Abstract
Sequencing the insertion sites of 8,865 Tn917 insertions in Enterococcus faecalis strain OG1RF identified a hot spot in the replication terminus region corresponding to 6% of the genome where 65% of the transposons had inserted. In E. faecalis, Tn917 preferentially inserted at a 29-bp consensus sequence centered on TATAA, a 5-bp sequence that is duplicated during insertion. The regional insertion site preference at the chromosome terminus was not observed in another low-G+C gram-positive bacterium, Listeria monocytogenes, although the consensus insertion sequence was the same. The 8,865 Tn917 insertion sites sequenced in E. faecalis corresponded to only ∼610 different open reading frames, far fewer than the predicted number of 2,400, assuming random insertion. There was no significant preference in orientation of the Tn917 insertions with either transcription or replication. Even though OG1RF has a smaller genome than strain V583 (2.8 Mb versus 3.2 Mb), the only E. faecalis strain whose sequence is in the public domain, over 10% of the Tn917 insertions appear to be in a OG1RF-specific sequence, suggesting that there are significant genomic differences among E. faecalis strains.
Many bacterial genetic studies involve the screening of random transposon mutant libraries. However, an ordered library offers several advantages over random libraries. First, a subset of the library containing one insertion in each nonessential open reading frame (ORF) can be assembled, significantly decreasing the number of mutants that need to be screened to reach saturation. Second, screening a library in which the mutated genes are known identifies genes that are not involved in a particular process as well as genes that are. Third, identification of mutants affecting a particular phenotype immediately identifies a set of relevant genes without further experimentation. Fourth, because the complete ordered library contains, on average, multiple insertions in most genes, several independent mutants can be tested to definitively correlate the inactivation of a particular gene and a phenotype. Finally, if a mutation appears to be in an operon, downstream insertions can be tested to determine if the original mutant's phenotype is the consequence of polarity of the transposon insertion.
In this study, we have attempted to construct an ordered transposon library of mutants for the human opportunistic pathogen Enterococcus faecalis, with the goal of obtaining an insertion in every nonessential ORF, analogous to a transposon library recently constructed for Pseudomonas aeruginosa (18). Transposon Tn917 insertion mutants were collected from E. faecalis strain OG1RF (25). Strain V583, the only strain with a publicly available sequence, was not selected, because unlike OG1RF, it has resistance to many antibiotics, including vancomycin, and therefore is difficult to genetically manipulate and poses a biohazard risk in the laboratory (33). OG1RF does not have these drawbacks and has been shown to successfully cause disease in a number of animal models. The replicative Tn3-like transposon Tn917 was chosen for the following reasons. First, it is the most commonly used gram-positive transposon and has been used successfully in Bacillus subtilis (43), E. faecalis (3), Staphylococcus aureus (4), and Streptococcus mutans (16) to construct stable insertion lines. Second, Tn917 is smaller and therefore easier to work with than Tn916 (5.2 kb versus 16 kb), and, unlike Tn916, it has not been shown to have a strong preference for intergenic regions (26). Third, although Tn4001 has successfully been used in Streptococcus pyogenes (22), pilot experiments with Tn4001 in E. faecalis indicated that it was prone to multiple insertions. Finally, and most importantly, several published reports that used a variety of gram-positive bacteria have purportedly shown that Tn917 insertion is sufficiently random to find mutations throughout the genome (3, 4, 16, 43). On the other hand, there have also been anecdotal reports that Tn917 does not insert randomly (see Discussion).
By sequencing a relatively large number of Tn917 insertions, we are able to report that Tn917 insertion in E. faecalis is far from random and is strongly biased towards the replication terminus. This study also generated interesting data with respect to whether Tn917 has a preferential target site and pinpointed significant differences between the OG1RF and V583 genomes.
MATERIALS AND METHODS
Transposon mutagenesis and sequencing.
The plasmid pTV1-OK, a temperature-sensitive plasmid with resistance to kanamycin that also contains the erythromycin resistance-encoding transposon Tn917 (16), was transformed into OG1RF, and transformants were selected on kanamycin (1,000 μg/ml) at 28°C to generate strain DAGF1-3. To generate transposants, single colonies of DAGF1-3 were used to inoculate 50 ml of brain heart infusion (BHI) medium containing 1,000 μg of kanamycin/ml. The culture was grown to mid-log phase (optical density of approximately 0.6), spun down, and brought up in 5 ml of medium plus 15% glycerol and either frozen or spread on a plate immediately. One milliliter of product was spread on a 230- by 230-mm square plate containing 250 ml of BHI agar plus 50 μg of erythromycin/ml and was incubated at 49.6°C for 5 to 6 days until transposants appeared. The colonies were picked robotically into 384-well plates containing 70 μl of BHI medium plus 15% glycerol and 50 μg of erythromycin/ml and were incubated at 37°C for 48 h without shaking to prepare glycerol stocks for freezing. The plates were stored at −80°C.
To determine the location of the transposon in each mutant in the library, we sequenced the flanking DNA by amplifying the region via PCR with a primer specific to one end of the Tn917 transposon and an arbitrary primer with a constant region at the 5′ end. In the second round of PCR, a specific primer, further nested, was used with a primer specific to the constant region of the arbitrary primer. The resulting product was sequenced by using a third nested primer specific to the transposon.
