Abstract
The light insensitive maize (Zea mays) mutant elongated mesocotyl1 (elm1) has previously been shown to be deficient in the synthesis of the phytochrome chromophore 3E-phytochromobilin (PΦB). To identify the Elm1 gene, a maize homolog of the Arabidopsis PΦB synthase gene AtHY2 was isolated and designated ZmHy2. ZmHy2 encodes a 297-amino acid protein of 34 kD that is 50% identical to AtHY2. ZmHY2 was predicted to be plastid localized and was targeted to chloroplasts following transient expression in tobacco (Nicotiana plumbaginifolia) leaves. Molecular mapping indicated that ZmHy2 is a single copy gene in maize that is genetically linked to the Elm1 locus. Sequence analysis revealed that the ZmHy2 gene of elm1 mutants contains a single G to A transition at the 3′ splice junction of intron III resulting in missplicing and premature translational termination. However, flexibility in the splicing machinery allowed a small pool of in-frame ZmHy2 transcripts to accumulate in elm1 plants. In addition, multiple ZmHy2 transcript forms accumulated in both wild-type and elm1 mutant plants. ZmHy2 splice variants were expressed in Escherichia coli and products examined for activity using a coupled apophytochrome assembly assay. Only full-length ZmHY2 (as defined by homology to AtHY2) was found to exhibit PΦB synthase activity. Thus, the elm1 mutant of maize is deficient in phytochrome response due to a lesion in a gene encoding phytochromobilin synthase that severely compromises the PΦB pool.
The quality and quantity of incident light provides the plant with a rich source of information to monitor a constantly changing environment (Neff et al., 2000). Light signals may trigger immediate adaptive responses or may be integrated over long periods of time to regulate seasonal changes in growth. Despite the variety of light-mediated responses, genetic analyses have identified a relatively modest number of primary photoreceptors. Among these, the best characterized are the phytochromes, a family of photoreceptors responsive to changes in the flux of red (R) and far-red (FR) light (Smith, 2000; Quail, 2002).
In Arabidopsis, the phytochrome family consists of five genes: PHYA, PHYB, PHYC, PHYD, and PHYE (Clack et al., 1994), while the grasses typically have three phytochromes: PhyA, PhyB, and PhyC (Mathews and Sharrock, 1996). In maize (Zea mays), an ancestral duplication of the genome has increased the total family size to six: PhyA1, PhyA2, PhyB1, PhyB2, PhyC1, and PhyC2. Each gene family member appears to encode a functional product that is expressed in several seedling tissues (Sheehan et al., 2004). These observations suggest that homologous Phy genes in maize may be functionally redundant.
Spectrally active plant phytochromes are conjugates of PHY apoprotein covalently attached to the linear tetrapyrrole chromophore 3E-phytochromobilin (PΦB; Lagarias and Rapoport, 1980). The light-dependent isomerization of the PΦB chromophore provides the basis of phytochrome action (Quail, 2002). Although phytochrome apoproteins are encoded by a multigene family, it is believed that all plant apophytochromes bind the same chromophore (Terry et al., 1993; Terry, 1997) and, therefore, the genetic disruption of PΦB synthesis offers a way to inactivate the entire phytochrome system. There are a number of known mutants in which linear tetrapyrrole synthesis is disrupted. These include the hy1 and hy2 mutants of Arabidopsis (Koornneef et al., 1980; Davis et al., 1999; Muramoto et al., 1999; Kohchi et al., 2001), the partially-etiolated-in-white-light1 (pew1) and pew2 mutants of tobacco (Nicotiana plumbaginifolia; Kraepiel et al., 1994), the phytochrome chromophore-deficient1 (pcd1) and pcd2 mutants of pea (Pisum sativum; Weller et al., 1996, 1997), the aurea (au) and yellow-green2 (yg-2) mutants of tomato (Lycopersicon esculentum; Koornneef et al., 1985; Terry and Kendrick, 1996), the photoperiodic sensitive5 (se5) mutant of rice (Oryza sativa; Yokoo and Okuno, 1993; Izawa et al., 2000), and the elongated mesocotyl1 (elm1) mutant of maize (Sawers et al., 2002).
PΦB is synthesized from 5-aminolevulinic acid, a precursor to both heme and chlorophyll (Elich and Lagarias, 1987). The first committed step in the synthesis of PΦB is the conversion of heme to biliverdin (BV) IXα by a ferredoxin-dependent heme oxygenase (Weller et al., 1996). BV IXα is then reduced to 3Z-PΦB by PΦB synthase and subsequently isomerized to 3E-PΦB (Terry et al., 1995). It is not known whether the isomerization of 3Z-PΦB to 3E- PΦB is enzyme-mediated or whether it occurs spontaneously. Of these three activities, genes encoding the first two have now been cloned (Davis et al., 1999; Muramoto et al., 1999; Kohchi et al., 2001). The HO1 (HY1) gene encodes heme oxygenase, which is targeted to the plastid (Muramoto et al., 1999). The AtHY2 gene encodes PΦB synthase, a ferredoxin-dependent biliverdin reductase, which is also plastid localized (Kohchi et al., 2001).
Characterization of the elm1 mutant strongly suggested that the mutant was deficient in a PΦB synthase activity (Sawers et al., 2002). Seedlings homozygous for the elm1 mutation lack photoactive phytochrome and responded only weakly to R and FR irradiation. HPLC analyses indicated that plastids isolated from elm1 mutant seedlings were unable to convert BV IXα to 3Z-PΦB. In this study, a maize homolog of the Arabidopsis AtHY2 (PΦB synthase) gene was isolated and designated Zea mays HY2 (ZmHy2). Comparative sequence analysis of the ZmHy2 gene between wild-type and elm1 plants revealed a single G to A transition that showed complete linkage to the elm1 mutant phenotype. The G to A transition present in elm1 mutants resulted in missplicing of ZmHy2 mRNA and premature translational termination. However, using a PCR assay, we were able to detect the low-level accumulation of in-frame ZmHy2 transcripts in elm1 plants that accumulate when a 3′ AA dinucleotide splice acceptor is utilized rather than the canonical AG. In addition, multiple ZmHy2 splice products were detected in both wild-type and elm1 mutant plants. ZmHY2 isoforms were expressed in E. coli and products assayed for activity. These results indicate that Elm1 encodes a PΦB synthase required for phytochrome activity in maize.
RESULTS
Isolation of the ZmHy2 Gene
A maize homolog of the Arabidopsis AtHY2 (PΦB synthase) gene was identified through TBLASTN searches of both private (Monsanto, St. Louis) and public DNA sequence databases. The search identified nine genomic sequences (Monsanto) and six expressed sequence tag (EST) sequences (5 Monsanto; 1 public, accession AI691542). The predicted gene was designated ZmHy2. The 891-bp coding region of ZmHy2 was amplified by reverse transcription (RT)-PCR from wild-type (W22) maize plants and 5′ RACE PCR was used to confirm the start of transcription. The EST AI691542 terminates with a poly(A+) tract and defines the 3′ untranslated region of ZmHy2. Genomic sequence recovered from database searches was used to generate a 1.6-kb contig encompassing the 3′ 666 bp of the gene. Additional 5′ genomic sequence was obtained using the Genome Walker PCR technique (see “Materials and Methods”). A 550-bp segment of intron I proved refractory to sequencing using standard protocols. Alignment of EST and genomic sequences predicted a gene structure of eight exons spanning an approximately 4.3-kb genomic region (Fig. 1).
