Abstract
The molecular motor myosin I is required for hyphal growth in the pathogenic yeast Candida albicans. Specific myosin I functions were investigated by a deletion analysis of five neck and tail regions. Hyphal formation requires both the TH1 region and the IQ motifs. The TH2 region is important for optimal hyphal growth. All of the regions, except for the SH3 and acidic (A) regions that were examined individually, were required for the localization of myosin I at the hyphal tip. Similarly, all of the domains were required for the association of myosin I with pelletable actin-bound complexes. Moreover, the hyphal tip localization of cortical actin patches, identified by both rhodamine-phalloidin staining and Arp3-green fluorescent protein signals, was dependent on myosin I. Double deletion of the A and SH3 domains depolarized the distribution of the cortical actin patches without affecting the ability of the mutant to form hyphae, suggesting that myosin I has distinct functions in these processes. Among the six myosin I tail domain mutants, the ability to form hyphae was strictly correlated with endocytosis. We propose that the uptake of cell wall remodeling enzymes and excess plasma membrane is critical for hyphal formation.
The pathogenic yeast Candida albicans can undergo a dramatic change in morphology, from spherical yeast cells to long filamentous hyphae. This extreme form of polarized growth requires continual remodeling of the cell wall at the hyphal tip (62). Both endocytosis and secretion contribute to balanced growth of the cell wall (55). These cellular processes are mediated by the actin cytoskeleton, which is comprised of the cortical actin patches and actin cables (3, 8, 17, 30, 31, 55, 56). Cortical actin patches are sites of endocytosis found at bud and hyphal tips, and actin cables are involved in the polarized secretion of vesicles (8, 17, 30). The myosin type I protein in C. albicans, CaMyo5p, is required for organizing the cortical actin patches at the hyphal tip and for hyphal formation (53), although a myosin I null mutant is capable of limited polarized growth and forms pseudohyphae, which are elongated cells that are separated by clear constrictions. Thus, the actin cytoskeleton and associated regulatory proteins are required for hyphal formation (1, 4, 32, 66, 68, 69).
All myosin heavy chains have a globular head domain, a neck, and a tail of various lengths. Class I myosin proteins function as a combination of a single heavy chain associated with one or more light chains (47). The force generated at the expense of ATP hydrolysis is catalyzed by the head domain, which is conserved among the different types of myosin (45). This “motor” domain has ATP and actin-binding sites as well as an actin-dependent Mg2+-ATPase activity. In Saccharomyces, Dictyostelium, and Acanthamoeba, this Mg2+-ATPase activity is modulated by the phosphorylation of a serine at the TEDS-rule site by kinases of the p21-activated kinase family (6, 9, 36, 70). In C. albicans, as in other organisms, this modulation of the motor activity is required for myosin I function, since mutations that prevent phosphorylation of the TEDS-rule site produce a null phenotype (33, 34, 52, 53, 65, 70, 71). In Aspergillus, replacement of the TEDS-rule site with alanine dramatically affected the Mg2+-ATPase activity of myosin I, whereas replacement with the phosphorylation mimic glutamic acid reduced but did not abolish this activity, suggesting that ATPase activity in MyoA is subject to modulation by phosphorylation of the TEDS-rule site. However, the significance of this phosphorylation is not clear because both of the myoA substitution mutants are equally functional in restoring the viability of the null mutant (40).
The N-terminal head domain is followed by a neck region consisting of IQ motifs, which are binding sites for the calmodulin light chain in type I myosins of yeasts and bullfrogs (10, 21, 47). The tail consists of several regions that are thought to specify the roles of myosin I in endocytosis and in organizing cortical actin. Myosin I binding to membranes, possibly through the TH1 region, may be required for endocytosis (13, 46, 47, 59, 64). The role of myosin I in organizing cortical actin is mediated by direct and indirect interactions with actin. First, the TH1 membrane-binding region has been shown to interact with F-actin in the presence of ATP (37, 39). Second, the TH2 region (ATP-independent actin-binding region, or GPA/Q domain) binds actin in vitro (27, 58) and in Saccharomyces cerevisiae assists in the formation of actin-patch-like structures on glass beads (25). Third, the SH3 domain within TH2 and the S. cerevisiae myosin I-specific C-terminal acidic (A) region are required for indirect interactions of myosin I with actin. The SH3 domain is required for the localization of myosin I and for interactions with verprolin (Vrp1) and Las17/Bee1 in S. cerevisiae (2, 18). The A region interacts with and activates the Arp2/3 complex to nucleate the assembly of highly branched actin filaments (18, 33, 38). Based on these multiple protein-protein interactions, myosin I is hypothesized to function in a large complex of cortical actin patch proteins. Many of these components were found in myosin I-containing complexes purified from S. cerevisiae and Dictyostelium discoideum (29, 61).
Genetic studies with various organisms have revealed that myosin I functions in actin-based processes, such as polarized growth, cell motility, phagocytosis, endocytosis, exocytosis, and contractile vacuolar activity (7, 14, 16, 20, 23, 28, 44, 45, 50, 51, 54, 56, 72). Despite extensive investigations of myosin I function at the biochemical level, much remains to be learned about its roles in cellular processes. The ability of C. albicans to switch from a budding to a hyphal form in response to specific environmental conditions is the subject of intense investigation, and many tools have become available to manipulate this process at the molecular level (11). Because myosin I is not essential yet plays a critical role in this morphogenesis (53), we have completed a thorough deletion analysis of the myosin I neck and tail domains. Specifically, we addressed the possible roles of each of these domains in myosin I intracellular localization, in the organization of the actin cytoskeleton, in hyphal formation, and in fluid-phase endocytosis and secretion.
MATERIALS AND METHODS
Strains.
The C. albicans strains used for this study are listed in Table 1. Transformation of C. albicans was done by the lithium acetate method (26). Mutant alleles of CaMYO5 were integrated into a Camyo5/Camyo5 Ura− mutant (53). First, plasmids containing these alleles were linearized with BglII and transformed into strain COU46. The integration of each at the CaMYO5::hisG locus was verified by Southern blot analysis (data not shown).
TABLE 1.
Strains
| Strain | Genotype | Reference |
|---|---|---|
| SC5314 | CaMYO5/CaMYO5 CaURA3/CaURA3 | 19 |
| CAI4 | ura3::imm434/ura3::imm434 | 19 |
| COU42 | CAI4 Camyo5::hisG/Camyo5::hisG-CaURA3-hisG | 51 |
| COU46 | CAI4 Camyo5::hisG/Camyo5::hisG | 51 |
| COU186 | CAI4 Camyo5::hisG/Camyo5 (CAMYO5-GFP) | 51 |
| COU298 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔIQ) | This study |
| COU304 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔA-GFP) | This study |
| COU316 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔSH3) | This study |
| COU325 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔA) | This study |
| COU329 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔSH3-GFP) | This study |
| COU338 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔIQ-GFP) | This study |
| COU362 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔTH2) | This study |
| COU368 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔTH1-GFP) | This study |
| COU370 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔTH1) | This study |
| COU389 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔTH2-GFP) | This study |
| COU406 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔSH3ΔA) | This study |
| COU410 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔSH3ΔA-GFP) | This study |
| COU493 | CAI4 (ARP3-GFP) | This study |
| COU503 | CAI4 Camyo5::hisG/Camyo5::hisG (ARP3-GFP) | This study |
| COU511 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔIQ) (ARP3-GFP) | This study |
| COU521 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔSH3) (ARP3-GFP) | This study |
| COU527 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔA) (ARP3-GFP) | This study |
| COU532 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔTH2) (ARP3-GFP) | This study |
| COU546 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔTH1) (ARP3-GFP) | This study |
| COU549 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔSH3ΔA) (ARP3-GFP) | This study |
| COU724 | CAI4 Camyo5::hisG/Camyo5 (Camyo5ΔC) | This study |
To obtain myosin I deletion mutants expressing ARP3-GFP, we first plated strains COU298 to COU370, which carried the different Camyo5 deletion alleles, on 5-fluoroorotic acid (Diagnostics Chemicals Ltd., Prince Edward Island, Canada) to select for Ura− colonies resulting from excision of the URA3 cassette by homologous recombination between the terminator sequences flanking the CaMYO5 locus. This was verified by Southern blot analysis and by PCR (51). Next, 10 μg of pU159 or pU165 linearized with BglII was transformed into the corresponding Ura− myosin I-deficient strains. Ura+ transformants were screened for the expression of Arp3-green fluorescent protein (GFP) by Western blot analysis (data not shown).
DNA manipulations.
