Abstract
Anthracnose is a destructive disease of the common bean (Phaseolus vulgaris L.). The Andean cultivar Hongyundou has been demonstrated to possess strong resistance to anthracnose race 81. To study the genetics of this resistance, the Hongyundou cultivar was crossed with a susceptible genotype Jingdou. Segregation of resistance for race 81 was assessed in the F2 population and F2:3 lines under controlled conditions. Results indicate that Hongyundou carries a single dominant gene for anthracnose resistance. An allele test by crossing Hongyundou with another resistant cultivar revealed that the resistance gene is in the Co-1 locus (therefore named Co-1HY). The physical distance between this locus and the two flanking markers was 46 kb, and this region included four candidate genes, namely, Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800. These candidate genes encoded serine/threonine-protein kinases. Expression analysis of the four candidate genes in the resistant and susceptible cultivars under control condition and inoculated treatment revealed that all the four candidate genes are expressed at significantly higher levels in the resistant genotype than in susceptible genotype. Phvul.001G243600 and Phvul.001G243700 are expressed nearly 15-fold and 90-fold higher in the resistant genotype than in the susceptible parent before inoculation, respectively. Four candidate genes will provide useful information for further research into the resistance mechanism of anthracnose in common bean. The closely linked flanking markers identified here may be useful for transferring the resistance allele Co-1HY from Hongyundou to elite anthracnose susceptible common bean lines.
Introduction
Common bean (Phaseolus vulgaris L.) is one of the most important and widely cultivated grain legumes, in China. Dry bean production in China in 2014 was 1.046 million tons from 936,000 ha harvested area [1]. In recent years, anthracnose caused by Colletotrichum lindemuthianum (Sacc. and Magn) Scrib has become a major cause of reduced dry bean production in China. Yield losses due to anthracnose may reach 100% in extreme cases [2]. Identifying anthracnose resistance genes and introgressing them into elite varieties is therefore desired to control anthracnose and reduce crop loss.
Currently, twenty-three related anthracnose resistance genes have been identified by the ‘Co’ symbol, including Co-1 to Co-7, Co-8, Co-9 to Co-15, Co-17, Co-u to Co-z, and CoPv09 [3–5]. Among these genes, Co-1, Co-x and Co-w belong to the Andean gene pool, and the others belong to the Mesoamerican gene pool. Different alleles have been described for the Co-1, Co-3 and Co-4 genes [4]. Specifically, Co-1 was identified as possessing the Co-1, Co-12, Co-13, Co-14, and Co-15 alleles [6–8]. Most of the anthracnose resistance genes have been located on common bean linkage maps, and flanking markers were determined. Three genes namely, Co-1, Co-x and Co-w, that mapped to linkage group Pv01 [9,10]. Further, Co-u mapped on Pv02 [11], and Co-13 and Co-17 mapped onto Pv03 [5,12]. The linkage group Pv04 includes six genes, namely: Co-3, Co-9, Co-y, Co-z, Co-10 and Co-15 [10,13–17]. Additionally, Co-5, Co-6 and Co-v were located on the Pv07 [18,19], Co-4 on the Pv08 linkage group and Co-2 on the Pv11 [16,20,21]. Recently, Campa et al. identified a new gene CoPv09 on Pv09 [3].
Genetic mapping analysis of anthracnose resistance genes indicated that some of these Co genes are not distributed randomly but tend to exist in complex clusters of race-specific resistance genes. Tight association of many Co-resistance genes have been well established on LGs Pv01, Pv02, Pv04, Pv07 and Pv11 [11,14,18,22,23] and correspond to the named clusters Co-1, Co-u, Co-3, Co-5 and Co-2, respectively.
At the molecular level, most cloned R genes encode proteins containing a nucleotide binding site (NBS) and a C-terminal leucine-rich repeat (LRR) domain. NBS-LRRs (NL) can be divided into two subclasses on the basis of their amino-terminal sequence. One encode an N-terminal domain with Toll/Interleukin-1 Receptor homology (TIR-NB-LRR, TNL), and the other one encode an N-terminal coiled-coil motif (CC-NB-LRR, CNL) [24]. Many clusters of anthracnose resistance genes have been located at the end of the pseudomolecules and correspond to NL clusters by genetic map and/or linked molecular markers. For example, an important number of NL clusters located at the ends of Pv04 and Pv11 containing R specificities against the fungi C. lindemuthianum, such as Co-3 and Co-2. Another example of large distal R cluster is the multi-resistance I cluster presenting R gene Co-u located at the end of Pv02 containing TNLs sequences. However, some atypical R genes have been observed, including: Co-1, Co-12, Co-13, Co-14, Co-15, Co-x, and Co-w located at the end of Pv01, and since they are located in a region that does not contain NL sequences [25].
In our previous research, an anthracnose resistance gene Co-2322 was identified from the common bean cultivar Hongyundou [26]. Based on the mapping of polymorphic markers identified by bulk segregant analysis, this gene maps to chromosome 1 and is flanked by a simple sequence repeat (SSR) marker, C871, and a cleaved amplified polymorphism sequence (CAPS) marker, g1224, at genetic distance of 3.58 cM and 3.81 cM, respectively. The Co-2322 locus is present in the Adean bean Hongyundou, with wildly planted in China. This locus is very valuable in the breeding of beans, particularly in those countries where race 81 of C.lindemuthianum predominate. Breeders in these countries should exercise caution in choosing resistance alleles at the Co-2322 locus, despite allelic differences in reaction to local races. The objectives of the present study were to (1) test allelism between the resistance gene Co-2322 from Hongyundou and the Co-1 locus, (2) develop SSR markers for the target region and construct a high-density genetic map of the Co-2322 region, (3) fine map the Co-2322 locus using a large F2 population from a cross between Hongyundou and Jingdou genotypes, and identify the candidate gene, and (4) identify the sequence polymorphism and expression pattern of the candidate gene.
Materials and Methods
Mapping population
Several common bean genotypes were used to analyze the inheritance and independence of the anthracnose resistance gene present in Hongyundou. The bean genotypes Hongyundou, Honghuayundou and Jingdou were obtained from National Gene Bank (China, Beijing). Other bean genotypes namely, G19833, Kaboon, Michigan Dark Red Kidney (MDRK) and Widusa are from the International Center for Tropical Agriculture (CIAT), Cali, Colombia. Hongyundou is a large-seed red Chinese landrace cultivar of Andean origin, and was used as the source of the resistance. Jingdou is middle-seed brown genotype of the Mesoamerican origin, and was used as the susceptible parent. The genome sequence of G19833 was acted as reference genome [27]. Kaboon carries the Co-12 gene, and Widusa has a unique Co-15 allele [6].
The anthracnose resistant genotype, Hongyundou, and the susceptible genotype, Jingdou, were crossed to produce F1 plants, F2 plants, and a F2:3 mapping population. A population of 1,092 F2:3 families as developed by self-pollination of the F2 plants. This population was used to fine map the anthracnose resistance locus Co-1HY (previously named Co-2322). Another cross was made in between two anthracnose resistant parents, Hongyundou and Michigan Dark Red Kidney (MDRK). The segregating F2 population of this cross was used to test allelism between the Co-1HY locus from Hongyundou and Co-1 from MDRK. The MDRK is a kidney red genotype of Andean origin, and it is thought that the resistance of MDRK to anthracnose (race 73) is conferred by Co-1 located on Pv01 and that MDRK is highly resistant to race 81 [28].
