Abstract
Background
Human obesity is due to a complex interaction among environmental, behavioral, developmental and genetic factors, including the interaction of leptin (LEP) and leptin receptor (LEPR). Several LEPR mutations and polymorphisms have been described in patients with early onset severe obesity and hyperphagic eating behavior; however, some contradictory findings have also been reported. In the present study we explored the association of six LEPR gene polymorphisms in patients with morbid obesity.
Findings
Twenty eight patients with morbid obesity and 56 non-obese Mexican Mestizo individuals were included. Typing of rs1137100, rs1137101, rs1805134, Ser492Thr, rs1805094 and rs1805096 LEPR polymorphisms was performed by PCR and allele specific hybridization. The LEPR Ser492Thr polymorphism was monomorphic with the presence of only the Ser492Thr-G allele. Allele C and genotype T/C for rs1805134 polymorphism were associated with susceptibility to morbid obesity (p = 0.02 and p = 0.03, respectively). No association was observed with any haplotype. Linkage disequilibrium (LD) showed that five polymorphisms (rs1137100, rs1137101, rs1805134, rs1805094 and rs1805096) were in absolute (D’ = 1) but none in perfect (r2 = 1) LD.
Conclusions
Our results suggest that rs1805134 polymorphism could be involved in the development of morbid obesity, whilst none of the alleles of the LEPR gene, rs1137100, rs1137101, rs1805094 and rs1805096 were associated as risk factors. However, more studies are necessary to confirm or reject this hypothesis.
Keywords: Genetic susceptibility, LEPR, Morbid obesity, Single nucleotide polymorphisms
Findings
Human obesity is due to a complex interaction among environmental, behavioral, developmental and genetic factors, the latter contributing to 40–70 % of the obese phenotype [1–3]. Leptin (LEP) is a hormone specifically produced by adipocytes, and its serum concentration is proportional to body fat mass which, in turn, has its amount regulated by the hypothalamic effects of LEP. Intravenous administration of LEP reduces appetite; while its deficiency increases food intake [4]. Its action occurs through the leptin receptor (LEPR), which is encoded by the LEPR gene. LEPR is a single-transmembrane-domain receptor of the cytokine-receptor family with widespread tissue distribution and several alternatively spliced isoforms [5].
Several LEPR mutations have been described in patients with early-onset of severe obesity and hyperphagic eating behavior [6, 7]. In contrast, a protective influence of two polymorphisms (rs1137100 and rs1137101) to higher blood pressure levels in men has been identified, increasing the protection when the carriers have the arginine allele in the two single nucleotide polymorphisms (SNPs) [8]. Thereby, several SNPs have been studied, and their replication in detail across different populations, for their potential association with obesity and its complication, has been stated. However, some contradictory findings have also been reported, adding the fact that there are scarce studies in non-Caucasian populations; therefore, the lack of data on this subject emphasizes the need for studies among and across different ethnic groups [9, 10]. In this work we explored the association of several LEPR gene polymorphisms with morbid obesity compared with non-obese Mexican Mestizo adults.
Blood samples were obtained from 28 patients with morbid obesity (mean age 39.6 ± 6.6 years, mean body mass index [BMI] 42.7 ± 6.5 Kg/m2) and 56 non-fat (mean age 32.7 ± 14.4, mean BMI 21.5 ± 1.5 Kg/m2) healthy unrelated volunteers. The sex of patients and controls was 89.7 and 93.3 % females, respectively. All participants were unrelated, with no consanguinity at all, and none couple was included. In addition to obesity, two patients presented hypothyroidism, one patient presented type 2 diabetes mellitus and arterial hypertension and one patient presented fat liver. This work complies with the current health laws of Mexico, and was approved by the Ethics and Research Committees of the Hospital General “Dr. Manuel Gea Gonzalez” with the reference number 04-92-2009. All participants were informed about the objectives of the study and were included only after providing written informed consent.
DNA was obtained from 10 ml of EDTA-peripheral blood using proteinase K and phenol/chloroform extraction [11]. The LEPR polymorphisms rs1137100 (Lys109Arg), rs1137101 (Gln223Arg), rs1805134 (Ser343Ser), Ser492Thr and rs1805096 (Pro1019Pro) were detected by PCR-restriction fragment length polymorphism (RFLP) technique, using primers described by Gotoda et al. [12] and Matsuoka et al. [13]. For allele determination of rs8179183 (Lys656Asn) SNP, a dot–blot format and the chemiluminescence method was used, employing the specific probes Lys656Asn-G: 5′-CTATGAAAAAGGAGAAAAATG-3' and Lys656Asn-C: 5'-CTATGAAAAACGAGAAAAATG-3′ ddUTP-Digoxigenin labelled [14, 15].