The oligonucleotides used for this study are listed in Table 2. The myosin I tail domains were deleted from CaMYO5 and CaMYO5-GFP and were verified by sequencing. The C-terminal GFP fusion construct was described by Oberholzer et al. (53). The SH3 domain (amino acids [aa] 1168 to 1216) was deleted from pU15 and pU88 (53) by fusion PCRs using the reverse primer with primer UO25 and primer UO24 with primer UO6. The 1.4- and 2.0-kb PCR products were subcloned as HindIII-XbaI fragments into pBluescript KS (Stratagene) to give pU49 and pU100, respectively. The A region (aa 1276 to 1302) was deleted from pU15 by fusion PCRs using the reverse primer with primer UO50 and primer UO49 with primer UO6. The A region was also deleted from pU88 by fusion PCRs using the reverse primer with primer UO60 and primer UO59 with primer UO6. The 1.5- and 2.2-kb PCR products were subcloned as HindIII-XbaI fragments into pKS, creating pU99 and pU107, respectively. The 1.4-, 1.5-, 2.0-, and 2.2-kb HindIII-XbaI fragments from pU49, pU99, pU100, and pU107, respectively, were subcloned together with a 3.8-kb HindIII-BamHI fragment from pU46A (53) into pVEC digested with BamHI and XbaI to generate pU79, pU111, pU112, and pU125, respectively. The Camyo5 and Camyo5-GFP alleles with SH3- and A-domain double deletions were also constructed by standard techniques. The 900-base NsiI-XbaI fragment from pU79 was replaced with the 820-base NsiI-XbaI fragment from pU99 and the 1.5-kb NsiI-XbaI fragment from pU107, both carrying the A region deletion, to generate pU151 and pU152, respectively. The two IQ motifs were also deleted. First, IQ1 (aa 740 to 751) was deleted from pU67 (53) by a fusion PCR using primer UO5 with primer UO56 and primer UO6 with primer UO55. The 5.3-kb PCR product was subcloned as a BamHI-XbaI fragment into pKS, creating pU113. IQ2 (aa 758 to 769) was deleted from pU113 by a fusion PCR using primer UO5 with primer UO58 and primer UO57 with primer UO6. The PCR product was subcloned as a 1.5-kb SalI-HindIII fragment together with a 2.3-kb BamHI-SalI fragment from pU14 (53) into pKS, creating pU123. The 3.8-kb BamHI-HindIII fragment from pU123 and the 1.5-kb HindIII-XbaI fragment from pU15 or the 2.2-kb HindIII-XbaI fragment from pU88 were cloned into pVEC digested with BamHI and XbaI to generate pU129 and pU132, respectively. The TH1 region (aa 775 to 985) was deleted from pU67 by a fusion PCR using primer UO5 with primer UO54 and primer UO53 with primer UO6. The PCR product was subcloned as a 940-bp SalI-HindIII fragment together with a 2.3-kb BamHI-SalI fragment from pU14 into pKS, creating pU135. The 3.2-kb BamHI-HindIII fragment from pU135 and the 1.5-kb HindIII-XbaI fragment from pU15 or the 2.2-kb HindIII-XbaI fragment from pU88 were cloned into pVEC digested with BamHI and XbaI to generate pU141 and pU143, respectively. The TH2 region (aa 1061 to 1167) was deleted from pU67 by a fusion PCR using primer UO5 with primer UO52 and primer UO6 with primer UO51. The PCR product was subcloned as a 1.6-kb SalI-NcoI fragment together with a 2.3-kb BamHI-SalI fragment from pU14 into pUCBM21 (Boehringer Mannheim), creating pU136. The 3.9-kb BamHI-NcoI fragment from pU136 and the 1.1-kb NcoI-XbaI fragment from pU15 or the 1.8-kb NcoI-BamHI fragment from pU88 were cloned into pVEC digested with BamHI and XbaI to generate pU142 and pU144, respectively. The C-terminal region, including the TH2, SH3, and A domains (aa 1061 to 1302), was deleted from pU67 by a PCR using primers UO5 and UO164. The PCR product was subcloned as a 1.5-kb SalI-PstI fragment into pU50, which has an engineered PstI site just before the stop codon (53), to generate pU193. The 2.3-kb SalI-XbaI fragment from pU193 together with the 2.3-kb BamHI-SalI fragment from pU67 were cloned into pVEC digested with BamHI and XbaI to generate pU195.
TABLE 2.
Oligonucleotides
| Oligonu-cleotide | Sequencea |
|---|---|
| UO5 | GCGGATCCCTGACCTTACACTGTCGTTGG |
| UO6 | GCTCTAGACATTGGCCACTTGTGGAGGTTC |
| UO24 | TTCCCTACATACAAGCTCAAAGAATGTTCAC |
| UO25 | GTGAACATTCTTTGAGCTTGTATGTAGGGAATTTAG |
| UO44 | AAACTGCAGTTATTTGTATAGTTCATCCATG |
| UO49 | CTACATTAGCGGGGTCATAGGTAAGAACTTG |
| UO50 | AACAAGTTCTTACCTATGACCCCGCTAATGTAG |
| UO51 | CAGTCACAACAACAAACAGCTATGTTTGATTATGATGG |
| UO52 | CCATCATAATCAAACATAGCTGTTTGTTGTTGTGACTG |
| UO53 | GGTAATCAATTTGAGCAATTCAAAAGACCAAGAGGGG |
| UO54 | AACCCCTCTTGGTCTTTTGAATTGCTCAAATTGATTACC |
| UO55 | AACATGGCAGCAAGAAAGGAAGATGCAGC |
| UO56 | TGCTGCATCTTCCTTTCTTGCTGCCATGTTATGC |
| UO57 | GATGCAGCAAAAACTAATCAATTTGAGCAATTCC |
| UO58 | TTGCTCAAATTGATTAGTTTTTGCTGCATCTTCC |
| UO59 | ACATTAGCGGGGTCAGCAAGCTTTATGAGTAAAGG |
| UO60 | ACTCATAAAGCTTGCTGACCCCGCTAATGTAG |
| UO92 | CCAAGAGACCTTTTTTCAGA |
| UO93 | CGACAAACCAATCAAACCGC |
| UO94 | TTATCTTCGTGGACTCCAAC |
| UO95 | ATGTATGTCATTTGCAACCA |
| UO96 | GTAAAGACGACGACGACAAAGAC |
| UO97 | TCATGGTGTTCAATTAATGA |
| UO105 | ACGCGTCGACATGAGTAAAGGAGAAGAACTTTTC |
| UO106 | AAGCCGGTTTTGCCGGTG |
| UO107 | TGGTGAACAATGGATGGAC |
| UO108 | GGTTGGATCCAATTGAAAC |
| UO109 | GCTCTAGATTCTGAGGACAAATCTCG |
| UO112 | CCGCTCGAGAGGTACTGAAAACAAAGAGAAGTTTC |
| UO113 | CCAATGCATTAAATATTTGTATAGTTTATTAAACAGAG |
| UO164 | AAAACTGCAGTGTTTGTTGTTGTGACTGTTGTGG |
| Reverse | GGAAACAGCTATGACCATG |
The underlined sequences correspond to restriction sites used for cloning.
The ARP3-GFP construct was made by PCR amplification of genomic DNA from SC5314, a wild-type strain, with primers UO108 and UO109. The 2.3-kb PCR product was cloned into pVEC as a BamHI-XbaI fragment, creating pU155. XhoI and NsiI restriction sites were added 5′ of the stop codon of ARP3 in pU155 by a PCR using primers UO112 and UO113. The GFP sequence (48) was PCR amplified with UO105 and UO44. The 700-base PCR product was cloned as a SalI-PstI fragment into the 8.6-kb PCR product consisting of ARP3 in pVEC to generate pU159. Several clones were verified by sequencing. The alanine at position 44 was replaced with a threonine in our cloned ARP3 sequence compared with the one available in the Stanford sequence database.
Phenotypic analyses.
The media used for hyphal induction were as follows: YPD (1% yeast extract, 2% Bacto peptone, 2% glucose, pH 6.5), YPD supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco BRL), 2% agar supplemented with 10% FBS, Spider medium (1% nutrient broth, 1% mannitol, 0.2% potassium phosphate [pH 7.2], 1.5% agar), and SD Ura− (0.67% yeast nitrogen base with ammonium sulfate, amino acids, adenine, tryptophan, and 2% glucose).
Strains were grown in YPD overnight, and 30 μl of a 10−5 dilution was spread on one-sixth of a plate (agar-FBS and Spider medium). Plates were incubated for 4 days at 37°C, and colonies were visualized with an inverted Nikon TMS microscope with a 2× objective and a 10× projection lens. Strains from overnight cultures grown in YPD were diluted 1:20 in YPD supplemented with 10% FBS and were incubated at 37°C for 2 h. The cells were visualized by Nomarski optics under an upright Leitz Aristoplan microscope with a 100× objective and a 10× projection lens.
Doubling times for each strain were determined for three independent colonies grown overnight in YPD and diluted to an optical density at 600 nm (OD600) of 0.1 at the beginning of the time course. The cultures were incubated at 30°C and the OD600 values were determined every hour for up to 7 h.
Northern blot analyses.
Total RNAs were extracted by the use of phenol and glass beads from wild-type and myosin I deletion strains grown to early log phase in YPD. Twenty micrograms of total RNA per sample was separated in a 7.5% formaldehyde-1% agarose gel, blotted onto a Zeta-Probe nylon membrane (Bio-Rad, Ontario, Canada) and probed with 32P-labeled DNAs specific for ACT1 (orf19.5007), DDR48 (orf19.4082), RTA2 (orf19.24), and orf19.675 as described previously (43). All Northern probes were PCR products that were subsequently labeled by random priming (Amersham Biosciences, Piscataway, N.J.). The oligonucleotides used were UO106 and UO107 (ACT1), UO92 and UO93 (orf19.675), UO94 and UO95 (RTA2), and UO96 and UO97 (DDR48) (Table 2).
Fluorescence microscopy.
