Abstract
The growth plate is a highly specialized and dynamic cartilage structure that serves many essential functions in skeleton patterning, growth and endochondral ossification in developing vertebrates. Major signaling pathways initiated by classical morphogens and by other systemic and tissue-specific factors are intimately involved in key aspects of growth plate development. As a corollary of these essential functions, disturbances in these pathways due to mutations or environmental factors lead to severe skeleton disorders. Here, we review these pathways and the most recent progress made in understanding their roles in chondrocyte differentiation in growth plate development and activity. Furthermore, we discuss newly uncovered pathways involved in growth plate formation, including mTOR, the circadian clock, and the COP9 signalosome.
Keywords: Chondrocytes, Osteoblasts, Transcriptional Regulator, Transdifferentiation, COP9 Signalosome
1. Introduction
The vertebrate skeleton has evolved as a dynamic system that serves numerous functions, such as protecting internal organs, creating attachment sites for muscles to produce locomotion, providing a reservoir for minerals, and serving as a hematopoietic niche. Many signaling pathways control the patterning, growth and maturation of skeletal structures from early development to adulthood, as demonstrated by the various skeletal diseases that occur in humans due to their dysregulation. The vertebrate bony skeleton is formed through two processes: most craniofacial bones form through intramembranous ossification, whereby mesenchymal condensations directly differentiate into bone-forming osteoblasts; whereas all other bones are created through endochondral ossification, whereby cartilage templates form first, and are replaced by bone later [1] [2] [3]. These cartilage templates are produced by chondrocytes that undergo successive, well-coordinated steps of differentiation (Figure 1A). Early chondrocytes actively proliferate and produce a highly specific cartilage extracellular matrix, and later become hypertrophic. Along with chondrocyte maturation, the cells present in the perichondrium flanking hypertrophic chondrocytes differentiate into osteoblasts to form a bone collar (interim bone cortex), wherein vascular invasion occurs. Following terminal maturation of hypertrophic chondrocytes, the mineralized cartilage matrix is invaded by blood vessels and associated pericytes, osteoclasts, and osteoblast progenitors. It is ultimately replaced by vascularized trabecular bone as well as bone marrow, forming the primary ossification center. A similar process occurs in epiphyses during the early postnatal stage, forming secondary ossification centers. The area between the primary and secondary ossification centers is the epiphyseal growth plate, which persists in humans until puberty to sustain longitudinal bone growth (Figure 1B).
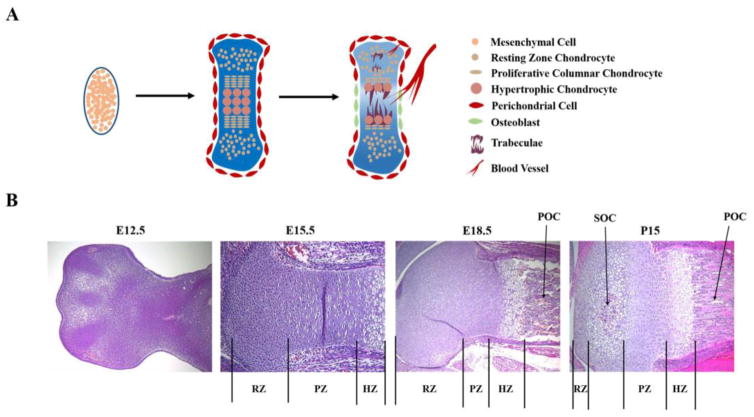
Fig. 1. Successive steps of endochondral ossification in mice.
A. Schematic representations of successive steps of cartilage growth plate formation. B. Hematoxylin and eosin staining of mouse limb growth plates at embryonic days E12.5, E15.5, E18.5, and post-natal days P15. Vertical lines indicate the Resting Zones (RZ), Proliferative Zones (PZ), and Hypertrophic Zones (HZ) at each stage. Arrows point to the Primary Ossification Center (POC) and the Secondary Ossification Center (SOC).
The epiphyseal growth plate is comprised of discrete zones representing chondrocytes undergoing stepwise maturation in a staggered manner, from resting, proliferative, and prehypertrophic stages, to hypertrophic and terminal stages (Figure 1B). In the reserve zone, chondrocytes are relatively inactive mitotically and express such markers as Col2a1 (encoding type II collagen). Underneath this zone, chondrocytes rapidly proliferate and highly express Col2a1 and Acan (aggrecan). Next, chondrocytes exit the cell cycle and become prehypertrophic, expressing Col10a1 (type X collagen) and Ihh (Indian Hedgehog) [3]. Prehypertrophic chondrocytes increase in size to become hypertrophic cells that stop expressing Col2a1 but continue expressing Col10a1. Hypertrophic chondrocytes eventually stop expressing Col10a1 and reach terminal stages, undergoing apoptosis, autophagic cell death, or transdifferentiation.
Not surprisingly, signaling cascades need to be finely tuned to ensure proper skeletal development. The complex regulatory system allows various developmental cues from the environment to transduce into proper spatial- and temporal-specific cellular responses. These signaling cascades converge on master chondrocyte transcription factors, including Sox9, Runx2, and Osterix. Here, we review critical signaling pathways involved in growth plate development and representative skeletal diseases associated with signaling dysregulation. For each signaling pathway, there is a considerable degree of combinatorial mixing and matching at the level of the ligands, receptors, antagonists, and downstream mediators. This produces a large diversity in transcriptional outputs and exerts precise context-dependent control. As of 2015, 436 genetic skeletal disorders have been recognized; as such, this topic has been intensively studied and reviewed [4, 5]. Thus, after a brief overview, we here focus only on the most recent progress regarding growth plate formation for each signaling pathway.
2. Transforming Growth Factor β (TGF-β)/Bone Morphogenetic Protein (BMP) Signaling
The TGF-β superfamily is a highly conserved group of more than 40 members, including TGF-βs, Activins, and BMPs, and many of them play an important role in skeletogenesis. The large number of TGF-β superfamily ligand-receptor combinations makes elucidating the role of each one very challenging. This difficult task is further compounded by functional redundancy among members and by the contextual-dependent nature of TGF-β signaling output [6]. Briefly, the binding of TGF-β superfamily ligands to single-pass transmembrane serine/threonine receptors results in the formation of a heteromeric receptor ligand complex, where the ligand-bound type II receptor phosphorylates the type I receptor. The type I receptor then recruits and phosphorylates regulatory Smad transcription factors (R-Smads), which form a complex with the coregulatory Smad (Co-Smad) Smad4. The complex accumulates in the nucleus and regulates gene transcription. TGF-β signaling primarily targets the R-Smads Smad2 and 3, whereas BMP signaling mostly mediates its response via Smad1, 5, and 8. Another group of Smads, the inhibitory Smads (I-Smads), act as negative regulators of R-Smads to inhibit TGF-β/BMP signaling.
Due to space limitations, we refer readers to a most recent review highlighting the well-established role of BMP signaling in skeletal development, particularly its role in bone and joint formation [7]. BMPs and their receptors are expressed in proliferating chondrocytes, hypertrophic chondrocytes, and perichondrium cells. BMP signaling is vital to endochondral bone formation through its roles in regulating chondrocyte proliferation and differentiation. Most importantly, BMP and TGF-β signaling are required for the expression and maintenance of SOX9, a master transcriptional regulator of chondrogenesis [8] [9] [10] and are thereby important inducers of chondrogenesis.
Mice with a global knockout of Bmp2 or Bmp4 die before E10.5, prior to the onset of skeletal development [11] [12]. In mice with Bmp2 deleted specifically in chondrocytes (using a Col2a1-CreER transgene), there was severe chondrodysplasia caused by impaired differentiation and maturation of chondrocytes. In contrast, Bmp4 deficiency (achieved using the same system) caused only a mild cartilage phenotype [13]. Thus, Bmp2 is a major ligand in chondrocytes. Interestingly, deletion of Bmp2 and Bmp4 in osteochondroprogenitors (Prx1-Cre) revealed that the genes are only required for mesenchymal condensations of the posterior digits in the forelimbs, possibly because other BMP genes are required for other condensations [14]. Accordingly, the overexpression of Noggin, a BMP inhibitor, blocked the formation of all prechondrogenic condensations [15, 16]. Noggin knockout mice have defects in all skeletal elements, resulting in shorter limbs, fused vertebrae, and fewer ribs [17]. On the other hand, when BMP2/4 was overexpressed in the developing chick limb, there was an increased number of chondrocytes and an increased production of cartilage extracellular matrix, and joints failed to form [18]. Recently, Dullard was discovered as a critical player in chondrogenesis. Although this phosphatase was originally identified as an inhibitor of BMPR1/2 in vitro [19], deletion of the gene Dullard using a Prx1-Cre driver resulted in a significant reduction in the proliferative and hypertrophic layers in growth plates of newborn mice, and these defects were associated with upregulated TGF-β signaling, but not BMP signaling [20]
Combined deletion of Bmpr1a (BMP Receptor Type IA) and Bmpr1b (BMP Receptor Type IB) in mouse chondrocytes resulted in a generalized chondrodysplasia with no growth plates [21]. These type I BMPR were shown to promote growth plate chondrocyte proliferation and late-stage differentiation by promoting Ihh expression and inhibiting Fgfr1 expression [22]. On the other hand, over-expression of a constitutively active form of Bmpr1a in mouse chondrocytes led to premature chondrocyte hypertrophy [23]. Another Type I receptor, Activin receptor IA/Activin-Like Kinase 2 (ACVR1/ALK2), binds to multiple BMP, TGF-β and Activin ligands. The activating mutations in ACVR1/ALK2 cause Fibrodysplasia Ossificans Progressiva (FOP), a rare and devastating genetic disorder characterized by endochondral ossification occurring at extra-skeletal sites, such as muscle and ligaments [24] [25]. Human mesenchymal stem cells overexpressing the ACVR1 activating mutation R206H, which occurs in 98% of FOP patients, were shown to cause ectopic bone formation in nude mice [26], and mice lacking Acvr1 in chondrocytes had significantly reduced BMP signaling, as well as craniofacial defects, axial skeleton defects, and thoracic kyphosis [27] [28].
The chondrocyte-specific deletion of Smad1/5/8 caused severe chondrodysplasia in mice [29]. These mice had small cartilage anlagen and reduced Sox9, Runx2, Col2a1 and Col10a1 expression. Growth plate chondrocytes displayed reduced proliferation and increased apoptosis, and lack of columnar organization and hypertrophy. While Smad4 expression was not altered in these mice, the expression of BMP receptor genes was lower, as was the expression of Tak1 and Mekk3, which encode kinases activating p38 MAPK through BMP signaling. Lastly, the Ihh/PTHrP feedback loop and functional antagonism between BMPs and FGFs were also disrupted [29]. There was a very similar lethal chondrodysplasia phenotype in the Smad1/5 double mutant mice, while Smad8-deficient mice had mostly normal growth plates and chondrogenesis [29]. Thus, these results point to a more important role of Smad1/5 over Smad8 in chondrocytes. Furthermore, the phenotype of Smad1/5 mutant mice was very similar to that of Bmpr1a/Bmpr1b receptor mutant mice [21], indicating that BMP signaling is mostly mediated through Smad1/5 in the growth plate. Interestingly, the chondrocyte-specific deletion of Smad4 using the same Col2a1 transgene resulted in a mild growth plate phenotype [7]. Thus, Smad1/5 likely may have many Smad4-independent actions in chondrocytes.
The inhibitory Smad6 and Smad7 add another layer of regulation to TGF-β/BMP signaling. In vitro, Smad7 can inhibit both TGF-β and BMP signaling, whereas Smad6 inhibits primarily BMP signaling [6]. Smad6/7 acts at multiple levels, including ubiquitin-mediated degradation of R-Smads and TGF-β receptors, competitive inhibition of Co-Smad4 binding, and binding of TGF-β receptors to prevent Smad phosphorylation. Both Smad6 and Smad7 play important roles in cartilage [30] [31]. Smad7-deficient mice are dwarfs, with delayed vascular invasion of growth plates and short hypertrophic zones. They show increased expression of Col2a1 and Col10a1, with a concomitant decrease in the expression of Mmp13, a terminal chondrocyte marker [31]. They have increased levels of phospho-Smad1/5/8, indicating enhanced BMP signaling in growth plates. Smad6-deficient mice displayed similar impairment of skeletal development [30]. They also showed an increase in BMP type 1 receptor expression, consistent with a study showing that Smad6 is responsible for the degradation of this receptor [30]. Therefore, Smad6/7’s control of chondrocyte proliferation and hypertrophy is at least in part through inhibiting BMP signaling.
