Abstract
Full genome microarrays were used to assess transcriptional responses of Arabidopsis seedlings to changing external supply of the essential macronutrient potassium (K+). Rank product statistics and iterative group analysis were employed to identify differentially regulated genes and statistically significant coregulated sets of functionally related genes. The most prominent response was found for genes linked to the phytohormone jasmonic acid (JA). Transcript levels for the JA biosynthetic enzymes lipoxygenase, allene oxide synthase, and allene oxide cyclase were strongly increased during K+ starvation and quickly decreased after K+ resupply. A large number of well-known JA responsive genes showed the same expression profile, including genes involved in storage of amino acids (VSP), glucosinolate production (CYP79), polyamine biosynthesis (ADC2), and defense (PDF1.2). Our findings highlight a novel role of JA in nutrient signaling and stress management through a variety of physiological processes such as nutrient storage, recycling, and reallocation. Other highly significant K+-responsive genes discovered in our study encoded cell wall proteins (e.g. extensins and arabinogalactans) and ion transporters (e.g. the high-affinity K+ transporter HAK5 and the nitrate transporter NRT2.1) as well as proteins with a putative role in Ca2+ signaling (e.g. calmodulins). On the basis of our results, we propose candidate genes involved in K+ perception and signaling as well as a network of molecular processes underlying plant adaptation to K+ deficiency.
Potassium (K+) is the most abundant inorganic cation in plants, comprising up to 10% of a plant's dry weight (Leigh and Jones, 1984). K+ is an important macronutrient for plants, which carries out vital functions in metabolism, growth, and stress adaptation. These functions can be classified into those that rely on high and relatively stable concentrations of K+ in certain cellular compartments and those that rely on K+ movement between different compartments, cells, or tissues. The first class of functions includes enzyme activation, stabilization of protein synthesis, and neutralization of negative charges on proteins (Marschner, 1995). The second class of functions of K+ is linked to its high mobility. This is particularly evident where K+ movement is the driving force for osmotic changes as, for example, in stomatal movement, light-driven and seismonastic movements of organs, or phloem transport. In other cases K+ movement provides a charge-balancing counter-flux essential for sustaining the movement of other ions. Thus, energy production through H+-ATPases relies on overall H+/K+ exchange (Tester and Blatt, 1989; Wu et al., 1991). The most general phenomenon that requires directed movement of K+ is growth. Accumulation of K+ (together with an anion) in plant vacuoles creates the necessary osmotic potential for rapid cell extension.
K+ deficiency is of great agricultural importance (Laegreid et al., 1999). This fact was recognized early in plant physiological research and has led to a good description of K+ starvation symptoms at the physiological level (Marschner, 1995). It is well established that K+ starvation leads to (1) growth arrest due to the lack of the major osmoticum, (2) impaired nitrogen and sugar balance due to inhibition of protein synthesis, photosynthesis, and long-distance transport, and (3) increased susceptibility to pathogens probably due to increased levels of low Mr nitrogen and sugar compounds. In a natural environment low-K+ conditions are often transient and therefore plants have developed mechanisms to adapt to short-term shortage of K+ supply. One important aspect of plant adaptation to K+ stress is cellular and tissue homeostasis of K+, which involves transport of K+ across various membranes in various tissues (Amtmann et al., 2004). K+ transport mechanisms have been studied extensively at the molecular level (Véry and Sentenac, 2003). Much less is known about the molecular nature of adaptive responses at the level of metabolism and development. Furthermore, it is completely unknown how plants sense external K+ concentration, how this information is communicated within the plant, and how physiological, biochemical, and molecular responses are integrated into a concerted adaptive response.
Microarray technology allows us to approach K+ deficiency again from a more integrative point of view considering all aspects of K+ management in plants, including processes related to growth, development, metabolism, and stress resistance. It therefore provides us for the first time with a molecular picture of how plants manage one of its most important nutrients under conditions of shortage in supply. Since the obtained picture is limited to transcripts, it will be far from complete. Nevertheless, the identified genes should provide a framework of molecular processes involved in K+ nutrition, which in future can be studied in more detail and complemented at the level of proteins and metabolites.
In this study we have used full genome microarrays to monitor the transcriptome of Arabidopsis seedlings exposed to long-term K+ starvation and short-term K+ resupply after starvation. Our experimental design ensures that we identify transcriptional responses that are specifically linked to the external K+ supply. Furthermore, we have employed new analysis methods to avoid determination of transcriptional changes based on arbitrary cut-off of fold-changes. Rank products (RP) is a novel nonparametric statistics test that assesses the consistency and magnitude of expression changes (Breitling et al., 2004b). We also employ a novel algorithm to identify groups of functionally related genes with significant transcriptional changes, which facilitates the biological interpretation of the obtained dataset (Breitling et al., 2004a). Our results indicate that changes in external K+ supply affect the transcription of genes within four major functional categories, namely genes related to the phytohormone jasmonic acid (JA), genes encoding cell wall proteins, genes involved in transport processes, and genes with possible function in cellular signaling.
Our study delineates a specific set of genes that respond rapidly and selectively to external K+. It is hoped that this will facilitate further studies of K+ signaling as screening for K+ perception mutants was previously hampered by the fact that K+-related physiological and morphological phenotypes appear late during the stress and are relatively unspecific. The identified set of specifically K+-regulated genes, which respond quickly to changes in external K+ supply, should be a useful tool for the further dissection of signaling pathways involved in perception or integration of K+ changes at the cellular and tissue level.
RESULTS
Experimental Design and Analysis
Design and Phenotypic Characterization of the Experimental System
Biological conditions and experimental design were tailored to characterize molecular responses to changes in the external supply of the macronutrient K+. To ensure that the biological material was sensitive to external K+, we used 2-week-old plants, which have low internal K+ storage capacity and exhibit high relative growth rate, thus requiring high rates of K+ uptake from the external medium. Long-term starvation responses were assessed by comparing plants grown on K+-free medium (K+ starved) with plants grown on K+-replete medium (control, 2 mm K+). Short-term K+ resupply responses were assessed by providing 10 mm KCl to K+-starved plants for 2 and 6 h (Fig. 1). This treatment was compared to two control experiments, i.e. resupply of K+-free solution (0) or resupply of 10 mm NaCl (Na). Transcriptional changes due to differences in ionic strength, osmotic potential, and chloride or sodium concentrations between the solutions were eliminated by identifying those genes that changed transcript levels upon K+ resupply with respect to both control treatments (see below).
Figure 1.
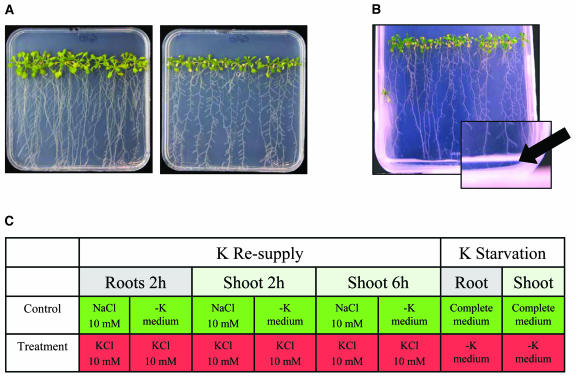
Biological system and experimental design. A, Phenotype of Arabidopsis seedlings grown vertically on petri dishes for 2 weeks after germination. Left, Complete medium (2 mm K+). Right, K+-free medium (−K). For composition of media see “Materials and Methods.” B, Resupply experiments; 2-week-old K-starved seedlings were provided with 5 mL of sterile −K medium supplemented with 10 mm KCl. Control plants received fresh −K medium with or without additional 10 mm NaCl. C, Overview of microarray experiments. Each column represents one array. Control samples are shown in green, treated samples in red. All eight comparisons were carried out for three independent biological replicates (24 arrays in total).
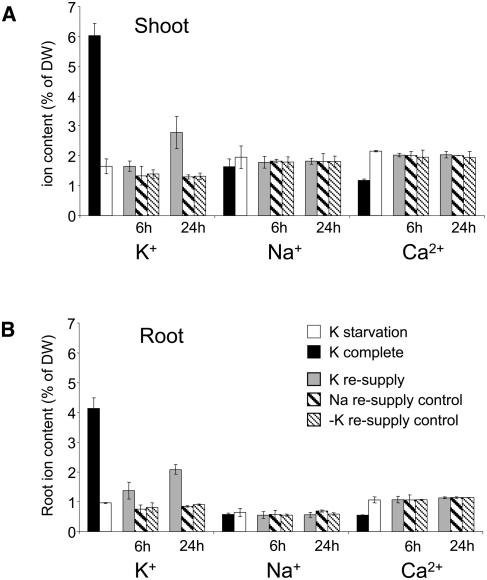
Plants grown on K+-free medium developed visible K+ starvation symptoms on day 10 (±2) after germination, which included chlorosis of older leaves and a typical growth arrest of lateral roots (Fig. 1A). Consequences of K+ starvation and resupply in terms of root and shoot ion contents were measured using inductively coupled plasma optical emission spectroscopy (ICP-OES). No significant changes in Mg, P, S, B, Fe, or Zn were observed during any of the treatments. K+, Na+, and Ca2+ tissue contents are shown in Figure 2. K+-starved seedlings contained about 4 times less K+ than K+-sufficient seedlings, in both roots and shoots. A concomitant doubling in tissue Ca2+ contents during K+ starvation was probably due to the increased Ca2+ concentration in the K+-free medium (see “Materials and Methods”). K+ resupply led to a slight but significant increase of root K+ content within the first 6 h. No significant increase of shoot K+ content was detected within this period of time, but after 24 h both tissues had regained a considerable amount of K+, i.e. 40% to 60% of K+ levels of plants grown in complete medium. Addition of the two control media (0 or Na) to K+-starved plants did not affect their ion contents, in particular no increase of tissue Na+ content was observed within 24 h after addition of 10 mm NaCl. We can conclude that our experimental design ensures that (1) responses to K+ starvation are assessed at a stage were K+ deficiency occurs but is not yet lethal, and that (2) responses to K+ resupply are monitored at a stage where external K+ is sensed by the plants but has not yet resulted in apparent morphological changes or a considerable increase of shoot K+ content.
Figure 2.
Seedling ion content after K+ starvation and resupply. Potassium, sodium, and calcium levels were measured with ICP-OES in shoots (A) and roots (B) of 2-week-old seedlings grown either on complete medium (black boxes) or on −K+ medium (white boxes). Ion content was also analyzed at 6 and 24 h after resupplying 10 mm KCl (gray boxes), 10 mm NaCl (bold dashed boxes), or a fresh K+-free-solution (light dashed boxes) to K+-starved plants. Fifty seedlings were pooled for each sample. Averages and ses of three independent experiments are shown.
Identification of Differentially Expressed Genes
Three biological replicates were examined for all experimental conditions (Fig. 1C). The statistical analysis was based on three replicates of the long-term starvation experiment and six replicates for K+ resupplied roots and shoots, merging the two replica sets obtained with 0 or Na+ control media at each time point. (It should be emphasized that the two sets of experiments differing in their control treatment were not treated as replicates in a narrow sense. Rather our analysis method [see below] identified genes that showed significant transcriptional responses that were consistent over the six arrays. Thus, the response of these genes was indeed due to a change in the external K+ concentration rather than to changes in Cl− or Na+ concentration, ionic strength, or osmotic potential.) For each treatment genes were sorted to produce a ranked list based on their fold-change (FC) compared to the control treatment (i.e. the gene with the highest FC is assigned rank 1 etc.). To identify genes with statistically significant expression changes, we used the RP test statistics (see “Materials and Methods;” Breitling et al., 2004b). We chose this recently developed, fully nonparametric test rather than a classical t test method (such as the one implemented in the Significance Analysis of Microarrays software; Tusher et al., 2001) because it does not require an estimate of measurement variation and is therefore more stable with regard to experimental noise and small data sets. Furthermore, it is applicable to low numbers of replicates, where traditional nonparametric methods (e.g. Wilcoxon rank test) fail due to the small number of possible permutations. Determination of expect-values and false discovery rate (E-values and FDR; see “Materials and Methods”) provided a well-defined significance estimate for the RP values obtained from replicated experiments. For each comparison the analysis resulted in two lists of genes sorted by their respective RP values, one for up- and one for down-regulated genes.
