Abstract
In recent years, studies have suggested that promoter methylation in human papilloma virus (HPV) positive head and neck squamous cell carcinoma (HNSCC) has a mechanistic role and has the potential to improve patient survival. The present study aimed to replicate key molecular findings from previous analyses of the methylomes of HPV positive and HPV negative HNSCC in an independent cohort, to assess the reliability of differentially methylated markers in HPV-associated tumors. HPV was measured using real-time quantitative PCR and the biological significance of methylation differences was assessed by Ingenuity Pathway Analysis (IPA). Using an identical experimental design of a 450K methylation platform, 7 of the 11 genes were detected to be significantly differentially methylated and all 11 genes were either hypo- or hypermethylated, which was in agreement with the results of a previous study. IPA's enriched networks analysis identified one network with msh homeobox 2 (MSX2) as a central node. Locally dense interactions between genes in networks tend to reflect significant biology; therefore MSX2 was selected as an important gene. Sequestration in the top four canonical pathways was noted for 5-hydroxytryptamine receptor 1E (serotonin signaling), collapsin response mediator protein 1 (semaphorin signaling) and paired like homeodomain 2 (bone morphogenic protein and transforming growth factor-β signaling). Placement of 9 of the 11 genes in highly ranked pathways and bionetworks identified key biological processes to further emphasize differences between HNSCC HPV positive and negative pathogenesis.
Keywords: differential methylation, pathway analysis, canonical pathways, networks
Introduction
The results from a number of studies involving next-generation sequencing sequence analysis, gene expression microarrays, whole-exome sequencing and genome-wide methylation studies support the observation that human papilloma virus (HPV) positive squamous head and neck cancer (HNSCC) is a distinct entity and therefore exhibits a particular set of somatic alterations (1–6). Data from epidemiological and clinical studies have also indicated that HPV positive HNSCC is clinically different from HPV negative HNSCC in terms of patient characteristics, sensitivity to treatment and molecular biology (7,8).
Global characterization of the HNSCC methylome demonstrates the different landscapes in HPV positive tumors compared with HPV negative tumors. CpG methylation is higher in HPV positive cells in both repetitive and non-repetitive regions (genic and non-genic) (9,10). Methylome analysis of HPV positive and HPV negative primary formalin-fixed paraffin-embedded tumors using the Illumina HumanMethylation450K Beadchip platform (2) have confirmed previous reports (9,11) that the DNA methylation signature of HPV positive HNSCC is different from that of HPV negative HNSCC and that HPV positive HNSCC has a strong tendency to undergo hypermethylation. A previous study (2) highlighted 11 genes: Chromosome 14 open reading frame 162/coiled-coil domain containing 177 (C14orf162/CCDC177), cadherin 8 (CDH8), collapsin response mediator protein 1 (CRMP1), engulfment and cell motility 1 (ELMO1), 5-hydroxytryptamine receptor 1E (HTR1E), meiotic double-stranded break formation protein 1 (MEI1), msh homeobox 2 (MSX2), protocadherin 10 (PCDH10), protocadherin β-11 (PCDHB11), paired like homeodomain 2 (PITX2) and synapsin II (SYN2) as undergoing significantly different methylation processes in HPV positive and HPV negative HNSCC (2) (Table I).
Table I.
Infinium HumanMethylation450 Beadchip assay data for HPV positive vs. HPV negative HNSCC samples for 11 genes.
| Gene name | Chromosome | HPV status | Mean β | SD | Ratio of HPV positive vs. HPV negative | P-value | Methylation |
|---|---|---|---|---|---|---|---|
| C14orf162/CCDC177 | 14 | Positive | 0.161 | 0.041 | 0.579 | 0.019a | Hypo |
| Negative | 0.278 | 0.034 | |||||
| CDH8 | 4 | Positive | 0.479 | 0.062 | 1.640 | 0.004a | Hyper |
| Negative | 0.292 | 0.055 | |||||
| CRMP1 | 4 | Positive | 0.208 | 0.058 | 1.155 | 0.749 | Hyper |
| Negative | 0.180 | 0.058 | |||||
| ELMO1 | 7 | Positive | 0.578 | 0.067 | 1.234 | 0.026a | Hyper |
| Negative | 0.468 | 0.033 | |||||
| HTR1E | 6 | Positive | 0.433 | 0.062 | 1.357 | 0.027a | Hyper |
| Negative | 0.319 | 0.060 | |||||
| MEI1 | 22 | Positive | 0.318 | 0.072 | 0.849 | 0.008a | Hypo |
| Negative | 0.531 | 0.087 | |||||
| MSX2 | 5 | Positive | 0.394 | 0.081 | 2.213 | 0.009a | Hyper |
| Negative | 0.178 | 0.065 | |||||
| PCDH10 | 4 | Positive | 0.333 | 0.103 | 1.524 | 0.166 | Hyper |
| Negative | 0.220 | 0.023 | |||||
| PCDHB11 | 5 | Positive | 0.548 | 0.074 | 1.302 | 0.042a | Hyper |
| Negative | 0.421 | 0.076 | |||||
| PITX2 | 4 | Positive | 0.416 | 0.112 | 1.386 | 0.138 | Hyper |
| Negative | 0.300 | 0.088 | |||||
| SYN2 | 3 | Positive | 0.399 | 0.115 | 1.013 | 0.781 | Hyper |
| Negative | 0.394 | 0.104 |
P<0.05, indicates a statistically significant difference. Cohort of 8 samples (4 HPV+HNSCC, 4 HPV-HNSCC). HNSCC, head and neck squamous cell carcinoma; HPV, human papilloma virus; chrom, chromosome; SD, standard deviation; meth, methylation status; hypo, hypomethylated; hyper, hypermethylated; C14orf162/CCDC177; chromosome 14 open reading frame 162/coiled-coil domain containing 177; CDH8, cadherin 8; CRMP1, collapsin response mediator protein 1; ELMO1, engulfment and cell motility 1; HTR1E, 5-hydroxytryptamine receptor 1E; MEI1, meiotic double-stranded break formation protein 1; MSX2, msh homeobox 2; PCDH10, protocadherin 10; PCDHB11, protocadherin β-11; PITX2, paired like homeodomain 2; SYN2, synapsin II.
