Abstract
The evolution of human immunodeficiency virus type 1 (HIV-1) coreceptor use has been described as the acquisition of CXCR4 use linked to accelerated disease progression. However, CXCR4-using virus can be isolated only from approximately one-half of individuals with progressive HIV-1 disease. The other half continue to yield only CCR5-using viruses (R5 phenotype) throughout the course of disease. In the present work, the use of receptor chimeras between CCR5 and CXCR4 allowed us to study the evolution of HIV-1 with the R5 phenotype, which was not revealed by studies of wild-type coreceptor use. All together, 246 isolates (173 with the R5 phenotype) from 31 individuals were tested for their ability to infect cells through receptor chimeras. R5narrow virus was able to use only wild-type CCR5, whereas R5broad(1) to R5broad(3) viruses were able to use one to three chimeric receptors, respectively. Broad use of chimeric receptors was interpreted as an increased flexibility in the mode of receptor use. R5broad isolates showed higher infectivity in cells expressing wild-type CCR5 than R5narrow isolates. Also, the increased flexibility of R5broad isolates was concomitant with a lower sensitivity to inhibition by the CC chemokine RANTES. Our results indicate a close relationship between HIV-1 phenotypic changes and the pathogenic process, since the mode and efficiency of CCR5 use as well as the decrease in the RANTES sensitivities of isolated viruses are significantly correlated with CD4+-T-cell decline in a patient. One possible explanation is that ligand competition at the CCR5 receptor or changed CCR5 availability may shape the outcome of HIV-1 infection.
The severity of human immunodeficiency virus type 1 (HIV-1) infection varies dramatically among different individuals. Many factors, including age, associated diseases, immune activation, viral load, and viral phenotype, have been shown to affect disease progression (6, 11, 37, 43). Conceivably, the final disease outcome is the result of interplay between several or all of these factors. Here we focus on viral phenotype, specifically, its changes over time, and consider mechanisms by which phenotypic evolution might be regulated.
One important phenotypic trait linked to HIV-1 pathogenesis is the type of coreceptor used by HIV-1 to enter cells (7, 8). The coreceptor is a chemokine receptor, CCR5 or CXCR4 (2, 12, 14, 18, 19, 24), which together with CD4 allows efficient uptake of HIV-1 into cells. CCR5-using viruses (the R5 phenotype) are present throughout all stages of HIV-1 infection (10, 13, 25-27, 33, 41, 42, 51, 62, 63, 65). The evolution of HIV-1 coreceptor use during progressive disease has been extensively studied in terms of both viral genetics and phenotype and involves change from CCR5 use to CXCR4 use, alone (the X4 phenotype) or in combination with the use of CCR5 (the R5X4 phenotype) and/or other less significant coreceptors (multitropic viruses) (10, 25, 28, 31, 51-53, 55). CXCR4 use is linked to an increased virulence of HIV-1, and CXCR4-using viruses can be isolated prior to or during progression to AIDS (32, 35, 57).
Even if the emergence of CXCR4-using HIV-1 has been linked to disease progression, it is isolated only from about one-half of AIDS patients (32, 35). Thus, progression to immunodeficiency may also occur in individuals in the absence of detectable CXCR4-using virus isolates. In these individuals the HIV-1 viral phenotype remains CCR5 dependent during the entire disease course (15, 29). However, R5 viruses from some AIDS patients develop increased resistance to inhibition by RANTES, i.e., the CC chemokine and natural ligand of CCR5 (29, 30, 34). It has been demonstrated that evolution to altered CCR5 usage can be induced in vitro, by exposing HIV-1 with the R5 phenotype to a small-molecule CCR5 antagonist (58). Recently, it was also shown that in individuals with progressive HIV-1 infection, R5 virus evolves to be more cytopathic over time, whereas this evolution is not seen in long-term asymptomatic individuals, indicating that this increasing cytopathicity of HIV-1 correlates with disease progression (36). Also, studies of macaques infected with different R5 clones implied that the fusogenicity of HIV-1 envelope is important to disease progression (21, 22). Thus, these observations suggest that R5 virus interaction with CCR5 may be altered both in vivo and in vitro.
In an attempt to document HIV biological variability during the pathogenic process, chimeric receptors between CCR5 and CXCR4 were used. Starting with the N terminus, parts of CCR5 were replaced with corresponding parts of CXCR4 (4). That study indicated that virus isolates from different individuals use CCR5 in different ways (33). In particular, the results hinted at differences in chimeric receptor use among viruses with the R5 phenotype. We therefore undertook a methodical study of sequentially collected HIV-1 isolates with the R5 phenotype and explored their ability to use chimeric receptors. Using a series of chimeric receptors as instruments, we here describe the in vivo evolution of the R5 phenotype to a phenotype in which the CCR5 receptor is used more flexibly and more efficiently. With parallel changes in the mode of CCR5 receptor use, isolated viruses become more resistant to inhibition by the CC chemokine RANTES. In association with declining CD4+-T-cell counts, changes in the mode of CCR5 use and the RANTES sensitivity of isolated viruses can be linked to pathogenesis.
MATERIALS AND METHODS
Patients and virus isolates.
The 31 patients studied were selected from a larger cohort of 53 HIV-1-infected individuals described previously (29, 32, 33). The patients were adult homo- or bisexual men living in Sweden whose median follow-up period was 103 months; follow-up included the counting of CD4 cells and viral isolations, and since 1996 plasma viral RNA loads have been determined at the South Hospital in Stockholm, Sweden. For the present study, the patients were selected on the basis of different rates of CD4 decline in the first 5 years of their infections and the difference in their virus biological phenotypes as assayed by syncytium induction on MT-2 cells and by coreceptor use on U87.CD4 and GHOST (3) cells. Accordingly, patients could be divided into two groups. Seventeen patients yielded non-syncytium-inducing virus that used CCR5 (the R5 phenotype) throughout the observation period (64 isolates). Fourteen patients initially yielded virus with the R5 phenotype but that later switched to CXCR4 use (also called syncytium-inducing viruses). In the patients with viruses that switched phenotypes, we tested 109 R5 isolates preceding the acquisition of CXCR4 use and, following the switch, 73 CXCR4-using isolates (the X4 and R5X4 or multitropic phenotypes).
Viruses were isolated from peripheral blood mononuclear cells (PBMC) by a standard procedure (50), and coreceptor use of sequential isolates was determined (29, 33). The evolutionary relationship between virus isolates of the same patients was studied by phylogenetic analysis of V3 sequences as previously described (39). Virus stocks were prepared by infecting 6 × 106 to 8 × 106 phytohemagglutinin (PHA) (Boule, Stockholm, Sweden)-stimulated PBMC from two healthy donors with 1.5 ml of supernatant from infected PBMC. Supernatants were harvested on days 7 and 10 or 11 after infection and stored at −80°C. The medium used was RPMI medium (Invitrogen, Lidingö, Sweden) containing 10% fetal calf serum (Invitrogen), 50 U of penicillin (Invitrogen) per ml, 50 U of streptomycin (Invitrogen) per ml, 2 μg of Polybrene (Sigma, Stockholm, Sweden) per ml, and 10 U of interleukin-2 (IL-2) (Amersham Pharmacia, Uppsala, Sweden) per ml.
