Abstract
Higher plants undergo a transition from a juvenile to an adult phase of vegetative development prior to flowering. Screens for mutants that undergo this transition precociously produced alleles of two genes required for posttranscriptional gene silencing (PTGS)—SUPPRESSOR OF GENE SILENCING3 (SGS3) and SUPPRESSOR OF GENE SILENCING2(SGS2)/SILENCING DEFECTIVE1 (SDE1)/RNA-DEPENDENT POLYMERASE6 (RDR6). Loss-of-function mutations in these genes have a phenotype similar to that of mutations in the Argonaute gene ZIPPY (ZIP). Epistasis analysis suggests that ZIP, SGS3, SGS2/SDE1/RDR6, and the putative miRNA export receptor, HASTY (HST), operate in the same pathway(s). Microarray analysis revealed a small number of genes whose mRNA is increased in ZIP, SGS3, and SGS2/SDE1/RDR6 mutants, as well as genes that are up-regulated in SGS3 and SGS2/SDE1/RDR6 mutants, but not in ZIP mutants. One of these latter genes (At5g18040) is silenced posttranscriptionally in trans by the sRNA255 family of endogenous, noncoding, small interfering RNAs (siRNAs). The increase in At5g18040 mRNA in SGS3 and SGS2/SDE1/RDR6 mutants is attributable to the absence of sRNA255-like siRNAs in these mutants. These results demonstrate a role for endogenous siRNAs in the regulation of gene expression, and suggest that PTGS plays a central role in the temporal control of shoot development in plants.
Keywords: siRNA, miRNA, vegetative phase change, posttranscriptional gene silencing, RNAi, heterochrony
The anatomy and morphology of a plant shoot changes over the course of its development. Although many traits (e.g., leaf size) vary gradually, other traits change abruptly at predictable times in shoot development (Hackett and Murray 1993; Greenwood 1995; Poethig 2003). Two such changes occur during the postembryonic growth of the shoot. The juvenile-to-adult transition (vegetative phase change) occurs early in the life of the shoot, and is marked by differences in the anatomy, morphology, and chemistry of leaves and internodes produced before and after this transition. The second transition (reproductive phase change) occurs during the adult phase, and results in the production of flowers and flower-bearing branches in place of vegetative shoots. Genetic analysis of floral induction in Arabidopsis has produced many genes affecting the transition from vegetative to reproductive growth, and significant progress has been made in understanding the molecular mechanism of this transition (Mouradov et al. 2002). In contrast, we are just beginning to understand the mechanism of vegetative phase change.
Most of the genes known to affect vegetative phase change in Arabidopsis have pleiotropic mutant phenotypes, indicating that they are involved in many different processes. This class includes SERRATE (Clarke et al. 1999; Prigge and Wagner 2001), SQUINT (SQN) (Berardini et al. 2001), and HASTY (HST) (Telfer and Poethig 1998; Bollman et al. 2003). In contrast, loss-of-function mutations of ZIPPY (ZIP) have a much more specific effect on vegetative phase change (Hunter et al. 2003). Mutations in this Argonaute-like gene (AGO7) (Fagard et al. 2000) accelerate vegetative phase change and produce slightly abnormal flowers, but have no other obvious effects on shoot morphology (Hunter et al. 2003). This specific phenotype, and the possibility that ZIP might be involved in a regulatory pathway involving microRNAs (miRNAs) and/or small interfering RNAs (siRNAs), encouraged us to search for other mutations that have this same vegetative phenotype.
Screens for mutations with a zip-like phenotype produced alleles of two genes—SUPPRESSOR OF GENE SILENCING3 (SGS3) and SUPPRESSOR OF GENE SILENCING2(SGS2)/SILENCING DEFECTIVE1(SDE1)/RNA-DEPENDENT POLYMERASE6 (RDR6)—required for posttranscriptional silencing (PTGS; also known as RNA interference, RNAi) of transgenes. Genes subject to PTGS are targeted for destruction by complementary 21-22-nt siRNAs (Elbashir et al. 2001; Tang et al. 2003). In plants, PTGS can be initiated by both single-stranded (ssRNA) and double-stranded RNA (dsRNA). SGS3 encodes a novel plant-specific protein and SGS2/SDE1/RDR6 encodes an RNA-dependent RNA polymerase (RdRP) required for silencing ssRNA. Both of these genes are believed to be involved in transforming ssRNA into the dsRNA precursors of siRNAs (Dalmay et al. 2000; Mourrain et al. 2000). Mutations in these genes confer sensitivity to cucumber mosaic virus (Beclin et al. 2002) and cabbage leaf curl virus (Muangsan et al. 2004), indicating that one of their normal functions is to provide resistance to viral infection. Two other RdRPs have been described in Arabidopsis. RNA-DEPENDENT RNA POLYMERASE2 (RDR2) is required for the production of siRNAs from endogenous transcripts, but is not required for virus resistance. RNA-DEPENDENT RNA POLYMERASE1 (RDR1) is induced upon viral infection and limits virus replication, but has no apparent role in the silencing of endogenous transcripts (Yu et al. 2003; Xie et al. 2004). RdRPs that participate in PTGS/RNAi have also been described in Caenorhabditis elegans (Smardon et al. 2000; Sijen et al. 2001; Simmer et al. 2002), Neurospora crassa (Cogoni and Macino 1999; Catalanotto et al. 2002; Shiu and Metzenberg 2002), Schizosaccharomyces pombe (Volpe et al. 2002, 2003) and Dictyostelium discoideum (Martens et al. 2002), and are required for a variety of different functions in the normal biology of these organisms. For example, ego-1 is required for germ-line development in C. elegans (Smardon et al. 2000), Rdp1 is necessary for centromere function in S. pombe (Volpe et al. 2003), and Sad-1 silences unpaired DNA during meiosis in N. crassa (Shiu and Metzenberg 2002).
Here we show that SGS3 and SGS2/SDE1/RDR6 act in the same pathway(s) as the Argonaute protein, ZIP, and the putative miRNA nuclear export receptor, HST, to regulate vegetative phase change and floral development in Arabidopsis. Expression profiling revealed that only a few genes are affected in common by mutations in ZIP, SGS3, and SGS2/SDE1/RDR6; these genes are excellent candidates for genes involved in vegetative phase change. In addition, we identified genes that are affected by mutations in SGS3 and SGS2/SDE1/RDR6, but not by a mutation in ZIP. One of the latter genes is silenced posttranscriptionally in trans by endogenous siRNAs derived from noncoding transcripts. These results reveal a developmental function for SGS3 and SGS2/SDE1/RDR6, and suggest possible targets of their regulation.
Results
Identification of zip-like mutants
In Arabidopsis, leaves produced at different times during shoot development can be distinguished from each other by their size, shape, and pattern of trichome distribution (Chien and Sussex 1996; Telfer et al. 1997; Tsukaya et al. 2000; Hunter et al. 2003). The first two leaves are small, round, relatively flat, have a smooth margin, and are incapable of producing trichomes on their abaxial surface. Subsequent leaves are larger, elliptical in shape, curl downward, have a serrated margin, and have the capacity to produce abaxial trichomes, although these are not usually produced until leaf 5 or 6.
Loss-of-function alleles of ZIP accelerate the expression of adult-phase vegetative traits, but have relatively little effect on other aspects of shoot development (Hunter et al. 2003). The phenotype of these mutations is most apparent in the first two leaves of the rosette, which are elongated and curl downward instead of being round and flat. In addition to this effect on leaf shape, zip mutations produce a forward shift in abaxial trichome production. zip does not affect the rate of leaf initiation or flowering time, but does have a weak effect on flower morphology. Mutant carpels typically have a split septum that produces ectopic stigmatic tissue, and flowers have a variably penetrant defect in stamen and carpel elongation, leading to poor seed set.
