Abstract
The heat-shock response is a complex cellular program that induces major changes in protein translation, folding and degradation to alleviate toxicity caused by protein misfolding. While heat-shock has been widely used to study proteostasis, it remained unclear how misfolded proteins are targeted for proteolysis in these conditions. We found that Rsp5 and its mammalian homologue Nedd4 are ones of the main E3-ligases responsible for the increased ubiquitination induced by heat-stress. We determined that Rsp5 ubiquitinates mainly cytosolic misfolded proteins upon heat-shock for proteasome degradation. We found that ubiquitination of heat-induced substrates requires the Hsp40 co-chaperone Ydj1 that is further associated with Rsp5 upon heat-shock. Additionally, ubiquitination is also promoted by PY Rsp5-binding motifs found primarily in the structured regions of stress-induced substrates, which can act as heat-induced degrons. Our results support a bipartite recognition mechanism combining direct and chaperone-dependent ubiquitination of misfolded cytosolic proteins by Rsp5.
Introduction
In eukaryotic cells, protein quality control (PQC) either promotes folding or eliminates misfolded polypeptides that threaten cell integrity due to their high propensity to aggregate1–3. The importance of PQC is underscored by the numerous conformational pathologies associated with protein misfolding or aggregation like Huntington’s disease. Several compartmentalized degradation PQC pathways have been identified in which E3 ubiquitin ligases selectively target misfolded proteins for degradation by the proteasome, often with the help of chaperones to mediate substrate recognition4, 5. These degradation pathways, especially in the cytosol, are often directly competing with other components of the folding machinery, and it is unclear how misfolded proteins are generally sorted by PQC.
The heat-shock (HS) response is a major system that protects the cell from perturbations causing protein misfolding6, 7. In addition to upregulating HS proteins (Hsp), pioneering work showed that HS also causes higher ubiquitination levels and increased proteasome degradation in eukaryotic cells8–10. While the Hul5 proteasome-associated ubiquitin ligase contributes to this response11, the major PQC pathway has remained elusive. More importantly, it is unclear how misfolded proteins that are destined for proteolysis are recognized and triaged under stress conditions, while most chaperone proteins are presumably sequestered by the large bulk of misfolded polypeptides.
In this study, we found that the yeast Rsp5 and its mammalian homologue Nedd4 are ones of the main ubiquitin ligases responsible for the increased ubiquitination upon heat-stress. We found that Rsp5 targets mainly cytosolic misfolded proteins for proteasome degradation upon HS. We also provide insight into the mechanism of how misfolded proteins are recognized.
Results
The RSP5 E3 ligase is required for the HS induced ubiquitination response
We sought to identify the main ubiquitin ligase that mediates the ubiquitination of cytosolic misfolded proteins upon HS. In addition to its roles in endocytosis, transcription and unsaturated fatty acid and sterol synthesis12–14, the yeast Rsp5 ubiquitin ligase targets misfolded plasma membrane proteins for lysosomal degradation15–17 and its overexpression increases the thermotolerance of S. cerevisiae cells18. We therefore tested whether Rsp5 could also target cytosolic misfolded proteins upon HS. We first assessed the rsp5-1 thermo-sensitive (ts) mutant allele, which has been widely used in the past19, by comparing levels of total ubiquitin conjugates before and after HS (45°C, 15min). We found that the HS-induced ubiquitination response was markedly reduced in rsp5-1 cells in comparison to wild-type (WT) cells when analyzed by western blot, by a quantitative dot-blot assay (Figures 1a, Supplementary Figure 1a), and when assessed at various temperatures or time points (Supplementary Figure 1b, c). We confirmed these data using two additional ts alleles of the essential RSP5 gene (rsp5-sm1 and rsp5-3), for which the HS-induced ubiquitination response was also significantly decreased (Supplementary Figure 1d).
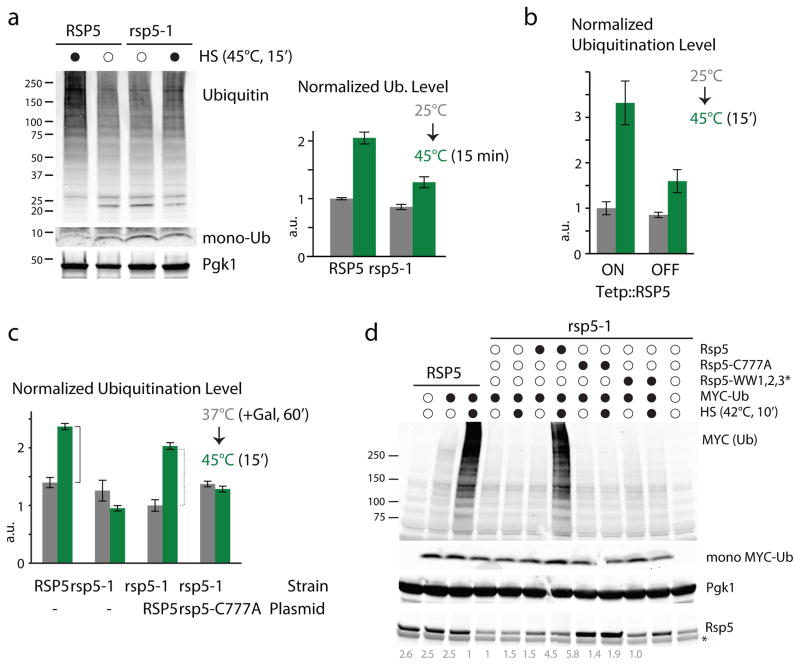
Figure 1. Rsp5 is a major E3 that mediates the increased ubiquitination upon HS.
a. Ubiquitination levels in both WT and rsp5-1 cells before (empty circle) and after a direct HS (black) analyzed by western blots and quantified by dot blot (right; see also Supplementary Figure 1a). Free mono-ubiquitin detected on the lower portion of the gel and Pgk1 loading-control are also shown. b. Normalized ubiquitination levels quantified by dot-blot in Tetp::RSP5 cells before and after HS (see also Supplementary Figure 1b). c. Normalized ubiquitination levels before and after HS in WT and rsp5-1 cells containing the indicated plasmids (- denotes presence of a control empty plasmid). Both RSP5 and rsp5-C777A were expressed under a Gal-promoter induced for 60min with 2% Galactose at 37°C. In a-c, a.u. denotes arbitrary units (each value is relative to the averaged value of the reference sample), n=3 (average±SD; source values are listed in Supplementary Table 5). d. In vitro ubiquitination performed with the indicated (black circles) recombinant proteins in WT and rsp5-1 cell extracts incubated at 25°C (empty circle) or 42°C for 10min (HS; black) and analyzed by western blot. Mono- MYC-ubiquitin, Rsp5 (with relative quantified revels) and Pgk1 loading control are also shown. The asterisk denotes an unspecific band. All uncropped images are in Supplementary Figure 8.
To further verify that RSP5 is involved in the HS ubiquitination response, we repeated our experiments by reducing RSP5 expression using a doxycycline-titratable promoter. As RSP5 is essential for its regulatory role in unsaturated fatty acid synthesis14, we supplemented cells with the oleic acid precursor TWEEN 80 to maintain cell viability during the RSP5 down-regulation. In these conditions, the increased ubiquitination level upon HS was also largely impaired in the absence of Rsp5 (Figures 1b & Supplementary Figure 1e).
To determine whether the ubiquitin ligase activity of Rsp5 is important, we performed an add-back experiment with WT RSP5 and the catalytic-inactive mutant rsp5-C777A. Whereas expression of RSP5 from a plasmid rescued the HS ubiquitination response in rsp5-1 cells, expression of rsp5-C777A did not (Figure 1c). These results indicate that a functional RSP5 ubiquitin ligase is required for the ubiquitination of proteins upon HS. As ubiquitin levels can be affected by the absence of Rsp5 in stressed cells20, we also verified that levels of free mono-ubiquitin were not significantly altered in rsp5-1 cells in our conditions (Figure 1a). In addition, overexpression of ubiquitin did not restore the increased ubiquitination in rsp5-1 cells (Supplementary Figure 1f), indicating that the observed impairment was unlikely due to reduced levels of free mono-ubiquitin.
Rsp5 directly ubiquitinates heat-induced misfolded proteins
We next sought to demonstrate that Rsp5 directly ubiquitinates proteins upon HS. One concern is that Rsp5 may indirectly affect ubiquitination levels, as it regulates the nuclear export of Hsf1 and Msn2/4 mRNAs, two major transcription factors of the HS response21, 22. We therefore developed an in vitro HS ubiquitination assay in cell extracts to monitor newly-catalyzed ubiquitination events. In these conditions, we found that there was an RSP5-dependent increase in poly-ubiquitination upon a short HS (10min) (Figure 1d, Supplementary Figure 1g). Importantly, the addition of WT recombinant Rsp5, but not the ligase-inactive mutant Rsp5-C777A, restored the increased poly-ubiquitination upon HS in extracts derived from rsp5-1 cells (Figure 1d). Rsp5 possesses three WW domains of approximately 35 amino acids each, which include two conserved tryptophan residues that bind predominately to substrates or substrate-adaptor proteins containing PY motifs23. Addition of the triple-WW-domain mutant Rsp5-WW1,2,3* failed to complement the lack of RSP5 activity in rsp5-1 cell extracts (Figure 1d). Our results indicate that Rsp5 ubiquitinates heat-induced misfolded proteins and that the recognition of these misfolded proteins is mediated by Rsp5-WW domains.