Cell lines.
Human glioma U87.CD4 cells, stably expressing CD4 and one of the chemokine receptors CCR5 and CXCR4, were previously described (14). Cells were grown in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal calf serum (Invitrogen) and antibiotics. Cultures were incubated in a humidified atmosphere with 5% CO2 at 37°C, detached by treatment with 5 mM (pH 8.0) EDTA (VWR International, Lund, Sweden), and subdivided 1:2 to 1:4 twice a week.
Chimeric receptors were constructed by replacing successively increasing portions of CCR5 with corresponding regions of CXCR4 by a modification of the single-overlap and extension PCR approach (4). The resulting six chimeric constructs and the wild-type receptors were stably expressed in U87.CD4 cells. Proper surface expression of the receptors was secured by flow cytometry with a monoclonal antibody (55B5 [4]) raised against CXCR4 and recognizing the N terminus of the receptor. The U87.CD4 cell lines carrying chimeric receptors were cultured in the presence of 300 μg of Geneticin (Invitrogen) per ml and 0.5 μg of puromycin (Sigma) per ml. Geneticin selects for cells expressing CD4, and puromycin selects for cells expressing the chemokine receptors. Parental U87.CD4 cells, engineered to express CD4 but no chemokine receptor, were also included in the experiments.
Infection of U87.CD4 cell lines.
Cells in 500 μl of medium per well were seeded into 48-well plates 1 or 2 days prior to infection to obtain a 50%-confluent cell layer by the time of infection. U87.CD4 cells with wild-type receptors were seeded in the same medium as described above for culturing. U87.CD4 cells with chimeric receptors were seeded in growth medium devoid of Geneticin and puromycin. When we infected the cells, the medium was removed and virus (at least 2 ng of the p24 antigen per ml according to an in-house p24 enzyme-linked immunosorbent assay [ELISA]) (56) was added to duplicate wells in a volume of 200 μl/well. Two hours after infection, medium with 2 μg of Polybrene (Sigma) per ml was added up to 500 μl/well. After an overnight incubation, cells were washed with PBS (0.12 M NaCl, 0.03 M phosphate [pH 7.2]), 1 ml of medium with Polybrene was added to each well, and the plates were further incubated. The cultures were kept for 7 days, and inspection for syncytium formation was performed daily. The supernatant for viral antigen detection by ELISA was removed on day 1, after being washed, and at the last day of infection. All supernatants were tested by the in-house p24 ELISA, and p24 production in cultures of cells expressing FC-1, FC-2, and FC-4b was also tested, using the commercially available Vironostika HIV Uni-Form II Ag/Ab Microelisa system (Biomérieux, Boxtel, The Netherlands), according to the protocol provided by the supplier. Viral antigen production was considered positive in the ELISA when the increase in absorbance between days 1 and 7 exceeded 0.2 in the in-house ELISA and 0.5 in the commercial ELISA.
Virus titration on U87.CD4 cells.
Virus titration by hematoxylin staining of U87.CD4 cells was as previously described (54). Briefly, U87.CD4-CCR5 cells were infected as described above, but with fivefold virus dilutions in triplicate wells, starting with 14 ng of p24 per ml, according to the protocol of the commercially available Vironostika HIV-1 antigen ELISA (Biomérieux, Boxtel, The Netherlands). The day after infection, the plates were washed once with PBS and 1 ml of new medium was added. On day 4, the cell monolayers were washed once with PBS and fixed with methanol-acetone (1:1) for 5 to 10 min. For hematoxylin staining, which visualizes cell nuclei, the fixed cells were incubated for 2 to 3 min with hematoxylin (Merck, Darmstadt, Germany), washed with tap water, and dried. The number of plaques (distinct groupings of syncytial cells) was counted under a light microscope. Virus titers were calculated as numbers of PFU per milliliter with the following equation: (average number of plaques in triplicate wells × virus dilution)/volume in the well.
RANTES inhibition assay.
RANTES inhibition of HIV-1 replication in PBMC cultures was done as described previously (29). In brief, PHA-stimulated donor PBMC (105 cells) were infected with HIV-1 primary isolates in the presence or absence of RANTES serially diluted in threefold steps starting from a final concentration of 600 ng/ml. At day 3 postinfection, the medium was changed and the RANTES content was restored. Supernatants were harvested at day 7 postinfection, and the 50% RANTES inhibitory concentration (IC50) was analyzed by determination of the HIV-1 p24 antigen concentration by an ELISA, the Vironostika HIV Uni-Form II Ag/Ab Microelisa system (Biomérieux, Boxtel, The Netherlands).
Statistical analyses.
The nonparametric Kruskal-Wallis test was used to compare groups of virus isolates with different R5 phenotypes {narrow (R5narrow), broad with the use of one to three chimeric receptors [R5broad(1), R5broad(2), and R5broad(3)]} based on sensitivity to RANTES and CD4+-T-cell counts at the time of virus isolation. To demonstrate evolution within the R5 phenotype over time, the first and last isolates from 26 patients were compared by the Wilcoxon signed-rank test. The same test was used to compare the infectivities of the different groups of R5 isolates. Use of the FC-4b chimeric receptor and the frequencies of different R5 phenotypes in different patient groups were compared by chi-square analysis. Spearman rank correlation was used to show an association between the CD4-T-cell count at the time of isolation and RANTES sensitivity (expressed as the IC50). Bivariate analysis was performed with a logistic regression model by calculating the odds ratio (OR) and its associated confidence-related interval (CI). In this model, CD4 status was treated as a categorical variable.
RESULTS
Defining the HIV-1 phenotype according to wild-type and chimeric coreceptor use.
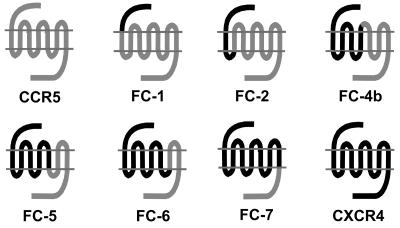
With the aim of studying a possible linkage between HIV pathogenesis and virus evolution in the context of host cell entry, we studied HIV-1 primary isolates obtained sequentially from 31 individuals with various levels of disease progression (a total of 246 isolates). In order to evaluate virus-coreceptor interactions, these isolates were tested on indicator U87.CD4 cells expressing CCR5, CXCR4, or one of the chimeric receptors FC-1, FC-2, FC-4b, FC-5, FC-6, and FC-7. As described previously (4), these chimeric receptors are hybrids of CCR5 and CXCR4 in which successively larger parts of CCR5 have been systematically replaced with corresponding parts of CXCR4 (Fig. 1). CXCR4-using viruses (with the X4, R5X4, and multitropic phenotypes) infected cells expressing all the different chimeric receptors, i.e., FC-4b, FC-5, FC-6, and FC-7, with the exception of FC-1 and FC-2 (33) (Table 1 and data not shown). Analysis of R5 isolates revealed that these viruses to various degrees used FC-1, FC-2, and FC-4b chimeras but did not use any other chimeric receptors or CXCR4. None of the viruses replicated in the parental U87.CD4 cells.