The details of this protocol are as follows. First, we replicated the library onto BHI agar plates with 50 μg of erythromycin/ml and then into liquid culture, all in a 96-well format. After overnight growth, 70 μl of the culture was heated to 99°C in a thermocycler for 10 min and then was spun down. Three microliters of the supernatant was added to a PCR mix containing 2.8 μl of 10× Taq buffer, 2.8 μl of dimethyl sulfoxide (DMSO), 1.4 μl of deoxynucleoside triphosphates (dNTPs) (5 mM) (Roche), 0.25 μl of Taq polymerase (Roche), 16.65 μl of sterilized, deionized water, and 0.55 μl of each of the following primers at a 50-ng/μl concentration: ARB1B, GGCCACGCGTCGACTAGTACNNNNNNNNNNGTAAT; and ODG29, GCAATAACCGTTACCTGTTTGTGC, which was specific to the left end of the transposon. The following cycling profile was used: 95°C for 5 min followed by 30 cycles of 95°C for 30 s, 42°C for 45 s, and 72°C for 1 min, followed by holding at 72°C for 5 min. The PCR was diluted 1:25 in sterile, deionized water, and 5 μl of the dilution was added to the second PCR, consisting of 2.5 μl of 10× Taq buffer, 2.5 μl of DMSO, 1.25 μl of dNTPs (5 mM) (Roche), 0.25 μl of Taq polymerase (Roche), 12.5 μl of sterile, deionized water, and 0.5 μl of the following primers at a 50-ng/μl concentration: ARB2, GGCCACGCGTCGACTAGTAC; and ODG30, GAAAACTGTACCACTAATAACTCACAATAGAGAGATGTC. The following cycling profile was used: 40 cycles of the 95°C for 30 s, 45°C for 30 s, and 72°C for 1 min, followed by 72°C for 5 min. Five microliters of the reaction product was added to 2 μl of ExoSAP-IT (USB), incubated at 37°C for 15 min, and then incubated at 80°C for 15 min to destroy the enzyme. The product was then sequenced using primer ODG31 (GATGTCACCGTCAAGTTAAATGTACAAAATAACAGCG).
A small pool of Tn917LTV3 insertion events was mapped from L. monocytogenes strain 10403S. The insertion pool was a kind gift from Helene Marquis (Cornell University, Ithaca, N.Y.) (2). Isolates from this pool were collected manually, and PCR was used to individually sequence these isolates according to the above-described protocol. The position of transposition events was determined by using the L. monocytogenes strain EGD-e genome sequence (14).
Determination of transposon-interrupted genes.
The analysis of the E. faecalis transposon insertion mutant database was accomplished by using an automated bioinformatic pipeline developed for the Pseudomonas aeruginosa Transposon Insertion Mutant Library (http://ausubellab.mgh.harvard.edu/enterococcus). Sequences generated by the Massachusetts General Hospital core sequencing facility were imported as ABI files corresponding to 96-well microtiter plates. Base calling and sequence quality estimation was accomplished with PHRED (http://www.phrap.org). The transposon sequence was located in these raw sequences through the use of a Smith-Waterman algorithm. Using the PHRED quality scores as a guide, low-quality sequence was trimmed off the 5′ and 3′ ends. The processed sequences were blasted against the V583 genome and the insertions, and the interrupted genes were determined with a high degree of confidence in regions common to V583 and OG1RF.
Determination of preferred insertion site consensus sequence.
Base frequency matrices and log-likelihood position-specific scoring matrices (PSSMs, weight matrices) were generated by methods analogous to those described previously (5). Two in-house-developed tools were used. MakeMatrices.pl (http://ausubellab.mgh.harvard.edu/enterococcus/files/makeMatrices.pl) was used to compute raw matrices, positional χ2 values, normalized matrices, and the normalized log-likelihood scoring matrices used in scoring matches to the Tn917 insert motif. Score Matrix.pl (http://ausubellab.mgh.harvard.edu/enterococcus/files/scoreMatrix.pl) was used for scoring matches to a matrix output by makeMatrices.pl.
PSSMs for the Tn917 insertion sequence were generated with two different sequence sets. For the purpose of identification of the chromosomal location of the insertion, flanking genomic sequence was obtained only from the left end of the transposon. Sequence quality was judged by the level of agreement between the known and the observed transposon sequence by using the Smith-Waterman algorithm. We obtained a PSSM for the left end by using 1,684 high-quality sequences for which the transposon sequence matched exactly. In order to characterize the right-end insertion consensus, we sequenced about 96 mutants from the right end of the transposon and obtained 74 right-end sequences that had three or fewer mismatches or gaps in the transposon sequence. In addition to allowing us to develop a PSSM specific to the right side of the transposon insertion, it verified that Tn917 inserts with a 5-base duplication.
In order to get additional information about the right side of the insertion site consensus, the V583 genome was used to retrieve sequences off the right end of the transposon, whereas only the left end had been sequenced. This was possible only in regions of the genome where there was perfect identity of the OG1RF transposon-flanking sequences with that of the V583 sequence. The consensus insertion sequence determined in this manner was found to agree closely with the consensus obtained when both sides were sequenced off the transposon.
To test whether or not the consensus sequence was symmetrical, we took the transposon insertion sequences, in both forward and reverse orientations, masked all but bases 3 through 7 (treating the middle base as position 0), and scored them against the original matrix. Sequences for which the reverse read scored higher than the forward read were reversed. The new sequence set containing some reversed sequences was then used to make a reoriented matrix. The reoriented matrix still showed signs of symmetry, suggesting that the consensus insertion site is at least partially symmetrical.
Determination of L. monocytogenes transposon-interrupted genes.
To investigate Tn917 target site preference in L. monocytogenes, individual insertions were mapped by using arbitrary PCR in strain 10403S (see above). As with E. faecalis, nonduplicate sequence reactions that could definitively call the actual base pair of transposon insertion were used to determine sequences flanking the point of insertion based on the sequenced genome (n = 12). The L. monocytogenes insertion sequences were aligned and compared with those determined for E. faecalis.
RESULTS
Creating an ordered insertion library in OG1RF.
We used the gram-positive transposon Tn917 to introduce disruptions into E. faecalis strain OG1RF and collected the mutants with the aid of a colony-picking robot to create a random insertion library in 384-well microtiter plates. We obtained DNA sequence flanking the transposon insertions in each of 8,865 mutants (see Materials and Methods) by amplifying the flanking region via PCR using a primer specific to one end of the Tn917 transposon and an arbitrary primer with a constant region at the 5′ end. In the second round of PCR, a specific primer, further nested, was used with a primer specific to the constant region of the arbitrary primer. The resulting product was sequenced with a third nested primer specific to the transposon.
