Abstract
The measles virus (MV) P gene codes for three proteins: P, an essential polymerase cofactor, and V and C, which have multiple functions but are not strictly required for viral propagation in cultured cells. V shares the amino-terminal domain with P but has a zinc-binding carboxyl-terminal domain, whereas C is translated from an overlapping reading frame. During replication, the P protein binds incoming monomeric nucleocapsid (N) proteins with its amino-terminal domain and positions them for assembly into the nascent ribonucleocapsid. The P protein amino-terminal domain is natively unfolded; to probe its conformational flexibility, we fused it to the green fluorescent protein (GFP), thereby also silencing C protein expression. A recombinant virus (MV-GFP/P) expressing hybrid GFP/P and GFP/V proteins in place of standard P and V proteins and not expressing the C protein was rescued and produced normal ratios of mono-, bi-, and tricistronic RNAs, but its replication was slower than that of the parental virus. Thus, the P protein retained nearly intact polymerase cofactor function, even with a large domain added to its amino terminus. Having noted that titers of cell-associated and especially released MV-GFP/P were reduced and knowing that the C protein of the related Sendai virus has particle assembly and infectivity factor functions, we produced an MV-GFP/P derivative expressing C. Intracellular titers of this virus were almost completely restored, and those of released virus were partially restored. Thus, the MV C protein is an infectivity factor.
Measles (MV) is an enveloped virus with a nonsegmented negative-strand RNA genome belonging to the Morbillivirus genus of the Paramyxoviridae family (37). The MV genome, as that of other Paramyxoviridae, including the model species Sendai virus (SeV), is tightly encapsidated by nucleocapsid (N) proteins, each one covering exactly 6 bp and forming a nuclease-resistant helical ribonucleocapsid (RNP) (19, 50). As initially inferred by analogy with SeV, the MV N protein is divided in a large well-conserved core domain, comprising most of the protein and supporting self-assembly and RNA binding (6), and a hypervariable small carboxyl-terminal tail that is intrinsically disordered (38). The N protein tail is also required for the RNP to act as a template for viral RNA synthesis (5, 15, 65).
Two others proteins are part of the RNP, the polymerase (L, for large) and the phosphoprotein P, a polymerase cofactor. The viral polymerase transcribes the six MV genes into mRNAs in a sequential and polar manner (26). At each gene junction transcription is terminated and reinitiated, leading to the production of monocistronic mRNAs. When the polymerase complex does not stop at gene junctions, bicistronic or polycistronic mRNAs are produced (11). In the replicative mode, the polymerase complex never stops at gene junctions, producing complete copies of the viral genome.
The P protein of Paramyxoviridae also has distinct functional domains (37). The carboxyl-terminal half is most conserved and contains regions required for transcription, in particular the polymerase-binding region (4, 13, 59) and sequences responsible for RNP binding (27, 54). In addition, it contains a coiled-coil motif responsible for oligomerization (14, 60); stable tetramers are the active form of SeV P for replication and transcription (36). The P protein extreme-carboxyl-terminal domain (amino acids 459 to 507) triggers an unstructured-to-structured transition of the N protein tail (38). The P protein amino-terminal domain (amino acids 1 to 231) is unstructured in solution and may undergo induced folding upon binding to monomeric N protein (34). This domain provides several additional functions required for replication and contains the conserved phosphorylation sites (37).
The MV P gene encodes the two additional proteins C and V from overlapping open reading frames (ORF) accessed by different mechanisms. The V protein shares the unstructured amino-terminal domain of the P protein, but its 68 carboxyl-terminal highly conserved amino acids forming a zinc-binding domain (47) are translated from a different ORF accessed by the cotranscriptional insertion of a pseudo-templated G residue (10). V proteins of Paramyxoviridae are dispensable for replication in cultured cells (17, 55) but are essential for virulence (18, 35, 55, 62). Their main function may be interference with host defense mechanisms, in particular interferon activation (46, 52, 53). The amino-terminal part of the SeV V protein regulates viral genome replication (29, 61).
The C protein, produced by translation of an ORF initiated 19 nucleotides downstream of the P/V start codon, colocalizes with the MV RNP in infected cells (2). Recombinant viruses defective for C protein expression grow well in certain cultured cells (48) but not in human peripheral blood mononuclear cells (20). The MV C protein reduces viral transcription in a chloramphenicol acetyltransferase reporter mini-genome assay (51) and inhibits the type I interferon response (56).
In the present study, we joined a reporter protein (green fluorescent protein [GFP]) to either end of the N or P protein and verified whether the hybrid proteins retained their functions. Remarkably, hybrid GFP/P and GFP/V proteins were able to functionally substitute standard P and V proteins even in a viral infection context. We also found that C protein expression silencing interfered with particle infectivity.
MATERIALS AND METHODS
Cells and viruses.
Vero (African green monkey kidney) cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, penicillin, and streptomycin at 37°C and 5% CO2. 293-3-46 helper cells (49) were grown in Dulbecco's modified Eagle's medium and 10% fetal bovine serum in the presence of 1.2 mg of G418/ml.
To prepare virus stocks, Vero cells were infected at a multiplicity of infection (MOI) of 0.05 50% tissue culture infective doses (TCID50)/cell with the relevant virus and incubated at 37°C. Cells were scraped in Opti-MEM (Gibco), and particles were released by two freeze-thaw cycles. Titers were determined by TCID50 titration on Vero cells according to the Spearman-Kärber method (32).
Virus growth kinetics were obtained by inoculating Vero cells at an MOI of 0.03 TCID50/cell. At the indicated time points, supernatants were clarified by centrifugation and cells were scraped in Opti-MEM (Gibco) and subjected to freeze-thaw cycles. Released and cell-associated viral titers were determined by TCID50 titration.
Plasmid construction.
The GFP ORF was fused upstream or downstream of the N or P ORFs. The two coding regions were connected through a flexible glycine-rich linker (5′-TCTGGAGGTTCCGGAGGTACCGGTGGA) containing the restriction site BspEI (underlined) to facilitate subsequent cloning. Two restriction sites were introduced upstream of the first ORF (SalI and MluI) or downstream of the second ORF (AatII and SphI) to facilitate cloning in the pCG intermediate vector with SalI and SphI. Then, the full-length MV plasmids were obtained by transfer of a MluI-AatII fragment in p(+)MV(IrGFP)P (63).
