Abstract
Axonal and synaptic degeneration is a hallmark of peripheral neuropathy, brain injury, and neurodegenerative disease. Axonal degeneration has been proposed to be mediated by an active autodestruction program, akin to apoptotic cell death; however, loss-of-function mutations capable of potently blocking axon self-destruction have not been described. Here, we show that loss of the Drosophila Toll receptor adaptor dSarm (sterile α/Armadillo/Toll-Interleukin receptor homology domain protein) cell-autonomously suppresses Wallerian degeneration for weeks after axotomy. Severed mouse Sarm1 null axons exhibit remarkable long-term survival both in vivo and in vitro, indicating that Sarm1 prodegenerative signaling is conserved in mammals. Our results provide direct evidence that axons actively promote their own destruction after injury and identify dSarm/Sarm1 as a member of an ancient axon death signaling pathway.
When axons are severed, the portion of the axon distal to the injury site undergoes extensive fragmentation. This process, termed Wallerian degeneration, was traditionally thought to result from passive wasting away of axons due to a lack of cell body–derived nutrients (1). However, this notion was challenged by the slow Wallerian degeneration (WldS) mutant mouse in which the distal portion of severed axons remained morphologically intact for weeks after axotomy (2, 3). The long-term survival of severed WldS+ axons raised the intriguing possibility that Wallerian degeneration might be driven by an active molecular program akin to apoptotic cell death (4, 5). However, the WldS phenotype results from a gain of function, likely neomorphic mutation that leads to neuronal overexpression of a chimeric fusion protein containing the nicotinamide adenine dinucleotide (NAD+) biosynthetic enzyme Nmnat1 (6, 7). Despite its ability to inhibit axonal degeneration, the gain-of-function nature of the WldS phenotype does not provide direct evidence supporting the existence of an axon death signaling pathway and may be unrelated to normal Nmnat1 function. Importantly, mutants reported to suppress Wallerian degeneration, such as wnd/DLK, delay the clearance of degenerating axons in Drosophila for only ~1 to 2 days, and mouse axons for several hours (8). This is quite weak suppression when compared with WldS. Thus, the existence of axon death pathways in Wallerian degeneration has remained largely speculative.
Wallerian degeneration appears to be molecularly distinct from apoptosis, because potent genetic or chemical inhibitors of cell death do not block axonal disintegration (9–11). We revisited this question and performed a comprehensive screen of existing mutants and dominant negative constructs for Drosophila genes affecting apoptosis, autophagy, or other defined cell degradative pathways, but these failed to suppress Wallerian degeneration (table S1).
