Abstract
Many intracellular pathogens are separated from the cytosol of their host cells by a vacuole membrane. This membrane serves as a critical interface between the pathogen and the host cell, across which nutrients are imported, wastes are excreted, and communication between the two cells takes place. Very little is known about the vacuole membrane proteins mediating these processes in any host-pathogen interaction. During a screen for monoclonal antibodies against novel surface or secreted proteins of Toxoplasma gondii, we identified ROP4, a previously uncharacterized member of the ROP2 family of proteins. We report here on the sequence, posttranslational processing, and subcellular localization of ROP4, a type I transmembrane protein. Mature, processed ROP4 is localized to the rhoptries, secretory organelles at the apical end of the parasite, and is secreted from the parasite during host cell invasion. Released ROP4 associates with the vacuole membrane and becomes phosphorylated in the infected cell. Similar results are seen with ROP2. Further analysis of ROP4 showed it to be phosphorylated on multiple sites, a subset of which result from the action of either host cell protein kinase(s) or parasite kinase(s) activated by host cell factors. The localization and posttranslational modification of ROP4 and other members of the ROP2 family of proteins within the infected cell make them well situated to play important roles in vacuole membrane function.
Toxoplasma gondii is an obligate intracellular parasite capable of invading nearly any nucleated cell within its vertebrate hosts. The process of host cell invasion is similar in all apicomplexan parasites, including T. gondii, Plasmodium spp. (causative agents of malaria), Eimeria tenella, and Cryptosporidium parvum. Internalized parasites undergo several rounds of replication within the parasitophorous vacuole (PV) and ultimately lyse the host cell. Repeated cycles of host cell invasion and lysis are directly responsible for the disease caused by T. gondii, which is particularly severe in immunocompromised individuals and in the congenitally infected fetus (19, 23, 33).
As with all apicomplexan parasites, a central feature of host cell invasion by T. gondii tachyzoites is the sequential release of proteins from apical secretory organelles. The tachyzoite secretes proteins first from the micronemes, then from the rhoptries, as it pulls itself into the host cell using a myosin-based motor complex (9, 12, 24). Proteins released from the micronemes include a variety of adhesins, which are thought to directly mediate binding to ligands on the host cell surface (reviewed in references 43 and 46). Rhoptry proteins are released concurrent with the formation of the PV and are thought to contribute to both the formation and functional properties of the PV membrane (PVM) (3, 9, 12, 41).
Host cell invasion by T. gondii tachyzoites is inhibited by cytochalasin D, while host cell attachment and rhoptry secretion remain unaffected (9, 14). Under these conditions, the contents of the rhoptries are directly injected into the host cell cytoplasm in the form of protein-rich vesicles called evacuoles (14). Like the PV itself, evacuoles avoid fusion with host cell endosomal and lysosomal compartments, and they associate with host cell mitochondria and the endoplasmic reticulum, suggesting a role for rhoptry proteins in one or more of these processes (14).
Of the nine rhoptry proteins identified with available antibodies, only ROP1 and ROP2 have been studied in any detail. ROP1 was initially implicated in invasion as an active component of the penetration-enhancing factor, a poorly characterized cellular fraction that increases the efficiency of host cell invasion by T. gondii (30, 31). Subsequent gene knockout studies revealed that ROP1 is not essential for either invasion or intracellular survival (21). Although its function remains unknown, ROP1 has been a useful model for studying rhoptry protein trafficking and processing (4-6, 44).
The ROP2 family of proteins (ROP2, ROP3/ROP8, and ROP4) was originally identified by cross-reacting monoclonal antibodies (MAbs) produced against a rhoptry-enriched fraction of T. gondii tachyzoites (36). Later studies demonstrated that the founding member of this family, ROP2, inserts into the PVM (2), where it mediates an association between the PV and host cell mitochondria (41). A recent study using antisense RNA suggested that ROP2 is important not only for the interaction between the PVM and host cell organelles but also for rhoptry biogenesis, parasite invasion, and intracellular replication (26).
In the course of a MAb screen for novel secretory or surface proteins of the T. gondii tachyzoite (47), we generated a MAb against a 60-kDa protein that we identify here as ROP4, a previously uncharacterized member of the ROP2 family of proteins. ROP4 is secreted from the rhoptries during or shortly after invasion and associates with the PVM, presumably through its predicted transmembrane domain. ROP4 becomes phosphorylated in the infected cell, by either host cell protein kinase(s) or parasite kinase(s) activated by host cell factors.
MATERIALS AND METHODS
Parasite culture.
T. gondii tachyzoites, RH(EP) strain unless otherwise noted, were cultured in human foreskin fibroblasts (HFFs) (CRL1634; American Type Culture Collection, Manassas, Va.) as previously described (35). Parasites were harvested from freshly lysed HFF monolayers and filtered through 3-μm-pore-size Nuclepore syringe filters (Whatman, Clifton, N.J.) to remove host cell debris prior to use. RHΔSAG3 strain parasites were a generous gift from Stanislas Tomavo.
MAbs.
MAbs C8.4 (against ROP4), A3.2 (against GRA8), and B3.90 (against AMA-1) were generated as previously described (8, 11, 47) and purified by ion exchange chromatography (10). MAb 88-70 was generated against an expressed fragment of cDNA encoding the carboxy-terminal 424 amino acids (aa) of ROP4 (rROP4). The cDNA fragment was cloned into the HindIII site of the pQE9 expression vector carrying the coding sequence for a six-histidine tag (QIAGEN, Valencia, Calif.) and expressed by inducing the transformed Escherichia coli strain M15[pREP4] with 1 mM isopropyl-β-d-1-thiogalactopyranoside (Sigma, St. Louis, Mo.) for 2 h at 30°C. The recombinant protein was enriched with bacterial protein extraction reagent, followed by the inclusion body solubilization reagent, in accordance with the manufacturer's instructions (Pierce, Rockford, Ill.). The proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to supported nitrocellulose (Schleicher & Schuell, Keene, N.H.), and stained with Ponceau S (Sigma). The band corresponding to rROP4 was excised from the blot; the nitrocellulose was scraped from the support, resuspended in phosphate-buffered saline (PBS), and sonicated with a probe sonicator until the suspension could pass through a 21-gauge needle. Immunizations with the sonicated antigen were performed as described previously (49). Hybridomas from the rROP4-immunized mice were generated and subcloned by Green Mountain Antibodies (Burlington, Vt.). Hybridoma supernatants were screened for reactivity by Western blotting against rROP4 and total tachyzoite extracts, and by immunofluorescence microscopy of extracellular and intracellular tachyzoites (47). MAb 88-70 was purified from hybridoma supernatant with a protein G-Sepharose column (Amersham Biosciences, Piscataway, N.J.), eluted from the column with 100 mM glycine, pH 2.5, and neutralized with 1/10 volume of 1 M Tris, pH 8.0. The antibody was dialyzed against PBS, pH 7.4, containing in 0.02% (vol/vol) sodium azide and stored at 4°C.
Polyclonal antibodies.
To prepare polyclonal antibodies specific to ROP4, unique peptides corresponding to residues 57 to 73 (acetyl-C-DKYSRDSTEGENTVSEG-amide) and 369 to 386 (acetyl-DMETFVDEIGRFPQEDRP-C-amide) were synthesized by the Biomolecular Resource Facility at the University of Texas Medical Branch (Galveston, Tex.). Rabbit polyclonal antibodies UVT68 (aa 369 to 386) and UVT70 (aa 57 to 73) were generated (Cocalico Biologicals, Reamstown, Pa.) and purified as previously described (11).
SDS-PAGE and Western blot analysis.
Proteins were resolved by SDS-PAGE in the presence (reducing) or absence (nonreducing) of 5% (vol/vol) β-mercaptoethanol, transferred to nitrocellulose, and Western blotted as previously described (47).
Immunofluorescence microscopy.
Confluent monolayers of HFFs on glass coverslips were infected with RH or RHΔSAG3 parasites approximately 24 h prior to use. All of the following manipulations were carried out on ice, with prechilled solutions. After cells were washed twice with PBS, pH 7.4, they were fixed and permeabilized with 100% methanol for 10 min, washed three times with PBS, and then blocked for 20 min with PBS containing 0.5% (wt/vol) fraction V bovine serum albumin (PBS-BSA; Fisher Scientific, Pittsburg, Pa.). Alternatively, infected monolayers were fixed for 30 min with PBS containing 2.5% (vol/vol) paraformaldehyde, washed three times with PBS, pH 7.4, and then blocked and permeabilized for 20 min with PBS-BSA containing 0.2% (vol/vol) Triton X-100 (TX-100). Fixed and permeabilized cells were incubated for 1 h with MAb C8.4, MAb 88-70, or UVT70 plus MAb 45.15 (directed against IMC1; K. L. Carey and G. E. Ward, unpublished data) diluted in PBS-BSA. Samples were washed three times with PBS-BSA and incubated for 45 min with PBS-BSA containing 5 μg of Alexa 488-conjugated goat anti-mouse immunoglobulin G (IgG; Molecular Probes, Eugene, Oreg.)/ml and/or Alexa 546-conjugated goat anti-rabbit IgG (Molecular Probes). Coverslips were washed four times with PBS, pH 7.4, and mounted on glass slides.
