Abstract
Ubiquitin-dependent degradation plays an important role in the negative regulation of TGF-β signaling. Here, we identify Tiul1 (for TGIF interacting ubiquitin ligase 1), a novel E3 ubiquitin ligase that inhibits TGF-β signaling by targeting both the activated receptor and Smad2 for degradation. Tiul1 associates constitutively with Smad7 and induces degradation of the activated type I receptor without affecting the expression levels of Smad7. Tiul1 can also interact with Smad2 and the nuclear corepressor TGIF upon activation of TGF-β signaling. Like Smad7, the steady-state levels of TGIF are not affected by Tiul1, but the interaction of Tiul1 with TGIF allows this ubiquitin ligase to target Smad2 for degradation. Consistent with this, overexpression of Tiul1 suppressed TGF-β-induced growth arrest and transcriptional responses. In addition, silencing of Tiul1 or TGIF genes by siRNA resulted in suppression of the TGF-β-dependent degradation of Smad2 and an enhancement of TGF-β-mediated gene expression. These results reveal a new role for TGIF as a component of a ubiquitin ligase complex that mediates the degradation of Smad2 in response to TGF-β signaling.
Keywords: Smad, TGF-β, TGIF, Tiul1, ubiquitin ligase
Introduction
Transforming growth factor-β (TGF-β) superfamily members regulate a wide variety of biological processes through two types of Ser/Thr transmembrane receptors, the type I (TβRI) and the type II receptor (TβRII) (Whitman, 1998; Massague and Chen, 2000). Upon ligand binding, the type I receptor activated by the type II receptor propagates signals to Smad proteins (Heldin et al, 1997; Derynck et al, 1998). Among three different classes of Smads, the receptor-regulated Smads (R-Smads) interact with the type I receptors and become activated through the phosphorylation on a C-terminal SSXS motif (Massague and Chen, 2000; Wrana, 2000). Smad2 and 3 are substrates for TGF-β and activin receptors, whereas Smad1, 5, and 8 are substrates for bone morphogenic protein (BMP) receptors. Phosphorylated R-Smads then form a complex with the common partner Smad, Smad4, and translocate into the nucleus, where they associate with DNA or DNA-binding proteins and act as transcriptional activators for TGF-β-responsive genes (Derynck et al, 1998; Massague and Chen, 2000). In contrast to R-Smads and Smad4, the antagonistic Smads, Smad6 and 7, appear to block signal transduction by associating stably with the activated type I receptors (Massague and Chen, 2000).
A growing body of evidence indicates that the activities of TGF-β signaling-related proteins are regulated by ubiquitin-dependent degradation (Mehra and Wrana, 2002). Ubiquitination of a protein is carried out by the action of E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme, and E3 ubiquitin ligases. Among these, the E3 ubiquitin ligases play a crucial role in determining substrate specificity and subsequent degradation by the 26S proteasome (Ciechanover et al, 2000). Recently, the E3 ubiquitin ligases Smurf1 and 2 were identified as Smad interacting proteins that target components of the TGF-β signaling pathways for degradation (Mehra and Wrana, 2002). Smurf1/2 contain the distinctive structural features of the HECT subclass of E3 ubiquitin ligases, including a phospholipid/calcium-binding C2 domain, WW domains that interact with PPXY (PY) motif on its substrates, and a catalytic HECT domain. Through its association with the Smad2 PY motif, Smurf2 can be recruited to the corepressor SnoN, thereby allowing Smurf2 to target SnoN for degradation (Bonni et al, 2001). By interacting with the PY motif of Smad7, Smurf1/2 can be recruited to the TGF-β receptors, leading to their degradation (Kavsak et al, 2000; Ebisawa et al, 2001). In addition to targeting TGF-β receptors, Smurf1 can interact with Smad1/5, and can mediate their degradation (Zhu et al, 1999). Smurf2 can also target Smad1 and 2 for degradation (Lin et al, 2000; Zhang et al, 2001).
The transcriptional function of Smad proteins can also be limited in the nucleus by TGIF, a transcriptional repressor that has been reported to interact with Smads on DNA and repress expression of TGF-β target genes (Wotton et al, 1999). Transcriptional repression by TGIF appears to act in multiple pathways through its ability to recruit divergent molecules (Wotton et al, 1999; Melhuish and Wotton, 2000; Pessah et al, 2001). In this report, we identify the novel ubiquitin ligase Tiul1, a member of the HECT subclass of E3 ubiquitin ligases, and show that Tiul1 interacts with TGIF to induce the degradation of Smad2. These results provide evidence for a novel function of TGIF that acts in partnership with the E3 ubiquitin ligase Tiul1 for the negative regulation of Smad signaling.
Results
Tiul1 interacts with Smad7
To identify new components of the TGF-β signaling pathway, we performed a yeast two-hybrid interaction assay using the entire mouse Smad7 as a bait. By screening 50 × 106 clones of a human placental cDNA expression library, we obtained 10 different cDNA species from 106 positive clones. The putative novel E3 ubiquitin ligase Tiul1 was one of the Smad7 interactors that exhibited specific and strong binding to Smad7 in the yeast two-hybrid system (Figure 1A and data not shown). Like Smurf1/2, Tiul1 has the distinctive structural features of the HECT subclass of E3 ubiquitin ligases, including a C2 domain, WW domains, and a HECT catalytic domain (Figure 1B). Comparison of the sequence of Tiul1 with those from Smurf1/2 revealed a divergent C2 domain (26 and 27%, respectively) and a very low degree of identity in the WW and HECT domains (36 and 39%). Furthermore, Tiul1 has four WW domains, whereas Smurf1/2 contain only three and two WW domains, respectively (Figure 1B).
Figure 1.
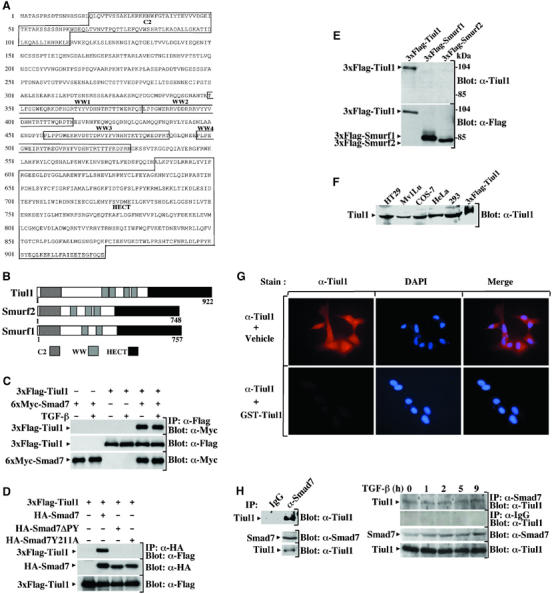
Smad7 interacts with Tiul1. (A) Amino-acid sequence of the human Tiul1. The conserved domains C2, WW, and HECT of Tiul1 are indicated. (B) Schematic representation of Tiul1, Smurf1, and Smurf2. (C) 293 cells were transfected with 6xMyc-Smad7 in the presence or absence of 3xFlag-Tiul1 and treated with or without TGF-βı (2 ng/ml) for 1 h. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody and immunoblotted (Blot) with anti-Myc antibody. (D) 293 cells were transfected with the indicated combinations of wild-type HA-Smad7, HA-Smad7ΔPY, HA-Smad7Y211A, and 3xFlag-Tiul1. The cell extracts were subjected to anti-HA immunoprecipitation followed by Flag immunoblotting. In each case, the total expression levels of transfected proteins were determined by immunoblotting with the indicated antibodies. (E) Extracts from 293 cells transfected with 3xFlag-Tiul1, 3xFlag-Smurf1, or 3xFlag-Smurf2 were immunoblotted with anti-Tiul1 antibody. The expression of 3xFlag-Tiul1, 3xFlag-Smurf1, and 3xFlag-Smurf2 was monitored by reprobing the same membrane with anti-Flag antibody. (F) Lysates from different cell lines were subjected to immunoblot analysis using anti-Tiul1 antibody. Lysates from 293 cells transfected with 3xFlag-Tiul1 was loaded as a positive control. (G) MDCK cells were stained with anti-Tiul1 antibody and DAPI, and the localization of Tiul1 was visualized under an immunofluorescence microscope. (H) Left panel, COS-7 cell lysates were immunoprecipitated with either normal goat IgG (IP: IgG) or anti-Smad7 antibody (IP: α-Smad7), and immunoblotted with anti-Tiul1 antibody. The total expression of endogenous proteins was monitored by direct immunoblotting. Right panel, Mv1Lu cells were exposed to TGF-βı (2 ng/ml) for the indicated times. In all, 1 mg of total cell lysates was subjected to immunoprecipitation with anti-Smad7 or goat IgG antibodies and immunoblotted with anti-Tiul1 antibody. The total expression of endogenous proteins was monitored by direct immunoblotting.
