Abstract
The pleckstrin homology (PH) domains of the homologous proteins Grp1 (general receptor for phosphoinositides), ARNO (Arf nucleotide binding site opener), and Cytohesin-1 bind phosphatidylinositol (PtdIns) 3,4,5-trisphosphate with unusually high selectivity. Remarkably, splice variants that differ only by the insertion of a single glycine residue in the β1/β2 loop exhibit dual specificity for PtdIns(3,4,5)P3 and PtdIns(4,5)P2. The structural basis for this dramatic specificity switch is not apparent from the known modes of phosphoinositide recognition. Here, we report crystal structures for dual specificity variants of the Grp1 and ARNO PH domains in either the unliganded form or in complex with the head groups of PtdIns(4,5)P2 and PtdIns(3,4,5)P3. Loss of contacts with the β1/β2 loop with no significant change in head group orientation accounts for the significant decrease in PtdIns(3,4,5)P3 affinity observed for the dual specificity variants. Conversely, a small increase rather than decrease in affinity for PtdIns(4,5)P2 is explained by a novel binding mode, in which the glycine insertion alleviates unfavorable interactions with the β1/β2 loop. These observations are supported by a systematic mutational analysis of the determinants of phosphoinositide recognition.
Keywords: ARNO, Cytohesin-1, Grp1, pleckstrin homology domain, phosphoinositide
Introduction
Phosphorylated derivatives of phosphatidylinositol (PtdIns) play a critical role in the subcellular localization and assembly of protein complexes implicated in signal transduction, membrane trafficking, and cytoskeletal dynamics (Corvera and Czech, 1998; Fruman et al, 1998; Leevers et al, 1999; Lemmon and Ferguson, 2000; Lemmon et al, 2002). Whereas several phosphoinositides, particularly PtdIns(3)P, PtdIns(4)P, and PtdIns(4,5)P2, are relatively abundant in quiescent cells, PtdIns(3,4)P2 and PtdIns(3,4,5)P3 are maintained at undetectable levels but accumulate transiently following stimulation (Auger and Cantley, 1991). The phosphorylation state of phosphoinositides is regulated by lipid kinases and phosphatases, including phosphoinositide 3-kinases, which have been implicated in cell survival and insulin action, and the PTEN tumor suppressor, which functions as a phosphoinositide 3-phosphatase (Maehama and Dixon, 1998; Cantley and Neel, 1999; Maehama et al, 2001; Vanhaesebroeck et al, 2001). PtdIns(4,5)P2 further serves as a substrate for the phospholipase C family of enzymes that generate diacyl glycerol, a potent activator of protein kinase C, and inositol trisphosphate, which stimulates Ca2+ release from internal stores (Tyers et al, 1988; Cifuentes et al, 1993).
As one of the largest domain families, with several hundred members in the human genome, pleckstrin homology (PH) domains occur frequently in modular signaling proteins (Lemmon and Ferguson, 2000; Cozier et al, 2004). Despite high sequence variability, PH domains preserve a common core structure consisting of a partly open β barrel capped at one end by a C-terminal α-helix (DiNitto et al, 2003). Many PH domains exhibit a dipolar electrostatic potential, with a basic surface at the open end of the β barrel, and are capable of interacting with membranes containing phosphoinositides (Macias et al, 1994; Kavran et al, 1998; Blomberg et al, 1999a, 1999b). In some cases, the interactions are best characterized as nonspecific, given that the partition coefficients correlate with the number of phosphate groups rather than the stereochemistry of phosphate substitution. However, a subset of PH domains have been shown to bind phosphoinositide head groups with sufficient affinity and specificity to provide the driving force for membrane localization (Rameh et al, 1997; Isakoff et al, 1998; Kavran et al, 1998). These include the PtdIns(4,5)P2-specific phospholipase Cδ (PLCδ) PH domain, the PtdIns(3,4,5)P3-specific PH domains of Bruton's tyrosine kinase (Btk) and general receptor for phosphoinositides (Grp1), as well as the PH domains of dual adaptor for phosphotyrosine and 3-phosphoinositides (Dapp1) and protein kinase B (PKB/Akt), which bind PtdIns(3,4)P2 and PtdIns(3,4,5)P3 with comparable affinity yet still discriminate against PtdIns(4,5)P2 (Fukuda et al, 1996; Salim et al, 1996; Frech et al, 1997; Klarlund et al, 1997; Rameh et al, 1997; Kavran et al, 1998; Venkateswarlu et al, 1998; Dowler et al, 1999).
Structures of PH domains with the head groups of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 have provided insight into the determinants of phosphoinositide recognition (DiNitto et al, 2003). A signature motif comprised of conserved basic residues from the β1 and β2 strands mediates critical interactions with the 1-, 4-, and 5-phosphates in the PLCδ PH domain and with the 1-, 3-, and 4-phosphates in the Btk, Grp1, and Dapp1 PH domains (Ferguson et al, 1995, 2000; Baraldi et al, 1999; Lietzke et al, 2000). The promiscuity of the Dapp1 PH domain for PtdIns(3,4)P2 and PtdIns(3,4,5)P3 can be explained by the absence of interactions with the 5-phosphate, which occupies a solvent exposed location (Ferguson et al, 2000). The high specificity of the Grp1 and Btk PH domains for PtdIns(3,4,5)P3 reflects additional interactions with three ‘specificity determining regions' (SDRs), also known as ‘variable loops' (VLs), corresponding to the β1/β2 loop (SDR1 or VL1), β3/β4 loop (SDR2 or VL2), and β6/β7 loop (SDR3 or VL3) (Baraldi et al, 1999; Ferguson et al, 2000; Lietzke et al, 2000). Despite a nearly identical head group orientation and an equivalent network of interactions with the signature residues, the Grp1 and Btk PH domains achieve selectivity for PtdIns(3,4,5)P3 through different structural mechanisms. In the Btk PH domain, the β1/β2 loop is sufficiently large to form a binding pocket for the 5-phosphate and contribute to a binding pocket for the 4-phosphate (Baraldi et al, 1999). In contrast, the six-residue β1/β2 loop in the Grp1 PH domain is too short to form a complete 5-phosphate binding pocket. However, a 20-residue insertion in the β6/β7 loop adopts a β hairpin structure that mediates interactions with both the 4- and 5-phosphates (Ferguson et al, 2000; Lietzke et al, 2000).
The highly homologous proteins Grp1, ARNO (Arf nucleotide binding site opener), and Cytohesin-1 define a functionally related family with a modular architecture consisting of an N-terminal heptad repeat, a Sec7 domain with exchange activity for Arf GTPases, a PH domain, and a C-terminal polybasic sequence. Splice variants of Grp1, ARNO, and Cytohesin-1 give rise to full-length proteins that differ only in the number of glycine residues at the N-terminus of the β1/β2 loop in the PH domain (Ogasawara et al, 2000). Whereas the ‘diglycine' (2G) variants exhibit strong selectivity for PtdIns(3,4,5)P3, the ‘triglycine' (3G) variants bind PtdIns(4,5)P2 and PtdIns(3,4,5)P3 with comparable affinity (Klarlund et al, 2000). The present study addresses the structural basis for these observations and provides general insights into the determinants of phosphoinositide recognition by PH domains.
Results
Structural basis for promiscuity in the 3G splice variants
The 3G ARNO PH domain was crystallized in complex with Ins(1,4,5)P3 and Ins(1,3,4,5)P4, which correspond to the head groups of PtdIns(4,5)P2 and PtdIns(3,4,5)P3, respectively. Crystals were also obtained for the 3G Grp1 PH domain in the absence of a ligand. The structures were solved by molecular replacement using a search model derived from the 2G Grp1 PH domain (Table I and Figure 1). Two sulfate ions from the crystallization medium occupy the phosphoinositide binding pocket in the structure of the 3G Grp1 PH domain. The significance of the latter observation is considered below.