For 15-min invasion assays, parasites were incubated with confluent monolayers of HFFs for 15 min at 4°C, followed by 15 min at 37°C, and processed for immunofluorescence microscopy as described above.
Evacuoles were generated in the presence of cytochalasin D as previously described (14). ROP1 and ROP4 were localized by dual-label immunofluorescence microscopy as described above using MAb Tg49 (anti-ROP1; a generous gift from Joseph Schwartzman) followed by Alexa 546-conjugated goat anti-mouse IgG (Molecular Probes) and MAb 88-70 directly conjugated to Alexa 488 (Molecular Probes).
Cells were imaged on a Nikon TE300 inverted microscope equipped with epifluorescence illumination and a 100× PlanApo objective (numerical aperture, 1.4). Digital images were captured with a SpotRT monochrome camera (Diagnostic Instruments Inc., Sterling Heights, Mich.) driven by QED imaging software (QED Imaging Inc., Pittsburg, Pa.) and processed with Photoshop, version 7.0 (Adobe Systems, Mountain View, Calif.).
Immunoelectron microscopy.
Extracellular tachyzoites were fixed in 0.1 M sodium cacodylate buffer containing 4% (vol/vol) paraformaldehyde and 0.1% (vol/vol) glutaraldehyde, dehydrated through a graded series of ethanol, and embedded in LR White resin (London Resin Co.). Ultrathin sections were cut with a Reichert Ultracut E and placed onto nickel grids. Grids were wet with PBS, blocked with goat blocking buffer (Aurion), and incubated with UVT68 or UVT70 in PBS containing acetylated BSA (Aurion). After a washing, grids were incubated with a secondary antibody (goat anti-rabbit IgG coupled to 6- or 10-nm gold beads; Aurion), washed, and fixed with 1% (vol/vol) glutaraldehyde. Samples were viewed on a JEOL 1200EX transmission electron microscope at the Albert Einstein College of Medicine Analytical Imaging Facility.
Immunoaffinity purification and sequence analysis.
An indirectly coupled immunoaffinity resin was prepared as previously described (8, 11, 17) with the following modifications. MAb C8.4 (10 mg) was dialyzed overnight against 100 mM sodium borate, pH 8.2, at 4°C. Rabbit anti-mouse IgG (9 mg; Jackson Laboratories, West Grove, Pa.) diluted to 10 ml with 100 mM sodium borate, pH 8.2, was incubated with prewashed protein A-agarose (1.5 ml of packed resin; Bio-Rad, Hercules, Calif.) for 40 min at 23°C with gentle rocking. The resin was washed four times with 100 mM sodium borate, pH 8.2, and then gently rocked with the dialyzed MAb C8.4 for 1 h at 23°C. After being cross-linked with 20 mM dimethylpimelidate, the resin was stored at 4°C in PBS containing 0.01% (vol/vol) sodium azide.
For immunoaffinity purification, tachyzoites were extracted at 2.5 × 108 parasites/ml for 30 min on ice in TX-100 lysis buffer (100 mM NaCl, 50 mM Tris [pH 7.4], 0.5% [vol/vol] TX-100, 1/100 [vol/vol] Sigma protease inhibitors [104 mM 4-{2-aminoethyl}benzenesulfonyl fluoride, 80 μM aprotinin, 2.1 mM leupeptin, 3.6 mM bestatin, 1.5 mM pepstatin A, and 1.4 mM E-64]). The extracts were pelleted for 5 min at 14,000 × g at 4°C to remove insoluble material. The MAb C8.4 affinity resin was washed twice with TX-100 lysis buffer, twice with 100 mM glycine, pH 3.0, and twice with TX-100 lysis buffer. Extract (500 μl) was added to prewashed resin (25 μl) and gently rotated for 2 h at 4°C, followed by four batchwise washes with TX-100 lysis buffer and one wash with 10 mM sodium phosphate, pH 6.8. Bound antigen was eluted by adding 100 mM glycine, pH 2.0, and gently rotating for 10 min at 23°C and then neutralized with 1/10 (vol/vol) 1 M Tris, pH 8.0. The antigen was resolved by SDS-PAGE, stained with Coomassie blue, excised, digested with trypsin, and analyzed by microcapillary reverse-phase high-performance liquid chromatography and nanoelectrospray tandem mass spectrometry as previously described (11).
5′ and 3′ RACE.
Total tachyzoite RNA was extracted with TRIZOL reagent (Invitrogen Life Technologies, Carlsbad, Calif.) according to the manufacturer's instructions. First-strand cDNA was synthesized with the Superscript preamplification system (Invitrogen). The 5′ and 3′ ends of the cDNA fragment were amplified by rapid amplification of cDNA ends (RACE) with gene-specific primers in accordance with the manufacturer's instructions (Invitrogen).
DNA sequencing and analysis.
All DNA sequencing was performed by dye terminator fluorescence-based analysis on an Applied Biosystems 373 automated sequencer with custom synthesized primers (Genosys Biotechnologies, Woodlands, Tex.). Signal sequence cleavage sites were predicted with SignalP, version 1.1 (http://www.cbs.dtu.dk/) (29), and transmembrane domains were predicted with TMpred (http://www.ch.embnet.org/).
Metabolic labeling and pulse-chase analysis.
Heavily infected HFF monolayers were rinsed with methionine- and cysteine-free Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10 mM HEPES, pH 7.0 (HyClone, Logan, Utah), and 1% (vol/vol) dialyzed fetal bovine serum (dFBS; Invitrogen). They were incubated in the same medium for 30 min at 37°C in a 5% CO2 incubator prior to the addition of 50 μCi of EXPRESS [35S]methionine-cysteine (Perkin-Elmer, Boston, Mass.)/ml. For pulse-chase experiments, the infected monolayers were labeled for 30 min, rinsed with complete DMEM (HyClone) containing 1% FBS (Invitrogen), and incubated at 37°C for various chase times prior to extraction and immunoprecipitation as described below.
[γ-32P]ATP labeling of cellular extracts.
Parasites from freshly lysed HFF monolayers were filtered, pelleted at 1,000 × g for 4 min, washed with phosphate-free DMEM (Invitrogen), and resuspended in intracellular buffer (120 mM KCl, 20 mM NaCl, 10 mM glucose, 20 mM HEPES [pH 7.0], 250 μM CaCl2, 0.5% [vol/vol] TX-100, 1 mg of DNase/ml, 0.5 mg of RNase/ml, phosphatase inhibitors [20 mM β-glycerophosphate, 5 mM sodium pyrophosphate, 20 mM sodium fluoride, 0.15 mM sodium orthovanadate], 1/100 [vol/vol] stock protease inhibitors [Sigma]). Uninfected HFF monolayers were scraped into phosphate-free DMEM, pelleted at 1,000 × g for 4 min, washed with phosphate-free DMEM, and resuspended in intracellular buffer. For combined parasite and HFF extracts, cells were washed as described above, pelleted together, and extracted in intracellular buffer. Cells were extracted for 1 h on ice and pelleted at 14,000 × g (10 min, 4°C). The clarified extract (100 μl) was supplemented with 100 μCi of [γ-32P]ATP (Perkin-Elmer), 20 μM ATP, and 1 mM MgCl2 and incubated for 30 min at 30°C. ROP4 was immunoprecipitated from the labeled extracts and processed as described below.
[32P]orthophosphate labeling of cells.
Monolayers of HFFs in T75 flasks were rinsed with phosphate-free DMEM (Invitrogen) containing 1/10 (vol/vol) complete DMEM (HyClone), 10 mM HEPES, pH 7.0, and 1% dFBS. They were incubated in the same medium (10 ml/flask) containing 250 μCi of [32P]orthophosphate (ICN Biomedicals, Irvine, Calif.)/ml for 8 h at 37°C in a 5% CO2 incubator. Tachyzoites (∼2 × 107 in 1 ml of phosphate-free DMEM) were then added to the 32P-labeled HFFs and incubated for 6 h at 37°C in a 5% CO2 incubator. Monolayers were rinsed with phosphate-free DMEM, scraped, and centrifuged at 1,000 × g for 4 min.
To label extracellular parasites, tachyzoites from freshly lysed infected monolayers were filtered, pelleted at 1,000 × g for 4 min, and resuspended in phosphate-free DMEM (∼5 × 107 tachyzoites in 10 ml) containing 250 μCi of [32P]orthophosphate (ICN Biomedicals)/ml. After being labeled for 1 to 6 h at 37°C in a 5% CO2 incubator, the parasites were pelleted for 4 min at 1,000 × g. Cell pellets were extracted and immunoprecipitated as described below.
Cell extraction and immunoprecipitation.