It has been established that patients with HPV positive HNSCC have better survival outcomes than those with HNSCC that are HPV negative, possibly due to improved responses to chemoradiation (12). However, it remains unclear why HPV positive patients experience improved prognoses-the underlying mechanisms responsible for these outcomes have not yet been investigated. A mechanistic role for promoter methylation is beginning to be established, with the aim of improving survival outcomes of patients with HPV positive HNSCC (13). The aim of the current study was to verify that the 11 aforementioned genes undergo significantly different methylation in HPV positive and HPV negative HNSCC tumor samples and to assess their biological significance using Ingenuity Pathway Analysis (IPA) for biomarker potential in HPV-associated HNSCC.
Materials and methods
DNA extraction and amplification
DNA from 4 HPV positive and 4 HPV negative freshly frozen (−80°C) primary HNSCC tumor samples, resected from patients admitted between December 2010 and January 2012 in the Department of Otolaryngology/Head and Neck Surgery (Henry Ford Hospital, Detroit, USA), were subjected to comprehensive genome-wide methylation profiling using the Infinium HumanMethylation450K BeadChip kit (Illumina, Inc., San Diego, CA, USA). Tumor site and demographic characteristics are presented in Table II. For the present study, patient informed consent for inclusion of surgically resected tissues was given, and the study was approved by the Henry Ford Health System Institutional Review Board committee.
Table II.
Patient cohort.
| Tissue ID | Site | Age | Ethnicity | Gender | HPV status |
|---|---|---|---|---|---|
| HFHS-HN19 | Oropharynx | 60 | AA | M | Positive |
| HFHS-HN28 | Oropharynx | 64 | CA | M | Positive |
| HFHS-HN30 | Oropharynx | 62 | AA | M | Positive |
| HFHS-HN48 | Tonsil | 53 | CA | M | Positive |
| HFHS-HN51 | Larynx | 49 | CA | M | Negative |
| HFHS-HN23 | Larynx | 67 | AA | M | Negative |
| HFHS-HN26 | Tongue | 71 | CA | M | Negative |
| HFHS-HN42 | Oropharynx | 66 | AA | F | Negative |
AA, African American; CA, caucasian; M, male; F, female; HPV, human papilloma virus.
DNA was extracted according to the manufacturer's protocol (Qiagen, Inc., Valencia, CA, USA). Tumor HPV DNA concentrations were measured using real-time quantitative PCR (qPCR) as previously described (14,15). Briefly, primers and probes to a housekeeping gene (β-globin) were run in parallel to standardize the input DNA. By using serial dilutions, standard curves were developed for the HPV viral copy number using CaSki (American Type Culture Collection, Manassas, VA, USA) cell line genomic DNA, which have 600 copies/genome equivalent (6.6 pg of DNA/genome). The cut-off value for HPV16 positive status was >0.03 (>3 HPV genome copy/100 cells) (14–16).
The Infinium HumanMethylation450 BeadChip kit includes 485,764 cytosine positions in the human genome and covers 99% of Reference Sequence genes. The Reference Sequence collection aims to provide an integrated, comprehensive, well-annotated non-redundant set of sequences, including genomic DNA, transcripts and proteins (17). The interrogated CpG sites are distributed among all 23 human chromosome pairs (17).
Processing samples for the Infinium HumanMethylation450
The Infinium HumanMethylation450 assays were performed at the Applied Genomics Technology Center, Wayne State University (Detroit, MI, USA). Following DNA checks of original DNA quality, quantification and bisulfite conversion, 4 µl of bisulfite-converted DNA was used for hybridization according to Illumina Infinium methylation protocols. Data were normalized using the Controls Normalization method (Illumina, Inc.). The methylation score for each CpG was represented as a beta (β) value according to the fluorescence intensity ratio and each β-value in the Infinium HumanMethylated450 BeadChip kit was accompanied by a detection P-value. β-values could take any value between 0 (non-methylated) and 1 (completely methylated) and were determined using the GenomeStudio® 2009.2, Methylation Module ver. 1.5.5., ver. 3.2 (Illumina, Inc.). Probes were discarded if P>0.05. All normalized and raw data were submitted to the GEO (Gene Expression Omnibus, NCBI) according to the instructions provided (GEO accession numbers: GSE67114). The resulting β-values were exported to Microsoft Excel, JMP and SAS (SAS Institute, Inc., Cary, NC, USA) for data analysis.
Pathway analysis
To determine the biological processes occurring within the 11 differentially methylated genes, they were uploaded to the Ingenuity pathway Analysis (IPA; Ingenuity Systems, Inc; Qiagen, Inc.). IPA integrates genes and molecules that are part of the same biological functions or regulatory networks interacting together.
Statistical methods
There was a priori evidence that the 11 genes were differentially methylated and each in a particular direction, therefore, P<0.05 was considered to indicate a statistically significant difference.