Allele frequencies (AF) and genotype frequencies (GF) were calculated by direct counting and were compared between patients and controls of each group. Chi-square analysis with Yate’s correction, considering p ≤ 0.05 as the minimum level of significance, was performed; exact Fisher test was used when appropriate. Relative risk was calculated as an odds ratio (OR). Ninety-five percent confidence intervals (95 % CI) were obtained by using Cornfield’s approximation. Haplotypes and linkage disequilibrium (LD) blocks were determined by confidence interval method using Haploview 4.2 software [16].
Table 1 summarizes the significant associations found between alleles, genotypes and haplotypes observed in patients with morbid obesity and in controls. Polymorphism Ser492Thr was monomorphic with the only presence of Ser492Thr-G allele. Allele C and genotype T/C for rs1805134 SNP were associated with susceptibility to morbid obesity (p = 0.02 and p = 0.03, respectively). Regarding the haplotype frequency, no association was observed; however, when shorter combinations were analyzed, haplotype rs1137101G-rs1805134C becomes associated with susceptibility (p = 0.036; OR [95 % CI] 3.4 [1.02–11.14]). Finally, according to LD plots generated in Haploview 4.2 (Fig. 1), the LEPR gene showed that five SNPs (rs1137100, rs1137101, rs1805134, rs1805094 and rs1805096) were in absolute but none in perfect LD.
Table 1.
Alleles, genotypes and haplotypes frequencies of LEPR SNPs in a Co-dominant model
| Patients (n = 28) | Controls (n = 56) | p valuea | OR (95 % CI)b | |
|---|---|---|---|---|
| Alleles | ||||
| rs1137100-C (Lys109Arg) | 0.45 | 0.43 | 0.99 | 1.08 (0.51–2.30) |
| rs1137100-T | 0.55 | 0.57 | 0.99 | 0.92 (0.43–1.96) |
| rs1137101-G (Gln223Arg) | 0.43 | 0.39 | 0.79 | 1.18 (0.58–2.39) |
| rs1137101-A | 0.57 | 0.61 | 0.79 | 0.85 (0.41–1.72) |
| rs1805134 - C (Ser343Ser) c | 0.17 | 0.06 | 0.02 | 3.41 (1.11–10.49) |
| rs1805134-T | 0.83 | 0.94 | 0.02 | 0.29 (0.09–0.90) |
| Ser492Thr-G | 1 | 1 | – | – |
| Ser492Thr-C | 0 | 0 | – | – |
| rs1805094-G (Lys656Asn) | 0.22 | 0.12 | 0.13 | 2.02 (0.89–4.59) |
| rs1805094-C | 0.78 | 0.88 | 0.13 | 0.49 (0.22–1.13) |
| rs1805096-G (Pro1019Pro) | 0.40 | 0.40 | 0.93 | 0.97 (0.48–1.95) |
| rs1805096-A | 0.60 | 0.60 | 0.93 | 1.03 (0.51–2.07) |
| Genotypes | ||||
| rs1137100 C/C | 0.05 | 0.10 | 0.87 | 0.66 (0.07–6.01) |
| rs1137100 C/T | 0.79 | 0.65 | 0.42 | 1.85 (0.19–0.59) |
| rs1137100 T/T | 0.16 | 0.25 | 0.65 | 0.64 (0.16–2.57) |
| rs1137101 A/A | 0.32 | 0.30 | 0.88 | 1.09 (0.38–3.16) |
| rs1137101 A/G | 0.50 | 0.61 | 0.54 | 0.65 (0.24–1.76) |
| rs1137101 G/G | 0.18 | 0.09 | 0.45 | 2.28 (0.55–9.43) |
| rs1805134 T/T d | 0.65 | 0.89 | 0.03 | 0.24 (0.07–0.83) |
| rs1805134 T/C d | 0.35 | 0.11 | 0.03 | 4.01 (1.20–13.41) |
| Ser492Thr G/G | 1.00 | 1.00 | – | – |
| rs1805094 C/C | 0.59 | 0.77 | 0.13 | 0.43 (0.17–1.13) |
| rs1805094 C/G | 0.38 | 0.22 | 0.17 | 2.19 (0.83–5.77) |
| rs1805094 G/G | 0.03 | 0.01 | 0.82 | 2.09 (0.13–34.61) |
| rs1805096 A/A | 0.33 | 0.33 | 0.84 | 1.05 (0.37–2.92) |
| rs1805096 A/G | 0.54 | 0.54 | 0.82 | 1.01 (0.38–2.66) |
| rs1805096 G/G | 0.13 | 0.13 | 0.80 | 0.99 (0.23–4.20) |
| Haplotypese | ||||
| CGTGCA | 0.266 | 0.271 | 0.94 | 0.97 (0.47–2.01) |