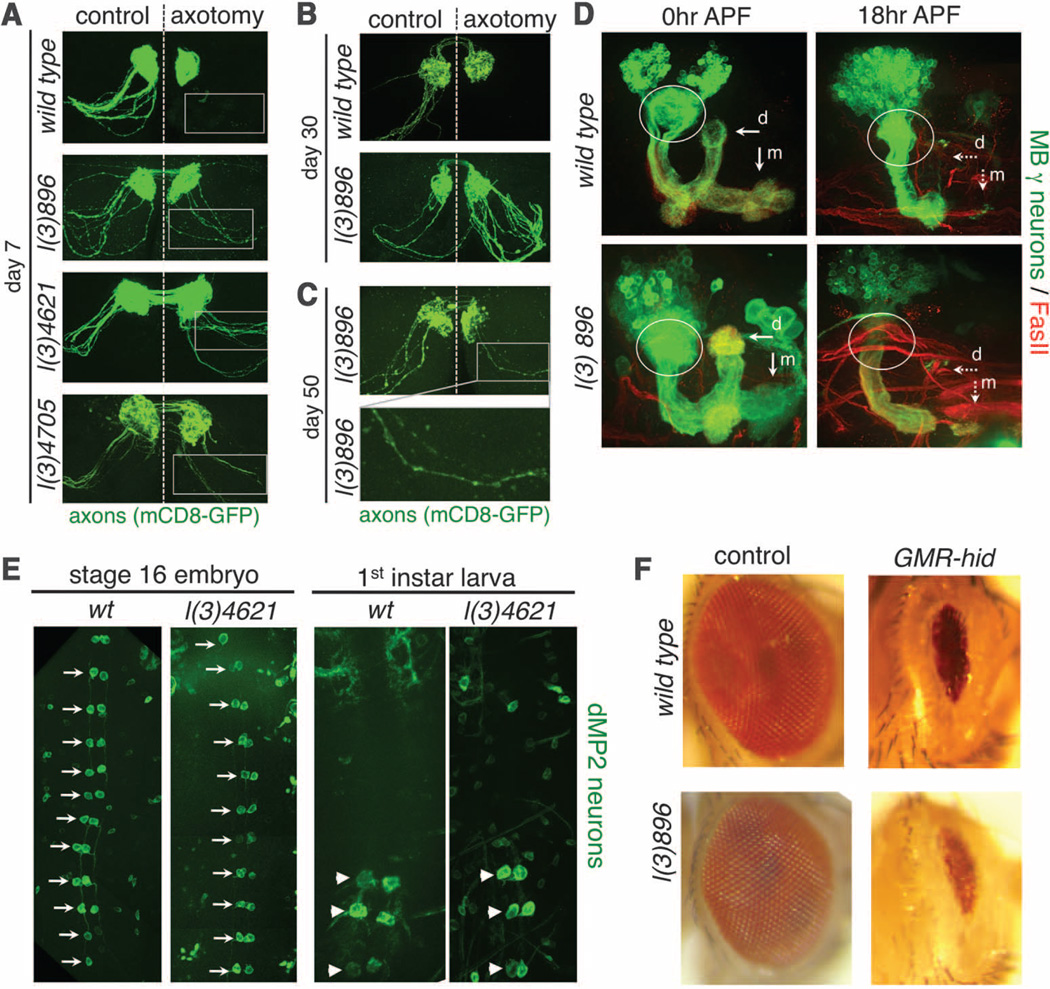
If Wallerian degeneration is indeed an active process, then loss-of-function mutants that exhibit-WldS-like protection of severed axons should exist. We therefore performed an F2 forward genetic screen in Drosophila for mutants that exhibited long-term survival of severed axons (fig. S1). Because genes required for Wallerian degeneration may be lethal when mutated, we designed our screen to allow for characterization of both viable and lethal mutants through mosaic analysis with a repressible cell marker (MARCM) clonal analysis (12). In control animals, severed olfactory receptor neuron (ORN) axons degenerated and were completely cleared from the antennal lobe 7 days after axotomy. We identified three lines, l(3)896, l(3)4621, and l(3)4705, in which severed homozygous mutant axons generated by MARCM remained intact 1 week after axotomy (Fig. 1A). Although the number of uninjured axons is slightly reduced in each mutant, 100% of green fluorescent protein (GFP)–labeled axons exhibited long-term preservation after injury (table S2). Mutant axons remained fully intact 30 days after injury (Fig. 1B), and a significant but reduced number remained even 50 days after injury (Fig. 1C). l(3)896, l(3)4621, and l(3)4705 therefore provide axonal preservation that rivals that of WldS in Drosophila, and lasts throughout the life span of the fly. Neuroprotection in these mutants extended to synapses: Synaptobrevin punctae localized to synaptic terminals even 30 days after axotomy (fig. S2, D and E). Neuronal morphology appeared normal in mutant animals (Fig. 1 and fig. S2B), indicating that these mutations do not grossly affect neuronal development.
Fig. 1.
Identification of mutations that suppress Wallerian degeneration. (A) ORN MARCM clones in control, l(3)896, l(3)4621, and l(3)4705. Right, axotomy; left, uninjured control. n ≥ 15 brains. (B) Control and l(3)896 brains 30 days after injury. n ≥ 10 brains. (C) l(3)896 clones 50 days after injury. n = 11 brains. (D) MARCM clones in mushroom body (MB) γ neurons in control and l(3)896 backgrounds at the indicated developmental stages. dorsal (d) and medial (m) axonal branches (arrows), and dendrites (circled). n ≥ 15 brains for all. (E) dMP2 neurons with GFP. Ventral views (anterior up) of stage 16 embryos (left) and first-instar larvae. dMP2 neurons before (arrows) and after (arrowheads) segment-specific apoptosis. n ≥ 20 flies at each time point. (F) Wild-type and l(3)896 mutant clones with (right) or without (left) ectopic expression of hid. n ≥ 20 flies.
