Abstract
Constructing containers with defined shape and size to load and protect therapeutics such as native RNA, DNA, or peptides, for controlled release in the human body has long been a dream. Here we report the fabrication of 3D RNA prisms for loading and protection of small molecules, proteins, small RNA molecules, and their subsequent release. RNA molecules loaded inside the prism container were protected from RNAse degradation for cell delivery. Utilizing planar geometry of the 3WJ motif from the pRNA of bacteriophage phi29 DNA packaging motor, the 10-nm prismoidal container was constructed and then characterized by gel electrophoresis, DLS, AFM, and Cryo-EM. To model small molecule drug encapsulation, malachite green dye was immobilized inside the nanoprism by binding to its RNA aptamer. Upon incubation with RNase, the half-life of the malachite green dye increased compared to unprotected constructs. This study demonstrates the concept of RNA nanoconstructs serving as containers for loading and protection of therapeutics in nanomedicine, drug delivery, and controlled release.
Keywords: RNA nanocontainer, pRNA 3WJ motif, RNA Drug Encapsulation, Nanotechnology, Nanobiotechnology
INTRODUCTION
In RNA Nanotechnology, canonical Watson-Crick pairs combined with non-canonical base pair interactions create a large variety of RNA structural building blocks or motifs such as multi-way junctions[1–5], kissing loops[6,6,6–11], and sticky ends[12,13,13] have been utilized to create two-dimensional (2D) RNA structures[14]. Combining diverse RNA connectivity into one nanostructure imparts complexity and allows for the addition of a third dimension in nanoparticle design. As a result, three-dimensional (3D) RNA nanoarchitectures have been explored[15–18]. 3D nanostructures are of particular interest as they hold tremendous potential for the encapsulation of small molecule drugs and therapeutic RNAs. Importantly, encapsulation may hold the distinct ability for controlled release of therapeutic modalities. Previously reported 3D DNA cages such as tetrahedrons, cubes, icosahedrons, and triangular prisms have been used as nano-containers to hold proteins[19,20], gold nanoparticles[21,22], as well as organic molecules[23]. Although construction of 3D RNA nanocages has been previously reported, their potential to encapsulate small reporter molecules, drugs, and proteins is yet to be realized.
RNA aptamers are important candidates for therapeutic and diagnostic applications as they display high affinity and specificity for their target molecules. Binding of malachite green (MG), a triphenylmethane dye, to its respective RNA aptamer, the malachite green aptamer (MGA), causes a drastic increase in MG fluorescence[24,25]. This fluorophore/RNA complex has been widely utilized as a marker in the construction of functional RNA nanoparticles[12,16,26–29]. However, the application of MGA in vivo is limited due to RNA specific exo- and endonucleases inside cells. To prevent this, ribose ring modification at 2’-C/U in RNA is a common strategy to enhance RNA’s nuclease resistance[30]. However, due to either structural hindrance or redistribution of the nearest neighbor landscape, 2’ ribose sugar or other chemical modifications can hamper the activities of aptamers selected with native RNA libraries. For example, 2'-fluorine (2’-F) modification to the MGA eliminates binding of the fluorophore, resulting in no fluorescence increase[26]. Protection of RNA aptamers would be beneficial as their lifetimes could be significantly increased, thereby increasing their utility in vivo. One possibility to extend their lives is encapsulation inside nuclease resistant nanocages.
One advancement utilized for constructing RNA architectures was the finding of an unusually thermostable three-way junction (3WJ) derived from the pRNA of bacteriophage phi29 DNA packaging motor[3]. The 3WJ can be used as a vehicle to integrate multiple functional RNA modules into one nanoparticle while retaining folding and function in vivo[3,8]. Utilizing the crystal structure of the 3WJ[31], various RNA nanoparticles including triangles, squares, and pentagons were constructed[32]. Upon systemic injection in mice, the RNA nanoparticles bind to xenograft and metastatic cells specifically and strongly with little or no accumulation in the liver or other healthy organs[33].
Herein, we describe a strategy to fabricate 3D RNA nanocages resembling triangular prisms based on pRNA-3WJ. Importantly, this 3D nanoparticle was used to demonstrate encapsulation of a small fluorescent dye, MG, while increasing its fluorescence half-life in an RNA unfriendly environment. The modularity of the triangular nanoprism design also allowed single strand oligonucleotide-drug conjugates to be positioned precisely inside the inner cavity of the RNA complex. The functional properties of the RNA nanocage to simultaneously carry 5 folate ligands for cell targeting as well as 6 DNA CpG motifs for cell immune response were further evaluated in vitro. This work paves the way towards the application of RNA based nanocaging as a delivery system for small reporter or therapeutic molecules.
RESULTS AND DISCUSSION
Computer-aided rational design and fabrication of 3D RNA nanoprism from planar pRNA 3WJ geometry
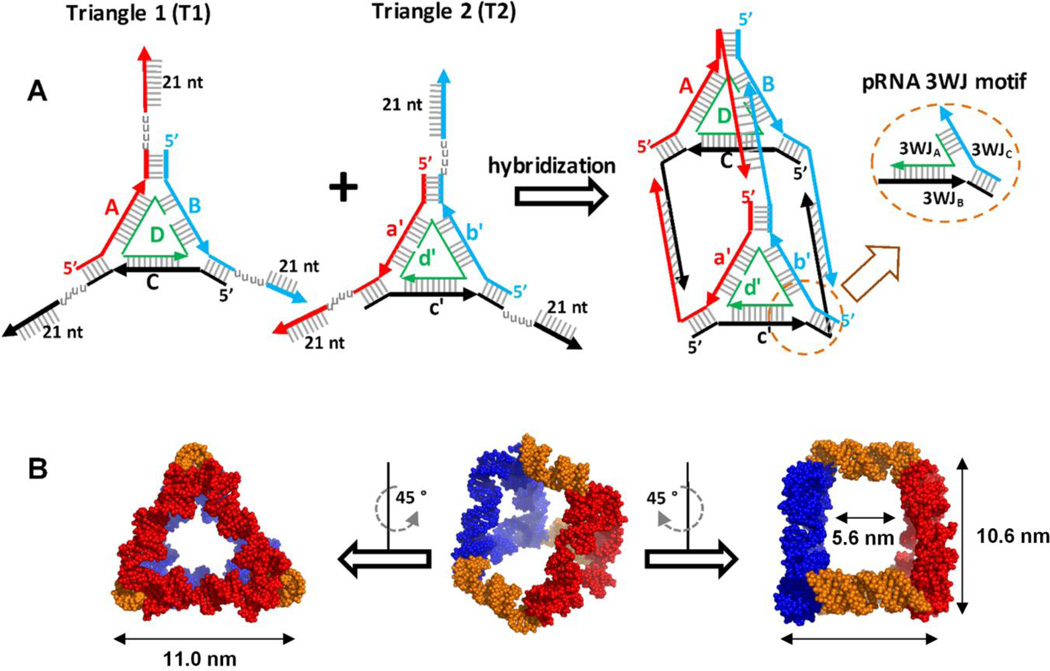
The triangular nanoprism was designed employing the structural information of pRNA 3WJ following a bottom-up approach[1]. As such, two helices of three adjacently arranged 3WJs were connected with duplex RNA to form flat triangular nanoparticles, a nanoparticle construction method devised previously[12]. To extend 2D triangle particles into a 3D RNA complex, two triangular nanoparticles (T1 and T2) containing 21 nucleotide (nt) single stranded (ss) complementary linker (L) regions on each vertex were tethered (Figure 1A, S1). Each ssL region was designed to interact specifically with its compliment strands L1-L1’, L2-L2’, L3-L3’ forming RNA-RNA duplex of 21 bp. To provide structural flexibility and allow the linkers to bend to a 90° angle necessary for triangular prism complex, poly uracils (poly-Us) were embedded between the ssL and triangle vertices. This resulted in the formation of a "face-to-face" dimer with a total of 8 RNA strands. The overall geometry was manually computed using SwissPDB Viewer software[34], the structure resembling triangular prism geometry with the pRNA-3WJ motif at the corners. The 3D RNA model displays a size of ~11 nm, measured from one vertex to an adjacent vertex, and a size of ~10 nm measured from one vertex to its nearest edge, as measured by the modeling software (Figure 1B). The largest diameter of the inner cavity of the prism was measured to be ~7 nm while the height was ~ 6 nm. Based on these dimensions, we assume that the central cavity of the prism could accommodate a spherical object of approximately 6 nm in diameter.
