Abstract
Treponema paraluiscuniculi, the etiologic agent of rabbit venereal syphilis, is morphologically indistinguishable from Treponema pallidum subsp. pallidum (T. pallidum), the human syphilis treponeme, and induces similar immune responses and histopathologic changes in the infected host. Because of their high degree of relatedness, comparative studies are likely to identify genetic determinants that contribute to pathogenesis or virulence in human syphilis. The tpr (Treponema pallidum repeat) genes are believed to code for potential virulence factors. In this study, we identified 10 tpr homologs in Treponema paraluiscuniculi Cuniculi A strain and determined their sequence architecture. Half of this group of paralogous genes were predicted to be nonfunctional due to the presence of frameshifts and premature stop codons. Furthermore, the immune response against the T. paraluiscuniculi Tpr homologs in long-term-infected rabbits was studied by enzyme-linked immunosorbent assay and lymphocyte proliferation assay, showing that TprK is the only target of the antibody and T-cell responses during experimental infection and emphasizing the importance of this putative virulence factor in venereal treponematosis.
The spirochetes are a small but diverse family of bacteria and show a wide range of host specificities. Examples of these are the easily cultivated free-living organisms (Spirochaeta spp.), commensal treponemes of the termite gut, the human and animal pathogens in the genera Borrelia, Leptospira, and Brachyspira, and the cultivable and noncultivatable treponemes of humans and animals. Of the five noncultivatable pathogenic treponemes, Treponema paraluiscuniculi naturally infects rabbits but is thought not to be infectious for humans (14). In contrast, the three subspecies of Treponema pallidum naturally infect humans but can also experimentally infect rabbits and other mammals. Treponema carateum naturally infects humans but is unable to multiply in rabbits or other nonprimates (28).
Treponema paraluiscuniculi is the etiologic agent of rabbit venereal syphilis. It was identified in 1913, only 8 years after Treponema pallidum subsp. pallidum (T. pallidum hereafter), the etiologic agent of human venereal syphilis, was discovered (3, 25). By dark-field microscopy, immunoassay, and electron microscopy, T. paraluiscuniculi and the other pathogenic treponemes are morphologically indistinguishable (17) and appear to induce similar immune responses and histopathologic changes. Furthermore, T. paraluiscuniculi-infected rabbits develop antibodies that can be detected with both treponemal and nontreponemal standard tests for syphilis (11, 12). Immunoblotting tests show only subtle differences between the specificities of T. paraluiscuniculi and T. pallidum antisera for T. pallidum antigens (2, 26). Despite this apparently high level of relatedness, the pathogenic treponemes have different host specificities and cause clinically distinct diseases. Infections with all these treponemal species are chronic, and while homologous immunity exists, there is no cross-protection between species of pathogenic treponemes and only partial cross-protection within species. These facts indicate that there are small but important differences between the virulence factors and antigens expressed by the different pathogenic treponemes.
Paralogous gene families arise during evolution by gene duplication and may yield new gene functions and new expression patterns (20-22). Initial analysis of the genome sequence of T. pallidum Nichols strain revealed 48 potential virulence genes, among them a novel family of 12 paralogous genes (tpr genes, for Treponema pallidum repeat) (13). The tpr genes account for ≈2% of the small T. pallidum genome. The family is divided into three subfamilies by their predicted amino acid homology: subfamily I (TprC, TprD, TprF, and TprI), subfamily II (TprE, TprG, and TprJ), and subfamily III (TprA, TprB, TprH, TprK, and TprL). The subfamily I and subfamily II proteins have conserved NH2- and COOH-terminal regions, whereas the central domains vary sequence and length. Within subfamily I, TprC and TprD are identical in the Nichols strain, and TprF lacks a central variable domain and the conserved COOH-terminal region due to a frameshift. Subfamily III members are comparatively less homologous to each other and to the other Tpr proteins. It has been shown that TprK, TprI, and TprF are the targets of a strong humoral and cellular immune response during syphilis infection in the rabbit model, and immunization with recombinant peptides significantly alters lesion development (4, 23, 27). In addition, TprK contains a cleavable leader sequence, possesses multiple alleles in T. pallidum isolates, and undergoes antigenic variation during syphilis infection (6, 7, 16). This suggests an important role for the Tpr antigens during syphilis infection.
Among the several approaches used to identify virulence factors are intra- and interspecies comparisons to determine which antigens are conserved and which are variable among different strains or species (30). Weinstock et al. reported that, while tpr genes are highly conserved among human syphilis strains, most of the differences between the T. pallidum subsp. pallidum and T. pallidum subsp. pertenue subspecies involved the tpr genes (30). Despite the high level of relatedness and the natural ability to infect rabbits, T. paraluiscuniculi is considered not to be infectious for humans (14). The host specificity shown by this microorganism in association with the similarities to syphilitic infection make T. paraluiscuniculi a unique comparison species for understanding which genetic determinants contribute to pathogenesis or virulence in human syphilis. For this reason, we studied the tpr gene family in the rabbit treponeme to determine the sequence architecture of and immune responses to these antigens. In this study, we characterized the tpr gene family of T. paraluiscuniculi, and here we describe distinctive features of several of its members.
MATERIALS AND METHODS
T. paraluiscuniculi propagation and DNA extraction.
T. paraluiscuniculi Cuniculi A strain was provided by Paul Hardy and Ellen Nell (Johns Hopkins University, Baltimore, Md.), propagated intratesticularly in New Zealand White rabbits and harvested as described elsewhere (19). Before infection, each rabbit had been serologically tested to rule out a naturally occurring infection with T. paraluiscuniculi. Treponemes were extracted from infected rabbit testes at peak orchitis or 3 weeks after the infection. Collected organisms were separated from host cellular gross debris by low-speed centrifugation (250 × g for 10 min at room temperature); the supernatants were spun in a microcentrifuge for 30 min at 12,000 × g at 4°C. The pellet was resuspended in 200 μl of 1× lysis buffer (10 mM Tris [pH 8.0] 0.1 M EDTA, 0.5% sodium dodecyl sulfate). DNA extraction was performed as previously described (5) with the QIAamp DNA minikit (Qiagen Inc., Chatsworth, Calif.), taking standard precautions to prevent cross-contamination between samples.
PCR amplification, cloning, sequencing, and sequence analysis.
The T. pallidum genome sequence (13) was used to design primers in the 5′- and 3′-flanking regions of the Nichols tpr genes to amplify the corresponding T. paraluiscuniculi DNA regions. The primers used for amplification are listed in Table 1. Each PCR was performed in a 50-μl final volume containing 200 μM deoxynucleoside triphosphates, 50 mM Tris-HCl (pH 9.0 at 20°C), 1.5 mM MgCl2, 20 mM NH4SO4, and 2.5 U of Taq polymerase (Promega, Madison, Wis.). Amplicons from two different PCRs with T. paraluiscuniculi DNA as the template were compared to minimize PCR errors. T. pallidum DNA was used as a positive control for each reaction. When required, a third amplification reaction followed by cloning and sequencing was used to confirm the data from the first two reactions (tprE and tprC). The cycling conditions were denaturation at 95°C for 2 min, followed by 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min for 35 cycles in total; the final extension was 72°C for 3 min. The products were separated in 1% agarose gels and cloned into the pCRII-Topo cloning vector (Invitrogen, Carlsbad, Calif.), according to the manufacturer's instruction.
TABLE 1.