In addition to the Smad-mediated canonical BMP pathway, BMPs also signal through noncanonical pathways in which BMP receptors initiate signaling via MAPKs, ultimately resulting in p38 MAPK family activation by downstream mediators including TGF-β activated kinase 1 (TAK1) [32]. TAK1 can be activated downstream of TGF-β and BMP4 in vitro. Both chondrocyte-specific and limb mesenchyme-specific deletion of Tak1 resulted in severe chondrodysplasia, with impaired formation of secondary ossification centers, absence of joints, and reduced BMP signaling [32] [33] [34]. Kindlins are a group of conserved cytoplasmic proteins involved in extracellular matrix adhesion, migration, and signaling. Knockout of Kindlin-2, a newly identified regulator of TGF-β signaling, in mice led to reduced Smad2 phosphorylation and neonatal lethal chondrodysplasia, disorganized chondrocyte columns, decreased chondrocyte proliferation, and increased apoptosis [35]. Lastly, Kartogenin, a small biomolecule, was able to stimulate limb skeletal growth and facilitate joint formation through the modulation of TGF-β and Ihh pathways [36]. All these latest results highlight that intricate TGF-β and BMP regulatory networks are directly involved in chondrogenesis and growth plate formation.
In humans, many skeletal dysplasias result from disrupted BMP signaling. In addition to FOP mentioned earlier, acromesomelic dysplasia is a group of rare osteochondrodysplasias characterized by short limbs and rudimentary fingers and toes. It results from mutations in BMPR1B or in the gene for the Growth and Differentiation Factor 5 (GDF5), a TGF-β superfamily member [4] [37]. Heterozygous mutations in the BMP antagonist NOGGIN results in multiple synostosis syndrome and proximal symphalangism, diseases characterized by failure to form joints in the hands and feet [38]. Moreover, many mutations in genes that alter BMP signaling through disrupting either ligand or receptor function can cause brachydactyly [5].
3. Fibroblast Growth Factor (FGF) Signaling
The FGF family is composed of at least 18 secreted ligands, which can bind to 4 different receptors (FGFR1–4). Many of the FGF ligands, including FGF4, FGF8, and FGF10, are responsible for limb bud formation and patterning [39]. Additionally, FGF9 and FGF18 are important regulators of endochondral bone formation [39]. FGF9 and FGF18 have both redundant and distinct functions. FGF18 has a biphasic effect, promoting chondrocyte proliferation and hypertrophy in early gestation (E14.5) and inhibiting them in late gestation (E16.5) [40] [41]. Fgf18−/− and Fgfr3−/− mice have similar growth plate phenotypes of expanded proliferating and hypertrophic zones, whereas Fgf9−/− mice have a normal proliferative zone. FGF9 and FGF18 promote and maintain Ihh and Runx2 expression [40]. Both are also involved in growth plate vascularization.
Alternative splicing creates FGFR isoforms with different specificities for FGF ligands. Upon ligand binding, FGFRs homodimerize and trans-autophosphorylate, leading to the phosphorylation of the docked adaptor protein FGFR substrate 2α(FRS2α). This allows the binding of other adaptor proteins and activation of different signaling cascades, involving MAPKs, PI3K, PLCγ, and JAK-STAT. FGFR3 signaling results in the activation of STAT1, ERK1/2, and p38 MAPK to regulate growth plate formation and endochondral ossification. FGFR3 can also indirectly control growth plate chondrocytes through modulating BMP, WNT, IHH, PTHrP and other developmental signaling pathways. The deletion of Fgfr3 in mice results in increased chondrocyte proliferation and an expansion of the hypertrophic zone, whereas its overexpression results in achondrodysplasia, a severe form of dwarfism due to very short growth plates [39]. In mice overexpressing Fgfr3 in limb bud mesenchyme, differentiation of chondrocytes from prehypertrophy to hypertrophy was blocked. Persistent Sox9 expression also occurred, possibly due to down-regulation of β-catenin [42].
Mutations in the FGF receptors lead to human skeletal dysplasias with variable severity. Activating mutations in FGFR3 cause achondroplasia, the most common form of dwarfism among live births. Some of these mutations, such as G380R, located in the FGFR3 transmembrane domain, stimulate FGF signaling in a ligand-dependent fashion. Others, like R248C in the extracellular domain or K650E in the intracellular domain, act in a ligand-independent manner and result in a more severe form of dwarfism, thanatophoric dysplasia [39]. The growth plates of humans with FGFR3 mutations show disrupted chondrocyte columns and reduced numbers of hypertrophic chondrocytes [39]. A recent study showed that the efficacy of five small molecule FGFR inhibitors, while effective in chondrocytes in vitro, did not improve skeletal growth in mice. Troublingly, they had no selectivity for FGFR3, but had significant off-target activity for IGFR1 [43]. These data show that new innovative approaches to treat achondrodysplasia are still required. Encouragingly, several candidate therapies are in progress [44]. These include competitive inhibitors such as a decoy receptor and a synthetic peptide that binds to FGFR3. Additionally, drugs such as statins decrease the activity of mutant FGFR3, whereas meclozine blocks MAPK and ERK signaling downstream of FGFR3. Both drugs increased the mouse Fgfr3 mutant limb length. The most promising therapy, an analog drug of C-type natriuretic peptide (CNP), is in phase II clinical trials, and was shown to increase growth velocity in children with achondrodysplasia through antagonism of MAPK signaling [44]. The tyrosine kinase inhibitor NVP-BGJ398, which is more selective for FGFR3 compared to the other FGF receptors, was shown to inhibit the hyperactivity of FGFR3 in a mouse model of achondrodysplasia and in human cartilage cells, as evidenced by restoration of chondrocyte proliferation and improvement of chondrocyte differentiation [45].
In addition to FGFR3, many human diseases are associated with loss and gain of function mutations in FGFR1 and FGFR2. Both activating and inactivating missense mutations in FGFR1 results in several different craniosynostosis syndromes, and missense mutations, deletions, and insertions in FGFR2 cause craniosynostosis syndromes, as well as defects in the appendicular skeleton and other organs [39]. FGFR1 and FGFR2 play many essential and mostly redundant roles during development, including growth plate formation. Fgfr2-deficient embryos fail to form limb buds [39]. On the other hand, overexpression of Fgfr1 in chondrocytes causes joint fusion. Deletion of both Fgfr1 and Fgfr2 in mice using an Osx-Cre driver caused a decreased length of the growth plate with a reduced number of proliferating chondrocytes [46]. Interestingly, the inactivation of Fgfr1/2 resulted in suppression of Ihh and Pthrp, and the growth plate phenotype was rescued by administration of PTH(1–34), suggesting an in vivo link between FGF signaling and IHH signaling in relaying information between growth plate chondrocytes [46].
MAPK/ERK Kinase 1 (MEK1) is activated by FGF signaling, and phosphorylates as well as activates ERK1/2 MAPKs (Extracellular signal-Regulated Kinase/Mitogen-Activated Protein Kinase). Constitutive activation of MEK1 in chondrocytes resulted in achondroplasia through inhibiting hypertrophic differentiation [47]. Erk1/2 inactivation in osteochondroprogenitor cells (Prx1-Cre) caused ectopic chondrocyte differentiation in the perichondrium, whereas expression of constitutively active MEK1 inhibited chondrocyte differentiation [48]. Furthermore, chondrocyte-specific deletion of Erk1/2 (Col2a1-Cre) caused severe chondrodysplasia [48], whereas Erk1/2 inactivation in hypertrophic chondrocytes (Osx-Cre) resulted in expansion of the hypertrophic zone and impairment of chondrocyte terminal differentiation [49]. MEK1 and ERK1/2 can also be activated by RAF kinases [50]. The deletion of a-Raf and b-Raf specifically in chondrocytes did not affect endochondral bone development [51]. However, chondrocyte-specific ablation of c-Raf caused expansion of the hypertrophic zone, decreased apoptosis of chondrocytes, and impaired vascular invasion of the growth plate concomitantly with reduced Vegf expression and decreased phospho-Erk1/2 levels in vivo [52]. These results indicate that c-Raf, which is the predominant isoform expressed by hypertrophic chondrocytes, plays a more important role in growth plate maturation than a- and b-Raf [52].
FGF and BMP signaling act antagonistically on chondrocyte proliferation and hypertrophic differentiation [1]. In a mouse model of achondrodysplasia, BMP2 treatment partially rescued the growth plate defect by increasing the size of the hypertrophic zone and increasing chondrocyte proliferation in limb explants [53]. BMP-mediated activation of a Msx2 promoter linked to a luciferase reporter was antagonized by FGF2 via the ERK/MAPK pathway. BMP induction of Ihh was also inhibited by ERK [29]. On the other hand, BMP signaling inhibits the activation of STAT and ERK1/2 MAPKs, which are key effectors of FGF signaling [22]. Interestingly, BMP and FGF signaling can also act synergistically to induce early chondrogenesis. Indeed, it was shown that FGF is necessary and sufficient to induce Sox9 expression and specification of a prechondrogenic identity in neural crest-derived mesenchymal cells. However, FGF was unable to maintain Sox9 expression and could not initiate the full Col2a1-expressing chondrogenic program. In contrast, BMP4 was able to maintain Sox9 expression, but only in combination with FGFs [54]. These findings highlight the complex nature of growth plate regulation, as many signaling pathways intersect in a delicate fashion.
4. WNT/β-Catenin Signaling
There are at least 19 WNT ligands and most signal through the canonical β-catenin pathway. When WNT ligands are absent, β-catenin is bound in a destruction complex consisting of glycogen synthase kinase 3β (GSK3β), axin, and adenomatosis polyposis coli (APC). GSK3β phosphorylates β-catenin and thereby triggers its ubiquitination and degradation. Wnt ligands bind to seven-pass transmembrane receptors Frizzled (Fzd) along with the co-receptors lipoprotein-related protein 5 (LRP-5) and LRP-6. Upon this interaction, GSK3β-mediated phosphorylation of β-catenin is inhibited, resulting in β-catenin stabilization. β-catenin then translocates to the nucleus and acts as a co-activator of TCF/LEF transcription factors. Members of the Dikkopf (DKK) family, secreted Frizzled-related proteins (sFRP), and Wnt inhibitory factor 1 (WIF-1), act as antagonists of Wnt signaling [55]. Mouse genetic studies have shown that Wnt signaling is intimately involved in the development of many tissues and organs, and both gain- and loss-of-function mutations in Wnt signaling cause various human diseases, including bone diseases [56] [57, 58].
WNT signaling is well known to regulate chondrocyte differentiation and cartilage development [3]. It is well established that β-catenin and SOX9 directly antagonize each other’s stability in differentiating chondrocytes [59] [60] [61] [62] [63]. The inactivation of β-catenin specifically in chondrocytes of mouse embryos led to severe dwarfism with decreased chondrocyte proliferation, delayed hypertrophy, and impaired endochondral bone formation accompanied by increased Sox9 expression [59]. The defect in chondrocyte proliferation and maturation was similar in another mouse model with deletion of β-catenin using a Dermo1-Cre driver [64]. Additionally, the deletion of β-catenin in hypertrophic chondrocytes (Col10a1-Cre) impaired chondrocyte maturation, as assessed by reduced expression of Vegf, Mmp13, Runx2, and Osx [65]. Further demonstrating the sensitivity of chondrocytes to β-catenin levels, β-catenin stabilization in chondrocytes of mouse embryos led to severe chondrodysplasia with decreased Sox9 expression [59]. Although Lrp5 and Lrp6 serve largely redundant functions, deletions of Lrp5 and Lrp6 using a Dermo1-Cre driver caused a severe shortening of the endochondral skeletal elements as a result of reduced hypertrophic chondrocytes and concomitant with a decreased expression of Col10a1 [66]. Additionally, the Col2a1-Cre-targeted deletion of Lrp5/6 resulted in lethality either in utero or shortly after birth, and analysis of E18.5 embryos showed abnormal growth plate development [67]. Inhibition of β-catenin in chondrocytes by expressing ICAT (Inhibitor of β-catenin and TCF), a Wnt inhibitor, resulted in delayed skeletal growth postnatally with decreased proliferative and hypertrophic zones [68]. Wnt signaling has also been shown to promote chondrocyte hypertrophy by inhibiting PTHrP activity [69].
Multiple Wnt ligands signal through non-canonical pathways, including the planar cell polarity (PCP) and the Wnt/Ca2+ pathways [57]. Non-canonical Wnt signaling has been implicated in promoting chondrocyte differentiation due to its ability to inhibit canonical Wnt signaling. The PCP pathway plays a unique role in inducing chondrocyte column formation in the growth plate [70]. Recently, time-lapse movies of live tissue revealed that chondrocyte columnar arrangement in the growth plate involves cadherin- and β-catenin-dependent expansion of surface adhesion [71].