Colletotrichum lindemuthianum inoculation procedure and disease screening
The Colletotrichum lindemuthianum race 81 was isolated from cultivated common bean in China and used to screen for anthracnose resistance in this study [29]. Race 81 was obtained from monosporic cultures maintained on fungus-colonized filter paper at -20°C for long-term storage. To obtain abundant sporulation, race 81 was grown at 19–21°C in darkness for approximately one week on Marthur’s agar [30]. Spore suspensions were prepared by flooding the plates with sterile distilled water and scraping the surface of the culture with a spatula. The two parental cultivars, F2 individuals and each of the F2:3 families were inoculated with the spore suspension at a final concentration of 2.0×106 spores/ml. Seven- to ten-day-old seedlings were inoculated by spraying with the aqueous conidial suspension and maintained at 20–22% and 95–100% humidity on a 12-h photoperiod in a climate controlled chamber [31]. The disease response of the plants was evaluated after 7–9 days and scored as either resistant (no symptoms) or susceptible (the seedlings died). For each of the F2:3 families, at least 30 seedlings were infected and grown as described by Marthur et al. [30], which enabled the classification of the F2 plants as homozygous resistant (when all F3 individuals were resistant), homozygous susceptible (when all F3 individuals were susceptible) or heterozygous (when F3 individuals were resistant and susceptible).
Marker development, bulked segregant analysis using SSRs and genetic mapping
The common bean G19833 reference genome sequence (www.phytozome.org) was used to identify and locate linkage markers. Primer3 software (version 4.0) was used to design PCR primers (http://frodo.wi.mit.edu/primer3/) (S1 Table).
Bulked segregant analysis (BSA) [32] was used to identify genomic SSR markers linked to Co-2322 (or Co-1HY). Two resistant and two susceptible bulks were prepared for SSR analysis. The resistant and susceptible bulks were obtained by pooling equivalent amounts of DNA from each of the 12 homozygous resistant and 12 homozygous susceptible F2 individuals.
Genomic DNA was isolated from lyophilized young leaves using 2×CTAB buffer (2% CTAB, 1.4 M NaCl, 100 mM, pH 8.0; Tris-HCl, 20 mM, pH 8.0 and EDTA) [33].
PCR was performed in a 15 μL mixture containing: 30 ng of DNA or cDNA template, 1.5 μL of 10× TransTaq HiFi buffer (including 2.0 mM MgSO4), 1.2 μL of 2.5 mM dNTPs, 0.3 μL of each primer at 10 μM and 1.0 U of TransTaq HiFi DNA polymerase (TransGen Biotech,China) using the standard PCR program: one cycle of 5 min at 94°C; 35 cycles of 30 s at 94°C, 30 s at 50–58°C, and 1–2 min at 72°C; and a final extension cycle of 10 min at 72°C. The PCR products were resolved on a 6% polyacrylamide gel or a 1% agarose gel. The polymorphic markers were considered putatively linked to the target gene and subsequently confirmed based on their co-segregation patterns in the F2 population.
A chi square test was used to test the goodness-of-fit of the observed to the expected ratios in the F2 Hongyundou×Jingdou population. The molecular marker data and corresponding individual phenotypes in the F2:3 population were analyzed using MAPMAKER Macintosh version 3.0 with a minimum logarithm of odds ratio (LOD) score of 3.0 [34]. The recombination values were converted into genetic map distances (cMs) via the Kosambi mapping function [35].
Sequence amplification and bioinformatics analysis
Gene-specific primers (S2 Table) were developed and used to amplify the genomic DNA (gDNA), complementary DNA (cDNA) and promoter sequences of the four genes from the resistant and susceptible parents. The P.vulgaris genome of G19833 genotype was used as the main reference genome [27] for assembly and annotation of candidate genes. Another BAT 93 genotype was used as an additional reference genome [36]. Structural and phylogenetic analysis of the four candidate genes from the common bean genome was performed by comparing them to several known CR4-related protein sequences from Zea mays (ZmCR4), Arabidopsis thaliana (ACR4 and AtCRR3) and Oryza sativa (OsCRR3). The GeneBank protein identification numbers are: ZmCR4, AAB09771.1; ACR4, CAB91612; AtCRR3, CAB87849; OsCRR3, EEE62193. BLAST analysis was used to anchor the mapped markers linked to the Co-1HY to the reference genomic sequence. The domains were predicted using Pfam 21.0 with an initial E-value cutoff of 0.1 [37]. The sequence alignment and phylogenetic analyses were performed with the DNAStar network service and ClustalW (European Bioinformatics Institute, EBI). Phylogenetic trees were constructed using the neighbor-joining method as implemented in MEGA 6.0 [38,39] with 1,000 bootstrap replicates. A stringency cutoff E value of <E−60 was used to identify similar sequences in other regions of the G19833 genome. The linkage map of homology genes was drawn with MapChart by Voorrips[40]
Real-time PCR (RT-PCR) analysis
The expression pattern of the four candidate genes, Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800, in response to inoculation with C. lindemuthianum race 81 was analyzed in the resistant (Hongyundou) and susceptible (Jingdou) genotypes. A comprehensive temporal expression analysis of the four genes was conducted at 6, 12, 24, 48, 72, 96, and 120 hours after-inoculation (HAI). The leaves from three different treatment and control plants were obtained and snap frozen in liquid nitrogen for RNA isolation. Total RNA was extracted with a GeneJETTM Plant RNA Purification Mini Kit according to the manufacturer’s instructions (Thermo Scientific, USA). cDNA synthesis was performed using a RevertAidTMFirst Strand cDNA Synthesis Kit (Thermo Scientific, USA) using 1 μg of total RNA after DNA enzyme digestion. The candidate gene-specific RT-PCR primers are listed in S2 Table. The primer pair Act-F 5'-GAAGTTCTCTTCCAACCATCC-3' and Act-R 5'-TTTCCTTGCTCATTCTGTCCG-3' was used to amplify the common bean actin gene, which was used as a reference gene. The qPCR was performed in a 20 μL reaction that consisted 2 μL of the cDNA, 0.4 μL each gene-specific forward and reverse primers with concentration 10μM, 10 μL of SYBR Premix Ex TaqTM, 0.4 μL ROX Reference Dye II (50×), and add distilled water to 20 μL (Takara Bio Inc., Beijing) was run on an ABI 7500 real-time PCR machine (Bio-Rad Corporation, USA) according to the manufacturer’s instructions. The gene expression data from three biological replicates and three technical replicates were processed and standardized according to the comparative 2-ΔΔCt method [41].
Results
Inheritance analysis and allele test of anthracnose resistance gene
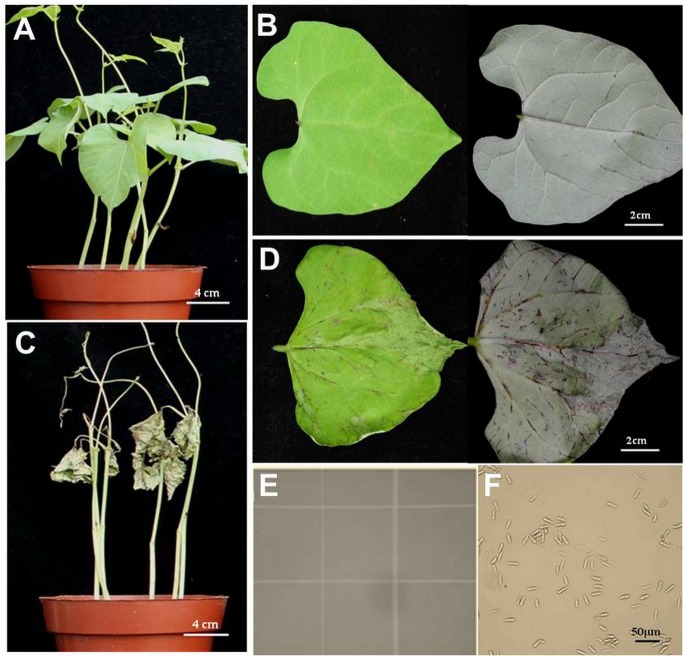
Seven days post-inoculation, the resistance plants grew normally and with no symptoms or small dark in the leaves (Fig 1A and 1B). However, the susceptible plants were severely wilting and the leaves exhibited largely collapsed lesions (Fig 1C and 1D). Sporulation never occurred from the inoculated leaves of the resistant plants, whereas abundant spores were collected from the inoculated leaves of the susceptible plants (Fig 1E and 1F).
