associated with active transcription (H3K27ac) and elongation (H2Bub1) (Supplementary Figure S2B–E) followed by high throughput sequencing (ChIP-seq). These studies revealed that differentiation-induced genes involved in the formation of bone phenotype as identified by GO analysis (Figure 2A) including decorin (DCN) and the odd-skipped related TF 2 (OSR2) display a marked increase in the occupancy of BRD4, H3K27ac, RNAPII and H2Bub1 during differentiation (Figure 2B). Consistently differentiation-induced genes showed the highest increase in BRD4 occupancy around their TSS (Figure 2C and G). Both RNAPII and H3K27ac levels similarly increased around the promoter regions of these genes while H2Bub1 occupancy increased on the transcribed regions (Figure 2E, F, H–J). In contrast, no notable change was observed on the set of un- and downregulated genes (Figure 2C–F, G–J). Altogether, consistent with a positive role in gene transcription, differentiation-induced changes in BRD4 recruitment around the TSS of genes positively correlated with the induction of lineage-specific gene expression.
Figure 2.
Differentiation-induced genes accumulate more BRD4 around TSS. (A) Functional classification summary of GO terms associated with genes upregulated during differentiation created by REVIGO(41). (B) Binding profiles for DCN and OSR2 identified from RNA-Seq as being upregulated during differentiation (DIF_UP). Each track represents normalized average occupancy (in RPKM) for BRD4, RNAPII, H3K27ac and H2Bub1 in undifferentiated (pink) and differentiated (cyan) hFOBs. For scaling, group autoscale was used for each condition. (C–F). Heatmaps depicting occupancy (RPKM values) of BRD4, RNAPII, H3K27ac and H2Bub1 in undifferentiated and differentiated hFOBs around the TSS (±3 KB) of genes upregulated (DIF_UP), unregulated (DIF_UN) and downregulated (DIF_DN) during differentiation. Each row of the heatmap represents one gene. The genes in heatmaps are sorted high to low based on BRD4 RPKM levels in differentiated cells. The order was maintained for RNAPII, H3K27ac and H2Bub1. (G–J). Average binding profiles (in RPKM values) of BRD4, RNAPII, H3K27ac and H2Bub1 in undifferentiated and differentiated hFOBs, respectively on the same genomic regions as used for heatmaps in (C–F). The profiles are depicted for differentiation upregulated (DIF_UP), unregulated (DIF_UN) and downregulated (DIF_DN) genes, separately in undifferentiated (UND) and differentiated (DIF) state.
BRD4-occupied genes differentially respond to JQ1 treatment
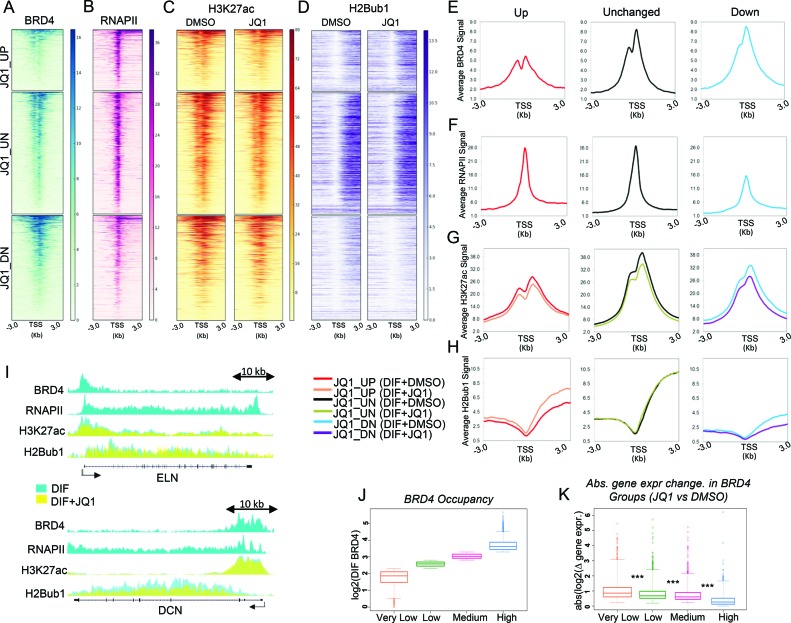
To understand the role of BRD4 in regulating gene expression and its association with active marks of transcription, we grouped genes as being upregulated, unregulated (unchanged) and downregulated based on their changes in mRNA expression following JQ1 treatment (Figure 3A–D). As expected, genes downregulated upon JQ1 treatment displayed proximal BRD4 and H3K27ac occupancy with summits shortly downstream of the TSS (Figure 3E and G). Interestingly, RNAPII and H2Bub1 occupancy at these genes were relatively lower when compared to the set of BRD4-independent genes (Figure 3F and H). Consistent with the gene expression change induced by JQ1, H2Bub1 levels dropped on JQ1-downregulated genes, whereas the JQ1-upregulated genes showed an increase in H2Bub1 occupancy (Figure 3D, H and I). Remarkably, JQ1 treatment resulted in a slight decrease in the overall H3K27ac levels across all genes (Figure 3C, G and I).
Figure 3.