Table I gives an overview of the number of genes whose expression was affected by K+- treatment according to significance levels expressed in terms of FDR (minimum FC are shown in brackets). The first point to note is that the number of genes showing significant transcriptional changes varied considerably depending on the applied treatment and the examined tissue. In roots, expression of a very large number of genes was significantly changed already after 2 h of K+ resupply (387 genes below 1% FDR, combining up and down-regulation). In shoots the same level of response was reached later (24 genes below 1% FDR at 2 h and 799 genes below 1% FDR at 6 h, combining up- and down-regulation). By contrast, 6 h incubation in K+ resupplied medium did not further increase the number of regulated root transcripts (as compared to 2 h), and we therefore restricted the analysis of early responses to K+ resupply in the roots to the 2 h time point. Expression level changes were stronger during starvation than after resupply (e.g. fold change of 2.14 or more at an FDR below 0.01%). The second point to make in relation to Table I is that our experimental design and analysis method allowed us to detect small transcriptional changes with high significance. For example, changes of only 1.3-fold were frequently detected at an FDR of 1% or lower.
Table I.
Significance analysis of genes differentially expressed in response K+ starvation and resupply
| FDR % | Down | Up | Down | Up |
|---|---|---|---|---|
| Starvation (Shoots, n = 3)
|
Starvation (Roots, n = 3)
|
|||
| <0.001 | 1 (2.91) | 4 (7.57) | 1 (3.08) | 1 (4.1) |
| <0.01 | 1 (2.91) | 19 (2.75) | 4 (2.14) | 11 (2.63) |
| <0.1 | 8 (1.62) | 45 (1.89) | 22 (1.72) | 35 (1.95) |
| <1 | 17 (1.45) | 99 (1.52) | 150 (1.49) | 149 (1.53) |
| <10 | 43 (1.33) | 224 (1.3) | 473 (1.30) | 417 (1.30) |
| Resupply (2 h Roots K(Na) and K(0), n = 6)
|
||||
| <0.001 | 23 (1.59) | 13 (1.51) | ||
| <0.01 | 40 (1.45) | 25 (1.45) | ||
| <0.1 | 72 (1.36) | 55 (1.40) | ||
| <1 | 177 (1.28) | 210 (1.21) | ||
| <10 | 658 (1.14) | 801 (1.10) | ||
| Resupply (2 h Shoots K(Na) and K(0), n = 6)
|
Resupply (6 h Shoot K(Na) and K(0), n = 6)
|
|||
| <0.001 | 5 (1.51) | 0 | 104 (1.55) | 50 (1.57) |
| <0.01 | 7 (1.45) | 0 | 152 (1.45) | 89 (1.44) |
| <0.1 | 14 (1.33) | 0 | 249 (1.37) | 175 (1.35) |
| <1 | 23 (1.29) | 1 (1.38) | 436 (1.27) | 363 (1.27) |
| <10 | 57 (1.18) | 5 (1.21) | 935 (1.18) | 934 (1.15) |
| Resupply (2 and 6 h Shoot K(Na) and K(0), n = 12)
|
||||
| <0.001 | 126 (1.31) | 64 (1.33) | ||
| <0.01 | 200 (1.25) | 122 (1.24) | ||
| <0.1 | 297 (1.18) | 222 (1.21) | ||
| <1 | 499 (1.16) | 507 (1.13) | ||
| <10 | 1020 (1.06) | 1217 (1.08) | ||
Cumulative numbers of differentially expressed genes according to FDRs ranging from <0.001% to <10%. FDRs were calculated by dividing E-values by the position of the gene in the RP list obtained from three (for K+ starvation) or six (for resupply) replicate experiments. E-values were assigned by comparing the RP of each gene to the RP calculated from 10,000 random permutations of the original dataset (see “Materials and Methods” for details). Numbers in brackets indicate the minimal FC at each level of significance.
Expression patterns of all genes listed in Table I with FDRs of up to 1% are available as supplemental material (www.plantphysiol.org). Furthermore, a fully annotated list of transcriptional changes with FDRs of up to 0.001%, sorted by putative physiological function, can be obtained from the web supplement. In both cases functional annotation was based on the Arabidopsis Information Resource (TAIR), Munich Information Center for Protein Sequences (MIPS), and The Institute for Genomic Research (TIGR). We also offer access to a searchable database of our results at http://www.brc.dcs.gla.ac.uk/∼rb106x/arabidopsis_results.htm.
Identification of Functional Categories of Differential Gene Expression
To detect physiologically relevant patterns in the gene lists resulting from RP analysis we employed iterative group analysis (iGA; Breitling et al., 2004a). In a traditional approach, such patterns are defined manually by looking for groups of genes that share a certain function or participate in a common process and are over-represented among a predefined list of differentially expressed genes. iGA encodes this procedure in an objective and statistically rigorous algorithm. Genes are automatically assigned to functional groups based on a wide variety of available annotations (gene names, gene families, Gene Ontology classes, etc.) and each of these groups is then tested for differential expression. In doing so, the iterative nature of the algorithm allows significance estimates even for imperfect annotations or when only some members of a functional group are changed. In addition, iGA does not rely on an arbitrarily restricted list of significantly changed genes, but rather determines the optimal cut-off for each particular group. The result is a list of groups, sorted by statistical significance (minimal P-value), and the corresponding member genes that show relevant expression changes (Tables II–V). Only groups up to an expected FDR of 10% are shown in Tables II to V.
Table II.
Groups of functionally related genes down-regulated in shoots after K+ resupply
| Groups | AGI | Code | Annotation | Rank | Category | |
|---|---|---|---|---|---|---|
| Myrosinase Binding Proteins | At1g54020 | Myrosinase-associated protein | 13 | JA-Related | ||
| Minimal P-value | Evidence | At1g52030 | MBP2 | MYROSINASE-BINDING PROTEIN 2 | 24 | JA-Related |
| 1.53E−08 | K | At1g52040 | MBP1 | MYROSINASE-BINDING PROTEIN 1 | 36 | JA-Related |
| At5g38540 | Myrosinase binding protein-like | 37 | JA-Related | |||
| At2g39330 | Putative myrosinase-binding protein | 71 | JA-Related | |||
| Lectins (Myrosine Binding Protein-Like) | At3g16470 | JR1 | Putative lectin (JR1) | 47 | JA-Related | |
| Minimal P-value | Evidence | At3g16400 | Putative lectin | 66 | JA-Related | |
| 7.15E−08 | G, K | At3g16410 | Putative lectin | 76 | JA-Related | |
| At3g16390 | Putative lectin | 82 | JA-Related | |||
| Vegetative Storage Proteins | ||||||
| Minimal P-value | Evidence | At5g24780 | VSP1 | VEGETATIVE STORAGE PROTEIN 1 | 2 | JA-Related |
| 1.40E−07 | K (2x), M | At5g24770 | VSP2 | VEGETATIVE STORAGE PROTEIN 2 | 3 | JA-Related |
| Jasmonic Acid Biosynthesis | At3g45140 | LOX2 | LIPOXYGENASE 2 | 11 | JA-Related | |
| Minimal P-value | Evidence | At5g42650 | AOS | ALLENE OXIDE SYNTHASE | 14 | JA-Related |
| 1.99E−06 | G | At3g25760 | AOC | ALLENE OXIDE CYCLASE | 87 | JA-Related |
| Tryptophan Catabolism/CYP79B | ||||||
| Minimal P-value | Evidence | At4g39950 | CYP79B2 | Cytochrome P450 (CYP79B2) | 57 | Stress |
| 6.44E−06 | G, F | At2g22330 | CYP79B3 | Cytochrome P450 (CYP79B3) | 61 | Stress |
| Trypsin Inhibitors | At2g43530 | Putative trypsin inhibitor | 38 | Stress | ||
| Minimal P-value | Evidence | At2g43510 | Putative trypsin inhibitor | 50 | Stress | |
| 8.59E−06 | K, G | At2g43520 | Putative trypsin inhibitor | 93 | Stress | |
| Degradation of Tyrosine | ||||||
| Minimal P-value | Evidence | At2g24850 | TAT3 | Tyrosine aminotransferase (TAT3) | 5 | JA-Related |
| 8.96E−06 | M (2x), K | At4g23600 | CORI3 | CORONATINE INDUCED 1 (CORI3) | 7 | JA-Related |
| Arginase/Biosynthesis of Polyamines | ||||||
| Minimal P-value | Evidence | At4g08870 | Putative arginase | 17 | JA-Related | |
| 1.62E−05 | K, M (2x) | At4g08900 | Arginase | 72 | JA-Related | |
| Phenylpropanoid Biosynthesis | ||||||
| Minimal P-value | Evidence | At3g53260 | PAL2 | Phenylalanine ammonia-lyase PAL2 | 127 | Stress |
| 4.27E−05 | G (2x) | At2g37040 | PAL1 | Phenylalanine ammonia-lyase PAL1 | 151 | Stress |
| Cold Acclimation | ||||||
| Minimal P-value | Evidence | At1g20440 | COR47 | COR47 | 40 | Stress |
| 7.47E−05 | G | At5g15960 | KIN1 | KIN1 | 65 | Stress |
| Dehydroascorbate Reductase | ||||||
| Minimal P-value | Evidence | At1g19570 | Dehydroascorbate reductase | 23 | Stress | |
| 9.11E−05 | K, M | At5g36270 | Dehydroascorbate reductase | 96 | Stress | |
| Sulfate Reduction, APS Pathway | ||||||
| Minimal P-value | Evidence | At3g22890 | APS1 | ATP sulfurylase | 85 | Sulfur |
| 1.16E−04 | G | At4g04610 | APR1 | 5-Adenylylsulfate reductase | 163 | Sulfur |
| Gluconolactonase Activity | ||||||
| Minimal P-value | Evidence | At5g24420 | 6-Phosphogluconolactonase-like protein | 1 | Growth | |
| 1.35E−04 | G | |||||
| Harpin-Induced Proteins | ||||||
| Minimal P-value | Evidence | At5g06320 | Harpin-induced protein-like | 42 | Stress | |
| 1.40E−04 | K | At2g35460 | Harpin-induced protein-like | 116 | Stress | |
| Other Extracellular Metabolism | At1g78660 | Gamma glutamyl hydrolase | 718 | Growth | ||
| Minimal P-value | Evidence | At1g78670 | Gamma glutamyl hydrolase | 746 | Growth | |
| 1.45E−04 | M | At1g78680 | Gamma glutamyl hydrolase | 928 | Growth | |
| S-Adenosyl-Methionine - Homocysteine Cycle | At4g01850 | S-Adenosylmethionine synthase 2 | 546 | Sulfur | ||
| Minimal P-value | Evidence | At2g36880 | Putative S-adenosylmethionine synthetase | 656 | Sulfur | |
| 1.87E−04 | M (2x) | At4g13940 | S-Adenosyl-l-homocysteinase | 967 | Sulfur | |
| At1g02500 | S-Adenosylmethionine synthetase | 1,206 | Sulfur | |||
| At5g57280 | Protein carboxyl methylase-like | 1,649 | Sulfur | |||
| At3g23810 | S-Adenosyl-l-homocysteinase | 2,868 | Sulfur | |||
| At3g17390 | Putative S-adenosylmethionine synthetase | 2,917 | Sulfur | |||
| Biosynthesis of Glycosides | ||||||
| Minimal P-value | Evidence | At5g28510 | Glycosyl hydrolase family 1 | 6 | ||
| 2.09E−04 | M | At1g52400 | BGL1 | Beta-Glucosidase | 9 | |
| Indoleacetic Acid Biosynthesis | At2g20610 | SUR1 | SUPERROOT 1 | 1,018 | ||
| Minimal P-value | Evidence | At3g44300 | NIT2 | NITRILASE 2 | 1,128 | |
| 2.28E−04 | G | At3g44310 | NIT1 | NITRILASE 1 | 1,350 | |
| At4g31500 | SUR2 | SUPERROOT 2 (CYP83B1) | 1,367 | |||
| Oxidoreductase Activity, Acting on Sulfur Group of Donors | ||||||
| Minimal P-value | Evidence | At4g21990 | APR3 | 5-Adenylylsulfate reductase PRH26 | 80 | Sulfur |
| 2.47E−04 | G | At4g04610 | APR1 | 5-Adenylylsulfate reductase | 163 | Sulfur |
K+-responsive groups of functionally related genes were identified by iGA based on RP lists of shoot genes sorted for down-regulation over 12 experiments (2 and 6 h K+ resupply, three replicates with two controls each). Significantly affected gene groups up to an FDR of 10% are shown. Some of the groups may overlap completely because genes are classified similarly by various annotations, and in these cases they are combined for the presentation in Tables II to V. For rank and evidence details, see footnote of Table IV.