Results
HumanMethylation450K BeadChip analysis
For all genes, 99.97% of the CpGs and 100% of the CpGs associated with the 11 genes of particular interest, met the detection threshold of P<0.05 and therefore were eligible for analysis.
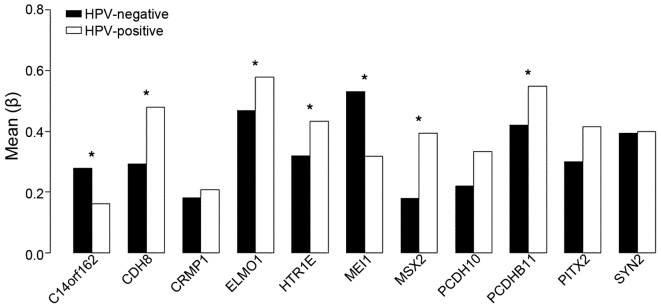
Of the 11 genes of interest, the methylation status of 7 differed significantly between HPV positive and HPV negative HNSCC (Table I). CDH8, PCDHB11, ELMO1, MSX2, and HTR1E were significantly hypermethylated and MEI1 and C14orf162/CCDC177 were significantly hypomethylated (Fig. 1). The methylation status of all 11 genes as either hypo- or hypermethylated was consistent with the results of a previous study by Lechner et al (2) even though the methylation of four genes did not differ significantly (P>0.05).
Figure 1.
Illumina 450K methylation beadchip beta values associated with HPV positive and HPV negative HNSCC. Of 11 differentially methylated genes, 7 demonstrate statistically significant differential methylation in HPV positive vs. HPV negative HNSCC (*P<0.05). For all 11 genes, hypermethylation and hypomethylation directional changes were concordant with the results from Lechner et al (2). Among these genes, MEI1 and C14orf162 were hypomethylated and the remaining 9 were hypermethylated. Differences in methylation in HPV negative and HPV positive HNSCC were significant in 7 of the genes analyzed. HPV, human papilloma virus; HNSCC, head and neck squamous cell carcinoma; C14orf162/CCDC177; chromosome 14 open reading frame 162/coiled-coil domain containing 177; CDH8, cadherin 8; CRMP1, collapsin response mediator protein 1; ELMO1, engulfment and cell motility 1; HTR1E, 5-hydroxytryptamine receptor 1E; MEI1, meiotic double-stranded break formation protein 1; MSX2, msh homeobox 2; PCDH10, protocadherin 10; PCDHB11, protocadherin β-11; PITX2, paired like homeodomain 2; SYN2, synapsin II.
Pathway analysis
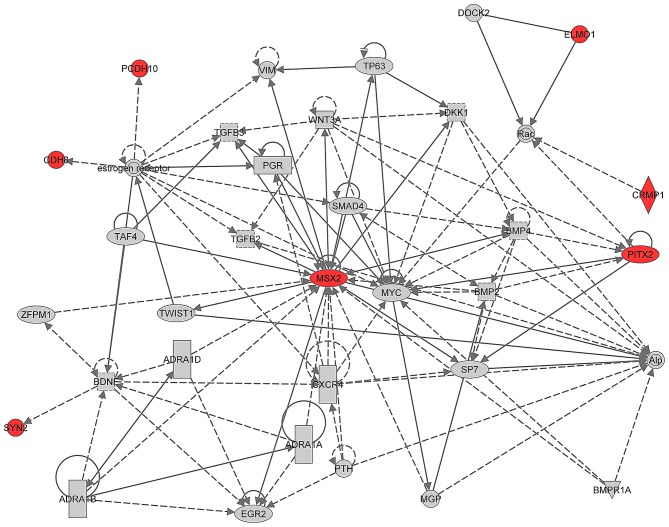
C14orf162/CCDC177 was excluded from the list of genes in IPA as it was unaccounted for in the IPA knowledge database. IPA connected 7/10 genes in a 35 gene network characterized by the following functions: Cellular Development, Skeletal and Muscular System Development and Function, and Embryonic Development as the sole enriched top network (Fig. 2). The seven genes exhibiting significantly different methylation were MSX2, CRMP1, ELMO1, CDH8, SYN2, PCDH10 and PITX2 (Fig. 1).
Figure 2.
Top network. IPA identified ‘Cellular Development, Skeletal and Muscular System Development and Function, Embryonic Development’ as the only enriched top network. This network is built with 35 genes from IPA's knowledgebase and includes 7 (MSX2, CRIMP1, ELMO1, CDH8, SYN2, PDCH10, PITX2) of the 11 differentially methylated genes (highlighted in red). MSX2, msh homeobox 2; CRIMP1, collapsing response mediator protein 1; ELMO1, engulfement and cell motility 1; CDH8, cadherin 8; SYN2, synapsin II; PDCH10, protocadherin 10; PITX2, paired like homeodomain 2.
The top five molecular and cellular functions included cell morphology, cellular assembly, cellular movement, cell-to-cell signaling and interaction and cell death and survival (Table III). ELMO1 was represented in all the top five functions followed by MSX2 and PTX2 (4/5); CRMP1 and SYN2 (3/5); PCDHB11 (2/5); and CDH8, HTR1E and PCDH10 (1/5).
Table III.