Of the 8,865 sequences we obtained, 2,628 of the sequences were found to be too short and/or of too poor quality to determine their origin. The remaining 6,237 were analyzed by comparison with the V583 genome database (www.tigr.org), using BLAST to determine the genomic location of each transposon insertion event (see Materials and Methods). Of these, 4,889 mutant sequences resulted in BLASTN alignments with the V583 genome having bit scores above a threshold of 60. There was a significant number of mutants that shared the same insertion location. Of the 4,889 good-quality mutants, 1,923 were unique but 2,869 had apparent siblings, and 97 could not be precisely located. These 2,869 nonunique mutants, representing 676 distinct positions, could have resulted from transposition events that were sequenced twice because of cell division following transposition. Alternatively, they could represent independent insertions. We did find 93 examples of mutants with insertions into the same base pair that could not be siblings, because they inserted in opposite directions. In total, transposition events were identified in 540 different genes that are also found in E. faecalis strain V583. This set of 540 mutants is available as an ordered library set from the Ausubel laboratory (http://ausubellab.mgh.harvard.edu/enterococcus). A description of the sequences that did not match our threshold is discussed later.
We estimate that the library of 540 mutants corresponds to 23% of the nonessential E. faecalis genes based on the following assumptions. The size of the chromosome of the sequenced strain of E. faecalis, V583, is 3.2 Mb and contains about 3,200 genes (28). Because OG1RF has a smaller genome of 2.8 Mb, we assumed that it has approximately 2,800 genes (24). Based on the number of nonessential genes in Escherichia coli (87%) (44), we calculated that there are approximately 2,400 nonessential genes in OG1RF.
Tn917 inserts preferentially at the chromosome terminus.
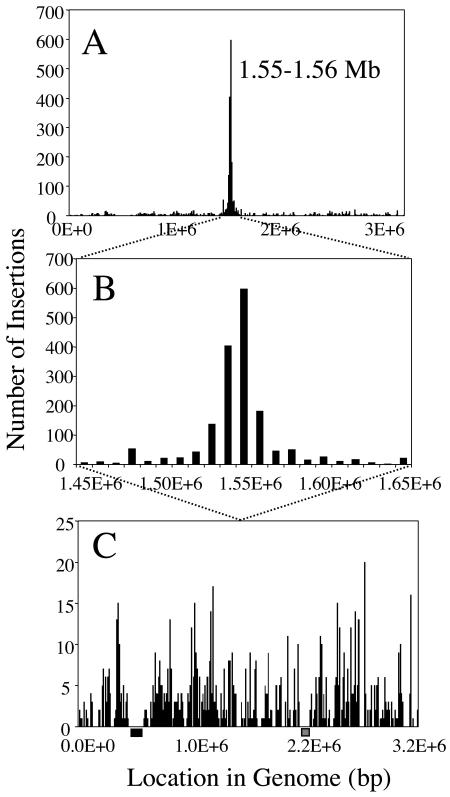
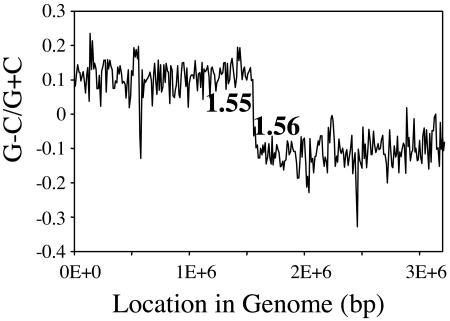
Assuming colinearity between the E. faecalis OG1RF and V583 genomes, we mapped the OG1RF Tn917 insertion events onto the V583 genome to identify any regional preferences, i.e., hot spots. There are several potential biases in our analysis. We cannot completely rule out the possibility that the sequences that did not amplify well and were discarded from our analysis were in particular regions of the chromosome. Double insertions in a large subset of our mutants could confuse our analysis, but by performing southern hybridization on a small subset of mutants we found that less than 5% contained double insertions (data not shown). Another potential bias is possible siblings, which we avoided by counting only once the mutants with insertions into the same base. As shown in Fig. 1, most of the mutants (65%) had insertions corresponding to a position in the V583 genome between 1.45 and 1.65 Mb, with the highest number of insertions occurring between 1.55 and 1.56 Mb (23%) (Fig. 1). This region corresponds to the terminus of the chromosome; it is directly opposite the gene encoding the DnaA protein, indicative of the origin of DNA replication. We performed G+C skew analysis with the V583 genome to identify the terminus, and as shown in Fig. 2, the region where the skew switches from positive to negative is between 1.55 and 1.56 Mb (Fig. 2). A cumulative skew analysis (15) pinpointed the location between 1.555 and 1.556 Mb (data not shown).
FIG. 1.
Genomic locations of Tn917 insertions, excluding siblings, in OG1RF relative to the V583 genome sequence, divided into 10-kb bins. (A) The number of hits throughout the genome. (B) The number of hits in the region immediately surrounding the terminus region. (C) The number of hits throughout the genome excluding the hot-spot region (14.5 to 16.5 Mb). The black box corresponds to the location of the putative V583 pathogenicity island, and the gray box corresponds to the region encoding vancomycin resistance (28), sequences not present in OG1RF.
FIG. 2.
Locating the terminus of replication by G/C skew analysis. The number of G's on the plus strand was subtracted by the number of C's and was divided by the total. The range where the outcome switches from positive to negative corresponds to the replication terminus.
Figure 1C shows the distribution of insertions, excluding the terminus region, to illustrate transposon insertion preference throughout the rest of the genome. As in the previous analysis, this assumes colinearity between the OG1RF and V583 genomes. Visual inspection of the figure suggests that insertions are not distributed randomly outside the terminus hot spot. There are some areas of V583 sequence that are devoid of transposition events, possible cold spots for transposition, while some areas have many insertions. Many cold spots were expected to occur, because the larger V583 genome (3.2 Mb) contains sequences absent in the smaller OG1RF genome (2.8 Mb). Indeed, some of these apparent cold spots include the region of vancomycin resistance (1.89 to 2.26 Mb) and the putative pathogenicity island located at 0.45 to 0.58 Mb (Fig. 1). OG1RF is not vancomycin resistant, and it does not carry the operon for cytolysin, which is found in the pathogenicity island of V583 (36). As detailed in the next section, there do seem to be additional genomic differences between OG1RF and V583, which may explain additional apparent cold spots. A full accounting of the apparent nonrandomness of Tn917 insertion outside the terminus hot spot awaits an OG1RF sequence.