Table III.
Groups of functionally related genes down-regulated in roots after K+ resupply
| Groups | AGI | Code | Annotation | Rank | Category | |
|---|---|---|---|---|---|---|
| Biosynthesis of Alkaloids; Fermentation; Linked; Oxidoreductase | At1g26420 | FAD-linked oxidoreductase family | 6 | Stress | ||
| Minimal P-value | Evidence | At1g26390 | FAD-linked oxidoreductase family | 7 | Stress | |
| 4.73E−11 | M (2x), K(2x) | At1g26400 | FAD-linked oxidoreductase family | 8 | Stress | |
| At1g26380 | FAD-linked oxidoreductase family | 15 | Stress | |||
| At4g20800 | Putative protein | 19 | Stress | |||
| At4g20830 | FAD-linked oxidoreductase family | 32 | Stress | |||
| At4g34900 | Xanthine dehydrogenase | 36 | Stress | |||
| At1g30730 | FAD-linked oxidoreductase family | 46 | Stress | |||
| Electron Transfer Flavoprotein | At4g20800 | Putative protein | 19 | Growth | ||
| Minimal P-value | Evidence | At2g46760 | Hypothetical protein common family | 79 | Growth | |
| 1.39E−06 | G | At2g46750 | Unknown protein | 99 | Growth | |
| At5g64250 | 2-Nitropropane dioxygenase-like protein | 129 | Growth | |||
| P-Type Pump | At1g27770 | ACA1/PEA1 | Calcium-transporting ATPase 1, PLM-type | 17 | Transport | |
| Minimal P-value | Evidence | At1g59820 | ALA3 | Chromaffin granule ATPase II homolog, putative | 212 | Transport |
| 2.50E−05 | T | At1g13210 | ALA11 | Potential phospholipid-transporting ATPase 11 | 216 | Transport |
| At4g29900 | ACA10 | Potential calcium-transporting ATPase 10, PLM-type | 321 | Transport | ||
| At4g30190 | AHA2 | Putative H+-transporting ATPase | 423 | Transport | ||
| At2g19110 | HMA4 | Putative cadmium-transporting ATPase | 425 | Transport | ||
| At4g30120 | HMA3 | Cadmium-transporting ATPase-like protein | 449 | Transport | ||
| At2g18960 | AHA1 | Plasma membrane proton ATPase (PMA) | 549 | Transport | ||
| At3g22910 | ACA13 | Potential calcium-transporting ATPase 13, PLM-type | 638 | Transport | ||
| Tryptophan Catabolism | ||||||
| Minimal P-value | Evidence | At4g39950 | CYP79B2 | Cytochrome P450-like protein | 23 | Stress |
| 2.68E−05 | G | At2g22330 | CYP79B3 | Putative cytochrome P450 | 116 | Stress |
| CYP79B | ||||||
| Minimal P-value | Evidence | At4g39950 | CYP79B2 | Cytochrome P450-like protein | 23 | Growth |
| 8.72E−05 | F | At2g22330 | CYP79B3 | Putative cytochrome P450 | 116 | Growth |
| EF-Hand Containing Proteins:Group IV | At2g41100 | CaM2 | Calmodulin-like protein, TOUCH3 | 14 | Calcium | |
| Minimal P-value | Evidence | At1g18210 | CML27 | Calcium-binding protein, putative | 60 | Calcium |
| 9.62E−05 | F | At3g56800 | CaM3 | Calmodulin-3 | 97 | Calcium |
| At3g43810 | CaM7 | Calmodulin 7 | 188 | Calcium | ||
| At2g41090 | CML10 | Calcium-binding protein (CaBP-22;CAM10) | 208 | Calcium | ||
| At4g03290 | CML6 | Putative calmodulin | 243 | Calcium | ||
| At3g51920 | CML9 | Putative calmodulin (CAM9) | 389 | Calcium | ||
| Arginine Decarboxylase Activity; Polyamine Biosynthesis | ||||||
| Minimal P-value | Evidence | At4g34710 | ADC2 | Arg decarboxylase SPE2 | 2 | JA-Related |
| 1.35E−04 | G (2x) | |||||
| Harpin | ||||||
| Minimal P-value | Evidence | At5g06320 | Harpin-induced protein-like | 13 | Stress | |
| 1.44E−04 | K | At2g35460 | Similar to harpin-induced protein hin1 from tobacco | 124 | Stress | |
| Stomach; Adrenal Gland; Catalase Reaction | At1g02920 | Glutathione transferase, putative | 56 | Stress | ||
| Minimal P-value | Evidence | At1g02930 | Glutathione transferase, putative | 96 | Stress | |
| 1.56E−04 | M (3x) | At2g30860 | Glutathione transferase, putative | 853 | Stress | |
| At1g69920 | Glutathione transferase, putative | 1,105 | Stress | |||
| Degradation of Met; S-Adenosyl-Methionine - Homocysteine Cycle; Transfer of Activated C-1 Groups | At2g36880 | Putative S-adenosylmethionine Synthetase | 292 | Sulfur | ||
| Minimal P-value | Evidence | At4g01850 | S-Adenosylmethionine synthase 2 | 358 | Sulfur | |
| 1.74E−04 | M (3x) | At1g02500 | S-Adenosylmethionine synthetase | 604 | Sulfur | |
| At3g23810 | S-Adenosyl-l-homocysteinas, putative | 797 | Sulfur | |||
| At5g57280 | Protein carboxyl methylase-like | 1,390 | Sulfur | |||
| At3g17390 | Putative S-adenosylmethionine synthetase | 2,185 | Sulfur | |||
| Phenylalanine Ammonia-Lyase Activity | ||||||
| Minimal P-value | Evidence | At3g53260 | PAL2 | Phenylalanine ammonia-lyase | 272 | Stress |
| 2.18E−04 | G | At2g37040 | PAL1 | Phenylalanine ammonia-lyase | 293 | Stress |
| Peroxidase | At5g64120 | AtP15 | Peroxidase, putative | 28 | Cell Wall | |
| Minimal P-value | Evidence | At2g41480 | Peroxidase, putative | 33 | Cell Wall | |
| 2.20E−04 | K | At5g39580 | AtP24 | Peroxidase, putative | 38 | Cell Wall |
| At1g14540 | AtP46 | Anionic peroxidase, putative | 83 | Cell Wall | ||
| 3′(2′),5′-Bisphosphate Nucleotidase Activity; Inositol/Phosphatidylinositol Phosphatase Activity | At5g64000 | SAL2 | 3(2),5-Bisphosphate nucleotidase (emb|CAB05889.1) | 146 | Growth | |
| Minimal P-value | Evidence | At5g63980 | FIERY 1 | 3(2),5-Bisphosphate nucleotidase | 326 | Growth |
| 2.51E−04 | G (2x) | |||||
K+-responsive groups of functionally related genes were identified by iGA based on RP lists of root genes sorted for down-regulation over six experiments (6 h K+ resupply, three replicates with two controls each). Significantly affected gene groups up to a FDR of 10% are shown. For rank and evidence details, see footnote of Table IV.
Table IV.
Groups of functionally related genes up-regulated in shoots after K+ resupply
| Groups | AGL | Code | Annotation | Ranka | Category | |
|---|---|---|---|---|---|---|
| Extensin | At2g43150 | Putative extensin | 21 | Cell wall | ||
| Minimal P-value | Evidenceb | At4g33970 | Extensin-like protein | 34 | Cell wall | |
| 1.22E−07 | K | At4g38080 | Extensin related | 90 | Cell wall | |
| At4g08370 | Extensin-like protein | 96 | Cell wall | |||
| At3g54590 | Extensin precursor-like protein | 115 | Cell wall | |||
| Aabinogalactan; Fasciclin | At5g44130 | FLA13 | Fasciclin-like arabinogalactan-protein, putative | 127 | Cell wall | |
| Minimal P-value | Evidence | At3g61640 | AGP20 | Arabinogalactan-protein | 133 | Cell wall |
| 1.10E−05 | K (2x) | At4g12730 | FLA2 | Fasciclin-like arabinogalactan-protein | 156 | Cell wall |
| At1g03870 | FLA9 | Fasciclin-like arabinogalactan-protein | 170 | Cell wall | ||
| At5g10430 | AGP4 | Arabinogalactan-protein | 182 | Cell wall | ||
| Aquaporin | At3g16240 | TIP2,1 | Delta tonoplast integral protein | 20 | Transport | |
| Minimal P-value | Evidence | At3g26520 | TIP1,2 | Gamma tonoplast intrinsic protein | 89 | Transport |
| 1.31E−05 | T | At4g35100 | PIP2,7 | Plasma membrane intrinsic protein | 106 | Transport |
| At4g01470 | TIP1,3 | Putative water channel protein | 513 | Transport | ||
| At1g52180 | TIP | Aquaporin, putative | 538 | Transport | ||
| At2g37170 | PIP2,2 | Aquaporin (plasma membrane intrinsic protein 2B) | 544 | Transport | ||
| At2g36830 | TIP1,1 | Putative aquaporin (tonoplast intrinsic protein gamma) | 703 | Transport | ||
| At3g04090 | SIP1,1 | Hypothetical protein | 712 | Transport | ||
| At4g00430 | PIP1,4 | Probable plasma membrane intrinsic protein 1c | 807 | Transport | ||
| At2g16850 | PIP2,8 | Putative plasma membrane intrinsic protein | 995 | Transport | ||
| At3g53420 | PIP2,1 | Plasma membrane intrinsic protein 2a | 1,217 | Transport | ||
| At2g25810 | TIP4,1 | Putative aquaporin (tonoplast intrinsic protein) | 1,357 | Transport | ||
| Cell Wall | At5g49080 | Putative protein | 28 | Cell wall | ||
| Minimal P-value | Evidence | At4g33970 | Extensin-like protein | 34 | Cell wall | |
| 2.31E−05 | M | At2g06850 | At-XTH4 | Xyloglucan endotransglycosylase (ext/EXGT-A1) | 50 | Cell wall |
| At5g06640 | Putative protein | 59 | Cell wall | |||
| At4g08370 | Extensin-like protein | 96 | Cell wall | |||
| Germin-Like Protein Family | Growth | |||||
| Minimal P-value | Evidence | At1g72610 | Germin-like protein | 31 | Growth | |
| 5.68E−05 | F | At5g20630 | Germin-like protein | 80 | Growth | |
| Plastidial Acyl Carrier Protein | At4g25050 | Acyl carrier-like protein | 58 | Growth | ||
| Minimal P-value | Evidence | At1g54580 | ACP2 | Acyl-carrier protein | 231 | Growth |
| 1.32E−04 | F | At1g54630 | ACP3 | Expressed protein | 486 | Growth |
| Plasma Membrane | At4g12420 | SKU5 | Pectinesterase (pectin methylesterase) family | 87 | ||
| Minimal P-value | Evidence | At4g02520 | Glutathione transferase, putative | 94 | ||
| 2.12E−04 | G | At4g35100 | PIP2,7 | Plasma membrane intrinsic protein (SIMIP) | 106 | |
| At5g65430 | GRF8 | 14-3-3 Protein GF14 kappa | 123 | |||
| Cell Wall (sensu Magnoliophyta) | Cell wall | |||||
| Minimal P-value | Evidence | At4g12420 | SKU5 | Pectinesterase (pectin methylesterase) family | 87 | Cell wall |
| 2.14E−04 | G | At5g65430 | GRF8 | 14-3-3 Protein GF14 kappa | 123 | Cell wall |
K+-responsive groups of functionally related genes were identified by iGA based on RP lists of shoot genes sorted for up-regulation over 12 experiments (2 and 6 h K+ resupply, three replicates with two controls each). Significantly affected gene groups up to a FDR of 10% are shown.
Rank within all time point and control RP gene list (n = 12 for shoot data, n = 6 for root data).
Evidence: K, Key words from gene names; T, transporters; M, MAtDB functional classification; F, TAIR gene family; G, GeneOntology annotation.
Table V.