Figure 1.
Alignment of ZmHY2 and related proteins. Alignment of ZmHY2 with the AtHY2 protein of Arabidopsis, a putative AtHY2 ortholog from rice (OsHY2) and the PebB protein of Synechococcus sp. WH8020. Residues present in two or more of the sequences define a consensus and are boxed. The structure of the ZmHy2 gene is shown above, introns as lines, exons as boxes. The positions of maize exon boundaries are shown on the alignment as filled triangles. The cleavage site of the chloroplast transit peptide is shown with an open triangle. The deleted region in one of the splice variants is also shown (Δ ELMC).
The ZmHy2 gene is predicted to encode a 297 amino acid protein of 34 kD. Figure 1 shows an alignment of the predicted ZmHy2 protein sequence with the Arabidopsis AtHY2 protein, a putative AtHY2 ortholog from rice, and the PebB protein of Synechococcus sp. WH8020. The degree of conservation between these sequences (maize to rice 79% residue identity; maize to Arabidopsis 50% residue identity) is consistent with the identification of ZmHY2 as a putative PΦB synthase. Sequence analysis revealed a putative chloroplast transit peptide in the 30 residues of the ZmHY2 N-terminal region (Bannai et al., 2002).
ZmHY2 Is Plastid Localized
To functionally test the predicted plastid localization of ZmHY2, green fluorescent protein (GFP) was fused to the C terminus of predicted full-length ZmHY2 protein and tobacco leaves infiltrated with an Agrobacterium culture containing the ZmHy2 expression construct (see “Materials and Methods”). A control plasmid carrying a 35S:GFP cassette resulted in predominantly cytosolic localization of GFP as shown by merged images of chlorophyll autofluorescence (magenta) and GFP fluorescence (green; Fig. 2, A–C). In contrast, the majority of ZmHY2:GFP protein was targeted to tobacco chloroplasts as shown by the overlay of chlorophyll (magenta) and GFP fluorescence images (yellow; Fig. 2, D–F). These data strongly suggest that ZmHY2 is plastid localized.
Figure 2.
Subcellular localization of ZmHY2. A to C, Transformed tobacco cells expressing nontargeted GFP (p35S:GFP) or D to E, expressing the ELM1 protein fused to GFP (p35S:ELM1-GFP). A and D, Chlorophyll autofluorescence; B and E, GFP fluorescence; C and F, merged images are shown.
ZmHy2 Is Genetically Linked to the Elm1 Locus
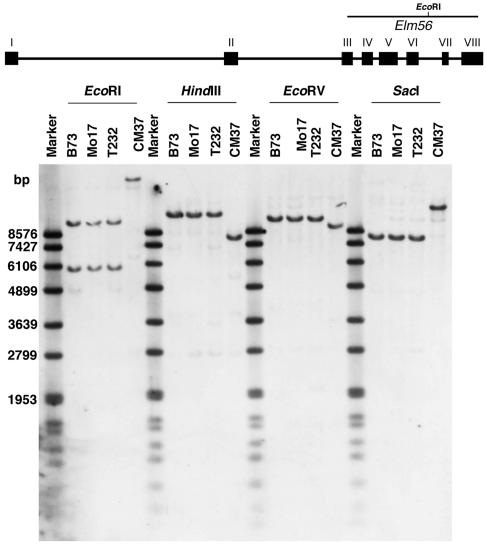
To genetically map the ZmHy2 gene, a 1,000-bp genomic fragment internal to ZmHy2 was labeled and hybridized to DNA isolated from 4 maize inbred lines that served as parents in generating the IBM94 (Lee et al., 2002) and BNL96 (Burr et al., 1988) recombinant inbred populations (Fig. 3). The ZmHy2 probe detected a single fragment in DNA digested with HindIII, EcoRV, and SacI and revealed polymorphisms that differentiated Elm1 alleles present in T232 and CM37. As expected, two bands were detected in EcoR1 digested DNA in inbreds B73, Mo17, and T232 DNA as the labeled DNA fragment spans an EcoRI cut site. The presence of a single EcoRI fragment detected in CM37 indicates that this restriction enzyme site is missing or methylated in the DNA from this inbred. The HindIII polymorphism detected between ZmHy2-T232 and ZmHy2-CM37 was used to map ZmHy2 on the BNL recombinant inbred population where it placed as a single locus on the long arm of chromosome 8 (8.06) flanked by markers umc48 and bng1240 (see “Materials and Methods”). The results of the DNA-blot analysis and mapping data indicate that ZmHy2 is a single copy gene in maize.
Figure 3.
DNA-blot analysis of ZmHy2. DNA isolated from maize inbred lines B73, Mo17, T232, and CM37 was digested with restriction enzymes shown and fractionated on 0.8% agarose gels prior to transfer to nylon membranes. DNA was hybridized to a digoxygenin-labeled probe generated using ZmHy2-specific primers Elm5 and Elm6.
DNA-blot analysis was also performed with elm1 mutants and near isogenic wild-type siblings to identify a potential lesion in the ZmHy2 gene of elm1 plants. Despite performing a number of restriction digests, no RFLPs were identified that distinguished ZmHy2 alleles from Elm1 and elm1 individuals. This indicated that the lesion in elm1 was not due to a large insertion or structural rearrangement of the ZmHy2 gene. However, small insertions or deletions in the ZmHy2 gene of elm1 plants would not be detectable using DNA-blot analysis. Therefore, to further investigate genetic linkage between elm1 and ZmHy2, an elm1 mutant [W22] was crossed to a wild-type plant from a different inbred background [H99]. The F1 plant was self-pollinated to generate a segregating F2 population. DNA was extracted from six phenotypically wild-type and six phenotypically mutant plants and digested with a restriction enzyme that distinguished the ZmHy2 alleles of W22 (ZmHy2-W22) and H99 (ZmHy2-H99). Filters were probed with the gene-specific ZmHy2 fragment, and complete linkage was observed between the W22 allele and the elm1 phenotype (data not shown), suggesting that the ZmHy2 gene was linked to the elm1 mutant phenotype.
The elm1 Mutant Is Defective in the Splicing of PΦB Synthase Gene Products
To precisely define the lesion in the ZmHy2 gene of elm1 plants, RT-PCR was used to amplify ZmHy2 cDNA products from Elm1 and elm1 plants. It is important to note that elm1 was identified as a spontaneous mutant derived from a highly inbred [W22] line. Thus, near isogenic comparisons can be made between Elm1 and elm1 plants. Sequencing of full-length cDNA products from elm1 seedlings identified a single bp deletion immediately 3′ to the junction of exons III and IV of the wild-type Elm1 transcript. The resulting frame shift creates an open reading frame of approximately 500 bp that diverges from the wild-type sequence and results in a premature stop codon. No other variations were detected between ZmHy2 sequences of Elm1 and elm1 siblings across the entire coding region of the gene.
