Abstract
A histone variant H2ABbd was recently identified, but its function is totally unknown. Here we have studied the structural and functional properties of nucleosome and nucleosomal arrays reconstituted with this histone variant. We show that H2ABbd can replace the conventional H2A in the nucleosome, but this replacement results in alterations of the nucleosomal structure. The remodeling complexes SWI/SNF and ACF are unable to mobilize the variant H2ABbd nucleosome. However, SWI/SNF was able to increase restriction enzyme access to the variant nucleosome and assist the transfer of variant H2ABbd–H2B dimer to a tetrameric histone H3–H4 particle. In addition, the p300- and Gal4-VP16-activated transcription appeared to be more efficient for H2ABbd nucleosomal arrays than for conventional H2A arrays. The intriguing mechanisms by which H2ABbd affects both nucleosome remodeling and transcription are discussed.
Keywords: histone transfer, histone variant H2Abbd, nucleosome remodeling, transcription
Introduction
Within the cell nucleus, DNA is packaged into chromatin. The basic unit of chromatin, the nucleosome, is composed of an octamer of the core histones (two each of H2A, H2B, H3 and H4) around which two superhelical turns of DNA are wrapped (van Holde, 1988). The nucleosome represents a barrier to both the accessibility of transcription factor to DNA and the passage of processive enzyme complexes (Beato and Eisfeld, 1997). To overcome the nucleosome barrier, the cell uses different strategies including post-translational histone modifications (Strahl and Allis, 2000), chromatin remodeling complexes (Langst and Becker, 2001) and the insertion of specific histone variants within the histone octamer (Malik and Henikoff, 2003). The first two mechanisms were the object of numerous studies and it is likely that they act in concert (Mizuguchi et al, 2001). The third mechanism is still poorly understood (Malik and Henikoff, 2003).
Different families of chromatin remodelers have been identified and isolated (for a review, see Becker, 2002). The chromatin remodelers are multiprotein assemblies, which contain an ATPase subunit, which is essential for their function. SWI/SNF is a chromatin remodeler able to both perturb the histone–DNA interaction and to mobilize the histone octamer (Peterson and Workman, 2000). ACF, an ISWI-containing remodeler, assists histone octamer sliding in an ATP-dependent manner (Ito et al, 1997). Acetylation of the tails of the core histones is directly associated with transcriptional regulation (reviewed in Strahl and Allis, 2000). In vitro and in vivo studies have shown that the HAT activity of the coactivators p300 and CBP is essential for the stimulation of transcription (Martinez-Balbas et al, 1998).
Histone variants are nonallelic isoforms of the conventional histones and they are present in the cell in a very low amount compared to their conventional counterparts (Tsanev et al, 1993). The primary structure of the histone variants shows a various degree of homology with the corresponding conventional histone (Malik and Henikoff, 2003). Incorporation of histone variants into the nucleosomes could result in the formation of nucleosome particles with different organization and novel functional properties. A typical example is the variant nucleosome containing the histone variant macroH2A (mH2A nucleosome). The mH2A nucleosomes exhibit an altered structure that interferes with both the binding of a transcription factor and its remodeling by the chromatin remodeling complexes SWI/SNF (Angelov et al, 2003).
Over the last few years, other histone variants have been shown to bring new properties to chromatin and participate in the regulation of gene expression. For example, the phosphorylation of H2AX is essential for maintaining genomic stability (Celeste et al, 2003). The histone variant H2A.Z is believed to participate in both gene activation (Santisteban et al, 2000) and gene silencing (Dhillon and Kamakaka, 2000). The histone variant H3.3, which differed at only four amino acids positions from the conventional H3, is involved in the replication-independent nucleosome assembly and marks active chromatin (Ahmad and Henikoff, 2002).
Recently, a novel histone variant, termed H2ABbd (Barr body deficient), was described (Chadwick and Willard, 2001). H2ABbd localizes to the nucleus and is excluded from the Barr body in interphase nuclei and from the inactive X chromosomes at metaphase. An overlapping between the nucleus distribution of H2ABbd and acetylated histone H4 was reported (Chadwick and Willard, 2001). H2ABbd shows a 48% identity to the conventional H2A and it is considerably shorter than the other proteins of the H2A family. Importantly, the residues that are targets in H2A for numerous post-translational modifications including acetylation, phosphorylation and ubiquitination are not conserved in H2ABbd. This suggests that H2ABbd could be regulated in a different fashion compared to the other members of the H2A family. This regulation as well as the function of H2ABbd is, however, completely unknown.
In this work, we have studied the structure of the nucleosomes containing H2ABbd and the functional consequences of the incorporation of this variant histone into the nucleosome for the interaction of transcription factor, the mobilization and remodeling of the variant nucleosome and the transcriptional capacity of H2ABbd nucleosomal templates. We have found that the H2ABbd nucleosome exhibits new properties different from those of conventional nucleosomes. The data suggest a distinct role of H2ABbd in both chromatin remodeling and transcriptional regulation.
Results
Presence of H2ABbd results in alterations of the nucleosome structure
The peculiar characteristics of the histone H2ABbd suggest that the variant H2ABbd nucleosome may exhibit a noncanonical structure. To test this, we have cloned, expressed and purified to homogeneity H2ABbd (Figure 1A, lane 2), as well as the other histones (lanes 1–5). An equimolar amount of this histone variant and of the conventional recombinant histones H2B, H3 and H4 (lane 7) were used to reconstitute nucleosomes on a 152 bp DNA fragment containing the Xenopus borealis somatic 5S RNA gene. In parallel, nucleosomes were also reconstituted on the same fragment of DNA, but using the conventional H2A (lane 6) and the other three remaining core histones. The electrophoretic mobility shift assay (EMSA) shows that the two proteins, conventional H2A and H2ABbd, are efficiently incorporated into the nucleosome: no or very little free DNA was detected on the gel (Figure 1B, lanes 1 and 2). These results are in agreement with the available data (Chadwick and Willard, 2001).
Figure 1.
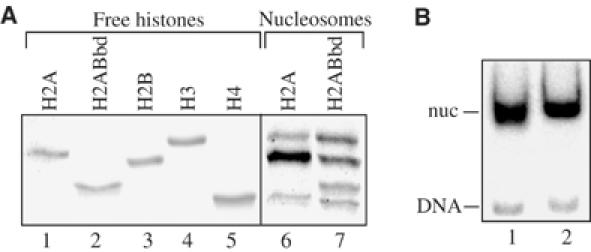
Reconstitution of conventional and H2ABbd nucleosomes. An equimolar mixture of recombinant core histones H2B, H3, H4 and either conventional H2A or H2ABbd and a 32P-end labeled 152 bp EcoRI–RsaI fragment comprising an X. borealis somatic 5S DNA gene was used for nucleosome reconstitution. (A) An 18% SDS–PAGE of the purified recombinant core histones (free histones) and the histone mixtures used for conventional H2A and H2ABbd nucleosome reconstitution (nucleosomes). Xenopus laevis H2A and H2B histones comigrate in this type of gel. (B) EMSA of the reconstituted conventional H2A (lane 1) and H2ABbd (lane 2) nucleosomes. The position of the free DNA (DNA) and that of the reconstituted nucleosomes (nuc) are indicated.