Groups of functionally related genes up-regulated in roots after K resupply
| Groups | AGI | Code | Annotation | Rank | Category | |
|---|---|---|---|---|---|---|
| Nitrate Transporter Activity | At1g08090 | NRT2.1 | High-affinity nitrate transporter NRT2 | 1 | Transport | |
| Minimal P-value | Evidence | At3g45060 | NRT2.6 | High-affinity nitrate transporter-like protein | 2 | Transport |
| 1.05E−10 | G (2x), K(3x), F, M (3x), T | At5g60780 | NRT2.3 | High-affinity nitrate transporter protein-like | 3 | Transport |
| Aquaporin; Water Channel Activity | At2g37170 | PIP2,2 | Aquaporin (plasma membrane intrinsic protein 2B) | 32 | Transport | |
| Minimal P-value | Evidence | At4g35100 | PIP2,7 | Plasma membrane intrinsic protein (SIMIP) | 68 | Transport |
| 1.53E−07 | T, G (3x), K, F | At3g26520 | TIP1,2 | Gamma tonoplast intrinsic protein | 73 | Transport |
| At3g53420 | PIP2,1 | Plasma membrane intrinsic protein 2a | 97 | Transport | ||
| At3g61430 | PIP1,1 | Plasma membrane intrinsic protein 1a | 143 | Transport | ||
| At2g34390 | NIP2,1 | Putative aquaporin (plasma membrane intrinsic protein) | 434 | Transport | ||
| At5g37820 | NIP4,2 | Membrane integral protein (MIP)-like | 643 | Transport | ||
| At4g00430 | PIP1,4 | Probable plasma membrane intrinsic protein 1c | 823 | Transport | ||
| At2g36830 | TIP1,1 | Putative aquaporin (tonoplast intrinsic protein gamma) | 847 | Transport | ||
| At2g16850 | PIP2,8 | Putative plasma membrane intrinsic protein | 951 | Transport | ||
| At4g01470 | TIP1,3 | Putative water channel protein | 954 | Transport | ||
| At1g52180 | TIP | Aquaporin, putative | 956 | Transport | ||
| At2g37180 | PIP2,3 | Aquaporin (plasma membrane intrinsic protein 2C) | 1,060 | Transport | ||
| At5g18290 | SIP1,2 | Putative protein | 1,276 | Transport | ||
| At3g04090 | SIP1,1 | Hypothetical protein | 1,313 | Transport | ||
| Peroxidase | At4g11290 | AtP19 | Peroxidase, putative | 10 | Cell wall | |
| Minimal P-value | Evidence | At5g19890 | AtPN | Peroxidase, putative | 21 | Cell wall |
| 8.75E−05 | K | At3g01190 | AtP12 | Peroxidase, putative | 25 | Cell wall |
| At4g30170 | AtP8 | Peroxidase, putative | 111 | Cell wall | ||
| At4g08770 | AtP38 | Peroxidase, putative | 151 | Cell wall | ||
| At5g66390 | AtP6 | Peroxidase, putative | 174 | Cell wall | ||
| Arabinogalactan | At4g26320 | AGP13 | Arabinogalactan-protein | 6 | Cell wall | |
| Minimal P-value | Evidence | At5g10430 | AGP4 | Arabinogalactan-protein | 11 | Cell wall |
| 9.37E−05 | K | At5g53250 | AGP22 | Arabinogalactan-protein, putative | 47 | Cell wall |
K+-responsive groups of functionally related genes were identified by iGA based on RP lists of root genes sorted for up-regulation over six experiments (6 h K+resupply, three replicates with two controls each). Significantly affected gene groups up to a FDR of 10% are shown. For rank and evidence details, see footnote of Table IV.
Together, the RP and iGA algorithms allowed us to obtain a dataset, which not only represents a highly structured overview of transcriptional responses to external K+, but also provides an estimate of the statistical significance of the observed patterns.
Biological Classification of K+-Responsive Genes
In this section we attempt to draw a comprehensive picture of the transcriptional adaptation occurring in response to changes in external K+ supply. For this purpose we used two different approaches. Firstly, iGA results were examined to obtain a general view of genes that share a common physiological role and were, as a group, transcriptionally affected by our treatments. From this analysis emerged several functional super-categories of genes that appear to play an important role in the plants adaptation to K+ stress. We then proceeded by looking at individual genes that showed highly significant transcriptional changes in response to K+ (i.e. FDR of 0.001% or lower) and identified those that are functionally linked to the iGA-based super-categories. This allowed us to include genes within the super-categories that are highly significant in their responsiveness to K+, but might have been overlooked by the iGA due to imperfect annotation.
K+-Responsive Functional Groups
The group lists resulting from iGA of the K+ resupply experiments revealed a number of recurring and statistically highly significant patterns (Tables II–V). Many of the functional groups identified in the K+ resupply experiments were also found when iGA was applied to the starvation experiments (with group members being regulated in the opposite direction; data not shown). The most prominent groups of genes down-regulated upon K+ resupply shared functional annotations related to either JA biosynthesis and known jasmonic acid-dependent processes (mostly in shoots; Table II) or synthesis of secondary metabolites (in both roots and shoots; Tables II and III). Furthermore, several gene groups encoding proteins functioning in sulfur metabolism (in particular linked to the homo-Cys cycle) were significantly down-regulated by K+ resupply and were detected as a group in both root and shoot datasets. K+ resupply also triggered down-regulation of genes known to be involved in stress adaptation (e.g. glutathione transferases, dehydroascorbate reductases, cold responsive genes, and polyamine synthesis). Another interesting result of the iGA was the detection of a number of genes encoding Ca2+-binding proteins (mainly calmodulins), which as a group showed a significant down-regulation in roots after resupply of K+. Significant groups among transcripts up-regulated upon K+ resupply overlapped between shoots and roots (Tables IV and V). In particular both tissues showed up-regulation of transporters (aquaporins) and cell wall related genes (arabinogalactans). By contrast, two groups of up-regulated genes linked to primary metabolism were shoot specific (Table IV).
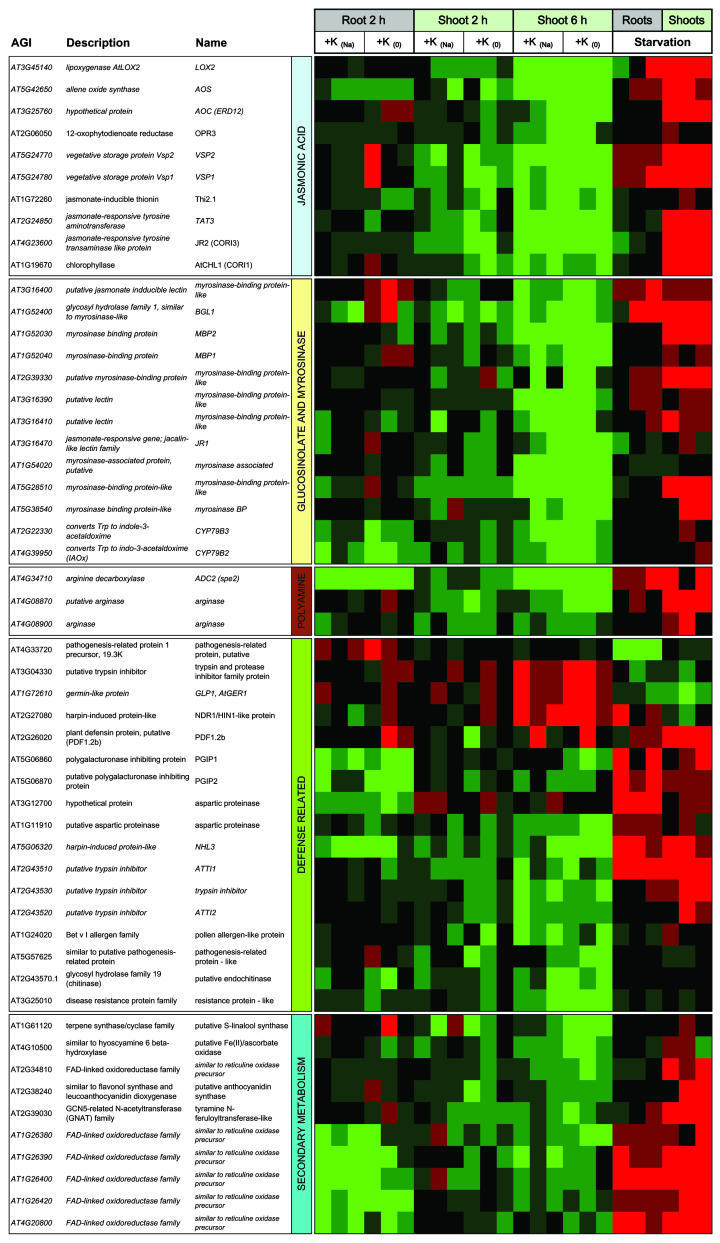
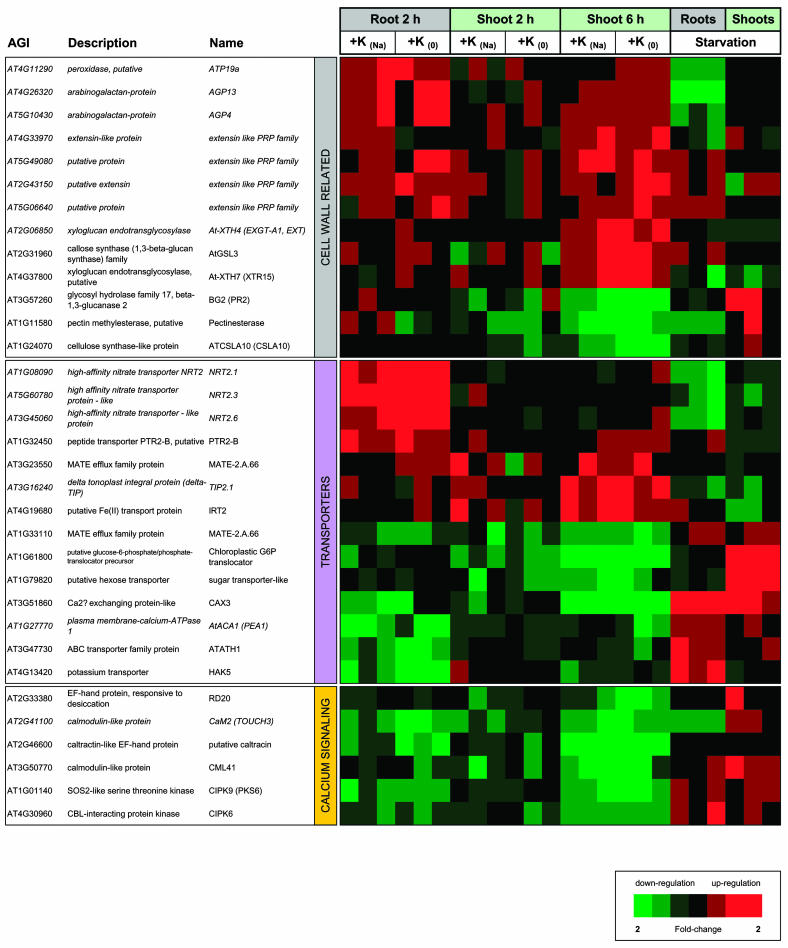
The iGA results highlighted four main functional super-categories: (1) genes related to the JA signaling pathway, (2) genes encoding cell wall proteins, (3) genes with a putative function in transport, and (4) genes encoding Ca2+-binding proteins. Figures 3 and 4 show expression profiles of all genes in these categories that were differentially expressed upon K+ resupply with high significance (FDR of 0.001% or lower). Note that while adding genes to the functional categories, the figures omit some of the group members identified by iGA. This has two reasons. Firstly, the chosen significance cut-off was extremely rigorous due to space limitations. Secondly, significant group responses can include changes in individual transcripts that have much lower significance when analyzed outside the group context. For comprehensive visualization, genes were sorted according to their expression profile.
Figure 3.
Expression profiles of highly significant K+-responsive genes related to JA-related genes were extracted from RP lists for K+ resupply experiments with FDR below 0.001%. On the left side AGI number, common name (if available) and a short description based on MIPS, TAIR, or TIGR are given for each gene. Genes identified by iGA are shown in italics. Functional super-categories extracted from iGA (Tables II–V) are given as vertical labels. On the right side expression profiles over all experimental conditions (compare Fig. 1C) are shown. Tissues and treatments are given on the top. Colors indicate change of transcript level in the treated samples with respect to the control samples (red for up-regulation, green for down-regulation, see color bar at the bottom of Fig. 4). For resupply treatments (+K+) respective controls are given in brackets (Na for supply of Na+ instead of K+, 0 for supply of K+-free medium). K+-starved plants were compared to plants grown on K+-sufficient medium. For each comparison data from three replicate experiments are shown.