Nedd4 is also required for the HS induced ubiquitination in mammalian cells
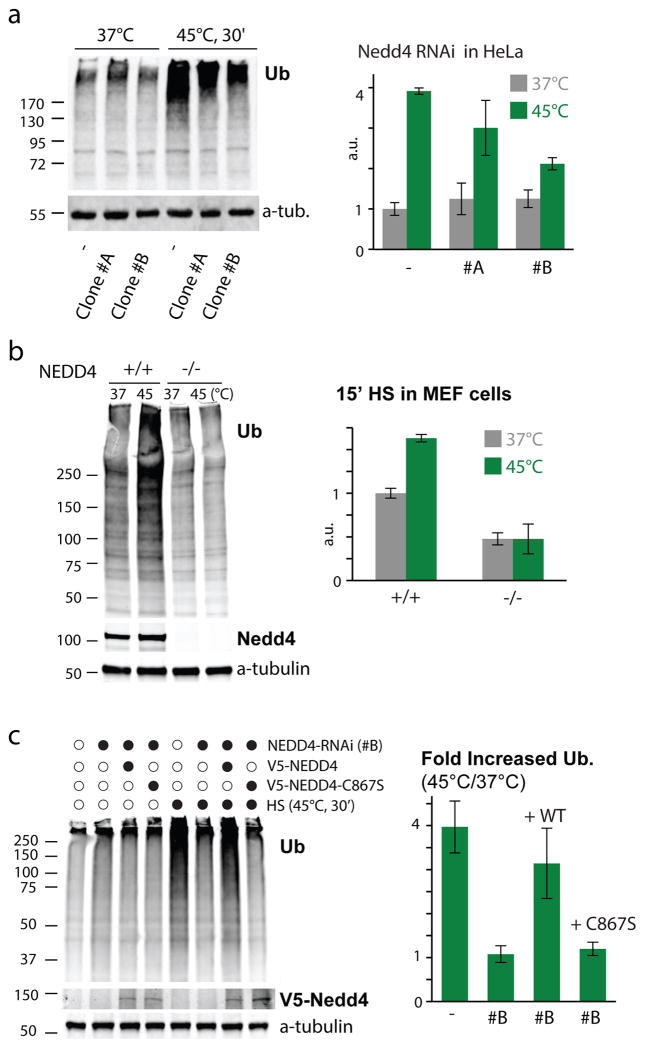
To determine whether the role of Rsp5 in HS-mediated ubiquitination is conserved in higher eukaryotes, we assessed its homologue Nedd4 in tissue culture cells. We first measured ubiquitination levels in two HeLa cell lines that were each stably transfected with a different shRNA targeting NEDD4. We found that the increased ubiquitination upon HS (45°C, 30min) was significantly reduced in these cells in comparison to the cells stably transfected with control shRNA (Figure 2a), and that the amplitude of the impairment correlated with Nedd4 levels in those cells (Supplementary Figure 2a), while levels of free mono-ubiquitin were not significantly altered in those same unstressed cells (Supplementary Figure 2b). The same results were obtained when we assessed ubiquitination levels after HS (45°C, 15 min) in MEF cells derived from WT or NEDD4−/− knock out mice (Figure 2b). Importantly, no reduced cell viability that could compromise the response was observed shortly after HS (Supplementary Figure 2c) and a similar increase in ubiquitination levels were obtained when cells were incubated at a lower temperature (42°C; Supplementary Figure 2d). To verify that Nedd4 (and its ligase activity) is required for the HS ubiquitination response, we performed add-back experiments in HeLa cells with NEDD4 stably knocked-down (targeting a non-coding region of the gene), choosing the clone with the lowest Nedd4 levels (clone #B)., We found that, following transient transfection, the expression of WT, but not the ligase-inactive Nedd4 (C867S), restored the increased ubiquitination levels upon HS (45°C, 30 min; Figure 2c). These results show that the role of Rsp5/Nedd4 in mediating the increased ubiquitination upon HS is conserved from yeast to mammalian cells.
Figure 2. The role of Rsp5/Nedd4 in the increased ubiquitination upon HS is conserved.
a. Ubiquitination levels in the indicated heat-stressed and unstressed HeLa cells stably knocked-down for Nedd4 (clones #A and #B), or scrambled control (−), as represented by the immunoblot in the left panel. Quantification of three experiments is provided in the right panel (average±SD; source values are listed in Supplementary Table 5). b. As in a, only Nedd4 knockout (−/−) or WT (+/+) MEFs were used instead of HeLa cells. c. As in a, but with the stably knocked-down HeLa cells re-transfected (or mock transfected) with WT NEDD4 or the catalytically-inactive NEDD4(C867S) for 24hrs prior to HS (45°C, 30min). All uncropped images are in Supplementary Figure 8.
Rsp5 ubiquitinates mainly heat-induced cytosolic misfolded proteins
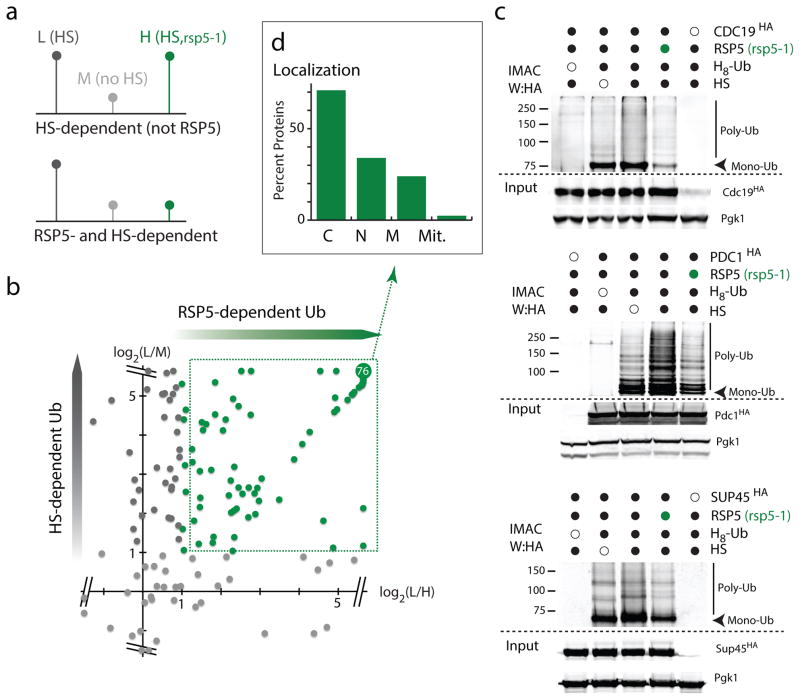
To then identify which proteins are ubiquitinated by Rsp5 upon HS, we used a proteomic approach in which we combined triple-SILAC (stable isotope labeling with amino acids in cell culture) analysis with antibody-based enrichment of diGly peptides (that correspond to ubiquitinated peptides)24, 25. In this experiment, we compared ubiquitinated proteins in WT cells that were subjected or not to a HS treatment (light and medium SILAC-labeled, respectively) and HS treated rsp5-1 cells (heavy-labeled) to unequivocally distinguish which proteins are ubiquitinated upon HS in an RSP5-dependent manner (Figure 3a). We found that about 80% (148/182 sites; Supplementary Table 1) of the diGly sites that were further enriched upon HS were affected by the rsp5 mutation (≥2 fold; Figures 3b, Supplementary Figure 3a). Analysis of the whole cell lysate also confirmed that the observed decrease in ubiquitination was not due to changes in corresponding proteins levels, except in one case (Supplementary Figure 3b). These results confirm that Rsp5 plays a major role in the heat-induced ubiquitination of proteins. To validate the proteomic analysis, we selected three Rsp5-candidate substrates: the Cdc19 pyruvate kinase, the Pdc1 pyruvate decarboxylase and the Sup45 translation release factor 1. We used an orthogonal approach, in which conjugated proteins were enriched from cells expressing the octo-histidine (H8) tagged ubiquitin by IMAC (Immobilized Metal Affinity Chromatography) for western blot analysis. We found that for all three substrates tested there was a marked increase in ubiquitination upon HS in WT cells, which was readily reduced in rsp5-1 cells (Figure 3c). We also confirmed that these proteins were poly-ubiquitinated in an RSP5-dependent manner upon HS by immunoprecipitation followed by ubiquitin immunoblotting (Supplementary Figure 3c; see also figure 4d, e). When assessing the proteomic data, we found that the majority of the 82 proteins ubiquitinated upon HS in an RSP5-dependent manner were cytosolic (Figure 3d). We confirmed this trend in a second independent experiment (Supplementary Figures 3a, d). Our results indicate that Rsp5 is a major E3 ligase that ubiquitinates mainly cytosolic misfolded proteins upon HS.
Figure 3. Rsp5 targets mainly cytosolic proteins upon HS.
a. Schematic representation of the triple SILAC experiment to identify HS-induced Rsp5 substrates (L:light; M:medium; and H: high labels). b. Plot of the log2 ratios of each quantified ubiquitinated peptide. In the y-axis, ratios of L versus M are reported to display sites affected by HS, and ratios of L versus H are reported in the x-axis to indicate sites affected by the absence of Rsp5 activity. RSP5-dependent ubiquitination sites are indicated in green. The are 76 overlapping sites with log2(ratios) ≥ 5.64. c. IMAC-enriched ubiquitin conjugates were analyzed by western blots. Samples were derived from indicated cells (rsp5-1 designated by green circles) was assed expressing endogenously tagged candidates (3xHA) and plasmid version of H8-Ubiquitin. All uncropped images are in Supplementary Figure 8. d. Localization of HS-induced Rsp5 substrates identified in b (C: Cytoplasm; N: Nuclear; M: Membrane; Mit: Mitochondria; source values are listed in Supplementary Table 5).
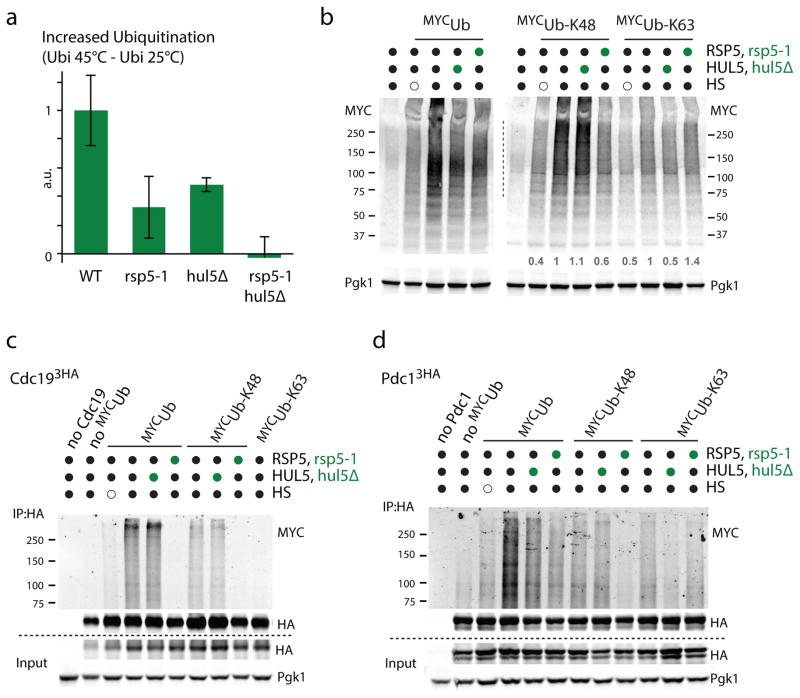
Figure 4. Both Rsp5 and Hul5 mostly target their substrates independently.
a. Increased ubiquitination levels quantified by dot-blot after a 15min HS at 45°C in the indicated cells. Experiment was done with three biological replicates (average±SD; source values are listed in Supplementary Table 5) and a.u. denotes arbitrary units. b. Western blot analysis of the indicated cells that expressed on a plasmid the indicated N-terminally MYC tagged ubiquitin constructs (K48 and K63 designate ubiquitin variants that only contain K48 and K63, respectively, while all other lysines are mutated to arginines) that were subjected or not to HS (45°C, 15min). c–d. C-terminally 3xHA tagged Cdc19 (c) and Pdc1 (d) expressed from their endogenous promoter on a plasmid were immunoprecipitated from the indicated cells (hul5Δ and rsp5-1 designated by green circles) expressing the indicated N-terminally MYC tagged ubiquitin constructs. Cells were HS or not for 20min at 45°C. Wild-type MYC tagged ubiquitin (MYCUb) was also expressed in the control cells used in lane 1. All uncropped images are in Supplementary Figure 8.