Screens for EMS, fast-neutron, and T-DNA-induced mutations that have a zip-like phenotype produced a number of phenotypically similar, recessive mutations in two complementation groups (Figs. 1, 2). We mapped a mutation in the first complementation group near SGS2/SDE1/RDR6 and a mutation in the second complementation group near SGS3. To confirm the identity of these new mutations, we performed complementation tests with sgs2-1 and sgs3-1, the alleles originally described by Mourrain et al. (2000). sgs2-1 and sgs3-1 were found to have a zippy-like phenotype that was not complemented by the mutations identified in our screen. The polymorphisms associated with these new alleles of SGS2/SDE1/RDR6 and SGS3 are described in Table 1. Two T-DNA insertions in SGS3 (SALK_001394 and SALK_039005) (Alonso et al. 2003) that have a morphological phenotype similar to these new alleles are also listed in this table. To simplify naming new alleles of SGS2/SDE1/RDR6 we have chosen to use the nomenclature (RDR6) suggested by Xie et al. (2004) for RdRP genes in Arabidopsis.
Figure 1.
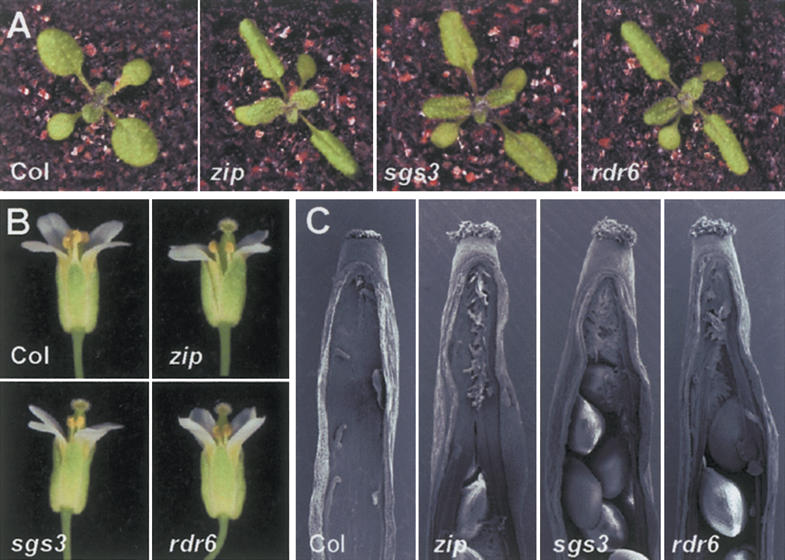
The morphology of wild-type Columbia (Col), zip-1, sgs3-11, and rdr6-11. (A) Thirteen-day-old plants. The first few leaves of mutant plants are elongated and curl downward. (B) Flowers. Mutant plants have variable seed sets because stamens frequently fail to contact the stigma. (C) Scanning electron micrographs of carpels with one valve and seeds removed. Mutant carpels have a split septum and produce stigmatic tissue in the middle of the septum at the apical end of the carpels.
Figure 2.
The morphology of the first two rosette leaves of wild-type (Col) and mutant plants.
Table 1.
New alleles of RDR6 and SGS3
| Allele | Mutagen | Mutationa |
|---|---|---|
| rdr6-11 | T-DNA | C → T (805), nonsense |
| rdr6-12 | Fast neutron | 7 bp del. (997-1003), frameshift |
| rdr6-13 | EMS | G → A (2597), missense |
| rdr6-14 | EMS | G → A (3539), nonsense |
| sgs3-11 | EMS | G → A (2283), splice site |
| sgs3-12 | EMS | G → A (719), nonsense |
| sgs3-13 | T-DNA | SALK_039005, insertion in exon 1 |
| sgs3-14 | T-DNA | SALK_001394, insertion in exon 1 |
Numbered according to the genomic sequence of At3g49500 (RDR6) and At5g23570 (SGS3) as described in TAIR (http://www.arabidopsis.org).
sgs3-11 and rdr6-11 prevent PTGS
The results presented here were obtained using sgs3-11 and rdr6-11. To determine whether these new mutations have an effect on transgene silencing like that of sgs3-1 and sgs2-1, we determined their effect on the expression of L1, a 35S::GUS transgene that is silenced posttranscriptionally in fully expanded leaves (Elmayan et al. 1998). Consistent with the phenotype of sgs3-1 and sgs2-1, we found that sgs3-11 and rdr6-11 prevented silencing of L1 in all leaves of the shoot (Fig. 3). Silencing of L1 occurred normally in zip-1, as previously observed for the related L2 transgene (Hunter et al. 2003).
Figure 3.
The effect of zip-1, sgs3-11, and rdr6-11 on posttranscriptional silencing of the L1 (35S::GUS) transgene. zip-1 does not interfere with the silencing of L1, but sgs3-11 and rdr6-11 prevent silencing in all fully expanded rosette leaves.
The phenotype of rdr6-11 and sgs3-11
An analysis of 25-35 plants of each genotype revealed that sgs3-11 and rdr6-11 have essentially the same phenotype as zip-1. All three mutants have elongated, down-wardly curled leaves (Figs. 1A, 2) and produce abaxial trichomes precociously (Fig. 4A). Abaxial trichomes appear at the base of leaf 3 in zip-1 and rdr6-11 and at the base of leaf 4 in sgs3-11, and gradually increase in extent over the next few leaves. This gradual increase in abaxial trichome production resembles the pattern in wild-type plants, and implies that these mutations accelerate an otherwise normal transition to the adult phase. Mutant and wild-type plants do not exhibit a major difference in either total rosette leaf number (Fig. 4B) or in flowering time (Fig. 4C). The carpels of zip-1, sgs3-11, and rdr6-11 typically have a split septum and produce stigmatic tissue at the center of the septum (Fig. 1C), although the expressivity of this floral phenotype varied among individual plants and along the length of the shoot. In addition, mutant plants often display a lack of coordination between stamen and carpel elongation, resulting in low seed set (Fig. 1B).
Figure 4.
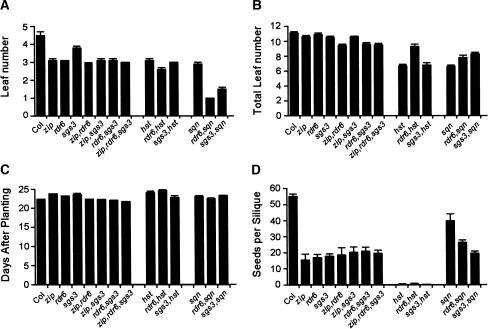
The phenotype of wild-type (Col) and mutant plants. (A) First leaf with abaxial trichomes. (B) Total number of leaves in the rosette. (C) Time to production of the first open flower. (D) Number of seeds per silique; 25-35 plants were measured for each genotype. Error bars indicate standard error of the mean.
SGS3 and RDR6 act in the same pathway as ZIP
To determine whether these proteins act in the same pathway, we examined the phenotype of plants homozygous for different combinations of zip-1, sgs3-11, and rdr6-11. We found that the seedling and floral morphology (Figs. 2, 5), abaxial trichome production, total leaf number, flowering time, and seed set (Fig. 4) of single, double, and triple mutants were not dramatically different. Double- and triple-mutant plants produced slightly fewer leaves and flowered earlier than single mutants, but these differences—although statistically significant—were quite small and are probably attributable to differences in the age or vigor of the seed stocks used in this experiment, because the flowering time of triple-mutant plants was identical to wild-type. These results support the hypothesis that ZIP, SGS3, and RDR6 act in the same pathway(s).
Figure 5.
The phenotype of zip-1, sgs3-11, rdr6-11, hst-1, and sqn-1, and plants homozygous for combinations of these mutations (13-day-old plants).