Figure 4.
Expression profiles of highly significant K+-responsive genes related to cell walls, transport, or calcium signaling. For explanation see Figure 3.
JA- and Defense-Related Genes
The largest functional category detected by iGA concerned genes related to the plant hormone JA. Our current knowledge of these genes derives mostly from work on defense mechanisms during pathogenesis and wounding. Transcriptional changes of JA-related genes in response to the external K+ supply were considerable in terms of both strength and statistical significance, thus pointing to a novel and exciting role of this plant hormone in mineral nutrition. Figure 3 gives an overview of K+-responsive genes, which are directly or indirectly linked to JA. The most prominent expression profile within this category consisted in up-regulation during starvation and down-regulation upon K+ resupply in shoots. Affected transcripts were related to (1) JA biosynthesis and known JA-downstream events, (2) glucosinolate synthesis and degradation, (3) polyamine metabolism, (4) defense mechanisms, and (5) production of secondary metabolites.
JA Biosynthesis and JA Markers
The first three steps of JA biosynthesis convert linolenic acid into 12-oxo-phytodienoic acid and are catalyzed by the enzymes lipoxygenase (LOX; Bell and Mullet, 1993), allene oxide synthase (AOS; Laudert and Weiler, 1998), and allene oxide cyclase (AOC; Weber, 2002). Genes for all three enzymes, i.e. LOX2, AOS, and AOC, were significantly up-regulated during K+ starvation and down-regulated after K+ resupply (the latter change was also observed for OPR3, encoding 12-oxo-phytodienoic acid reductase; Sanders et al., 2000). An increase of JA levels in response to K+-starvation and a rapid decrease after K+ resupply was also indicated by the concomitant regulation of well-known JA-responsive transcripts encoding vegetative storage protein (VSP1, VSP2; Utsugi et al., 1998), thionin (THI2.1; Epple et al., 1995), and chlorophyllase (CORI1/CHL1; Tsuchiya et al., 1999). These transcripts, as well as several plant defensin transcripts (PDF1.2a, b, c; PDF1.3) showed the highest average fold change (5–10) in shoots of K+-starved plants (see supplemental material). Two transcripts encoding a Tyr aminotransferase are also included in this group as TAT3 is annotated as JA-responsive (TAIR; Titarenko et al., 1997).
Glucosinolate Synthesis and Degradation
A second gene category reported to be regulated by JA contains genes involved in the glucosinolate-myrosinases system (for reviews, see Rask et al., 2000; Wittstock and Halkier, 2002). Transcript levels of the methylthioalkylmalate synthase-like gene (MAM-L, At5g23020) but not the methylthioalkylmalate synthase1 (MAM1, At5g23010; Kroymann et al., 2001) were reduced in shoots upon starvation. These enzymes are implicated in early synthesis steps of chain-elongated Met derived glucosinolates. Genes encoding cytochrome-P450-dependent mono-oxygenases of the CYP79 family are involved in the second step of glucosinolate synthesis by catalyzing the conversion of Trp to indole-3-acetaldoxime (Hull et al., 2000; Zhao et al., 2002). In our experiments, CYP79B2 and CYP79B3 were down-regulated upon K+ resupply in roots and shoots. CYP79B2 and CYP79B3 are also involved in the Trp-dependent auxin biosynthesis (Wittstock and Halkier, 2002; Zhao et al., 2002; Mikkelsen et al., 2003). A possible involvement of auxin in the response to external K+ supply was also pointed out by iGA with a minimal P-value of 2.28e−04 for a group of genes involved in auxin biosynthesis (NIT1, NIT2, SUR1, and SUR2; Table II). The counterpart to the synthesis of glucosinolates is their specific degradation by thioglucoside glucohydrolase also known as myrosinases. None of the three myrosinase genes characterized to date (TGG1, At5g26000; TGG2, At5g25980; pseudogene TGG3, At5g48375; Husebye et al., 2002) changed expression in our experiments. However, two myrosinase-like genes (BGL1, At1g52400 and At5g28510) were strongly up-regulated during K+ starvation (average fold change of 8.6 and 7.6, respectively) and down-regulated upon K+ resupply. Furthermore, several myrosinase-binding protein, homologs of which are induced by wounding and exogenous JA treatment in Brassica napus (Taipalensuu et al., 1997), showed the same expression profile.
Polyamine Metabolism
The third group of known JA-responsive genes showing the typical expression profile of up-regulation during K+ starvation and down-regulation upon K+ resupply concerned genes involved in polyamine synthesis. High levels of the polyamine putrescine are well known to be associated with K+ deficiency in plants (Watson and Malmberg, 1996). The strongest response in terms of strength and significance was found for AtADC2 (spe2, At4g34710), which encodes an Arg decarboxylase. ADC2 induction has been reported for various stresses (Urano et al., 2003) and various growth regulators including JA (Perez-Amador et al., 2002). By contrast, transcript levels of AtADC1 (spe1, At2g16500), which is not regulated by JA (Perez-Amador et al., 2002), remained unchanged in our treatments. Transcripts of the two arginase genes implicated in Orn production (usually a polyamine precursor) responded to K+ treatments in Arabidopsis (compare Tables II and III).
Defense Mechanisms and Production of Secondary Metabolites
As JA is involved in signaling events related to pathogenesis and wounding (McConn et al., 1997; Leon et al., 2001), K+-responsive transcripts implicated in defense and secondary metabolism were also included in Figure 3. Within theses categories, several different expression profiles emerged. Most genes showed the typical response of up-regulation during starvation and down-regulation upon resupply. Strikingly strong regulation was observed for genes encoding polygalacturonase inhibiting proteins (PGIP1, PGIP2; Ferrari et al., 2003), aspartic proteinases, protease-inhibitors, and several FAD-related oxidoreductases. A second pattern consisted of a strong up-regulation in roots and/or in shoots after K+ resupply with weak or nondetectable change during long-term starvation. Transcripts presenting this profile encoded protease-inhibitors, germin-like proteins, and a harpin-induced protein.
In summary, a large number of K+-responsive transcripts are linked to JA either as components of the JA biosynthesis pathway or through well-known JA-dependent processes. A clear relation with JA was evident in several cases where out of two closely related genes only the known JA-dependent isoform was affected by the K+ treatment (e.g. CHL1 versus CHL2; Tsuchiya et al., 1999; ADC2 versus ADC1; Perez-Amador et al., 2002).
Cell Wall Related Genes
Cell wall related transcripts constituted another large category of K+-responsive genes pointed out by iGA. Within this category, transcripts for (1) extensins, (2) xyloglucan glucosyltransferases, (3) arabinogalactans (AGPs), and (4) peroxidases showed highly significant changes (Fig. 4). Their main expression profiles were characterized by up- or down-regulation upon K+ resupply. Most cell wall related genes did not show a strong response to long-term starvation (compare supplemental material).
Extensins
Most extensin and extensin-like transcripts identified by our analysis were up-regulated upon resupply. These proteins are generally associated with cell wall reinforcement (for review, see Kieliszewski and Lamport, 1994; Cassab, 1998; Sommer-Knudsen et al., 1998). Up-regulation of extensin encoding transcripts was first observed in roots (within 2 h of K+ resupply), whereas increase of transcript levels in shoots appeared with a delay. Extensin expression has also been shown to be increased in response to wounding, pathogen infection, and exogenous application of methyl JA and other hormones (Merkouropoulos and Shirsat, 2003).
Xyloglucan Glucosyltransferases
A second group of cell wall related transcripts showing a strong but delayed up-regulation in shoots encoded group-1 xyloglucan endotransglycosidases-hydrolases, namely At-XTH4 and At-XTH7. These enzymes are involved in the rearrangement of the network formed by association of xyloglucans with cellulose microfibrils (Cosgrove, 1997; Perrin et al., 1999). Although contrasting correlations between XTH and cell expansion have been reported, the overall available evidence argues for a functional role for XTH in primary cell wall enlargement (Rose et al., 2002). Besides its structural function, xyloglucans are also a source of growth-regulating oligosaccharides, which might play role in signaling (Perrin et al., 1999; Pilling and Hofte, 2003).
AGPs
Another well-represented group of K+-responsive cell wall transcripts is the AGP family of highly glycosylated Hyp-rich glycoproteins (compare Tables IV and V). AGPs are localized in the cell wall and the plasma membrane and have been implicated in various processes of plant growth and development (Schultz et al., 2000; Gaspar et al., 2001; Showalter, 2001). Transcripts for AGP4 and AGP13 were down-regulated in roots during starvation and up-regulated upon K+ reresupply. As most AGPs, all K+-responsive AGPs contain a glycosylphosphatidylinositol-anchor, which provide an alternative to transmembrane domains for anchoring proteins to the cell surface (Udenfriend and Kodukula, 1995). As for animal cells, involvement of AGPs in signal transduction has been proposed for plants (Schultz et al., 1998).
Peroxidases
A member of the peroxidase gene family, ATP19a, known to be predominantly expressed in roots (Welinder et al., 2002), was strongly down-regulated in K+-starved plants and derepressed upon K+ resupply (Fig. 4). Many other members of this family were highlighted by iGA (Tables III and V). Peroxidases are involved in oxidative cross-linking of cell wall components (e.g. lignification) and in H2O2 detoxification. Secreted peroxidases are also involved in generating reactive oxygen species coupled to oxidation of hormones and defense compounds such as indole-3-acetic acid and salicylic acid (SA; Kawano, 2003).
Genes Encoding Membrane Transporters
K+ Transporters
Changes in K+ supply affected a large number of membrane transporters, but surprisingly only two of the transcripts belonged to a gene family with known function in K+ transport. The HAK5 gene, a member of the KUP/HAK family of K+ transporters (13 members in Arabidopsis), was up-regulated during K+ starvation and quickly down-regulated after K+ resupply. The response occurred only in the roots. Similar results have been obtained with mature plants using real time PCR (Ahn et al., 2004). Another member of the same family, KUP12, was down-regulated in shoots after K+ resupply (E-value 0.0079, FDR <0.004%; see supplemental material). None of the KUP/HAK genes that have been functionally characterized to date seem to be involved in K+ nutrition although many of them transport K+ when expressed in heterologous systems (Fu and Luan, 1998; Kim et al., 1998; Rubio et al., 2000), but some of them produce developmentally impaired phenotypes when knocked-out in Arabidopsis (Rigas et al., 2001; Elumalai et al., 2002).
Nitrate Transporters
Significant transcriptional changes were pointed out by iGA for nitrate transporters, aquaporins, and P-type pumps (Tables III–V). The strongest changes were observed for three members of the NRT2 family of high-affinity nitrate transporters. In contrast to HAK5, NRT2.1, NRT2.3, and NRT2.6 were down-regulated during starvation and quickly up-regulated after K+ resupply (Fig. 4). This response was specific for roots.
Aquaporins
Another group of genes that responded strongly to changes in external K+ supply belonged to the family of aquaporins, which contains 38 genes in Arabidopsis. IGA identified subsets of 15 aquaporin genes in roots and 12 genes in shoots (9 common genes; Tables IV and V), which as a group were significantly up-regulated after resupply of K+. Up-regulation of TIP2.1 in shoots upon K+ resupply was particularly significant and is shown in Figure 4. Aquaporins are membrane integral proteins that facilitate the transport of water across the plasma membrane and internal membranes (Chrispeels et al., 2001). Some aquaporins have been shown to transport small molecules such as urea, ammonia, and CO2 in addition to water (Liu et al., 2003).
Pumps and Antiporters
A set of P-type pumps showed high group ranking among genes, which responded to K+ resupply with a decrease of transcript level in roots (Table III), and included members of different functional classes such as plasma membrane proton pumps, known and putative Ca2+-pumps, and heavy metal pumps (Axelsen and Palmgren, 2001). The strongest individual response was shown by the Ca2+ pump ACA1, which in addition to responding quickly to K+ resupply was also up-regulated in roots during long-term K+ starvation (Fig. 4). A possible link of K+ nutrition and Ca2+ homeostasis was also evident in the transcriptional regulation of CAX3 by external K+. CAX3, a tonoplast located Ca2+/H+ antiporter (Pittman et al., 2002), displayed strongly increased mRNA levels during K+ starvation in both roots and shoots and was quickly down-regulated after K+ resupply (Fig. 4).
