Abstract
Estrogen Receptor-β (ERβ) has been implicated in many cancers. In prostate and breast cancer its function is controversial, but genetic studies implicate a role in cancer progression. Much of the confusion around ERβ stems from antibodies that are inadequately validated, yet have become standard tools for deciphering its role. Using an ERβ-inducible cell system we assessed commonly utilized ERβ antibodies and show that one of the most commonly used antibodies, NCL-ER-BETA, is non-specific for ERβ. Other antibodies have limited ERβ specificity or are only specific in one experimental modality. ERβ is commonly studied in MCF-7 (breast) and LNCaP (prostate) cancer cell lines, but we found no ERβ expression in either, using validated antibodies and independent mass spectrometry-based approaches. Our findings question conclusions made about ERβ using the NCL-ER-BETA antibody, or LNCaP and MCF-7 cell lines. We describe robust reagents, which detect ERβ across multiple experimental approaches and in clinical samples.
Keywords: Estrogen receptor beta, Prostate, Breast, Cancer, Antibody
Highlights
-
•
ERβ is important in prostate and breast cancer, but its role is controversial.
-
•
ERβ antibodies are problematic, with varying specificity.
-
•
We tested a panel of ERβ antibodies and show the most commonly used is non-specific.
-
•
Two antibodies were validated across multiple experimental approaches.
-
•
Using multiple techniques, we show cell lines used to study ERβ lack its expression.
1. Introduction
Estrogen receptor beta (ERβ) was first discovered in the rat prostate (Kuiper et al., 1996). Since then, there has been considerable interest in understanding its role in both breast and prostate cancer. Despite a large body of literature, the function of ERβ in these two cancers remains unclear (Haldosen et al., 2014, Nelson et al., 2014). Most authors agree that ERβ has a predominantly antiproliferative, pro-apoptotic and tumor-suppressive role (Attia and Ederveen, 2012, Bottner et al., 2014, Chang and Prins, 1999, Ellem and Risbridger, 2007, Horvath et al., 2001, Madak-Erdogan et al., 2013, McPherson et al., 2010, Muthusamy et al., 2011, Nakajima et al., 2011, Rizza et al., 2014, Ruddy et al., 2014, Zhu et al., 2004), however ERβ has also been implicated as an oncogene. This is particularly in the context of Castrate Resistant Prostate Cancer (CRPC) where it has been proposed as a driver of androgen receptor (AR)-dependent gene transcription (Yang et al., 2012, Yang et al., 2015), along with a potential role in mediating the transition from hormone-sensitive to CRPC (Zellweger et al., 2013). In breast cancer, it has been suggested that ERβ may have a ‘bi-faceted role’ and should not simply be considered a tumor-suppressor (Jonsson et al., 2014). ERβ has been reported to ‘cross-talk’ with androgen receptor-positive breast cancer (Rizza et al., 2014) and may be an important factor in ERα-negative breast cancer (Gruvberger-Saal et al., 2007, Smart et al., 2013).
Inconsistencies in the reported expression of ERβ in breast and prostate cancers as determined by immunohistochemistry (IHC) have contributed to this uncertainty. In prostate, most data support the conclusion that ERβ is highly expressed in benign epithelial cells, with expression declining in cancer development and inversely correlating with increasing Gleason grade (Asgari and Morakabati, 2011, Attia and Ederveen, 2012, Dey et al., 2014, Horvath et al., 2001, Leav et al., 2001, Risbridger et al., 2007). However, it has also been reported that ERβ expression is high in bone and lymph node metastases (Bouchal et al., 2011, Zhu et al., 2004) and that high ERβ expression correlates with poor clinical prognosis (Horvath et al., 2001, Zellweger et al., 2013). In breast cancer, high ERβ expression has been described both as a poor (Guo et al., 2014a, Guo et al., 2014b) and favorable (Esslimani-Sahla et al., 2004, Gruvberger-Saal et al., 2007, Hieken et al., 2015, Leygue and Murphy, 2013, Myers et al., 2004, Omoto et al., 2002, Roger et al., 2001) prognostic marker, with others finding no association between clinico-pathological parameters and ERβ expression (Umekita et al., 2006).
It is recognized that there is wide variability in the sensitivity and specificity of ERβ antibodies, which may contribute to the uncertainties surrounding its molecular action and tissue expression (Choi et al., 2001, Hartman et al., 2012, Skliris et al., 2002, Weitsman et al., 2006, Wu et al., 2012). Previous ERβ antibody validation studies have been published (Carder et al., 2005, Choi et al., 2001, Skliris et al., 2002, Weitsman et al., 2006, Wu et al., 2012), however some of them are limited by reliance on two key assumptions. Firstly, that when assessing an antibody by Western blotting in a cell line model, the factor of interest is expressed and secondly, when assessing an antibody's specificity by IHC in tissue, the tissue expression of the factor has been well characterized. In the case of ERβ, these assumptions are problematic, as its expression in commonly used cell line models and in tissues is not universally accepted (Al-Bader et al., 2011, Asgari and Morakabati, 2011, Attia and Ederveen, 2012, Bouchal et al., 2011, Dey et al., 2014, Gruvberger-Saal et al., 2007, Guo et al., 2014a, Guo et al., 2014b, Hieken et al., 2015, Holbeck et al., 2010, Horvath et al., 2001, Leav et al., 2001, Nakajima et al., 2011, Omoto et al., 2002, Risbridger et al., 2007, Shaaban et al., 2003, Skliris et al., 2002, Umekita et al., 2006, Zellweger et al., 2013, Zhou et al., 2012, Zhu et al., 2004).
In light of this, we sought to test and validate six commonly used, commercially available ERβ antibodies and two non-commercially available ERβ antibodies (Choi et al., 2001, Wu et al., 2012) in a systematic manner that addresses these assumptions. To achieve this, we employed a number of assays for antibody validation, including a novel proteomic-based pull down method called Rapid Immunopreciptation Mass spectrometry of Endogenous protein (RIME) (Mohammed et al., 2013). We then applied successfully validated antibodies to cell line models of breast and prostate cancer commonly used for studies of ERβ to assess them for ERβ expression. ERβ expression in the cell lines was validated by a non-antibody dependent, targeted proteomics method known as Parallel Reaction Monitoring (PRM) (Gallien et al., 2012). Finally, benign and malignant prostate and breast tissues were stained with the validated ERβ antibody to assess tissue expression of ERβ by IHC.
2. Materials and methods
2.1. Cell culture
The cancer cell line MDA-MB-231 with doxycycline-inducible ERβ expression (MDA-MB-231-ERβ) (Reese et al., 2014) was cultured in Dulbeccos Modified Eagle Medium with F12 supplement (DMEM/F12) with 10% heat-inactivated tetracycline-free fetal bovine serum (FBS) (Fisher-Scientific), 2 mM L-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin, 5 μg/ml blasticidin S (Invivogen) to select for the tetracycline repressor and 500 μg/ml zeocin (Invitrogen) to select for the ERβ expression vector. To induce ERβ expression in MDA-MB 231-ERβ cells, 15 cm2 plates were seeded with 5 × 106 cells and doxycycline added at either 0.1 μg/ml (for Western blot, real-time polymerase chain reaction (qRT-PCR) and PRM) or 0.5 μg/ml (for RIME) for 24 h. The MCF-7 breast cancer cell line was cultured in Dulbecco's Modified Eagle Medium (DMEM) with 10% heat-inactivated FBS (Fisher-Scientific), 2 mM L-glutamine, 50 U/ml penicillin and 50 μg/ml streptomycin. The LNCaP prostate cancer cell line was cultured in RPMI 1640 with 10% heat-inactivated FBS (Fisher-Scientific), 2 mM L-glutamine, 50 U/ml penicillin and 50 μg/ml streptomycin. All cells were incubated at 37 °C with 5% CO2 and cultured to 80–90% confluence. LNCaP and MCF-7 cell lines were obtained from ATCC (Middlesex, UK) and validated by STR genotyping.
2.2. Preparation of mRNA and qRT-PCR
MDA-MB-231-ERβ+, MDA-MB-231-ERβ–, MCF-7 and LNCaP cells were harvested for collection of mRNA using the RNEasy Mini Kit (Qiagen, California USA). On-column DNase digestion was performed to remove contaminating genomic DNA. RNA was quantified with the NanoDrop 8000 (Thermo Scientific, Delaware USA). Samples containing 250 ng random primers, 1 μg RNA, 1 μl 10 mM dNTP mix and water to a total volume of 13 μl were heated to 65 °C for 5 min, followed by 1 min incubation on ice. To each sample 4 μl 5X First-strand buffer, 1 μl 0.1 M DTT, 1 μl RNaseOUT and 1 μl SuperScript III reverse transcriptase (RT) (Thermofisher Scientific, Leicestershire, UK) were added and incubated at 25 °C for 5 min then 50 °C for 60 min followed by heating at 70 °C for 15 min qRT-PCR primers for wild type ERβ (Table 1) were designed based on published sequence of ESR2 (available from USCS genome browser at http://genome.ucsc.edu/) using the Primer3 software package (Koressaar and Remm, 2007, Untergasser et al., 2012) available at http://bioinfo.ut.ee/primer3-0.4.0/primer3/. UBC primers (SY121212648) were obtained from Sigma-Aldrich (Dorset, UK). Each qRT-PCR reaction contained 7.5 μl Power SYBR Green PCR Master Mix (Applied Biosystems, California USA), 0.5 μl of 10 μM primer mix, 2 μl of a 1:5 dilution of cDNA and nuclease-free water to a final volume of 15 μl. Reactions were performed with the Stratagene Mx3005P RealTime machine in triplicate. Hot-start Taq polymerase was heat-activated at 95 °C for 10 min followed by 40 cycles of 15 s at 95 °C and 30 s at 60 °C. Fluorescence was read in each cycle and a melting curve constructed as the temperature was increased from 65 °C to 95 °C with continuous fluorescence readings. UBC was used as a control gene to normalize between the samples and relative expression determined using the delta-delta Ct method (Livak and Schmittgen, 2001).
Table 1.
Sequence of ERβ mRNA primers used in qRT-PCR validation of the MDA-MB-231-ERβ cell line. These primer sequences flank a region spanning exons 2 and 3, which is common to wild type ERβ and ERβ isoforms.
| Primer | Sequence |
|---|---|
| ERβ – fwd | 5′ AAAACCGGCGCAAGAGCTG 3′ |
| ERβ – rev | 3′ TGCTCGTCGGCACTTCTCTG 5′ |
2.3. Western blotting
MDA-MB-231-ERβ+, MDA-MB-231-ERβ–, MCF-7 and LNCaP cells were harvested for nuclear extract using the Ne-Per nuclear extraction kit (Thermo Scientific Pierce, Rockford IL USA) according to the manufacturer's instructions. Extracted protein was quantified using the Direct Detect system (Merrick Millipore, Massachusetts USA). Nuclear extracts were prepared with 4X protein sample loading buffer (LI-COR Biosciences, USA), 10X NuPage sample reducing agent (Thermofisher Scientific, Leicestershire, UK) and water, and 15 μg protein per lane loaded into Bolt 4–12% Bis-Tris gels (Thermofisher Scientific, Leicestershire, UK). Gels were run with MOPS running buffer for 30 min at 60 V followed by 30 min at 120 V. Western transfer was performed using the iBlot system (Invitrogen, Paisley, UK) according to the manufacturer's instructions. Odyssey blocking buffer (LI-COR Biosciences, USA) was added to membranes for one hour at room temperature. Primary antibodies (Table 2 and Supplementary Fig. 1) were added at the following dilutions and incubated overnight at 4 °C: Novocastra-ER-beta (EMR02-NCL-ER-BETA) (Leica Biosystems, Newcastle, UK) 1:100, ERβ1 PPG5/10 (MAI-81281) (Thermo Scientific Pierce, Rockford IL USA) 1:100, ERβ-antibody H150 (sc8974) (Santa Cruz Biotechnology, Dallas TX, USA) 1:200, CWK-F12, USA) (Choi et al., 2001) 1:200, MC10 (Wu et al., 2012) 1:300, GeneTex ERβ 70182 (Irvine, CA, USA) 1:200, ERβ 06-629 (Merck Millipore, Watford, UK), 1:500, Abcam 288 [14C8] (Cambridge, UK) 1:500. The following were used as loading controls: rabbit anti-beta actin (ab8227) (Abcam, Cambridge, UK) 1:5000 or mouse anti-beta actin [AC-15](ab6276) 1:1000 according to the species of the ERβ antibody. The membranes were washed three times with PBS/0.1% tween and incubated with secondary antibodies for one hour at room temperature: Goat anti-mouse (green) 1:5000 with Goat anti-rabbit (red) 1:20000 or Goat anti-rabbit (green) 1:5000 with Goat anti-mouse (red) 1:20000 according to the species of the ERβ antibody. Membranes were imaged using the Li-Cor Odyssey fluorescent imaging system (LI-COR Biosciences, USA).