Fig 1. The anthracnose resistant (Hongyundou) and susceptible (Jingdou) plants following C. lindemuthianum race 81 inoculation.
(A) The Hongyundou plants grew normally and were completely asymptomatic. (B) The Jingdou plants began to wilt seven days after inoculation. (C) Sporadic disease spots on the front side and underside of an inoculated Hongyundou leaf. (D) The Jingdou plants exhibited large lesions on the inoculated leaves and collapsed with severe anthracnose symptoms. (E) No spores could be extracted from the inoculated leaves of the Hongyundou plants. (F) Abundant spores were detected from the inoculated leaves of the Jingdou plants.
In addition, we tested the reaction of the offspring to races 81. The F1 plants exhibited similar phenotype as that of the resistant parent, indicating that the resistance was dominant. Initial analysis of the 182 F2 plants identified resistant and susceptible individuals. The segregation of the resistant and susceptible plants was consistent with that expected for segregation at a single locus. The F2 population segregated as 3 resistant: 1 susceptible, and the F2:3 lines segregated as 247 homozygous resistant:549 segregating:296 homozygous susceptible, which was also expected for single gene segregation (χ21:2:1 = 4.43<χ20.05 = 5.99; Table 1).
Table 1. The anthracnose resistance of the parents and F2:3 families.
| Parent or cross | No. of plants or lines | Expected ratio | Χ2 | P-value | |||
|---|---|---|---|---|---|---|---|
| Resistant | Segregating | Susceptible | |||||
| Hongyundou | P1 | 30 | |||||
| Jingdou | P2 | 30 | |||||
| Hongyundou × Jingdou | F1 | 50 | |||||
| Hongyundou × Jingdou | F2 | 138 | 44 | 3:1 | 0.029 | 0.864 | |
| Hongyundou × Jingdou | F2:3 | 247 | 549 | 296 | 1:2:1 | 4.43 | 0.109 |
The values for the significance of the χ2 tests at P = 0.05 were 3.83 for 1 df and 5.99 for 2 df.
The resistance gene in Hongyundou was previously mapped onto chromosome 1 [26]. To identify the relationship between Co-2322 and the Co-1 locus, an allele test were carried out between the resistance gene in Hongyundou and the Co-1 locus. The result showed that after inoculation with race 81, the parent plants of Hongyundou and MDRK grew normally without any leaf symptoms, and the 605 F2 plants crossed by Hongyundou and MDRK presented the same phenotypic character, remarkable resistance to race 81. This suggests that Co-2322 may be a member of the Co-1 allelic series. Therefore, we propose the use of symbol Co-1HY for the major dominant gene in Hongyundou.
Identification of markers linked with the Co-1HY locus
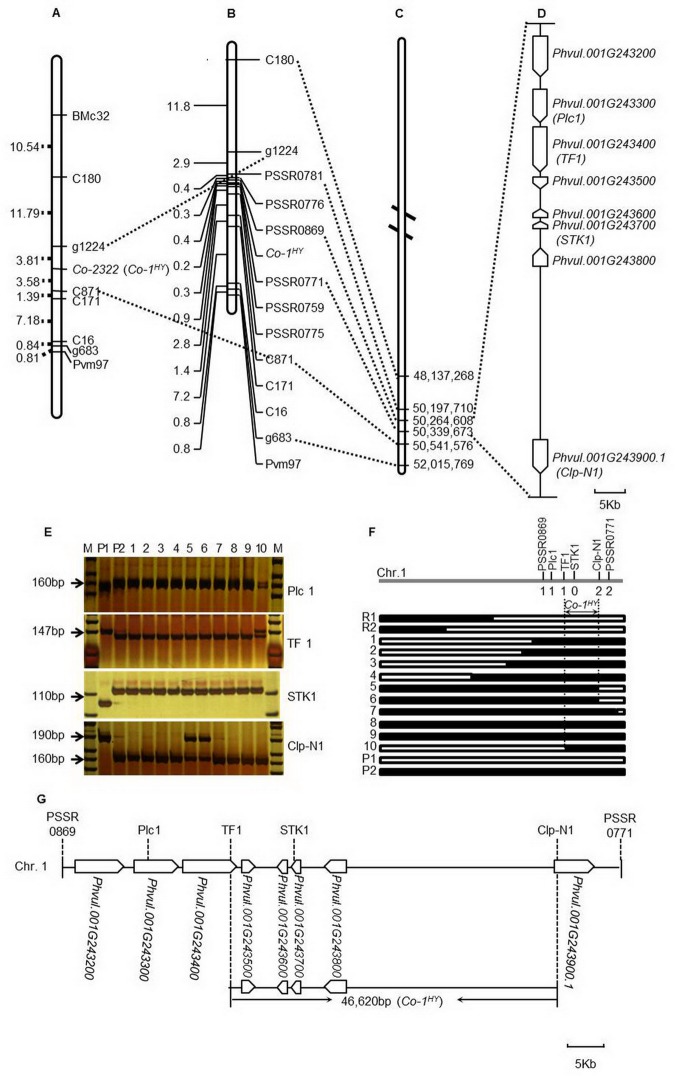
In our previous study, we mapped the Co-1HY to chromosome 1 at a locus flanked by a marker, C871, and a marker, g1224, at genetic distance of 3.58 cM and 3.81 cM, respectively [26] (Fig 2A). The C871 was located at 50,541,576 bp while g1224 could not be mapped, maybe it's because there have some difference between the sequences of g1224 maker and reference genome sequence. Therefore, we selected another linkage marker C180 as an anchor as it was 11.79 cM away from g1224. The C180 marker was anchored at 48,137,268 bp. We then utilized the genome sequence information of the 2.4 Mb region between the nucleotides at 48,137,268 and at 50,541,576. We identified 282 SSR markers from this region that were used to fine map the Co-1HY locus. Of all of the markers chosen for initial screening, 32 (11.35%) were polymorphic between the parental lines. Ultimately, the genomic SSR markers PSSR0771 and PSSR0869 were polymorphic between the contrasting parents and bulks, which indicated that PSSR0771 and PSSR0869 were tightly linked to the Co-1HY (Fig 2B). A single recombinant event was detected between PSSR0771 and Co-1HY, whereas two recombinant events were detected between PSSR0869 and Co-1HY. Based on these results, we suggest that Co-1HY is located between the markers PSSR0771 and PSSR0869. BLAST analysis allowed us to anchor the markers PSSR0771 and PSSR0869 within approximately 75kb region that is located between the 50,339,673 and 50,264,608 bp of the common bean genome (Fig 2C). Subsequently, we used the annotation data of the G19833 reference genome to identify genes located within this 75kb region. We identified eight predicted genes namely, Phvul.001G243200, Phvul.001G243300, Phvul.001G243400, Phvul.001G243500, Phvul.001G243600, Phvul.001G243700, Phvul.001G243800 and Phvul.001G243900 (Fig 2D).
Fig 2. Positional cloning of the resistance gene Co-1HY.
(A) The primary genetic linkage map of Co-2322 gene constructed by Chen et al. [26]. (B) The fine mapping of the Co-2322 (or Co-1HY) gene and associated molecular markers. Recombination distances are indicated on the left side of the linkage group in centiMorgans (cM), and the marker names are shown on the right side. (C) The physical map showing the locations of the Co-1HY locus and SSR markers. The Co-1HY region was positioned between 50,264,608 bp and 50,339,673 bp in an approximately 75 kb interval of chromosome 1 between the SSR markers PSSR0869 and PSSR0771. The dotted lines indicate common markers. (D) The predicted genes flank the target 75-kb region. Fifteen ORFs are shown as open arrows, and the parentheses show PCR markers Plc1, TF1, STK1 and Clp-N1 that were developed with reference to the predicted genes. (E) The amplification of the F2 population with the PCR markers Plc1, TF1, STK1 and Clp-N1. P1: resistant parent; P2: susceptible parent; 1–10: susceptible individuals of the F2 population; individual 10 in the markers of Plc1 and TF1 and individuals 5 and 6 in Clp-N1 are recombined. (F) Diagrammatic representation of the Co-1HY mapping. The distribution of recombination events that were detected by high-resolution mapping is depicted. The hatched bar represents the region of the Hongyundou genome, and the open bar represents the region of the Jingdou genome. The number below the markers PSSR0869, Plc1, TF1, STK1, Clp-N1 and PSSR0771 represents the recombinant. Genotyping of the homozygous progeny delimited the Co-1HY locus to an approximately 46 kb stretch flanked by Clp-N1 and TF1. P1: resistant parent; P2: susceptible parent; 1–10: susceptible individuals of the F2 population; R1, R2: resistant individuals of the F2 population. (G) Annotation of the 46-kb candidate region. The pentagons represent the predicted genes with the sizes indicated in the lengths of each gene, and the arrows indicate the relative orientation.