Genes Related to Intracellular Ca2+ Signaling
A last category of K+-responsive transcripts identified by iGA included genes encoding proteins with known or predicted Ca2+-binding properties, such as calmodulins (Table III). Additional genes for calcium-binding proteins as well as interacting kinases appeared in the list of individual genes with highly significant transcriptional response to K+ resupply and are shown in Figure 4. Similarly to the group of genes identified by iGA, they were all down-regulated by K+ resupply; in two cases the response was specific for shoots. Very few of the Ca2+-related genes showed long-term responses during starvation. A transient response of these genes to changes in the environment is in accordance with their putative function in cellular signaling.
Other K+-Responsive Genes
While iGA is a powerful tool to detect coherent groups of significantly affected genes, it may miss single genes with specialized functions or incomplete annotation. Thus, other genes that showed particularly strong significant changes (FDR <0.001%) to external K+ supply are listed in the supplemental material. Among these are genes encoding transcription factors (bH2H, WRKY, NAM, AP2, E2F) as well as putative signal transduction components such as protein kinases (MAPK, receptor-like, and other types of protein kinases), ubiquitin-related proteins, and those containing putative F-box domains. Several transcript encoding stress responsive genes, GSH-dependent dehydroascorbate reductase (oxidative stress), COR47, COR6.6 (cold stress), and ERD1, dehydrin (dehydration stress) were mostly down-regulated upon K+ resupply and some of them also up-regulated during starvation. The same expression profile was also found for several transcripts encoding methyltransferase or methyltransferase-like proteins. Transfer of methyl groups through S-adenosyl-methyltransferase occurs in several metabolic and hormonal pathways including the JA pathway. Transcripts such as SYP81, COPII, and AtMEMB12 encoding trafficking components were up-regulated upon K+ resupply. No such uniform expression profile was found for cytoskeleton-related transcripts. Finally a large number of K+-responsive transcripts can be linked to primary metabolism. Among those, a particularly strong response in terms of fold change and reproducibility both during K+ starvation (down) and after resupply (up) was found in a gene (At5g24420) with homology to 6-phosphogluconolactonase, the second enzyme of the pentose-phosphate pathway. Other primary-metabolism linked K+-responsive transcripts are involved in sulfur assimilation (APS1, APR3), sugar metabolism (starch synthase, Glc-6-phosphate dehydrogenase), lipid synthesis (MGDG synthase AtMGD3, acyl-carrier proteins), and organic nitrogen metabolism (Glu dehydrogenase, Ala:glyoxylate aminotransferase 2).
DISCUSSION
As existing information on individual K+-responsive genes has already been given in the Results section, the discussion focuses on three main questions: Which signaling pathways are involved in the perception of external K+ and in the integration of responses at the whole plant level? Which membrane transporters are involved in nutrient allocation during K+ deficiency? And, can we assign general physiological functions to K+-responsive genes?
The K+-Responsive Signaling Network
The response of JA to changes in external K+ supply was the most conspicuous event observed in our study and therefore JA is likely to play a prominent role in plant responses to external K+. Several transcripts for proteins involved in the biosynthesis of JA responded to K+ starvation and resupply indicating that JA levels increase during starvation and rapidly decrease after K+ resupply. The earliest steps of JA biosynthesis take place in chloroplasts (Schaller, 2001), and the transcriptional response of JA biosynthesis genes was indeed limited to the shoot with the exception of AOS, which was also regulated in the roots (Fig. 3). Since JA appears to be an important component of a K+ signaling network, we compared our results with known JA responses, in particular those related to wounding and pathogen attack.
Signaling pathways leading to defense mechanisms are well characterized and involve positive and negative cross talks between JA, SA, and ethylene (ET; Devoto and Turner, 2003). This hormonal network has been progressively dissected using reporter genes for specific pathways as well as hormone signaling mutants and transcriptomic approaches (Berger, 2002; Van Zhong and Burns, 2003). ET production is unlikely to occur during long-term K+ starvation since ET-biosynthesis and ET-responsive genes such as ACO2 and ERS1 (Gomez-Lim et al., 1993; Hua et al., 1998) were not changed in our treatments. Furthermore, VSP2, whose transcript level is strongly decreased in response to exogenous ET (Rojo et al., 1999; Van Zhong and Burns, 2003), was up-regulated during K+ starvation. Similarly, searching our data for known SA-responsive genes such as PR1-like (At2g14610; Uknes et al., 1992; Laird et al., 2004), PR5 (At1g75040), NPR1 (At1g64280; Cao et al., 1997), and WRKY18 (At4g31800; Yu et al., 2001) revealed no indication for increased SA production during K+ starvation. Cross-talk between SA, JA, and ET has been modeled from gene expression data obtained from Pseudomonas syringae infected signaling-defective Arabidopsis mutants (using the Affymetrix 8K microarray; Glazebrook et al., 2003). The authors classified JA, SA, and ET signaling pathway on the basis of four specific responsive gene clusters (note that these clusters were only based on partial genome coverage). Comparing our results (restricted to <0.001% FDR gene list of all treatments and tissues) with these clusters revealed an overlap of K+-responsive transcripts mainly with cluster D, representing COI1-mediated JA-dependent gene expression (37% overlap; Glazebrook et al., 2003). Very few K+-responsive transcripts were found in ET, SA, and combined clusters. We conclude that signaling pathways involved in plant responses to long-term starvation and short-term resupply of K+ share signaling components with defense processes mainly through the JA pathway. This, however, does not preclude a role of ET in early responses to K+ deprivation as recently proposed by Shin and Schachtman (2004).
A role of JA in nutrient signaling might not be restricted to potassium. Many JA-related genes including JA biosynthesis genes were also found to be induced in response to sulfur starvation (Hirai et al., 2003; Nikiforova et al., 2003) as well as in sel1-10 mutants affected in the high-affinity sulfate transporter SULTR1;2 (Maruyama-Nakashita et al., 2003). By contrast, in nitrogen and phosphorus starved plants, no JA-responsive genes were identified by expression profiling of up to 8,000 genes, although differential regulation of several defense-related transcripts was observed (Wang et al., 2000, 2003; Hammond et al., 2003; Wu et al., 2003). To further assess this issue oxylipin profiling of plants exposed to various nutrient stresses is currently underway in our laboratory.
Whereas our microarray data revealed many downstream targets of JA signaling, identification of putative up-stream elements, in particular of those involved in early perception and signaling in roots, is more challenging. Several K+-responsive peroxidases (Table III) and transcripts encoding enzymes related to oxidative stress were identified in our study supporting a role of reactive oxygen species in early K+ signaling (Shin and Schachtman, 2004). H2O2 and other reactive oxygen species are also important signals in ion channel regulation (Demidchik et al., 2003; Kohler et al., 2003) as well as in early host-pathogen interactions (Lamb and Dixon, 1997).
Other candidates for early events in K+-perception in roots include rapidly induced plasma membrane anchored cell wall proteins such as AGPs, known to participate in signaling events in animal cells (Tables IV and V; Fig. 4). Furthermore, transcriptional regulation of several Ca2+-related proteins was detected in response to external K+. This hints toward an involvement of cytoplasmic Ca2+ in K+ signaling. Cytoplasmic Ca2+ was found to transiently increase after many abiotic stress treatments (Knight et al., 1997) but has not yet been assessed in response to changing nutrient supply.
Interestingly, many JA-related shoot genes responded to K+ resupply before any measurable change of shoot K+ had taken place (i.e. within 6 h, compare Fig. 2). It is therefore likely that they respond to an early root-shoot signal rather than to a rise of shoot K+ content. The microarray data gave no obvious clues as to what might be the signal initiating the JA response. No genes involved in the synthesis of other hormones such as ABA, ET, or brassinosteroids responded to our K+ treatments and the observed response of CYP79 genes with possible function in auxin biosynthesis is likely to be located downstream of JA rather than upstream.
K+-Responsive Membrane Transporters
The finding that very few known K+ transporters featured in the list of K+ responsive genes is remarkable considering that K+ homeostasis under varying K+ supply requires reallocation of K+ between different cellular compartments and tissues (Amtmann et al., 2004). The result is however in agreement with a recent microarray study on roots of mature, K+-starved plants (Maathuis et al., 2003) as well as a northern-blot analysis of all Shaker K+ channel genes (Pilot et al., 2003) and a real-time PCR analysis of all KUP/HAK/KT genes (Ahn et al., 2004). It appears that at the transcriptional level K+ transporters are in general little responsive to external K+. The strong transcriptional response to external K+ of HAK5, a member of the KUP/HAK/KT family of putative high-affinity K+ transporters, is an exception and might indicate a role in plant K+ nutrition under low K+ conditions. This will require further investigation using reverse genetic approaches.
The strong response of aquaporins to changing external K+ supply is not surprising since K+ is the main osmoticum and its uptake will be accompanied by an adjustment of water flux through aquaporins. Aquaporins might also contribute to ion homeostasis at the whole plant level, i.e. differential activity of tissue and membrane specific aquaporins will affect the ratio of apoplastic/symplastic water flow and thus direct solute flux through plant tissues.
A particularly strong response was found for three genes of the NRT2 family of nitrate transporters, which were all down-regulated during starvation and up-regulated after K+ resupply in roots. Furthermore, the nitrate inducible gene NRT1.1 (CHL1, At1g12110), encoding a dual-affinity nitrate transporter, was up-regulated with a 3-time fold change in roots during starvation (E-value 0.0020; FDR <0.013%; supplemental material). These results are interesting with respect to the well-known inter-dependence between nitrogen and potassium fertilization (Gething, 1993; Laegreid et al., 1999). Regulation of several nitrate transporter genes by changes in external K+ was also reported for tomato (Wang et al., 2002). Although direct regulation of these genes by external K+ cannot be excluded, this response might be related to changes in internal nitrate level or nitrogen metabolites (Forde, 2002). Changes in nitrogen assimilation during K+ starvation were indicated by the up-regulation of characterized nitrogen assimilatory genes (Wang et al., 2003) such as GS-GLN1-1 (At5g37600; E-value 0.0027; FDR <0.01%) and GS-GLN1-2 (At1g66200; E-value 0.0001; FDR <0.005%) in roots and AS-ASN1 (At3g47340; E-value 0.008; FDR <0.027%) in shoots of K+-starved plants (see supplemental material).
Finally, transcriptional regulation of genes encoding Ca2+ pumps and Ca2+ transporters (e.g. ACA1 and CAX3) was observed during K+ starvation and resupply. Ca2+ can act as an osmoticum in fully expanded cells but due to its low mobility cannot replace K+ in its osmotic function in fast growing tissues. Together with the elevated Ca2+ concentrations during K+ starvation, up-regulation of Ca2+ transporters in K+-starved plants could illustrate a preferential uptake of Ca2+ into the vacuoles of older tissues thus freeing up K+ for transport to expanding tissues. The fast down-regulation of ACA1 and CAX3 upon K+ resupply, which did not involve a change in external Ca2+ concentrations, suggests that active Ca2+ homeostasis is indeed a component of plant K+ adaptation.