Tn917 has a slight preference for noncoding regions.
We examined whether or not Tn917 has a preference for noncoding regions. Other transposons have been shown to have a strong preference for intergenic regions, which is potentially a survival strategy. For example, 90% of Tn916 insertions were found to be in noncoding regions (26). To not bias this analysis, we only chose a single representative sequence from a given genomic position when more than one mutant had an insertion in that position. In addition, we also only selected insertions that had a strong match to the V583 genome (bit score of greater than 200) to ensure that we would be able to easily determine whether or not the transposon was in an ORF or an intergenic region. Of the 2,522 sequences that matched the criteria, we found that 30% had insertions in intergenic regions. Only 13% of the E. faecalis chromosome is made up of intergenic space, suggesting that there is a preference for noncoding DNA, albeit not as dramatic a bias as that for Tn916.
Consensus sequence for Tn917 insertion site.
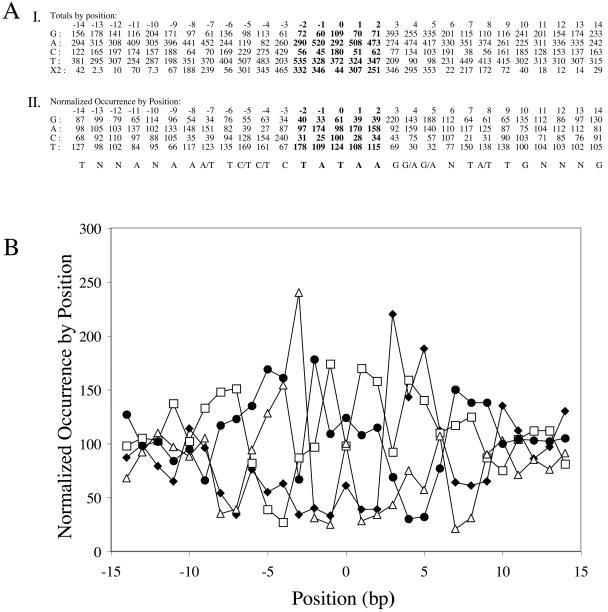
We examined the sequences flanking the transposon insertion sites for commonalities that would define a preferential insertion sequence. We included only sequences that were a perfect match to the V583 genome in the first 60 bp flanking the end of the transposon and included redundant insertions only once. We sequenced off the end of the transposon closest to the erythromycin resistance-encoding gene (erm), known as the erm-proximal end. After assigning the location, we used the V583 sequence database to predict the sequence on the erm-distal side of each transposon insertion. To assess these predictions, and also to confirm the presence of the 5-bp duplication that is a hallmark of Tn917 transposition (29), we sequenced the flanking regions on the erm-distal side of about 100 insertions. When we compared these sequences to our virtual sequences we did not find any significant differences (data not shown). Therefore, using the erm-proximal end sequence and erm-distal end virtual sequence, the frequency of A, T, G, and C at each position was tabulated to create a base frequency matrix. The matrix was normalized to reflect differences in nucleotide occurrence frequencies in the E. faecalis genome (a low-G+C gram-positive bacterium), and a log-likelihood PSSM was generated. The χ2 test was used to determine the statistical significance of deviations from the expected frequency of occurrence of each nucleotide at each position. Based on this analysis, a 29-bp motif was identified, characterized by a central A/T-rich region that consists of the residues that make up the 5-bp duplication. Flanking this central sequence is a preference for G/A residues on the erm-proximal side (right side in Fig. 3) and C/T residues on the erm-distal side (left side in Fig. 3). In particular, a guanine is strongly favored at position 3 and a cytidine is strongly favored at position −3. The 29-bp sequence appears somewhat symmetric, suggesting that the insertion site is palindromic. However, an alignment of randomly oriented sequences could artificially produce a seemingly palindromic insertion site. Following a previously published method (5), we reoriented the sequenced insertion sites so that the sides that best match bases 3 through 7 of the palindrome were on the same side. This corresponds to the region of the insertion site that directly abuts the duplication. In contrast to what was previously observed with Tn3 (5), such an analysis resulted in an insertion sequence that was nearly identical to the original, strongly suggesting that the palindromy is not artifactual (data not shown).
FIG. 3.
Identifying the preferred insertion sequence motif. (A) Section I shows the number of each nucleotide observed at each position flanking the transposon and tabulated into a matrix. Positions −2 to +2 (in boldface) represent the duplicated 5-bp target site. The χ2 test was used to detect deviations from the expected frequency. The 95% confidence level was greater than 14.07, and the 99% confidence level was greater than 18.48. Section II shows the calculated and the preferred sequence motif, based on the overall nucleotide frequency in the V583 genome (63% A/T, 37% G/C), the normalized occurrence by position (normal equals a value of 100). The nucleotide designation was based on a normalized occurrence of 120 or greater. (B) We used the numbers generated in matrix II to graphically represent the deviation in nucleotide frequencies of the region directly flanking the site of Tn917 insertion. G, solid diamond; A, open square; C, open triangle; T, closed circle.
Tn917 insertion orientation is not strongly correlated with the direction of transcription or replication.
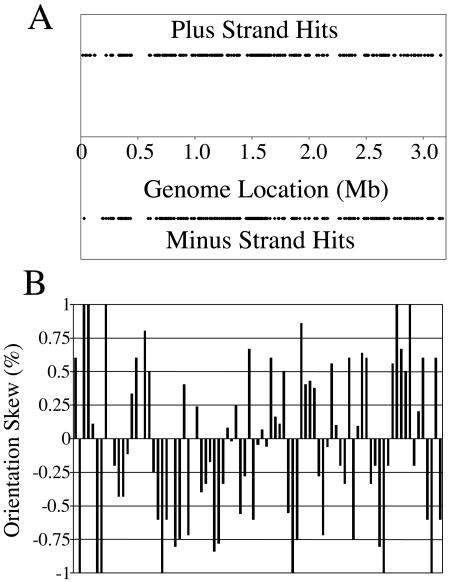
We also checked for a correlation between Tn917 insertion orientation and the directions of transcription and replication. If insertion orientation correlated with replication direction, then we would expect one orientation to predominate before the replication terminus with a reversal to the other orientation after the replication terminus. But our data showed equal numbers of insertions in both directions on both sides of the terminus region, arguing against such a correlation (Fig. 4A). These data also argue against an orientation preference related to transcription, because many bacteria, including E. faecalis, display a preference for the transcriptional orientation of genes consistent with the direction of replication. To more directly address any possible influence of transcription on the insertion orientation, we checked the orientation of each insertion with the direction of transcription at the insertion site and saw a weak bias against transcription; 60% of the insertions were oriented against the direction of transcription (data not shown).