FIG. 1.
Schematic pictures of the chemokine receptors CCR5 and CXCR4 and the chimeric receptors FC-1, FC-2, FC-4b, FC-5, FC-6 and FC-7, where successively larger parts of CCR5 have been replaced with corresponding regions of CXCR4.
TABLE 1.
Use of chimeric receptors FC-1, FC-2, and FC-4b
| Viral phenotypea | No. of isolates | % of replicating isolates using:
|
|||||
|---|---|---|---|---|---|---|---|
| FC-1
|
FC-2
|
FC-4b
|
|||||
| p24 onlyb | p24 + syncc | p24 only | p24 + sync | p24 only | p24 + sync | ||
| R5 nonswitch | 64 | 20 | 2 | 3 | 63 | 22 | 17d |
| R5 switch | 109 | 37 | 8 | 6 | 79 | 17 | 63 |
| CXCR4 using | 73 | 14 | 51 | 7 | 89 | 0 | 100 |
R5 nonswitch, R5 isolates from patients with CCR5-using virus throughout the study; R5 switch, R5 isolates preceding the acquisition of CXCR4-using virus in patients with a detected switch to X4, R5X4, or multitropic virus; CXCR4 using, X4, R5X4, or multitropic isolates from the patients in the R5 virus switch group.
p24 only, p24 antigen production but no syncytium formation.
p24 + sync, syncytium induction and p24 antigen production.
Significantly lower numbers and percentages of replicating R5 isolates were obtained from nonswitch virus patients than from R5 switch virus patients when two isolates from each patient were analyzed as described for Table 2 (P = 0.01, chi-square analysis).
On the basis of chimeric receptor use, R5 viruses could be subdivided into two groups: those with the R5narrow phenotype and those with the R5broad phenotype. The R5narrow phenotype is shown by viruses that use wild-type CCR5 but no chimeric receptors, whereas viruses using at least one chimeric receptor in addition to CCR5 are designated R5broad viruses. Depending on the number of chimeric receptors used, the R5broad viruses could be further divided into R5broad(1), R5broad(2), and R5broad(3) phenotypes. The 38 isolates in the R5broad(1) category used either FC-2 or FC-4b (71 or 29%, respectively). Among the 64 isolates in the R5broad(2) category, the combination of FC-2 and FC-4b was the most frequent (77%). As previously described (33), the FC-1 chimera appeared to have the most restricted coreceptor function, since few of the isolates that were able to use the FC-2 or FC-4b receptor could use FC-1 for cell entry. Restriction is likely to be the result of an altered receptor conformation structure in which only the external part of the CXCR4 N terminus is anchored to CCR5, as with FC-1 but not with FC-2, as earlier suggested (4, 33).
The results indicate that HIV-1 isolates with the R5 phenotype may differ in their modes of CCR5 coreceptor use. Some viruses require the N terminus of CCR5 (R5narrow), while others can bind to the coreceptor without the CCR5 N terminus (R5broad). Within the latter group, exchange of the first extracellular loop of CCR5 to CXCR4 introduces an alternative receptor conformation. In spite of this, some of the R5 viruses can utilize chimeric receptor FC-4b when only the second and the third extracellular loops are derived from CCR5. Thus, these isolates appear not to depend on the N terminus and the first extracellular loop of CCR5 for host cell entry. However, the third extracellular loop of CCR5, represented by chimeras FC-5 and FC-6, is not enough to allow infection by any of the 173 R5 viruses that thus far have been tested (reference 33 and data not shown), indicating that these viruses depend on the second extracellular loop of CCR5 for infection.
The evolution of the R5 phenotype is related to pathogenesis.
The first question we asked was whether the R5narrow and R5broad phenotypes indicate evolutionary steps of R5 virus. The phenotypes of the first and last R5 isolates from 26 patients (patients with two R5 isolates obtained at least 18 months apart) were compared (Table 2). Sixteen patients showed changes over time as follows: R5narrow→R5broad(1)→R5broad(2)→R5broad(3). These results established that the increasing capacity to use chimeric receptors indicates the evolution of the mode of CCR5 receptor use (P = 0.004, Wilcoxon signed-rank test). Thus, the phenotypic changes resulting in a broader ability to use chimeric receptors indeed represent sequential steps in R5 virus evolution.
TABLE 2.
Changes within the R5 phenotype over time
| Patienta | Patient codeb | No. of R5 isolates | Time between first and last R5 isolate (mo) | CD4 cell count (106 cells/liter) at the time of isolation
|
Phenotype of R5 isolatec
|
Antiretroviral therapy at time of isolatione
|
|||
|---|---|---|---|---|---|---|---|---|---|
| First | Last | First | Last | First | Last | ||||
| R5 nonswitch | |||||||||
| 292 | I | 2 | 41 | 140 | 90 | Narrow | Broad(2) | AZT | AZT + ddI |
| 435 | 10 | 54 | 730 | 462 | Narrow | Narrow | |||
| 451 | J | 2 | 31 | 220 | 20 | Broad(1) | Broad(1) | AZT | ddI |
| 1047 | 14 | 99 | 630d | 360 | Broad(1) | Broad(1) | |||
| 1276 | 9 | 80 | 320 | 40 | Narrow | Broad(2) | ddI | ||
| 1838 | 4 | 63 | 455 | 290 | Narrow | Broad(1) | |||
| 1703 | F | 2 | 27 | 310 | 190 | Broad(1) | Narrow | AZT | AZT |
| 2010 | G | 2 | 35 | 260 | 5 | Broad(2) | Broad(3) | AZT | ddI |
| 2061 | L | 2 | 44 | 220 | 13 | Broad(3) | Broad(3) | ||
| 2146 | M | 2 | 74 | 750 | 20 | Broad(2) | Broad(3) | AZT | |
| 2216 | H | 2 | 33 | 290 | 6 | Broad(2) | Broad(3) | ||
| 2245 | N | 3 | 64 | 310 | 280 | Broad(2) | Broad(1) | ||
| 3408 | O | 3 | 77 | 270 | 140 | Broad(1) | Broad(2) | HAART | |
| 4021 | Q | 3 | 58 | 240 | 220 | Broad(1) | Broad(1) | AZT | HAART |
| 4468 | R | 2 | 18 | 200 | 9 | Broad(1) | Broad(3) | AZT | |
| R5 switch | |||||||||
| 958 | 5 | 26 | 300d | 30 | Narrow | Broad(2) | AZT | ||
| 965 | 11 | 41 | 310 | 360d | Narrow | Broad(2) | AZT | ||
| 1023 | 4 | 36 | 500 | 500 | Broad(2) | Broad(3) | |||
| 1679 | 23 | 26 | 230 | 81 | Broad(2) | Broad(3) | AZT | ||
| 1991 | 13 | 39 | 260 | 160 | Broad(2) | Broad(3) | AZT | ||
| 2112 | 6 | 42 | 340d | 338 | Broad(1) | Broad(3) | AZT | ||
| 2239 | 2 | 20 | 730 | 410 | Broad(1) | Broad(1) | |||
| 2242 | 18 | 49 | 260 | 119 | Broad(2) | Broad(3) | AZT | ||
| 2282 | 7 | 34 | 350 | 163 | Broad(3) | Broad(2) | AZT | ||
| 2289 | 10 | 48 | 490d | 250 | Broad(2) | Broad(3) | AZT | ||
| 3382 | 3 | 38 | 150 | 160 | Broad(2) | Broad(1) | |||
R5 nonswitch, R5 phenotype was maintained throughout the study period; R5 switch, later acquisition of CXCR4-using virus.