Additional evidence for this conclusion is provided by the interaction between these mutations and hst-1 and sqn-1. HST is the ortholog of the pre-miRNA nuclear export receptor exportin 5 (Bollman et al. 2003; Yi et al. 2003; Bohnsack et al. 2004; Lund et al. 2004), whereas SQN is the ortholog of cyclophilin 40 (Berardini et al. 2001). Loss-of-function mutations in these genes resemble zip-1, sgs3-11, and rdr6-11 in that they accelerate the juvenile-to-adult transition. However, hst and sqn mutations have a much stronger effect on the expression of phase-specific traits and are more pleiotropic than these three mutations. Previously, we found that zip-1 interacts additively with sqn-1, and that hst-1 is epi-static to zip-1 (Hunter et al. 2003). sgs3-11 and rdr6-11 interact with sqn-1 and hst-1 in the same fashion. sgs3-11 hst-1 and rdr6-11 hst-1 were morphologically indistinguishable from hst-1 (Figs. 2, 5) and had the same pattern of trichome production as hst-1 plants (Fig. 4A). Although rdr6-11 hst-1 plants had two more leaves (Fig. 4B) and flowered 2 d earlier than hst-1 (Fig. 4C), we do not believe that this difference is biologically meaningful, because this difference is the opposite of the normal relationship between leaf number and flowering time. Furthermore, the expressivity of these two traits varies significantly in hst-1 even under fairly uniform conditions (Telfer and Poethig 1998). In contrast, sgs3-11 sqn-1 and rdr6-11 sqn-1 plants had an additive phenotype. The first two leaves of these double mutants were intermediate in morphology (Figs. 2, 5) and produced abaxial trichomes (Fig. 4A); abaxial trichomes were never seen earlier than leaf 3 in single mutants. Furthermore, the siliques of double mutants had multiple carpels, completely lacked a septum, and had a very reduced seed set (Fig. 4D). These results support the conclusion that RDR6 and SGS3 are functionally related to ZIP, and that all three of these proteins probably operate in the same pathway (or on the same substrates) as HST. The additive interaction between these mutations and sqn-1 may mean that SQN operates in a completely different regulatory pathway, or that SQN and these three proteins have related but nonoverlapping functions.
Genes regulated by ZIP, SGS3, and RDR6
In order to identify genes regulated by ZIP, SGS3, and RDR6, we performed a microarray analysis of gene expression in wild-type and mutant plants. RNA was isolated from shoot apices of 2-week-old plants grown under short days (10 h light:14 h dark) in order to delay flowering and thus limit the analysis to genes involved in vegetative development. Hybridizations to the Affymetrix ATH1 array (22,800 genes) were performed in duplicate, using RNA isolated from two sets of plants grown at different times in the same growth chamber.
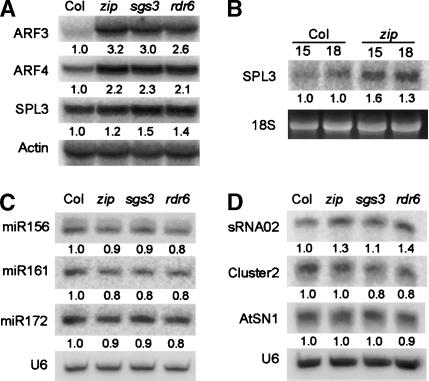
This analysis produced 17 genes whose expression was increased in all three mutants in both replicates by an average of 1.3-fold or greater, but no genes that were down-regulated in a similar fashion (Table 2). We chose nine genes from this group for further analysis based on their level of expression or potential role in phase change. Northern analysis of RNA isolated from additional samples confirmed these microarray results in the case of ARF3/ETTIN, ARF4, and SPL3 (Fig. 6A,B). However, we were unable to confirm these microarray results for six other genes, implying that most of the 17 genes listed in Table 2 are false positives.
Table 2.
Microarray analysis of genes up-regulated in 2-week-old zip-1, sgs3-11, and rdr6-11 plants grown in short days
| Fold increasea
|
|||||
|---|---|---|---|---|---|
| Gene | Annotation | zip | sgs3 | rdr6 | Conf.b |
| At2g33860 | auxin response transcription factor 3 (ETTIN/ARF3) | 3.1 | 3.6 | 4.2 | yes |
| At5g60450 | auxin response transcription factor (ARF4) | 2.7 | 2.8 | 2.5 | yes |
| At1g08830 | copper/zinc superoxidase dismutase (CSD1) | 2.0 | 2.5 | 2.3 | no |
| At1g62480 | vacuolar calcium-binding protein-related | 2.2 | 2.1 | 1.7 | no |
| At4g35770 | senescence-associated protein (SEN1) | 2.0 | 1.7 | 1.9 | n.d. |
| At1g64660 | methionine/cystathionine gamma lyase, putative | 2.0 | 1.7 | 1.6 | n.d. |
| At1g62500 | lipid-transfer protein family | 1.6 | 1.9 | 1.8 | n.d. |
| At1g73500 | MAP kinase, putative | 1.7 | 1.8 | 1.8 | no |
| At5g14780 | formate dehydrogenase | 1.8 | 1.9 | 1.4 | n.d. |
| At3g05900 | similar to anion exchange proteins | 1.9 | 1.8 | 1.4 | n.d. |
| At5g14920 | gibberellin-regulated protein family | 1.7 | 1.6 | 1.5 | no |
| At1g12780 | uridine diphosphate glucose epimerase | 1.7 | 1.6 | 1.5 | no |
| At4g15800 | rapid alkalinization factor (RALF) family protein | 1.5 | 1.6 | 1.4 | no |
| At2g38530 | lipid-transfer protein 2 (LTP2) | 1.4 | 1.6 | 1.6 | n.d. |
| At3g23030 | auxin-inducible gene (IAA2) | 1.4 | 1.4 | 1.7 | n.d. |
| At2g33810 | squamosa-promoter binding protein (SPL3) | 1.3 | 1.5 | 1.5 | yes |
| At1g11260 | glucose transporter | 1.4 | 1.4 | 1.3 | n.d. |
Relative to wild-type Columbia.
Confirmed by Northern analysis of mRNA prepared from a separately harvested sample. (n.d.) Not determined.
Figure 6.
The effect of zip-1, sgs3-11, and rdr6-11 on the accumulation of mRNAs, miRNAs, and siRNAs. (A) mRNA of ARF3/ETTIN, ARF4, and SPL3 accumulates in zip, sgs3-11, and rdr6-11. Total RNA from 2-week-old rosettes of plants grown in short days was hybridized with probes to the indicated genes. Actin was used as a loading control. (B) SPL3 mRNA is elevated in zip-1 at 15 and 18 d after planting. Blot of total RNA from shoot apices of wild-type and zip-1 rosettes hybridized with a probe to SPL3. 18S ribosomal RNA was used as a loading control. (C) The accumulation of miRNAs in zip-1, sgs3-11, and rdr6-11. Blots of low-molecular-weight RNA from 2-week-old mutant and wild-type plants hybridized with probes complementary to the functional strand of the indicated miRNAs. (D) The effect of zip-1, sgs3-11, and rdr6-11 on the level of endogenous sRNAs. Blots of low-molecular-weight RNA from 2-week-old mutant and wild-type plants hybridized with probes to the indicated sRNAs. U6 was used as a loading control. The intensity of each signal was measured using NIH Image; the ratio of this signal relative to wild-type is indicated below each image.
SPL3 is a potential target of miR156 (Rhoades et al. 2002). To determine whether the increase in SPL3 mRNA could be attributed to a defect in miRNA biogenesis, we examined the accumulation of miR156 and two additional miRNAs in 2-week-old mutant and wild-type plants. Little or no difference in the levels of these miRNAs was observed (Fig. 6C), confirming the results obtained for miR171 (Boutet et al. 2003). This result suggests that ZIP, SGS3, and RDR6 are not involved in miRNA biogenesis. To test the hypothesis that ZIP, SGS3, and RDR6 are required for PTGS of ARF3/ETTIN, ARF4, and SPL3, we searched for siRNAs derived from these genes. Blots of low-molecular-weight RNA were hybridized with cDNAs complementary to the coding region of these genes; however, we were unable to detect hybridization to these probes. We detected a weak signal with a probe for the SPL3 3′ UTR but not with a 3′ UTR probe containing seven mutations in the miR156 cognate site, indicating that this signal probably represents cross-hybridization with miR156. The basis for the accumulation of ARF3/ETTIN, ARF4, and SPL3 mRNA in these mutants therefore remains to be determined.