FIG. 4.
Direction of Tn917 orientation is biased locally but not globally. (A) The insertion of Tn917 into the plus strand (positive orientation) or the minus strand (negative orientation) was noted for each mutant relative to the transposon's location in the genome. (B) The orientation bias of 1,000-bp regions which contained five or more insertions were analyzed by subtracting the number of insertions on the minus strand from the number on the plus strand and dividing by the total. Insertions that occurred multiple times in the same base were counted only once.
Tn917 orientation is biased within small regions.
In contrast to our global analysis of Tn917 insertion orientation, within relatively small regions of the chromosome (on the order of 100 to 1,000 bp) many of the insertions were oriented in the same direction. To look at this more closely, we examined successive 1,000-bp windows for frames that contained five or more insertions and then analyzed the orientation bias by subtracting the number of insertions on the minus strand from the number on the plus strand and dividing by the total (Fig. 4B). An equal number of insertions in both strands would result in a ratio of 0.0, whereas all the insertions being in one strand or the other would result in a ratio of −1.0 or 1.0. As in the analysis above, insertions that had occurred multiple times at the same base position were only counted once to avoid possibly including siblings in this analysis. We considered a ratio between −0.25 and 0.25 (i.e., close to 0) to have relatively little bias, which was the case in 25 out of 85 cases (29%). In contrast, a random transposon would have a ratio between −0.25 and 0.25 62% of the time (data not shown). This suggests that Tn917 insertion orientation is biased on a local level for reasons unassociated with the direction of transcription. Regional orientation biases have also been found in another element, IS903. Interestingly, this regional orientation bias was found to be dependent on the host-encoded nucleoid protein H-NS (38). IS903 also has a preference for certain hot spots and is sensitive to a signal associated with replication termination (see Discussion) (17).
Tn917 insertion preferences in L. monocytogenes.
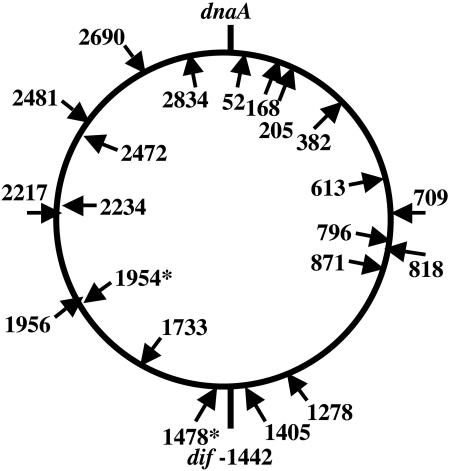
To determine if Tn917's strong bias for the terminus also occurs in a different low-G+C gram-positive species, we used arbitrary PCR to map a small collection of Tn917 insertions in the chromosome of L. monocytogenes strain 10403S (n = 24) (2). A survey of the literature reveals anecdotal accounts of favored hot spots for Tn917 transposition (see Discussion). We used the publicly available DNA sequence from strain EGD-e (14) to determine the location of insertions in 10403S, as was done for E. faecalis. All but one of the sequences used as insertion sites in strain 10403S matched the EGD-e genome; sequencing revealed that one apparent transposon insertion resulted from integration of the entire delivery vector or reversion of its temperature-sensitive origin of DNA replication. Both L. monocytogenes strains EGD-e and 10403S are of the same serotype, but a genome size estimate has not been determined for 10403S. We found that the sequences did not cluster in the terminus; rather, they appeared to be scattered relatively evenly around the chromosome (Fig. 5). As with E. faecalis, some duplicate insertions were also identified (Fig. 5).
FIG. 5.
Location of sequenced Tn917 insertions in L. monocytogenes. The predicted location of Tn917 insertions (arrows) and a putative dif site are indicated in kilobase pairs on the L. monocytogenes strain EGD-e genome. The orientation of individual insertion events is indicated by placement of the arrow inside (left to right) or outside (right to left) the circle. Duplicate insertions are indicated by asterisks, and the position of the dnaA gene is noted. Sequencing revealed that one apparent transposon insertion resulted from integration of the entire delivery vector or a reversion event.
The insertion site preference for L. monocytogenes was similar to what we had observed for E. faecalis. The likelihood matrix was very similar, though χ2 values were not significant due to our small sample size (data not shown). The L. monocytogenes sequences were scored against the E. faecalis matrix. The average log odds score for L. monocytogenes insertion site sequences was 1.59, compared to 2.59 for the average of the E. faecalis insertion site sequences (Material and Methods). By comparison, random sequences from the L. monocytogenes genome have an average log odds score of −1.96, suggesting that Tn917 prefers an insertion sequence in L. monocytogenes similar to that of E. faecalis. Also like E. faecalis, the transposon orientation did not seem to be strongly affected by the direction of replication (Fig. 5).
Genomic differences between OG1RF and V583.
Recall that of the 6,237 sequences that were of sufficient quality and length to merit further analysis, only 4,889 had matches to the V583 strain sequence by BLAST analysis (see Materials and Methods). We hypothesized that some of the Tn917 insertions, which occurred in DNA sequences that did not match the V583 sequence, might be vector sequences due to insertion of the entire shuttle vector into the chromosome, but in only a few mutants did we find significant homology to the delivery vector. Therefore, we examined these 1,348 sequences more closely.