Pellets were extracted in either TX-100 lysis buffer or SDS lysis buffer (100 mM NaCl, 50 mM Tris [pH 7.4], 0.5% [wt/vol] SDS, phosphatase inhibitors, 1/100 [vol/vol] protease inhibitor stocks [Sigma]). For TX-100 extractions, pellets were resuspended in lysis buffer, incubated for 30 min on ice, and then centrifuged for 10 min at 14,000 × g at 4°C. For SDS extractions, pellets were boiled in lysis buffer for 10 min, cooled to 23°C, and then supplemented with 1/100 (vol/vol) protease inhibitor stocks (Sigma) and 2% (vol/vol) TX-100, followed by a 30-min incubation on ice and centrifugation at 14,000 × g (10 min, 4°C).
For immunoprecipitations, antibodies against ROP4 (MAb C8.4, UVT68, or UVT70) or ROP2/ROP3/ROP4 (MAb T34A7, a generous gift from Jean-Francois Dubremetz) were incubated with 0.5 ml of extract for 1.5 h at 4°C with gentle rocking, followed by the addition of prewashed protein A-Sepharose (25 μl packed; Zymed, San Francisco, Calif.) and another 1.5-h incubation at 4°C with gentle rocking. After the resin was washed five times with TX-100 lysis buffer, the antigen was eluted by boiling in sample buffer, separated by SDS-PAGE, transferred to nitrocellulose (Schleicher & Schuell) or Immobilon-P (Millipore, Bedford, Mass.), and detected by autoradiography and/or Western blotting.
Phosphoamino acid analysis and two-dimensional phosphopeptide mapping.
For phosphoamino acid analysis, 32P-labeled ROP4 was immunoaffinity purified with the MAb C8.4 resin, resolved by SDS-PAGE, transferred to Immobilon-P (Millipore), and detected by autoradiography. The ROP4 band was excised, rinsed several times with double-distilled H2O, and hydrolyzed in 200 μl of constant boiling 6 N HCl (Pierce) for 1 h at 110°C. The hydrolysis products were dried under vacuum and dissolved in thin-layer chromatography (TLC) buffer (5 parts isobutyric acid, 3 parts 0.5 M NaOH). Three microliters of a mixture of phosphoserine, phosphothreonine, and phosphotyrosine (each at 1 mg/ml in water) was added to 3 μl of the sample, which was then spotted onto a cellulose thin-layer plate (Chromagram 13255; 20 by 20 cm; Eastman Kodak, Rochester, N.Y.). The samples were resolved by chromatography in TLC buffer. Plates were dried, sprayed with 0.2% ninhydrin in ethanol (Sigma), and developed at 65°C. The phosphoamino acid standards were visualized directly; 32P-labeled amino acids were visualized by autoradiography.
Two-dimensional tryptic peptide mapping of gel-purified ROP4 was performed on Chromagram 13255 cellulose thin-layer plates as previously described (48).
Nucleotide sequence accession number.
The complete 1,734-bp open reading frame of the ROP4 gene has been assigned GenBank accession number AY662677.
RESULTS
MAb C8.4 recognizes both a 60-kDa apical antigen and the 43-kDa surface protein SAG3.
In experiments designed to identify membrane proteins on the surface or within the secretory organelles of T. gondii tachyzoites, a MAb (C8.4) that recognized a 60-kDa protein on Western blots of total tachyzoite extracts run under reducing conditions was generated (Fig. 1A, left, lane RH). By immunofluorescence microscopy, the antigen localized to the surface of nonpermeabilized extracellular tachyzoites (data not shown) and to the surfaces and internal apical structures of detergent-permeabilized extracellular and intracellular tachyzoites (Fig. 1B, a, and data not shown).
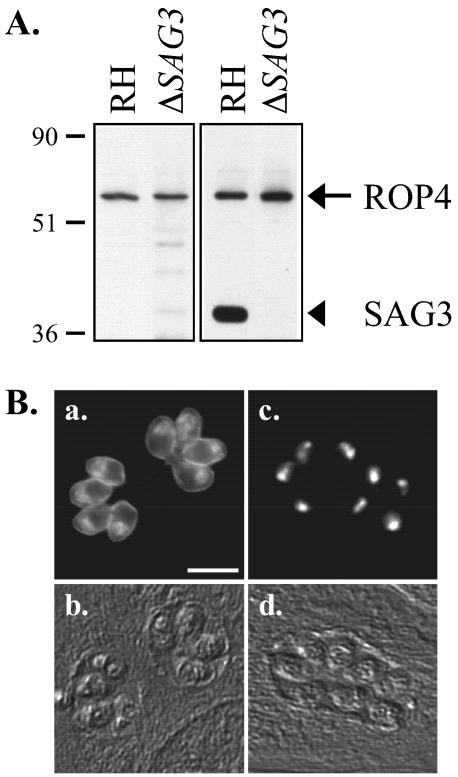
FIG. 1.
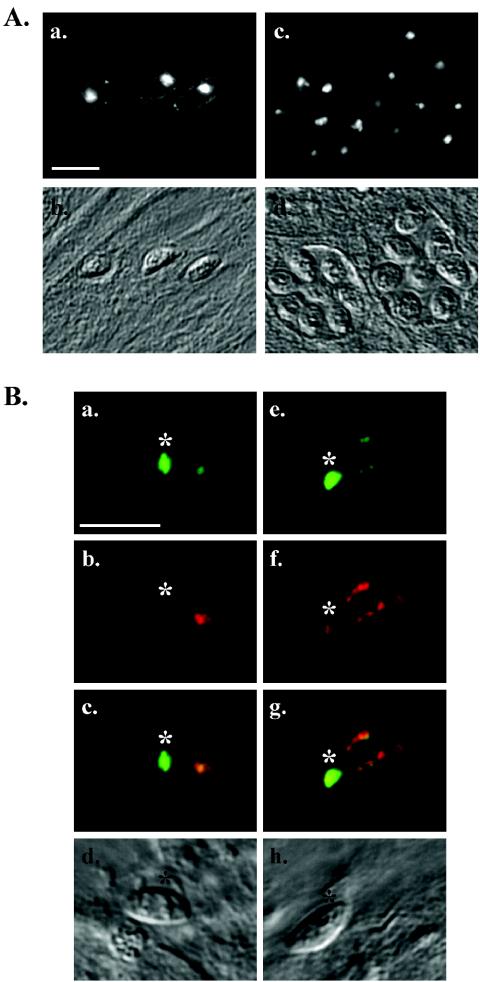
Western blot analysis and localization of the antigen(s) recognized by MAb C8.4. (A) Lysates of RH and RHΔSAG3 parasites were resolved by SDS-PAGE under either reducing (left) or nonreducing (right) conditions and Western blotted with MAb C8.4. Numbers on left indicate molecular masses in kilodaltons. (B) Indirect immunofluorescence analysis with MAb C8.4 of methanol-fixed HFF cells 24 h after infection with RH (a) or RHΔSAG3 (c) parasites; (b and d) corresponding differential interference contrast images. Bar, 10 μm.
On Western blots of gels run under nonreducing conditions, MAb C8.4 recognized both the 60-kDa antigen and a band at 43 kDa (Fig. 1A, right, lane RH). The 60- and 43-kDa antigens were also both immunoprecipitated by MAb C8.4 (data not shown). The 43-kDa protein was identified as SAG3, a member of the immunodominant, glycosylphosphatidylinositol-anchored surface protein family of T. gondii tachyzoites (13, 20), by Western blotting with an anti-SAG3 antibody (data not shown). This result was confirmed by Western blotting a parasite strain lacking SAG3 (13), extracted under both reducing and nonreducing conditions, with MAb C8.4 (Fig. 1A, lanes ΔSAG3).
Immunofluorescence with RHΔSAG3 strain parasites demonstrated that the surface labeling observed with MAb C8.4 (Fig. 1B, a) was due to its cross-reactivity with SAG3. In the absence of SAG3, MAb C8.4 localizes to the apical ends of permeabilized tachyzoites (Fig. 1B, c) with no detectable peripheral staining.
Identification of the 60-kDa antigen as ROP4.
The 60-kDa antigen recognized by MAb C8.4 was purified by immunoaffinity chromatography and SDS-PAGE. Tryptic peptides were analyzed by microcapillary reverse-phase high-performance liquid chromatography and nanoelectrospray tandem mass spectrometry. The fragmentation spectra matched four expressed sequence tags (ESTs) in the T. gondii EST database (data not shown). A search of the T. gondii EST database identified a 656-bp contig containing three of the ESTs but lacking the 5′ and 3′ ends of the sequence. By 5′- and 3′ RACE, the complete 1,734-bp open reading frame was determined. The corresponding genomic DNA was sequenced and found to contain no introns.
The predicted protein sequence of the 60-kDa antigen (578 aa) exhibits significant sequence similarity to the T. gondii rhoptry proteins ROP2 and ROP8. Based on its sequence (Fig. 2), the 60-kDa antigen was identified as ROP4, a previously sequenced (GenBank accession number Z71787) but otherwise uncharacterized rhoptry protein (J. F. Dubremetz, personal communication). The ROP4 cDNA and genomic sequences presented here differ from the previously published sequence. The nucleotides corresponding to positions 490, 563, and 566 in our sequence are missing in the previously published sequence, resulting in two frameshifts within the predicted amino acid sequence (Fig. 2). Restriction digests of ROP4 with NotI (data not shown) were consistent with our sequence (two predicted cleavage sites) and inconsistent with the previously published sequence (one predicted cleavage site). Furthermore, tryptic digestion of the purified antigen followed by mass spectrometry identified a peptide within this region with a mass predicted by our sequence (Fig. 2) but not by the previously published sequence.