Consistent with a role in targeting cytosolic proteins, we found the addition of a nuclear localization signal to Rsp5 impaired the increased ubiquitination in cells upon HS (Supplementary Figure 3e). Furthermore, we found that the rapid HS ubiquitination response mediated by Rsp5 is distinct from the plasma membrane surveillance system that relies on a network of arrestin-related trafficking adaptor (ART) proteins to mediate the ubiquitin-dependent endocytosis and lysosome degradation of misfolded plasma membrane proteins15. Deletion of all yeast ART proteins (art1-10Δ) does not inhibit the increase in levels of ubiquitin conjugates upon HS (Supplementary Figure 3f). Furthermore, deletion of the N-terminal C2 domain of Rsp5, which is required for Rsp5’s function in sorting cargo into MVB vesicles26, does not impair RSP5 function in the HS ubiquitination response (Supplementary Figure 3g). These data indicate that the role of Rsp5 in the increased ubiquitination induced by HS is a novel function of this ubiquitin ligase distinct from its role at the plasma membrane.
Rsp5 and Hul5 ubiquitin ligase mainly target their heat-induced substrates independently
We previously found that the HECT ligase Hul5 also ubiquitinates a fraction of cytosolic misfolded proteins11. It was proposed that Hul5 is an E4-elongating enzyme that further extends ubiquitin chains on conjugated substrates27, 28. One possibility is that the main role of Rsp5 is to prime misfolded proteins that would then be further processed by Hul5. In this case, both ligases would mainly target the same pool of substrates. However, we found that the defect of the HS-response in the double hul5 and rsp5 mutant (hul5Δ, rsp5-1) was additive compared to the single mutants and, strikingly, caused a complete abolition of the accumulation of ubiquitin conjugates upon HS (Figure 4a, Supplementary Figure 4a). These results suggest that a subset of Hul5 substrates is not targeted by Rsp5. For instance, ubiquitination of Lsm7, a previously identified substrate of Hul511, was not impaired in rsp5-1 cells (Supplementary Figure 4b). In addition, we found that the accumulation of K48-linked chains upon HS was impaired in rsp5-1 but not in hul5Δ cells (Figure 4b). In contrast, the HS-induced increase of K63-linked chains was mostly impaired in hul5Δ but not in rsp5-1 cells (Figure 4b). If Hul5 was mostly further conjugating substrates mono-ubiquitinated by Rsp5, then a greater decrease of K48-link chains should have also been observed in hul5Δ cells. To confirm these data, we assessed the ubiquitination levels of the three Rsp5 substrates in hul5Δ cells. While ubiquitination of Pdc1 was also HUL5 dependent, the ubiquitination of both Cdc19 and Sup45 was not affected by HUL5 deletion (Figure 4c, d, Supplementary Figure 4c). Importantly, while there was a loss of K48-linked chains conjugated on Cdc19 and Pdc1 in rsp5-1 cells, deletion of HUL5 only affected the conjugation of K63 chains on Pdc1 that was affected to a much lesser extent in rsp5-1 cells (Figure 4c, d). These results indicate that Rsp5 and Hul5 are the two main ubiquitin ligases responsible for the accumulation of ubiquitin conjugates upon HS, and imply that both ubiquitin ligases most likely target their substrates independently of each other.
Rsp5 is required for the degradation of cytosolic misfolded proteins
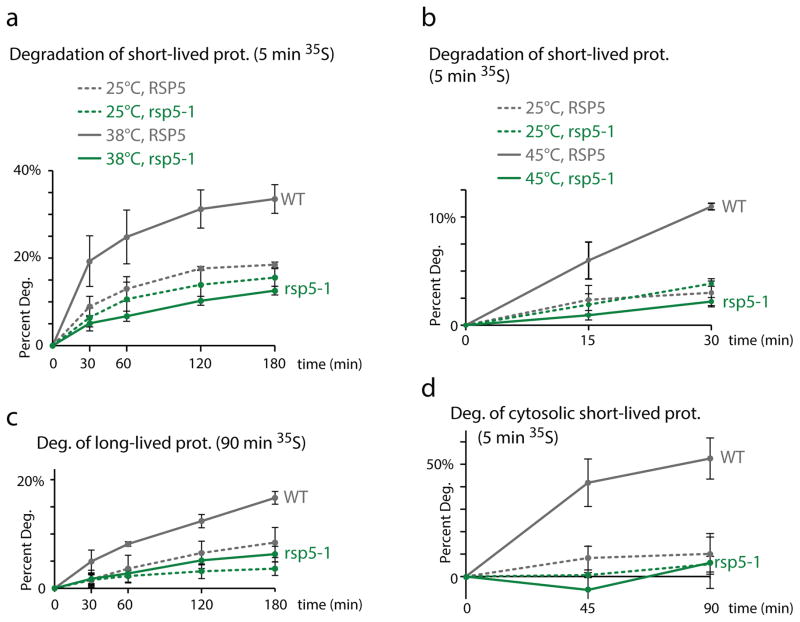
We next sought to identify the role of Rsp5-mediated ubiquitination of cytosolic misfolded proteins. Degradation of short-lived proteins by the proteasome is markedly augmented upon increased misfolding due to a mild HS10. We found that the increased degradation rate of short-lived proteins induced in these mild HS conditions was abolished in rsp5-1 cells (Figure 5a). We confirmed that, in our experimental conditions, the increased degradation of 35S pulsed-labeled proteins upon HS was proteasome-dependent in RSP5 cells and not affected by the deletion of two major lysosome proteases (Supplementary Figure 5a, b). We also verified that RSP5 is required for the heat-induced degradation of short-lived proteins when shifting the cells to a higher temperature (45°C; Figure 5b). While cell viability was not affected by a 30min HS at 45°C in both WT and rsp5-1 cells, longer incubations reduced viability (data not shown). We therefore performed subsequent turnover experiments at lower temperatures. In addition to short-lived proteins, we also observed that the increased degradation of long-lived proteins (90min labeling) upon HS was abrogated in rsp5-1 cells (Figure 5c). Consistent with a role in targeting long-lived proteins, the increased ubiquitination upon HS in cells pre-treated with cycloheximide (that blocks translation, thereby allowing the depletion of short-lived proteins) was also affected in rsp5-1 cells (Supplementary Figure 5c). To further demonstrate a role for Rsp5 in targeting cytosolic misfolded proteins for degradation, we isolated a cytosolic fraction prior to quantification of radio-labeled proteins using a lysis buffer lacking detergent to deplete membrane associated-proteins (Supplementary Figure 5d). In these conditions, the degradation of these mostly cytosolic pulse-labeled proteins was abrogated in rsp5-1 cells (Figure 5d). These results indicate that RSP5 is essential for the HS-induced proteasome degradation of cytosolic proteins.
Figure 5. Rsp5 is required for the degradation of cytosolic misfolded proteins.
a–d. Degradation of 35S labeled proteins in WT (grey) and rsp5-1 (green) cells at 25°C (dotted lines) or upon HS (solid lines; 38°C in a, c and d and 45°C in b). The portion of proteins degraded after the chase at the indicated times was measured for short-lived (a, b) and long-lived (c) proteins. The portion of proteins that mainly correspond to cytosolic proteins degraded after the chase was measured at the indicated times in (d). All experiments were done in three independent experiments (average±SD; source values are listed in Supplementary Table 5).