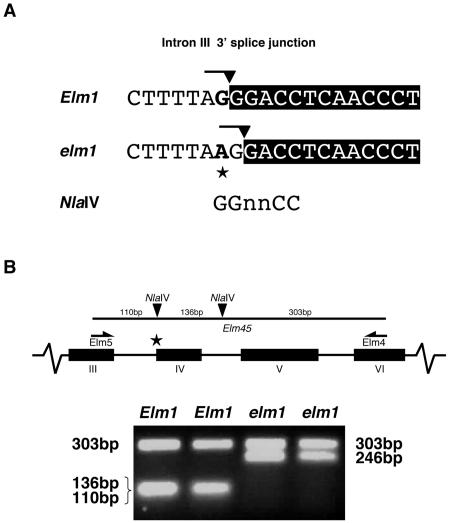
To investigate the putative splice junction lesion of ZmHy2 in elm1 mutants, PCR was used to amplify across intron III from Elm1 and elm1 genomic DNA. Cloning and sequence analysis of ZmHy2 genomic PCR products revealed a G to A transition at the 3′ splice acceptor site of intron III (Fig. 4A). The dinucleotide AG is normally essential to define the 3′ splice site, and alterations to this guanidine have previously been reported to result in missplicing and the creation of frameshift mutations (Brown, 1996). The presence of a G at the first position in the adjacent exon IV creates a novel 3′ splice junction one nucleotide downstream in elm1 mutants and results in splicing at a position +1 nt relative to wild type.
Figure 4.
Identification of elm1 mutant allele. A, Sequence of the ZmHy2 intron III 3′ splice site from Elm1 and elm1. Intron sequence and adjoined boxed exon IV sequence is shown. The AG dinucleotide defining the junction is marked with a line and the splice site with a filled arrow. The G to A transition in elm1 is marked with a star. The recognition site of the NlaIV restriction enzyme is shown and is ablated in elm1 mutants. B, PCR genotyping of ZmHy2 alleles. Top, Schematic showing primer binding sites (Elm5 and Elm4) for amplification of a genomic fragment that includes introns III, IV, and V. NlaIV cut sites are shown as black arrows. In the elm1 mutant, a substitution at the 3′ end of intron III (★) results in the ablation of the first NlaIV site. Bottom, Ethidium bromide stained agarose gel of digested products from a representative assay. DNA from two samples of three pooled wild-type (Elm1) and mutant (elm1) plants. Products were amplified with primers Elm4 and Elm5 and digested with NlaIV. Two NlaIV cut sites in wild-type fragments produced fragments of 303 bp, 136 bp, and 110 bp, whereas a single NlaIV cut site in elm1 mutants resulted in fragments of 303 bp and 246 bp.
A PCR-based assay was developed to confirm the association of this nucleotide change with the elm1 phenotype (Fig. 4B). PCR primers designed to exon III and exon IV were used to amplify ZmHy2 genomic sequence from wild-type and elm1 plants. The resulting products were digested with the enzyme NlaIV and analyzed by gel electrophoresis. An NlaIV site 110 bp into the fragment was ablated by the A to G transition mutation in the elm1 mutant allele, allowing the presence of the single nucleotide polymorphism to be easily assayed. A second NlaIV site 246 bp into the fragment provided a control for complete digestion. Figure 4B shows the results obtained using DNA extracted from two independent pools of wild-type Elm1 and mutant elm1 seedlings.
A Small Pool of ZmHy2 Transcripts Are Spliced In-Frame in elm1 Mutants
The presence of a 3′ terminal AG is highly conserved in introns of many plant and animal nuclear genes. The elm1 mutation changes the authentic 3′ splice sequence from AG:GG to AA:GG such that splicing is expected to occur at the new +1 AG dinucleotide (AAG:G) producing an out-of-frame mRNA. However, a number of reports have described splicing events at an AA dinucleotide, including at least one instance in maize (Aroian et al., 1993; Brown et al., 1995; Lal et al., 1999).
In light of these data, an RT-PCR-based assay was used to investigate the possibility that in elm1 mutants a proportion of ZmHy2 transcripts may be spliced at the authentic position, using the AA as the 3′ splice site and generating in-frame mRNAs. A primer (RS302) was designed that would only form a perfect complement to ZmHy2 transcripts that were spliced in-frame either in wild-type Elm1 transcripts or in elm1 transcripts using the AA as 3′ splice site. The use of the +1 AG as a splice acceptor would delete a single G residue from the mature transcript and result in noncomplementarity at the 3′ end of the primer (Fig. 5A). When used in conjunction with a primer designed to exon II (Pcb10), a 199-bp product was amplified from plasmid DNA containing in-frame product (Fig. 5B). However, very little product was obtained when using plasmid containing the mismatched base (+1 AG), indicating that this PCR assay can differentiate between these two splicing events.
Figure 5.
ZmHy2 splicing assay. A, Junction of exon III and exon IV showing site of primer RS302. When splicing of ZmHy2 is in-frame, primer RS302 is completely complementary. In elm1 mutants deletion of a G from the mature transcript results in a 3′ terminal mismatch between primer and template (white box). The positions of primers used in the assay are shown on a schematic of the ZmHy2 genomic locus. B, DNA gel-blot analysis of RT-PCR products amplified from wild-type (Elm1) and mutant (elm1) plants (see “Materials and Methods”). Product sizes predicted from known sequence are shown in bp.
To estimate the relative product abundance of ZmHy2 splice products in wild-type and elm1 plants, a semiquantitative PCR assay was developed. A primer designed to exon IV (Elm4) was used in conjunction with Pcb10 to assess differences in template concentration. RNA was extracted from wild-type and mutant seedling leaf tissue, and cDNA products were amplified by RT-PCR using Pcb10/RS302 and Pcb10/Elm4 primer pairs. Interestingly, primer pair Pcb10/RS302 amplified a 199-bp product from both wild-type and mutant cDNA (Fig. 5B), although the yield was much lower for elm1 plants. These data suggest that a small amount of ZmHy2 product is spliced using the AA dinucleotide as the 3′ splice site in elm1 mutants. Primer pair Pcb10/Elm4 amplified similar yields of products from wild type and mutants, thus confirming an approximately equal concentration of template. In addition to the 199-bp product, a 283-bp product was amplified from mutant elm1 cDNA using primer pair Pcb10/RS302. This 283-bp product was sequenced and found to include intron III but not intron II of the unprocessed transcript. The absence of intron II in these products suggests that they are not amplified from contaminating genomic DNA but are derived from partially spliced transcripts.
Multiple ZmHy2 Transcripts Accumulate in Elm1 and elm1 Plants
In the analysis of ZmHy2 RT-PCR products, it became apparent that multiple transcripts of varying size were being amplified from Elm1 and elm1 cDNA. Comparisons of RT-PCR products revealed that all deleted regions began immediately downstream of an exon boundary and terminated with an AG dinucleotide. These observations suggested that multiple ZmHy2 transcripts were differentially spliced products of a single gene. The cloning and sequencing of 28 ZmHy2 RT-PCR products (14 each from wild-type and elm1 samples) identified multiple 3′ splice acceptor sites in introns III and IV (Fig. 6).