FIG. 2.
Nucleotide and amino acid sequences of ROP4. The open reading frame of ROP4 encodes a polypeptide of 578 aa. Arrow, predicted signal peptide cleavage site; dashed underline, putative transmembrane domain; single solid underlines, peptides used to prepare ROP4-specific polyclonal antibodies (UVT70, aa 57 to 73; UVT68, aa 369 to 386). Shaded nucleotides are absent in the published ROP4 sequence (GenBank accession number Z71787), resulting in two frameshifts within the boxed amino acid sequence. A tryptic peptide whose mass matches precisely the mass of the double-underlined peptide was identified by mass spectrometry analysis.
The ROP4 primary sequence contains several features of note, including two hydrophobic regions: an N-terminal signal sequence, with a predicted cleavage site between residues 33 and 34, and a potential C-terminal transmembrane domain (Fig. 2). BLAST searches of the T. gondii genomic database with the ROP4 amino acid sequence yielded the two known rhoptry proteins, ROP2 (37% identity) and ROP8 (39% identity), and an unidentified protein with even greater sequence identity (77%). Protein kinases from a variety of organisms were identified by BLAST searches of the National Center for Biotechnology Information database. A putative serine/threonine protein kinase domain in the C-terminal half of the ROP4 sequence was identified by a PROSITE search (16), although this domain lacks a conserved aspartic acid in the catalytic loop that is critical for phosphotransferase activity. The ROP4 sequence also contains a potential cleavage site for the serine protease TgSUB2, which may function in ROP4 processing by removing the prodomain (25), and a YXXφ motif within the C-terminal tail that may function in trafficking to the rhoptries (18, 28). The epitope on SAG3 that cross-reacts with MAb C8.4 is likely to be conformational rather than linear, as it is sensitive to reduction and there are no stretches of significant sequence identity between ROP4 and SAG3.
Generation and localization of ROP4-specific antibodies.
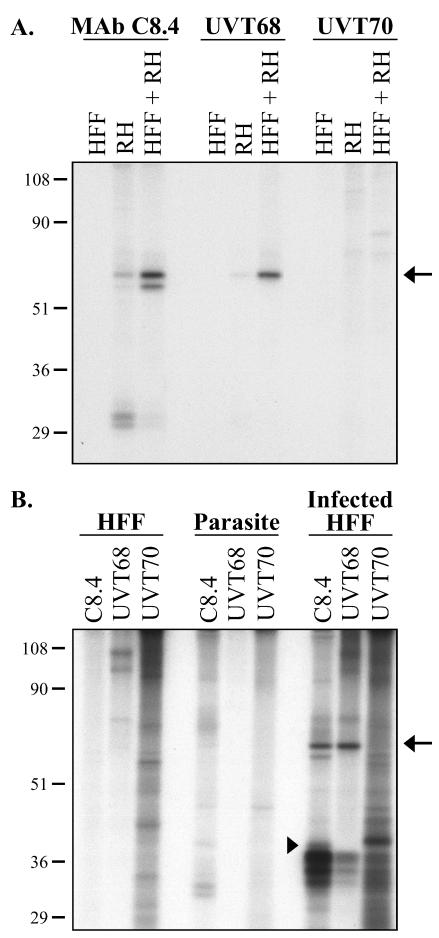
To eliminate the problem of cross-reactivity of MAb C8.4 with SAG3, new ROP4-specific antibodies were generated. MAbs against the C-terminal 424 aa of recombinant histidine-tagged ROP4 were generated. One of these MAbs, 88-70 (Fig. 3A), recognized ROP4 (60 kDa) on Western blots of total T. gondii extracts run under both reducing and nonreducing conditions (Fig. 3B). Polyclonal antibodies against synthetic peptides corresponding to ROP4 sequences absent in the other known ROP2 family members were also prepared (Fig. 2 and 3A). One of these polyclonal antibodies, UVT68, recognized ROP4 (60 kDa) on Western blots of total T. gondii extracts run under both reducing and nonreducing conditions (Fig. 3B). The lower-molecular-weight 43- to 45-kDa proteins recognized by MAb 88-70 and UVT68 under reducing conditions may be degradation products. Neither MAb 88-70 nor any of the polyclonal antipeptide antibodies show any detectable cross-reactivity with SAG3.
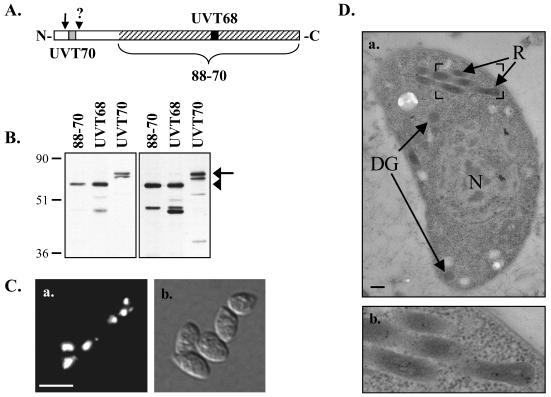
FIG. 3.
Western blot analysis and immunolocalization using ROP4-specific antibodies. (A) Schematic representation of regions recognized by the ROP4-specific antibodies. Hatched region, C-terminal 424 aa of ROP4 used as the immunogen for mouse MAb 88-70; gray and black boxes, synthetic peptides used to prepare rabbit polyclonal antibodies UVT70 and UVT68, respectively; arrow, predicted signal peptide cleavage site; arrowhead, approximate site of prodomain cleavage (see text). (B) Total RH parasite extracts were resolved by SDS-PAGE under either reducing (left) or nonreducing (right) conditions and Western blotted with MAb 88-70, UVT68, or UVT70. Arrow, pro-ROP4 (68 kDa), recognized by UVT70; arrowhead, mature ROP4 (60 kDa), recognized by MAb 88-70 and UVT68. On longer exposures, pro-ROP4 (68 kDa) was also recognized by MAb 88-70 and UVT68 (data not shown). Numbers on left indicate molecular masses in kilodaltons. (C) Indirect immunofluorescence microscopy with MAb 88-70 of methanol-fixed extracellular parasites (a) and the corresponding differential interference contrast image (b). Bar, 10 μm. (D) Immunoelectron microscopy using polyclonal antibody UVT68. ROP4 localizes specifically to the rhoptries (R), as is evident from the distribution of the 10-nm immunogold particles. N, nucleus; DG, dense granules. Bar, 200 nm. Panel b shows an enlargement of the boxed region in panel a.
Immunofluorescence microscopy with the ROP4-specific antibodies localized ROP4 to discrete foci at the apical end extracellular tachyzoites (Fig. 3C, a). This apical distribution was indistinguishable from that seen with MAb C8.4 on the RHΔSAG3 parasites (Fig. 1B, c). It was also indistinguishable from that of ROP2 but different from the staining pattern of the dense granule and microneme markers GRA8 and TgAMA-1 (data not shown). Immunoelectron microcopy confirmed that the apical compartment labeled by the ROP4-specific antibodies is indeed the rhoptries (Fig. 3D).
ROP4 processing.
An additional polyclonal antibody, UVT70, reacts with a 68-kDa doublet in parasite lysates, but not the 60-kDa band recognized by MAb C8.4, MAb 88-70, or UVT68 (Fig. 3B). UVT70 was generated against an N-terminal peptide of ROP4 (aa 57 to 73; Fig. 2 and 3A). Previous studies have shown that rhoptry proteins can undergo multiple processing events to remove the signal peptide and an N-terminal prodomain (4, 36, 44). It therefore seems likely that the peptide recognized by UVT70 lies within the ROP4 prodomain and that cleavage of the prodomain converts the 68-kDa proprotein to the mature, 60-kDa ROP4. The second band of the 68-kDa doublet recognized by UVT70 may correspond to either an additional processed form of ROP4 or to the unidentified ROP4 homolog referred to above that is predicted to be 77% identical to ROP4.
To confirm that the peptide recognized by UVT70 lies within the cleaved prodomain of ROP4, pulse-chase experiments were performed. Intracellular parasites were labeled with [35S]methionine and [35S]cysteine, and ROP4 was immunoprecipitated with the different ROP4 antibodies. With UVT68, the 68-kDa form of ROP4 was detected first, followed by the 60-kDa form (Fig. 4, left) indicating a precursor-product relationship. The half-life of the 68-kDa precursor protein is ∼60 min, longer than the ∼15-min half-life previously observed for ROP1 (44).
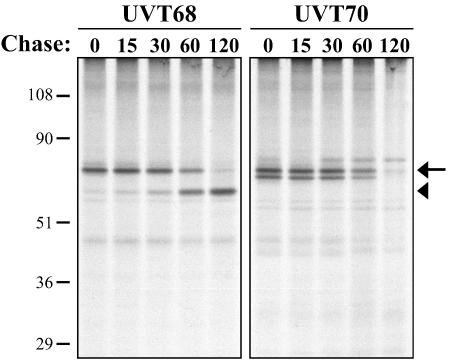
FIG. 4.