Primers in the 5′- and 3′-flanking regions of the tpr genes
| Subfamily and gene | Sense primer (5′-flanking region) | Antisense primer (3′-flanking region) | Amplicon size (bp)a |
|---|---|---|---|
| Subfamily I | |||
| tprC | 5′-GGGGGTGAGGTAGAAGTGAGA | 5′-CCCTCATCCGAGACAAAAT | 1,916 |
| tprD | 5′-AAGAGGTTCAGGAAGCAACG | 5′-ACTTCGTAGGAGCAGCAGGA | 2,792 |
| Subfamily II | |||
| tprG1 | 5′-CAGGATTTTCCGGTTCATTG | 5′-TCACGCGTTTAATGTTCTGC | 3,257 |
| tprG2 | 5′-AAGTTTGCTTTCAGAT | 5′-GTACTACCTTCCCCCGGTCT | 3,018 |
| tprG/I hybrid | 5′-CGCGTACCCACTTCTCTCTC | 5′-GTACTACCTTCCCCCGGTCT | 2,459 |
| Subfamily III | |||
| tprA | 5′-CGTATGCTTTTACCCGCTGT | 5′-TGCAACCATCTTCGATTACG | 2,795 |
| tprB | 5′-CTTCGCGCCTAAGTTAATGC | 5′-TACTCATGCGGATCCCCTAC | 3,011 |
| tprK | 5′-TCCCCCAGTTGCAGCACTAT | 5′-TCGCGGTAGTCAACAATACCA | 2,083-2,133b |
| tprL | 5′-TTCTGAGCAGCTAGGCATTG | 5′-ACGGCGTCTTTCTGTACGTG | 1,876 |
| tprH | 5′-ACGTGGCAGAGGAGTTTTTG | 5′-TTCCTCAGGAGTTTGGAACG | 3,197 |
The size refers to amplicons obtained with T. paraluiscuniculi DNA as the template.
Size range of 10 tprK amplicons.
Plasmid DNA from colonies containing inserts was extracted with the Qiagen plasmid minikit (Qiagen), and at least two clones, each one from a different amplification reaction, were sequenced in both directions with the Applied Biosystems dye terminator sequencing kit (Perkin-Elmer, Foster City, Calif.) with the primer walking approach (unpublished data). Sequences were aligned with the Multiple Alignment Program (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html), and shading of identical bases was done with the Boxshade 3.21 program (http://www.ch.embnet.org/software/BOX_form.html). Open reading frames (ORFs) and reading frames were determined with the NCBI ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) and the Six Frames Translation (http://searchlauncher.bcm.tmc.edu/seq-util/seq-util.html) programs. Supplemental data is available at http://depts.washington.edu/treplab/iai72_11.htm.
Infection with T. paraluiscuniculi.
Approximately 107 organisms were used to infect six New Zealand White rabbits intratesticularly. Each rabbit had tested seronegative for prior T. paraluiscuniculi infection by the Venereal Disease Research Laboratory and fluorescent treponemal antibody absorption tests. To test antibody responses against recombinant Tprs, sera were collected at day 0, day 30 postinfection, and approximately every 30 days thereafter until day 270. At 270 days after infection, the rabbits were sacrificed and splenocytes were harvested to perform the lymphocyte proliferation assay.
Recombinant and synthetic peptides.
The recombinant Tprs used to perform the enzyme-linked immunosorbent assays (ELISAs) and lymphocyte proliferation assays are listed in Table 2. The partial open reading frames coding for these peptides have been cloned from T. pallidum Nichols strain genomic DNA. Expression and purification by affinity chromatography on nickel agarose of the 6-His-tagged proteins were performed as previously described (4). TprL could not be expressed. With the exception of TprF and the peptides labeled as being from conserved regions, each recombinant peptide represents the unique central region, which contains sequences that differentiate the individual Tpr proteins; only small portions of the sequence that is conserved among members of the same subfamily are present in the peptides.
TABLE 2.
Recombinant peptides used for antibody response and lymphocyte proliferation assays
| Subfamily and peptide | Amino acid sequence limits | Size (no. of residues) | Identitya with T. paraluiscuniculi predicted ORFs |
|---|---|---|---|
| Subfamily I | |||
| 5′-subfamily 1 | YAGVLT. . . . . QHQDPA | 267 | 98.6% identity with TprC and TprD before frameshift |
| TprF (N-terminal and central region) | YAGVLT. . . . . GTGGAC | 349 | 98.6% identity with TprC and TprD before frameshift |
| TprD2-central | TSLGD. . . . . WLQFTY | 90 | 96% with TprC and TprD |
| 3′-subfamily I (A)b | GFLRLE. . . . . TKSGDP | 86 | 81% with TprC and TprD |
| 3′-subfamily I (B)b | RIPGLS. . . . . IAESIW | 76 | 82% with TprC and TprD |
| TprI-central | RLTLEP. . . . . YTHLLT | 211 | 80% with TprG/I hybrid |
| Subfamily II | |||
| TprG-central | RLTLEP. . . . . DLIPKT | 219 | 85.3% with TprG1 and TprG2 |
| 3′-Subfamily II | LFTAQW. . . . . GVTLSV | 202 | 89.3% with TprG1; 89.6% with TprG2 |
| TprE-central | RLTLEP. . . . . QQTVAA | 191 | 9.5% with TprG1 and TprG2 |
| TprJ-central | RLTLEP. . . . . MRTEIT | 226 | 56.6% with TprG1 and TprG2 |
| Subfamily III | |||
| TprA (N-terminus) | MRLVLG. . . . . NCGTRR | 253 | 91.6% |
| TprA (C-terminus) | MGLVVT. . . . . GCKITW | 368 | 90% |
| TprB-central | RLTLSP. . . . . SLSKLV | 194 | 99.4% |
| TprH-central | RITLTP. . . . . YTHLID | 180 | 98.8% |
| TprK-centralc | IEGYAE. . . . . DTSFLE | 315 | 84% |
| TprLd | NA | NA | NA |
To define identity, T. pallidum recombinant peptide sequences were aligned with their T. paraluiscuniculi homologues and the value was calculated as (number of identical amino acids in identical position/recombinant peptide size) × 100.
3′-subfamily I A and B are partially overlapping peptides encompassing positions 313 to 395 (peptide A) and 379 to 454 (peptide B) of T. paraluiscuniculi TprC and TprD (B ORF, Fig. 1), respectively.
The TprK 1.1 clone was used for this calculation.
Recombinant peptide not available (NA).
The subfamily III recombinant proteins (TprA N-terminal and C-terminal, TprB-central, TprH-central, and TprK-central) have virtually no overlap, while Tpr I-central, TprE-central, TprG-central, and TprJ-central each contain 23 amino acids of the NH2-terminal conserved region; TprI-central also contains 21 amino acids of the COOH-terminal conserved region. TprF, in contrast, contains 262 amino acids of the NH2-terminal constant region of subfamily I in addition to 87 amino acids representing the central region. TprD2-central is an allele-specific peptide. The peptides representing the conserved amino terminus and carboxyl terminus of subfamilies I and II are labeled 5′-conserved and 3′-conserved, respectively. We were unable to produce the 5′-conserved peptide for subfamily II, and the 3′-conserved peptide of subfamily I had to be produced in two smaller fragments, as expression of full-length peptide appeared to be toxic to Escherichia coli. The percent identity between the Nichols-based recombinant antigens and T. paraluiscuniculi Tprs is indicated in Table 2.
Overlapping 20-amino-acid-long synthetic peptides were synthesized based on the Seattle Nichols strain TprK sequence (GenBank accession number AF194369) as described elsewhere (24). Thirty-nine peptides with a 10-amino-acid overlap, spanning the entire mature TprK sequence (amino acids 30 through 349) (16) (see Fig. 6 and data not shown) were used in lymphocyte proliferation assays. Additionally, the conserved carboxyl terminus (amino acids 425 to 478, listed as peptide 40) was cloned and expressed as a recombinant protein as described previously (24).
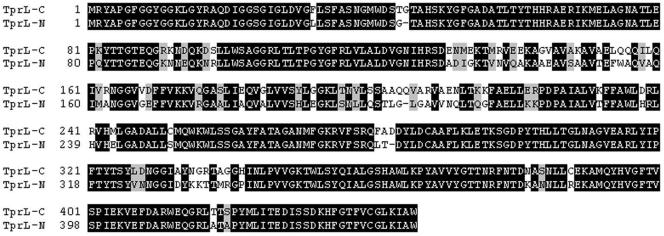
FIG. 6.
Alignment of TprL homologues in T. paraluiscuniculi and T. pallidum subsp. pallidum. TprL-C, T. paraluiscuniculi sequence; TprL-N, T. pallidum Nichols strain sequence.
ELISAs.
The appropriate purified recombinant Tpr antigen (0.5 μg per well) in phosphate-buffered saline (PBS) containing 0.1% sodium azide and 0.1% sodium dodecyl sulfate was used to coat the wells of a 96-well ELISA plate (EIA II Plus Microplate; ICN, Irvine, Calif.). The plates were incubated at 37°C for 2 h and subsequently at 4°C overnight. The wells were then washed three times with PBS, blocked by incubation for 1 h at room temperature with 200 μl of 3% nonfat milk-PBS/well, and washed again.