To produce the fusion proteins N/GFP and P/GFP, cDNAs encoding N, P, or GFP ORFs were obtained by PCR using the following primers (restriction sites used are underlined in the primer sequences): 5′-GCAGTCGACGCGTCATGGCCACACTTTTAAGG (SalI-MluI) and 5′-GTACCTCCGGAACCTCCTGAGTCTAGAAGATTTCTG (BspEI) for the N ORF; 5′-GCAGTCGACGCGTCATGGCAGAAGAGCAGG (SalI-MluI) and 5′-GTACCTCCGGAACCTCCAGACTTGATTATTATCTTCATCA (BspEI) for the P ORF; 5′-GAGGTTCCGGAGGTACCGGTGGAGTGAGCAAGGGCGAG (BspEI) and 5′-GCAGCATGCGACGTCGTTACTTGTACAGCTCGTCCA (SphI-AatII) for the GFP ORF.
To produce the fusion proteins GFP/N and GFP/P, cDNAs encoding N, P, or GFP ORFs were obtained by PCR using the following primers (restriction sites as above): 5′-GAGGTTCCGGAGGTACCGGTGGAGCCACACTTTTAAGGAG and 5′-GCAGCATGCGACGTCGTCAGTCTAGAAGATTTCTGTC for the N ORF; 5′-GAGGTTCCGGAGGTACCGGTGGAGCAGAAGAGCAGGCAC and 5′-GCAGCATGCGACGTCGCTACTTCATTATTATCTTCA for the P ORF; 5′-GCAGTCGACGCGTCATGGTGAGCAAGGGCG and 5′-GTACCTCCGGAACCTCCAGACTTGTACAGCTCGTCCA for the GFP ORF.
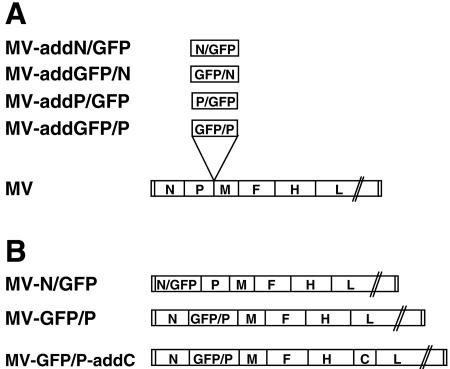
For each construct, the two ORFs were linked in a three-way ligation and cloned in the intermediate pCG vector by using the SalI-BspEI-SphI restriction sites, thus yielding the four plasmids pCG-N/GFP, pCG-P/GFP, pCG-GFP/N, and pCG-GFP/P. Full-length plasmids were obtained by transferring the MluI and AatII fragments in the additional transcription unit located between P and M in the full-length MV genome p(+)MV(IrGFP)P, resulting in the following four plasmids: p(+)MVaddN/GFP, p(+)MVaddP/GFP, p(+)MVaddGFP/N, and p(+)MVaddGFP/P (Fig. 1A). The integrity of all constructs was verified by sequencing.
FIG. 1.
Genomes of recombinant MVs. (A) MVs expressing hybrid proteins in addition to the standard N or P proteins. The MV genome is represented with its 3′ end on the left, and its six genes are indicated by capital letters. The ATUs coding for the hybrid proteins are inserted between the P and M genes. (B) MVs expressing hybrid proteins in place of the standard proteins.
To produce full-length plasmids with the N or P genes replaced by a hybrid GFP gene, site-directed mutagenesis was performed using the QuikChange system (Stratagene). To obtain p(+)MV-N/GFP, a BsiWI restriction site was introduced downstream of the N gene stop codon in pCG-N-P [intermediate plasmid containing the N ORF and part of the P ORF, fragment ScaI-StuI from p(+)MV-NSe (58)]. The same restriction site (underlined) was introduced downstream of the GFP stop codon in pCG-N/GFP, using primers 5′-GTGTACAATGACAGAAATCTTCTAGACTAGCGTACGGTGCGAGAGGCCGAGGACCAG and 5′-GCATGGACGAGCTGTACAAGTAGCGACGTACGATGCAAGCTGATCTTTTTCCCTCTGCC. After mutagenesis, the BstXI-BsiWI fragment from pCG-N/GFP, containing the N/GFP coding sequence was transferred to pCG-N-P in exchange for the corresponding fragment containing only N. Then, p(+)MV-N/GFP was obtained by transfer of a SfiI-SacII fragment in p(+)MV-NSe.
To obtain p(+)MV-GFP/P, the MluI restriction site (underlined) was introduced upstream the ATG of the P gene in pCG-N-P using the following primer: 5′-CCATCCACTCCCACGATTGGAGCAGACACGCGTCTAGATGGCAGAAGAGCAGGCACGCC. A MluI-SacII fragment was transferred from pCG-GFP/P in the MluI-mutagenized pCG-N-P. Then, p(+)MV-GFP/P was obtained by transfer of a SfiI-SacII fragment in p(+)MV-NSe. The integrity of all constructs was verified by sequencing.
Finally, to obtain a virus expressing GFP/P as well as the C protein, the C gene ORF was amplified by PCR using the following primers: 5′-CAGCCCATCAACGCGTATGTCAAAAACGGACTGGAATGC and 5′-TCGATCGCGAGACGTCTCATCAGGAGCTCGTGGATCTCC. The MluI and AatII sites (underlined) were introduced upstream or downstream of the C gene, respectively, to allow cloning in the additional transcription unit of p(+)MV(IrGFP)H, resulting in p(+)MV-addC. p(+)MV-GFP/P-addC was generated by transfer of the PacI-SpeI fragment from p(+)MV-addC to p(+)MV-GFP/P.
Recombinant virus recovery.
Recombinant MVs were generated essentially as described previously by Radecke et al. (49). Briefly, the helper cell line 293-3-46 stably expressing MV N, MV P, and T7 polymerase was transfected by calcium phosphate precipitation using the ProFection kit (Promega) with two plasmids, one coding for the relevant MV genome and the other for the MV polymerase (pEMCLa). Three days after transfection, the helper cells were overlaid on Vero cells, appearance of infectious centers was monitored, and then single syncytia were picked and propagated on Vero cells.