Pulse-chase analysis of metabolically labeled ROP4. Intracellular parasites were labeled for 30 min with [35S]methionine and [35S]cysteine, followed by chase intervals of 0 to 120 min in medium lacking 35S. ROP4 was immunoprecipitated from SDS-extracted parasites with either UVT68 (left) or UVT70 (right), resolved by SDS-PAGE, transferred to Immobilon-P, and detected by autoradiography. UVT68 recognizes both the proprotein (68 kDa; arrow) and the mature protein (60 kDa; arrowhead), while UVT70 recognizes the proprotein (and a second, closely migrating band [see text]), but not the mature form. Numbers on the left indicate molecular masses in kilodaltons.
A similar turnover rate for the 35S-labeled 68-kDa form of ROP4 was observed with UVT70. However, the 60-kDa form was not immunoprecipitated, providing direct evidence that the epitope recognized by UVT70 is removed during cleavage of the prodomain (Fig. 4, right). Attempts to identify the prodomain cleavage site of affinity-purified 60-kDa ROP4 by amino-terminal sequencing were unsuccessful, most likely a result of a blocked amino terminus.
In immunofluorescence studies, a relatively low percentage of fixed and permeabilized intracellular parasites were labeled by UVT70. In these parasites, the antibody localized to a few discrete internal foci, anterior to the nucleus but posterior to the rhoptries (Fig. 5B). This staining pattern is reminiscent of the distribution of N-terminally epitope-tagged ROP1 (44). The compartment labeled by UVT70 may therefore correspond to the nascent rhoptries of daughter parasites (44), in which ROP1 and ROP4 are processed.
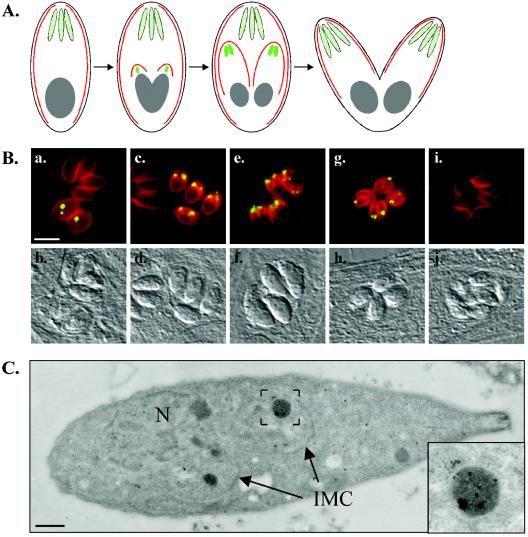
FIG. 5.
Polyclonal antibody UVT70 labels a prerhoptry compartment in developing daughter parasites. (A) Schematic representation of T. gondii endodyogeny, a process in which two daughter parasites are formed within an intact mother parasite. Red, inner membrane complex of the mother and daughter parasites; light green, rhoptries of the mother parasite; bright green, daughter rhoptries. (B) Dual-label immunofluorescence microscopy. The inner membrane complex (MAb 45.15) of the mother and daughter parasites is red, and the UVT70-positive compartment is green in the merged images. The compartment labeled by UVT70 is located immediately anterior to the nucleus and posterior to the rhoptries in developing daughter parasites from the initial stages (a) to very late in endodyogeny (g). UVT70 labeling disappears immediately prior to the budding of the daughters from the mother (i), and no labeling is detectable in mature, extracellular parasites (not shown). Corresponding differential interference contrast images are shown below. Bar, 10 μm. (C) Immunoelectron microscopy with polyclonal antibody UVT70. In parasites undergoing endodyogeny, UVT70 localizes to electron-dense vesicles in the daughter parasites, as is evident from the distribution of the 6-nm immunogold particles. N, mother nucleus; IMC, daughter inner-membrane complexes. Bar, 500 nm. Inset, enlargement of the boxed region.
T. gondii divides by endodyogeny, a process in which two daughter parasites are formed within an intact mother parasite (Fig. 5A). The daughter parasites are surrounded by an inner membrane complex that grows from the anterior region to the posterior of the daughter as endodyogeny proceeds, and within which daughter organelles either assemble (e.g., the rhoptries) or are partitioned from the mother (e.g., the nucleus). Dual labeling with MAb 45.15 (which recognizes IMC1, a major inner membrane complex protein; Carey and Ward, unpublished) and UVT70 confirms that the subset of parasites that are labeled with UVT70 are undergoing endodyogeny, as UVT70 labeling is only observed within emerging 45.15-positive daughter parasites (Fig. 5B). Parasites are UVT70 positive from the initial stages of daughter formation (Fig. 5B, a) until just prior to budding from the mother cell (Fig. 5B, g). Immunoelectron microscopy with UVT70 revealed that the antigen is concentrated in electron-dense vesicles within the forming daughter parasites (Fig. 5C).
Secretion of ROP4 into the PV and evacuoles.
In infected cells, mature ROP4 is found within both the rhoptries of the intracellular parasites and the PVM (Fig. 6A, a). ROP4 is detectable in the PVM within 15 min after the addition of parasites to the host cells (Fig. 6A, a). As the parasites divide and the vacuole grows, ROP4 becomes more difficult to visualize, probably a result of either ROP4 turnover or dilution in the expanding PVM (Fig. 6A, c).
FIG. 6.
Localization of ROP4 in infected cells and secretion into evacuoles. (A) Immunofluorescence microscopy showing that MAb 88-70 labels both the rhoptries and the PVM 15 min (a) or 24 h (c) postinfection. (b and d) Corresponding differential interference contrast (DIC) images. Bar, 10 μm. (B) Parasites were incubated with HFF cells in the presence of cytochalasin D as previously described (14). Evacuoles within the host cell were visualized by dual-label immunofluorescence microscopy with antibodies against ROP4 (a and e; MAb 88-70; green) and ROP1 (b and f; MAb Tg49; red). (c and g) Merged images; (d and h) corresponding DIC images. *, residual rhoptry staining within the attached tachyzoites. Bar, 10 μm.
Parasites pretreated with cytochalasin D for 10 min prior to incubation with HFF cells are able to attach and secrete the contents of their rhoptries but are unable to invade. As previously shown for ROP1 and ROP2 (14), ROP4 is injected into the host cell by cytochalasin D-arrested parasites and can be localized in the resultant evacuoles (Fig. 6B). Interestingly, dual labeling with antibodies against ROP4 and ROP1 shows that these two rhoptry proteins are secreted into the evacuoles to different extents: more ROP4 is detected in the rhoptries of the attached parasites than in the evacuoles, while the opposite is observed for ROP1 (Fig. 6B).
Phosphorylation of ROP4.
Although ROP4 may not be an active protein kinase itself, since it lacks a critical aspartic acid within its degenerate protein kinase domain (16), it was found to be a substrate for protein kinase activity(ies) present in parasite extracts. When such extracts are incubated with [γ-32P]ATP, a weakly 32P-labeled 60-kDa band is immunoprecipitated with either MAb C8.4 or UVT68 (Fig. 7A, lanes RH). No labeled bands are immunoprecipitated with these antibodies from [γ-32P]ATP-labeled HFF cell extracts (Fig. 7A, lanes HFF). When extracts of parasites and HFF cells are mixed and then labeled with [γ-32P]ATP, ROP4 incorporates significantly more 32P (Fig. 7A, lanes HFF + RH) than is incorporated in parasite extracts alone. Western blotting confirmed that the increase in 32P labeling was not due to the recovery of more ROP4 from the mixed extracts (data not shown). Interestingly, the 68-kDa form of ROP4 immunoprecipitated by UVT70 incorporates no detectable 32P, suggesting that prodomain cleavage may be a necessary prerequisite for ROP4 phosphorylation in cellular extracts (Fig. 7A).
FIG. 7.
Phosphorylation of ROP4 in cell extracts and in intact cells. (A) Cell extracts. HFF cell extracts (lanes HFF), tachyzoite extracts (lanes RH), and HFF extracts combined with tachyzoite extracts (lanes HFF + RH) were labeled with [γ-32P]ATP for 30 min at 30°C and immunoprecipitated with MAb C8.4, UVT68, or UVT70. The immunoprecipitates were resolved by SDS-PAGE, transferred to Immobilon-P, and visualized by autoradiography. The identity of the 60-kDa 32P-labeled band (arrow) as ROP4 was confirmed by Western blotting (data not shown). Numbers on the left indicate molecular masses in kilodaltons. (B) Intact cells. HFF cells, tachyzoites, and tachyzoite-infected HFF cells were labeled with [32P]orthophosphate, extracted, and immunoprecipitated with MAb C8.4, UVT68, or UVT70. Proteins were separated by SDS-PAGE, transferred to Immobilon-P, and detected by autoradiography. The mature ROP4 protein (60 kDa), immunoprecipitated by MAb C8.4 and UVT68, was phosphorylated in infected HFF cells (arrow); pro-ROP4 (68 kDa), immunoprecipitated by UVT70, was not. SAG3, precipitated with MAb C8.4, was also phosphorylated (arrowhead), as previously reported (45). The identities of the heavily phosphorylated 33- to 43-kDa proteins precipitated by all three antibodies are unknown. Numbers on the left indicate molecular masses in kilodaltons.