Top molecular and cellular functions.
| Name | P-value | Differentially methylated genes |
|---|---|---|
| Cell-to-cell signalling and interaction | <0.0001–0.028a | PCDHB11, HTR1E, CDH8, PITX2, ELMO1, MSX2, SYN2 |
| Cellular movement | <0.0001–0.048a | CRMP1, PCDH10, PITX2, ELMO1, MSX2 |
| Cellular assembly and organization | <0.001–0.027a | PCDHB11, CRMP1, ELMO1, SYN2 |
| Cell death and survival | <0.001–0.038a | PITX2, ELMO1, MSX2 |
| Cell morphology | <0.001–0.04a | CRMP1, PITX2, SYN2, ELMO1, MSX2 |
P<0.05; indicates a statistically significant result. C14orf162/CCDC177; chromosome 14 open reading frame 162/coiled-coil domain containing 177; CDH8, cadherin 8; CRMP1, collapsin response mediator protein 1; ELMO1, engulfment and cell motility 1; HTR1E, 5-hydroxytryptamine receptor 1E; MEI1, meiotic double-stranded break formation protein 1; MSX2, msh homeobox 2; PCDH10, protocadherin 10; PCDHB11, protocadherin β-11; PITX2, paired like homeodomain 2; SYN2, synapsin II.
The highly ranked canonical pathways (P<0.05) indicate sequestration of HTR1E in serotonin receptor signaling, CRMP1 in sematophorin signaling, and PITX2 in bone morphogenic protein (BMP) and transforming growth factor (TGF) β signaling (Table IV).
Table IV.
Top canonical pathways.
| Pathway | P-value | (Name) of differentially methylated genes/total pathway genes |
|---|---|---|
| Serotonin receptor signaling | 0.02a | 1 (HTR1E)/33 |
| Semaphorin signaling in neurons | 0.03a | 1 (CRMP1)/52 |
| BMP signaling | 0.04a | 1 (PITX2)/80 |
| TGF-β signaling | 0.056 | 1 (PITX2)/89 |
P<0.05; HTR1E, 5-hydroxytryptamine receptor 1E; CRMP1, collapsin response mediator protein 1; PITX2, paired like homeodomain 2; BMP, bone morphogenic protein; TGF-β, transforming growth factor β.
Discussion
Squamous cell carcinomas primarily develop in the larynx, pharynx and oral cavity, and make up the vast majority of mucosal head and neck cancer (18). It has been challenging to accurately and reliably stratify HNSCC to predict outcomes, primarily due to the numerous anatomic sites and subsites from which tumors can arise. Globally, 600,000 people are annually diagnosed with HNSCC (19). The number of cases of HNSCC has gradually increased over the past three decades and currently account for 5% of all malignancies (20).
Just two risk factors, tobacco and alcohol (21) are responsible for 72% of HNSCC cases (22). However, previous studies have also implicated HPV status as a possible cause of certain HNSCC cases (23) and an independent risk factor for oropharyngeal cancer (OPSCC) (24). The biological significance of HPV as an additional independent risk factor is highlighted by the improved prognosis of patients with HPV positive HNSCC compared to HPV negative HNSCC (23,24), which may partly be due to improved therapeutic responses to chemoradiotherapy (12).
It has been demonstrated that HPV positive OPSCC is a distinct type of HNSCC characterized by overexpression of cyclin-dependent kinase inhibitor 2A (CDKN2A), improved patient prognoses, nonkeratinizing histology and a high prevalence of HPV infection (25). At present, HPV status is the most robust and valid molecular diagnostic and prognostic biomarker for HNSCC (26). The expression of E6 and E7 HPV oncoproteins may influence the genetic profile of HPV positive HNSCC (23). HPV positive tumors are characterized by E6 suppression of wild-type p53 function (27), retinoblastoma pathway inactivation by E7 and overexpression of wild-type CDKN2A (28), and infrequent amplification of cyclin D (29), whereas for HPV-HNSCC, the opposite occurs (30). Tumor protein 53 mutations are detected in all HPV negative cases and in the majority of HPV negative tumors, downregulation of CDKN2A and/or cyclin D1 amplification occur (1). The improved prognosis of HPV positive patients compared to HPV negative HNSSC patients has been confirmed (12,16,24), however the mechanistic explanation for this phenomenon remains elusive.
Continuing to adapt the idea of assessing the methylation status of patients with HPV positive HNSCC compared to those with HPV negative HNSCC, Lechner et al (2), analyzed 32 cases of HNSCC (18 HPV positive and 14 HPV negative), detected 11 genes that were methylated differently between HPV positive and HPV negative HNSCC and analyzed mRNA expression for two genes: CDH8 and PCDH10. The mRNA expression data for the remaining 9 genes were obtained from a different study by Pyeon et al (6). The study also confirmed previous reports (9,11) that HPV positive patients have a distinct DNA methylation signature from their HPV negative HNSCC counterparts.
The present study, in an independent HNSCC sample set using an identical experimental design of the 450K methylation platform, confirmed that 7 of the 11 aforementioned genes were significantly differentially methylated. Additionally, replication of complete concordance of methylation direction (hypo- vs. hypermethylated) was observed for all 11 genes. A recent analysis demonstrated that 9 genes, CDH8, CRMP1, ELMO1, HTR1E, MEI1, MSX2, PCDHB11, PITX2, SYN2, were observed to be significantly differentially methylated in HPV positive compared to HPV negative HNSCC (31).
Differentially methylated genes can be investigated by the pathway analysis framework, which is able to identify distinct signaling pathway networks and therefore may provide a biological basis for further exploration of methylated genes as differential targets in HPV positive and HPV negative HNSCC. In this type of analysis, a biological system is surveyed in the context of a specific phenotype for example, the context of disease, to identify gene groups associated with biological systems (32,33). The pairing of biological relationships allows for a strategic knowledge base approach and has been used to improve understanding of the systems biology of disease processes and more intuitively identify potential therapeutic targets (34–36).