Figure 1. Triangular Nanoprism design and 3D model structure.
A. Fabrication of the triangle nanoprism from two planar equilateral RNA triangles. The triangle prism nanostructure forms by hybridization of the flexible 21 nt ssRNA “arms” of each triangles. The overall prismoidal construct contains the preserved geometry of the pRNA 3WJ structural building block. B. Computer model structure of the triangular nanoprism demonstrating its average dimension in 3D space.
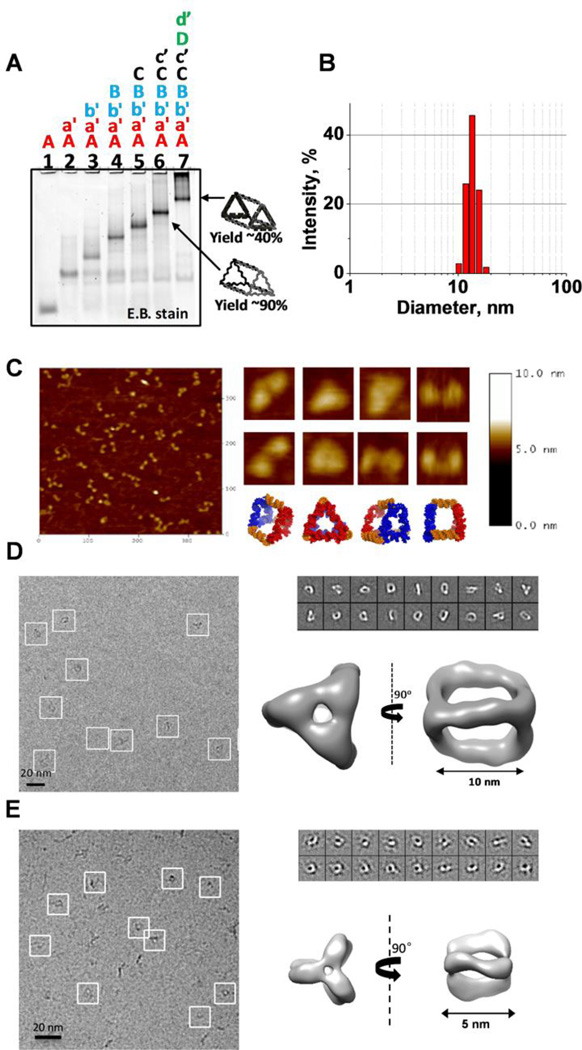
Assembly efficiency of the RNA strands into the desired conformation was first assayed by 6% native polyacrylamide gel electrophoresis (PAGE). Upon addition of each of eight component strands, a step-wise decrease in electrophoretic migration can be clearly seen, indicating association of each complimentary ssRNA oligomer into the complex RNA structure (Figure 2A). The distinct band in lane 7 indicates the formation of a stable RNA complex containing all 8 RNA strands. Integration of the intensity of gel bands results in an estimated assembly yield of 43.0 ± 3.1%. A large fraction of RNA aggregates are localized in the top of lane 7. Presumably, this is due to non-specific interactions between the 21nt RNA single stranded sticky ends. Interestingly, the RNA complex with no D and d’ stands has much higher assembly efficiency with estimated yield of 90% (Figure 2A lane 6).
Figure 2. RNA Triangular nanoprism self-assembly and characterization.
A. Assembly efficiency of the RNA triangle nanoprism complex evaluated by 6% native polyacrylamide gel electrophoresis (PAGE). Step-wise association of the 8 RNA strands (0.5 µM each) into the final complex was conducted in 1 × TMS buffer using one-pot assembly. B. Typical dynamic light scattering (DLS) data demonstrating average hydrodynamic diameter of the prism. C. Atomic force microscopy (AFM) images of the triangle nanoprism (5 nM) taken in air. D. CryoEM image of the large (10 nm) triangular RNA prism. Each white box indicates an individual RNA complex. Class averages of RNA nanoprisms as observed by cryo-EM. Reconstructed three-dimensional nanoprism at 2.5 nm resolution. E. CryoEM image of the small (5 nm) triangular RNA prism. Each white box indicates an individual RNA complex. Class averages of RNA nanoprisms as observed by cryo-EM. Reconstructed three-dimensional nanoprism at 2.2 nm resolution.
DLS was utilized to measure the apparent dimensions of the purified RNA prism. As measured by DLS, the hydrodynamic diameter of the RNA complex is 11.5 ± 1.6 nm, consistent with the 3D model structure of the nanoprism (Figure 2B). AFM imaging of the pure RNA complex has revealed that the particles do in fact display prismoidal-like architecture (Figure 2C). AFM is 2D observation of the 3D RNA prism and cannot be used to conclusively indicate the formation of the triangular prism geometry, nevertheless, the particles do resemble the designed triangular prism in two dimensional projections.
Cryo-EM imaging studies revealed triangular prism geometry
To address the concern whether the folding of 8 RNA stands resulted in 3D geometry, the purified RNA complex was visualized by single-particle cryo-electron microscopy (cryo-EM) (Figure 2D, 2E). Cryo-electron microscopy is a useful method to show the true 3D nature of the RNA nanoprisms, as AFM analysis causes distortion of 3D nanoparticles imaged in 2D[35]. The 3D reconstruction of the RNA nanoparticles was achieved implementing single-particle reconstruction using the EMAN2 approach as described in the methods section. Reconstruction of the 3D structure of the RNA complex were determined at a resolution of 2.5 nm (10 nm RNA prism) and 2.2 nm (5nm RNA prism), and demonstrated that the RNA particles have an average sizes of 10 nm and 5nm, respectively, in agreement with DLS and AFM results. More importantly, the overall geometry is similar to the computer modeled 3D RNA prism. It is important to note that the reconstruction of the RNA prism by Cryo-EM clearly demonstrates the presence of an inner cavity. Collectively, the data from native PAGE, DLS, AFM, and Cryo-EM clearly indicate the formation of closed and compact RNA nanoprisms.