| TATGCG | 0.173 | 0.223 | 0.47 | 0.74 (0.32–1.69) |
| TATGCA | 0.108 | 0.106 | 0.98 | 1.01 (0.36–2.88) |
| CATGCG | 0.078 | 0.117 | 0.44 | 0.64 (0.20–2.00) |
| TATGGA | 0.084 | 0.088 | 0.92 | 0.94 (0.30–3.01) |
| TGCGCA | 0.073 | 0.058 | 0.70 | 1.28 (0.36–4.62) |
| CATGCA | 0.037 | 0.041 | 0.91 | 0.91 (0.17–4.92) |
| TGTGCG | 0.037 | 0.029 | 0.79 | 1.27 (0.22–7.53) |
| TATGGG | 0.047 | 0.024 | 0.39 | 2.12 (0.37–12.03) |
| TACGGG | 0.032 | 0.005 | 0.13 | 7.97 (0.34–185.4) |
| TGTGCA | 0.005 | 0.016 | 0.56 | 0.34 (0.01–15.99) |
* p value with Yates correction; bOdds ratio (95 % confidence interval); cStatistical power = 0.632; dStatistical power = 0.679; e LEPR haplotypes: rs1137100-rs1137101-rs1805134-Ser492Thr-rs1805094-rs1805096. Characters in bold and italics indicate statistically significant values
Fig. 1.
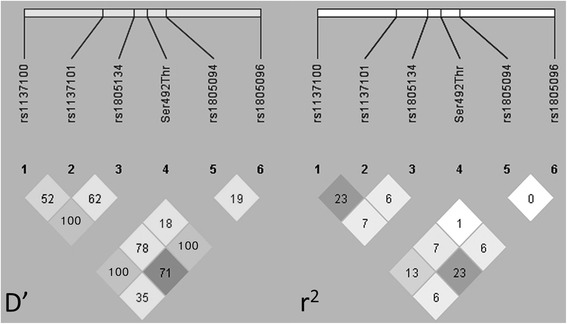
Linkage disequilibrium (LD) in LEPR polymorphisms between obese and control groups. The pairwise LD plot was created by Haploview 4.2. Within each diamond the pairwise standardized coefficient of LD (D’) or the correlation coefficient (r2) is presented in percentage. Standard color coding was used for the Haploview LD plots with the confidence bounds color scheme, for the D’ plot, numbers are representative LD and the logarithm of the odds are in color: white = strong evidence of recombination; light grey = uninformative; dark grey = strong evidence of LD; for r2 LD plots white (r2 = 0), shades of grey (0 < r2 < 1), black (r2 = 1). A marker pair is said to show moderate or usable LD if |D’| is between 0.33 and 0.5, and strong LD if |D’| is 0.5 or above (i.e. at least half the maximum value)
There are data pointing to the fact that the Hispanic population exhibits predisposition towards obesity ([17, 18], http://www.who.int/topics/obesity/en/). Therefore, it is important to study the genetic mechanisms that predispose individuals to this pathology. In the present study, of the six SNPs analyzed, only allele C and genotype T/C for rs1805134 (Ser343Ser) SNP was associated with morbid obesity. Interestingly, this polymorphism has been scanty studied in obesity; when association between serum lipids and LEP/LEPR gene polymorphisms in obese Japanese children was studied, the rs1805134 SNPs showed a significant relationship with serum lipid profile, since lower triglyceride levels were obtained in rs1805134 C/C homozygotes [19]. In another study performed in Spanish adults with obesity, no significant case–control differences were found in allele/genotype frequencies for rs1137100, rs1137101, rs1805134 and rs8179183 SNPs [20]. Since rs1805134 SNP addresses a synonymous amino acid change and, according to our results, it was in absolute LD with both flanking SNPs (rs1137100 and rs1805096) in the LEPR gene, it suggests that SNPs have not been affected by recombination events, which is in concordance with the literature. The present observations allow us to speculate about the presence of others polymorphisms, probably located in introns, close to rs1805134 SNP in absolute and perfect LD, which might regulate the expression of exon 9 of LEPR gene, as it has been described for other Eukaryotes genes [21].