Purification of viral particles.
Vero cells (5 × 106) were infected at an MOI of 0.05 and incubated at 32°C until 80 to 90% of all nuclei were found in syncytia. Before the start of syncytium lysis, supernatants were collected and clarified by centrifugation at 20,000 × g for 20 min at 4°C. The supernatant was then transferred into a 12-ml polypropylene tube (Sorvall) and pelleted by velocity centrifugation (100,000 × g for 90 min at 4°C) through 20% sucrose onto a 60% sucrose cushion prepared in TNE buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA). The virus-containing interphase fraction was harvested, diluted to less than 30% sucrose by addition of TNE buffer, and pelleted by 90 min of centrifugation at 100,000 × g. All liquid was carefully removed, and the pellet was dissolved in 50 μl of urea buffer (200 mM Tris-HCl [pH 6.8], 8 M urea, 0.1 mM EDTA [pH 8], 5% sodium dodecyl sulfate [SDS], 0.03% bromophenol blue) containing 1.5% dithiothreitol and denatured for 10 min at 90°C for fractionation on SDS-polyacrylamide gels as described below.
Western blot analysis of cell extracts.
Cells (4 × 105) were infected with an MOI of 0.1 and, 36 h later or when 70 to 80% of the nuclei were found in syncytia, washed in phosphate-buffered saline (PBS), incubated for 10 min at 4°C in lysis buffer (50 mM Tris [pH 8.0], 62.5 mM EDTA, 0.4% deoxycholate, 1% Igepal [Sigma]) containing protease inhibitors (Complete mix [Roche]) and 1 mM phenylmethylsulfonylfluoride, and centrifuged at 5,000 × g for 10 min at 4°C. The supernatants were denatured with urea buffer for 10 min at 90°C. Samples were fractionated on SDS-polyacrylamide gels, blotted to polyvinylidene difluoride membranes, and subjected to enhanced chemiluminescence detection using antibodies specific for MV N, P, H, F, V, C, or GFP as indicated.
Antibodies.
For N and P detection, mouse monoclonal anti-N antibody (cl25) (23) and an anti-P rabbit polyclonal serum were kindly provided by D. Gerlier. For F and H protein detection, rabbit antipeptide antisera recognizing the 14 carboxyl-terminal F protein amino acids or the 14 amino-terminal H protein amino acids were used (8). For V and C protein detection, similar rabbit antipeptide antisera recognizing the respective carboxyl-terminal amino acids were generated. For this endeavor, the peptide (C)DTRIWHHNLPEIPE (the amino-terminal cysteine was added for coupling purposes), corresponding to MV V amino acids 286 to 299, and the peptide CKLQKEGRSTSS, corresponding to MV C amino acids 176 to 186, were synthesized and coupled to keyhole limpet hemocyanin. These conjugates were used to produce a rabbit antiserum. A commercially available monoclonal antibody (Boehringer) was used to detect the GFP-tagged proteins.
Total RNA extraction and Northern blot analysis.
Vero cells (5 × 105) on six-well plates were infected and incubated at 37°C until extensive cytopathic effect was observed. Total RNA was isolated using the RNeasy mini kit (QIAGEN). Northern blot analysis was performed as described previously (11). Briefly, total RNA was separated by electrophoresis on a 1% agarose formaldehyde gel, transferred to a nylon membrane (Amhersham), and analyzed with a digoxigenin (DIG)-labeled probe (DIG RNA labeling kit; Boehringer [42]) corresponding to 851 bp of MV N mRNA or 787 bp MV H mRNA (11).
Confocal microscopy.
Infected Vero cells (5 × 103 cells in chamber slides [Lab Tec]) were fixed with PBS-2% paraformaldehyde at room temperature. Cells were washed with PBS and then fixed with the ProLong antifade kit (Molecular Probes) and analyzed with a Zeiss LSM 510 confocal microscope.
RESULTS
Production of recombinant MV expressing a GFP hybrid protein in addition to the N or P protein.
We generated hybrid proteins in which the GFP coding region was fused in frame with either end of the N or P coding regions. A flexible linker (SGGSGGTGG [single-letter amino acid code]) was added between the domains to increase the probability of the independent folding function. This linker contributes to preserve GFP function in other contexts (41). After transfection of the plasmids expressing hybrid proteins N/GFP, GFP/N, P/GFP, or GFP/P in Vero cells, proteins of the expected molecular mass were detected (data not shown).
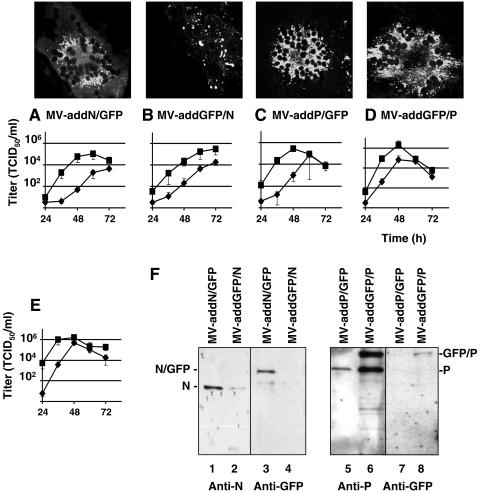
The hybrid genes N/GFP, GFP/N, P/GFP, and GFP/P were then subcloned into a plasmid providing the MV transcription regulatory elements in the form of an additional transcription unit (ATU) located in the 3′-untranslated region of the P gene (Fig. 1A). The structure of the four recombinant MV genomes with an ATU expressing an additional hybrid protein is shown in Fig. 1A. Virus rescue was attempted on helper 293-3-46 cells (49). All four viruses were rescued, and efficiently propagated through the monolayer, and they expressed GFP (Fig. 2A to D, upper panels), indicating that interference effects from the hybrid proteins, if any, were small.
FIG. 2.
Characteristics of the viruses expressing additional hybrid proteins. Vero cells were infected with recombinant viruses expressing hybrid proteins in addition to the viral proteins MV-addN/GFP (A), MV-addGFP/N (B), MV-addP/GFP (C), or MV-addGFP/P (D). (A to D) Expression of the hybrid GFP proteins in infected cells (top panels) and titers of cell-associated (squares) or released (diamonds) virus (bottom graphs). All titers were determined in triplicate. Means and standard deviations are indicated. Small standard deviation intervals are occasionally covered by the symbols. (E) Titers of control MV virus. (F) Western blot of purified virus particles. The viruses used for infection are indicated above each lane, the antibodies used for analysis are shown below the gels, and the proteins detected are shown on the sides.