The 152 bp 5S DNA fragment contains a strong positioning signal, which allows the reconstitution of precisely positioned and well-defined nucleosome particles (Thiriet and Hayes, 1998). The DNA organization within such conventional and variant nucleosomes can be visualized at 1 bp resolution by footprinting techniques. A combination of hydroxyl radical and DNase I footprinting was used to study the structural consequences of the incorporation of the histone variant H2ABbd into the reconstituted nucleosomes (Figure 2). The two types of reconstituted nucleosomes exhibit identical hydroxyl radical cleavage pattern with peaks separated by 10 bp intervals (Figure 2A, lanes 3 and 4, and Figure 2B, lanes 3 and 4). This evidences for a lack of steric hindrance to the hydroxyl radical and a wrapping of the nucleosomal DNA around the histone octamer in the reconstituted nucleosomes (Hayes and Lee, 1997). The DNase I footprinting pattern was, however, different for the two types of particles (Figure 2). Indeed, significant alterations were detected in both the top (Figure 2A, compare lanes 1 and 2) and bottom (Figure 2B, compare lanes 1 and 2) strands of the H2ABbd nucleosome. These alterations are observed at different positions along the nucleosomal DNA. Interestingly, a very pronounced modification in the accessibility of DNase I is detected around the dyad axis of the bottom strand of the variant H2ABbd particle (Figure 2B, lane 2).
Figure 2.
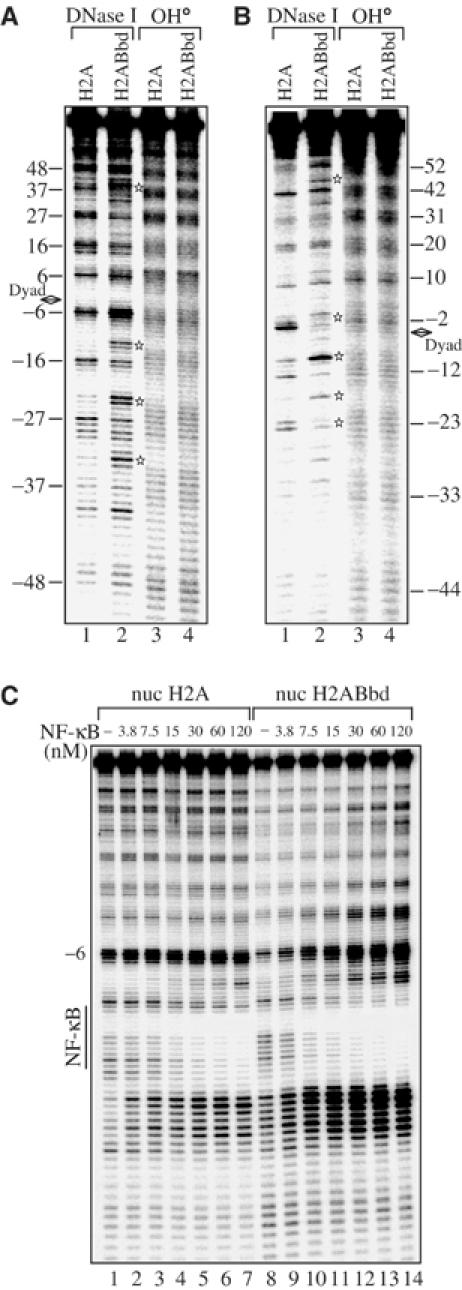
The presence of H2ABbd induces alterations in the structure of the H2ABbd nucleosome and increases the accessibility of NF-κB to nucleosomal DNA. (A) Reconstituted H2A or H2ABbd nucleosomes were treated with either DNase I (lanes 1 and 2) or hydroxyl radical (OH·, lanes 3 and 4) and the digestion pattern was analyzed on an 8% sequencing gel. The bottom strand of the nucleosomal DNA was 32P-end labeled. (B) Same as (A), except for 5′-end-labeled DNA (top strand). The thick black arrow shows the nucleosome dyad axis. The stars designate alterations in the nucleosome structure. (C) Nucleosomes were reconstituted using conventional H2A or H2ABbd proteins and a 152 bp fragment comprising the 5S DNA X. borealis gene with inserted recognition sequence of the transcription factor NF-κB in the vicinity of the dyad axis. An increasing amount of NF-κB (as indicated in the figure) was allowed to interact with the reconstituted nucleosomes and the binding was assessed by DNase I footprinting. The bottom strand of DNA was labeled. The vertical lines indicate the binding sequence of NF-κB.
Remodeling of variant H2ABbd nucleosomes by SWI/SNF
The footprinting data show alterations in the structure of H2ABbd. Analogous alterations were found for the macroH2A nucleosomes, which were intimately related with the lack of transcription factors accessibility to macroH2A nucleosomes and the inability of SWI/SNF to remodel such variant nucleosomes (Angelov et al, 2003). To test if this is also the case for the H2ABbd nucleosomes, the following experiments were carried out. Within the 152 bp DNA fragment comprising the sequence of the X. borealis 5S gene, a single recognition site for the transcription factor NF-κB was inserted close to the dyad axis at positions −16 to −26 (Angelov et al, 2003), where the major alterations of the H2ABbd nucleosomal structure were observed. This fragment was used to reconstitute conventional and H2ABbd nucleosomes, and the binding of NF-κB to these two types of nucleosomes was studied by DNase I footprinting (Figure 2C). To determine precisely the efficiency of binding of NF-κB to H2ABbd compared to conventional nucleosomes, the DNase I footprinting was performed after the interaction of increasing amounts of NF-κB with the nucleosomal templates. A very clear footprinting of NF-κB was detected for both nucleosomes. The titration experiment indicates a slight preference of NF-κB for the H2ABbd nucleosomes compared to H2A nucleosomes (not more than two-fold). Thus, in contrast to macroH2A, the presence of H2ABbd does not impede the binding of NF-κB to the variant nucleosomes but instead seems to favor the access of the transcription factor to its nucleosomal binding site.
The binding of the remodeling complex SWI/SNF and of the ATPase ISWI subunit were also not affected by the presence of the histone variant H2ABbd (Figure 3A and B). Indeed, the EMSA experiments show that incubation of the conventional and H2ABbd nucleosomes with increasing amounts of SWI/SNF or ISWI results in a shift of the nucleosomal particles with similar efficiency (Figure 3A and B). Importantly, the ATPase activity of SWI/SNF in the presence of each type of nucleosome was essentially the same (Figure 3C). Only a very slight decrease (not exceeding 5–10%) of the SWI/SNF ATPase activity was observed in the presence of H2ABbd nucleosomes (Figure 3C). The incorporation of conventional H2A and H2ABbd histones into the histone octamer has, however, a completely different effect on the ability of SWI/SNF to remodel nucleosomes (Figure 3D). Incubation of the conventional nucleosomes with increasing amounts of SWI/SNF in the presence of ATP results in an efficient remodeling (Figure 3D, compare lane 1 with lanes 2–4). Indeed, the addition of a low amount (0.8 μl) of SWI/SNF was sufficient to alter the pattern of DNase I digestion (lanes 2–4). In contrast, SWI/SNF was unable to efficiently remodel H2ABbd nucleosomes. Although some sites of hypersensitivity become visible in the presence of high amounts of SWI/SNF (compare lane 8 with 5), the clear 10 bp DNase I cleavage pattern observed in the absence of the remodeler was unaffected (Figure 3D, compare lane 5 with 8).
Figure 3.