5. Hedgehog Signaling and Parathyroid related Peptide (PTHrP) Signaling
Hedgehog (Hh) signaling is involved in many developmental processes. Mutations in hedgehog genes cause various congenital limb deformities [5]. Vertebrates produce three Hh ligands: Sonic (Shh), Indian (Ihh), and Desert Hedgehog (Dhh). If Hh ligands are absent, the G protein-coupled receptor Smoothened (Smo) is inhibited catalytically by Patched (Ptc). Upon Hh ligand binding to Ptc, the inhibition of Smo is released and Gli transcription factors are activated [72] [73]. The two Patched homologues, Patched1 and Patched2, were recently found to play overlapping and complementary roles in limb development [74]. Shh-deficient mouse embryos die at birth with holoprosencephaly and oligodaactyly, a disorder caused in humans by SHH mutations, whereas Shh overexpression causes polydactyly [75] [76]. Ihh is produced by prehypertrophic chondrocytes and plays a crucial role in chondrocyte differentiation. Ihh–deficient mouse embryos display severe skeletal growth retardation. They show reduced chondrocyte proliferation, and premature and abnormal hypertrophy [77] [78] [79]. Scube2 encodes a secreted and cell membrane-bound protein that mediates long-range Ihh signaling, probably through increasing Ihh solubility by binding to its cholesterol moiety. Genetic ablation of Scube2 led to defective chondrocyte differentiation and proliferation, manifested by short growth plates [80]. Thus, post-translational modifications play an important role in modulating Ihh activity.
Ihh plays a crucial role in chondrocyte hypertrophy. In a negative feedback loop, Ihh promotes the expression of the gene for the parathyroid hormone-related peptide (PTHrP), which maintains growth plate chondrocytes in a proliferative state. Overall, the concerted actions of Ihh and PTHrP regulate chondrocyte proliferation and maturation. Pthrp is expressed in resting chondrocytes and maintains chondrocyte proliferation, whereas chondrocytes farther away from the resting zone in the growth plate do not receive PTHrP signals, and undergo hypertrophy. Indeed, Pthrp-deficient mice had accelerated hypertrophic differentiation and premature mineralization [81].
PTHrP signals through parathyroid hormone type I receptor (PTH1R), a G-protein coupled receptor, to control skeletogenesis and Pth1r knockout mice also showed accelerated hypertrophy [82]. Disruption of PTH1R signaling is associated with a number of osteochondrodysplasias, such as brachydactyly type E, caused by a mutation in PTHrP, and metaphyseal dysplasia and Eiken dysplasia, which are rare skeletal disorders caused by mutations in PTH1R [82].
6. Notch Signaling
Notch signaling is another evolutionarily conserved pathway involved in cell fate determination, differentiation, proliferation, and apoptosis [5]. It occurs when the Notch receptor is activated by binding to Delta or Jagged, ligands anchored on adjacent cells as type 1 transmembrane proteins [83]. The Notch receptor then undergoes cleavage to generate a Notch Extracellular Truncation (NEXT) fragment. This fragment is further cleaved by γ-secretase proteinase to produce the Notch Intracellular Domain (NICD). NICD translocates to the cell nucleus to interact with the transcription factor RBPJ and the transcriptional coactivator Mastermind-like 1 (MAML1). The ternary complex promotes the expression of basic helix-loop-helix (bHLH) transcription factors of HES and HEY families, which then transcriptionally repress downstream target genes [83].
Notch signaling plays important roles in bone and cartilage formation. Mouse genetic deletions and human genetic mutations in Notch signaling are associated with a variety of defects in patterning, axial skeleton development, and skeletal diseases [83, 84]. Notch signaling is important for chondrocyte hypertrophy in part via regulating Sox9 expression [83] [85]. Mouse models with loss and gain of function in Notch signaling have revealed that Notch acts upstream of Sox9 and Runx2 in the regulation of osteochondro-progenitor cell proliferation and lineage specification. In Prx1-Cre targeted osteochondral progenitor cells, the loss of Notch enhanced chondrocytic lineage commitment but impaired fracture healing due to depletion of the bone marrow stromal cell pool, whereas gain of Notch function suppressed chondrocyte differentiation and proliferation [86] [87]. Additionally, the overexpression of NICD specifically in chondrocytes (Col2a1Cre and Col2a1CreER) caused skeletal malformations with decreased proliferation and impaired chondrocyte hypertrophy, whereas the loss of Notch function in these cells increased the hypertrophic zone size [88] [89, 90]. Notch negatively regulates chondrocyte differentiation in the axial skeleton by suppressing Sox9 expression [89]. A RBPjκ-dependent pathway mediates the chondrocyte maturation, whereas a RBPjk-independent pathway is incriminated in the suppression of chondrocyte proliferation, survival and columnar organization, as well as impaired responsiveness to Ihh signaling [90]. Again, the role of Notch signaling in growth plates is likely to be even more complex as Notch signaling was recently shown to regulate chondrocyte hypertrophy in part in a Sox9-independent manner [85]. Furthermore, Prx1-Cre deletion of Hes1 and Hes5, the key targets of RBPjκ-dependent Notch signaling, confirms that deletion of Notch effectors in osteochondral progenitors accelerates chondrocyte differentiation [91]. While Hes5 alone controls Sox9 transcription, both Hes1 and Hes5 are required to suppress chondrogenesis and promote the onset of hypertrophy. Additionally, the overexpression of Hes1 in osteochondral progenitor cells delays chondrocyte hypertrophy and reduces proliferation. On the other hand, the HEY family of transcription factors play no role in chondrogenesis or matrix catabolism. However, this contrasts with an earlier study of combined deletion of Hes1 and Hes5 using a Col2a1-Cre driver showed that they are dispensable for endochondral ossification in chondrocytes [92]. This again highlights the stage-specific role that Notch signaling plays in growth plate homeostasis. Another new study has shown that Notch may inhibit chondrogenic differentiation of limb bud mesenchyme by upregulating the expression of Twist1, which encodes a bHLH transcription factor involved in skeletal formation [93]. These data show that a key action of Notch signaling in chondrogenesis and growth plate formation is to control the expression of such master cell fate regulators as Sox9 and Twist1, and understanding of the downstream Notch targets provides insight into normal development and the role of Notch in skeletal disorders.
7. Retinoic Acid Signaling
Retinoic acid (RA), the main metabolite of vitamin A, elicits a key pathway involved in growth plate regulation. Signaling occurs when RA binds to RA Receptors (RARα/β/γ), which will then form a heterodimer with Retinoid X Receptors (RXRα/β/γ), and bind to RA Response Elements in DNA to directly regulate gene transcription. Importantly, the RAR and RXR heterodimers can also act as transcriptional repressors in the absence of ligand through the recruitment of co-repressors and histone deacetylase complexes to promote cartilage growth and chondrocyte differentiation [94]. The amount of RA available to a cell is dependent on the balance between the RA synthesizing enzymes retinol dehydrogenases (RDH) and retinaldehyde dehydrogenases (RALDH), and cytochrome P450 26 (CYP26), which degrades RA [94]. The genetic deletion of any single member of the RAR family (RARα, RARβ, RARγ) in mice results in a minor phenotype, while deletions of multiple RAR receptors (e.g. RARα/γ or RARβ/γ) results in perinatal lethality with severe skeletal abnormalities [95] [96]. While most RAR and RXR genes are functionally redundant, RXRα-deficient mice die in utero [94].
RA signaling regulates anteroposterior and proximodistal patterning in early limb development [94]. RA signaling in the proximal limb promotes the expression of transcription factors Meis1/2 in the limb bud, whereas Fgf8 signaling in the distal limb represses the expression of Meis1/2 to promote proper proximodistal patterning [94]. Furthermore, in mouse and zebrafish models deficient for Raldh2, the enzyme catalyzing the conversion of retinaldehyde to RA, there is a severely reduced expression of T-box 5 (Tbx5), a transcription factor expressed in the lateral plate mesoderm and important for forelimb initiation [94]. Indeed, the deletion of Raldh2 in mice causes the absence of forelimb bud development.
Elucidating the role of RA signaling in the growth plate has been challenging since activating as well as arresting RA signaling can result in similar phenotypes. Multiple animal models, including quail, zebrafish, chick and murine models, show that RA is important in forelimb development and nearly dispensable in the hindlimb [94]. Furthermore, exogenous treatment with RA has a teratogenic effect on chondrocyte differentiation in murine limb development. Indeed, increasing RA signaling through global and Prx1-Cre-mediated deletion of Cyp26b1 in mice resulted in truncated limbs and abnormal digit formation. Furthermore, the addition of a RARα antagonist in culture stimulates chondrogenesis, whereas a RARγ agonist can inhibit BMP signaling response in chondrogenesis. Thus, RA signaling generates an important signaling cascade for limb bud development, and interacts with many pathways involved in control of growth plate homeostasis, including FGF, WNT, BMP, and SHH [94]. For example, BMP signaling inhibits the expression of Aldh1a2, an enzyme that synthesizes RA [97]. Furthermore, inhibition of RA signaling in primary cultures of mouse limb mesenchyme repressed Sox9 expression, and p38 MAPK and PKA signaling is activated when RA signaling is inhibited [98]. Interestingly, global deletion of RA Receptor γ (RARγ) in Cyp26b1-deficient mice only partially rescues limb morphology, indicating that there are two distinct effects of RA signaling in the limb, a teratogenic effect and a patterning effect [99]. Additionally, deletion of RARα/γ or RARβ/γ in chondrocytes (Col2a1-Cre) caused severe growth retardation by 3 weeks of age, with shorter growth plates, reduced proliferating chondrocytes and decreased Aggrecan expression, whereas RARα/β-deficient mice are essentially normal [96]. In contrast, RA treatment of mouse epiphyseal chondrocytes in culture stimulates Wnt signaling and enhances the inhibitory role of Wnt signaling on matrix accumulation [100]. Therefore, the RA signaling pathway is also likely to be context-dependent, and further study is warranted to better understand the mechanism with which RA metabolism controls growth plate development.
8. mTOR Signaling
The mammalian Target of Rapamycin (mTOR) pathway is a ubiquitous, central regulator of cell metabolism, survival and proliferation and is thereby critical for organismal growth and homeostasis. This pathway integrates various types of extracellular and intracellular signals. It can be activated by various growth factors, such as epidermal growth factor and insulin-like growth factor, which activate Akt/PI3K signaling, as well as nutrients such as amino acids, glucose, and oxygen levels. It can be inhibited by AMP-activated protein Kinase (AMPK) in response to the absence of nutrients in the cell [101].
The importance of mTOR in the cartilage growth plate has only recently started to emerge. mTOR is a highly conserved serine/threonine kinase that takes part in two distinct complexes: mTORC1 and mTORC2. These complexes share several common protein subunits in addition to mTOR, and differ by other components, namely RAPTOR for mTORC1 and RICTOR for mTORC2 [101]. The deletion of mTOR specifically in limb bud mesenchyme (Prx1-Cre) resulted in impaired embryonic skeletal growth [102]. Mutant mice died at birth due to exencephaly, and displayed smaller appendicular bones, although the authors did not describe any growth plate abnormalities in these mutants. The disruption of mTORC1 specifically by deleting Raptor (Prx1-Cre) also resulted in neonatal lethality [102]. Cartilage elements showed reduced production of cartilage extracellular matrix due to deficient protein synthesis. There was no effect on growth plate chondrocyte proliferation or survival, but chondrocyte hypertrophy was delayed and cells showed limited enlargement [102]. The disruption of mTORC2 specifically by deleting Rictor (mTORC2) in limb bud mesenchyme caused a similar phenotype, with shorter skeletal elements and delayed chondrocyte hypertrophy. However, these mutants had no defect in cartilage matrix production and although the hypertrophic zone was shortened, there was no change in hypertrophic chondrocyte cell size [103].
Interestingly, a recent study demonstrated that mTORC1 promotes early stage chondrocyte proliferation and prevents later stage terminal differentiation [104]. Indeed, mice treated with rapamycin showed thinner growth plates and a reduced number of proliferating and prehypertrophic chondrocytes. Furthermore, the deletion of TSC1, an upstream negative regulator of mTORC1, specifically in chondrocytes (Col2a1-Cre) delays postnatal endochondral bone formation and causes chondrodysplasia and dwarfism. The mutants had enhanced chondrocyte proliferation but impaired chondrocyte hypertrophy and terminal differentiation. This phenotype was rescued by rapamycin treatment, indicating there is an uncoupling of the proliferation and differentiation program in the growth plate as a result of mTOR signaling disruption. mTORC1 activity is also crucial for PTHrP transcription by phosphorylating and promoting the nuclear accumulation of Gli2, a Hedgehog signaling mediator, in a smoothened-independent manner. [104]. Finally, the deletion in mice of Lkb1, a serine/threonine kinase inhibitor of mTOR signaling, in chondrocytes (Col2a1-Cre) resulted in marked growth retardation in postnatal mice, concurrent with a disorganized growth plate that had increased proliferating chondrocytes and ectopic hypertrophic chondrocytes [105]. Thus, mTOR signaling is an important coordinator of chondrocyte proliferation and hypertrophy. mTORC1 and mTORC2 play important and distinct roles in the growth plate and the disruption of mTOR signaling might affect growth plate homeostasis through disrupting other signaling pathways. However, much more work is still needed to further elucidate how mTOR signaling regulates the growth plate.