Rabbit sera collected from six animals prior to infection and at days 30, 60, 90, 130, 160, 190, 230, and 270 postinfection were pooled for each time point and adsorbed with a lysate from the E. coli strain used to express the recombinant antigens. This lysate of the E. coli strain expressing an unrelated Trypanosoma cruzi recombinant protein (SA85 1.1) was used to remove cross-reacting antibody against possible E. coli contaminants and the histidine tag added to the recombinant peptides during expression (27). Ten microliters of each adsorbed pool was diluted 1:20 in 1% nonfat milk-PBS plus 0.05% Tween 20 (Sigma) prior to being dispensed into wells. Only one concentration of serum was tested. One hundred microliters of diluted primary antibody was incubated for 1 h at room temperature, and then the wells were washed three times with PBS. One hundred microliters of secondary antibody solution, alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Sigma, St. Louis, Mo.), diluted 1:2,000 in 1% nonfat milk-PBS-0.05% Tween 20, was then added to each well, and the plates were incubated for 1 h at room temperature before repeating the washing step. Finally, the plates were developed for 45 min after addition of 50 μl of 1-mg/ml para-nitrophenyl phosphate (Sigma) to each well and read at 405 nm on a Multiskan MC plate reader (Titertek, Huntsville, Ala.). The mean of background readings (from preimmune rabbit sera) was subtracted from the mean of triplicate experimental wells for each antigen. The reported data are the mean ± standard error of the mean for each antigen per time point.
Lymphocyte proliferation assays.
Three uninfected and five Cuniculi A strain-infected rabbits (day 270 postinfection) were sacrificed, and splenic lymphocytes were harvested and cultured as previously described (1, 19) in RPMI medium supplemented with glutamine (2 mM final concentration), penicillin (100 U/ml final concentration), streptomycin (100 μg/ml final concentration), and heat-inactivated normal rabbit serum (1% final concentration). Approximately 5 × 105 cells in 200 μl of culture medium were plated in quadruplicate for each animal and each antigen in flat-bottomed 96-well plates (Costar, Cambridge, Mass.) and incubated at 37°C in a 5% CO2 atmosphere. Recombinant proteins were added at 2 and 10 μg/ml final concentrations, and synthetic peptides were added at a 25 μg/ml final concentration; 10 μl/well of a sonicated suspension of T. pallidum was used as the treponeme-specific positive control, and 0.5 μg/well (as determined by titration) of concanavalin A (ICN) used as the positive T-cell control. PBS was used to measure background reactivity. Three days after exposure to recombinant peptide or controls, cells were pulsed with 0.5 μCi of [3H]thymidine per well and harvested after 24 h to measure tritiated thymidine incorporation. The geometric mean of quadruplicate wells with no antigens (determined as background value) was subtracted from the geometric means of the quadruplicate experimental wells. The reported data represent the geometric means (± standard error) of values for each antigen for five rabbits.
Nucleotide sequence accession numbers.
The sequences identified in this study can be found at GenBank under accession numbers AY685232 to AY685240 (tprA, tprB, tprH, tprL, tprC, tprD, tprG/I, tprG1, and tprG2, respectively) and AY685241 to AY685250 for tprK (clones 1.1 to 1.5 and 2.1 to 2.5, respectively).
RESULTS
Identification of tpr homologs in T. paraluiscuniculi.
All tpr loci defined in the Nichols genome sequence were analyzed in the corresponding chromosomal locations in T. paraluiscuniculi Cuniculi A strain. The DNA and predicted amino acid sequences were aligned with the T. pallidum Nichols strain homologs, followed by BLAST searches with the T. paraluiscuniculi sequences as the query to find additional regions of homology with other tpr sequences available in the databases. Overall, we identified three major subfamilies and one hybrid ORF. These are described in Table 3.
TABLE 3.
Characteristics of T. paraluiscuniculi tpr genes and predicted proteins
| T. paraluiscuniculi Tpr | Closest Nichols strain homologe | Primer seta | ORF lengthb (bp) | Predicted size (kDa) | Signal peptidec | Predicted cellular locationd |
|---|---|---|---|---|---|---|
| TprG/I hybrid | tprG (sense), tprF (antisense) | |||||
| Reading frame A | TprG (amino-terminal region) | 777 | 27.7 | U | IM | |
| Reading frame B | Tprl (central and carboxyl-terminal regions) | 1,344 | ||||
| Subfamily I | ||||||
| TprC | tprC | |||||
| Reading frame A | TprD2e | 591 | 21.4 | C | P/OM | |
| Reading frame B | TprD2 | 1,413 | ||||
| TprD | tprD | |||||
| Reading frame A | TprD2 | 591 | 21.4 | C | P/OM | |
| Reading frame B | TprD2 | 1,413 | ||||
| Subfamily II | ||||||
| TprG1 | tprE | |||||
| Reading frame A | TprG | 753 | 27.7 | U | IM | |
| Reading frame B | 1,752 | |||||
| TprG2 | tprJ | |||||
| Reading frame A | TprG | 753 | 27.7 | U | IM | |
| Reading frame B | TprG | 1,752 | ||||
| Subfamily III | ||||||
| TprA | TprA | tprA | 1,797 | 65.9 | U | IM |
| TprB | TprB | tprB | 1,935 | 69.5 | N | Cy |
| TprH | TprH | tprH | 2,082 | 76.0 | N | Cy |
| TprKf | TprK | tprK | 1,518 | 57.7 | C | P/OMg |
| TprL | TprL | tprL | 1,341 | 48.8 | N | Cy |
Indicates which tpr gene was targeted for amplification. In the Nichols strain of T. pallidum, each set recognizes the flanking regions of the gene indicated. Reading frames A and B indicate the two ORFs generated by the frameshift. No predictions (size, signal peptide, and cellular localization) are reported for the B ORFs due to the lack of evidence that these regions are actually transcribed or translated.
Total length includes stop codon.
Signal peptide predicted by Psort (http://psort.nibb.ac.jp/). U, uncleavable; C, cleavable; N, none.
Location of the predicted protein in the bacterial cell, predicted by Psort. OM, outer membrane; IM, inner membrane; P, periplasmic; Cy, cytoplasmic.
The tprD2 allele is not present in the Nichols strain genome. However, it was reported in several strains of T. pallidum subsp. pallidum (8).
The TprK 1.1 clone was used for this prediction.
Subfamily I in T. pallidum is composed of four members, tprC, tprD, tprF, and tprI. In the Cuniculi A strain, however, we identified only the tprC and tprD alleles. These loci contain identical sequences (Fig. 1), which are highly homologous to the T. pallidum tprD2 allele described previously (8). However, these ORFs do not encode full-length TprD2 amino acid sequences. Instead, there is an identical frameshift in each ORF due to the presence of an extra guanosine at position 439 of the coding sequence (amino acid position 147 of the Cuniculi A TprC and TprD ORFs; Fig. 1). This introduces a premature termination, which generates a putative protein sequence of only 196 amino acids instead of 597 (expected in the absence of a frameshift). However, both amino acid sequences (upstream and downstream of the frameshift) are included in the alignments for comparison purposes. The region corresponding to the Nichols tprI locus is absent in T. paraluiscuniculi, but its 3′-flanking region is nonetheless preserved. A hybrid ORF containing the central domain and carboxyl terminus of a tprI-like sequence was found in the region of the Nichols tprF locus (described below as a tprG/I hybrid).
FIG. 1.
Comparison of the TprC and TprD loci protein sequences of T. paraluiscuniculi with the TprD2 sequence of T. pallidum. TprC-C, TprD-C indicate the TprC and TprD alleles of T. paraluiscuniculi, respectively. A, NH2-terminal region; B. central unique and COOH-terminal regions. Identical residues are indicated by black shading, and nonsynonymous changes are indicated in gray or no shading, respectively. *, frameshift position in T. paraluiscuniculi Tprs (TprC-C and TprD-C A ORFs, amino acid position 147); TprD2, sequence from GenBank (Accession number AF217541).