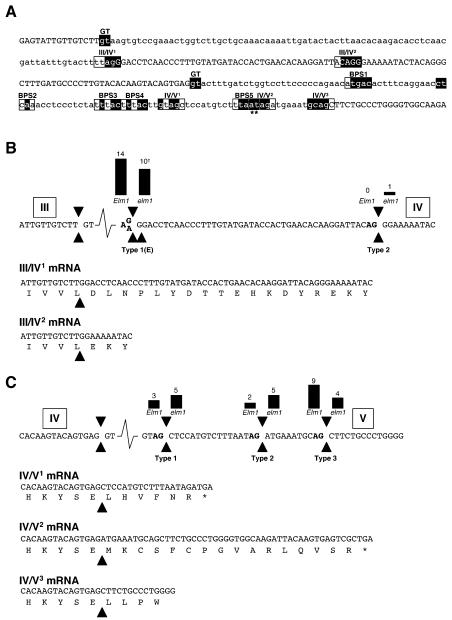
Figure 6.
Splicing events in the ZmHy2 gene. A, Genomic sequence of ZmHy2 from the end of exon III to the start of exon V. Exon sequence shown in uppercase. Intron sequence shown in lowercase. The 5′ splice sites of introns III and IV are boxed and labeled GT. 3′ splice sites of introns III and IV are boxed. Five putative BPSs within intron IV are boxed and labeled BPS1-5. Shaded nucleotides within boxed sequence match consensus BPS or 3′ splice site sequences as described in the text. The pair of nucleotides labeled with an asterisk match the branch point consensus in BPS5 but do not match the 3′ splice site consensus in the IV/V2 3′ splice site. B, Splicing of intron III. Top, Genomic sequence of ZmHy2. Exon numbers are shown boxed above sequence. Type 1, type 1E, and type 2 splice sites are indicated by black arrows. AG dinucleotides are shown in bold. The G to A transition in elm1 is also indicated. Bars and numbers above splice junctions indicate the observed frequencies of these events. †, A single type 1 event was identified from elm1 mutants resulting in an in-frame transcript. Bottom, mRNAs arising from type 1 and type 2 splicing. The predicted protein sequence is shown below. C, Splicing of intron IV. Top, Genomic sequence of ZmHy2. Exons are shown boxed above sequence. Type 1, type 2, and type 3 cleavage sites are indicated by black arrows. Bars and numbers above splice junctions indicate the observed frequencies of these events. Bottom, mRNAs formed by type 1, type 2, or type 3 splicing. The predicted protein sequence is shown below.
All ZmHy2 mRNA species identified from wild-type seedlings utilized the same 3′ splice acceptor in intron III (Fig. 6A). This event was designated type 1 splicing at the boundary of exon III and exon IV and the resulting mRNA denoted III/IV1. In elm1 mutants the AG dinucleotide defining this site is mutated, resulting in the creation of a novel splice acceptor site one nucleotide downstream and designated III/IV1E. In 10 of 14 sequences from elm1 seedlings, the type 1E pattern of splicing was observed (Fig. 6B). However, one sequence was found to contain a 45-bp deletion (relative to III/IV1), corresponding to the presence of an alternative 3′ splice site AG present 45 nt downstream of the exon III/IV boundary. This event was designated type 2 splicing at the boundary of exon III and exon IV. In contrast to III/IV1E mRNAs, III/IV2 mRNAs remain in-frame with the predicted full-length wild-type ZmHy2, although they contain an internal deletion corresponding to the loss of 15 amino acid residues (Fig. 1). An additional AG dinucleotide is present 36 nt downstream of the type 1 splice site, but no PCR products were amplified corresponding to this transcript length. Two sequences from elm1 were amplified in which intron III was not spliced from the mature transcript, and a single product was amplified from elm1 that was spliced in-frame. These latter observations are consistent with the RT-PCR results shown in Figure 5, suggesting that some in-frame ZmHy2 transcripts are generated in the elm1 mutants.
The use of multiple splice acceptor sites was also observed at the junction of intron IV and exon V. RT-PCR products were amplified from wild-type and elm1 cDNAs corresponding to splicing at three different positions at the 3′ end of ZmHy2 intron IV. These species were designated type 1, type 2, and type 3 splicing at the boundary of exon IV and exon V (Fig. 6C). All three splice sites were defined by the presence of an AG dinucleotide (Fig. 6A). The observed frequencies of these events are shown in Figure 6C. In both vertebrate and plant introns, the 3′ splice site AG is found within the 5 nt consensus GCAG:G (highly conserved AG motif is underlined and the cleavage site is denoted by a colon; Kramer, 1996; Brown and Simpson, 1998). If multiple AGs lie in close proximity to each other, sequence context can affect selection of the preferred AG (Smith et al., 1993). The position of an upstream branch point sequence (BPS) further influences the selection of the 3′ splice site (Simpson et al., 1996). The BPS is defined by the consensus sequence CURAY (most highly conserved underlined) and is generally located 18 to 40 nt upstream of the 3′ splice site. Five putative BPSs were identified upstream of the 3′ end of intron IV (Fig. 6A). Interestingly, ZmHy2 IV/V1 and IV/V2 mRNAs contain frameshifts, which may give rise to truncated proteins. Only ZmHy2 IV/V3 mRNA is predicted to encode a full-length protein, as defined by homology with AtHY2, thereby suggesting that the type 3 AG defines the authentic splice site.
Recombinant ZmHY2 Exhibits PΦB Synthase Activity
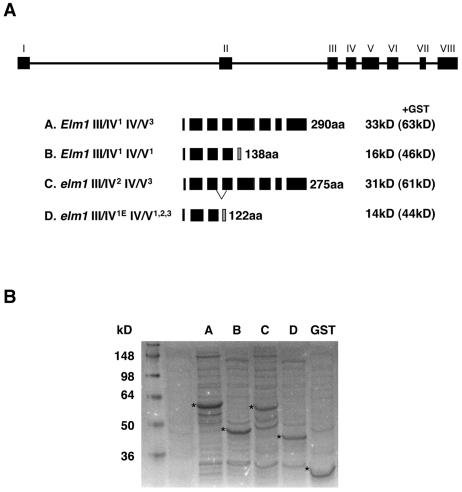
To assay the potential function of multiple ZmHy2 mRNA species in Elm1 and elm1 plants, four ZmHY2 derivatives were expressed in E. coli as fusions to glutathione S-transferase (GST). The constructs designated ELMA-D are illustrated in Figure 7A. All four lacked the N-terminal 38 residues predicted to constitute the chloroplast transit peptide (as determined by homology to Arabidopsis AtHY2). ELMA encodes the predicted mature full-length ZmHY2 protein. ELMB encodes a truncated ZmHY2 protein predicted to be the product of the IV/V1 splicing event. ELMC encodes a protein containing an internal deletion predicted to be the product of III/IV2 splicing in elm1 mutants. ELMD encodes a truncated protein resulting from III/IV1E splicing in elm1 mutants.
Figure 7.