Only the N/GFP and GFP/P proteins are incorporated in viral particles.
We then addressed the question of whether the intracellular distributions of the hybrid proteins were compatible with their functions. This analysis indicated that N/GFP, P/GFP, and GFP/P were excluded from the nuclei and concentrated in the perinuclear region as the other viral proteins (Fig. 2A, C, and D, upper panels). This localization was similar to that of unmodified N or P proteins (8). In contrast, GFP/N was concentrated in cytoplasmic patches (Fig. 2B, upper panel), thus suggesting exclusion from replication centers.
Growth kinetics of the four recombinant viruses indicated that the maximal cell-associated virus titers were similar (Fig. 2A to D, bottom panels) and only slightly lower than those of the control virus (1.8 × 106 TCID50/ml) (Fig. 2E). The control virus was derived from p(+) MV-NSe (58). In particular, MV-addP/GFP (Fig. 2C, bottom panel) reached almost identical titers with a similar kinetics as the parental control. Maximal supernatant titers of the four recombinant viruses (Fig. 2A to D, bottom panels) were in a similar but slightly lower range than the control and were reached at later time points. These data suggest that certain hybrid proteins may have a slight inhibitory effect on viral replication and particle release but are not dominant negative.
To investigate whether the hybrid proteins retained functional interactions with the unmodified N and P proteins, we asked whether they were incorporated in viral particles isolated from the supernatants of infected cells. The Western blots in Fig. 2F document hybrid protein incorporation in particles of two recombinant viruses: in MV-addN/GFP and MV-addGFP/P, an anti-GFP antibody detected the hybrid proteins (Fig. 2F, lanes 3 and 8, respectively). The GFP/P protein was detected at similar levels as P by using an anti-P antibody (Fig. 2F, lane 6), whereas N/GFP was not detected with an anti-N antibody (Fig. 2F, lane 1) and, thus, its incorporation levels must be much lower than those of N. The GFP/N and P/GFP hybrid proteins were excluded from viral particles (Fig. 2F, lanes 2 and 4 and lanes 5 and 7, respectively).
A recombinant MV expressing GFP/P in place of P can be rescued but replicates inefficiently.
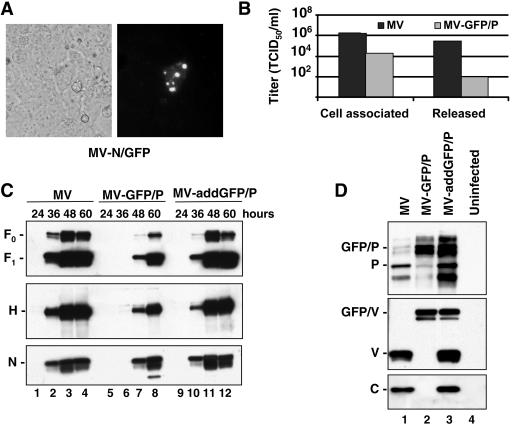
Having shown that the two hybrid proteins N/GFP and GFP/P are incorporated into viral particles, albeit with different efficiencies, we asked whether they could functionally replace the standard proteins. To this purpose we replaced the standard genes in the infectious MV cDNA with the genes coding for the hybrid proteins, producing the recombinant genomes MV-N/GFP and MV-GFP/P (Fig. 1B). We failed to rescue the MV-N/GFP virus, in spite of consistent monitoring of isolated green cells 3 days after transfection of the full-length genome in the helper cell line (Fig. 3A). This suggests that RNA replication and transcription did initially occur, possibly based on the endogenous N protein present in the helper cell line, but that N/GFP was then unable to support efficient virus replication. In contrast, MV-GFP/P was rescued, although it propagated on Vero cells only to low titers (Fig. 3B): maximum titers of cell-associated and released viruses were reduced by about 100 and 3,000 times, respectively, compared to the control virus (Fig. 3B).
FIG. 3.
Characteristics of the viruses expressing hybrid proteins in place of N or P. (A) Rescue cell line 293-3-46 transfected with full-length MV-N/GFP analyzed by fluorescence microscopy. (B) Titers of cell-associated or released virus produced during infection of Vero cells with MV (black columns) or MV-GFP/P (gray columns). Titers were determined based on the TCID50. (C) Western blot analysis of Vero cells infected with the viruses indicated at the top at different time points (24, 36, 48, and 60 h) using anti-Fcyt (top panel), anti-Hcyt (center panel), or anti-N (bottom panel) antibodies. (D) Western blot analysis of Vero cells infected with MV (lane 1), MV-GFP/P (lane 2), or MV-addGFP/P (lane 3) or uninfected (lane 4). Membranes were blotted with antibodies against P (top), V (center), or C (bottom).
MV-GFP/P produces a balanced ratio of all proteins but no C protein.
Having shown that infectious viral particle production by MV-GFP/P is strongly reduced, we sought to characterize the cause of this effect. Suspecting defective polymerase processivity resulting in transcription and protein expression unbalance, we analyzed the amount of N, hemagglutinin (H), and fusion (F) proteins available at 24, 36, 48, and 60 h after infection with the three viruses at the same MOI (Fig. 3C). A significant delay in the production of these three viral proteins was observed for MV-GFP/P compared to MV (Fig. 3C, compare lanes 6 to 8 with lanes 2 to 4). MV-addGFP/P protein expression kinetics was similar to that of the control MV (Fig. 3C, compare lanes 10 to 12 with lanes 2 to 4). Thus, this analysis indicated a significant delay in MV-GFP/P protein expression without detecting an unbalance in viral protein synthesis.
We then analyzed the expression of the P gene products using antibodies specific for the P, V, or C proteins (Fig. 3D, top, center, or bottom panel, respectively) in Western blot assays. As expected, P and V proteins of standard molecular mass were detected in cells infected with the parental MV (lanes 1), whereas larger P and V proteins were seen in MV-GFP/P-infected cells (lane 2) and a mixture of the two forms was found in MV-addGFP/P-infected cells (lane 3). C protein expression was not detectable in MV-GFP/P-infected cells (Fig. 3D, bottom panel, lane 2). The C protein is encoded by an ORF for which the start codon is moved from 19 to more than 700 nucleotides downstream of the P/V ORF AUG upon insertion of the GFP ORF. As a result, C protein expression is silenced.