We next asked whether l(3)896 was broadly required for neuron pruning or apoptotic cell death. We examined dendritic and axonal pruning in MARCM clones in Drosophila mushroom body γ neurons during metamorphosis (13). In both control and l(3)896 animals, mushroom body γ neuron axons and dendrites were pruned normally (Fig. 1D). During early embryogenesis, dMP2 neurons are present in each segment, but by late embryogenesis, all but the posterior three pairs undergo developmentally programmed apoptosis (14). We found that dMP2 neurons were generated normally in l(3)4621 animals, and the appropriate subset of neurons underwent apoptosis (Fig. 1E). Finally, we expressed the proapoptotic gene hid in the Drosophila visual system (15) to induce widespread apoptotic death in cells of the developing eye disc. We found that l(3)896 mutant clones failed to suppress activation of cell death (Fig. 1F).
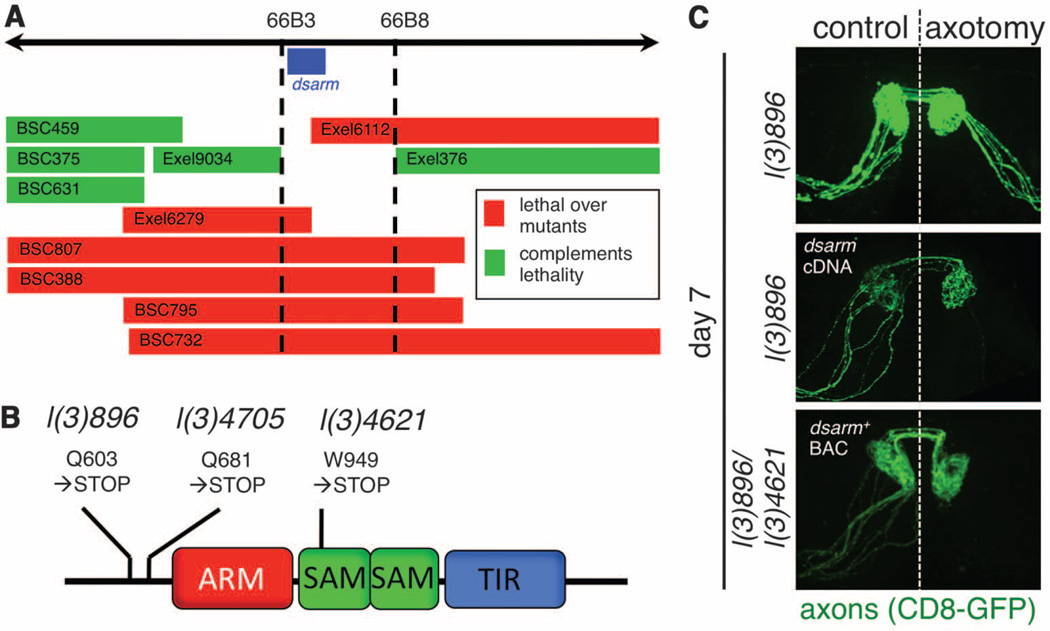
These mutants were all recessive (fig. S2A) in axon degenerative phenotypes and fell into a single lethal complementation group; therefore, each line represented an independently isolated lethal mutation in the same gene. To identify the gene mutated in l(3)896, l(3)4621, and l(3)4705, we took two complementary approaches. First, we mapped the lethality of each mutant using small chromosome deficiencies and identified a single gene at cytogenetic region 66B (Fig. 2A). We also used next-generation sequencing technology to resequence mutant genomes to a read depth of 70× (table S3). We identified a single gene affected in all three mutants, which resided in cytogenetic region 66B: ect4, which we refer to as dsarm (Drosophila sterile alpha and Armadillo motif). The dsarm gene encodes a protein with an Armadillo/HEAT (ARM) domain, two sterile alpha motifs (SAM), and a Toll/interleukin-1 receptor homology (TIR) domain. Each identified dsarm allele contained a unique premature stop codon in dsarm open reading frame (Fig. 2B and table S4). From these data we conclude that l(3)896, l(3)4621, and l(3)4705 are loss-of-function alleles of dsarm. Consistent with this interpretation, expression of full-length dsarm cDNA using the postmitotic OR22a-Gal4 driver in l(3)896 mutant clones was sufficient to fully revert the suppression of axonal degeneration observed in dsarm mutants (Fig. 2C). In addition, we rescued both the lethality and suppression of Wallerian degeneration phenotypes of l(3)896/l(3)4621 and l(3)896/Df(3L)BSC795 trans-heterozygous animals with a BAC clone containing the dsarm gene (fig. S2C). Together, these data indicate that dSarm function is necessary in postmitotic neurons to drive axonal destruction after axotomy.