We hypothesized that the 1,348 sequences that did not match the V583 genome might be bacterial genes contained in OG1RF but not the sequenced strain, V583. We compared these sequences against the National Center for Biotechnology Information (NCBI) microbial database and indeed found that many of these sequences were homologous to bacterial genes found in related species. We grouped overlapping non-V583 sequences by assembling them with PHRAP (8, 9) and in this way identified a total of 48 stretches of contiguous DNA sequence that could be assembled from overlapping transposon sequences (contigs). One-hundred twenty of the 1,348 sequences did not match the V583 genome and did not overlap with other sequences, but these singlet sequences had at least 100 bp of sequence adjacent to the transposon such that we could include them in our analysis. Twenty of the contigs and 58 of the singlets had homology to known bacterial genes in the GenBank bacterial database (gbbct) as judged by the BLAST algorithm, corresponding to approximately 69 different genes in total. No significant hits were identified for the remaining 18 contigs and 62 singlets. Ten of the sequences that matched other sequences found in the NCBI database were examined more closely by designing primers specific to the OG1RF sequence, generating labeled probes, and hybridizing against genomic DNA from Enterococcus spp. strains OG1RF, V583, FA2-2, E002, E006, and E007. As expected, all the probes hybridized to OG1RF but none hybridized to V583, confirming that these sequences are indeed present in strain OG1RF but not strain V583. Some of the sequences were found in FA2-2 (10), another commonly used laboratory strain of clinical origin, and in E006, a clinical isolate. None were present in E002, another clinical isolate, or in E007, which is an E. faecium clinical isolate (Table 1) (12).
TABLE 1.
Prevalence of OG1RF-specific genes in other Enterococcus spp. strainsa
| Gene and description | Score (bits) | Strainb
|
||
|---|---|---|---|---|
| OG1RF | FA2-2 | E006 | ||
| AAK4953 (AY033764); histidine kinase VanSc3 (F. flavescens) | 109 | X | X | |
| JC5217; site-specific DNA-methyltransferase (adenine-specific) (EC2.1.1.72) HsdM (validated); (Pasteurella haemolytica) | 33 | X | X | |
| NP_266583; beta-glucoside-specific PTS system; IIABC component (Lactococcus lactis subsp. lactis) | 68.6 | X | ||
| NP_244418.1; integrase (phage-related protein) (B. halodurans) | 45.8 | X | ||
| NP_815574.1; FtsK/SpoIIIE family protein (E. faecalis V583) | 67 | X | ||
| ZP_00124788.1 COG0673; predicted dehydrogenases and related proteins (Pseudomonas syringae pv. syringae B728a) | 104 | X | X | X |
| ZP_00037078.1 COG4932; predicted membrane protein (E. faecium) | 48.1 | X | ||
| CAD97577.1; putative histidine kinase (Clostridium beijerinckii) | 30.8 | X | X | |
| ZP_00037043.1 COG1131; ABC-type multidrug transport system, ATPase component (E. faecium) | 67 | X | X | X |
| NP_350277.1; uncharacterized conserved membrane protein, YUEB B. subtilis homolog (Clostridium acetobutylicum) | 49.3 | X | X | |
Genomic DNA was prepared from the strains, digested, run on a gel, blotted, and then probed for the OG1RF genes that were not found in the V583 genome. The X's indicate a positive signal.
Strains V583, E002, and E007 had no positive signals.
We estimate that 10 to 15% of the OG1RF genome may consist of sequences not found in V583. We base this estimate on the fact that we identified approximately 69 genes in OG1RF that were not present in V583 and 540 that were present. This figure is likely an underestimate, because it does not include the possibility that the sequences flanking the element did not result in a match to the NCBI database; the identification of presumptive OG1RF-specific genes is based on sequencing regions flanking transposon insertions, a procedure which does not always result in high sequence quality. Careful characterization of these putatively identified genes and a full understanding of the global genomic differences between these different E. faecalis strains await comprehensive sequencing of the OG1RF genome.
DISCUSSION
In this work, we generated an ordered insertion library of transposon mutants in E. faecalis strain OG1RF corresponding to disruptions in approximately 25% of the nonessential genes. In the process we characterized aspects of Tn917 insertion and genomic differences between OG1RF and V583.
The distribution of Tn917 insertions in E. faecalis prompted us to reexamine published accounts from previous mutant screens that utilized Tn917. Multiple examples exist in the literature where certain genes were found to be highly overrepresented in collections of Tn917 transposition events. The most notable is the finding of hot spots in the gltAB genes in B. subtilis, which are located near the replication terminus (35, 39, 43). However, when these genes were replaced with DNA from another strain of B. subtilis the bias became much weaker, suggesting that this hot spot did not solely result from its location near the terminus (39).
Our data show that both sequence and regional biases strongly affect the composition of collections of Tn917 insertions in E. faecalis. However, caution is warranted in extending this finding to bacteria other than E. faecalis. Any regional bias will need to be determined on a case-by-case basis given our results with L. monocytogenes. In the small collection of Tn917 insertions in L. monocytogenes we sequenced, there was no strong bias for transposition into any region of the chromosome. It is true, however, that a strong insertion bias for certain DNA sequences may still limit the use of Tn917 in many gram-positive organisms, and there is a need for the development of new tools for these medically and industrially important bacteria.
The hint of fairly significant genomic differences between OG1RF and V583 is perhaps not surprising, considering that a vast array of mobile genetic elements has been found to be characteristic of E. faecalis. It is worth noting, however, that OG1RF and V583 were also isolated under very different circumstances. V583 was isolated from a blood culture (33), while the parent of OG1RF, OG1, is thought to have been isolated from dental caries (27), very different environments that may require unique subsets of genes. Significant differences between strains have also been noted for E. coli (41), Helicobacter pylori (34), and S. aureus (11), while considerably less heterogeneity has been observed in Vibrio cholerae (7) and P. aeruginosa (42).