To confirm the interaction of Tiul1 with Smad7 in mammalian cells, we expressed a Flag-Tiul1 with Myc-Smad7 in 293 cells. Immunoprecipitation of cell lysates with anti-Flag antibody revealed the presence of Myc-Smad7, which was absent in a control transfection in which only Smad7 was transfected (Figure 1C). Interestingly, exposure of 293 cells to TGF-β had no effect on the association between Tiul1 and Smad7 (Figure 1C), suggesting that the interaction between Tiul1 and Smad7 can occur in a manner independent of TGF-β signaling. A similar conclusion could be drawn from COS-7 cells transfected with Flag-Tiul1 and Myc-Smad7, together with a constitutively activated form of TβRI (data not shown) (Wieser et al, 1995).
Previous studies have shown that Smurf1/2 associate with Smad7 through interactions between WW domains of Smurf1/2 and the PY motif in Smad7 protein (Kavsak et al, 2000; Ebisawa et al, 2001). To determine the role of the Smad7 PY motif in mediating binding to Tiul1, we employed two Smad7 PY motif mutants, one (Smad7PY) with the PPPY sequence deleted, and another (Smad7Y211A) with the conserved Tyr residue in the PY motif mutated to Ala. In contrast to wild-type Smad7, the Smad7 PY motif mutants failed to interact with Tiul1, indicating that the PY motif in Smad7 is critical for its interaction with Tiul1 (Figure 1D).
To determine whether the association of Tiul1 with Smad7 occurs with physiological levels of these proteins, a polyclonal antibody against Tiul1 was raised. This antibody recognizes full-length Flag-Tiul1, but not Flag-Smurf1 or Flag-Smurf2 (Figure 1E). Use of this antibody for Western blot analysis of extracts from various cell lines identified a specific band of approximately 95 kDa, which is the expected size for Tiul1 (Figure 1F). We also performed indirect immunofluorescence to investigate the subcellular localization of endogenous Tiul1 and found that the anti-Tiul1 antibody can detect a protein that is distributed both in the nucleus and the cytoplasm in MDCK (Figure 1G) and Mv1Lu cells (data not shown). The specificity of this immunoreactivity of Tiul1 was evident, since we were unable to detect a signal when the anti-Tiul1 antibody was saturated by preincubation with an excess of GST-Tiul1 protein (Figure 1G). To characterize the endogenous association of Tiul1 with Smad7, Smad7 was immunoprecipitated from COS-7 extracts with an anti-Smad7 antibody and associated Tiul1 was visualized by immunoblotting with anti-Tiul1 antibody. In immunoprecipitates prepared with preimmune antisera, no Tiul1 was detectable (Figure 1H, left panel). However, in the anti-Smad7 immunoprecipitates, we clearly observed Tiul1 co-precipitating with Smad7 (Figure 1H, left panel). Analysis of cell extracts from Mv1Lu cells treated with TGF-β for various time periods confirmed that the association of endogenous Smad7 and Tiul1 occurred in a manner independent of TGF-β signaling (Figure 1H, right panel).
Tiul1 associates with Smad7 to induce the degradation of the TGF-β type I receptor
Previous studies have reported that Smad7 inhibits TGF-β signaling through interaction with the activated type I receptor (Hayashi et al, 1997; Nakao et al, 1997). To determine whether Tiul1 could affect the ability of Smad7 to bind TβRI, we expressed in 293 cells Smad7 together with wild-type or constitutively activated TβRI in either the presence or absence of Tiul1. Consistent with previous reports (Hayashi et al, 1997), we observed a strong interaction between the constitutively activated TβRI and Smad7 (Figure 2A). However, the amount of activated TβRI bound to Smad7 was significantly decreased in the presence of wild-type Tiul1, correlating with a decrease in the expression of the activated TβRI, but not in the expression of Smad7 (Figure 2A). In contrast to wild-type Tiul1, the level of activated TβRI bound to Smad7 was not changed in the presence of a catalytically inactive mutant of Tiul1 (Figure 2A), Tiul1-C890A, in which the Cys that is believed to conjugate ubiquitin was replaced to Ala. The ability of Tiul1 to induce degradation of the activated TβRI without affecting the expression levels of Smad7 is specific, because we found that Smurf1/2 can mediate degradation of both Smad7 and the activated TβRI (Figure 2A), consistent with previous observations (Kavsak et al, 2000; Ebisawa et al, 2001). A similar effect of Tiul1 was observed in 293 cells transfected with wild-type TβRI and treated with TGF-β (Figure 2B).
Figure 2.
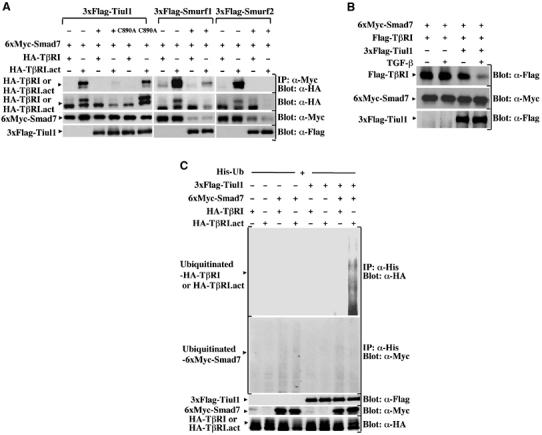
Tiul1 associates with Smad7 to induce the degradation of TβRI. (A) 293 cells were transfected with the indicated combinations of wild-type 3xFlag-Tiul1, 3xFlag-Tiul1-C890A, 3xFlag-Smurf1, 3xFlag-Smurf2, 6xMyc-Smad7, HA-TβRI, and HA-TβRI.act. Cell lysates were immunoprecipitated (IP) with anti-Myc antibody and immunoblotted (Blot) with anti-HA antibody. (B) 293 cells were transfected with 6xMyc-Smad7 and Flag-wild-type TβRI in the presence or absence of 3xFlag-Tiul1 and treated with or without TGF-βı for 4 h. The total expression levels of transfected proteins were determined by immunoblotting with the indicated antibodies. (C) 293 cells were transfected with His-Ub, together with the indicated combinations of 3xFlag-Tiul1, 6xMyc-Smad7, HA-TβRI, and HA-TβRI.act. Before lysis, cells were incubated with the proteasome inhibitor MG132 (10 μM) for 6 h to prevent degradation. Cell lysates were immunoprecipitated with anti-His antibody and immunoblotted with anti-HA or anti-Myc antibodies.
To determine whether Tiul1 acts in partnership with Smad7 to induce the ubiquitination of the activated TβRI, we expressed His-ubiquitin with wild-type or constitutively activated TβRI in the presence or absence of Tiul1 and Smad7 in 293 cells and examined ubiquitination in the presence of the proteasome inhibitor MG132, which allowed us to visualize ubiquitinated species of the activated TβRI by blocking its degradation (Figure 2C). Under these conditions, the ubiquitination of the type I receptor was not detected by the expression of Tiul1 or Smad7 alone. However, when Tiul1 was coexpressed along with Smad7, we observed a strong increase in the ubiquitination of the activated TβRI (Figure 2C). A similar analysis of Smad7 revealed that neither coexpression of Tiul1 alone nor together with the activated TβRI significantly alter the ubiquitination of Smad7 (Figure 2A and B). Taken together, these results suggest that Tiul1 requires Smad7 for efficient ubiquitination of the activated TβRI, with subsequent degradation.