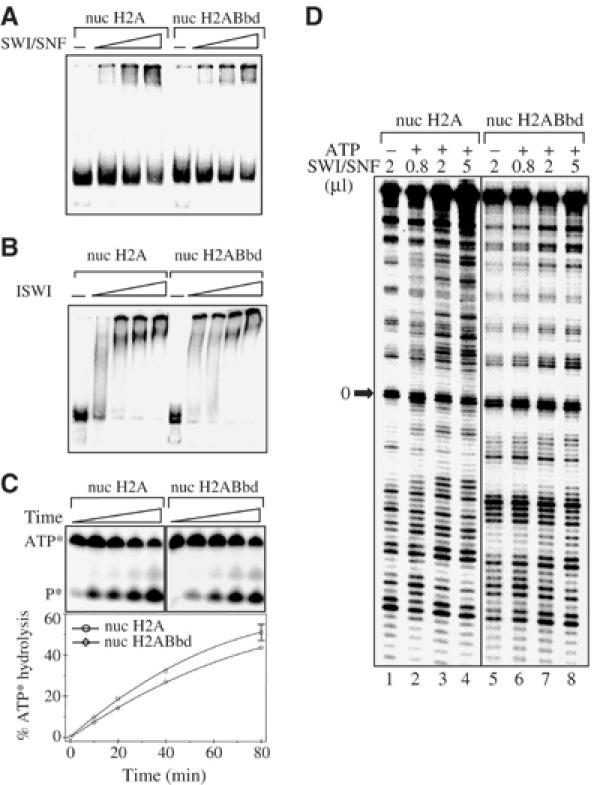
SWI/SNF is unable to remodel H2ABbd nucleosomes. (A) SWI/SNF efficiently binds H2ABbd nucleosomes. The nucleosomes were reconstituted on the 152 bp fragment, comprising the X. borealis 5S DNA gene. An increasing amount of SWI/SNF was added to either conventional H2A or H2ABbd nucleosomes in a buffered solution containing 1 mM ATP and 2.5 mM MgCl2. After incubation for 10 min at 30°C, the reactions were analyzed by EMSA. (B) ISWI binding to the H2A and H2ABbd nucleosomes. Same as (A), except that the end-positioned 241 bp 601 nucleosomes and ISWI were used. (C) H2ABbd does not interfere with the ATPase activity of SWI/SNF. The kinetics of the SWI/SNF ATP hydrolysis were analyzed on 15% denaturing polyacrylamide gels in the presence of conventional and H2ABbd nucleosomes. The positions of ATP and the released phosphate are designated (upper panel). The lower panel shows the quantification of the ATP hydrolysis. (D) Remodeling of conventional H2A and H2ABbd particles. Increasing amounts of SWI/SNF were added to either conventional or H2ABbd nuclesomes with a 32P-end–labeled DNA (the bottom strand) in a buffered solution containing ATP and the reaction mixtures were incubated for 45 min at 30°C. The remodeling of conventional H2A (lanes 2–4) and H2ABbd (lanes 6–8) was visualized by DNase I footprinting. Lanes 1 (conventional nucleosomes) and 5 (H2ABbd nucleosomes) show the DNase I digestion pattern of the control reactions carried out in the absence of ATP.
The nucleosome mobilization experiments further confirmed this conclusion (Figure 4). These experiments were first carried out with conventional and H2ABbd nucleosomes reconstituted on a 248 bp mouse rDNA fragment (Langst et al, 1999). Since SWI/SNF promotes histone octamer sliding from the center to the end of DNA (Becker, 2002), in order to study this process one needs only centrally positioned nucleosomes. Centrally positioned conventional and H2ABbd nucleosomes were isolated and incubated with increasing amounts of SWI/SNF in the presence of ATP and the mobilization of the octamers was assessed on a 5.5% native polyacrylamide gel (Figure 4A). A very clear SWI/SNF-dependent sliding is observed for conventional nucleosomes (Figure 4A, lanes 1–5), whereas SWI/SNF was unable to induce mobilization of the H2ABbd octamers, since even at the highest amount of SWI/SNF no band corresponding to the end-positioned nucleosome was detected (Figure 4A, compare lane 6 with 10). To determine if this lack of nucleosome sliding was dependent on the DNA sequence used, we repeated these experiments with the 601 positioning sequence (Figure 4B) (Lowary and Widom, 1998). Reconstitution of conventional nucleosomes on this sequence gives rise to centrally positioned nucleosomes (Kagalwala et al, 2004). Incubation of the 601 reconstituted nucleosomes (nuc H2A and nuc H2ABbd) with increasing amounts of SWI/SNF clearly indicates that conventional H2A nucleosomes slide efficiently as expected (Kagalwala et al, 2004), whereas H2ABbd nucleosomes are not mobilized (Figure 4B, lanes 6–10).
Figure 4.
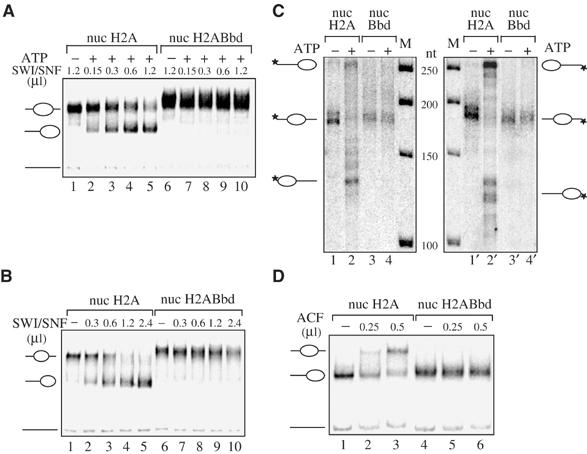
SWI/SNF and ACF mobilization of conventional H2A and variant H2ABbd nucleosomes. (A) A 248 bp mouse rDNA fragment was used for reconstitution of the two types of nucleosomes. After gel purification, conventional and variant H2ABbd nucleosomes that are centrally positioned were incubated for 45 min at 30°C with increasing amounts of SWI/SNF in the presence of ATP and the sliding of the histone octamers was assessed on native 5.5% polyacrylamide gels. The center- and end-positioned nucleosomes and free DNA are indicated on the left side of the figure. (B) Same as (A), except that nucleosomes were reconstituted on the 601 DNA fragment (Kagalwala et al, 2004). (C) Exonuclease III digestion of conventional and variant nucleosomes reconstituted on the 601 DNA templates in the presence or absence of active SWI/SNF as indicated in the figure. Mapping on the top strand (lanes 1–4) and bottom strand (lanes 1′–4′) is shown. (D) ACF mobilization of H2A and H2ABbd nucleosomes reconstituted on an end-positioned 601 DNA sequence (Kagalwala et al, 2004). End-positioned nucleosomes were incubated with increasing amounts of ACF for 45 min at 30°C in the presence of ATP, and the sliding was assessed on a 5.5% polyacrylamide gel.
One possible explanation of the absence of sliding of H2ABbd nucleosomes in the presence of SWI/SNF might be that these variant nucleosomes reconstitute preferentially on the 601 DNA fragment in an end position. To determine the starting and final positions of the H2A and H2ABbd nucleosomes, we performed exonuclease III digestion mapping of these particles before and after SWI/SNF remodeling (Figure 4C). These data clearly indicate that the starting central position is the same for H2A and H2ABbd nucleosomes (compare lanes 1 and 3, or 1′ and 3′). After SWI/SNF remodeling of the H2A nucleosomes, the protection from exonuclease digestion indicates that conventional nucleosomes move as expected from the central to the end positions (lanes 2 and 2′). In contrast, H2ABbd nucleosomes do not move after incubation with SWI/SNF (lanes 4 and 4′). To further characterize the mobility properties of nuc H2ABbd nucleosomes, we used the ACF remodeling factor, which promotes histone octamer sliding from the end to the center of DNA (Eberharter et al, 2001) (Figure 4D). Addition of an increasing amount of ACF moves efficiently conventional nucleosomes (lanes 1–3), whereas H2ABbd nucleosomes are not mobilized (lane 4–6). Altogether, these experiments show that independently of the DNA sequence used and of the starting position of the nucleosomes, H2ABbd nucleosomes cannot be efficiently mobilized by the SWI/SNF and ACF machineries.