Fig. 2.
Mutations in dsarm block Wallerian degeneration. (A) The lethality of l(3)896, l(3)4621, and l(3)4705 was mapped to region 66B using Exel or Bloomington Stock Center deficiencies. (B) Dsarm protein domains, positions, and effect of predicted point mutations. (C) UAS-dsarm in l(3)896 mutant clones or dsarm+ bacterial artificial chromosome rescue axonal degeneration defects in l(3)896/l(3)4621 flies. n = 12 flies.
Based on RNA in situ hybridizations to embryos, larval brains, and adult brains, reverse transcription polymerase chain reaction from dissected neural tissues, and analysis of a dsarm-Gal4 driver line, dsarm is widely expressed in the Drosophila nervous system (fig. S3, A to E). These data raise the possibility that dSarm may be broadly required to promote Wallerian degeneration in the nervous system.
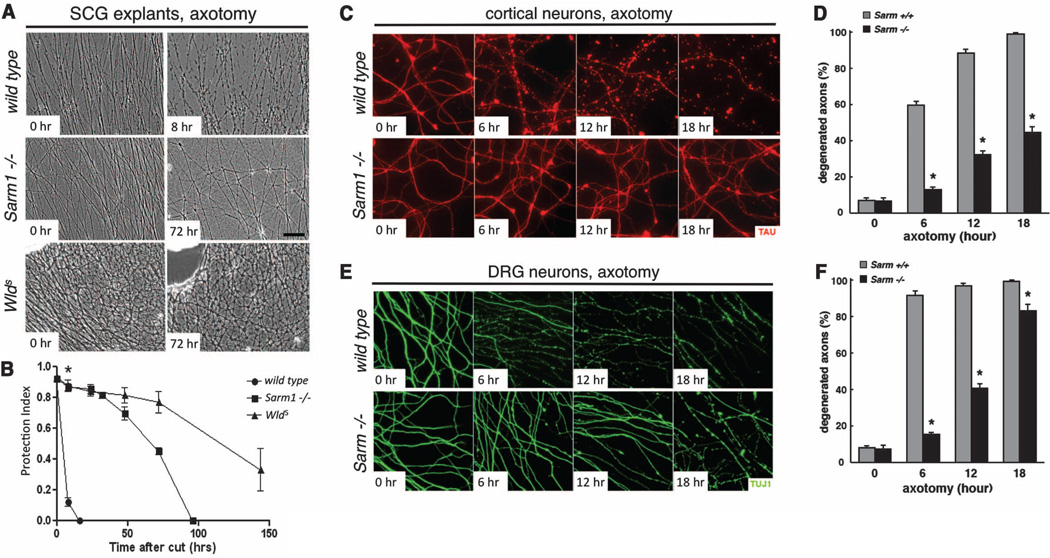
We next assayed Wallerian degeneration in null mutants for the mouse ortholog of dsarm, Sarm1. We grew 5-day cultures of superior cervical ganglia (SCG) from wild-type, Sarm1−/− (16) and WldS mice, severed axons, and scored axonal integrity over the next week. Sarm1−/− SCG explants exhibited robust protection from degeneration up to 72 hours after axotomy, similar to what is observed with WldS SCG neurons, whereas wild-type axons degenerated within 8 hours (Fig. 3, A and B). This robust protection was also seen in cultured Sarm1−/− cortical neuron axons (Fig. 3, C and D, and fig. S5C) and dorsal root ganglia (DRG) (Fig. 3, E and F, and fig. S5D). Notably, Sarm1−/− DRG explants were not protected from nerve growth factor (NGF) deprivation (fig. S5, A and B), a model for developmental axon pruning (4, 13, 17, 18), suggesting that Sarm1 protection is specific to injury-induced axon degeneration.