Regarding rs1137100, rs1137101 and rs8179183 SNPs, our data are similar with those reported in others Mexican populations; Guizar-Mendoza et al. [22] assessed rs1137101 and rs1805096 LEPR polymorphisms in Mexican adolescents from Guanajuato state, finding no differences in the genotype frequencies of these SNPs between obese and non-obese participants. Furthermore, another study in Mexican children and adolescents from Colima state, analyzed rs1137100, rs1137101 and rs8179183 SNPs, and no statistically significant association with obesity was found in any of the alleles [18].
The main limitation of the present study was the small sample size; nevertheless we were able to demonstrate statistically significant differences with an adequate power between case and control groups. Rosmond [10] has made harsh criticisms on the association with risk factors of case–control genetic studies, highlighting that the current literature linking central obesity to genetic variants has many reports of associations that either cannot be reproduced or corroborated. The present study, as opposed to Rosmond’s criticism, does replicate the findings reported by other researchers in the same ethnic group, supporting the consistency of allele and genotype frequency for some LEPR polymorphisms in a specific group of participants; therefore, we propose that the rs1805134 SNP is interesting and might be involved in morbid obesity; however, more studies are necessary to confirm or reject this hypothesis.
Acknowledgments
The authors would like to thank Rocio Jimenez-Lucio and David Sierra-Barrera for technical support.
Funding
No financial support was received during the present study.
Abbreviations
- LEP
leptin
- LEPR
leptin receptor
- SNP
single nucleotide polymorphism
- BMI
body mass index
- DNA
deoxyribonucleic acid
- EDTA
ethylenediaminetetraacetic acid
- PCR
polymerase chain reaction
- RFLP
restriction fragment length polymorphism
- AF
allele frequency
- GF
genotype frequency
- LD
linkage disequilibrium
- OR
odds ratio
- CI
confidence interval
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MER-R and JLB-H selected the patients and collected the samples and obtained the written informed consent from each participant before their recruitment. BZ-V, MR-V and AO-D performed the PCR and dot-blot assays. MER-R, JLB-H and AO-D formulated the idea and obtained the authorizations by the Institutional Committees for Research and Ethics in Research. PM, MR-V and AO-D improved the analysis of data and performed critical comments. All authors participated during the discussion and writing of the manuscript and approved its final version.
Contributor Information
Martin Edgardo Rojano-Rodriguez, Email: drrojano@icloud.com.
Jose Luis Beristain-Hernandez, Email: jlberistain@yahoo.com.
Beatriz Zavaleta-Villa, Email: bezavi@yahoo.com.mx.
Pablo Maravilla, Email: maravillap@yahoo.com.
Mirza Romero-Valdovinos, Email: mirzagrv@yahoo.com.
Angelica Olivo-Diaz, Phone: +5255-5528-4228, Email: aolivod@yahoo.com.
References
- 1.Guh DP, Zhang W, Bansback N, Amarsi Z, Birmingham CL, Anis AH. The incidence of co-morbidities related to obesity and overweight: a systematic review and meta-analysis. BMC Public Health. 2009;9:88. doi: 10.1186/1471-2458-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malis C, Rasmussen EL, Poulsen P, Petersen I, Christensen K, Beck-Nielsen H, et al. Total and regional fat distribution is strongly influenced by genetic factors in young and elderly twins. Obes Res. 2005;13:2139–45. doi: 10.1038/oby.2005.265. [DOI] [PubMed] [Google Scholar]
- 3.Marti A, Martínez-González MA, Martínez JA. Interaction between genes and lifestyle factors on obesity. Proc Nutr Soc. 2008;67:1–8. doi: 10.1017/S002966510800596X. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–32. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 5.Stefan N, Vozarova B, Del Parigi A, Ossowski V, Thompson DB, Hanson RL, et al. The Gln223Arg polymorphism of the leptin receptor in Pima Indians: influence on energy expenditure, physical activity and lipid metabolism. Int J Obes Relat Metab Disord. 2002;26:1629–32. doi: 10.1038/sj.ijo.0802161. [DOI] [PubMed] [Google Scholar]
- 6.Clément K, Vaisse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392:398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 7.Farooqi IS, Wangensteen T, Collins S, Kimber W, Matarese G, Keogh JM, et al. Clinical and molecular genetic spectrum of congenital deficiency of the leptin receptor. N Engl J Med. 2007;356:237–47. doi: 10.1056/NEJMoa063988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosmond R, Chagnon YC, Holm G, Chagnon M, Pérusse L, Lindell K, et al. Hypertension in obesity and the leptin receptor gene locus. J Clin Endocrinol Metab. 2000;85:3126–31. doi: 10.1210/jcem.85.9.6781. [DOI] [PubMed] [Google Scholar]
- 9.Fan SH, Say YH. Leptin and leptin receptor gene polymorphisms and their association with plasma leptin levels and obesity in a multi-ethnic Malaysian suburban population. J Physiol Anthropol. 2014;33:15. doi: 10.1186/1880-6805-33-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosmond R. Association studies of genetic polymorphisms in central obesity: a critical review. Int J Obes Relat Metab Disord. 2003;27:1141–51. doi: 10.1038/sj.ijo.0802397. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fitsch EF, Maniatis T. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; New York 2001.
- 12.Gotoda T, Manning BS, Goldstone AP, Imrie H, Evans AL, Strosberg AD, et al. Leptin receptor gene variation and obesity: lack of association in a white British male population. Hum Mol Genet. 1997;6:869–76. doi: 10.1093/hmg/6.6.869. [DOI] [PubMed] [Google Scholar]
- 13.Matsuoka N, Ogawa Y, Hosoda K, Matsuda J, Masuzaki H, et al. Human leptin receptor gene in obese Japanese subjects: evidence against either obesity-causing mutations or association of sequence variants with obesity. Diabetologia. 1997;40:1204–10. doi: 10.1007/s001250050808. [DOI] [PubMed] [Google Scholar]
- 14.Bignon JD, Fernández-Viña M, Arnaiz-Villena A. Technical handbook of twelfth international histocompatibility workshop. 1. France: HLA et Medicine Paris; 1996. [Google Scholar]
- 15.Olivo-Diaz A, Romero-Valdovinos M, Gudiño-Ramirez A, Reyes-Gordillo J, Jimenez-Gonzalez DE, Ramirez-Miranda ME, et al. Findings related to IL-8 and IL-10 gene polymorphisms in a Mexican patient population with irritable bowel syndrome infected with Blastocystis. Parasitol Res. 2012;111:487–91. doi: 10.1007/s00436-012-2830-0. [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 17.Wauters M, Mertens I, Rankinen T, Chagnon M, Bouchard C, Van Gaal L. Leptin receptor gene polymorphisms are associated with insulin in obese women with impaired glucose tolerance. J Clin Endocrinol Metab. 2001;86:3227–32. [DOI] [PubMed]
- 18.Angel-Chávez LI, Tene-Pérez CE, Castro E. Leptin receptor gene K656N polymorphism is associated with low body fat levels and elevated high-density cholesterol levels in Mexican children and adolescents. Endocr Res. 2012;37:124–34. doi: 10.3109/07435800.2011.648360. [DOI] [PubMed] [Google Scholar]
- 19.Okada T, Ohzeki T, Nakagawa Y, Sugihara S, Arisaka O, Study Group of Pediatric Obesity and Its related Metabolism Impact of leptin and leptin-receptor gene polymorphisms on serum lipids in Japanese obese children. Acta Paediatr. 2010;99:1213–7. doi: 10.1111/j.1651-2227.2010.01778.x. [DOI] [PubMed] [Google Scholar]
- 20.Marti A, Santos JL, Gratacos M, Moreno-Aliaga MJ, Maiz A, Martinez JA, et al. Association between leptin receptor (LEPR) and brain-derived neurotrophic factor (BDNF) gene variants and obesity: a case–control study. Nutr Neurosci. 2009;12:183–8. doi: 10.1179/147683009X423355. [DOI] [PubMed] [Google Scholar]
- 21.Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, Wellinger RJ, et al. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell. 2011;147:320–31. doi: 10.1016/j.cell.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 22.Guízar-Mendoza JM, Amador-Licona N, Flores-Martínez SE, López-Cardona MG, Ahuatzin-Trémary R, Sánchez-Corona J. Association analysis of the Gln223Arg polymorphism in the human leptin receptor gene, and traits related to obesity in Mexican adolescents. J Hum Hypertens. 2005;19:341–6. doi: 10.1038/sj.jhh.1001824. [DOI] [PubMed] [Google Scholar]