Table 1.
Structure determination and refinement
| PH domain | 3G ARNO | 3G ARNO | 3G Grp1 |
|---|---|---|---|
| Ligand | Ins(1,4,5)P3 | Ins(1,3,4,5)P4 | Unliganded |
| Resolution range (Å) | 20–1.8 | 20–2.3 | 20–1.8 |
| Rsyma,b | 0.037 (0.242) | 0.063 (0.339) | 0.066 (0.340) |
| 〈I/σ〉a | 27.0 (7.0) | 25.1 (3.1) | 20.1 (4.5) |
| Redundancy | 6 | 7 | 5 |
| # Reflections | 12 727 | 5239 | 14 899 |
| Completeness (%)a | 99.8 (99.5) | 99.0 (99.2) | 98.8 (96.9) |
| R factor | 0.229 | 0.231 | 0.218 |
| Rfreec | 0.262 | 0.288 | 0.240 |
| r.m.s. deviations | |||
| Bond angle (deg) | 1.2 | 1.4 | 1.1 |
| Bond length (Å) | 0.009 | 0.008 | 0.009 |
| Mean B factor | |||
| Protein (Å2) | 28 | 57 | 25 |
| Head group (Å2) | 27 | 70 | |
| Sulfate ions 1/2 (Å2) | 31/50 | ||
| aValues within parentheses represent the highest resolution shell. | |||
| bRsym=∑h∑j∣Ij(h)−〈(h)〉∣/∑h∑j Ij(h). | |||
| cR-value for a 5% subset of reflections selected at random and omitted from refinement. | |||
Figure 1.
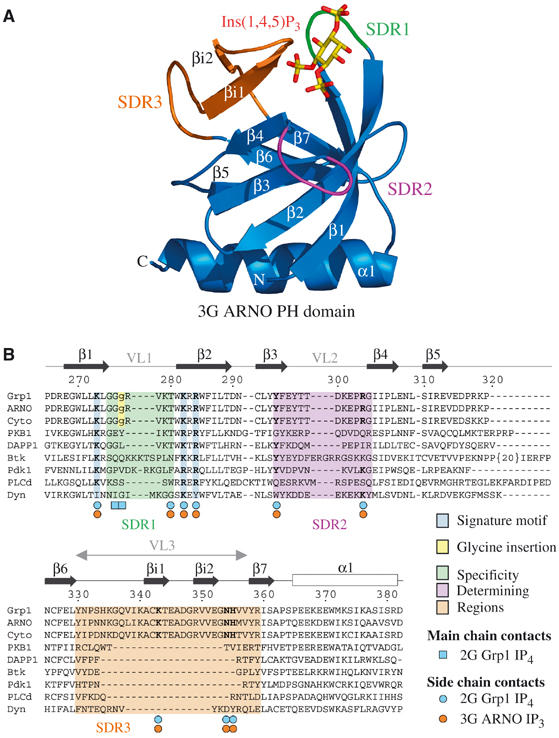
Structure of the 3G ARNO and 3G Grp1 PH domains. (A) Ribbon representation of the 3G ARNO PH domain bound to Ins(1,4,5)P3. Regions involved in specificity determination are highlighted in green (β1/β2 loop), magenta (β3/β4 loop), and orange (β6/β7 loop). Ins(1,4,5)P3 is shown in yellow (carbon and phosphorus atoms) and red (oxygen atoms). (B) Structure-based alignment of PH domain sequences with residue numbers and secondary structure corresponding to 2G Grp1.
The 3G ARNO PH domain binds Ins(1,3,4,5)P4 in an orientation indistinguishable from that of the 2G Grp1 PH domain (Figure 2). This orientation preserves interactions with the 3-, 4-, and 5-phosphates mediated by the signature residues as well as nonconserved residues from the β3/β4 and β6/β7 loops. In contrast, hydrogen bonds between the 5-phosphate and backbone NH groups at the N-terminus of the β1/β2 loop are disrupted in the 3G ARNO PH domain as are hydrogen bonds between the 1-phosphate and the side-chain hydroxyl of a threonine residue at the C-terminus of the β1/β2 loop. The loss of backbone interactions with the 1- and 5-phosphates reflects a local structural perturbation induced by the insertion of a glycine residue into the β1/β2 loop (Figure 1B) and accounts for the lower affinity of the 3G variants for PtdIns(3,4,5)P3. Thus, despite significant conformational differences in the β1/β2 loop, the mode of PtdIns(3,4,5)P3 head group binding resembles that observed for other PH domains (Baraldi et al, 1999; Ferguson et al, 2000; Lietzke et al, 2000; Thomas et al, 2002).
Figure 2.
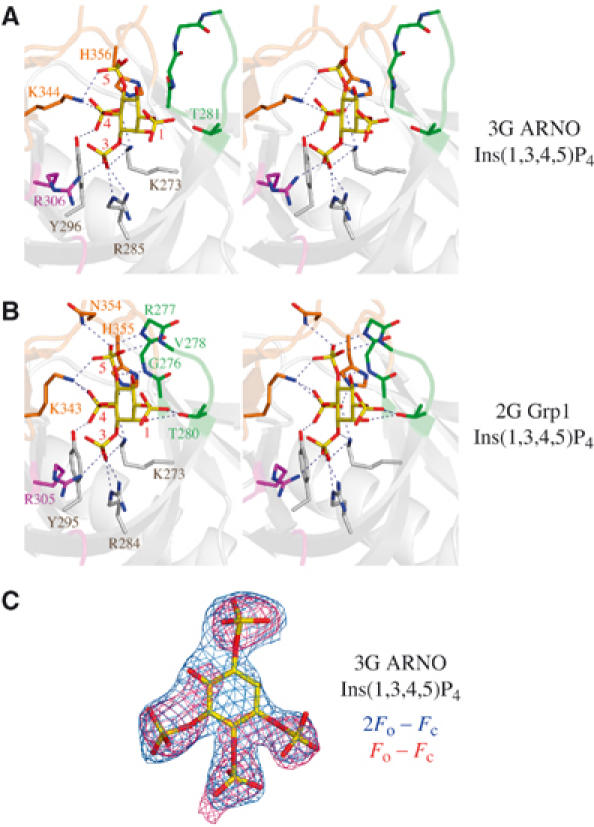
Mode of PtdIns(3,4,5)P3 binding by the 3G ARNO PH domain. (A) Stereo-pair depicting direct interactions between the 3G ARNO PH domain and Ins(1,3,4,5)P4. Residues from the signature motif are colored blue, whereas residues from the β1/β2 loop, β3/β4 loop, and β6/β7 loop are colored green, magenta, and orange, respectively. To simplify comparison, Grp1 residue numbering is used for ARNO residues in all figures. The actual ARNO residue numbers can be obtained by subtracting 5. (B) Stereo-pair depicting direct interactions between the 2G Grp1 PH domain and Ins(1,3,4,5)P4 (PDB ID code: 1FGY). Residues are colored and numbered as in (A). (C) Ins(1,3,4,5)P4 from the refined complex with the 3G ARNO PH domain is overlaid with the corresponding electron density from σA weighted Fo–Fc (red) and 2Fo–Fc (blue) maps contoured at 2.5σ and 1.0σ, respectively. The maps were generated after multiple rounds of simulated annealing refinement of the protein model with an initial temperature of 5000 K, but prior to inclusion of the head group.