Patient code used by Jansson and coauthors (29).
Narrow, virus using wild-type CCR5 but no chimeric receptor; Broad(1) to Broad(3), virus able to use one to three chimeric receptors in addition to CCR5.
CD4+-T-cell count was not available at the exact time point; the closest previous value is used.
AZT, zidovudine; ddI, didanosine; HAART, highly active antiretroviral therapy.
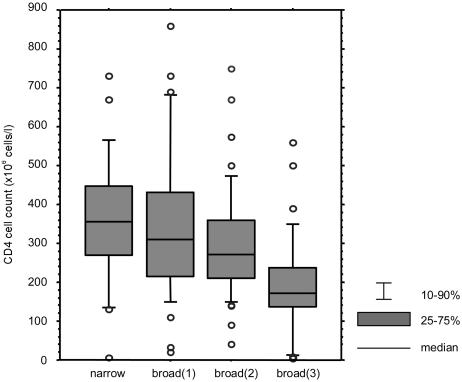
Next we asked whether the R5 phenotype correlated with the degree of immune suppression of the patients from whom the virus was isolated. The results showed that viruses with the R5broad(3) phenotype were more often isolated from patients with low CD4 counts than from patients with high CD4 counts (Fig. 2). For the sake of statistical analysis, two isolates from each patient (the first and last as presented in Table 2) were considered (P = 0.04, Kruskal-Wallis test). For further analysis, our patient group of 31 HIV-1-infected individuals was divided into two groups on the basis of the phenotypes of viruses isolated: 17 patients retained virus with the R5 phenotype throughout the study period, and 14 patients acquired CXCR4-using virus. Accordingly, R5 viruses derived from these two groups of patients were designated R5 nonswitch and R5 switch viruses, respectively. When these two groups were analyzed separately, the correlation between viral phenotype and CD4 count was statistically significant in the nonswitch virus patient group (P = 0.008, Kruskal-Wallis test). These results indicate that during the HIV-1 pathogenic process, the loss of CD4+ T cells parallels the evolution of the R5 phenotype within nonswitch virus patients.
FIG. 2.
Correlation between different R5 phenotypes, i.e., the R5narrow, R5broad(1), R5broad(2), and R5broad(3) phenotypes, and CD4-T-cell counts. CD4+-T-cell counts in the four different groups are significantly different when two isolates are analyzed from each patient as in Table 2 (P = 0.04, Kruskal-Wallis test). Determination of coreceptor use was based on p24 antigen production. We used R5 isolates from both nonswitch and switch virus patients for whom CD4+ T cells were counted on the same day as virus isolation (a total of 142 isolates).
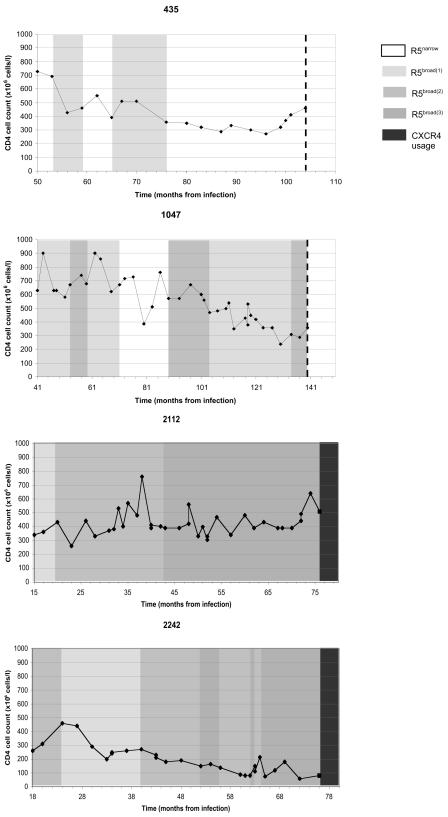
While the evolution of receptor use from the R5narrow to the R5broad phenotype was significantly associated with CD4 decline, evolution in single patients appeared to be discontinuous and was associated with a striking fluctuation of virus variants able to use one (usually FC-2) or several chimeric receptors. These patterns of R5 virus evolution are illustrated for four patients, two of whom were infected with virus that maintained the R5 phenotype during the entire disease course and two who later acquired CXCR4-using isolates (Fig. 3).
FIG. 3.
CD4+-T-cell counts and use of chimeric receptors of R5 viruses for four patients. Patients 435 and 1047 are nonswitch virus patients and yielded virus with the R5 phenotype throughout the study (10 and 14 isolates tested, respectively). The dashed line denotes the end of the study. Patients 2112 and 2242 are switch virus patients who later acquired CXCR4-using virus. Each virus was tested twice. The left axis and line show the CD4+-T-cell counts as numbers of cells (106) per liter of blood. The four different shades of grey and white display changes in viral phenotypes over time, with white indicating the R5narrow phenotype; increasing darkness indicating the R5broad(1), R5broad(2), and R5broad(3) phenotypes; and the darkest grey indicating CXCR4-using virus. The infection date was calculated as the midpoint between the last negative and the first positive sample.
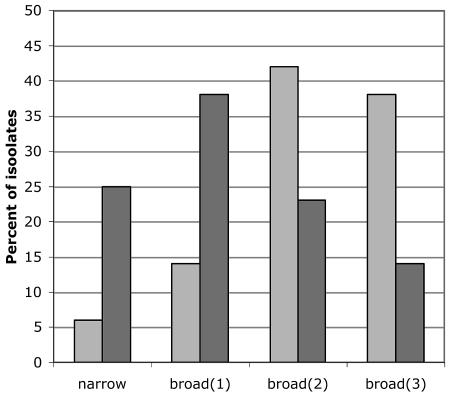
The R5narrow phenotype as well as the different categories of the R5broad phenotype occurred in both the nonswitch and switch virus groups. However, differences in frequencies of the various phenotypes in the two groups were observed (Fig. 4). The majority of viruses (63%) in the R5 nonswitch group had the R5narrow or R5broad(1) phenotype, and conversely, as many as 80% of the viruses with the R5broad(2) and R5broad(3) phenotypes occurred in the R5 switch group (OR = 6.0; 95% CI, 1.5 to 23.7; P = 0.01) when the R5 switch group was compared with the R5 nonswitch group and adjustment was made for CD4 status (logistic regression), using two isolates from each patient as described in Table 2. This finding suggests that while evolution of the R5 phenotype appears to be a general phenomenon, evolution is more pronounced in patients whose virus later switches to CXCR4 use.