A previous study by our group (5) demonstrated that genes involved in signal transduction pathways of HPV positive HNSCC tumor genomes exhibited a predominant hypermethylation profile. In the current study, 9 of the 11 genes that were represented in IPA's top functions, pathways and networks were hypermethylated. Hypomethylated C14orf162 was not registered in IPA's knowledge database and thus was not included in pathway analyses. Hypomethylated MEI1 did not place among significantly ranked pathways/functions/networks. C14orf162/CCDC177, also known as myelin proteolipid protein-like protein, is encoded by a gene that maps to human chromosome 14q24.1. MEI1 is expressed almost exclusively in the gonads and is required for normal vertebrate meiotic chromosome synapsis (37). Its role in cancer has not yet been investigated and the results of the present study suggest opportunities for additional studies to further investigate the mechanism by which MEI1 operates in cancer.
The IPA network generation algorithm iteratively constructs networks that strive for optimization of the number of focus genes (10 in the current study) and interconnectivity, under the constraint of a maximum network size of 35 genes. The former identifies how user-specified genes interact with each other or with neighboring genes, thus highlighting the molecular biology implied in a dataset. In the present study, IPA's enriched networks analysis produced only one network: Cellular development, skeletal and muscular system development and function and embryonic development. This network included 7/10 focus genes, MSX2, CRMP1, ELMO1, CDH8, SYN2, PCDH10, PITX2 that completed the 35 gene network from IPA's knowledgebase. Locally dense interactions between genes tend to reflect significant biology therefore, the role of the MSX2 gene in particular has been highlighted as significant.
MSX2 is a homeobox gene implicated in bone metabolism, organ development, and breast and colorectal tumorigenesis (38–41). Its involvement in HPV-associated HNSCC has been previously documented (2) and was confirmed in the present independent study, which presents additional rationale for further investigations into the role of MSX2 in HNSCC.
ELMO1 was represented in all top five functions, followed by MSX2 and PTX2 (4/5), CRMP1 and SYN2 (3/5), PCDHB11 (2/5), and CDH8, HTR1E, and PCDH10 (1/5) (Table III). ELMO1 is an evolutionarily conserved cytoplasmic engulfment protein that promotes actin-dependent phagocytosis and cell migration in a number of invertebrate models and immortalized mammalian cell lines (42). Its role in tumorigenesis and increased invasiveness has been investigated in glioma cells (43), esophageal adenocarcinoma (44) and breast cancer (45). Furthermore, it has been demonstrated that hypermethylated ELMO1 was part of one of the three methylation epigenotypes for colorectal cancer (46).
The top four canonical pathways indicated the inclusion of HTR1E, CRMP1 and PITX2. HTR1E is one of the 33 genes in the highest ranked serotonin receptor signaling canonical pathway (Table IV). Serotonin (5-HT) is a growth factor and a cell cycle mediator that regulates DNA synthesis (47). In the mammalian nervous system, 5-HT receptors are part of the superfamily of G-protein-coupled receptors. Treatment of tumor cells with EGF (Epidermal Growth Factor) and 5-HT may decrease the number of G0/G1 cells that are dormant, resulting in more active, dividing cells that consequently have increased sensitivity to chemotherapeutic treatment (48). Differential methylation of the promoters of HTR1E has been detected in patients with schizophrenia (49).
CRMPs are cytosolic phosphoproteins involved in axonal guidance and neuronal differentiation (50). CRMP1 is one of 52 genes in the second ranked semaphorin signaling canonical pathway implicated in semaphorin-induced growth cone collapse during neural development and axonal guidance (50). Semaphorin signaling affects focal adhesion assembly/disassembly and induces cytoskeletal remodeling, which consequently impacts cell motility, cell shape, cell migration and attachment to the extracellular matrix (51). Oncology studies are thus increasingly focused on axon guidance molecules.
CRMP1 has been characterized as a novel invasion-suppression gene. It has been demonstrated that low expression of CRMP1 mRNA in lung cancer tissue is significantly associated with lymph node metastasis, early post-operative relapse, advanced disease and shorter patient survival times (52). How CRMP1 inhibits cell invasion remains unclear, however, DNA methylation in the context of a hypermethylated CRMP1 suggests low expression and therefore implicates epigenetic mechanisms in HNSCC tumorigenesis.
The PITX2 gene was placed in 4/5 of the top molecular and cellular functions (Table III) and two of the four top canonical pathways (Table IV). PITX2 is 1 of 80 genes in the BMP and 1 of 89 genes in the TGF-β signaling pathways. TGF-β/BMP signaling pathways are involved in bone formation during mammalian development with various regulatory functions (53) and their disruptions have been implicated in multiple bone diseases, including tumor metastasis (54). PITX2 encodes a transcription factor of the paired-like homeodomain protein family, a crucial component during normal embryonic development (55) that has diverse roles in cell differentiation, proliferation, organogenesis and hematopoiesis (56,57). PITX2 is downstream of TGF-β and fibroblast growth factor (FGF) signaling and may regulate cell proliferation by activating the expression of cyclins D1 and D3 (58). It has been demonstrated that methylation of PITX2 in human tumorigenesis occurs in several types of human cancer, including colon, breast, ovarian and prostate cancer (59–61). A previous study indicated that PITX2 downregulation with associated promoter hypermethylation predicted good clinical outcomes following radical prostatectomy (61).