Tn917 is in the Tn3 transposon family, and its transposase shares 80% similarity to the Tn3 transposase (37). Previous work looked at the sequences at the site of Tn917 insertion and, as was the case with Tn3, identified a 5-bp duplication (29). We have confirmed this 5-bp duplication in our examination of about a hundred clones for which we obtained sequences from both sides of the insertion. Like Tn3, the preferred 5-bp target is AT rich, followed by a G or a C. But unlike Tn3, for which the target sequence was determined to be asymmetric, upon careful analysis the preferred insertion sequence for Tn917 is strongly palindromic. It was concluded in previous work on Tn3 that the primary sequence structure is necessary, but not sufficient, for the creation of a transposition hot spot and that primary sequence is not a factor in the creation of a cold spot. The authors hypothesized that some other factor, such as DNA secondary structure, might be involved. More recent work has shown that the deformability of certain sequences can be of prime importance for transposable elements from highly divergent organisms (21, 40). Molecular dissection of Tn10 target site selection has revealed that the ability of the transposase to alter a potential target sequence can be a critical component of target site selection (32). We have found that primary sequence cannot explain Tn917's penchant for the terminus region. There is no difference in the preferred sequence within the hot spot region compared to that used in the rest of the genome, and examination of the OG1RF sequence does not indicate that there are more preferred sequence positions in the terminus (data not shown). We hypothesize that there must be some aspect to the terminus that attracts transposon insertion by Tn917 in E. faecalis. We cannot formally rule out the possibility that Tn917 insertions are preferentially isolated in terminus regions because they are fitter; however, insertions outside the terminus region did not give any obvious growth disadvantage based on colony size (personal observation). It is very unlikely that the tight grouping of insertions to such a small area of the chromosome is driven by a lack of essential genes in this region, given that a relatively small number of genes are believed to be essential in the laboratory in other bacteria, such as E. coli and B. subtilis (13, 20).
What is it about the terminus that so strongly attracts Tn917 transposition events? Perhaps Tn917 is attracted to a certain accessory protein or DNA secondary structure associated with replication termination. Accessory-targeting proteins are sometimes element encoded, as in the case of Tn7, which uses the transposon-encoded protein TnsE to target the terminus region and other DNA replication-associated targets (30). However, in the case of Tn7, targeting is not as precise as it is in the case of Tn917 (31). Additionally, Tn917 does not have any obvious element-encoded accessory proteins that would suggest a targeting mechanism. This does not rule out the possibility that host-encoded accessory proteins are used to direct transposition events preferentially to the terminus region. It is clear that host-encoded accessory proteins are important for other transposable elements that show strong regional biases in transposition. For example, in yeast, Ty1 and Ty3 use transcription factors to target upstream of RNA polymerase III-transcribed genes (6, 19) and Ty5 uses SIR4 to target silent DNA (45).
Tn917 could target a replication termination apparatus in E. faecalis. In B. subtilis and E. coli, DNA replication forks are impeded by one-way terminators of DNA replication progression, called ter sites, through the use of a trans-acting protein called Tus or RTP (1). It has been previously shown that Tn7 preferentially targets the terminus region and that this process is altered, although it is not eliminated, in E. coli Tus− cells (31). The transposable element IS903 also seems to have a bias for places where replication fork progression can be inhibited, but not necessarily in the terminus of DNA replication where opposing replication forks meet (38). When IS903 inserts into the chromosome it preferentially inserts into eight hot spot regions. While five of these hot spots are within 30 kb of ter sites, only one of the IS903 hot spots is in the terminus region where DNA replication forks likely meet. Unfortunately, a termination apparatus has not been identified in E. faecalis. We have looked but have been unable to find a Tus or RTP homolog and have not been able to find ter sequences. It is possible that E. faecalis uses an undefined mechanism of replication termination which could be a target for Tn917 transposition. It is also possible that E. faecalis completely lacks a mechanism, as replication termination is not an essential process, at least in laboratory grown bacteria.
Another phenomenon that occurs in the terminus region that could attract Tn917 transposition events is dif-mediated site-specific recombination. Site-specific recombination at dif allows the resolution of dimer chromosomes, which are formed via homologous recombination between sister chromosomes in circular bacterial chromosomes after DNA replication. In E. coli and in B. subtilis, tyrosine-recombinase proteins XerC/CodV and XerD/RipX resolve dimer chromosomes at a specific site in the terminus region called dif. We have identified a putative dif site in the terminus region centered at 1,565,836 based on homology to dif sites found in the chromosomes of E. coli and B. subtilis. The putative dif site is within 10,000 bp of the point where G-C/G+C shifts in the E. faecalis chromosome. XerC/CodV and XerD/RipX homologs exist in the E. faecalis V583 genome. Precedent for transposons targeting site-specific recombination systems exists. The Tn5053 family of transposons selectively directs transposition into the region of resolution used by site-specific serine recombinases when the transposons are located on plasmids (23). There are significant differences between the targeting found with Tn5053 elements and that with Tn917. One difference is that Tn5053 does not appear to target the dif region in E. coli; however, only two Tn5053 insertions were mapped in the E. coli chromosome (200 and 1,241 kb from dif) (23). A second difference between the Tn5053 and Tn917 insertion patterns concerns how close they occur to the point of resolution in the two systems. Tn5053 insertions are focused primarily over a few base pairs within the region of dimer resolution, while Tn917 insertions occur primarily over a region of many thousands of base pairs unequally distributed around the putative dif site identified in E. faecalis.
In addition to the above examples, many other structures or complexes could be imagined that might occur in the terminus region and provide an attractive target for Tn917. Whatever factor results in the frequent isolation of Tn917 insertion events in the terminus region of E. faecalis, this highly preferred target does not appear to exist in L. monocytogenes. Much remains unknown about the factors that contribute to the acquisition and integration of mobile DNA. It is intriguing that the regional bias for Tn917 seems so different between E. faecalis and L. monocytogenes. It will be interesting to learn if certain hosts have developed adaptations to direct mobile DNA to certain regions of the genome in an attempt to protect essential genes. Further research will be needed to determine if a host mechanism specific to E. faecalis mediates this process and how widespread this hypothetical system might be in bacteria.
Acknowledgments
This work was supported by grants from Aventis, SA, and NHLBI grant UO1 HL66678 to F.M.A. and NSF grant MCB-0315316 to J.E.P.
We thank Helene Marquis for providing us with the pool of L. monocytogenes Tn917 mutants.