SWI/SNF-dependent transfer of histone H2ABbd–H2B dimers
Recently, it was reported that the remodeling complexes RSC and SWI/SNF, but not ISWI, were able to induce transfer of conventional H2A–H2B dimers to H3–H4 tetrameric particles only from remodeled nucleosomes (Bruno et al, 2003). Since SWI/SNF was unable to mobilize the H2ABbd nucleosomes, it was of interest to study the capacity of this remodeler to promote transfer of the variant H2ABbd–H2B dimer (Figure 5). To this end, we have reconstituted conventional H2A and H2ABbd nucleosomes onto a 205 bp DNA fragment containing the X. borealis 5S RNA gene by using radiolabeled H2B and the remaining nonlabeled histones. In addition, we have reconstituted H3–H4 tetramer particles on the 147 bp 5S DNA fragment using nonlabeled histones H3 and H4. Centrally positioned, labeled H2B nucleosomes were used in the transfer experiments. These nucleosomes were mixed with the nonlabeled tetramers in a buffered solution containing ATP and SWI/SNF, and the transfer of the labeled dimers to the H3–H4 tetramer particles was carried out at 23°C. Under these conditions, the incubation of particles containing radioactively labeled H3 in the presence of tetrameric particles shows no SWI–SNF-dependent transfer of label (Figure 5A, compare lane 9 with 10), demonstrating a lack of transfer of the H3–H4 tetramer. In the absence of SWI/SNF, only a small amount (not exceeding 2%) of conventional H2A–H2B dimer is transferred (Figure 5A, compare lane 1 with 3). In contrast, for the H2ABbd–H2B dimer, a significant transfer (>5%) to the unlabeled H3–H4 tetramer is observed (Figure 5A, compare lanes 5 and 7). The kinetics of the transfer of the heterotypic conventional and variant dimers support this observation (Figure 5B, compare lanes 1–5 of H2A top panel with lanes 1–5 of Bbd bottom panel). In the absence of SWI/SNF, the amount of transferred conventional H2A–H2B dimers increases with time but with a transfer efficiency about two to three times smaller than that of the variant H2ABbd dimers (Figure 5B, see the quantification of the data). Both these equilibrium and kinetics data suggest that within the nucleosome particle, the H2ABbd–H2B dimer exhibits a weaker interaction with the H3–H4 tetramer compared to that of the conventional H2A–H2B one, which allows a more efficient spontaneous transfer of the variant H2ABbd–H2B dimers.
Figure 5.
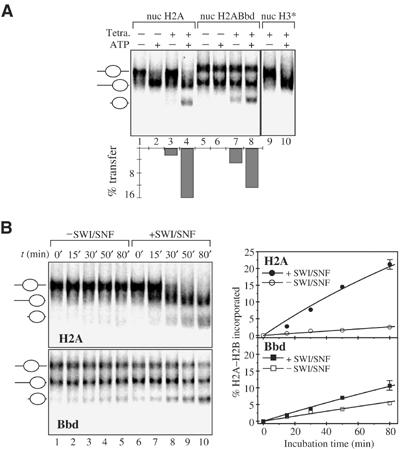
Spontaneous and SWI/SNF-dependent transfer of conventional H2A–H2B and variant H2ABbd–H2B dimers. H2B was radioactively labeled and used to reconstitute conventional and H2ABbd nucleosomes on an unlabeled 205 bp fragment derived from plasmid pXp10 containing the X. borealis 5S DNA gene. On this sequence, the conventional octamer is essentially centrally positioned (panel A, lane 1) while the variant H2ABbd octamer is both end and centrally positioned (panel A, lane 5). Histone H3–H4 tetrameric particles were reconstituted using the 152 bp fragment containing the X. borealis 5S gene. For the histone H3–H4 tetramer transfer studies (lanes 9 and 10), histone H3, instead of histone H2B, was radioactively labeled. (A) H2ABbd–H2B dimers exhibit higher spontaneous and lower SWI/SNF-dependent transfer compared to that of the H2A–H2B dimers. Conventional (lanes 1–4) and H2ABbd (lanes 5–8) nucleosomes reconstituted on the 205 bp 5S DNA fragment were incubated for 60 min at 23°C in the presence or absence of a 2.6-fold molar excess of tetrameric particles. An equal amount of SWI/SNF was present in all reactions. In the presence of ATP, the conventional (lane 2) but not the H2ABbd nucleosomes are mobilized (lane 6). A smaller amount of H2A–H2B (not exceeding 2%, lane 3) compared to that of variant H2A–H2B (∼5%, lane 7) is spontaneously transferred in the absence of ATP to the tetrameric particles. The total (the sum of spontaneous and SWI/SNF-dependent) amount of conventional heterotypic dimers (lane 4) and variant H2ABbd–H2B dimers (lane 8) differed slightly, however. The quantification of the data is shown on the lower part of the panel. The incubation of particles containing radioactively labeled H3 in the presence of tetrameric particles shows no transfer of the label (compare lane 9 with 10), demonstrating a lack of transfer of the H3–H4 tetramer. (B) Kinetics of spontaneous and SWI/SNF-dependent transfer of H2A–H2B and H2ABbd–H2B dimers. A 20 μl portion of conventional or H2ABbd nucleosome reconstituted with radioactively labeled H2B on the 205 bp DNA fragment was incubated at 23°C in the presence or absence of SWI/SNF. A 2.6-fold molar excess of the acceptor 152 bp H3–H4 tetrameric particle and 1 mM ATP were present in all reactions. After incubation for the indicated times, the reactions were stopped and the material was used for EMSA. The quantification of the data is presented on the right part of the figure. Each point of the curves representing the SWI/SNF-dependent transfer was obtained by subtracting the value of the spontaneous transfer from the respective value of the measured total (the sum of spontaneous and SWI/SNF-dependent) transfer of the heterotypic dimers.
The presence of SWI/SNF mobilizes the conventional nucleosome and induces an additional (about 10-fold) SWI–SNF-dependent high-efficiency transfer of the H2A–H2B dimer to the tetramer particle (Figure 5A, compare lane 3 with 4). This is in agreement with the recent report of Bruno et al (2003). In contrast, the presence of SWI/SNF increases the transfer of H2ABbd–H2B to the H3–H4 tetramer particle only by about 2.5-fold (Figure 5A, lanes 7 and 8). The kinetics data presented in Figure 5B support these results. The SWI/SNF-dependent efficiency for the transfer of the H2ABbd–H2B dimer was found to be about three- to four-fold smaller than that of the conventional H2A–H2B dimer (compare lanes 6–10 of the top panel H2A with lanes 6–10 of the bottom panel Bbd and see the quantification panel). This evidences that SWI/SNF, which cannot remodel detectably the variant nucleosome, is however able to induce a low-efficiency transfer of the H2ABbd–H2B dimer to the tetrameric H3–H4 particle. This indicates that SWI/SNF has some destabilization effects on the H2ABbd nucleosome, which promote the transfer of the H2ABbd–H2B dimer.