Confirmation of hypermethylation of CDH8 and PCDHB11 in the independent sample set of the current study is consistent with findings from previous studies demonstrating that cadherin genes are targets for transcriptional silencing in HNSCC and HPV-mediated hypermethylation (2). Cadherins are targets of Polycomb repressive complex 2 and are involved in different types of cancer and cancer specific processes (62) including the epithelial to mesenchymal transition, a process that facilitates cell invasion and metastasis (63). Polycomb group target genes are more likely to contain cancer-specific promoter DNA hypermethylation than other genes (64). This supports the theory that cancer originates from the stem cells, where permanent silencing replaces reversible gene expression thus locking the cell into a perpetual state of self-renewal that predisposes it to malignant transformation (65).
In conclusion, the current study investigated an independent HNSCC sample set. The concordance of methylation direction (hypo- vs. hypermethylated) from previous analyses of the methylomes of HPV positive and HPV negative HNSCC was replicated for all 11 genes. Of the 11 genes, 9 were represented in IPA's top functions, pathways and network, and 7 genes were reported as undergoing significantly different methylation. The present study suggests that cadherins CDH8 and PCDHB11, together with C14orf162/CCDC177, ELMO1, HTR1E MEI1 and MSX2, are biomarkers for HPV-associated HNSCC. Cell signaling events are critical in the execution of key biological functions and insights into how complex cellular signaling cascades and networks are programmed may be important in aiding the development of novel biological agents with multiple targets. Placement of 9 of the 11 genes in key pathways and biological processes focuses attention on the unique biological processes in HPV-associated HNSCC tumors and identify the differences between the pathogenesis of HPV positive and HPV negative HNSCC at the epigenetic level.
Acknowledgements
The current study was supported by Susan G Komen Race for the Cure® (grant no., KG110218 to Dr M. J. Worsham).
References
- 1.Lechner M, Frampton GM, Fenton T, Feber A, Palmer G, Jay A, Pillay N, Forster M, Cronin MT, Lipson D, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV- tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lechner M, Fenton T, West J, Wilson G, Feber A, Henderson S, Thirlwell C, Dibra HK, Jay A, Butcher L, et al. Identification and functional validation of HPV-mediated hypermethylation in head and neck squamous cell carcinoma. Genome Med. 2013;5:15. doi: 10.1186/gm419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Worsham MJ, Chen KM, Ghanem T, Stephen JK, Divine G. Epigenetic modulation of signal transduction pathways in HPV-associated HNSCC. Otolaryngol Head Neck Surg. 2013;149:409–416. doi: 10.1177/0194599813490895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillison ML. Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol. 2004;31:744–754. doi: 10.1053/j.seminoncol.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen NP, Chi A, Nguyen LM, Ly BH, Karlsson U, Vinh-Hung V. Human papillomavirus-associated oropharyngeal cancer: A new clinical entity. QJM. 2010;103:229–236. doi: 10.1093/qjmed/hcp176. [DOI] [PubMed] [Google Scholar]
- 9.Sartor MA, Dolinoy DC, Jones TR, Colacino JA, Prince ME, Carey TE, Rozek LS. Genome-wide methylation and expression differences in HPV(+) and HPV(−) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics. 2011;6:777–787. doi: 10.4161/epi.6.6.16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards KL, Zhang B, Baggerly KA, Colella S, Lang JC, Schuller DE, Krahe R. Genome-wide hypomethylation in head and neck cancer is more pronounced in HPV-negative tumors and is associated with genomic instability. PLoS One. 2009;4:e4941. doi: 10.1371/journal.pone.0004941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lleras RA, Smith RV, Adrien LR, Schlecht NF, Burk RD, Harris TM, Childs G, Prystowsky MB, Belbin TJ. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin Cancer Res. 2013;19:5444–5455. doi: 10.1158/1078-0432.CCR-12-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fakhry C, Westra WH, Li S, Cmelak A, Ridge JA, Pinto H, Forastiere A, Gillison ML. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst. 2008;100:261–269. doi: 10.1093/jnci/djn011. [DOI] [PubMed] [Google Scholar]
- 13.Gubanova E, Brown B, Ivanov SV, Helleday T, Mills GB, Yarbrough WG, Issaeva N. Downregulation of SMG-1 in HPV-positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin Cancer Res. 2012;18:1257–1267. doi: 10.1158/1078-0432.CCR-11-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephen JK, Chen KM, Shah V, Havard S, Lu M, Schweitzer VP, Gardner G, Worsham MJ. Human papillomavirus outcomes in an access-to-care laryngeal cancer cohort. Otolaryngol Head Neck Surg. 2012;146:730–738. doi: 10.1177/0194599811434482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stephen JK, Divine G, Chen KM, Chitale D, Havard S, Worsham MJ. Significance of p16 in site-specific HPV positive and HPV negative head and neck squamous cell carcinoma. Cancer Clin Oncol. 2013;2:51–61. doi: 10.5539/cco.v2n1p51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worsham MJ, Stephen JK, Chen KM, Mahan M, Schweitzer V, Havard S, Divine G. Improved survival with HPV among African Americans with oropharyngeal cancer. Clin Cancer Res. 2013;19:2486–2492. doi: 10.1158/1078-0432.CCR-12-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 18.Horner MJ, Ries LAG, Krapcho M, Neyman N, Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A, Miller BA, Lewis DR, Eisner MP, Stinchcomb DG, Edwards BK, editors. Based on November 2008 SEER data submission, posted to the SEER web site. National Cancer Institute; Bethesda, MD: 2009. SEER cancer statistics review, 1975–2006, section 16: Melanoma of the Skin. Retrieved March 29, 2010, from. 2009. [Google Scholar]