Table 2.
Details of ERβ antibodies validated. Application details are as recommended by the manufacturer. IHC, immunohistochemistry; WB, western blot; IF, immunofluorescence; ChIP, chromatin immunoprecipitation; ELISA, enzyme-linked immunosorbent assay; Flow cyt, flow cytometry; ICC, immunocytochemistry; IP, immunoprecipitation; Wt, wild type; NTD, N terminal domain; LBD, ligand binding domain.
| Antibody | Immunogen | Host species | Class | Binding region | Application |
|---|---|---|---|---|---|
| NCL-ER-BETA | Recombinant protein. Wt ERβ. C terminus | Mouse | Monoclonal | C terminus | IHC, WB |
| PPG5/10 | Synthetic peptide C terminus of wt ERβ | Mouse | Monoclonal | C terminus | IF, IHC, WB |
| GeneTex 70182 | Amino acids 1-153 of human ERβ expressed in E.coli | Mouse | Monoclonal | N terminus | IP, WB, ChIP |
| Millipore 06-629 | Amino acids 46-63 of human ERβ | Rabbit | Polyclonal | NTD | WB, IHC |
| Santa cruz sc8974 | Amino acids 1-150 of human ERβ | Rabbit | Polyclonal | N terminus | WB, ChIP, IF, ELISA |
| Abcam 288 [14C8] | Recombinant fusion protein. Amino acids 1-153 of human ERβ in E.coli | Mouse | Monoclonal | N terminus | WB, Flow cyt, IHC, ICC, ChIP |
| CWK-F12 | Recombinant protein. Amino acids 272-285 of human wt ERβ | Mouse | Monoclonal | LBD | WB, IP, IHC |
| MC10 | Fusion protein. Amino acids 1-140 of human ERβ in E.coli | Mouse | Monoclonal | N terminus | IHC, IP, WB, IF |
2.4. Immunohistochemistry (IHC)
Formalin-fixed, paraffin-embedded MDA-MB-231-ERβ– and MDA-MB-231-ERβ+ cell pellets were generated, with ∼2 × 107 cells per pellet. ERβ expression was induced with 0.5 μg/ml doxycycline for 24 h. Antigen retrieval was achieved by incubating in citrate-based retrieval solution for 20 min. Sections were stained using CWK-F12 ERβ antibody, diluted 1:250 in standard Bond diluent using Leica's Polymer Refine Kit (Catalogue No: DS9800) on the automated Bond platform (Leica Biosystems Newcastle Ltd, Newcastle UK). Images were captured using Aperio® software (Leica Biosystems Newcastle Lt, Newcastle UK).
A prostate tissue microarray (TMA) was created from a random selection of prostate cancers, including a range of different tumor grades, and benign prostatic tissue (10 cancer, 5 benign in total) (ethical approval: ProMPT study MREC/01/4/061). The areas to be sampled from the formalin-fixed and paraffin embedded tissue blocks were marked on the corresponding Haematoxylin and Eosin stained paraffin sections. Each block was assessed to ensure that there was an adequate amount of tissue for sampling, and cores of tissue punched from the selected area of the block using 5 mm skin biopsy punches. Each core was re-embedded into a new recipient paraffin block and its position in the block recorded on a TMA map. Cores of pig kidney were used as orientation markers.
The breast TMA was constructed using the Chemicon Advanced Tissue Arrayer (Merck Millipore, Germany) according to the manufacturer's instructions. This contained 30 benign samples, 56 grade I, 55 grade II and 57 grade III ER alpha positive tumors. An additional TMA was constructed from 10 invasive carcinomas and 10 non-malignant tissues for optimisation of antibody staining. To ensure adequate representation of the tissue, core size of 1 mm was selected and cores arranged in duplicate with liver and spleen as orientation cores. The study protocol for tissue collection was approved by the University of Adelaide Human Research Ethics Committee (#s H-2005-065).
For the prostate IHC, 3.5 μm sections were cut and mounted onto charged slides, dried and sealed with paraffin. The CWK-F12 ERβ antibody was further optimized to the clinical samples and diluted at 1:200 in diluent consisting of 1% donkey serum, 0.05% Tween20 in 300 mM TBS to reduce background staining. Antigen retrieval was achieved by incubating in Tris EDTA for 20 min at 100 °C. Images were captured at 250 × magnification using Image Pro-Insight (Media Cybernetics. Rockville, MD. USA).
For the breast IHC, 4 μm sections were cut and adhered to Superfrost UltraPLUS slides (Thermo-Fisher Scientific #1014356190). Slides were dewaxed in xylene followed by 100% EtOH and then PBS. Endogenous peroxidase was quenched with 0.3% hydrogen peroxide (Ajax Finchem ##7722-84-1). Antigen retrieval was performed in 10 mM Citric acid buffer (pH 6.0) within a decloaking chamber (Biocare Medical #DC2012), for 5 min at 120 °C. Slides were blocked in 5% normal goat serum (Sigma-Aldrich #G9023) in PBS for 30 min at room temperature. CWK-F12 antibody was added at a dilution of 1:100 and incubated overnight at 4 °C. A second section of TMA tissue that received buffer in the absence of primary antibody served as a negative control. Secondary antibody (biotinylated anti-mouse antibody (Dako #E0433) diluted in PBS with 5% normal goat serum was added and incubated for 60 min at room temperature. Sections were washed twice in PBS followed by addition of HRP-conjugated streptavidin (Dako #P0397). Tissue was counterstained with haematoxylin and mounted under DPX mountant (Sigma #06522). Slides were scanned on a Nanozoomer slide scanner (Hamamatsu #C9600).
2.5. Rapid immunoprecipitation and Mass Spectrometry of Endogenous Protein (RIME)
RIME experiments were conducted as previously described (Mohammed et al., 2013). Briefly, MDA-MB-231-ERβ+, MDA-MB-231-ERβ– (2 × 107 cells per condition for antibody evaluation), LNCaP and MCF-7 cells (4 × 107 cells per condition for cell line characterization) were grown in 15 cm2 plates to 90% confluency. Cells were crosslinked with media containing 1% EM grade formaldehyde (TEBU biosciences, Peterborough UK) for 8 min and the formaldehyde quenched with 0.1 M glycine. Cells were washed, harvested and pelleted in cold PBS. To enrich the nuclear fraction the cell pellet was suspended in 10 ml of lysis buffer 1 (50 mM HEPES-KOH [pH 7.5], 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40 or Igepal CA-630, and 0.25% Triton X-100) for 10 min at 4 °C. Cells were pelleted and resuspended in lysis buffer 2 (10 mM Tris-HCL [pH 8.0], 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA) for five minutes at 4 °C. Cells were pelleted and resuspended in 300 μl of lysis buffer 3 (10 mM Tris-HCl [pH 8], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-deoxycholate, and 0.5% N-lauroylsarcosine) and sonicated (Diagenode bioruptor. Diagenode, Seraing Belgium) for 45 min 30 μl of 10% Triton-X was added and the sonicated lysate centrifuged at 17,000G for 10 min to remove cell debris. The supernatant was incubated with 100 μl of magnetic beads (Dynabeads®, Thermo Fisher Scientific, Waltham MA USA) pre-bound with antibody.
For evaluation of the 8 ERβ antibodies, immunoprecipitations (IP) were set up each for MDA-MB-231-ERβ– and MDA-MB-231-ERβ+ cells using 10 μg of antibody (NCL-ER-BETA, GeneTex 70182, Millipore 06-629, Abcam 288 [14C8], MC10, CWK-F12, sc8974 and PPG5/10). For characterization of LNCaP and MCF-7 cells, 20 μg of MC10 ERβ antibody was used in each IP. In all cases, 10 μg of E2F1-C20 IP was used as a positive control (Sc-193, Santa Cruz Biotechnology, Dallas TX, USA) and species-specific IgG used to detect non-specific pull-down (Mouse sc2025 or Rabbit sc2027, Santa Cruz Biotechnology, Dallas TX, USA). Samples were incubated overnight at 4 °C. Beads were washed 10 times in 1 ml RIPA buffer (50 mM HEPES pH 7.6, 1 mM EDTA, 0.7% Na deoxycholate, 1% NP-40, 0.5 M LiCL) and twice in 100 mM ammonium hydrogen carbonate (AMBIC) solution. Dry, frozen beads were submitted for tryptic digestion of bead-bound protein, and peptides pulled down by IP identified by mass-spectrometry (LTQ Velos-Orbitrap MS, Thermo Fisher Scientific, Waltham MA USA). Raw MS data files were processed using Proteome Discoverer v.1.3 (Thermo Scientific). Processed files were searched against the SwissProt human database using the Mascot search engine version 2.3.0 with a false discovery rate (FDR) of less than 1%. For each ERβ antibody tested, the resulting list of purified peptides identified was filtered against the corresponding IgG control to remove non-specific proteins pulled down. Mean percentage ERβ peptide coverage, and mean number of unique ERβ peptides identified in biological duplicate experiments were calculated.
2.6. Parallel Reaction monitoring (PRM)
Nuclear pellets of MDA-MB-231-ERβ+, MDA-MB-231-ERβ–, LNCaP and MCF-7 cells were prepared using the Panomics nuclear extraction kit (Affymetrix, CA USA) as per the manufacturer's provided instructions. Nuclear pellets were lysed in 8 M Urea, 0.1% SDS in 50 mM TEAB by sonication twice, each for 5 min. After protein estimation 20 μg of protein was taken for tryptic digestion. 50 mM of TEAB (pH = 8) was added up to a total volume of 100 μl. Cysteines were reduced in 0.1 mM DTT for 1 h at room temperature and alkylated in 0.1 mM IAA for 30 min at room temperature in the dark. Alkylation was quenched by adding 0.1 mM DTT for 15 min. Trypsin (Promega trypsin (V5111)) was added in a 1:100 trypsin:protein ratio for 1 h at room temperature. Another batch of trypsin (1:100 ratio) was added to have a final ratio of 1:50 for incubation overnight. Samples were acidified to a final concentration of 1% formic acid (FA) and cleaned over C18 spin columns (Harvard apparatus C18 Micro SpinColumn™). After elution from the columns samples were lyophilized in a speedvac and resolubilized in 0.1% FA, 5% ACN to a final peptide concentration of 1 μg/μl. Samples were subjected to liquid chromatography-electrospray ionization in an Orbitrap nano-ESI Q-Exactive mass spectrometer (Thermo Scientific), coupled to a nanoLC (Dionex Ultimate 3000 UHPLC). Samples were trapped on a 100 μm × 2 cm, C18, 5 μm, 100 trapping column (Acclaim PepMap 100) in μL-pickup injection mode at 4 μL/min flow rate for 10 min. Samples were loaded on a Rapid Separation Liquid Chromatography, 75 μm × 25 cm nanoViper C18 3 μm 100 column (Acclaim, PepMap) retrofitted to an EASY-Spray source with a flow rate of 300 nL/min (buffer A, HPLC H2O, 0.1% FA; buffer B, 100% ACN, 0.1% FA; 60-min gradient; 0–5 min: 5% buffer B, 5–45 min: 5 to >56% buffer B, 45.1–50 min: 56% to >95% buffer B, 50.1–60 min, 5% buffer B). Peptides were transferred to the gaseous phase with positive ion electrospray ionization at 1.8 kV. Precursors were targeted in a 2Th selection window around the m/z of interest. Precursors were fragmented in high-energy collisional dissociation mode with normalized collision energy dependent on the target peptide. The first mass analysis was performed at a 70,000 resolution, an automatic gain control target of 3 × 106, and a maximum C-trap fill time of 200 ms; MS/MS was performed at 35,000 resolution, an AGC target of 5 × 104, and a maximum C-trap fill time of 100 ms. Spectra were analyzed using Skyline with manual validation.