In addition to miRNAs, Arabidopsis possesses many 20-26-nt RNAs (i.e., small RNAs, sRNAs) derived from transposons and intergenic sequences (Llave et al. 2002a; Xie et al. 2004). sRNAs derived from several of these sequences require the RdRP, RDR2, and the Dicer-like protein, DCL3, for their biogenesis (Xie et al. 2004). To determine whether ZIP, SGS3, and RDR6 play a role in the biogenesis of these sRNAs, we examined the effect of zip-1, sgs3-11, and rdr6-11 on the accumulation of sRNAs derived from the SINE retroelement AtSN1, and the sRNA02 and Cluster02 loci, all of which are 24 nt in size. Previous investigators have reported that rdr6 and sgs3 mutations have no effect on the accumulation of AtSN1 (Hamilton et al. 2002). We confirmed this result and found that these mutations also do not affect the accumulation of sRNA02 and Cluster02 sRNAs (Fig. 6D).
SGS3 and RDR6 are required for the biogenesis of trans-acting siRNAs
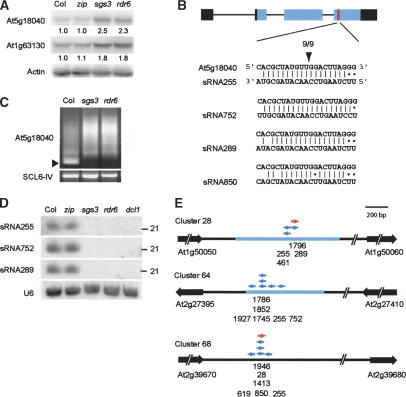
Along with genes whose mRNA accumulates in all three mutants, we identified two genes (At1g63130 and At5g18040) whose mRNA accumulates in sgs3-11 and rdr6-11, but not in zip-1 (Fig. 7A). At1g63130 encodes a pentratricopeptide repeat (PPR) protein, whereas At5g18040 encodes a protein of unknown function. A BLAST search of the Small RNA Database (http://cgrb.orst.edu/smallRNA) (Xie et al. 2004) revealed that several endogenous 21-nt sRNAs are partially complementary to a site in the coding region of At5g18040 (Fig. 7B). Using a modified 5′ RACE protocol (Llave et al. 2002b), we found that At5g18040 mRNA is cleaved in the middle of this complementary sequence (Fig. 7B) and that the 3′ product of this cleavage reaction is reduced or absent in rdr6-11 and sgs3-11 plants (Fig. 7C). To determine the basis for this effect, we examined the expression of four sRNAs that are complementary to this site (sRNA255, sRNA752, sRNA289, and sRNA850) in 2-week-old plants mutant for ZIP, SGS3, RDR6, or DCL1. We were unable to detect expression of sRNA850 (data not shown). However, Northern analysis revealed that sRNA255, sRNA289, and sRNA752 are present in wild-type and zip-1 2-week-old rosettes, but are absent in rdr6-11, sgs3-11, and dcl1-7 (Fig. 7D). It should be noted that because of their sequence similarity we cannot rule out the possibility that most, if not all, of the hybridization observed in these experiments corresponds to a single one of these sRNAs. These observations suggest that the increase in At5g18040 mRNA in rdr6-11 and sgs3-11 is attributable to a reduction in mRNA cleavage resulting from a defect in the biogenesis of one or more of these sRNAs.
Figure 7.
SGS3 and RDR6 repress gene expression by promoting the production of trans-acting siRNAs. (A) Genes affected by sgs3-11 and rdr6-11, but not by zip-1. Total RNA from 2-week-old rosettes of plants grown in short days hybridized with probes to the indicated genes. Actin was used as a loading control. The intensity of each signal was measured using NIH Image; the ratio of this signal relative to wild-type is indicated below each image. (B) The genomic structure of At5g18040 and the location and sequence of the cognate site of sRNA255, sRNA752 and sRNA289, and sRNA850. UTRs are black, the coding region is blue, and the sRNA255 cognate sequence is red. Sequencing of nine 5′ RACE clones revealed that At5g18040 is cleaved in the middle of this cognate site. (C) 5′RACE products from Col, sgs3-11, and rdr6-11 2-week-old rosettes. This reaction gives one major product in Col (arrow-head) that is missing in sgs3-11 and rdr6-11. 5′RACE of SCL6-IV, which is cleaved by miR171, was used as a control. (D) sgs3-11, rdr6-11, and dcl1-7 block the production of sRNA255-like RNAs. Blot of low-molecular-weight RNA from 2-week-old plants hybridized with probes complementary to the indicated sRNAs. (E) sRNA255-like RNAs are found in three clusters on two different chromosomes. Arrows indicate the 5′-to-3′ orientation of the sRNA sequence. Red arrows are in direct orientation and blue arrows are in reverse orientation to the genomic DNA sequence.
sRNA255, sRNA752, sRNA289, and sRNA850 sRNAs are found at three sites in the genome (clusters 24, 64, and 68) in association with a number of other sRNAs (Table 3; Fig. 7E) (http://cgrb.orst.edu/smallRNA). Because sRNA255 is present at all three sites, we refer to this class of sRNAs as sRNA255-like sRNAs. The RNAs in cluster 28 are contained within an ∼900-nt clone isolated from a root cDNA library (RAFL14-47-C13; GenBank AU226586 and AI997465), whereas cluster 64 sRNAs are present in an ∼650-nt clone isolated from a cDNA library of senescing leaves of Landsberg erecta (Ler) (GenBank CD534180, CD534192). The sequences of sRNA255 and sRNA1745 are conserved in this Ler sequence, but sRNA752 and sRNA1927 are polymorphic between Ler and Col. The existence of these cDNAs suggests that the sRNAs in cluster 28 and cluster 64 derive from single transcripts. The precursor(s) of cluster 68 has not been identified, but the proximity of these sRNAs strongly suggests that they arise from a common transcript as well. sRNAs complementary to sRNA289 in cluster 28 and to sRNA28/850/1413 in cluster 68 have been identified, suggesting that these molecules derive from a dsRNA precursor (Table 3; Fig. 7E). This prediction is further supported by the observation that these complementary pairs of sRNAs (sRNA289/sRNA1796 and sRNA1413/sRNA1946) are predicted to form dsRNAs that have one or two 3′ unpaired nucleotides at each end—a feature characteristic of the products of Dicer-mediated cleavage (Elbashir et al. 2001). Based on these observations, and the fact the production of sRNA255-like sRNAs depends on SGS3 and RDR6, we conclude that the sRNAs in these clusters are siRNAs. Biogenesis of sRNA255-like sRNAs also requires the Dicer-like protein, DCL1, as these sRNAs are absent in dcl1-7 mutants (Fig. 7D). DCL1 is required for miRNA biogenesis (Park et al. 2002; Reinhart et al. 2002), but is not required for PTGS initiated by a hairpin transgene (Finnegan et al. 2003) or for the production of endogenous 24-nt sRNAs (Xie et al. 2004). This result was therefore unexpected, and may reveal that DCL1 has a broader array of substrates than has previously been suspected.