Expression of ZmHY2 and derivatives in E. coli. A, Schematic illustrating the proteins expressed by the four constructs ELMA, B, C, and D. Black boxes represent exon sequence. Shaded boxes show regions of divergent protein sequence arising from frame shifts. Lines in ELMC mark the region of deletion. Predicted mass is shown in kilodaltons. Mass in brackets is predicted size of protein variant:GST fusion. B, SDS-PAGE fractionation of bacterial extracts following induction of ELMA-D or GST alone. Accumulation of fusion protein is marked with an asterisk. Molecular mass is shown in kilodaltons.
A coupled holophytochrome assembly assay was used to determine whether ZmHY2 or derivatives possessed PΦB synthase activity. ELMA-D proteins were purified as GST-fusions by affinity chromatography and incubated in a reaction mixture containing BV IXα and cyanobacterial apophytochrome (Cph1). The phytochrome difference spectrum obtained following incubation with ELMA had a maximum at 676 nm and a minimum at 724 nm, which is consistent with a PΦB-Cph1 adduct (Kohchi et al., 2001) and suggested that ZmHY2 possesses PΦB synthase activity (Fig. 8). Difference spectra obtained following incubation with ZmHY2 derivatives ELMB-D (Fig. 8) did not differ from a no protein control. These data indicate that ELMB-D are not active in vitro.
Figure 8.
Phytochrome difference spectra of Cph1 incubated with BV metabolites. Recombinant ZmHY2 and derivatives were assayed for PΦB synthase activity as described in “Materials and Methods.” Cph1 was incubated with bilin reaction mixture and a phytochrome difference spectrum obtained. Traces are displaced vertically to allow comparison. Traces were obtained following incubation with ELMA-D as labeled. The upper trace is a no protein control.
DISCUSSION
ZmHy2 is a maize homolog of the Arabidopsis HY2 gene, which encodes a PΦB synthase (Kohchi et al., 2001). PΦB synthase catalyses the ferredoxin-dependent reduction of BV IXα to 3Z-PΦB, a precursor of the phytochrome chromophore (Kohchi et al., 2001; McDowell and Lagarias, 2001). Although highly divergent in the amino-terminal regions of the proteins, AtHY2 and ZmHY2 are both predicted to be targeted to the plastid and are localized to chloroplasts in transient expression assays (Kohchi et al., 2001). Comparison of AtHY2 and ZmHY2 mature protein sequences shows a degree of conservation that is consistent with common enzymatic activity. Unfortunately, little is known about the domain structure of AtHY2, making it difficult to speculate on the functional significance of divergence seen between AtHY2 and potential orthologs. Comparison of plant PΦB synthase proteins with cyanobacterial bilin reductases may identify certain residues essential to function (Fig. 1). However, as has been previously noted, conserved residues may be plesiomorphies shared by this large protein family and do not necessarily define regions essential to PΦB synthase activity per se (Kohchi et al., 2001). Recombinant AtHY2 and ZmHY2 both exhibit PΦB synthase activity in an in vitro coupled holophytochrome assembly assay (Kohchi et al., 2001).
The elm1 mutant of maize has previously been characterized as severely deficient in active phytochrome pools (Sawers et al., 2002). Furthermore, plastids isolated from elm1 mutants do not support the reduction of exogenously supplied BV IXα to 3Z-PΦB, and it was proposed that the elm1 mutation is a lesion in a PΦB synthase gene. Sequencing of the ZmHy2 gene from Elm1 and elm1 plants revealed a G to A transition disrupting the 3′ splice site of intron III in elm1 mutants. The dinucleotide AG is essential to the definition of the 3′ splice junction of most eukaryotic introns, and lesions of the 3′ AG dinucleotide of plant introns have previously been reported to generate frameshift mutations (Umen and Guthrie, 1995; Brown, 1996). The maize allele shrunken2-intermediate (sh2-i) contains an A to G transition in a splice acceptor site but mutant plants are able to accumulate correctly spliced sh2-i transcript as the result of splicing at the noncanonical dinucleotide AA (Lal et al., 1999). Evidence presented here suggests that a small percentage of ZmHy2 message may similarly be correctly spliced in elm1 mutants as a result of the use of the dinucleotide AA as the 3′ splice acceptor site. Our previous characterization of elm1 noted that mutant seedlings were weakly responsive to R and that mature plants differ only subtly from wild-type siblings (Sawers et al., 2002). It is possible that a low level of in-frame ZmHy2 transcript may allow partial recovery over the life of elm1 plants. The accumulation of correctly spliced sh2-i transcript is tissue specific (Lal et al., 1999). Similar context-dependent splicing at AA may permit the accumulation of different amounts of correctly spliced ZmHy2 in different tissues and at different stages of plant growth.
Missplicing of intron III of ZmHy2 in elm1 plants leads to the accumulation of two species of ZmHy2 mRNA. The predominant mRNA species encodes a truncated protein (ELMD), while a minor form encodes a protein containing an internal deletion (ELMC). The potential accumulation of ELMC and ELMD offers a further explanation of the partial light responsiveness of elm1 mutant plants. However, when tested in a coupled holophytochrome assembly assay, neither ELMC nor ELMD exhibited measurable PΦB synthase activity.
In both wild-type and elm1 backgrounds, intron IV of the ZmHy2 gene is spliced at multiple 3′ splice sites. Splicing is a two step cleavage-ligation reaction catalyzed by the machinery of the spliceosome (Kramer, 1996; Brown and Simpson, 1998; Lorkovic et al., 2000). In the first step, transesterification results in cleavage at the 5′ splice site and formation of an intron lariat with an adenosine residue within the BPS located upstream of the 3′ splice site. A second transesterification results in 3′ cleavage, exon ligation, and the release of the intron lariat. Prevailing models of vertebrate intron splicing maintain that the first AG dinucleotide downstream from the BPS is selected as the 3′ splice site by scanning of the spliceosome from the BPS (Smith et al., 1989). As detailed above and in Figure 6A, the 3′ splice site AG and the branch point adenosine are further defined by sequence context. Splicing of intron IV at the type 3 site produces transcript-encoding full-length protein and the type 3 AG may be considered to define the authentic splice junction. Based upon a scanning model for AG selection, the sequence TTAAT (BPS5 in Fig. 6A) is the best candidate for the BPS used in conjunction with the type 3 AG. However, the normally observed distance between the BPS and 3′ splice site AG is 18 to 40 nt (Simpson et al., 1996, 2002) and therefore larger than the 16 nt between BPS5 and the type 3 AG. The suboptimal placement of the intron IV type 3 AG relative to BPS5 may promote the use of the additional type 1 and type 2 sites. The use of type 1 and type 2 3′ splice sites requires the use of BPSs other than BPS5. Four additional candidate BPSs (BPS1–4) are shown in Figure 6A. The observed multiple splicing forms of ZmHy2 may result from competition among BPS/AG combinations for a pool of splicing factors that define the 3′ junction. The functional role of IV/V1 and IV/V2 mRNAs is unclear. In vitro analysis of ELMB suggested that the N-terminal half of ZmHY2 alone is unable to catalyze the reduction of BV IXα to PΦB. Nevertheless, it is possible that truncated forms of ZmHY2 play a role in sequestering BV IXα and in the regulation of tetrapyrrole metabolism. However, no such truncated proteins have been identified as the products of Arabidopsis AtHY2 (Kohchi et al., 2001); thus, the multiple splicing events at intron IV of ZmHy2 may simply be the result of poor splice site definition.