BRD4 bound genes display different patterns in gene expression upon JQ1 treatment. (A–D) Heatmaps depicting occupancy (in RPKM) of BRD4 and RNAPII in differentiated hFOBs and occupancy of H3K27ac and H2Bub1 in differentiated hFOBs with DMSO or JQ1 treatment. Each row represents ±3 KB around the TSS of genes upregulated (JQ1_UP), unregulated (JQ1_UN) and downregulated (JQ1_DN) upon JQ1 treatment. Genes are sorted from high to low based on their RPKM values for BRD4 in differentiated cells. The order was maintained for RNAPII, H3K27ac and H2Bub1 profiles. (E–H) Average occupancy profiles (in RPKM) of BRD4, RNAPII in differentiated hFOBs and of H3K27ac and H2Bub1 in differentiated hFOBs with DMSO or JQ1 treatment on the same genomic regions as used for heatmaps in (A–D). The profiles are depicted for JQ1 upregulated (JQ1_UP), unregulated (JQ1_UN) and downregulated (JQ1_DN) genes, separately for differentiated control (DIF_DMSO) and JQ1-treated (DIF_JQ1) states (the last only for H3K27ac and H2Bub1 profiles). (I). The binding profiles for ELN and DCN genes identified by RNA-seq as JQ1-downregulated genes (JQ1_DN). Each track represents normalized average occupancy (in RPKM) for BRD4, RNAPII, H3K27ac and H2Bub1 in differentiated hFOBs treated with DMSO (cyan) or JQ1 (yellow). For scaling, group autoscale was used for each condition. (J) Boxplots showing log2 RPKM values of BRD4 ±500 bp around the TSS of genes in differentiated hFOBs. Based on the BRD4 values genes were grouped in four classes: ‘Very Low’, ‘Low’, ‘Medium’ and ‘High’ having log2 BRD4 RPKM < 2.3; 2.3 < log2 BRD4 RPKM < 2.8; 2.8 < log2 BRD4 RPKM < 3.3 and log2 BRD4 RPKM > 3.3, respectively. (K). Boxplots showing log2 fold gene expression change induced by JQ1 in differentiated hFOBs within ‘Very Low’, ‘Low’, ‘Medium’ and ‘High’ BRD4 groups. For statistical analysis Wilcoxon Rank Sum test was performed where ***P < 0.001, **P < 0.01, *P < 0.05.
Notably, we observed that genes unaffected by JQ1 show a high occupancy of BRD4 and RNAPII as well as the active histone modifications H3K27ac and H2Bub1 (Figure 3 A–D, E–H). Moreover, genes downregulated by JQ1 treatment displayed a wide range of BRD4 occupancy around their TSS (see the BRD4 heatmap for JQ1_DN genes, Figure 3A). To further investigate the correlation between BRD4 occupancy and gene expression, we classified genes based on BRD4 occupancy into 4 groups referred to as ‘Very Low’, ‘Low’, ‘Medium’ and ‘High’ (Figure 3J). RNAPII, H3K27ac and H2Bub1 levels correlated with BRD4 within each group where they displayed lowest occupancy around the genes of ‘Very Low’ group and increased incrementally in the ‘Low’, ‘Medium’ and ‘High’ groups (Supplementary Figure S3A–C). Similarly, gene expression levels also correlated with BRD4 occupancy (Supplementary Figure S3D). Strikingly, the absolute gene expression change following JQ1 treatment was highest for the ‘Very Low’ group (Figure 3K), implying that low BRD4-marked and lowly expressed genes were more sensitive to inhibition of BRD4. In fact, almost a third of the JQ1 downregulated genes belonged to the ‘Very Low’ group (Supplementary Figure S3E) while the rest of the genes were scattered across the other three groups. Altogether, these results indicate that TSS-associated BRD4 occupancy generally correlates with gene expression levels but only partially explains the extent of its requirement for gene expression, suggesting that additional mechanisms of transcriptional control may play a more significant role in determining the effects of BRD4 inhibition or loss.
BRD4 is enriched on osteoblast-specific putative enhancers
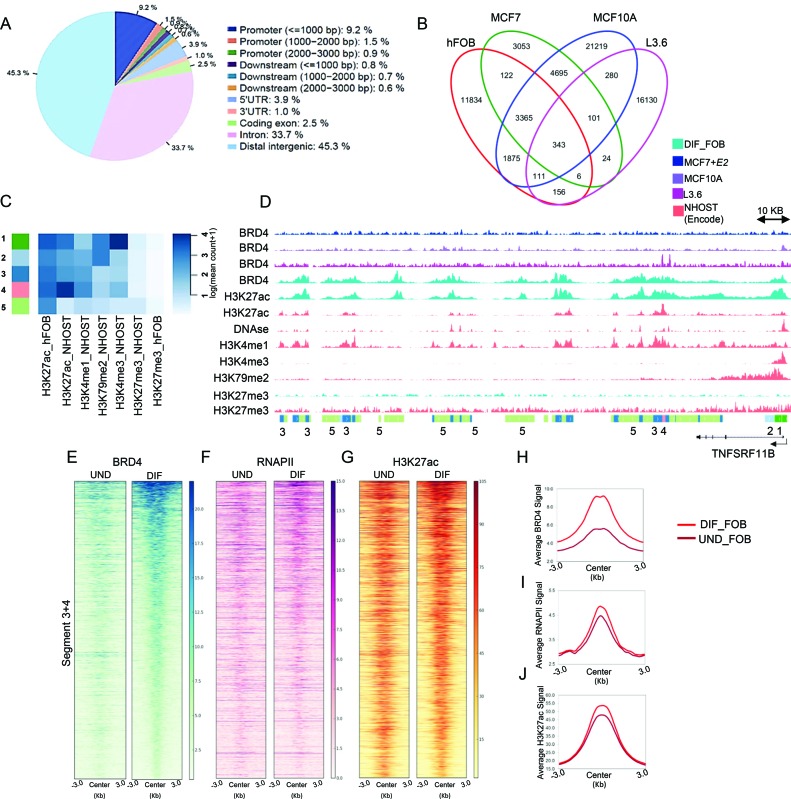
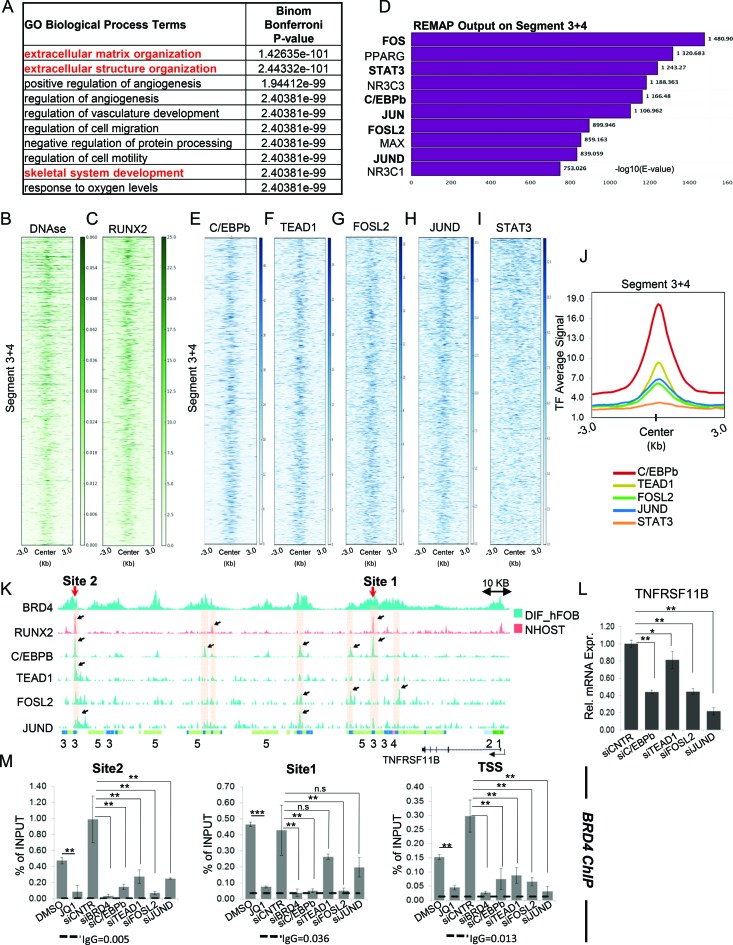