2.7. Statistics
Differences in ERβ mRNA levels observed in MDA-MB-231-ERβ– and MDA-MB-231-ERβ+ conditions were analyzed using unpaired t-tests. Differences were considered statistically significant at p ≤ 0.05. Data presented are mean of technical triplicate experiments ± standard deviation. Analysis was performed in GraphPad Prism version 6.
3. Results
3.1. ERβ antibody validation
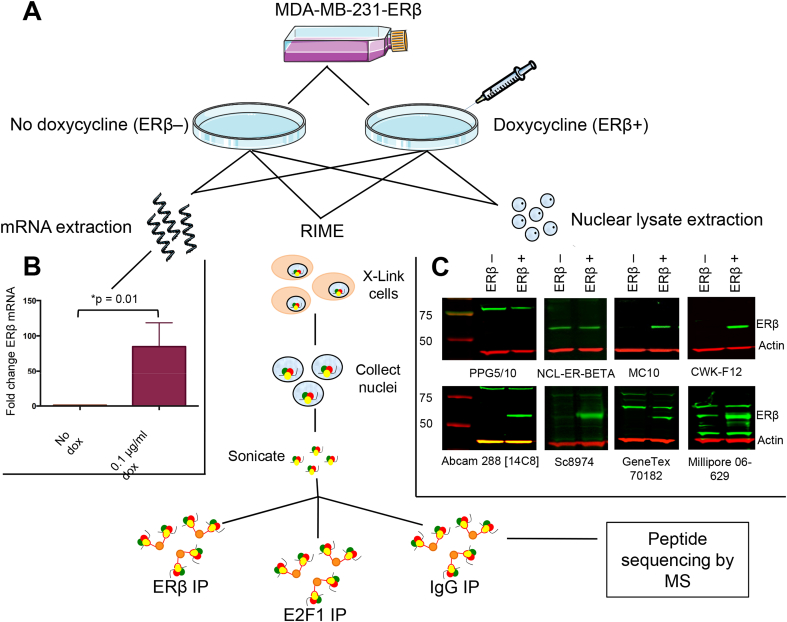
Given the confusion in the ERβ field and the concern associated with variable and potentially non-specific reagents, we sought to extensively validate commonly used ERβ antibodies in a systematic manner that does not rely upon a priori assumptions regarding ERβ expression in cell line models or in tissues. As a control, we employed a cell line system with doxycycline-inducible expression of the ERβ protein, allowing us to assess antibodies in ERβ negative and matched ERβ positive conditions (Fig. 1A). One hundred-fold induction of ERβ mRNA in MDA-MB-231-ERβ cells treated with doxycycline 0.1 μg/ml for 24 h (p = 0.01) was confirmed by qRT-PCR (Fig. 1B).
Fig. 1.
Validation of ERβ antibodies using doxycycline-inducible MDA-MB-231-ERβ cells. (A) MDA-MB-231-ERβ cells were treated with doxycycline to induce ERβ expression. Untreated cells provided an ERβ-negative control. Messenger RNA was extracted for qRT-PCR and protein for Western blotting. MDA-MB-231-ERβ+ and MDA-MB-231-ERβ– cells were crosslinked and immunoprecipitated with antibody for RIME. (B) qRT-PCR confirmed 100-fold induction of ERβ mRNA in MDA-MB-231-ERβ+ cells versus untreated MDA-MB-231-ERβ– cells. Data are mean ± S.D. of technical triplicate experiments. (C) Western blots of MDA-MB-231-ERβ+ and MDA-MB-231-ERβ– cells with the 8 antibodies undergoing assessment. The MC10, CWK-F12, Abcam 288[14C8] and sc8974 antibodies detected bands of 59 kDa, with differential signal in the ERβ+ versus ERβ– conditions, indicating specificity to ERβ. GeneTex 70182 detected ERβ, although there was non-specific signal at 65 kDa. Millipore 06-629 appears to detect ERβ, although there is also a 59 kDa band in the ERβ– condition. Review of the RIME data suggests this may be cross-reactivity with LACTB. NCL-ER-BETA, the most commonly used ERβ antibody, gives bands of the correct size for ERβ, but there is no difference between ERβ– and ERβ+ conditions, confirming that this antibody is not specific to ERβ.
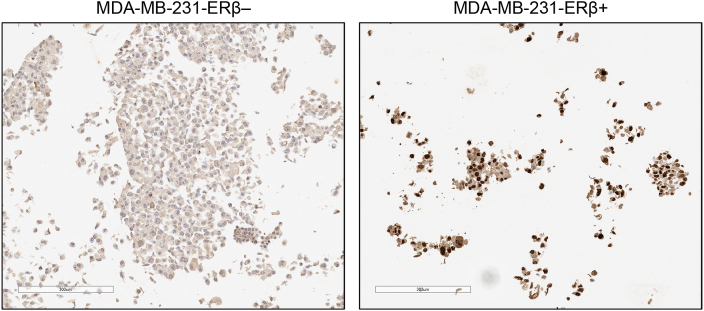
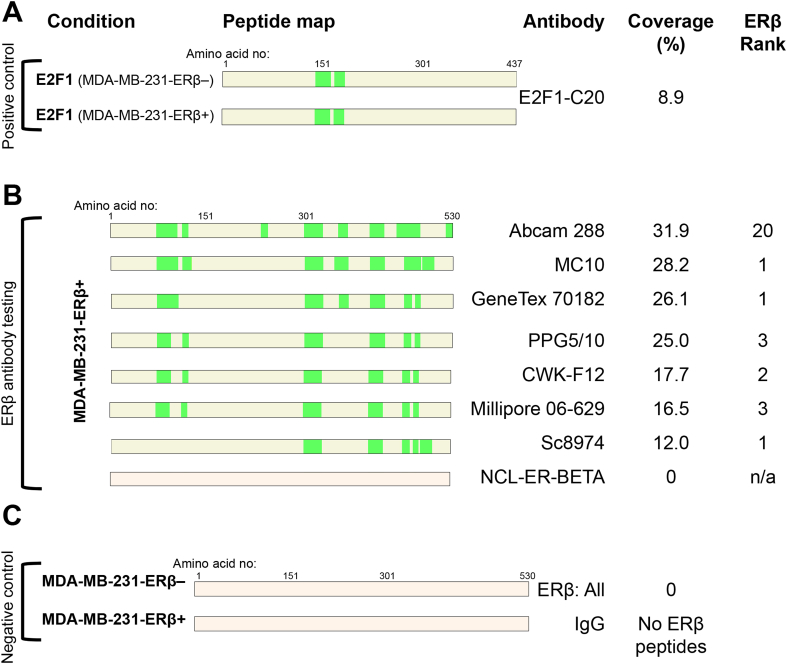
Western blots of MDA-MB-231-ERβ+ and MDA-MB-231-ERβ– cell lysates with 8 different ERβ antibodies were performed (Fig. 1C). Six commonly used antibodies in the literature were included; PPG5/10 (Asgari and Morakabati, 2011, Carder et al., 2005, Ciucci et al., 2014, Shaaban et al., 2003, Wimberly et al., 2014), NCL-ER-BETA (Ellem et al., 2014, Hussain et al., 2012, McPherson et al., 2007; 2010; Morais-Santos et al., 2015, Oliveira et al., 2007, Umekita et al., 2006, Yang et al., 2015, Zellweger et al., 2013), Genetex 70182 (Celhay et al., 2010, Madak-Erdogan et al., 2013, Mak et al., 2013, Mak et al., 2015a, Mak et al., 2015b, ; Nakajima et al., 2011), Millipore 06-629 (Bouchal et al., 2011, Chen et al., 2009, Grubisha et al., 2012), Abcam 288 [14C8] (Abd Elmageed et al., 2013, Carder et al., 2005, Colciago et al., 2014, Cotrim et al., 2013, Dey et al., 2012, Dey et al., 2014, Setlur et al., 2008, Shaaban et al., 2003, Vivar et al., 2010, Yang et al., 2012) and Santa Cruz 8974 (Al-Bader et al., 2011, Foryst-Ludwig et al., 2008, Han et al., 2015, Rossi et al., 2011, Zhou et al., 2012) antibodies. The PPG5/10 antibody detected a protein band of 77 kDa with no difference between ERβ+ or ERβ– conditions, suggesting it is recognizing a non-specific protein. Similarly, the NCL-ER-BETA antibody detected a band of ∼59 kDa, which is the correct size for ERβ however, there was no difference between ERβ+ or ERβ– conditions implying that this band was not ERβ. The GeneTex 70182 antibody detected a band of 59 kDa with differential signal between ERβ+ and ERβ– conditions, and a non-specific band was present at around 65 kDa. The Millipore 06-629 antibody detected a band of 59 kDa in both ERβ+ and ERβ– conditions, however the band was stronger in the ERβ+ condition, suggesting that the antibody could be cross-reacting with another protein of 59 kDa in addition to detecting ERβ. MC10, CWK-F12, Abcam 288 [14C8] and sc8974 ERβ antibodies all detected protein bands of 59 kDa with differential signal between ERβ+ and ERβ– conditions, confirming their specificity for ERβ by Western blotting. Further confirmation of the specificity of CWK-F12 to ERβ was demonstrated by IHC of MDA-MB-231-ERβ+ and MDA-MB-231-ERβ– cell pellets (Fig. 2), showing differential nuclear staining between the two conditions. The 8 ERβ antibodies were then assessed by an independent method called RIME, which uses an antibody-based purification followed by mass spectrometry (MS) to identify enriched peptides. We conducted RIME in MDA-MB-231-ERβ– and MDA-MB-231-ERβ+ cells using all 8 antibodies. E2F1 antibody was included in parallel as a positive control since E2F1 is a ubiquitous protein (Fig. 3A) and an IgG was used as a negative control (Fig. 3C). In MDA-MB-231-ERβ– cells, no ERβ peptides were purified by any of the ERβ antibodies, confirming the ERβ negative status of the uninduced MDA-MB-231-ERβ cell line (Fig. 3C). Following ERβ induction, RIME revealed diverse coverage of the ERβ protein by the different antibodies. The percent coverage of the ERβ protein following purification with each of the ERβ antibodies, and the location of the peptide fragments identified by MS are shown in Fig. 3B. To provide an indication of the specificity of each antibody, we ranked all the proteins purified by the IP and identified by MS according to the number of unique peptides (confirmed with a false discovery rate (FDR) of <1%). We hypothesized that the higher the ranking of ERβ, the greater the specificity of the antibody. Hence, if ERβ has the greatest number of unique peptides relative to all other proteins, it is ranked 1st.
Fig. 2.