Ligand-dependent association of Tiul1 with R-Smads
Next, we examined whether Tiul1 may function to target other Smads for degradation. To approach this question, we first examined the interaction of Tiul1 with different Smads in the presence of wild-type or constitutively activated TβRI. As shown in Figure 3A, Tiul1 interacted not only with Smad7 but also with Smad2, 3, 4, and 6. However, when Smad1 was used in this assay, we could not detect any interaction with Tiul1, indicating that the interactions that we observed between Tiul1 and Smad2, 3, 4, 6, and 7 were specific (Figure 3A). More importantly, we observed that the associations of Tiul1 with the R-Smads, Smad2 and 3, but not Smad4, 6, and 7, were increased in response to TGF-β signaling (Figure 3A).
Figure 3.
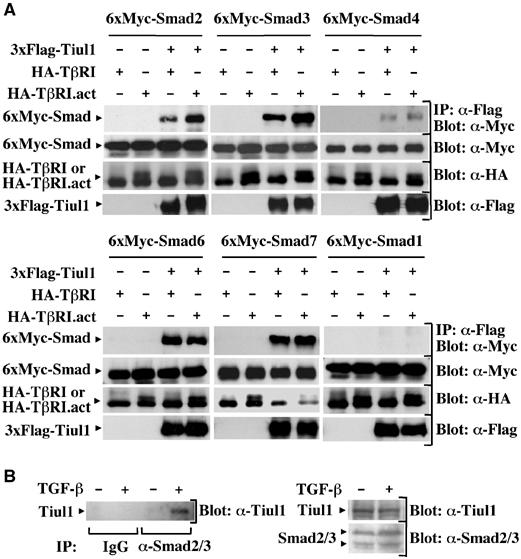
Tiul1 interacts with Smads but does not induce their degradation. (A) 293 cells were transfected with 3xFlag-Tiul1, 6xMyc-Smads, HA-TβRI, and HA-TβRI.act as indicated and cell lysates were immunoprecipitated (IP) with anti-Flag antibody and immunoblotted (Blot) with anti-Myc antibody. The total expression levels of transfected proteins were determined by immunoblotting with the indicated antibodies. (B) Lysates from 293 cells untreated or treated with TGF-βı (2 ng/ml) for 1 h were immunoprecipitated with either normal goat IgG (IP: IgG) or anti-Smad2/3 antibody (IP: α-Smad2/3), and immunoblotted with anti-Tiul1 antibody (α-Tiul1).
To provide further evidence that TGF-β signaling can induce the association of Tiul1 with Smad2/3, we examined the endogenous association of Tiul1 with Smad2/3 in 293 cells using the anti-Tiul1 antibody and a polyclonal antibody that recognizes both Smad2 and 3. We observed that Tiul1 co-precipitated with Smad2/3 only in 293 cells that had been exposed to TGF-β (Figure 3B), indicating that Tiul1 can associate with Smad2 and Smad3 at the endogenous levels in the presence of TGF-β signaling.
We next examined whether Tiul1 decreases the steady-state levels of these Smads. As shown in Figure 3A, Tiul1 did not significantly alter the steady-state levels of Smads 2, 3, 4, or 7 even in the presence of the constitutively activated TβRI. Under these experimental conditions, we confirmed that Tiul1 acts in association only with Smad7 to target the activated TβRI for degradation (Figure 3A). These results indicate that the interaction of Tiul1 with Smads is not sufficient for Tiul1 to target these proteins for degradation.
Tiul1 interacts with TGIF
Our previous observations indicate that Tiul1 acts in association with Smad7 to trigger the ubiquitin-dependent degradation of the activated TβRI. In an attempt to explore whether Tiul1 could use additional mechanisms to target other components of the TGF-β signaling pathway, we tested whether Smad2 might function to recruit Tiul1 to TGIF, a transcriptional corepressor that associates with R-Smad in the nucleus and represses its ability to mediate TGF-β-dependent transcription (Wotton et al, 1999). As a first step to approach this question, we looked for possible interactions between Tiul1 and TGIF. Immunoprecipitation of cell lysates from transfected 293 cells with anti-Flag antibody directed against Tiul1 revealed the presence of HA-TGIF, which was absent in a control experiment in which only HA-TGIF was transfected (Figure 4A). Interestingly, treatment of cells with TGF-β enhanced the interaction between Tiul1 and TGIF (Figure 4A).
Figure 4.
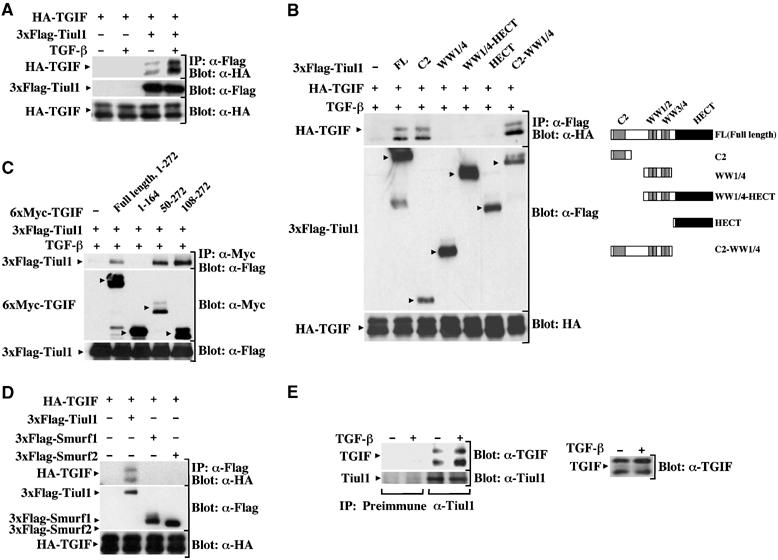
Tiul1 interacts with TGIF. (A) 293 cells were transfected with HA-TGIF either alone or together with 3xFlag-Tiul1. Cells were treated with or without TGF-βı (2 ng/ml) for 1 h and cell lysates were immunoprecipitated (IP) with anti-Flag antibody and immunoblotted (Blot) with anti-HA antibody. (B) 293 cells were transfected with HA-TGIF either alone or together with wild-type 3xFlag-Tiul1 or 3xFlag-Tiul1 fragments as indicated. Cells were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-HA antibody. (C) 293 cells were transfected with 3xFlag-Tiul1 either alone or together with wild-type 6xMyc-TGIF or deletion mutants of TGIF as indicated. Cells were immunoprecipitated with anti-Myc antibody and immunoblotted with anti-Flag antibody. (D) 293 cells were transfected with HA-TGIF, 3xFlag-Tiul1, 3xFlag-Smurf1, and 3xFlag-Smurf2 as indicated. The cell extracts were subjected to anti-Flag immunoprecipitation followed by anti-HA immunoblotting. (E) Cell lysates from Mv1Lu cells untreated or treated with TGF-β (2 ng/ml) for 1 h were immunoprecipitated with either rabbit preimmune serum as a negative control (IP: Preimmune) or anti-Tiul1 antibody (IP: α-Tiul1), and immunoblotted with anti-TGIF antibody (Blot: α-TGIF). The membrane was reprobed with anti-Tiul1 antibody (Blot: α-Tiul1) to see the expression of Tiul1. The expression of TGIF was monitored by direct immunoblotting.
To analyze in more detail the interaction between Tiul1 and TGIF, various Flag-tagged Tiul1 constructs were transfected into 293 cells and tested for interaction with cotransfected HA-TGIF. As shown in Figure 4B, a Tiul1 fragment corresponding to the C2 domain was sufficient for interaction with HA-TGIF. A similar analysis of N-terminal truncations revealed that deletion of the C2 domain of Tiul1 abrogated binding to TGIF. We also generated a series of deletion mutants of TGIF and tested for their interaction with Tiul1. We observed that the Tiul1-interacting region of TGIF maps to the C-terminus domain of TGIF between residues 108 and 272 (Figure 4C). To establish the specificity for the interaction between TGIF and Tiul1, cell extracts from 293 cells transfected with HA-TGIF, together with Flag-Smurf1, Flag-Smurf2, or Flag-Tiul1, were subjected to immunoprecipitation with anti-Flag antibody, followed by immunoblotting with anti-HA antibody. In contrast to Tiul1, we detected no or only weak interaction between Smurf1/2 and TGIF (Figure 4D).