The transcriptional regulation of a gene is achieved by a cooperation of promoter- and enhancer-mediated events. Consistent with a proposed function at both enhancers and promoter proximal regions, nearly half of BRD4-enriched regions (45.3%) were localized to distal intergenic regions (Figure 4A). Moreover, BRD4 occupancy markedly correlated with RNAPII and H3K27ac on all the genomic regions (Supplementary Figure S3F). These findings support the notion that distal BRD4-bound regions might serve as transcribed enhancers responsible for the establishment and maintenance of an osteoblast-specific transcriptional program. In order to identify putative osteoblast-specific enhancer regions we performed differential analysis of BRD4-bound regions among four diverse cell systems including hFOB, MCF7 (estrogen-receptor-positive breast cancer cell line) (20), MCF10A (normal mammary epithelial cell line) and L3.6 (pancreatic cancer cell line). Strikingly, the fraction of commonly shared BRD4-occupied sites was lower between these cells than the number of cell type-specific regions for each of them (Figure 4B). 7822 regions were found to be specifically enriched for BRD4 occupancy in hFOBs (FDR < 0.05). Since active enhancers can be identified based on the combination of active histone marks we performed chromatin segmentation analysis (50) to further characterize osteoblast-specific BRD4-enriched regions. Based on the high degree of epigenetic similarity of hFOB cells and NHOST cells (Supplementary Figure S4A and B), we also utilized available H3K4me3, H3K4me1, H3K79me2, H3K27me3 and H3K27ac occupancy profiles from NHOST cells along with H3K27ac and H3K27me3 profiles from hFOBs to identify chromatin states which subdivide osteoblast-specific BRD4-bound regions (Figure 4C and Supplementary Figure S4C). All these states were positive for H3K27ac and negative for H3K27me3, confirming the association of BRD4 with active chromatin regions. States 1 and 2 were most likely associated with actively transcribed promoter regions as revealed by the combination of H3K27ac, H3K4me3 and H3K79me2 occupancy, where H3K4me3 is particularly associated with TSS-proximal regions and H3K79me2 with the proximal transcribed region of active genes (Figure 4D and Supplementary Figure S4E). States 3 and 4 appear to represent putative enhancers (Supplementary Table S5) based on the presence of both H3K27ac and H3K4me1 marks, both known to be associated with active enhancer regions. State 5 was only positive for H3K27ac, and specific for hFOBs, possibly indicating that these regions may be specific for the hFOB system and therefore less informative about general mechanisms related to osteoblast lineage-specification. Consistently, the majority of ‘Segment 3+4’ constituted distal intergenic regions or introns (Supplementary Figure S4D), where some were in close proximity to osteoblast-specific genes as seen on the example of the TNFRSF11B and COL1A1 genes (Figure 4D and Supplementary Figure S4E).
Figure 4.
BRD4 occupancy increases on putative enhancers upon differentiation. (A) Distribution of BRD4 occupied regions over the genome in differentiated hFOBs. The figure was obtained with the program CEAS (78) with default options. (B) The Venn diagrams depicting overlap of BRD4 peaks among four cell lines used in the study: differentiated hFOBs (DIF_FOB), MCF10A, MCF7 treated with estradiol (+E2) and L3.6 cells. (C) hFOB-specific regions displaying differential BRD4 occupancy were subjected to EpiCSeg segmentation analysis. Five histone modifications from NHOST cells (H3K27ac, H3K4me1, H3K79me2, H3K4me3 and H3K27me3) and two from hFOBs (H3K27ac and H3K27me3) were used to identify chromatin states associated with selected BRD4 regions. The heatmap shows five chromatin states which were identified based on the combination of normalized counts of the histone modifications datasets. (D) IGV profile showing the distribution of segments around the osteoblast-specific TNFRSF11B gene. Each lane represents normalized occupancy (from top to low) of BRD4 in MCF7+E2 (darkblue), in MCF10A (purple), in L3.6 (magenta); BRD4 and H3K27ac in differentiated hFOBs (cyan); H3K27ac, H3K4me1, DNAse-Seq, H3K4me3, H3K79me2 in NHOST (coral); H3K27me3 in differentiated hFOBs (cyan) and NHOST (coral). The coverage signals were autoscaled by the software. (E–G) Heatmaps showing BRD4, RNAPII, H3K27ac occupancy in undifferentiated and differentiated hFOBs ±3 KB around the center of ‘Segment 3+4’ identified from EpiCSeg. Segments in heatmaps were ordered from high to low based on BRD4 RPKM values in differentiated cells. (H–J). Average binding profiles (in RPKM) of BRD4, RNAPII, H3K27ac in undifferentiated and differentiated hFOBs on the same genomic regions as heatmaps in (E–G).