Despite the absence of a well-defined BPS and 3′ splice junction at intron IV, ZmHy2 IV/V3 RT-PCR products were clearly the dominant form isolated from wild-type plants (chi-square test of observed against equal frequency at all sites; χ2 = 46, P < 0.005). However, it should be noted that the assay used does not distinguish between differences in splicing efficiencies and differential stability of the resulting mRNA products. Both ZmHy2 IV/V1 and IV/V2 mRNAs are potential targets of the nonsense-mediated decay pathway, a system characterized in yeast, invertebrates, mammals, and plants that functions to rapidly degrade mRNAs containing premature stop codons (Pulak and Anderson, 1993; Bhattacharya et al., 2000; Ruiz-Echevarria and Peltz, 2000, Isshiki et al., 2001). It is interesting to note that the observed frequencies of type 1, type 2, and type 3 splicing at intron IV are much more similar to each other in the elm1 background and are consistent with equal splicing at the three sites (chi-square test of observed against equal frequency at all sites; χ2 = 0.16, P > 0.05). Although apparent differences in intron IV splicing patterns between wild type and elm1 may result directly from differential splicing triggered by events in intron III, such a possibility is not consistent with existing models of intronic or exonic definition. Alternatively, differences in intron IV splicing variant accumulation between wild type and elm1 may be the result of changes in mRNA stability. The presence of a premature stop codon in exon IV of the majority of ZmHy2 mRNAs in the elm1 background makes all such messages potential targets of the nonsense-mediated decay pathway irrespective of splicing events at intron IV. Consequently, the equal frequencies of ZmHy2 IV/V1, IV/V2, and IV/V3 products observed in elm1 may be a truer reflection of the relative efficiencies of splicing events at these sites.
The elm1 mutant is the first phytochrome-deficient mutant of maize to be characterized, and it has proven a useful resource in beginning to understand the role of phytochrome signaling in maize development and growth (e.g. Sawers et al., 2002; Markelz et al., 2003). However, interpretation of elm1 physiology has been hindered by limited information concerning the molecular nature of the lesion. The cloning of ZmHy2 has identified the elm1 mutation as a ZmHy2 allele and provided evidence that elm1 mutant plants have the capacity for partial recovery due to low-level accumulation of full-length in-frame ZmHy2 transcripts. Characterization of chromophore-deficient mutants from a number of species has suggested that a complete loss of PΦB synthase activity may have severe and pleiotropic phenotypic consequences (Terry, 1997). Thus, null alleles of PΦB synthase-encoding genes may be difficult to isolate and of limited use in the characterization of mature plant phenotypes. Indeed, screens of reverse genetics resources in maize (e.g. MTMdB, http://mtm.cshl.org/ and PML, http://chloroplast.uoregon.edu/) have failed to result in the recovery of additional elm1 alleles. Thus, the elm1 mutant allele described here offers the most direct method to understand the role of phytochrome in maize development and to determine the extent, and potential agronomic importance, of light-regulated phenotypic plasticity.
MATERIALS AND METHODS
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Plant Material and Growth Conditions
Segregating elm1 mutant stocks was as previously described (Sawers et al., 2002). All material was greenhouse grown in winter, Ithaca, NY (supplemental lighting provided by 400-W metal halide lamps, 28°C) and seedlings harvested at 10 d after planting. RNA was extracted from approximately 0.75 g of second/third leaves pooled from three individuals.
Sequence Analysis
Genomic sequence not present in database searches was amplified using the Genome Walker kit (CLONTECH, Palo Alto, CA) according to the manufacturer's protocol. The nested ZmHy2-specific primers PCB2.1 (5′-GTCAAGCTCCGAAGCAGTCTAATTTTGG-3′) and PCB2.2 (5′-CATTGAGAACGGTGTTATCCTCATTTGC-3′) were used for amplification. Sequence alignments and gene assemblies were performed with Sequencher (Gene Codes, Ann Arbor, MI) and DNAstar (DNAstar, Madison, WI) software.
Transcript Analysis
RNA isolation and RT-PCR was as previously described (Sawers et al., 2002; Singh et al., 2003). The full-length coding region of ZmHy2 was amplified using primers RS301 (5′-TGCCTCTCTGCTCTTCCCAAGCTC-3′) and ELMSALIREV (5′-CTTGTCGACTATCCATCCACAGCGTCGCCGACGAA-3′). RACE PCR was performed using the GeneRacer kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The nested ZmHy2-specific primers ELMRACE3 (5′-CACTTGTAATCTTGCCACCCCAGGGCAGAA-3′) and ELMRACE4 (5′-AGCGCGAAGTGGACGAACTTCTGGT-3′) were used for amplification. A semiquantitative PCR assay was used to examine splice site selection in wild-type and elm1 mutants. cDNA was synthesized from wild-type and mutant leaf tissue and low-cycle PCR amplification performed using PCB10 (5′-ACCGTTCTCAATGCCTTGTC-3′) and RS302 (5′-CATACAAAGGGTTGAGGTCC-3′) or PCB10 and ELM4 (5′-ACTTTCGCGAACTGCTCCGTCC-3′) primer sets. Products were transferred to nylon membrane and detected using an [α-32P]CTP-labeled fragment of ZmHy2. The ZmHy2 fragment was generated from PCR amplification of a cDNA clone (pPCB1013) using primers PCB10 and PCB13 (5′-GTCGGGTTCCAAAGGACTTC-3′). The fragment was labeled using random hexamers as previously described (Sheehan et al., 2004).
Recombinant Inbred Mapping
A HindIII restriction site polymorphism was used to map ZmHy2 in the BNL96 population (Burr et al., 1988). A digoxygenin-labeled DNA fragment was generated as previously described Singh et al. (2003) using primers ELM5 (5′-CCCCTGCACGGAGTATTGTTGTC-3′) and ELM6 (5′-CGCCATGCTCCGCTTCTT-3′).
PCR-Based Genotyping of Elm1 and elm1 Plants
Primers ELM5 and ELM4 (5′-ACTTTCGCGAACTGCTCCGTCC-3′) were used to amplify a 549-bp genomic region spanning introns III, IV, and V. PCR products were digested with the restriction enzyme NlaIV, fractionated on 2% (w/v) agarose gels, and detected using ethidium bromide. The presence of the G to A transition at the 5′ end of intron III was detected by the ablation of an NlaIV site. See Figure 4B for details.