Table 3.
sRNAs in Cluster 28, Cluster 64, and Cluster 68a
| Cluster | sRNA | Sequence | Location | Length | Chromosome |
|---|---|---|---|---|---|
| 28 | 255 | UUCUAAGUCCAACAUAGCGUA | 18553232-18553212 | 21 | 1 |
| 461 | UUCUAAGUCCAACAUAGCGUAA | 18553232-18553213 | 22 | 1 | |
| 289 | UUCUAAGUCCAACAUAGCAUA | 18553253-18553233 | 21 | 1 | |
| 1796 | UGCUAUGUUGGACUUAGAAU | 18553235-18553254 | 20 | 1 | |
| 64 | 1927 | UUUCUUUAAAAUUGUUUCCGUGUA | 11728892-11728869 | 24 | 2 |
| 1786 | UGAUAUUUGUAGUAAUGGCG | 11729020-11729001 | 20 | 2 | |
| 1852 | AUGAUAUUUGUAGUAAUGGCG | 11729021-11729001 | 21 | 2 | |
| 1745 | AAUGAUAUUUGUAGUAAUGGCG | 11729022-11729001 | 22 | 2 | |
| 255 | UUCUAAGUCCAACAUAGCGUA | 11729063-11729043 | 21 | 2 | |
| 752 | UCCUAAGUCCAACAUAGCGUU | 11729147-11729127 | 21 | 2 | |
| 68 | 619 | AUAUUCCAGGAUAUGCAAAAG | 16544872-16544852 | 21 | 2 |
| 1946 | UCGAUAUGUUGAACUUAGAAUA | 16544895-16544916 | 22 | 2 | |
| 28 | UUCUAAGUUCAACAUAUCGA | 16544914-16544895 | 20 | 2 | |
| 850 | UUCUAAGUUCAACAUAUCGAC | 16544914-16544894 | 21 | 2 | |
| 1413 | UUCUAAGUUCAACAUAUCGACG | 16544914-16544893 | 22 | 2 | |
| 255 | UUCUAAGUCCAACAUAGCGUA | 16544935-16544915 | 21 | 2 |
Sequences and nomenclature according to the Small RNA Database (http://cgrb.orst.edu/smallRNA/db).
Discussion
The mechanism of PTGS and its function in plant biology are only beginning to be deciphered. In plants, PTGS is involved in transgene silencing, virus resistance, the production of siRNAs from endogenous genes, and the suppression of gene expression by miRNAs. SGS3 and RDR6 are known to be involved in two of these processes: transgene silencing and virus resistance (Dalmay et al. 2000; Mourrain et al. 2000; Beclin et al. 2002; Vaistij et al. 2002; Himber et al. 2003; Muangsan et al. 2004). Here we demonstrate that SGS3 and RDR6 also play a role in normal development, and show that these genes are required for the production of endogenous siRNAs that act in trans to repress gene expression by an RNAi-like mechanism.
The function of SGS3 and RDR6
We found that loss-of-function mutations of SGS3 and RDR6 have a precocious vegetative phenotype and floral phenotype nearly identical to that of mutations in the Argonaute-like gene ZIP. In addition, we found that plants homozygous for any combination of these three mutations are not significantly different from single mutant plants, strongly suggesting that these genes operate in the same genetic pathway(s). The observation that sgs3 and rdr6 mutants have a defect in transgene silencing that is not shared by zip implies that these proteins have distinct functions. Argonaute proteins are associated with siRNAs and miRNAs in the RNA-induced silencing complex in animals (Hammond et al. 2001; Mourelatos et al. 2002; Hutvagner et al. 2004; Shi et al. 2004) and fungi (Catalanotto et al. 2002), and are required for the activity of siRNAs and miRNAs in fungi, plants, and animals (Carmell et al. 2002). SGS3 and RDR6 are required for the synthesis of 21-nt siRNAs from transgenes that produce single-stranded transcripts (Dalmay et al. 2000; Mourrain et al. 2000; Beclin et al. 2002; Boutet et al. 2003; Himber et al. 2003), but are not required for PTGS initiated by hairpin transgenes (Beclin et al. 2002).
ZIP, SGS3, and RDR6 could regulate the biogenesis or function of endogenous small RNAs in several ways. One possibility is that ZIP is required for the activity of miRNAs or siRNAs that initiate transitive PTGS. In this scenario, ZIP would initiate miRNA-directed or siRNA-directed mRNA cleavage, and SGS3 and RDR6 would amplify this effect by producing dsRNA from untargeted regions of the mRNA, as RDR6 has been shown to do in the case of transgenes (Vaistij et al. 2002; Himber et al. 2003). Although there is as yet no evidence that miRNAs initiate transitive gene silencing, we believe it is reasonable to entertain this possibility because of the way in which these mutations interact with hst. Plants doubly mutant for any of these genes and hst are phenotypically identical to hst, implying that HST operates in the same pathway(s) as these genes. HST is the Arabidopsis ortholog of the miRNA export receptor, exportin 5 (Bollman et al. 2003; Yi et al. 2003; Lund et al. 2004), and mutations in HST reduce the levels of most miRNAs in Arabidopsis (M.-Y. Park and R.S. Poethig, unpubl.). The interaction between these mutations therefore suggests that miRNAs play important roles in the pathway(s) in which ZIP, SGS3, and RDR6 operate. This hypothesis also receives some support from the identity of the genes whose mRNA accumulates in zip, sgs3, and rdr6 mutants. We identified three such genes by microarray analysis: ARF3/ETTIN, ARF4, and SPL3. ARF3/ETTIN and ARF4 encode members of a large family of auxin-regulated transcription factors, several of which are potential targets of miRNAs (Park et al. 2002; Rhoades et al. 2002); whether ARF3/ETTIN and ARF4 are subject to miRNA regulation is unknown. SPL3 encodes a member of the Squamosa Promoter Binding family of transcription factors (Cardon et al. 1999), 10 of which possess a cognate site for miR156 in either their coding region or 3′ UTR (Rhoades et al. 2002). SPL3 is of particular interest because its mRNA increases during vegetative development (Cardon et al. 1997, 1999), making it an excellent candidate for a gene involved in vegetative phase change. Furthermore, overexpression of SPL3 in transgenic plants causes early flowering (Cardon et al. 1997). We were unable to detect siRNAs derived from the coding region of these genes by Northern analysis, and thus have no evidence that these genes are actually subject to transitive PTGS. However, the possibility that these siRNAs may be present at too low a level to be detected by this approach has not been eliminated.
Although we are intrigued by the hypothesis that SGS3 and RDR6 are involved in miRNA-initiated transitive PTGS, Tang et al. (2003) noted that this phenomenon is inconsistent with the observation that mutations in miRNA cognate sites are typically dominant to the wild-type allele (McConnell et al. 2001; Palatnik et al. 2003; Vaucheret et al. 2004). If miRNAs are capable of initiating secondary siRNA production from untargeted regions of a transcript, the wild-type allele would be a source of such siRNAs in heterozygous plants, and these siRNAs should be capable of silencing both the wild-type and mutant alleles. Thus, if SGS3 and RDR6 are indeed involved in miRNA-initiated transitivity, it is likely that this process only occurs in a few cases. Another possibility is that the function that SGS3 and RDR6 perform in miRNA-directed gene silencing does not involve dsRNA synthesis, and thus is different from the function they perform in transgene silencing. For example, these genes may be required for miRNA-directed cleavage of target mRNAs. This function is suggested by the observation that the C. elegans RdRP, rrf-1, is likely to have a function beyond its role in dsRNA synthesis (Sijen et al. 2001).
A third hypothesis is that SGS3 and RDR6 are involved in the biogenesis of endogenous siRNAs that mediate PTGS in association with ZIP or another Argonaute protein. This hypothesis is suggested by our observation that SGS3 and RDR6 are required for the accumulation of a family of siRNAs that mediate the cleavage of At5g18040 mRNA. Although we have no evidence that ZIP is required for this particular cleavage event, it is not unreasonable to believe that it could be involved in the biogenesis or function of other SGS3/RDR6-dependent siRNAs that silence genes involved in vegetative phase change.
The function of PTGS in vegetative phase change
What is the role of ZIP, SGS3, and RDR6 in vegetative phase change? The precocious loss-of-function phenotype of these genes demonstrates that they normally promote the expression of the juvenile phase, presumably by repressing the expression of adult-promoting genes during this phase. Therefore, one way in which phase change might be regulated is by a decrease in the activity of these genes late in shoot development. The available information about the expression of these genes does not support this hypothesis, however. ZIP mRNA actually increases, rather than decreases, during shoot development (Hunter et al. 2003). Although the expression pattern of SGS3 and RDR6 has not been characterized, we found that mutations in these genes interfere with PTGS throughout shoot development, implying that they are expressed constitutively. These data suggest that the juvenile-to-adult transition is regulated by a change in the transcription of regulatory RNAs whose subsequent processing or function is dependent on ZIP, SGS3, and RDR6, rather than by a change in the abundance or activity of these three proteins.