Physiological Role of K+-Responsive Genes
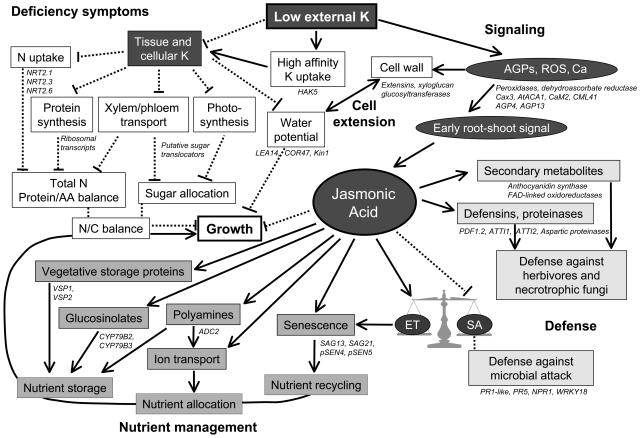
This study has revealed a framework of physiological and metabolic processes occurring during plant adaptation to K+ starvation. Based on K+ regulated transcripts we developed a working model for future verification with other techniques such as reverse genetics and metabolic profiling (Fig. 5). Well-described symptoms of K+ deficiency (Marschner, 1995) were reflected in many K+-regulated genes, for example genes for nitrate transporters (reduced nitrogen levels), ribosomal proteins (inhibition of protein synthesis), and sugar translocators (impaired photosynthesis and long-distance transport). To compensate for decreased K+ availability plants increase K+ absorption through induction of high-affinity K+ transport systems such as HAK5. However, during long-term starvation this strategy is not sufficient and the plant needs to engage in a proper acclimation response to complete its life cycle. This process requires perception and signaling of the plant K+ status. Putative components of early signaling events emerging from our study include ROS, cytoplasmic Ca2+, and cell wall proteins. JA was identified as the main player in the further integration of adaptive responses. Integrated nutrient management could involve storage of nutrients in energy-rich compounds, reallocation of nutrients via regulation of transporters, and recovery of nutrients from senescent leaves. K+-regulated genes encoding vegetative storage proteins and enzymes involved in the biosynthesis and degradation of glucosinolates and polyamines support the notion of altered storage strategies for nitrogen and carbon in K+-deficient plants (Staswick, 1984; Rask et al., 2000; Kakkar and Sawhney, 2002). JA-dependent management of nitrogen allocation is in agreement with recent findings that JA treatment of tomato plants decreased nitrogen uptake and altered nitrogen partitioning toward root nitrogen storage (Meuriot et al., 2004). Regulation of K+ channels by JA (Evans, 2003; Suhita et al., 2003) and polyamines (Brüggemann et al., 1998; Liu et al., 2000) has been reported and might contribute to K+ reallocation between cellular compartments and tissues (Amtmann et al., 2004). Nutrient recycling is the purpose of the reversible stage of leaf senescence. Senescence leads to the induction of specific transcripts of so-called senescence-associated genes (Gepstein et al., 2003), several of which were reversibly up-regulated by K+ starvation. Transfer of nutrients from senescing leaves to sink organs has been clearly demonstrated (Himelblau and Amasino, 2001; for review, see Quirino et al., 2000; Buchanan-Wollaston et al., 2003; Yoshida, 2003) and several lines of evidence support an involvement of JA (Park et al., 1998; Weaver et al., 1998; He et al., 2002). Finally, JA is well known for its role in plant defense responses against insect herbivores and (mainly necrotrophic) fungi, which are the most relevant enemies of K+-starved plants (Perrenoud, 1990; Kessler and Baldwin, 2002; Kunkel and Brooks, 2002). One might hypothesize that the observed increase in JA serves to protect K+-deficient plants against herbivore and fungal attack, while not entirely compensating for increased feeding and pathogen development. Conversely, constitutively increased JA levels in K+-starved plants might interfere with inducible defense responses. JA, ET, and SA are all involved in pathogen responses, acting either synergistically or antagonistically on a partly overlapping range of pathogens and abiotic stresses (Turner et al., 2002). Elevated JA levels in K+-starved plants might not only specifically inhibit some SA-dependent responses, but also generally interfere with the highly dynamic nature of an integrated stress response to pathogens.
Figure 5.
Model of molecular processes underlying plant adaptation to K+ deficiency. Putative components of K+ deficiency and adaptive responses are shown in boxes. Connecting lines are based on K+-responsive genes identified in this study (shown in italics) and published information (see text). Black arrows indicate stimulation, dashed lines inhibition. Known K+ deficiency symptoms are shown in white boxes. Putative components of signaling events are indicated in dark gray. Lighter gray shading marks different JA-dependent processes potentially leading to adaptive nutrient management and defense responses. For further discussion see text.
CONCLUSIONS
Our study provides for the first time, to our knowledge, a comprehensive insight into molecular processes in growing seedlings induced by varying K+ supply. The plant material used in the experiments was characterized with respect to morphology and tissue ion contents, and a thorough experimental design allowed us to assess both short-term and long-term effects and to compare them to several control treatments. We furthermore used new analysis tools for functional data mining that led to the discovery of four main super-categories of K+-responsive genes namely genes related to the phytohormone JA, cell wall, ion transport, and Ca2+ signaling. On the basis of these findings, we propose a model of molecular mechanisms for nutrient stress adaptation. The large number of K+-responsive genes identified here provides a platform for future analysis of their physiological roles as well as further dissection of signaling pathways involved in K+ perception and nutrient homeostasis.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seeds were surface sterilized (2.5% sodium hypochlorite; 0.1% Tween 20) for 5 min, rinsed 5 times with sterile water, and placed in darkness at 4°C for 3 to 4 d to synchronize germination. Seeds were then sown in 120 × 120 mm square petri dishes (approximately 15 seeds/plate) containing 70 mL of nutrient medium with 3% Suc and 1% agar Type A (Sigma, Poole, UK) added. The control nutrient medium contained 1.25 mm KNO3, 0.5 mm Ca(NO3)2, 0.5 mm MgSO4, 42.5 μm FeNaEDTA, 0.625 mm KH2PO4, 2 mm NaCl, and micronutrients (see Maathuis et al., 2003) at pH 5.6. In the K+-free medium KNO3 was replaced by Ca(NO3)2, KH2PO4 by NaH2PO4, and NaCl was lowered to 1.375 mm. Final ion concentrations in the two media, control (versus K+-free), were 1.875 (0) mm K+, 0.5 (1) mm Ca2+, 1.25 (1) mm NO3− and 2 (1.375) mm Cl− (all other ions unchanged). Petri dishes were sealed with parafilm and placed vertically under the light source (16 h at 100 μE) at 22°C. Resupply experiments were carried out with 2-week-old seedlings and consisted in replacing the condensed solution at the bottom of the petri dishes with liquid K+-free medium supplemented or not (−K+ control) with 10 mm KCl (+K+ treatment) or 10 mm NaCl (+Na+ control).
Ion Content Determination
For ion content analysis, seedlings from three to four petri dishes (approximately 50 seedlings) were pooled. A total of three seedling batches were grown and treated independently for replicates. Roots were briefly rinsed in ice-cold water and blotted dry before determination of fresh and dry weight. Ions were acid extracted in 2 m HCl overnight, diluted 50 times with distilled water, and analyzed by ICP-OES using an Optima 4300 DV Optical Emission spectrometer (Perkin Elmer Instruments, Wellesley, MA).
RNA Extraction, cDNA Synthesis, and Labeling
Shoot and root material were frozen and ground in liquid nitrogen. Total RNA was extracted using a guanidinium-based buffer and purified through a 5.7-m CsCl cushion as described by Chomczynski and Sacchi (1987). One hundred micrograms of total RNA were concentrated to a volume of 20 μL using a microcon column YM-30 (Millipore, Bedford, MA), denatured at 65°C for 10 min and cooled down to room temperature with 50 ng of a polyT20 primer (MWG, Ebersberg, Germany). Labeling was carried out during reverse transcription adding 500 μm dATP, dGTP, dTTP, 200 μm dCTP, and 45 μm of either Cy3-dCTP or Cy5-dCTP (Amersham, Little Chalfont, UK) for synthesis of “control cDNA” and “treated cDNA,” respectively, to the reaction mix, which also contained 500 units of Superscript II RT (INVITROGEN, Paisley, UK), 10 mm dithiothreitol, and 1× Superscript II reverse transcriptase buffer (final volume of 40 μL). Labeled cDNA was purified using a PCR product purification kit (Qiagen, Crawley, UK) according to the manufacturer instructions.
Microarray Preparation and Hybridization
Microarrays spotted with the Arabidopsis Genome Oligo Set version 1.0. (Qiagen) were obtained from D. Galbraith (University of Arizona, http://www.ag.arizona.edu/microarray/). Slides were rehydrated and UV-cross linked according to the supplier's web page with an additional step consisting of a 45-min incubation in a blocking solution (5× SSC, 0.1% SDS, 1% bovine serum albumin) followed by 5 rinses with double-distilled water. Excess water was drained using a 1,000 rpm spin for 5 min. Cy3- and Cy5-labeled cDNA were speed-vacuum concentrated, resuspendend, and combined in a final volume of 36 μL formamide-based hybridization buffer (MWG, Ebersberg, Germany), deposited onto the array, and covered with hydrophobic coverslips (Sigma). Hybridization took place overnight at 42°C in a hybridization chamber. Arrays were then washes for 5 min at room temperature (2× SSC, 0.1%SDS; 1× SSC and 0.5× SSC solutions) and scanned immediately (ScanArray Express scanner and software suite; Perkin Elmer, Warrington, UK). Signal quantification was carried out using the fixed circle method as defined in QuantArray software.
Microarray Data Analysis
Normalization
For each chip, Cy3 and Cy5 hybridization signals were quantile normalized (Bolstad et al., 2003), so that the distribution of signal intensities in both channels became identical. Expression ratios were calculated directly from these data without background subtraction to obtain a minimal dependence of measurement variance on signal intensity. This procedure is a variation of the started-log procedure of Rocke and Durbin (2003) and led to excellent variance stabilization for our data (not shown).
Detection of Differentially Expressed Genes
Probes were sorted by their normalized expression ratio for each chip. Two sorted lists were produced for each chip, sorted in ascending and descending order, respectively. RPs were calculated for each gene according to Breitling et al., 2004b. RPs were compared to the RPs of 10,000 random permutations of the same data to assign E-values. To correct for the multiple testing problem inherent in microarray experiments we employed the FDR (Storey, 2003), i.e. we divided the E-value of each gene by its position in the list of changed transcripts. An FDR of ≤1% means that only 1% or less of the genes up to this position is expected to be observed by chance (false positives), the remaining 99% being genes that are indeed significantly affected (true positives).
Detection of Differentially Expressed Gene Groups
Classifications of genes into functional groups were obtained from various sources: gene ontology classifications and gene family assignments from TAIR (http://www.arabidopsis.org), functional categories from MAtDB (http://mips.gsf.de/proj/thal/db/index.html), and membrane transporter families from the annotation of the Arabidopsis Membrane Transporters microarrays (Maathuis et al., 2003). In addition, genes were classified according to the shared occurrence of keywords (strings of more than three nonnumerical characters, followed by any number of alphanumerical characters) in the gene names and functional annotation included in the microarray description files (http://www.ag.arizona.edu/microarray/). For each of these classification schemes we performed iGA (Breitling et al., 2004a), which determines the functional classes that are most enriched at the top of the gene lists sorted by rank products (separately for up- and down-regulation). The iGA procedure is based on calculating P-values using the hypergeometric distribution. For each functional class it iteratively finds the subset of members that minimizes this P-value. Only genes that were annotated in a specific classification scheme were considered for the calculations. As in the case of single genes, we face a multiple testing problem, because so many groups are examined simultaneously. To correct for this, we again used the FDR. An approximate FDR was estimated by comparing these results to those obtained for 100 randomly permuted lists for each type of classification.
Supplementary Material
Acknowledgments
The authors thank Pawel Herzyk for advices in bioinformatics, Bo Wang for helping with the ICP-OES, Mike Blatt for fruitful discussions, Giorgia Riboldi-Tunnicliffe at the MBSU facility of the SHWFGF for providing scanner facilities, and David Galbraith for supplying microarrays.
This work was supported by the Biotechnology and Biological Science Research Council (grant nos. 17/P17237 and 17/G17989) and the Nuffield Foundation (grant no. NAL/00562/G).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046482.