Evidence for a destabilization effect of SWI/SNF on H2ABbd nucleosomes can be obtained by assessing the accessibility of nucleosomal DNA by restriction enzyme (Sengupta et al, 2001) as described in Figure 6A. Conventional and H2ABbd nucleosomes reconstituted on a labeled 152 bp 5S DNA fragment were incubated in the presence or absence of SWI/SNF and ATP for 45 min, and then ATP was depleted in some reactions before the addition of the restriction enzyme EcoRV, so that transient or more stable changes causing access to nucleosomal DNA could be detected (Figure 6B and C). The EcoRV restriction cut site at +33 (109 bp from the 3′-labeled bottom strand) is located within the nucleosome-bound region of DNA. The small amount of DNA cut within the first minute after the addition of EcoRV corresponds to the free or incorrectly packed DNA present in the sample (lane 2) and has been subtracted from the normalized data. The time course of the incubation of H2ABbd and conventional nucleosomes with EcoRV shows that the DNA from the variant nucleosome is more accessible, since EcoRV cleaves the H2ABbd nucleosome with a better efficiency (two-fold) (Figure 6C). Analysis of EcoRV cleavage after SWI/SNF remodeling (lanes 7–11) shows that the accessibility of the DNA from the H2A nucleosomes is significantly enhanced, whereas the accessibility of DNA from the variant H2ABbd nucleosome is very similar to the control, that is, incubation of H2ABbd nucleosome with SWI/SNF does not significantly enhance the accessibility of the DNA. These stationary data are in complete agreement with the results of the DNase I footprinting analysis shown in Figure 3. In contrast, when the accessibility of both nucleosomes is assessed during the remodeling process (lanes 12–16), a significant increase of DNA accessibility is observed for both nucleosomes (about a four-fold increase). These data indicate that SWI/SNF is able to induce transient destabilization effects on the H2ABbd nucleosomes, which can be detected by the restriction enzyme assay. This destabilization could explain the transfer of the H2ABbd–H2B dimer to the H3–H4 tetrameric particle induced by SWI/SNF (Figure 5).
Figure 6.
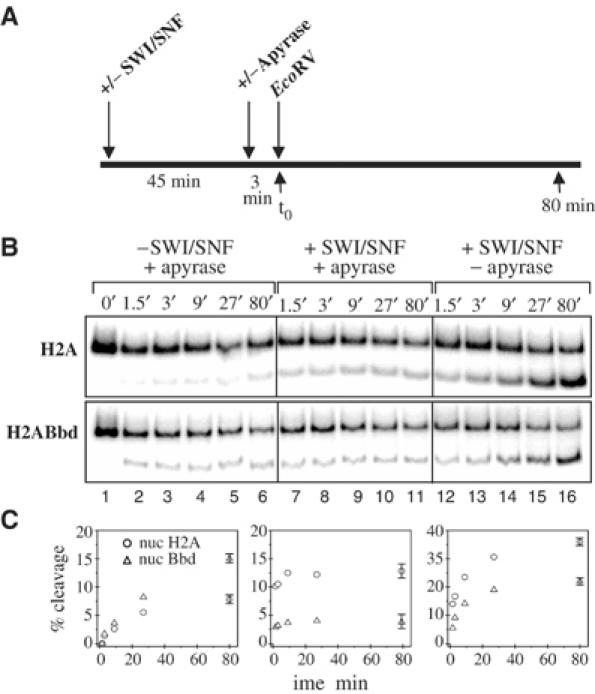
Restriction enzyme assay of DNA accessibility in H2A and H2ABbd nucleosomes. (A) Scheme of the experiment. Nucleosomes were preincubated for 45 min in the presence or absence of SWI/SNF, and then some reactions were depleted of ATP by apyrase treatment before the addition of EcoRV. Samples were taken at different time points, and then DNA was extracted and analyzed on an 8% native PAGE. (B) Nucleosomes were reconstituted on a 152 bp 5S DNA fragment using the H2A or H2ABbd histones and were incubated with EcoRV in the presence or absence of SWI/SNF or apyrase as indicated in the figure. (C) Quantification of the data. The DNA cleaved within the first minute was likely to represent free DNA (Sengupta et al, 2001) (lane 2) and was therefore subtracted from all other time points. To quantify and represent in the figure the effect of SWI/SNF on the efficiency of DNA accessibility by EcoRV, the amount of nucleosomal DNA cleaved in the absence of SWI/SNF (lanes 3–6) was subtracted from the amount of nucleosomal DNA cleaved in the presence of the remodeler (lanes 7–16).
Effect of H2ABbd on the p300-dependent transcription and histone acetylation of nucleosome arrays
In order to shed further light on the functional consequence of the incorporation of H2ABbd in the nucleosome, we have analyzed the p300-dependent transcription and histone acetylation of chromatin templates. Chromatin was assembled on a pG5ML array plasmid according to the procedure of Ito et al (1999). Drosophila recombinant Acf1, ISWI, nucleosome assembly protein-1 (NAP-1), the H2B, H3 and H4 histones and either the variant H2ABbd or the conventional H2A histone were used for the assembly. The reconstituted chromatin was analyzed by microccocal nuclease digestion (Figure 7A). The nucleosome digestion pattern of chromatin templates assembled with conventional H2A and variant H2ABbd histones shows a clear 200 bp repeat, evidencing for a proper structural organization (Figure 7A, lanes 2–5). These two chromatin templates were further used in transcription assays. Basal (Gal4-VP16 independent) transcription was not detected for both templates (data not shown). The addition of Gal4-VP16 to the reactions results in both cases in small, but detectable, transcription (Figure 7B, lanes 1 and 3). The presence of p300 leads to a dramatic increase in the transcription from the conventional and variant chromatin templates (Figure 7B, compare lane 1 with 2, and 3 with 4). Interestingly, the transcription efficiency from the H2ABbd templates was found to be slightly higher than that of the conventional H2A templates (compare lane 2 with 4). This moderate effect (not exceeding two-fold) was reproducible in the whole set of experiments carried out.
Figure 7.
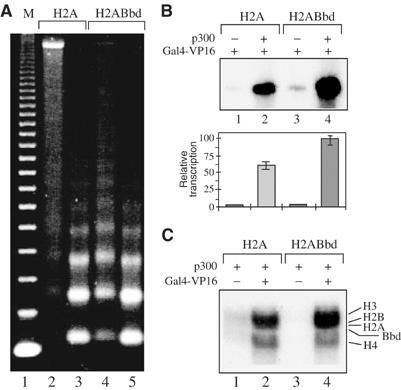
p300- and Gal4-VP16-dependent transcription from H2ABbd chromatin templates. (A) Micrococcal nuclease (MNase) digestion of the assembled chromatin. Chromatin was assembled on the pG5ML array templates using recombinant Drosophila Acf1, ISWI and NAP-1 and either conventional H2A (lanes 2 and 3) or variant H2ABbd (lanes 4 and 5) and the three remaining core histones H2B, H3 and H4. The assembled chromatin was digested with 0.1 mU MNase (lanes 2 and 4) or with 0.5 mU MNase (lanes 3 and 5) for 10 min at 22°C. Then, DNA was extracted and analyzed on a 1.2% agarose gel. Lane 1, a 123 bp DNA ladder marker. (B) Transcription from chromatin templates assembled with conventional H2A (lanes 1 and 2) or H2ABbd (lanes 3 and 4). Templates were incubated with Gal4-VP16 alone or together with p300 as indicated. The lower panel shows the quantification of the data from three independent experiments. (C) Differential effect of H2ABbd on the p300-mediated histone acetylation in the assembled nucleosomal arrays. HAT assays with chromatin templates reconstituted with conventional H2A (lanes 1 and 2) or with the variant histone H2ABbd (lanes 3 and 4). All reactions contained p300. Reactions 2 and 4, in addition to p300, also contained Gal4-VP16.