Interestingly, upon differentiation, BRD4 occupancy substantially increased on ‘Segment 3+4’ regions, implying a potential function of these enhancers in controlling osteoblast-specific gene expression (Figure 4E and H). Consistently, H3K27ac and RNAPII occupancy also slightly increased on these regions after differentiation (Figure 4F, G, I and J). Thus, by combining differential binding and chromatin segmentation analysis, we were able to successfully identify putative osteoblast-specific enhancers potentially involved in lineage determination.
A distinct set of tissue-specific transcription factors co-localizes with BRD4 on putative enhancers
To identify possible target genes associated with the putative enhancers from ‘Segment 3+4’ we used ‘Genomic Regions Enrichment of Annotations Tool’ (GREAT) (52). Interestingly, the majority of these enhancers were found to be 50–500 kb away from known gene-specific TSS regions (Supplementary Figure S5A). Consistent with their specific enrichment in hFOB cells, these genes were enriched for pathways associated with skeletal development and extracellular matrix organization (Figure 5A). Importantly, many of these genes were differentially regulated during differentiation or following BETi treatment (Supplementary Figure S5B and C), supporting a role for BRD4 function in osteoblast lineage determination.
Figure 5.
BRD4-bound putative enhancers show C/EBPb, AP1 and TEAD1 binding. (A) Biological process gene ontology (GO.BP) analysis of genes associated with putative enhancers from ‘Segment 3+4’ as identified by GREAT analysis. (B and C). Heatmaps showing DNAse-Seq signal and RUNX2 occupancy in NHOST ± 3 KB around the center of regions identified from EpiCSeg ‘Segment 3+4’. The ordering of segments in heatmaps is based on BRD4 RPKM values levels in differentiated hFOB cells from high to low. (D) Top 10 hits according to ReMap results performed on ‘Segment 3+4’. The bars represent the significance values for intersected regions of identified factors. (E–I) Heatmaps depicting occupancy (in RPKM) of C/EBPb, TEAD1, FOSL2, JUND and STAT3 ±3 KB around the center of ‘Segment 3+4. Each row represents one segment and is ordered from high to low based on BRD4 RPKM values levels in differentiated hFOB cells. (J) Average binding intensity (RPKM) of C/EBPb, TEAD1, FOSL2, JUND and STAT3 ±3 KB around the center of ‘Segment 3+4’. (K) IGV profile depicting binding of C/EBPb, TEAD1, FOSL2 and JUND in differentiated hFOBs (cyan) and RUNX2 in NHOST (coral) in the proximity of the TNFRSF11B gene. Two potential enhancer sites 1 and 2 are indicated. (L). The relative expression of TNFRSF11B in differentiated hFOBs following siRNA-mediated knockdown of C/EBPb, TEAD1, FOSL2 and JUND evaluated via qRT-PCR and normalized to expression level of RPLP0 and the expression in the control (siCNTR) state. Mean ± SD, n = 3. One-way ANOVA followed by Dunnett's test were performed for statistical analysis where ***P < 0.001, **P < 0.01, *P < 0.05. (M) BRD4 occupancy was evaluated by ChIP at potential enhancer sites 1 and 2 indicated in (K) and the TSS of the TNFRSF11B gene and measured via qPCR in differentiated hFOBs following JQ1 treatment or siRNA-mediated knockdown of BRD4, C/EBPb, TEAD1, FOSL2 and JUND. For background estimation IgG ChIP was used. The ChIP efficiency is displayed as enrichment over Input in percent. Mean ± SD, n = 2. Student's t-test (for DMSO versus JQ1 comparison) and one-way ANOVA followed by Dunnett's test (for the rest) were performed for statistical analysis where ***P < 0.001, **P < 0.01, *P < 0.05.
The tissue-specific function of an enhancer is dictated by the panel of TFs bound to it at a given time. The TFs define the composition and hence the outcome of enhancer-promoter interactions during gene regulation. Notably, ‘Segment 3+4’ displayed DNase hypersensitivity in NHOST cells (Figure 5B), suggesting that these regions may be bound by specific TFs. One of the main TFs involved in the osteoblast differentiation is the Runt-related TF 2 (RUNX2) (57). Consistently, analysis of RUNX2 occupancy in NHOST cells revealed a prominent enrichment of RUNX2 on ‘Segment 3+4’ (Figure 5C). To identify additional TF binding motifs enriched in these regions, we performed differential motif analysis of ‘Segment 3+4’ versus the ‘Segments 1+2+5’. We observed an enrichment of consensus sequences for the AP1 TF family as well as the Hippo/YAP-regulated Transcriptional enhancer factor TEF-1 (TEAD1) within the osteoblast-specific BRD4-bound regions of ‘Segment3+4’ (Supplementary Table S4). Moreover, an enrichment analysis within ‘Segment 3+4’ of a large number of publicly available datasets for the occupancy of various TFs and cofactors was performed using ReMap (53). These analyses confirmed the significant enrichment of AP1-bound regions on ‘Segment 3+4’ (Figure 5D) and further identified a significant overlap with additional TFs including Signal Transducer and Activator of Transcription 3 (STAT3) and CCAAT/Enhancer-Binding Protein-β (C/EBPb). Notably, based on RNA-seq data, all the identified TFs are expressed in hFOBs and only RUNX2 and Fos-related antigen 2 (FOSL2) expression were dependent on BRD4 (Supplementary Figure S5D), suggesting that these factors may serve as central mediators to facilitate BRD4 recruitment to osteoblast-specific enhancer regions. ChIP-qPCR analyses on the overlapping regions identified by ReMap showed a significant enrichment for C/EBPb, TEAD1, STAT3, JUND and FOSL2 (Supplementary Figure S5E–H). Thus, we performed additional genome-wide occupancy studies of all five TFs in hFOB cells to examine their overlap with osteoblast-specific BRD4-enriched regions. Interestingly, these TFs displayed differential binding patterns in hFOBs, where a significant number of C/EBPb-bound regions were largely devoid of other factors (Supplementary Figure S5I). A number of sites displayed similar TEAD1, JUND and FOSL2 occupancy, while strongly enriched STAT3-bound regions lacked binding of the other factors (Supplementary Figure S5J–M). Notably, BRD4 exhibited differential binding profiles at the TF bound regions, where C/EBPb-, JUND- and FOSL2-enriched sites displayed higher binding of BRD4, compared to TEAD1- and STAT3-enriched regions, which showed moderate and low levels of BRD4 occupancy, respectively (Supplementary Figure S5N). Remarkably, ‘Segment 3+4’ displayed substantial binding of C/EBPb, TEAD1, FOSL2 and JUND (Figure 5E–H, J) whereas almost no STAT3 signal was detected on these regions (Figure 5I and J). Interestingly, some ‘Segment 3+4’ regions showed co-occupancy of all TFs (Supplementary Figure S6A) while some osteoblast-specific BRD4 target genes such as TNFRSF11B (Figure 1C), ELN and DCN (Supplementary Figure S5C) showed distinct occupancy of the identified TFs at putative enhancer sites located within 50 kb of the genes (Figure 5K; Supplementary Figure S6B and E).