IHC validation of CWK-F12 ERβ antibody in MDA-MB-231-ERβ cell pellets. Nuclear staining is evident in MDA-MB-231-ERβ+ cells and absent from the MDA-MB-231-ERβ– control, confirming the specificity of CWK-F12 to ERβ.
Fig. 3.
RIME demonstrates specificity and peptide coverage of ERβ antibodies. Eight ERβ antibodies were assessed by RIME in MDA-MB-231-ERβ+/– cells. Coverage of the protein relates to green areas on the peptide maps, indicating peptides identified by MS with false discovery rate of ≤1% (mean of 2 biological replicates). (A) E2F1 antibody was applied to MDA-MB-231-ERβ– and MDA-MB-231-ERβ+ conditions as a positive control, as E2F1 is a ubiquitously expressed protein. (B) ERβ antibody tests: ‘ERβ ranking’ indicates where ERβ features in a list of proteins purified by the antibody, ranked by number of unique peptides identified in MS, giving an indication of antibody specificity. NCL-ER-BETA failed to identify ERβ. (C) Negative controls: All of the ERβ antibodies were tested in MDA-MB-231-ERβ– cells, to confirm absence of ERβ expression in the non-induced condition. Mouse IgG antibodies were used to identify non-specific peptides pulled down by the IP. None of the IgG antibodies purified ERβ.
NCL-ER-BETA did not purify any ERβ peptides (Fig. 3B), which is consistent with the lack of specificity identified from the Western blot result (Fig. 1C). The Millipore 06-629 antibody positively pulled down ERβ in the test condition, although coverage and ranking were not as favorable as compared with some of the other antibodies. Interestingly, LACTB, a 60 kDa protein was purified by Millipore 06-629 in both ERβ+ and ERβ– conditions (data not shown), which may explain the ∼60 kDa band identified from Western blotting. Whilst the PPG5/10 did not detect ERβ by Western blotting, by RIME it detected ERβ with 25% coverage, with ERβ ranking 3rd in the list of identified peptides, suggesting differences in the specificity of this antibody from one experimental assay to another. PPG5/10 has been previously validated for IHC in a doxycycline-inducible U2OS-ERβ cell line, developed using the same plasmids as the MDA-MB-231-ERβ cell line (Wu et al., 2012). The Abcam 288 [14C8] antibody is a very commonly used ERβ antibody (Abd Elmageed et al., 2013, Colciago et al., 2014, Cotrim et al., 2013, Dey et al., 2012, Dey et al., 2014, Setlur et al., 2008, Shaaban et al., 2003, Vivar et al., 2010, Yang et al., 2012), which performed well by Western blotting, and also had the best antibody coverage by RIME (31.9%). However ERβ ranked 20th in the list of identified peptides when using Abcam 288 [14C8], suggesting that this antibody might also be purifying additional non-specific proteins. The MC10 antibody had the second-greatest coverage (28.2%) with ERβ ranking 1st in the list of identified peptides. In view of this finding, along with the positive Western blot result (Fig. 1), the MC10 antibody was carried forward into the RIME experiments for the cell line characterization. The CWK-F12 antibody had 17.7% coverage, with ERβ ranking 2nd in the list of purified peptides. As the CWK-F12 antibody produced very clean results by Western blotting, IHC and ranked ERβ second in the list of purified proteins, it was used for Western blotting in the cell line characterization and directly compared against the non-specific NCL-ER-BETA antibody. The goal was to use independent validated ERβ antibodies and additional independent methods to assess whether the most commonly studied breast and prostate cancer cell line models express ERβ.
3.2. Characterization of LNCaP and MCF-7 cell lines for ERβ expression
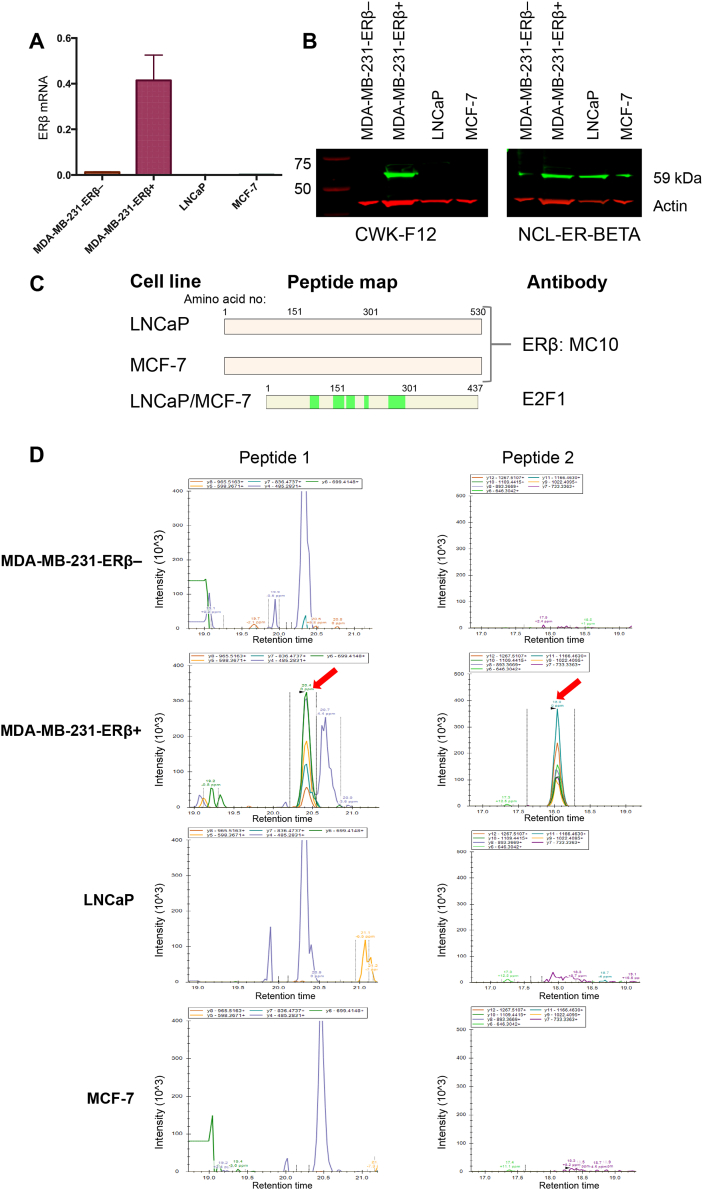
Given the wealth of publications assessing ERβ in breast (MCF-7) and prostate (LNCaP) cancer cell lines (Abd Elmageed et al., 2013, Al-Bader et al., 2011, Bouchal et al., 2011, Chen et al., 2009, Dey et al., 2014, Ellem et al., 2014, Fuqua et al., 1999, Hinsche et al., 2015, Kim et al., 2002a, Kim et al., 2002b, Lau et al., 2000, Mak et al., 2013, Shaaban et al., 2003, Skliris et al., 2002, Weng et al., 2013, Yang et al., 2012, Yang et al., 2015, Zhou et al., 2012), we sought to investigate the expression of ERβ in these models, using the newly validated ERβ antibodies. Protein lysate and RNA was collected from LNCaP and MCF-7 cells. Using primers validated in the inducible MDA-MB-231-ERβ cell line, which binds to sequence common to wild type (wt) ERβ and its isoforms (Fig. 1B), LNCaP and MCF-7 were shown to express no detectable levels of ERβ mRNA (Fig. 4A). Using the validated CWK-F12 ERβ antibody, ERβ protein was undetectable by Western blotting in these cells. By way of contrast, using the NCL-ER-BETA antibody on the same cell lysates, we detected a protein band of approximately 59 kDa in all conditions tested, including the MDA-MB-231-ERβ– cell line, confirming the non-specificity of this antibody to ERβ (Fig. 4B). Importantly, this demonstrates that the NCL-ER-BETA antibody is not detecting ERβ in either LNCaP or MCF-7 cancer cell line models and is instead identifying a non-specific protein of similar molecular weight.
Fig. 4.
Multimodal characterization of LNCaP and MCF-7 cell lines confirms absence of ERβ expression. LNCaP and MCF-7 are cell lines commonly used to study ERβ. We detected no ERβ expression in either cell line at mRNA level by qRT-PCR (A) or at protein level by western blot (B) or RIME (C) using validated CWK-F12 and MC10 antibodies respectively. Western blot of the same cell lysates using the NCL-ER-BETA antibody clearly shows how some of the conflicting data in the literature has arisen, as this antibody shows bands of the correct size for ERβ in all conditions including MDA-MB-231-ERβ– negative control. (D) PRM confirms, using an antibody-independent technique, the absence of ERβ protein expression in LNCaP and MCF-7 cells. The ERβ peptides (peptide 1 is SLEHTLPVNR and peptide 2 is SSITGSECSPAEDSK) identified in the MDA-MB-231-ERβ+ positive control (red arrows) are absent in the other cell lines. Data shown are representative of 2 independent biological replicates.
Furthermore, RIME analysis of LNCaP and MCF-7 cells using the validated MC10 ERβ antibody did not purify any ERβ peptides by MS (Fig. 4C). This result was confirmed by an antibody-independent approach known as Parallel Reaction Monitoring (PRM), which demonstrated that no ERβ peptides were present in either of these cell lines (Fig. 4D). As such, our early passage LNCaP and MCF-7 cell line models are ERβ negative and these cancer models should not be used for the analysis of this protein.