9. Insulin-like Growth Factor (IGF) Signaling
Insulin-like growth factor signaling has key roles in endochondral ossification. It consists of two ligands (IGF1 and IGF2), two receptors (IGFR1 and IGFR2), and six IGF-binding proteins (IGFBP1 to 6). IGF1 is the main signaling ligand. IGF1 binds to IGFR1 with 20 times more affinity than IGF2. However, IGF1 barely binds IGFR2. Furthermore, IGFR1 is a receptor with tyrosine kinase activity that is highly homologous to the insulin receptor, whereas IGFR2 is a single pass transmembrane receptor with no kinase activity.
The binding of IGFs or insulin to IGFR1 initiates PI3K/AKT and RAS/RAF/MEK/ERK signaling to control growth and survival through activation of the mTOR pathway, inactivation of caspase-mediated cell death, and induction of cell cycle progression. IGFBPs act extracellularly as endocrine factors, although they can also be found intracellularly, to regulate IGF half-life or to modulate its signaling [106]. The global deletion of Igf1 resulted in shortening of the skeleton and dwarfism, and Igfr1 deletion caused a more severe growth reduction and neonatal lethality [107]. Igf1 deletion is associated with reduced proliferation, decreased survival, and impaired maturation of growth plate chondrocytes [108]. Moreover, Igfr1 deficiency specifically in chondrocytes (Col2a1-Cre) resulted in neonatal lethality, and the growth plates of these mutants showed disorganized chondrocyte columns and delayed ossification and vascularization [109]. Mutant chondrocytes exhibited decreased proliferation and increased apoptosis, as well as increased Pthrp expression. Mice with the deletion of Igfr1 in prehypertrophic chondrocytes and osteoblasts (Osx-Cre) were viable, but resulted in impaired postnatal growth [108, 110]. The morphology of growth plates was abnormal, and chondrocytes showed decreased proliferation and delayed maturation. Interestingly, Wnt-induced secreted protein 3 (WISP3), a member of the CCN family of secreted growth factors, inhibits IGF1 signaling by binding to IGF1 and preventing its secretion. This results in impaired chondrocyte hypertrophy, with reduced Col10a1 expression [111]. WISP3 mutations in humans results in the autosomal recessive skeletal disease progressive pseudorheumatoid dysplasia (PPD), a spondylo-epiphyseal dysplasia characterized by cartilage loss and joint failure by early adulthood [112]. However, its deletion or overexpression in mice does not result in an apparent phenotype [112]. Therefore, more study is needed to elucidate its role in the growth plate.
While IGF1 signaling mostly predominates IGF2 signaling, IGF2 does play a role in growth plate chondrocyte differentiation. For example, treatment of mouse E18.5 limb explant cultures with IGF2 and BMP9 led to increased endochondral ossification with an expanded hypertrophic zone [113]. IGF2 synergistically acted with BMP9 in activating the Smad pathway, and this effect was antagonized by IGFBP3 and IGFBP4. Additionally, in fibroblasts isolated from patients with 3-M Syndrome, a human disorder characterized by pre and postnatal growth impairment, there was reduced expression of IGF2 [114].
In humans, homozygous deletions of IGF1 result in severe growth retardation in utero and postnatally, accompanied with severe osteopenia [107]. Various other IGF1 mutations have also been reported, including a partial deletion of exons 4 and 5, a homozygous missense mutation, and a nucleotide substitution. These IGF1 mutations caused various degrees of retarded skeletal maturation, microcephaly, deaf-mutism, and mental retardation [107]. Furthermore, humans with heterozygous mutations in IGFR1 display a similar phenotype to IGF1 mutations, underlying the importance of IGF1-IGFR1 signaling axis in human skeletal development [107].
10. The Circadian Clock
Another important emerging area of growth plate research is the regulation of endochondral ossification by the circadian clock and its effect on developmental signaling pathways. The circadian clock is an endogenous time-keeping system present in nearly all organisms. It is involved in the regulation of gene expression, physiology, and behavior with an approximately 24-hour periodicity. It is composed of a positive and negative regulatory feedback loop that is entrained by external zeitgebers (temporal cues), such as the sun, with the positive arm consisting of the transcription factors BMAL1 and CLOCK, and the negative arm consisting of the transcription factors PERIODS (PER) and CRYPTOCHROMES [115]. Increasing evidence indicates that circadian clocks also regulate skeletal homeostasis [116] [117]. Interestingly, some chondrocyte-specific genes, such as Col2a1 and Col10a1, are expressed in a diurnal pattern along with the core clock genes in chondrogenic ATDC5 cells as well as in articular cartilage and growth plates [118] [119]. Mouse Per1 and Per2 expression is increased by PTH through PKA-CREB signaling [120]. Indeed, PTH can reset the circadian clock in femurs in a time- and dose-dependent manner [121] [122]. Thus, there might exist a chondrocyte-specific peripheral clock that is critical to growth plate development. In Bmal1-deficient mice, the femora and tibiae were significantly shorter, with impaired Ihh-mediated chondrocyte differentiation [123] [124]. More importantly, the reduced bone length and decreased Ihh expression persisted when Bmal1 was deleted specifically in chondrocytes, confirming that the effect is likely to be cell-autonomous [124]. Moreover, the chondrocyte-specific ablation of Bmal1 (Col2a1-Cre) also resulted in severe cartilage degeneration by 3 months of age due to dysregulated TGF-β signaling [125]. Given that the circadian clock signaling pathway is implicated in many physiological processes, it will be interesting to understand the detailed mechanism of circadian control of the musculoskeletal system, especially in regards to its interaction with other signaling pathways.
11. Novel Transcriptional Co-Factors involved in Growth Plate Signaling
The effects of diverse signaling pathways reviewed above all ultimately impinge on a multi-layered transcriptional network in which key transcription factors, such as Sox9, Runx2 and Osterix, have been extensively studied and their role in growth plates has been well established. On the other hand, some transcriptional cofactors, such as β-catenin, CBFβ, JAB1, and YAP1, have been shown in recent years to also play a key role in growth plate formation. Such transcriptional cofactors, although they do not have identifiable DNA-binding domains themselves, can interact with key transcription factors such as Sox9, Runx2, and key signaling downstream effectors, such as Smads, to regulate chondrocyte differentiation and skeletal development in a spatio-temporal manner. In this regard, β-catenin serves as a prime example as a crucial transcriptional co-activator that plays essential roles at multiple steps of growth plate formation [57]. Overall, various transcription co-factors often interpret various developmental signaling pathways in an exquisite fashion, serving as a crucial bridge between upstream signaling cues and downstream events in the nucleus [1]. We refer the readers to the review by Liu and colleagues in the same journal issue for a review on master transcription factors involved in chondrocyte specification and differentiation. We here below review very recent studies on two novel, and crucial transcriptional co-factors, the COP9 signalosome and YAP1
11.1. The COP9 signalosome
In recent years, chondrocyte differentiation and growth plate formation have been shown to be regulated by a novel transcriptional co-factor Jab1 (c-Jun activation domain binding protein-1). Jab1, also known as Cops5 and Csn5, is the fifth member of the COP9 Signalosome complex (CSN), an evolutionarily conserved proteolysis regulator consisting of 8 subunits and playing an essential role in the regulation of development in mammals, plants and flies [126]. The CSN has the deneddylase activity that regulates the stability of the SCF E3 ubiquitin ligase complex, and is involved in the regulation of protein stability, the control of protein subcellular localization, cell cycle control, DNA repair and many developmental processes [127]. Jab1 interacts with many master transcription factors and key signaling pathways, and is also overexpressed in various cancers [127] [128]. Jab1 directly interacts with the TGF-β/BMP pathway. Jab1 can bind Smad4 in vitro and induce Smad4 degradation, and this ultimately attenuates TGF-β signaling [129]. However, Jab1 was also shown to interact with and induce the degradation of Smad7, resulting in increased TGF-β signaling [130]. Interestingly, Jab1 can interact with Smad5 to inhibit BMP signaling in human chondrocyte cultures [131]. Thus, the role of Jab1 in TGF-β/BMP signaling is very likely to be highly context-dependent.
Interestingly, in both human and mouse growth plates, Jab1 is highly expressed in the resting zone and proliferative zone, but not in the hypertrophic zone, suggesting a critical role for Jab1 in controlling chondrocyte hypertrophy. However, global knockouts of Jab1 in mice resulted in embryonic lethality by E8.5 prior to skeletogenesis [132] [133]. To better understand the physiological role of Jab1 in growth plate development, using a Col2a1-Cre driver, we have recently shown that Jab1 plays a crucial role in chondrocyte differentiation in vivo, most likely as a negative regulator of BMP signaling [134]. These mutant mice died at birth with severe chondrodysplasia, and had defects in the development of both the appendicular and axial skeleton, resulting in severe dwarfism [134]. Chondrocytes from these mice had altered BMP signaling, with increased BMP downstream effector Smad1/5 phosphorylation, enhanced response of primary Jab1 knockout chondrocytes to BMP treatment, and increased expression of BMP ligands and target genes, including Runx2 [134]. Furthermore, we showed that Jab1 is also essential for early limb development in a mouse model using a Prx1-Cre driver to delete Jab1 in osteochondral progenitor cells. The mutant embryos all displayed drastically shorter limbs that progressively worsen with age, with smaller cartilage elements, fewer hypertrophic chondrocytes, and a reduced or absent secondary ossification centers [135]. Interestingly, in micromass culture experiments, BMP response was decreased in the mutants, thus highlighting the differential role of Jab1 at different stages of BMP-mediated growth plate development. Moreover, in Jab1 knockout mutants, Sox9 expression and activity was severely compromised, suggesting Jab1 positively regulates Sox9 in early growth plate development. Interestingly, in a recent Sox9 ChIP-seq study in E12.5 limb buds, Jab1 was identified as a putative key downstream target for Sox9, suggesting a possible Jab1-Sox9 positive feedback loop in early limb development [136]. Thus, it is essential to further elucidate the underlying mechanism of the interaction between Jab1 and master transcription factors such as Sox9 and Runx2 in growth plate development.
11.2. YAP1
The transcriptional co-factor Yes-Associated Protein 1 (YAP1) is a key effector of the Hippo signaling, which is primarily involved in the regulation of organ size and tissue regeneration. YAP1 and closely related TAZ regulate downstream target genes by interacting with transcription factors such as TEAD. The evolutionarily conserved Hippo-YAP1/TAZ signaling pathway functions in a unique transduction manner, and we direct readers to a very recent comprehensive overview [137]. Briefly, when the tumor suppressor Hippo pathway is inactive, oncogenic YAP1 and TAZ are unphosphorylated and located in the nucleus. YAP1 and TAZ interact with transcription factors TEADs to promote the expression of target genes involved in survival, proliferation, differentiation, and migration/invasion [137]. When the Hippo pathway is activated by TAO kinases, MTS1/2 kinases are phosphorylated and activated, which subsequently phosphorylate LATS1/2 kinases. LATS1/2 kinases then phosphorylate and inactivate YAP1 and TAZ. As a result, YAP1 and TAZ are retained in the cytoplasm and are inactivated [137]. YAP1 serves as a key transcriptional regulator to integrate multiple signaling cascades, including Wnt, TGF-β/BMP, and hedgehog [138]. While a role for TAZ in Runx2-mediated osteogenesis has been well-established, most recently the role of YAP1 in chondrocyte differentiation has also begun to emerge. YAP1 can inhibit chondrogenic differentiation through downregulating BMP signaling by suppressing Smads [139]. Furthermore, a most recent mouse genetic study showed that Yap1 promoted chondrocyte proliferation, but inhibited hypertrophy during endochondral ossification and fracture healing [138]. The study shows that Yap1 expression is high in early chondrocyte differentiation and progressively decreases. Mechanism-wise, the high-level expression of Yap1 in early chondrogenesis promotes chondrocyte proliferation, at least in part, through inducing the expression of Sox6, a major downstream target of Sox9 during chondrogenesis. On the other hand, lower Yap1 expression in hypertrophic chondrocytes relieved its inhibition of Runx2-mediated Col10a1 expression [138]. This provides another example of a transcriptional co-factor that has differential roles at different stages of growth plate development through its interaction with key transcription factors.
