Abstract
Molecular and genetic studies have demonstrated that members of the Toll-like receptor (TLR) family are critical innate immune receptors. TLRs are recognition receptors for a diverse group of microbial ligands including bacteria, fungi, and viruses. This study demonstrates that distinct TLRs are responsible for the recognition of Helicobacter lipopolysaccharide (LPS) versus intact Helicobacter bacteria. We show that the cytokine-inducing activity of Helicobacter LPS was mediated by TLR4; i.e., TLR4-deficient macrophages were unresponsive to Helicobacter pylori LPS. Surprisingly, the cytokine response to whole Helicobacter bacteria (H. pylori, H. hepaticus, and H. felis) was mediated not by TLR4 but rather by TLR2. Studies of HEK293 transfectants revealed that expression of human TLR2 was sufficient to confer responsiveness to intact Helicobacter bacteria, but TLR4 transfection was not sufficient. Our studies further suggest that cag pathogenicity island genes may modulate the TLR2 agonist activity of H. pylori as cagA+ bacteria were more active on a per-cell basis compared to cagA mutant bacteria for interleukin-8 (IL-8) cytokine secretion. Consistent with the transfection studies, analysis of knockout mice demonstrated that TLR2 was required for the cytokine response to intact Helicobacter bacteria. Macrophages from both wild-type and TLR4-deficient mice produced a robust cytokine secretion response (IL-6 and MCP-1) when stimulated with intact Helicobacter bacteria. In contrast, macrophages from TLR2-deficient mice were profoundly unresponsive to intact Helicobacter stimulation, failing to secrete cytokines even at high (100:1) bacterium-to-macrophage ratios. Our studies suggest that TLR2 may be the dominant innate immune receptor for recognition of gastrointestinal Helicobacter species.
Helicobacter pylori is a gram-negative, spiral-shaped bacterium that infects half of the world's population (29). While infection with H. pylori invariably leads to a chronic inflammatory response (chronic active gastritis), most infected patients remain asymptomatic, with only minimal inflammation (62). However, a significant percentage of patients do progress to more serious outcomes, which include peptic ulcer disease, gastric lymphoma, and gastric cancer (12, 22, 42, 57). On the basis of its strong link to gastric cancer, H. pylori has been classified by the International Agency for Research on Cancer (a branch of the World Health Organization) as a class I carcinogen (29). In both animal models (40, 79) and human studies (14, 15), progression of H. pylori disease from superficial gastritis to gastric cancer appears to be related to the severity of the host inflammatory response. The identification of H. pylori components and host factors that contribute to the inflammatory response may lead to important insights into the mechanism of peptic ulcer disease and/or gastric malignancy.
Although H. pylori induces chronic mucosal inflammation to some degree in all infected patients, the organism does not appear to invade the gastric epithelium (reviewed in reference 27). Gastric epithelial cells and macrophages are considered to be the main sources of proinflammatory cytokines and key components of innate immunity. With respect to H. pylori, the gastric epithelial cell layer is thought to represent the first line of defense and the initial trigger for host innate and inflammatory responses (8, 19, 65). H. pylori has been shown to activate intracellular signaling in gastric epithelial cells, leading to transcriptional responses. Epithelial cells release a variety of proinflammatory mediators including both cytokines and chemokines, leading to the subsequent attraction of monocytes/macrophages. In addition, both epithelial cells and macrophages appear to recognize microbial pathogens by sampling the environment with a family of receptors that discriminate between pathogens and self, pattern recognition receptors known as the Toll-like receptor (TLR) family (8, 19, 65).
Several animal models of Helicobacter-induced gastrointestinal inflammation and gastric cancer have been developed (40, 79, 80). H. felis is a mouse pathogen, and infection of C57BL6 mice mimics many of the pathogenic changes commonly found in humans infected with H. pylori (40, 80). H. pylori from human isolates is not usually a mouse pathogen; however, a mouse-adapted strain, H. pylori SS1, has proven very useful in modeling gastric disease and cancer progression in mice (41, 75). H. hepaticus is an endogenous mouse pathogen. H. hepaticus infection produces a typhlocolitis in mice (16, 17). Recently, H. hepaticus infection has been shown to lead to the development of colon cancer in Rag-2-deficient mice (18). The receptors involved in the inflammatory response to H. hepaticus are largely unknown, although a prominent role for NF-κB has been demonstrated in studies by Erdman et al. (16).
In this study, we have investigated the role of TLRs in the recognition of H. pylori, H. felis, and H. hepaticus. With normal human monocytes and macrophages, transfected cell lines, and genetically deficient animals, we demonstrate that TLR2 is an important cytokine signaling receptor for H. pylori, H. felis, and H. hepaticus. Further, we demonstrate that although the lipopolysaccharide (LPS) of H. pylori is recognized by TLR4, the major TLR for intact Helicobacter bacteria is TLR2, not TLR4.
MATERIALS AND METHODS
Human cells and cell lines.
Human embryonic kidney (HEK293) cells (American Type Culture Collection, Manassas, Va.) were grown in RPMI 1640 medium or Dulbecco modified Eagle medium (Gibco BRL, Grand Island, N.Y.) supplemented with 10% heat-inactivated fetal bovine serum (Atlanta Biologicals, Norcross, Ga.). HEK293 cells stably expressing human TLR2, TLR4, MD2, and/or CD14 were engineered as previously described (38). Peripheral blood mononuclear cells (PBMC) were isolated from normal human donors by Ficoll-Hypaque gradient centrifugation (Lymphocyte Separation Medium; Mediatech, Herndon, Va.). Monocytes were isolated by countercurrent centrifugal elutriation of mononuclear leukocyte-enriched cell preparations from leukapheresis donors. Macrophages were differentiated from blood monocytes by culturing the cells in the presence of 1,000 U of macrophage colony-stimulating factor (R&D Systems, Minneapolis, Minn.) per ml for 2 days and in culture medium (Dulbecco modified Eagle medium plus 10% fetal bovine serum) alone for an additional 8 to 14 days (76).
PECs.
Murine peritoneal exudate cells (PECs) were harvested from mice injected intraperitoneally with 1 ml of 4% thioglycolate after 4 days by peritoneal lavage with Ca2+- and Mg2+-free sterile saline. TLR2-deficient mice were the gift of S. Akira (Osaka, Japan) (24). TLR4-deficient C57BL/10ScN mice were obtained from the National Cancer Institute. C57BL/6, C57BL/6 × 129Sv F2 (B6129F2), and C57BL/10SnJ wild-type control mice were purchased from The Jackson Laboratory (Bar Harbor, Maine).
Cells were cultured in 24-well culture dishes at densities of 105 to 106 cells per well in 1 ml of medium. HEK293 cells at 105/ml, PBMC at 106/ml, monocytes and macrophages at 105/ml, and PECs at 105 to 106/ml were used. Cells were stimulated with LPS (1 to100 ng/ml), Pam3CSK4 (10 to100 ng/ml), or bacteria (103 to 109 CFU/ml). For analysis of cytokine secretion, culture supernatants were harvested 18 h after stimulation and cytokine levels were determined by enzyme-linked immunosorbent assay (ELISA) (OptEIA; BD-Pharmingen). For analysis of cytokine gene transcription, cells were stimulated with bacteria or medium alone for 2 h. The cells were harvested and lysed, and RNA was prepared with a commercial RNA extraction kit (RNeasy; QIAGEN, Valencia, Calif.). Total mRNA (5 μg per blot) was reverse transcribed, labeled with biotin, and hybridized to a Mouse Inflammatory Cytokine SuperArray in accordance with the manufacturer's (SuperArray Inc., Frederick, Md.) instructions. Chemiluminescent images on film were transilluminated, scanned, and analyzed with GEArray software.
LPS preparations.