To provide further evidence that Tiul1 associates with TGIF, we examined the endogenous association of these proteins in Mv1Lu cells using specific antibodies to Tiul1 and TGIF. Similar to our previous results in 293 cells (Figure 4A), some constitutive interactions between TGIF and Tiul1 could be detected in unstimulated cells and activation of TGF-β signaling enhanced this interaction (Figure 4E), indicating that the TGF-β-inducible Tiul1/TGIF complex can occur with physiological levels of these proteins.
TGIF binds to Tiul1 independently of Smad2
To examine the possibility that Smad2 may function to recruit Tiul1 to TGIF, we made use of a mutant form of TGIF (TGIF 1–148:177–262) lacking the Smad-interacting domain (Figure 5A, left panel; Wotton et al, 1999). Interestingly, we observed that this mutant form of TGIF retained its ability to interact with Tiul1 (Figure 5A, right panel), suggesting that the interaction of TGIF with Smad2 is not required for its association with Tiul1.
Figure 5.
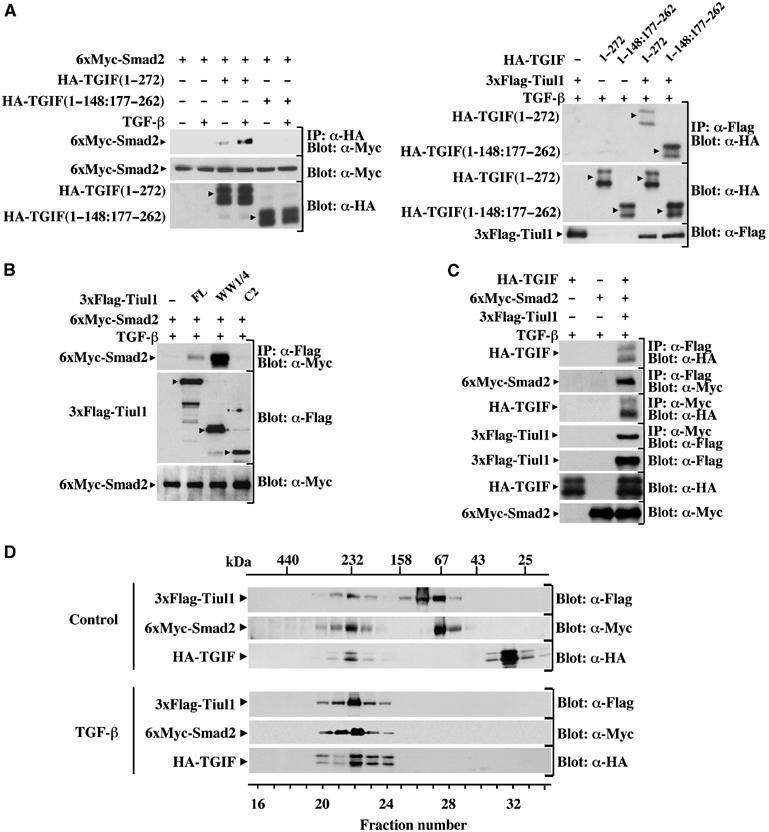
TGIF binding to Tiul1 is independent of Smad2. (A) 293 cells were transfected with the indicated combinations of wild-type HA-TGIF (1–272), the mutant of TGIF (1–148:177–262), 6xMyc-Smad2, and 3xFlag-Tiul1. Cells were treated with or without TGF-βı (2 ng/ml) for 1 h and cell lysates were immunoprecipitated (IP) with anti-HA (left panel) or anti-Flag (right panel) antibodies and immunoblotted (Blot) with anti-Myc (left panel) or anti-HA (right panel) antibodies. (B) 293 cells were transfected with wild-type 3xFlag-Tiul1 or the indicated deletion mutants fused to 3xFlag and 6xMyc-Smad2 as indicated. Cells were immunoprecipitated with anti-Flag antibody and immunoblotted with anti-Myc antibody. (C) 293 cells were transfected with HA-TGIF, 3xFlag-Tiul1, and 6xMyc-Smad2 as indicated and treated with TGF-βı for 1 h. The cell extracts were subjected to anti-Flag or anti-Myc immunoprecipitation, followed by anti-HA, anti-Myc, or anti-Flag immunoblotting. (D) 293 cells were transfected with 3x-Flag-Tiul1, HA-TGIF, and 6xMyc-Smad2 and treated with or without TGF-βı for 1 h. Cell lysates were loaded onto a Superdex 200 gel filtration column and fractions were analyzed for the presence of Tiul1, TGIF, and Smad2 by Western blotting.
To provide direct evidence that the interaction of TGIF with Tiul1 occurs independently of Smad2, we expressed our different constructs of Tiul1 with Smad2 in 293 cells. As shown in Figure 5B, the C2 domain mutant, which strongly associates with TGIF (Figure 4B), was defective in its ability to interact with Smad2. Conversely, we could not detect any interaction between TGIF and the WW1/4 mutant of Tiul1 (Figure 4B), which strongly interacts with Smad2 (Figure 5B). Thus, it is likely that Tiul1 contains discrete binding sites for Smad2 and TGIF.
Since our previous results indicate that Tiul1 can interact with both Smad2 and TGIF, we wanted to know whether Smad2 and TGIF could coexist with Tiul1 or whether Smad2–Tiul1 and TGIF–Tiul1 complexes are mutually exclusive. Cotransfection of 293 cells with Flag-Tiul1, Myc-Smad2, and HA-TGIF revealed that TGIF and Smad2 were clearly detectable in complexes precipitated via the Flag epitope present on Tiul1 (Figure 5C). We also analyzed the interactions of TGIF and Tiul1 with Smad2 by immunoprecipitation of the same cell lysates with anti-Myc antibody directed towards Smad2. We observed that both TGIF–Smad2 and Tiul1–Smad2 complexes are present (Figure 5C). These results suggest that Smad2, TGIF and, Tiul1 might interact with each other at the same time to form a complex. To examine this directly, cell lysates from transfected 293 cells were subjected to size fractionation on a Superdex 200 column. In unstimulated cells, Tiul1, TGIF, and Smad2 were eluted from the gel filtration column at their respective size (see Supplementary Figure, available at the EMBO Journal Online) and in a peak with an apparent molecular mass of ∼230 kDa, close to the expected size of the Tiul1–TGIF–Smad2 complex (∼200 kDa) (Figure 5D). Following TGF-β treatment, the majority of Tiul1, TGIF, and Smad2 were coeluted in the ∼230 kDa peak (Figure 5D). Taken together, these data indicate that Tiul1, TGIF, and Smad2 can all coexist in the same complex, the level of which can be enhanced by activation of the TGF-β signaling pathway.
Tiul1 degrades Smad2 in the presence of TGIF
Since Tiul1 associates with Smad2 as well as with TGIF upon activation of TGF-β signaling, we tested whether coexpression of Tiul1 together with Smad2 and TGIF might alter the steady-state levels of TGIF and/or Smad2. As shown in Figure 6A, neither coexpression of Tiul1 alone nor together with Smad2 significantly alters the TGIF expression levels both in the presence and absence of TGF-β signaling. Consistent with this, we did not detect any effect of Tiul1 on the ubiquitination of TGIF in the presence of Smad2 and TGF-β signaling (see Figure 6F). Pulse-chase analysis also confirmed that the turnover of TGIF was unaffected by coexpression of Tiul1 and Smad2 (see Figure 6E). In contrast, when TGIF was coexpressed with Tiul1 and Smad2, we observed that exposure of cells to TGF-β induces a significant decrease in the amount of Smad2 protein expressed in cells (Figure 6A), suggesting that the association of TGIF and Tiul1 may enhance the degradation of Smad2. To examine whether coexpression of TGIF might enhance the ability of Tiul1 to induce ubiquitin-dependent degradation of Smad2, we expressed Smad2 with Tiul1 or Tiul1-C890A in the presence or absence of TGIF. As expected, coexpression of TGIF with Tiul1 strongly decreased the steady-state levels of Smad2 (Figure 6B). In contrast, coexpression of TGIF with Tiul1-C890A did not yield a decrease in the steady-state levels of Smad2 (Figure 6B). A similar degradation by Tiul1 and TGIF was observed for Smad3, but not for Smad7 (Figure 6C), suggesting that TGIF might act in partnership with Tiul1 to degrade the R-Smads, Smad2 and 3. This degradation is specifically mediated by Tiul1 and TGIF, since we were unable to detect a significant degradation of Smad2 by overexpression of Tiul1 together with SnoN (Figure 6D), which, like TGIF, can act as a repressor of Smad signaling (Mehra and Wrana, 2002). In these experiments, we observed that Smurf2 can degrade both SnoN and Smad2 (Figure 6D), similar to previous observations (Bonni et al, 2001). Taken together, these results suggest that TGIF might selectively associate with Tiul1 to target Smad2 for degradation.