Fine mapping and physical localization of the Co-1HY locus
To accurately define the Co-1HY locus relative to the closely linked flanking markers according to the position on the chromosome, we selected four of the predicted genes (i.e., Phvul.001G243300, Phvul.001G243400, Phvul.001G243700 and Phvul.001G243900) and PCR amplified them from the resistant and susceptible parents using primers based on the 5' UTR and 3' UTR sequences. A comparison of DNA sequences from the resistant (Hongyundou) and susceptible (Jingdou) parents revealed a number of single nucleotide polymorphisms (SNPs) and insertions or deletions (InDels) (S1 Fig). Based on these nucleotide differences in the parents, we developed four InDel markers, namely, Plc1, TF1, STK1, and Clp-N1 (Fig 2D;S1 Fig; S3 Table). Finally, the four InDel markers were amplified from the mapping population that included 296 homozygous susceptible F2:3 families from the 1,092 F2:3 lines (Fig 2E).
We detected two recombinants for the Clp-N1 marker, only one recombinant each for the TF1 and Plc1 markers, and no recombinant for the STK1 marker. These results indicate that STK1 co-segregated with the target gene Co-1HY. We therefore suggest that the Co-1HY is located between the TF1 and Clp-N1 markers (Fig 2E and 2F), and includes a genomic region that is 46,620 bp long and is located between the 50,286,325 and 50,332,945 bp of the G19833 reference genome. There are four potential genes within this 46,620 bp region (Fig 2G). Using the annotation information of the reference genome from the database, all the four predicted genes namely, Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800 are classified as ‘similar to CRINKLY4 related 3’ from A. thaliana (S4 Table). These four genes, were considered as the candidate genes for the Co-1HY locus (Fig 2G).
Analysis of genetic structure and sequence polymorphism of the four candidate genes
The amplification of the four candidate genes reveals that Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800 were 1,470 bp, 1,107 bp, 1,233 bp and 2,421 bp long, respectively in the resistant genotype Hongyundou, with no intron and kinase catalytic domain. Further BLASTP analysis of the predicted protein sequences of the candidate genes showed Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800 from Hongyundou encoded proteins with 489 AA, 368 AA, 410 AA and 806 AA, respectively. These predicted proteins matched with the entries PHASIBEAM10F002206T1, PHASIBEAM10F002205T1, PHASIBEAM10F002204T1 and PHASIBEAM10F002203T1, respectively from the BAT93 database. All the four predicted proteins shared sequence similarities with A. thaliana CRINKLY4-related 3 (CRR3, AT3G55950).
Structural and phylogenetic analyses indicated all the four candidate genes were more similar to AtCRR3 (Arabidopsis thaliana) than other CR4-related genes such as ZmCR4 (Zea mays), ACR4 (Arabidopsis thaliana) and OsCRR3 (Oryza sativa). The Phvul.001G243800 encodes a full-length AtCRR3 protein with 97% query coverage. It has 58% identity and 71% similarity. Further, the proteins encoded by Phvul.001G243500, Phvul.001G243600 and Phvul.001G243700 only included the C-terminal part of the kinase, with 38–40% amino acid query coverage, 52%-58% identities and 63%-69% similarities. All of the four predicted proteins lacked crinkly repeats but contained kinase domains (S2 Fig). We further determined the phylogenetic relationships between these four genes. The four proteins fell into 2 major clusters (S3 Fig). Phvul.001G243500, Phvul.001G243600 and Phvul.001G243700 were more closely related to each other than Phvul.001G243800. Interestingly, Phvul.001G243800 and AtCRR3 formed a clade and appeared to be potential orthologs.
All the candidate genes had single polymorphic differences in the genomic, cDNA and amino acid sequences in parents. The sequences of Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800 revealed SNPs corresponding to one amino acid variation in Phvul.001G243500 and Phvul.001G243800, and two amino acid variations in Phvul.001G243600 (S4 Fig). Interestingly, compared to the resistant parent, Phvul.001G243700 contained ten SNPs and three missing nucleotides near the termination codon of the cDNA sequences that resulted in six polymorphic sites and one missing amino acid (arginine) in the susceptible parent (S4 Fig). To corroborate the role of these polymorphisms in resistance or susceptibility to anthracnose, we sequenced the Phvul.001G243700 homologs from four different anthracnose resistant cultivars (G19833, Kaboon, MDRK, Honghuayundou) and one other susceptible cultivar (Widusa). These sequences were aligned to the Phvul.001G243700 homologs from the Hongyundou and Jingdou cultivars. Astonishingly, the Phvul.001G243700 from Kaboon, MDRK, Honghuayundou and Hongyundou were 100% identical, whereas the Phvul.001G243700 from G19833 and Hongyundou were 99% identical. Remarkably, the susceptible cultivars Widusa and Jingdou had same four amino acid sites among the six polymorphic sites (S5 Fig).
To further examine the transcriptional regulation of these four genes, we analyzed the upstream promoter sequences of the genes using the PlantCARE database. The BLAST searches revealed that the amplified sequence included essential cis-regulatory elements of the promoters such as the core promoter element TATA and the common element CAAT that were highly conserved across the investigated species (S6 Fig). Moreover, other specific cis-acting elements were also detected in all those predicted genes. For example, TC-rich repeats involved in defense and stress responsiveness were detected in Phvul.001G243500, Phvul.001G243700 and Phvul.001G243800; a CGTCA motif involved in Me-JA responsiveness was detected in Phvul.001G243600; and the 5UTR Py-rich stretch element that confers high transcription levels was detected only in Phvul.001G243700. The Phvul.001G243800 also contained the TA-rich region (enhancer), Box-W1 (the fungal elicitor responsive element), P-box (the gibberellin-responsive element) and others. There were nearly no differences between the promoter sequences of Phvul.001G243500 and Phvul.001G243600 from the Hongyundou and Jingdou genotypes. We obtained about 1.5kb sequence for the Phvul.001G243700 gene. Interestingly, at approximately 600 bp upstream of the start codon, a sequence of approximately 130 bp was deleted from the Hongyundou, whilst at approximately 800 bp upstream of the start codon, there was an approximately 60 bp insertion. Quite remarkably, the 130 bp missing sequence and 60 bp insertion sequence in Hongyundou all include a TATA and CAAT box that are known to be essential transcription regulators in eukaryotes (S6 Fig).