LPS from Escherichia coli serotype O111:B4 was purchased from Sigma (St. Louis, Mo.). Phenol extraction of the LPS to remove contaminating lipopeptides was performed as described elsewhere (26). The following clinical isolates of H. pylori were also used as sources of LPS: AM1, isolated from a patient with chronic gastritis; AM2 and AM3, from patients with superficial gastritis; AM4, from a patient with chronic atrophic gastritis; AM5, from a patient with intestinal metaplasia; and AM6, from a patient with gastric adenocarcinoma. Similarly, LPS was obtained from clinical isolates of E. coli and Salmonella enterica (55).
Bacterial biomass was obtained by growth of H. pylori strains on blood agar under microaerobic conditions (54). H. pylori LPS was obtained from this biomass by phenol-water extraction and subsequent enzymatic purification with RNase A, DNase II, and proteinase K, as well as by ultracentrifugation as described previously (55). The H. pylori LPS obtained was essentially free of proteins (<0.1%) and nucleic acids (<0.1%) and had an electrophoretic profile similar to that reported previously for the high-molecular-mass LPS of other H. pylori strains (54). Moreover, in the Limulus amebocyte lysate assay and for induction of tumor necrosis factor alpha, the LPS exhibited bioactivities identical to those reported previously (63). High-molecular-mass LPS was produced by these strains when they were grown either on solid medium or in liquid culture. Importantly, the H. pylori LPS used in these studies was free of contaminating peptidoglycan as determined in biochemical assays (A. P. Moran, unpublished results), and furthermore, the bioactivities of these preparations was unaffected by treatment with lysozyme, which removes trace peptidoglycan contamination (data not shown).
Helicobacter strains.
H. pylori SS1, H. pylori ATCC 43504 (cagA+ vacA+), H. pylori Astra 244 (cagA− vacA+), H. hepaticus (ATCC 51449), and H. felis (ATCC 49179) were grown on tryptic soy broth-blood agar (Becton Dickinson, Cockeysville, Md.), harvested, and washed, and the optical density at 600 nm and CFU counts were determined (28). Bacteria were stored in aliquots at −70°C. On the day of assay, the bacteria were washed in sterile saline, recovery was checked by determining the optical density at 600 nm, and the bacteria were resuspended to 109/ml in sterile saline. Serial dilutions of the bacteria were prepared in culture medium immediately prior to addition to the cell cultures. Cells and bacteria were incubated together in tissue culture medium for 18 h at 37°C, and culture supernatants were harvested for cytokine analysis. (Experiments with bacteria grown in liquid culture showed the same results. Data are representative of at least three independent experiments.)
RESULTS
H. pylori activates the innate immune response in human cells.
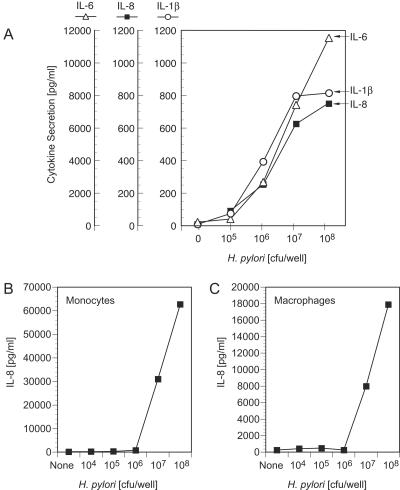
Normal human peripheral blood cells incubated with H. pylori SS1 bacteria secreted inflammatory cytokines, including interleukin-1β (IL-1β), IL-6, IL-8, and tumor necrosis factor alpha (Fig. 1). Cytokines were produced in a dose-dependent manner by PBMC upon incubation with H. pylori SS1 bacteria (Fig. 1A). Purified human monocytes and monocyte-derived human macrophages exhibited similar dose-dependent cytokine responses to H. pylori bacteria (Fig. 1B and C). Optimal cytokine secretion was observed at a dose of approximately 108 H. pylori SS1 CFU/105 monocytes (Fig. 1B) or macrophages (Fig. 1C).
FIG. 1.
Human monocytes and macrophages challenged with whole H. pylori bacteria secrete inflammatory cytokines. Human PBMC (A), purified monocytes (B), or monocyte-derived macrophages (C) were stimulated with various numbers of H. pylori SS1 bacteria or medium alone. Culture supernatants were harvested following an 18-h incubation, and IL-6, IL-8, and IL-1β levels were determined by cytokine-specific ELISA.
Human monocytes and macrophages express innate immune receptors, including TLR2, TLR4, and the pattern recognition receptor CD14. These receptors participate in the inflammatory response to a variety of bacteria and bacterial products, including LPS from gram-negative bacteria, peptidoglycan from gram-positive bacteria, and yeast zymosan. Therefore, we determined if these receptors are also important for the response to H. pylori.
Human TLR2 expression is sufficient to confer responsiveness to H. pylori bacteria.
We investigated the role of TLRs in the response to H. pylori in gain-of-function studies with stably transfected HEK293 cells. Transfection of HEK293 cells with human TLR4 confers responsiveness to the TLR4 ligand E. coli LPS (10, 38). Likewise, transfection with human TLR2 confers responsiveness to TLR2 ligands, such as yeast zymosan and S.aureus peptidoglycan (38, 44). CD14 expression enhances the response of HEK293 cells to both TLR2 and TLR4 ligands (10, 38, 44).
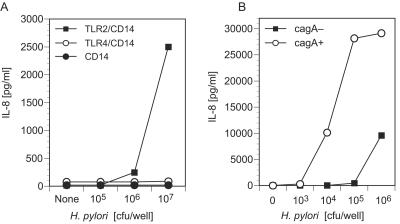
We examined the response of HEK293 cells stably transfected with CD14 alone or in combination with TLR2 or TLR4 to H. pylori. Intact H. pylori bacteria induced IL-8 secretion from cells expressing TLR2 and CD14 but not from cells expressing TLR4 and CD14 or CD14 alone (Fig. 2A). The IL-8 response of TLR2- and CD14-expressing cells to H. pylori SS1 bacteria was dose dependent (Fig. 2A and B). These results suggested that although H. pylori is a gram-negative bacterium, the cytokine secretion response to whole H. pylori bacteria occurs by TLR2, rather than TLR4, receptor stimulation.
FIG. 2.
Whole H. pylori bacteria are potent stimulators of HEK293 cells expressing TLR2 but not TLR4. (A) HEK293 cell clones stably expressing CD14, TLR2 and CD14, or TLR4 and CD14 were stimulated with H. pylori SS1 bacteria or medium alone. (B) TLR2- and CD14-expressing HEK293 cell clones were stimulated with various numbers of cagA+ or cagA mutant H. pylori bacteria or medium alone. Culture supernatants were harvested after an 18-h incubation, and IL-8 levels were determined by ELISA.
cagA+ H. pylori stimulation of TLR2-expressing cells.
Studies from several laboratories have shown that a set of H. pylori genes in the cag pathogenicity island influences the cytokine-stimulating activity of H. pylori. To address the role of the cag region genes in the TLR2 agonist activity of intact H. pylori bacteria, cagA+ and cagA mutant H. pylori bacteria were used to stimulate TLR2- and CD14-expressing HEK293 cells (Fig. 2B). Both strains of bacteria stimulated IL-8 secretion from the TLR2- and CD14-expressing cells, indicating that cagA is not required for a TLR2-mediated cytokine response. Nevertheless, cagA+ bacteria were 2 logs more potent stimulators of IL-8 secretion than cagA mutant bacteria and the response to cagA+ bacteria was dependent on TLR2 expression, suggesting that cagA (or genes associated with cagA in the cag pathogenicity island) augments the TLR2 response either directly or by regulating the expression of other TLR2 target molecules on H. pylori.