FIG. 4.
Distribution of R5 isolates from nonswitch virus patients (dark gray, 64 isolates) and switch virus patients (light gray, 109 isolates) in the different R5 phenotypes; levels of viruses with the R5narrow, R5broad(1), R5broad(2), and R5broad(3) phenotypes are shown as percentages of replicating isolates. The majority of viruses in the nonswitch virus group had the R5narrow or R5broad(1) phenotype, and conversely, most of the isolates in the switch virus group had the R5broad(2) or R5broad(3) phenotype. Statistical analysis was carried out with two isolates from each patient (compare Table 2) after adjustment for CD4 status (logistic regression) (OR = 6.0; 95% CI, 1.5 to 23.7; P = 0.01).
Considering the use of FC-4b only, R5 viruses from switching individuals may more frequently utilize this chimeric receptor than R5 viruses from those not switching (P = 0.01, chi-square test) (Table 1). This suggests that R5 switch isolates are more flexible in their use of the CCR5 receptor than R5 nonswitch isolates since the second and third extracellular loops together are sufficient for infection to take place.
Infectivities of R5 viruses with different phenotypes at the wild-type CCR5 receptor.
In order to clarify whether the ability to use a broad range of chimeric receptors also influences infectivity, we compared the infectious titers of viruses with the R5narrow and R5broad phenotypes in a plaque assay using U87.CD4-CCR5 cells (54). A pairwise comparison of infectious titers of sequential R5 viruses with different phenotypic characteristics, derived from individual patients, was performed. A titer comparison, illustrated in Fig. 5 with ratios of R5broad to R5narrow viruses isolated sequentially from seven individuals, showed that R5broad isolates had significantly higher infectious titers than R5narrow isolates (P = 0.04, Wilcoxon signed-rank test). Thus, viruses with a broad capacity to use CCR5 and CXCR4 chimeric receptors also showed increased infectivity on CCR5-expressing cells.
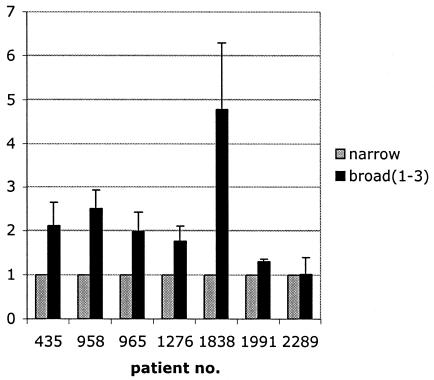
FIG. 5.
Comparison of the infectivities of sequential R5narrow and R5broad(1) to R5broad(3) isolates from seven patients. A plaque assay with U87.CD4-CCR5 cells was used, and titers were determined at the same time with the same amount of p24 antigen per volume of virus supernatant. A pairwise comparison of sequential isolates with different R5 phenotypes was performed. Results are shown as ratios of numbers of plaque-forming units of R5broad(1), R5(broad2), and R5(broad3) isolates to R5narrow isolates per milliliter, where the infectivity of R5narrow was set at 1. Error bars show intra-assay variation. Infectious titers of R5broad(1), R5(broad2), and R5(broad3) isolates were significantly higher than those of isolates with the R5narrow phenotype (P = 0.04, Wilcoxon signed-rank test).
Resistance to inhibition by RANTES is related to chimeric receptor use.
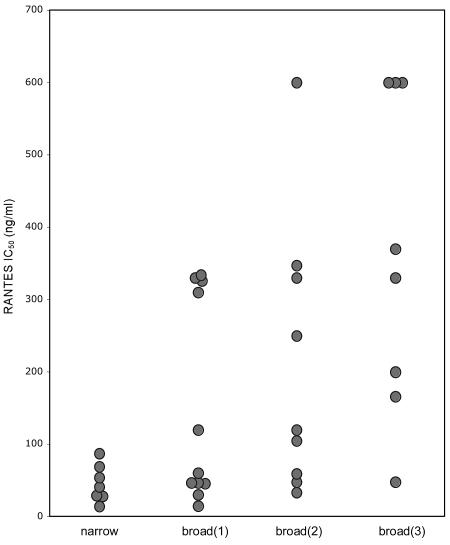
In an earlier work (29) 24 isolates from 12 patients in the R5 nonswitch virus group were tested on PBMC for sensitivity to inhibition by the CC chemokine RANTES. The results showed that R5 viruses with reduced RANTES sensitivity could be isolated from five patients with severe immune suppression. With this in mind, we asked if there is any relationship between RANTES sensitivity and chimeric receptor usage. We plotted RANTES IC50 data previously obtained (29), together with the results of RANTES inhibitions performed with 11 isolates from an additional four patients in the R5 nonswitch virus group (a total of 35 R5 isolates from 16 patients) against the use of chimeric receptors (Fig. 6). This analysis revealed a significant correlation between the evolution of the R5 phenotype and sensitivity to inhibition by RANTES (P = 0.01, Kruskal-Wallis test). Viruses with the R5narrow phenotype had the lowest RANTES IC50s, while R5broad(3) viruses had the highest (Fig. 6). Thus, the mode of the R5 virus-coreceptor interaction, monitored through chimeric receptor use, translated into RANTES inhibition sensitivity. Further analysis also showed a significant inverse correlation (P = 0.04 according to Spearman rank correlation analysis) between CD4-T-cell counts at the time of R5 virus isolation and the RANTES sensitivity of corresponding isolates (data not shown). Our observations suggest that during HIV-1 disease progression, an increased resistance to inhibition by RANTES is concomitant with the evolution of the R5 phenotype.
FIG. 6.
Correlation between chimeric receptor use and RANTES sensitivity of 35 R5 isolates from patients maintaining the R5 phenotype during the entire disease course. A broader use of chimeric CCR5 and CXCR4 receptors correlated with reduced RANTES sensitivity (P = 0.01, Kruskal-Wallis test). RANTES sensitivity is indicated with IC50s.
DISCUSSION
The use of receptor chimeras between CXCR4 and CCR5 allowed us to gain further insight into HIV biological variation. We found that HIV-1 isolates able to use CCR5 but not CXCR4 (R5 phenotype) vary in their capacity to use chimeric receptors. The change from an exclusive use of wild-type CCR5 (R5narrow) to the use of increasing numbers of chimeric receptors [R5broad(1) to R5(broad3)] over time is interpreted as the evolution to extended flexibility in the use of the CCR5 receptor. Viruses with flexible CCR5 use had higher infectivity at the wild-type CCR5 than isolates with the R5narrow phenotype. Moreover, a broadened R5 phenotype also correlated with a reduced number of CD4+ T cells within nonswitch virus patients, indicating that the increased flexibility of R5 isolates to use CCR5 seen in vitro is also relevant in vivo and may influence disease progression.