To determine if ROP4 is phosphorylated in intact parasites, extracellular tachyzoites were incubated with [32P]orthophosphate, extracted, and immunoprecipitated with ROP4 antibodies. Under these conditions, ROP4 does not incorporate detectable levels of 32P, although many other parasite proteins are 32P labeled (Fig. 7B and data not shown).
To determine if ROP4 is phosphorylated in infected HFF cells, HFF monolayers were incubated with [32P]orthophosphate, infected with parasites, and extracted 6 h later. The 60-kDa form of ROP4, immunoprecipitated with either MAb C8.4 or UVT68, is clearly phosphorylated under these conditions (Fig. 7B). Western blotting confirmed that the increased amount of 32P incorporation in ROP4 from infected monolayers compared to that in ROP4 from extracellular parasites is not due to the presence of more ROP4 in the infected-cell samples (data not shown). SAG3 (Fig. 7B) is also phosphorylated, as previously observed (45). The identities of the other heavily phosphorylated 33- to 43-kDa bands in the immunoprecipitates are not known. These proteins are not detectable by silver staining or by 35S autoradiography in immunoprecipitates of metabolically labeled parasites, indicating that they are heavily phosphorylated but otherwise minor components of the immunoprecipitate. They are not present in precipitates in the absence of the primary antibody or in immunoprecipitates prepared with an unrelated MAb directed against the dense granule protein GRA8 (data not shown). Immunoprecipitation of the 32P-labeled proteins with three different ROP4 antibodies (Fig. 7B) makes it unlikely that their presence is due to antibody cross-reactivity. They are not found in the same fractions as ROP4 on sucrose density gradients (data not shown), suggesting that they are not physically complexed with ROP4.
ROP4 is phosphorylated on multiple sites.
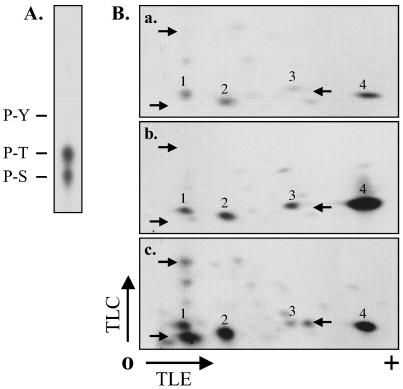
ROP4 phosphorylation in infected HFF cells occurs on both serine and threonine (Fig. 8A). Two-dimensional phosphopeptide mapping was used to determine the number of phosphorylated peptides and to compare the phosphorylation patterns under different labeling conditions. Four phosphopeptides were reproducibly detected in [γ-32P]ATP-labeled parasite extracts (Fig. 8B, a, spots 1 to 4). In mixed parasite-HFF extracts, the same four phosphopeptides were detected (Fig. 8B, b) but the amount of radiolabel associated with two of these spots increased dramatically compared to the amount in parasite extracts (Fig. 8B, a and b, spots 3 and 4). These same four peptides are phosphorylated in intact, infected HFF cells (Fig. 8B, c); however, three additional peptides, phosphorylated in infected cells, are not observed in [γ-32P]ATP-labeled extracts (Fig. 8B, c). Taken together, these results show that (i) ROP4 is phosphorylated on multiple serine and threonine residues in infected HFF cells, (ii) phosphorylation of these sites can be partially reproduced in cell extracts, and (iii) a subset of the sites are phosphorylated either by host cell protein kinase(s) or by parasite kinase(s) activated by host cell factors.
FIG. 8.
Phosphoamino acid analysis and peptide mapping of phosphorylated ROP4. (A) Phosphoamino acid analysis. Purified ROP4 from [32P]orthophosphate-labeled infected HFF cells was hydrolyzed with 6 N HCl and analyzed by TLC. 32P-labeled spots comigrating with ninhydrin-stained phosphoserine (P-S), phosphothreonine (P-T), and phosphotyrosine (P-Y) standards were detected by autoradiography. (B) Phosphopeptide mapping. ROP4 was immunoprecipitated with MAb C8.4 from parasite extracts labeled with [γ-32P]ATP (a), combined HFF and parasite extracts labeled with [γ-32P]ATP (b), or extracts prepared from [32P]orthophosphate-labeled infected HFF cells (c). Proteins were resolved by SDS-PAGE, transferred to Immobilon-P, and detected by autoradiography. In each case, the band corresponding to ROP4 was excised, digested with TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-trypsin, and spotted onto the origin (O) of a cellulose TLC plate. Tryptic peptides were separated by electrophoresis in the first dimension and chromatography in the second. The labeling intensity of two of the phosphopeptides increased dramatically in the combined host cell-parasite extract compared to that in the extract of parasites alone (a and b, spots 3 and 4). Several additional phosphopeptides were detected in the infected HFF cells (c, arrows) that were not phosphorylated in cell extracts (a and b, arrows). TLE, thin-layer electrophoresis.
Phosphorylation of ROP2.
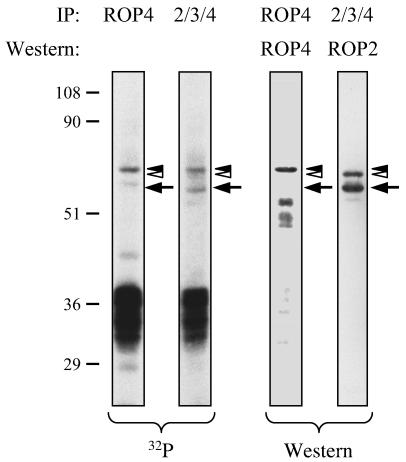
Given the sequence similarity between ROP4 and ROP2, we investigated whether ROP2 is also phosphorylated in infected host cells. Confluent monolayers of HFF cells were incubated with [32P]orthophosphate and then infected with parasites for 6 h. After extraction, MAb T34A7 (22) was used to immunoprecipitate ROP2, ROP3, and ROP4. Two 32P-labeled proteins of 60 and 55 kDa were recovered in the immunoprecipitates; these proteins were confirmed by Western blotting to be ROP4 and ROP2 (Fig. 9), respectively. The same heavily phosphorylated 33- to 43-kDa proteins seen in the ROP4 immunoprecipitates (Fig. 7) were also immunoprecipitated by the ROP2/ROP3/ROP4 antibody.
FIG. 9.
Phosphorylation of ROP2 in infected cells. Infected HFF cells were labeled with [32P]orthophosphate, extracted, and immunoprecipitated with MAb C8.4 (ROP4) or MAb T34A7 (ROP2/ROP3/ROP4). Proteins were separated by SDS-PAGE, transferred to Immobilon-P, and detected by autoradiography and Western blotting. Two of the phosphorylated proteins immunoprecipitated by MAb T34A7 corresponded to ROP4 (black arrowhead) and ROP2 (arrow), as determined by Western blotting with UVT68 and anti-ROP2 polyclonal antisera. The protein indicated by the white arrowhead on the ROP2 Western blot was also recognized on Western blots probed with MAb T34A7 and is presumed to be ROP3. This protein incorporates little, if any, 32P. Numbers on the left indicate molecular masses in kilodaltons.
DISCUSSION
In this report, we have identified and characterized T. gondii ROP4, a new member of the ROP2 family of proteins. Previous studies have demonstrated that rhoptry proteins are synthesized as proproteins, which are proteolytically processed to their mature forms en route to the rhoptries (44). Rhoptry protein processing has been studied primarily with ROP1. ROP1 cleavage does not appear to be required for targeting to the rhoptries, suggesting that it may instead be involved in folding or function (6). Brefeldin A blocks ROP1 processing, indicating that processing occurs downstream of the endoplasmic reticulum (44). Immunofluorescence microscopy localized the N-terminal epitope tag on recombinant myc-ROP1 to an anterior compartment distinct from mature rhoptries in the subset of parasites undergoing replication (44). These data suggested that the N-terminal prodomain of ROP1 is removed late in the secretory pathway within an anterior “prerhoptry” compartment.
We show here that ROP4 is similarly processed at its N terminus to a mature 60-kDa polypeptide, with a half-life of ∼60 min. The immunofluorescence data with UVT70 demonstrate that the propeptide is removed from ROP4 en route to the mature rhoptries. The discrete compartment labeled by UVT70 in daughter parasites during endodyogeny—anterior to the nucleus and posterior to the rhoptries—may represent the site of ROP4 processing. UVT70 reacts with native ROP4, rather than a recombinant construct, and may be a useful reagent for characterizing rhoptry protein processing and the compartment in which processing occurs.
Pharmacological inhibition of either cysteine or serine protease activity results in disruption of rhoptry formation during parasite replication (38). Two candidate target enzymes have recently been identified and localized to the rhoptries: TgSUB2, a subtilisin-like serine protease, and Toxopain-1, a cathepsin B cysteine protease (25, 32). The amino acid sequence of ROP4 contains a potential TgSUB2 cleavage site immediately C-terminal to the epitope recognized by polyclonal antibody UVT70, and preliminary data suggest that ROP4 is indeed a substrate for TgSUB2 in vivo (K. Kim, unpublished observation).