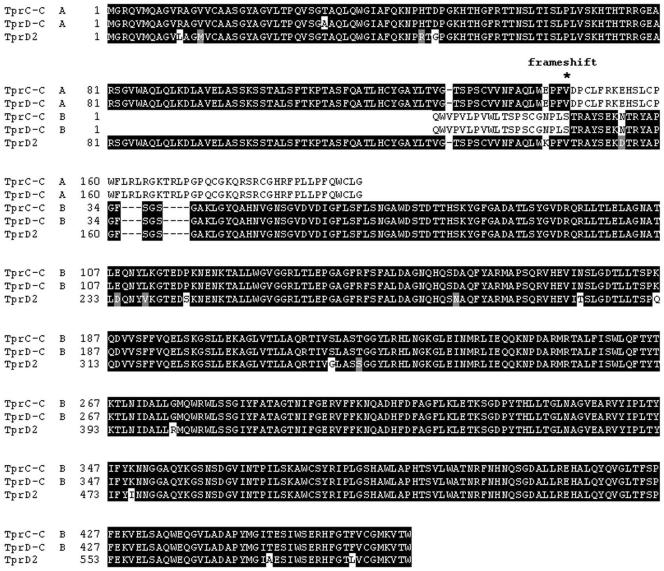
The DNA regions in Cuniculi A corresponding to the Nichols tprE and tprJ loci were found to contain two identical sequences with a complex pattern of homology to the Nichols subfamily II tprs (Fig. 2 and Fig. 4A). Their 5′ end aligns with the conserved 5′ end of the subfamily II tprs (tprE, tprG, tprJ); the central regions align with the central portion of the Nichols tprG and their 3′ end with the corresponding fragment of the Nichols tprJ (Fig. 2 and 4B). Despite the complex nature of the sequence composition, they show the highest homology to Nichols tprG. For this reason, they are called here tprG1 and tprG2. Similar to the tprC and tprD alleles, these two ORFs have an extra guanosine in position 653 of the coding sequence, which also creates a frameshift (amino acid position 217 of Cuniculi A TprG1 and TprG2 ORFs; Fig. 2) and a premature stop in the predicted amino acid sequences. There is no tprE homolog.
FIG. 2.
Alignment of the subfamily II Tpr homologs of T. paraluiscuniculi and T. pallidum. TprG1-C and TprG2-C, T. paraluiscuniculi sequences. TprG-N, TprJ-N, and TprE-N, T. pallidum Nichols strain sequences. A, open reading frame A (NH2-terminal region); B, open reading frame B (central unique and COOH-terminal regions). *, frameshift position in T. paraluiscuniculi Tprs (TprG1-C and TprG2-C A ORFs, amino acid position 217).
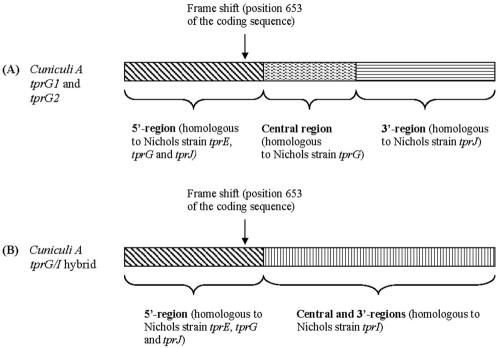
FIG. 4.
Schematic representation of T. paraluiscuniculi subfamily II members found in loci analogous to Nichols tprE (tprG1) and tprJ (tprG2) (A), and tprG/I hybrid (B) found in the locus analogous to Nichols tprG and tprF.
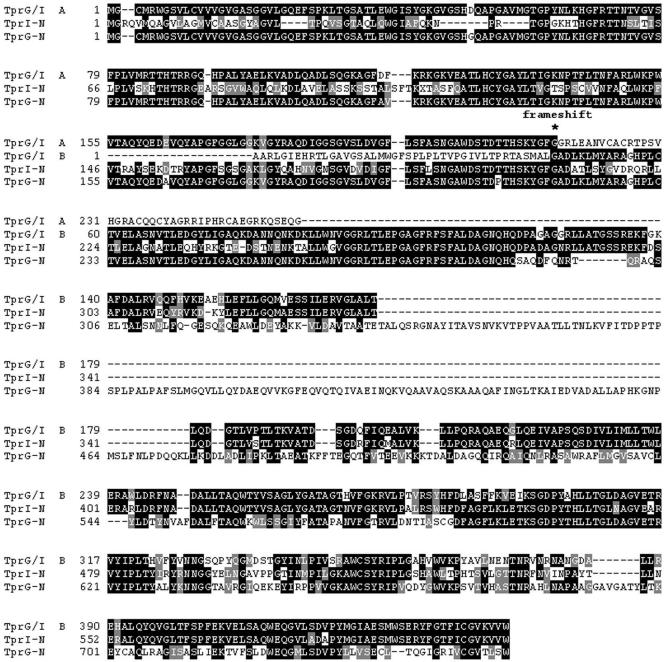
In T. paraluiscuniculi, a single tprG/I hybrid ORF replaces the loci occupied by tprG and tprF in the Nichols strain (Fig. 3). It is unique in that the 5′ end is homologous to the Nichols subfamily II 5′ conserved region, and the central unique region and the 3′ end contain a tprI-like sequence (Fig. 4B). The presence of a complete central region and a 3′ end with a stop codon as predicted in the Nichols tprI ORF rules out the possibility that the sequence in the G/I hybrid is tprF (tprF contains a large deletion in the central region, a frameshift and a premature stop). Interestingly, as seen in the Cuniculi A tprD2 alleles and in the tprG1 and tprG2 ORFs, this hybrid molecule also has a frameshift at nucleotide position 653 (amino acid position 217 of TprG/I, A ORF; Fig. 3), which introduces a premature stop in the predicted protein sequence (Fig. 3).
FIG. 3.
Alignment of the T. paraluiscuniculi TprG/I hybrid and the TprG and TprI homologs of T. pallidum. TprG/I, T. paraluiscuniculi TprG/I hybrid. TprG-N, TprI-N, T. pallidum Nichols strain sequences. A, open reading frame A (NH2-terminal region); B, open reading frame B (central unique and COOH-terminal regions). *, frameshift position in T. paraluiscuniculi TprG/I (amino acid position 217 of TprG/I A ORF).
As in the human treponeme, Cuniculi A subfamily III is composed of five members (tprA, tprB, tprH, tprK, and tprL) with poor homology to each other and to the other predicted Cuniculi A tprs. tprH and tprB are virtually identical to their T. pallidum homologs except for scattered nonsynonymous single-nucleotide polymorphisms which introduce four amino acid changes in TprB and five in TprH (unpublished data). tprA in the Nichols strain contains an authentic frameshift at nucleotide number 712 (amino acid position 237 of Nichols TprA ORF; Fig. 5) located in a region containing three dinucleotide repeats (CT) at positions 706 to 711 in the ORF. In contrast, the tprA ORF in T. paraluiscuniculi contains four CT dinucleotide repeats, which revert the frameshift and generate a sequence encoding a full-length ORF.
FIG. 5.
Alignment of TprA homologs in T. paraluiscuniculi and T. pallidum. TprA-N, T. pallidum Nichols strain sequence; TprA-C, T. paraluiscuniculi sequence. A, open reading frame A (NH2-terminal region); B, open reading frame B (central unique and COOH-terminal regions). *, frameshift position in T. pallidum TprA (amino acid position 237 of TprA-N, A ORF).
The presence of dinucleotide repeats is frequently associated with regulatory mechanisms of gene expression, where the number of repeats varies according to an on or off state to achieve phase variation of gene expression. To see whether variation in the number of dinucleotide repeats is due to such a mechanism as an explanation for the absence of a frameshift and premature termination in Cuniculi A tprA, we sequenced 15 more clones of a DNA region of approximately 800 bp containing the dinucleotide repeats (data not shown). All sequences confirmed the absence of a frameshift in the rabbit treponeme tprA homolog. We are aware, however, that changes in the number of repeats that occur at low frequencies may not be detected with this approach. Alignment of the Nichols and Cuniculi A tprL homologs showed conserved 5′ and 3′ ends with a different central region of 366 nucleotides (122 amino acids, positions 131 to 252 of Cuniculi A TprL; Fig. 6). This unique region in Cuniculi A has no sequence homology in the T. pallidum databases.