- 19.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 20.Brockstein B, Haraf DJ, Rademaker AW, Kies MS, Stenson KM, Rosen F, Mittal BB, Pelzer H, Fung BB, Witt ME, et al. Patterns of failure, prognostic factors and survival in locoregionally advanced head and neck cancer treated with concomitant chemoradiotherapy: A 9-year, 337-patient, multi-institutional experience. Ann Oncol. 2004;15:1179–1186. doi: 10.1093/annonc/mdh308. [DOI] [PubMed] [Google Scholar]
- 21.Hashibe M, Brennan P, Benhamou S, Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova E, Fernandez L, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers and the risk of head and neck cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99:777–789. doi: 10.1093/jnci/djk179. [DOI] [PubMed] [Google Scholar]
- 22.Hashibe M, Brennan P, Chuang SC, Boccia S, Castellsague X, Chen C, Curado MP, Dal Maso L, Daudt AW, Fabianova E, et al. Interaction between tobacco and alcohol use and the risk of head and neck cancer: Pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. Cancer Epidemiol Biomarkers Prev. 2009;18:541–550. doi: 10.1158/1055-9965.EPI-08-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillison ML, Lowy DR. A causal role for human papillomavirus in head and neck cancer. Lancet. 2004;363:1488–1489. doi: 10.1016/S0140-6736(04)16194-1. [DOI] [PubMed] [Google Scholar]
- 24.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marur S, D'Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: A virus-related cancer epidemic. Lancet Oncol. 2010;11:781–789. doi: 10.1016/S1470-2045(10)70017-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mydlarz WK, Hennessey PT, Califano JA. Advances and perspectives in the molecular diagnosis of head and neck cancer. Expert Opin Med Diagn. 2010;4:53–65. doi: 10.1517/17530050903338068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafkamp HC, Speel EJ, Haesevoets A, Bot FJ, Dinjens WN, Ramaekers FC, Hopman AH, Manni JJ. A subset of head and neck squamous cell carcinomas exhibits integration of HPV 16/18 DNA and overexpression of p16INK4A and p53 in the absence of mutations in p53 exons 5–8. Int J Cancer. 2003;107:394–400. doi: 10.1002/ijc.11389. [DOI] [PubMed] [Google Scholar]
- 28.Smith EM, Wang D, Kim Y, Rubenstein LM, Lee JH, Haugen TH, Turek LP. P16INK4a expression, human papillomavirus and survival in head and neck cancer. Oral Oncol. 2008;44:133–142. doi: 10.1016/j.oraloncology.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Ragin CC, Taioli E, Weissfeld JL, White JS, Rossie KM, Modugno F, Gollin SM. 11q13 amplification status and human papillomavirus in relation to p16 expression defines two distinct etiologies of head and neck tumours. Br J Cancer. 2006;95:1432–1438. doi: 10.1038/sj.bjc.6603394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gillison ML, D'Souza G, Westra W, Sugar E, Xiao W, Begum S, Viscidi R. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008;100:407–420. doi: 10.1093/jnci/djn025. [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Network, corp-author. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lum PY, Chen Y, Zhu J, Lamb J, Melmed S, Wang S, Drake TA, Lusis AJ, Schadt EE. Elucidating the murine brain transcriptional network in a segregating mouse population to identify core functional modules for obesity and diabetes. J Neurochem. 2006;97:S50–S62. doi: 10.1111/j.1471-4159.2006.03661.x. (Suppl 1) [DOI] [PubMed] [Google Scholar]
- 33.Nilsson EE, Savenkova MI, Schindler R, Zhang B, Schadt EE, Skinner MK. Gene bionetwork analysis of ovarian primordial follicle development. PLoS One. 2010;5:e11637. doi: 10.1371/journal.pone.0011637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gargalovic PS, Imura M, Zhang B, Gharavi NM, Clark MJ, Pagnon J, Yang WP, He A, Truong A, Patel S, et al. Identification of inflammatory gene modules based on variations of human endothelial cell responses to oxidized lipids. Proc Natl Acad Sci USA. 2006;103:12741–12746. doi: 10.1073/pnas.0605457103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horvath S, Zhang B, Carlson M, Lu KV, Zhu S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA. 2006;103:17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Libby BJ, Reinholdt LG, Schimenti JC. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc Natl Acad Sci USA. 2003;100:15706–15711. doi: 10.1073/pnas.2432067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davideau JL, Demri P, Hotton D, Gu TT, MacDougall M, Sharpe P, Forest N, Berdal A. Comparative study of MSX-2, DLX-5, and DLX-7 gene expression during early human tooth development. Pediatr Res. 1999;46:650–656. doi: 10.1203/00006450-199912000-00015. [DOI] [PubMed] [Google Scholar]
- 39.Malewski T, Milewicz T, Krzysiek J, Gregoraszczuk EL, Augustowska K. Regulation of Msx2 gene expression by steroid hormones in human nonmalignant and malignant breast cancer explants cultured in vitro. Cancer Invest. 2005;23:222–228. doi: 10.1081/CNV-200055958. [DOI] [PubMed] [Google Scholar]