MV-GFP/P produces normal ratios of mono- and polycistronic RNAs but replicates slowly.
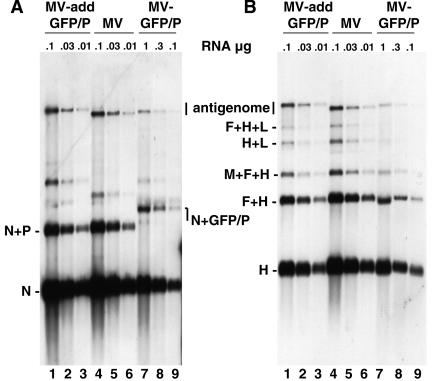
All three P gene proteins have been assigned functions in transcription or the control of replication. To analyze viral transcription in MV-GFP/P-infected Vero cells, we extracted total RNA and performed Northern blotting using either N or H probes (Fig. 4). In control MV-infected cells, in addition to the monocistronic transcripts, polycistronic transcripts were produced when the viral polymerase failed to recognize an intercistronic stop-start signal (Fig. 4A and B, lanes 4 to 6). In MV-GFP/P- or MV-addGFP/P-infected cells (Fig. 4A and B, lanes 7 to 9 or 1 to 3, respectively) the ratios of the monocistronic and polycistronic RNAs, as well as the ratio between different monocistronic mRNAs, were similar to those observed in MV-infected cells. This indicated that in spite of the presence of an additional GFP domain in the P protein and the absence of the C protein, mRNA termination, polyadenylation by repeated copying of a short poly(U) stretch, and reinitiation are not impaired in MV-GFP/P.
FIG. 4.
MV-GFP/P transcription is delayed but not qualitatively altered. Shown is a Northern blot analysis on total RNA purified from Vero cells infected with the viruses indicated at the top. Membranes were incubated with DIG-labeled N (A) or H (B) RNA probes. The positions of the mono-, di-, and selected tricistronic mRNAs and of the antigenomes are indicated on the side. Note that 10 times more total RNA from cells infected with MV-GFP/P than from cells infected with the other viruses was loaded, as indicated above the gels.
Remarkably, the amount of each viral mRNA species detected in infections with MV-GFP/P was about 30 times lower than that detected in MV infections started with the same MOI (Fig. 4, both panels, compare lanes 7 to 9 and 4 to 6; note that 10 times more total RNA from cells infected with MV-GFP/P than from cells infected with the other viruses was loaded). We suspect that the GFP domain added to P slows down transcription and replication of MV-GFP/P.
C protein expression partially restores MV-GFP/P infectivity.
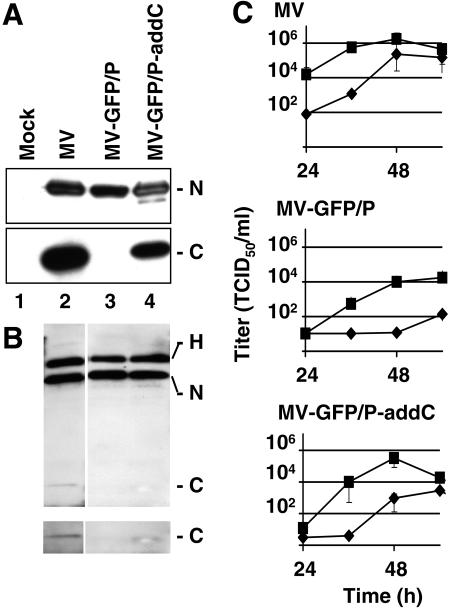
Since MV-GFP/P transcription was slower but otherwise not qualitatively impaired, it seemed possible that the main reason for reduced production of infectious particles was C protein expression silencing. We thus attempted to compensate these defects by adding to the MV-GFP/P genome a transcription unit expressing C (Fig. 1B, bottom). The corresponding virus, named MV-GFP/P-addC, was rescued and expression of the C protein was confirmed by Western blotting (Fig. 5A, lane 4). Other viral proteins were also analyzed and were found to be produced at similar levels as those of the parental virus (MV-GFP/P) (Fig. 5A, top panel, N in lanes 3 and 4, and data not shown).
FIG. 5.
C protein expression partially restores infectivity. (A) Western blot analysis of cells infected with the indicated viruses. Membranes were blotted with antibodies against N (top) or C (bottom). (B) Western blot analysis of purified viral particles. Membranes were blotted with antibodies against H, N, and C. The bottom panel is a longer exposure of the lower portion of the gel shown in the top panel. (C) Growth curves of MV (upper panel), MV-GFP/P (middle panel), or MV-GFP/P-addC (lower panel) in Vero cells infected at an MOI of 0.03. Titers of cell-associated (squares) or released (diamonds) virus are indicated. All titrations were done in triplicate. Means and standard deviations are indicated. Small standard deviation intervals are sometimes covered by symbols.
We then measured the kinetics and maximum titer of cell-associated and released virus produced in MV-GFP/P-addC infections (Fig. 5C, lower panel). Maximum cell-associated virus titers were about 50 times higher than in MV-GFP/P infections (Fig. 5C, center panel) but remained about three times lower than in MV infections (Fig. 5C, upper panel); the kinetics of MV-GFP/P-add C infectious particle production was intermediate between those of MV-GFP/P and MV. Titers of released MV-GFP/P-addC were about 20 times higher than those of MV-GFP/P but remained about 100 times lower than those of MV.
The kinetics of viral protein expression in MV-GFP/P-addC infections (data not shown) was similar to that with MV-GFP/P (Fig. 3C). Thus, C protein expression restoration did not improve transcription or replication. On the other hand, it resulted in 50-times-higher titers of cell-associated virus (Fig. 5C). We conclude that the main role of the C protein in this system is to support the production of infectious virus particles.