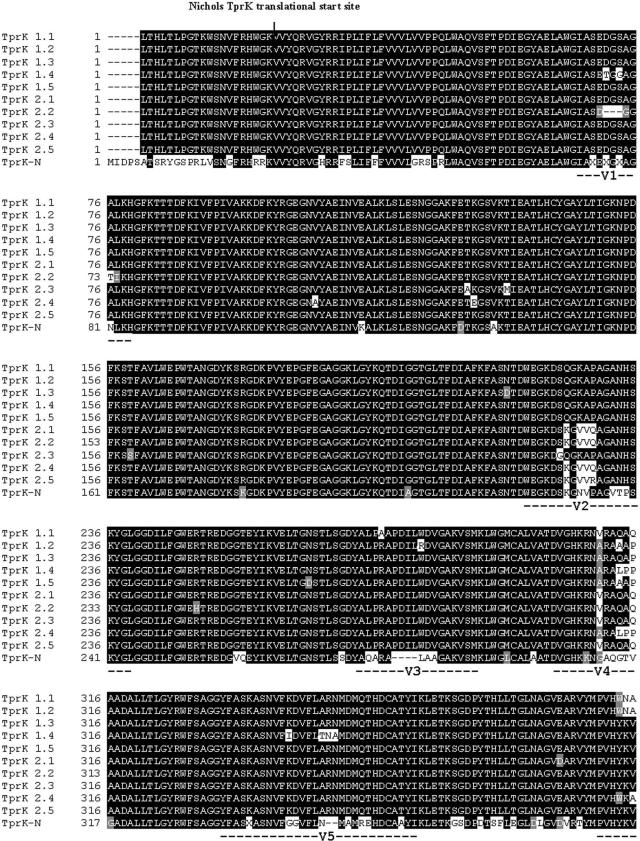
We have shown previously that the TprK antigen in T. pallidum is heterogeneous within and among T. pallidum isolates. The heterogeneity is localized in seven discrete variable regions (V1 to V7) flanked by highly conserved sequences (6). In the Cuniculi A strain of the rabbit treponeme, however, heterogeneity was identified in only a subset of V regions (Fig. 7), corresponding to T. pallidum V1, V2, V4, V6, and V7; V3 is not variable, at least in this isolate of T. paraluiscuniculi, and V5 shows very limited sequence diversity. The Cuniculi A V regions differ in their degree of heterogeneity, but diversity is, overall, less impressive than in the human treponeme. As in T. pallidum, V6 is the most diverse (eight sequences in 10 clones).
FIG. 7.
Alignment of TprK deduced amino acid sequences in T. paraluiscuniculi and T. pallidum Nichols strain. TprK-N, T. pallidum Nichols strain sequence. Amino acid sequences upstream of the Nichols TprK translational start site are included for comparison (4, 15). TprK 1.1 to 1.5 and TprK 2.1 to 2.5, T. paraluiscuniculi sequences from two independent PCRs; V1 to V7, TprK variable regions (6).
In addition to the differences in the number of V regions between the rabbit and human treponemes, there were 27 additional amino acid differences in the conserved regions compared with the sequence of the Nichols strain. Most were randomly dispersed throughout the conserved regions except for a region of 20 amino acids located immediately upstream of V6. This peptide sequence (SGDPYTHLLTGLNAGVEARV, amino acid positions 367 to 387 of tprK-C clones; Fig. 7) differs significantly from the corresponding TprK sequence in the human treponemes and has no homology to any of the seven V regions. Further analysis of this peptide sequence revealed that it matched a highly conserved region of TprL in both the Nichols and Cuniculi A strains and was also present in the carboxyl terminus of the Nichols subfamily I and II antigens.
Antibody and T-cell responses.
Pooled sera collected from six T. paraluiscuniculi-infected rabbits at nine different time points (day 0 to day 270) were tested by ELISA against recombinant peptides based on the Nichols strain Tpr sequences (Table 2). Sera collected prior to infection showed no specific reactivity against any of the recombinant antigens tested. ELISA background absorbance readings ranged from 0.001 to 0.03 units. Immune sera raised against each of the recombinants were used as positive controls and to assess binding of the antigen to the plate well (data not shown). All sera tested (pre- and postinfection and positive-control sera) were not reactive against the unrelated SA85 1.1 Trypanosoma cruzi recombinant protein (24) used as a control antigen to assess cross-reactivity (data not shown).
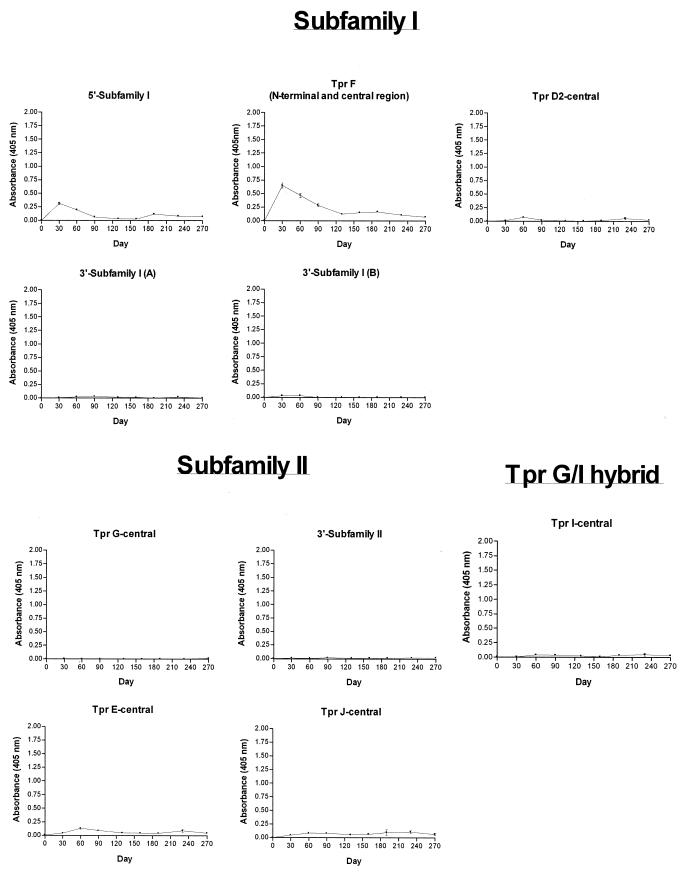
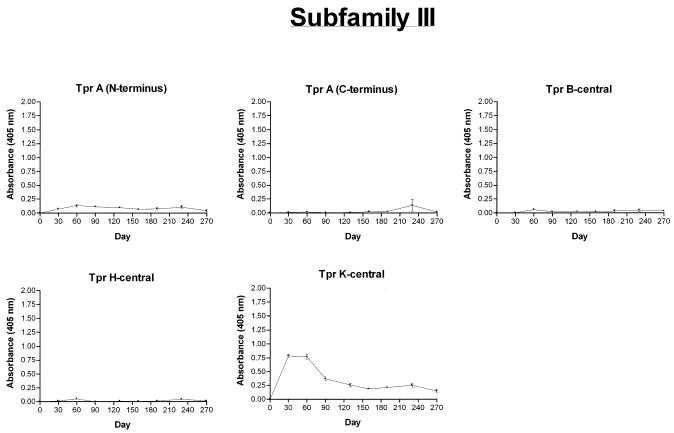
Antibody responses to Cuniculi A strain subfamily I members (TprC and TprD) were determined with five recombinant peptides (5′-subfamily I, TprF, TprD2-central, and 3′-subfamily I A/B; Fig. 8). TprF and the 5′-subfamily I recombinant share a sequence of 267 amino acids (the conserved amino terminus of subfamily I). The strongest reactivity peak against these two recombinants was detected at day 30, decreasing gradually over time, with virtually no detectable reactivity by day 190. We interpret this antibody reactivity in infected rabbits as the immune response against the amino terminus of the Cuniculi A TprD2-like peptides due to sequence conservation in this region in all members of subfamily I. Although the Cuniculi A tprC and tprD loci contain frameshifts which cause premature stops, it is still possible that the coding region upstream of the frameshift could be translated. The TprD2-central and the 3′-subfamily I A/B recombinant peptides were not recognized by the antisera (Fig. 8), consistent with the hypothetical partial translation of TprC and TprD.
FIG. 8.