We next examined the functional importance of each of the TFs identified to be differentially enriched at the osteoblast-specific BRD4-enriched putative enhancers within ‘Segment 3+4’ for the expression of the associated target genes. Consistent with reduced recruitment of BRD4 at both the TSS as well as the putative enhancer sites (Site 1 and 2, Figure 5K) following the depletion of C/EBPb, TEAD1, FOSL2 and JUND in differentiated hFOBs (Supplementary Figure S6C and D), the expression of the TNFRSF11B gene was decreased in each of the knockdown conditions, albeit to a varying degrees (Figure 5L and M). Notably, the expression of BRD4 was partially dependent on JUND expression (Supplementary Figure S6D). Likewise, BRD4 occupancy both at TSS as well as at a potential enhancer site (Site 1, Supplementary Figure S6E) upstream of the ELN gene was markedly decreased following the knockdown of each of the four TFs (Supplementary Figure S6F). However, the expression of elastin (ELN) was significantly affected only upon TEAD1 and JUND knockdown (Supplementary Figure S6G), suggesting a unique and, perhaps, hierarchical contribution pattern of TFs to BRD4-dependent gene expression regulation. Thus, the predictive value of utilizing a combination of chromatin segmentation of differentially bound osteoblast-specific BRD4-enriched regions, motif analyses and integration of publically available TF genome occupancy datasets, could be confirmed by genome-wide occupancy studies for four out of five predicted TFs (C/EBPb, TEAD1, JUND and FOSL2). Moreover, the association of these putative enhancers with osteoblast-related genes and the dependence of these genes on functional activity of BRD4 support a role for BRD4 in regulating gene expression and controlling lineage determination.
DISCUSSION
Cellular differentiation is a complex process involving the coordination of multiple TFs and epigenetic modifiers acting on both cis- and trans-regulatory elements to selectively regulate lineage-specific gene expression. In this study, we sought to elucidate the context-specific function of BRD4 in regulating gene expression by exploring its function and localization during osteoblast differentiation. In fact, we show that the function of BRD4 is indispensable for osteoblast lineage commitment at early as well as later stages of osteoblast differentiation. Although global mRNA or protein levels of BRD4 do not significantly change during differentiation there is a considerable increase of BRD4 occupancy around the TSS of genes and the change is markedly higher on genes upregulated during differentiation.
Importantly, pharmacological inhibition of BET proteins was shown to be a promising strategy in the treatment of bone-associated tumors and inflammatory bone resorption (58,59). Consistently, in addition to inhibition of tumor growth or osteoclastogenesis, BETi inhibited the differentiation of osteoblasts. However, BETi injections into healthy mice did not lead to any significant changes in bone formation rate or bone morphometric parameters, possibly due to the adult age of the animals used in the studies. In fact, among other developmental defects, heterozygous BRD4 null mice were reported to display malformations of skull bones, possibly supporting the importance of BRD4 for bone formation (60).