12. Chondrocyte Terminal Fates and Transdifferentiation
The precise fates of hypertrophic chondrocytes and the origins of trabecular-forming osteoblasts during endochondral ossification had eluded us for decades, It is conventionally assumed that the hypertrophic chondrocytes, both in the cartilage anlagen prior to the establishment of growth plates and those at the chondro-osseous junction of growth plates, are terminally differentiated and are ultimately eliminated through programmed cell death, by either apoptosis (type I programed cell death) or autophagy (type II programed cell death) [140, 141]. Indeed, many studies in various organisms have detected evidences of apoptosis or autophagy in growth plate hypertrophic chondrocytes either under physiological condition or in response to various stimuli, including altered extracellular matrix and hypoxia in the microenvironment [142] [143] [144] [145] [146] [147, 148] [149]. The findings of these studies vary quite extensively in terms of type and percent of cells involved in programed cell death. For example, Farnum and Wilsman [144, 150] observed only about 12% of the hypertrophic chondrocytes with condensed morphology, indicative of apoptotic cells, in the lower lacunae adjacent to the cartilage–bone marrow interface in Yucatan swine. However, Gibson [145] reported that more than 50% of hypertrophic chondrocytes displayed characteristics of apoptosis at the chondro-osseous interface in the chick sterna. Similarly, studies with the TUNEL method also yielded divergent results, from as low as 9% of TUNEL-positive cells of hypertrophic chondrocytes isolated from the chick growth plate [151], to as much as 44% of TUNEL-positive cells found in the hypertrophic zone [152].
Unlike cells in other tissues, most of growth plate chondrocytes are well embedded within cartilage matrix and cannot be phagocytosed by macrophages as apoptotic cells in other tissues. While lacking typical apoptotic morphological changes in the terminal hypertrophic chondrocytes, there was the presence of autophagic vacuoles and the expression of autophagy-regulating genes by growth plate chondrocytes, suggesting that these cells may undergo processes more similar to autophagy than apoptosis [140, 144, 153]. Indeed, the autophagy pathway promotes cell survival in the mouse growth plate. The chondrocyte-specific deletion of Autophagy-related (Atg) genes, Atg5 and Atg7, resulted in reduced proliferation and enhanced cell death through caspase-dependent mechanisms [154]. Another study showed that autophagy in growth plates regulated the secretion of type II collagen through Fgf18-Fgfr4 signaling via JNK-dependent activation of the VPS34-Beclin1 [155].
Evidently, there are a significant number of hypertrophic chondrocytes that are not accounted for by programed cell death. As autophagy can lead to cell survival as well as cell death, some autophagy might also lead to reprogramed progenies. Indeed, the idea that some of these hypertrophic chondrocytes, rather than dying, can transdifferentiate into osteoblasts, was suggested several decades ago, and is now regaining traction [156]. The fact that both osteoblasts and chondrocytes are derived from the same Sox9-expressing osteochondro-progenitors hints at their potential common gene regulatory networks. Indeed, the transcription factors involved in chondrocyte and osteoblast differentiation serve complementary functions in the development of cartilage and bone [157]. In addition to their own specific markers such as Col10a1, hypertrophic chondrocytes also express many osteoblast markers, including Alp, SPARC, BGLAP, Opn, and Bsp, as well as Osx and Runx2, albeit at relatively lower levels compared with their expressions in osteoblasts [158, 159]. In fact, earlier morphological, histological [147], and cell culture experiments [158, 160–162] all observed the presence of potential chondrocyte-derived osteoblasts and likely transdifferntiation events. It was suggested that hypertrophic chondrocytes to osteoblasts transdifferentiation might have occurred through either asymmetrical division or direct transdifferentiation [147] [162] [163] [164]. However, most of such evidence came from in vitro studies and was not fully conclusive.
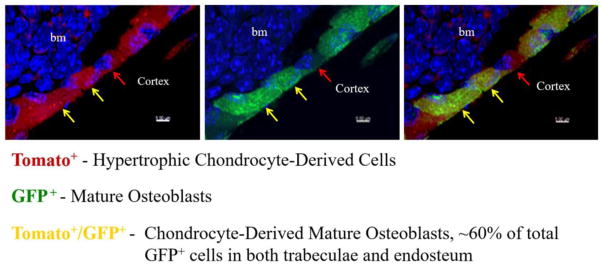
Recently, several groups independently reported the findings of their Cre/reporter lineage tracing studies in mice [165] [166, 167]. In these studies, chondrocytes or hypertrophic chondrocytes were labelled with reporter genes driven by chondrocyte or hypertrophic chondrocyte-specific promotors either constitutively or in an inducible manner. Collectively, their findings provide strong in vivo evidence that a significant number of hypertrophic chondrocytes indeed become osteoblasts, indicating that hypertrophic chondrocytes are a direct major source of osteoblasts in endochondral bone formation [165]. Zhou et al. showed that the Col10a1-Cre marked reporter+ cells started to appear in the primary ossification center coincident with the onset of primary ossification [165]. These cells were gradually increased in numbers and were distributed throughout the primary spongiosa. They were located in the bone marrow, on trabecular surfaces, and embedded within bone matrix (Figure 2). In postnatal mice, these Col10a1-Cre marked reporter+ cells were also found on endosteum and within cortex. Zhou et al. further showed that approximately 60% of the total mature osteoblasts on both trabecular and endosteum surfaces were derived from hypertrophic chondrocytes [165]. In addition to hypertrophic chondrocyte-derived osteoblasts and osteocytes, there were abundant numbers of Osx-expressing reporter+ bone marrow cells, which did not express Col1a1 and Osteocalcin but displayed skeletal progenitor characteristics, implying that hypertrophic chondrocytes may be the source of some Osx-expressing skeletal progenitors [168, 169].
Fig. 2. Mouse hypertrophic chondrocytes can transdifferentiate into mature osteoblasts.
The triple transgenic Col10a1-Cre;2.3Col1-GFP;ROSA-tdTomato mice express tomato (red) upon Cre recombination in hypertrophic chondrocytes, express Enhanced Green Fluorescent Protein (EGFP) in 2.3Col1-GFP+ mature osteoblasts, and a Tomato Red/EGFP overlay creates yellow, indicating chondrocyte-derived mature osteoblasts. Blue indicates nuclei. Image shows a magnified region of the cortical bone. bm: bone marrow. The image is adapted from reference 165.
These studies also found that embryonic hypertrophic chondrocyte-derived cells persisted well into adulthood, as long term chasing showed the labeled cells were present as osteoblasts, osteocytes and stromal cells in the metaphysis after birth [167]. It is worth noting that the Cre-induced reporter+ cells were not found in the perichondrium and periosteum, in which Osx is highly expressed, indicating that the chondrocyte-derived osteoblasts represent a sub-population of Osx-expressing osteoblasts [165]. Bone fracture healing occurs mostly through processes similar to developmental endochondral ossification. Recent studies also demonstrated that chondrocytes in the repair callus were a direct source of osteoblasts responsible for bone formation during fracture healing, as hypertrophic chondrocyte-derived osteoblasts were found in the fracture callus [165] [167]. It is rational to speculate that chondrocyte-derived osteoblasts may also be involved in other endochondral ossification processes, such as osteophyte formation in osteoarthritis and heterotopic ossification in FOP patients. It will be important to dissect the underlying mechanism of transdifferentiation, as these results raise many key questions for future studies, such as what determines the final fate of hypertrophic chondrocytes as to undergo apoptosis, autophagy, or transdifferentiation. Hopefully, in the near future, this hypertrophic chondrocyte-to-osteoblast trans-differentiation notion will be more firmly established with more advanced technology, such as better live imaging techniques and more sophisticated cell lineage tracing genetic tools. Nevertheless, the very fact that these chondrocyte-targeted Cre recombinases are capable of marking abundant numbers of osteoblasts, osteocytes and osteo-progenitors should be taken into consideration in our interpretation of bone phenotypes in genetic mouse models when a given gene is specifically deleted in chondrocytes.
10. Concluding remarks
In summary, growth plate formation is a dynamic, well-coordinated, tightly controlled process. Many signaling pathways interact with each other to exert essential roles during the process. The large number of skeletal diseases that exist in humans, and the limited number and efficacy of currently available treatment options mean that much research remains to be done. Newer methodologies, including Next Generation Sequencing (NGS), the Clustered regularly-interspaced short palindromic repeats CRISPR/Cas9 genome editing system, and induced pluripotent stem cells, will accelerate our uncovering of novel signaling transduction mechanisms involved in the formation and activity of cartilage growth plates [170–172]. Indeed, rapid progress in NGS allows single cell analyses to be performed, and has already provided insightful results. For example, a single cell analysis study showed that the expression patterns of chondrocyte differentiation markers are heterogeneous amongst cells, and do not correlate with functional capacity, challenging the widely-held notion that a marker gene expression profile correlates with a cellular phenotype [173]. Furthermore, a new bioinformatics pipeline, which analyzed single cell RNA-seq data among 217 single cells that represent various stages of growth plate development, allowed spatiotemporal analysis and identification of genes that are involved in regulatory networks controlling growth plate development [174]. In addition to validating previously identified transcription factors involved in growth plate development, this study also identified novel factors, including the Krüppel-like family and Fos family, as having high regulatory potential [174]. Notably, this study shows the potential power of bioinformatics and the ability to practically trace single cells from early to late stages during growth plate development. In the future, the acquisition of detailed, specific knowledge of genes and pathways involved in the control of the growth plate should allow for the development of new therapies and design of new drugs to directly and effectively target culprit signaling pathways or stimulate alternative pathways, and thereby achieve marked efficacy and reduce side effects in musculoskeletal diseases.