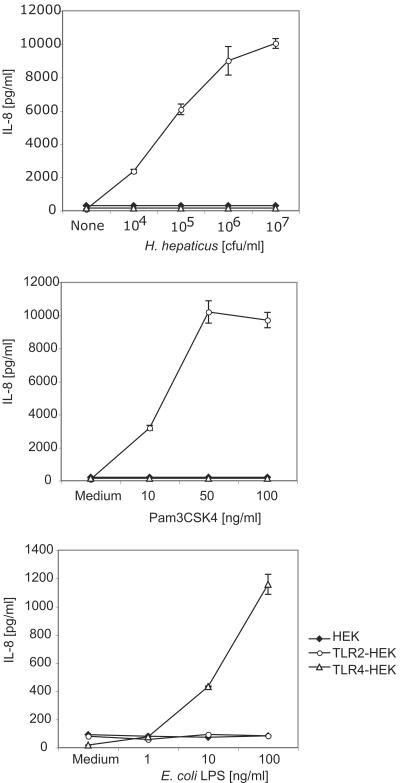
H. hepaticus bacteria activate cytokine secretion via TLR2.
Similar to the response seen with H. pylori, whole H. hepaticus bacteria stimulated IL-8 secretion (Fig. 3) from HEK293 cells transfected with human TLR2. The response was dose dependent, and IL-8 secretion was detectable at bacterium-to-cell ratios as low as 0.5:1 (Fig. 3). In contrast, neither H. hepaticus nor H. pylori bacteria activated TLR4 (and MD2)-expressing HEK293 cells, although the TLR4-expressing HEK293 cells responded to an LPS challenge (Fig. 2 and 3).
FIG. 3.
Whole H. hepaticus bacteria activate TLR2-expressing but not TLR4-expressing HEK cells. HEK293 clones stably expressing TLR2 and CD14, TLR4 and MD2, or CD14 alone (control) were stimulated with H. hepaticus bacteria (upper panel), Pam3CSK4 (TLR2 ligand, middle panel), E. coli LPS (TLR4 ligand, lower panel), or medium alone. Culture supernatants were harvested after an 18-h incubation, and IL-8 levels were determined by ELISA.
H. pylori bacteria activate inflammatory cytokine gene expression and secretion from murine macrophages.
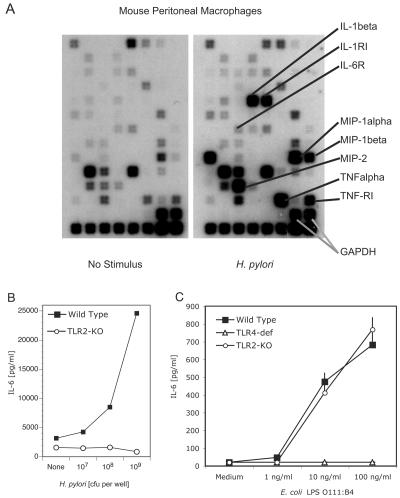
Like human monocytes and macrophages, mouse peritoneal macrophages incubated with whole H. pylori bacteria exhibited a robust inflammatory cytokine response (Fig. 4). Gene array analysis of peritoneal macrophages demonstrated upregulation of a large number of inflammatory cytokine and chemokine genes within 2 h of stimulation of the macrophages with whole H. pylori bacteria (Fig. 4A). This increase in gene expression was reflected at the protein level; H. pylori induced a dose-dependent increase in the synthesis and secretion of inflammatory cytokines, including IL-6 and MCP-1, from mouse macrophages (Fig. 4B).
FIG. 4.
Murine macrophages are activated by whole H. pylori SS1 bacteria in a TLR2-dependent manner. (A) Wild-type mouse peritoneal exudate macrophages were stimulated with medium (left panel) or H. pylori SS1 bacteria (right panel). Following a 2-h incubation, RNA was extracted, labeled, and hybridized to a mouse SuperArray blot. The positions of inflammatory cytokine and cytokine receptor spots are indicated. (Bottom two rows) Negative controls included blanks and pUC18. Positive controls included the gene for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and housekeeping genes (those for cyclophilin A and actin). (B) Wild-type and TLR2 knockout (KO) mouse peritoneal exudate macrophages were stimulated with medium (None) or H. pylori SS1 bacteria (107 to 109 per well). Supernatants were harvested 18 h later, and IL-6 levels were determined by ELISA. (C) Wild-type, TLR2 knockout, or TLR4-deficient (def) ScN mouse peritoneal exudate macrophages were stimulated with medium alone or E. coli LPS (TLR4 ligand). Supernatants were harvested 18 h later, and IL-6 levels were determined by ELISA.
TLR2 knockout macrophages are unresponsive to H. pylori bacteria, while TLR4-deficient macrophages respond normally.
The gain-of-function studies of transfected cell lines indicated that TLR2 plays an important role in the response to whole H. pylori bacteria. We next determined if the targeted deletion of TLR2 in knockout mice would affect the inflammatory response to H. pylori bacteria. Peritoneal macrophages from wild-type and TLR2−/− mice were cultured with H. pylori bacteria, and the release of cytokines was measured. Wild-type macrophages secreted IL-6 in response to H. pylori bacteria, while TLR2−/− macrophages failed to respond to H. pylori (Fig. 4B), even at very high doses of bacteria (109 CFU/106 macrophages). Control cultures demonstrated a robust cytokine response of wild-type and TLR2−/− macrophages to E. coli LPS (Fig. 4C) and to IL-1β (data not shown). As expected, TLR2−/− macrophages were unresponsive when challenged with the TLR2 ligands (i.e., zymosan and peptidoglycan) (11, 70; data not shown), whereas TLR4-deficient macrophages were unresponsive to LPS (Fig. 4C).
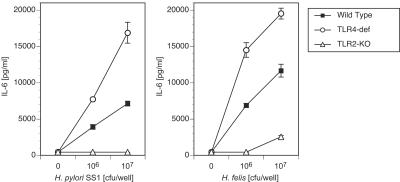
In a similar experiment, we compared wild-type and TLR2- and TLR4-deficient macrophage responses to H. pylori SS1 and H. felis (Fig. 5). We wanted to determine whether (i) the failure of TLR2−/− mice to respond to H. pylori SS1 was peculiar to this species of Helicobacter or whether responses to other gastric Helicobacter species were also deficient in these animals and (ii) whether TLR4 is necessary for the cytokine response to Helicobacter bacteria. These studies demonstrate that TLR2−/− macrophages failed to respond to either Helicobacter species (Fig. 5A and B). In contrast, TLR4-deficient mice had an enhanced response to both Helicobacter species compared to the response of control (wild-type) animals (Fig. 5A and B). (The basis for the hyperresponsiveness of TLR4-deficient macrophages to Helicobacter bacteria is not clear, but it may reflect higher levels of TLR2 expression on these macrophages or increased availability of intracellular adaptor or signaling components in the absence of TLR4 expression.) Thus, TLR2 but not TLR4 was necessary and sufficient for the cytokine response to intact Helicobacter bacteria.
FIG. 5.
H. pylori SS1 and H. felis both activate murine macrophages in a TLR2-dependent, TLR4-independent manner. Wild-type, TLR2-knockout (KO), and TLR4-deficient (def) mouse peritoneal exudate macrophages were stimulated with medium alone or with H. pylori SS1 (left panel) or H. felis (right panel) bacteria. Supernatants were harvested 18 h later, and IL-6 levels were determined by ELISA.