The identification of osteoblast-specific BRD4-bound putative enhancers and TFs associated with these regions further strengthens the role of BRD4 in lineage determination. Previous studies established important functions of C/EBPb, AP1 and STAT3 TFs in regulating osteoblast differentiation (61–64). Although the specific role of TEAD1 in the context of bone formation has not yet been studied, its importance can be implied based on its interaction and dependence on the transcriptional co-activator with PDZ-binding motif (TAZ) (65). Specifically, TAZ was shown to regulate osteoblast differentiation and mediate the effects of canonical Wnt signaling, a central regulator of osteoblast differentiation (66,67). Although the RUNX2 binding motif was not specifically identified by motif discovery, analysis of RUNX2 in NHOST cells suggests its co-localization on osteoblast-specific BRD4-enriched regions. Consistently, RUNX2 was previously reported to interact with both AP1 factors as well as C/EBPb to regulate osteoblast differentiation (61,68). Thus, it is likely that BRD4 localization to ‘Segment 3+4’ may be mediated by functional cooperation between RUNX2 and other DNA binding TFs. In general, the distinct binding pattern of the identified TFs at osteoblast-specific genes suggests that each might play a specific role in coordinating BRD4 function during the establishment of osteoblast-specific transcriptional programs. The cluster of active enhancers marked by TFs, the mediator complex and a combination of histone modifications are frequently found near genes associated with cell fate specification (12). However, the functional importance of each of the enhancer units within these clusters might vary greatly (69,70). Whereas some of the enhancers might display critical regulatory functions, the function of others is dispensable for the activation of the target genes. The functional hierarchy of individual enhancers likely reflects the complexity of TF cooperativity involved in enhancer activation (70). Moreover, the three-dimensional organization of the chromatin and the looping of several enhancers with the target promoter within one topologically associated domain may enable multiple enhancers to promote target gene transcription in an additive manner (71). Hence, given the (at least partial) additive and redundant function of enhancers, it is not surprising that depletion of the identified TFs in this study resulted in differential regulation of BRD4-target genes despite similar effects on BRD4 recruitment at individual potential enhancer sites and TSS of these genes. Moreover, whether each of the putative enhancers located in the proximity of a specific gene serve as regulatory elements controlling the expression of this gene requires further gene editing approaches to definitively establish their role. Futhermore, although BRD4 recruitment was dependent on each of the identified TFs, whether these TFs directly interact with BRD4 in a bromodomain-dependent manner or indirectly by means of additional co-factors needs further investigation. The strong association of p300 with BRD4 specifically on enhancers as well as with C/EBPb, AP1, TEAD1 and STAT proteins suggests a possible model where the identified TFs could utilize p300 to recruit BRD4 onto tissue-specific enhancers (65,72–75). This assumption can further be supported by synergistic effects of CBP/p300 inhibitors with BETi (76). Moreover, some of the TFs may themselves serve as acetylation targets for p300 (4,77), offering an additional, more direct mode of mediating BRD4 recruitment to enhancers.
Although BRD4 was reported as a ubiquitously expressed protein that serves as a general facilitator of transcriptional activation, an increasing body of evidence indicates that it regulates gene expression in a context-specific manner where the specificity is defined by the repertoire of TFs and co-factors expressed. Together, our findings support the role of BRD4 as a central intermediary in the induction of gene expression in response to external signaling via its recruitment to both promoter and enhancer regions bound by a subset of TFs. Thus, these findings begin to provide an explanation of the intricacies and diversity of BETi-mediated transcriptional effects elicited in different tissues and cell lines. A further understanding of the context-dependent functions of BET proteins will be essential for the efficient and correct clinical application of BET inhibitors in various diseases.
Acknowledgments
The authors thank M. Thomas-Chollier and D. Thieffry for guiding the work of R.T.M.; C. Hernandez and S. Collombet for useful discussions and invaluable assistance on the data analysis; G. Salinas-Riester, B. Downey, F. Ludewig and T. Lingner for excellent support with next generation sequencing; and C.-M. Chiang for kindly providing the BRD4 antibody.
Author contributions: The experiments were designed by S.A.J. and Z.N. R.T.M. contributed to bioinformatic analyses including HTSeq, DESeq2, GO analysis, REVIGO, DiffBind, motif discovery (RSAT & ReMap) and chromatin segmentation (EpiCSeg) analysis. Z.N. performed all the in vitro experiments in hFOBs and hMSCs, generated all the ChIP-Seq and mRNA-Seq data from hFOBs. The analysis on the Galaxy platform were performed by Z.N. M.S. and J.R.H. conducted the experiments with IDG-SW3 cells and the qPCR gene expression analysis from these cells were performed by G.S. S.J.B. assisted with hFOB experiments and T.H. assisted with transcription factor ChIP experiments. S.N., V.K.M. and U.B. generated the BRD4 ChIP-Seq datasets from MCF7, L3.6 and MCF10A cells, respectively. S.K. synthesized JQ1. The manuscript was written by Z.N. and S.A.J. All authors read and corrected the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
German Academic Exchange Service (DAAD) (to S.N.); EpiGeneSys consortium travel grant (to R.T.M.); Erasmus Mundus External Cooperation Window EURINDIA and the Göttingen Graduate School for Neurosciences, Biophysics and Molecular Biosciences (to U.B.); German Research Foundation (DFG) [JO 815/3-1]; Deutsche Krebshilfe [111600 to S.A.J.]; German Ministry for Science and Education (BMBF) and Agence Nationale de la Recherche-funded iBONE consortium [01KU1401A to S.A.J., 01KU1401B to E.H., ANR-14-CE11-0006 to M. Thomas-Chollier]; German Research Foundation [HE 5208/2-1 to E.H.]; NIH [DE14036 to J.R.H., M.S.]. Funding for open access charge: Internal house funding/university funding.
Conflict of interest statement. None declared.