Association of the Hsp40 Ydj1 to Rsp5 mediates the ubiquitination of Rsp5 substrates upon HS
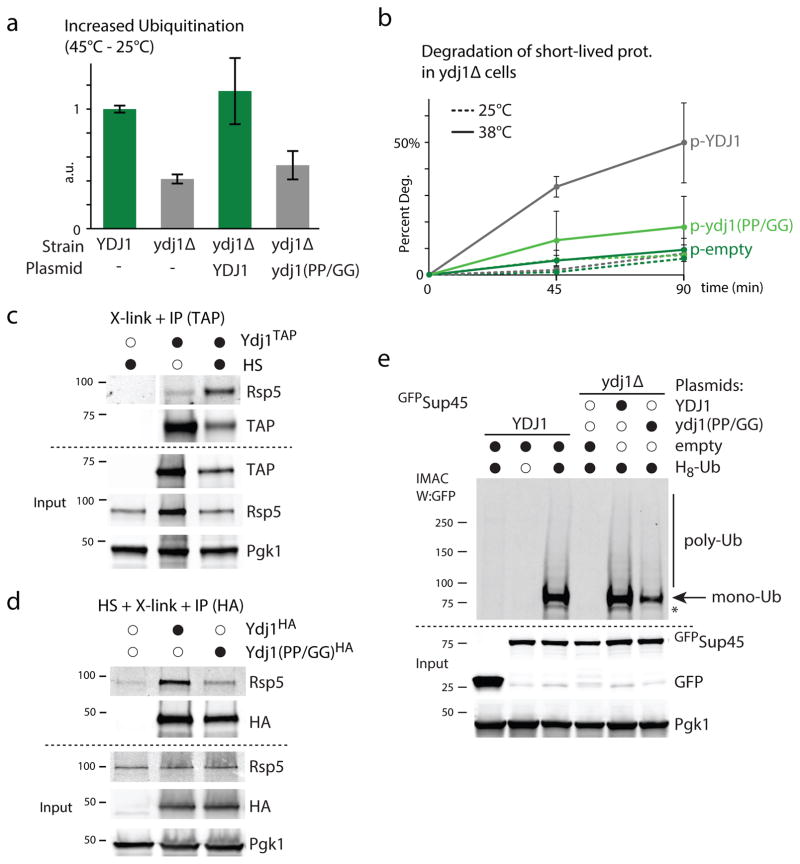
We next sought to determine how Rsp5 recognizes cytosolic misfolded proteins to target them for proteasome degradation. We first hypothesized that a chaperone could mediate the recognition of a wide-range of heat-induced misfolded proteins. Similarly to other Rsp5 substrate adaptor proteins, several chaperone proteins contain PY or PY-like motifs including Ydj1, which contains a putative PY sequence (PIPKY) in its C-terminal region. Ydj1 is a type I Hsp40 co-chaperone of the DnaJ family, which both stimulates Hsp70 ATPase activity and mediates selectivity of Hsp70 client proteins29. Interestingly, YDJ1 was also shown to be required for the ubiquitination and degradation of certain misfolded proteins in earlier studies30, 31. However, the putative role of Ydj1 in the proteolysis of cytosolic proteins remained unclear. We posit that Ydj1 may associate with Rsp5 to facilitate substrate recognition. Indeed, we found that there was a significant enrichment of proteins containing short sequences predicted to mediate binding with Ydj1 upon misfolding among HS-induced Rsp5 substrates (Supplementary Figure 6a).
We first assessed the overall role of Ydj1 in the increase of ubiquitination upon HS and found that the deletion of YDJ1 impaired the ubiquitination response (Figure 6a, Supplementary Figure 6b). Correspondingly, absence of YDJ1 also prevented the degradation of 35S-labeled short-lived proteins upon HS (Figure 6b). We next determined whether Ydj1 and Rsp5 interact in HS conditions by performing co-immunoprecipitations (IP) in extracts derived from cells crossed-linked prior to lysis (to preserve a possible heat-induced interaction during IP). Under these conditions, we observed an increased interaction between Rsp5 and Ydj1 upon HS (Figure 6c). Importantly, the HS-induced Rsp5 interaction was mostly abrogated upon the mutagenesis of the putative PY motif of Ydj1 (PIPKY to GIGKY; Figure 6d). Accordingly, the increased ubiquitination was restored upon the expression of the WT YDJ1 but not after the expression of the ydj1 (PP/GG) mutant (Figure 6a, Supplementary Figure 6b). We verified this result by assessing the ubiquitination levels of Sup45, a heat-induced substrate of Rsp5. We found that the deletion of YDJ1 led to the abolition of Sup45 ubiquitination in HS conditions, which can be fully restored upon the expression of WT YDJ1 but not of the ydj1 (PP/GG) mutant (Figure 6e). Similarly, expression of the ydj1 (PP/GG) mutant failed to restore degradation of short-lived proteins upon HS (Figure 6b). Our results indicate that the association of Ydj1 with Rsp5 plays a major role in the HS-induced ubiquitination and degradation of Rsp5 substrates.
Figure 6. An Ydj1-adaptor mediates ubiquitination of misfolded proteins upon HS.
a. Increased ubiquitination levels quantified by dot-blot after HS (45°C, 15min) in the indicated cells that carried a LEU2 plasmid that was empty (−) or with YDJ1 or ydj1 (PP/GG) expressed from the YDJ1 promoter. Three biological replicates were assessed (average±SD; source values are listed in Supplementary Table 5). b. Degradation of short-lived 35S-labeled (5 min) proteins in ydj1Δ cells that carried a plasmid expressing YDJ1 (grey) or the ydj1 (PP/GG) mutant (light green) from the YDJ1 promoter or an empty plasmid (dark green) at 25°C (dotted lines) or 38°C (solid lines). Three independent experiments were performed (average±SD; source values are listed in Supplementary Table 5). c. TAP immunoprecipitation (IP) was performed after in vivo cross-linking with 1% formaldehyde in cells grown at 25°C (empty circle) or during a HS (45°C, 20min; black). The endogenous YDJ1 was C-terminally TAP-tagged in the indicated lanes. The IP samples were analyzed by western blot (the - TAP control was analyzed on the same membrane but not adjacent to the other two lanes). d. HA IP was performed after in vivo cross-linking with 1% formaldehyde in ydj1Δ cells subjected to a HS (45°C, 20min) and that carried a LEU2 plasmid that was empty or contained C-terminally tagged (3xHA) YDJ1 or ydj1 (PP/GG) expressed from the YDJ1 promoter. e. IMAC from YDJ1 or ydj1Δ cells expressing when indicated H8-Ubiquitin from a first URA3 plasmid, Sup45 fused N-terminally to GFP or GFP alone from a second HIS3 plasmid, and YDJ1 or ydj1 (PP/GG) under the YDJ1 promoter from third LEU2 plasmid. All uncropped images are in Supplementary Figure 8.
Heat-induced Rsp5 substrates contain PY motifs preferably embedded in structured regions
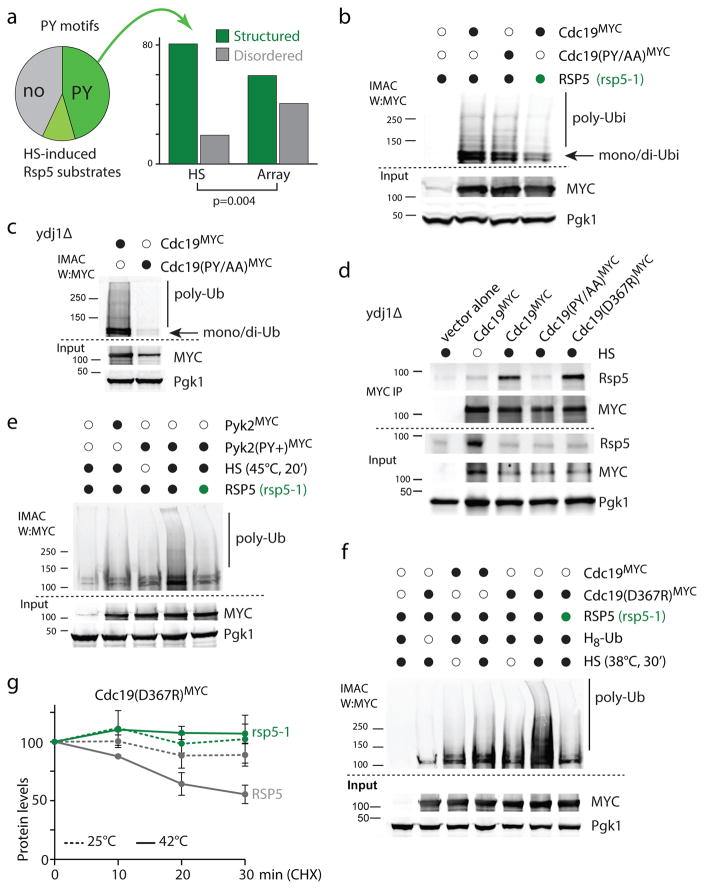
We reasoned that, in addition to Ydj1-binding, substrates themselves could contain elements that directly promote their recognition by Rsp5. Indeed, we observed that the heat-induced ubiquitination of Cdc19, another Rsp5 substrate upon HS, was not fully abrogated in ydj1Δ cells (Supplementary Figure 6c), a similar observation was obtained for Pdc1 (see also Supplementary Figure 7). These results suggest that, in some cases, a bipartite mechanism could mediate the recognition of misfolded proteins by Rsp5. Accordingly, we found a large portion of HS-induced substrates with PY or PY-like motifs (Figure 7a, Supplementary Table 2). Note that not all Rsp5 candidate substrates contain obvious PY-like motifs (e.g., Sup45 which was entirely ubiquitinated in a YDJ1-dependent manner in Figure 6e). Interestingly, the Rsp5 canonical PPxY motif was more prevalent among candidate Rsp5 substrates identified using a protein array32, while slight variations of the PxY motif such as, SPxF and VPxF (referred to as PY-like) were significantly enriched among HS-induced candidate substrates (Supplementary Table 2). We also found that PY and PY-like motifs among HS-induced Rsp5 candidate-substrates had significantly a lower frequency of locating in regions predicted to be disordered in comparison to other Rsp5 candidate substrates identified in the protein array (Figure 7a). The higher prevalence of PY motifs in disordered regions among substrates identified in non-HS conditions is consistent with the required accessibility of the motif to mediate binding with one of the Rsp5 WW domains under normal conditions (i.e., no HS). One possibility is that the HS-induced substrates are mainly recognized by Rsp5 upon misfolding, which exposes otherwise shielded motifs. In agreement with this idea, PY and PY-like motifs of Rsp5 candidate-substrates induced by HS were predicted to be less accessible in comparison to the other candidate substrates, when considering motifs in structured regions (Supplementary Figure 7a).
Figure 7. PY-motifs on substrates promote RSP5-dependent ubiquitination upon HS.
a. The pie chart (left) indicates which portion of HS induced Rsp5 candidate substrates identified in the two mass spectrometry experiments contains a proline-containing motifs ([PLSV]Px[YF], or PPPP; dark green), additional PxY motifs (light green) or no obvious PY motif (grey). × denotes any residue. The histogram (right) indicates whether [PLSV]Px[YF] or PPPP motifs are located in regions predicted disordered or not among Rsp5 candidate substrates induced by HS or identified in a protein array (Array). Assigned PY motifs are listed in Supplementary Table 5. b, c, e, f. IMAC analyzed by western blots. Samples were derived from cells expressing H8-ubiquitin (from a URA3 plasmid) and the indicated wild-type or mutants Cdc19 and Pyk2 (fused C-terminally with 13xMYC and expressed from a HIS3 plasmid). Py+ designates the A365P/L366N mutations in Pyk2. RSP5 (black) or rsp5-1 (green) cells were analyzed in b, e and f, and ydj1Δ cells in c. A HS at 45°C for 20min was applied in b and c, and in e when indicated (black); cells were subjected to a 38°C HS for 30min in f when indicated (black). d. MYC immunoprecipitation (IP) was performed after in vivo cross-linking with 1% formaldehyde (10 min) in cells grown at 25°C (empty circle) or during a HS (40°C, 20min; black). The indicated Cdc19 (fused C-terminally with 13xMYC) were expressed (GDP promoter) in ydj1Δ cells. The IP and input samples were analyzed by western blots as indicated. All uncropped images are in Supplementary Figure 8. g. Levels of Cdc19(D367R) C-terminally tagged with 13×MYC was determined by quantitative western blotting after incubating cells (wild type in grey and rsp5-1 in green) in the presence of 100μg/ml cycloheximide at 25°C (dotted lines) or 42°C (solid lines) at the indicated times in three independent experiments (average±SEM; source values are listed in Supplementary Table 5; representative images are shown in Supplementary Figure 7f).