Homology search in P. vulgaris genome sequence
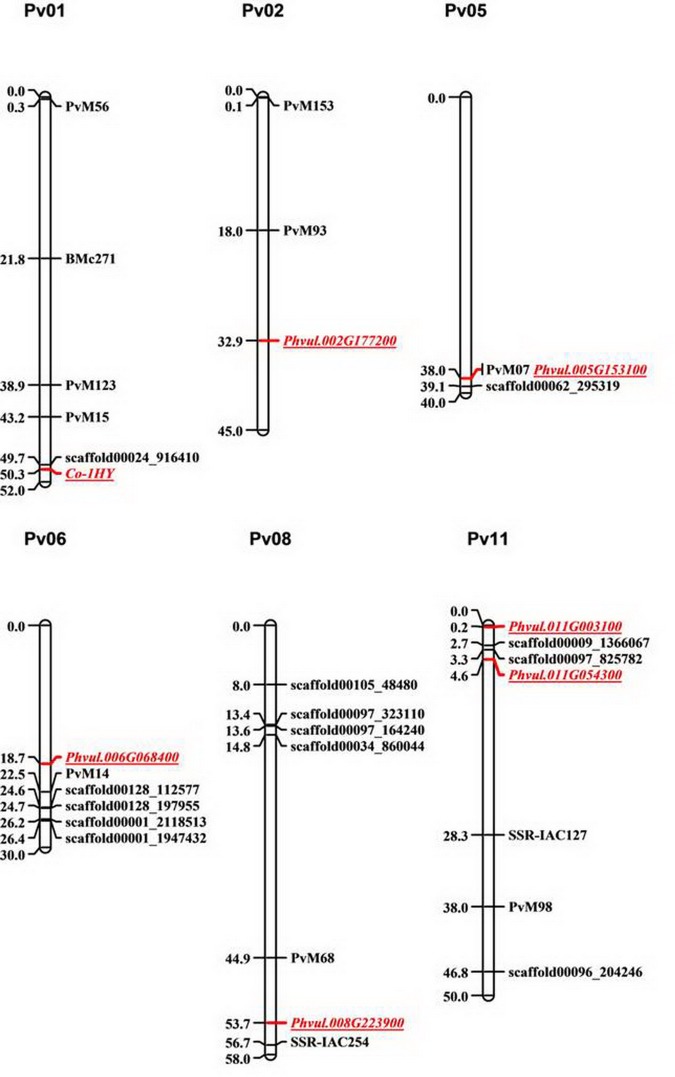
Homologous sequences were accessed of each four candidate gene within the G19833 reference genome (Fig 3). The representative markers by Perseguini are associated with anthracnose resistance [42].In addition to the candidate genes identified in the 46 kb Co-1HY target region, Phvul.001G243500 identified 5 additional specific genes located on Pv05, Pv06, Pv08 and Pv011 with Kinase domains. These are Phvul.005G153100, Phvul.006G068400, Phvul.008G223900, Phvul.011G003100 and Phvul.011G054300. Furthermore, Phvul.001G243600 identified 2 homology genes on Pv06 (Phvul.006G068400) and Pv08 (Phvul.008G223900). Phvul.001G243700 identified 3 homology genes on Pv06 (Phvul.006G068400), Pv08 (Phvul.008G223900) and Pv11 (Phvul.011G003100) apart from Co-1HY location. Besides the same location as identified by Phvul.001G243500, Phvul.001G243800 also had a single homology gene Phvul.002G177200 located on Pv02. However, Phvul.001G243700 and Phvul.001G243800 all did not identify sequence similarity to Phvul.001G243500 based on the stringency criteria used. These homology genes can be divided into two categories according to the G19833 annotation. The Phvul.002G177200 gene belongs to the leucine repeat protein kinase (LRR-kinase) family while others have a similar protein domain as the four candidate genes on Pv01. For example, Phvul.005G153100 and Phvul.011G054300 have a CRINKLY 4 (CR4) protein kinase, whilst Phvul.006G068400 and Phvul.008G223900 had a near identical region with CRINKLY 4 related 3 (CRR3) protein, and Phvul.011G003100 had high sequence similarity to CRINKLY 4 related 4 (CRR4) proteins (Table 2). It is noted that the homology genes have characterized by SSR and SNP markers associated with anthracnose (Fig 3).
Fig 3. The eight homologous genes were positioned on the common bean linkage map.
Representative markers in black are associated with anthracnose resistance and those in red genes homologous to four candidate gene identified from the Co-1HY region. The marker positions are represented in Mb.
Table 2. Gene homologous with candidate genes Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800. The G19833 genome sequence was used to identify homologous genes (www.phytozome.org).
| Homology gene | G19833 genome location | Number of exon | Coding sequence length (bp) | E-value | Predicted putative annotation |
|---|---|---|---|---|---|
| Phvul.002G177200 | Chr02:32,856,832..32,862,036 | 15 | 2808 | 7.2E-61 | Leucine-rich protein kinase/PTK |
| Phvul.005G153100 | Chr05:38,001,220..38,004,640 | 1 | 2721 | 6.2E-64 | CRINKLY4 protein/ CR4 |
| Phvul.006G068400 | Chr06:18,689,709..18,692,381 | 1 | 2397 | 0.0 | CRINKLY4 Related 3 protein/ CRR3 |
| Phvul.008G223900 | Chr08:53,741,844..53,744,583 | 1 | 2391 | 0.0 | CRINKLY4 Related 3 protein/ CRR3 |
| Phvul.011G003100 | Chr11:237,814..240,513 | 1 | 2409 | 2.5E-128 | CRINKLY4 Related 4 protein/ CRR4 |
| Phvul.011G054300 | Chr11:4,602,210..4,605,788 | 1 | 2760 | 5.1E-66 | CRINKLY4 protein/ CR4 |
Expression analysis of the candidate genes
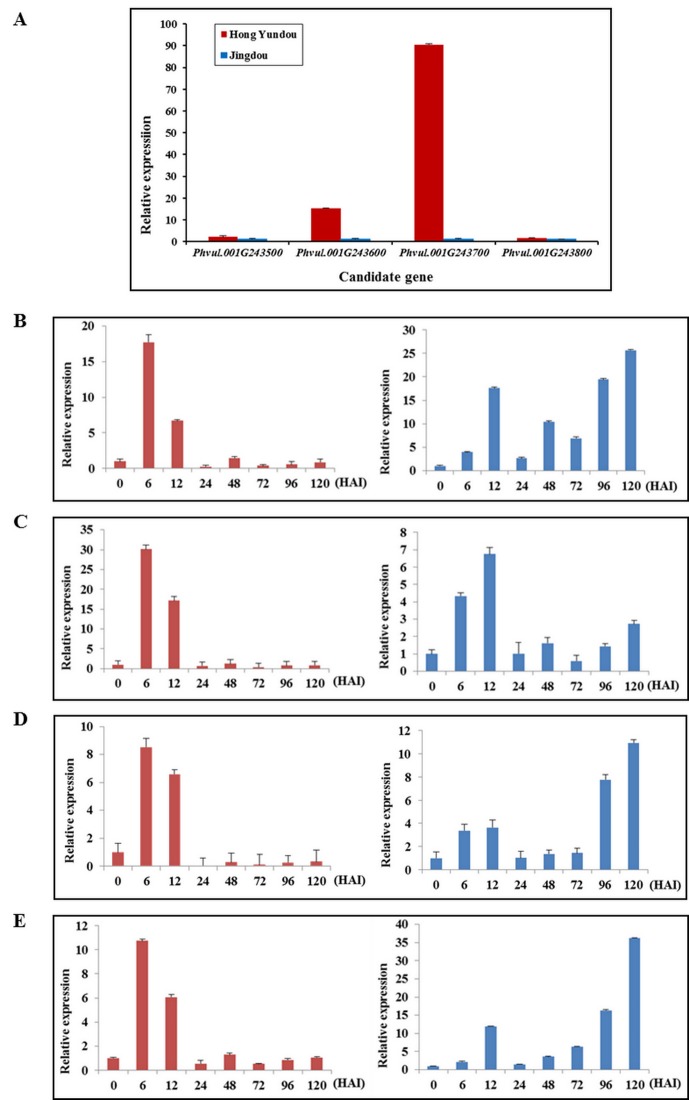
The results revealed that the expression levels of the four genes, i.e., Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800 were higher in the resistant parent than in the susceptible parent by 2.0, 15, 90, and 1.6 fold, respectively (Fig 4A). To further ascertain the roles of these candidate genes in anthracnose resistance, we preformed expression analyses of these genes in the leaves of the resistant and susceptible parents at ternate compound leaf stage, using qRT-PCR with gene-specific primers. Strikingly, temporal gene expression analysis showed that the four candidate genes were induced in anthracnose race 81 infected Hongyundou and Jingdou plants. Interestingly, the expression pattern of the four genes was almost consistently different between resistant and susceptible genotypes. In the resistant genotype, four genes were significantly induced early at 6 HAI, followed by slight reduction in expression at 12 HAI (Fig 4B–4E). Thereafter the expression of these genes in the resistant genotype dropped significantly to almost no noticeable expression from 24 to 120 HAI (Fig 4B–4E). Comparatively, the expression of Phvul.001G243600 peaked at 12 HAI in the susceptible genotype, whilst the expression of other 3 candidates peaked only at 120 HAI. Further, an apparent pattern of some expression at 6 HAI, slightly increased expression at 12 HAI, significantly reduced expression between 24–72 HAI, followed by significantly increased expression at 96 and 120 HAI was observed in the susceptible genotype (Fig 4B–4E). This indicates that the resistant genotype responds very quickly with expression of the four genes peaking at 6 HAI whereas the susceptible genotype with early expression at 6 HAI, manages to lift expression somewhat at 12 HAI, but gives up between 24–72 hours, only to increase expression at a very later stage (96–120 HAI). This suggests that the susceptible genotype misses the critical window of early expression (0–6 HAI), which possibly allows the fungi to penetrate the leaves and may be even multiply within the host (as suggested by increased defense response at later time points). However, low/insignificant gene expression is observed in the susceptible genotype between 24–72 HAI.