H. pylori LPS stimulates cytokine secretion via TLR4.
We next determined if the LPS component of H. pylori could stimulate an inflammatory response similar to that seen with whole bacteria. LPS from gram-negative bacteria, such as E. coli, stimulates the innate immune response through TLR4. This response is independent of TLR2 expression (23, 26, 77). In contrast to E. coli, the innate immune response to H. pylori was dependent on TLR2 but not TLR4. To determine if differences in the structure of the LPS of E. coli compared to that of H. pylori LPS could account for differences in TLR activation, we compared the E. coli and H. pylori LPSs as stimulants of wild-type, TLR2−/−, and TLR4-deficient macrophages. The LPSs of both E. coli and H. pylori activated macrophages in a dose-dependent (Fig. 6) and TLR4-dependent (Fig. 7) manner. Thus, H. pylori LPS preparations triggered cytokine secretion from wild-type and TLR2−/− macrophages, as well as from TLR4-transfected HEK293 cells (Fig. 7A, B, and D). In contrast, H. pylori LPS failed to activate TLR4−/− macrophages or control HEK293 cells (Fig. 7C and data not shown).
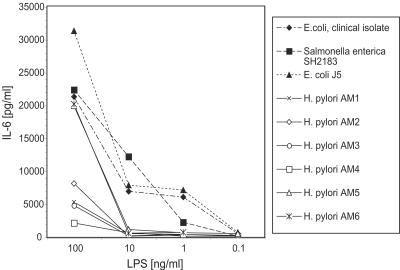
FIG. 6.
Dose-dependent activation of cytokine secretion and comparison of LPS preparations purified from E. coli with those from clinical isolates of H. pylori. Wild-type peritoneal exudate macrophages were stimulated with medium alone or with various doses of LPS isolated from E. coli, S. enterica, or clinical isolates of H. pylori (designated AM1 to -6). Following an 18-h incubation, supernatants were harvested and IL-6 levels were determined by ELISA.
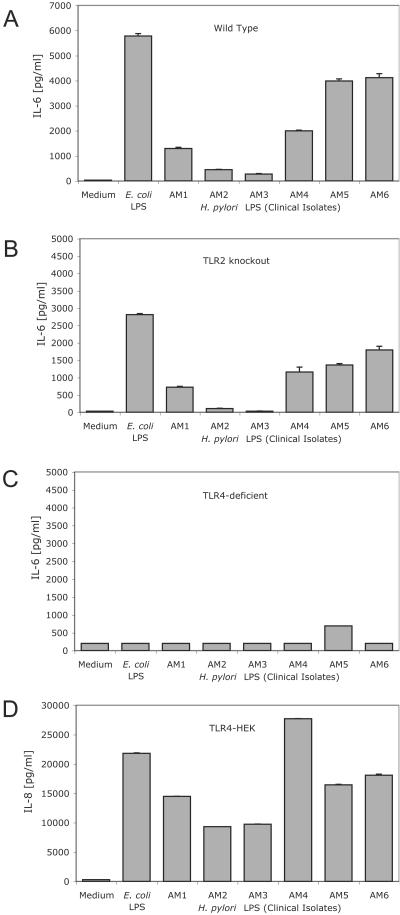
FIG. 7.
LPS preparations from clinical isolates of H. pylori are TLR4 ligands. Wild-type peritoneal exudate macrophages (A), TLR2 knockout macrophages (B), TLR4-deficient macrophages (C), or TLR4-expressing HEK293 cells (D) were stimulated with medium alone, LPS isolated from E. coli O11B4, or LPS from clinical isolates of H. pylori. All LPS stimulations were done with protein-free LPS re-extracted with 10 ng of phenol/ml. H. pylori LPS was prepared from six different clinical isolates (designated AM1 to -6) as detailed in Materials and Methods. Following an 18-h incubation, supernatants were harvested and IL-6 (macrophages) or IL-8 (TLR4-expressing HEK293 cells) levels were determined by ELISA.
As noted in other studies, LPS prepared from clinical isolates of H. pylori varies substantially in its cytokine-stimulating activity (5, 31, 35). We observed that LPS from isolates AM5 and AM6 induced a robust IL-6 cytokine response (Fig. 6). LPS from isolates AM1, -2, -3, and -4 induced lesser levels of IL-6 (Fig. 6). These differences in activity may reflect variations in the degree of acylation and/or phosphorylation of the LPS in these different clinical isolates (Moran, unpublished). Differences in the core and O chains of the LPS from the individual isolates are also observed that could perhaps modulate the cytokine-inducing activity of the lipid A component of LPS (Moran, unpublished).
Overall, the H. pylori LPS preparations were 1 to 2 logs less stimulatory than either E. coli or Salmonella LPS similarly prepared from clinical isolates (Fig. 6). Nevertheless, all of the H. pylori LPS preparations tested that were active in assays with wild-type cells (i.e., AM1, AM4, AM5, and AM6) displayed the same TLR4-dependent, TLR2-independent pattern of response (Fig. 7). Similarly, differences in the structure of the LPS derived from H. pylori compared to that of the LPS from E. coli cannot account for the TLR2 dependence of the response to intact H. pylori bacteria (53, 56). Taken together, our studies suggest that TLR4 expression was both necessary and sufficient for a robust cytokine secretion response to H. pylori LPS, whereas TLR2 expression was required for responsiveness to intact Helicobacter bacteria.
Intact E. coli bacteria stimulate both TLR2- and TLR4-expressing cells.
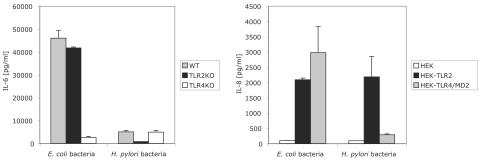
We directly compared the TLR2 and TLR4 dependence of cytokine secretion induced by intact E. coli and H. pylori bacteria (Fig. 8). E. coli bacteria induced IL-6 secretion from both wild-type and TLR2 knockout peritoneal macrophages but only weakly activated TLR4 knockout cells (Fig. 8). In contrast, H. pylori bacteria activated wild-type and TLR4 knockout cells but not TLR2 knockout cells. H. pylori bacteria were weak inducers of cytokine secretion compared to E. coli bacteria on a per-cell basis (Fig. 8 and data not shown). Although the response of peritoneal macrophages to E. coli bacteria was largely TLR4 dependent, both TLR2- and TLR4-expressing HEK293 cells secreted cytokines in response to E. coli bacteria, indicating that both receptors potentially contributed to the response to intact E. coli (Fig. 8). In contrast, H. pylori bacteria activated TLR2-expressing, but not TLR4-expressing, HEK293 cells (Fig. 8). Thus, intact H. pylori activated TLR2, while intact E. coli activated both TLR2 and TLR4, although TLR4 was the predominant activating receptor for E. coli on normal macrophages.
FIG. 8.
E. coli bacteria activate cytokine secretion via both TLR2 and TLR4. Wild-type (WT), TLR2 knockout (KO), and TLR4 knockout mouse peritoneal exudate macrophages were stimulated with E. coli or H. pylori bacteria (2.5 × 105 CFU/well, left panel). Supernatants were harvested 18 h later, and IL-6 levels were determined by ELISA. (IL-6 levels were less than 200 pg/ml in medium controls.) HEK293 cells transfected with TLR2 or TLR4 were stimulated with E. coli or H. pylori bacteria (106 CFU/well, right panel). Supernatants were harvested 18 h later, and IL-8 levels were determined by ELISA. (IL-8 levels in medium controls were less than 200 pg/ml in HEK and HEK-TLR2 cells and 350 pg/ml in HEK-TLR4 cells.)
DISCUSSION
TLRs are transmembrane proteins that function as pattern recognition receptors for the detection and response to microbial ligands (reviewed in references 30, 32, and 37). To date, 10 TLRs have been identified in humans and natural or synthetic ligands for at least 9 TLRs have been identified (9, 30, 32, 33, 37, 52, 67). The extracellular regions of TLRs are diverse but contain variable numbers of leucine-rich repeat regions and conserved cysteine domains that are thought to contribute to receptor structure and function. All of the TLRs have a signature intracellular signaling motif in common with the IL-1 receptor, called the TIR (Toll-IL-1-R) domain (6, 9, 52, 67). Activation of TLRs results in the recruitment of adaptor proteins, including MyD88 and Mal/TIRAP, to the TIR domain. A series of phosphorylation-recruitment-activation events leads to the activation and translocation of NF-κB to the nucleus and the transcription of inflammatory cytokine genes (1, 30, 50, 51, 59).
Ligands recognized by TLRs can be found among many genera of bacteria, viruses, and fungi (3, 7, 44, 49, 77, 78, 82). These include LPS from gram-negative bacteria (TLR4 ligand), peptidoglycan and lipoteichoic acid from gram-positive bacteria (TLR2 ligands), and zymosan from yeast (TLR2 ligand). Recent data also indicate that TLRs are important in the innate immune response to bacterial DNA (TLR9 [24]) and viral RNA (TLR3 [2]), as well as intact bacteria and viruses (TLR2 and TLR4 [11, 39]).
Our data demonstrate that TLR2 is a critical receptor for the recognition of intact H. pylori bacteria. This is remarkable since most gram-negative bacteria preferentially activate TLR4 by the interaction of the potent TLR4 ligand with LPS in their outer membrane (reviewed in reference 43). Although the LPS produced by most gram-negative bacteria activates via TLR4, a reported notable exception is the LPS of Porphyromonas gingivalis, which is a ligand for TLR2 but not TLR4 (48). Our studies of the LPS derived from different H. pylori isolates indicate that H. pylori LPS is a TLR4 ligand similar to the LPS of other gram-negative strains, such as E. coli and Salmonella spp. Thus, the TLR2 dependence of the response to intact H. pylori cannot be explained by the expression of an unusual LPS.
Consistent with our finding, the previously published studies of Kawahara et al. (34, 36) demonstrated that the response to H. pylori LPS occurs via TLR4 in guinea pig gastric pit cells. In contrast, Smith et al. (74) recently reported that H. pylori LPS activated via TLR2, not TLR4. The reason for the discrepancy between their study and the present report (as well as the studies of Kawahara et al. [34, 36]) is not clear. However, by using the same experimental approach as Smith et al. (74), i.e., transfected HEK293 cells, we have found that H. pylori LPS from clinical isolates signals preferentially via TLR4. Furthermore, with knockout macrophages we found that H. pylori LPS does not elicit a cytokine response from TLR4-deficient macrophages; the response of TLR4-deficient macrophage to TLR2 ligands such as zymosan, peptidoglycan, and lipopeptides was indistinguishable from the response of wild-type macrophages, indicating that the TLR4-deficient macrophages had an intact TLR2-signaling capacity. One possible explanation for the discrepancy between our results (as well as the studies of Kawahara et al. [36]) and those of Smith et al. (74) is the strain of bacteria from which the LPS was isolated. Smith et al. (74) used only one H. pylori strain (26695), a heavily passaged laboratory strain, to prepare their LPS. In the present studies, we used a number of clinical isolates of H. pylori for LPS purification. It has been previously established that clinical isolates and laboratory strains of H. pylori have very different LPS molecules and that passage of H. pylori and culture conditions can induce variation in the LPS structures expressed (54, 55, 71).
It is also noteworthy that the dosages of H. pylori LPS used for cytokine stimulation in the present study were much lower than the dosages at which Smith et al. detect TLR2 agonist activity in their H. pylori LPS (i.e., nanograms per milliliter compared to micrograms per milliliter) (74). A potential difference between the H. pylori LPS studied by Smith et al. (74) and the LPS preparations in the present study may be the level of trace contaminants such as peptidoglycan or lipopeptides, both potent TLR2 agonists (26, 38, 72).
Despite the capacity of H. pylori LPS to activate via TLR4, the response to intact H. pylori bacteria was dependent on TLR2, and not TLR4, expression. This suggests that a non-LPS component of the bacterium is the major cytokine-activating molecule. It also raises the question of why it is difficult to detect a TLR4-dependent response to H. pylori LPS on the intact bacterium. One reason may be that H. pylori LPS is only weakly active as a cytokine inducer, even under ideal conditions, i.e., when it is added in a purified form to cultured cells. We and others (Fig. 7 and references 5, 58, 63, and 64) have noted that H. pylori LPS is 100- to 10,000-fold less active than the LPS of other gram-negative bacteria, such as E. coli. It is estimated that E. coli bacteria yield 10 ng of LPS per 106 CFU of bacteria (66). Given its weak intrinsic activity (1,000-fold lower than the activity of E. coli LPS), a response to H. pylori LPS on intact bacteria would be difficult to detect with challenge doses of less than 108 to 109 CFU even if all of the H. pylori LPS were exposed and accessible for interaction with TLR4.
Although the LPS of H. pylori is apparently not the major determinant of the response to intact H. pylori bacteria, the LPS-TLR4 interaction may, nevertheless, play a role in disease pathogenesis. This may be particularly important in the stomach, where we and others have failed to detect TLR2 expression (60; unpublished observations). In contrast, TLR4 has been detected on gastric pit cells, where it may sample the stomach environment for pathogens (34). In the absence of TLR2, the interaction of H. pylori LPS with TLR4 may be critical to the early detection of H. pylori infection or colonization. Several previous studies have suggested that H. pylori may activate the innate immune response through interactions between its LPS and TLR4. Initial work by Sakagami et al. (68) demonstrated that C3H/He mice (with an intact TLR4-encoding gene) show severe atrophic gastritis in response to H. felis infection, whereas C3H/HeJ mice (which have a mutated TLR4-encoding gene and are LPS nonresponders) show heavy colonization but minimal atrophic gastritis with a much reduced macrophage infiltration of the lamina propria. Recently, Panthel et al. (61) showed that, in short-term colonization studies, TLR2 knockout mice had reduced colonization compared to C57BL/6J mice, but the degree of gastric inflammation was not reported. In addition, strain differences play an important role in colonization, as C57BL/6 mice are reported to have lower levels of colonization than either C3H/HeN or C3H/HeJ mice (46). The importance of TLR4 in the H. pylori response has been supported by work by another group that has reported that guinea pig gastric pit cells express TLR4 and show significant responses to H. pylori LPS (34, 36). Another group has suggested that H. pylori NF-κB activation in macrophages also involves TLR4 and CD14 (45).
Nevertheless, H. pylori LPS has been shown to be a weak inducer of TLR4 activation with a potency that is 1,000- to 10,000-fold less than that of LPS from E. coli or S. enterica (5, 58, 63, 64). The lower bioactivity of H. pylori LPS compared to that of E. coli LPS may reflect a lower interaction with TLR4 and/or MD2. The biochemical basis of the lower activity of H. pylori LPS compared to that of E. coli LPS is the underphosphorylation of lipid A, as well as the presence of longer-chain fatty acids than normally encountered with E. coli LPS (53, 56). In addition, H. pylori LPS generally contains only four fatty acids (56) rather than the six-fatty-acid architecture seen in E. coli and other bioactive LPS molecules (53). Given its relatively weak bioactivity, H. pylori LPS may not contribute to the overall profile of bacterial stimulation or signaling seen with intact H. pylori bacteria. Furthermore, it has also been shown that H. pylori can activate cells by an LPS-independent and TLR4-independent mechanism (4, 47). The importance of the latter pathway relative to the LPS-dependent pathway in macrophages has not been examined.
Our studies reveal a role for genes in the cag pathogenicity island. In humans the induction of cytokine gene expression in infected stomach tissue has been linked to the level of colonization with cagA+ H. pylori bacteria (73, 81). cagA+ H. pylori bacteria were more potent activators of TLR2-expressing cells than cagA mutant H. pylori, suggesting that cagA or other associated cag pathogenicity island genes may themselves stimulate cytokine secretion by activating TLR2 and CD14 receptors. Alternatively, cag pathogenicity island-encoded genes may regulate the expression levels of H. pylori components that signal through TLR2 receptors. (Although cagA is a useful marker for the cag pathogenicity island, cagA+ strains of H. pylori SS1 may have an incomplete cag region [13, 69].) In previous studies (reviewed in 21), cagA has been linked to IL-8 secretion from epithelial cells and is proposed to involve the translocation of the CagA protein into epithelial cells and its subsequent intracellular phosphorylation and activation of IL-8-encoding gene expression (20, 25). Our studies suggest that cagA-induced IL-8 secretion from transfected HEK cells is a TLR2-dependent process, since HEK cells lacking TLR2 failed to secrete IL-8 when challenged with cagA+ or cagA mutant H. pylori bacteria. Nevertheless, cagA expression was not required for TLR2-induced cytokine secretion, since cagA mutant strains of H. pylori, as well as H. felis and H. hepaticus, which do not express cagA, all induced cytokine secretion via TLR2.
In conclusion, our studies demonstrate that TLR2 is both necessary and sufficient for responses to whole bacteria of several Helicobacter species. A major, nonredundant role for TLR2 in the recognition of intact Helicobacter bacteria was seen both with cell lines stably transfected with TLRs and with normal cells from wild-type, TLR2 knockout, and TLR4 knockout animals. Moreover, TLR4 was neither necessary nor sufficient for responses to intact Helicobacter bacteria. In contrast to the TLR2-dependent response to intact Helicobacter bacteria, LPS as a pure lipid product extracted from the outer membrane of Helicobacter bacteria and depleted of lipoproteins and glycan cell wall components was a ligand for TLR4 but its activity was variable. We demonstrated this with both knockout animals and transfected cell lines. Thus, TLR4 (but not TLR2) was both necessary and sufficient for response to the isolated bacterial lipid, while TLR2 was required for response to intact Helicobacter bacteria. (Control cultures demonstrated that intact E. coli bacteria activated cytokine secretion via both TLR2 and TLR4 and that TLR4 was the dominant receptor on macrophages, consistent with the potent agonist activity of E. coli LPS.)
We hypothesize that, upon initial infection in the stomach, intact H. pylori bacteria may be weakly recognized by interaction of its LPS with TLR4. However, a substantial inflammatory cytokine response to H. pylori may only develop after the recruitment and accumulation of TLR2-expressing cells, such as infiltrating blood leukocytes (polymorphonuclear granulocytes and monocytes) in the stomach (38). Thus, H. pylori may escape detection and elimination by the immune system because it colonizes a TLR2-deficient environment (i.e., the stomach) and at the same time expresses an LPS with very weak TLR4 agonist activity.
Acknowledgments
This work was supported by grants from the National Institutes of Health (RO1 AI51415 [to E.K.J.], RO1 CA93405 [to T.C.W.], RO1 AI37750, and RO1 CA67529 [to J.G.F.]) and the Irish Health Research Board (to A.P.M.).
Editor: F. C. Fang
REFERENCES
- 1.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Aliprantis, A. O., R. B. Yang, M. R. Mark, S. Suggett, B. Devaux, J. D. Radolf, G. R. Klimpel, P. Godowski, and A. Zychlinsky. 1999. Cell activation and apoptosis by bacterial lipoproteins through Toll-like receptor 2. Science 285:736-739. [DOI] [PubMed] [Google Scholar]
- 4.Backhed, F., B. Rokbi, E. Torstensson, Y. Zhao, C. Nilsson, D. Seguin, S. Normark, A. M. Buchan, and A. Richter-Dahlfors. 2003. Gastric mucosal recognition of Helicobacter pylori is independent of Toll-like receptor 4. J. Infect. Dis. 187:829-836. [DOI] [PubMed] [Google Scholar]
- 5.Bliss, C. M., Jr., D. T. Golenbock, S. Keates, J. K. Linevsky, and C. P. Kelly. 1998. Helicobacter pylori lipopolysaccharide binds to CD14 and stimulates release of interleukin-8, epithelial neutrophil-activating peptide 78, and monocyte chemotactic protein 1 by human monocytes. Infect. Immun. 66:5357-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowie, A., and L. A. O'Neill. 2000. The interleukin-1 receptor/Toll-like receptor superfamily: signal generators for pro-inflammatory interleukins and microbial products. J. Leukoc. Biol 67:508-514. [DOI] [PubMed] [Google Scholar]
- 7.Brightbill, H. D., D. H. Libraty, S. R. Krutzik, R. B. Yang, J. T. Belisle, J. R. Bleharski, M. Maitland, M. V. Norgard, S. E. Plevy, S. T. Smale, P. J. Brennan, B. R. Bloom, P. J. Godowski, and R. L. Modlin. 1999. Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science 285:732-736. [DOI] [PubMed] [Google Scholar]
- 8.Cario, E., G. Gerken, and D. K. Podolsky. 2002. “For whom the bell tolls!”—innate defense mechanisms and survival strategies of the intestinal epithelium against lumenal pathogens. Z. Gastroenterol. 40:983-990. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhary, P. M., C. Ferguson, V. Nguyen, O. Nguyen, H. F. Massa, M. Eby, A. Jasmin, B. J. Trask, L. Hood, and P. S. Nelson. 1998. Cloning and characterization of two Toll/interleukin-1 receptor-like genes, TIL3 and TIL4: evidence for a multi-gene receptor family in humans. Blood 91:4020-4027. [PubMed] [Google Scholar]
- 10.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor 4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 11.Compton, T., E. A. Kurt-Jones, K. W. Boehme, J. Belko, E. Latz, D. T. Golenbock, and R. W. Finberg. 2003. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J. Virol. 77:4588-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Correa, P. 1992. Human gastric carcinogenesis: a multistep and multifactorial process—first American Cancer Society award lecture on cancer epidemiology and prevention. Cancer Res. 52:6735-6740. [PubMed] [Google Scholar]
- 13.Crabtree, J. E., R. L. Ferrero, and J. G. Kusters. 2002. The mouse colonizing Helicobacter pylori strain SS1 may lack a functional cag pathogenicity island. Helicobacter 7:139-140. (Author reply, 7:140-141.) [DOI] [PubMed] [Google Scholar]
- 14.El-Omar, E. M., M. Carrington, W. H. Chow, K. E. McColl, J. H. Bream, H. A. Young, J. Herrera, J. Lissowska, C. C. Yuan, N. Rothman, G. Lanyon, M. Martin, J. F. Fraumeni, Jr., and C. S. Rabkin. 2000. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 404:398-402. [DOI] [PubMed] [Google Scholar]
- 15.El-Omar, E. M., C. S. Rabkin, M. D. Gammon, T. L. Vaughan, H. A. Risch, J. B. Schoenberg, J. L. Stanford, S. T. Mayne, J. Goedert, W. J. Blot, J. F. Fraumeni, Jr., and W. H. Chow. 2003. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 124:1193-1201. [DOI] [PubMed] [Google Scholar]
- 16.Erdman, S., J. G. Fox, C. A. Dangler, D. Feldman, and B. H. Horwitz. 2001. Typhlocolitis in NF-κB-deficient mice. J. Immunol. 166:1443-1447. [DOI] [PubMed] [Google Scholar]
- 17.Erdman, S. E., T. Poutahidis, M. Tomczak, A. B. Rogers, K. Cormier, B. Plank, B. H. Horwitz, and J. G. Fox. 2003. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am. J. Pathol. 162:691-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erdman, S. E., V. P. Rao, T. Poutahidis, M. M. Ihrig, Z. Ge, Y. Feng, M. Tomczak, A. B. Rogers, B. H. Horwitz, and J. G. Fox. 2003. CD4+ CD25+ regulatory lymphocytes require interleukin 10 to interrupt colon carcinogenesis in mice. Cancer Res. 63:6042-6050. [PubMed] [Google Scholar]
- 19.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. [DOI] [PubMed] [Google Scholar]
- 20.Fischer, W., J. Puls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Haas. 2001. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 21.Graham, D. Y., and Y. Yamaoka. 2000. Disease-specific Helicobacter pylori virulence factors: the unfulfilled promise. Helicobacter 5(Suppl. 1):S3-S9. (Discussion, 5:S27-S31.) [DOI] [PubMed] [Google Scholar]
- 22.Hansson, L. E., O. Nyren, A. W. Hsing, R. Bergstrom, S. Josefsson, W. H. Chow, J. F. Fraumeni, Jr., and H. O. Adami. 1996. The risk of stomach cancer in patients with gastric or duodenal ulcer disease. N. Engl. J. Med. 335:242-249. [DOI] [PubMed] [Google Scholar]
- 23.Heine, H., C. J. Kirschning, E. Lien, B. G. Monks, M. Rothe, and D. T. Golenbock. 1999. Cutting edge: cells that carry a null allele for Toll-like receptor 2 are capable of responding to endotoxin. J. Immunol. 162:6971-6975. [PubMed] [Google Scholar]
- 24.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 25.Higashi, H., R. Tsutsumi, S. Muto, T. Sugiyama, T. Azuma, M. Asaka, and M. Hatakeyama. 2002. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295:683-686. [DOI] [PubMed] [Google Scholar]
- 26.Hirschfeld, M., Y. Ma, J. H. Weis, S. N. Vogel, and J. J. Weis. 2000. Cutting edge: repurification of lipopolysaccharide eliminates signaling through both human and murine Toll-like receptor 2. J. Immunol. 165:618-622. [DOI] [PubMed] [Google Scholar]
- 27.Houghton, J., J. G. Fox, and T. C. Wang. 2002. Gastric cancer: laboratory bench to clinic. J. Gastroenterol. Hepatol. 17:495-502. [DOI] [PubMed] [Google Scholar]
- 28.Houghton, J. M., L. M. Bloch, M. Goldstein, S. Von Hagen, and R. M. Korah. 2000. In vivo disruption of the fas pathway abrogates gastric growth alterations secondary to Helicobacter infection. J. Infect. Dis. 182:856-864. [DOI] [PubMed] [Google Scholar]
- 29.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. 1994. Schistosomes, liver flukes and Helicobacter pylori. IARC Monogr. Eval. Carcinog. Risks Hum. 61:1-241. [PMC free article] [PubMed] [Google Scholar]
- 30.Imler, J. L., and J. A. Hoffmann. 2001. Toll receptors in innate immunity. Trends Cell Biol. 11:304-311. [DOI] [PubMed] [Google Scholar]
- 31.Janvier, B., B. Grignon, C. Audibert, L. Pezennec, and J. L. Fauchere. 1999. Phenotypic changes of Helicobacter pylori components during an experimental infection in mice. FEMS Immunol. Med. Microbiol. 24:27-33. [DOI] [PubMed] [Google Scholar]
- 32.Kaisho, T., and S. Akira. 2000. Critical roles of Toll-like receptors in host defense. Crit. Rev. Immunol. 20:393-405. [PubMed] [Google Scholar]
- 33.Kaisho, T., and S. Akira. 2002. Toll-like receptors as adjuvant receptors. Biochim. Biophys. Acta 1589:1-13. [DOI] [PubMed] [Google Scholar]
- 34.Kawahara, T., Y. Kuwano, S. Teshima-Kondo, T. Kawai, T. Nikawa, K. Kishi, and K. Rokutan. 2001. Toll-like receptor 4 regulates gastric pit cell responses to Helicobacter pylori infection. J. Med. Investig. 48:190-197. [PubMed] [Google Scholar]
- 35.Kawahara, T., Y. Kuwano, S. Teshima-Kondo, T. Sugiyama, T. Kawai, T. Nikawa, K. Kishi, and K. Rokutan. 2001. Helicobacter pylori lipopolysaccharide from type I, but not type II, strains stimulates apoptosis of cultured gastric mucosal cells. J. Med. Investig. 48:167-174. [PubMed] [Google Scholar]
- 36.Kawahara, T., S. Teshima, A. Oka, T. Sugiyama, K. Kishi, and K. Rokutan. 2001. Type I Helicobacter pylori lipopolysaccharide stimulates Toll-like receptor 4 and activates mitogen oxidase 1 in gastric pit cells. Infect. Immun. 69:4382-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopp, E. B., and R. Medzhitov. 1999. The Toll-receptor family and control of innate immunity. Curr. Opin. Immunol. 11:13-18. [DOI] [PubMed] [Google Scholar]
- 38.Kurt-Jones, E. A., L. Mandell, C. Whitney, A. Padgett, K. Gosselin, P. E. Newburger, and R. W. Finberg. 2002. Role of Toll-like receptor 2 (TLR2) in neutrophil activation: GM-CSF enhances TLR2 expression and TLR2-mediated interleukin 8 responses in neutrophils. Blood 100:1860-1868. [PubMed] [Google Scholar]
- 39.Kurt-Jones, E. A., L. Popova, L. Kwinn, L. M. Haynes, L. P. Jones, R. A. Tripp, E. E. Walsh, M. W. Freeman, D. T. Golenbock, L. J. Anderson, and R. W. Finberg. 2000. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 1:398-401. [DOI] [PubMed] [Google Scholar]
- 40.Lee, A., J. G. Fox, G. Otto, and J. Murphy. 1990. A small animal model of human Helicobacter pylori active chronic gastritis. Gastroenterology 99:1315-1323. [DOI] [PubMed] [Google Scholar]
- 41.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386-1397. [DOI] [PubMed] [Google Scholar]
- 42.Levi, S., K. Beardshall, G. Haddad, R. Playford, P. Ghosh, and J. Calam. 1989. Campylobacter pylori and duodenal ulcers: the gastrin link. Lancet i:1167-1168. [DOI] [PubMed] [Google Scholar]
- 43.Lien, E., and R. R. Ingalls. 2002. Toll-like receptors. Crit. Care Med. 30:S1-S11. [PubMed] [Google Scholar]
- 44.Lien, E., T. J. Sellati, A. Yoshimura, T. H. Flo, G. Rawadi, R. W. Finberg, J. D. Carroll, T. Espevik, R. R. Ingalls, J. D. Radolf, and D. T. Golenbock. 1999. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Biol. Chem. 274:33419-33425. [DOI] [PubMed] [Google Scholar]
- 45.Maeda, S., M. Akanuma, Y. Mitsuno, Y. Hirata, K. Ogura, H. Yoshida, Y. Shiratori, and M. Omata. 2001. Distinct mechanism of Helicobacter pylori-mediated NF-κB activation between gastric cancer cells and monocytic cells. J. Biol. Chem. 276:44856-44864. [DOI] [PubMed] [Google Scholar]
- 46.Mahler, M., C. Janke, S. Wagner, and H. J. Hedrich. 2002. Differential susceptibility of inbred mouse strains to Helicobacter pylori infection. Scand. J. Gastroenterol. 37:267-278. [DOI] [PubMed] [Google Scholar]
- 47.Mai, U. E., G. I. Perez-Perez, L. M. Wahl, S. M. Wahl, M. J. Blaser, and P. D. Smith. 1991. Soluble surface proteins from Helicobacter pylori activate monocytes/macrophages by lipopolysaccharide-independent mechanism. J. Clin. Investig. 87:894-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin, M., J. Katz, S. N. Vogel, and S. M. Michalek. 2001. Differential induction of endotoxin tolerance by lipopolysaccharides derived from Porphyromonas gingivalis and Escherichia coli. J. Immunol. 167:5278-5285. [DOI] [PubMed] [Google Scholar]
- 49.Means, T. K., E. Lien, A. Yoshimura, S. Wang, D. T. Golenbock, and M. J. Fenton. 1999. The CD14 ligands lipoarabinomannan and lipopolysaccharide differ in their requirement for Toll-like receptors. J. Immunol. 163:6748-6755. [PubMed] [Google Scholar]
- 50.Medzhitov, R., and C. Janeway, Jr. 2000. Innate immune recognition: mechanisms and pathways. Immunol. Rev. 173:89-97. [DOI] [PubMed] [Google Scholar]
- 51.Medzhitov, R., and C. A. Janeway, Jr. 1998. An ancient system of host defense. Curr. Opin. Immunol. 10:12-15. [DOI] [PubMed] [Google Scholar]
- 52.Medzhitov, R., P. Preston-Hurlburt, and C. A. Janeway, Jr. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394-397. [DOI] [PubMed] [Google Scholar]
- 53.Moran, A. 1998. The products of Helicobacter pylori that induce inflammation. Eur. J. Gastroenterol. Hepatol. 10(Suppl. 1):S3-S8. [Google Scholar]
- 54.Moran, A. P., I. M. Helander, and T. U. Kosunen. 1992. Compositional analysis of Helicobacter pylori rough-form lipopolysaccharides. J. Bacteriol. 174:1370-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moran, A. P., Y. A. Knirel, S. N. Senchenkova, G. Widmalm, S. O. Hynes, and P. E. Jansson. 2002. Phenotypic variation in molecular mimicry between Helicobacter pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. Acid-induced phase variation in Lewis(x) and Lewis(y) expression by H. pylori lipopolysaccharides. J. Biol. Chem. 277:5785-5795. [DOI] [PubMed] [Google Scholar]
- 56.Moran, A. P., B. Lindner, and E. J. Walsh. 1997. Structural characterization of the lipid A component of Helicobacter pylori rough- and smooth-form lipopolysaccharides. J. Bacteriol. 179:6453-6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulholland, G., J. E. Ardill, D. Fillmore, R. S. Chittajallu, G. M. Fullarton, and K. E. McColl. 1993. Helicobacter pylori related hypergastrinaemia is the result of a selective increase in gastrin 17. Gut 34:757-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muotiala, A., I. M. Helander, L. Pyhala, T. U. Kosunen, and A. P. Moran. 1992. Low biological activity of Helicobacter pylori lipopolysaccharide. Infect. Immun. 60:1714-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Neill, L. 2000. The Toll/interleukin-1 receptor domain: a molecular switch for inflammation and host defence. Biochem. Soc. Trans. 28:557-563. [DOI] [PubMed] [Google Scholar]
- 60.Ortega-Cava, C. F., S. Ishihara, M. A. Rumi, K. Kawashima, N. Ishimura, H. Kazumori, J. Udagawa, Y. Kadowaki, and Y. Kinoshita. 2003. Strategic compartmentalization of Toll-like receptor 4 in the mouse gut. J. Immunol. 170:3977-3985. [DOI] [PubMed] [Google Scholar]
- 61.Panthel, K., G. Faller, and R. Haas. 2003. Colonization of C57BL/6J and BALB/c wild-type and knockout mice with Helicobacter pylori: effect of vaccination and implications for innate and acquired immunity. Infect. Immun. 71:794-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parsonnet, J., R. A. Harris, H. M. Hack, and D. K. Owens. 1996. Modelling cost-effectiveness of Helicobacter pylori screening to prevent gastric cancer: a mandate for clinical trials. Lancet 348:150-154. [DOI] [PubMed] [Google Scholar]
- 63.Pece, S., D. Fumarola, G. Giuliani, E. Jirillo, and A. P. Moran. 1995. Activity in the Limulus amebocyte lysate assay and induction of tumour necrosis factor by diverse Helicobacter pylori lipopolysaccharide preparations. J. Endotoxin Res. 2:455-462. [Google Scholar]
- 64.Perez-Perez, G. I., V. L. Shepherd, J. D. Morrow, and M. J. Blaser. 1995. Activation of human THP-1 cells and rat bone marrow-derived macrophages by Helicobacter pylori lipopolysaccharide. Infect. Immun. 63:1183-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Podolsky, D. K. 1999. Mucosal immunity and inflammation. V. Innate mechanisms of mucosal defense and repair: the best offense is a good defense. Am. J. Physiol. 277:G495-G499. [DOI] [PubMed] [Google Scholar]
- 66.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rock, F. L., G. Hardiman, J. C. Timans, R. A. Kastelein, and J. F. Bazan. 1998. A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 95:588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakagami, T., J. Vella, M. F. Dixon, J. O'Rourke, F. Radcliff, P. Sutton, T. Shimoyama, K. Beagley, and A. Lee. 1997. The endotoxin of Helicobacter pylori is a modulator of host-dependent gastritis. Infect. Immun. 65:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandor, F., E. Latz, F. Re, L. Mandell, G. Repik, D. T. Golenbock, T. Espevik, E. A. Kurt-Jones, and R. W. Finberg. 2003. Importance of extra- and intracellular domains of TLR1 and TLR2 in NFκB signaling. J. Cell Biol. 162:1099-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scherer, C., K. D. Muller, P. M. Rath, and R. A. Ansorg. 2003. Influence of culture conditions on the fatty acid profiles of laboratory-adapted and freshly isolated strains of Helicobacter pylori. J. Clin. Microbiol. 41:1114-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by Toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 73.Shimoyama, T., S. M. Everett, M. F. Dixon, A. T. Axon, and J. E. Crabtree. 1998. Chemokine mRNA expression in gastric mucosa is associated with Helicobacter pylori cagA positivity and severity of gastritis. J. Clin. Pathol. 51:765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith, M. F., Jr., A. Mitchell, G. Li, S. Ding, A. M. Fitzmaurice, K. Ryan, S. Crowe, and J. B. Goldberg. 2003. Toll-like receptor (TLR) 2 and TLR5, but not TLR4, are required for Helicobacter pylori-induced NF-κB activation and chemokine expression by epithelial cells. J. Biol. Chem. 278:32552-32560. [DOI] [PubMed] [Google Scholar]
- 75.Sutton, P., J. Wilson, and A. Lee. 2000. Further development of the Helicobacter pylori mouse vaccination model. Vaccine 18:2677-2685. [DOI] [PubMed] [Google Scholar]
- 76.Swingler, S., A. Mann, J. Jacque, B. Brichacek, V. G. Sasseville, K. Williams, A. A. Lackner, E. N. Janoff, R. Wang, D. Fisher, and M. Stevenson. 1999. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat. Med. 5:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 78.Underhill, D. M., A. Ozinsky, A. M. Hajjar, A. Stevens, C. B. Wilson, M. Bassetti, and A. Aderem. 1999. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401:811-815. [DOI] [PubMed] [Google Scholar]
- 79.Wang, T. C., C. A. Dangler, D. Chen, J. R. Goldenring, T. Koh, R. Raychowdhury, R. J. Coffey, S. Ito, A. Varro, G. J. Dockray, and J. G. Fox. 2000. Synergistic interaction between hypergastrinemia and Helicobacter infection in a mouse model of gastric cancer. Gastroenterology 118:36-47. [DOI] [PubMed] [Google Scholar]
- 80.Wang, T. C., J. R. Goldenring, C. Dangler, S. Ito, A. Mueller, W. K. Jeon, T. J. Koh, and J. G. Fox. 1998. Mice lacking secretory phospholipase A2 show altered apoptosis and differentiation with Helicobacter felis infection. Gastroenterology 114:675-689. [DOI] [PubMed] [Google Scholar]
- 81.Yamaoka, Y., T. Kodama, M. Kita, J. Imanishi, K. Kashima, and D. Y. Graham. 1999. Relation between clinical presentation, Helicobacter pylori density, interleukin 1β and 8 production, and cagA status. Gut 45:804-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoshimura, A., E. Lien, R. R. Ingalls, E. Tuomanen, R. Dziarski, and D. Golenbock. 1999. Cutting edge: recognition of gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 163:1-5. [PubMed] [Google Scholar]