To assess the role of these PY and PY-like motifs in mediating substrate recognition by Rsp5 upon HS, we mutated the putative Rsp5-binding motif of Cdc19 (LPNY to LANA), which we previously identified as an Rsp5 substrate. This putative motif is located, based on a crystal structure, close to the interaction interface between two monomers (Supplementary Figure 7b) and is thereby not readily accessible for Rsp5 binding. We found that the ubiquitination of the Cdc19 (PY/AA) mutant upon HS was reduced in comparison to the WT Cdc19. (Figure 7b). When we repeated the same analysis in ydj1Δ cells, we found that the Cdc19 (PY/AA) mutant was no longer ubiquitinated upon HS (Figure 7c). Similarly, mutation of either the LPTF motif (to LATA) or LPVF motif (to LAVA) of Pdc1 reduced ubiquitination of this Rsp5 substrate in ydj1Δ cells (Supplementary Figure 7c) but not in YDJ1 cells (Supplementary Figure 7d). As well, while crossed-linked Rsp5 was specifically co-immunoprecipitated with Cdc19 in ydj1Δ cells upon HS, it was not with Cdc19(PY/AA) (Figure 7d). These results suggest that, in addition to Ydj1-mediated recognition, PY or PY-like motifs on substrates also promote HS-induced Rsp5-interaction and ubiquitination. To further test this idea, we determined whether the addition of a PY motif would be sufficient to mediate the Rsp5-dependent ubiquitination upon HS. Pyk2 is a Cdc19 paralog that arose from the whole genome duplication. Despite good conservation between both proteins (71% identity), the PY motif is not conserved in Pyk2, which is also less abundant in the cell in standard growth conditions33. Remarkably, substitution of two amino acids in Pyk2 to create the same PY motif as in Cdc19 (LALY to LPNY; Pyk2(PY+)) was sufficient to cause an RSP5-dependent ubiquitination of the mutated Pyk2(PY+) upon HS (Figure 7e). Based on the homology model of Pyk2, the generation of the PY motif was not predicted to destabilized the structure of the mutated protein (see Methods) and Pyk2, Pyk2(PY+) and Cdc19, under the same promoter, were expressed at similar levels (Figure 7e), indicating that the mutant protein was likely expressed and folded properly in unstressed conditions. We then mutated a Cdc19 residue near the PY-motif (D367R) that we predicted to likely disrupt the homo-dimer formation (Supplementary Figure 7b). We reasoned that this mutation may further expose the PY motif and thereby promote Cdc19 ubiquitination. Indeed, while a 38°C HS led to a mild increased ubiquitination of Cdc19, Cdc19-D367R was readily ubiquitinated in a RSP5-dependent manner in these conditions (Figure 7f). Consistent, with these results, cross-linked Rsp5 was more readily co-immunoprecipitated with Cdc19-D367R upon HS (Figure 7d). Accordingly, Cdc19-D367R was less stable than wild-type Cdc19 in stressed conditions (Supplementary Figure 7e). In this case, degradation of the mutated Cdc19 was HS-, RSP5- and proteasome-dependant, and accordingly was not affected by deletion of HUL5 (Figure 7g, Supplementary Figure 7f–h). These results indicate that PY motifs can act as a heat-induced degron to mediate ubiquitination of misfolded proteins by Rsp5 under stress conditions.
Discussion
Here, we discovered a conserved PQC degradation pathway in which Rsp5/Nedd4 are ones of the main ubiquitin ligases that target misfolded proteins upon HS. Using both genetic and biochemical approaches, we show that Rsp5 ubiquitinates mainly cytosolic misfolded proteins upon HS to target them for proteasome degradation. We previously found that the Hul5 ubiquitin ligase, which has been proposed to be an E4 enzyme, also conjugates cytosolic misfolded protein upon HS11. The absence of Hul5 impaired the accumulation of K63-linked ubiquitin chains upon HS, consistent with previous data27. In contrast, mainly the accumulation of K48-linked chains was affected in rsp5-1 mutant cells, indicating that each ligase conjugates its substrates independently. Because Rsp5 preferentially catalyzes K63-linked ubiquitin chains34, it will be important to determine whether the RSP5-dependent accumulation of K48 chains upon HS is directly mediated by Rsp5 or with the help of another ligase and/or deubiquitinase.
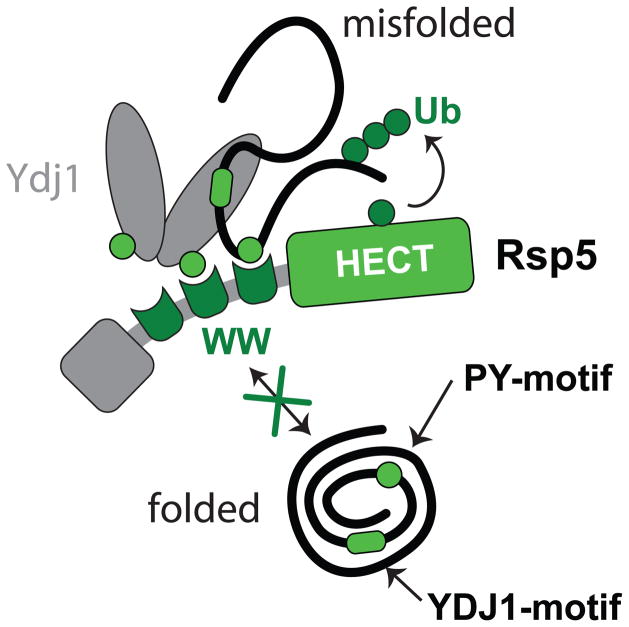
We propose that Ydj1 acts as a substrate-adaptor protein to promote the ubiquitination of its misfolded client proteins either alone or in conjunction with exposed PY-motifs on the substrate (Figure 8). Buried PY motifs in proteins could signal misfolding by mediating Rsp5 recognition when exposed. Consistent with this misfolding inducible degron model, PY motifs are widely distributed in the proteome and primarily found in structured regions (Supplementary Figure 7i).
Figure 8. Bipartite model for the recognition of cytosolic misfolded proteins by Rsp5.
Schematic of the proposed bipartite model in which misfolded substrates are recruited by Ydj1 Hsp40 and/or directly recognized by Rsp5.
Interestingly, the Hsp40 co-chaperone Sis1, but not Ydj1, is sequestered in protein aggregates35. Sis1 can cooperate with the Ubr1 ubiquitin ligase to target a misfolded model substrate for degradation36, and Sis1 sequestration in protein aggregates inhibits the degradation of misfolded proteins35. One possibility is that Ydj1 and Rsp5 participate in a major PQC pathway that is activated upon major misfolding stresses when other PQC components like Sis1 are sequestered by the abundant load of misfolded proteins.
Nedd4/Rsp5 were recently shown to potentially play a major role in α-synuclein proteopathies. The mammalian Nedd4 targets α-synuclein for degradation37 and the NAB2 compound that reduces α-synuclein toxicity in mammalian and yeast cells targets Rsp538. More recently, Rsp5 was also shown to be required for the degradation of aggregation-prone proteins by macro-autophagy39. It will be important to determine the interplay between these pathways and whether a similar mechanism for the recognition of misfolded proteins upon heat-shock is used in other instances.
Online Methods
Strains, Plasmids and Reagents
All Saccharomyces cerevisiae strains (S288C background) and plasmids are listed in Supplementary Tables 3 and 4, respectively. Temperature sensitive (ts) mutant strains rsp5-1, rsp5-sm1, rsp5-3 and rpt6-20 were generously provided by Dr. Charlie Boone from the yeast ts conditional mutants collection40. The strain in which Rsp5 expression is controlled by the Tet-Promoter (Tetp::RSP5) was purchased from the Yeast Tet-Promoter Hughes Collection distributed by Open Biosystems (Thermo Scientific)41. All the single deletion strains were from the Yeast Knockout (YKO) Collection (Open Biosystems, Thermo Scientific) and kindly provided by Dr. Michel Roberge. The art1-10Δ and art1-8,10Δ deletions strains in the BY4741 background were kindly provided by Dr. Pelham42. All the double mutant strains were generated by mating and tetrad dissection. All SILAC mass spectrometry experiments were done in BY4742 background cells. YDJ1-TAP was kindly provided by Dr. Elizabeth Conibear from the Yeast TAP-Tagged ORFs Collection (Open Biosystems, Thermo Scientific). CDC19-3HA, PDC1-3HA and SUP45-3HA were generated by inserting the 3xHA tag at the 3′ end of the endogenous genes by homologous recombination43.