ROP4 is secreted during or shortly after invasion and associates with the PVM, most likely through its predicted transmembrane domain. If parasite internalization is blocked with cytochalasin D, ROP4 is secreted from the parasite into the host cell cytoplasm, where it can be detected in evacuoles, poorly defined structures that are thought to function in the delivery of rhoptry proteins to the PVM (14). Surprisingly, we found that ROP4 and ROP1 appear to be secreted into evacuoles to different extents. This may indicate that different rhoptry proteins are stored in different rhoptries or rhoptry subcompartments, the secretion of which is differentially regulated. Alternatively, different rhoptry proteins may be modified postsecretion in ways that differentially affect antibody binding.
A search for functional domains in the amino acid sequence of ROP4 identified a degenerate protein kinase domain. All members of the ROP2 family contain at least some of the consensus residues that define the kinase domain, but only ROP2 encodes the conserved aspartic acid in the catalytic loop critical to phosphotransferase activity (16). Members of the ROP2 family may therefore have diverged from an ancestral protein kinase; whether ROP2 has retained protein kinase activity is unknown, as are the functions of the other members of the ROP2 family. Attempts to determine whether immunoprecipitated ROP4 or ROP2 or both are capable of autophosphorylation were complicated by the presence of additional proteins in the immunoprecipitates (Fig. 7 and 9 and data not shown).
Regardless of whether or not ROP4 and ROP2 are themselves active kinases, we show here that both of them are substrates for kinase activity(ies) present in infected cells. Further analysis of ROP4 showed it to be phosphorylated on several serine or threonine residues both in parasite extracts and in intact infected cells. ROP4 is not phosphorylated in intact extracellular parasites, indicating that (i) ROP4, the ROP4 kinase(s), and/or the labeled ATP pool are compartmentalized from one another within the parasite; (ii) the conformation of ROP4 within the rhoptries somehow precludes its phosphorylation; or (iii) the parasite ROP4 kinase(s) is activated during invasion. The observation that pro-ROP4 (68 kDa) is not phosphorylated in parasite extracts, in contrast to mature ROP4 (60 kDa), suggests that the conformation of the protein may indeed affect its ability to be phosphorylated. The increased phosphorylation of specific peptides in combined host cell-parasite extracts compared to that in extracts of parasites alone suggests that these sites may be targets for host cell kinases and/or parasite kinases that are activated by host cell factors. The observation that additional sites are phosphorylated in infected host cells compared to the sites phosphorylated in combined extracts indicates either that the conditions in our extracts are suboptimal for reconstituting full ROP4 phosphorylation or that these additional phosphopeptides are targets for host or parasite kinases that are activated in response to invasion. Identification of the kinase(s) that phosphorylates the various sites on ROP4 will be an important area for future investigation.
It was recently reported that parasites deficient in ROP4 show no obvious growth or invasion defect in cell culture (7). The high degree of sequence similarity that exists among members of the ROP2 family of proteins may indicate some degree of functional redundancy. The generation of parasites that are simultaneously deficient in multiple members of the ROP2 family and/or the analysis of mutant strains in animal models may ultimately be required to elucidate the function of ROP4 and other uncharacterized members of the ROP2 family. The recent development of RNA-based methods for controlling gene expression in T. gondii (1, 39) may enable the simultaneous downregulation of genes that have significant sequence identity, such as those of the ROP2 family.
The behavior of ROP4 described here is strikingly similar to that of Chlamydia psittaci IncA. IncA is secreted by C. psittaci into its host cell, where it associates with the vacuole membrane and is phosphorylated on multiple serine and threonine residues by host cell kinases(s) (34). The functions of ROP4 and IncA and their intracellular phosphorylations are currently unknown. In any intracellular parasite surrounded by a vacuolar membrane, the membrane serves as a critical functional interface between the parasite and the host cell cytoplasm (reviewed in reference 40). For T. gondii, nutrient import and waste disposal occur across the PVM, host mitochondria and the endoplasmic reticulum are recruited and physically tethered to the PVM, the host cell's intermediate filament network is reorganized around the PVM, and the intracellular parasite is capable of inhibiting host cell apoptosis from within the PVM (15, 27, 37, 42). As predicted transmembrane proteins of the PVM that are also posttranslationally modified within the infected cell, ROP2, ROP4, and other members of the ROP2 family are well situated to play an important role(s) in PVM function. Indeed, ROP2 appears to function in the recruitment of host cell mitochondria and the endoplasmic reticulum to the PVM in infected cells (41). Given the molecular genetic tools available for studying protein function in Toxoplasma (24, 35), future studies in which the phosphorylation sites of individual ROP2 family members are disrupted may reveal important new insights into the function of this family of vacuolar proteins and their posttranslational modification within the host cell.
Acknowledgments
We thank Stanislas Tomavo for providing RHΔSAG3 parasites; Joseph Schwartzman, Con Beckers, and Jean-Francois Dubremetz for providing antibodies; Murry Stein and Daniel Zurawski for assistance with preparation of MAb 88-70; and the Vermont Cancer Center DNA sequencing facility for the sequencing services provided. We thank the Albert Einstein College of Medicine Analytical Imaging Facility (supported by NIH Cancer Center grant P30CA13330) for technical assistance with immunoelectron microscopy. We thank J.-F. Dubremetz for helpful discussions and Mariana Matrajt and members of the Ward laboratory for comments on the manuscript.
This work was supported by PHS grants AI42355 and AI01719 (G.E.W.).
REFERENCES
- 1.Al-Anouti, F., T. Quach, and S. Ananvoranich. 2003. Double-stranded RNA can mediate the suppression of uracil phosphoribosyltransferase expression in Toxoplasma gondii. Biochem. Biophys. Res. Commun. 302:316-323. [DOI] [PubMed] [Google Scholar]
- 2.Beckers, C. J., J. F. Dubremetz, O. Mercereau-Puijalon, and K. A. Joiner. 1994. The Toxoplasma gondii rhoptry protein ROP 2 is inserted into the parasitophorous vacuole membrane, surrounding the intracellular parasite, and is exposed to the host cell cytoplasm. J. Cell Biol. 127:947-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckers, C. J., T. Wakefield, and K. A. Joiner. 1997. The expression of Toxoplasma proteins in Neospora caninum and the identification of a gene encoding a novel rhoptry protein. Mol. Biochem. Parasitol. 89:209-223. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, P. J., and J. C. Boothroyd. 1999. Identification of the pro-mature processing site of Toxoplasma ROP1 by mass spectrometry. Mol. Biochem. Parasitol. 100:103-109. [DOI] [PubMed] [Google Scholar]
- 5.Bradley, P. J., and J. C. Boothroyd. 2001. The pro region of Toxoplasma ROP1 is a rhoptry-targeting signal. Int. J. Parasitol. 31:1177-1186. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, P. J., C. L. Hsieh, and J. C. Boothroyd. 2002. Unprocessed Toxoplasma ROP1 is effectively targeted and secreted into the nascent parasitophorous vacuole. Mol. Biochem. Parasitol. 125:189-193. [DOI] [PubMed] [Google Scholar]
- 7.Bradley, P. J., N. Li, and J. C. Boothroyd. 2004. A GFP-based motif-trap reveals a novel mechanism of targeting for the Toxoplasma ROP4 protein. Mol. Biochem. Parasitol. 137:111-120. [DOI] [PubMed] [Google Scholar]
- 8.Carey, K. L., C. G. Donahue, and G. E. Ward. 2000. Identification and molecular characterization of GRA8, a novel, proline-rich, dense granule protein of Toxoplasma gondii. Mol. Biochem. Parasitol. 105:25-37. [DOI] [PubMed] [Google Scholar]
- 9.Carruthers, V. B., and L. D. Sibley. 1997. Sequential protein secretion from three distinct organelles of Toxoplasma gondii accompanies invasion of human fibroblasts. Eur. J. Cell Biol. 73:114-123. [PubMed] [Google Scholar]