Antibody responses of T. paraluiscuniculi-infected rabbits to Tpr recombinant peptides through day 270 postinfection as measured by ELISA. Peptides were designed based on the Nichols strain Tpr sequences. The reported values (mean OD405 ± standard error of the mean) represent three replicates of pooled sera from six Cuniculi A strain-infected rabbits.
The TprG-central and 3′-subfamily II recombinant peptides were used to detect antibody responses against Cuniculi A TprG1 and TprG2. These two identical homologs encode hybrid proteins which contain the following Nichols fragments: the amino terminus of subfamily II (containing a frameshift), the unique central region of TprG, and the carboxyl terminus of TprJ (Fig. 2 and Fig. 4A). The antisera showed no reactivity (Fig. 8), most likely because these recombinants represent proteins that are not translated due to the frameshift in the Cuniculi A coding sequences. No recombinant peptide was available to investigate the antibody response against the NH2-terminal portion of the subfamily II Tprs. Antisera were also tested against two antigens (TprE-central and TprJ-central) representing the central regions of Nichols subfamily II members TprE and TprJ. These sequences have a very low degree of homology with any of the Cuniculi A antigens described (Table 2), but they were tested to rule out expression of unidentified Tpr antigens with significant homology to these two Tprs elsewhere in the chromosome. No significant reactivity against them was detected (Fig. 8). Antibodies against the Cuniculi A TprG/I hybrid were investigated with the TprI-central peptide, corresponding to the hybrid's central unique region, but no reactivity was found (Fig. 8). As in subfamilies I and II, this suggests no translation due to the frameshift upstream of this region.
Five peptides (TprA N-terminal, TprA C-terminal, TprB-central, TprH-central, and TprK-central) were tested to assess reactivity against subfamily III members. Antisera reacted against TprK, with the highest and strongest reactivity at day 30 postinfection (Fig. 7). No significant antibody reactivity was detectable against TprA N-terminal, TprA C-terminal, TprB-central, and TprH-central.
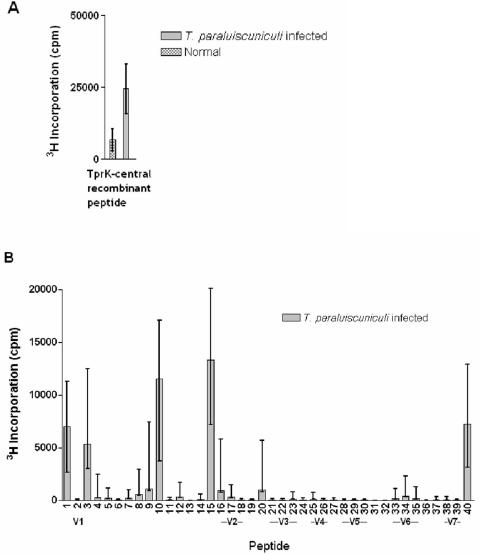
T-cell responses against the T. paraluiscuniculi Tprs were assessed with the same recombinants used for the ELISA tests (Table 2) plus 40 additional peptides (39 synthetic and 1 recombinant) spanning the entire Nichols TprK sequence (unpublished data). Concanavalin A-stimulated splenic lymphocytes from infected and uninfected rabbits showed strong proliferative responses, while no significant proliferation was detected in the absence of antigen (data not shown). Splenocytes from T. paraluiscuniculi-infected rabbits had significant [3H]thymidine incorporation when stimulated with T. pallidum sonicate and, in contrast, splenocytes from uninfected rabbits failed to proliferate in the presence of T. pallidum sonicates (data not shown). Of all the recombinant peptides listed in Table 2, only the TprK-central recombinant elicited a significant response in infected rabbits (Fig. 9A); no response was found to any other recombinant peptides (data not shown). Proliferation was seen in response to synthetic peptides 1, 3, 10, and 15 and recombinant peptide 40, all of which carry highly conserved TprK regions. None of the other peptides induced significant T-cell responses (Fig. 9B).
FIG. 9.
T-cell epitopes. Splenocytes from five rabbits infected for 270 days with T. paraluiscuniculi Cuniculi A strain were tested for their ability to proliferate when stimulated with recombinant TprK-central peptide (A) and synthetic peptides (B). All peptides were designed according to the Nichols strain Tpr sequences. A. TprK-central (315 amino acids, Table 2) encompasses the sequences of peptides 2 to 30 and partially those of peptides 1 and 31. The concentration of antigen was 10 μg/ml. B. Among the peptides encompassing sequences in the conserved regions of TprK, T-cell response was confined to peptides 1, 3, 10, 15, and 40 during T. paraluiscuniculi experimental infection. V1 to V7 indicate peptides containing amino acid sequences found in the variable regions (unpublished data).
The tpr gene family in T. paraluiscuniculi is composed of at least 10 paralogs; six of them are single-copy genes, and two have identical alleles in two different loci. These characteristics are summarized in Table 3, along with prediction of cellular location, molecular weight, and presence of a signal peptide.
Sequence analysis predicts that half of these genes are nonfunctional due to the presence of frameshifts and premature stop codons; only the subfamily III ORFs are presumably fully functional. Antibody and T-cell assays show, however, that only TprK is recognized by the humoral and cellular immune response, suggesting that the other subfamily III members may not be expressed. The diversity in Cuniculi A TprK resembles that observed in T. pallidum and suggests the presence of an antigenic variation system similar to the one described for the human treponeme (6, 7).
DISCUSSION
The Tpr antigen families of T. pallidum and T. paraluiscuniculi, pathogenic spirochetes which cause human and rabbit venereal syphilis, respectively, were the focus of this comparative study. Both spirochetes cause persistent infections, are transmitted from host to host by a similar route, and are indistinguishable in morphology and the immunopathological changes induced. However, the human venereal syphilis spirochete has the ability to infect both humans and rabbits, while the rabbit treponeme has a more restricted host specificity, being unable to cause active infection in humans (14).
Morphological, genetic, and immunological analyses of T. paraluiscuniculi and the other pathogenic treponemes indicate that these organisms are highly related. It has been postulated that the pathogenic treponemes arose originally from free-living treponemes in mud (29). These organisms came to be carried commensally by animals and humans and then developed the ability to cause disease as natural selection ensured the optimal survival of mutants best suited for transmission from host to host in the prevailing environment (31). It is most likely that the rabbit spirochete and the other treponemes evolved with mechanisms similar to those of other organisms, by acquisition or loss of new traits resulting in differences in host specificity and mechanisms of pathogenesis.
The tpr gene family likely arose by gene duplication events in the distant past. Duplication of an ancestor tpr gene during evolution may have generated this paralogue family, whose members then underwent further differential evolution. Some might have been randomly silenced, while others might have undergone neofunctionalization (one copy acquires a novel, beneficial function and becomes preserved by natural selection, while the other copy retains its original function) (20, 22) or subfunctionalization (complementary loss of function of both copies to the level of the single-copy ancestral gene, resulting in the preservation of both loci) (21).
All the subfamily I (tprC, tprD, tprF, and tprI) and II (tprE, tprG, and tprJ) tprs in the human syphilis treponeme Nichols strain are thought to be transcribed and translated (4, 27; unpublished data) and, except for tprF, do not contain frameshifts and early termination. In contrast to T. pallidum, all subfamily I (tprC and tprD), II (tprG1 and tprG2), and the I/II hybrid (tprG/I) ORFs identified in T. paraluiscuniculi are predicted to encode truncated proteins due to frameshifts caused by a single extra guanosine in their coding sequences. This predicts that these genes are not essential for survival in the rabbit. Sequences obtained from two independent PCRs and from shorter amplicons containing the frameshift regions (data not shown), obtained with primers internal to the ones used for amplification of the whole ORFs, ruled out the possibility that the extra G nucleotide was due to PCR error. In addition, it is highly unlikely that PCR errors would be introduced at the same nucleotide position for tprC and tprD and, for the tprG1 and tprG2 alleles, in two independent PCRs for each template. It is also possible, however, that the frameshifts might be corrected in a small proportion of mRNAs during transcription or that ribosomal slippage occurs during translation, permitting translation of a complete product.