- 40.Satoh K, Ginsburg E, Vonderhaar BK. Msx-1 and Msx-2 in mammary gland development. J Mammary Gland Biol Neoplasia. 2004;9:195–205. doi: 10.1023/B:JOMG.0000037162.84758.b5. [DOI] [PubMed] [Google Scholar]
- 41.Liu F, Ji F, Ji Y, Jiang Y, Sun X, Lu Y, Zhang L, Han Y, Liu X. Dissecting the mechanism of colorectal tumorigenesis based on RNA-sequencing data. Exp Mol Pathol. 2015;98:246–253. doi: 10.1016/j.yexmp.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 42.Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/S0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- 43.Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, Haney LB, Ravichandran KS, Nishikawa R, Cheng SY. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Yang L, Fu H, Yan J, Wang Y, Guo H, Hao X, Xu X, Jin T, Zhang N. Association between Gαi2 and ELMO1/Dock180 connects chemokine signalling with Rac activation and metastasis. Nat Commun. 2013;4:1706. doi: 10.1038/ncomms2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yagi K, Akagi K, Hayashi H, Nagae G, Tsuji S, Isagawa T, Midorikawa Y, Nishimura Y, Sakamoto H, Seto Y, et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res. 2010;16:21–33. doi: 10.1158/1078-0432.CCR-09-2006. [DOI] [PubMed] [Google Scholar]
- 47.Vicaut E, Laemmel E, Stücker O. Impact of serotonin on tumour growth. Ann Med. 2000;32:187–194. doi: 10.3109/07853890008998826. [DOI] [PubMed] [Google Scholar]
- 48.Hambek M, Werner C, Baghi M, Gstöttner W, Knecht R. Prestimulation of head and neck cancer cells with growth factors enhances treatment efficacy. Anticancer Res. 2006;26:1091–1095. [PubMed] [Google Scholar]
- 49.Nishioka M, Bundo M, Koike S, Takizawa R, Kakiuchi C, Araki T, Kasai K, Iwamoto K. Comprehensive DNA methylation analysis of peripheral blood cells derived from patients with first-episode schizophrenia. J Hum Genet. 2013;58:91–97. doi: 10.1038/jhg.2012.140. [DOI] [PubMed] [Google Scholar]
- 50.Deo RC, Schmidt EF, Elhabazi A, Togashi H, Burley SK, Strittmatter SM. Structural bases for CRMP function in plexin-dependent semaphorin3A signaling. EMBO J. 2004;23:9–22. doi: 10.1038/sj.emboj.7600021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gelfand MV, Hong S, Gu C. Guidance from above: Common cues direct distinct signaling outcomes in vascular and neural patterning. Trends Cell Biol. 2009;19:99–110. doi: 10.1016/j.tcb.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shih JY, Lee YC, Yang SC, Hong TM, Huang CY, Yang PC. Collapsin response mediator protein-1: A novel invasion-suppressor gene. Clin Exp Metastasis. 2003;20:69–76. doi: 10.1023/A:1022598604565. [DOI] [PubMed] [Google Scholar]
- 53.Katagiri T, Takahashi N. Regulatory mechanisms of osteoblast and osteoclast differentiation. Oral Dis. 2002;8:147–159. doi: 10.1034/j.1601-0825.2002.01829.x. [DOI] [PubMed] [Google Scholar]
- 54.Siegel PM, Massagué J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 55.Cox CJ, Espinoza HM, McWilliams B, Chappell K, Morton L, Hjalt TA, Semina EV, Amendt BA. Differential regulation of gene expression by PITX2 isoforms. J Biol Chem. 2002;277:25001–25010. doi: 10.1074/jbc.M201737200. [DOI] [PubMed] [Google Scholar]
- 56.Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, et al. Identification of a Wnt/Dvl/beta-Catenin ->Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/S0092-8674(02)01084-X. [DOI] [PubMed] [Google Scholar]
- 57.Liu W, Selever J, Lu MF, Martin JF. Genetic dissection of Pitx2 in craniofacial development uncovers new functions in branchial arch morphogenesis, late aspects of tooth morphogenesis and cell migration. Development. 2003;130:6375–6385. doi: 10.1242/dev.00849. [DOI] [PubMed] [Google Scholar]
- 58.Iwata J, Tung L, Urata M, Hacia JG, Pelikan R, Suzuki A, Ramenzoni L, Chaudhry O, Parada C, Sanchez-Lara PA, Chai Y. Fibroblast growth factor 9 (FGF9)-pituitary homeobox 2 (PITX2) pathway mediates transforming growth factor β (TGFβ) signaling to regulate cell proliferation in palatal mesenchyme during mouse palatogenesis. J Biol Chem. 2012;287:2353–2363. doi: 10.1074/jbc.M111.280974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hirose H, Ishii H, Mimori K, Tanaka F, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. The significance of PITX2 overexpression in human colorectal cancer. Ann Surg Oncol. 2011;18:3005–3012. doi: 10.1245/s10434-011-1653-z. [DOI] [PubMed] [Google Scholar]
- 60.Fung FK, Chan DW, Liu VW, Leung TH, Cheung AN, Ngan HY. Increased expression of PITX2 transcription factor contributes to ovarian cancer progression. PLoS One. 2012;7:e37076. doi: 10.1371/journal.pone.0037076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vinarskaja A, Schulz WA, Ingenwerth M, Hader C, Arsov C. Association of PITX2 mRNA down-regulation in prostate cancer with promoter hypermethylation and poor prognosis. Urol Oncol. 2013;31:622–627. doi: 10.1016/j.urolonc.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 62.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 64.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 65.Widschwendter M, Apostolidou S, Raum E, Rothenbacher D, Fiegl H, Menon U, Stegmaier C, Jacobs IJ, Brenner H. Epigenotyping in peripheral blood cell DNA and breast cancer risk: A proof of principle study. PLoS One. 2008;3:e2656. doi: 10.1371/journal.pone.0002656. [DOI] [PMC free article] [PubMed] [Google Scholar]