Acknowledgments
This work was supported in part by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health under Award Numbers R01 AR068361 to GZ and AR049072 08 to XZ, by a Rolanette and Berdon Lawrence Texas Bone Disease Program Award to XZ, and by the National Cancer Center under Award Number R03 CA175874 to GZ. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. We also apologize to the many investigators whose important studies could not be cited directly here owing to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kozhemyakina E, Lassar AB, Zelzer E. A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development. 2015;142:817–31. doi: 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berendsen AD, Olsen BR. Bone development. Bone. 2015;80:14–8. doi: 10.1016/j.bone.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long F, Ornitz DM. Development of the endochondral skeleton. Cold Spring Harb Perspect Biol. 2013;5:a008334. doi: 10.1101/cshperspect.a008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonafe L, Cormier-Daire V, Hall C, Lachman R, Mortier G, Mundlos S, et al. Nosology and classification of genetic skeletal disorders: 2015 revision. Am J Med Genet A. 2015;167:2869–92. doi: 10.1002/ajmg.a.37365. [DOI] [PubMed] [Google Scholar]
- 5.Baldridge D, Shchelochkov O, Kelley B, Lee B. Signaling pathways in human skeletal dysplasias. Annu Rev Genomics Hum Genet. 2010;11:189–217. doi: 10.1146/annurev-genom-082908-150158. [DOI] [PubMed] [Google Scholar]
- 6.Akhurst RJ, Padgett RW. Matters of context guide future research in TGFbeta superfamily signaling. Sci Signal. 2015;8:re10. doi: 10.1126/scisignal.aad0416. [DOI] [PubMed] [Google Scholar]
- 7.Salazar VS, Gamer LW, Rosen V. BMP signalling in skeletal development, disease and repair. Nat Rev Endocrinol. 2016;12:203–21. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–28. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22:85–9. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 10.Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 1997;17:2336–46. doi: 10.1128/mcb.17.4.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Bradley A. Mice deficient for BMP2 are nonviable and have defects in amnion/chorion and cardiac development. Development. 1996;122:2977–86. doi: 10.1242/dev.122.10.2977. [DOI] [PubMed] [Google Scholar]
- 12.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995;9:2105–16. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 13.Shu B, Zhang M, Xie R, Wang M, Jin H, Hou W, et al. BMP2, but not BMP4, is crucial for chondrocyte proliferation and maturation during endochondral bone development. J Cell Sci. 2011;124:3428–40. doi: 10.1242/jcs.083659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bandyopadhyay A, Tsuji K, Cox K, Harfe BD, Rosen V, Tabin CJ. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006;2:e216. doi: 10.1371/journal.pgen.0020216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pizette S, Niswander L. BMPs are required at two steps of limb chondrogenesis: formation of prechondrogenic condensations and their differentiation into chondrocytes. Dev Biol. 2000;219:237–49. doi: 10.1006/dbio.2000.9610. [DOI] [PubMed] [Google Scholar]
- 16.Zimmerman LB, De Jesus-Escobar JM, Harland RM. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell. 1996;86:599–606. doi: 10.1016/s0092-8674(00)80133-6. [DOI] [PubMed] [Google Scholar]
- 17.Brunet LJ, McMahon JA, McMahon AP, Harland RM. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–7. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- 18.Duprez D, Bell EJ, Richardson MK, Archer CW, Wolpert L, Brickell PM, et al. Overexpression of BMP-2 and BMP-4 alters the size and shape of developing skeletal elements in the chick limb. Mech Dev. 1996;57:145–57. doi: 10.1016/0925-4773(96)00540-0. [DOI] [PubMed] [Google Scholar]
- 19.Satow R, Kurisaki A, Chan TC, Hamazaki TS, Asashima M. Dullard promotes degradation and dephosphorylation of BMP receptors and is required for neural induction. Dev Cell. 2006;11:763–74. doi: 10.1016/j.devcel.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Hayata T, Ezura Y, Asashima M, Nishinakamura R, Noda M. Dullard/Ctdnep1 regulates endochondral ossification via suppression of TGF-beta signaling. J Bone Miner Res. 2015;30:318–29. doi: 10.1002/jbmr.2343. [DOI] [PubMed] [Google Scholar]
- 21.Yoon BS, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. Bmpr1a and Bmpr1b have overlapping functions and are essential for chondrogenesis in vivo. Proc Natl Acad Sci U S A. 2005;102:5062–7. doi: 10.1073/pnas.0500031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon BS, Pogue R, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, et al. BMPs regulate multiple aspects of growth-plate chondrogenesis through opposing actions on FGF pathways. Development. 2006;133:4667–78. doi: 10.1242/dev.02680. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi T, Lyons KM, McMahon AP, Kronenberg HM. BMP signaling stimulates cellular differentiation at multiple steps during cartilage development. Proc Natl Acad Sci U S A. 2005;102:18023–7. doi: 10.1073/pnas.0503617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pignolo RJ, Shore EM, Kaplan FS. Fibrodysplasia ossificans progressiva: clinical and genetic aspects. Orphanet J Rare Dis. 2011;6:80. doi: 10.1186/1750-1172-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatsell SJ, Idone V, Wolken DM, Huang L, Kim HJ, Wang L, et al. ACVR1R206H receptor mutation causes fibrodysplasia ossificans progressiva by imparting responsiveness to activin A. Sci Transl Med. 2015;7:303ra137. doi: 10.1126/scitranslmed.aac4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Duffhues G, Fotsis T, Dijke PT. Signal Transduction: Gain of Activin Turns Muscle into Bone. Curr Biol. 2015;25:R1136–8. doi: 10.1016/j.cub.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 27.van Dinther M, Visser N, de Gorter DJ, Doorn J, Goumans MJ, de Boer J, et al. ALK2 R206H mutation linked to fibrodysplasia ossificans progressiva confers constitutive activity to the BMP type I receptor and sensitizes mesenchymal cells to BMP-induced osteoblast differentiation and bone formation. J Bone Miner Res. 2010;25:1208–15. doi: 10.1359/jbmr.091110. [DOI] [PubMed] [Google Scholar]
- 28.Rigueur D, Brugger S, Anbarchian T, Kim JK, Lee Y, Lyons KM. The type I BMP receptor ACVR1/ALK2 is required for chondrogenesis during development. J Bone Miner Res. 2015;30:733–41. doi: 10.1002/jbmr.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Retting KN, Song B, Yoon BS, Lyons KM. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development. 2009;136:1093–104. doi: 10.1242/dev.029926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estrada KD, Retting KN, Chin AM, Lyons KM. Smad6 is essential to limit BMP signaling during cartilage development. J Bone Miner Res. 2011;26:2498–510. doi: 10.1002/jbmr.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Estrada KD, Wang W, Retting KN, Chien CT, Elkhoury FF, Heuchel R, et al. Smad7 regulates terminal maturation of chondrocytes in the growth plate. Dev Biol. 2013;382:375–84. doi: 10.1016/j.ydbio.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shim JH, Greenblatt MB, Xie M, Schneider MD, Zou W, Zhai B, et al. TAK1 is an essential regulator of BMP signalling in cartilage. EMBO J. 2009;28:2028–41. doi: 10.1038/emboj.2009.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunnell LM, Jonason JH, Loiselle AE, Kohn A, Schwarz EM, Hilton MJ, et al. TAK1 regulates cartilage and joint development via the MAPK and BMP signaling pathways. J Bone Miner Res. 2010;25:1784–97. doi: 10.1002/jbmr.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao L, Sheu TJ, Dong Y, Hoak DM, Zuscik MJ, Schwarz EM, et al. TAK1 regulates SOX9 expression in chondrocytes and is essential for postnatal development of the growth plate and articular cartilages. J Cell Sci. 2013;126:5704–13. doi: 10.1242/jcs.135483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C, Jiao H, Lai Y, Zheng W, Chen K, Qu H, et al. Kindlin-2 controls TGF-beta signalling and Sox9 expression to regulate chondrogenesis. Nat Commun. 2015;6:7531. doi: 10.1038/ncomms8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Decker RS, Koyama E, Enomoto-Iwamoto M, Maye P, Rowe D, Zhu S, et al. Mouse limb skeletal growth and synovial joint development are coordinately enhanced by Kartogenin. Dev Biol. 2014;395:255–67. doi: 10.1016/j.ydbio.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stange K, Desir J, Kakar N, Mueller TD, Budde BS, Gordon CT, et al. A hypomorphic BMPR1B mutation causes du Pan acromesomelic dysplasia. Orphanet J Rare Dis. 2015;10:84. doi: 10.1186/s13023-015-0299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Serra R, Chang C. TGF-beta signaling in human skeletal and patterning disorders. Birth Defects Res C Embryo Today. 2003;69:333–51. doi: 10.1002/bdrc.10023. [DOI] [PubMed] [Google Scholar]
- 39.Ornitz DM, Marie PJ. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015;29:1463–86. doi: 10.1101/gad.266551.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hung IH, Schoenwolf GC, Lewandoski M, Ornitz DM. A combined series of Fgf9 and Fgf18 mutant alleles identifies unique and redundant roles in skeletal development. Dev Biol. 2016;411:72–84. doi: 10.1016/j.ydbio.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z, Lavine KJ, Hung IH, Ornitz DM. FGF18 is required for early chondrocyte proliferation, hypertrophy and vascular invasion of the growth plate. Dev Biol. 2007;302:80–91. doi: 10.1016/j.ydbio.2006.08.071. [DOI] [PubMed] [Google Scholar]
- 42.Shung CY, Ota S, Zhou ZQ, Keene DR, Hurlin PJ. Disruption of a Sox9-beta-catenin circuit by mutant Fgfr3 in thanatophoric dysplasia type II. Hum Mol Genet. 2012;21:4628–44. doi: 10.1093/hmg/dds305. [DOI] [PubMed] [Google Scholar]
- 43.Gudernova I, Vesela I, Balek L, Buchtova M, Dosedelova H, Kunova M, et al. Multikinase activity of fibroblast growth factor receptor (FGFR) inhibitors SU5402, PD173074, AZD1480, AZD4547 and BGJ398 compromises the use of small chemicals targeting FGFR catalytic activity for therapy of short-stature syndromes. Hum Mol Genet. 2016;25:9–23. doi: 10.1093/hmg/ddv441. [DOI] [PubMed] [Google Scholar]
- 44.Klag KA, Horton WA. Advances in treatment of achondroplasia and osteoarthritis. Hum Mol Genet. 2016;25:R2–8. doi: 10.1093/hmg/ddv419. [DOI] [PubMed] [Google Scholar]
- 45.Komla-Ebri D, Dambroise E, Kramer I, Benoist-Lasselin C, Kaci N, Le Gall C, et al. Tyrosine kinase inhibitor NVP-BGJ398 functionally improves FGFR3-related dwarfism in mouse model. J Clin Invest. 2016;126:1871–84. doi: 10.1172/JCI83926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karuppaiah K, Yu K, Lim J, Chen J, Smith C, Long F, et al. FGF signaling in the osteoprogenitor lineage non-autonomously regulates postnatal chondrocyte proliferation and skeletal growth. Development. 2016;143:1811–22. doi: 10.1242/dev.131722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murakami S, Balmes G, McKinney S, Zhang Z, Givol D, de Crombrugghe B. Constitutive activation of MEK1 in chondrocytes causes Stat1-independent achondroplasia-like dwarfism and rescues the Fgfr3-deficient mouse phenotype. Genes Dev. 2004;18:290–305. doi: 10.1101/gad.1179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matsushita T, Chan YY, Kawanami A, Balmes G, Landreth GE, Murakami S. Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Mol Cell Biol. 2009;29:5843–57. doi: 10.1128/MCB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Z, Yue SX, Zhou G, Greenfield EM, Murakami S. ERK1 and ERK2 regulate chondrocyte terminal differentiation during endochondral bone formation. J Bone Miner Res. 2015;30:765–74. doi: 10.1002/jbmr.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cseh B, Doma E, Baccarini M. “RAF” neighborhood: protein-protein interaction in the Raf/Mek/Erk pathway. FEBS Lett. 2014;588:2398–406. doi: 10.1016/j.febslet.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Provot S, Nachtrab G, Paruch J, Chen AP, Silva A, Kronenberg HM. A-raf and B-raf are dispensable for normal endochondral bone development, and parathyroid hormone-related peptide suppresses extracellular signal-regulated kinase activation in hypertrophic chondrocytes. Mol Cell Biol. 2008;28:344–57. doi: 10.1128/MCB.00617-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu ES, Raimann A, Chae BT, Martins JS, Baccarini M, Demay MB. c-Raf promotes angiogenesis during normal growth plate maturation. Development. 2016;143:348–55. doi: 10.1242/dev.127142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minina E, Kreschel C, Naski MC, Ornitz DM, Vortkamp A. Interaction of FGF, Ihh/Pthlh, and BMP signaling integrates chondrocyte proliferation and hypertrophic differentiation. Dev Cell. 2002;3:439–49. doi: 10.1016/s1534-5807(02)00261-7. [DOI] [PubMed] [Google Scholar]