Plasmids expressing RSP5 (BPM587) and rsp5-C777A (BPM588) were obtained by subcloning the GALp into pRS426 (URA3, 2μ) using EcoRI and NaeI, and HA-RSP5 and HA-rsp5-C777A that were generously provided by Dr. Jon Huibregtse using NaeI and NotI. The GST-RSP5 (BPM98) in pGEX-6P-1 was previously described44. GST-rsp5-C777A (BPM514) was generated by subcloning rsp5-C777A to replace a C-terminus fragment of RSP5 in BPM98 using NotI and AfeI restriction sites. rsp5-WW1,2,3* (in which all three WW domains are mutated)45 was kindly provided by Dr. Teresa Zodalek and subcloned with primers containing EcoRI and SalI restriction sites to replace RSP5 in BPM98 to generate GST-Rsp5-WW1,2,3* (BPM518). The human Nedd4 plasmids in pcDNA3.1N-V5 (Invitrogen) were previously described46. The MYC tagged (1 epitope) ubiquitin expressing constructs were generating by PCR amplification of the wild-type or K0 ubiquitin47 that were subcloned after a GPD promoter and before the PGK1 terminator sequence in pRS313 (BPM590 for WT). K48only (BPM591) and K63 only (BPM592) were then generated by site directed mutagenesis. The YDJ1 plasmid (BPM390) was obtained from Elizabeth Craig, which was then modified by site-directed mutagenesis to mutate P317 and P319 to glycine residues to generate ydj1-PY/GG (BPM569). The magaprimer method used to generate HA-tagged YDJ1 and ydi1-PY/GG was adapted from a previously descripted method48. Briefly, the 3xHA sequence was PCR amplified using pFA6a-3xHA-His3MX6 to generate the megaprimer, which is then used to PCR amplify BPM390, BPM569 prior to DpnI digestion and bacteria transformation. The LSM7-TAP, His8-ubiquitin plasmid (BPM297) was generated by PCR amplifying LSM7 with its endogenous promoter (+570bp) and C-terminal TAP sequence and inserted into pRS316-GPDp-His8-ubiquitin-PGKt (BPM30) using NotI site. The GFP-SUP45 (BPM580) was obtained by PCR amplifying SUP45 from genomic DNA and inserted into pRS313-GPDp-EGFP-PGKt (BPM519). The CDC19-3HA (BPM635) and PDC1-3HA (BPM636) were PCR amplified from endogenously C-terminally tagged genes (3xHA; YTM1308 and 1295) with their own promoters (−600bp) into pRS316. The CDC19-13MYC (BPM563), PYK2-13MYC (BPM603) and PDC1-13MYC (BPM565) plasmids were generated by PCR amplifying the endogenous genes from genomic DNA and inserting them into a pRS313 plasmid with GPD promoter, 13×MYC tag and PGK terminator sequence (BPM173) using BamHI and XmaI restriction sites. The cdc19-PY/AA-13MYC plasmid (BPM564) was obtained by site-directed mutagenesis of BPM563 so that P363 and Y365 were mutated to alanine residues. pdc1-PY1-13MYC (BPM566), pdc1-PY2-13MYC (BPM567) and pdc1-PY1/2-13MYC (BPM568) were similarly generated to mutate P500, Y502 (PY1) and P541, Y543 (PY2) to alanine residues. cdc19-D367R (BPM625) was obtained by site-directed mutagenesis of BPM563 so that D367 was mutated to an arginine residue. The pyk2-PY(+)-13MYC (BPM604) was obtained by site-directed mutagenesis of BPM603, so that A365 and L366 were mutated to proline and asparagine residues, respectively, to regenerate the same PY motif (LPNY) present in CDC19.
Mouse monoclonal anti-ubiquitin antibody MAB1510 (Millipore, 1:3,000) was used for assessing yeast ubiquitin levels and the mouse monoclonal P4G7 antibody (Santa Cruz, 1:1,000) was used for mammalian ubiquitin. Goat anti-Rsp5 antibody (sc-26193, 1:1,000) from Santa Cruz, rabbit anti-hsNedd4 monoclonal antibody (3607S, 1:1,000) from Cell Signaling and mouse anti-mmNedd4 monoclonal antibody (15/Nedd4, 1:2,000) from BD Transduction were used, as well as rabbit polyclonal anti-Pgk1 antibodies (AP21371AF-N, 1:10,000) from Acris Antibodies, anti-α-tubulin antibodies (sc-12462-R, 1:1,000) from Santa Cruz, mouse monoclonal anti-MYC (9E10, 1:7,000) and anti-HA (12CA5, 1:2,000) antibodies from the AbLab UBC in-house facility, anti-V5 antibody (R960-25, 1:10,000) form Invitrogen, anti-GFP antibody (11814460001, 1:1,000) from Roche, and polyclonal anti-TAP (CAB1001, 1:1,000) from Pierce (Thermo Scientific). Anti-mouse-800, anti-rabbit-700 and anti-goat-800 fluorescent secondary antibodies (1:10,000; LI-COR) were detected with an Odyssey Infrared Imaging System (LiCor) or with an Odyssey Fc System (LiCor) for Figure 2a, 2b and S2c. In Supplementary Figure 2a, hsNedd4 was detected using Perkin Elmer Western Lightning Plus-ECL reagents. Protease inhibitor cocktail was purchased from Roche and all other reagents were purchased from Sigma unless specified.
Heat-shock assays in yeast cells
To quantify ubiquitination levels after HS, yeast cells were collected and processed as previously described11. For direct HS experiments, overnight-saturated cultures were diluted and grown to exponential phase in YPD at 253°C to an A600 of 1–1.5, about 2 ml of cells were then incubated at 45°C in a thermomixer for the indicated times (typically 153min). For some temperature sensitive strains, a pre-incubation at 37°C was included when indicated. In this case, overnight-saturated cultures were diluted and grown to exponential phase in YPD at 253°C to an A600 of 1–1.5 before pre-incubation at 37°C for 30min followed by heat-shock at 453°C for 15min. For add-back experiments using the GALp-controlled expression plasmids, cells were grown in SD-URA media with 2% raffinose and 0.05% dextrose until the A600 reached 1, and 2% galactose was added for 1 hour at 37°C to induce expression of Rsp5 or Rsp5-C777A prior to heat-shock at 45°C for 15min. For regular HS assay, Tetp::RSP5 cells were grown in YPD supplemented with 0.5% Tween-80 with/without 100μg/ml doxycycline at 25°C until saturated and then diluted into the same media. This process was repeated three times before the last dilution, and cells were then grown to mid-log phase for a direct heat-shock assay as above. For IP experiments, Tetp::RSP5 cells were grown in synthetic selection media at 25°C until saturated, which were then diluted into 150ml of the same media containing 0.2% Tween-80 with/without 100μg/ml doxycycline and grown to mid-log phase. For western and dot blots, cell pellets were snap-frozen in liquid nitrogen after treatment and washed twice with cold 1×TBS (50mM Tris, 150mM, NaCl, pH 7.5) before lysis. Lysis was carried out with glass beads in a Precellys 24 tissue homogenizer (Precellys) in pre-warmed 1×SDS-PAGE Laemmli sample buffer without reducing agent and dye. All samples were normalized using a Bradford assay (BioRad). For the dot blot assay, 3μl of each normalized sample (5–10μg proteins) was spotted and dried overnight on a nitrocellulose membrane. Membranes were rehydrated with 1×TBS and processed as other western blots. All quantitative dot blots were performed by measuring signals from three biological replicates. For each sample, the ubiquitin signal was normalized using the Pgk1 signal. When reporting the normalized ubiquitination levels (e.g., 25°C and 45°C), the signals were normalized to the averaged ubiquitination level in the reference sample (typically wild-type at 25°C) that was set to the arbitrary value of 1 and then averaged. When reporting the increase in ubiquitination, the difference of normalized signals between two temperatures for each sample (e.g., 45°C-25°C) was calculated, then averaged across the three replicates and normalized to the averaged difference in the reference sample (set to the arbitrary value of 1).
In vitro heat shock ubiquitination assay in yeast cell extracts
Recombinant Rsp5, Rsp5-C777A and Rsp5-ww1,2,3* were purified from BL21 (DE3) bacteria cells. After a three-hour induction with 1mM IPTG at 25°C, cells were lysed in cold lysis buffer (1xPBS pH7.3, 1mM EDTA, 10% glycerol, 1mM DTT, 0.5% Triton-X100, 1mM PMSF, 1×Protease inhibitor cocktail) by sonication. Purification procedures were following the manufacturer’s protocol using Glutathione Sepharose 4B resin (GE healthcare). Rsp5, Rsp5-C777A or Rsp5-ww1,2,3* were cleaved from the GST tag with the PreScission Protease (GE healthcare) in cleavage buffer (50mM Tris-HCl pH7.0, 150mM NaCl, 1mM EDTA, 1mM DTT, 10% glycerol, 0.01% Triton-X100) at 4°C for 5 hours. Cleaved proteins were obtained from the flow-through and stored at −80°C.
Mid-log phase cells of both WT and rsp5-1 grown in YPD were subjected to 37°C pre-incubation for 30min to inactivate rsp5-1. Frozen cell pellets were lysed in native lysis buffer (30mM HEPES pH7.4, 150mM KOAc, 250mM Sorbitol, 7.5mM MgCl2, 1mM DTT, 1mM EDTA, 1mM PMSF, 1× protease inhibitors cocktail, 1mM sodium orthovandate, 2.5mM sodium pyrophosphate and 1mM β-glycerophosphate) using mortar and pestle. Clarified lysates were aliquoted and stored in liquid nitrogen. For the heat-shock ubiquitination assay, cell lysates were warmed up to room temperature. 2mM ATP, and if indicated around 0.05μg (in 0.2μl) of Rsp5, Rsp5-C777A, Rsp5-ww1,2,3* and 2μl of 1μg/μl MYC-ubiquitin (BostonBiochem) were added to 20μl of cell lysate right before incubating the samples at the indicated temperatures for 10min. Samples were mixed with 3× SDS-PAGE Laemmli sample buffer to stop the reaction and processed for western blot analysis.
Heat-shock assays in mammalian cells
HeLa cells were grown in DMEM media supplemented with 10% FBS, penicillin, streptomycin, and fungizone. Nedd4 (+/+) or (−/−) MEFs (mouse embryonic fibroblasts) were grown in the same media further supplemented with L-glutamine. All cell media was purchased from Life Technologies. The generation of stably transfected HeLa cells, in which Nedd4 (also called Nedd4-1) was stably knocked-down, is described elsewhere49. As a control an empty shRNA (pGIPZ, Open Biosystems) was used and two different sequences complementary to Nedd4 were used in clone #A and #B. Generation of the MEF cells was previously described50, in which β-gal cDNA was inserted between exons 6 and 7, and embryonic (13.5–14.5 days) cells were immortalized by serial passages. Cells at approximately 90% confluency in 6 cm diameter plates were replenished with fresh media (4mL) 2–3 hrs prior to HS, then were transferred to an incubator pre-warmed at 45°C for the indicated times. For add-back experiments, the indicated DNA was transfected using PolyJet (SignaGen) according to the manufacturer’s instructions 24hrs prior to HS. Mock transfection without DNA was performed for control cells. After HS, cells were washed twice with cold 1xPBS and lysed in 50mM Tris-HCl pH 7.5, 150mM NaCl, 1% NP-40, 0.1% SDS, 1mM PMSF, 1× Protease inhibitor cocktail by cell scraping. Protein concentrations were measured using a Bradford assay (BioRad) before mixing with SDS sample buffer (final 2%SDS). All samples were boiled at 96°C for 15–20min with constant vortexing to reduce the viscosity prior to western blot analysis. Ubiquitination levels were normalized to α-tubulin levels. For the add-back experiments, ubiquitination levels were first compared on each membrane between heat-shock and unstressed cells for each replicate then averaged between the three experiments. This method was preferred due to variability of the ubiquitin signals from one membrane to another.