Although the loss of interactions with the 5-phosphate is consistent with the decreased affinity of the 3G variants for PtdIns(3,4,5)P3, it does not explain the increased affinity for PtdIns(4,5)P2 (Klarlund et al, 2000). If both head groups are bound in a similar orientation, the absence of the 3-phosphate coupled with the loss of interactions with the 5-phosphate should result in significantly lower affinity for PtdIns(4,5)P2. This conundrum is resolved by the structure of the 3G ARNO PH domain in complex with Ins(1,4,5)P3, which reveals a novel head group orientation distinct from either the common orientation for Ins(1,3,4,5)P4 or the ‘flipped' orientation observed for Ins(1,4,5)P3 bound to the PLCδ PH domain (Ferguson et al, 1995; Baraldi et al, 1999; Ferguson et al, 2000; Lietzke et al, 2000; Thomas et al, 2002). Compared with Ins(1,3,4,5)P4, the inositol ring of Ins(1,4,5)P3 in the ARNO PH domain is rotated by ∼60° such that the 4-phosphate occupies the highly basic ‘3-phosphate' pocket while the 5-phosphate is located near the ‘4-phosphate' pocket (Figure 3). Consequently, the detailed network of hydrogen bonding/electrostatic interactions differs markedly, although the residues involved in head group recognition are largely the same. Despite these obvious differences, the general mode of interaction with the 4- and 5-phosphates is remarkably well preserved, provided the identity of the individual phosphate groups and orientation of the pseudosymmetrical inositol ring are neglected (see below). Furthermore, in all of the observed binding modes, the terminal oxygens of the 1-phosphate group extend from the pocket such that there is room to accommodate a diacyl glycerol moiety.
Figure 3.
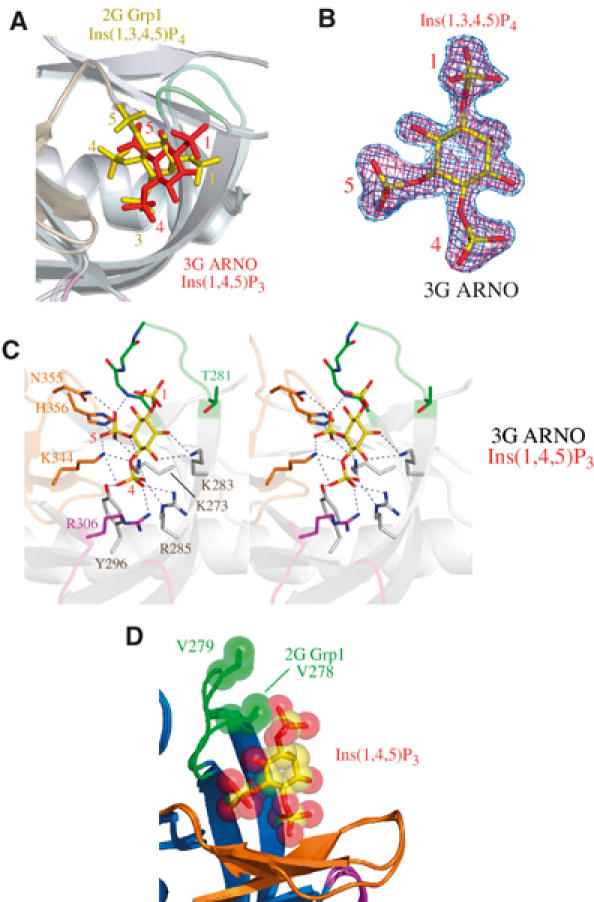
Novel mode of PtdIns(4,5)P2 head group binding by the 3G ARNO PH domain. (A) Comparison of head group binding modes for the 2G Grp1 and 3G ARNO PH domains following superposition of Cα atoms. Ins(1,3,4,5)P4 (2G Grp1) is colored yellow, while Ins(1,4,5)P3 (3G ARNO) is colored red. The 3G ARNO and 2G Grp1 structures are rendered with the same color scheme with the exception that the 3G ARNO PH domain is semitransparent. (B) Ins(1,4,5)P3 from the refined complex with the 3G ARNO PH domain is overlaid with the corresponding density from σA weighted Fo–Fc and 2Fo–Fc maps contoured at 3.0σ and 1.0σ, respectively. The maps were generated as in Figure 2C. (C) Stereo-pair depicting direct interactions between the 3G ARNO PH domain and Ins(1,4,5)P3. Residues are colored and numbered as in Figure 2A. (D) Comparison of the β1/β2 loops of the 3G ARNO and 2G Grp1 PH domains following superposition of Cα atoms. Also depicted is the Ins(1,4,5)P3 ligand from the complex with the 3G ARNO PH domain. The head group and a valine residue in the β1/β2 loop (Val 278 in 2G Grp1 and Val 279 in 3G ARNO) are rendered as spheres.
Superposition of the Ins(1,4,5)P3-bound 3G ARNO PH domain with the Ins(1,3,4,5)P4-bound 2G Grp1 PH domain suggests a mechanism whereby the 2G variants may selectively hinder PtdIns(4,5)P2 binding (Figure 3D). As a consequence of the rotated binding mode for the head group of PtdIns(4,5)P2, the 1-phosphate is displaced towards the β1/β2 loop. This displacement is readily accommodated by the longer β1/β2 loop in the 3G ARNO PH domain. However, the nonpolar side chain of Val 278 in the 2G Grp1 PH domain occupies a spatial location that is compatible with the PtdIns(3,4,5)P3 head group binding mode, but would result in a steric clash with the 1-phosphate of PtdIns(4,5)P2. In the structure of the unliganded 2G Grp1 PH domain, residues 278VKT280 at the C-terminus of the β1/β2 loop adopt a similar conformation, albeit with higher B factors indicative of increased mobility (Lietzke et al, 2000). In the 3G ARNO structures, a crystal contact involving Arg 278 influences the conformation of the β1/β2 loop, which is likely to be mobile in solution. In contrast, the β1/β2 loop interacts directly with the head group in the 2G Grp1 structure with Ins(1,3,4,5)P4 and is consequently well ordered despite the absence of crystal contacts. These observations could explain the small increase in PtdIns(4,5)P2 affinity for the 3G variant of Grp1 (Klarlund et al, 2000).
Determinants of phosphoinositide recognition in the 2G and 3G variants
To gain further insight into the relative contributions of the signature motif and SDRs, residues 277RVKT280 in the β1/β2 loop, as well as the other residues that mediate head group contacts, were replaced with alanine and/or glycine in both 2G Grp1 and 3G ARNO. Wild-type and mutant PH domains were expressed as GST fusions and purified over glutathione sepharose. All of the mutants expressed in a soluble form at wild-type levels with no indication of instability or degradation. The ability of the wild-type and mutant proteins to bind the head groups and soluble dioctanoyl forms of PtdIns(4,5)P2 and/or PtdIns(3,4,5)P3 was analyzed by isothermal titration microcalorimetry (ITC) (Figure 4 and Supplementary Tables I and II). The wild-type 2G Grp1 PH domain fused to GST binds to the head groups of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 with dissociation constants (Kd) of 6 μM and 34 nM, respectively, and molar enthalpies (ΔH) of −8 and −21 kcal/mol, respectively. These values agree well with those reported previously for the untagged 2G Grp1 PH domain (Kavran et al, 1998).
Figure 4.