Design and construction of RNA aptamer encapsulated nanoparticle
The MGA is a commonly used tool in biochemistry for structure-function verification [36–38]. This is attributed to its unique properties as free MG dye in solution displays little to no fluorescent signal, yet when bound to MGA the photoemission increases more than 2000-fold[39,40]. However, fluorescence can only occur if the RNA MGA folds into its authentic 3D conformation. Previous attempts to achieve fluorescence using 2’-F modified RNA bases resulted in little to no fluorescence [26]. Therefore, for RNA MGA to remain a useful tool within a cellular environment, it must be placed into a container that will protect the RNA from a degrading environment. This is a common problem among many small RNAs and small molecule drugs with therapeutic utility. Using 2’-F modified RNA to construct the prism’s frame (2’-fluoro uridine (2’-F-U) and 2’-fluoro cytosine (2’-F-C)), which are known to display resistance to a degrading environment [41], we hypothesize that the RNA MGA will be protected when positioned inside the 2’-F modified nanocage.
As a proof-of-principle, we employed the MGA RNA sequence by embedding it within the "D" and "d’" strands at their 3' ends. The schematic encapsulation design is summarized in Figure 3A. Throughout this paper, we refer to the stable RNA complex formed by six 2’-F modified RNA strands without D or d' strands as a "frame". It was interesting to find that this complex is highly stable with apparent melting temperature (TM) of 74 °C, with the assembly efficiency being Mg2+ dependent (Figure S2).
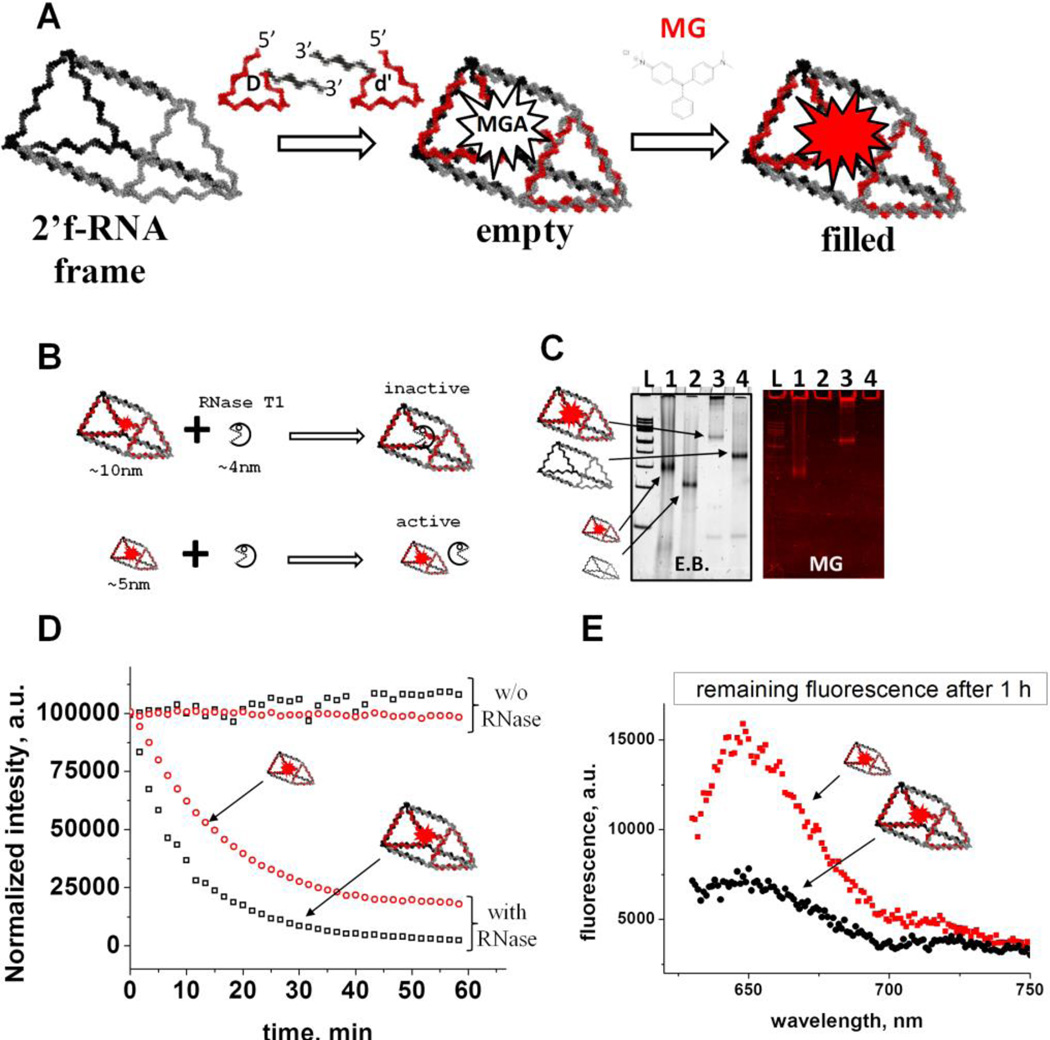
Figure 3. Encapsulation of the RNA MGA and fluorescence assay.
A. Schematic design approach for the RNA MGA encapsulation. The RNA MGA sequence is embedded within the 3’-end of D and d’ strands. Upon hybridization MGA folds into native conformation. B. Schematic representation of the RNase T1 protection experiment demonstrating the relative dimensions of RNase T1 and nanoprisms. C. Assembly efficiency and relative migration of the triangle prisms on native 6% PAGE. The smaller RNA prism migrates much faster compared to the regular prism and emits fluorescence signal indicating proper folding of the functional RNA complex. Lane “L” is 100 bp DNA ladder (Thermo Scientific). D. Time-dependent fluorescence emission profile of the 0.1 µM RNA MGA aptamer in the presence of 500U of RNase T1 and its absence (control). E. The remaining fluorescence signal of the triangle prisms after one hour of RNase T1 treatment.