35S-labeling and protein turnover assays
Quantification of the degradation of 35S-labeled proteins was performed as previously described10. Yeast cells grown in SC medium with additional tyrosine (20μg/ml) to an A600 of 0.8–1, washed and incubated in SD-Met media with additional tyrosine (20μg/ml) for 50 min51. EXPRE35S35S protein labeling mix (50μCi/m1, PerkinElmer) was added for 5min or 90min prior to washing cells with ice-cold SC-chase media containing cysteine (0.5mg/ml), methionine (6mg/ml) and cycloheximide (0.5mg/ml). Cells were collected at the indicated times to be then mixed to a final concentration of 10% TCA. After an overnight incubation at 4°C, radioactivity in both TCA-soluble and -insoluble fractions was measured in a MicroBeta2 radiometric detector (PerkinElmer). The percentage of protein degradation was calculated by subtracting the signal from TCA-soluble at time 0 from indicated time that was then divided by the signal in the TCA-insoluble fraction at time 0. To assess turnover of soluble proteins, around 5 A600 of radiolabeled cells were lyzed in 100μl native buffer (50mM Tris-HCL pH 7.5, 150mM NaCl, 2mM MgCl2, 1mM PMSF, 103mM chloroacetamide, 1mM phenanthroline, and 1× protease inhibitors cocktail) by glass beating. After a 15min centrifugation at 16,000rpm in a microfuge, samples were normalized to same concentration in 90μl. One fraction of soluble proteins (70μl) was precipitated by TCA, while the other fraction (20μl) was analyzed by SDS-PAGE. Radioactivity for each time point was measured in the TCA-soluble fraction and normalized to the amount of proteins detected by Coomassie on the protein gel. All experiments were performed in three independent replicates.
Microscopy
Cells were grown to an A600 of 1 at 25°C and subjected or not to a 40°C heat-shock for 2hrs. In the last 30min of incubation, 2.5μg/ml of Hoechst 33342 (Invitrogen) was added to each culture for DNA staining. Live cells were mounted on a slide with 1×PBS and immediately imaged with an inverted Zeiss Axio Observer microscope. One image plane per field (225μm2) was acquired with a 63×oil objective and processed with Zeiss Zen software.
DiGly peptide enrichment for triple SILAC mass spectrometry analysis
Enrichment of diGly containing peptides was performed using the PTMScan kit (Cell Signaling) with minor modifications from previous description52. Only lysine (K) labeling was used for the diGly SILAC experiments. RSP5 cells labeled with light-lysine (K0) and rsp5-1 cells labeled with heavy-lysine (K8) were subjected to 20min heat-shock at 45°C prior to lysis, while medium-labeled RSP5 cells (K4) were kept at 25°C for control. Equal amounts of lysate from differentially labeled cells were mixed together to obtain ~30mg of proteins in total. The PTMScan Ubiquitin Remnant Motif Kit (Cell Signaling Technology) was used to immuno-precipitate (IP) diGly peptides according to the manufacturer’s protocol except beads were cross-linked prior to the IP53 and 1/8 of the recommended amount of beads was used per IP instead. Bound peptides were eluted with 50μl 0.15 % (vol/vol) Trifluoroacetic acid at 25°C for 5min before being cleaned up by C18 stage tips without fractionation54. An aliquot of the whole cell lysate (WCL) was also analyzed after C18 stage tipping. For the first experiment, WCL peptides were also separated in 6 fractions by strong cationic exchange (SCX).
Mass spectrometry analysis
The instrument methods below are adapted from a previous study 55. Purified peptides were analyzed using a LTQ-Obitrap Velos (ThermoFisher Scientific) coupled to an Agilent 1290 Series HPLC using a nanospray ionization source (ThermoFisher Scientific). The LTQ-Orbitrap Velos was set with the following parameters: full-range scan at 60,000 resolution from 350 to 1600Th in the Orbitrap, fragmentation of top 5 ions by HCD in each cycle in the LTQ (minimum intensity 1000 counts), exclusion of singly charged ions and previously analyzed for 30 sec. The Orbitrap was continuously recalibrated using lock-mass function56. Mass accuracy: error of mass measurement is typically within 5ppm and is not allowed to exceed 10ppm.
Centroided fragment peak lists were processed to Mascot generic format using Proteome Discoverer (PD, 1.2). Fragment spectra were searched using the Mascot algorithm (2.3.0) against the Saccharomyces Genome Database (SGD-05Jan2012 with 6147 protein sequences and 6147 randomized sequences). The cut-off FDR of glygly peptides was set below 1% (0.01). Similar to previous studies, we removed peptides with a C-terminus GlyGly and considered peptides with log2 (ratios) ≥ 157, 58. For the peptide quantitation, missing values were replaced with default minimum ion intensity. For each dataset, peptide ratios were normalized to the median protein ratio obtained from an aliquot of the input whole cell lysate sample. The ratios of diGly peptides independently quantified several times were averaged.
IMAC—purification of ubiquitinated proteins
In all experiments, about 1μl of MagneHis (Promega) was used per 200μg of protein extract. For HA-tagged substrate validation, 150ml of cells carrying His8-ubiquitin plasmid (BPM30)59 or corresponding empty plasmid for control were grown in SD-URA media at 25°C. Cells with or without 20min heat-shock treatment at 45°C were washed twice with ice-cold 1xTBS and cell pellets snap frozen in liquid nitrogen. Thawed cells were lysed in HU buffer (8M urea, 100mM HEPES at pH38, 0.05% SDS, 10mM chloroacetamide, 1mM PMSF, 10mM imidazole and protease inhibitors cocktail) by glass beads. Following 90min incubation with cell extracts at ambient temperature, nickel beads were washed three times in HU buffer with 1% SDS. Bound proteins were eluted by incubating the beads in one volume of 8M HU and one volume of 2M imidazole for 10min at ambient temperature with shaking. One volume of 3×SDS-PAGE Laemmli sample buffer was added and samples were heated at 70°C for 10min before western blot analysis.
In vivo cross-linking and co-immunoprecipitation experiments
Cells were grown at 25°C to an A600 of 1 and then heat shocked at 40°C or 45°C for 10min before adding 1% formaldehyde and incubating for a further 10min. The cross-linking reaction was quenched with an excess of glycine (250mM) for five minutes at 4°C. The samples were centrifuged, washed twice with 1×TBS, and then frozen in liquid nitrogen. Cells were lysed with modified RIPA buffer (50mM Tris-HCl pH7.5, 150mM NaCl, 1% Triton X-100, 0.5%SDS, 1mM PMSF, 103mM chloroacetamide, 1mM phenanthroline, and 1× protease inhibitor cocktail) using glass beads. For the TAP tagged proteins IP, Tris-HCl was replaced by 50mM HEPES at pH8. For MYC tagged proteins, 25mM Tris-HCl pH7.6, 1% NP40 were used instead of 50mM and 1% Triton X-100, respectively. Lysates were then slowly diluted to 0.1% SDS final for IP. Cell pellets were also resolubilized in lysis buffer containing 0.1% SDS at 4°C for thirty minutes and mixed with corresponding lysates, as some additional precipitations occurred in the presence of 0.5% SDS. Sample concentrations were measured by Bradford assay and normalized before incubation with beads overnight at 4°C. Beads were washed six times in lysis buffer with 500mM NaCl and eluted with 1×SDS buffer without reducing agent to avoid elution of cross-linked IgG. Reducing agent was added and samples were boiled prior to SDS-PAGE. For detecting presence of ubiquitin on immuno-precipitated proteins, cells were lysed and processed as for MYC IP (lysis in 0.5% SDS before dilution to 0.1% for IP), but without protein cross-linking. IgG coupled Dynal (Invitrogen), anti-MYC (9E10; OriGene) and anti-HA (Pierce) magnetic beads were used for TAP, MYC and HA co-IP experiments, respectively.
Computational analyses of Rsp5 candidate substrates
Localization of Rsp5 candidate substrates was assigned according to a previous systems-wide localization analysis60. Candidate Rsp5 substrates induced by heat-shock (combining both proteomic experiments; 112 proteins) and high-confidence Rsp5 substrates identified in a protein array (41 proteins)61 were used to search for PY motifs ([PLSV]Px[YF], and PPPP) or PXY using an in-house python script. Enrichment of a given motif among Rsp5 candidates relative to its prevalence in the whole proteome was tested using Fisher’s exact test. We determined whether PY motifs ([PLSV]Px[YF], and PPPP) are located in the disordered regions using Disopred262, 63. Solvent accessibility of motifs was predicted using Sable64, 65 for the four individual residues in the PY motifs and was then averaged. The Ydj1 binding motif was previously defined as GX[LMQ]{P}X{P}{CIMPVW}66. Fisher’s exact test was used to determine if proteins containing at least one, two or three Ydj1 motifs were enriched in the HS dataset compared to the genome. To identify which Cdc19 residue mutation may enhance ubiquitination, X-ray crystallography structure (1A3W67) was repaired using the repairPDB function of FoldX. Using the BuildModel function of FoldX, residue D367 was mutated to Arg. The interaction energies of the wild-type structure and of the mutant were measured using the AnalyseComplex function of FoldX. A similar approach was used to assess other residues located in the binding interface; the D367R mutation was selected as it is near the PY motif and was predicted to more strongly affect the homo-dimer interaction compared to other mutations (ranked 5th among 45 mutations assessed). We verified that the addition of a PY motif on Pyk2 and the mutation destabilizing the Cdc19 dimer were not predicted to destabilize the structure of Pyk2 and Cdc19, respectively, by using the Stability function of Foldx. For Pyk2, the Modbase-generated homology model68 was used.
Datasets
All raw proteomics data have been deposited to the ProteomeXchange Consortium69 via the PRIDE partner repository with the identifier PXD001214.