3.3. ERβ expression in prostate and breast tissue
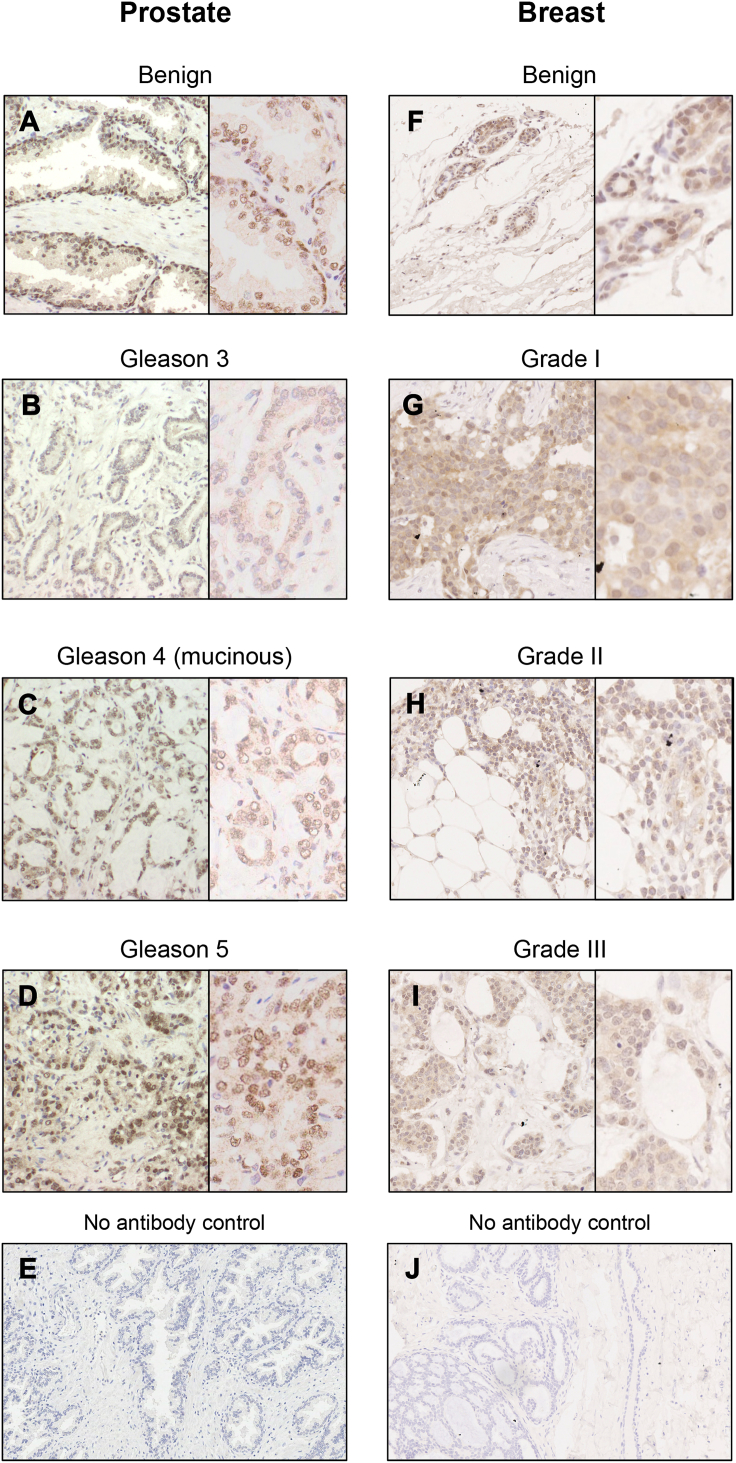
Importantly, whilst the LNCaP and MCF-7 cell-line models do not express ERβ, application of the validated CWK-F12 ERβ antibody to prostate and breast cancer TMAs demonstrated variable ERβ expression in differing cancer grades. In prostate tissue, previous reports have described an inverse correlation between ERβ expression and increasing Gleason grade of prostate cancer (Asgari and Morakabati, 2011, Attia and Ederveen, 2012, Dey et al., 2014, Horvath et al., 2001, Leav et al., 2001, Risbridger et al., 2007), whereas others have reported an association between increased ERβ expression and higher Gleason grade (Zellweger et al., 2013) or increased expression of ERβ in bone and lymph node metastases (Bouchal et al., 2011, Zhu et al., 2004). In our prostate TMA (Fig. 5A–D) we observed high expression of ERβ in the basal epithelium of benign glands, with no expression in Gleason grade 3 cancer. Gleason grade 4 cancer showed weak nuclear staining of ERβ and in areas of Gleason grade 5 cancer, ERβ nuclear expression was of moderate intensity. In breast tissue, previous studies have shown greatest ERβ expression in benign tissue, with a gradual decrease in expression associated with increasing cancer grade (Guo et al., 2014a, Guo et al., 2014b, Omoto et al., 2002). Conversely, a non-statistically significant trend towards higher ERβ expression in Grade 3 tumors has also been reported (Myers et al., 2004). In our breast TMA (Fig. 5F–I), we observed greatest expression of ERβ in benign epithelium, with a trend towards decreasing ERβ expression associated with increasing cancer grade.
Fig. 5.
IHC of prostate and breast tissue with validated CWK-F12 ERβ antibody. Demonstration of variable ERβ expression in differing grades of prostate (A–D) and breast (F–I) cancer. In prostate, ERβ was highly expressed in basal and luminal epithelial cells of benign glands (A), whereas there was no nuclear staining in Gleason grade 3 cancer (B). In Grade 4 mucinous tumor (C) and high grade tumor (D) nuclei showed weak to moderate expression of ERβ. In breast, ERβ expression was greatest in nuclei of benign epithelial cells (F), which was observed to decrease with increasing grade of cancer (G, H and I – Grade 1, 2 and 3 respectively). The greatest difference in expression was observed between benign (F) and Grade 3 cancer (I). E and I - no primary antibody negative controls.
One potential explanation for the inconsistencies in ERβ tissue expression is the presence of ERβ splice-variant isoforms, which are fully conserved in exons 1–6, but have differing C-terminal domains (Leung et al., 2006). Different antibodies that bind either to the conserved regions or only to the C-terminal domain of the full-length ERβ protein may therefore give differing results (Supplementary Fig. 1). This may particularly be the case in prostate cancer, where it has been reported that ERβ isoform expression increases with the development of CRPC (Dey et al., 2012, Leung et al., 2010). Whilst this is likely to have an impact, our data suggest that some of the differing conclusions around ERβ expression in primary tissues are a direct result of certain investigations utilizing non-specific reagents that lack specificity for ERβ.
4. Discussion
Despite a large body of published literature, the role of ERβ in cancers of the prostate and breast is not clear. Contradictions between IHC findings and antibody-dependent molecular biology methods have contributed to this uncertainty, particularly the lack of clear consensus regarding correlation between tissue expression of ERβ and clinico-pathological parameters (Asgari and Morakabati, 2011, Attia and Ederveen, 2012, Bouchal et al., 2011, Dey et al., 2014, Esslimani-Sahla et al., 2004, Guo et al., 2014a, Guo et al., 2014b, Hieken et al., 2015, Horvath et al., 2001, Leav et al., 2001, Leygue and Murphy, 2013, Myers et al., 2004, Omoto et al., 2002, Risbridger et al., 2007, Roger et al., 2001, Umekita et al., 2006, Zellweger et al., 2013, Zhu et al., 2004).
Our results have demonstrated marked variation in the ability of commonly used commercially available ERβ antibodies to accurately detect ERβ by Western blotting and protein purification-MS based methods. Most notably, NCL-ER-BETA, a commonly used antibody (Ellem et al., 2014, Hussain et al., 2012, McPherson et al., 2007; 2010; Morais-Santos et al., 2015, Oliveira et al., 2007, Umekita et al., 2006, Yang et al., 2015, Zellweger et al., 2013) did not detect ERβ by any methodological approach. This antibody consistently yields bands on Western blots of the appropriate size for ERβ (59 kDa) in all tested conditions (Figs. 1C and 3B), but we have confirmed that this protein band is not ERβ through the use of the MDA-MB-231-ERβ inducible cell line system and the RIME technique. As such, this non-specific ∼59 kDa band is likely to be the source of much of the controversy and confusion surrounding the study and characterization of ERβ. The PPG5/10 antibody targets the C-terminus of wt ERβ, and as such may be useful for distinguishing wt ERβ from expression of ERβ isoforms. PPG5/10 identified ERβ in the MDA-MB-231-ERβ cell line by RIME, and has previously been shown to be ERβ-specific by IHC in both an inducible cell line model (Wu et al., 2012) and in breast tissue (Carder et al., 2005). However, in our study this antibody did not show specificity by Western blot analysis (Fig. 1C). In their antibody validation study, Carder et al. also assessed the Abcam 288[14C8] antibody and found it to be ERβ-specific for IHC in tissue (Carder et al., 2005). Whilst our Western blotting data support this assertion (Fig. 1C), our RIME data suggest that this antibody also purifies additional, non-specific peptides, and as such should be used with caution for IP-based methods (Fig. 3B). Taken together, these findings reassert the importance of validating antibodies for individual experimental approaches, rather than assuming general applicability across methodological platforms (Baker, 2015, Bordeaux et al., 2010).
RIME was initially developed as a discovery tool to study the interacting proteomes of transcription factors in an unbiased manner (Mohammed et al., 2013). The advantage of using RIME in antibody validation arises from being able to identify specific, named peptides purified by an antibody, rather than relying on the presence of a protein band of approximate size on a Western blot. This is typified by the NCL-ER-BETA antibody, which gave bands on Western blot in both ERβ– and ERβ+ conditions and no ERβ peptides identified by RIME. Taken together, these data confirm that this antibody is not specific to ERβ. The non-commercially available ERβ antibodies tested (MC10 and CWK-F12) have been previously validated by other approaches (Choi et al., 2001, Wu et al., 2012) and our results add further confidence in their specificity using multiple independent assays. By comparing the peptide coverage of each antibody along with the ERβ ranking (as a surrogate of specificity) RIME facilitated an informed decision-making process in selecting which antibody to carry forward to the cell-line characterization. Our multi-modal approach to cell-line characterization using both antibody-dependent (Western blotting and RIME) and antibody-independent (qRT-PCR and PRM) approaches has shown that low-passage, genotyped LNCaP and MCF-7 cell lines do not express detectable ERβ, despite numerous publications making conclusions about ERβ biology using these cell line models (Abd Elmageed et al., 2013, Al-Bader et al., 2011, Bouchal et al., 2011, Chen et al., 2009, Dey et al., 2014, Ellem et al., 2014, Fuqua et al., 1999, Hinsche et al., 2015, Kim et al., 2002a, Kim et al., 2002b, Lau et al., 2000, Mak et al., 2013, Weng et al., 2013, Yang et al., 2012, Yang et al., 2015). Whilst we acknowledge that immortalized cell lines may have variable expression of certain factors across passage numbers and laboratories (Masters, 2000), our data suggest the need for caution in making this assumption with respect to ERβ. Reassuringly, we have confirmed expression of ERβ in prostate and breast tissue using the validated CWK-F12 antibody. Our IHC study is not intended to be an exhaustive analysis of ERβ expression in prostate and breast tissue, and we acknowledge the limitations presented by our small sample size and lack of statistical correlation with clinico-pathological parameters. We have however, demonstrated that the CWK-F12 ERβ antibody is validated for IHC and in principle can be used for larger scale assessment of ERβ expression in tissue.
Epidemiological evidence suggests that estrogen and its receptors have important roles in the development and progression of prostate cancer. Japanese men are known to have a very low incidence of prostate cancer (Ross et al., 1992), and it has been proposed that their traditional diet, which is high in ERβ selective phytoestrogens may exert a protective role (Andres et al., 2011, Attia and Ederveen, 2012, Hori et al., 2011, Messina, 2010, Reiter et al., 2011, Shen et al., 2000, Stettner et al., 2007, Thelen et al., 2007, Thelen et al., 2005, Wuttke et al., 2002). Further evidence from studies of ERβ knockout mice (βERKO) shows a clear phenotype and tumor-suppressive effect of ERβ (Ricke et al., 2008). However, clinical trials of agents seemingly effective in vitro have demonstrated no clinical benefit of estrogen-selective agents in prostate cancer (Bergan et al., 1999, Kim et al., 2002a, Kim et al., 2002b). There are numerous explanations as to why this might be, for example, expression of ERβ in non-epithelial cell types (Gargett et al., 2002, Pierdominici et al., 2010) modulating the tissue response to these agents, but in light of our findings we would suggest that use of poorly validated reagents and inadequately characterized cancer cell line models is an important contributing factor.
In the presented study, detailed validation of commonly used ERβ antibodies has demonstrated that some of these reagents either detect ERβ in specific experimental conditions only or lack any specificity for ERβ across multiple assays. ERβ has been investigated in numerous cancers including prostate, breast, kidney (Yu et al., 2013), colon (Dey et al., 2013), endometrium (Han et al., 2015), ovary (Ciucci et al., 2014, Suzuki et al., 2008) bladder (Hsu et al., 2013) and non-small cell lung cancer (He et al., 2015, Luo et al., 2015) but in many cases, the findings are predicated on non-specific reagents. As such, a re-evaluation of ERβ expression and biology is needed using reliable, specific reagents. Our determination of ERβ antibody specificity will contribute towards clarifying existing, conflicting data on the role of ERβ in these diverse cancers and provide the necessary, validated tools with which to move forward our understanding of ERβ biology.
Ethics
Prostate tissue included in the tissue microarray was obtained under approval granted for the ProMPT study (MREC/01/4/061). Breast tissue collection and assessment was approved by the University of Adelaide Human Research Ethics Committee (#s H-2005-065).
Financial support
AWN is supported by The Medical Research Council (MR/L00156X/1) and The Urology Foundation Scholarship (RESCH1302). WDT is supported by the National Health and Medical Research Council of Australia (ID 1008349 and 1084416), Cancer Australia/National Breast Cancer Foundation (ID 1043497), National Breast Cancer Foundation Pilot Study (PS-15-041) and a Prostate Cancer Research Program Transformative Impact Award from the US Department of Defense (W81XWH-13-2-0093). BSK receives a grant from the Breast Cancer Research Foundation. JSC is funded by Cancer Research UK and an ERC Consolidator award.