REFFERENCES
- 1.Filippakopoulos P., Picaud S., Mangos M., Keates T., Lambert J.-P., Barsyte-Lovejoy D., Felletar I., Volkmer R., Müller S., Pawson T., et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang M.K., Mochizuki K., Zhou M., Jeong H.-S., Brady J.N., Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z., Yik J.H.N., Chen R., He N., Jang M.K., Ozato K., Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 4.Roe J.-S., Mercan F., Rivera K., Pappin D.J., Vakoc C.R. BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia. Mol. Cell. 2015;58:1028–1039. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi J., Wang Y., Zeng L., Wu Y., Deng J., Zhang Q., Lin Y., Li J., Kang T., Tao M., et al. Disrupting the interaction of BRD4 with di-acetylated twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown J.D., Lin C.Y., Duan Q., Griffin G., Federation A., Paranal R.M., Bair S., Newton G., Lichtman A., Kung A., et al. NF-kB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang B., Yang X.-D., Zhou M.-M., Ozato K., Chen L.-F. Brd4 coactivates transcriptional activation of NF-κB via specific binding to acetylated RelA. Mol. Cell. Biol. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart H.J.S., Horne G.A., Bastow S., Chevassut T.J.T. BRD4 associates with p53 in DNMT3A-mutated leukemia cells and is implicated in apoptosis by the bromodomain inhibitor JQ1. Cancer Med. 2013;2:826–835. doi: 10.1002/cam4.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahman S., Sowa M.E., Ottinger M., Smith J.A., Shi Y., Harper J.W., Howley P.M. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3 ▿. Mol. Cell. Biol. 2011;31:2641–2652. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W., Ma Q., Wong K., Li W., Ohgi K., Zhang J., Aggarwal A., Rosenfeld M.G. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visel A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ong C.-T., Corces V.G. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat. Rev. Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Santa F., Barozzi I., Mietton F., Ghisletti S., Polletti S., Tusi B.K., Muller H., Ragoussis J., Wei C.-L., Natoli G. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hah N., Murakami S., Nagari A., Danko C.G., Kraus W.L. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim T.-K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D.A., Laptewicz M., Barbara-Haley K., Kuersten S., et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Notani D., Ma Q., Tanasa B., Nunez E., Chen A.Y., Merkurjev D., Zhang J., Ohgi K., Song X., et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y., Wang L., Ren S., Wang L., Blackburn P.R., McNulty M.S., Gao X., Qiao M., Vessella R.L., Kohli M., et al. Activation of P-TEFb by androgen receptor-regulated enhancer RNAs in castration-resistant prostate cancer. Cell Rep. 2016;15:599–610. doi: 10.1016/j.celrep.2016.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanno T., Kanno Y., LeRoy G., Campos E., Sun H.-W., Brooks S.R., Vahedi G., Heightman T.D., Garcia B.A., Reinberg D., et al. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol. 2014;21:1047–1057. doi: 10.1038/nsmb.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagarajan S., Hossan T., Alawi M., Najafova Z., Indenbirken D., Bedi U., Taipaleenmäki H., Ben-Batalla I., Scheller M., Loges S., et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell Rep. 2014;8:460–469. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Micco R., Fontanals-Cirera B., Low V., Ntziachristos P., Yuen S.K., Lovell C.D., Dolgalev I., Yonekubo Y., Zhang G., Rusinova E., et al. Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep. 2014;9:234–247. doi: 10.1016/j.celrep.2014.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang W., Prakash C., Sum C., Gong Y., Li Y., Kwok J.J.T., Thiessen N., Pettersson S., Jones S.J.M., Knapp S., et al. Bromodomain-containing Protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J. Biol. Chem. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coudé M.-M., Braun T., Berrou J., Dupont M., Bertrand S., Masse A., Raffoux E., Itzykson R., Delord M., Riveiro M.E., et al. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget. 2015;6:17698–17712. doi: 10.18632/oncotarget.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Z., Gong Y., Ma Y., Lu K., Lu X., Pierce L.A., Thompson R.C., Muller S., Knapp S., Wang J. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin. Cancer Res. 2013;19:1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lockwood W.W., Zejnullahu K., Bradner J.E., Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mertz J.A., Conery A.R., Bryant B.M., Sandy P., Balasubramanian S., Mele D.A., Bergeron L., Sims R.J. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Asangani I.A., Dommeti V.L., Wang X., Malik R., Cieslik M., Yang R., Escara-Wilke J., Wilder-Romans K., Dhanireddy S., Engelke C., et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuber J., Shi J., Wang E., Rappaport A.R., Herrmann H., Sison E.A., Magoon D., Qi J., Blatt K., Wunderlich M., et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpiuk O., Najafova Z., Kramer F., Hennion M., Galonska C., König A., Snaidero N., Vogel T., Shchebet A., Begus-Nahrmann Y., et al. The histone H2B monoubiquitination regulatory pathway is required for differentiation of multipotent stem cells. Mol. Cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Liu W., Stein P., Cheng X., Yang W., Shao N.-Y., Morrisey E.E., Schultz R.M., You J. BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ. 2014;21:1950–1960. doi: 10.1038/cdd.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu T., Pinto H.B., Kamikawa Y.F., Donohoe M.E. The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression. Stem Cell Rep. 2015;4:390–403. doi: 10.1016/j.stemcr.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris S.A., Enger R.J., Riggs B.L., Spelsberg T.C. Development and characterization of a conditionally immortalized human fetal osteoblastic cell line. J. Bone Miner. Res. 1995;10:178–186. doi: 10.1002/jbmr.5650100203. [DOI] [PubMed] [Google Scholar]
- 33.Woo S.M., Rosser J., Dusevich V., Kalajzic I., Bonewald L.F. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J. Bone Miner. Res. 2011;26:2634–2646. doi: 10.1002/jbmr.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Håkelien A.-M., Bryne J.C., Harstad K.G., Lorenz S., Paulsen J., Sun J., Mikkelsen T.S., Myklebost O., Meza-Zepeda L.A. The regulatory landscape of osteogenic differentiation. Stem Cells. 2014;32:2780–2793. doi: 10.1002/stem.1759. [DOI] [PubMed] [Google Scholar]
- 36.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:1–13. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goecks J., Nekrutenko A., Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:1–13. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:1–12. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Liaw W.H.A., Lumley T., Maechler M., Magnusson A., Moeller S., Schwartz M., et al. gplots: various R programming tools for plotting data. 2015. R Package 2.