To examine C protein incorporation, we analyzed the protein composition of released viral particles concentrated on sucrose gradients. When similar amounts of N protein (corresponding to different virus titers) were loaded on each lane (Fig. 5B), roughly similar amounts of H protein were detected (lanes 2 to 4) in MV, MV-GFP/P, and MV-GFP/P-addC particles. Thus, lack of C protein did not cause a marked N-H incorporation unbalance. C protein was detected at low levels in the MV control particles (Fig. 5B, lane 2, bottom panel) and at levels only slightly above background in MV-GFP/P-addC particles (Fig. 5B, lane 4, bottom panel). Thus, MV C, as for SeV C (28), is a low-abundance structural protein that enhances particle assembly and stabilizes infectivity.
DISCUSSION
We have studied selected functions of the proteins encoded by the MV P gene. We demonstrated that the P protein accurately functions as a polymerase cofactor, even when GFP is attached to its amino terminus. In addition, we showed that the C protein enhances particle assembly and functions as an infectivity factor.
Conformational flexibility of the P and V protein common amino-terminal domain.
A nearly perfect cylindrical domain 42 Å long and 24 Å in diameter (45) was added to the common amino terminus of the P and V proteins without significantly compromising function. In contrast, when the same GFP beta-can domain was added to either the N protein termini or to the P protein carboxyl terminus, it strongly impaired their stability or their functions.
When GFP was added to the N protein amino terminus, which is part of the well-conserved core supporting RNP self-assembly, it caused a change in protein localization and reduced protein stability, possibly due to steric hindrance. When added to the N protein carboxyl terminus, which is intrinsically disordered and exposed on the RNP outer surface, it did not cause protein instability but allowed only inefficient RNP incorporation and also slightly slowed down virus replication (Fig. 2A). Moreover, N/GFP did not functionally replace N. Thus, we suspect that GFP added to the N protein amino terminus interferes with replication and transcription to a point impeding virus rescue.
The hybrid P protein with GFP added to its carboxyl terminus was expressed, but it was not incorporated in virus particles and may not be functional. Since the extreme carboxyl-terminal domain is the region of P responsible for the interaction and the induced folding of the N protein carboxyl-terminal tail (3, 33, 38), it is not surprising that GFP addition to this terminus interferes with its function. On the other hand, the P protein amino-terminal domain is natively unfolded (34) and, therefore, we suspected that it would be more tolerant to modification.
Two P protein domains have been characterized structurally: the tetramerization domain (a coiled-coil; amino acids 320 to 446) (60) and the 60-amino-acid carboxyl-terminal N-binding domain (3, 30). In the case of SeV, the tetrameric P protein carboxyl-terminal half has been modeled into a 160-Å-long rod with a radius of gyration of 48 Å (3). The P protein coiled-coil has been proposed to act as a molecular axle that allows tetrameric P protein rotation while L slides along N proteins arranged in the RNP (36). In this model the carboxyl terminus protrudes from one end of the P tetramer and makes contacts with the N protein carboxyl-terminal tail on the parental RNP. During replication the P amino terminus would recognize incoming monomeric N proteins, positioning them for assembly into the nascent RNP (36).
Our data suggest that four 42-Å-long, 24-Å-diameter GFP cylinders protruding from one end of the tetrameric rod do not interfere with the initial monomeric N protein binding and with their subsequent delivery to the nascent RNP. Indeed MV-addGFP/P particles incorporated similar levels of P and GFP/P (Fig. 2F, lane 6), an observation consistent with structural and functional equivalence between the unmodified and the hybrid protein.
The number of P proteins incorporated in Paramyxoviridae particles is probably in the hundreds range (37). Since each P protein in a recombinant MV-GFP/P is fluorescent, this should allow visualization of single viral particles by microscopic analysis. Indeed, this is the case (data not shown), but the heterogeneous nature of MV particles (50) and the high particle-to-infectivity ratio complicate the analysis of productive viral entry. Nevertheless, the availability of recombinant viruses expressing GFP/P in place of P does allow the real-time study of the cellular compartmentalization of MV replication and transcription.
C protein as infectivity factor.
The P gene of Paramyxoviridae codes for P/V or C on overlapping ORFs. Most Paramyxoviridae express a single C protein (37, 43), but SeV produces a set of four proteins with staggered amino termini. Paramyxoviridae C proteins have been assigned three different functions: transcriptional regulators, infectivity factors, and host control. C proteins have been recognized as down-modulators of mRNA synthesis and genome amplification using synthetic mini-genomes as templates (7, 16, 51) and have also been implicated in viral assembly: a SeV mutant defective for the expression of all four C proteins variants produces particles that are largely noninfectious and have anomalous morphologies and a broad sedimentation profile (28).
C proteins also antagonize the antiviral action of interferons (21). The interactions of the SeV and MV C proteins with the interferon system are complex and involve different STAT proteins (22, 24, 25, 43, 56). While the MV C protein is not essential for virus propagation in Vero cells (48), probably because these cells don't have a functional interferon system, it is required in human peripheral blood mononuclear cells or in HeLa cells to support high viral titers (20, 56).
Here we report that a C-protein-defective MV expressing hybrid GFP/P and GFP/V proteins produces particles that have strongly reduced infectivity and that particles of a derivative of this virus expressing C have partially restored infectivity. It was previously shown that a C-defective MV retains intracellular titers similar to those of the parental strain expressing C protein (48). We have confirmed this observation but have also monitored titers of released virus approximately 100 times lower than those of the parental virus (data not shown). Taken together, these data suggest that the MV C protein is an infectivity factor.
The human immunodeficiency virus virion infectivity factor (Vif) is a low-abundance structural protein (31, 44) that sequesters a cellular RNA-modifying cytidine deaminase (39, 40, 57, 64). It is interesting that infection of MV and other negative-strand RNA viruses can be restricted by another RNA-modifying enzyme, an adenosine deaminase (1, 9, 12). However, not all negative-strand RNA viruses whose genomes were modified by adenosine-to-inosine hypermutation have a C protein homologue that could be inactivated. The possible role of MV C in stabilizing virus particles is currently under investigation.
Acknowledgments
We thank Denis Gerlier and Sonia Longhi for providing the anti-N and anti-P antibodies and reviewing the manuscript, Beda Brun Del Re for expression plasmids, and Sompong Vongpunsawad for excellent technical support.
This work was supported by grants of the Mayo and Siebens Foundations.