- 54.Kumar M, Ray P, Chapman SC. Fibroblast growth factor and bone morphogenetic protein signaling are required for specifying prechondrogenic identity in neural crest-derived mesenchyme and initiating the chondrogenic program. Dev Dyn. 2012;241:1091–103. doi: 10.1002/dvdy.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J Cell Sci. 2003;116:2627–34. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 56.Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 57.Maupin KA, Droscha CJ, Williams BO. A Comprehensive Overview of Skeletal Phenotypes Associated with Alterations in Wnt/beta-catenin Signaling in Humans and Mice. Bone Res. 2013;1:27–71. doi: 10.4248/BR201301004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Amerongen R, Berns A. Knockout mouse models to study Wnt signal transduction. Trends Genet. 2006;22:678–89. doi: 10.1016/j.tig.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 59.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, et al. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–87. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo X, Day TF, Jiang X, Garrett-Beal L, Topol L, Yang Y. Wnt/beta-catenin signaling is sufficient and necessary for synovial joint formation. Genes Dev. 2004;18:2404–17. doi: 10.1101/gad.1230704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–50. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 62.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–38. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 63.Bhattaram P, Penzo-Mendez A, Kato K, Bandyopadhyay K, Gadi A, Taketo MM, et al. SOXC proteins amplify canonical WNT signaling to secure nonchondrocytic fates in skeletogenesis. J Cell Biol. 2014;207:657–71. doi: 10.1083/jcb.201405098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu H, Hilton MJ, Tu X, Yu K, Ornitz DM, Long F. Sequential roles of Hedgehog and Wnt signaling in osteoblast development. Development. 2005;132:49–60. doi: 10.1242/dev.01564. [DOI] [PubMed] [Google Scholar]
- 65.Golovchenko S, Hattori T, Hartmann C, Gebhardt M, Gebhard S, Hess A, et al. Deletion of beta catenin in hypertrophic growth plate chondrocytes impairs trabecular bone formation. Bone. 2013;55:102–12. doi: 10.1016/j.bone.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 66.Joeng KS, Schumacher CA, Zylstra-Diegel CR, Long F, Williams BO. Lrp5 and Lrp6 redundantly control skeletal development in the mouse embryo. Dev Biol. 2011;359:222–9. doi: 10.1016/j.ydbio.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schumacher CA, Joiner DM, Less KD, Drewry MO, Williams BO. Characterization of genetically engineered mouse models carrying Col2a1-cre-induced deletions of Lrp5 and/or Lrp6. Bone Res. 2016;4:15042. doi: 10.1038/boneres.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen M, Zhu M, Awad H, Li TF, Sheu TJ, Boyce BF, et al. Inhibition of beta-catenin signaling causes defects in postnatal cartilage development. J Cell Sci. 2008;121:1455–65. doi: 10.1242/jcs.020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guo X, Mak KK, Taketo MM, Yang Y. The Wnt/beta-catenin pathway interacts differentially with PTHrP signaling to control chondrocyte hypertrophy and final maturation. PLoS One. 2009;4:e6067. doi: 10.1371/journal.pone.0006067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y, Dudley AT. Noncanonical frizzled signaling regulates cell polarity of growth plate chondrocytes. Development. 2009;136:1083–92. doi: 10.1242/dev.023820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Romereim SM, Conoan NH, Chen B, Dudley AT. A dynamic cell adhesion surface regulates tissue architecture in growth plate cartilage. Development. 2014;141:2085–95. doi: 10.1242/dev.105452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12:393–406. doi: 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 73.Lee RT, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367–72. doi: 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- 74.Zhulyn O, Nieuwenhuis E, Liu YC, Angers S, Hui CC. Ptch2 shares overlapping functions with Ptch1 in Smo regulation and limb development. Dev Biol. 2015;397:191–202. doi: 10.1016/j.ydbio.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 75.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, et al. Identification of Sonic hedgehog as a candidate gene responsible for holoprosencephaly. Nat Genet. 1996;14:353–6. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 76.Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, et al. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–13. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- 77.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–86. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Amano K, Densmore MJ, Lanske B. Conditional Deletion of Indian Hedgehog in Limb Mesenchyme Results in Complete Loss of Growth Plate Formation but Allows Mature Osteoblast Differentiation. J Bone Miner Res. 2015;30:2262–72. doi: 10.1002/jbmr.2582. [DOI] [PubMed] [Google Scholar]
- 79.Long F, Chung UI, Ohba S, McMahon J, Kronenberg HM, McMahon AP. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development. 2004;131:1309–18. doi: 10.1242/dev.01006. [DOI] [PubMed] [Google Scholar]
- 80.Lin YC, Roffler SR, Yan YT, Yang RB. Disruption of Scube2 Impairs Endochondral Bone Formation. J Bone Miner Res. 2015;30:1255–67. doi: 10.1002/jbmr.2451. [DOI] [PubMed] [Google Scholar]
- 81.Vortkamp A, Lee K, Lanske B, Segre GV, Kronenberg HM, Tabin CJ. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–22. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 82.Cheloha RW, Gellman SH, Vilardaga JP, Gardella TJ. PTH receptor-1 signalling-mechanistic insights and therapeutic prospects. Nat Rev Endocrinol. 2015;11:712–24. doi: 10.1038/nrendo.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Engin F, Lee B. NOTCHing the bone: insights into multi-functionality. Bone. 2010;46:274–80. doi: 10.1016/j.bone.2009.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen S, Lee BH, Bae Y. Notch signaling in skeletal stem cells. Calcif Tissue Int. 2014;94:68–77. doi: 10.1007/s00223-013-9773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kohn A, Rutkowski TP, Liu Z, Mirando AJ, Zuscik MJ, O’Keefe RJ, et al. Notch signaling controls chondrocyte hypertrophy via indirect regulation of Sox9. Bone Res. 2015;3:15021. doi: 10.1038/boneres.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dong Y, Jesse AM, Kohn A, Gunnell LM, Honjo T, Zuscik MJ, et al. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137:1461–71. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang C, Inzana JA, Mirando AJ, Ren Y, Liu Z, Shen J, et al. NOTCH signaling in skeletal progenitors is critical for fracture repair. J Clin Invest. 2016;126:1471–81. doi: 10.1172/JCI80672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mead TJ, Yutzey KE. Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc Natl Acad Sci U S A. 2009;106:14420–5. doi: 10.1073/pnas.0902306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen S, Tao J, Bae Y, Jiang MM, Bertin T, Chen Y, et al. Notch gain of function inhibits chondrocyte differentiation via Rbpj-dependent suppression of Sox9. J Bone Miner Res. 2013;28:649–59. doi: 10.1002/jbmr.1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kohn A, Dong Y, Mirando AJ, Jesse AM, Honjo T, Zuscik MJ, et al. Cartilage-specific RBPjkappa-dependent and -independent Notch signals regulate cartilage and bone development. Development. 2012;139:1198–212. doi: 10.1242/dev.070649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rutkowski TP, Kohn A, Sharma D, Ren Y, Mirando AJ, Hilton MJ. HES factors regulate specific aspects of chondrogenesis and chondrocyte hypertrophy during cartilage development. J Cell Sci. 2016;129:2145–55. doi: 10.1242/jcs.181271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Karlsson C, Brantsing C, Kageyama R, Lindahl A. HES1 and HES5 are dispensable for cartilage and endochondral bone formation. Cells Tissues Organs. 2010;192:17–27. doi: 10.1159/000280416. [DOI] [PubMed] [Google Scholar]
- 93.Tian Y, Xu Y, Fu Q, Chang M, Wang Y, Shang X, et al. Notch inhibits chondrogenic differentiation of mesenchymal progenitor cells by targeting Twist1. Mol Cell Endocrinol. 2015;403:30–8. doi: 10.1016/j.mce.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cunningham TJ, Duester G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat Rev Mol Cell Biol. 2015;16:110–23. doi: 10.1038/nrm3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dranse HJ, Sampaio AV, Petkovich M, Underhill TM. Genetic deletion of Cyp26b1 negatively impacts limb skeletogenesis by inhibiting chondrogenesis. J Cell Sci. 2011;124:2723–34. doi: 10.1242/jcs.084699. [DOI] [PubMed] [Google Scholar]
- 96.Williams JA, Kondo N, Okabe T, Takeshita N, Pilchak DM, Koyama E, et al. Retinoic acid receptors are required for skeletal growth, matrix homeostasis and growth plate function in postnatal mouse. Dev Biol. 2009;328:315–27. doi: 10.1016/j.ydbio.2009.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoffman LM, Garcha K, Karamboulas K, Cowan MF, Drysdale LM, Horton WA, et al. BMP action in skeletogenesis involves attenuation of retinoid signaling. J Cell Biol. 2006;174:101–13. doi: 10.1083/jcb.200604150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weston AD, Chandraratna RA, Torchia J, Underhill TM. Requirement for RAR-mediated gene repression in skeletal progenitor differentiation. J Cell Biol. 2002;158:39–51. doi: 10.1083/jcb.200112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pennimpede T, Cameron DA, MacLean GA, Petkovich M. Analysis of Cyp26b1/Rarg compound-null mice reveals two genetically separable effects of retinoic acid on limb outgrowth. Dev Biol. 2010;339:179–86. doi: 10.1016/j.ydbio.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 100.Yasuhara R, Yuasa T, Williams JA, Byers SW, Shah S, Pacifici M, et al. Wnt/beta-catenin and retinoic acid receptor signaling pathways interact to regulate chondrocyte function and matrix turnover. J Biol Chem. 2010;285:317–27. doi: 10.1074/jbc.M109.053926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Eltschinger S, Loewith R. TOR Complexes and the Maintenance of Cellular Homeostasis. Trends Cell Biol. 2016;26:148–59. doi: 10.1016/j.tcb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 102.Chen J, Long F. mTORC1 signaling controls mammalian skeletal growth through stimulation of protein synthesis. Development. 2014;141:2848–54. doi: 10.1242/dev.108811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chen J, Holguin N, Shi Y, Silva MJ, Long F. mTORC2 signaling promotes skeletal growth and bone formation in mice. J Bone Miner Res. 2015;30:369–78. doi: 10.1002/jbmr.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yan B, Zhang Z, Jin D, Cai C, Jia C, Liu W, et al. mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat Commun. 2016;7:11151. doi: 10.1038/ncomms11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lai LP, Lilley BN, Sanes JR, McMahon AP. Lkb1/Stk11 regulation of mTOR signaling controls the transition of chondrocyte fates and suppresses skeletal tumor formation. Proc Natl Acad Sci U S A. 2013;110:19450–5. doi: 10.1073/pnas.1309001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Baxter RC. IGF binding proteins in cancer: mechanistic and clinical insights. Nat Rev Cancer. 2014;14:329–41. doi: 10.1038/nrc3720. [DOI] [PubMed] [Google Scholar]
- 107.Agrogiannis GD, Sifakis S, Patsouris ES, Konstantinidou AE. Insulin-like growth factors in embryonic and fetal growth and skeletal development (Review) Mol Med Rep. 2014;10:579–84. doi: 10.3892/mmr.2014.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Y, Nishida S, Sakata T, Elalieh HZ, Chang W, Halloran BP, et al. Insulin-like growth factor-I is essential for embryonic bone development. Endocrinology. 2006;147:4753–61. doi: 10.1210/en.2006-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y, Cheng Z, Elalieh HZ, Nakamura E, Nguyen MT, Mackem S, et al. IGF-1R signaling in chondrocytes modulates growth plate development by interacting with the PTHrP/Ihh pathway. J Bone Miner Res. 2011;26:1437–46. doi: 10.1002/jbmr.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Y, Menendez A, Fong C, ElAlieh HZ, Kubota T, Long R, et al. IGF-I Signaling in Osterix-Expressing Cells Regulates Secondary Ossification Center Formation, Growth Plate Maturation, and Metaphyseal Formation During Postnatal Bone Development. J Bone Miner Res. 2015;30:2239–48. doi: 10.1002/jbmr.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Repudi SR, Patra M, Sen M. WISP3-IGF1 interaction regulates chondrocyte hypertrophy. J Cell Sci. 2013;126:1650–8. doi: 10.1242/jcs.119859. [DOI] [PubMed] [Google Scholar]
- 112.Nakamura Y, Weidinger G, Liang JO, Aquilina-Beck A, Tamai K, Moon RT, et al. The CCN family member Wisp3, mutant in progressive pseudorheumatoid dysplasia, modulates BMP and Wnt signaling. J Clin Invest. 2007;117:3075–86. doi: 10.1172/JCI32001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen L, Jiang W, Huang J, He BC, Zuo GW, Zhang W, et al. Insulin-like growth factor 2 (IGF-2) potentiates BMP-9-induced osteogenic differentiation and bone formation. J Bone Miner Res. 2010;25:2447–59. doi: 10.1002/jbmr.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Murray PG, Hanson D, Coulson T, Stevens A, Whatmore A, Poole RL, et al. 3-M syndrome: a growth disorder associated with IGF2 silencing. Endocr Connect. 2013;2:225–35. doi: 10.1530/EC-13-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–9. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reale ME, Webb IC, Wang X, Baltazar RM, Coolen LM, Lehman MN. The transcription factor Runx2 is under circadian control in the suprachiasmatic nucleus and functions in the control of rhythmic behavior. PLoS One. 2013;8:e54317. doi: 10.1371/journal.pone.0054317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gafni Y, Ptitsyn AA, Zilberman Y, Pelled G, Gimble JM, Gazit D. Circadian rhythm of osteocalcin in the maxillomandibular complex. J Dent Res. 2009;88:45–50. doi: 10.1177/0022034508328012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gossan N, Zeef L, Hensman J, Hughes A, Bateman JF, Rowley L, et al. The circadian clock in murine chondrocytes regulates genes controlling key aspects of cartilage homeostasis. Arthritis Rheum. 2013;65:2334–45. doi: 10.1002/art.38035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Okubo N, Minami Y, Fujiwara H, Umemura Y, Tsuchiya Y, Shirai T, et al. Prolonged bioluminescence monitoring in mouse ex vivo bone culture revealed persistent circadian rhythms in articular cartilages and growth plates. PLoS One. 2013;8:e78306. doi: 10.1371/journal.pone.0078306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hinoi E, Ueshima T, Hojo H, Iemata M, Takarada T, Yoneda Y. Up-regulation of per mRNA expression by parathyroid hormone through a protein kinase A-CREB-dependent mechanism in chondrocytes. J Biol Chem. 2006;281:23632–42. doi: 10.1074/jbc.M512362200. [DOI] [PubMed] [Google Scholar]