Fig. 3.
Sarm1−/− neurons in primary culture are protected from Wallerian degeneration. (A) Phase contrast images of SCG explant cultures from wild-type (top), Sarm−/− (middle), and WldS-expressing (bottom) animals at the indicated time after axotomy. (B) Quantification from (A). Mean T SEM, *P < 0.01. (C) Axon preservation at the indicated time points in cortical neuron cultures from E16.5 mouse embryos. α-Tau, red. (D) Quantification from (C). Mean T SEM, *P < 0.01. (E) Axon preservation at the indicated time points in DRG cultures from E13.5 mouse embryos. α-TUJI, green. (F) Quantification from (E). Mean T SEM, *P < 0.01.
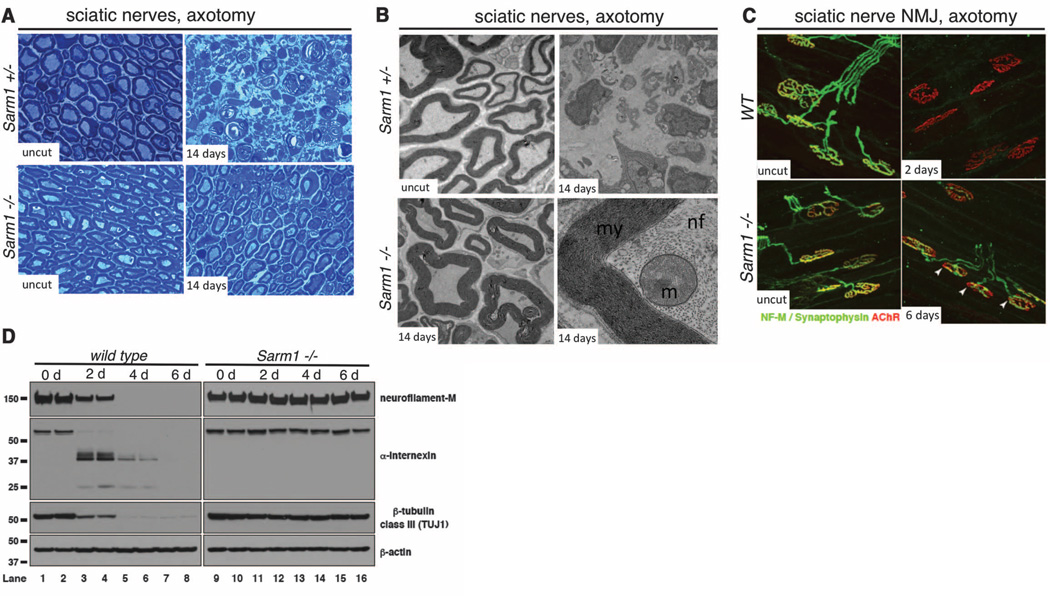
To determine whether Sarm1 is required for Wallerian degeneration in vivo, we performed sciatic nerve lesions of the right hind limb of Sarm1−/− mice and controls. Whereas Sarm1+/− controls exhibited a dramatic breakdown of the axon and myelin sheath within 3 days of injury, 61.2% of lesioned Sarm1−/− axons were protected from degeneration at least 14 days after injury (P = 0.002) (Fig. 4A). Ultrastructural analysis revealed a remarkable preservation of the Schwann cell and myelin sheath, axonal neurofilaments, and axonal mitochondria at this time point (Fig. 4B and fig. S4). Western blots of sciatic nerve with antibodies to neurofilament-M (NF-M), α-Internexin, and β-tubulin class III (TUJI) confirmed that the axonal cytoskeleton was robustly preserved in severed Sarm1−/− axons (Fig. 4D). We further note preservation of the NF-M signal by immunofluorescent staining nerves in Sarm1−/− mutants (fig. S6, A and E).
Fig. 4.