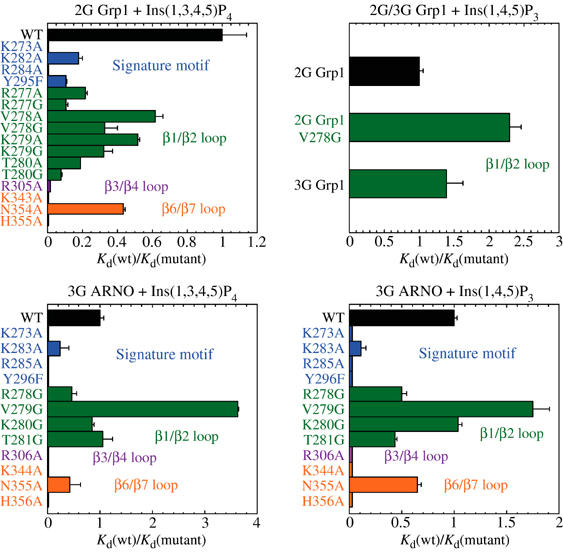
Mutational analysis of Grp1 and ARNO mutants. Relative Kd values for the binding of Ins(1,3,4,5)P4 and Ins(1,4,5)P3 to wild-type (WT) and mutants of the 2G Grp1, 3G Grp1, and 3G ARNO PH domains fused to GST. Kd and ΔH values for head group and dioctanoyl–phosphoinositide binding to the wild-type and mutant PH domains (listed in Supplementary Tables I and II) were determined at 25°C using isothermal titration calorimetry. The effects of mutations within the signature motif are indicated in blue, whereas mutations within the β1/β2, β3/β4, and β6/β7 loops are indicated in green, magenta, and orange, respectively.
PH domains known to bind polyphosphoinositides with high affinity share a signature motif of conserved basic residues, K–Xm–(K/R)–X–R, where the initial lysine residue occupies the penultimate position of the β1 strand, whereas the (K/R)–X–R sequence spans positions 2–4 of the β2 strand (Isakoff et al, 1998; Lietzke et al, 2000). Alanine substitution of the lysine residue at the N-terminus of β2 (Lys 282) exhibits a five-fold decrease in Kd, whereas alanine mutants of the terminal lysine (Lys 273) and arginine (Arg 284) of the signature motif show no detectable binding. Substitution of the conserved tyrosine residue near the C-terminus of the β3 strand with phenylalanine leads to an order of magnitude decrease in affinity. The severity of the mutations correlates well with the stereochemistry and physiochemical properties of the interactions. For example, Lys 273 and Arg 284 mediate buried, multivalent, stereochemically optimal electrostatic/hydrogen bonding interactions with the 3- and 4-phosphate groups. On the other hand, Tyr 295 contributes a single buried hydrogen bonding interaction with the 4-phosphate, whereas the solvent-exposed side chain of Lys 282 contributes to the basic electrostatic potential near the 1-phosphate.
Following similar arguments reflecting the stereochemistry and physiochemical properties of the interactions observed in the crystal structures, it would be expected that nonconserved basic residues would also contribute significantly. As with the signature residues, alanine mutations in all three SDRs exhibit moderate to severe defects (Figure 4). No detectable binding to Ins(1,3,4,5)P4 was observed for the alanine mutants of Lys 343 and His 355 in the β6/β7 loop. Although detectable, the affinity of the alanine mutant of Arg 305 in the β3/β4 loop was diminished nearly 100-fold. In contrast, two- to six-fold effects were observed for alanine mutants in the β1/β2 loop, with the most significant effects resulting from alanine substitutions of Arg 277 and Thr 280. Arg 277 contributes to the positive electrostatic potential in the 5-phosphate binding pocket, whereas the polar side chain of Thr 280 engages in a bifurcated hydrogen bonding interaction with the 1-phosphate. Alanine mutants of Asn 354, which mediates a solvent-exposed hydrogen bonding interaction with the 5-phosphate, as well as residues that do not contact the head group (Val 278 and Lys 279) show relatively minor (<3-fold) effects. Finally, insertion of a glycine residue at the N-terminus of the β1/β2 loop to generate the 3G variant of Grp1 results in an order of magnitude decrease in affinity, consistent with the loss of backbone contacts with the 5-phosphate and/or the displacement of Arg 278 observed in the structure of the 3G variant of the ARNO PH domain in complex with Ins(1,3,4,5)P4. Thus, the affinity for the head group of PtdIns(3,4,5)P3 reflects contributions from all three specificity-determining regions in addition to the signature motif, consistent with the distribution of stereochemically specific electrostatic/hydrogen bonding interactions observed in the crystal structure.
The binding of the head groups and soluble dioctanoyl phosphoinositides to the 3G ARNO PH domain is sensitive to mutation of conserved residues in the signature motif as well as nonconserved residues in the VLs. In general, residues that mediate critical interactions with Ins(1,3,4,5)P4 in the 2G Grp1 PH domain also mediate critical interactions with both Ins(1,4,5)P3 and Ins(1,3,4,5)P4 in the 3G ARNO PH domain, with the exception of residues in the β1/β2 loop. In the 3G ARNO PH domain, glycine substitutions of Arg 278, Val 279, Lys 280, or Thr 281 result in small changes in head group affinity, consistent with the absence of direct hydrogen bonding interactions with these residues in the crystal structures. Thus, the β1/β2 loop contributes significantly to the affinity and specificity of the 2G Grp1 PH domain for the head group of PtdIns(3,4,5)P3 but relatively little to the interaction of the 3G ARNO PH domain with the head group of PtdIns(4,5)P2 or PtdIns(3,4,5)P3.
In addition to a local structural perturbation, it is possible that the glycine insertion in the 3G splice variants influences phosphoinositide affinity indirectly through increased flexibility of the β1/β2 loop. To assess the potential contribution of increased flexibility, residues in the β1/β2 loop of the 2G Grp1 PH domain were substituted with glycine. The glycine mutants exhibit defects that are consistently two- to three-fold more severe than the corresponding alanine substitutions when analyzed with respect to the affinity for Ins(1,3,4,5)P4. This effect can be attributed to the predictable increase in main-chain conformational entropy and the resulting penalty for ordering the β1/β2 loop in order to facilitate backbone contacts with the 5-phosphate. In contrast, the same glycine substitutions result in negligible or small increases in affinity for Ins(1,4,5)P3, with the most significant increase observed for the V278G mutant. These results are consistent with the head group orientations and interactions observed in the structures of the 2G Grp1 and 3G ARNO PH domains.
In general, the effects of the mutants observed for the dioctanoyl–phosphoinositides parallel those of the head group. Notably, however, the R277A substitution has little effect on affinity for dioctanoyl PIP3, despite a six-fold reduction in affinity for the head group. This discrepancy may reflect decreased accessibility of the head group in the context of the phosphoinositide. However, a caveat with the experiments involving dioctanoyl phosphoinositides is a tendency for the complexes to aggregate at concentrations in excess of 5 μM. Conversely, the head group complexes show no indication of aggregation. Consequently, the dissociation constants and thermodynamic parameters are better estimated for the head group than for the dioctanoyl phosphoinositides, particularly for the experiments with the 3G ARNO PH domain.
Despite weak homology and distinct head group orientations, superposition of the 3G ARNO and PLCδ PH domains reveals a fundamental relationship in the mode of interaction with Ins(1,4,5)P3. As shown in Figure 5A, the 4- and 5-phosphates in the complex with the 3G ARNO PH domain overlap the 5- and 4-phosphates, respectively, in the corresponding complex with the PLCδ PH domain, thereby preserving both the stereochemistry and electrostatic character of the interactions with conserved residues. Thus, the two binding modes can be regarded as pseudosymmetric, such that the orientation of Ins(1,4,5)P3 in the 3G ARNO PH domain can be approximately derived from that in the PLCδ PH domain by a 180° rotation about an axis bisecting the C1–C2 and C4–C5 bonds followed by 120° rotations about the C4–P and C5–P bonds to alternative rotomer conformations (Figure 5B). Furthermore, the location of the 4- and 5-phosphate groups closely approximates that of two inorganic sulfate ions or two inorganic phosphate ions in the unliganded structures of the 2G/3G Grp1 and Dapp1 PH domains, respectively (see, for example, Figure 5C).
Figure 5.