Although there is no evidence that temporal changes in the expression of ZIP, SGS3, and RDR6 contribute to vegetative phase change, this may not be true for other components of gene-silencing pathways. Temporal variation in gene silencing has been described for transposons (Banks et al. 1988; Fedoroff and Banks 1988; Martienssen et al. 1990) and the Pl-Bh mutation (Cocciolone and Cone 1993; Hoekenga et al. 2000) in maize, as well as for several transgenes in Arabidopsis (Elmayan et al. 1998; Glazov et al. 2003; Vaucheret et al. 2004). As a rule, silencing increases along the length of the shoot; genes subject to silencing are typically expressed at a higher level in organs produced early in development (cotyledons, juvenile leaves) than in organs produced later in shoot development (adult leaves, flowers). In maize, this temporal decrease in gene expression or transposon activity has been associated with increased DNA methylation (Fedoroff and Banks 1988; Hoekenga et al. 2000; Martienssen et al. 1990). This phenomenon suggests that temporal variation in gene-silencing pathways not only regulates vegetative phase change, but may also be controlled by this developmental transition. Determining the endogenous targets of these gene-silencing pathways is an important objective for future research.
The function of endogenous small RNAs
The first endogenous small RNAs to be described in animals (Lee et al. 1993; Lau et al. 2001; Lee and Ambros 2001; Lagos-Quintana et al. 2002) and plants (Llave et al. 2002a; Park et al. 2002; Rhoades et al. 2002) were miRNAs. These 20-22-nt RNAs are derived from hairpin precursors and repress the expression of mRNAs that have complementary sequences by directing the cleavage of these mRNAs, or repressing their translation. Other types of small (20-26-nt) noncoding RNAs have since been described in C. elegans (Ambros et al. 2003b) and Arabidopsis (Llave et al. 2002a; Xie et al. 2004). In C. elegans, these RNAs are called “tiny noncoding RNAs” (tncRNAs) (Ambros et al. 2003b). Some of these sRNAs and tncRNAs are fragments of mRNAs, whereas others arise from intergenic regions of the genome. tncRNAs and sRNAs do not appear to derive from hairpin precursors, although most require Dicer for their biogenesis and thus probably arise from dsRNA precursors. Several Arabidopsis sRNAs have been shown to promote heterochromatin formation (Zilberman et al. 2003; Xie et al. 2004). The function of tncRNAs in C. elegans is unknown.
The evidence presented here suggests that some sRNAs in Arabidopsis act in trans to cleave mRNAs. This conclusion is based on the effect of sgs3-11 and rdr6-11 on the level of sRNA255-like sRNAs and their putative target, At5g18040. We found that At5g18040 is cleaved in the middle of a sequence that is complementary to sRNA255-like sRNAs, and that this process is defective in sgs3-11 and rdr6-11 because sRNA255-like sRNAs are absent in these mutants. sRNA255-like sRNAs arise from noncoding transcripts. Although the precursor of sRNAs in cluster 64 can be folded to produce a hairpin containing sRNA752, several observations indicate that these sRNAs are siRNAs, not miRNAs. The most compelling evidence for this conclusion is the observation that RDR6 is required for their biogenesis. By definition, miRNAs arise from precursor molecules that are capable of forming dsRNA by intra-molecular pairing (Ambros et al. 2003a). There is no obvious reason why an RdRP should be required for miRNA biogenesis and, indeed, we found no evidence that rdr6-11 has a major effect on the accumulation of miRNAs. It is also significant that sRNA255-like sRNAs are produced from precursors that are the source of multiple sRNAs, some of which are complementary to each other. This observation suggests that sRNA255-like sRNAs arise from long, perfectly paired dsRNA precursors produced by SGS3/RDR6-mediated reverse transcription. One argument against this hypothesis is that the biogenesis of sRNA255-like sRNAs also requires DCL1, a protein that was shown to be required for miRNA biogenesis (Park et al. 2002; Reinhart et al. 2002), but not for the biogenesis of endogenous siRNAs (Xie et al. 2004). This result may reveal a role for DCL1 in the generation of a subset of endogenous siRNAs (see below). An alternative possibility is that sRNA255 precursors are cleaved once by DCL1 (because they resemble pre-miRNAs?) and are then transformed into siRNAs by a mechanism that is dependent on SGS3, RDR6, and another Dicer-like protein.
Two types of siRNAs are produced from transgenes undergoing PTGS: short (21-22-nt) siRNAs and long (23-26-nt) siRNAs (Hamilton et al. 2002; Klahre et al. 2002; Himber et al. 2003; Tang et al. 2003). Short siRNAs are associated with sequence-specific mRNA degradation, whereas long siRNAs are correlated with systemic silencing and DNA methylation (Hamilton et al. 2002). Endogenous sRNAs in Arabidopsis can be grouped into similar size and functional categories. Arabidopsis sRNAs range in size from 20-26 nt, with long (23-26-nt) sRNAs being the predominant class (Hamilton et al. 2002; Xie et al. 2004). Long sRNAs (e.g., AtSN1, Cluster02, and sRNA02) require RDR2 and DCL3 for their biogenesis and direct heterochromatin formation (Xie et al. 2004). sRNA255-like sRNAs are short (20-22-nt) sRNAs. These sRNAs resemble miRNAs and transgene-derived siRNAs in size, in their requirement for, respectively, DCL1 and RDR6, and in their ability to direct mRNA cleavage (Hamilton et al. 2002; Llave et al. 2002a,b; Park et al. 2002; Reinhart et al. 2002; Himber et al. 2003; Kasschau et al. 2003). This observation suggests that miRNAs and short sRNAs/siRNAs share components of their biosynthetic pathways and have similar regulatory functions. Hundreds of short sRNAs have been identified in Arabidopsis (Llave et al. 2002a; Xie et al. 2004). Even if only a small fraction of these short sRNAs target coding transcripts, their regulatory potential could be enormous.
Materials and methods
Plant material and growth conditions
All genetic stocks used in this study were in a Columbia (Col) genetic background. Sequence-indexed T-DNA insertions in sgs3 (SALK_001394 and SALK_039005) were generated by Jose Alonso and Joe Ecker (Salk Institute) (Alonso et al. 2003) and were obtained from the Arabidopsis Biological Resource Center. Seeds of the L1 line, sgs2-1, and sgs3-1 were provided by H. Vaucheret (INRA). Seeds were grown on Metromix 200 (Scotts) and placed at 4°C for 2 d before transfer to growth chambers (Conviron). For phenotypic analysis, plants were grown in 96-well flats under constant fluorescent light (120 μE/min/m2; Sylvania VHO) at 22°C. High humidity was maintained during germination and early growth by covering flats with transparent plastic lids. The lids were removed after about 10 d.
Abaxial trichomes were scored 2-3 wk after planting with the aid of a stereomicroscope. After flowering, leaves were removed, attached to cardboard with double-sided tape, and then scanned in a digital scanner. Scanning electron microscopy was performed on siliques that had been fixed in formalin-acetic acid, dehydrated in ethanol, dried, and coated with palladium.
Genetic analysis
New zip-like mutations were mapped using an F2 population from a cross to Ler. Once their identity had been determined, PCR markers useful for the following mutant alleles in genetic crosses were generated by taking advantage of the nucleotide changes corresponding to these mutations. sgs3-11 was identified using a PCR primer pair (5′-CAAAAAACCTGTGGTGGTCTGCA-3′ and 5′-ACAACCTTGGCACGTTCCTGC-3′) that incorporate a PstI polymorphism at the site of the mutation. rdr6-11 was identified by amplifying DNA with the primers 5′-TACTGTCCCTGGCGATCTCT-3′ and 5′-CCACCTCACACGTTCCTCTT-3′, followed by cleavage with Taq1, and zip-1 was identified by performing PCR with 5′-CTGTACTTTGACAGCGGAAACC-3′ and 5′-ACTGGCTTGGACTTTCTACTAGGTTC-3′, followed by digestion with BsaI.