Sarm1 is required for Wallerian degeneration in mice in vivo. (A) Sciatic nerve distal to the injury site stained with Toludine blue. Time points and genotypes as indicated. (B) Ultrastructural analysis of Sarm+/− and Sarm−/− axons before or 14 days after axotomy. Higher magnification image (lower right) is shown in its entirety in fig. S4. my, myelin; nf, neurofilaments; m, mitochondrion. (C) Neuromuscular junction (NMJ) preservation at tibialis anterior muscles. red, AChR (postsynapse/muscle); green, NF-M/synpatophysin (presynapse). Arrows, motor end plates. (D) Immunoblot analysis of distal injured nerve segment. n = 4 mice at each time point and genotype.
Synaptic integrity was scored by colocalization of presynaptic marker (NF-M/synaptophysin) with the postsynaptic acetylcholine receptor (AChR) in tibialis anterior muscles after sciatic nerve transection. In wild-type animals, motor end plate denervation was complete by 2 days after axotomy. However, in Sarm1−/− animals, most synaptic terminals were partially innervated even at 6 days after injury (Fig. 4C and fig. S5D). Consistent with a strong preservation of nerve integrity, macrophage/monocyte infiltration of lesioned nerves was suppressed in Sarm1 knockout animals (fig. S6, B, C, and F). Taken together, our results indicate that Sarm1−/− mutations strongly preserve sciatic nerves in vivo from initial axonal cytoskeletal breakdown to recruitment of macrophages for myelin clearance.
To help define potential sites of action, we assayed localization of dSarm and Sarm1 in neurons. Expression of dSarm::GFP in Drosophila larval neurons resulted in punctate localization in neuronal cell bodies and broad localization to neurites in vivo (fig. S7, A and B). Immunostaining with antibodies to Sarm1 of in vitro cultured mouse neurons showed a similar pattern, and we note that endogenous Sarm1 did not show preferential localization with a mitochondrial marker (fig. S7, C and D), although mitochondrial localization has been reported for overexpressed Sarm1::GFP (16). Thus, dSarm/Sarm1 appears to be broadly localized in the axonal compartment.
Our analysis of dSarm/Sarm1 provides evidence that Wallerian degeneration is driven by an ancient, conserved axonal death program. Intriguingly, its Caenorhabditis elegans homolog, Tir-1, was recently implicated in nonapoptotic cell death (19), suggesting mechanistic links between these nonapoptotic degenerative mechanisms. Influx of extracellular calcium is known to be necessary and sufficient to trigger Wallerian degeneration (20). Tir-1 functions downstream of the Ca2+-CaM kinase signaling pathway in C. elegans (21, 22); therefore, dSarm/Sarm1 may respond directly to axonal calcium increases after axotomy. Sarm1−/− neurons in slice cultures exhibit reduced cell death in response to oxygen/glucose deprivation (18); whether this is an indirect result or reflects axonal preservation remains untested. Pharmacological inhibition of Sarm1 may represent a promising therapeutic avenue for patients with axonal loss, particularly if Sarm1 deletion is shown to benefit animal models of neurodegenerative disease.
Supplementary Material
Acknowledgments
We thank J. Gilley for preliminary work with Sarm1 primary cultures and Y. Hsueh for the Sarm1 antibody. We thank J. Patrick, D. Cameron, and D. Weaver for assistance with mouse work. This work was supported by the U.S. Department of Defense grant BC093796 (to T.M.R.); National Institute for Neurological Diseases and Stroke (NINDS) grants R01NS072248 and U54NS065712 (to S.Z.); an Alzheimer’s Research UK Grant ART/PG2009/2, a Biotechnology and Biological Sciences Research Council Institute Strategic Programme Grant (to M.C.); NIH grant AI030165 to A.D.; support from Rockefeller University (J.Y. and M.T.-L.); and NINDS grant R01NS059991 and the ALS Therapy Alliance (to M.R.F.). M.R.F. is an Early Career Scientist with the Howard Hughes Medical Institute. M.R.F. and S.Z. are inventors on a patent applied for by University of Massachusetts Medical School, University of Miami, and the Howard Hughes Medical Institute covering therapeutic applications that target Sarm1 to block axonal and synaptic degeneration. R.H.B. was supported by NINDS5RO1-NS050557-05, NINDS ARRA Award RC2-NS070-342, the Angel Fund, the ALS Association, P2ALS, Project ALS, Pierre L. de Bourgknecht ALS Research Foundation, and the ALS Therapy Alliance. R.H.B. is cofounder of AviTx, an ALS research company, and serves as a consultant to BiogenIdec and Iperion, is President of the ALS Therapy Alliance, and serves on the advisory boards of Project ALS, P2ALS, and the Angel Fund for ALS research.
