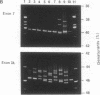
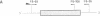Abstract
Hemophilia A is an X chromosome-linked disorder resulting from deficiency of factor VIII, an important protein in blood coagulation. A large number of disease-producing mutations have been reported in the factor VIII gene. However, a comprehensive analysis of the mutations has been difficult because of the large gene size, its many scattered exons, and the high frequency of de novo mutations. Recently, we have shown that nearly all mutations resulting in mild-to-moderate hemophilia A can be detected by PCR and denaturing gradient gel electrophoresis (DGGE). In this study, we attempted to discover the mutations causing severe hemophilia A by analyzing 47 unselected patients, 30 of whom had severe hemophilia and 17 of whom had mild-to-moderate disease. Using DGGE as a screening method, we analyzed 99% of the coding region, 94% of the splice junctions, the promoter region, and the polyadenylylation site of the gene. We found the mutation in 16 of 17 (94%) patients with mild-to-moderate disease but in only 16 of 30 (53%) patients with severe hemophilia A. Since DGGE after computer analysis appears to detect all mutations in a given fragment, the lower-than-expected yield of mutations in patients with severe disease is likely not due to failure of the detection method; it is probably due to the presence of mutations in DNA sequences outside the regions studied. Such sequences may include locus-controlling regions, other sequences within introns or outside the gene that are important for its expression, or another gene involved in factor VIII expression that is very closely linked to the factor VIII gene.
Full text
PDF
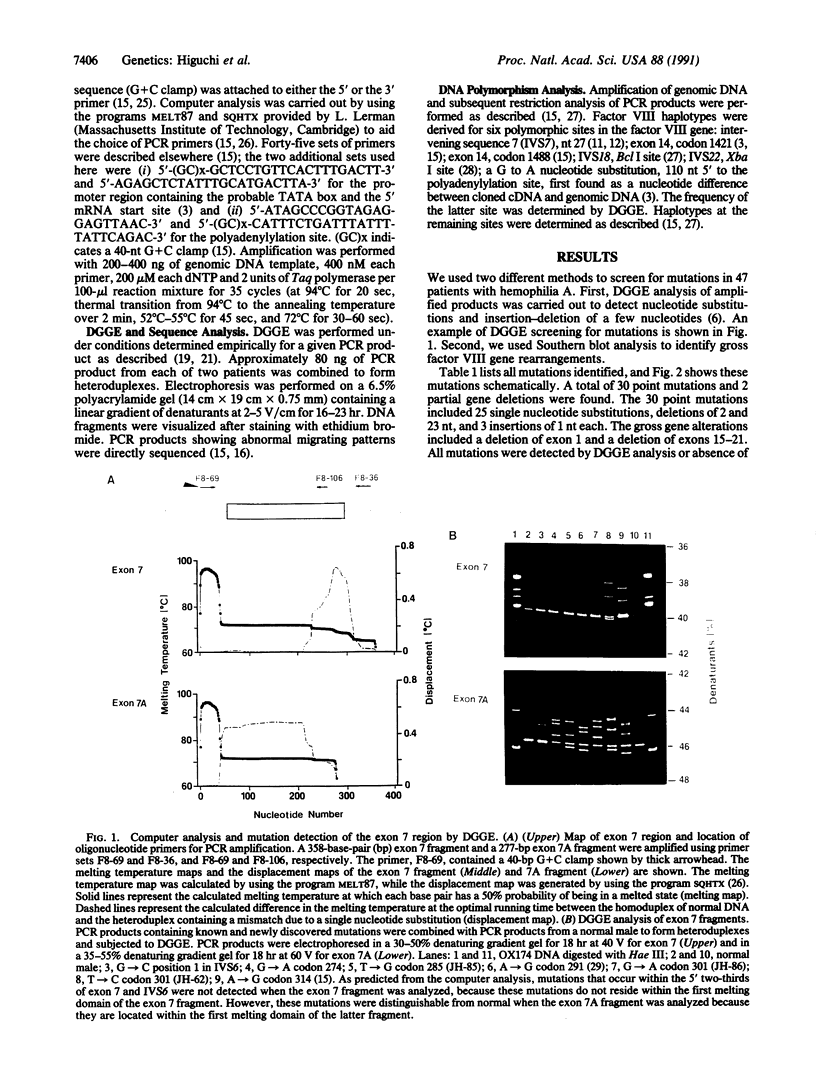


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antonarakis S. E., Kazazian H. H., Jr The molecular basis of hemophilia A in man. Trends Genet. 1988 Aug;4(8):233–237. doi: 10.1016/0168-9525(88)90156-4. [DOI] [PubMed] [Google Scholar]
- Arai M., Higuchi M., Antonarakis S. E., Kazazian H. H., Jr, Phillips J. A., 3rd, Janco R. L., Hoyer L. W. Characterization of a thrombin cleavage site mutation (Arg 1689 to Cys) in the factor VIII gene of two unrelated patients with cross-reacting material-positive hemophilia A. Blood. 1990 Jan 15;75(2):384–389. [PubMed] [Google Scholar]
- Arai M., Inaba H., Higuchi M., Antonarakis S. E., Kazazian H. H., Jr, Fujimaki M., Hoyer L. W. Direct characterization of factor VIII in plasma: detection of a mutation altering a thrombin cleavage site (arginine-372----histidine). Proc Natl Acad Sci U S A. 1989 Jun;86(11):4277–4281. doi: 10.1073/pnas.86.11.4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M. V., Hirst M. C., Nakahori Y., MacKinnon R. N., Roche A., Flint T. J., Jacobs P. A., Tommerup N., Tranebjaerg L., Froster-Iskenius U. Physical mapping across the fragile X: hypermethylation and clinical expression of the fragile X syndrome. Cell. 1991 Feb 22;64(4):861–866. doi: 10.1016/0092-8674(91)90514-y. [DOI] [PubMed] [Google Scholar]
- Cotton R. G., Rodrigues N. R., Campbell R. D. Reactivity of cytosine and thymine in single-base-pair mismatches with hydroxylamine and osmium tetroxide and its application to the study of mutations. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4397–4401. doi: 10.1073/pnas.85.12.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll M. C., Dobkin C. S., Alter B. P. Gamma delta beta-thalassemia due to a de novo mutation deleting the 5' beta-globin gene activation-region hypersensitive sites. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7470–7474. doi: 10.1073/pnas.86.19.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Fischer S. G., Lerman L. S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitschier J., Drayna D., Tuddenham E. G., White R. L., Lawn R. M. Genetic mapping and diagnosis of haemophilia A achieved through a BclI polymorphism in the factor VIII gene. 1985 Apr 25-May 1Nature. 314(6013):738–740. doi: 10.1038/314738a0. [DOI] [PubMed] [Google Scholar]
- Gitschier J., Kogan S., Levinson B., Tuddenham E. G. Mutations of factor VIII cleavage sites in hemophilia A. Blood. 1988 Sep;72(3):1022–1028. [PubMed] [Google Scholar]
- Gitschier J., Wood W. I., Goralka T. M., Wion K. L., Chen E. Y., Eaton D. H., Vehar G. A., Capon D. J., Lawn R. M. Characterization of the human factor VIII gene. Nature. 1984 Nov 22;312(5992):326–330. doi: 10.1038/312326a0. [DOI] [PubMed] [Google Scholar]
- Gitschier J., Wood W. I., Tuddenham E. G., Shuman M. A., Goralka T. M., Chen E. Y., Lawn R. M. Detection and sequence of mutations in the factor VIII gene of haemophiliacs. 1985 May 30-Jun 5Nature. 315(6018):427–430. doi: 10.1038/315427a0. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Wong C., Kochhan L., Olek K., Aronis S., Kasper C. K., Kazazian H. H., Jr, Antonarakis S. E. Characterization of mutations in the factor VIII gene by direct sequencing of amplified genomic DNA. Genomics. 1990 Jan;6(1):65–71. doi: 10.1016/0888-7543(90)90448-4. [DOI] [PubMed] [Google Scholar]
- Kazazian H. H., Jr The thalassemia syndromes: molecular basis and prenatal diagnosis in 1990. Semin Hematol. 1990 Jul;27(3):209–228. [PubMed] [Google Scholar]
- Koeberl D. D., Bottema C. D., Ketterling R. P., Bridge P. J., Lillicrap D. P., Sommer S. S. Mutations causing hemophilia B: direct estimate of the underlying rates of spontaneous germ-line transitions, transversions, and deletions in a human gene. Am J Hum Genet. 1990 Aug;47(2):202–217. [PMC free article] [PubMed] [Google Scholar]
- Kogan S. C., Doherty M., Gitschier J. An improved method for prenatal diagnosis of genetic diseases by analysis of amplified DNA sequences. Application to hemophilia A. N Engl J Med. 1987 Oct 15;317(16):985–990. doi: 10.1056/NEJM198710153171603. [DOI] [PubMed] [Google Scholar]
- Kogan S., Gitschier J. Mutations and a polymorphism in the factor VIII gene discovered by denaturing gradient gel electrophoresis. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2092–2096. doi: 10.1073/pnas.87.6.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman L. S., Silverstein K. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:482–501. doi: 10.1016/0076-6879(87)55032-7. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Larin Z., Maniatis T. Detection of single base substitutions by ribonuclease cleavage at mismatches in RNA:DNA duplexes. Science. 1985 Dec 13;230(4731):1242–1246. doi: 10.1126/science.4071043. [DOI] [PubMed] [Google Scholar]
- Myers R. M., Maniatis T., Lerman L. S. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- Orita M., Iwahana H., Kanazawa H., Hayashi K., Sekiya T. Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2766–2770. doi: 10.1073/pnas.86.8.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattinson J. K., Millar D. S., McVey J. H., Grundy C. B., Wieland K., Mibashan R. S., Martinowitz U., Tan-Un K., Vidaud M., Goossens M. The molecular genetic analysis of hemophilia A: a directed search strategy for the detection of point mutations in the human factor VIII gene. Blood. 1990 Dec 1;76(11):2242–2248. [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sheffield V. C., Cox D. R., Lerman L. S., Myers R. M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989 Jan;86(1):232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima M., Ware J., Yoshioka A., Fukui H., Fulcher C. A. An arginine to cysteine amino acid substitution at a critical thrombin cleavage site in a dysfunctional factor VIII molecule. Blood. 1989 Oct;74(5):1612–1617. [PubMed] [Google Scholar]
- Tantravahi U., Murty V. V., Jhanwar S. C., Toole J. J., Woozney J. M., Chaganti R. S., Latt S. A. Physical mapping of the factor VIII gene proximal to two polymorphic DNA probes in human chromosome band Xq28: implications for factor VIII gene segregation analysis. Cytogenet Cell Genet. 1986;42(1-2):75–79. doi: 10.1159/000132255. [DOI] [PubMed] [Google Scholar]
- Toole J. J., Knopf J. L., Wozney J. M., Sultzman L. A., Buecker J. L., Pittman D. D., Kaufman R. J., Brown E., Shoemaker C., Orr E. C. Molecular cloning of a cDNA encoding human antihaemophilic factor. Nature. 1984 Nov 22;312(5992):342–347. doi: 10.1038/312342a0. [DOI] [PubMed] [Google Scholar]
- Traystman M. D., Higuchi M., Kasper C. K., Antonarakis S. E., Kazazian H. H., Jr Use of denaturing gradient gel electrophoresis to detect point mutations in the factor VIII gene. Genomics. 1990 Feb;6(2):293–301. doi: 10.1016/0888-7543(90)90569-g. [DOI] [PubMed] [Google Scholar]
- Vincent A., Heitz D., Petit C., Kretz C., Oberlé I., Mandel J. L. Abnormal pattern detected in fragile-X patients by pulsed-field gel electrophoresis. Nature. 1991 Feb 14;349(6310):624–626. doi: 10.1038/349624a0. [DOI] [PubMed] [Google Scholar]
- Wion K. L., Tuddenham E. G., Lawn R. M. A new polymorphism in the factor VIII gene for prenatal diagnosis of hemophilia A. Nucleic Acids Res. 1986 Jun 11;14(11):4535–4542. doi: 10.1093/nar/14.11.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Dowling C. E., Saiki R. K., Higuchi R. G., Erlich H. A., Kazazian H. H., Jr Characterization of beta-thalassaemia mutations using direct genomic sequencing of amplified single copy DNA. 1987 Nov 26-Dec 2Nature. 330(6146):384–386. doi: 10.1038/330384a0. [DOI] [PubMed] [Google Scholar]
- Youssoufian H., Antonarakis S. E., Aronis S., Tsiftis G., Phillips D. G., Kazazian H. H., Jr Characterization of five partial deletions of the factor VIII gene. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3772–3776. doi: 10.1073/pnas.84.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Kazazian H. H., Jr, Phillips D. G., Aronis S., Tsiftis G., Brown V. A., Antonarakis S. E. Recurrent mutations in haemophilia A give evidence for CpG mutation hotspots. 1986 Nov 27-Dec 3Nature. 324(6095):380–382. doi: 10.1038/324380a0. [DOI] [PubMed] [Google Scholar]
- Youssoufian H., Wong C., Aronis S., Platokoukis H., Kazazian H. H., Jr, Antonarakis S. E. Moderately severe hemophilia A resulting from Glu----Gly substitution in exon 7 of the factor VIII gene. Am J Hum Genet. 1988 Jun;42(6):867–871. [PMC free article] [PubMed] [Google Scholar]