The histone NH2-terminal tails play a selective, acetylation-dependent role in transcriptional activation by p300 (An et al, 2002). Since the changes in the status of acetylation of the NH2-termini are associated with transcriptional activation (An et al, 2002), we have studied the acetylation level of both conventional H2A and H2ABbd variant chromatin templates induced by p300. In the absence of Gal4-VP16, very faint acetylation of the histones is observed (Figure 7C, lanes 1 and 3). The addition of Gal4-VP16 results in a drastic increase in the level of histone acetylation within both templates (Figure 7C, lanes 2 and 4). Interestingly, this increase in the acetylation histone level is higher for the H2ABbd chromatin (Figure 7C, compare lane 2 with 4). Thus, the incorporation of H2ABbd into the nucleosomes leads to better efficiency of acetylation of the histone tails by p300.
The moderate stimulatory effect of H2ABbd on transcription may be either due to higher initiation of transcription or easier elongation (more efficient passage of RNA polymerase II through the H2ABbd nucleosome) or both. To differentiate between these possibilities, we have studied the efficiency of transcription through conventional and H2ABbd nucleosomes (Figure 8). These templates were transcribed using Pol II elongation complexes immobilized on beads and ligated to the respective mononucleosomes (Kireeva et al, 2002). The nascent RNA was visualized by pulse labeling. The transcription reactions were carried out at three different concentrations of KCl: 40, 300 and 800 mM (Kireeva et al, 2002). In agreement with our earlier observations (Kireeva et al, 2002), approximately 20% of the templates (roughly corresponding to the amount of free DNA present in the nucleosomes; Figure 8A) were transcribed at 40 mM KCl to completion (Figure 8B, lanes 7 and 11). Strong nucleosome-specific pausing was apparent during transcription at 40 mM KCl. Thus, both the conventional and the variant nucleosomes represent a strong barrier to the passage of the polymerase at 40 mM KCl. Increasing the ionic strength to 300 and 800 mM KCl results in some nucleosome destabilization and, as expected (Kireeva et al, 2002), in a much higher efficiency of transcription, very similar for the conventional and variant nucleosomes (Figure 8B, lanes 8 and 9, and 12 and 13). The data suggest that the incorporation of H2ABbd within the nucleosome has no detectable effect on the rate of passage of the Pol II through it. We conclude that the moderate stimulatory effect of H2ABbd on transcriptional activation is likely to be exerted at the level of initiation of transcription.
Figure 8.
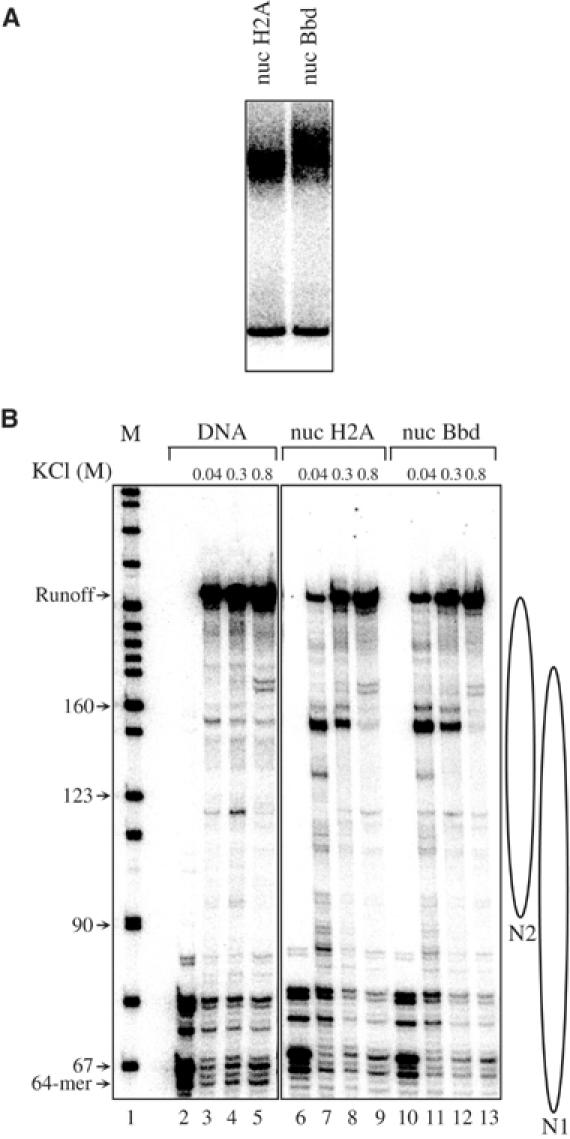
Transcription through conventional and H2ABbd nucleosomes. Nucleosomal transcription templates containing either H2A or H2ABbd were reconstituted on a 245 bp DNA fragment (Kireeva et al, 2002). The templates are a mixed population of two positioned mononucleosomes (N1 and N2). The nucleosomal templates were ligated to Pol II elongation complex immobilized on beads and the RNA was pulse labeled by the ‘walking' RNA polymerase. The transcription was carried out in the presence of either 40, 300 or 800 mM NaCl. Then, RNA was extracted and separated on an 8% denaturing polyacrylamide gel. (A) EMSA of the reconstituted conventional (lane 1) and H2ABbd (lane 2) nucleosomes. (B) Pol II transcribes conventional and H2ABbd nucleosomal templates with similar efficiency. Denaturing PAGE of the RNA extracted from the transcription reactions performed with naked DNA (lanes 3–5), conventional (lanes 7–9) and variant H2ABbd (lanes 11–13) nucleosomal templates. Lanes 2, 6 and 10 show the transcription from the preformed stalled elongation complexes. A marker for the molecular mass of the transcripts is shown on the left site of the figure. The arrows indicate the length of the transcripts. The positions of the N1 and N2 nucleosomes are shown on the right part of the figure.
Discussion
This work is focused on the effect of histone variant H2ABbd on the structural and functional properties of the H2ABbd nucleosomes. It is shown that H2ABbd can replace the conventional H2A within the nucleosomal particle. The variant H2ABbd nucleosome exhibits, however, numerous structural perturbations compared to the conventional nucleosome: clear alterations in the DNase I footprinting pattern were observed all along the H2ABbd nucleosomal DNA. In addition, the H2ABbd nucleosome shows an easier spontaneous transfer of the H2ABbd–H2B dimer to the H3–H4 tetramer particle, testifying for disturbed interactions of this variant dimer with the histone H3–H4 tetramer within the H2ABbd nucleosome. All these data suggest a weaker thermodynamic stability of H2ABbd nucleosome. Interestingly, SWI/SNF was unable to induce detectable mobilization of the variant particle. This could be viewed as an example of a lack of a direct relationship between the stability of a nucleosome and the capacity of a remodeling machine to stably remodel it.