Our results show that the evolution of HIV-1 coreceptor use in the course of disease progression not only encompasses a switch to CXCR4 use but is a process that involves the evolution of the R5 phenotype as well. R5narrow isolates were preferentially isolated from patients with the highest CD4+-T-cell counts, while R5broad(3) isolates were most frequently found during severe immunodeficiency. Viruses evolved to a more flexible and more efficient CCR5 use even if a switch to CXCR4 use occurred later during the observation period. However, the broader R5 phenotypes were more common in patients whose viruses switched phenotypes than in patients who retained R5 phenotype virus throughout the course of disease.
Previous studies using mutagenesis and biochemistry have suggested that CCR5 binding and host cell infection of R5 HIV-1 variants depend on the highly acidic and tyrosine-rich amino-terminal region of CCR5 (20, 23). However, other regions of CCR5, such as the second extracellular loop, have also been implied to play a role in infection by R5 viruses (38, 44). Similar observations were noted in studies using various types of receptors (1, 9, 17, 46, 49). In a recent study by Platt et al., the passage of R5 virus on cells expressing CCR5 containing amino-terminal mutations resulted in adapted viruses with mutations in the gp120 V3 loop (47). Adaptation led to enhanced fusogenicity and was interpreted by those authors such that the viruses either were not dependent on the affinity between gp120 and CCR5 or showed enhanced affinity to the coreceptor. Interestingly, the apparent increased affinity of gp120 to CCR5 depended on interaction with the second extracellular loop and not the N terminus. It is likely also in our study that the interaction of R5 viruses with receptor chimeras occurs through the second extracellular loop and that this confers increased receptor affinity and the capacity to yield higher infectious titers at wild-type CCR5. Indeed, it was previously reported that gp120s of different R5 viruses may bind CCR5 with different affinities (16, 64). In the present study, we noted a striking difference in the use of one chimeric receptor, FC-4b, by R5 isolates derived from patients later yielding CXCR4-using isolates and R5 isolates from those patients continuing to yield only R5 isolates. Thus, these results suggest that R5 variants with the ability to use FC-4b contain envelope structures that are more prone to mutate towards CXCR4 usage.
What is the selective pressure that drives HIV-1 to a flexible and more efficient use of CCR5? Our results show that resistance to inhibition by the CCR5 ligand RANTES parallels the evolution of CCR5 coreceptor use by the virus and disease progression in the patient. We suggest that HIV-1 variants that develop RANTES resistance may have an advantage and successfully compete for the receptor even in the presence of the natural ligand(s). It is possible that the selection is driven by RANTES or any other CCR5 ligand(s). We previously reported on the evolution of R5 isolates in relation to reduced RANTES sensitivity at the time of severe immunosuppression within patients maintaining CCR5-dependent isolates during the entire disease course (29, 30). This observation has recently also been confirmed by Koning and coauthors (34). Thus, in relation to chimeric receptor use, it is intriguing to note that among isolates from patients infected with a virus that maintains the R5 phenotype, we found that broadness in chimeric receptor use correlated with resistance to RANTES inhibition. Taken together, these observations suggest that virus-coreceptor interactions may be altered also within infected individuals who develop AIDS while continuing to yield truly CCR5-dependent isolates. Experimentally, it has been shown that in vitro passage of an R5 virus in the presence of a CCR5 antagonist selected for a highly resistant escape mutant still dependent on CCR5 for host cell entry (58). Variation in affinity has also been shown to translate into divergence in R5 virus sensitivity to entry inhibitors such as T20 and TAK-779 (48). Thus, our findings of R5 variants with RANTES resistance and broadened flexibility during coreceptor binding, emerging concomitantly with disease progression, may be important in the optimal design of new, effective HIV-1 entry inhibitors.
An alternative mechanism for the selection of R5 variants with an altered receptor interaction may be a reduced expression of CCR5 on target cells. Down-regulation of CCR5 as a consequence of CCR5-ligand binding or altered cytokine balance has been suggested to occur during HIV-1 disease progression (3, 59). Several cytokines, such as IL-4, IL-7, and IL-10, have been shown to down-regulate CCR5, and increased expression of these cytokines has also been implied in the triggering of the HIV-1 coreceptor switch (40, 45, 60). The selective loss of certain CD4+-cell subsets during HIV-1 disease progression has also been implied to occur in the selection of certain virus variants (5, 61). In line with this, we believe that R5 variants with an altered CCR5 engagement also may emerge as a consequence of changed CCR5 availability.
In summary, we show that HIV-1 evolves in vivo towards a more efficient use of CCR5. It is likely that this is a selection process that favors the replication of virus variants that bind CCR5 in an alternative way, or with higher affinity, as a consequence of competition with the natural ligand or changed CCR5 availability.
Acknowledgments
The expert technical assistance of Elzbieta Vincic, Margareta Pusch, and Kajsa Aperia and statistical advice by Håkan Lövkvist, Jonas Björk, and Birgitta Holmgren are greatly appreciated.
This work was supported by the Swedish Research Council, the Swedish International Development Agency/Department for Research Cooperation (SIDA/SAREC), and the Network for Inflammation Research, funded by the Swedish Foundation for Strategic Research. Grants were also received from GS Development, Ingabritt and Arne Lundberg's Foundation, Alfred Österlund's Foundation, the Crafoord Foundation, Magn Bergvall's Foundation, the Lars Hiertas Minne Foundation, and the Royal Physiographic Society in Lund, Sweden.