Conflicts of interest
The authors confirm there are no conflicts of interest to disclose.
Acknowledgements
AWN is an Honorary Research Fellow of the Royal College of Surgeons of England and acknowledges their support. The authors are grateful to Dr. Rasmus Siersbaek for providing MCF-7 cells, Ms. Marie Pickering for her assistance with IHC, and the staff of the Cambridge Institute's core facilities, particularly Biorepository, Proteomics, Histopathology and Research Core Instrumentation Facility. The authors acknowledge the Breast Cancer Research Foundation, Cancer Research UK, ERC Consolidator award (grant number 646876), Cambridge Biomedical Research Campus and Cambridge Cancer Centre, which fund the tissue bank and the Urology Biorepository.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.mce.2016.11.016.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
ERβ Antibody binding sites. Schematic of the full length, wild type (wt) ERβ protein, indicating the binding sites of the 8 antibodies evaluated. NTD, N terminal domain; DBD, DNA-binding domain; LBD, ligand-binding domain.
References
- Abd Elmageed Z.Y., Moroz K., Srivastav S.K., Fang Z., Crawford B.E., Moparty K., Thomas R., Abdel-Mageed A.B. High circulating estrogens and selective expression of ERbeta in prostate tumors: implications for racial disparity of prostate Cancer. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bader M., Ford C., Al-Ayadhy B., Francis I. Analysis of estrogen receptor isoforms and variants in breast cancer cell lines. Exp. Ther. Med. 2011;2:537–544. doi: 10.3892/etm.2011.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres S., Abraham K., Appel K.E., Lampen A. Risks and benefits of dietary isoflavones for cancer. Crit. Rev. Toxicol. 2011;41:463–506. doi: 10.3109/10408444.2010.541900. [DOI] [PubMed] [Google Scholar]
- Asgari M., Morakabati A. Estrogen receptor beta expression in prostate adenocarcinoma. Diagn Pathol. 2011;6:61. doi: 10.1186/1746-1596-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia D.M., Ederveen A.G. Opposing roles of ERalpha and ERbeta in the genesis and progression of adenocarcinoma in the rat ventral prostate. Prostate. 2012;72:1013–1022. doi: 10.1002/pros.21507. [DOI] [PubMed] [Google Scholar]
- Baker M. Reproducibility crisis: blame it on the antibodies. Nature. 2015;521:274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- Bergan R.C., Reed E., Myers C.E., Headlee D., Brawley O., Cho H.K., Figg W.D., Tompkins A., Linehan W.M., Kohler D., Steinberg S.M., Blagosklonny M.V. A Phase II study of high-dose tamoxifen in patients with hormone-refractory prostate cancer. Clin. Cancer Res. 1999;5:2366–2373. [PubMed] [Google Scholar]
- Bordeaux J., Welsh A., Agarwal S., Killiam E., Baquero M., Hanna J., Anagnostou V., Rimm D. Antibody validation. Biotechniques. 2010;48:197–209. doi: 10.2144/000113382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottner M., Thelen P., Jarry H. Estrogen receptor beta: tissue distribution and the still largely enigmatic physiological function. J. Steroid Biochem. Mol. Biol. 2014;139:245–251. doi: 10.1016/j.jsbmb.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Bouchal J., Santer F.R., Hoschele P.P., Tomastikova E., Neuwirt H., Culig Z. Transcriptional coactivators p300 and CBP stimulate estrogen receptor-beta signaling and regulate cellular events in prostate cancer. Prostate. 2011;71:431–437. doi: 10.1002/pros.21257. [DOI] [PubMed] [Google Scholar]
- Carder P.J., Murphy C.E., Dervan P., Kennedy M., McCann A., Saunders P.T., Shaaban A.M., Foster C.S., Witton C.J., Bartlett J.M., Walker R.A., Speirs V. A multi-centre investigation towards reaching a consensus on the immunohistochemical detection of ERbeta in archival formalin-fixed paraffin embedded human breast tissue. Breast Cancer Res. Treat. 2005;92:287–293. doi: 10.1007/s10549-004-4262-8. [DOI] [PubMed] [Google Scholar]
- Celhay O., Yacoub M., Irani J., Dore B., Cussenot O., Fromont G. Expression of estrogen related proteins in hormone refractory prostate cancer: association with tumor progression. J. Urol. 2010;184:2172–2178. doi: 10.1016/j.juro.2010.06.089. [DOI] [PubMed] [Google Scholar]
- Chang W.Y., Prins G.S. Estrogen receptor-beta: implications for the prostate gland. Prostate. 1999;40:115–124. doi: 10.1002/(sici)1097-0045(19990701)40:2<115::aid-pros7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Chen M., Ni J., Chang H.C., Lin C.Y., Muyan M., Yeh S. CCDC62/ERAP75 functions as a coactivator to enhance estrogen receptor beta-mediated transactivation and target gene expression in prostate cancer cells. Carcinogenesis. 2009;30:841–850. doi: 10.1093/carcin/bgn288. [DOI] [PubMed] [Google Scholar]
- Choi I., Ko C., Park-Sarge O.K., Nie R., Hess R.A., Graves C., Katzenellenbogen B.S. Human estrogen receptor beta-specific monoclonal antibodies: characterization and use in studies of estrogen receptor beta protein expression in reproductive tissues. Mol. Cell Endocrinol. 2001;181:139–150. doi: 10.1016/s0303-7207(01)00492-0. [DOI] [PubMed] [Google Scholar]
- Ciucci A., Zannoni G.F., Travaglia D., Petrillo M., Scambia G., Gallo D. Prognostic significance of the estrogen receptor beta (ERbeta) isoforms ERbeta1, ERbeta2, and ERbeta5 in advanced serous ovarian cancer. Gynecol. Oncol. 2014;132:351–359. doi: 10.1016/j.ygyno.2013.12.027. [DOI] [PubMed] [Google Scholar]
- Colciago A., Ruscica M., Mornati O., Piccolella M., Montagnani-Marelli M., Eberini I., Festuccia C., Magni P., Motta M., Negri-Cesi P. Vitro chronic administration of ERbeta selective ligands and prostate Cancer cell growth: hypotheses on the selective role of 3beta-Adiol in AR-positive RV1 cells. Biomed. Res. Int. 2014;2014:801473. doi: 10.1155/2014/801473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotrim C.Z., Fabris V., Doria M.L., Lindberg K., Gustafsson J.A., Amado F., Lanari C., Helguero L.A. Estrogen receptor beta growth-inhibitory effects are repressed through activation of MAPK and PI3K signalling in mammary epithelial and breast cancer cells. Oncogene. 2013;32:2390–2402. doi: 10.1038/onc.2012.261. [DOI] [PubMed] [Google Scholar]
- Dey P., Barros R.P., Warner M., Strom A., Gustafsson J.A. Insight into the mechanisms of action of estrogen receptor beta in the breast, prostate, colon, and CNS. J. Mol. Endocrinol. 2013;51:T61–T74. doi: 10.1530/JME-13-0150. [DOI] [PubMed] [Google Scholar]
- Dey P., Jonsson P., Hartman J., Williams C., Strom A., Gustafsson J.A. Estrogen receptors beta1 and beta2 have opposing roles in regulating proliferation and bone metastasis genes in the prostate Cancer cell line PC3. Mol. Endocrinol. 2012 doi: 10.1210/me.2012.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P., Strom A., Gustafsson J.A. Estrogen receptor beta upregulates FOXO3a and causes induction of apoptosis through PUMA in prostate cancer. Oncogene. 2014;33:4213–4225. doi: 10.1038/onc.2013.384. [DOI] [PubMed] [Google Scholar]
- Ellem S.J., Risbridger G.P. Treating prostate cancer: a rationale for targeting local oestrogens. Nat. Rev. Cancer. 2007;7:621–627. doi: 10.1038/nrc2174. [DOI] [PubMed] [Google Scholar]
- Ellem S.J., Taylor R.A., Furic L., Larsson O., Frydenberg M., Pook D., Pedersen J., Cawsey B., Trotta A., Need E., Buchanan G., Risbridger G.P. A pro-tumourigenic loop at the human prostate tumour interface orchestrated by oestrogen, CXCL12 and mast cell recruitment. J. Pathol. 2014;234:86–98. doi: 10.1002/path.4386. [DOI] [PubMed] [Google Scholar]
- Esslimani-Sahla M., Simony-Lafontaine J., Kramar A., Lavaill R., Mollevi C., Warner M., Gustafsson J.A., Rochefort H. Estrogen receptor beta (ER beta) level but not its ER beta cx variant helps to predict tamoxifen resistance in breast cancer. Clin. Cancer Res. 2004;10:5769–5776. doi: 10.1158/1078-0432.CCR-04-0389. [DOI] [PubMed] [Google Scholar]
- Foryst-Ludwig A., Clemenz M., Hohmann S., Hartge M., Sprang C., Frost N., Krikov M., Bhanot S., Barros R., Morani A., Gustafsson J.A., Unger T., Kintscher U. Metabolic actions of estrogen receptor beta (ERbeta) are mediated by a negative cross-talk with PPARgamma. PLoS Genet. 2008;4:e1000108. doi: 10.1371/journal.pgen.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua S.A., Schiff R., Parra I., Friedrichs W.E., Su J.L., McKee D.D., Slentz-Kesler K., Moore L.B., Willson T.M., Moore J.T. Expression of wild-type estrogen receptor beta and variant isoforms in human breast cancer. Cancer Res. 1999;59:5425–5428. [PubMed] [Google Scholar]
- Gallien S., Duriez E., Crone C., Kellmann M., Moehring T., Domon B. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol. Cell Proteomics. 2012;11:1709–1723. doi: 10.1074/mcp.O112.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargett C.E., Bucak K., Zaitseva M., Chu S., Taylor N., Fuller P.J., Rogers P.A. Estrogen receptor-alpha and -beta expression in microvascular endothelial cells and smooth muscle cells of myometrium and leiomyoma. Mol. Hum. Reprod. 2002;8:770–775. doi: 10.1093/molehr/8.8.770. [DOI] [PubMed] [Google Scholar]
- Grubisha M.J., Cifuentes M.E., Hammes S.R., Defranco D.B. A local paracrine and endocrine network involving TGFbeta, Cox-2, ROS, and estrogen receptor beta influences reactive stromal cell regulation of prostate cancer cell motility. Mol. Endocrinol. 2012;26:940–954. doi: 10.1210/me.2011-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruvberger-Saal S.K., Bendahl P.O., Saal L.H., Laakso M., Hegardt C., Eden P., Peterson C., Malmstrom P., Isola J., Borg A., Ferno M. Estrogen receptor beta expression is associated with tamoxifen response in ERalpha-negative breast carcinoma. Clin. Cancer Res. 2007;13:1987–1994. doi: 10.1158/1078-0432.CCR-06-1823. [DOI] [PubMed] [Google Scholar]
- Guo L., Zhang Y., Zhang W., Yilamu D. Correlation between estrogen receptor beta expression and the curative effect of endocrine therapy in breast cancer patients. Exp. Ther. Med. 2014;7:1568–1572. doi: 10.3892/etm.2014.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Zhu Q., Yilamu D., Jakulin A., Liu S., Liang T. Expression and prognostic value of estrogen receptor beta in breast cancer patients. Int. J. Clin. Exp. Med. 2014;7:3730–3736. [PMC free article] [PubMed] [Google Scholar]
- Haldosen L.A., Zhao C., Dahlman-Wright K. Estrogen receptor beta in breast cancer. Mol. Cell Endocrinol. 2014;382:665–672. doi: 10.1016/j.mce.2013.08.005. [DOI] [PubMed] [Google Scholar]