Figure 6.
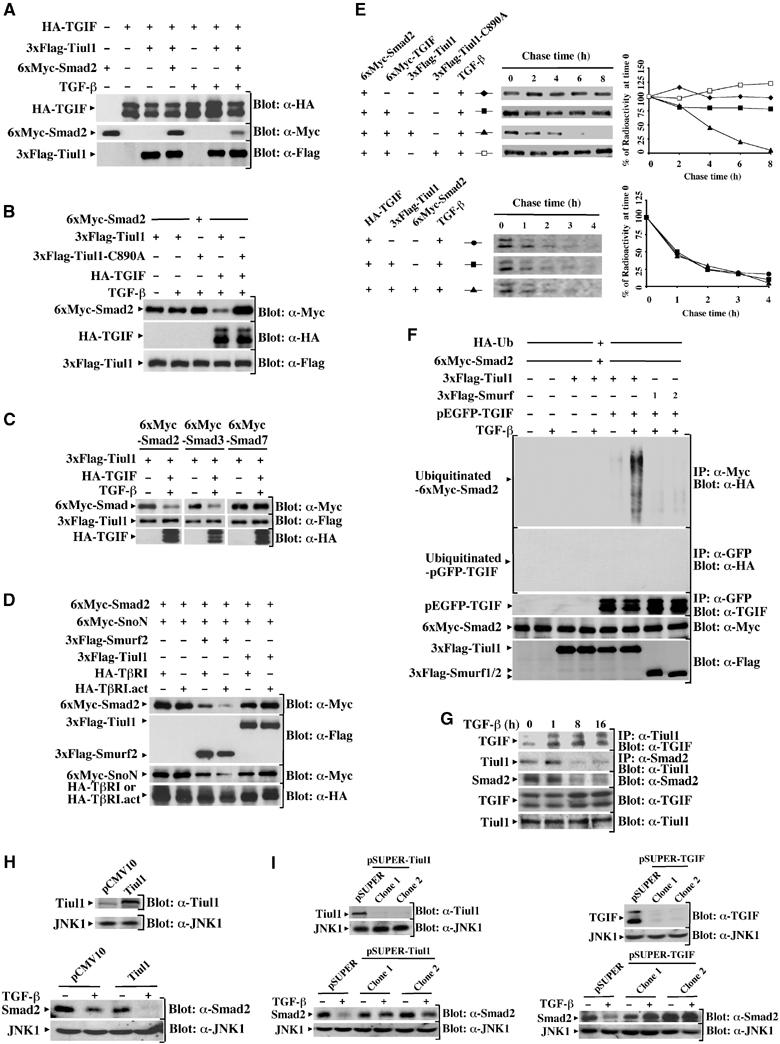
Tiul1 induces ubiquitin-dependent degradation of Smad2 in the presence of TGIF. (A) 293 cells were transfected with the indicated combinations of 3xFlag-Tiul1, HA-TGIF, and 6xMyc-Smad2. Cells were treated with or without TGF-βı for 1 h and cell lysates were subjected to immunoblotting (Blot) with the indicated antibodies. (B) 293 cells were transfected with the indicated combinations of 3xFlag-Tiul1, 3xFlag-Tiul1-C890A, 6xMyc-Smad2, and HA-TGIF. Cells were treated with or without TGF-βı (2 ng/ml) for 1 h and cell lysates were analyzed by immunoblotting with the indicated antibodies. (C) 293 cells were transfected with 3xFlag-Tiul1, HA-TGIF, 6xMyc-Smad2, 6xMyc-Smad3, and 6xMyc-Smad7 as indicated and treated with or without TGF-βı (2 ng/ml) for 1 h. The total expression levels of transfected proteins were determined by immunoblotting with the indicated antibodies. (D) 293 cells were transfected with 3xFlag-Tiul1, 3xFlag-Smurf2, 6xMyc-Smad2, 6xMyc-SnoN, HA-TβRI, and HA-TβRI.act as indicated. The total expression levels of transfected proteins were determined by immunoblotting with the indicated antibodies. (E) 293 cells were transfected with the indicated combinations of 3xFlag-Tiul1, 3xFlag-Tiul1-C890A, 6xMyc-Smad2, 6xMyc-TGIF, and HA-TGIF. Cells were treated with TGF-βı (2 ng/ml) and cell lysates were immunoprecipitated with anti-Myc (Smad2; upper panel) or anti-HA (TGIF; lower panel) antibodies before being visualized and quantified by phosphoimager. Labeled Smad2 (upper panel) or TGIF (lower panel) is plotted at each time point as the percentage of the amount present at time 0. (F) 293 cells were transfected with HA-Ub, together with the indicated combinations of 3xFlag-Tiul1, 3xFlag-Smurf1, 3xFlag-Smurf2, 6xMyc-Smad2, and pEGFP-TGIF. Before lysis, cells were incubated with the proteasome inhibitor MG132 (10 μM) for 6 h. Cells were treated with or without TGF-βı (2 ng/ml) for 1 h and cell lysates were immunoprecipitated with anti-Myc or anti-GFP antibodies before being analyzed by immunoblotting with anti-HA antibody. (G) Mv1Lu cells were treated with TGF-βı (2 ng/ml) for the indicated times and cell extracts were immunoprecipitated with anti-Tiul1 or anti-Smad2 antibodies and immunoblotted with anti-TGIF or anti-Tiul1 antibodies. (H) MDCK cells stably expressing empty vector or 3xFlag-Tiul1 (Blot: α-Tiul1) were treated with or without TGF-βı (2 ng/ml) for 16 h and cell extracts were immunoblotted with anti-Smad2 antibody (Blot: α-Smad2). In all cases, the membranes were reprobed with anti-JNK1 antibody (Blot: α-JNK1) to show that equal amounts of proteins were loaded in the gel. (I) Top, MDCK cells were stably cotransfected with the pEMP4 vector containing a hygromycin resistance gene and empty vector (pSUPER), pSUPER-Tiul1, or pSUPER-TGIF. Clones with reduced expression of Tiul1 or TGIF were identified by immunoblotting total cell lysates with anti-Tiul1 (α-Tiul1; left panel) or anti-TGIF (blot: α-TGIF; right panel) antibodies. Bottom, cells were treated with or without TGF-βı (2 ng/ml) for 16 h and total cell extracts were analyzed by immunoblotting with anti-Smad2 antibody (blot: α-Smad2).
We next analyzed the effects of TGIF and Tiul1 on the turnover of Smad2 using pulse-chase experiments. As shown in Figure 6E, expression of TGIF alone had no effect on the turnover of Smad2 even in the presence of TGF-β signaling. However, when TGIF was coexpressed with wild-type Tiul1, the turnover rate of Smad2 was significantly increased in response to TGF-β signaling (Figure 6E). In contrast to wild-type Tiul1, expression of Tiul1-C890A along with TGIF had no effect on the turnover of Smad2 (Figure 6E). Therefore, we concluded that TGIF acts in partnership with Tiul1 to accelerate the turnover of Smad2.