Fig 4. Expression pattern of the four candidate genes with gene-specific primers and the bean actin gene was used as an internal control for qRT-PCR.
(A) Expression pattern of the four candidate genes in the leaves of the parental Hongyundou and Jingdou plants without inoculation. The graph's horizontal axis indicates the predicted genes (Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800), and the vertical axis indicates the relative expression levels. (B)-(E) Expression analysis of Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800, respectively, in the leaves of Hongyundou and Jingdou at different times (0, 6, 12, 24, 48, 72, 96 and 120 hours after inoculation (HAI) with Colletotrichum lindemuthianum race 81). Plot with the time after inoculation on the horizontal axis and the relative expression level on the vertical axis. The filled red bars in (A), (B), (C), (D) and (E) refers to Hongyundou, and the blue bars refer to Jingdou. The means were generated from three independent measurements, and the bars indicate the standard errors.
Discussion
Our results reveal that Co-1HY is a potential member of the Co-1 allelic series [6,8]. So far, seven alleles related to anthracnose resistance have been reported from the Co-1 locus on chromosome 1. They are Co-1 in MDRK, Co-12 in Kaboon, Co-13 in Perry Marrow [8], Co-14 in AND277 [13], Co-15 in Widusa [6], and Co-w and Co-x in JaloEEP558 [9]. Here, the allelism test for independence of the single dominant resistant gene in Hongyundou was performed on a single population Hongyundou×MDRK with one race (race81). Moreover, the Widusa (Co-15), after inoculation with Colletotrichum lindemuthianum, was highly susceptible to race 81, which indicates that the resistant gene in Hongyundou has a different allele from Co-15. This is consistent with previous reports that Co-15 is a special gene that is different from other reported alleles on the Co-1 locus [6]. However, further analysis is required to determine if the resistance gene in Hongyundou is different from other alleles that have been previously reported for the Co-1 locus (example, Co-12, Co-13 and Co-14). Nonetheless, the results support the presence, in Hongyundou, of a resistance allele at the Co-1 locus.
To date, no anthracnose resistant gene has been cloned from common bean, several complex disease resistance gene clusters have been mapped on the specific linkage that contain classical nucleotide-binding site leucine-rich repeat (NBS-LRR) sequence and protein kinases such as Co-2, Co-3, Co-4, Co-9, Co-10, Co-y and Co-z [11,43,44,45]. The Co-u has been localized to the vicinity of TNL sequences [46]. However, fine mapping of the common bean Co-1HY locus reveals a resistance candidate that encodes a subfamily of CR4 (CRR3) protein, which belong to RLK family. In plant, RLKs are reported to involve in plant defense responses [47,48],such as, Xa21 and Xa3 in rice [49,50], Rpg1 in barley [51], RFo1 and FLS2 in Arabidopsis [52,53]. However, there are no reports documenting the seemingly direct relevance of the CR4 to the resistance to anthracnose or other diseases in plants. Previous studies reported that cr4 gene regulated leaf epidermal cell differentiation and aleurone endosperm cell specialization, AtCRR1, AtCRR2, AtCRR3, and AtCRK1, were identified to play important roles in the protodermal cells of the embryos and shoots and OsCR4 affect rice seeds in terms of the functions of the palea and lemma during epidermal cell differentiation. [54,55,56,57]. In this paper, the expression level of Phvul.001G243600 and Phvul.001G243700 in the resistant genotype were higher than in the susceptible genotype. Notably, there have some difference in the sequence of these genes between the resistant and susceptible genotype. We also noticed that Phvul.001G243600 contained a CGTCA motif involved in Me-JA response. As Me-JA response is critical to plant defense response [58], the early and significant induction in the resistant genotype along with 2 amino acid difference, makes it a more likely candidate.
Our results suggested that the candidate genes Phvul.001G243500, Phvul.001G243600, Phvul.001G243700 and Phvul.001G243800 maybe have new functions. Here, the four candidate genes related to Co-1HY were more similar to AtCRR3 than to the other CR4 members based on the phylogenetic analysis. These findings suggest that the CRR3 protein may have significant role in plant disease resistance, at least in the common bean. However, Richard et al. [59] noted that Co-x is located in the region flanked by two markers, P05 and K06, which are located at nucleotide positions 50,264,307–50,265,284 and 50,320,965–50,322,583 of chromosome 1, respectively. However, the Co-1HY gene was located between the markers TF1 and Clp-N1 with nucleotide position of 50,286,325 and 50,332,737, respectively. These results indicate that the Co-x and Co-1HY genes are likely located in the same locus. In the Co-x region, three phosphoinositide-specific phospholipases C (PI-PlC), one zinc finger protein, and four kinases were predicted to be present [59]. The authors speculated the three PI-PLC members as potential Co-x candidate genes. Our results reveal that four kinases Phvul.001G243500, Phvul.001G243600, Phvul.001G243700, and Phvul.001G243700 may be more likely the potential Co-1HY candidate genes. Further studies using for example, a knock-out approach like virus-induced gene silencing are required to ascertain which one of these genes is most important for anthracnose resistance [60].
Supporting Information
The white rectangle indicates the insertion deletion length polymorphism.
(TIF)
The protein sequences of the candidate gens were amplified from Hongyundou. The following are indicated with symbols and shadings: identical (black shading), 80% identity (grey shading) and 60% identity (light grey shading) residues; the seven ‘crinkly’ repeats (solid bars); the predicted transmembrane domain of ZmCR4 (dotted line labeled TM); and the twelve conserved kinase subdomains (solid bars with round ends).
(PDF)
The utilized multiple sequence alignment is the same as that in A. The results are displayed graphically using NJ-plot from MEGA version 6.0 [38,39]. The bootstrap values from 1,000 bootstrap replicates that were used to assess the robustness of the tree are shown at the nodes.
(TIF)
The black shading indicates polymorphic amino acids.
(PDF)
Resistance cultivars: a) G19833, b) Kaboon, c) MDRK, d) Honghuayundou, e) Hongyundou. Susceptible cultivars: f) Jingdou and g) Widusa. Rectangle boxes in solid line reveal the efficient SNPs between resistance cultivars and susceptible cultivars while boxes in dotted line present the meaningless SNPs.
(PDF)
The following elements are marked: initiation codon ATG (solid-line arrows); TATA box (grey shading); CAAT box (light grey shading); TC-rich repeats (green shading); TCA-element (red shading); TC-rich repeats (olive green shading); BoxW1 (blue shading); skn-1 motif (blue ash shading); and P-box (clear blue shading).