Cultures of the various strains grown overnight in YPD were diluted 1:20 in either YPD or SD Ura− supplemented with 10% FBS and then grown for 1 to 2 h at 37°C. Cells expressing GFP-tagged myosin I were washed once in phosphate-buffered saline and then mounted on slides for visualization with an appropriate filter at room temperature by epifluorescence microscopy under an upright Leitz Aristoplan microscope with a 100× immersion oil objective and a 10× projection lens. Pictures were acquired with a Princeton Instruments RTE/CCD camera and Northern Eclipse software (Empix Imaging). Cells expressing GFP-tagged Arp3 were mounted directly on slides and were visualized at room temperature by epifluorescence microscopy under a Leica-DM-IRB inverted microscope with a 63× objective and a 10× projection lens. Pictures were acquired with a Sensys charge-coupled device camera and Openlab 3.1 software. Rhodamine-phalloidin (Molecular Probes, Eugene, Oreg.), 4′,6′-diamidino-2-phenylindole (DAPI; Sigma), and Calcofluor White (Sigma) staining was done as described by Oberholzer et al. (53).
Video microscopy.
Hyphal growth rates were measured for strains SC5314, COU316, COU325, COU362, and COU406 by video microscopy as described previously (51).
Protein extracts and Western blot analysis.
Overnight cultures of the different strains expressing GFP-tagged proteins were diluted 1:100 in fresh YPD and grown to late exponential phase (OD600 of 2 to 3). The cells were lysed as described previously (51). Equal amounts of protein per sample were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto nitrocellulose membranes, and analyzed by Western blotting for myosin I-GFP and Hog1 protein levels with anti-GFP (a generous gift from B. Massie) and anti-p38 (Santa Cruz Biotechnology) antibodies.
Subcellular fractionation of myosin I mutants.
Cells expressing CaMYO5-GFP (WT), CaMYO5ΔIQ-GFP (ΔIQ), CaMYO5ΔTH2-GFP (ΔTH2), or CaMYO5ΔAΔSH3-GFP (ΔAΔSH3) were grown in YPD to late exponential phase. Cell equivalents of 250 OD600 units were harvested by centrifugation, washed with ice-cold extraction buffer (buffer A) containing 50 mM HEPES (pH 7.5), 10% glycerol, 100 mM KCl, 1 mM dithiothreitol, 2 mM MgCl2, 0.25 mM ATP, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, and protease inhibitor cocktail tablets (Roche Diagnostics GmbH), and disrupted in a French press. The homogenates were cleared of intact cells and debris at 1,000 × g for 5 min at 4°C and then were centrifuged for 30 min at 100,000 × g at 4°C. The supernatant (S) fraction was removed, and the pellet fraction (P) was washed with extraction buffer and centrifuged as described above. Aliquots of all of the fractions were adjusted with SDS sample buffer to equivalent final volumes, and the samples were subjected to SDS-PAGE, transferred to nitrocellulose, and processed for Western blot analysis.
For alkaline phosphatase treatment, the pellet was resuspended in extraction buffer supplemented with 0.1% Triton X-100 (buffer B), and pelletable myosin I-GFP was immunoprecipitated with the mouse monoclonal 3E6 anti-GFP antibody coupled to protein G-Sepharose beads (Amersham Pharmacia Biotech) for 1 h. The beads were extensively washed with the extraction buffer, and the immunoprecipitates were treated with 10 U of alkaline phosphatase (New England Biolabs) and resuspended in a reaction buffer containing 50 mM Tris-HCl (pH 9.0), 100 mM KCl, 10 mM MgCl2, 10 μM ZnCl2, and 1 mM dithiothreitol or in the same buffer lacking phosphatase for 30 min. Proteins were eluted from the beads with 0.5 M acetic acid, pH 3.4. The eluates were then lyophilized and resuspended in SDS sample buffer. The proteins were separated by SDS-PAGE and analyzed by Western blotting.
For extraction experiments, the pellets were resuspended in 500 μl of extraction buffer A. Another 500 μl of extraction buffer A containing 1 M NaCl was added to the tubes and incubated for 15 min. The extracts were centrifuged at 100,000 × g for 30 min. Proteins in the supernatants were precipitated with 10% trichloroacetic acid, washed with ice-cold acetone, and resuspended in SDS sample buffer. The remaining pellets were washed with buffer A and extracted with buffer A containing 1% Triton X-100 for another 15 min. The extracts were processed as described above. Pellet and supernatant fractions were analyzed by Western blotting.
For sucrose gradient fractionation, the cell lysates were centrifuged for 15 min at 10,000 × g at 4°C. The supernatant aliquots, yielding 50 OD280 units, were loaded on an eight-step (25 to 60% [wt/wt]) sucrose gradient over a 65% (wt/wt) 2-ml sucrose pad, with each step prepared in 50 mM HEPES, pH 7.5, and 2 mM MgCl2. The gradients were centrifuged at 38,000 rpm in an SW41 swinging-bucket rotor (Beckman) at 4°C for 12 h. Fractions (0.85 ml) were collected from the tip. Proteins were precipitated with 10% trichloroacetic acid, and the protein pellets were washed with ice-cold acetone, resuspended in SDS sample buffer, and processed for Western blot analysis. GFP fusion proteins were probed with the mouse monoclonal 11E5 anti-GFP antibody. Rvs167 and cofilin were probed with rabbit polyclonal antibodies against these yeast proteins (generous gifts from Brenda Andrews and David Drubin, respectively). Actin was probed with the mouse monoclonal mAB150 antibody against mouse actin (Chemicon International). Finally, calmodulin was detected with a rabbit polyclonal CaM I antibody against rabbit calmodulin (Santa Cruz Biotechnology). Detection was done by enhanced chemiluminescence.
LY and FM4-64 uptake assay and secretion assay.
Lucifer Yellow (LY) uptake was measured essentially as described by Basrai et al. (5) except that the cells were harvested by centrifugation at 1,500 × g. Dead cells were identified by propidium iodide staining (10 μg/ml), and the fraction of cells that internalized LY into their vacuoles was determined by counting. The procedure for visualizing FM4-64 uptake was done as described by Vida and Emr (67). The secretion of secreted aspartyl proteases was monitored as described by Lee et al. (35).
RESULTS
IQ and TH1 are essential for proper growth rates.
We previously identified a single gene encoding myosin I in C. albicans and generated a null mutant. The resulting Δ/Δ Camyo5 strain was unable to make hyphae (53). To identify specific domains of the neck and C-terminal tail of myosin I that are required for hyphal formation and for the intracellular localization of myosin I and cortical actin patches, we generated a series of mutants that lacked the IQ motifs, the TH1 domain, the large N-terminal TH2 region, the SH3 domain, and the A region (Fig. 1). We introduced these mutant alleles into the Δ/Δ Camyo5 mutant. Phenotypic comparisons of the mutants were made with the SC5314 and reintegrant COU73 (53) wild-type strains. Because these wild-type strains displayed identical behaviors in all processes examined, only data for the SC5314 strain are shown here. The morphology and doubling times of the mutants relative to the wild type and the myosin I-deficient strain under yeast growth conditions were determined. As observed previously, the Δ/Δ Camyo5 null mutant formed very round cells and grew in clumps, probably due to a defect in cytokinesis (53). Deletion of the IQ motifs and the TH1 domain resulted in similar spherical cells that grew in clumps, whereas deletion of the TH2, SH3, and A regions or a double deletion of the SH3 and A domains did not affect the cellular morphology (data not shown). Deletion of either the TH1 domain (ΔTH1) or the IQ motifs (ΔIQ) affected the growth rate to the same extent as a complete myosin I deficiency (compare strains COU370 and COU298 with strain COU42 in Table 3). In contrast, deletion of the TH2, SH3, or A region (ΔTH2, ΔSH3, or ΔA [strains COU362, COU316, and COU325, respectively]) did not affect the growth rates of cells compared to that of the wild-type strain SC5314, while the growth rate was moderately affected by a double deletion of the SH3 and A regions (ΔSH3ΔA [COU406]). These results suggest that the TH1 region and the IQ motifs are essential for myosin I function.
FIG. 1.
Deletion analysis of myosin I tail domains of C. albicans. The black lines correspond to the deletion alleles used in this study. The numbers above the deletion constructs correspond to the amino acids deleted based on the wild-type sequence.
TABLE 3.
Doubling times
| Strain | Myosin I allele | Doubling time (h)a |
|---|---|---|
| SC5314 | CaMYO5 | 1.04 ± 0.03 |
| COU42 | None | 1.51 ± 0.14 |
| COU298 | Camyo5ΔIQ | 1.39 ± 0.15 |
| COU316 | Camyo5ΔSH3 | 1.06 ± 0.05 |
| COU325 | Camyo5ΔA | 1.05 ± 0.06 |
| COU362 | Camyo5ΔTH2 | 1.13 ± 0.08 |
| COU370 | Camyo5ΔTH1 | 1.50 ± 0.19 |
| COU406 | Camyo5ΔSH3ΔA | 1.25 ± 0.15 |
Data are means ± SD calculated from three independent colonies per strain.
The TH1 and IQ regions are essential for hyphal growth.