We also examined whether TGIF enhances the ability of Tiul1 to induce the ubiquitination of Smad2. In the absence of proteasome inhibitors, Smad2 level is strongly reduced by Tiul1 and TGIF, precluding analysis of Smad2 ubiquitination. Therefore, to visualize ubiquitinated species of Smad2, we inhibited the proteasome using MG132 (Figure 6F). Consistent with our analysis of the turnover and the steady-state levels of Smad2, Tiul1 did not induce a detectable ubiquitination of Smad2 in the absence of TGIF (Figure 6F). However, when Tiul1 was coexpressed with TGIF, a significant increase in high molecular weight of ubiquitin-conjugated Smad2 was detected upon activation of TGF-β signaling (Figure 6F). Unlike Tiul1, overexpression of TGIF together with Smurf1/2 failed to induce a significant ubiquitination of Smad2 (Figure 6F). Together, these data demonstrate that TGIF plays an important role in the ubiquitin-dependent degradation of Smad2 by Tiul1.
To determine whether endogenous Tiul1 and TGIF could regulate the TGF-β-dependent degradation of Smad2, we first investigated the kinetics of Tiul1 and TGIF interactions in response to TGF-β. Analysis of Tiul1 or TGIF revealed that treatment of Mv1Lu cells with TGF-β did not appreciably affect the expression levels of Tiul1 and TGIF (Figure 6G). However, exposure of cells to TGF-β caused a significant and persistent increase in the association of Tiul1 and TGIF (Figure 6G). A TGF-β-inducible interaction of Smad2 and Tiul1 was also clearly detectable after 1 h treatment of TGF-β (Figure 6G). In contrast to the Tiul1–TGIF complexes, the interaction of Smad2 with Tiul1 appeared to decrease within 8 h of TGF-β treatment, and this decrease correlated with a decrease in the expression levels of Smad2 (Figure 6G). Taken together, these results suggest the possibility that the association of Tiul1 with both TGIF and Smad2 might trigger Smad2 for degradation.
Our results, and those of others, indicate that TGF-β signaling can induce the degradation of Smad2 (Figure 6G; Lo and Massague, 1999). To investigate whether the TGF-β-dependent degradation of Smad2 might occur through Tiul1, we generated pools of MDCK cells stably overexpressing Tiul1. As shown in Figure 6H, expression of Tiul1 enhanced the ability of TGF-β to induce the degradation of endogenous Smad2. In a second experimental approach, we used the RNA interference approach to generate stable MDCK cell lines in which the expression of endogenous Tiul1 was reduced by 80–90% compared to control cells. Interestingly, the reduction in the level of endogenous Tiul1 suppressed TGF-β-mediated degradation of Smad2 (Figure 6I). In the course of these analyses, we also established stable MDCK cell lines in which the expression of TGIF is knocked down by siRNA. Here again, we found that expression of siRNA to TGIF abolished the ability of TGF-β to induce the degradation of Smad2 (Figure 6I). Similar results were obtained with pools of MDCK cells stably expressing a second interfering sequence to Tiul1 or TGIF (data not shown), confirming that TGF-β-induced degradation of Smad2 is mediated by endogenous Tiul1 and TGIF.
Tiul1 inhibits TGF-β-mediated transcriptional responses and growth arrest
To examine the role of Tiul1 in the regulation of TGF-β-mediated responses, we used the (CAGA)9-Lux reporter construct, which contains concatamerized CAGA elements previously shown to bind complexes of Smad3 and 4 and to be transactivated by both TGF-β and Smad3/4 expression (Dennler et al, 1998). As shown in Figure 7A, TGF-β-induced expression of the reporter gene was markedly inhibited by Tiul1 or TGIF expression in 293 cells. Interestingly, we observed that Tiul1 can synergize with TGIF to suppress TGF-β-mediated luciferase activity (Figure 7A). A similar inhibition was also observed in cells transfected with FAST1 and the ARE-Lux construct (Figure 7A), which contains three copies of the activin response element from the Mix.2 promoter, known to bind the FAST1–Smad2–Smad4 complexes (Whitman, 1998).
Figure 7.
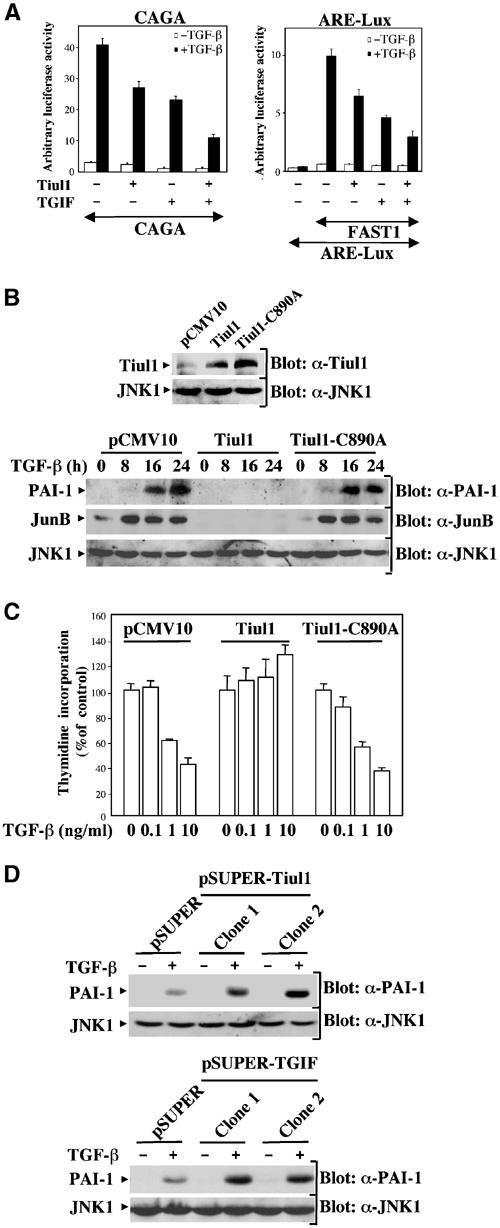
Tiul1 inhibits TGF-β-mediated growth arrest and transcriptional responses. (A) 293 cells were transfected with the reporter constructs (CAGA)9-Lux (left panel) or ARE-Lux, together with FAST1 (right panel) and the indicated combinations of Tiul1 and TGIF. In all cases, cells were treated with (black bars) or without (open bars) TGF-βı (2 ng/ml) for 16 h prior to lysis and then analyzed for luciferase activity. Luciferase activity was normalized to β-galactosidase activity and was expressed as the mean±s.d. of triplicates from a representative experiment performed at least three times. (B) MDCK cells stably expressing the empty vector, 3xFlag-Tiul1, or 3xFlag-Tiul1-C890A (blot: α-Tiul1) were treated with or without TGF-βı (2 ng/ml) for the indicated times and cell extracts were immunoblotted with anti-PAI-1 (blot: α-PAI-1) or anti-JunB (Blot: α-JunB) antibodies. (C) MDCK cells stably expressing empty vector, 3xFlag-Tiul1, or 3xFlag-Tiul1-C890A were treated with the indicated concentrations of TGF-β for 48 h and the rate of cell proliferation was determined by the thymidine incorporation method. Data are expressed as percentages of the relative radioactivity in the absence of TGF-β. (D) Stable MDCK cells with reduced expression of Tiul1 (pSUPER-Tiul1) or TGIF (pSUPER-TGIF) were treated with or without TGF-βı (2 ng/ml) for 16 h and cell extracts were immunoblotted with anti-PAI-1 or anti-JNK1 antibodies.
Next we generated pools of MDCK cells overexpressing wild-type Tiul1 or Tiul1-C890A (Figure 7B) to investigate whether Tiul1 could block the ability of Smad proteins to mediate TGF-β-induced expression of endogenous genes. We chose to focus our analysis on plasminogen activator inhibitor-1 (PAI-1) gene as a target of Smad signaling pathway because activation of PAI-1 promoter by TGF-β requires the formation of a Smad3–Smad4 complex that binds to the CAGA sequence (Dennler et al, 1998). Western blotting analysis with a specific anti-PAI-1 antibody demonstrated that stable expression of Tiul1, but not that of Tiul1-C890A, can block the TGF-β-dependent transcription of the PAI-1 gene (Figure 7B). This inhibitory effect of Tiul1 was also observed on endogenous JunB (Figure 7B), the expression of which is upregulated by TGF-β through a mechanism similar to that of PAI-1 (Jonk et al, 1998).