- 44.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramírez F., Dündar F., Diehl S., Grüning B.A., Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mammana A., Chung H.-R. Chromatin segmentation based on a probabilistic model for read counts explains a large portion of the epigenome. Genome Biol. 2015;16:1–12. doi: 10.1186/s13059-015-0708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Griffon A., Barbier Q., Dalino J., van Helden J., Spicuglia S., Ballester B. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res. 2014;43:1–14. doi: 10.1093/nar/gku1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Medina-Rivera A., Defrance M., Sand O., Herrmann C., Castro-Mondragon J.A., Delerce J., Jaeger S., Blanchet C., Vincens P., Caron C., et al. RSAT 2015: Regulatory Sequence Analysis Tools. Nucleic Acids Res. 2015;43:W50–W56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas-Chollier M., Darbo E., Herrmann C., Defrance M., Thieffry D., van Helden J. A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat. Protoc. 2012;7:1551–1568. doi: 10.1038/nprot.2012.088. [DOI] [PubMed] [Google Scholar]
- 56.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Otto F., Thornell A.P., Crompton T., Denzel A., Gilmour K.C., Rosewell I.R., Stamp G.W.H., Beddington R.S.P., Mundlos S., Olsen B.R., et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 58.Lamoureux F., Baud'huin M., Rodriguez Calleja L., Jacques C., Berreur M., Rédini F., Lecanda F., Bradner J.E., Heymann D., Ory B. Selective inhibition of BET bromodomain epigenetic signalling interferes with the bone-associated tumour vicious cycle. Nat. Commun. 2014;5:3511. doi: 10.1038/ncomms4511. [DOI] [PubMed] [Google Scholar]
- 59.Park-Min K.-H., Lim E., Lee M.J., Park S.H., Giannopoulou E., Yarilina A., van der Meulen M., Zhao B., Smithers N., Witherington J., et al. Inhibition of osteoclastogenesis and inflammatory bone resorption by targeting BET proteins and epigenetic regulation. Nat. Commun. 2014;5:5418. doi: 10.1038/ncomms6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Houzelstein D., Bullock S.L., Lynch D.E., Grigorieva E.F., Wilson V.A., Beddington R.S.P. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4†. Mol. Cell. Biol. 2002;22:3794–3802. doi: 10.1128/MCB.22.11.3794-3802.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gutierrez S., Javed A., Tennant D.K., van Rees M., Montecino M., Stein G.S., Stein J.L., Lian J.B. CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 2002;277:1316–1323. doi: 10.1074/jbc.M106611200. [DOI] [PubMed] [Google Scholar]
- 62.Nicolaidou V., Wong M.M., Redpath A.N., Ersek A., Baban D.F., Williams L.M., Cope A.P., Horwood N.J. Monocytes induce STAT3 Activation in human mesenchymal stem cells to promote osteoblast formation. PLoS One. 2012;7:e39871. doi: 10.1371/journal.pone.0039871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou H., Newnum A.B., Martin J.R., Li P., Nelson M.T., Moh A., Fu X.-Y., Yokota H., Li J. Osteoblast/osteocyte-specific inactivation of Stat3 decreases load-driven bone formation and accumulates reactive oxygen species. Bone. 2011;49:404–411. doi: 10.1016/j.bone.2011.04.020. [DOI] [PubMed] [Google Scholar]
- 64.Sabatakos G., Sims N.A., Chen J., Aoki K., Kelz M.B., Amling M., Bouali Y., Mukhopadhyay K., Ford K., Nestler E.J., et al. Overexpression of ΔFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat. Med. 2000;6:985–990. doi: 10.1038/79683. [DOI] [PubMed] [Google Scholar]
- 65.Stein C., Bardet A.F., Roma G., Bergling S., Clay I., Ruchti A., Agarinis C., Schmelzle T., Bouwmeester T., Schübeler D., et al. YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet. 2015;11:e1005465. doi: 10.1371/journal.pgen.1005465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hong J.-H., Hwang E.S., McManus M.T., Amsterdam A., Tian Y., Kalmukova R., Mueller E., Benjamin T., Spiegelman B.M., Sharp P.A., et al. TAZ, a transcriptional modulator of mesenchymal stem cell differentiation. Science. 2005;309:1074–1078. doi: 10.1126/science.1110955. [DOI] [PubMed] [Google Scholar]
- 67.Byun M.R., Hwang J.-H., Kim A.R., Kim K.M., Hwang E.S., Yaffe M.B., Hong J.-H. Canonical Wnt signalling activates TAZ through PP1A during osteogenic differentiation. Cell Death Differ. 2014;21:854–863. doi: 10.1038/cdd.2014.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.D'Alonzo R.C., Selvamurugan N., Karsenty G., Partridge N.C. Physical interaction of the activator protein-1 factors c-Fos and c-Jun with Cbfa1 for collagenase-3 promoter activation. J. Biol. Chem. 2002;277:816–822. doi: 10.1074/jbc.M107082200. [DOI] [PubMed] [Google Scholar]
- 69.Shin H.Y., Willi M., Yoo K.H., Zeng X., Wang C., Metser G., Hennighausen L. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016;48:904–911. doi: 10.1038/ng.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang J., Liu X., Li D., Shao Z., Cao H., Zhang Y., Trompouki E., Bowman T.V., Zon L.I., Yuan G.-C., et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev. Cell. 2016;36:9–23. doi: 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kieffer-Kwon K.-R., Tang Z., Mathe E., Qian J., Sung M.-H., Li G., Resch W., Baek S., Pruett N., Grøntved L., et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paulson M., Pisharody S., Pan L., Guadagno S., Mui A.L., Levy D.E. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 73.Schwartz C., Beck K., Mink S., Schmolke M., Budde B., Wenning D., Klempnauer K.-H. Recruitment of p300 by C/EBPβ triggers phosphorylation of p300 and modulates coactivator activity. EMBO J. 2003;22:882–892. doi: 10.1093/emboj/cdg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arias J., Alberts A.S., Brindle P., Claret F.X., Smeal T., Karin M., Feramisco J., Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 75.Mink S., Haenig B., Klempnauer K.H. Interaction and functional collaboration of p300 and C/EBPbeta. Mol. Cell. Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Picaud S., Fedorov O., Thanasopoulou A., Leonards K., Jones K., Meier J., Olzscha H., Monteiro O., Martin S., Philpott M., et al. Generation of a selective small molecule inhibitor of the CBP/p300 bromodomain for leukemia therapy. Cancer Res. 2015;75:5106–5119. doi: 10.1158/0008-5472.CAN-15-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ni J., Shen Y., Wang Z., Shao D., Liu J., Kong Y., Fu L., Zhou L., Xue H., Huang Y., et al. P300-dependent STAT3 acetylation is necessary for angiotensin II-induced pro-fibrotic responses in renal tubular epithelial cells. Acta Pharmacol. Sin. 2014;35:1157–1166. doi: 10.1038/aps.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]