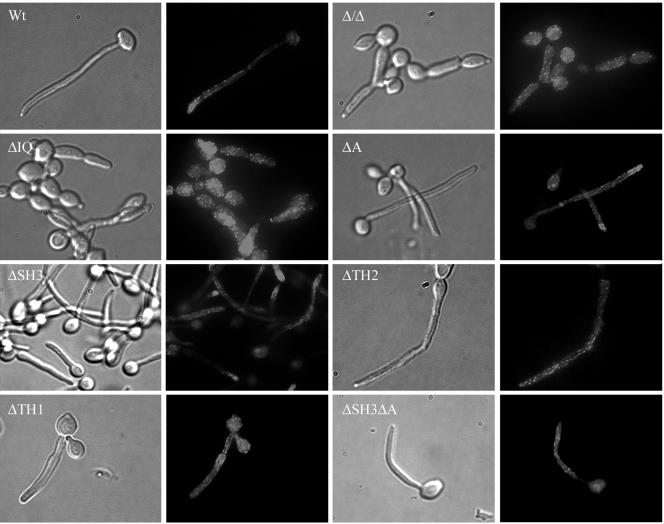
The myosin I mutants were tested for the ability to form hyphae. First, cells from the mutant strains and the wild-type strain SC5314 were incubated in a liquid hypha-inducing medium. The ΔIQ and ΔTH1 mutants rarely formed germ tubes and failed to form true hyphae. Instead, they formed pseudohyphal cells similar to those of the Δ/Δ Camyo5 mutant (Fig. 2a). Results on solid medium were similar; only the ΔIQ motif or the ΔTH1 region mutants did not form hyphae (Fig. 2b and c). However, the ΔA, ΔTH2, and ΔSH3ΔA mutants exhibited a less extensive hyphal network than did colonies of the wild type. Deletion of the C-terminal region (ΔC), which removed the TH2, SH3, and A regions, affected the ability of cells to form hyphae in liquid and solid media, although some true hyphae could be observed in liquid medium (Fig. 2a). Overall, these observations suggested that these regions of the C terminus together play a role in hyphal formation and therefore that the IQ motifs and TH1 region are not sufficient for hyphal formation.
FIG. 2.
Hyphal formation in wild-type and myosin I mutant strains. (a) Hyphal formation in liquid YPD supplemented with 10% FBS was determined at 37°C after 2 h of incubation. Bar = 5 μm. Hyphal formation on solid media was determined by plating single colonies of each strain on 10% FBS (b) or Spider medium (c) plates. Plates were incubated for 4 days at 37°C. Bar = 1 mm.
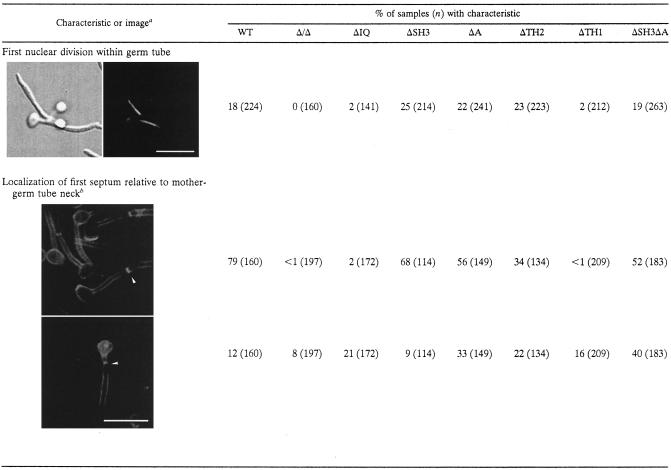
True hyphal cells form filaments of uniform width and present other characteristics regarding cellular division that differentiate them from pseudohyphal cells (63). To determine the extent of true hyphal formation, we quantified nuclear migration into germ tubes and measured the distance between the first septum and the mother-germ tube neck (Table 4). A first characteristic of true hyphae is the migration and division of the nuclei in the germ tubes 100 min after hyphal induction (63). Nuclei in pseudohyphal cells divide across the mother cell and germ tube neck and do not migrate into the hyphae (63). Of the ΔIQ and ΔTH1 mutant elongated cells, 2% had nuclei that had entered into the germ tube and divided compared to between 18 and 25% for wild-type and other mutant strains (Table 4). A second characteristic that is indicative of true hyphal formation is a distance of 5 to 15 μm between the first septum and the mother-germ tube neck, while in pseudohyphae the first septum is located at the neck of the mother and the daughter cell (63). According to these criteria, the ΔIQ and ΔTH1 strains had fewer than 2% true hyphal cells, compared to between 34 and 79% in wild-type and other mutant strains (Table 4). However, the myosin I deletion, ΔIQ, and ΔTH1 strains had a significant proportion of cells in which the first septum was >0 but <5 μm from the mother-germ tube neck, suggesting that in these mutants hyphal growth was initiated but not maintained. Together with the cellular morphologies of the mutants, these data show that the ΔA, ΔSH3, ΔTH2, and ΔSH3ΔA mutants can form true hyphae and that the Δ/Δ, ΔIQ, and ΔTH1 strains cannot.
TABLE 4.
True hyphal characteristics
The top image was stained with DAPI and the bottom images were stained with calcofluor. Bar, 10 μm.
The top image is for septa of >5 μm, and the bottom image is for septa of >0 and <5 μm.
Hyphal growth rates were also measured for the wild-type and mutant strains that could form hyphae (Table 5). In all but the ΔTH2 mutant, the hyphal growth rates were higher than 20 μm/h and were not significantly different from the wild-type growth rate. In ΔTH2 cells, hyphal growth rates were reduced to 16 μm/h (P < 10−8), suggesting that TH2 is required for optimal hyphal growth.
TABLE 5.
Hyphal growth rates
| Strain | Myosin I allele | Growth rate (μm/h)a | No. of hyphae |
|---|---|---|---|
| SC5314 | CaMYO5 | 23.2 ± 4.4 | 37 |
| COU316 | Camyo5ΔSH3 | 23.9 ± 3.9 | 36 |
| COU325 | Camyo5ΔA | 24.2 ± 5.2 | 34 |
| COU362 | Camyo5ΔTH2 | 16.5 ± 3.7b | 36 |
| COU406 | Camyo5ΔSH3ΔA | 21.4 ± 4.5c | 32 |
Data are means ± SD.
The difference in the means for hyphal growth rates for COU362 and SC5314 is significant with a P value of <10−8.
The difference in the means for COU406 and SC5314 is not significant (P = 0.09).
Transcript levels of DDR48, RTA2, and a hypothetical open reading frame (ORF) are upregulated in hypha-defective myosin I mutants.
Preliminary DNA microarray analyses of transcript levels showed that the expression of several genes involved in different processes is induced in a myosin I null mutant (data not shown). The levels of the three most upregulated transcripts, DDR48, RTA2, and orf19.675, the last of which encodes a protein of unknown function, were determined for the myosin I deletion strains by Northern blot analyses (Fig. 3). Transcript levels for all three genes were elevated relative to those of ACT1 in the hypha-defective Δ/Δ, ΔIQ, and ΔTH1 mutants. They were also elevated, but to a lesser extent, in the ΔTH2 mutant, which also exhibited more severe hyphal growth defects than the ΔA, ΔSH3, and ΔSH3ΔA mutant strains. The expression of the stress response gene DDR48 is activated during the yeast-to-hypha transition (49), indicating that the elevated transcript levels observed are probably not the cause of the hyphal defects in these mutants. Rather, these elevated transcript levels may reflect the physiological state of stressed cells.
FIG. 3.

DDR48, RTA2, and orf19.675 transcript levels in wild-type and mutant strains. Equal amounts of total RNA from the wild-type and mutant strains were separated by electrophoresis, blotted onto nylon membranes, and probed with 32P-labeled DNA probes specific for DDR48, RTA2, orf19.675, and ACT1 (actin). The transcript levels were quantified with a phosphorimager and normalized relative to the actin transcript levels. The numbers below each panel correspond to the fold expression relative to the wild type.
Localization of myosin I correlates with cortical actin patch distribution.
To identify the region(s) responsible for proper myosin I distribution, we determined the localization of mutant myosin I proteins fused to GFP at the C terminus and expressed in a strain that is otherwise deficient for myosin I (i.e., the Δ/Δ Camyo5 background) under hypha-inducing conditions. Western blot analysis showed that the mutant myosin I-GFP fusion proteins were expressed at similar levels to that of wild-type myosin I-GFP, except in the ΔSH3 and ΔSH3ΔA mutants, in which lower levels of expression were observed (Fig. 4A). The wild-type myosin I-GFP protein was able to fully complement the myosin I deficiency and was localized in patches at the hyphal tips (53). The mutant myosin-GFP-tagged alleles generated similar hyphal phenotypes to those of the corresponding nontagged alleles (data not shown). Deletion of the SH3 or A regions did not affect myosin I localization in patches at the hyphal tips (Fig. 4B). However, deletion of these domains together had a dramatic effect on localization. While the majority of wild-type hyphae (70%; n = 140) showed myosin I-GFP localization in patches at the tips, this was observed in only 7% of ΔSH3ΔA mutant hyphae (n = 95), and most of the signal was cytoplasmic and diffuse. Deletion of the IQ motifs resulted in punctate or patch-like myosin I localization throughout the cell surface (Fig. 4B). The absence of the TH1 and TH2 regions resulted in a uniform cytoplasmic distribution in pseudohyphal and hyphal cells.
FIG. 4.
Localization of wild-type and mutant CaMyo5-GFP. (A) Equal amounts of protein extracts (25 μg) prepared from cells expressing CaMYO5-GFP (WT), CaMYO5ΔIQ-GFP (ΔIQ), CaMYO5ΔA-GFP (ΔA), CaMYO5ΔSH3-GFP (ΔSH3), CaMYO5ΔTH2-GFP (ΔTH2), CaMYO5ΔTH1-GFP (ΔTH1), or CaMYO5ΔAΔSH3-GFP (ΔAΔSH3) grown in YPD to late exponential phase were separated by SDS-PAGE and analyzed by Western blotting with anti-GFP and anti-p38 antibodies. Molecular weight markers (M; in thousands) are indicated on the right. (B) Cells induced with 10% FBS in YPD at 37°C for 2 h were visualized for GFP fluorescence. Bar = 5 μm.