ZmHY2 Cellular Localization
To generate a translation protein fusion between ZmHY2 and GFP, the full-length ORF of ZmHY2 was amplified by PCR and introduced into the binary vector pGWB5 (a gift of Dr. Tsuyoshi Nakagawa, Shimane University, Japan) using the GATEWAY in vitro site-specific recombination (Invitrogen). This binary construct contains a cauliflower mosaic virus 35S promoter to confer constitutive expression of the ZmHY2:GFP introduced into Agrobacterium tumefaciens strain GV3101 (pMP90; Koncz et al., 1984) by electroporation. A control vector expressing a nontargeted GFP (pBIN35S:GFP) was kindly provided by Dr. Ian Moore, Oxford University. Control A. tumefaciens-mediated transient expression was carried out in tobacco (Nicotiana tabacum) SR1 (cv Petit Havana) as described by Batoko et al. (2000) with some modifications. A single colony of the transformed A. tumefaciens was used to inoculate 5 mL of yeast extract medium containing beef extract, supplemented with 100 μg/mL kanamycin and 10 μg/mL gentamycin. The bacterial culture was incubated at 28°C overnight, and the bacteria were pelleted by centrifugation at 2,200g for 15 s in a microcentrifuge at room temperature. The pellet was washed three times with 1 mL of the infiltration buffer (50 mm MES, pH 5.6, 2 mm Na3PO4, 0.5% Glc [w/v], and 100 μm acetosyringone; Sigma-Aldrich, St. Louis) and then resuspended in 1 mL of the same buffer. The bacterial suspension was diluted with infiltration buffer to adjust the inoculum concentration to 0.05 OD600 value. The inoculum was delivered to the lamina tissues of tobacco leaves by gentle pressure infiltration through a small needle puncture created in the lower epidermis. The plant was incubated under normal growing conditions for 48 h before confocal microscopy analysis.
Construction of pGEX-ZmHY2 Expression Vectors
Vectors were constructed to express four species of ZmHy2 products. Transcripts encoding mature peptides without predicted chloroplast transit sequences were amplified using the primers ELMBGLIIFWD (5′-GGCAGATCTGGGTCCGGCTGCTCGTACCAG-3′) and ELMSALIREV (5′-CTTGTCGACTATCCATCCACAGCGTCGCCGACGAA-3′), which contained BglII and SalI sites (underlined) respectively, and cloned into the pGEMT-EZ vector for sequence analysis. Cloned fragments representative of different species of ZmHy2 were excised using BglII and SalI and subcloned into the Escherichia coli expression vector pGEX-6-P1 (Amersham Pharmacia Biotech, Piscataway, NJ) using BamHI and SalI sites. The constructed vectors contained mature ZmHy2 sequence fused 3′ to the GST gene of Schistosoma japonicum under the control of the Ptac promoter. Constructs are detailed in Figure 7.
Expression and Purification of Recombinant ZmHY2
Overexpression of Elm-GST constructs was performed in E. coli BL21 DE3 cells containing a plasmid encoding the trxA gene for E. coli thioredoxin (Yasukawa et al., 1995) to enhance protein solubility. One-liter cell cultures containing 100 mg L−1 ampicillin and 34 mg L−1 chloramphenicol were incubated at 18°C for 16 to 20 h before protein expression was induced with 0.5 mm isopropylthio-β-galactoside. Cultures were incubated for a further 6 h at 18°C and then lysed by sonication in buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.05% [v/v] Triton X-100, 1 mm dithiothreitol (DTT), Complete Protease Inhibitor; F. Hoffman La Roche, Basel) using six 20-s pulses. Lysate was cleared at 45,000gmax for 60 min. Protein purification was performed using columns with a 1.5-mL bed of Glutathione Sepharose 4B resin (Amersham Biosciences, Buckinghamshire, UK) according to the manufacturer's protocol. Removal of the GST protein domain with site-specific protease was not performed due to low expression level.
PΦB Synthase Activity Assay
Phytochromobilin synthesizing capacity for each purified protein fraction was determined in 1-mL assay mixes as described in Kohchi et al. (2001). Assays were started by the addition of purified recombinant protein, and incubations were performed in darkness at 28°C for 60 min. Samples were analyzed directly following incubation. Cph1 apophytochrome from Synecchocystis sp. PCC6803 was expressed in E. coli M15 [pREP4]. A crude lysate was prepared by sonication (6 times 10 s) in Cph1 lysis buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 5 mm EDTA, 1 mm DTT, 5% glycerol) and cleared by centrifugation at 11,600gmax for 10 min. Lysate was kept on ice. Phytochromobilin synthesis assay product (500 μL) plus 0.5 mm DTT was mixed with 10 μL Cph1 lysate and incubated on ice for 5 min. Absorption spectra (450–850 nm) were taken in dark then following 60 s R illumination and 60 s FR illumination. For spectrophotometer and filter details, see Kami et al. (2004). Difference spectra were obtained by subtraction of R spectra from FR spectra.
Sequence data from this article have been deposited with the GenBank data library under accession numbers AY560384 and AI691542. Database accession numbers used in sequence alignments were BAB33374 (AtHY2), AK101395 (OsHY2), and Q02190 (PebB).
Acknowledgments
We thank Drs. J.W.S. Brown and C.G. Simpson (Scottish Crop Research Institute, Invergowrie, Dundee, Scotland) for discussions and comments on splicing of the ZmHy2 gene and Ms. Katya Anufrikova (Boyce Thompson Institute, Ithaca, NY) and Ms. Ling Bai (Cornell University, Ithaca, NY) for excellent technical assistance. We also thank Drs. Michael Edgerton (Monsanto, St. Louis), Paul Chomet (Monsanto/Mystic Research, Mystic, CT), and Ms. Moira Sheehan (Cornell University, Ithaca, NY) for many helpful discussions and Dr. Ben Burr (Brookhaven National Laboratories, Upton, NY) for contributing the mapping data.
This work was supported by the National Science Foundation (grant no. IBN 0110297 to T.P.B.) and in part by the Biotechnology and Biological Sciences Research Council International Scientific Interchange Scheme (award no. ISIS 982 to M.J.T. and T.K.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046417.