References
- Ahn SJ, Shin R, Schachtman DP (2004) Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 134: 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amtmann A, Armengaud P, Volkov V (2004) Potassium nutrition and salt stress. In MR Blatt, ed, Membrane Transport in Plants. Blackwell Publishing, Oxford (in press)
- Axelsen KB, Palmgren MG (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol 126: 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell E, Mullet JE (1993) Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol 103: 1133–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S (2002) Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214: 497–504 [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193 [DOI] [PubMed] [Google Scholar]
- Breitling R, Amtmann A, Herzyk P (2004. a) Iterative Group Analysis (iGA): a simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinformatics 5: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P (2004. b) Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Brüggemann LI, Pottosin II, Schonknecht G (1998) Cytoplasmic polyamines block the fast-activating vacuolar cation channel. Plant J 16: 101–105 [Google Scholar]
- Buchanan-Wollaston V, Earl S, Harrison E, Mathas E, Navabpour S, Page T, Pink D (2003) The molecular analysis of leaf senescence—a genomic approach. Plant Biotechnol J 1: 3–22 [DOI] [PubMed] [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Cassab GI (1998) Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol 49: 281–309 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate- phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Morillon R, Maurel C, Gerbeau P, Kjellbom P, Johansson I (2001) Aquaporins of plants: Structure, function, regulation, and role in plant water relations. In S Hohmann, P Agre, S Nielsen, eds, Aquaporins, Vol 51. Academic Press, San Diego, pp 277–334
- Cosgrove DJ (1997) Assembly and enlargement of the primary cell wall in plants. Annu Rev Cell Dev Biol 13: 171–201 [DOI] [PubMed] [Google Scholar]
- Demidchik V, Shabala SN, Coutts KB, Tester MA, Davies JM (2003) Free oxygen radicals regulate plasma membrane Ca2+- and K+-permeable channels in plant root cells. J Cell Sci 116: 81–88 [DOI] [PubMed] [Google Scholar]
- Devoto A, Turner JG (2003) Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann Bot (Lond) 92: 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elumalai RP, Nagpal P, Reed JW (2002) A mutation in the Arabidopsis KT2/KUP2 potassium transporter gene affects shoot cell expansion. Plant Cell 14: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H (1995) An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that for pathogenesis-related proteins. Plant Physiol 109: 813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NH (2003) Modulation of guard cell plasma membrane potassium currents by methyl jasmonate. Plant Physiol 131: 8–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15: 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG (2002) Local and long-range signaling pathways regulating plant responses to nitrate. Annu Rev Plant Biol 53: 203–224 [DOI] [PubMed] [Google Scholar]
- Fu HH, Luan S (1998) AtKuP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell 10: 63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ (2001) The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Mol Biol 47: 161–176 [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp M-J, Hajouj T, Nesher MFO, Yariv I, Dor C, Bassani M (2003) Large-scale identification of leaf senescence-associated genes. Plant J 36: 629–642 [DOI] [PubMed] [Google Scholar]
- Gething PA (1993) The Potassium-Nitrogen Partnership. International Potash Institute, Basel
- Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, Metraux JP, Zhu T, Katagiri F (2003) Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J 34: 217–228 [DOI] [PubMed] [Google Scholar]
- Gomez-Lim MA, Valdes-Lopez V, Cruz-Hernandez A, Saucedo-Arias LJ (1993) Isolation and characterization of a gene involved in ethylene biosynthesis from Arabidopsis thaliana. Gene 134: 217–221 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fukushige H, Hildebrand DF, Gan S (2002) Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol 128: 876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E, Amasino M (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158: 1317–1323 [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai H, Nourizadeh S, Chen QG, Bleecker AB, Ecker JR, Meyerowitz EM (1998) EIN4 and ERS2 are members of the putative ethylene receptor gene family in Arabidopsis. Plant Cell 10: 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull AK, Vij R, Celenza JL (2000) Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc Natl Acad Sci USA 97: 2379–2384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husebye H, Chadchawan S, Winge P, Thangstad OP, Bones AM (2002) Guard cell- and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol 128: 1180–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakkar RK, Sawhney VK (2002) Polyamine research in plants—a changing perspective. Physiol Plant 116: 281–292 [Google Scholar]
- Kawano T (2003) Roles of the reactive oxygen species-generating peroxidase reactions in plant defense and growth induction. Plant Cell Rep 21: 829–837 [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53: 299–328 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DT (1994) Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J 5: 157–172 [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kwak JM, Uozumi N, Schroeder JI (1998) AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10: 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1997) Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J 12: 1067–1078 [DOI] [PubMed] [Google Scholar]
- Kohler B, Hills A, Blatt MR (2003) Control of guard cell ion channels by hydrogen peroxide and abscisic acid indicates their action through alternate signaling pathways. Plant Physiol 131: 385–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroymann J, Textor S, Tokuhisa JG, Falk KL, Bartram S, Gershenzon J, Mitchell-Olds T (2001) A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. Plant Physiol 127: 1077–1088 [PMC free article] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM (2002) Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol 5: 325–331 [DOI] [PubMed] [Google Scholar]
- Laegreid M, Bockman OC, Kaarstad O (1999) Agriculture, Fertilizers and the Environment. CABI, Oxon, UK
- Laird J, Armengaud P, Giuntini P, Laval V, Milner JJ (2004) Inappropriate annotation of a key defence marker in Arabidopsis: Will the real PR-1 please stand up? Planta (in press) [DOI] [PubMed]
- Lamb C, Dixon RA (1997) The oxidative burst in plant disease resistance. Annu Rev Plant Physiol Plant Mol Biol 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Laudert D, Weiler EW (1998) Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signalling. Plant J 15: 675–684 [DOI] [PubMed] [Google Scholar]
- Leigh RA, Jones RGW (1984) A hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant-cell. New Phytol 97: 1–13 [Google Scholar]
- Leon J, Rojo E, Sanchez-Serrano JJ (2001) Wound signalling in plants. J Exp Bot 52: 1–9 [DOI] [PubMed] [Google Scholar]
- Liu K, Fu HH, Bei QX, Luan S (2000) Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiol 124: 1315–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L-H, Ludewig U, Gassert B, Frommer WB, von Wiren N (2003) Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiol 133: 1220–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJ, Filatov V, Herzyk P, Krijger GC, Axelsen KB, Chen S, Green BJ, Li Y, Madagan KL, Sanchez-Fernandez R, et al (2003) Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J 35: 675–692 [DOI] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants, Ed 2. Academic Press, London
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H (2003) Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol 132: 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkouropoulos G, Shirsat AH (2003) The unusual Arabidopsis extensin gene atExt1 is expressed throughout plant development and is induced by a variety of biotic and abiotic stresses. Planta 217: 356–366 [DOI] [PubMed] [Google Scholar]
- Meuriot F, Noquet C, Avice J-C, Volenec JJ, Cunningham SM, Sors TG, Caillot S, Ourry A (2004) Methyl jasmonate alters N partitioning, N reserves accumulation and induces gene expression of a 32-kDa vegetative storage protein that possesses chitinase activity in Medicago sativa taproots. Physiol Plant 120: 113–123 [DOI] [PubMed] [Google Scholar]
- Mikkelsen MD, Petersen BL, Glawischnig E, Jensen AB, Andreasson E, Halkier BA (2003) Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 131: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33: 633–650 [DOI] [PubMed] [Google Scholar]
- Park JH, Oh SA, Kim YH, Woo HR, Nam HG (1998) Differential expression of senescence-associated mRNAs during leaf senescence induced by different senescence-inducing factors in Arabidopsis. Plant Mol Biol 37: 445–454 [DOI] [PubMed] [Google Scholar]
- Perez-Amador MA, Leon J, Green PJ, Carbonell J (2002) Induction of the arginine decarboxylase ADC2 gene provides evidence for the involvement of polyamines in the wound response in Arabidopsis. Plant Physiol 130: 1454–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrenoud S (1990) Potassium and Plant Health. International Potash Institute, Basel
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K (1999) Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Pilling E, Hofte H (2003) Feedback from the wall. Curr Opin Plant Biol 6: 611–616 [DOI] [PubMed] [Google Scholar]
- Pilot G, Gaymard F, Mouline K, Cherel I, Sentenac H (2003) Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol Biol 51: 773–787 [DOI] [PubMed] [Google Scholar]
- Pittman JK, Sreevidya CS, Shigaki T, Ueoka-Nakanishi H, Hirschi KD (2002) Distinct N-terminal regulatory domains of Ca(2+)/H(+) antiporters. Plant Physiol 130: 1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quirino BF, Noh YS, Himelblau E, Amasino RM (2000) Molecular aspects of leaf senescence. Trends Plant Sci 5: 278–282 [DOI] [PubMed] [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J (2000) Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol 42: 93–113 [PubMed] [Google Scholar]
- Rigas S, Debrosses G, Haralampidis K, Vicente-Agullo F, Feldmann KA, Grabov A, Dolan L, Hatzopoulos P (2001) TRH1 encodes a potassium transporter required for tip growth in Arabidopsis root hairs. Plant Cell 13: 139–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocke DM, Durbin B (2003) Approximate variance-stabilizing transformations for gene-expression microarray data. Bioinformatics 19: 966–972 [DOI] [PubMed] [Google Scholar]
- Rojo E, Leon J, Sanchez-Serrano JJ (1999) Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J 20: 135–142 [DOI] [PubMed] [Google Scholar]
- Rose JK, Braam J, Fry SC, Nishitani K (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43: 1421–1435 [DOI] [PubMed] [Google Scholar]
- Rubio F, Santa-Maria GE, Rodriguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109: 34–43 [Google Scholar]
- Sanders PM, Lee PY, Biesgen C, Boone JD, Beals TP, Weiler EW, Goldberg RB (2000) The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12: 1041–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller F (2001) Enzymes of the biosynthesis of octadecanoid-derived signalling molecules. J Exp Bot 52: 11–23 [PubMed] [Google Scholar]
- Schultz C, Gilson P, Oxley D, Youl J, Bacic A (1998) GPI-anchors on arabinogalactan-proteins: implications for signalling in plants. Trends Plant Sci 3: 426–431 [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A (2000) The classical Arabinogalactan protein gene family of Arabidopsis. Plant Cell 12: 1751–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Schachtman DP (2004) Hydrogen peroxide mediates plant root cell response to nutrient deprivation. Proc Natl Acad Sci USA 101: 8827–8832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM (2001) Arabinogalactan-proteins: structure, expression and function. Cell Mol Life Sci 58: 1399–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer-Knudsen J, Bacic A, Clarke AE (1998) Hydroxyproline-rich plant glycoproteins. Phytochemistry 47: 483–497 [Google Scholar]
- Staswick PE (1984) Novel regulation of vegetative storage protein genes. Plant Cell 2: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD (2003) The positive false discovery rate: a Bayesian interpretation and the q-value. Ann Stat 31: 2013–2035 [Google Scholar]
- Suhita D, Kolla VA, Vavasseur A, Raghavendra AS (2003) Different signaling pathways involved during the suppression of stomatal opening by methyl jasmonate or abscisic acid. Plant Sci 164: 481–488 [Google Scholar]
- Taipalensuu J, Andreasson E, Eriksson S, Rask L (1997) Regulation of the wound-induced myrosinase-associated protein transcript in Brassica napus plants. Eur J Biochem 247: 963–971 [DOI] [PubMed] [Google Scholar]
- Tester M, Blatt MR (1989) Direct measurement of K+ channels in thylakoid membranes by incorporation of vesicles into planar lipid bilayers. Plant Physiol 91: 249–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titarenko E, Rojo E, Leon J, Sanchez-Serrano JJ (1997) Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol 115: 817–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, Takamiya K (1999) Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA 96: 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell 14 (Suppl): S153–S164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udenfriend S, Kodukula K (1995) How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem 64: 563–591 [DOI] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J (1992) Acquired resistance in Arabidopsis. Plant Cell 4: 645–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K, Yoshiba Y, Nanjo T, Igarashi Y, Seki M, Sekiguchi F, Yamaguchi-Shinozaki K, Shinozaki K (2003) Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ 26: 1917–1926 [Google Scholar]
- Utsugi S, Sakamoto W, Murata M, Motoyoshi F (1998) Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol Biol 38: 565–576 [DOI] [PubMed] [Google Scholar]
- Van Zhong G, Burns JK (2003) Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol Biol 53: 117–131 [DOI] [PubMed] [Google Scholar]
- Véry AA, Sentenac H (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54: 575–603 [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12: 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YH, Garvin DF, Kochian LV (2002) Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol 130: 1361–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson MB, Malmberg RL (1996) Regulation of Arabidopsis thaliana (L.) Heynh arginine decarboxylase by potassium deficiency stress. Plant Physiol 111: 1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver LM, Gan S, Quirino B, Amasino RM (1998) A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol Biol 37: 455–469 [DOI] [PubMed] [Google Scholar]
- Weber H (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7: 217–224 [DOI] [PubMed] [Google Scholar]
- Welinder KG, Justesen AF, Kjaersgard IV, Jensen RB, Rasmussen SK, Jespersen HM, Duroux L (2002) Structural diversity and transcription of class III peroxidases from Arabidopsis thaliana. Eur J Biochem 269: 6063–6081 [DOI] [PubMed] [Google Scholar]
- Wittstock U, Halkier BA (2002) Glucosinolate research in the Arabidopsis era. Trends Plant Sci 7: 263–270 [DOI] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Peters J, Berkowitz GA (1991) Surface charge-mediated effects of Mg2+ and K+ flux across the chloroplast envelope are associated with regulation of stroma pH and photosynthesis. Plant Physiol 97: 580–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S (2003) Molecular regulation of leaf senescence. Curr Opin Plant Biol 6: 79–84 [DOI] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL (2002) Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16: 3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.