REFERENCES
- 1. Bussiere, D. E., and D. Bastia. 1999. Termination of DNA replication of bacterial and plasmid chromosomes. Mol. Microbiol. 31:1611-1618. [DOI] [PubMed] [Google Scholar]
- 2. Camilli, A., A. Portnoy, and P. Youngman. 1990. Insertional mutagenesis of Listeria monocytogenes with a novel Tn917 derivative that allows direct cloning of DNA flanking transposon insertions. J. Bacteriol. 172:3738-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, A. E. Franke, Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coulter, S. N., W. R. Schwan, E. Y. Ng, M. H. Langhorne, H. D. Ritchie, S. Westbrock-Wadman, W. O. Hufnagle, K. R. Folger, A. S. Bayer, and C. K. Stover. 1998. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol. Microbiol. 30:393-404. [DOI] [PubMed] [Google Scholar]
- 5.Davies, C. J., and C. A. Hutchison, III. 1995. Insertion site specificity of the transposon Tn3. Nucleic Acids Res. 23:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devine, S. E., and J. D. Boeke. 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10:620-633. [DOI] [PubMed] [Google Scholar]
- 7.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 9.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, G. F., and D. B. Clewell. 1985. A conjugative transposon (Tn919) in Streptococcus sanguis. Infect. Immun. 47:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. Garcia-del Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 15.Grigoriev, A. 1998. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26:2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutierrez, J. A., P. J. Crowley, D. P. Brown, J. D. Hillman, P. Youngman, and A. S. Bleiweis. 1996. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 178:4166-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, W. Y., and K. M. Derbyshire. 1998. Target choice and orientation preference of the insertion sequence IS903. J. Bacteriol. 180:3039-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirchner, J., C. M. Connolly, and S. B. Sandmeyer. 1995. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science 267:1488-1491. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao, G. C., E. J. Rehm, and G. M. Rubin. 2000. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97:3347-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyon, W. R., C. M. Gibson, and M. G. Caparon. 1998. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 17:6263-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Minakhina, S., G. Kholodii, S. Mindlin, O. Yurieva, and V. Nikiforov. 1999. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol. Microbiol. 33:1059-1068. [DOI] [PubMed] [Google Scholar]
- 24.Miranda, A. G., K. V. Singh, and B. E. Murray. 1991. DNA fingerprinting of Enterococcus faecium by pulsed-field gel electrophoresis may be a useful epidemiologic tool. J. Clin. Microbiol. 29:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray, B. E., F. Y. An, and D. B. Clewell. 1988. Plasmids and pheromone response of the beta-lactamase producer Streptococcus (Enterococcus) faecalis HH22. Antimicrob. Agents Chemother. 32:547-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, K. E., D. L. Richardson, and B. A. Dougherty. 1997. Tn916 transposition in Haemophilus influenzae Rd: preferential insertion into noncoding DNA. Microb. Comp. Genomics 2:313-321. [DOI] [PubMed] [Google Scholar]
- 27.Oliver, D. R., B. L. Brown, and D. B. Clewell. 1977. Analysis of plasmid deoxyribonucleic acid in a cariogenic strain of Streptococcus faecalis: an approach to identifying genetic determinants on cryptic plasmids. J. Bacteriol. 130:759-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 29.Perkins, J. B., and P. J. Youngman. 1984. A physical and functional analysis of Tn917, a Streptococcus transposon in the Tn3 family that functions in Bacillus. Plasmid 12:119-138. [DOI] [PubMed] [Google Scholar]
- 30.Peters, J. E., and N. L. Craig. 2001. Tn7 recognizes transposition target structures associated with DNA replication using the DNA-binding protein TnsE. Genes Dev. 15:737-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters, J. E., and N. L. Craig. 2000. Tn7 transposes proximal to DNA double-strand breaks and into regions where chromosomal DNA replication terminates. Mol. Cell. 6:573-582. [DOI] [PubMed] [Google Scholar]
- 32.Pribil, P. A., and D. B. Haniford. 2003. Target DNA bending is an important specificity determinant in target site selection in Tn10 transposition. J. Mol. Biol. 330:247-259. [DOI] [PubMed] [Google Scholar]
- 33.Sahm, D. F., J. Kissinger, M. S. Gilmore, P. R. Murray, R. Mulder, J. Solliday, and B. Clarke. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33:1588-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandman, K., R. Losick, and P. Youngman. 1987. Genetic analysis of Bacillus subtilis spo mutations generated by Tn917-mediated insertional mutagenesis. Genetics 117:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shankar, N., A. S. Baghdayan, and M. S. Gilmore. 2002. Modulation of virulence within a pathogenicity island in vancomycin-resistant Enterococcus faecalis. Nature 417:746-750. [DOI] [PubMed] [Google Scholar]
- 37.Shaw, J. H., and D. B. Clewell. 1985. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J. Bacteriol. 164:782-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swingle, B., M. O'Carroll, D. Haniford, and K. M. Derbyshire. 2004. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 52:1055-1067. [DOI] [PubMed] [Google Scholar]
- 39.Vandeyar, M. A., and S. A. Zahler. 1986. Chromosomal insertions of Tn917 in Bacillus subtilis. J. Bacteriol. 167:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vigdal, T. J., C. D. Kaufman, Z. Izsvak, D. F. Voytas, and Z. Ivics. 2002. Common physical properties of DNA affecting target site selection of sleeping beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323:441-452. [DOI] [PubMed] [Google Scholar]
- 41.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfgang, M. C., B. R. Kulasekara, X. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youngman, P. J., J. B. Perkins, and R. Losick. 1983. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc. Natl. Acad. Sci. USA 80:2305-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu, B. J., B. H. Sung, M. D. Koob, C. H. Lee, J. H. Lee, M. S. Kim, and S. C. Kim. 2002. Minimization of the Eschericdhia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 20:1018-1023. [DOI] [PubMed] [Google Scholar]
- 45.Zhu, Y., J. Dai, P. G. Fuerst, and D. F. Voytas. 2003. Controlling integration specificity of a yeast retrotransposon. Proc. Natl. Acad. Sci. USA 100:5891-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]