- 121.Hanyu R, Hayata T, Nagao M, Saita Y, Hemmi H, Notomi T, et al. Per-1 is a specific clock gene regulated by parathyroid hormone (PTH) signaling in osteoblasts and is functional for the transcriptional events induced by PTH. J Cell Biochem. 2011;112:433–8. doi: 10.1002/jcb.22957. [DOI] [PubMed] [Google Scholar]
- 122.Okubo N, Fujiwara H, Minami Y, Kunimoto T, Hosokawa T, Umemura Y, et al. Parathyroid hormone resets the cartilage circadian clock of the organ-cultured murine femur. Acta Orthop. 2015;86:627–31. doi: 10.3109/17453674.2015.1029393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Samsa WE, Vasanji A, Midura RJ, Kondratov RV. Deficiency of circadian clock protein BMAL1 in mice results in a low bone mass phenotype. Bone. 2016;84:194–203. doi: 10.1016/j.bone.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Takarada T, Kodama A, Hotta S, Mieda M, Shimba S, Hinoi E, et al. Clock genes influence gene expression in growth plate and endochondral ossification in mice. J Biol Chem. 2012;287:36081–95. doi: 10.1074/jbc.M112.408963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dudek M, Gossan N, Yang N, Im HJ, Ruckshanthi JP, Yoshitane H, et al. The chondrocyte clock gene Bmal1 controls cartilage homeostasis and integrity. J Clin Invest. 2016;126:365–76. doi: 10.1172/JCI82755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bashur L, Chen D, Zhou G. The Cop9 Signalosome, a Novel, Essential Regulator of Skeletal Development and Tumorigenesis. Case Orthop J. 2011;8:45–50. [PMC free article] [PubMed] [Google Scholar]
- 127.Kato JY, Yoneda-Kato N. Mammalian COP9 signalosome. Genes Cells. 2009;14:1209–25. doi: 10.1111/j.1365-2443.2009.01349.x. [DOI] [PubMed] [Google Scholar]
- 128.Shackleford TJ, Claret FX. JAB1/CSN5: a new player in cell cycle control and cancer. Cell Div. 2010;5:26. doi: 10.1186/1747-1028-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wan M, Cao X, Wu Y, Bai S, Wu L, Shi X, et al. Jab1 antagonizes TGF-beta signaling by inducing Smad4 degradation. EMBO Rep. 2002;3:171–6. doi: 10.1093/embo-reports/kvf024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim BC, Lee HJ, Park SH, Lee SR, Karpova TS, McNally JG, et al. Jab1/CSN5, a component of the COP9 signalosome, regulates transforming growth factor beta signaling by binding to Smad7 and promoting its degradation. Mol Cell Biol. 2004;24:2251–62. doi: 10.1128/MCB.24.6.2251-2262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Haag J, Aigner T. Jun activation domain-binding protein 1 binds Smad5 and inhibits bone morphogenetic protein signaling. Arthritis Rheum. 2006;54:3878–84. doi: 10.1002/art.22261. [DOI] [PubMed] [Google Scholar]
- 132.Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY. Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J Biol Chem. 2004;279:43013–8. doi: 10.1074/jbc.M406559200. [DOI] [PubMed] [Google Scholar]
- 133.Tian L, Peng G, Parant JM, Leventaki V, Drakos E, Zhang Q, et al. Essential roles of Jab1 in cell survival, spontaneous DNA damage and DNA repair. Oncogene. 2010;29:6125–37. doi: 10.1038/onc.2010.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen D, Bashur LA, Liang B, Panattoni M, Tamai K, Pardi R, et al. The transcriptional co-regulator Jab1 is crucial for chondrocyte differentiation in vivo. J Cell Sci. 2013;126:234–43. doi: 10.1242/jcs.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bashur LA, Chen D, Chen Z, Liang B, Pardi R, Murakami S, et al. Loss of jab1 in osteochondral progenitor cells severely impairs embryonic limb development in mice. J Cell Physiol. 2014;229:1607–17. doi: 10.1002/jcp.24602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Garside VC, Cullum R, Alder O, Lu DY, Vander Werff R, Bilenky M, et al. SOX9 modulates the expression of key transcription factors required for heart valve development. Development. 2015;142:4340–50. doi: 10.1242/dev.125252. [DOI] [PubMed] [Google Scholar]
- 137.Meng Z, Moroishi T, Guan KL. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Deng Y, Wu A, Li P, Li G, Qin L, Song H, et al. Yap1 Regulates Multiple Steps of Chondrocyte Differentiation during Skeletal Development and Bone Repair. Cell Rep. 2016;14:2224–37. doi: 10.1016/j.celrep.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 139.Karystinou A, Roelofs AJ, Neve A, Cantatore FP, Wackerhage H, De Bari C. Yes-associated protein (YAP) is a negative regulator of chondrogenesis in mesenchymal stem cells. Arthritis Res Ther. 2015;17:147. doi: 10.1186/s13075-015-0639-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roach HI, Clarke NM. Physiological cell death of chondrocytes in vivo is not confined to apoptosis. New observations on the mammalian growth plate. J Bone Joint Surg Br. 2000;82:601–13. doi: 10.1302/0301-620x.82b4.9846. [DOI] [PubMed] [Google Scholar]
- 141.Ahmed YA, Tatarczuch L, Pagel CN, Davies HM, Mirams M, Mackie EJ. Physiological death of hypertrophic chondrocytes. Osteoarthritis Cartilage. 2007;15:575–86. doi: 10.1016/j.joca.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 142.Shapiro IM, Adams CS, Freeman T, Srinivas V. Fate of the hypertrophic chondrocyte: microenvironmental perspectives on apoptosis and survival in the epiphyseal growth plate. Birth Defects Res C Embryo Today. 2005;75:330–9. doi: 10.1002/bdrc.20057. [DOI] [PubMed] [Google Scholar]
- 143.Bronckers AL, Goei W, Luo G, Karsenty G, D’Souza RN, Lyaruu DM, et al. DNA fragmentation during bone formation in neonatal rodents assessed by transferase-mediated end labeling. J Bone Miner Res. 1996;11:1281–91. doi: 10.1002/jbmr.5650110913. [DOI] [PubMed] [Google Scholar]
- 144.Farnum CE, Wilsman NJ. Condensation of hypertrophic chondrocytes at the chondro-osseous junction of growth plate cartilage in Yucatan swine: relationship to long bone growth. The American journal of anatomy. 1989;186:346–58. doi: 10.1002/aja.1001860404. [DOI] [PubMed] [Google Scholar]
- 145.Gibson GJ, Kohler WJ, Schaffler MB. Chondrocyte apoptosis in endochondral ossification of chick sterna. Dev Dyn. 1995;203:468–76. doi: 10.1002/aja.1002030409. [DOI] [PubMed] [Google Scholar]
- 146.Gibson G. Active role of chondrocyte apoptosis in endochondral ossification. Microscopy research and technique. 1998;43:191–204. doi: 10.1002/(SICI)1097-0029(19981015)43:2<191::AID-JEMT10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 147.Roach HI. New aspects of endochondral ossification in the chick: chondrocyte apoptosis, bone formation by former chondrocytes, and acid phosphatase activity in the endochondral bone matrix. J Bone Miner Res. 1997;12:795–805. doi: 10.1359/jbmr.1997.12.5.795. [DOI] [PubMed] [Google Scholar]
- 148.Roach HI, Aigner T, Kouri JB. Chondroptosis: a variant of apoptotic cell death in chondrocytes? Apoptosis : an international journal on programmed cell death. 2004;9:265–77. doi: 10.1023/b:appt.0000025803.17498.26. [DOI] [PubMed] [Google Scholar]
- 149.Zenmyo M, Komiya S, Kawabata R, Sasaguri Y, Inoue A, Morimatsu M. Morphological and biochemical evidence for apoptosis in the terminal hypertrophic chondrocytes of the growth plate. The Journal of pathology. 1996;180:430–3. doi: 10.1002/(SICI)1096-9896(199612)180:4<430::AID-PATH691>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 150.Farnum CE, Wilsman NJ. Morphologic stages of the terminal hypertrophic chondrocyte of growth plate cartilage. The Anatomical record. 1987;219:221–32. doi: 10.1002/ar.1092190303. [DOI] [PubMed] [Google Scholar]
- 151.Hatori M, Klatte KJ, Teixeira CC, Shapiro IM. End labeling studies of fragmented DNA in the avian growth plate: evidence of apoptosis in terminally differentiated chondrocytes. J Bone Miner Res. 1995;10:1960–8. doi: 10.1002/jbmr.5650101216. [DOI] [PubMed] [Google Scholar]
- 152.Ohyama K, Farquharson C, Whitehead CC, Shapiro IM. Further observations on programmed cell death in the epiphyseal growth plate: comparison of normal and dyschondroplastic epiphyses. J Bone Miner Res. 1997;12:1647–56. doi: 10.1359/jbmr.1997.12.10.1647. [DOI] [PubMed] [Google Scholar]
- 153.Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vuppalapati KK, Bouderlique T, Newton PT, Kaminskyy VO, Wehtje H, Ohlsson C, et al. Targeted Deletion of Autophagy Genes Atg5 or Atg7 in the Chondrocytes Promotes Caspase-Dependent Cell Death and Leads to Mild Growth Retardation. J Bone Miner Res. 2015;30:2249–61. doi: 10.1002/jbmr.2575. [DOI] [PubMed] [Google Scholar]
- 155.Cinque L, Forrester A, Bartolomeo R, Svelto M, Venditti R, Montefusco S, et al. FGF signalling regulates bone growth through autophagy. Nature. 2015;528:272–5. doi: 10.1038/nature16063. [DOI] [PubMed] [Google Scholar]
- 156.Yeung Tsang K, Wa Tsang S, Chan D, Cheah KS. The chondrocytic journey in endochondral bone growth and skeletal dysplasia. Birth Defects Res C Embryo Today. 2014;102:52–73. doi: 10.1002/bdrc.21060. [DOI] [PubMed] [Google Scholar]
- 157.Gomez-Picos P, Eames BF. On the evolutionary relationship between chondrocytes and osteoblasts. Front Genet. 2015;6:297. doi: 10.3389/fgene.2015.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bianco P, Cancedda FD, Riminucci M, Cancedda R. Bone formation via cartilage models: the “borderline” chondrocyte. Matrix biology : journal of the International Society for Matrix Biology. 1998;17:185–92. doi: 10.1016/s0945-053x(98)90057-9. [DOI] [PubMed] [Google Scholar]
- 159.Gerstenfeld LC, Shapiro FD. Expression of bone-specific genes by hypertrophic chondrocytes: implication of the complex functions of the hypertrophic chondrocyte during endochondral bone development. J Cell Biochem. 1996;62:1–9. doi: 10.1002/(SICI)1097-4644(199607)62:1%3C1::AID-JCB1%3E3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 160.Erenpreisa J, Roach HI. Epigenetic selection as a possible component of transdifferentiation. Further study of the commitment of hypertrophic chondrocytes to become osteocytes. Mechanisms of ageing and development. 1996;87:165–82. doi: 10.1016/0047-6374(96)01703-4. [DOI] [PubMed] [Google Scholar]
- 161.Kirsch T, Swoboda B, von der Mark K. Ascorbate independent differentiation of human chondrocytes in vitro: simultaneous expression of types I and X collagen and matrix mineralization. Differentiation; research in biological diversity. 1992;52:89–100. doi: 10.1111/j.1432-0436.1992.tb00503.x. [DOI] [PubMed] [Google Scholar]
- 162.Roach HI, Erenpreisa J, Aigner T. Osteogenic differentiation of hypertrophic chondrocytes involves asymmetric cell divisions and apoptosis. J Cell Biol. 1995;131:483–94. doi: 10.1083/jcb.131.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Roach HI, Erenpreisa J. The phenotypic switch from chondrocytes to bone-forming cells involves asymmetric cell division and apoptosis. Connect Tissue Res. 1996;35:85–91. doi: 10.3109/03008209609029178. [DOI] [PubMed] [Google Scholar]
- 164.Wlodarski KH, Wlodarski PK, Brodzikowska A. Metaplasia of chondrocytes into osteoblasts. Folia Biol (Krakow) 2006;54:75–80. doi: 10.3409/173491606778557491. [DOI] [PubMed] [Google Scholar]
- 165.Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B. Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice. PLoS Genet. 2014;10:e1004820. doi: 10.1371/journal.pgen.1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Park J, Gebhardt M, Golovchenko S, Perez-Branguli F, Hattori T, Hartmann C, et al. Dual pathways to endochondral osteoblasts: a novel chondrocyte-derived osteoprogenitor cell identified in hypertrophic cartilage. Biol Open. 2015;4:608–21. doi: 10.1242/bio.201411031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang L, Tsang KY, Tang HC, Chan D, Cheah KS. Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation. Proc Natl Acad Sci U S A. 2014;111:12097–102. doi: 10.1073/pnas.1302703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mizoguchi T, Pinho S, Ahmed J, Kunisaki Y, Hanoun M, Mendelson A, et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev Cell. 2014;29:340–9. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ono N, Ono W, Nagasawa T, Kronenberg HM. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. 2014;16:1157–67. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Goodwin S, McPherson JD, McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet. 2016;17:333–51. doi: 10.1038/nrg.2016.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Dominguez AA, Lim WA, Qi LS. Beyond editing: repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat Rev Mol Cell Biol. 2016;17:5–15. doi: 10.1038/nrm.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xu Y, Zhang M, Li W, Zhu X, Bao X, Qin B, et al. Transcriptional Control of Somatic Cell Reprogramming. Trends Cell Biol. 2016;26:272–88. doi: 10.1016/j.tcb.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 173.Cote AJ, McLeod CM, Farrell MJ, McClanahan PD, Dunagin MC, Raj A, et al. Single-cell differences in matrix gene expression do not predict matrix deposition. Nat Commun. 2016;7:10865. doi: 10.1038/ncomms10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li J, Luo H, Wang R, Lang J, Zhu S, Zhang Z, et al. Systematic Reconstruction of Molecular Cascades Regulating GP Development Using Single-Cell RNA-Seq. Cell Rep. 2016;15:1467–80. doi: 10.1016/j.celrep.2016.04.043. [DOI] [PubMed] [Google Scholar]