REFERENCES
- 1.Bass, B. L., and H. Weintraub. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55:1089-1098. [DOI] [PubMed] [Google Scholar]
- 2.Bellini, W. J., G. Englund, S. Rozenblatt, H. Arnheiter, and C. D. Richardson. 1985. Measles virus P gene codes for two proteins. J. Virol. 53:908-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanchard, L., N. Tarbouriech, M. Blackledge, P. Timmins, W. P. Burmeister, R. W. Ruigrok, and D. Marion. 2004. Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution. Virology 319:201-211. [DOI] [PubMed] [Google Scholar]
- 4.Bowman, M. C., S. Smallwood, and S. A. Moyer. 1999. Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J. Virol. 73:6474-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchholz, C. J., C. Retzler, H. E. Homann, and W. J. Neubert. 1994. The carboxy-terminal domain of Sendai virus nucleocapsid protein is involved in complex formation between phosphoprotein and nucleocapsid-like particles. Virology 204:770-776. [DOI] [PubMed] [Google Scholar]
- 6.Buchholz, C. J., D. Spehner, R. Drillien, W. J. Neubert, and H. E. Homann. 1993. The conserved N-terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J. Virol. 67:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cadd, T., D. Garcin, C. Tapparel, C. R. Pringle, M. Itho, M. Homma, L. Roux, J. Curran, and D. Kolakofsky. 1996. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter specific fashion. J. Virol. 70:5067-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cattaneo, R. 1994. Biased (A→I) hypermutation of animal RNA virus genomes. Curr. Opin. Genet. Dev. 4:895-900. [DOI] [PubMed] [Google Scholar]
- 10.Cattaneo, R., K. Kaelin, K. Baczko, and M. A. Billeter. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759-764. [DOI] [PubMed] [Google Scholar]
- 11.Cattaneo, R., G. Rebmann, A. Schmid, K. Baczko, V. ter Meulen, and M. A. Billeter. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cattaneo, R., A. Schmid, D. Eschle, K. Baczko, V. ter Meulen, and M. A. Billeter. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curran, J. 1996. Reexamination of the Sendai virus P protein domains required for RNA synthesis: a possible supplemental role for the P protein. Virology 221:130-140. [DOI] [PubMed] [Google Scholar]
- 14.Curran, J., R. Boeck, N. Lin-Marq, A. Lupas, and D. Kolakofsky. 1995. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 214:139-149. [DOI] [PubMed] [Google Scholar]
- 15.Curran, J., H. Homann, C. Buchholz, S. Rochat, W. Neubert, and D. Kolakofsky. 1993. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J. Virol. 67:4358-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curran, J., J. B. Marq, and D. Kolakofsky. 1992. The Sendai virus nonstructural C proteins specifically inhibit mRNA synthesis. Virology 189:647-656. [DOI] [PubMed] [Google Scholar]
- 17.Delenda, C., S. Hausmann, D. Garcin, and D. Kolakofsky. 1997. Normal cellular replication of Sendai virus without the trans-frame, nonstructural V protein. Virology 228:55-62. [DOI] [PubMed] [Google Scholar]
- 18.Delenda, C., G. Taylor, S. Hausmann, D. Garcin, and D. Kolakofsky. 1998. Sendai viruses with altered P, V, and W protein expression. Virology 242:327-337. [DOI] [PubMed] [Google Scholar]
- 19.Egelman, E. H., S. S. Wu, M. Amrein, A. Portner, and G. Murti. 1989. The Sendai virus nucleocapsid exists in at least four different helical states. J. Virol. 63:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escoffier, C., S. Manie, S. Vincent, C. P. Muller, M. Billeter, and D. Gerlier. 1999. Nonstructural C protein is required for efficient measles virus replication in human peripheral blood cells. J. Virol. 73:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcin, D., J. B. Marq, L. Strahle, P. le Mercier, and D. Kolakofsky. 2002. All four Sendai Virus C proteins bind Stat1, but only the larger forms also induce its mono-ubiquitination and degradation. Virology 295:256-265. [DOI] [PubMed] [Google Scholar]
- 23.Giraudon, P., M. F. Jacquier, and T. F. Wild. 1988. Antigenic analysis of African measles virus field isolates: identification and localisation of one conserved and two variable epitope sites on the NP protein. Virus Res. 10:137-152. [DOI] [PubMed] [Google Scholar]
- 24.Gotoh, B., T. Komatsu, K. Takeuchi, and J. Yokoo. 2003. The C-terminal half-fragment of the Sendai virus C protein prevents the gamma-activated factor from binding to a gamma-activated sequence site. Virology 316:29-40. [DOI] [PubMed] [Google Scholar]
- 25.Gotoh, B., K. Takeuchi, T. Komatsu, and J. Yokoo. 2003. The STAT2 activation process is a crucial target of Sendai virus C protein for the blockade of alpha interferon signaling. J. Virol. 77:3360-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin, D. E. 2001. Measles virus, p. 1401-1441. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 27.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863-2867. [DOI] [PubMed] [Google Scholar]
- 28.Hasan, M. K., A. Kato, M. Muranaka, R. Yamaguchi, Y. Sakai, I. Hatano, M. Tashiro, and Y. Nagai. 2000. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J. Virol. 74:5619-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horikami, S. M., S. Smallwood, and S. A. Moyer. 1996. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology 222:383-390. [DOI] [PubMed] [Google Scholar]
- 30.Johansson, K., J. M. Bourhis, V. Campanacci, C. Cambillau, B. Canard, and S. Longhi. 2003. Crystal structure of the measles virus phosphoprotein domain responsible for the induced folding of the C-terminal domain of the nucleoprotein. J. Biol. Chem. 278:44567-44573. [DOI] [PubMed] [Google Scholar]
- 31.Kao, S., M. A. Khan, E. Miyagi, R. Plishka, A. Buckler-White, and K. Strebel. 2003. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 77:11398-11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kärber, G. 1931. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Arch. Exp. Pathol. Pharmakol. 162:480-483. [Google Scholar]
- 33.Karlin, D., F. Ferron, B. Canard, and S. Longhi. 2003. Structural disorder and modular organization in Paramyxovirinae N and P. J. Gen. Virol. 84:3239-3252. [DOI] [PubMed] [Google Scholar]