To determine whether the tail subregions are required for the myosin I-dependent localization of cortical actin patches, we incubated mutant and wild-type strains under hypha-inducing conditions and visualized cortical actin patches by rhodamine-phalloidin staining and fluorescence microscopy. Whereas in the wild type, cortical actin patches were primarily polarized to the hyphal tips, deletion of the IQ motifs, the TH1 region, or the TH2 region depolarized the distribution of cortical actin patches in pseudohyphal and hyphal cells to extents similar to the mislocalization observed in the myosin I null mutant (Fig. 5). The cortical actin patch distribution patterns were quantified for mutants that were able to make hyphae. In hyphae of the ΔTH2 mutant, cortical actin patches were mostly depolarized, similar to those of the phosphorylation-mimetic mutant, S366D (Table 6) (53). Deletion of the A region did not alter the polarized distribution of cortical actin patches. Deletion of the SH3 domain reduced the number of hyphal cells with polarized cortical actin patches (Table 6). However, deletion of the SH3 and A regions together led to a significant decrease in hyphal cells with a polarized distribution of cortical actin patches and to a concomitant increase in the proportion of hyphal cells with an abnormal distribution pattern of these patches (Table 6). These results suggest that the SH3 and A regions function together to organize cortical actin. Moreover, among the myosin I mutants, there was a close correlation between the severity of the defects in both myosin I localization and the polarized localization of cortical actin patches at hyphal tips.
FIG. 5.
Cortical actin cytoskeleton in wild-type and myosin I mutants. Hyphal formation was induced with 10% FBS at 37°C for 1 h 30 min. The cells were then fixed and stained with rhodamine-phalloidin (RP). The Nomarski and corresponding rhodamine-phalloidin images are shown for each strain. Bar = 5 μm.
TABLE 6.
Cortical actin patch patternsa
 |
Polarized, cortical actin patches localized exclusively at the hyphal tip; loose, cortical actin patches localized preferentially at the tip but also throughout the hyphae; abnormal, depolarized distribution of cortical actin patches, and in the ΔSH3ΔA strain cortical actin patches were reduced in number.
Bar = 5 μm.
Structural determinants of myosin I localization.
Since myosin I is known to localize to cortical actin patches (53), the patterns of localization of the mutant myosin I-GFP fusion proteins might reflect their ability to associate with these cortical patches. This hypothesis was tested by comparing the distributions of the wild-type and mutant myosin I-GFP fusion proteins in subcellular fractions. To separate supernatant (S) and pellet (P) pools of myosin I, we subjected the cellular extracts to high-speed centrifugation. The majority of actin was pelleted from wild-type and mutant extracts under these conditions (Fig. 6A, actin; also data not shown). As shown in Fig. 6A, the amounts of pelletable myosin I were reduced by the ΔTH1 and ΔSH3ΔA mutations. We observed differences in the mobility of myosin I in the soluble and pelletable fractions during SDS-PAGE. The wild-type myosin I protein in the P fraction was mostly represented by a lower-mobility species than that in the S fraction (Fig. 6B). Myosin I is known to contain consensus phosphorylation sites for p21-activated kinase and other kinases (53; U. Oberholzer and M. Whiteway, unpublished observations). To determine whether the lower-mobility isoforms were due to protein phosphorylation, we immunoprecipitated the fusion proteins with anti-GFP antibodies and then treated them with alkaline phosphatase. As shown in Fig. 6B, phosphatase treatment increased the mobility of wild-type myosin I-GFP in the P fraction to that of the faster migrating isoform in the S fraction, indicating that the former is phosphorylated. We determined that deletion of the TH2 region did not significantly affect this phosphorylation. In contrast, the mutant GFP-fusion protein lacking the IQ motifs, the TH1 region, or the SH3 and A regions did not show the lower-mobility isoform or produced a reduced mobility shift in the pelletable fractions, suggesting that these domains are required for phosphorylation (Fig. 6A and B).
FIG. 6.
Subcellular fractionation of myosin I-GFP-containing protein complexes. (A) Lack of TH1 and TH2 regions and simultaneous deletion of SH3 and A domains increases solubility of myosin I. CaMYO5-GFP (WT)-, CaMYO5ΔIQ-GFP (ΔIQ)-, CaMYO5ΔTH1-GFP (ΔTH1)-, CaMYO5ΔTH2-GFP (ΔTH2)-, or CaMYO5ΔAΔSH3-GFP (ΔAΔSH3)-expressing cells were extracted, as described in Materials and Methods, and centrifuged for 30 min at 100,000 × g at 4°C. The supernatant (S) and pellet (P) fraction aliquots were subjected to SDS-PAGE. Myosin I-GFP (CaMyo5-GFP) was detected with an anti-GFP 11E5 monoclonal antibody. Actin was detected in wild-type (shown here) and mutant (data not shown) fractions with anti-mouse actin mAB150. (B) Wild-type and mutant myosin I-GFP in pellet fractions were treated with (+) or without (−) alkaline phosphatase (AP) as described in Materials and Methods. (C) Sucrose velocity centrifugation at 10,000 × g. Supernatants of the cell extracts obtained from either yeast or hyphal wild-type cells were loaded on a 25 to 60% (wt/wt) sucrose gradient and centrifuged at 38,000 rpm for 12 h. Fractions were collected from the top. Proteins were analyzed by Western blotting using monoclonal antibodies directed against GFP and actin, and rabbit polyclonal anti-Rvs167, anti-cofilin, and anti-calmodulin antibodies. (D) Sequential extraction of pelletable myosin I. Pelleted proteins were sequentially extracted with 500 mM NaCl (NaCl) and then with 1% Triton X-100 (TX). S corresponds to the 100,000 × g soluble fraction, NaCl and TX correspond to solubilized proteins after sequential extraction of the pellet, and P corresponds to 100,000 × g pelletable proteins that were not extracted with NaCl and TX.
To confirm that pelletable myosin I is indeed associated with cortical actin patches, we determined its distribution in sucrose density gradients. Extracts were prepared from both the yeast and hyphal forms of the strain expressing the wild-type myosin I-GFP fusion protein. In each case, myosin I was represented by two separable pools: a light pool, which remained within the few fractions at the top of the gradient, and a heavy pool, which sedimented closer to the bottom (Fig. 6C). The distribution of myosin I within the gradient was compared to that of actin and other cortical actin patch components by Western blot analysis. The heavy pool of myosin I largely cosedimented with actin, Rvs167, cofilin, and calmodulin (Fig. 6D). The higher extractability of all of the mutant forms of myosin I indicated that their association with pelletable actin was compromised. Sequential extraction of the mutant proteins with 500 mM NaCl followed by 1% Triton X-100 (TX) resulted in a complete (for ΔIQ and ΔSH3ΔA) or nearly complete (for ΔTH1 and ΔTH2) shift of myosin I to the supernatant, while the full-length protein was poorly extractable under these conditions (Fig. 6C). Interestingly, the ΔIQ mutant was more readily extracted with detergent than with salt.
Arp3-GFP tip localization is dependent on myosin I.
Evangelista et al. (18) showed that the A domain interacts with components of the Arp2/3 complex in S. cerevisiae. To determine if myosin I contributes to Arp2/3 localization, we introduced Arp3-GFP into the myosin I-deficient and wild-type strains. A Western blot analysis revealed that the resulting strains, except for the ΔSH3 and ΔTH2 mutants, expressed similar levels of a GFP fusion protein of the expected molecular mass (73 kDa) (Fig. 7A). Three observations indicated that this Arp3-GFP fusion protein is associated with cortical actin patches, as in S. cerevisiae (40). First, the Arp3-GFP signal was localized in patches at the hyphal tips of the wild-type strain (Fig. 7B). Second, a treatment of cells with latrunculin A (200 μM), which causes disassembly of actin filaments, also produced depolarization and the fragmentation of Arp3-GFP patches, suggesting that Arp3-GFP localization is F-actin dependent (Fig. 7B). Third, in the Δ/Δ Camyo5 strain, which has mislocalized cortical actin patches, Arp3-GFP was also present in the cytoplasm and in mislocalized patches (Fig. 7C). Thus, myosin I is required for Arp3 localization.
FIG. 7.
Arp3-GFP localization in wild-type and myosin I mutant cells. Arp3-GFP was introduced into Ura− strains of CAI4, Δ/Δ Camyo5, ΔIQ, ΔA, ΔSH3, ΔTH1, ΔTH2, and ΔAΔSH3. (A) Western blot analysis of the levels of Arp3-GFP in wild-type and myosin I mutants. Equal amounts of protein extracts prepared from each strain were separated by SDS-PAGE and analyzed by Western blotting. (B) Wild-type hyphal cells expressing Arp3-GFP treated with (+LatA) or without (−LatA) 200 μM latrunculin A. (C) Arp3-GFP localization in wild-type and mutant cells incubated at 37°C in SD-URA containing 10% FBS for 1 h 30 min (WT, ΔA, ΔSH3, ΔTH2, and ΔAΔSH3) or 2 h 30 min (Δ/Δ Camyo5, ΔIQ, and ΔTH1) was visualized by epifluorescence microscopy. Arrowheads indicate the Arp3-GFP signal at hyphal tips of WT, ΔSH3, and ΔA cells. Bars = 10 μm.