References
- Aroian RV, Levy AD, Koga M, Ohshima Y, Kramer JM, Sternberg PW (1993) Splicing in Caenorhabditis elegans does not require an AG at the 3′ splice acceptor site. Mol Cell Biol 13: 626–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannai H, Tamada Y, Maruyama O, Nakai K, Miyano S (2002) Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18: 298–305 [DOI] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12: 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW (1996) Arabidopsis intron mutations and pre-mRNA splicing. Plant J 10: 771–780 [DOI] [PubMed] [Google Scholar]
- Brown JW, Simpson CG (1998) Splice site selection in plant pre-mRNA splicing. Annu Rev Plant Physiol Plant Mol Biol 49: 77–95 [DOI] [PubMed] [Google Scholar]
- Brown WR, Kacskovics I, Amendt BA, Blackmore NB, Rothschild M, Shinde R, Butler JE (1995) The hinge deletion allelic variant of porcine IgA results from a mutation at the splice acceptor site in the first C alpha intron. J Immunol 154: 3836–3842 [PubMed] [Google Scholar]
- Burr B, Burr FA, Thompson KH, Albertson MC, Stuber CW (1988) Gene mapping with recombinant inbreds in maize. Genetics 118: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD (1999) The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci USA 96: 6541–6546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elich T, Lagarias JC (1987) Phytochrome chromophore biosynthesis: Both 5-minolevulinic acid and biliverdin overcome inhibition by gabaculine in etiolated Avena sativa L. seedlings. Plant Physiol 84: 304–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki M, Yamamoto Y, Satoh H, Shimamoto K (2001) Nonsense-mediated decay of mutant waxy mRNA in rice. Plant Physiol 125: 1388–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izawa T, Oikawa T, Tokutomi S, Okuno K, Shimamoto K (2000) Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J 22: 391–399 [DOI] [PubMed] [Google Scholar]
- Kami C, Mukougawa K, Muramoto T, Yokota A, Shinomura T, Lagarias JC, Kohchi T (2004) Complementation of phytochrome chromophore-deficient Arabidopsis by expression of phycocyanobilin:ferredoxin oxidoreductase. Proc Natl Acad Sci USA 101: 1099–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC (2001) The Arabidopsis Hy2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13: 425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Kreuzaler F, Kalman Z, Schell J (1984) A simple method to transfer, integrate and study expression of foreign genes, such as chicken ovalbumin and alpha-actin in plant tumors. EMBO J 3: 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Cone JW, Dekens RG, O'Herne-Robers EG, Spruitt CJP, Kendrick RE (1985) Photomorphogenic responses of long-hypocotyl mutants of tomato. J Plant Physiol 120: 153–165 [Google Scholar]
- Koornneef M, Rolff E, Spruitt CJP (1980) Genetic control of light-induced hypocotyl elongation in Arabidopsis thaliana L. Z Pflanzenphysiol 100: 147–160 [Google Scholar]
- Kraepiel Y, Jullien M, Cordonnier-Pratt MM, Pratt L (1994) Identification of two loci involved in phytochrome expression in Nicotiana plumbaginifolia and lethality of the corresponding double mutant. Mol Gen Genet 242: 559–565 [DOI] [PubMed] [Google Scholar]
- Kramer A (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem 65: 367–409 [DOI] [PubMed] [Google Scholar]
- Lagarias JC, Rapoport H (1980) Chromopeptides from phytochrome: the structure and linkage of the Pr form of the phytochrome chromphore. J Am Chem Soc 102: 4821–4828 [Google Scholar]
- Lal S, Choi JH, Curtis Hannah L (1999) The AG dinucleotide terminating introns is important but not always required for pre-mRNA splicing in the maize endosperm. Plant Physiol 120: 65–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Sharopova N, Beavis WD, Grant D, Katt M, Blair D, Hallauer A (2002) Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol Biol 48: 453–461 [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Wieczorek Kirk DA, Lambermon MH, Filipowicz W (2000) Pre-mRNA splicing in higher plants. Trends Plant Sci 5: 160–167 [DOI] [PubMed] [Google Scholar]
- Markelz NH, Costich DE, Brutnell TP (2003) Photomorphogenic responses in maize seedling development. Plant Physiol 133: 1578–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews S, Sharrock RA (1996) The phytochrome gene family in grasses (Poaceae): a phylogeny and evidence that grasses have a subset of the loci found in dicot angiosperms. Mol Biol Evol 13: 1141–1150 [DOI] [PubMed] [Google Scholar]
- McDowell MT, Lagarias JC (2001) Purification and biochemical properties of phytochromobilin synthase from etiolated oat seedlings. Plant Physiol 126: 1546–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto T, Kohchi T, Yokota A, Hwang I, Goodman HM (1999) The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11: 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J (2000) Light: an indicator of time and place. Genes Dev 14: 257–271 [PubMed] [Google Scholar]
- Pulak R, Anderson P (1993) mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev 7: 1885–1897 [DOI] [PubMed] [Google Scholar]
- Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3: 85–93 [DOI] [PubMed] [Google Scholar]
- Ruiz-Echevarria MJ, Peltz SW (2000) The RNA binding protein Pub1 modulates the stability of transcripts containing upstream open reading frames. Cell 101: 741–751 [DOI] [PubMed] [Google Scholar]
- Sawers RJ, Linley PJ, Farmer PR, Hanley NP, Costich DE, Terry MJ, Brutnell TP (2002) Elongated mesocotyl1, a phytochrome-deficient mutant of maize. Plant Physiol 130: 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MJ, Farmer PR, Brutnell TP (2004) Structure and expression of maize phytochrome family homeologs. Genetics 167: 1395–1405 [DOI] [PMC free article] [PubMed]
- Simpson CG, Clark G, Davidson D, Smith P, Brown JW (1996) Mutation of putative branchpoint consensus sequences in plant introns reduces splicing efficiency. Plant J 9: 369–380 [DOI] [PubMed] [Google Scholar]
- Simpson CG, Thow G, Clark GP, Jennings SN, Watters JA, Brown JW (2002) Mutational analysis of a plant branchpoint and polypyrimidine tract required for constitutive splicing of a mini-exon. RNA 8: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Lewis PE, Hardeman K, Bai L, Rose JK, Mazourek M, Chomet P, Brutnell TP (2003) Activator mutagenesis of the pink scutellum1/viviparous7 locus of maize. Plant Cell 15: 874–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Chu TT, Nadal-Ginard B (1993) Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol 13: 4939–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Porro EB, Patton JG, Nadal-Ginard B (1989) Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature 342: 243–247 [DOI] [PubMed] [Google Scholar]
- Smith H (2000) Phytochromes and light signal perception by plants—an emerging synthesis. Nature 407: 585–591 [DOI] [PubMed] [Google Scholar]
- Terry MJ (1997) Phytochrome chromophore-deficient mutants. Plant Cell Environ 20: 740–745 [Google Scholar]
- Terry MJ, Kendrick RE (1996) The aurea and yellow-green-2 mutants of tomato are deficient in phytochrome chromophore synthesis. J Biol Chem 271: 21681–21686 [DOI] [PubMed] [Google Scholar]
- Terry MJ, McDowell MT, Lagarias JC (1995) (3Z)- and (3E)-phytochromobilin are intermediates in the biosynthesis of the phytochrome chromophore. J Biol Chem 270: 11111–11118 [DOI] [PubMed] [Google Scholar]
- Terry MJ, Wahleithner JA, Lagarias JC (1993) Biosynthesis of the plant photoreceptor phytochrome. Arch Biochem Biophys 306: 1–15 [DOI] [PubMed] [Google Scholar]
- Umen JG, Guthrie C (1995) The second catalytic step of pre-mRNA splicing. RNA 1: 869–885 [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Terry MJ, Rameau C, Reid JB, Kendrick RE (1996) The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXa. Plant Cell 8: 55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Terry MJ, Reid JB, Kendrick RE (1997) The phytochrome-deficient pcd2 mutant of pea is unable to convert biliverdin IXa to 3Z-phytochromobilin. Plant J 11: 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasukawa T, Kanei-Ishii C, Maekawa T, Fujimoto J, Yamamoto T, Ishii S (1995) Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J Biol Chem 270: 25328–25331 [DOI] [PubMed] [Google Scholar]
- Yokoo T, Okuno K (1993) Genetic analysis of earliness mutations induced in the rice cultivar Norin 8. Jpn J Breed 43: 1–11 [Google Scholar]