Double-mutant plants were identified in the F2 progeny from intercrosses between zip-1, sgs3-11, and rdr6-11 using the PCR assays described above. Plants homozygous for all three mutations were identified in the F2 progeny of a cross between zip-1 sgs3-11 and rdr6-11. Plants homozygous for these mutations and the L1 transgene were generated by backcrossing F1 progeny to the L1 line. Progeny of this backcross were allowed to self-pollinate, and these families were then screened on kanamycin to identify families homozygous for L1 (kanR/kanR) and segregating for the zip-like mutation. Mutant plants in these families were stained for GUS activity as described (Hunter et al. 2003).
Microarray analysis
RNA was harvested from shoot apices of 2-week-old Col, zip-1, sgs3-11, and rdr6-11 plants grown under short-day conditions (10 h light:14 h dark) at 22°C. All leaves greater than 5 mm in length were removed prior to freezing the samples in liquid nitrogen. Total RNA was extracted with Trizol reagent (Invitrogen) from frozen tissue disrupted in a bead-beater. The extracted RNA was further purified with RNAeasy columns (QIAGEN). Synthesis of biotin-labeled cRNA, hybridization to the Affymetrix ATH1 Genome Array, and scanning was carried out by the University of Pennsylvania Microarray Facility. Microarray Suite 5.0 was used for image analysis.
Northern analysis
For Northern analysis, total RNA (20 μg) extracted from 2-week-old seedlings with Trizol reagent as described above was transferred to Hybond-N+ membranes (Amersham Biosciences). Probes were generated from Col genomic DNA or cDNA by PCR using the following primers: ARF3/ETTIN (5′-AGAGGCCGCTTCAAAGGAACAGAA-3′ and 5′-TGCATAGATGTCCCTTCCTTGCGA-3′), ARF4 (5′-GTTTCCAAGGGTCTTGCAAGGTCA-3′ and 5′-TTTGCTCGAGCTTTGCGGCTTAGA-3′), and SPL3 (5′-ACGAGAGAAGGCGGAAAAGCACAA-3′ and 5′-CGGGATCCCTAAGTCTCAATGCATTTAT-3′), and amplified fragments were sequenced after purification with the QIAquick Gel Extraction kit (QIAGEN). Hybridization was carried out at 68°C using PerfectHyb Plus buffer (Sigma). Probes were labeled with 32P-dCTP using a Prime-it II Random Primer Labeling kit (Stratagene). Blots were washed once in 2× SSC and 0.1% SDS for 5 min at room temperature, twice in 0.5× SSC and 0.1% SDS for 20 min at 68°C, and once in 0.1× SSC 0.1% SDS for 20 min at 68°C. The hybridization signal was detected with a Storm 860 (Molecular Dynamics), and contrast was adjusted with Photoshop 7.
Low-molecular-weight RNA was purified from total RNA of 2-week-old seedlings or floral buds, and 10-μg or 20-μg aliquots were then resolved on 15% denaturing gel and electrically transferred to Hybond-N+ membranes following Dalmay et al. (2000). Oligonucleotide probes whose sequences are complementary to individual miRNAs were 32P-labeled with T4 poly-nucleotide kinase (New England Biolabs). Probes to sRNA02 and Cluster02 sRNA were made as described (Xie et al. 2004). The AtSN1 probe was prepared as described by Zilberman et al. (2003), except that its fragment was cloned onto pGEM-T Easy vector (Promega) and SP6 RNA polymerase (Invitrogen) was used for in vitro transcription. Hybridization was carried out at 38°C using ULTRAhyb-oligo hybridization buffer (Ambion). Blots were washed twice in 2× SSC and 0.5% SDS for 30 min at 38°C. The hybridization signal was detected with a Storm 860, and contrast was adjusted with Photoshop 7.
5′ RACE
Total RNA was extracted from 2-week-old rosettes as described above. Poly(A)+ mRNA was purified from total RNA using the Oligotex mRNA Mini kit (QIAGEN). 5′ RACE was carried out using the GeneRacer Kit (Invitrogen). The GeneRacer RNA Oligo adapter was directly ligated to mRNA (100 ng) without calf intestinal phosphatase and tobacco acid pyrophosphatase treatment. The GeneRacer oligo dT primer was used for cDNA synthesis. Initial PCR was carried out using the GeneRacer 5′ Primer and At5g18040 RACE1 (5′-GTGGGATACAGAAGTCAACAAGCAGACC-3′) or SCL6-IV-1152R, as described by Llave et al. (2002b). Nested PCR was carried out using 1 μL of the initial PCR reaction, GeneRace 5′ nested primer, and At5g18040 RACE2 (5′-GTGGGATACAGAAGTCAACAAGCAGACC-3′). RACE fragments were cloned and sequenced after gel purification.
Acknowledgments
We are grateful to Christine Hunter and Matthew Willmann for their comments on this manuscript, to John Tobias for assistance with the analysis of microarray data, and to Herve Vaucheret and the Arabidopsis Biological Resource Center for seed stocks used in this study. Support for this work was provided by a National Institutes of Health training grant to A.P., a fellowship from the National Institute of Agrobiological Sciences (Japan) to M.Y., and an NIH grant (RO1 GM51893) to R.S.P.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1231804.
References
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. [DOI] [PubMed] [Google Scholar]
- Ambros V., Bartel, B., Bartel, D.P., Burge, C.B., Carrington, J.C., Chen, X., Dreyfuss, G., Eddy, S.R., Griffiths-Jones, S., Marshall, M., et al. 2003a. A uniform system for microRNA annotation. RNA 9: 277-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V., Lee, R.C., Lavanway, A., Williams, P.T., and Jewell, D. 2003b. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13: 807-818. [DOI] [PubMed] [Google Scholar]
- Banks J.A., Masson, P., and Fedoroff, N. 1988. Molecular mechanisms in the developmental regulation of the maize Suppressor-mutator transposable element. Genes & Dev. 2: 1364-1380. [DOI] [PubMed] [Google Scholar]
- Beclin C., Boutet, S., Waterhouse, P., and Vaucheret, H. 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12: 684-688. [DOI] [PubMed] [Google Scholar]
- Berardini T.Z., Bollman, K., Sun, H., and Poethig, R.S. 2001. Regulation of vegetative phase change in Arabidopsis thaliana by cyclophilin 40. Science 291: 2405-2407. [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T., Czaplinski, K., and Gorlich, D. 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollman K.M., Aukerman, M.J., Park, M.Y., Hunter, C., Berardini, T.Z., and Poethig, R.S. 2003. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130: 1493-1504. [DOI] [PubMed] [Google Scholar]
- Boutet S., Vazquez, F., Liu, J., Beclin, C., Fagard, M., Gratias, A., Morel, J.B., Crete, P., Chen, X., and Vaucheret, H. 2003. Arabidopsis HEN1. A genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13: 843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon G.H., Hohmann, S., Nettesheim, K., Saedler, H., and Huijser, P. 1997. Functional analysis of the Arabidopsis thaliana SBP-box gene SPL3: A novel gene involved in the floral transition. Plant J. 12: 367-377. [DOI] [PubMed] [Google Scholar]
- Cardon G., Hohmann, S., Klein, J., Nettesheim, K., Saedler, H., and Huijser, P. 1999. Molecular characterisation of the Arabidopsis SBP-box genes. Gene 237: 91-104. [DOI] [PubMed] [Google Scholar]
- Carmell M.A., Xuan, Z., Zhang, M.Q., and Hannon, G.J. 2002. The Argonaute family: Tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumori-genesis. Genes & Dev. 16: 2733-2742. [DOI] [PubMed] [Google Scholar]
- Catalanotto C., Azzalin, G., Macino, G., and Cogoni, C. 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes & Dev. 16: 790-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien J.C. and Sussex, I.M. 1996. Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L) Heynh. Plant Physiol. 111: 1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J.H., Tack, D., Findlay, K., Van Montagu, M., and Van Lijsebettens, M. 1999. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 20: 493-501. [DOI] [PubMed] [Google Scholar]
- Cocciolone S.M. and Cone, K.C. 1993. Pl-Bh, an anthocyanin regulatory gene of maize that leads to variegated pigmentation. Genetics 135: 575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C. and Macino, G. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166-169. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543-553. [DOI] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21 and 22-nucleotide RNAs. Genes & Dev. 15: 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., et al. 1998. Arabidopsis mutants impaired in cosuppression. Plant Cell 10: 1747-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. 2000. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. 97: 11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N.V. and Banks, J.A. 1988. Is the Suppressor-mutator element controlled by a basic developmental regulatory mechanism? Genetics 120: 559-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E.J., Margis, R., and Waterhouse, P.M. 2003. Posttranscriptional gene silencing is not compromised in the Arabidopsis CARPEL FACTORY (DICER-LIKE1) mutant, a homolog of Dicer-1 from Drosophila. Curr. Biol. 13: 236-240. [DOI] [PubMed] [Google Scholar]
- Glazov E., Phillips, K., Budziszewski, G.J., Meins, F., and Levin, J.Z. 2003. A gene encoding an RNase D exonuclease-like protein is required for post-transcriptional silencing in Arabidopsis. Plant J. 35: 342-349. [DOI] [PubMed] [Google Scholar]
- Greenwood M.S. 1995. Juvenility and maturation in conifers: Current concepts. Tree Physiol. 15: 433-438. [DOI] [PubMed] [Google Scholar]
- Hackett W.P. and Murray, J.R. 1993. Maturation and rejuvenation in woody species. In Micropropagation of woody plants (ed. M.R. Ahuja), pp. 93-105. Kluwer, Amsterdam.