The distribution of the Arp3-GFP fusion protein was also determined for the myosin I deletion mutants. In the ΔSH3 and ΔA deletion strains, Arp3-GFP patches were localized to the hyphal tips of 58% (n = 130) and 66% (n = 167) of the cells, respectively, compared to 87% of the wild-type hyphae (n = 116). In the ΔSH3ΔA mutant, only 18% (n = 151) of the hyphae showed a polarized localization of Arp3-GFP, suggesting that these domains function together in localizing Arp2/3 to hyphal tips. In the other myosin I deletion strains examined, the Arp3-GFP patches were also mislocalized (Fig. 7C). These results suggest that the IQ, TH1, and TH2 domains of myosin I are required for the polarized distribution of Arp3-GFP patches.
Endocytosis, but not secretion, is compromised in myosin I mutants that are defective for hyphal formation.
Increasing evidence points to a role for the cortical actin patches in endocytosis (17, 30, 55, 60). Because some of the myosin I mutants displayed defects in cortical actin patch distribution, we examined their ability to perform endocytosis. The uptake of a low-molecular-weight dye, LY, into vacuoles occurs by fluid-phase endocytosis and can be readily visualized by microscopy (5). Dead Δ/Δ Camyo5 cells could be stained with LY, so propidium iodide was used to discriminate between cells that were alive and had taken up LY and those that were dead (12). Of the wild-type cells, 32% incorporated LY (Table 7), while only 4% of Δ/Δ Camyo5 mutant cells did so. Between 3 and 6% of cells of the ΔIQ, ΔTH1, and ΔTH2 strains incorporated LY, which is comparable to the case for the myosin I null mutant. Of the ΔSH3 or ΔA mutant cells, 22% incorporated LY. A double deletion of the SH3 and A domains did not have a major effect on LY uptake. To confirm these results, we also examined the uptake of the vacuolar membrane dye FM4-64 (67). We found that the deletion of myosin I or of its IQ or TH1 domains dramatically reduced the uptake of FM4-64 in budding and filamentous cells compared to that in wild-type cells (Fig. 8; data not shown). Deletion of TH2 partially reduced FM4-64 uptake, whereas deletion of the SH3 and A regions had no significant effect on FM4-64 uptake in yeast and hyphal cells (Fig. 8; data not shown). These results suggest that the IQ motifs and the TH1 region are required for myosin I function in fluid-phase endocytosis, whereas the TH2 region is only partially required.
TABLE 7.
LY uptake
| Strain | Myosin I allele | % of cells with LY uptake | % of cells with propidium iodide uptake | Total no. of cells |
|---|---|---|---|---|
| SC5314 | CaMYO5 | 37 | 1 | 615 |
| COU42 | None | 4 | 12 | 480 |
| COU298 | Camyo5ΔIQ | 3 | 9 | 266 |
| COU316 | Camyo5ΔSH3 | 22 | 1 | 389 |
| COU325 | Camyo5ΔA | 22 | <0.5 | 445 |
| COU362 | Camyo5ΔTH2 | 6 | 3 | 365 |
| COU370 | Camyo5ΔTHI | 3 | 12 | 294 |
| COU406 | Camyo5ΔSH3ΔA | 17 | 3 | 661 |
FIG. 8.
Fluid-phase endocytosis in wild-type and myosin I mutant mutants. FM4-64 uptake in cells of wild-type and myosin I mutants is shown. Cells were incubated with 20 μM FM4-64 for 5 min, chased for 1 h 30 min, and visualized by epifluorescence microscopy. Bar = 5 μm.
To determine if myosin I is also required for secretion in C. albicans, we examined the ability of the myosin I-deficient and wild-type strains to secrete aspartyl proteases (Saps) by using an assay that monitors the degradation of bovine serum albumin (BSA) in the medium (35). We found no significant difference between the wild type and the myosin I mutants (Fig. 9; data not shown). In a positive control experiment, we found that the complete degradation of BSA was dependent on the secretion of Sap4, Sap5, and Sap6 since an ∼45-kDa BSA degradation product remained intact in the supernatant of sap4 sap5 sap6 triple null cells for up to 6 h of growth. Thus, myosin I is not required for secretion of the Sap4, Sap5, and Sap6 aspartyl proteases.
FIG. 9.

Secretion of aspartyl proteases (SAP) in wild-type and mutant strains. 10 OD units of wild-type, myosin I, and sap4 sap5 sap6 null mutant cells were incubated in medium containing 2% BSA. Two microliters of medium was loaded and separated in SDS-8% polyacrylamide gels stained with Coomassie brilliant blue.
DISCUSSION
We have characterized the functions of individual neck and tail regions of C. albicans myosin I during hyphal formation, the organization of the cortical actin cytoskeleton, and endocytosis. Based on the known functions of the myosin I regions, we deleted regions that are known to be required for (i) interactions of myosin I with actin filaments (TH2 and TH1), (ii) intracellular targeting (TH1, SH3, and A), and (iii) regulation (IQ). The most striking phenotypes were associated with deletion of the IQ motifs and the TH1 region. The deletion of these regions rendered myosin I nonfunctional, and the corresponding mutants were indistinguishable from the null mutant. These regions have also been found to be essential for function and/or viability in S. cerevisiae and Aspergillus nidulans (21, 73), suggesting a common mechanism for myosin I activity in fungal organisms. The results for the other regions examined (TH2, SH3, and A) suggest that they are required primarily for the organization of cortical actin patches within cells, a function that is distinct and separable from endocytosis. Our results also suggest that the SH3 and A regions function together in activating the Arp2/3 actin-nucleating complex at the hyphal tips during hyphal development. Deletion of either domain alone did not greatly affect the polarization of cortical actin patches to the hyphal tip, as visualized by rhodamine-phalloidin staining or the use of Arp3-GFP. However, we found that the organization of the cortical actin patches was significantly different in cells of the double mutant ΔSH3ΔA strain from that in cells of either a single deletion or wild-type strain. It is possible that the double deletion of the SH3 and A regions would affect myosin I function similarly in A. nidulans, even if single deletions do not apparently affect hyphal growth (73). In S. cerevisiae, activation of the Arp2/3 complex involves interactions between the myosin I SH3 and A regions and other proteins (41). The SH3 domain interacts with Las17/Bee1, a WASP-like protein that activates the Arp2/3 complex (18, 42). The A domain of S. cerevisiae myosin I interacts with Arc40 and Arc19, which are components of the Arp2/3 complex, and thereby also contributes to its activation (18). In addition, the activation of Arp2/3 by the A region may require the WH2 domain of Vpr1, which also binds to the SH3 domain of myosin I (2, 41). In Dictyostelium, the A region is absent. Instead, the CARMIL protein interacts with the SH3 domain to activate the Arp2/3 complex (29). Thus, our finding that the A and SH3 regions share redundant functions for cortical actin patch organization is consistent with findings for other organisms.
If cortical actin patches are not required for polarized growth, what is? Endocytosis and hyphal formation in C. albicans.
Phenotypic analyses of myosin I deletion mutants indicated that (i) cortical actin patch distribution per se is not required for hyphal formation but that (ii) endocytosis correlates with hyphal formation. A striking observation of this study was the ability of mutant C. albicans to form hyphae that elongate at the same rate and with the same characteristics as wild-type hyphae despite the dramatic depolarization of cortical actin patch distribution. Perhaps cortical actin patches in wild-type hyphae play a minor role in the process of hyphal elongation while actin cables and polarized secretion are more important for polarized growth at the hyphal tips. However, the myosin I null mutant, which has intact actin cables (data not shown), is not able to develop true hyphae. Because cortical actin patches are also depolarized in this mutant, it is possible that endocytosis-defective cortical actin patches distinguish mutants who are not able to form hyphae from those who can.
Both endocytosis and secretion are crucial for polarized growth in S. cerevisiae and filamentous fungi (8, 24, 55). Here we report a correlation in the abilities of myosin I mutants to form hyphae and to perform endocytosis (Table 7; Fig. 8). Myosin I, specifically its IQ motifs and the TH1 region, is required for fluid-phase endocytosis and hyphal formation, whereas the SH3 and A regions combined are not. The TH2 region is partially required for hyphal growth and fluid-phase endocytosis. Thus, an inability to form true hyphae may result from a diminished capacity to endocytose. Yet endocytosis may not be completely abolished in these mutants, because they can still grow as pseudohyphae.
Our finding that endocytosis correlates with hyphal growth in C. albicans supports a current model in S. cerevisiae whereby endocytosis retrieves cell-wall-modifying enzymes, the plasma membrane, and associated proteins to maintain polarized growth (55). These enzymes and membrane proteins can be either rapidly redirected to the growing site via the early endosome and secretory pathway or sent to the vacuole for degradation (57). An inability to do so may lead to isotropic slower cellular growth. Interestingly, the deletion of WAL1, the C. albicans homologue of LAS17, leads to endocytosis and hyphal defects that are strikingly similar to those of the myosin I null mutant (68).
Our results show that myosin I does not have a major role in secretion. In particular, myosin I mutants are not affected in the secretion of aspartyl proteases (Fig. 9). The activities of several other secreted enzymes, as measured in the culture medium, were also not significantly different between wild-type and myosin I null strains (15). Invertase secretion was reported to be slightly slower in the myosin I mutant of S. cerevisiae (2), suggesting that myosin I plays a minor role in secretion. However, this defect could be caused indirectly from a primary endocytic defect, as suggested by Pruyne and Bretscher (55).
The observation that transcript levels of several genes, including the stress response gene DDR48, inversely correlate with the ability to perform endocytosis is intriguing. We are currently investigating the cause of the altered gene expression profiles observed for several endocytosis-defective mutants.