Using SwissPDB Viewer, we first modeled the complex structure of the RNA nanocage with encapsulated MGA by positioning the 3D structure of the MGA inside the inner cavity formed by the Nanoprism. This allowed us to estimate the distance constraint in base pairs required to mount the RNA MGA (extracted from PDB ID: 1F1T [24]) into the inner cavity of the prism. The complexation of the RNA MGA within the nanocage was evaluated on native PAGE (Figure S3). The apparent gel shift of the band localized in lane 4 (2’-F-RNA frame with MGA) compared to lane 1 (2’-F-RNA frame only) shows complexation of the MGA sequences with the frame structure detected by total RNA stain with Ethidium Bromide (EB). Staining of the same gel in the presence of MG dye resulted in strong fluorescence emission from the distinct band on lane 4, indicating correct folding of the RNA MGA (Figure S3). We performed additional control experiments based on migration properties of the closed (compact) and open (relaxed) 3D RNA complexes as it is generally accepted that more compact RNA structures migrate faster in comparison to relaxed RNA conformations. To fabricate the open 3D prism conformation, the interaction of one of the complementary sticky ends linking two RNA triangle together were intentionally disrupted though sequence design (Table S1). Comparing migration of the open RNA prism to the closed version revealed slight differences in migration distance (Figure S3: lanes 4 and 5; Figure S4). This indicates that the prism is closed and that the MGA was located, presumably, within the inner cavity of the nanocage. Data obtained from the mobility shift assay alone do not provide a concrete conclusion whether the RNA MGA formed inside of the closed cage; additional data supporting our assumption comes from RNase T1 cleavage experiments as described below.
RNase T1 protection assay confirmed the encapsulation of the MG RNA aptamer by the nanocage
Encapsulation of an RNA module inside the negatively charged RNA nanoprism is challenging due to the repulsion forces caused by the negatively charged phosphodiester backbone of both the inner cavity of the RNA cage and the RNA MGA. Development of a method to overcome this challenge would be exceptionally beneficial as the activity of functional RNA modules could be retained by protecting them from enzymatic degradation in vivo. To further confirm that the RNA MGA was localized inside the cavity of the nanocage, we treated the RNA complex with endonuclease RNase T1, which specifically cleaves unpaired guanosine (Gs) nucleotide[42]. While the single stranded Gs are absent from the 2’-F-RNA prismoidal construct, there are several ss Gs present in the RNA MGA itself that are essential for MG binding. Thus, we hypothesize that RNase T1 will cleave the exposed Gs of the MGA sequence resulting in the loss of fluorescence. However, if the cage were to restrict RNase T1 access to the MG aptamer within the nanoprism, fluorescence emission would be retained or decay at a slower rate.
According to the modeled 3D structure of the prism, the estimated average dimensions of the inner cavity of the prism could accommodate a spherical object of about 6 nm. The average size of the RNase T1 structure was measured (PDB ID: 1YGW) to be 3.7 nm in diameter [43] (Figure S5) allowing RNase T1 to easily access the inner cavity and quench the fluorescence of the MGA. To address this obstacle, a nanoprism having a smaller cavity size of 3 nm was designed by decreasing the length of the RNA prism helical regions by one half (Supplemental Table 1). The schematic design of the experiment is summarized in Figure 3B. The smaller prism is expected to insulate the MGA from RNase T1 cleavage, thus extending fluorescence activity.
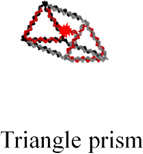
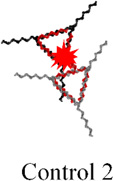
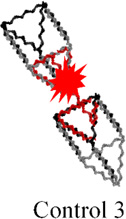
The assembly of the small prism is highly efficient, and RNA MGA is functional according to native PAGE analysis (Figure 3C, lanes 1 and 2). Next, we carried out time-dependent MG fluorescence decay in the presence of RNase T1 to test the activity of the MGA embedded inside the large and small nanoprisms. Figure 3D shows the fluorescence exponential decay profiles of the large (black squares) and small (red circles) nanoprisms. The calculated lifetimes (τ) were found to be 445.5 sec and 781.4 sec for the large and small nanoprisms respectively. The small prism displayed a half-life almost double that of the large prism, suggesting that RNase T1 has limited access to MGA encapsulated inside of the small nanoprism. Complete protection against RNase T1 has not been observed presumably due to the dynamic nature of the interacting linker arms of two triangles. This is also supported by AFM images where significant fractions of prisms are in an open state, which allows RNase T1 to digest the MGA. Notably, fluorescence emission of the small prism was still observed after 1 hour of incubation with RNase T1 while fluorescence in the large prism was negligible (Figure 3E). Additional control experiments were performed using a variety of other RNA control complexes containing RNA MGA. The calculated values of τ are summarized in Table 1. Mean lifetime comparison of the small RNA prism to the controls demonstrates that no control construct has greater τ than the small prism. Thus, the RNase T1 cleavage assay indicates the successful encapsulation of MGA encapsulated in both the small and large nanocages. More importantly, we have shown that by tuning the size of the inner cavity, fluorescence half-life can be significantly increased. This is synonymous with increasing the release time of a drug that can be potentially used an in vivo system.
Table 1.
Fluorescence life times of different RNA constructsa
| Nanoparticle type and structure |
 |
 |
 |
 |
 |
|---|---|---|---|---|---|
| Life time τ, secb |
445.5 ± 50.0 | 352.3± 47.8 | 273.2 ± 39.3 | 386.5 ± 45.3 | 781.4 ± 43.0 |
The analysis of the electrophoretic migration of the Control 2 and Control 3 is shown in Figure S3.
The standard error of the mean obtained from at least three independent experiments.
Encapsulation of oligonucleotide-drug conjugates inside the nanocage
Taking advantage of the programmability of the D and d’ prism strands, we next demonstrated the encapsulation of single stranded oligodeoxynucleotides (ssODN) containing functional moieties such as fluorescence reporters and drugs. To this end, one long ssRNA was designed to connect D and d’ strands through a 22 nt linker. The resulting RNA sequence will be referred as a “core” RNA strand. While the 5’- and 3’-ends of the core RNA strand interact with the triangular edges of the prism, its single stranded middle region is designed to localize inside the nanocage. The portion of the core strand inside the prism was utilized for encapsulation of either ssDNA or ssRNA carrying different functional groups via base pair hybridizations (Figure S6A). Hence, we anticipate that the encapsulation of the functional moieties will prevent them from enzymatic degradation and increase their cellular life-time. Alternatively, this principle can be applied as a safety mechanism. For instance, carrying highly toxic compounds that should be released only upon reaching its target.
Due to the programmable nature of the RNA prism, the size of the nano-container can be adjusted by simply increasing or decreasing the sides of the prism by complete helical turns as shown previously [41,44]. This is advantageous as the capacity of the RNA nano-container can be controlled enabling encapsulation of diverse size biomolecules or inorganic nanoparticles, as well as increase the payload of a single nano-container for increased detection sensitivity and increased efficacy in the treatment of disease. Moreover, from a medical point of view, size alternation of the nano-prism imparts differences in toxicity, cellular binding and internalization, renal clearance, physicochemical features, as well as pharmacokinetics and pharmacodynamics profiles. As such, the prismoidal nano-container provides a unique system to further exploit these factors.