Footnotes
www.sciencemag.org/cgi/content/full/science.1223899/DC1
Materials and Methods
Figs. S1 to S7
Tables S1 to S4
References (23–31)
References and Notes
- 1.Waller A. Philos. Trans. R. Soc. London Ser. B Biol. Sci. 1850;140:423. [Google Scholar]
- 2.Lunn ER, Perry VH, Brown MC, Rosen H, Gordon S. Eur. J. Neurosci. 1989;1:27. doi: 10.1111/j.1460-9568.1989.tb00771.x. [DOI] [PubMed] [Google Scholar]
- 3.Glass JD, Brushart TM, George EB, Griffin JW. J Neurocytol. 1993;22:311. doi: 10.1007/BF01195555. [DOI] [PubMed] [Google Scholar]
- 4.Raff MC, Whitmore AV, Finn JT. Science. 2002;296:868. doi: 10.1126/science.1068613. [DOI] [PubMed] [Google Scholar]
- 5.Coleman MP, Perry VH. Trends Neurosci. 2002;25:532. doi: 10.1016/s0166-2236(02)02255-5. [DOI] [PubMed] [Google Scholar]
- 6.Mack TG, et al. Nat. Neurosci. 2001;4:1199. doi: 10.1038/nn770. [DOI] [PubMed] [Google Scholar]
- 7.Coleman MP, Freeman MR. Annu. Rev. Neurosci. 2010;33:245. doi: 10.1146/annurev-neuro-060909-153248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller BR, et al. Nat. Neurosci. 2009;12:387. doi: 10.1038/nn.2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deckwerth TL, Johnson EM., Jr Dev. Biol. 1994;165:63. doi: 10.1006/dbio.1994.1234. [DOI] [PubMed] [Google Scholar]
- 10.Finn JT, et al. J Neurosci. 2000;20:1333. doi: 10.1523/JNEUROSCI.20-04-01333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitmore AV, Lindsten T, Raff MC, Thompson CB. Cell Death Differ. 2003;10:260. doi: 10.1038/sj.cdd.4401147. [DOI] [PubMed] [Google Scholar]
- 12.Lee T, Luo L. Trends Neurosci. 2001;24:251. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- 13.Luo L, O’Leary DD. Annu. Rev. Neurosci. 2005;28:127. doi: 10.1146/annurev.neuro.28.061604.135632. [DOI] [PubMed] [Google Scholar]
- 14.Miguel-Aliaga I, Thor S. Development. 2004;131:6093. doi: 10.1242/dev.01521. [DOI] [PubMed] [Google Scholar]
- 15.Grether ME, Abrams JM, Agapite J, White K, Steller H. Genes Dev. 1995;9:1694. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y, et al. J Exp. Med. 2007;204:2063. doi: 10.1084/jem.20070868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacInnis BL, Campenot RB. Mol. Cell. Neurosci. 2005;28:430. doi: 10.1016/j.mcn.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Buss RR, Sun W, Oppenheim RW. Annu. Rev. Neurosci. 2006;29:1. doi: 10.1146/annurev.neuro.29.051605.112800. [DOI] [PubMed] [Google Scholar]
- 19.Blum ES, Abraham MC, Yoshimura S, Lu Y, Shaham S. Science. 2012;335:970. doi: 10.1126/science.1215156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.George EB, Glass JD, Griffin JW. J Neurosci. 1995;15:6445. doi: 10.1523/JNEUROSCI.15-10-06445.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chuang CF, Bargmann CI. Genes Dev. 2005;19:270. doi: 10.1101/gad.1276505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang C, Hsieh YW, Lesch BJ, Bargmann CI, Chuang CF. Development. 2011;138:3509. doi: 10.1242/dev.069740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.