- Han S.J., Jung S.Y., Wu S.P., Hawkins S.M., Park M.J., Kyo S., Qin J., Lydon J.P., Tsai S.Y., Tsai M.J., DeMayo F.J., O'Malley B.W. Estrogen receptor beta modulates apoptosis complexes and the inflammasome to drive the pathogenesis of endometriosis. Cell. 2015;163:960–974. doi: 10.1016/j.cell.2015.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman J., Strom A., Gustafsson J.A. Current concepts and significance of estrogen receptor beta in prostate cancer. Steroids. 2012;77:1262–1266. doi: 10.1016/j.steroids.2012.07.002. [DOI] [PubMed] [Google Scholar]
- He Y.F., Luo H.Q., Wang W., Chen J., Yao Y.W., Cai S.B., He J., Yan Y., Wu S.S., Hu X.X., Ke L.H., Niu J.Y., Li H.M., Ji C.S., Hu B. Clinical features and prognosis-associated factors of non-small cell lung cancer exhibiting symptoms of bone metastasis at the time of diagnosis. Oncol. Lett. 2015;9:2706–2712. doi: 10.3892/ol.2015.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieken T.J., Carter J.M., Hawse J.R., Hoskin T.L., Bois M., Frost M., Hartmann L.C., Radisky D.C., Visscher D.W., Degnim A.C. ERbeta expression and breast Cancer risk prediction for women with atypias. Cancer Prev. Res. (Phila) 2015;8:1084–1092. doi: 10.1158/1940-6207.CAPR-15-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinsche O., Girgert R., Emons G., Grundker C. Estrogen receptor beta selective agonists reduce invasiveness of triple-negative breast cancer cells. Int. J. Oncol. 2015;46:878–884. doi: 10.3892/ijo.2014.2778. [DOI] [PubMed] [Google Scholar]
- Holbeck S., Chang J., Best A.M., Bookout A.L., Mangelsdorf D.J., Martinez E.D. Expression profiling of nuclear receptors in the NCI60 cancer cell panel reveals receptor-drug and receptor-gene interactions. Mol. Endocrinol. 2010;24:1287–1296. doi: 10.1210/me.2010-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S., Butler E., McLoughlin J. Prostate cancer and diet: food for thought? BJU Int. 2011;107:1348–1359. doi: 10.1111/j.1464-410X.2010.09897.x. [DOI] [PubMed] [Google Scholar]
- Horvath L.G., Henshall S.M., Lee C.S., Head D.R., Quinn D.I., Makela S., Delprado W., Golovsky D., Brenner P.C., O'Neill G., Kooner R., Stricker P.D., Grygiel J.J., Gustafsson J.A., Sutherland R.L. Frequent loss of estrogen receptor-beta expression in prostate cancer. Cancer Res. 2001;61:5331–5335. [PubMed] [Google Scholar]
- Hsu I., Vitkus S., Da J., Yeh S. Role of oestrogen receptors in bladder cancer development. Nat. Rev. Urol. 2013;10:317–326. doi: 10.1038/nrurol.2013.53. [DOI] [PubMed] [Google Scholar]
- Hussain S., Lawrence M.G., Taylor R.A., Lo C.Y., Frydenberg M., Ellem S.J., Furic L., Risbridger G.P. Estrogen receptor beta activation impairs prostatic regeneration by inducing apoptosis in murine and human stem/progenitor enriched cell populations. PLoS One. 2012;7:e40732. doi: 10.1371/journal.pone.0040732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson P., Katchy A., Williams C. Support of a bi-faceted role of estrogen receptor beta (ERbeta) in ERalpha-positive breast cancer cells. Endocr. Relat. Cancer. 2014;21:143–160. doi: 10.1530/ERC-13-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I.Y., Kim B.C., Seong D.H., Lee D.K., Seo J.M., Hong Y.J., Kim H.T., Morton R.A., Kim S.J. Raloxifene, a mixed estrogen agonist/antagonist, induces apoptosis in androgen-independent human prostate cancer cell lines. Cancer Res. 2002;62:5365–5369. [PubMed] [Google Scholar]
- Kim I.Y., Seong D.H., Kim B.C., Lee D.K., Remaley A.T., Leach F., Morton R.A., Kim S.J. Raloxifene, a selective estrogen receptor modulator, induces apoptosis in androgen-responsive human prostate cancer cell line LNCaP through an androgen-independent pathway. Cancer Res. 2002;62:3649–3653. [PubMed] [Google Scholar]
- Koressaar T., Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kuiper G.G., Enmark E., Pelto-Huikko M., Nilsson S., Gustafsson J.A. Cloning of a novel receptor expressed in rat prostate and ovary. Proc. Natl. Acad. Sci. U. S. A. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau K.M., LaSpina M., Long J., Ho S.M. Expression of estrogen receptor (ER)-alpha and ER-beta in normal and malignant prostatic epithelial cells: regulation by methylation and involvement in growth regulation. Cancer Res. 2000;60:3175–3182. [PubMed] [Google Scholar]
- Leav I., Lau K.M., Adams J.Y., McNeal J.E., Taplin M.E., Wang J., Singh H., Ho S.M. Comparative studies of the estrogen receptors beta and alpha and the androgen receptor in normal human prostate glands, dysplasia, and in primary and metastatic carcinoma. Am. J. Pathol. 2001;159:79–92. doi: 10.1016/s0002-9440(10)61676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Y.K., Lam H.M., Wu S., Song D., Levin L., Cheng L., Wu C.L., Ho S.M. Estrogen receptor beta2 and beta5 are associated with poor prognosis in prostate cancer, and promote cancer cell migration and invasion. Endocr. Relat. Cancer. 2010;17:675–689. doi: 10.1677/ERC-09-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Y.K., Mak P., Hassan S., Ho S.M. Estrogen receptor (ER)-beta isoforms: a key to understanding ER-beta signaling. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13162–13167. doi: 10.1073/pnas.0605676103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leygue E., Murphy L.C. A bi-faceted role of estrogen receptor beta in breast cancer. Endocr. Relat. Cancer. 2013;20:R127–R139. doi: 10.1530/ERC-12-0389. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo Z., Wu R., Jiang Y., Qiu Z., Chen W., Li W. Overexpression of estrogen receptor beta is a prognostic marker in non-small cell lung cancer: a meta-analysis. Int. J. Clin. Exp. Med. 2015;8:8686–8697. [PMC free article] [PubMed] [Google Scholar]
- Madak-Erdogan Z., Charn T.H., Jiang Y., Liu E.T., Katzenellenbogen J.A., Katzenellenbogen B.S. Integrative genomics of gene and metabolic regulation by estrogen receptors alpha and beta, and their coregulators. Mol. Syst. Biol. 2013;9:676. doi: 10.1038/msb.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P., Chang C., Pursell B., Mercurio A.M. Estrogen receptor beta sustains epithelial differentiation by regulating prolyl hydroxylase 2 transcription. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4708–4713. doi: 10.1073/pnas.1221654110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P., Li J., Samanta S., Chang C., Jerry D.J., Davis R.J., Leav I., Mercurio A.M. Prostate tumorigenesis induced by PTEN deletion involves estrogen receptor beta repression. Cell Rep. 2015;10:1982–1991. doi: 10.1016/j.celrep.2015.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak P., Li J., Samanta S., Mercurio A.M. ERbeta regulation of NF-kB activation in prostate cancer is mediated by HIF-1. Oncotarget. 2015;6:40247–40254. doi: 10.18632/oncotarget.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters J.R. Human cancer cell lines: fact and fantasy. Nat. Rev. Mol. Cell Biol. 2000;1:233–236. doi: 10.1038/35043102. [DOI] [PubMed] [Google Scholar]
- McPherson S.J., Ellem S.J., Simpson E.R., Patchev V., Fritzemeier K.H., Risbridger G.P. Essential role for estrogen receptor beta in stromal-epithelial regulation of prostatic hyperplasia. Endocrinology. 2007;148:566–574. doi: 10.1210/en.2006-0906. [DOI] [PubMed] [Google Scholar]
- McPherson S.J., Hussain S., Balanathan P., Hedwards S.L., Niranjan B., Grant M., Chandrasiri U.P., Toivanen R., Wang Y., Taylor R.A., Risbridger G.P. Estrogen receptor-beta activated apoptosis in benign hyperplasia and cancer of the prostate is androgen independent and TNFalpha mediated. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3123–3128. doi: 10.1073/pnas.0905524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina M. Insights gained from 20 years of soy research. J. Nutr. 2010;140:2289S–2295S. doi: 10.3945/jn.110.124107. [DOI] [PubMed] [Google Scholar]
- Mohammed H., D'Santos C., Serandour A.A., Ali H.R., Brown G.D., Atkins A., Rueda O.M., Holmes K.A., Theodorou V., Robinson J.L., Zwart W., Saadi A., Ross-Innes C.S., Chin S.F., Menon S., Stingl J., Palmieri C., Caldas C., Carroll J.S. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3:342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais-Santos M., Nunes A.E., Oliveira A.G., Moura-Cordeiro J.D., Mahecha G.A., Avellar M.C., Oliveira C.A. Changes in estrogen receptor ERbeta (ESR2) expression without changes in the estradiol levels in the prostate of aging rats. PLoS One. 2015;10:e0131901. doi: 10.1371/journal.pone.0131901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy S., Andersson S., Kim H.J., Butler R., Waage L., Bergerheim U., Gustafsson J.A. Estrogen receptor beta and 17beta-hydroxysteroid dehydrogenase type 6, a growth regulatory pathway that is lost in prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 2011;108:20090–20094. doi: 10.1073/pnas.1117772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E., Fleming F.J., Crotty T.B., Kelly G., McDermott E.W., O'Higgins N.,J., Hill A.D., Young L.S. Inverse relationship between ER-beta and SRC-1 predicts outcome in endocrine-resistant breast cancer. Br. J. Cancer. 2004;91:1687–1693. doi: 10.1038/sj.bjc.6602156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y., Akaogi K., Suzuki T., Osakabe A., Yamaguchi C., Sunahara N., Ishida J., Kako K., Ogawa S., Fujimura T., Homma Y., Fukamizu A., Murayama A., Kimura K., Inoue S., Yanagisawa J. Estrogen regulates tumor growth through a nonclassical pathway that includes the transcription factors ERbeta and KLF5. Sci. Signal. 2011;4 doi: 10.1126/scisignal.2001551. ra22. [DOI] [PubMed] [Google Scholar]
- Nelson A.W., Tilley W.D., Neal D.E., Carroll J.S. Estrogen receptor beta in prostate cancer: friend or foe? Endocr. Relat. Cancer. 2014;21:T219–T234. doi: 10.1530/ERC-13-0508. [DOI] [PubMed] [Google Scholar]
- Oliveira A.G., Coelho P.H., Guedes F.D., Mahecha G.A., Hess R.A., Oliveira C.A. 5alpha-Androstane-3beta,17beta-diol (3beta-diol), an estrogenic metabolite of 5alpha-dihydrotestosterone, is a potent modulator of estrogen receptor ERbeta expression in the ventral prostrate of adult rats. Steroids. 2007;72:914–922. doi: 10.1016/j.steroids.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Omoto Y., Kobayashi S., Inoue S., Ogawa S., Toyama T., Yamashita H., Muramatsu M., Gustafsson J.A., Iwase H. Evaluation of oestrogen receptor beta wild-type and variant protein expression, and relationship with clinicopathological factors in breast cancers. Eur. J. Cancer. 2002;38:380–386. doi: 10.1016/s0959-8049(01)00383-5. [DOI] [PubMed] [Google Scholar]