- Hamilton A., Voinnet, O., Chappell, L., and Baulcombe, D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146-1150. [DOI] [PubMed] [Google Scholar]
- Himber C., Dunoyer, P., Moissiard, G., Ritzenthaler, C., and Voinnet, O. 2003. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 22: 4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekenga O.A., Muszynski, M.G., and Cone, K.C. 2000. Developmental patterns of chromatin structure and DNA methylation responsible for epigenetic expression of a maize regulatory gene. Genetics 155: 1889-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C., Sun, H., and Poethig, R.S. 2003. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Curr. Biol. 13: 1734-1739. [DOI] [PubMed] [Google Scholar]
- Hutvagner G., Simard, M.J., Mello, C.C., and Zamore, P.D. 2004. Sequence-specific inhibition of small RNA function. PLoS Biol. 2: 465-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau K.D., Xie, Z., Allen, E., Llave, C., Chapman, E.J., Krizan, K.A., and Carrington, J.C. 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev. Cell 4: 205-217. [DOI] [PubMed] [Google Scholar]
- Klahre U., Crete, P., Leuenberger, S.A., Iglesias, V.A., and Meins Jr., F. 2002. High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. 99: 11981-11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735-739. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858-862. [DOI] [PubMed] [Google Scholar]
- Lee R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862-864. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with anti-sense complementarity to lin-14. Cell 75: 843-854. [DOI] [PubMed] [Google Scholar]
- Llave C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. 2002a. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002b. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053-2056. [DOI] [PubMed] [Google Scholar]
- Lund E., Guttinger, S., Calado, A., Dahlberg, J.E., and Kutay, U. 2004. Nuclear export of microRNA precursors. Science 303: 95-98. [DOI] [PubMed] [Google Scholar]
- Martens H., Novotny, J., Oberstrass, J., Steck, T.L., Postlethwait, P., and Nellen, W. 2002. RNAi in Dictyostelium: The role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell 13: 445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R., Barkan, A., Taylor, W.C., and Freeling, M. 1990. Somatically heritable switches in the DNA modification of Mu transposable elements monitored with a suppressible mutant in maize. Genes & Dev. 4: 331-343. [DOI] [PubMed] [Google Scholar]
- McConnell J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411: 709-713. [DOI] [PubMed] [Google Scholar]
- Mouradov A., Cremer, F., and Coupland, G. 2002. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell (Suppl.) 14: S111-S130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Jouette, D., Lacombe, A.M., Nikic, S., Picault, N., et al. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533-542. [DOI] [PubMed] [Google Scholar]
- Muangsan N., Beclin, C., Vaucheret, H., and Robertson, D. 2004. Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 38: 1004-1014. [DOI] [PubMed] [Google Scholar]
- Palatnik J.F., Allen, E., Wu, X., Schommer, C., Schwab, R., Carrington, J.C., and Weigel, D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425: 257-263. [DOI] [PubMed] [Google Scholar]
- Park W., Li, J., Song, R., Messing, J., and Chen, X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S. 2003. Phase change and the regulation of developmental timing in plants. Science 301: 334-336. [DOI] [PubMed] [Google Scholar]
- Prigge M.J. and Wagner, D.R. 2001. The Arabidopsis SERRATE gene encodes a zinc-finger protein required for normal shoot development. Plant Cell 13: 1263-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110: 513-520. [DOI] [PubMed] [Google Scholar]
- Shi H., Djikeng, A., Tschudi, C., and Ullu, E. 2004. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: Control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell. Biol. 24: 420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu P.K. and Metzenberg, R.L. 2002. Meiotic silencing by unpaired DNA: Properties, regulation and suppression. Genetics 161: 1483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H., and Fire, A. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465-476. [DOI] [PubMed] [Google Scholar]
- Simmer F., Tijsterman, M., Parrish, S., Koushika, S.P., Nonet, M.L., Fire, A., Ahringer, J., and Plasterk, R.H. 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317-1319. [DOI] [PubMed] [Google Scholar]
- Smardon A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., and Maine, E.M. 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10: 169-178. [DOI] [PubMed] [Google Scholar]
- Tang G., Reinhart, B.J., Bartel, D.P., and Zamore, P.D. 2003. A biochemical framework for RNA silencing in plants. Genes & Dev. 17: 49-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A. and Poethig, R.S. 1998. HASTY: A gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125: 1889-1898. [DOI] [PubMed] [Google Scholar]
- Telfer A., Bollman, K.M., and Poethig, R.S. 1997. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645-654. [DOI] [PubMed] [Google Scholar]
- Tsukaya H., Shoda, K., Kim, G.T., and Uchimiya, H. 2000. Heteroblasty in Arabidopsis thaliana (L.) Heynh. Planta 210: 536-542. [DOI] [PubMed] [Google Scholar]
- Vaistij F.E., Jones, L., and Baulcombe, D.C. 2002. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez, F., Crata, P., and Bartel, D.P. 2004. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes & Dev. 18: 1187-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833-1837. [DOI] [PubMed] [Google Scholar]
- Volpe T., Schramke, V., Hamilton, G.L., White, S.A., Teng, G., Martienssen, R.A., and Allshire, R.C. 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11: 137-146. [DOI] [PubMed] [Google Scholar]
- Xie Z., Johansen, L.K., Gustafson, A.M., Kasschau, K.D., Lellis, A.D., Zilberman, D., Jacobsen, S.E., and Carrington, J.C. 2004. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2: 642-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R., Qin, Y., Macara, I.G., and Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Dev. 17: 3011-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Fan, B., MacFarlane, S.A., and Chen, Z. 2003. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact. 16: 206-216. [DOI] [PubMed] [Google Scholar]
- Zilberman D., Cao, X., and Jacobsen, S.E. 2003. ARGO-NAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716-719. [DOI] [PubMed] [Google Scholar]