The equilibrium and the kinetics data demonstrate that the presence of SWI/SNF induces a clear increase of the transfer of both the conventional H2A–H2B and the variant H2ABbd–H2B dimers. This effect was found to be three- to four-fold higher for the conventional H2A–H2B dimer, which, as suggested by Bruno et al (2003), could be associated with the remodeling of the particle by SWI/SNF. Although the H2ABbd nucleosome cannot be mobilized by SWI–SNF (Figures 3, 4 and 5), transfer of the dimer H2ABbd–H2B to H3–H4 tetramer templates can be stimulated by SWI–SNF (Figure 5). To explain this apparent discrepancy, we should keep in mind that SWI–SNF interacts with the H2ABbd nucleosomes with the same efficiency as that for the canonical nucleosome (Figure 3A). This interaction might be sufficient for the generation of some transient structural alteration sufficient to further destabilize the H2ABbd–H2B contacts with the H3–H4 tetramer. The half-life of the destabilization state being very small, the destabilization can be observed only in the presence of the competitor H3–H4 tetrameric particle, which in this case acts as a trap for the destabilization state and allows its detection. This hypothesis is in complete agreement with the EcoRV enzyme restriction accessibility assay (Figure 6). Indeed, the data from the EcoRV enzyme restriction assay clearly demonstrate that during the remodeling process (in the presence of SWI/SNF and ATP) the accessibility of nucleosomal DNA for EcoRV is increased (Figure 6). Therefore, H2ABbd nucleosome variants possess new properties, which allow to disconnect effects of remodeling complexes on mobilization and on transfer or access to restriction enzymes.
The p300 transcriptional assay reflects the specific structural properties of the H2ABbd nucleosome. Indeed, H2ABbd exerted a moderate stimulatory effect on the p300-dependent transcriptional activation. This stimulatory effect seems to be the consequence of the level of transcriptional initiation rather than transcriptional elongation, since the measured efficiency of the passage of Pol II through the H2ABbd nucleosome was found to be identical to that of the conventional H2A nucleosome (Figure 8). This H2ABbd effect on transcription is surprising, since the H2ABbd chromatin template could not be mobilized by such a strong remodeler as SWI/SNF. This lack of H2ABbd nucleosome mobilization would intuitively suggest that the presence of H2ABbd might negatively interfere with transcription and not stimulate it. Then, how can the stimulatory effect of H2Abbd be explained? Different hypotheses could be proposed. First, the nuclear extract used for the transcription assay may contain a faint amount of a remodeling activity specific for H2ABbd nucleosomes. Such activity, although present in low amounts, may strongly remodel some H2ABbd template promoters and render them completely accessible to the transcriptional machinery, thus allowing an efficient transcription. This suggestion seems plausible since the existence of a remodeler that was specific for the histone variant H2A.Z and was able to catalyze the exchange of the histone variant H2A.Z was recently demonstrated (Mizuguchi et al, 2004). A second scenario could be proposed in light of the enhanced histone acetylation by p300 in the presence of H2ABbd nucleosomes. The H2ABbd noncanonical structure of nucleosomes may allow a better accessibility of p300 to the histone tails, which would result in the observed in vitro-dependent transcriptional activation. In this context, it is worthwhile to recall that H2ABbd was detected in vivo mainly in chromatin regions containing hyperacetylated histones (Chadwick and Willard, 2001). Alternatively, a better accessibility of a transcription factor, as suggested by our data (Figure 2), may contribute to the increase of transcription of chromatin templates.
This report on the novel properties of chromatin templates that contained the histone variant H2ABbd is another example of how the cell can use the diversity of histone variants to regulate gene expression. The variant H2A.Z seemed to participate in both gene activation (Santisteban et al, 2000) and gene silencing (Dhillon and Kamakaka, 2000), macroH2A is rather associated with the repression of transcription (Perche et al, 2000), and we show here that H2ABbd is likely to be associated with gene activation.
Materials and methods
Preparation of DNA probes
The 205 bp fragment comprising the sequence of the X. borealis 5S RNA gene was prepared from the plasmid pXP10 by PCR amplification with appropriate primers. The 152 bp fragment was obtained by digestion of the PCR products with EcoRI and RsaI (Lee and Hayes, 1998). The recognition sequence of NF-κB (GGGGATTCCCC) was inserted at positions −16 to −26 by site-directed mutagenesis as described earlier (Angelov et al, 2003). Polynucleotide kinase and [γ-32P]ATP or klenow enzyme and [α-32P]ATP were used to radioactively label the 5′ or 3′ ends of the EcoRI site, respectively. The plasmids pGEM3Z-601 and p199-1 (a kind gift from Drs J Widom and B Bartholomew) were used for the preparation of the 255 and 241 bp DNA fragments containing the strongly nucleosome positioning sequence 601 at the middle or at one extremity, respectively (Lowary and Widom, 1998). The plasmid PMR974 was used for preparation of the 248 bp mouse rDNA fragment containing the sequence −232 to +16 (Langst et al, 1999).
Protein expression and purification
Appropriate primers were used for PCR-amplifying H2ABbd cDNA from testis Marathon-ready cDNA library. The cDNAs were further cloned in a pet3d vector. After expression in Escherichia coli, the recombinant H2ABbd was purified to homogeneity. Conventional recombinant X. laevis histones were expressed in bacteria and purified according to the protocol of Luger et al (1999). SWI/SNF was prepared as described earlier (Shen et al, 2000). The procedures of Ito et al (1999) and Kraus and Kadonaga (1998) were used to express and purify recombinant Acf1, ISWI, NAP-1, Flag-tagged Gal4-VP16 and human His6-tagged p300.
Reconstitution of nucleosomes
The nucleosome reconstitution was carried out as described previously (Mutskov et al, 1998; Angelov et al, 2003). If some free DNAs were present in the nucleosome preparations, they were further purified on a 5–30% sucrose gradient (Mutskov et al, 1998).
The mobilization experiments were performed with centrally positioned nucleosomes reconstituted on either a 248 bp mouse rDNA fragment or a 255 bp 601 DNA fragment (Lowary and Widom, 1998). To isolate centrally positioned nucleosomes, after reconstitution the centrally positioned nucleosomes were separated from the end-positioned ones on a preparative 5.5% native polyacrylamide gel, excised from the gel and eluted. End-positioned nucleosomes used in ACF mobilization experiments were reconstituted using the 241 bp 601 DNA fragment. The binding of NF-κB to the conventional and variant nucleosomes was carried out as described (Angelov et al, 2003).
EMSA, DNase I and hydroxyl radical footprinting
EMSA was carried out in 4 or 5.5% (w/v) polyacrylamide gel (acrylamide to bisacrylamide, 29:1) and 0.3 × TBE at 4°C (Angelov et al, 2003). DNase I and hydroxyl radical footprinting were performed as described (Hayes and Lee, 1997; Angelov et al, 2003).
SWI/SNF nucleosome remodeling
The ATPase activity of SWI/SNF was measured as described previously (Angelov et al, 2003). The nucleosome remodeling reactions were performed essentially as described by Angelov et al (2003).
SWI/SNF and ACF nucleosome mobilization experiments were carried out essentially as described by Angelov et al (2003) and Kagalwala et al (2004), respectively.