The principle behind chemical drug encapsulation relies on the conjugation of a drug at the 3'/5'-end or at both ends of DNA /RNA oligonucleotides, followed by hybridization of this conjugate inside the cavity of the RNA nanoprism. To demonstrate this in practice, short DNA sequences (22nt) labeled at the 5’-end with different chemicals including folic acid (FA), cyclodextrin, taxol, Cy5 fluorophore, and biotin. To verify the formation of the RNA nanoparticle with the conjugated DNA sequence, prism strands and corresponding DNA oligonucleotides were assembled in one-pot by mixing at equimolar concentrations, followed by native PAGE analysis (Figure S6B). Formation of distinct bands corresponding to the frame structure and frame containing the core RNA sequence are shown in Figure S6, lanes 1 and 2, respectively. The additional lanes correspond to loaded nanocage with the 22nt DNA containing a variety of small molecules. Based on the slower rate and almost identical distance of all five loaded cargoes as compared to the bands in lanes 1 and 2, we can conclude that the conjugated ssDNA were successfully hybridized inside the nanocage structure. Additionally, co-migration of Cy5 and EB signal of Cy5 loaded nanocage indicates correct assembly.
To demonstrate incorporation and function of the DNA-biotin conjugate within the RNA 3D cage, biotin-streptavidin binding was utilized[45](Figure S6C). To separately visualize the biotin-nanoprism and the streptavidin, the DNA-biotin conjugate was tagged with Cy5 and the streptavidin with Cy3. The ensuing gel shift assay demonstrated complexation between the encapsulated biotin and streptavidin as the complex migrates much slower than the biotin-nanoprism or streptavidin alone (Figure S6C, lane 1 vs 2 and 5). Colocalization of EB (RNA), Cy3 (streptavidin), and Cy5 (DNA-biotin) signals in the distinct band in lane 1 reveals the functional properties of the encapsulated molecule (Figure S6C). Although PAGE results cannot directly confirm whether the functionalities were located inside the nanocage, results based on computer modeling and fluorescence functional assays are a strong indication of the encapsulation mechanism. Utilizing the programmability of RNA D and d’ strands as well as modularity of the ssDNA-drug conjugates we were able to demonstrate feasibility of the encapsulation approach, in addition to RNA MGA aptamer encapsulation, for caging of small molecules.
Varying RNA Chemical Modifications Allows Control over Nanoprism Serum Stability and Drug Release
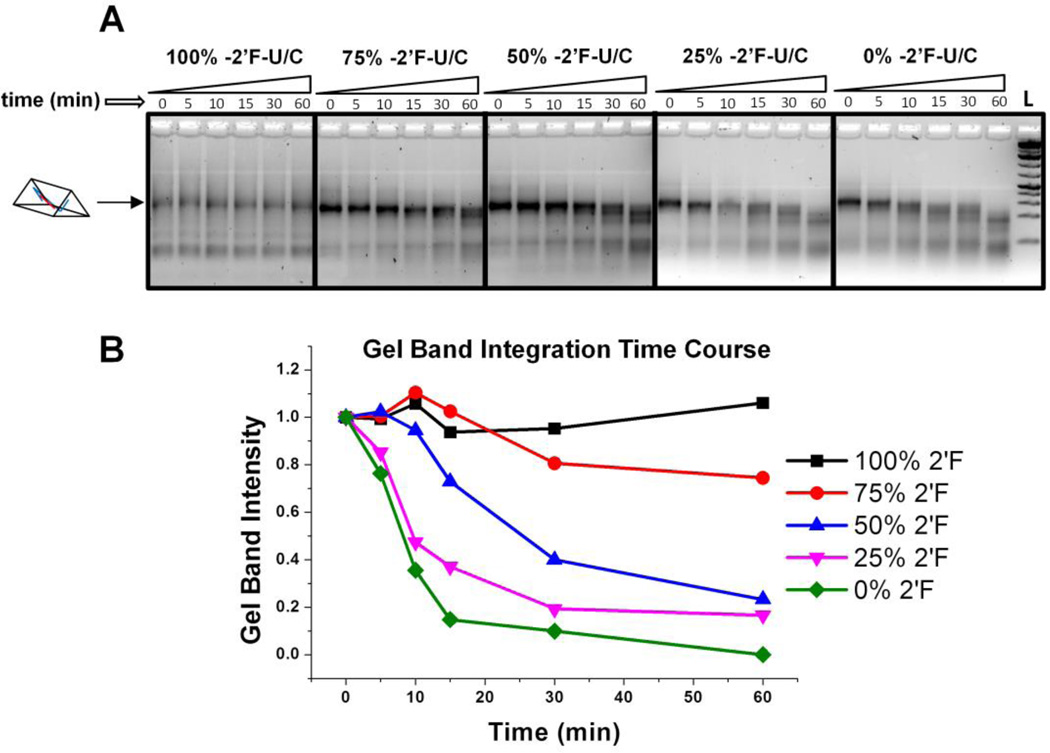
One potential advantage of RNA 3D nanocontainers is the ability to safely deliver and release encapsulated drugs in a fine-tunable fashion. Development of controlled release is extremely challenging yet highly desirable in RNA nanotechnology[1]. To release the encapsulated drug(s), one approach is to use the help of naturally occurring nucleases to break down RNA 3D nanoscaffold. It was our hypothesis that changing the percentage of 2-’F nucleotides in the nanoprism scaffold would result in diverse degradation profiles. To change the amount of 2’-F nucleotides in the nanoprisms, the ratio of 2’-F cytidine phosphate (CTP) and uridine phosphate (UTP) nucleotides to unmodified cytidine phosphate uridine phosphate were adjusted during in vitro transcription reactions to 4:0, 3:1, 2:2, 1:3, and 0:4, respectively. After transcription and purification, five different nanoprisms were assembled with 2’-F-C/U modifications of approximately 100%, 75%, 50%, 25%, and 0%. The degradation profiles of each nanoprism were observed over the course of one hour in 2% fetal bovine serum (FBS) solution at a final concentration of 200 nM and analyzed on 3% agarose gel (Figure 4A). Gel band intensity was integrated using ImageJ software and intensity at each time point was compared to initial gel band intensity for each individual nanoprism (Figure 4B). As shown by the time course plot, decreasing the percentage of 2’-F nucleotides increases the digestion rate of the nanoprism scaffold. This approach shows feasibility of controlled drug release by degradation of the partially 2’-F modified nano-container, which can be fine-tuned to the scale of minutes.
Figure 4. Tuning degradation profile of nanoprism in fetal bovine serum.
A. Gel images of nanoprisms with different percentages of 2’-F modified pyrimidines used during assembly. Nanoprisms of 200 nM concentration were incubated in 2% FBS solution and analyzed by 3% agarose gel. B. Prism relative gel band intensity plotted versus time. The intensity of each band was compared to the zero time point for each nanoprism.
In vitro cell binding studies revealed high binding affinity to cancer cells by RNA nanocage carrying ssDNA-Cy5
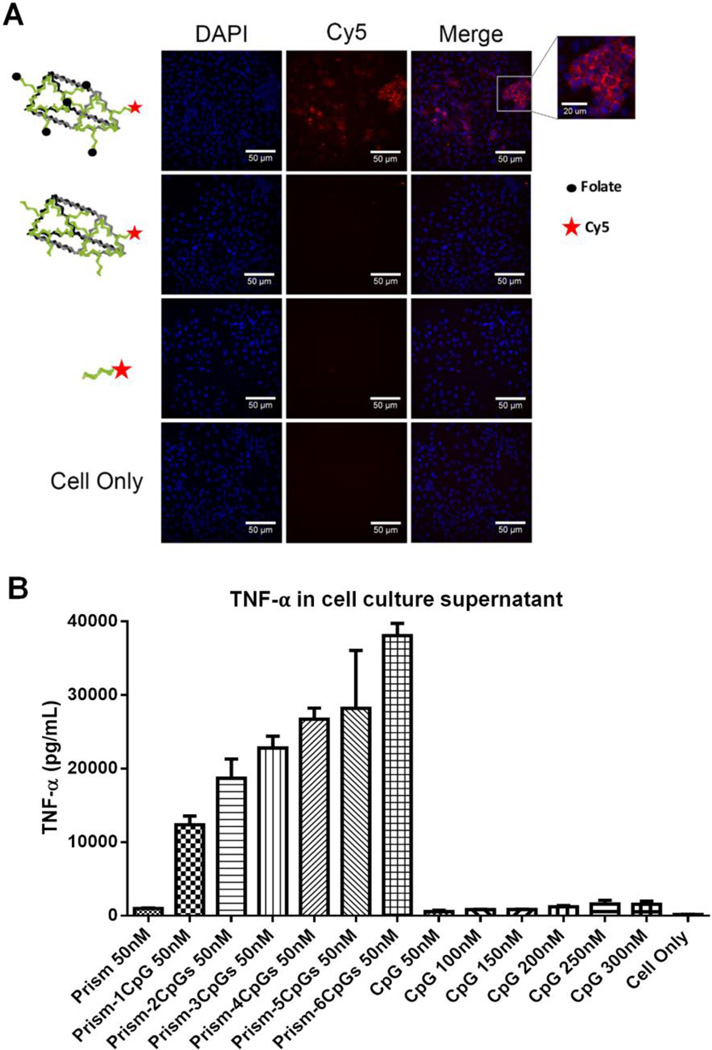
The ability of the RNA nano-container to successfully bind and deliver drugs or reporter molecules into cancer cells would tremendously benefit the advancement of nanomedicine. To investigate whether the RNA nanocage can serve as a container to deliver small molecules, the 2’-F RNA frame structure was decorated with 5 copies of DNA oligonucleotide-folate conjugates and 1 copy of DNA-Cy5, as shown in Figure 5A (Assembly shown in Figure S7). Folate-containing nanoconstructs are known to have a strong affinity to cancer cells of epithelial origin as these types of cancer cells overexpress folate-binding protein receptors [46]. After incubation of the folate functionalized RNA nanoprism with KB cells, strong binding of the RNA complex with the cells was observed (Figure 5A) in comparison to control complex having the same RNA 3D nanostructure but lacking folate molecules. This result demonstrates applicability of the 3D RNA system for detection and delivery of the reporter molecules.
Figure 4. Evaluation of the RNA triangle prism to carry functional moieties.
A. Confocal microscopy images of KB cells incubated with RNA nanoprism decorated with 5 folates as targeting modules. B. TNF-a cytokine induction by the RNA nanoprism harboring different number of CpG -ODN per nanoparticle. Experiments conducted using macrophage-like RAW264.7 cells showing that cytokine secretion increases with the number of CpG payload per nanovehicles. The error bar obtained through three independent experiments.
The 3D RNA triangular nanoprism possess elevated immunostimulatory activity in vitro
CpG oligodeoxynucleotide (ODN) is a short synthetic DNA sequence containing unmethylated C and G nucleotides that has recently gained attention in immunology as it has shown to be useful as a vaccine adjuvant or immunotherapy reagent[37,47,48]. It was of particular importance to further investigate whether the unique 3D conformation of the RNA triangular nanoprism can be used to enhance the immunomodulation effect in vitro. Six CpG oligonucleotides were incorporated onto the 2'-F-RNA frame construct (Figure S8). The immunostimulatory efficacy of the nanoprism was evaluated by measuring the release of cytokines TNF-α after addition to mouse macrophage-like RAW264.7 cells (Figure 5B), as previously described [37]. The results demonstrate that upon increasing the number of CpG induced a stronger immune response, observed by measuring the level of the induction of TNF-α. The results suggest that the cytokine released by the CpG prism complex significantly increases the immunostimulatory activity compared to CpG alone (Figure 5B). Interestingly, the same observation was obtained by previous studies using RNA polygonal nanoconstructs [37], as with more CpG per nanoparticle a higher level of host immune response can be triggered. This data combined with cell binding studies demonstrates that the 3D RNA nanoprism can be readily utilized in cell studies to effectively carry functional moieties. This paves the way to further in vivo applications.
CONCLUSION
The proof-of-concept for a technique to construct 3D RNA nanocontainers to encapsulate small RNA molecules and a method for loading the nanocontainers with model drugs was demonstrated. The assembly of RNA triangular nanoprism utilizes pRNA 3WJ motifs positioned at the prism vertices, imparting stability into the system and allowing for future in vivo delivery. Loading the container with model drugs (MG) was accomplished via diffusion of the model drug into the container with subsequent immobilization by RNA receptors (MGA) pre-encapsulated within the nanoprism. The concept of RNA aptamer encapsulation is a promising platform for exploring new applications in pharmaceutical and materials science. The RNA-directed triangular prism brings together the functionality, biocompatibility, and self-assembly potential with fine control of geometry, positioning, and the responsive characteristic of RNA nanoparticles.
Experimental Section
RNA sequence design and 3D modeling
RNA sequences were optimized using RNA 2D folding software Mfold (www.mfold.bioinfo.rpi.edu) prior to synthesis. Computer models of the RNA prism were assembled from two previously constructed RNA triangles [12]. The vertices of the triangles were assembled by overlapping helical domains of the triangular "arms" to position them at the correct angle and distance using the 'autofit' tool in Swiss-PDB Viewer (www.spdbv.vital-it.ch) and manual adjustment. The detailed procedure for the triangle design can be found in our previous reports [12]. Each triangle vertex contains a 21 nt ssRNA tethered to the 3’-end using a poly-uracil (poly-U) linker. The poly-U is inserted to provide a hinge allowing for the flexibility of the ssRNA when hybridized with its complementary sequence of another triangle. The resulting 21 bp RNA duplexes were chosen to have similar lowest free-energy secondary structures (calculated ΔG =−39.0 ± 2.0 kcal/mol). The sequence of the RNA MGA module was extracted from the PDB databank (www.pdb.org), PDB ID: 1F1T [24]. Detailed 2D structures and sequence of the nanoprisms as well as synthetic oligonucleotides containing conjugates are provided in Supplementary Table 1.
RNA synthesis, purification and assembly
Templates for RNA transcription were prepared by standard protocols described in detail elsewhere[49]. Briefly, PCR was used to prepare transcription templates using PCR reaction kit (Promega Corporation, GoTaq® FlexiDNA Polymerase) for the amplification of DNA primers (IDT). Following purification of DNA templates on spin columns (QIAGEN), the RNA strands or 2’-F-U/C modified RNA strands were transcribed using home-made T7 RNA polymerase. The RNA was precipitated with cold ethanol, then the pellets dried and resuspended in ddH2O. Homogeneity of individual RNA strands is checked by denaturing 10% PAGE. For assembly assays, equimolar RNA strands (0.5 µM each) were mixed in 1× TMS (40 mM TRIS-HCl pH = 8.0, 100 mM NaCl and 10 mM MgCl2) buffer. The RNA mixture was heated to 80 °C for 5 min and slowly cooled to 4 °C at the rate of 2 °C/min in a PCR thermocycler. RNA assembly products were checked on native 6% or 7% PAGE (29:1 acrylamide: bis-acrylamide ratio) in 1 × TBM (89 mM TRIS-Borate pH = 8.0, 5 mM MgCl2) running buffer. All native PAGE experiments were run at 4°C at constant 90 V for 2 hours. Gels were stained in Ethidium Bromide buffer for total RNA strand visualization or in MG binding buffer (20 µM MG dye, 10 mM HEPES pH = 7.4, 100 mM KCl, 5mM MgCl2) for detection of MG aptamer signal as previously reported[37].
Cryo-electron microscopy
2 µl of RNA triangular prism nanoparticle solution (1 mg/ml) was applied onto a glow-discharged 200-mesh R1.2/1.3 Quantifoil grid. The grids were blotted for 3 s and rapidly frozen in liquid ethane using a Vitrobot Mark IV (FEI). Then the girds were transferred to JEM2200FS cryo-electron microscope (JEOL) operated at 200 kV with in-column filter for screening. Micrographs of 10 nm RNA prism were recorded with a direct detection device (DDD) (DE-20 4k×5k camera, Direct Electron, LP) operating in movie mode at a recording rate of 25 raw frames per second at 25,000×microscope magnification (corresponding to a calibrated sampling of 2.51 Å per pixel) and a dose rate of ~20 electrons per second per Å2 with a total exposure time of 3 s. Micrographs of 5 nm RNA prism were recorded with a 4k × 4k CCD (Gatan) at 80,000× microscope magnification (corresponding to a calibrated sampling of 1.36 Å per pixel) and a dose rate of ~20 electrons per second per Å2 with a total exposure time of 3 s. A total 52 images of 10 nm RNA prism and 30 images of 5 nm RNA prism were collected with a defocus range of 2~4 µm.
Single particle image processing and 3D reconstruction
The image processing software package EMAN2 was used for the micrograph evaluation, particle picking, CTF correction, 2-D reference-free class averaging, initial model building and 3-D refinement of the cryo-electron microscopy data. We boxed total 2340 particles for 10 nm RNA prism and 2206 particles for 5 nm RNA prism to generate the 2D class averages for building the initial models. Finally, 1514 particles for the 10 nm RNA prism and 1648 particles for the 5 nm RNA prism were used for final refinement, applying the D3 symmetry. The resolution for the final maps was estimated by the 0.143 criterion of FSC curve without any mask. 25 Å and 22 Å Gauss low-pass filter were applied to the final 3D maps displayed in the Chimera software package.
AFM images
For all samples, specially modified mica surfaces (APS mica) were used. The APS mica was obtained by incubation of freshly cleaved mica in 167nM 1-(3-aminopropyl) silatrane following previously reported protocol [50]. The RNA samples were diluted with 1 × TMS buffer to a final concentration of 3 – 5 nM. Then, 5−10 µL was immediately deposited on APS mica. After 2min incubation on the surface, excess samples were washed with DEPC treated water and dried under a flow of Argon gas. AFM images in air were acquired using MultiMode AFM NanoScope IV system (Veeco/Digital Instruments, Santa Barbara, CA) operating in tapping mode. Two types of AFM probes were used for tapping mode imaging in air: (1) regular tapping Mode Silicon Probes (Olympus from Asylum Research, Santa Barbara, CA) with a spring constant of about 42N/m and a resonant frequency between 300 and 320kHz. (2) Non-contact NSG01 DLC probes (K-Tek Nanotechnology, Wilsonville, OR) with a spring constant of about 5.5N/m and a resonance frequency between 120 and 150kHz.
Dynamic light scattering
The hydrodynamic diameter of the RNA nanoprisms was determined at a concentration of 10 µM in 50µL TMS buffer using a Zetasizer nano-ZS (Malvern Instrument, LTD) at 25°C.
Prism_MG degradation assay
The function of the MGA within the triangular prism nanoparticles was assayed by mixing the RNA complex at a concentration of 0.1 µM with 1µM MG dye in 1× TMS buffer. The solutions were allowed to equilibrate at room temperature for 10 minutes. The fluorescence emission spectrum (recorded from 630 – 750 nm) of the complexes was measured using a fluorospectrophotometer (Horiba Jobin Yvon) with an excitation wavelength positioned at 615 nm. 1 µL of 500 U/µL RNase T1 (Thermo Scientific) was added to 99 µL of the above RNA-MG complex directly in a fluorometer cuvette. Fluorescence at λmax = 650 nm was immediately recorded and monitored every 1 sec for a total time of 3600 sec. The time resolved fluorescence life time measurements were performed by the software integrated within the fluorometer. The obtained data were fit using exponential decay function I(t) = I0 e(−t/τ), where I0 is the initial intensity (at time zero) and τ is the mean life time defined as the time for the intensity to drop by 1/e or to ~37%. All fluorescence decay experiments were conducted in triplicate with indication of ± for mean standard error (SEM).
Supplementary Material
Acknowledgments
This research was supported by NIH grants R01EB019036, and U01CA151648 to PG. Funding to P.G.'s Endowed Chair position is by CM Chen Foundation. PG is a consultant of Oxford Nanopore. His inventions at the University of Kentucky have been licensed to the Matt Holding and RNA Nanobio, Ltd.
Footnotes
Supporting Information
Supporting information is available online from the Wiley Online Library or from the author.