We also investigated whether stable expression of Tiul1 in MDCK cells might interfere with TGF-β-mediated growth inhibition. As shown in Figure 7C, the growth-inhibitory effect of TGF-β is largely lost in MDCK cells stably expressing Tiul1, but not Tiul1-C890A (Figure 7C). Thus, in stably transfected cells and in transient transfection assays, the presence of Tiul1 caused not only a loss of Smad-dependent transcriptional responses but also the antiproliferative responses to TGF-β.
To provide direct evidence that endogenous Tiul1 acts as an inhibitor of TGF-β-activated transcription, we took advantage of the availability of our MDCK clones in which the expression of endogenous Tiul1 or TGIF was knocked down by siRNA (Figure 6I). As expected, the reduction in the level of endogenous Tiul1 and TGIF increased the sensitivity of the cells to TGF-β-mediated transcriptional activation from endogenous PAI-1 promoter (Figure 7D). Similarly, we observed an increase in the sensitivity of cells to TGF-β-mediated expression of JunB and growth arrest using MDCK cells with reduced expression of Tiul1 or TGIF (data not shown). Thus, reducing the level of endogenous Tiul1 or its partner TGIF results in a concomitant increase in TGF-β signaling, which is consistent with the notion that Tiul1 and TGIF can act as inhibitors in the TGF-β signaling pathway.
Discussion
In this study, we report on a new member of the HECT subclass of E3 ubiquitin ligases that we call Tiul1, and show that Tiul1 can function as a negative regulator in the TGF-β signaling pathway. We demonstrate that Tiul1 degrades not only the activated type I receptor in association with Smad7 but also Smad2 in association with TGIF. Consistent with this, we observed that Tiul1 is localized both in the cytoplasm and the nucleus, suggesting that this distribution may allow Tiul1 to target the degradation of TβRI at the membrane or in vesicles and Smad2 in the nucleus. Taken together, the results outlined in the present study provide evidence for the existence of a novel mechanism for the negative regulation of TGF-β signaling pathway by E3 ubiquitin ligases through degradation of active components of TGF-β signaling molecules.
Tiul1 has significant homology to the HECT subclass E3 ubiquitin ligases, which selectively target protein for ubiquitination and degradation by the 26S proteasome (Ciechanover et al, 2000). Other members of this class include Smurf1/2, which share with Tiul1 distinctive structural features, including a phospholipid/calcium-binding C2 domain, WW protein–protein interaction domains, and a HECT domain that catalyzes ubiquitination on target proteins. Recent findings have suggested that Smurf1/2 interact with Smad7 and TGF-β signaling results in Smad7-dependent recruitment of Smurf1/2 to TGF-β receptors (Kavsak et al, 2000; Ebisawa et al, 2001). The Smad7-dependent recruitment of Smurf1/2 to TGF-β receptors resulted in proteasome-mediated degradation of the Smad7/TGF-β receptor complexes, thereby allowing interruption of TGF-β-mediated responses. A remarkable difference between Smurf1/2 and Tiul1 is that ectopic expression of Tiul1 induces degradation of the activated type I receptor in the presence of Smad7 but has no effect on the expression levels of Smad7. At present, it is unclear why Tiul1 selectively induces degradation of the type I receptor. The expression of Smad7 is increased in response to TGF-β, providing a mechanism for negative feedback regulation of the Smad signaling pathway (Nakao et al, 1997). The expression of Smad7 can also be induced by interferon-γ (IFN-γ) (Ulloa et al, 1999) and by tumor necrosis factor-α (TNF-α) (Bitzer et al, 2000), raising the interesting possibility that Smad7 may fulfill other cellular functions independent of its inhibitory role in TGF-β signaling. Since Tiul1 functions to limit TGF-β-mediated responses, one possibility is that Tiul1 might provide additional mechanisms to allow quantitative inhibition by Smad7 in the presence of high levels of TGF-β signaling. Alternatively, the ubiquitin-dependent degradation of the activated type I receptor might selectively suppress TGF-β signaling, which will allow Smad7 to control other biological functions in response to other signal inputs.
The ability of Smads to modulate transcription in response to ligand results from a functional cooperativity with several nuclear partners (Massague and Chen, 2000), and these interacting proteins could potentially be targeted by Tiul1 for ubiquitin-mediated degradation. TGIF is one of the characterized R-Smad interacting proteins, which is believed to repress expression of TGF-β-responsive genes by assembling a repressor complex (Wotton et al, 1999). In this study, we demonstrated that TGIF interacts with Tiul1, in order to trigger the ubiquitination-dependent degradation of Smad2. The association of TGIF and Tiul1 is apparently of physiological significance, since coimmunoprecipitation experiments using specific antibodies have identified endogenous Tiul1-TGIF complexes, the level of which can be enhanced by activation of the TGF-β signaling pathway. We have investigated the basis of this interaction in transfected cells and found that Tiul1 associates with TGIF through interactions between the C2 domain of Tiul1 and the C-terminal part of TGIF. Analysis of truncation mutants that removed the C2 domain or the WW1/4 domain showed that the C2 domain mediates interaction with TGIF, whereas the WW1/4 domain mediates interaction with Smad2, thereby indicating that the interaction of Tiul1 and TGIF occurs independently of the association of TGIF with Smad2.
The findings outlined in the present study are consistent with a model in which the corepressor TGIF functions to enhance the ability of Tiul1 to degrade Smad2 through ubiquitination. Furthermore, we observed that Tiul1 is localized both in the cytoplasm and the nucleus. Since the ubiquitin-dependent degradation of Smad2 appears to take place in the nucleus (Lo and Massague, 1999), one attractive possibility it that TGIF might facilitate the Tiul1-dependent degradation of Smad2 by controlling the nuclear localization of Tiul1. Another alternative possibility is that TGIF might associate with Tiul1 to increase the specificity of the interaction between Smad2 and Tiul1 or to stabilize the Smad2–Tiul1 complexes, thereby allowing Tiul1 to induce ubiquitin-dependent degradation of Smad2. However, we were unable to detect that TGIF stabilizes the association of Smad2 with Tiul1 or Tiul1-C890A, which is defective in its ability to induce the degradation of Smad2 (data not shown). In any case, our findings that TGIF associates with Tiul1 to facilitate the degradation of Smad2 is consistent with the traditional function of TGIF as an active repressor in TGF-β signaling, and provides a novel mechanism by which ubiquitin ligases function to limit TGF-β signaling.
Materials and methods
Yeast two-hybrid screen
DNA encoding full-length Smad7 was cloned into the yeast GAL4 DNA-binding domain vector pGBT10. The resulting plasmid was used as a bait in two-hybrid screens of a human placental cDNA library fused to the GAL4 activation domain in the pGAD-10 plasmid. Positive clones were selected by prototrophy for His and expression of β-galactosidase. Several cDNAs displaying similarity to a putative ubiquitin ligase WWP1 (accession number: NM007013) with unknown function were isolated and one cDNA was used to screen a λ-ZAP11 human placental cDNA library (Clontech) to obtain a full-length Tiul1 cDNA clone. We renamed this protein Tiul1 (for TGIF-interacting ubiquitin ligase 1).
Expression vectors
Expression vectors for Myc-Smurf1, 6xMyc-SnoN, 6xMyc-Smad1, 6xMyc-Smad2, 6xMyc-Smad3, 6xMyc-Smad4, 6xMyc-Smad6, and 6xMyc-Smad7 were a gift from Dr K Miyazono. The expression vector for Smurf2 was kindly provided by Dr XH Feng. Expression vectors for (CAGA)9-Lux, ARE-Lux, FAST1, HA-TβRI or HA-TβRI.act were described previously (Pessah et al, 2001; Prunier et al, 2001). For Tiul1 expression constructs, the open reading frame was amplified from the λ-ZAP11 clone by PCR and subcloned into p3xFlag-CMV-10 expression vector (Sigma). A similar approach was used to clone Smurf1/2 in p3xFlag-CMV-7.1 and TGIF fragments in pcDNA-3-6xMyc. A PCR-based procedure was employed to generate the catalytically inactive ubiquitin ligase mutant of Tiul1-C890A. For stable expression of short interfering RNAs to Tiul1 or TGIF in MDCK cells, oligonucleotides containing sequences from target genes (Tiul1: GGCACGAATGGAATAGATA; TGIF: ATCTGGACCAAGTACGAAT) were inserted into the pSUPER vector as described previously (Brummelkamp et al, 2002).