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant No. 31301387), the Ministry of Agriculture of China [the earmarked fund for China Agriculture Research System (CARS-09)], Program of International Science & Technology Cooperation, Cooperation Research on Collecting Techniques and Practice in Crop Genebank between China and United States of America (2014DFG31860) and the Agricultural Science and Technology Innovation Program (ASTIP) of CAAS.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (grant No. 31301387), the Ministry of Agriculture of China [the earmarked fund for China Agriculture Research System (CARS-09)], Program of International Science & Technology Cooperation, Cooperation Research on Collecting Techniques and Practice in Crop Genebank between China and United States of America (2014DFG31860), the National Key Technology Research and Development Program of the Ministry of Science and Technology of China (2013BAD01B03-18a) and the Agricultural Science and Technology Innovation Program (ASTIP) of CAAS. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.FAO (2015) Food and agriculture organization of the United Nations Statistical database. http://faostat3.fao.org/browse/Q/QC/E
- 2.Singh SP, Schwartz HF (2010) Breeding common bean for resistance to diseases: a review. Crop Sci 50: 2199–2223. [Google Scholar]
- 3.Campa A, Rodríguez-Suárez C, Giraldez R, Ferreira JJ (2014) Genetic analysis of the response to eleven Colletotrichum lindemuthianum races in a RIL population of common bean (Phaseolus vulgaris L.). BMC Plant Biol 14: 115 10.1186/1471-2229-14-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira JJ, Campa A, Kelly JD (2013) Organization of genes conferring resistance to anthracnose in common bean. Translational Genomics for Crop Breeding: Biotic Stress 1: 151–181. [Google Scholar]
- 5.Trabanco N, Campa A, Ferreira JJ (2015) Identification of a new chromosomal region involved in the genetic control of resistance to anthracnose in common bean. Plant Genome [DOI] [PubMed] [Google Scholar]
- 6.Gonçalves-Vidigal MC, Kelly JD (2006) Inheritance of anthracnose resistance in the common bean cultivar Widusa. Euphytica 151: 411–419. [Google Scholar]
- 7.Gonçalves-Vidigal MC, Cruz AS, Garcia A, Kami J, Vidigal Filho PS, Sousa LL, et al. (2011) Linkage mapping of the Phg-1 and Co-14 genes for resistance to angular leaf spot and anthracnose in the common bean cultivar and 277. Theor Appl Genet 122: 893–903. 10.1007/s00122-010-1496-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melotto M, Kelly JD (2008) An allelic series at the Co-1 locus conditioning resistance to anthracnose in common bean of Andean origin. Euphytica 116: 143–149. [Google Scholar]
- 9.Geffroy V, Sévignac M, Billant P, Dron M, Langin T (2008) Resistance to Colletotrichum lindemuthianum in Phaseolus vulgaris: a case study for mapping two independent genes. Theor Appl Genet 116: 407–415. 10.1007/s00122-007-0678-y [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez-Suárez C, Ferreira JJ, Campa A, Paňeda A, Giraldez R (2008) Molecular mapping and intra-cluster recombination between anthracnose race-specific resistance genes in the common bean differential cultivars Mexico 222 and Widusa. Theor Appl Genet 116: 807–814. 10.1007/s00122-008-0714-6 [DOI] [PubMed] [Google Scholar]
- 11.Geffroy V, Macadré C, David P, Pedrosa-Harand A, Sévignac M, Dauga C, et al. (2009) Molecular analysis of a large subtelomeric nucleotide-binding-site-leucine-rich-repeat family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics 181: 405–419. 10.1534/genetics.108.093583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonçalves-Vidigal MC, Filho PS, Medeiros AF, Pastor-Corrales MA (2009) Common bean Landrace Jalo Listras Pretas is the source of a new Andean anthracnose resistance gene. Crop Sci 49: 133–138. [Google Scholar]
- 13.Alzate-Marin AL, Costa MR, Arruda KM, Gonçalves de Barros EG, Alves Moreira MA (2003) Characterization of the anthracnose resistance gene present in Ouro Negro (Honduras 35) common bean cultivar. Euphytica 133;165–169. [Google Scholar]
- 14.Geffroy V, Sicard D, de Oliveira JCF, Sévignac M, Cohen S, Gepts P, et al. (1999) Identification of an ancestral resistance gene cluster involved in the coevolution process between Phaseolus vulgaris and its fungal pathogen Colletotrichum lindemuthianum. Mol Plant Microbe Interact 12: 774–784. 10.1094/MPMI.1999.12.9.774 [DOI] [PubMed] [Google Scholar]
- 15.Méndez-Vigo B, Rodríguez-Suárez C, Paňeda A, Ferreira JJ, Giraldez R (2005) Molecular markers and allelic relationships of anthracnose resistance gene cluster B4 in common bean. Euphytica 141, 237–245. [Google Scholar]
- 16.Rodríguez-Suárez C, Méndez-Vigo B, Pañeda A, Ferreira JJ, Giraldez R (2007) A genetic linkage map of Phaseolus vulgaris L. and localization of genes for specific resistance to six races of anthracnose (Colletotrichum lindemuthianum). Theor Appl Genet 114: 713–722. 10.1007/s00122-006-0471-3 [DOI] [PubMed] [Google Scholar]
- 17.Sousa LL, Gonçalves-Vidigal MC, Gonçalves AO, Vidigal Filho PS, Awale H, Kelly JD (2013) Molecular mapping of the anthracnose resistance gene co-15 in the common bean cultivars Corinthiano. Ann Rep Bean Improv Coop 56: 45–46. [Google Scholar]
- 18.Campa A, Giraldez R, Ferreira JJ (2009) Genetic dissection of the resistance to nine anthracnose races in the common bean differential cultivars MDRK and TU. Theor Appl Genet 119: 1–11. 10.1007/s00122-009-1011-8 [DOI] [PubMed] [Google Scholar]
- 19.Geffroy V (1997) Dissection génétique de la résistance à Colletotrichum lindemuthianum, agent de l'anthracnose, chezdeux génotypes représentatifs des pools géniques de Phaseolus vulgaris L. Paris-Grignon: PhD Thesis. Inst Natl Agron.
- 20.Melotto M, Coelho MF, Pedrosa-Harand A, Kelly JD, Camargo L (2004) The anthracnose resistance locus co-4 of common bean is located on chromosome 3 and contains putative disease resistance-related genes. Theor Appl Genet 109: 690–699. 10.1007/s00122-004-1697-6 [DOI] [PubMed] [Google Scholar]
- 21.Adam-Blondon AF, Sevignac M, Dron M, Bannerot H (1994) A genetic map of common bean to localize specific resistance genes against anthracnose. Genome 37: 915–924. [DOI] [PubMed] [Google Scholar]
- 22.Kelly JD, Vallejo VA (2004) A comprehensive review of the major genes conditioning resistance to anthracnose in common bean. Hort Sci 39: 1196–1207. [Google Scholar]
- 23.Miklas PN, Kelly JD, Beebe SE, Blair MW (2006) Common bean breeding for resistance against biotic and abiotic stresses: from classical to MAS breeding. Euphytica 147: 105–131. [Google Scholar]
- 24.Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341: 746–751. 10.1126/science.1236011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meziadi C, Richard MMS, Derquennes A, Thareau V, Blanchet S, Gratias A, et al. (2016) Development of molecular markers linked to disease resistance genes in common bean based on whole genome sequence. Plant Science 242: 351–357. 10.1016/j.plantsci.2015.09.006 [DOI] [PubMed] [Google Scholar]
- 26.Chen ML, Wang LF, Wang XM, Zhang XY, Wang SM (2011) Mapping of gene conferring resistance to anthracnose in common bean (Phaseolus vulgaris L.) by molecular markers. Acta Agronomica Sinica 37: 2130–2135. [Google Scholar]
- 27.Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, et al. (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46: 707–713. 10.1038/ng.3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McRostie GP (1919) Inheritance of anthracnose resistance as indicated by a cross between a resistant and a susceptible bean. Phytopathology 9: 141–148. [Google Scholar]
- 29.Wang K, Wang X-M, Zhu Z-D, Zhang X-Y, Wang S-M (2008) Identification of Colletotrichum lindemuthianum races and bean germplasm evaluation for anthracnose resistance. J Plant Genet Resour 9(2):168–172 [Google Scholar]
- 30.Marthur RS, Barnett HL, Lilly VG (1950) Sporulation of Colletotrichum lindemuthianum in culture. Phytopathol 40: 104–114. [Google Scholar]
- 31.Balardin RS, Pastor-Corrales MA, Otoya MM (1990) Variabilidade patogênica de Colletotrichum lindemuthianum no Estado de Santa Catarina. Fitopatol Bras 15: 243–245. [Google Scholar]
- 32.Quarrie SA, Lazić-Jančić V, Kovačević D, Steed A. Pekić S (1999) Bulk segregant analysis with molecular markers and its use for improving drought resistance in maize. J Exp Bot 50: 1299–1306. [Google Scholar]
- 33.Stewart CN Jr, Via LE (1993) A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14(5): 748–750. [PubMed] [Google Scholar]
- 34.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, et al. (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1(2):174–181. [DOI] [PubMed] [Google Scholar]
- 35.Kosambi DD (1943) The estimation of map distances from recombination values. Ann of Eugenics 12(1): 172–175. [Google Scholar]
- 36.Vlasova A, Capella-Gutiérrez S, Rendón-Anaya M, Hernández-Oñate M, Minoche AE, Erb I, et al. (2016) Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome biology 17(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, et al. (2004) The Pfam protein families database. Nucleic Acids Res 32(Suppl. 1): D138–D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Bio Evol 4(4): 406–425. [DOI] [PubMed] [Google Scholar]
- 40.Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J of Hered 93(1): 77–78. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4): 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 42.Perseguini JMKC, Oblessuc PR, Rosa JRBF, Gomes KA, Chiorato AF, Carbonell SAM, et al. (2016) Genome-Wide association studies of anthracnose and angular leaf spot resistance in common bean (Phaseolus vulgaris L.). PloS ONE 11(3): e0150506 10.1371/journal.pone.0150506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David P, Chen NWG, Pedrosa-Harand A, Thareau V, Sévignac M, Cannon SB, et al. (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol 151: 1048–1065. 10.1104/pp.109.142109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrier-Cana E, Geffroy V, Macadré C, Creusot F, Imbert-Bolloré P, Sévignac M, et al. (2003) Characterization of expressed NBS-LRR resistance gene candidates from common bean. Theor Appl Genet 106: 251–261. 10.1007/s00122-002-1032-z [DOI] [PubMed] [Google Scholar]
- 45.Ferrier-Cana E, Macadré C, Sévignac M, David P, Langin T, Geffroy V (2005) Distinct post-transcriptional modifications result into seven alternative transcripts of the CC-NBS-LRR gene JA1tr of Phaseolus vulgaris. Theor Appl Genet 110: 895–905. 10.1007/s00122-004-1908-1 [DOI] [PubMed] [Google Scholar]
- 46.Vallejos CE, Astua-Monge G, Jones V, Plyler TR, Sakiyama NS, Mackenzie SA (2006) A genetic and molecular characterization of the I locus of Phaseolus vulgaris. Genetics 172: 1229–1242. 10.1534/genetics.105.050815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afzal AJ, Wood AJ, Lightfoot DA (2008) Plant receptor-like serine threonine kinase: Roles in signaling and plant defense. Mol Plant Microbe Interact 21:507–517. 10.1094/MPMI-21-5-0507 [DOI] [PubMed] [Google Scholar]
- 48.Sekhwal MK, Li PC, Lam I, Wang X, Cloutier S, You FM (2015) Disease resistance gene analogs (RGAs) in plants. Int J Mol Sci 16: 19248–19290. 10.3390/ijms160819248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, et al. (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270: 1804–1806. [DOI] [PubMed] [Google Scholar]
- 50.Sun X, Cao Y, Wang S (2006) Point mutations with positive selection were a major force during the evolution of a receptor-kinase resistance gene family of rice. Plant Physiol. 140: 998–1008. 10.1104/pp.105.073080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brueggeman R, Rostoks N, Kudrna D, Kilian A, Han F, Chen J, et al. (2002) The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc. Natl. Acad. Sci. USA 99: 9328–9333. 10.1073/pnas.142284999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diener AC, Ausubel FM (2005) Resistance to fusarium oxysporum1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171: 305–321. 10.1534/genetics.105.042218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tabata S, Kaneko T, Nakamura Y, Kotani H, Kato T, Asamizu E, et al. (2000) Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 408: 823–826. 10.1038/35048507 [DOI] [PubMed] [Google Scholar]
- 54.Becraft PW, Stinard PS, McCarty DR (1996) CRINKLY4: a TNFR-like receptor kinase involved in maize epidermal differentiation. Science 273: 1406–1409. [DOI] [PubMed] [Google Scholar]
- 55.Becraft PW, Kang SH, Suh SG (2001) The maize CRINKLY4 receptor kinase controls a cell-autonomous differentiation response. Plant Physiol 127:486–496. [PMC free article] [PubMed] [Google Scholar]
- 56.Cao X, Li K, Suh SG, Guo T, Becraft PW (2005) Molecular analysis of the CRINKLY4 gene family in Arabidopsis thaliana. Planta 220: 645–657. 10.1007/s00425-004-1378-3 [DOI] [PubMed] [Google Scholar]
- 57.Pu CX, Ma Y, Wang J, Zhang YC, Jiao XW, Hu YH, et al. (2012) Crinkly4 receptor-like kinase is required to maintain the interlocking of the palea and lemma, and fertility in rice, by promoting epidermal cell differentiation. Plant J 70: 940–953. 10.1111/j.1365-313X.2012.04925.x [DOI] [PubMed] [Google Scholar]
- 58.Xu Y, Chang PFL, Liu D, Narasimhan ML, Raghothama KG, Hasegawa PM,et al. (1994) Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 6:1077–1085. 10.1105/tpc.6.8.1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richard MM, Pflieger S, Sévignac M, Thareau V, Blanchet S, Li Y, et al. (2014) Fine mapping of Co-x, an anthracnose resistance gene to a highly virulent strain of Colletotrichum lindemuthianum in common bean. Theor Appl Genet 127:1653–1666. 10.1007/s00122-014-2328-5 [DOI] [PubMed] [Google Scholar]
- 60.Pflieger S, Richard MM, Blanchet S, Meziadi C, Geffroy V (2013) VIGS technology: an attractive tool for functional genomics studies in legumes. Funct Plant Biol 40: 1234–1248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The white rectangle indicates the insertion deletion length polymorphism.
(TIF)
The protein sequences of the candidate gens were amplified from Hongyundou. The following are indicated with symbols and shadings: identical (black shading), 80% identity (grey shading) and 60% identity (light grey shading) residues; the seven ‘crinkly’ repeats (solid bars); the predicted transmembrane domain of ZmCR4 (dotted line labeled TM); and the twelve conserved kinase subdomains (solid bars with round ends).
(PDF)
The utilized multiple sequence alignment is the same as that in A. The results are displayed graphically using NJ-plot from MEGA version 6.0 [38,39]. The bootstrap values from 1,000 bootstrap replicates that were used to assess the robustness of the tree are shown at the nodes.
(TIF)
The black shading indicates polymorphic amino acids.
(PDF)
Resistance cultivars: a) G19833, b) Kaboon, c) MDRK, d) Honghuayundou, e) Hongyundou. Susceptible cultivars: f) Jingdou and g) Widusa. Rectangle boxes in solid line reveal the efficient SNPs between resistance cultivars and susceptible cultivars while boxes in dotted line present the meaningless SNPs.
(PDF)
The following elements are marked: initiation codon ATG (solid-line arrows); TATA box (grey shading); CAAT box (light grey shading); TC-rich repeats (green shading); TCA-element (red shading); TC-rich repeats (olive green shading); BoxW1 (blue shading); skn-1 motif (blue ash shading); and P-box (clear blue shading).
(PDF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.