The T. paraluiscuniculi tprD2-like sequences found in the tprC and tprD loci are virtually identical to the tprD2 allele present in other T. pallidum isolates. Approximately 50% of human syphilis isolates contain the tprD2 gene; some humans with venereal syphilis make antibodies to a TprD2-specific peptide (8). tprD2 is also found in T. pallidum subsp. pertenue and T. pallidum subsp. endemicum. The TprD2 antigen is predicted to be an outer membrane antigen and may provide functional diversity to individual isolates. It is notable that both tprD2-like alleles in the rabbit treponeme contain authentic frameshifts introducing early terminations and generating a short predicted peptide sequence of only 196 amino acids (instead of 597). The frameshift is due to an extra G located at exactly the same nucleotide position in both loci, suggesting that the frameshifts are not artifactual and that these two alleles are nonfunctional. In contrast, the T. pallidum tprD2 homologue has been retained intact during evolution, and in the presence of selective advantage, it might have undergone neofunctionalization. This could provide advantages to particular T. pallidum isolates, such as adaptation to specific tissues in the human host.
Due to the presence of frameshifts and premature stops in the subfamily I Tprs, we reasoned that only the NH2-terminal portion of these genes is likely expressed and that the central and COOH-terminal regions are not translated. This is supported by our ELISA findings, where antibody reactivity was seen against the conserved subfamily I amino terminus and no reactivity was detected against the central region (TprD2-central) or the carboxyl terminus (3′-subfamily I peptides A and B). Alternative explanations for the lack of reactivity against the central and COOH-terminal regions are that these regions are not the target of the humoral antibody responses and that they are expressed at a very low level during infection. However, the recent demonstration that human syphilitic sera react against a 216-amino-acid peptide of the subfamily I Tprs carboxyl terminus (V. Sambri et al., unpublished data) supports the idea that the absence of reactivity to this particular region in the Cuniculi A strain is due not to lack of antigenicity but to lack of translation. This hypothesis is equally supported by the absence of detectable reactivity against the central region and carboxyl terminus of Cuniculi A subfamily II members (TprG1 and TprG2) and the TprG/I hybrid, with these regions being coded by sequences downstream of the frameshift position in their respective genes. No recombinant antigens were available to test reactivity against the amino terminus of these antigens due to difficulties in expressing this particular peptide region.
Based on the analysis of the predicted protein sequences, it appears that only the subfamily III Tpr proteins may be functional molecules in T. paraluiscuniculi. There is a very high degree of conservation between the T. paraluiscuniculi and T. pallidum tprB and tprH genes, which are virtually identical. In contrast, there are significant sequence differences in their tprA, tprK, and tprL genes. tprA is an intact ORF in the rabbit treponeme but not in T. pallidum Nichols strain, where this ORF has an authentic frameshift due to the presence of only three CT repeats instead of four. In absence of full-length functional tprC, tprD, tprF, tprI, tprG, tprJ, and tprE genes, one could assume that tprA and the other subfamily III members having intact full-length coding sequences are playing an important role in T. paraluiscuniculi biology. Although this assumption may seem obvious from sequence analysis, ELISA antibody reactivity and proliferation assays do not entirely support this hypothesis.
Overall, antibody responsiveness to subfamily III Tpr antigens in T. pallidum isolates has been shown to vary in specificity and temporal pattern of reactivity elicited against individual Tpr proteins during infection (18). Of particular interest is the Nichols strain, in which the expression level of most subfamily III Tprs (except for TprK) seems to be lower than in other T. pallidum isolates. It has been suggested that this may reflect a high degree of adaptation to the rabbit, which by analogy could be extrapolated to T. paraluiscuniculi as well. The lack of antibody specificities during T. paraluiscuniculi infection is similar to that seen against subfamily III members in the Nichols strain, in which TprK is the only apparent target of the immune response. T-cell proliferation data confirm these observations, with TprK being the only target of the cellular immune response. As in T. pallidum (24), Cuniculi A T-cell epitopes were found in the TprK conserved regions, with the same peptides being recognized in both infections. The lack of antibody and T-cell responses to the other subfamily III Tprs (in the presence of intact ORFs) may reflect different expression patterns (very low levels of expression throughout infection) or lack of antigenicity of these proteins, or may indicate that these ORFs are actually be pseudogenes.
The tprK locus is of particular interest. It has been shown in T. pallidum that the TprK antigen contains seven variable regions targeted by the antibody response of the host and that it undergoes antigenic variation during infection with donor sites located in a ≈4-kb region flanking the tprD gene (24, 7). As in T. pallidum, Cuniculi A TprK is the target of an early and strong immune response. The heterogeneity in the V regions of the tprK alleles in the Cuniculi A isolate suggests that its TprK may undergo antigenic variation too. Interestingly, a significant difference is the presence of limited diversity in the V regions in the rabbit treponeme, with the observed heterogeneity not being as impressive as in the human isolates. It has been postulated that the extensive diversity in the TprK antigen may represent mechanisms for immune evasion and adaptation to a changing environment in the host during infection. In this context, the overall lower diversity in the Cuniculi A TprK antigen might imply that T. paraluiscuniculi uses a smaller subset of TprK variants to achieve successful adaptation in its host.
So far, we have been able to obtain only a fragment of about 600 nucleotides from the corresponding Cuniculi A donor regions flanking the tprD gene by PCR amplification. We do not know at this time whether this is due to lack of binding sites for the primers, fragmentation, and different localization of the donor sites in the chromosome, or smaller donor regions in T. paraluiscuniculi than in T. pallidum.
The successful amplification of the tpr ORFs in T. paraluiscuniculi with primers based on the Nichols genome sequence of the flanking regions for each locus suggests sequence conservation at the primer binding sites and similar organizations of the ORFs (tprs and flanking ORFs) in the amplified regions of these two species. We sequenced a large number of tpr ORFs from several T. pallidum isolates and observed variations in sequence composition in the coding regions but not in their chromosomal locations, supporting earlier morphological and immunopathological observations that these are highly related organisms. However, we cannot completely rule out the presence of additional paralogues in the Cuniculi A chromosome. Alternative approaches, such as Southern blot analysis or the construction of genomic, subgenomic, and lambda libraries, have already been considered. Although technically feasible, T. paraluiscuniculi grows very slowly with very low bacterial yields, making it impractical to collect Percoll-purified treponemes (free of most contaminating rabbit DNA) in sufficient numbers to create these libraries. It is more likely that other approaches that require minimal amounts of DNA (such as whole-genome long-PCR fingerprinting) might yield more information without the need for large amounts of treponemal DNA.
It is intriguing to find five apparently nonfunctional tpr genes in T. paraluiscuniculi due to frameshifts and premature stops (tprC, tprD, tprG1, tprG2, and the hybrid tprG/I). It is likely that these genes were silenced during evolution after gene duplication generated the tpr family in an ancestor treponeme. One hypothesis is that the human and rabbit treponemes arose from a common predecessor containing a set of fully functional tpr genes. Later on, during adaptation to the genetic background of the rabbit, the subfamily I and II genes were then silenced by single-base insertions. However, to explain the presence of identical alleles in the tprC and tprD loci as well as in the tprG1 and tprG2 loci containing frameshifts at exactly the same positions, we need to assume that the original set of genes were composed of a smaller number of single-copy genes, some of which underwent further duplication after the frameshifts were introduced. The lack of functionality of these genes suggests that they are not essential for survival in the rabbit but may represent pathogenicity factors in T. pallidum. As mentioned above, we have not completely ruled out the possibility of other tpr genes elsewhere in the Cuniculi A chromosome, and ongoing studies in our laboratory will determine whether second functional copies of these genes are present in this strain and, also, whether other T. paraluiscuniculi strains have similar sets of genes and similar patterns of expression.
Besides the most obvious differences between the T. pallidum and T. paraluiscuniculi homologues presented here, small numbers of single synonymous and nonsynonymous nucleotide mutations can also be identified in the tpr sequences. The amino acid changes due to single-nucleotide mutations that might influence protein function can be observed throughout some Tpr sequences, suggesting selective evolutionary divergence. Nevertheless, silent mutations are also known to influence protein function by mediating in vivo protein folding changes or regulating gene transcription (9, 10). Although the sequence organization of and host immune responses to the Tpr antigens are being unraveled, the function of these proteins and the role in antigenicity or protein structure of the observed diversity due to single-nucleotide changes or more extensive sequence modifications still remain unclear.
Acknowledgments
We are grateful to Heidi Pecoraro for manuscript preparation.