Cell lines
All cell lines used in this study were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). For stable transfectants, cells were transfected with expression vectors by the LipofectAMINE method (GIBCO/BRL) and selected in G418 or hygromycin-containing growth medium.
[3H]thymidine incorporation
[3H]thymidine incorporation was performed as described previously (Prunier et al, 2001). Briefly, cells were seeded in six-well plates and then incubated for 48 h with various concentrations of TGF-β1 in DMEM containing 0.2% FCS. Cells were labeled with 1 μCi/ml [3H]thymidine for the last 4 h of incubation and cell extracts were collected and counted in a liquid scintillation spectrometer.
Immunoprecipitation and immunoblotting
Cells were transfected with expression vectors by the LipofectAMINE method. After 48 h, cells were resuspended in TNMG buffer (Prunier et al, 2001) and lysed by sonication. Cell lysates were subjected to immunoprecipitation with anti-Flag M2 (Sigma), anti-c-Myc (9E10) (Santa Cruz), anti-His (Roche), anti-HA (Roche), anti-Smad2/3 (Santa Cruz), anti-Smad2 (Zymed), anti-Smad7 (Santa Cruz), anti-GFP (Clontech), or anti-Tiul1 (see below) antibodies for 2 h, followed by adsorption to sepharose-coupled protein G for 1 h. For the anti-Tiul1 antibody, rabbits were immunized with bacterially produced GST-Tiul1-encoding amino acids 121–340, and the serum was affinity purified using standard methods. Immunoprecipitates were separated by SDS–PAGE and analyzed by immunoblotting with the indicated antibodies. In all cases, the total expression levels of transfected or endogenous proteins were determined by immunoblotting with the appropriate antibodies.
Gel filtration
Cell lysates from transfected 293 cells with Flag-Tiul1, HA-TGIF, and Myc-Smad2 were fractionated on a Superdex 200 column-fast protein liquid chromatography system with elution buffer containing 20 mM HEPES (pH 7.5), 250 mM NaCl, 1 mM EDTA, 0.1% β-mercaptoethanol, and 0.01% Tween 20. Column calibration was performed under the same conditions with ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), and chymotrypsin (25 kDa). Fractions of 0.5 ml were collected and analyzed by immunoblotting with anti-Flag, anti-HA, or anti-Myc antibodies.
Immunofluorescence
The distribution of endogenous Tiul1 in MDCK cells was visualized by immunofluorescence using the rabbit anti-Tiul1 antibody and Texas Red-conjugated goat anti-rabbit IgG. Image was visualized using a fluorescence microscope.
Pulse-chase analysis
Transfected 293 cells were preincubated for 1 h in Met/Cys-free DMEM, and labeled for 10 min with 100 μCi/ml [35S]-Met/Cys in Met/Cys-free DMEM. The cells were chased for the indicated time periods and the cell lysates were subjected to immunoprecipitation. The immunoprecipitates were resolved by SDS–PAGE, visualized by phosphoimager, and quantified.
Gene expression analysis
For the reporter gene assays, 293 cells were transfected with expression vectors using FuGENE. Cells were subsequently treated with or without TGF-β1 (2 ng/ml) (Sigma) for 16 h. Luciferase activity was measured by the luciferase assay system (Promega) and was normalized for transfection efficiency using a β-galactosidase expressing vector (pCMV5.LacZ) and the Galacto-Star system (Perkin-Elmer).
GenBank Accession Number
Tiul1 sequence data have been submitted to the GenBank database under the accession number AY345857.
Supplementary Material
Supplementary Figure
Acknowledgments
We thank Drs J-M Gauthier, XH Feng, J Massagué, K Miyazono, and J Wrana for providing plasmids. This work was supported by INSERM, CNRS, la Ligue contre le Cancer Comité de Paris, and Association pour la Recherche sur le Cancer (ARC). Su Ryeon Seo was a recipient of a fellowship from la Ligue contre le Cancer.
References
- Bitzer M, von Gersdorff G, Liang D, Dominguez-Rosales A, Beg AA, Rojkind M, Bottinger EP (2000) A mechanism of suppression of TGF-β/SMAD signaling by NF-kappa B/RelA. Genes Dev 14: 187–197 [PMC free article] [PubMed] [Google Scholar]
- Bonni S, Wang HR, Causing CG, Kavsak P, Stroschein SL, Luo K, Wrana JL (2001) TGF-β induces assembly of a Smad2-Smurf2 ubiquitin ligase complex that targets SnoN for degradation. Nat Cell Biol 3: 587–595 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Orian A, Schwartz AL (2000) Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays 22: 442–451 [DOI] [PubMed] [Google Scholar]
- Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM (1998) Direct binding of Smad3 and Smad4 to critical TGF-β-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J 17: 3091–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang Y, Feng XH (1998) Smads: transcriptional activators of TGF-β responses. Cell 95: 737–740 [DOI] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, Miyazono K (2001) Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem 276: 12477–12480 [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA Jr, Wrana JL, Falb D (1997) The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell 89: 1165–1173 [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature 390: 465–471 [DOI] [PubMed] [Google Scholar]
- Jonk LJ, Itoh S, Heldin CH, ten Dijke P, Kruijer W (1998) Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-beta, activin, and bone morphogenetic protein-inducible enhancer. J Biol Chem 273: 21145–21152 [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL (2000) Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF β receptor for degradation. Mol Cell 6: 1365–1375 [DOI] [PubMed] [Google Scholar]
- Lin X, Liang M, Feng XH (2000) Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J Biol Chem 275: 36818–36822 [DOI] [PubMed] [Google Scholar]
- Lo RS, Massague J (1999) Ubiquitin-dependent degradation of TGF-β-activated Smad2. Nat Cell Biol 1: 472–478 [DOI] [PubMed] [Google Scholar]
- Massague J, Chen YG (2000) Controlling TGF-β signaling. Genes Dev 14: 627–644 [PubMed] [Google Scholar]
- Mehra A, Wrana JL (2002) TGF-β and the Smad signal transduction pathway. Biochem Cell Biol 80: 605–622 [DOI] [PubMed] [Google Scholar]
- Melhuish TA, Wotton D (2000) The interaction of the carboxyl terminus-binding protein with the Smad corepressor TGIF is disrupted by a holoprosencephaly mutation in TGIF. J Biol Chem 275: 39762–39766 [DOI] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P (1997) Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature 389: 631–635 [DOI] [PubMed] [Google Scholar]
- Pessah M, Prunier C, Marais J, Ferrand N, Mazars A, Lallemand F, Gauthier JM, Atfi A (2001) c-Jun interacts with the corepressor TG-interacting factor (TGIF) to suppress Smad2 transcriptional activity. Proc Natl Acad Sci USA 98: 6198–6203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier C, Ferrand N, Frottier B, Pessah M, Atfi A (2001) Mechanism for mutational inactivation of the tumor suppressor Smad2. Mol Cell Biol 21: 3302–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa L, Doody J, Massague J (1999) Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature 397: 710–713 [DOI] [PubMed] [Google Scholar]
- Whitman M (1998) Smads and early developmental signaling by the TGFβ superfamily. Genes Dev 12: 2445–2462 [DOI] [PubMed] [Google Scholar]
- Wieser R, Wrana JL, Massague J (1995) GS domain mutations that constitutively activate TβR-I, the downstream signaling component in the TGF-β receptor complex. EMBO J 14: 2199–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D, Lo RS, Lee S, Massague J (1999) A Smad transcriptional corepressor. Cell 97: 29–39 [DOI] [PubMed] [Google Scholar]
- Wrana JL (2000) Regulation of Smad activity. Cell 100: 189–192 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R (2001) Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA 98: 974–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400: 687–693 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure