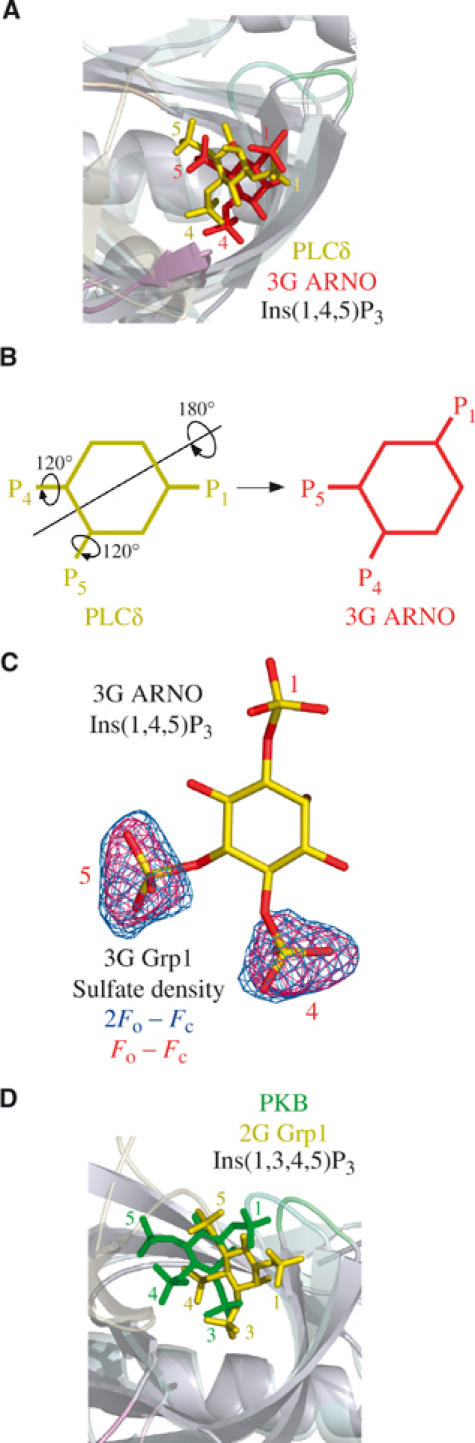
Comparison with other PH domain structures. (A) Overlay of the 3G ARNO PH domain (semitransparent) and PLCδ PH domain (PDB ID code: 1MAI), both in complex with Ins(1,4,5)P3, following superposition of Cα atoms. Ins(1,4,5)P3 is depicted in red (3G ARNO) and yellow (PLCδ). (B) Schematic diagram illustrating the approximate rigid body and torsion angle rotations that transform Ins(1,4,5)P3 from the orientation in the PLCδ PH domain to that in the 3G ARNO PH domain. (C) Overlay of Ins(1,4,5)P3 bound to the 3G ARNO PH domain with the electron density corresponding to inorganic sulfate ions from the unliganded 3G Grp1 PH domain following superposition of Cα atoms. The electron density is from σA weighted Fo–Fc and 2Fo–Fc maps contoured at 3.0σ and 1.2σ, respectively. The maps were generated as in Figure 2C. (D) Overlay of the 2G Grp1 PH domain (semitransparent) and the PKB PH domain (PDB ID code: 1H10), both in complex with Ins(1,3,4,5)P4, following superposition of Cα atoms. Ins(1,3,4,5)P4 is depicted in yellow (2G Grp1) and green (PKB).
Discussion
In both fetal and adult brain, 2G Grp1, 3G ARNO, and 3G Cytohesin-1 represent the most abundant variants (Ogasawara et al, 2000). Whereas the PH domains of the 2G variants bind selectively to PtdIns(3,4,5)P3 with sufficient affinity to drive the plasma membrane recruitment of the full-length proteins (Klarlund et al, 1997; Kavran et al, 1998; Venkateswarlu et al, 1998, 1999), the lower affinity of the promiscuous 3G variants, although necessary, is not sufficient (Klarlund et al, 2000). In this case, weak but essential determinants outside the PH domain contribute to the localization of the full-length proteins. These include the C-terminal polybasic region as well as unidentified determinants N-terminal to the PH domain (Nagel et al, 1998; Klarlund et al, 2000; Macia et al, 2000). With respect to the latter, the interaction of the Sec7 domain with Arf GTPases and/or the interaction of the N-terminal heptad repeat with other proteins could enhance membrane targeting (Nevrivy et al, 2000; Klarlund et al, 2001; Mansour et al, 2002; Tang et al, 2002; Venkateswarlu, 2003).
In this study, the 2G and 3G variants were shown to bind the head group of PtdIns(3,4,5)P3 in a common orientation that preserves interactions with both signature and variable residues, with the exception of the hydrogen bonds with the 1- and 5-phophates donated by residues in the β1/β2 loop, which are disrupted in the 3G variant as a consequence of a local structural perturbation resulting from the insertion of a glycine residue. The results of the mutational analysis support the conclusion that the large decrease in PtdIns(3,4,5)P3 affinity in the 3G variants is due primarily to the loss of contacts with the head group. Secondary effects, such as increased flexibility of the β1/β2 loop or changes in the local electrostatic environment resulting from displacement of basic residues in the β1/β2 loop, contribute to the observed differences, although to a lesser extent.
A key insight into the promiscuity of the 3G variants, as well as the differential effects of the glycine insertion on the affinity for PtdIns(4,5)P2 and PtdIns(3,4,5)P3, is provided by the observation that the head group of PtdIns(4,5)P2 binds in a novel orientation that is distinct from that of either the 2G variants or the PH domain of PLCδ (Ferguson et al, 1995). Remarkably, the same residues are critical for binding to both PtdIns(3,4,5)P3 and PtdIns(4,5)P2 even though the detailed network of interactions is quite different. The structural basis of the promiscuity in the 3G variants, which reflects alternative binding modes with similar affinities, differs fundamentally from that proposed for the PH domains of Dapp1 and PKB (Ferguson et al, 2000; Thomas et al, 2002). For the Dapp1 and PKB PH domains, which bind with equivalent affinity to PtdIns(3,4)P2 and PtdIns(3,4,5)P3, the structural basis for promiscuous recognition can be adequately explained in terms of the absence of potential contacts with the 5-phosphate rather than alternative binding modes.
PH domains that bind phosphoinositides with moderate to high affinity typically contain a signature motif that encodes conserved basic residues located at the C- and N-termini of the β1 and β2 strands, respectively (Rameh et al, 1997; Isakoff et al, 1998; Kavran et al, 1998). Crystallographic and mutational studies have highlighted the critical requirement for the conserved basic residues in this motif (DiNitto et al, 2003). The structural studies also reveal a number of additional interactions with basic and polar residues from the three VL regions that flank the binding site. In the present study, we have systematically assessed the relative contribution of the signature as well as variable residues that mediate interactions with the head groups of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in splice variants of the Grp1 and ARNO PH domains. Consistent with earlier work, the first and last basic residues of the signature motif are essential for phosphoinositide binding (Salim et al, 1996; Yagisawa et al, 1998; Klarlund et al, 2000). In addition, three nonconserved basic residues from the variable β3/β4 and β6/β7 loops were found to be critical determinants. Moreover, the effects of the alanine and glycine substitutions are strongly correlated with the structural observations. For example, the most critical determinants correspond to basic residues that mediate solvent excluded, electrostatically complementary, hydrogen bonding interactions with either the 3-, 4-, or 5-phosphate groups. Likewise, polar residues that form buried hydrogen bonds with the head group also contribute significantly. Conversely, basic or polar residues that mediate exposed hydrogen bonding and/or nonstereospecific electrostatic interactions contribute little to head group affinity.
A significant proportion (30–40%) of the mammalian PH domains in the PFAM and SMART databases conserve the first and third basic residues of the signature motif that were found to be essential for phosphoinositide binding in the present study and in earlier work on other PH domains (Fukuda et al, 1996; Salim et al, 1996; Rameh et al, 1997; Isakoff et al, 1998; Yagisawa et al, 1998). The side chains of these residues extend from the structurally conserved core to form the most buried and positively charged region of the head group binding site. These residues further contribute the majority of the interactions with the 3-phosphate in the structures of PH domains bound to the head group of PtdIns(3,4,5)P3 and the 4- or 5-phosphate of the PtdIns(4,5)P2 head group in the 3G ARNO and PLCδ PH domains, respectively (Baraldi et al, 1999; Ferguson et al, 2000; Lietzke et al, 2000; Thomas et al, 2002). Despite critical contributions from nonconserved residues in the SDRs and large differences in the orientation of the head group, fundamental similarities are evident in the various modes of polyphosphoinositide recognition, which further resemble the binding of inorganic sulfate or phosphate ions to the unliganded Grp1 and Dapp1 PH domains. These observations support a generalized view of phosphoinositide recognition in which the number of potential binding modes is limited to those that dispose adjacent phosphate groups so as to mediate stereochemically and electrostatically analogous interactions with conserved basic residues from the signature motif (Figure 6). Favorable as well as unfavorable interactions with the variable SDRs determine the relative affinity of each potential binding mode and, consequently, the specificity for phosphoinositides.
Figure 6.
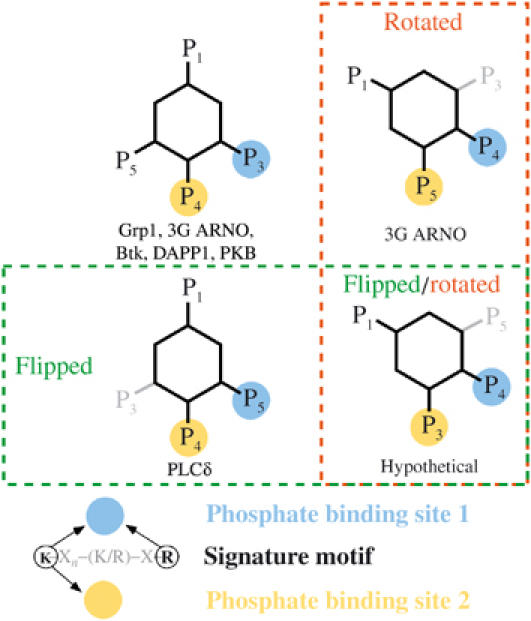
Observed and hypothetical modes of polyphosphoinositide recognition. Schematic diagram depicting potential modes of polyphosphoinositide recognition by PH domains. The orientation of the inositol ring and disposition of phosphate groups are categorized with respect to the location of inorganic sulfate/phosphate ions observed in the unliganded structures of the 2G Grp1 and Dapp1 PH domains. Blue circles represent the phosphate binding site corresponding to the most buried and electropositive region of the head group binding site formed primarily by the N-terminal lysine and C-terminal arginine residue of the signature motif. Yellow circles represent the phosphate binding site comprised of the N-terminal lysine residue from the signature motif as well as basic and/or polar residues from the variable SDRs. In this classification of binding modes, rotational and/or translational displacements of the inositol ring that are not sufficiently large to alter the network of interactions with conserved residues are neglected as are the specific rotomer conformations of the phosphate groups.
A recent genome-wide analysis provides insight into the lipid binding properties and localization of yeast PH domains (Yu et al, 2004). Of the 36 yeast PH domains, only eight were found to bind phosphoinositides, with the majority exhibiting little or no specificity for particular phosphoinositide head groups. Interestingly, all eight of these PH domains conserve the first and third basic residues of the signature motif, underscoring the role of the core basic residues as general determinants of phosphoinositide binding but not specificity. Thus, for all of these cases, degenerate head group binding modes such as those categorized in Figure 6 could explain the predominately nonspecific interaction with polyphosphoinositides and, in principle, monophosphoinositides. Together, these observations support the conclusion that selective phosphoinositide binding represents a rare property of PH domains acquired at a late stage in the evolution of eukaryotic organisms through accessory interactions with the VLs that increase the affinity for a particular binding mode while selecting against alternate modes.
A characteristic property of PH domains that bind phosphoinositides, with either low or high affinity, is a strong positive electrostatic potential at the open end of the central β barrel. Furthermore, the β1/β2 loops of 3-phosphoinositide-specific PH domains contain a higher content of basic residues (Ferguson et al, 2000). In the case of the 2G and 3G splice variants, alanine substitutions involving the basic residues in the β1/β2 loop have relatively modest effects on the affinity for phosphoinositides, consistent with the absence of direct interactions with the head group in the crystal structures. Although it is possible that electrostatic interactions with head group would be enhanced in the physiological environment of the plasma membrane, the proximal location of the β1/β2 loop with respect to the 1-phosphate suggests that residues in the β1/β2 loop are likely to penetrate into the interfacial region of the bilayer. The contribution of nonspecific interactions mediated by nonpolar and basic residues in loop regions flanking phosphoinositide binding sites has been well documented for other membrane targeting modules, including FYVE and PX domains (Dumas et al, 2001; Kutateladze and Overduin, 2001; Karathanassis et al, 2002; Diraviyam et al, 2003; Stahelin et al, 2003; Hayakawa et al, 2004; Kutateladze et al, 2004). Basic residues outside the phosphoinositide binding site have also been implicated in the membrane targeting of PLCδ as well as the weak nonspecific interaction of many PH domains with acidic membranes (Yagisawa et al, 1998; Diraviyam et al, 2003). These observations suggest that the bias in the composition of the β1/β2 loop, in favor of basic and nonpolar residues, is likely to reflect nonspecific interactions with the plasma membrane. The extent to which such interactions contribute to membrane targeting in the 2G and 3G splice variants remains to be determined.
Materials and methods
Materials and constructs
Phosphoinositides and head groups were obtained from Cell Signals. Constructs corresponding to residues 261–385 of the 3G Grp1 PH domain and 260–378 of the 3G ARNO PH domain were amplified using Vent polymerase and subcloned into a modified pET15b vector using BamHI/SalI restriction sites. The resulting constructs contain an N-terminal 6 × His tag (MGHHHHHHGS). Site-specific mutations were generated using the Quick Change mutagenesis kit (Stratagene). Wild-type and mutant constructs were sequenced from 3′ and 5′ directions.
Expression and purification
BL21(DE3) cells transformed with modified pET15b plasmids containing the 3G Grp1 or 3G ARNO PH domain constructs were grown at 22°C to an OD600 of 0.3–0.4 and induced with 50 μM IPTG for 16 h. For purification, the cells were suspended in buffer (50 mM Tris, pH 8.0, 0.1 M NaCl, 0.1% β-mercaptoethanol, 0.1 mM PMSF, 1 mg/ml lysozyme) and disrupted by sonication. Triton X-100 was added to a final concentration of 0.5% and the crude cell lysates were centrifuged at 35 000 g for 40 min. The supernatants were loaded onto an NiNTA–agarose column (Qiagen). After washing with 10 column volumes of buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 10 mM imidazole, 0.1% mercaptoethanol), the 6x His fusion proteins were eluted with a gradient of 10–150 mM imidazole (10–500 mM imidazole for 10x His fusions). Subsequent ion exchange chromatography on Source Q (Pharmacia) followed by gel filtration chromatography over Superdex-75 (Pharmacia) resulted in preparations that were >99% pure as judged by SDS–PAGE.
For ITC experiments, wild-type and mutant PH domains were expressed as GST fusions in pGEX-6p1. BL21(DE3)-RIL cells expressing the GST-PH domains were suspended in buffer (50 mM Na/K phosphate, pH 8.0, 150 mM NaCl, 0.1% β-mercaptoethanol, 0.1 mM PMSF, 1 mg/ml lysozyme, 10 μg/ml DNase I) and lysed by sonication. Triton X-100 was then added to 0.5% followed by centrifigation at 32 000 g for 45 min. Clarified lysates were added to glutathione–sepharose beads and mutated for 1 h at 4°C. The beads were washed extensively with buffer and the fusion proteins were eluted with 10 mM reduced glutathione. The GST fusion proteins were >95% pure as determined by SDS–PAGE.