This work was supported by NIH grants AI42143 (S.A.L.), AI34616 (S.A.L.), and AI43456 (W.V.V.) and the University of Washington Royalty Research Fund Award (A.C.L.). E.S.S. was supported by NIH institutional training grant AI07140 and a fellowship from the Achievement Rewards for College Scientists Foundation.
Editor: A. D. O'Brien
REFERENCES
- 1.Arroll, T. W., A. Centurion-Lara, S. A. Lukehart, and W. C. Van Voorhis. 1999. T-cell responses to Treponema pallidum subsp. pallidum antigens during the course of experimental syphilis infection. Infect Immun. 67:4757-4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker-Zander, S. A., and S. A. Lukehart. 1984. Antigenic cross-reactivity between Treponema pallidum and other pathogenic members of the family Spirochaetaceae. Infect Immun. 46:116-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayon, H. 1913. A new species of Treponema found in the genital sores of rabbits. Br. Med. 2:1159. [Google Scholar]
- 4.Centurion-Lara, A., C. Castro, L. Barrett, C. Cameron, M. Mostowfi, W. C. Van Voorhis, and S. A. Lukehart. 1999. Treponema pallidum major sheath protein homologue Tpr K is a target of opsonic antibody and the protective immune response. J. Exp. Med. 189:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centurion-Lara, A., C. Castro, R. Castillo, J. M. Shaffer, W. C. Van Voorhis, and S. A. Lukehart. 1998. The flanking region sequences of the 15-kDa lipoprotein gene differentiate pathogenic treponemes. J. Infect. Dis. 177:1036-1040. [DOI] [PubMed] [Google Scholar]
- 6.Centurion-Lara, A., C. Godornes, C. Castro, W. C. Van Voorhis, and S. A. Lukehart. 2000. The tprK gene is heterogeneous among Treponema pallidum strains and has multiple alleles. Infect Immun. 68:824-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centurion-Lara, A., R. E. LaFond, K. Hevner, C. Godornes, B. J. Molini, W. C. Van Voorhis, and S. A. Lukehart. 2004. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol. Microbiol. 52:1579-1596. [DOI] [PubMed] [Google Scholar]
- 8.Centurion-Lara, A., E. S. Sun, L. K. Barrett, C. Castro, S. A. Lukehart, and W. C. Van Voorhis. 2000. Multiple alleles of Treponema pallidum repeat gene D in Treponema pallidum isolates. J. Bacteriol. 182:2332-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortazzo, P., C. Cervenansky, M. Marin, C. Reiss, R. Ehrlich, and A. Deana. 2002. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem. Biophys. Res. Commun. 293:537-541. [DOI] [PubMed] [Google Scholar]
- 10.Deana, A., R. Ehrlich, and C. Reiss. 1996. Synonymous codon selection controls in vivo turnover and amount of mRNA in Escherichia coli bla and ompA genes. J. Bacteriol. 178:2718-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiGiacomo, R. F., S. A. Lukehart, C. D. Talburt, S. A. Baker-Zander, W. E. Giddens, Jr., J. Condon, and C. W. Brown. 1985. Chronicity of infection with Treponema paraluis-cuniculi in New Zealand White rabbits. Genitourin. Med. 61:156-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiGiacomo, R. F., C. D. Talburt, S. A. Lukehart, S. A. Baker-Zander, and J. Condon. 1983. Treponema paraluis-cuniculi infection in a commercial rabbitry: epidemiology and serodiagnosis. Lab. Anim. Sci. 33:562-566. [PubMed] [Google Scholar]
- 13.Fraser, C. M., S. J. Norris, G. M. Weinstock, O. White, G. G. Sutton, R. Dodson, M. Gwinn, E. K. Hickey, R. Clayton, K. A. Ketchum, E. Sodergren, J. M. Hardham, M. P. McLeod, S. Salzberg, J. Peterson, H. Khalak, D. Richardson, J. K. Howell, M. Chidambaram, T. Utterback, L. McDonald, P. Artiach, C. Bowman, M. D. Cotton, J. C. Venter, et al. 1998. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281:375-388. [DOI] [PubMed] [Google Scholar]
- 14.Graves, S., and J. Downes. 1981. Experimental infection of humans with rabbit-virulent Treponema paraluis-cuniculi. Br. J. Vener. Dis. 57:7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazlett, K. R., D. L. Cox, R. A. Sikkink, F. Auch'ere, F. Rusnak, and J. D. Radolf. 2002. Contribution of neelaredoxin to oxygen tolerance by Treponema pallidum. Methods Enzymol. 353:140-156. [DOI] [PubMed] [Google Scholar]
- 16.Hazlett, K. R. O., T. J. Sellati, T. T. Nguyen, D. L. Cox, M. L. Clawson, M. J. Caimano, and J. D. Radolf. 2001. The TprK protein of Treponema pallidum is periplasmic and is not a target of opsonic antibody or protective immunity. J. Exp. Med. 193:1015-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hougen, K. H., A. Birch-Andersen, and H. J. Jensen. 1973. Electron microscopy of Treponema cuniculi. Acta Pathol. Microbiol. Scand. 81:15-28. [DOI] [PubMed] [Google Scholar]
- 18.Leader, B. T., K. Hevner, B. J. Molini, L. K. Barrett, W. C. Van Voorhis, and S. A. Lukehart. 2003. Antibody responses elicited against the Treponema pallidum repeat proteins differ during Infection with different isolates of Treponema pallidum subsp. pallidum. Infect. Immun. 71:6054-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukehart, S. A., S. A. Baker-Zander, and S. Sell. 1980. Characterization of lymphocyte responsiveness in early experimental syphilis. I. In vitro response to mitogens and Treponema pallidum antigens. J. Immunol. 124:454-460. [PubMed] [Google Scholar]
- 20.Lynch, M., and J. S. Conery. 2000. The evolutionary fate and consequences of duplicate genes. Science 290:1151-1155. [DOI] [PubMed] [Google Scholar]
- 21.Lynch, M., and A. Force. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154:459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch, M., M. O'Hely, B. Walsh, and A. Force. 2001. The probability of preservation of a newly arisen gene duplicate. Genetics 159:1789-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morgan, C. A., S. A. Lukehart, and W. C. Van Voorhis. 2003. Protection against syphilis correlates with specificity of antibodies to the variable regions of Treponema pallidum repeat protein K. Infect. Immun. 71:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan, C. A., B. J. Molini, S. A. Lukehart, and W. C. Van Voorhis. 2002. Segregation of B and T cell epitopes of Treponema pallidum repeat protein K to variable and conserved regions during experimental syphilis infection. J. Immunol. 169:952-957. [DOI] [PubMed] [Google Scholar]
- 25.Ross, E. H. 1912. An intracellular parasite developing into spirochaetes. Br. Med. J. 2:1651-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schell, R. F., and D. M. Musher (ed.). 1983. Pathogenesis and immunology of treponemal infection. Marcel Dekker, Inc., New York, N.Y.
- 27.Sun, E. S., B. J. Molini, L. K. Barrett, A. Centurion-Lara, S. A. Lukehart, and W. C. Van Voorhis. 2004. Subfamily I Treponema pallidum repeat protein family: sequence variation and immunity. Microbes Infect. 6:725-737. [DOI] [PubMed] [Google Scholar]
- 28.Turner, T. B., and D. H. Hollander. 1957. Biology of the treponematoses. World Health Organization, Geneva, Switzerland.
- 29.Veldkamp, H. 1960. Isolation and characteristics of Treponema zuelzerae nov. spec., and anaerobic, free-living spirochete. Antonie van Leeuwenhoek 26:103-125. [DOI] [PubMed] [Google Scholar]
- 30.Weinstock, G. M., S. J. Norris, E. J. Sodergren, and D. Smajs. 2002. Identification of virulence genes in silico: infectious disease genomics, p. 251-261. In K. A. Brogden, J. A. Roth, T. B. Stanton, C. A. Bolin, F. C. Minion, and M. J. Wannemuehler (ed.), Virulence mechanisms of bacterial pathogens, 3rd ed. ASM Press, Washington, D.C.
- 31.Willcox, R. R. 1972. The treponemal evolution. Trans. St. Johns Hosp. Dermatol. Soc. 58:21-37. [PubMed] [Google Scholar]