Histone transfer experiments
For the histone transfer experiment, a wild-type H3 and swapped tail H3–H2B mutant histones were used. The N-terminal tail of histone H2B was replaced with the N-terminus of histone H3 within the H2B mutant protein by using standard molecular biology approaches. This allows the mutant H2B to be radioactively labeled at serine 10 by the Aurora A kinase (Scrittori et al, 2001) and its transfer to the H3–H4 tetrameric particle to be studied. Labeled mutant H2B or H3 histones were mixed with equimolar amounts of the remaining three histones in 10 mM HCl and dialyzed overnight against histone folding buffer. Nucleosomes were reconstituted as described above. Acceptor tetramers were reconstituted on the 152 bp 5S DNA fragment with an equimolar mixture of the H3 and H4 histones at 0.4:1 w:w H3–H4/DNA ratio.
For the transfer experiments, 14 ng of histone-labeled nucleosomes (reconstituted on the 205 bp 5S DNA) was mixed in remodeling buffer together with a 2.6-fold molar excess of tetrameric H3–H4 particles and 2 μl of SWI/SNF in 10 μl final volume. For the time-course experiments, the reactions were proportionally scaled and aliquots were taken at the time indicated. Reactions were stopped with 1 μg of plasmid DNA, 0.1 U of apyrase and 7.5 mM EDTA and stored on ice until loading on the gel. Gels were run at 4°C.
Restriction enzyme accessibility and exonuclease III assay
H2A and H2ABbd nucleosomes reconstituted on the 152 bp 5S DNA were incubated for 45 min at 28°C in 0.8 × EcoRV buffer supplemented with 1 mM ATP and 100 μg/ml BSA in the presence or absence of 2 μl SWI/SNF (final volume 30 μl). Then, 0.5 μl of apyrase (0.1 U/μl) was added in some tubes and incubated for 3 min at 28°C, and EcoRV (30 U) was added. Aliquots were taken at the indicated time and the reactions were immediately stopped by adding 2 μl of 4 × stop/loading buffer: 0.8% SDS, 0.5 μg/μl proteinase K, 60 mM EDTA, 10% glycerol. DNA fragments were analyzed on 8% native PAGE.
For the exonuclease assay on the nucleosomes incubated in the presence of SWI/SNF, the remodeling was arrested by the addition of 0.1 U of apyrase and 1 μg of plasmid DNA. Then, 5 U of exonuclease III was added and incubated for 15 min at 30°C. The digestion was stopped by the addition of SDS (0.15% final concentration), 1 μg proteinase K, EDTA (10 mM final concentration) and a further incubation of 15 min at 50°C. DNA was extracted and then analyzed on an 8% acrylamide–8 M urea denaturing gel. The experiment was performed with the labeled top or bottom strands.
Transcription experiments
For the p300- and Gal4-VP16-mediated transcription experiments, the chromatin arrays containing either conventional or H2ABbd histones were assembled using the pG5ML array template and recombinant Acf1, ISWI and NAP-1 as described (Ito et al, 1999; An et al, 2002). The p300 and Gal4-VP16 transcription and the HAT assays were carried out according to previously described protocols (An et al, 2002).
Nucleosome reconstitution, Pol II elongation complex assembly and its ligation to either the DNA or the nucleosome and the Pol II transcription analysis were carried out as described previously (Kireeva et al, 2002).
Acknowledgments
This work was supported by CNRS, INSERM, Région Rhône-Alpes and a grant from the ministère de la Recherche (ACI Biologie cellulaire Moléculaire et Structurale, BCM0070). DA is on leave from the Institute of Solid State Physics, BAS, Sofia.
References
- Ahmad K, Henikoff S (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 9: 1191–1200 [DOI] [PubMed] [Google Scholar]
- An W, Palhan VB, Karymov MA, Leuba SH, Roeder RG (2002) Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol Cell 9: 811–821 [DOI] [PubMed] [Google Scholar]
- Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S (2003) The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome Remodeling. Mol Cell 11: 1033–1041 [DOI] [PubMed] [Google Scholar]
- Beato M, Eisfeld K (1997) Transcription factor access to chromatin. Nucleic Acids Res 25: 3559–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB (2002) Nucleosome sliding: facts and fiction. EMBO J 21: 4749–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T (2003) Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell 12: 1599–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A (2003) H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114: 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF (2001) A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J Cell Biol 152: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N, Kamakaka RT (2000) A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell 6: 769–780 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB (2001) Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J 20: 3781–3788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JJ, Lee KM (1997) In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins. Methods 12: 2–9 [DOI] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT (1997) ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155 [DOI] [PubMed] [Google Scholar]
- Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT (1999) ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev 13: 1529–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B (2004) Topography of the ISW2–nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J 23: 2092–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM (2002) Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol Cell 9: 541–552 [DOI] [PubMed] [Google Scholar]
- Kraus WL, Kadonaga JT (1998) p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev 12: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Becker PB (2001) Nucleosome mobilization and positioning by ISWI-containing chromatin-remodeling factors. J Cell Sci 114: 2561–2568 [DOI] [PubMed] [Google Scholar]
- Langst G, Bonte EJ, Corona DF, Becker PB (1999) Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97: 843–852 [DOI] [PubMed] [Google Scholar]
- Lee K-M, Hayes JJ (1998) Linker DNA and H1-dependent reorganization of histone–DNA interactions within the nucleosome. Biochemistry 37: 8622–8628 [DOI] [PubMed] [Google Scholar]
- Lowary PT, Widom J (1998) New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol 276: 19–42 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol 119: 1–16 [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S (2003) Phylogenomics of the nucleosome. Nat Struct Biol 10: 882–891 [DOI] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Bannister AJ, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T (1998) The acetyltransferase activity of CBP stimulates transcription. EMBO J 17: 2886–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C (2004) ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Vassilev A, Tsukiyama T, Nakatani Y, Wu C (2001) ATP-dependent nucleosome remodeling and histone hyperacetylation synergistically facilitate transcription of chromatin. J Biol Chem 276: 14773–14783 [DOI] [PubMed] [Google Scholar]
- Mutskov V, Gerber D, Angelov D, Ausio J, Workman J, Dimitrov S (1998) Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol Cell Biol 18: 6293–6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perche P, Vourch C, Souchier C, Robert-Nicoud M, Dimitrov S, Khochbin C (2000) Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr Biol 10: 1531–1534 [DOI] [PubMed] [Google Scholar]
- Peterson CL, Workman JL (2000) Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr Opin Genet Dev 10: 187–192 [DOI] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM (2000) Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103: 411–422 [DOI] [PubMed] [Google Scholar]
- Scrittori L, Hans F, Angelov D, Charra M, Prigent C, Dimitrov S (2001) pEg2 aurora-A kinase, histone H3 phosphorylation, and chromosome assembly in Xenopus egg extract. J Biol Chem 276: 30002–30010 [DOI] [PubMed] [Google Scholar]
- Sengupta SM, VanKanegan M, Persinger J, Logie C, Cairns BR, Peterson CL, Bartholomew B (2001) The interactions of yeast SWI/SNF and RSC with the nucleosome before and after chromatin remodeling. J Biol Chem 276: 12636–12644 [DOI] [PubMed] [Google Scholar]
- Shen X, Mizuguchi G, Hamiche A, Wu C (2000) A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Thiriet C, Hayes JJ (1998) Functionally relevant histone–DNA interactions extend beyond the classically defined nucleosome core region. J Biol Chem 273: 21352–21358 [DOI] [PubMed] [Google Scholar]
- Tsanev R, Russev G, Pashev I, Zlatanova J (1993) Replication and Transcription of Chromatin. Boca Raton, FI: CRC Press [Google Scholar]
- van Holde K (1988) Chromatin. Berlin, Germany: Springer-Verlag KG [Google Scholar]