Crystallization, data collection, and refinement
Crystals of the 3G ARNO PH domain bound to Ins(1,4,5)P3 were grown at 20°C in microseeded hanging drops containing 10 mg/ml of the complex, 0.5–2% PEG-4000, 50 mM HEPES, pH 7.0, 10% glycerol, and a 1.2-fold stoichiometric excess of Ins(1,4,5)P3. The crystals are in the primitive orthorhombic space group P212121, with cell constants a=43.0 Å, b=56.4 Å, and c=57.1 Å. Crystals were soaked at 20°C in a cryoprotectant-stabilizer solution (10% PEG-4000, 10% MPD, 10% glycerol, 50 mM HEPES, pH 7.0) prior to flash freezing in a liquid nitrogen cryostream at 100 K. Crystals of the 3G ARNO PH domain bound to Ins(1,3,4,5)P4 were grown at 20°C in microseeded hanging drops containing 1.5–3.0% PEG-4000, 50 mM sodium MES, pH 6.0, and 10% glycerol. The crystals are in the primitive orthorhombic space group P212121, with cell constants a=41.9 Å, b=56.0 Å, and c=59.6 Å. Crystals of the unliganded 3G Grp1 PH domain were obtained at 20°C in microseeded hanging drops containing 10 mg/ml protein, 6–7% PEG-4000, 50 mM sodium acetate, pH 4.2, 0.1 M Li2SO4, and 10% glycerol. The crystals are in the space group P21212 with unit-cell dimensions of a=40.8 Å, b=107.5 Å, and c=37.4 Å. Diffraction data were collected on a Rigaku RUH3R generator using Mar 30 cm (3G Grp1 and 3G ARNO-Ins(1,4,5)P3) or R-axis IV (3G ARNO-Ins(1,3,4,5)P4) image plate detectors, both equipped with Osmic mirrors, processed with DENZO, and scaled with Scalepack (Otwinowski and Minor, 1994). The structures were solved by molecular replacement with AmoRe (Collaborative Computational Project Number 4, 1994). The initial models were improved by iterative refinement using Arp/wArp and Refmac5 (3G Grp1 and 3G ARNO-Ins(1,4,5)P3) or CNS (3G ARNO-Ins(1,3,4,5)P4) combined with rebuilding in O (Jones et al, 1991; Collaborative Computational Project Number 4, 1994; Brunger et al, 1998). Structural figures were rendered with PyMOL (DeLano, 2002).
Isothermal titration microcalorimetry
ITC experiments were performed at 25°C using a VP-ITC isothermal titration calorimeter (Microcal). GST fusions of the Grp1 and ARNO PH domains were dialyzed against 50 mM MOPS (pH 7.5), and 100 mM NaCl. Protein concentrations were determined from the absorbance at 280 nm using extinction coefficients derived from amino-acid analysis. All solutions were degassed prior to the experiment. The GST fusions at a concentration of 5 μM (Grp1) or 20 μM (ARNO) were titrated with 100 μM (Grp1) or 400 μM (ARNO) head group or diC8 phosphoinositide dissolved in the same buffer as the protein. Heats of dilution obtained from the control titrations into buffer were incorporated into the analysis. Raw data were corrected for baseline drift and the total heat released during each injection (ΔQi) determined by integrating over the injection period. The resulting data were fit with a 1:1 binding model:
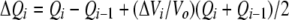 |
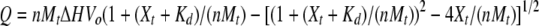 |
where Kd is the dissociation constant, n is the number of binding sites, ΔH is the molar enthalpy of binding, ΔHm is the molar enthalpy of mixing, and Vo is the volume of the sample chamber. Mt and Xt represent the bulk concentration of the macromolecule and ligand, respectively, following correction for dilution:
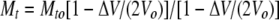 |
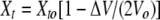 |
where ΔV is the cumulative injection volume and Mto and Xto represent the uncorrected bulk concentrations. To improve the accuracy of parameter estimation, the conventional χ2 merit function was replaced by the generalized maximum-likelihood function:
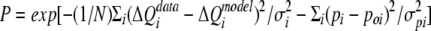 |
where N is the number of injections, σi is the uncertainty in the data, pi is the value of the ith parameter, poi is the expected parameter value, and σpi is the uncertainty in the estimate of poi. Kd and ΔH were treated as freely adjustable parameters, whereas n, ΔHm, Mto, and Xto were restrained near their estimated values by the maximum-likelihood function. Agreement between the model and data was improved by minimizing the negative logarithm of the maximum-likelihood function using the Marquardt and Simplex algorithms. Systematic analysis of model data sets indicates that the generalized maximum-likelihood approach, as implemented here, reduces the systematic error in estimates of Kd and ΔH compared with the conventional χ2 parameter estimation.
PDB accession codes
Coordinates for the unliganded 3G Grp1 PH domain (PDB ID code: 1U2B) and the 3G ARNO PH domain in complex with Ins(1,4,5)P3 (PDB ID code: 1U29) and Ins(1,3,4,5)P4 (PDB ID code: 1U27) have been deposited.
Supplementary Material
Supplementary Tables I and II
Acknowledgments
This work was supported by a grant from the National Institutes of Health (DK 60564).
References
- Auger KR, Cantley LC (1991) Novel polyphosphoinositides in cell growth and activation. Cancer Cells 3: 263–270 [PubMed] [Google Scholar]
- Baraldi E, Carugo KD, Hyvonen M, Surdo PL, Riley AM, Potter BV, O'Brien R, Ladbury JE, Saraste M (1999) Structure of the PH domain from Bruton's tyrosine kinase in complex with inositol 1,3,4,5-tetrakisphosphate. Struct Fold Des 7: 449–460 [DOI] [PubMed] [Google Scholar]
- Blomberg N, Baraldi E, Nilges M, Saraste M (1999a) The PH superfold: a structural scaffold for multiple functions. Trends Biochem Sci 24: 441–445 [DOI] [PubMed] [Google Scholar]
- Blomberg N, Gabdoulline RR, Nilges M, Wade RC (1999b) Classification of protein sequences by homology modeling and quantitative analysis of electrostatic similarity. Proteins 37: 379–387 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Cantley LC, Neel BG (1999) New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA 96: 4240–4245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes ME, Honkanen L, Rebecchi MJ (1993) Proteolytic fragments of phosphoinositide-specific phospholipase C-delta 1. Catalytic and membrane binding properties. J Biol Chem 268: 11586–11593 [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4, C (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Corvera S, Czech MP (1998) Direct targets of phosphoinositide 3-kinase products in membrane traffic and signal transduction. Trends Cell Biol 8: 442–446 [DOI] [PubMed] [Google Scholar]
- Cozier GE, Carlton J, Bouyoucef D, Cullen PJ (2004) Membrane targeting by pleckstrin homology domains. Curr Top Microbiol Immunol 282: 49–88 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA. http://www.pymol.org [Google Scholar]
- DiNitto JP, Cronin TC, Lambright DG (2003) Membrane recognition and targeting by lipid-binding domains. Sci STKE 2003, Dec 16; 2003(213):re16 [DOI] [PubMed]
- Diraviyam K, Stahelin RV, Cho W, Murray D (2003) Computer modeling of the membrane interaction of FYVE domains. J Mol Biol 328: 721–736 [DOI] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Downes CP, Alessi DR (1999) DAPP1: a dual adaptor for phosphotyrosine and 3-phosphoinositides. Biochem J 342 (Part 1): 7–12 [PMC free article] [PubMed] [Google Scholar]
- Dumas JJ, Merithew E, Sudharshan E, Rajamani D, Hayes S, Lawe D, Corvera S, Lambright DG (2001) Multivalent endosome targeting by homodimeric EEA1. Mol Cell 8: 947–958 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA (2000) Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell 6: 373–384 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1995) Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell 83: 1037–1046 [DOI] [PubMed] [Google Scholar]
- Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA (1997) High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J Biol Chem 272: 8474–8481 [DOI] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC (1998) Phosphoinositide kinases. Annu Rev Biochem 67: 481–507 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kojima T, Kabayama H, Mikoshiba K (1996) Mutation of the pleckstrin homology domain of Bruton's tyrosine kinase in immunodeficiency impaired inositol 1,3,4,5-tetrakisphosphate binding capacity. J Biol Chem 271: 30303–30306 [DOI] [PubMed] [Google Scholar]
- Hayakawa A, Hayes SJ, Lawe DC, Sudharshan E, Tuft R, Fogarty K, Lambright D, Corvera S (2004) Structural basis for endosomal targeting by FYVE domains. J Biol Chem 279: 5958–5966 [DOI] [PubMed] [Google Scholar]
- Isakoff SJ, Cardozo T, Andreev J, Li Z, Ferguson KM, Abagyan R, Lemmon MA, Aronheim A, Skolnik EY (1998) Identification and analysis of PH domain-containing targets of phosphatidylinositol 3-kinase using a novel in vivo assay in yeast. EMBO J 17: 5374–5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Karathanassis D, Stahelin RV, Bravo J, Perisic O, Pacold CM, Cho W, Williams RL (2002) Binding of the PX domain of p47(phox) to phosphatidylinositol 3,4-bisphosphate and phosphatidic acid is masked by an intramolecular interaction. EMBO J 21: 5057–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA (1998) Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem 273: 30497–30508 [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP (1997) Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science 275: 1927–1930 [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Holik J, Chawla A, Park JG, Buxton J, Czech MP (2001) Signaling complexes of the FERM domain-containing protein GRSP1 bound to ARF exchange factor GRP1. J Biol Chem 276: 40065–40070 [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Tsiaras W, Holik JJ, Chawla A, Czech MP (2000) Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J Biol Chem 275: 32816–32821 [DOI] [PubMed] [Google Scholar]
- Kutateladze T, Overduin M (2001) Structural mechanism of endosome docking by the FYVE domain. Science 291: 1793–1796 [DOI] [PubMed] [Google Scholar]
- Kutateladze TG, Capelluto DG, Ferguson CG, Cheever ML, Kutateladze AG, Prestwich GD, Overduin M (2004) Multivalent mechanism of membrane insertion by the FYVE domain. J Biol Chem 279: 3050–3057 [DOI] [PubMed] [Google Scholar]
- Leevers SJ, Vanhaesebroeck B, Waterfield MD (1999) Signalling through phosphoinositide 3-kinases: the lipids take centre stage. Curr Opin Cell Biol 11: 219–225 [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM (2000) Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J 350 (Part 1): 1–18 [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS (2002) Pleckstrin homology domains and the cytoskeleton. FEBS Lett 513: 71–76 [DOI] [PubMed] [Google Scholar]
- Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG (2000) Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell 6: 385–394 [DOI] [PubMed] [Google Scholar]
- Macia E, Paris S, Chabre M (2000) Binding of the PH and polybasic C-terminal domains of ARNO to phosphoinositides and to acidic lipids. Biochemistry 39: 5893–5901 [DOI] [PubMed] [Google Scholar]
- Macias MJ, Musacchio A, Ponstingl H, Nilges M, Saraste M, Oschkinat H (1994) Structure of the pleckstrin homology domain from beta-spectrin. Nature 369: 675–677 [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 273: 13375–13378 [DOI] [PubMed] [Google Scholar]
- Maehama T, Taylor GS, Dixon JE (2001) PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem 70: 247–279 [DOI] [PubMed] [Google Scholar]
- Mansour M, Lee SY, Pohajdak B (2002) The N-terminal coiled coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with the scaffolding protein CASP. J Biol Chem 277: 32302–32309 [DOI] [PubMed] [Google Scholar]
- Nagel W, Schilcher P, Zeitlmann L, Kolanus W (1998) The PH domain and the polybasic c domain of cytohesin-1 cooperate specifically in plasma membrane association and cellular function. Mol Cell Biol 9: 1981–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevrivy DJ, Peterson VJ, Avram D, Ishmael JE, Hansen SG, Dowell P, Hruby DE, Dawson MI, Leid M (2000) Interaction of GRASP, a protein encoded by a novel retinoic acid-induced gene, with members of the cytohesin family of guanine nucleotide exchange factors. J Biol Chem 275: 16827–16836 [DOI] [PubMed] [Google Scholar]
- Ogasawara M, Kim SC, Adamik R, Togawa A, Ferrans VJ, Takeda K, Kirby M, Moss J, Vaughan M (2000) Similarities in function and gene structure of cytohesin-4 and cytohesin-1, guanine nucleotide-exchange proteins for ADP-ribosylation factors. J Biol Chem 275: 3221–3230 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1994) Processing of X-ray diffraction data in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Rameh LE, Arvidsson A, Carraway KL III, Couvillon AD, Rathbun G, Crompton A, VanRenterghem B, Czech MP, Ravichandran KS, Burakoff SJ, Wang DS, Chen CS, Cantley LC (1997) A comparative analysis of the phosphoinositide binding specificity of pleckstrin homology domains. J Biol Chem 272: 22059–22066 [DOI] [PubMed] [Google Scholar]
- Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CI, Driscoll PC, Waterfield MD, Panayotou G (1996) Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J 15: 6241–6250 [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Burian A, Bruzik KS, Murray D, Cho W (2003) Membrane binding mechanisms of the PX domains of NADPH oxidase p40phox and p47phox. J Biol Chem 278: 14469–14479 [DOI] [PubMed] [Google Scholar]
- Tang P, Cheng TP, Agnello D, Wu CY, Hissong BD, Watford WT, Ahn HJ, Galon J, Moss J, Vaughan M, O'Shea JJ, Gadina M (2002) Cybr, a cytokine-inducible protein that binds cytohesin-1 and regulates its activity. Proc Natl Acad Sci USA 99: 2625–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CC, Deak M, Alessi DR, van Aalten DM (2002) High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr Biol 12: 1256–1262 [DOI] [PubMed] [Google Scholar]
- Tyers M, Rachubinski RA, Stewart MI, Varrichio AM, Shorr RG, Haslam RJ, Harley CB (1988) Molecular cloning and expression of the major protein kinase C substrate of platelets. Nature 333: 470–473 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD (2001) Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem 70: 535–602 [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K (2003) Interaction protein for cytohesin exchange factors 1 (IPCEF1) binds cytohesin 2 and modifies its activity. J Biol Chem 278: 43460–43469 [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Gunn-Moore F, Tavare JM, Cullen PJ (1999) EGF-and NGF-stimulated translocation of cytohesin-1 to the plasma membrane of PC12 cells requires PI 3-kinase activation and a functional cytohesin-1 PH domain. J Cell Sci 112 (Part 12): 1957–1965 [DOI] [PubMed] [Google Scholar]
- Venkateswarlu K, Oatey PB, Tavare JM, Cullen PJ (1998) Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol 8: 463–466 [DOI] [PubMed] [Google Scholar]
- Yagisawa H, Sakuma K, Paterson HF, Cheung R, Allen V, Hirata H, Watanabe Y, Hirata M, Williams RL, Katan M (1998) Replacements of single basic amino acids in the pleckstrin homology domain of phospholipase C-delta1 alter the ligand binding, phospholipase activity, and interaction with the plasma membrane. J Biol Chem 273: 417–424 [DOI] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA (2004) Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13: 677–688 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables I and II