Myosin I subcellular fractionation.
Fluorescence microscopy and subcellular fractionation experiments revealed that all of the regions are required for the association of myosin I with cortical actin patches. Notably, the SH3 domain or the A region by itself is not required for the localization of myosin I, in accordance with observations in Dictyostelium and Aspergillus (52, 73). Together, these regions appear to play an important role in myosin I localization, which may also be the case in Aspergillus. However, we cannot exclude the possibility that the C-terminal GFP fusion to the ΔSH3ΔA mutant may have negatively affected its overall structure and protein levels and hence biased our interpretation of its localization. Fractionation experiments indicated that the TH1 region and the SH3 and A regions together are required for the association of myosin I with pelletable actin (Fig. 6A). These domains are known to be responsible for tail-ligand interactions and for intracellular targeting of various isoforms of myosin I (2, 18, 29, 33, 38, 41). In addition, TH1 has been shown to associate with acidic phospholipid membranes (13). Our data suggest that in the absence of these multiple protein-protein interactions, as in the ΔSH3ΔA mutant, or with an inability to associate with lipids, as in the ΔTH1 mutant, CaMyo5 fails to be recruited to cortical actin patches. In contrast, the IQ motifs and the TH2 region were required for the association with pelletable actin only when myosin I complexes were subjected to agents that disrupt protein-protein and protein-membrane associations (Fig. 6D). Deletion of the same N-terminal portion of TH2 in S. cerevisiae myosin I did not result in reduced binding to actin, whereas a deletion of SH3 and the C-terminal portion of TH2 had dramatic effects on actin binding and other myosin I functions (22). Thus, the complete deletion of TH2 may affect actin-binding properties of C. albicans myosin I to a greater extent. Further experiments are required to determine the differences between the mutant and wild-type complexes.
Our results suggest that the phosphorylation state of CaMyo5 correlates with its association with cortical actin patches. First, the majority of myosin I was present as a phosphorylated isoform in the pelletable and high-molecular-weight fractions (Fig. 6A and C). Second, mutants that had a reduced phosphorylation-dependent mobility shift also showed reduced protein levels in the pelletable fraction (Fig. 6A, compare the ΔSH3ΔA and ΔTH1 mutants with the wild type). The ΔTH2 mutant showed intermediate effects on the phosphorylation of myosin I and its association with pelletable actin (Fig. 6A and D). The exception to this correlation was the ΔIQ mutant, which appeared to be underphosphorylated in the actin-bound pellet fraction. It remains unclear whether the IQ motifs play a direct role in CaMyo5 phosphorylation. One possibility is that IQ-dependent binding to calmodulin (data not shown) is a prerequisite for this phosphorylation. In addition to the previously identified Ser366 (53), other phosphorylation sites of myosin I may be required for the observed mobility shift. The identification of the kinase(s) and the residues that are subject to phosphorylation will help to elucidate the role of phosphorylation in myosin I regulation.
Acknowledgments
We thank Brenda Andrews, David Drubin, and Bernard Massie for providing antibodies. We also thank André Nantel for help with statistical analyses of the data. We thank Catherine Bachewich and Bill Zerges for critical readings of the manuscript.
U.O. was a recipient of a Swiss National Foundation fellowship. T.I. was supported by CIHR grant number MOP85769 to D.Y.T. and M.W.
Footnotes
This is NRC publication number 46 222.
REFERENCES
- 1.Akashi, T., T. Kanbe, and K. Tanaka. 1994. The role of the cytoskeleton in the polarized growth of the germ tube in Candida albicans. Microbiology 140:271-280. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, B. L., I. Boldogh, M. Evangelista, C. Boone, L. A. Greene, and L. A. Pon. 1998. The Src homology domain 3 (SH3) of a yeast type I myosin, Myo5p, binds to verprolin and is required for targeting to sites of actin polarization. J. Cell Biol. 141:1357-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, J. M., and D. R. Soll. 1986. Differences in actin localization during bud and hypha formation in the yeast Candida albicans. J. Gen. Microbiol. 132:2035-2047. [DOI] [PubMed] [Google Scholar]
- 4.Asleson, C. M., E. S. Bensen, C. A. Gale, A. S. Melms, C. Kurischko, and J. Berman. 2001. Candida albicans INT1-induced filamentation in Saccharomyces cerevisiae depends on Sla2p. Mol. Cell. Biol. 21:1272-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basrai, M. A., F. Naider, and J. M. Becker. 1990. Internalization of lucifer yellow in Candida albicans by fluid phase endocytosis. J. Gen. Microbiol. 136:1059-1065. [DOI] [PubMed] [Google Scholar]
- 6.Bement, W. M., and M. S. Mooseker. 1995. TEDS rule: a molecular rationale for differential regulation of myosins by phosphorylation of the heavy chain head. Cell Motil. Cytoskeleton 31:87-92. [DOI] [PubMed] [Google Scholar]
- 7.Bose, A., A. Guilherme, S. I. Robida, S. M. C. Nicoloro, Q. L. Zhou, Z. Y. Jiang, D. P. Pomerleau, and M. P. Czech. 2002. Glucose transporter recycling in response to insulin is facilitated by Myo1c. Nature 420:821-824. [DOI] [PubMed] [Google Scholar]
- 8.Bretscher, A. 2003. Polarized growth and organelle segregation in yeast: the tracks, motors, and receptors. J. Cell Biol. 160:811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brzeska, H., and E. D. Korn. 1996. Regulation of class I and class II myosins by heavy chain phosphorylation. J. Biol. Chem. 271:16983-16986. [DOI] [PubMed] [Google Scholar]
- 10.Cyr, J. L., R. A. Dumont, and P. G. Gillepsie. 2002. Myosin-1c interacts with hair-cell receptors through its calmodulin-binding IQ domains. J. Neurosci. 22:2487-2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Backer, M. D., P. T. Magee, and J. Pla. 2000. Recent developments in molecular genetics of Candida albicans. Annu. Rev. Microbiol. 54:463-498. [DOI] [PubMed] [Google Scholar]
- 12.de la Fuente, J. M., A. Alvarez, C. Nombela, and M. Sanchez. 1992. Flow cytometric analysis of Saccharomyces cerevisiae autolytic mutants and protoplasts. Yeast 8:39-45. [DOI] [PubMed] [Google Scholar]
- 13.Doberstein, S. K., and T. D. Pollard. 1992. Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J. Cell Biol. 117:1241-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doberstein, S. K., I. C. Baines, G. Wiegand, E. D. Korn, and T. D. Pollard. 1993. Inhibition of contractile vacuole function in vivo by antibodies against myosin-I. Nature 365:841-843. [DOI] [PubMed] [Google Scholar]
- 15.Dunphy, G. B., U. Oberholzer, M. Whiteway, R. J. Zakarian, and I. Boomer. 2003. Virulence of Candida albicans mutants toward larval Galleria mellonella (Insecta, Lepidoptera, Galleridae). Can. J. Microbiol. 49:514-524. [DOI] [PubMed] [Google Scholar]
- 16.Durrbach, A., K. Collins, P. Matsudaira, D. Louvard, and E. Coudrier. 1996. Brush border myosin-I truncated in the motor domain impairs the distribution and the function of endocytic compartments in an hepatoma cell line. Proc. Natl. Acad. Sci. USA 93:7053-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engqvist-Goldstein, A. E. Y., and D. G. Drubin. 2003. Actin assembly and endocytosis: from yeast to mammals. Annu. Rev. Cell Dev. Biol. 19:287-332. [DOI] [PubMed] [Google Scholar]
- 18.Evangelista, M., B. M. Klebl, A. H. Tong, B. A. Webb, T. Leeuw, E. Leberer, M. Whiteway, D. Y. Thomas, and C. Boone. 2000. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J. Cell Biol. 148:353-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geli, M. I., and H. Riezman. 1996. Role of type I myosins in receptor-mediated endocytosis in yeast. Science 272:533-535. [DOI] [PubMed] [Google Scholar]
- 21.Geli, M. I., A. Wesp, and H. Riezman. 1998. Distinct functions of calmodulin are required for the uptake step of receptor-mediated endocytosis in yeast: the type I myosin Myo5p is one of the calmodulin targets. EMBO J. 17:635-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geli, M. I., R. Lombardi, B. Schmezl, and H. Riezman. 2000. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 19:4281-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodson, H. V., B. L. Anderson, H. M. Warrick, L. A. Pon, and J. A. Spudich. 1996. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J. Cell Biol. 133:1277-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta, G. D., and I. B. Heath. 2002. Predicting the distribution, conservation, and functions of SNARES and related proteins in fungi. Fungal Genet. Biol. 36:1-21. [DOI] [PubMed] [Google Scholar]
- 25.Idrissi, F.-Z., B. L. Wolf, and M. I. Geli. 2002. Cofilin, but not profilin, is required for myosin-I-induced actin polymerization and the endocytic uptake in yeast. Mol. Biol. Cell 21:4074-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito, H., Y. Fukuda, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung, G., and J. A. Hammer III. 1994. The actin binding site in the tail domain of Dictyostelium myosin IC (myoC) resides within the glycine- and proline-rich sequence (tail homology region 2). FEBS Lett. 342:197-202. [DOI] [PubMed] [Google Scholar]
- 28.Jung, G., X. Wu, and J. A. Hammer III. 1996. Dictyostelium mutants lacking multiple classic myosin I isoforms reveal combinations of shared and distinct functions. J. Cell Biol. 133:305-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung, G., K. Remmert, X. Wu, J. M. Volosky, and J. A. Hammer III. 2001. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J. Cell Biol. 153:1479-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karksonen, M., Y. Sun, and D. G. Drubin. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell 115:475-487. [DOI] [PubMed] [Google Scholar]
- 31.Karpova, T. S., S. L. Reck-Peterson, N. B. Elkind, M. S. Mooseker, P. J. Novick, and J. A. Cooper. 2000. Role of actin and Myo2p in polarized secretion and growth of Saccharomyces cerevisiae. Mol. Biol. Cell 11:1727-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurischko, C., and R. K. Swoboda. 2000. Cytoskeletal proteins and morphogenesis in Candida albicans and Yarrowia lipolytica, p. 173-184. In J. F. Ernst and A. Schmidt (ed.), Dimorphism in human pathogenic and apathogenic yeasts, vol. 5. Karger, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 33.Lechler, T., A. Shevchenko, and R. Li. 2000. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J. Cell Biol. 148:363-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lechler, T. A., G. A. Jonsdottir, S. K. Klee, D. Pellman, and R. Li. 2001. A two-tiered mechanism by which Cdc42 controls the localization and activation of an Arp2/3-activating motor complex in yeast. J. Cell Biol. 155:261-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee, S. A., Y. Mao, Z. Zhang, and B. Wong. 2001. Overexpression of a dominant negative allele of YPT1 inhibits growth and aspartyl protease secretion in Candida albicans. Microbiology 147:1961-1970. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S.-F., and G. P. Côté. 1995. Purification and characterization of a Dictyostelium protein kinase required for the actin-activated MgATPase activity of Dictyostelium myosin ID. J. Biol. Chem. 270:11776-11782. [DOI] [PubMed] [Google Scholar]
- 37.Lee, W.-L., E. M. Ostap, H. G. Zot, and T. D. Pollard. 1999. Organization and ligand binding properties of the tail of Acanthamoeba myosin-IA. J. Biol. Chem. 49:35159-35171. [DOI] [PubMed] [Google Scholar]
- 38.Lee, W.-L., M. Bezanilla, and T. D. Pollard. 2000. Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J. Cell Biol. 151:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, X., H. Brzeska, and E. D. Korn. 2000. Functional analysis of tail domains of Acanthamoeba myosin IC by characterization of truncation and deletion mutants. J. Biol. Chem. 275:24886-24892. [DOI] [PubMed] [Google Scholar]
- 40.Liu, X., N. Osherov, R. Yamashita, H. Brzeska, E. D. Korn, and G. S. May. 2001. Myosin I mutants with only 1% of wild-type actin-activated MgATPase activity retain essential in vivo function(s). Proc. Natl. Acad. Sci. USA 98:9122-9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Machesky, L. M. 2000. The tails of two myosins. J. Cell Biol. 148:219-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madania, A., P. Dumoulin, S. Grava, H. Kitamoto, C. Shärer-Brodbeck, A. Soulard, V. Moreau, and B. Winsor. 1999. The Saccharomyces cerevisiae human homologue Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell 10:3521-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martchenko, M., A.-A. Alarco, D. Harcus, and M. Whiteway. 2004. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hypha-induced SOD5 gene. Mol. Biol. Cell 15:456-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGoldrick, C. A., C. Gruver, and G. S. May. 1995. myoA of Aspergillus nidulans encodes an essential myosin I required for secretion and polarized growth. J. Cell Biol. 128:577-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mermall, V., P. L. Post, and M. S. Mooseker. 1998. Unconventional myosins in cell movement, membrane traffic, and signal transduction. Science 279:527-533. [DOI] [PubMed] [Google Scholar]
- 46.Miyata, H., B. Bowers, and E. D. Korn. 1989. Plasma membrane association of Acanthamoeba myosin I. J. Cell Biol. 109:1519-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mooseker, M. S., and R. E. Cheney. 1995. Unconventional myosins. Annu. Rev. Cell Dev. Biol. 11:633-675. [DOI] [PubMed] [Google Scholar]
- 48.Morschhauser, J., S. Michel, and J. Hacker. 1998. Expression of a chromosomally integrated, single-copy GFP gene in Candida albicans, and its use as a reporter of gene regulation. Mol. Gen. Genet. 257:412-420. [DOI] [PubMed] [Google Scholar]
- 49.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuhaus, E. M., and T. Soldati. 2000. A myosin I is involved in membrane recycling from early endosomes. J. Cell Biol. 150:1013-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novak, K. D., M. D. Peterson, M. C. Reedy, and M. A. Titus. 1995. Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J. Cell Biol. 131:1205-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novak, K. D., and M. A. Titus. 1998. The myosin I SH3 domain and TEDS rule phosphorylation site are required for in vivo function. Mol. Biol. Cell 9:75-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oberholzer, U., A. Marcil, E. Leberer, D. Y. Thomas, and M. Whiteway. 2002. Myosin I is required for hyphal formation in Candida albicans. Eukaryot. Cell 1:213-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ostap, E. M., P. Maupin, S. K. Doberstein, I. C. Baines, E. D. Korn, and T. D. Pollard. 2003. Dynamic localization of myosin-I to endocytic structures in Acanthamoeba. Cell Motil. Cytoskeleton 54:29-40. [DOI] [PubMed] [Google Scholar]
- 55.Pruyne, D., and A. Bretscher. 2000. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J. Cell Sci. 113:571-585. [DOI] [PubMed] [Google Scholar]
- 56.Pruyne, D. W., D. H. Schott, and A. Bretscher. 1998. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J. Cell Biol. 143:1931-1945. [DOI] [PubMed] [Google Scholar]
- 57.Raposo, G., M. N. Cordonnier, D. Tenza, B. Menichi, A. Durrbach, D. Louvard, and E. Coudrier. 1999. Association of myosin I alpha with endosomes and lysosomes in mammalian cells. Mol. Biol. Cell 10:1477-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenfeld, S. S., and B. Rener. 1994. The GPQ-rich segment of Dictyostelium myosin IB contains an actin binding site. Biochemistry 33:2322-2328. [DOI] [PubMed] [Google Scholar]
- 59.Senda, S., and M. A. Titus. 2000. A potential mechanism for regulating myosin I binding to membranes in vivo. FEBS Lett. 484:125-128. [DOI] [PubMed] [Google Scholar]
- 60.Shaw, J. D., K. B. Cummings, G. Huyer, S. Michaelis, and B. Wendland. 2001. Yeast as a model system for studying endocytosis. Exp. Cell Res. 271:1-9. [DOI] [PubMed] [Google Scholar]
- 61.Soulard, A., T. Lechler, V. Spiridonov, A. Shevchenko, A. Shevchenko, R. Li, and B. Winsor. 2002. Saccharomyces cerevisiae Bzz1p is implicated with type I myosins in actin patch polarization and is able to recruit actin-polymerizing machinery in vitro. Mol. Cell. Biol. 22:7889-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Staebell, M., and D. R. Soll. 1985. Temporal and spatial differences in cell wall expansion during bud and mycelium formation in Candida albicans. J. Gen. Microbiol. 131:1467-1480. [DOI] [PubMed] [Google Scholar]
- 63.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19-31. [DOI] [PubMed] [Google Scholar]
- 64.Tang N., T. Lin, and E. M. Ostap. 2002. Dynamics of myo1c (myosin-ibeta) lipid binding and dissociation. J. Biol. Chem. 277:42763-42768. [DOI] [PubMed] [Google Scholar]
- 65.Toya, M., F. Motegi, K. Nakano, I. Mabuchi, and M. Yamamoto. 2001. Identification and functional analysis of the gene for type I myosin in fission yeast. Genes Cells 6:187-199. [DOI] [PubMed] [Google Scholar]
- 66.Ushinsky, S. C., D. Harcus, J. Ash, A. Marcil, J. Morchhauser, D. Y. Thomas, M. Whiteway, and E. Leberer. 2002. CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryot. Cell 1:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vida, T. A., and S. D. Emr. 1995. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128:779-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walther, A., and J. Wendland. 2004. Polarized hyphal growth in Candida albicans requires the Wiskott-Aldrich syndrome protein homolog Wal1p. Eukaryot. Cell 3:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woo, M., K. Lee, and K. Song. 2003. MYO2 is not essential for viability, but is required for polarized growth and dimorphic switching in Candida albicans. FEMS Microbiol. Lett. 218:195-202. [DOI] [PubMed] [Google Scholar]
- 70.Wu, C., S. F. Lee, E. Furmaniak-Kazmierczak, G. P. Côté, D. Y. Thomas, and E. Leberer. 1996. Activation of myosin-I by members of the Ste20p protein kinase family. J. Biol. Chem. 271:31787-31790. [DOI] [PubMed] [Google Scholar]
- 71.Wu, C., V. Lytvyn, D. Y. Thomas, and E. Leberer. 1997. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J. Biol. Chem. 272:30623-30626. [DOI] [PubMed] [Google Scholar]
- 72.Yamashita, R. A., and G. S. May. 1998. Constitutive activation of endocytosis by mutation of myoA, the myosin I gene of Aspergillus nidulans. J. Biol. Chem. 273:14644-14648. [DOI] [PubMed] [Google Scholar]
- 73.Yamashita, R. A., N. Osherov, and G. S. May. 2000. Localization of wild type and mutant class I myosin proteins in Aspergillus nidulans using GFP-fusion proteins. Cell Motil. Cytoskeleton 45:163-172. [DOI] [PubMed] [Google Scholar]