- Pierdominici M., Maselli A., Colasanti T., Giammarioli A.M., Delunardo F., Vacirca D., Sanchez M., Giovannetti A., Malorni W., Ortona E. Estrogen receptor profiles in human peripheral blood lymphocytes. Immunol. Lett. 2010;132:79–85. doi: 10.1016/j.imlet.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Reese J.M., Suman V.J., Subramaniam M., Wu X., Negron V., Gingery A., Pitel K.S., Shah S.S., Cunliffe H.E., McCullough A.E., Pockaj B.A., Couch F.J., Olson J.E., Reynolds C., Lingle W.L., Spelsberg T.C., Goetz M.P., Ingle J.N., Hawse J.R. ERbeta1: characterization, prognosis, and evaluation of treatment strategies in ERalpha-positive and -negative breast cancer. BMC Cancer. 2014;14:749. doi: 10.1186/1471-2407-14-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter E., Gerster P., Jungbauer A. Red clover and soy isoflavones–an in vitro safety assessment. Gynecol. Endocrinol. 2011;27:1037–1042. doi: 10.3109/09513590.2011.588743. [DOI] [PubMed] [Google Scholar]
- Ricke W.A., McPherson S.J., Bianco J.J., Cunha G.R., Wang Y., Risbridger G.P. Prostatic hormonal carcinogenesis is mediated by in situ estrogen production and estrogen receptor alpha signaling. FASEB J. 2008;22:1512–1520. doi: 10.1096/fj.07-9526com. [DOI] [PubMed] [Google Scholar]
- Risbridger G.P., Ellem S.J., McPherson S.J. Estrogen action on the prostate gland: a critical mix of endocrine and paracrine signaling. J. Mol. Endocrinol. 2007;39:183–188. doi: 10.1677/JME-07-0053. [DOI] [PubMed] [Google Scholar]
- Rizza P., Barone I., Zito D., Giordano F., Lanzino M., De Amicis F., Mauro L., Sisci D., Catalano S., Wright K.D., Gustafsson J.A., Ando S. Estrogen receptor beta as a novel target of androgen receptor action in breast cancer cell lines. Breast Cancer Res. 2014;16(R21) doi: 10.1186/bcr3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger P., Sahla M.E., Makela S., Gustafsson J.A., Baldet P., Rochefort H. Decreased expression of estrogen receptor beta protein in proliferative preinvasive mammary tumors. Cancer Res. 2001;61:2537–2541. [PubMed] [Google Scholar]
- Ross R.K., Bernstein L., Lobo R.A., Shimizu H., Stanczyk F.Z., Pike M.C., Henderson B.E. 5-alpha-reductase activity and risk of prostate cancer among Japanese and US white and black males. Lancet. 1992;339:887–889. doi: 10.1016/0140-6736(92)90927-u. [DOI] [PubMed] [Google Scholar]
- Rossi V., Bellastella G., De Rosa C., Abbondanza C., Visconti D., Maione L., Chieffi P., Della Ragione F., Prezioso D., De Bellis A., Bellastella A., Sinisi A.A. Raloxifene induces cell death and inhibits proliferation through multiple signaling pathways in prostate cancer cells expressing different levels of estrogen receptor alpha and beta. J. Cell Physiol. 2011;226:1334–1339. doi: 10.1002/jcp.22461. [DOI] [PubMed] [Google Scholar]
- Ruddy S.C., Lau R., Cabrita M.A., McGregor C., McKay B.C., Murphy L.C., Wright J.S., Durst T., Pratt M.A. Preferential estrogen receptor beta ligands reduce Bcl-2 expression in hormone-resistant breast cancer cells to increase autophagy. Mol. Cancer Ther. 2014;13:1882–1893. doi: 10.1158/1535-7163.MCT-13-1066. [DOI] [PubMed] [Google Scholar]
- Setlur S.R., Mertz K.D., Hoshida Y., Demichelis F., Lupien M., Perner S., Sboner A., Pawitan Y., Andren O., Johnson L.A., Tang J., Adami H.O., Calza S., Chinnaiyan A.M., Rhodes D., Tomlins S., Fall K., Mucci L.A., Kantoff P.W., Stampfer M.J., Andersson S.O., Varenhorst E., Johansson J.E., Brown M., Golub T.R., Rubin M.A. Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J. Natl. Cancer Inst. 2008;100:815–825. doi: 10.1093/jnci/djn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban A.M., O'Neill P.A., Davies M.P., Sibson R., West C.R., Smith P.H., Foster C.S. Declining estrogen receptor-beta expression defines malignant progression of human breast neoplasia. Am. J. Surg. Pathol. 2003;27:1502–1512. doi: 10.1097/00000478-200312000-00002. [DOI] [PubMed] [Google Scholar]
- Shen J.C., Klein R.D., Wei Q., Guan Y., Contois J.H., Wang T.T., Chang S., Hursting S.D. Low-dose genistein induces cyclin-dependent kinase inhibitors and G(1) cell-cycle arrest in human prostate cancer cells. Mol. Carcinog. 2000;29:92–102. doi: 10.1002/1098-2744(200010)29:2<92::aid-mc6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Skliris G.P., Parkes A.T., Limer J.L., Burdall S.E., Carder P.J., Speirs V. Evaluation of seven oestrogen receptor beta antibodies for immunohistochemistry, western blotting, and flow cytometry in human breast tissue. J. Pathol. 2002;197:155–162. doi: 10.1002/path.1077. [DOI] [PubMed] [Google Scholar]
- Smart E., Hughes T., Smith L., Speirs V. Estrogen receptor beta: putting a positive into triple negative breast cancer? Horm. Mol. Biol. Clin. Investig. 2013;16:117–123. doi: 10.1515/hmbci-2013-0042. [DOI] [PubMed] [Google Scholar]
- Stettner M., Kaulfuss S., Burfeind P., Schweyer S., Strauss A., Ringert R.H., Thelen P. The relevance of estrogen receptor-beta expression to the antiproliferative effects observed with histone deacetylase inhibitors and phytoestrogens in prostate cancer treatment. Mol. Cancer Ther. 2007;6:2626–2633. doi: 10.1158/1535-7163.MCT-07-0197. [DOI] [PubMed] [Google Scholar]
- Suzuki F., Akahira J., Miura I., Suzuki T., Ito K., Hayashi S., Sasano H., Yaegashi N. Loss of estrogen receptor beta isoform expression and its correlation with aberrant DNA methylation of the 5'-untranslated region in human epithelial ovarian carcinoma. Cancer Sci. 2008;99:2365–2372. doi: 10.1111/j.1349-7006.2008.00988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen P., Peter T., Hunermund A., Kaulfuss S., Seidlova-Wuttke D., Wuttke W., Ringert R.H., Seseke F. Phytoestrogens from Belamcanda chinensis regulate the expression of steroid receptors and related cofactors in LNCaP prostate cancer cells. BJU Int. 2007;100:199–203. doi: 10.1111/j.1464-410X.2007.06924.x. [DOI] [PubMed] [Google Scholar]
- Thelen P., Scharf J.G., Burfeind P., Hemmerlein B., Wuttke W., Spengler B., Christoffel V., Ringert R.H., Seidlova-Wuttke D. Tectorigenin and other phytochemicals extracted from leopard lily Belamcanda chinensis affect new and established targets for therapies in prostate cancer. Carcinogenesis. 2005;26:1360–1367. doi: 10.1093/carcin/bgi092. [DOI] [PubMed] [Google Scholar]
- Umekita Y., Souda M., Ohi Y., Sagara Y., Rai Y., Takahama T., Yoshida H. Expression of wild-type estrogen receptor beta protein in human breast cancer: specific correlation with HER2/neu overexpression. Pathol. Int. 2006;56:423–427. doi: 10.1111/j.1440-1827.2006.01983.x. [DOI] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B.C., Remm M., Rozen S.G. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivar O.I., Zhao X., Saunier E.F., Griffin C., Mayba O.S., Tagliaferri M., Cohen I., Speed T.P., Leitman D.C. Estrogen receptor beta binds to and regulates three distinct classes of target genes. J. Biol. Chem. 2010;285:22059–22066. doi: 10.1074/jbc.M110.114116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitsman G.E., Skliris G., Ung K., Peng B., Younes M., Watson P.H., Murphy L.C. Assessment of multiple different estrogen receptor-beta antibodies for their ability to immunoprecipitate under chromatin immunoprecipitation conditions. Breast Cancer Res. Treat. 2006;100:23–31. doi: 10.1007/s10549-006-9229-5. [DOI] [PubMed] [Google Scholar]
- Weng C., Cai J., Wen J., Yuan H., Yang K., Imperato-McGinley J., Zhu Y.S. Differential effects of estrogen receptor ligands on regulation of dihydrotestosterone-induced cell proliferation in endothelial and prostate cancer cells. Int. J. Oncol. 2013;42:327–337. doi: 10.3892/ijo.2012.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly H., Han G., Pinnaduwage D., Murphy L.C., Yang X.R., Andrulis I.L., Sherman M., Figueroa J., Rimm D.L. ERbeta splice variant expression in four large cohorts of human breast cancer patient tumors. Breast Cancer Res. Treat. 2014;146:657–667. doi: 10.1007/s10549-014-3050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Subramaniam M., Negron V., Cicek M., Reynolds C., Lingle W.L., Goetz M.P., Ingle J.N., Spelsberg T.C., Hawse J.R. Development, characterization, and applications of a novel estrogen receptor beta monoclonal antibody. J. Cell Biochem. 2012;113:711–723. doi: 10.1002/jcb.23443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuttke W., Jarry H., Westphalen S., Christoffel V., Seidlova-Wuttke D. Phytoestrogens for hormone replacement therapy? J. Steroid Biochem. Mol. Biol. 2002;83:133–147. doi: 10.1016/s0960-0760(02)00259-5. [DOI] [PubMed] [Google Scholar]
- Yang L., Ravindranathan P., Ramanan M., Kapur P., Hammes S.R., Hsieh J.T., Raj G.V. Central role for PELP1 in nonandrogenic activation of the androgen receptor in prostate cancer. Mol. Endocrinol. 2012;26:550–561. doi: 10.1210/me.2011-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Wang J., Wang L., Shen C., Su B., Qi M., Hu J., Gao W., Tan W., Han B. Estrogen induces androgen-repressed SOX4 expression to promote progression of prostate cancer cells. Prostate. 2015;75:1363–1375. doi: 10.1002/pros.23017. [DOI] [PubMed] [Google Scholar]
- Yu C.P., Ho J.Y., Huang Y.T., Cha T.L., Sun G.H., Yu D.S., Chang F.W., Chen S.P., Hsu R.J. Estrogen inhibits renal cell carcinoma cell progression through estrogen receptor-beta activation. PLoS One. 2013;8:e56667. doi: 10.1371/journal.pone.0056667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger T., Sturm S., Rey S., Zlobec I., Gsponer J.R., Rentsch C.A., Terracciano L.M., Bachmann A., Bubendorf L., Ruiz C. Estrogen receptor beta expression and androgen receptor phosphorylation correlate with a poor clinical outcome in hormone-naive prostate cancer and are elevated in castration-resistant disease. Endocr. Relat. Cancer. 2013;20:403–413. doi: 10.1530/ERC-12-0402. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Zhou J., Du Y. Estrogen receptor alpha interacts with mitochondrial protein HADHB and affects beta-oxidation activity. Mol. Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.011056. M111 011056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Leav I., Leung Y.K., Wu M., Liu Q., Gao Y., McNeal J.E., Ho S.M. Dynamic regulation of estrogen receptor-beta expression by DNA methylation during prostate cancer development and metastasis. Am. J. Pathol. 2004;164:2003–2012. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ERβ Antibody binding sites. Schematic of the full length, wild type (wt) ERβ protein, indicating the binding sites of the 8 antibodies evaluated. NTD, N terminal domain; DBD, DNA-binding domain; LBD, ligand-binding domain.