Reference List
- 1.Guo P. Nature Nanotechnology. 2010;5:833. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 3.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Nature Nanotechnology. 2011;6:658. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bindewald E, Hayes R, Yingling YG, Kasprzak W, Shapiro BA. Nucleic Acids Res. 2008;36:D392–D397. doi: 10.1093/nar/gkm842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leontis NB, Westhof E. RNA. 2001;7:499. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Zhang C, Guo P. RNA. 1999;5:805. doi: 10.1017/s1355838299990350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pattabiraman Nagarajan, Martinez Hugo M, Shapiro Bruce A. Journal of Bimolecular Structure & Dynamics. 2002;20:397. doi: 10.1080/07391102.2002.10506858. [DOI] [PubMed] [Google Scholar]
- 8.Guo P, Zhang C, Chen C, Trottier M, Garver K. Mol. Cell. 1998;2:149. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Sheng S, Shao Z, Guo P. J Biol Chem. 2000;275(23):17510. doi: 10.1074/jbc.M909662199. [DOI] [PubMed] [Google Scholar]
- 10.Jaeger L, Leontis NB. Angew. Chem Int. Ed Engl. 2000;39:2521. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 11.Westhof E, Masquida B, Jaeger L. Folding & Design. 1996;1:R78–R88. doi: 10.1016/S1359-0278(96)00037-5. [DOI] [PubMed] [Google Scholar]
- 12.Khisamutdinov EF, Jasinski DL, Guo P. ACS Nano. 2014;8:4771. doi: 10.1021/nn5006254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu D, Moll WD, Deng Z, Mao C, Guo P. Nano Lett. 2004;4:1717. doi: 10.1021/nl0494497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Lee T, Dziubla T, Pi F, Guo S, Xu J, Li C, Haque F, Liang X, Guo P. Nano Today. 2015;10:631. doi: 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. Nat. Chem. 2010;2:772. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. Nat. Nanotechnol. 2010;5:676. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao C, Li X, Tian C, Jiang W, Wang G, Mao C. Nat. Commun. 2014;5:3890. doi: 10.1038/ncomms4890. [DOI] [PubMed] [Google Scholar]
- 18.Yu JW, Liu ZY, Jiang W, Wang GS, Mao CD. Nature Communications. 2015;6:5724. doi: 10.1038/ncomms6724. [DOI] [PubMed] [Google Scholar]
- 19.Erben CM, Goodman RP, Turberfield AJ. Angewandte Chemie-International Edition. 2006;45:7414. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 20.Juul S, Iacovelli F, Falconi M, Kragh SL, Christensen B, Frohlich R, Franch O, Kristoffersen EL, Stougaard M, Leong KW, Ho YP, Sorensen ES, Birkedal V, Desideri A, Knudsen BR. ACS Nano. 2013;7:9724. doi: 10.1021/nn4030543. [DOI] [PubMed] [Google Scholar]
- 21.Lo PK, Karam P, Aldaye FA, McLaughlin CK, Hamblin GD, Cosa G, Sleiman HF. Nature Chemistry. 2010;2:319. doi: 10.1038/nchem.575. [DOI] [PubMed] [Google Scholar]
- 22.Zhang C, Li X, Tian C, Yu GM, Li YL, Jiang W, Mao CD. ACS Nano. 2014;8:1130. doi: 10.1021/nn406039p. [DOI] [PubMed] [Google Scholar]
- 23.Edwardson TGW, Carneiro KMM, McLaughlin CK, Serpell CJ, Sleiman HF. Nature Chemistry. 2013;5:868. doi: 10.1038/nchem.1745. [DOI] [PubMed] [Google Scholar]
- 24.Baugh C, Grate D, Wilson C. J Mol. Biol. 2000;301:117. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen DH, Defina SC, Fink WH, Dieckmann T. J Am. Chem. Soc. 2002;124:15081. doi: 10.1021/ja027635d. [DOI] [PubMed] [Google Scholar]
- 26.Reif R, Haque F, Guo P. Nucleic Acid Ther. 2013;22(6):428. doi: 10.1089/nat.2012.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haque F, Shu D, Shu Y, Shlyakhtenko L, Rychahou P, Evers M, Guo P. Nano Today. 2012;7:245. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Nano Lett. 2011;11:878. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu W, Lu Y. Anal. Chem. 2010;82:574. doi: 10.1021/ac9018473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu Y, Shu D, Diao Z, Shen G, Guo P. IEEE/NIH Life Science Systems and Applications Workshop. 2009:9. doi: 10.1109/LISSA.2009.4906696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Endrizzi JA, Shu Y, Haque F, Sauter C, Shlyakhtenko LS, Lyubchenko Y, Guo P, Chi YI. RNA. 2013;19:1226. doi: 10.1261/rna.037077.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haque F, Wang S, Stites C, Chen L, Wang C, Guo P. Biomaterials. 2015;53:744. doi: 10.1016/j.biomaterials.2015.02.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shu Y, Cinier M, Fox SR, Ben-Johnathan N, Guo P. Molecular Therapy. 2011;19:1304. doi: 10.1038/mt.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 35.Veneziano R, Ratanalert S, Zhang K, Zhang F, Yan H, Chiu W, Bathe M. Science. 2016;352:1534. doi: 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shu Y, Haque F, Shu D, Li W, Zhu Z, Kotb M, Lyubchenko Y, Guo P. RNA. 2013;19:766. doi: 10.1261/rna.037002.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khisamutdinov E, Li H, Jasinski D, Chen J, Fu J, Guo P. Nucleic Acids Res. 2014;42:9996. doi: 10.1093/nar/gku516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shu D, Khisamutdinov E, Zhang L, Guo P. Nucleic Acids Res. 2013;42:e10. doi: 10.1093/nar/gkt885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolpashchikov DM. J Am. Chem. Soc. 2005;127:12442. doi: 10.1021/ja0529788. [DOI] [PubMed] [Google Scholar]
- 40.Babendure JR, Adams SR, Tsien RY. J Am. Chem. Soc. 2003;125:14716. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 41.Jasinski D, Khisamutdinov EF, Lyubchenko YL, Guo P. ACS Nano. 2014;8:7620. doi: 10.1021/nn502160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikehara M, Ohtsuka E, Tokunaga T, Nishikawa S, Uesugi S, Tanaka T, Aoyama Y, Kikyodani S, Fujimoto K, Yanase K, Fuchimura K, Morioka H. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:4695. doi: 10.1073/pnas.83.13.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeiffer S, KarimiNejad Y, Ruterjans H. J Mol Biol. 1997;266:400. doi: 10.1006/jmbi.1996.0784. [DOI] [PubMed] [Google Scholar]
- 44.Li H, Zhang K, Pi F, Guo S, Shlyakhtenko L, Chiu W, Shu D, Guo P. Adv. Mater. 2016 doi: 10.1002/adma.201601976. Accepted. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Green NM, Toms EJ. Biochem. J. 1973;133:687. doi: 10.1042/bj1330687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leamon CP, Low PS. Drug Discovery Today. 2001;6:44. doi: 10.1016/s1359-6446(00)01594-4. [DOI] [PubMed] [Google Scholar]
- 47.Hartmann G, Weeratna RD, Ballas ZK, Payette P, Blackwell S, Suparto I, Rasmussen WL, Waldschmidt M, Sajuthi D, Purcell RH, Davis HL, Krieg AM. J Immunol. 2000;164:1617. doi: 10.4049/jimmunol.164.3.1617. [DOI] [PubMed] [Google Scholar]
- 48.Weiner GJ, Liu HM, Wooldridge JE, Dahle CE, Krieg AM. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:10833. doi: 10.1073/pnas.94.20.10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shu Y, Shu D, Haque F, Guo P. Nat Protoc. 2013;8:1635. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lyubchenko YL, Gall AA, Shlyakhtenko LS, Harrington RE, Jacobs BL, Oden PI, Lindsay SM. J Biomol Struct Dyn. 1992;10:589. doi: 10.1080/07391102.1992.10508670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.