REFERENCES
- 1.Alkhatib, G., S. S. Ahuja, D. Light, S. Mummidi, E. A. Berger, and S. K. Ahuja. 1997. CC chemokine receptor 5-mediated signaling and HIV-1 co-receptor activity share common structural determinants. Critical residues in the third extracellular loop support HIV-1 fusion. J. Biol. Chem. 272:19771-19776. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib, G., M. Locati, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1997. HIV-1 coreceptor activity of CCR5 and its inhibition by chemokines: independence from G protein signaling and importance of coreceptor downmodulation. Virology 234:340-348. [DOI] [PubMed] [Google Scholar]
- 4.Antonsson, L., A. Boketoft, A. Garzino-Demo, B. Olde, and C. Owman. 2003. Molecular mapping of epitopes for interaction of HIV-1 as well as natural ligands with the chemokine receptors, CCR5 and CXCR4. AIDS 17:2571-2579. [DOI] [PubMed] [Google Scholar]
- 5.Auewarakul, P., K. Sangsiriwut, S. Suwanagool, and C. Wasi. 2001. Target cell populations of human immunodeficiency virus type 1 in peripheral blood lymphocytes with different chemokine receptors at various stages of disease progression. J. Virol. 75:6384-6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babiker, A. G., T. Peto, K. Porter, A. S. Walker, and J. H. Darbyshire. 2001. Age as a determinant of survival in HIV infection. J. Clin. Epidemiol. 54(Suppl. 1):S16-S21. [DOI] [PubMed] [Google Scholar]
- 7.Berger, E. A., R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391:240. [DOI] [PubMed] [Google Scholar]
- 8.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 9.Bieniasz, P. D., R. A. Fridell, I. Aramori, S. S. Ferguson, M. G. Caron, and B. R. Cullen. 1997. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 16:2599-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjorndal, A., H. Deng, M. Jansson, J. R. Fiore, C. Colognesi, A. Karlsson, J. Albert, G. Scarlatti, D. R. Littman, and E. M. Fenyo. 1997. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J. Virol. 71:7478-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanchard, A., L. Montagnier, and M. L. Gougeon. 1997. Influence of microbial infections on the progression of HIV disease. Trends Microbiol. 5:326-331. [DOI] [PubMed] [Google Scholar]
- 12.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 13.Connor, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 15.de Roda Husman, A. M., R. P. van Rij, H. Blaak, S. Broersen, and H. Schuitemaker. 1999. Adaptation to promiscuous usage of chemokine receptors is not a prerequisite for human immunodeficiency virus type 1 disease progression. J. Infect. Dis. 180:1106-1115. [DOI] [PubMed] [Google Scholar]
- 16.Doranz, B. J., S. S. Baik, and R. W. Doms. 1999. Use of a gp120 binding assay to dissect the requirements and kinetics of human immunodeficiency virus fusion events. J. Virol. 73:10346-10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doranz, B. J., Z. H. Lu, J. Rucker, T. Y. Zhang, M. Sharron, Y. H. Cen, Z. X. Wang, H. H. Guo, J. G. Du, M. A. Accavitti, R. W. Doms, and S. C. Peiper. 1997. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J. Virol. 71:6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 19.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 20.Dragic, T., A. Trkola, S. W. Lin, K. A. Nagashima, F. Kajumo, L. Zhao, W. C. Olson, L. Wu, C. R. Mackay, G. P. Allaway, T. P. Sakmar, J. P. Moore, and P. J. Maddon. 1998. Amino-terminal substitutions in the CCR5 coreceptor impair gp120 binding and human immunodeficiency virus type 1 entry. J. Virol. 72:279-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etemad-Moghadam, B., D. Rhone, T. Steenbeke, Y. Sun, J. Manola, R. Gelman, J. W. Fanton, P. Racz, K. Tenner-Racz, M. K. Axthelm, N. L. Letvin, and J. Sodroski. 2001. Membrane-fusing capacity of the human immunodeficiency virus envelope proteins determines the efficiency of CD+ T-cell depletion in macaques infected by a simian-human immunodeficiency virus. J. Virol. 75:5646-5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etemad-Moghadam, B., Y. Sun, E. K. Nicholson, M. Fernandes, K. Liou, R. Gomila, J. Lee, and J. Sodroski. 2000. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J. Virol. 74:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farzan, M., H. Choe, L. Vaca, K. Martin, Y. Sun, E. Desjardins, N. Ruffing, L. Wu, R. Wyatt, N. Gerard, C. Gerard, and J. Sodroski. 1998. A tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J. Virol. 72:1160-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 25.Fenyo, E. M. 2001. The role of virus biological phenotype in human immunodeficiency virus pathogenesis. AIDS Rev. 3:157-168. [Google Scholar]
- 26.Fiore, J. R., A. Bjorndal, K. A. Peipke, M. Di Stefano, G. Angarano, G. Pastore, H. Gaines, E. M. Fenyo, and J. Albert. 1994. The biological phenotype of HIV-1 is usually retained during and after sexual transmission. Virology 204:297-303. [DOI] [PubMed] [Google Scholar]
- 27.Fiore, J. R., G. Buccoliero, P. Pezzotti, G. Rezza, A. Saracino, G. Pastore, E. M. Fenyo, and G. Angarano. 1999. HIV-1 disease progression in women: role of the viral phenotype of the HIV-positive steady partner. AIDS 13:1801-1802. [DOI] [PubMed] [Google Scholar]
- 28.Hu, Q. X., A. P. Barry, Z. X. Wang, S. M. Connolly, S. C. Peiper, and M. L. Greenberg. 2000. Evolution of the human immunodeficiency virus type 1 envelope during infection reveals molecular corollaries of specificity for coreceptor utilization and AIDS pathogenesis. J. Virol. 74:11858-11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansson, M., E. Backstrom, A. Bjorndal, V. Holmberg, P. Rossi, E. M. Fenyo, M. Popovic, J. Albert, and H. Wigzell. 1999. Coreceptor usage and RANTES sensitivity of non-syncytium-inducing HIV-1 isolates obtained from patients with AIDS. J. Hum. Virol. 2:325-338. [PubMed] [Google Scholar]
- 30.Jansson, M., M. Popovic, A. Karlsson, F. Cocchi, P. Rossi, J. Albert, and H. Wigzell. 1996. Sensitivity to inhibition by beta-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc. Natl. Acad. Sci. USA 93:15382-15387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jensen, M. A., F. S. Li, A. B. van't Wout, D. C. Nickle, D. Shriner, H. X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karlsson, A., K. Parsmyr, E. Sandstrom, E. M. Fenyo, and J. Albert. 1994. MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J. Clin. Microbiol. 32:364-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karlsson, I., L. Antonsson, Y. Shi, A. Karlsson, J. Albert, T. Leitner, B. Olde, C. Owman, and E. M. Fenyo. 2003. HIV biological variability unveiled: frequent isolations and chimeric receptors reveal unprecedented variation of coreceptor use. AIDS 17:2561-2569. [DOI] [PubMed] [Google Scholar]
- 34.Koning, F. A., D. Kwa, B. Boeser-Nunnink, J. Dekker, J. Vingerhoed, H. Hiemstra, and H. Schuitemaker. 2003. Decreasing sensitivity to RANTES (regulated on activation, normally T cell-expressed and -secreted) neutralization of CC chemokine receptor 5-using, non-syncytium-inducing virus variants in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 188:864-872. [DOI] [PubMed] [Google Scholar]