- 34.Karlin, D., S. Longhi, V. Receveur, and B. Canard. 2002. The N-terminal domain of the phosphoprotein of morbilliviruses belongs to the natively unfolded class of proteins. Virology 296:251-262. [DOI] [PubMed] [Google Scholar]
- 35.Kato, A., K. Kiyotani, Y. Sakai, T. Yoshida, and Y. Nagai. 1997. The paramyxovirus, Sendai virus, V protein encodes a luxury function required for viral pathogenesis. EMBO J. 16:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolakofsky, D., P. Le Mercier, F. Iseni, and D. Garcin. 2004. Viral RNA polymerase scanning and the gymnastics of Sendai virus RNA synthesis. Virology 318:463-473. [DOI] [PubMed] [Google Scholar]
- 37.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxovirus: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 38.Longhi, S., V. Receveur-Brechot, D. Karlin, K. Johansson, H. Darbon, D. Bhella, R. Yeo, S. Finet, and B. Canard. 2003. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 278:18638-18648. [DOI] [PubMed] [Google Scholar]
- 39.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 40.Marin, M., K. M. Rose, S. L. Kozak, and D. Kabat. 2003. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9:1398-1403. [DOI] [PubMed] [Google Scholar]
- 41.Miesenbock, G., D. A. De Angelis, and J. E. Rothman. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192-195. [DOI] [PubMed] [Google Scholar]
- 42.Mrkic, B., J. Pavlovic, T. Rulicke, P. Volpe, C. J. Buchholz, D. Hourcade, J. P. Atkinson, A. Aguzzi, and R. Cattaneo. 1998. Measles virus spread and pathogenesis in genetically modified mice. J. Virol. 72:7420-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagai, Y., and A. Kato. 2004. Accessory genes of the Paramyxoviridae, a large family of nonsegmented negative strand RNA viruses, as a focus of active investigation by reverse genetics. Curr. Top. Microbiol. Immunol. 283:197-248. [DOI] [PubMed] [Google Scholar]
- 44.Ohagen, A., and D. Gabuzda. 2000. Role of Vif in stability of human immunodeficiency virus type 1 core. J. Virol. 74:11055-11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ormo, M., A. B. Cubitt, K. Kallio, L. A. Gross, R. Y. Tsien, and S. J. Remington. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science 273:1392-1395. [DOI] [PubMed] [Google Scholar]
- 46.Palosaari, H., J. P. Parisien, J. J. Rodriguez, C. M. Ulane, and C. M. Horvath. 2003. STAT protein interference and suppression of cytokine signal transduction by measles virus V protein. J. Virol. 77:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 48.Radecke, F., and M. A. Billeter. 1996. The nonstructural C protein is not essential for multiplication of Edmonston B strain measles virus in cultured cells. Virology 217:418-421. [DOI] [PubMed] [Google Scholar]
- 49.Radecke, F., P. Spielhofer, H. Schneider, K. Kaelin, M. Huber, C. Dotsch, G. Christiansen, and M. A. Billeter. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773-5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rager, M., S. Vongpunsawad, W. P. Duprex, and R. Cattaneo. 2002. Polyploid measles virus with hexameric genome length. EMBO J. 21:2364-2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reutter, G. L., C. Cortese-Grogan, J. Wilson, and S. A. Moyer. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285:100-109. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez, J. J., J. P. Parisien, and C. M. Horvath. 2002. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 76:11476-11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez, J. J., L. F. Wang, and C. M. Horvath. 2003. Hendra virus V protein inhibits interferon signaling by preventing STAT1 and STAT2 nuclear accumulation. J. Virol. 77:11842-11845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryan, K. W., E. M. Morgan, and A. Portner. 1991. Two noncontiguous regions of Sendai virus P protein combine to form a single nucleocapsid binding domain. Virology 180:126-134. [DOI] [PubMed] [Google Scholar]
- 55.Schneider, H., K. Kaelin, and M. A. Billeter. 1997. Recombinant measles viruses defective for RNA editing and V protein synthesis are viable in cultured cells. Virology 227:314-322. [DOI] [PubMed] [Google Scholar]
- 56.Shaffer, J. A., W. J. Bellini, and P. A. Rota. 2003. The C protein of measles virus inhibits the type I interferon response. Virology 315:389-397. [DOI] [PubMed] [Google Scholar]
- 57.Sheehy, A. M., N. C. Gaddis, and M. H. Malim. 2003. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9:1404-1407. [DOI] [PubMed] [Google Scholar]
- 58.Singh, M., and M. A. Billeter. 1999. A recombinant measles virus expressing biologically active human interleukin-12. J. Gen. Virol. 80:101-106. [DOI] [PubMed] [Google Scholar]
- 59.Smallwood, S., K. W. Ryan, and S. A. Moyer. 1994. Deletion analysis defines a carboxyl-proximal region of Sendai virus P protein that binds to the polymerase L protein. Virology 202:154-163. [DOI] [PubMed] [Google Scholar]
- 60.Tarbouriech, N., J. Curran, R. W. Ruigrok, and W. P. Burmeister. 2000. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7:777-781. [DOI] [PubMed] [Google Scholar]
- 61.Tober, C., M. Seufert, H. Schneider, M. A. Billeter, I. C. Johnston, S. Niewiesk, V. ter Meulen, and S. Schneider-Schaulies. 1998. Expression of measles virus V protein is associated with pathogenicity and control of viral RNA synthesis. J. Virol. 72:8124-8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valsamakis, A., H. Schneider, P. G. Auwaerter, H. Kaneshima, M. A. Billeter, and D. E. Griffin. 1998. Recombinant measles viruses with mutations in the C, V, or F gene have altered growth phenotypes in vivo. J. Virol. 72:7754-7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang, Z., L. Hangartner, T. I. Cornu, L. R. Martin, A. Zuniga, M. A. Billeter, and H. Y. Naim. 2001. Recombinant measles viruses expressing heterologous antigens of mumps and simian immunodeficiency viruses. Vaccine 19:2329-2336. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, X., C. Glendening, F. Linke, C. L. Parks, C. Brooks, S. A. Udem, and M. Oglesbee. 2002. Identification and characterisation of a regulatory domain on the carboxyl terminus of the measles virus nucleocapsid protein. J. Virol. 76:8737-8746. [DOI] [PMC free article] [PubMed] [Google Scholar]