Supplementary Material
Acknowledgments
We thank Dr. N. Stoynov for his support for the mass spectrometry analyses, all colleagues cited in Methods who provided reagents and a previous anonymous reviewer who suggested to investigate Rsp5. This work was supported by a CIHR grant. RJD is an HHMI investigator and TM is CIHR and MSFHR new investigator.
Footnotes
Contributions
N.N.F. designed most of the experiments through discussions with T.M. and additional inputs from J.G., R.J.D. and D.R.; N.N.F. carried out most experiments; G.T.C. carried out the computational analyses with additional participation from J.G.; M.Z. and S.A.C. prepared several plasmids and strains; A.P. and N.N.F carried out the mammalian cells experiments together. D.R. provided reagents. Development of the conjugate assay was initiated in R.J.D.’s lab. T.M. supervised the study. N.N.F. and T.M. wrote the paper and all other authors edited or commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 2.Powers ET, Balch WE. Diversity in the origins of proteostasis networks--a driver for protein function in evolution. Nat Rev Mol Cell Biol. 2013;14:237–248. doi: 10.1038/nrm3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol. 2011;3:a004374. doi: 10.1101/cshperspect.a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comyn SA, Chan GT, Mayor T. False start: Cotranslational protein ubiquitination and cytosolic protein quality control. J Proteomics. 2013 doi: 10.1016/j.jprot.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vabulas RM, Raychaudhuri S, Hayer-Hartl M, Hartl FU. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb Perspect Biol. 2010;2:a004390. doi: 10.1101/cshperspect.a004390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morimoto RI. The heat shock response: systems biology of proteotoxic stress in aging and disease. Cold Spring Harb Symp Quant Biol. 2011;76:91–99. doi: 10.1101/sqb.2012.76.010637. [DOI] [PubMed] [Google Scholar]
- 8.Carlson N, Rogers S, Rechsteiner M. Microinjection of ubiquitin: changes in protein degradation in HeLa cells subjected to heat-shock. J Cell Biol. 1987;104:547–555. doi: 10.1083/jcb.104.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parag HA, Raboy B, Kulka RG. Effect of heat shock on protein degradation in mammalian cells: involvement of the ubiquitin system. EMBO J. 1987;6:55–61. doi: 10.1002/j.1460-2075.1987.tb04718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medicherla B, Goldberg AL. Heat shock and oxygen radicals stimulate ubiquitin-dependent degradation mainly of newly synthesized proteins. J Cell Biol. 2008;182:663–673. doi: 10.1083/jcb.200803022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011;13:1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galan JM, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 13.Huibregtse JM, Yang JC, Beaudenon SL. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoppe T, et al. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Macgurn JA, Liu M, Emr S. The ART-Rsp5 ubiquitin ligase network comprises a plasma membrane quality control system that protects yeast cells from proteotoxic stress. Elife. 2013;2:e00459. doi: 10.7554/eLife.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keener JM, Babst M. Quality control and substrate-dependent downregulation of the nutrient transporter Fur4. Traffic. 2013;14:412–427. doi: 10.1111/tra.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crapeau M, Merhi A, Andre B. Stress Conditions Promote Yeast Gap1 Permease Ubiquitylation and Downregulation via the Arrestin-like Bul and Aly Proteins. J Biol Chem. 2014 doi: 10.1074/jbc.M114.582320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shahsavarani H, Sugiyama M, Kaneko Y, Chuenchit B, Harashima S. Superior thermotolerance of Saccharomyces cerevisiae for efficient bioethanol fermentation can be achieved by overexpression of RSP5 ubiquitin ligase. Biotechnol Adv. 2012;30:1289–1300. doi: 10.1016/j.biotechadv.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Wang G, Yang J, Huibregtse JM. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krsmanovic T, Kolling R. The HECT E3 ubiquitin ligase Rsp5 is important for ubiquitin homeostasis in yeast. FEBS Lett. 2004;577:215–219. doi: 10.1016/j.febslet.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Haitani Y, Takagi H. Rsp5 is required for the nuclear export of mRNA of HSF1 and MSN2/4 under stress conditions in Saccharomyces cerevisiae. Genes Cells. 2008;13:105–116. doi: 10.1111/j.1365-2443.2007.01154.x. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez MS, Gwizdek C, Haguenauer-Tsapis R, Dargemont C. The HECT ubiquitin ligase Rsp5p is required for proper nuclear export of mRNA in Saccharomyces cerevisiae. Traffic. 2003;4:566–575. doi: 10.1034/j.1600-0854.2003.00115.x. [DOI] [PubMed] [Google Scholar]
- 23.Zarrinpar A, Lim WA. Converging on proline: the mechanism of WW domain peptide recognition. Nat Struct Biol. 2000;7:611–613. doi: 10.1038/77891. [DOI] [PubMed] [Google Scholar]
- 24.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111.013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunn R, Klos DA, Adler AS, Hicke L. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J Cell Biol. 2004;165:135–144. doi: 10.1083/jcb.200309026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crosas B, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 28.Aviram S, Kornitzer D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol. 2010;30:985–994. doi: 10.1128/MCB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee DH, Sherman MY, Goldberg AL. Involvement of the molecular chaperone Ydj1 in the ubiquitin-dependent degradation of short-lived and abnormal proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4773–4781. doi: 10.1128/mcb.16.9.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta R, et al. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol. 2007;3:116. doi: 10.1038/msb4100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 34.Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol. 2009;29:3307–3318. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park SH, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 36.Summers DW, Wolfe KJ, Ren HY, Cyr DM. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 2013;8:e52099. doi: 10.1371/journal.pone.0052099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tofaris GK, et al. Ubiquitin ligase Nedd4 promotes alpha-synuclein degradation by the endosomal-lysosomal pathway. Proc Natl Acad Sci U S A. 2011;108:17004–17009. doi: 10.1073/pnas.1109356108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tardiff DF, et al. Yeast Reveal a “Druggable” Rsp5/Nedd4 Network that Ameliorates α-Synuclein Toxicity in Neurons. Science. 2013 doi: 10.1126/science.1245321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu K, Psakhye I, Jentsch S. Autophagic Clearance of PolyQ Proteins Mediated by Ubiquitin-Atg8 Adaptors of the Conserved CUET Protein Family. Cell. 2014;158:549–563. doi: 10.1016/j.cell.2014.05.048. [DOI] [PubMed] [Google Scholar]
- 40.Li Z, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mnaimneh S, et al. Exploration of essential gene functions via titratable promoter alleles. Cell. 2004;118:31–44. doi: 10.1016/j.cell.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Nikko E, Pelham HR. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic. 2009;10:1856–1867. doi: 10.1111/j.1600-0854.2009.00990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 44.Saeki Y, Isono E, Toh-E A. Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 2005;399:215–227. doi: 10.1016/S0076-6879(05)99014-9. [DOI] [PubMed] [Google Scholar]
- 45.Gajewska B, et al. WW domains of Rsp5p define different functions: determination of roles in fluid phase and uracil permease endocytosis in Saccharomyces cerevisiae. Genetics. 2001;157:91–101. doi: 10.1093/genetics/157.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Persaud A, et al. Comparison of substrate specificity of the ubiquitin ligases Nedd4 and Nedd4–2 using proteome arrays. Mol Syst Biol. 2009;5:333. doi: 10.1038/msb.2009.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 48.Miyazaki K. MEGAWHOP cloning: a method of creating random mutagenesis libraries via megaprimer PCR of whole plasmids. Methods Enzymol. 2011;498:399–406. doi: 10.1016/B978-0-12-385120-8.00017-6. [DOI] [PubMed] [Google Scholar]
- 49.Persaud A, et al. Tyrosine phosphorylation of NEDD4 activates its ubiquitin ligase activity. Sci Signal. 2014 doi: 10.1126/scisignal.2005290. In press. [DOI] [PubMed] [Google Scholar]
- 50.Fouladkou F, et al. The ubiquitin ligase Nedd4–1 is dispensable for the regulation of PTEN stability and localization. Proc Natl Acad Sci U S A. 2008;105:8585–8590. doi: 10.1073/pnas.0803233105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller MJ, Xuong NH, Geiduschek EP. A response of protein synthesis to temperature shift in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979;76:5222–5225. doi: 10.1073/pnas.76.10.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ng AH, Fang NN, Comyn SA, Gsponer J, Mayor T. System-wide analysis reveals intrinsically disordered proteins are prone to ubiquitylation after misfolding stress. Mol Cell Proteomics. 2013;12:2456–2467. doi: 10.1074/mcp.M112.023416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Udeshi ND, et al. Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics. 2013;12:825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 55.Kristensen AR, Gsponer J, Foster LJ. A high-throughput approach for measuring temporal changes in the interactome. Nat Methods. 2012;9:907–909. doi: 10.1038/nmeth.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 57.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111.013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007;6:1885–1895. doi: 10.1074/mcp.M700264-MCP200. [DOI] [PubMed] [Google Scholar]
- 60.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 61.Gupta R, et al. Ubiquitination screen using protein microarrays for comprehensive identification of Rsp5 substrates in yeast. Mol Syst Biol. 2007;3:116. doi: 10.1038/msb4100159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 63.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–8. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner M, Adamczak R, Porollo A, Meller J. Linear regression models for solvent accessibility prediction in proteins. J Comput Biol. 2005;12:355–369. doi: 10.1089/cmb.2005.12.355. [DOI] [PubMed] [Google Scholar]
- 65.Adamczak R, Porollo A, Meller J. Combining prediction of secondary structure and solvent accessibility in proteins. Proteins. 2005;59:467–475. doi: 10.1002/prot.20441. [DOI] [PubMed] [Google Scholar]
- 66.Kota P, Summers DW, Ren HY, Cyr DM, Dokholyan NV. Identification of a consensus motif in substrates bound by a Type I Hsp40. Proc Natl Acad Sci U S A. 2009;106:11073–11078. doi: 10.1073/pnas.0900746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jurica MS, et al. The allosteric regulation of pyruvate kinase by fructose-1,6-bisphosphate. Structure. 1998;6:195–210. doi: 10.1016/s0969-2126(98)00021-5. [DOI] [PubMed] [Google Scholar]
- 68.Pieper U, et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2011;39:D465–74. doi: 10.1093/nar/gkq1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vizcaino JA, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.