- 10.Church, W. R., S. A. Brown, and A. B. Mason. 1988. Monoclonal antibodies to the amino- and carboxyl-terminal domains of ovotransferrin. Hybridoma 7:471-484. [DOI] [PubMed] [Google Scholar]
- 11.Donahue, C. G., V. B. Carruthers, S. D. Gilk, and G. E. Ward. 2000. The Toxoplasma homologue of Plasmodium apical membrane antigen-1 (AMA-1) is a microneme protein secreted in response to elevated intracellular calcium levels. Mol. Biochem. Parasitol. 111:15-30. [DOI] [PubMed] [Google Scholar]
- 12.Dubremetz, J. F., A. Achbarou, D. Bermudes, and K. A. Joiner. 1993. Kinetics and pattern of organelle exocytosis during Toxoplasma gondii/host-cell interaction. Parasitol. Res. 79:402-408. [DOI] [PubMed] [Google Scholar]
- 13.Dzierszinski, F., M. Mortuaire, M. F. Cesbron-Delauw, and S. Tomavo. 2000. Targeted disruption of the glycosylphosphatidylinositol-anchored surface antigen SAG3 gene in Toxoplasma gondii decreases host cell adhesion and drastically reduces virulence in mice. Mol. Microbiol. 37:574-582. [DOI] [PubMed] [Google Scholar]
- 14.Hakansson, S., A. J. Charron, and L. D. Sibley. 2001. Toxoplasma evacuoles: a two-step process of secretion and fusion forms the parasitophorous vacuole. EMBO J. 20:3132-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halonen, S. K., and E. Weidner. 1994. Overcoating of Toxoplasma parasitophorous vacuoles with host cell vimentin type intermediate filaments. J. Eukaryot. Microbiol. 41:65-71. [DOI] [PubMed] [Google Scholar]
- 16.Hanks, S., and T. Hunter. 1995. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576-596. [PubMed] [Google Scholar]
- 17.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Hoppe, H. C., H. M. Ngo, M. Yang, and K. A. Joiner. 2000. Targeting to rhoptry organelles of Toxoplasma gondii involves evolutionarily conserved mechanisms. Nat. Cell Biol. 2:449-456. [DOI] [PubMed] [Google Scholar]
- 19.Israelski, D. M., and J. S. Remington. 1993. Toxoplasmosis in patients with cancer. Clin. Infect. Dis. 17(Suppl. 2):S423-S435. [DOI] [PubMed] [Google Scholar]
- 20.Jacquet, A., L. Coulon, J. De Neve, V. Daminet, M. Haumont, L. Garcia, A. Bollen, M. Jurado, and R. Biemans. 2001. The surface antigen SAG3 mediates the attachment of Toxoplasma gondii to cell-surface proteoglycans. Mol. Biochem. Parasitol. 116:35-44. [DOI] [PubMed] [Google Scholar]
- 21.Kim, K., D. Soldati, and J. C. Boothroyd. 1993. Gene replacement in Toxoplasma gondii with chloramphenicol acetyltransferase as selectable marker. Science 262:911-914. [DOI] [PubMed] [Google Scholar]
- 22.Leriche, M. A., and J. F. Dubremetz. 1991. Characterization of the protein contents of rhoptries and dense granules of Toxoplasma gondii tachyzoites by subcellular fractionation and monoclonal antibodies. Mol. Biochem. Parasitol. 45:249-259. [DOI] [PubMed] [Google Scholar]
- 23.Luft, B. J., and J. S. Remington. 1992. Toxoplasmic encephalitis in AIDS. Clin. Infect. Dis. 15:211-222. [DOI] [PubMed] [Google Scholar]
- 24.Meissner, M., D. Schluter, and D. Soldati. 2002. Role of Toxoplasma gondii myosin A in powering parasite gliding and host cell invasion. Science 298:837-840. [DOI] [PubMed] [Google Scholar]
- 25.Miller, S. A., V. Thathy, J. W. Ajioka, M. J. Blackman, and K. Kim. 2003. TgSUB2 is a Toxoplasma gondii rhoptry organelle processing proteinase. Mol. Microbiol. 49:883-894. [DOI] [PubMed] [Google Scholar]
- 26.Nakaar, V., H. M. Ngo, E. P. Aaronson, I. Coppens, T. T. Stedman, and K. A. Joiner. 2003. Pleiotropic effect due to targeted depletion of secretory rhoptry protein ROP2 in Toxoplasma gondii. J. Cell Sci. 116:2311-2320. [DOI] [PubMed] [Google Scholar]
- 27.Nash, P. B., M. B. Purner, R. P. Leon, P. Clarke, R. C. Duke, and T. J. Curiel. 1998. Toxoplasma gondii-infected cells are resistant to multiple inducers of apoptosis. J. Immunol. 160:1824-1830. [PubMed] [Google Scholar]
- 28.Ngo, H. M., M. Yang, K. Paprotka, M. Pypaert, H. Hoppe, and K. A. Joiner. 2003. AP-1 in Toxoplasma gondii mediates biogenesis of the rhoptry secretory organelle from a post-Golgi compartment. J. Biol. Chem. 278:5343-5352. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 30.Norrby, R., and E. Lycke. 1967. Factors enhancing the host-cell penetration of Toxoplasma gondii. J. Bacteriol. 93:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ossorio, P. N., J. D. Schwartzman, and J. C. Boothroyd. 1992. A Toxoplasma gondii rhoptry protein associated with host cell penetration has unusual charge asymmetry. Mol. Biochem. Parasitol. 50:1-15. [DOI] [PubMed] [Google Scholar]
- 32.Que, X., H. Ngo, J. Lawton, M. Gray, Q. Liu, J. Engel, L. Brinen, P. Ghosh, K. A. Joiner, and S. L. Reed. 2002. The cathepsin B of Toxoplasma gondii, toxopain-1, is critical for parasite invasion and rhoptry protein processing. J. Biol. Chem. 277:25791-25797. [DOI] [PubMed] [Google Scholar]
- 33.Remington, J. S., R. McLeod, and G. Desmonts. 1995. Toxoplasmosis, p. 140-267. In J. S. Remington and J. O. Klein (ed.), Infectious diseases of the fetus and the newborn infant, 4th ed. W. B. Saunders Company, Philadelphia, Pa..
- 34.Rockey, D., D. Grosenbach, D. Hruby, M. Peacock, R. Heinzen, and T. Hackstadt. 1997. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol. Microbiol. 24:217-228. [DOI] [PubMed] [Google Scholar]
- 35.Roos, D. S., R. G. Donald, N. S. Morrissette, and A. L. Moulton. 1994. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45:27-63. [DOI] [PubMed] [Google Scholar]
- 36.Sadak, A., Z. Taghy, B. Fortier, and J. F. Dubremetz. 1988. Characterization of a family of rhoptry proteins of Toxoplasma gondii. Mol. Biochem. Parasitol. 29:203-211. [DOI] [PubMed] [Google Scholar]
- 37.Schwab, J. C., C. J. Beckers, and K. A. Joiner. 1994. The parasitophorous vacuole membrane surrounding intracellular Toxoplasma gondii functions as a molecular sieve. Proc. Natl. Acad. Sci. USA 91:509-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw, M. K., D. S. Roos, and L. G. Tilney. 2002. Cysteine and serine protease inhibitors block intracellular development and disrupt the secretory pathway of Toxoplasma gondii. Microbes Infect. 4:119-132. [DOI] [PubMed] [Google Scholar]
- 39.Sheng, J., F. Al-Anouti, and S. Ananvoranich. 2004. Engineered delta ribozymes can simultaneously knock down the expression of the genes encoding uracil phosphoribosyltransferase and hypoxanthine-xanthine-guanine phosphoribosyltransferase in Toxoplasma gondii. Int. J. Parasitol. 34:253-263. [DOI] [PubMed] [Google Scholar]
- 40.Sinai, A. P., and K. A. Joiner. 1997. Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu. Rev. Microbiol. 51:415-462. [DOI] [PubMed] [Google Scholar]
- 41.Sinai, A. P., and K. A. Joiner. 2001. The Toxoplasma gondii protein ROP2 mediates host organelle association with the parasitophorous vacuole membrane. J. Cell Biol. 154:95-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinai, A. P., P. Webster, and K. A. Joiner. 1997. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. J. Cell Sci. 110:2117-2128. [DOI] [PubMed] [Google Scholar]
- 43.Soldati, D., J. F. Dubremetz, and M. Lebrun. 2001. Microneme proteins: structural and functional requirements to promote adhesion and invasion by the apicomplexan parasite Toxoplasma gondii. Int. J. Parasitol. 31:1293-1302. [DOI] [PubMed] [Google Scholar]
- 44.Soldati, D., A. Lassen, J. F. Dubremetz, and J. C. Boothroyd. 1998. Processing of Toxoplasma ROP1 protein in nascent rhoptries. Mol. Biochem. Parasitol. 96:37-48. [DOI] [PubMed] [Google Scholar]
- 45.Tomavo, S., A. Martinage, and J. F. Dubremetz. 1992. Phosphorylation of Toxoplasma gondii major surface antigens. Parasitol. Res. 78:541-544. [DOI] [PubMed] [Google Scholar]
- 46.Tomley, F., and D. Soldati. 2001. Mix and match modules: structure and function of microneme proteins in apicomplexan parasites. Trends Parasitol. 17:81-88. [DOI] [PubMed] [Google Scholar]
- 47.Ward, G. E., and K. L. Carey. 1999. 96-well plates providing high optical resolution for high-throughput, immunofluorescence-based screening of monoclonal antibodies against Toxoplasma gondii. J. Immunol. Methods 230:11-18. [DOI] [PubMed] [Google Scholar]
- 48.Ward, G. E., and M. W. Kirschner. 1990. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell 61:561-577. [DOI] [PubMed] [Google Scholar]
- 49.Zurawski, D., and M. Stein. 2003. SseA acts as the chaperone for the SseB component of the Salmonella pathogenicity island 2 translocon. Mol. Microbiol. 47:1341-1351. [DOI] [PubMed] [Google Scholar]