- 35.Koot, M., I. P. Keet, A. H. Vos, R. E. de Goede, M. T. Roos, R. A. Coutinho, F. Miedema, P. T. Schellekens, and M. Tersmette. 1993. Prognostic value of HIV-1 syncytium-inducing phenotype for rate of CD4+ cell depletion and progression to AIDS. Ann. Intern. Med. 118:681-688. [DOI] [PubMed] [Google Scholar]
- 36.Kwa, D., J. Vingerhoed, B. Boeser, and H. Schuitemaker. 2003. Increased in vitro cytopathicity of CC chemokine receptor 5-restricted human immunodeficiency virus type 1 primary isolates correlates with a progressive clinical course of infection. J. Infect. Dis. 187:1397-1403. [DOI] [PubMed] [Google Scholar]
- 37.Lawn, S. D., S. T. Butera, and T. M. Folks. 2001. Contribution of immune activation to the pathogenesis and transmission of human immunodeficiency virus type 1 infection. Clin. Microbiol. Rev 14:753-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, B., M. Sharron, C. Blanpain, B. J. Doranz, J. Vakili, P. Setoh, E. Berg, G. Liu, H. R. Guy, S. R. Durell, M. Parmentier, C. N. Chang, K. Price, M. Tsang, and R. W. Doms. 1999. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J. Biol. Chem. 274:9617-9626. [DOI] [PubMed] [Google Scholar]
- 39.Leitner, T., G. Korovina, S. Marquina, T. Smolskaya, and J. Albert. 1996. Molecular epidemiology and MT-2 cell tropism of Russian HIV type 1 variant. AIDS Res. Hum. Retrovir. 12:1595-1603. [DOI] [PubMed] [Google Scholar]
- 40.Llano, A., J. Barretina, A. Gutierrez, J. Blanco, C. Cabrera, B. Clotet, and J. A. Este. 2001. Interleukin-7 in plasma correlates with CD4 T-cell depletion and may be associated with emergence of syncytium-inducing variants in human immunodeficiency virus type 1-positive individuals. J. Virol. 75:10319-10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long, E. M., S. M. Rainwater, L. Lavreys, K. Mandaliya, and J. Overbaugh. 2002. HIV type 1 variants transmitted to women in Kenya require the CCR5 coreceptor for entry, regardless of the genetic complexity of the infecting virus. AIDS Res. Hum. Retrovir. 18:567-576. [DOI] [PubMed] [Google Scholar]
- 42.McNearney, T., Z. Hornickova, R. Markham, A. Birdwell, M. Arens, A. Saah, and L. Ratner. 1992. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc. Natl. Acad. Sci. USA 89:10247-10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mellors, J. W., A. Munoz, J. V. Giorgi, J. B. Margolick, C. J. Tassoni, P. Gupta, L. A. Kingsley, J. A. Todd, A. J. Saah, R. Detels, J. P. Phair, and C. R. Rinaldo, Jr. 1997. Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann. Intern. Med. 126:946-954. [DOI] [PubMed] [Google Scholar]
- 44.Olson, W. C., G. E. Rabut, K. A. Nagashima, D. N. Tran, D. J. Anselma, S. P. Monard, J. P. Segal, D. A. Thompson, F. Kajumo, Y. Guo, J. P. Moore, P. J. Maddon, and T. Dragic. 1999. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J. Virol. 73:4145-4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patterson, B. K., M. Czerniewski, J. Andersson, Y. Sullivan, F. Su, D. Jiyamapa, Z. Burki, and A. Landay. 1999. Regulation of CCR5 and CXCR4 expression by type 1 and type 2 cytokines: CCR5 expression is downregulated by IL-10 in CD4-positive lymphocytes. Clin. Immunol. 91:254-262. [DOI] [PubMed] [Google Scholar]
- 46.Picard, L., G. Simmons, C. A. Power, A. Meyer, R. A. Weiss, and P. R. Clapham. 1997. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J. Virol. 71:5003-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Platt, E. J., S. E. Kuhmann, P. P. Rose, and D. Kabat. 2001. Adaptive mutations in the V3 loop of gp120 enhance fusogenicity of human immunodeficiency virus type 1 and enable use of a CCR5 coreceptor that lacks the amino-terminal sulfated region. J. Virol. 75:12266-12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rucker, J., M. Samson, B. J. Doranz, F. Libert, J. F. Berson, Y. Yi, R. J. Smyth, R. G. Collman, C. C. Broder, G. Vassart, R. W. Doms, and M. Parmentier. 1996. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell 87:437-446. [DOI] [PubMed] [Google Scholar]
- 50.Scarlatti, G., V. Lombardi, A. Plebani, N. Principi, C. Vegni, G. Ferraris, A. Bucceri, E. M. Fenyo, H. Wigzell, P. Rossi, et al. 1991. Polymerase chain reaction, virus isolation and antigen assay in HIV-1-antibody-positive mothers and their children. AIDS 5:1173-1178. [DOI] [PubMed] [Google Scholar]
- 51.Scarlatti, G., E. Tresoldi, A. Bjorndal, R. Fredriksson, C. Colognesi, H. K. Deng, M. S. Malnati, A. Plebani, A. G. Siccardi, D. R. Littman, E. M. Fenyo, and P. Lusso. 1997. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat. Med. 3:1259-1265. [DOI] [PubMed] [Google Scholar]
- 52.Shankarappa, R., P. Gupta, G. H. Learn, Jr., A. G. Rodrigo, C. R. Rinaldo, Jr., M. C. Gorry, J. I. Mullins, P. L. Nara, and G. D. Ehrlich. 1998. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 241:251-259. [DOI] [PubMed] [Google Scholar]
- 53.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi, Y., J. Albert, G. Francis, H. Holmes, and E. M. Fenyo. 2002. A new cell line-based neutralization assay for primary HIV type 1 isolates. AIDS Res. Hum. Retrovir. 18:957-967. [DOI] [PubMed] [Google Scholar]
- 55.Singh, A., and R. G. Collman. 2000. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J. Virol. 74:10229-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sundqvist, V. A., J. Albert, E. Ohlsson, J. Hinkula, E. M. Fenyo, and B. Wahren. 1989. Human immunodeficiency virus type 1 p24 production and antigenic variation in tissue culture of isolates with various growth characteristics. J. Med. Virol. 29:170-175. [DOI] [PubMed] [Google Scholar]
- 57.Tersmette, M., R. A. Gruters, F. de Wolf, R. E. de Goede, J. M. Lange, P. T. Schellekens, J. Goudsmit, H. G. Huisman, and F. Miedema. 1989. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J. Virol. 63:2118-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trkola, A., S. E. Kuhmann, J. M. Strizki, E. Maxwell, T. Ketas, T. Morgan, P. Pugach, S. Xu, L. Wojcik, J. Tagat, A. Palani, S. Shapiro, J. W. Clader, S. McCombie, G. R. Reyes, B. M. Baroudy, and J. P. Moore. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trkola, A., W. A. Paxton, S. P. Monard, J. A. Hoxie, M. A. Siani, D. A. Thompson, L. Wu, C. R. Mackay, R. Horuk, and J. P. Moore. 1998. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J. Virol. 72:396-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valentin, A., W. Lu, M. Rosati, R. Schneider, J. Albert, A. Karlsson, and G. N. Pavlakis. 1998. Dual effect of interleukin 4 on HIV-1 expression: implications for viral phenotypic switch and disease progression. Proc. Natl. Acad. Sci. USA 95:8886-8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Rij, R. P., H. Blaak, J. A. Visser, M. Brouwer, R. Rientsma, S. Broersen, A. M. de Roda Husman, and H. Schuitemaker. 2000. Differential coreceptor expression allows for independent evolution of non-syncytium-inducing and syncytium-inducing HIV-1. J. Clin. Investig. 106:1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Investig. 94:2060-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]
- 64.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]